Abstract
Microbiota-derived metabolites that elicit DNA damage can contribute to intestinal tumorigenesis. However, the full spectrum of genotoxic chemicals produced by indigenous gut microbes remains to be defined. We established a pipeline to systematically evaluate the genotoxicity of a large collection of human gut commensals and identified isolates from divergent phylogenies whose small molecule metabolites caused DNA damage. Using comparative metabolomics and bioactivity-guided natural product-discovery techniques, we discovered a previously undescribed family of genotoxic metabolites—termed the indolimines—produced by the colorectal cancer (CRC)-associated species Morganella morganii. Finally, we found that M. morganii exacerbated CRC in gnotobiotic mice. These studies reveal the existence of a previously unexplored universe of genotoxic small molecules from the human microbiome and imply a broader role for microbiota-derived genotoxins in CRC.
One Sentence Summary:
Functional screening of human commensal bacteria identifies diverse genotoxic microbes that may contribute to intestinal tumorigenesis.
Colorectal cancer (CRC) is the third most common malignancy and the second leading cause of cancer death worldwide (1). Two-thirds of all CRC cases occur in individuals without a family history of CRC or inherited CRC-predisposing genetic mutations (2). Thus, environmental risk factors that promote the acquisition and accumulation of somatic genetic and epigenetic aberrations are chief contributors to CRC development. The gut microbiota can regulate intestinal tumorigenesis through diverse mechanisms (3–5) such as short-chain fatty acid-producing clostridia species that induce regulatory T cells as well as temper inflammation-induced tumorigenesis (6) and Fusobacterium nucleatum strains that enhance tumor growth by inducing epithelial proliferation through FadA-mediated engagement of E-cadherin and activation of Wnt/β-catenin signaling (7). Microbial products can also trigger DNA modifications in intestinal epithelial cells (8). For example, the 20 kDa Bacteroides fragilis toxin induces DNA damage through induction of reactive oxygen species (ROS) (9), while cytolethal distending toxin (CDT) from pathogenic Gram-negative bacteria has direct DNase activity (10).
Small molecule metabolites from the microbiome are also known to influence CRC risk by directly causing DNA damage. Select Escherichia coli strains produce the reactive small molecule genotoxin colibactin, which directly alkylates and crosslinks DNA, triggering double-stranded DNA breaks (DSBs) and facilitating intestinal tumorigenesis in mouse models (11–14). The colibactin biosynthetic machinery is encoded by a 54 kb hybrid polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS) gene cluster referred to as the pks or clb locus (11), and the mature chemical structure of colibactin responsible for the pathway’s DNA interstrand crosslinking activity was recently determined (15, 16). Human CRC tumors also contain mutational signatures consistent with colibactin-induced DNA damage, implicating colibactin in human CRC development (17, 18).
The colibactin paradigm illustrates the importance of microbiota metabolite-induced DNA damage in human CRC. However, aside from colibactin, the role of microbiota-derived small molecule genotoxins in CRC initiation or progression remains mostly unexplored. Given the enormous complexity and diversity of metabolites produced by bacteria (19), we hypothesized that diverse taxa from the human gut microbiome may produce previously undiscovered or unappreciated small molecules that cause DNA damage in intestinal epithelial cells and contribute to the development of CRC. Here, we established a pipeline to evaluate the genotoxicity of small molecule metabolites derived from over 100 phylogenetically diverse human gut microbes which were isolated from the microbiomes of inflammatory bowel disease (IBD) patients (20). We identified a diverse set of microbes that produced genotoxic small molecule metabolites, including the Gram-positive bacteria Clostridium perfringens and Clostridium ramosum, and the CRC-associated Gram-negative species Morganella morganii. However, none of these isolates produced known genotoxins, such as colibactin, or encoded known genotoxin-producing biosynthetic gene clusters. We combined untargeted metabolomics and bioactivity-guided natural product discovery techniques, to isolate, characterize, and synthesize a family of previously undescribed genotoxic metabolites—termed the indolimines—from M. morganii under in vitro and in vivo culture conditions. Finally, genotoxic M. morganii exacerbated intestinal tumorigenesis in a gnotobiotic mouse model of CRC. These studies thus uncover an expanded universe of genotoxic gut microbes and metabolites that may critically influence CRC initiation and progression.
Establishing a pipeline to identify genotoxic bacteria from the human gut microbiota
We established a pipeline to screen diverse human gut microbes based on their ability to directly damage DNA. We then applied this pipeline to a gut microbiota culture collection assembled by anaerobic culturomics of stool samples from 11 IBD patients (20), as IBD patients are considered to be at a significantly increased risk of developing CRC (21). This collection consists of 122 unique bacterial isolates that span 5 phyla, 9 classes, 10 orders, and 18 families, as well as multiple strains that were assigned to the same species (Fig. 1A).
Figure 1. Establishing a pipeline to identify genotoxic bacteria from the human gut microbiota.
