Abstract
Differential chromatin structure is one of the hallmarks distinguishing active and inactive genes. For the X-linked human hypoxanthine phosphoribosyltransferase gene (HPRT), this difference in chromatin structure is evident in the differential general DNase I sensitivity and hypersensitivity of the promoter regions on active versus inactive X chromosomes. Here we characterize the nucleosomal organization responsible for the differential chromatin structure of the active and inactive HPRT promoters. The micrococcal nuclease digestion pattern of chromatin from the active allele in permeabilized cells reveals an ordered array of translationally positioned nucleosomes in the promoter region except over a 350-bp region that is either nucleosome free or contains structurally altered nucleosomes. This 350-bp region includes the entire minimal promoter and all of the multiple transcription initiation sites of the HPRT gene. It also encompasses all of the transcription factor binding sites identified by either dimethyl sulfate or DNase I in vivo footprinting of the active allele. In contrast, analysis of the inactive HPRT promoter reveals no hypersensitivity to either DNase I or a micrococcal nuclease and no translational positioning of nucleosomes. Although nucleosomes on the inactive promoter are not translationally positioned, high-resolution DNase I cleavage analysis of permeabilized cells indicates that nucleosomes are rotationally positioned over a region of at least 210 bp on the inactive promoter, which coincides with the 350-bp nuclease-hypersensitive region on the active allele, including the entire minimal promoter. This rotational positioning of nucleosomes is not observed on the active promoter. These results suggest a model in which the silencing of the HPRT promoter during X chromosome inactivation involves remodeling a transcriptionally competent, translationally positioned nucleosomal array into a transcriptionally repressed architecture consisting of rotationally but not translationally positioned nucleosomal arrays.
Differential chromatin structure and accessibility, particularly at the promoter, have long been recognized as characteristics that distinguish active from inactive genes. Active genes are in general more accessible to regulatory factors than inactive genes, as indicated by nuclease sensitivity. In addition, the promoters of active genes often exhibit marked DNase I hypersensitivity, especially in the vicinity of transcription factor binding sites (10, 20). This hypersensitivity is postulated to be due to changes in the chromatin architecture of the promoter and may represent nucleosomal remodeling or displacement, stretches of single-stranded DNA, torsionally stressed DNA, or other distortions in chromatin structure arising from factor binding (18, 20, 62).
The functional effect of nucleosomes on transcription initiation is thought to be repressive since in vitro assembly of nucleosomal arrays on DNA templates drastically reduces the capacity of these templates to support basal transcription (25, 35, 36, 57). Furthermore, the differential accessibility and transcriptional potential of chromatin structure in active versus inactive promoters are often associated with differential nucleosomal organization (3, 5, 23, 30, 46, 62). Thus, remodeling the nucleosomal architecture of a promoter is likely to be an integral feature of mechanisms of gene activation and/or silencing, which may involve histone acetylation and chromatin-remodeling complexes such as SWI/SNF (15, 20, 60).
The organization of genomic DNA into nucleosomal arrays is defined by both the translational position of the nucleosome relative to the linear nucleotide sequence and the rotational orientation of the DNA helix relative to the surface of the histone octamer. The translational position of nucleosomes on a DNA template (i.e., the linear position of the nucleosome relative to the DNA sequence [46]) has been shown to affect the accessibility of cis-acting elements in the promoter to various transcription factors as well as the basal transcription complex. Whether a transcription factor binding site is incorporated into a nucleosomal core or is in the linker region between nucleosomes can dramatically affect its accessibility to its cognate binding protein(s) in vitro (25, 58). However, the extent to which incorporating a transcription factor binding site into a nucleosomal core reduces the accessibility of that site can vary widely among transcription factors (4, 25). The rotational orientation of the DNA helix wound around a nucleosomal core also affects the accessibility of that DNA to transcription factors (25). For instance, the rotational orientation of the TATA box, the glucocorticoid response element, and the thyroid response element within a nucleosome strongly affects the accessibility of these elements to their cognate binding factors (14, 24, 59). These findings suggest that both the translational position and rotational orientation of cis-acting regulatory elements relative to those of the histone octamer can significantly affect their accessibility to regulatory factors and therefore their function in the context of nucleosomal arrays.
In addition to their repressive effects on transcription, nucleosomal positioning and orientation appear, in some cases, to modulate transcriptional activation. In particular, translational positioning of nucleosomes can mediate the cooperative binding of transcription factors such as Gal4 (48, 54) and pho4 (53), allowing rapid transcriptional induction. Furthermore, the nucleosomal organization of chromatin may establish a sequential hierarchy of transcription factor binding, with those factors that can bind to their cognate sites in nucleosomes binding first, leading to remodeling of adjacent nucleosomes so other factors can subsequently bind and activate transcription (21, 35, 48). In addition, precisely positioned nucleosomes can juxtapose linearly distant transcription factor binding sites, allowing the factors bound to these sites to interact (29, 45, 46). Alternatively, it has been suggested that the curvature of DNA along the surface of the histone octamer may reduce steric hindrance, which would otherwise occur between adjacent factors bound to closely spaced binding sites on linear DNA (30, 52). Together, these studies strongly suggest that examination of the nucleosomal organization of endogenous promoters is vital to the understanding of their regulation.
Here, we examine the nucleosomal structure of the promoter of the X-linked human hypoxanthine phosphoribosyltransferase gene (HPRT) on the active and inactive X chromosomes. The HPRT gene is subject to X chromosome inactivation, a process that leads to the transcriptional silencing of genes on one of the two X chromosomes in each female somatic cell (9). This results in the presence of both a transcriptionally active and a transcriptionally inactive HPRT allele within each female nucleus. The HPRT promoter lies within a CpG island, lacks a TATA box, and contains a potential AP-2 binding site, a cluster of five GC boxes, a potential initiator element, and a region of multiple transcription initiation sites (13, 16, 32, 42).
Several epigenetic characteristics distinguish the active and inactive HPRT alleles, including differential DNA methylation, general DNase I sensitivity, and DNase I hypersensitivity. On the active allele, the promoter region in vivo is unmethylated (11), is relatively sensitive to DNase I (26), and contains a DNase I-hypersensitive site that maps to the 5′ flanking region (13, 26, 56). Multiple transcription factor binding sites, including the potential AP-2 binding site, all five GC boxes, and the potential initiator element, have been identified in the active promoter by dimethyl sulfate (DMS) in vivo footprinting (13). In contrast, the inactive promoter is densely methylated (11), is resistant to DNase I, and does not exhibit DNase I hypersensitivity (26) or detectable transcription factor binding in vivo (13).
To examine the nucleosomal organization of the HPRT promoter, we investigated both the translational and rotational positioning of nucleosomes in the active and inactive HPRT promoter regions in permeabilized cells. Micrococcal nuclease (MNase) analysis showed that the active promoter was assembled into an ordered array of translationally positioned nucleosomes which was interrupted over a 350-bp region that showed increased accessibility to MNase and DNase I and that contained the entire functional promoter. In contrast, the inactive promoter was relatively inaccessible to both nucleases and transcription factors and was not assembled into a translationally positioned nucleosomal array. However, high-resolution DNase I cleavage analysis revealed rotational positioning of nucleosomes over a 210-bp region on the inactive promoter coincident with the 350-bp nuclease-accessible region on the active promoter. This differential organization of nucleosomes between the active and inactive HPRT promoters suggests that the transcriptional silencing of the HPRT promoter by X inactivation involves reconfiguration of the translationally positioned nucleosomal array in the active promoter to rotationally, but not translationally positioned nucleosomal arrays on the inactive promoter. Furthermore, it suggests that this rotational positioning of nucleosomes may be involved in the transcriptional repression of the HPRT promoter.
MATERIALS AND METHODS
Cell lines.
Two cell lines, 8121 and 4.12, described previously (13), were used for analysis of the active and inactive human HPRT promoter regions. 8121 is a human-hamster hybrid containing an inactive human X chromosome, whereas 4.12 is a human-hamster hybrid containing an active human X chromosome. 4.12 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, and 1× hypoxanthine-aminopterin-thymidine (Gibco/BRL) supplement. 8121 cells were maintained in DMEM supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, and 1× 6-thioguanine (2-amino-6-mercaptopurine; final concentration, 5 μg/ml; Sigma). Cells were maintained in culture at 37°C in 5% CO2.
MNase treatment of permeabilized cells
MNase treatment of permeabilized cells was based on a protocol for DNase I treatment of permeabilized cells modified from that described by Rincon Limas et al. (41). Cells were grown to confluence in T-150 culture flasks, trypsinized, combined in a 50-ml conical tube, and gently pelleted at ∼500 × g in a tabletop centrifuge. They were then washed gently once in solution A (150 mM sucrose, 80 mM KCl, 35 mM HEPES [pH 7.4], 5 mM K2HPO4, 5 mM MgCl2, 0.5 mM CaCl2) and once in solution B (150 mM sucrose, 80 mM KCl, 35 mM HEPES [pH 7.4], 5 mM K2HPO4, 5 mM MgCl2, 2 mM CaCl2) and were then resuspended at 500 μl per T-150 flask in solution B. For each MNase treatment, 500 μl of cells was combined with 500 μl of solution B plus 0.4% NP-40 (Sigma) and the desired amount (0 to 100 U [see Fig. 2B] or 0 to 200 U [see Fig. 2A]) of MNase (resuspended at 20 U/μl; Sigma), gently mixed, and then incubated at room temperature for 2 min. The digestion was stopped by adding 4 ml of DNA lysis buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 25 mM EDTA [pH 8.0], 0.5% sodium dodecyl sulfate [SDS], 300 μg of proteinase K/ml), and the lysate was then incubated overnight at room temperature. MNase-treated DNA was then isolated by standard phenol extraction and ethanol precipitation techniques (43), resuspended at 1 μg/μl in TE (10 mM Tris, 1 mM EDTA [pH 8.0]), and stored at 4°C.
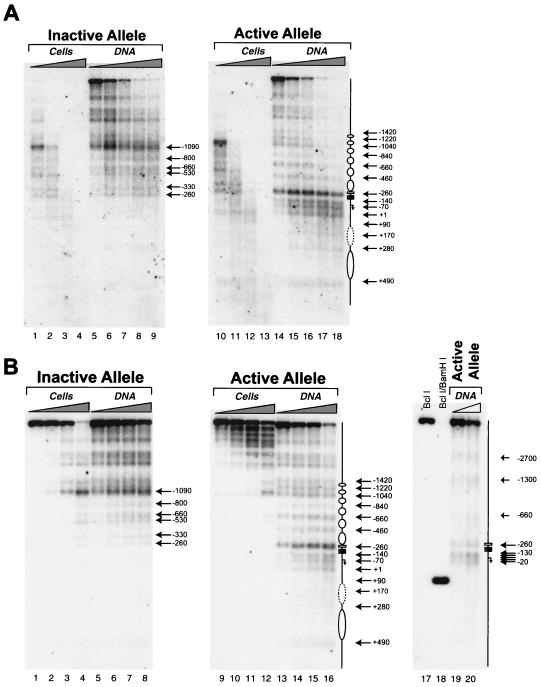
FIG. 2.