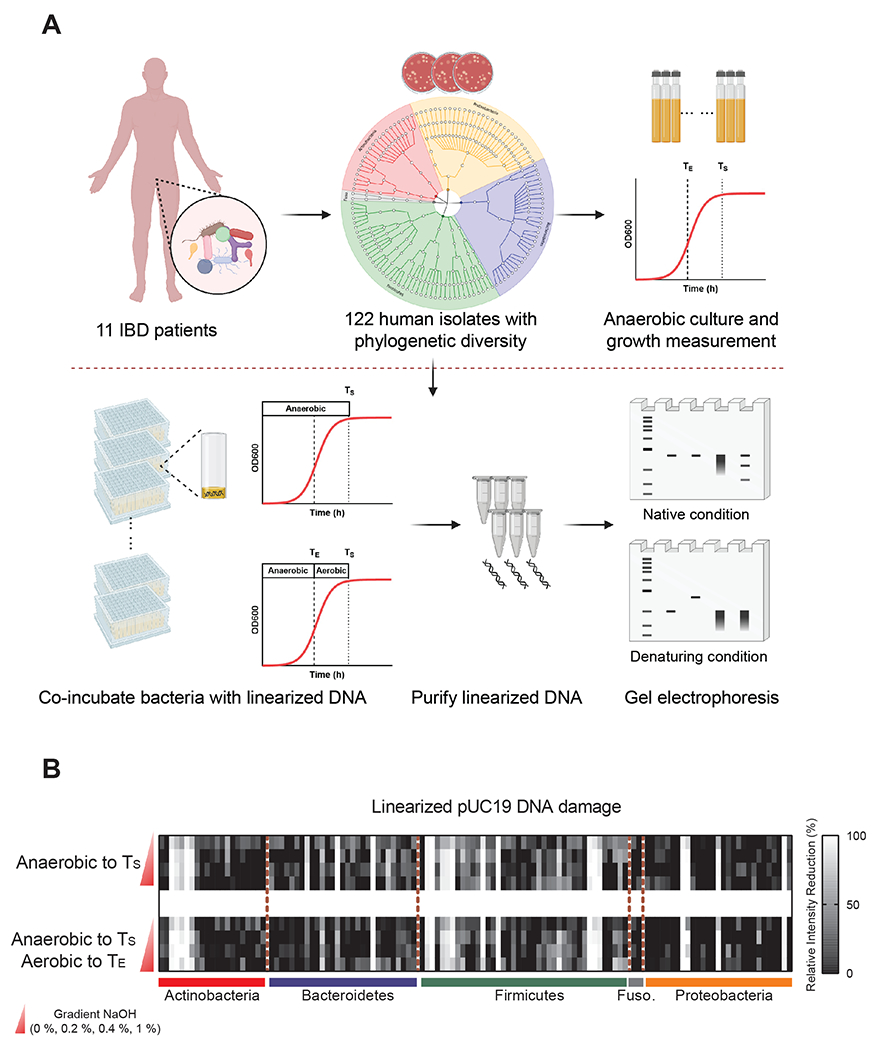
(A) Overview of functional screening of gut microbes for direct genotoxicity. 122 phylogenetically diverse bacterial isolates from 11 IBD patients (shaded based on phylum: Red, Actinobacteria; Blue, Bacteroidetes; Orange, Proteobacteria; Gray, Fusobacteria) were evaluated for genotoxicity via co-incubation with plasmid DNA followed by gel electrophoresis. Bacterial growth curves for all isolates were determined via OD600 and individual isolates were co-cultured with linearized plasmid DNA under anaerobic conditions to stationary phase (TS, light dashed line), or co-cultured under anaerobic conditions to exponential phase (TE, bold dashed line), and then under aerobic conditions to stationary phase. After co-incubation, DNA damage was assessed via gel electrophoresis of purified plasmid DNA under native (top) or denaturing (bottom) conditions.
(B) Diverse human gut bacteria exhibit direct DNA damaging activities. Bacterial genotoxicity was determined by calculating the relative intensity reduction (RIR, %) of linearized pUC19 DNA bands after co-incubation with 122 diverse human gut bacteria (as outlined in A) as compared to medium only controls. pUC19 DNA was then purified via column purification and treated with or without gradient NaOH (0 %, 0.2 %, 0.4 %, 1 %) before evaluating DNA integrity via gel electrophoresis. Heatmap columns represent 122 phylogenetically diverse isolates and rows represent native or denaturing conditions (0 %, 0.2 %, 0.4 %, or 1 % NaOH).
To probe for genotoxicity, we evaluated the activity of each isolate in a plasmid DNA damage assay. As microbiome metabolites such as colibactin can be recalcitrant to isolation (15), we focused our primary studies on co-incubation of individual bacterial isolates with linearized pUC19 plasmid DNA (Fig. 1A). This assay is based on the principle that the extent and modes of DNA damage can be assessed by electrophoresis under native and denaturing conditions (22, 23): The intact linearized pUC19 DNA forms a clear band at ~2.7 kb under native conditions, while denaturation with increasing concentrations of NaOH reveals a ~1.3 kb band of single-stranded DNA. The formation of DNA interstrand cross-links (ICLs) prevents unwinding under denaturing conditions, thereby resulting in slower migration of duplexed DNA. Alkylation at many of the sites in DNA bases is known to decrease the stability of the glycoside bonds, resulting in deglycosylation and fragmentation, which is detectable as smaller fragments of higher mobility following electrophoresis (24). Extensive DNA damage, for example, by DNase-mediated degradation, results in a loss of DNA even under native conditions. Finally, DNA damage caused by restriction enzyme-like molecules produces multiple bands under native conditions and even smaller fragments under denaturing conditions.
We confirmed that plasmid DNA was stable under diverse anaerobic cultivation conditions including incubation in Gifu medium, which supports the growth of all isolates in our collections (Fig. S1). To minimize the damage caused by bacterial DNases that are often produced in the stationary phase of bacterial growth, we measured growth curves for all 122 isolates in our collection and established a TE (time point of exponential phase) and TS (time point of stationary phase) for each isolate (Table S1). The isolates were then clustered into 7 groups that exhibited similar growth dynamics (Fig. S2). We selected two culture conditions for the initial screening: anaerobic co-incubation with DNA to TS; or anaerobic co-incubation with DNA to TE which was followed by aerobic co-incubation to TS to approximate the oxygen stress encountered in an inflammatory gut environment. Finally, we purified the linearized pUC19 DNA from the bacterial cultures via column purification and performed gel electrophoresis under native and gradient denaturing conditions (0 %, 0.2 %, 0.4 % and 1 % NaOH) (Fig. 1A, Fig. S3A–G).
We used the relative intensity reduction (RIR, %) of DNA after co-incubation with bacteria as a general measure of bacterially-induced DNA damage (Fig. 1B, Table S2). We found that diverse gut microbes exhibited DNA damaging activities, which suggests that microbiota-mediated genotoxicity may be more widespread than previously appreciated. While previously described microbiota-derived genotoxins discovered in a case-by-case manner are primarily produced by Gram-negative bacteria (e.g., E. coli, B. fragilis and K. oxytoca) (25, 26), we observed that multiple Gram-positive microbes from the phyla Actinobacteria and Firmicutes also caused significant DNA damage. DNA damaging activity was largely independent of culture conditions, although a few microbes displayed slightly varied genotoxicity in the presence or absence of oxygen stress (Fig. 1B, Table S2).