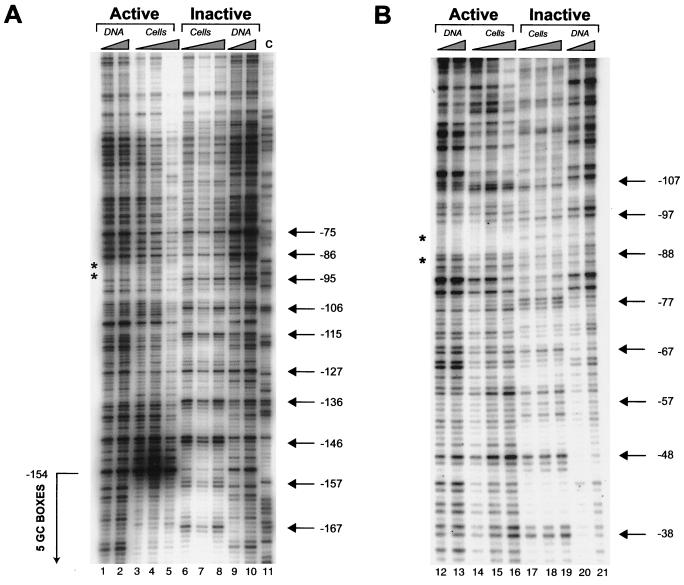
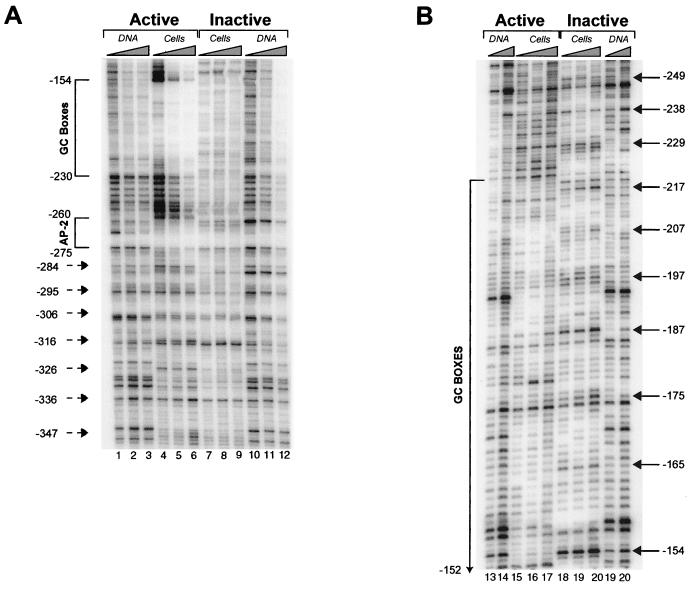
Mapping of MNase cleavage sites and DNase I-hypersensitive sites on the active and inactive HPRT promoters in vivo. (A) Mapping of nucleosome positions on the inactive and active HPRT promoters by MNase digestion. All lanes are from the same gel and autoradiogram and are aligned accordingly. All designations and symbols are described below. (B) Mapping of DNase I-hypersensitive sites relative to MNase cleavage sites. All lanes are from the same gel and autoradiogram and are aligned accordingly. Lanes 1 to 16, MNase-treated samples; lanes 17 to 20, DNase I-treated samples. MNase-treated samples in panel B were digested with lower concentrations of MNase than were samples in panel A. Band BclI (lane 17), position of the full-length genomic BclI fragment containing the HPRT promoter in purified genomic DNA; band BclI/BamHI (lane 18), relative position of a BamHI site 100 bp downstream of the translation initiation site within the full-length BclI genomic DNA fragment (Fig. 1). Lanes 19 and 20 (active allele), relative positions of DNase I-hypersensitive sites on the active HPRT allele; the diagram to the right indicates the positions of the DNase I-hypersensitive sites relative to those of the transcription factor binding sites (small boxes) and the major transcription initiation sites of the HPRT promoter (bent arrow) and the direction of transcription (Fig. 1). All bands in the Southern blots (A and B) were visualized by indirect labeling with a radiolabeled 400-bp probe located just upstream of a reference BclI site 838 bp downstream of the translation initiation site of the HPRT gene (Fig. 1). The diagrams to the right of lanes from the active allele (A, lanes 10 to 18, and B, lanes 9 to 16) indicate the translational positions of nucleosomes (ovals) on the active HPRT promoter relative to transcription factor binding sites (small boxes) as determined by the in vivo MNase cleavage pattern. The dashed oval indicates that the first downstream nucleosome may be modified, shifted, or absent in a subpopulation of cells since an “intranucleosomal” MNase cleavage occurs at position +170 (see text). Numbers to the right of the autoradiogram indicate the positions of MNase or DNase I cleavage sites relative to the translation initiation site. Active allele, samples from 4.12 cells containing an active HPRT gene on the active human X chromosome; inactive allele, samples from 8121 cells containing an inactive HPRT gene on the inactive human X chromosome; cells, DNA from permeabilized cells; DNA, naked DNA treated with MNase. All position numbers are relative the translation initiation site of the HPRT gene.
DNase I treatment of permeabilized cells for mapping DNase I-hypersensitive sites.
Cells were grown to confluence in a T-75 culture flask and then washed with phosphate-buffered saline and trypsinized. They were pelleted by centrifugation at 500 × g in a tabletop centrifuge and then washed once with solution A and resuspended in 500 μl of solution B. Then 500 μl of solution B containing 0.4% NP-40 and 20 or 40 μg of DNase I (1 mg/ml; Worthington) was added to the cell suspension, and the suspension was mixed and incubated for 2 min at 37°C. The digestion was stopped with 4 ml of lysis buffer (see above), and the suspension was incubated overnight at room temperature. DNA was subsequently isolated by standard techniques (43), resuspended at 1 μg/μl in TE, and stored at 4°C.
MNase treatment of naked DNA.
Ten micrograms of purified genomic DNA was digested to completion with BclI for each MNase concentration point. The BclI-digested DNA was then extracted with phenol-chloroform, precipitated with ethanol, and resuspended in 100 μl of Ex50 buffer (10 mM HEPES [pH 7.6], 60 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA [pH 8.0], 10% glycerol, 10 mM glycerol phosphate) (1) plus 5 mM CaCl2. Then 0.125 to 1 U (see Fig. 2B) or 1 to 8 U (see Fig. 2A) of MNase was added, and the reaction mixture was allowed to incubate for 2 min at room temperature. The reaction was stopped by adding 50 μl of stop solution (2.5% Sarkosyl, 100 mM EDTA [pH 8.0]). The treated DNA was extracted with phenol-chloroform, precipitated with ethanol, resuspended at 1 μg/μl in TE, and stored at 4°C.
Southern transfer, probe preparation, and hybridization.
MNase-treated DNA samples from permeabilized cells and naked DNA were digested to completion with BclI, size-fractionated on a 1.4% agarose gel at 60 V for 12 h, stained with ethidium bromide, and transferred to Hybond N+ by capillary transfer. Indirect end labeling was performed using a 400-bp hybridization probe located just upstream of a BclI restriction site in intron I of HPRT (see Fig. 1). This probe was amplified by PCR from a plasmid containing a BamHI/PstI restriction fragment from intron 1 that includes a previously described single-copy region (26) using the following primer set: MnaseBcl400 (GTTTGGGGTGCGATGGTGAGG) and MnaseBcldownstream (CAGAACGGTTGAGGAGGGAGGCCA). The PCR product was gel purified using the Qiagen gel extraction kit and radiolabeled by random priming. Hybridization was performed as described by Hornstra and Yang (12) at 65°C overnight in 4 ml of hybridization solution (0.25 M Na2HPO4 [pH 7.2] with o-phosphoric acid, 7% SDS, 1% bovine serum albumin [fraction V], 1 mM EDTA [pH 8.0]). The blot was then washed at 65°C in wash solution (20 mM Na2HPO4 [pH 7.2; with o-phosphoric acid], 1% SDS, 1 mM EDTA) and exposed to Kodak MR film for 4 to 5 days with intensifying screens at −80°C.
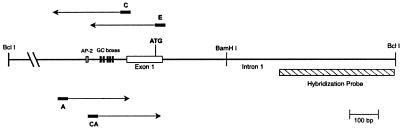
FIG. 1.
Locations of probes and primers for analysis of the HPRT promoter region. Horizontal line bounded by BclI sites, 4.3-kb BclI fragment containing the HPRT promoter; gray box, potential AP-2 site; five black boxes, cluster of GC boxes in the HPRT promoter; white box, first exon of the HPRT gene including the region of multiple transcription initiation sites in the promoter; ATG, translation initiation site; BamHI, position of a reference BamHI site in the first intron 100 bp downstream of the translation initiation site; hatched box, position of the 400-bp hybridization probe used to map DNase I and MNase cleavage sites in the HPRT promoter by indirect end labeling; black rectangles above and below the line, positions of the LMPCR primer sets used to map the high-resolution DNase I cleavage pattern of the HPRT minimal promoter; arrows extending from the black boxes, strand and region analyzed with each primer set.
Reconstitution of nucleosomes onto the HPRT promoter in vitro.
The DNA template used for in vitro reconstitution of chromatin was the pBS HPRT 1.8-kb plasmid, which is a pBluescript-derived plasmid containing a 1.8-kb EcoRI-to-BamHI fragment that includes the entire HPRT promoter. DNA methylation at CpG dinucleotides in vitro was performed using HpaII, HhaI, and SssI methylases (New England Biolabs) essentially as described by the supplier. The methylated DNA was then extracted with phenol-chloroform, precipitated, and resuspended at 100 ng/μl in TE. Completeness of methylation was assayed by digestion of in vitro-methylated templates with HpaII and HhaI. Chromatin reconstitution was performed with a Drosophila melanogaster chromatin assembly extract essentially as described by Becker et al. (1). The reconstituted chromatin (total volume = 70 μl) was mixed with 100 μl of a preassembled MNase mixture (94 μl of Ex50 buffer, 5 μl of 100 mM CaCl2, 1 μl of MNase [50 U/μl; Sigma]) and incubated at room temperature. At 2 and 5 min, 40 μl of this digestion mixture was removed and mixed with 20 μl of stop solution (2.5% Sarkosyl, 100 mM EDTA [pH 8.0]) to stop the reaction. To remove RNA and proteins, 1 μl of RNase cocktail (Ambion), 8 μl of 2% SDS, and 5 μl of proteinase K (resuspended at 10 mg/ml; Gibco/BRL) were added at each time point and the mixtures were incubated at 37°C overnight. The digested DNA (approximately 150 ng per time point) was extracted with phenol-chloroform, precipitated, and resuspended in 10 μl of TE (pH 8.0). The purified DNA at each MNase digestion time point was digested to completion with BamHI. The digested DNA was size-fractionated on a 1.6% agarose gel at 100 V for 4 h and then transferred to Hybond N+ by capillary transfer. The blot was hybridized overnight at 40°C with radiolabeled oligonucleotide BamHINuc1Probe (5′-GCCCTGAGGCGCGGGATC-3′), which corresponds to the sequence immediately upstream of the BamHI site in the first intron of the HPRT promoter, and visualized by exposure to Kodak AR film at room temperature for 1 h.