Small molecule metabolites produced by human gut microbes induced DNA damage
From our primary screening, we selected 24 bacterial isolates that exhibited strong DNA damaging activity and 18 phylogenetically-related non-genotoxic isolates for evaluation in a secondary screening (Fig. 2A). We re-established precise growth curves for each isolate (Table S1) and re-screened all 42 isolates under four distinct culture conditions, including co-incubation of DNA with bacterial supernatants collected from anaerobic cultures at TS (Fig. S3H–J, Table S2). The vast majority of isolates that caused DNA damage in our primary screening also exhibited genotoxicity upon secondary screening (Fig. 2B). Moreover, for 18 of these isolates, the level of DNA damage induced by clarified bacterial supernatants was comparable to that derived by direct incubation with live bacterial cultures (Fig. 2B).
Figure 2. Small molecule metabolites produced by human gut microbes induced DNA damage.
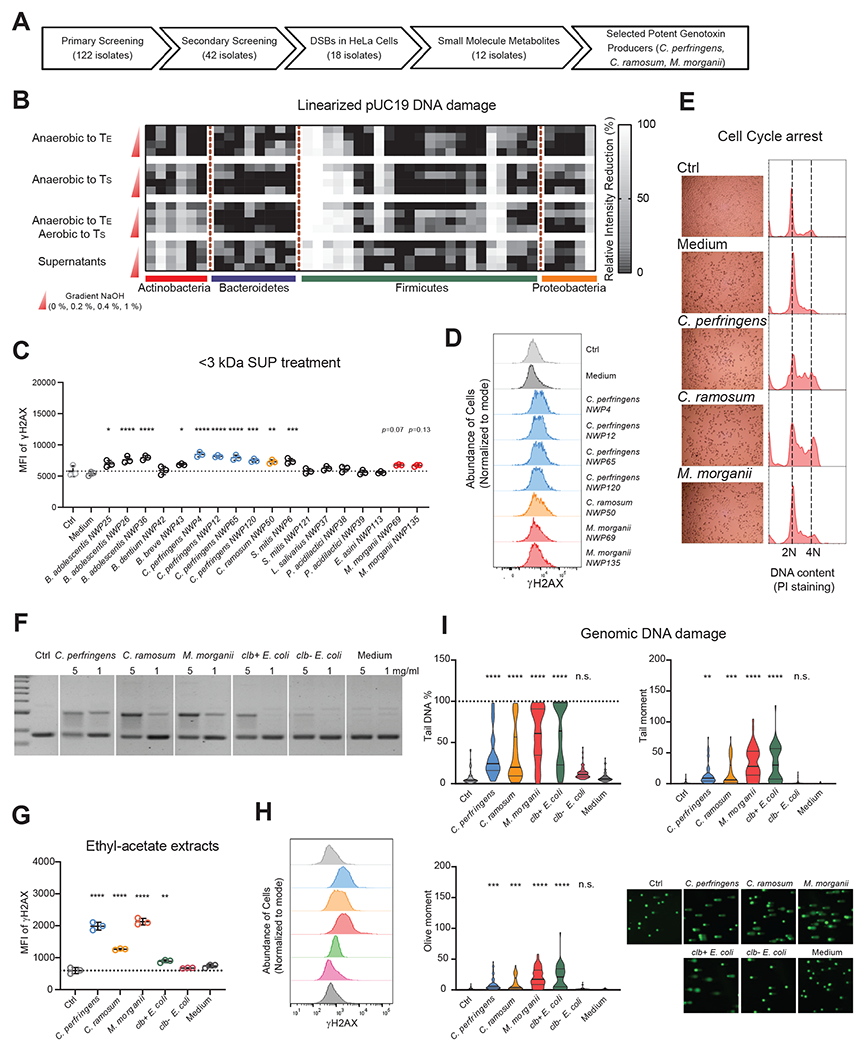
(A) Sequential screening of diverse human gut microbes for genotoxicity using orthogonal methodologies identifies human gut bacteria that produce small molecule genotoxins.
(B) Relative intensity reduction (RIR, %) of linearized pUC19 DNA bands in a secondary screening of 42 putative genotoxic and non-genotoxic isolates selected based on primary screening results (Fig. 1). Linearized pUC19 DNA was co-incubated with select isolates anaerobically to TS or TE, anaerobically to TE and then aerobically to TS, or with bacterial supernatants from isolates cultured anaerobically to TS for 4 h. pUC19 DNA was isolated via column purification after co-incubation and treated with or without NaOH (0 %, 0.2 %, 0.4 %, 1 %) before evaluating DNA integrity via gel electrophoresis. Columns represent 42 isolates and rows represent native or denaturing conditions. Relative intensity reduction (RIR, %) was calculated based on normalization to control samples incubated in Gifu medium alone.
(C) MFI (geometric mean fluorescence intensity) of γ-H2AX staining of HeLa cells treated with 40 % (v/v) PBS (Ctrl) or <3 kDa SUP (small-molecule bacterial supernatants) for 5-6 h.
(D) Representative histograms of γ-H2AX staining of HeLa cells treated with <3 kDa SUP from C. perfringens, C. ramosum or M. morganii isolates.
(E) HeLa cells were treated with 40 % (v/v) PBS or <3 kDa SUP from medium, C. perfringens, C. ramosum or M. morganii isolates for 24 h. Cell cycle arrest was evaluated by propidium iodide (PI) staining based on flow cytometry.
(F-I) Assessment of DNA damage induced by ethyl-acetate extracts of C. perfringens, C. ramosum or M. morganii supernatants. Evaluation of nicking of circular pUC19 DNA (top band = nicked DNA) after co-incubation for 5-6 h. Ctrl, control pUC19 DNA in TE buffer. (F); γ-H2AX staining (G and H) and genomic DNA comets (I) in HeLa cells after treatment with 5 mg/ml extracts or without (Ctrl) for 5-6 h. n = 49 for comet assay analysis, n.s., not significant; * p<0.05; ** p<0.01; *** p<0.001; **** p<0.0001, one-way ANOVA.