DNase I treatment of permeabilized cells for high-resolution DNase I cleavage analysis.
Cells were grown to 80% confluence in T-75 culture flasks. They were then washed once with phosphate-buffered saline, once with solution A, and once with solution B. One milliliter of solution B containing 0.2% NP-40 and 0 to 100 μg of DNase I (Worthington) was gently distributed over the monolayer of cells in each T-75 flask, and the cells were incubated at 37°C for 2 min. The DNase I digestion was then stopped by adding 4 ml of lysis buffer (see above), and the mixture was incubated overnight at room temperature. DNA was then isolated by standard phenol extraction and ethanol precipitation techniques (43), resuspended at 1 μg/μl in TE, and stored at 4°C.
DNase I treatment of naked DNA.
Fifty micrograms of genomic DNA was first digested to completion with EcoRI for each DNase I concentration point. The EcoRI-digested DNA was then extracted with phenol-chloroform, precipitated with ethanol, and then resuspended in a solution containing 100 μl of distilled water and 200 μl of solution B. DNase I (1 mg/ml; Worthington) was added to a final concentration of 0.0125 to 0.05 μg/ml, and the DNA was digested for 2 min at room temperature. The reaction was then stopped by adding 12 μl of 0.5 M EDTA (pH 8.0) and 3 μl of 20% SDS. The DNA was extracted with phenol-chloroform, precipitated with ethanol, resuspended in TE, and stored at 4°C.
LMPCR.
Amplification of DNase I-treated DNA was accomplished by ligation-mediated PCR (LMPCR) essentially as described by Hornstra and Yang (12), with an extension product capture modification described by Tormanen et al. (49) to reduce background. Primer extension was performed using Vent DNA polymerase (New England Biolabs), and PCR amplification was performed using Taq DNA polymerase (Gibco/BRL). The E1/E2 LMPCR primer set (13) was used to examine the upper strand in the minimal promoter, whereas the CA1/CA2 primer set (CCTAGTGAGCCTGCAAACTG/AAACTGGTAGGCGCCGGCGTAGG) was used to examine the lower strand. Additional upper- and lower-strand LMPCR primer sets C1/C2 and A1/A2, respectively (13), were used to analyze the region immediately upstream of the minimal promoter. Primer extension was performed essentially as described by Hornstra and Yang (12) using the same annealing temperature (45°C) and ramping parameters for all primer sets. For the PCR amplification, an annealing temperature of 64°C was used for all primers sets. Visualization of the footprint was achieved by size-fractionation of the PCR products on a DNA sequencing gel, followed by electrotransfer, hybridization, and autoradiography, essentially as described by Hornstra and Yang (12) using a radiolabeled single-strand-specific probe synthesized from single-stranded M13 templates containing the upper or lower strand of the HPRT promoter, as appropriate.
RESULTS
To determine the nucleosomal organization of the HPRT promoter region on the active and inactive X chromosomes and its relationship to transcription factor binding sites and nuclease hypersensitivity, the translational and rotational positioning of nucleosomes and the pattern of transcription factor binding in the HPRT promoter were examined. To examine translational positioning in vivo, NP-40-permeabilized cells were treated with MNase and then the MNase cleavage sites in the active and inactive HPRT promoters were mapped by Southern blotting and indirect end labeling. To identify regions of rotational nucleosomal positioning in the active and inactive promoters in vivo, NP-40-permeabilized cells were treated with DNase I and the high-resolution DNase I cleavage pattern of the promoter was determined by LMPCR. These high-resolution DNase I cleavage patterns were also used to delineate regions within the promoter that are occupied and footprinted in vivo by sequence-specific DNA-binding factors on the active and inactive HPRT alleles.
Translational positioning of nucleosomes in the HPRT promoter region in vivo.
To examine the translational positioning of nucleosomes in the human HPRT promoter region, 4.12 and 8121 cells (human-hamster hybrid cell lines containing either the active or the inactive human X chromosome, respectively) were permeabilized using detergent NP-40 and then treated with increasing concentrations of MNase. MNase preferentially cleaves DNA within the linker region between nucleosomal cores and, in conjunction with Southern blotting and indirect end labeling, can detect the presence and positions of translationally positioned nucleosomes within a given region of chromatin. The positions of the MNase cleavages within chromatin of the HPRT promoter region of permeabilized cells relative to a downstream BclI site in the first intron of the HPRT gene were mapped using a 400-bp hybridization probe located just upstream of the BclI site (Fig. 1). To determine the positions of DNase I-hypersensitive sites relative to MNase cleavage sites, NP-40-permeabilized cells containing the active HPRT allele were treated with increasing concentrations of DNase I and the DNase I-hypersensitive sites in chromatin of the HPRT promoter relative to the same BclI site were also mapped by indirect end labeling using the same hybridization probe.