To determine whether the putative genotoxic bacterial isolates we identified via electrophoresis-based screening (Fig. S4A) produce genotoxic small molecules that cause DNA damage in human cells, we separated their supernatants (SUP) into small- (<3 kDa SUP) and large- (>3 kDa SUP) molecular weight fractions and evaluated the relative genotoxicity of these fractions in HeLa cells. We found that small molecules from 12 isolates enhanced the expression of γ-H2AX, a marker of DNA double-strand breaks (DSBs) (27), in HeLa cells (Fig. 2C, Fig. S4B–F): two of three genotoxic isolates of Bifidobacterium adolescentis and one isolate of Streptococcus mitis induced DSBs through both small and large molecule metabolites; one isolate of Bifidobacterium breve, four isolates of Clostridium perfringens, one isolate of Clostridium ramosum and two isolates of Morganella morganii induced DSBs through small molecule metabolites only; and the remaining six isolates exhibited no detectable DNA damaging activity in this assay. The DSBs induced by these small molecule metabolites were not due to alterations in cell viability (Fig. S4B), as only C. perfringens supernatants and large molecules induced significant cell death (Fig. S4C–F) which is likely due to the production of general cytotoxins that do not directly induce DNA damage (28).
Recent meta-analyses have identified cross-cohort microbial signatures associated with CRC. As Clostridiaceae, Erysipelotrichaceae, and M. morganii are prevalent members of the CRC-associated microbiome (29, 30), we focused our subsequent studies on isolates from each of these taxonomic groups that exhibited genotoxicity in our initial screens: C. perfringens, C. ramosum, and M. morganii. Consistent with their ability to induce γ-H2AX (Fig. 2C–D), we found that small molecule metabolites from C. perfringens, C. ramosum, and M. morganii also induced cell cycle arrest in HeLa cells (Fig. 2E), further implicating these taxa as potential genotoxin producers. Next, to enrich for genotoxic small molecules, we performed ethyl-acetate extractions using supernatants from one representative genotoxic isolate from each species, as well as control colibactin-producing E. coli strains (K-12 BW25113 with clb locus, designated as clb+ E. coli) and non-colibactin-producing E. coli strains (K-12 BW25113 with empty bacterial artificial chromosome, designated as clb− E. coli). We found that ethyl-acetate extracts from C. perfringens, C. ramosum, M. morganii, and clb+ E. coli cultures nicked circular pUC19 plasmid DNA, while extracts from clb− E. coli or Gifu medium alone had negligible impacts on DNA integrity (Fig. 2F). Similarly, ethyl-acetate extracts from genotoxic species induced γ-H2AX expression (Fig. 2G–H) and tailing in an alkaline comet unwinding assay that measures DNA DSBs at the single-cell level (Fig. 2I) in HeLa cells. Taken together, these data reveal that diverse taxa from the human gut microbiota produce small molecule genotoxins.
M. morganii produces a previously uncharacterized genotoxin that is distinct from colibactin
The biosynthetic machinery involved in the production of microbial metabolites, including previously characterized small molecule genotoxins, is often encoded by biosynthetic gene clusters (BGCs) (31). For example, colibactin production is encoded by a multimodular PKS-NRPS pathway in E. coli, and tilimycin and tilivalline are encoded by an NRPS pathway in Klebsiella oxytoca (26, 32). However, BGC analyses of genotoxic C. perfringens, C. ramosum and M. morganii using antiSMASH (33) failed to detect any known genotoxin-encoding BGCs (Fig. S5A, Table S3). While M. morganii harbors one NRPS/PKS gene cluster, this BGC is entirely distinct from the clb genomic island. In fact, M. morganii lacks key genes involved in colibactin synthesis, such as clbI and clbP (Fig. S5B) (34, 35). This is consistent with recent analyses of 69 publicly available Morganella genomes, which suggests that clb genes are absent in Morganella spp. (36). The genotoxicity caused by M. morganii is also distinct from that caused by colibactin—while clb+ E. coli caused DNA crosslinking, DNA exposed to M. morganii-derived metabolites displayed a smearing pattern under both native and denaturing conditions (Fig. S5C). Finally, as previously reported, colibactin-induced γ-H2AX may require cell-to-cell contact between bacterial and host cells as both separation via a filter abrogates activity and supernatants from clb+ E. coli failed to induce dramatic increases in γ-H2AX in cell lines, likely due to documented colibactin instability (11, 37, 38). By contrast, M. morganii supernatants and small molecule metabolites elicited significant increases in γ-H2AX (Fig. S5D–G). Together, these data suggest that M. morganii produces a previously uncharacterized genotoxin(s) that is distinct from colibactin and is readily diffusible.
Isolation and identification of a previously undescribed genotoxic metabolite derived from M. morganii
To identify specific genotoxin(s) produced by M. morganii, we employed a combination of ultra-performance liquid chromatography quadrupole time-of-flight mass spectrometry (UPLC-QTOF-MS)-based untargeted metabolomics and bioactivity-guided fractionation using small-scale cultures, followed by large-scale cultivation and isolation for unambiguous structure elucidation and genotoxicity analyses (Fig. 3A). We generated an initial candidate ion list of the most abundant M. morganii-derived metabolites relative to Gifu medium control (~100 ion features, Table S4) via comparative metabolomics. We then performed two rounds of activity-guided fractionation using preparative high-performance liquid chromatography (HPLC) and circular pUC19 plasmid-based genotoxicity assays, then profiled the resulting fractions and subfractions using UPLC-QTOF-MS-based metabolomics (Fig. S6A–B). To identify potential genotoxins, we excluded ions present in inactive fractions from the aforementioned initial ion list and ultimately identified 4 ion features (I–IV) as potential genotoxic hits (Fig. 3B, Table S4). To enable structural elucidation and genotoxic activity assessment for these compounds, we performed a large-scale cultivation (18 liters) and ethyl acetate extraction of M. morganii supernatant based on previously observed retention times that imply relatively low polarity of the compounds of interest. This crude extract was subjected to two rounds of HPLC to generate four semi-pure fractions enriched in the four target ion features (F1-F4 enriched in I–IV, respectively). One of these fractions (F2) exhibited dose-dependent genotoxicity in circular pUC19 plasmid-based genotoxicity assays (Fig. 3C). Based on UPLC-QTOF-MS analyses, F2 was primarily comprised of two previously undescribed metabolites with m/z values of 215.1543 (indolimine-214, compound 1, target ion feature II) and 234.1852 (compound 2, a M. morganii-derived metabolite that was absent from the initial ion list, but was enriched during extraction and fractionation) at a ratio of 4:6. This fraction was recalcitrant to further purification by preparative HPLC using diverse combinations of stationary and mobile phases. Thus, the chemical structures of the two components were characterized as a mixture using one- and two-dimensional nuclear magnetic resonance (NMR) spectroscopy analyses (Fig. 3D, Fig. S7). To confirm which compound exerted the observed genotoxicity, we synthesized both indolimine-214 (1) and the phenethylamine-derived compound 2.