Figure 2 shows the Southern blot analysis of the DNase I and MNase cleavage patterns on the inactive and active HPRT promoters in permeabilized 8121 and 4.12 cells, respectively. The MNase cleavage pattern of the inactive HPRT promoter in permeabilized 8121 cells (Fig. 2A, lanes 5 to 9, and B, lanes 5 to 8) was essentially identical to that of MNase-treated naked DNA (Fig. 2A, lanes 1 to 4, and B, lanes 1 to 4), although DNA from permeabilized 8121 cells exhibited a significantly wider distribution of sizes at any single MNase concentration. For naked DNA, Fig. 2A, lanes 1 to 4, shows the lower-molecular-weight bands, whereas Fig. 2B, lanes 1 to 4, shows the higher-molecular-weight bands. The similarity between the MNase cleavage pattern of the inactive allele in permeabilized 8121 cells and the cleavage pattern in naked DNA (albeit at much higher MNase concentrations for the permeabilized cells) indicates that the MNase cleavage pattern of the inactive allele is largely determined by the underlying DNA sequence rather than the nucleosomal organization. Because the inactive promoter region in permeabilized cells was more nuclease resistant (relative to the active promoter), it is likely that nucleosomes are present on the inactive promoter. However, the lack of an ∼200-bp periodicity in MNase cleavage and the similarity between the MNase cleavage pattern in permeabilized 8121 cells (Fig. 2A, lanes 5 to 9, and B, lanes 5 to 8) and that in naked DNA (Fig. 2A, lanes 1 to 4, and B, lanes 1 to 4) suggest that nucleosomes on the inactive promoter region are not translationally positioned. Instead, this cleavage pattern is consistent with random translational positioning of nucleosomes in the inactive promoter region, where MNase cleavage in linker regions of randomly positioned nucleosomes occurs uniformly throughout the region in a population of cells (each with a differently positioned nucleosomal array), yielding a pattern of cleavage dependent only on sites of preferential MNase cleavage in naked DNA.
In contrast, the active HPRT promoter in permeabilized 4.12 cells exhibited a pattern of strong MNase cleavages with a periodicity of approximately 200 bp, as represented by cleavages at positions −260, −460, −660, −840, −1040, −1220, and −1420 relative to the translation initiation site (Fig. 2A, lanes 14 to 18, and B, lanes 13 to 16). This ∼200-bp periodicity is consistent with the potential unit length of DNA in a mammalian nucleosome, suggesting that six distinct translationally positioned nucleosomes are positioned immediately upstream of the potential AP-2 site (positions −264 to −272) on the active HPRT promoter (Fig. 1 and schematic diagrams in Fig. 2A and B). Two additional translationally positioned nucleosomes are suggested by the presence of a second ∼200-bp cleavage periodicity consisting of three weak MNase cleavage sites at positions +90, +280, and +490 in the first intron of the HPRT gene (Fig. 2A, lanes 14 to 18, and B, lanes 13 to 16). While these downstream cleavage sites were relatively weak, they were consistently reproducible in multiple independent MNase assays. This pattern of periodic 200-bp cleavages observed in the chromatin of the active allele was not observed on MNase-treated naked DNA through a range of MNase concentrations that eventually achieve complete digestion of the parental 4.3-kb BclI band (Fig. 2A, lanes 10 to 13, and B, lanes 9 to 12). This difference between the MNase cleavage patterns of permeabilized 4.12 cells (Fig. 2A, lanes 14 to 18, and B, lanes 13 to 16) and naked DNA (Fig. 2A, lanes 10 to 13, and B, lanes 9 to 12) suggests that the periodic cleavages observed in permeabilized cells are due to nucleosomal organization rather than the underlying DNA sequence.
An additional weak MNase cleavage site was observed in permeabilized cells within the first downstream nucleosome at position +170 in the active promoter (Fig. 2A, lanes 14 to 18). Since the MNase cleavages at +90 and +280 suggest that a nucleosome is in fact positioned within this region, the cleavage site at +170 may simply indicate an intranucleosomal MNase cleavage site similar to those previously seen at high resolution in the pS2 promoter by Sewack and Hansen (46). Alternatively, the cleavage site at +170 could represent (i) a modified nucleosome that is inherently more susceptible to nuclease attack (e.g., the “split” nucleosomes observed by Lee and Garrard [22]), (ii) the linker region of an alternatively positioned downstream nucleosomal array in a subpopulation of cells, or (iii) the absence of a nucleosome between +90 and +280.
Between the upstream and downstream nucleosomal arrays on the active allele in permeabilized cells was an approximately 350-bp region that exhibited multiple MNase cleavages at nonnucleosomal intervals at positions −140, −70, and +1 (Fig. 2A, lanes 14 to 18, and B, lanes 13 to 16). MNase cleavage at position −260 was also greatly enhanced relative to that at all other sites on the active allele (in cells as well as in naked DNA). The presence of cleavages at positions −140, −70, and +1 suggests that nucleosomes are not translationally positioned over this region and, in conjunction with the strong MNase cleavage at position −260, indicates an increased accessibility to nucleases, consistent with the absence (or modification) of nucleosomes in this region. This highly nuclease-accessible 350-bp region from positions −260 to +90 contains the entire functional promoter including the known transcription factor binding sites as well as the multiple transcription initiation sites of the HPRT gene (Fig. 2, diagrams). The absence of MNase cleavage at positions −140, −70, and +1 in naked DNA from 4.12 cells (Fig. 2A, lanes 10 to 13, and B, lanes 9 to 12), even at higher MNase concentrations (Fig. 2A, lanes 10 to 13), suggests that these three cleavages in permeabilized cells are dictated by the chromatin structure of the active allele rather than the inherent susceptibility of the underlying DNA to MNase cleavage. Significant cleavage within this 350-bp region was also absent in permeabilized 8121 cells (Fig. 2A, lanes 5 to 9, and B, lanes 5 to 8) and naked DNA from 8121 cells (Fig. 2A, lanes 1 to 4, and B, lanes 1 to 4), further supporting the notion that DNA in this region is highly accessible in vivo on the active HPRT promoter.
DNase I hypersensitivity is characteristic of the HPRT promoter on the active X chromosome, whereas the promoter on the inactive X chromosome does not contain DNase I-hypersensitive sites (26, 61). To examine the basis of the hypersensitivity of the active promoter, we mapped DNase I-hypersensitive sites in the promoter relative to the translationally positioned nucleosomes. Figure 2B (lanes 19 and 20) shows two major and three minor DNase I-hypersensitive sites in chromatin of the active HPRT promoter. All three minor hypersensitive sites, centered at positions −660, −1300, and −2700, comapped to complete or partial Alu repeat elements upstream of the functional promoter. However, both of the major DNase I-hypersensitive sites, centered at positions −260 and −70, map to the MNase-accessible 350-bp region corresponding to the functional promoter on the active allele. The major DNase I-hypersensitive site centered at position −260 mapped between the AP-2 site and the cluster of CG boxes in the active HPRT promoter. The other major DNase I-hypersensitive site, which spanned the region from position −20 to −130, mapped immediately downstream of the CG boxes and encompasses the major transcription initiation sites as well as the potential initiator element (Fig. 1 and 2B, lanes 19 and 20). On the active promoter, the presence of both DNase I hypersensitivity and increased MNase accessibility within the 350-bp region suggests that this region is preferentially accessible to nucleases and transcription factors and may be devoid of nucleosomes. Furthermore, the locations of both the DNase I-hypersensitive sites (at positions −260 and −70) and the MNase cleavages (at positions −260, −140, and −70) between transcription factor binding sites suggest that transcription factor binding protects the underlying DNA from nuclease cleavage but may also distort the adjacent DNA, making it more accessible for nuclease cleavage (20). In contrast, both DNase I hypersensitivity (26) and MNase cleavage (Fig. 2A, lanes 5 to 9, and B, lanes 5 to 8) are markedly absent within this 350-bp region on the inactive HPRT allele, consistent with the nuclease-inaccessible and transcriptionally silent nature of the inactive promoter.
Overall, the data in Fig. 2 suggest that the promoter region of the transcriptionally active HPRT allele is assembled into an ordered array of translationally positioned nucleosomes interrupted by a highly nuclease-accessible 350-bp interval that contains the known functional elements of the HPRT promoter including the AP-2 site, the cluster of GC boxes, the potential initiator element, and the region of multiple transcription initiation sites. Furthermore, three CpG dinucleotides, at positions −97, −54, and −48, whose methylation appears to be critical for transcriptional repression of the HPRT promoter (6) all lie within this 350-bp region. In contrast, nucleosomes on the inactive HPRT promoter region do not appear to be translationally positioned, and both hypersensitivity to DNase I and accessibility to MNase within this 350-bp region are reduced or absent.
Effects of DNA methylation on the translational positioning of nucleosomes on the HPRT promoter in vitro.
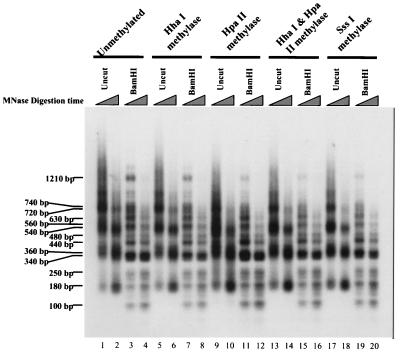