Figure 3. Isolation and identification of a previously undescribed genotoxic metabolite derived from M. morganii.
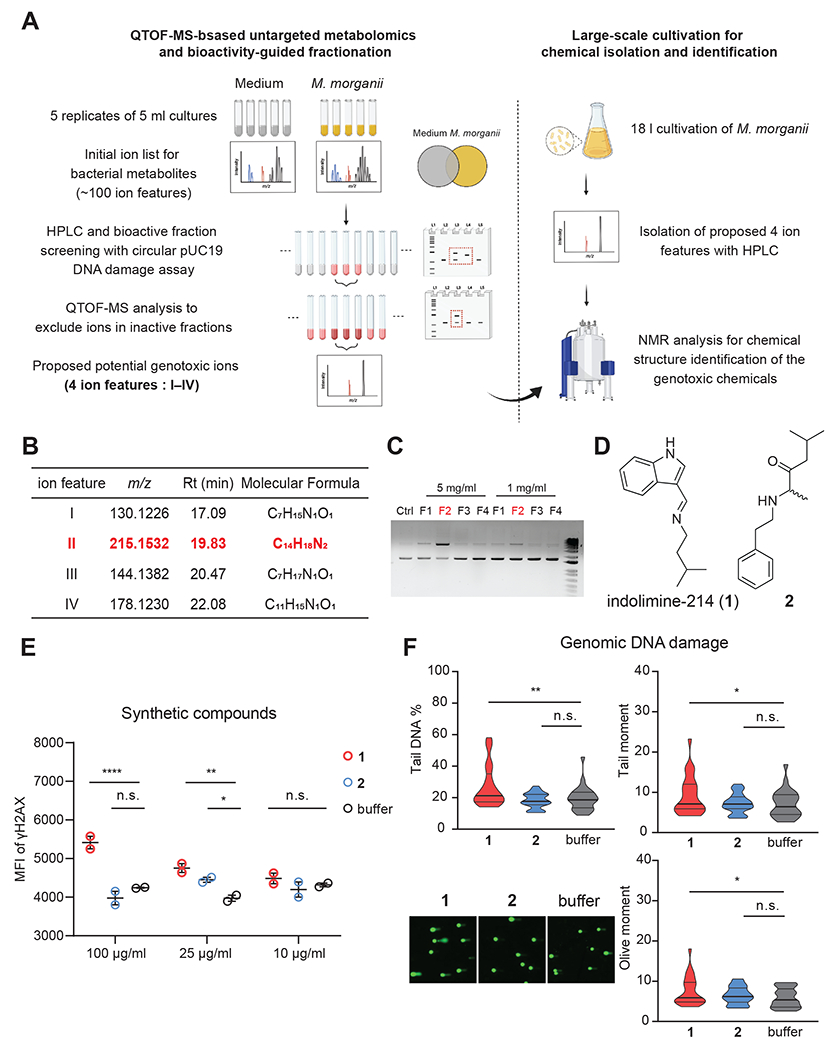
(A) Overview of isolation and identification of genotoxins derived from M. morganii.
(B) Proposed 4 candidate ion features initially detected from M. morganii cultures. Rt, retention time.
(C) Evaluation of nicking of circular pUC19 DNA (top band = nicked DNA) after co-incubation overnight with F1–F4 fractions enriched with ion features I–IV, respectively. Ctrl, control pUC19 DNA in TE buffer.
(D) Chemical structures of compounds indolimine-214 (1) and 2.
(E) MFI of γ-H2AX staining for HeLa cells treated with synthetic compounds at indicated concentrations for 5 h. n.s., not significant; * p<0.05; ** p<0.01; *** p<0.001, two-way ANOVA.
(F) Genomic DNA comets in HeLa cells after treatment with 100 μg/ml synthetic compounds for 5-6 h. n = 25 for comet assay analysis, n.s., not significant; * p<0.05; ** p<0.01; *** p<0.001; **** p<0.0001, one-way ANOVA.
Synthetic indolimine-214 (1) was unstable due to the reversible nature of its imine bridge. Therefore, we fractionated fresh synthetic material and assessed the purity of each fraction using 1H NMR to secure pure compounds for genotoxicity analysis (Fig. S6C). Nonetheless, neither synthetic indolimine-214 (1) nor the phenethylamine-derived metabolite 2 induced DNA damage in a circular pUC19 plasmid-based genotoxicity assay even at high concentrations (1 mg/ml). However, the mixture of indolimine-214 (1) and compound 2 elicited dose-dependent DNA damage at the experimentally observed isolation ratio of 4:6 (F2, Fig. S6D), suggesting that compound 2 may facilitate indolimine-214-induced DNA damage in cell-free assays. We next assessed potential genotoxicity of the synthetic compounds individually in HeLa cells and found that indolimine-214 (1) alone, but not compound 2, triggered increased γ-H2AX in a dose-dependent manner (Fig. 3E) and induced tailing in an alkaline comet unwinding assay (Fig. 3F). Furthermore, the genotoxicity of synthetic indolimine-214 (1) correlated directly with its purity (i.e., in relation to its hydrolytic degradation products, Fig. S6E). Thus, we defined metabolite indolimine-214 (1) as a previously undescribed genotoxic small molecule metabolite produced by M. morganii.
M. morganii produces multiple genotoxic indolimines in vivo and exacerbates CRC in gnotobiotic mice
M. morganii is enriched in fecal samples from IBD- and CRC-patients (29) and in CRC tumors (39). As such, we chose to examine the potential impact of M. morganii on CRC in vivo as compared to a phylogenetically-related non-genotoxic control. The Enterobacterales strain E. coli NC101 exhibits genotoxicity in vitro (40) and exacerbates CRC in AOM/Il10−/− gnotobiotic mice via the production of colibactin (12). However, an isogenic ΔclbP E. coli mutant (NC101 mut) with a disrupted clbP gene failed to induce γ-H2AX in HeLa cells (Fig. S5E–F) or CRC in AOM/Il10−/− gnotobiotic mice (12). UPLC-QTOF-MS-based quantification of indolimine-214 (1) with reference to its synthetic standard confirmed that M. morganii produces high levels of this metabolite in vitro (~40 μg/ml, Fig. 4A), which are comparable to the concentrations of synthetic compound sufficient to induce genotoxicity in HeLa cells (Fig. 3E–F). By contrast, indolimine-214 (1) was undetectable in supernatants from colibactin-producing E. coli strains, clb+ E. coli and NC101 wt, and isogenic non-genotoxic controls, clb− E. coli and NC101 mut (Fig. 4A). Correspondingly, M. morganii supernatants and small molecule metabolites induced γ-H2AX in HeLa cells, while clb− E. coli strains failed to induce detectable increases in γ-H2AX (Fig. 4B, Fig. S5G).
Figure 4. M. morganii produces multiple genotoxic indolimines in vivo and exacerbates CRC in gnotobiotic mice.
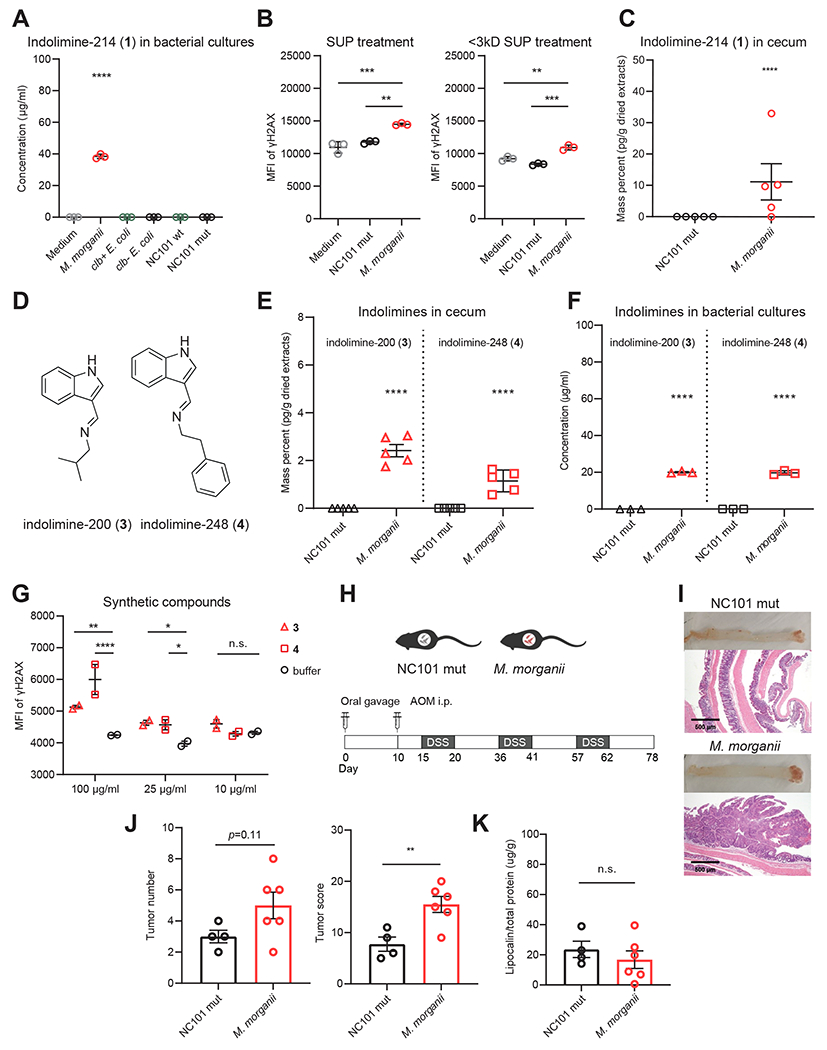
(A) UPLC-QTOF-MS quantification of indolimine-214 (1) in medium or bacterial supernatants of M. morganii, clb+ E. coli, clb− E. coli, NC101 wt E. coli or NC101 mut E. coli. **** p<0.0001, one-way ANOVA.
(B) MFI of γ-H2AX in HeLa cells treated with 40 % (v/v) SUP or <3 kDa SUP from medium, M. morganii, or NC101 mut E. coli for 5-6 h. ** p<0.01; *** p<0.001, one-way ANOVA.
(C) QTOF-MS quantification of indolimine-214 (1) in cecal contents of gnotobiotic mice colonized by M. morganii, or NC101 mut E. coli. **** p<0.0001, Student’s t-test.
(D) Chemical structures of compounds indolimine-200 (3) and indolimine-248 (4).
(E) QTOF-MS quantification of indolimine-200 (3) and indolimine-248 (4) in cecal contents of gnotobiotic mice colonized by M. morganii, or NC101 mut E. coli. **** p<0.0001, Student’s t-test.
(F) UPLC-QTOF-MS quantification of indolimine-200 (3) and indolimine-248 (4) in bacterial supernatants of M. morganii or NC101 mut E. coli. **** p<0.0001, Student’s t-test.
(G) MFI of γ-H2AX staining for HeLa cells treated with synthetic compounds at indicated concentrations for 5 h. n.s., not significant; * p<0.05; ** p<0.01; **** p<0.0001, two-way ANOVA.
(H) Schematic of experimental design for CRC induction in age-matched gnotobiotic mice colonized with M. morganii or NC101 mut E. coli.
(I-K) Representative colon tissue and histology images (I), tumor number and tumor score (J), and fecal lipocalin 2 levels (K; Day 78) in gnotobiotic mice colonized with M. morganii or NC101 mut E. coli. Each dot represents one mouse (n = 4-6 per group), n.s., not significant; ** p<0.01, Student’s t-test.
To assess genotoxic indolimine production in vivo, we colonized germ-free mice with either M. morganii or NC101 mut. Cecal contents from mice colonized with M. morganii contained high levels of genotoxic indolimine-214 (1), but this compound was undetectable in mice colonized with NC101 mut (Fig. 4C). Moreover, in the process of quantifying cecal indolimine-214 (1), we observed the production of two additional previously undescribed indolimines. As indolimine-214 (1) consists of a conjugation of indole-3-carboxaldehyde and the leucine-derived metabolite 3-methylbutan-1-amine, we speculated that M. morganii may also produce related indole conjugates. Indeed, we found that cecal contents from mice colonized with M. morganii, but not mice colonized with E. coli, contained two additional indolimines: conjugates of indole-3-carboxaldehyde with 2-methyl-propan-1-amine (indolimine-200, compound 3, m/z 201.1386) or phenylethylamine (indolimine-248, 4, m/z 249.1386) (Fig. 4D–E, Fig. S8). These additional indolimines were also detected in in vitro M. morganii bacterial cultures, but not E. coli cultures, at slightly lower abundance (~20 μg/ml) than indolimine-214 (1), as confirmed by synthetic standards (Fig. 4F). Finally, synthetic indolimine-200 (3) and indolimine-248 (4) also triggered increased γ-H2AX in a dose-dependent manner (Fig. 4G) Thus, M. morganii produced multiple genotoxic indolimines both in vitro and in vivo.
Finally, to elucidate the potential effects of M. morganii on CRC, we measured the induction of colorectal tumors in gnotobiotic mice colonized with genotoxin-producing M. morganii or non-genotoxin-producing E. coli (NC101 mut) after repeated administration of azoxymethane (AOM) and dextran sulfate sodium (DSS) (Fig. 4H). We observed that M. morganii colonized mice developed more adenomatous polyps and invasive adenocarcinomas (Fig. 4I), and exhibited increased tumor numbers and overall tumor burden as compared to mice colonized with the NC101 mut (Fig. 4J). However, both groups of mice displayed similar levels of overt intestinal inflammation, as measured by fecal lipocalin 2, suggesting that M. morganii-induced exacerbation of CRC is independent of inflammation (Fig. 4K). Overall, these data suggest that M. morganii-derived genotoxins can exacerbate CRC.
Discussion
Aside from a small number of case studies (26, 41), the taxonomic distribution and repertoire of small molecule genotoxins produced by the microbiota remain mostly unexplored. Here, we undertook a systematic evaluation of the carcinogenic potential of a diverse selection of human gut microbes based on the reasoning that somatic mutations resulting from DNA damage are critical mediators of tumor initiation and progression (42). We found that diverse taxa from the human gut microbiota exhibited genotoxicity, identified and characterized a previously undiscovered family of genotoxic M. morganii-derived small molecules termed the indolimines, and determined that colonization with genotoxin-producing M. morganii exacerbated CRC in gnotobiotic mice.
By revealing the existence of a previously uncharted universe of microbiota-derived genotoxins and defining the indolimines as a novel family of bioactive microbiota-derived small molecules, these studies imply an expanded role for genotoxic metabolites in CRC. More broadly, novel genotoxins, including the indolimines, may also impact diverse aspects of host biology beyond tumor initiation in the gut. Indeed, in addition to their direct DNA damaging activities, microbiota-derived genotoxins can potentially initiate or exacerbate inflammatory processes (32, 43, 44), thus contributing to a vicious cycle of inflammation and DNA damage in IBD patients that culminates in tumorigenesis (45). Bacterial production of genotoxins can also enhance competitive fitness for species that thrive in inflammatory microenvironments and thereby shape microbiota composition (46, 47), which may select for the evolution or maintenance of DNA-damaging compounds by individual commensal species. Notably, somatic mutations can be detected in colonic epithelial cells even in early life, which suggests persistent mutagenesis throughout the lifespan of an individual (48). Furthermore, while CRC patients display increased carriage of clb+ E. coli, clb+ taxa (including E. coli relatives, such as Klebsiella species) are also found in healthy individuals (49). Altogether, these observations support a model whereby genotoxic gut microbes contribute to CRC development by persistently inducing DNA damage in host epithelial cells, which synergizes with chronic inflammation in the gut microenvironment, along with additional environmental factors, and eventually facilitates the initiation and progression of CRC.
Finally, these studies underscore the power of function-based assessments of the microbiome to provide new insights into the diverse impacts of indigenous microbes on host biology and disease susceptibility. Recent illumination of the ‘tumor microbiome’ beyond the gut highlights an additional potential role for microbial genotoxins in tumor initiation and progression (50). Furthermore, the broad distribution of genotoxicity across diverse gut species suggests that resident microbes from other mucosal tissues may also produce previously undiscovered genotoxins. Thus, in addition to revealing an expanded diversity and significance of microbiota-derived genotoxins in CRC, these studies provide a roadmap for future identification and characterization of novel microbiota-derived genotoxins across diverse tissues and disease states.
Supplementary Material
Funding:
This work was supported by a Yale Cancer Center Team Challenge award funded by the National Cancer Institute of the National Institutes of Health under award number 3P30CA016359 (to N.W.P, S.B.H and J.M.C.) as well as R01CA215553 to S.B.H. and J.M.C. Approximately 35% of the funding for this research project (~$200,000) was financed with NIH funds; the remainder was financed by nongovernmental sources. N.W.P. also gratefully acknowledges support from the Common Fund of the National Institutes of Health (DP2DK125119), Leona M. and Henry B. Helmsley Charitable Trust (3083), Chan Zuckerberg Initiative, Michael J. Fox Foundation for Parkinson’s Research, Ludwig Family, Mathers Foundation, Pew Charitable Trust, NIA and NIGMS (R01AG068863 and RM1GM141649), and F. Hoffmann-La Roche Ltd. The funders of this work had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Competing interests:
N.W.P. is a co-founder of Artizan Biosciences, Inc. and Design Pharmaceuticals, Inc. N.W.P. has received research funding for unrelated studies from Artizan Biosciences, Inc. and F. Hoffmann-La Roche AG.
Data and materials availability:
All data is available in the main text or in the supplementary materials.
References
- 1.Keum N, Giovannucci E, Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol 16, 713–732 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Jasperson KW, Tuohy TM, Neklason DW, Burt RW, Hereditary and familial colon cancer. Gastroenterology 138, 2044–2058 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brennan CA, Garrett WS, Gut Microbiota, Inflammation, and Colorectal Cancer. Annu Rev Microbiol 70, 395–411 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tilg H, Adolph TE, Gerner RR, Moschen AR, The Intestinal Microbiota in Colorectal Cancer. Cancer Cell 33, 954–964 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Garrett WS, Cancer and the microbiota. Science 348, 80–86 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F, From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 165, 1332–1345 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Rubinstein MR et al. , Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe 14, 195–206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen J, Sears CL, Impact of the gut microbiome on the genome and epigenome of colon epithelial cells: contributions to colorectal cancer development. Genome Med 11, 11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodwin AC et al. , Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc Natl Acad Sci U S A 108, 15354–15359 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buc E et al. , High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS One 8, e56964 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nougayrede JP et al. , Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313, 848–851 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Arthur JC et al. , Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cougnoux A et al. , Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut 63, 1932–1942 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Cuevas-Ramos G et al. , Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci U S A 107, 11537–11542 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xue M et al. , Structure elucidation of colibactin and its DNA cross-links. Science 365, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang YD et al. , Reactivity of an Unusual Amidase May Explain Colibactin’s DNA Cross-Linking Activity. Journal of the American Chemical Society 141, 11489–11496 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pleguezuelos-Manzano C et al. , Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nature, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dziubanska-Kusibab PJ et al. , Colibactin DNA-damage signature indicates mutational impact in colorectal cancer. Nat Med 26, 1063–1069 (2020). [DOI] [PubMed] [Google Scholar]
- 19.da Silva RR, Dorrestein PC, Quinn RA, Illuminating the dark matter in metabolomics. Proc Natl Acad Sci U S A 112, 12549–12550 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palm NW et al. , Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adami HO et al. , The continuing uncertainty about cancer risk in inflammatory bowel disease. Gut 65, 889–893 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue M et al. , Characterization of Natural Colibactin-Nucleobase Adducts by Tandem Mass Spectrometry and Isotopic Labeling. Support for DNA Alkylation by Cyclopropane Ring Opening. Biochemistry 57, 6391–6394 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bossuet-Greif N et al. , The Colibactin Genotoxin Generates DNA Interstrand Cross-Links in Infected Cells. mBio 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gates KS, An Overview of Chemical Processes That Damage Cellular DNA: Spontaneous Hydrolysis, Alkylation, and Reactions with Radicals. Chemical Research in Toxicology 22, 1747–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dejea CM et al. , Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359, 592–597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Unterhauser K et al. , Klebsiella oxytoca enterotoxins tilimycin and tilivalline have distinct host DNA-damaging and microtubule-stabilizing activities. P Natl Acad Sci USA 116, 3774–3783 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan J, Adamski R, Chen J, Focus on histone variant H2AX: to be or not to be. Febs Lett 584, 3717–3724 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freedman JC, Shrestha A, McClane BA, Clostridium perfringens Enterotoxin: Action, Genetics, and Translational Applications. Toxins (Basel) 8, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas AM et al. , Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nature Medicine 25, 667-+ (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wirbel J et al. , Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nature Medicine 25, 679-+ (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shine EE, Crawford JM, Molecules from the Microbiome. Annual review of biochemistry 90, 789–815 (2021). [DOI] [PubMed] [Google Scholar]
- 32.Schneditz G et al. , Enterotoxicity of a nonribosomal peptide causes antibiotic-associated colitis. P Natl Acad Sci USA 111, 13181–13186 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blin K et al. , antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47, W81–W87 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trautman EP et al. , Domain-Targeted Metabolomics Delineates the Heterocycle Assembly Steps of Colibactin Biosynthesis. J Am Chem Soc 139, 4195–4201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zha L et al. , Colibactin assembly line enzymes use S-adenosylmethionine to build a cyclopropane ring. Nature Chemical Biology 13, 1063-+ (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wami H et al. , 10.1101/2021.01.22.427840v1 (2021). [DOI] [Google Scholar]
- 37.Shine EE et al. , Model Colibactins Exhibit Human Cell Genotoxicity in the Absence of Host Bacteria. ACS Chem Biol 13, 3286–3293 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Healy AR et al. , Synthesis and reactivity of precolibactin 886. Nat Chem 11, 890–898 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen W et al. , Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One 7, e39743 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang-Fichaux M et al. , The Polyphosphate Kinase of Escherichia coli Is Required for Full Production of the Genotoxin Colibactin. mSphere 5, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dougherty MW, Jobin C, Shining a Light on Colibactin Biology. Toxins (Basel) 13, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martincorena I, Campbell PJ, Somatic mutation in cancer and normal cells. Science 349, 1483–1489 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Tse H et al. , A tricyclic pyrrolobenzodiazepine produced by Klebsiella oxytoca is associated with cytotoxicity in antibiotic-associated hemorrhagic colitis. Journal of Biological Chemistry 292, 19503–19520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zechner EL, Inflammatory disease caused by intestinal pathobionts. Curr Opin Microbiol 35, 64–69 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Olafsson S et al. , Somatic Evolution in Non-neoplastic IBD-Affected Colon. Cell, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tronnet S et al. , The Genotoxin Colibactin Shapes Gut Microbiota in Mice. mSphere 5, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Silpe JE et al. , 10.1101/2021.05.24.445430v3 (2021). [DOI] [Google Scholar]
- 48.Lee-Six H et al. , The landscape of somatic mutation in normal colorectal epithelial cells. Nature 574(7779), 532–537 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Putze J et al. , Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. Infect Immun 77, 4696–4703 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nejman D et al. , The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 368, 973-+ (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vizcaino MI, Crawford JM, The colibactin warhead crosslinks DNA. Nat Chem 7, 411–417 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dodd D et al. , A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648-+ (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tautenhahn Ralf et al. XCMS Online: a web-based platform to process untargeted metabolomic data. Analytical chemistry 84, 11 5035–5039 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen H et al. , A Forward Chemical Genetic Screen Reveals Gut Microbiota Metabolites That Modulate Host Physiology. Cell 177, 1217–1231 e1218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bolger AM, Lohse M, Usadel B, Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nurk S et al. , Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20, 714–737 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is available in the main text or in the supplementary materials.