To examine the possibility that the differential methylation of the active versus inactive HPRT promoters (11) might mediate the differences in their nucleosomal organizations, a Drosophila chromatin assembly extract (1) was used to assemble nucleosomal arrays onto methylated and unmethylated supercoiled human HPRT promoter templates. Templates were methylated in vitro with HhaI, HpaII, SssI, or HhaI plus HpaII methylase prior to chromatin assembly. The reconstituted chromatin was then digested with MNase, and the positions of MNase cleavage sites relative to a BamHI site within the first intron of the HPRT gene were mapped by Southern blotting and indirect end labeling using a radiolabeled oligonucleotide probe (BamHINuc1Probe; see Materials and Methods) corresponding to the sequence immediately upstream of the BamHI site (Fig. 1). When the reconstituted chromatin was treated with MNase and not subsequently digested with BamHI (Fig. 3, lanes 1, 2, 5, 6, 9, 10, 13, 14, 17, and 18, labeled uncut), an approximately 180-bp ladder (i.e., 180, 360, 540 bp, etc.) of MNase-cleaved fragments was observed for all templates, indicating that all templates formed regularly spaced nucleosomal arrays. However, when these reconstituted MNase-treated templates were subsequently digested with BamHI to map the positions of the MNase cleavage sites relative to the intronic BamHI site (Fig. 3, lanes 3, 4, 7, 8, 11, 12, 15, 16, 19, and 20, labeled BamHI), this 180-bp ladder disappeared in all templates regardless of the methylation status of the template. This loss of the MNase-generated 180-bp ladder in samples digested with BamHI suggests that nucleosomes in the reconstituted arrays are not translationally positioned (i.e., phased) in the same manner on each template molecule. This is in direct contrast to the strong translational positioning of nucleosomes observed on the active HPRT promoter in vivo (i.e., in permeabilized cells; Fig. 2). Since the cleavage pattern generated by MNase treatment followed by BamHI digestion is unaffected by the methylation status of the template (Fig. 3, lanes labeled BamHI), DNA methylation does not appear to directly affect the translational positioning of nucleosomes on the HPRT promoter in vitro. This suggests that differential DNA methylation is unlikely to play a major role in the differential translational positioning of nucleosomes on the active versus inactive promoters in vivo. Furthermore, the absence of a detectable 180-bp ladder of fragments in in vitro-reconstituted samples that were MNase treated and BamHI digested suggests that the HPRT promoter region does not contain a strong nucleosome-positioning sequence. Interestingly, the positions of the MNase cleavages represented by the 100-, 180-, 250-, and 340-bp fragments seen in vitro (Fig. 3, lanes labeled BamHI) coincide with the major hypersensitive MNase cleavage sites (at positions +1, −70, −140, and −260) in the 350-bp nuclease-accessible region on the active HPRT promoter in permeabilized cells (Fig. 2A, lanes 14 to 18, and Fig. 2B, lanes 13 to 16).
FIG. 3.
Effects of DNA methylation on the translational positioning of nucleosomes on the human HPRT promoter in vitro. Nucleosomes were assembled in vitro onto methylated and unmethylated DNA templates containing the human HPRT promoter. HpaII methylase, HhaI methylase, and SssI methylase, DNA methyltransferases used to methylate each template; uncut, reconstituted chromatin that was digested with MNase but not BamHI; BamHI, reconstituted chromatin that was digested with MNase, purified, and then digested with BamHI. All samples were probed with BamHINuc1Probe, an 18-mer oligonucleotide immediately upstream of the BamHI site in the first intron of the HPRT gene. Triangles indicate increasing MNase digestion times used to cleave the reconstituted chromatin. Numbers to the left, approximate sizes of the bands.
DNase I in vivo footprinting of the human HPRT promoter.
To further examine the correlation between the locations of the nuclease cleavage sites and the positions of transcription factor binding sites in the active functional HPRT promoter in permeabilized cells, DNase I in vivo footprint analysis was performed, concentrating on the region containing the DNase I-hypersensitive sites. While the pattern of DMS in vivo footprints (i.e., the pattern of guanine contacts in DNA by sequence-specific DNA-binding factors) in the HPRT promoter has been previously determined (13), DNase I in vivo footprinting allowed examination of the extent of DNA-protein contacts at each transcription factor binding site and permitted detection of the binding of additional transcription factors that do not interact with DNA at guanine residues. In vivo DNase I footprint analysis was performed by treating permeabilized 4.12 and 8121 cells with increasing concentrations of DNase I, purifying the DNase I-treated genomic DNA, and examining the DNase I cleavage pattern in the region of interest by LMPCR. The strand and region covered by each of the LMPCR primer sets used in this analysis are shown in Fig. 1.
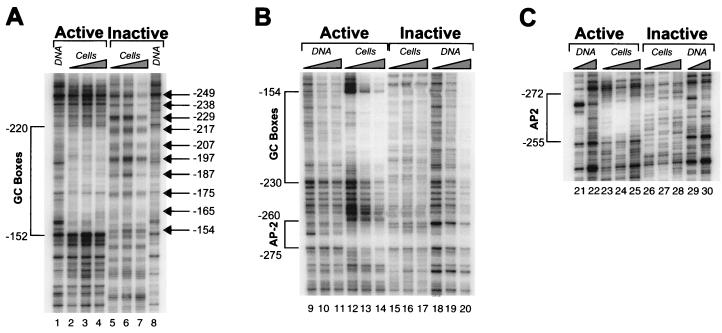
DNase I footprint analysis of the minimal promoter in permeabilized cells identified two major footprints on the active HPRT allele, which corresponded to the potential AP-2 binding site and the cluster of GC boxes (Fig. 4). The footprint over the GC boxes extended from position −152 to −220 on the upper strand (Fig. 4A, lanes 2 to 4) and from position −154 to −230 on the lower strand (Fig. 4B, lanes 12 to 14). The footprint over the AP-2 binding site spanned positions −255 to −272 on the upper strand (Fig. 4C, lanes 23 to 25) and −260 to −275 on the lower strand (Fig. 4B, lanes 12 to 14). This study complements previous DMS in vivo footprinting studies (13), which identified footprints over the AP-2 binding site and the cluster of GC boxes, by further defining the region of DNA occupied and protected by the bound proteins. It also confirms that DNase I hypersensitivity and increased MNase cleavage appear to occur between rather than within transcription factor binding sites in this region. DNase I in vivo footprinting identified no new footprints in the functional promoter that were not previously detected by DMS in vivo footprinting over the region analyzed (from positions −10 to −350).
FIG. 4.
DNase I in vivo footprint analysis of the human HPRT promoter. Active, samples from cells containing an active HPRT gene on the active human X chromosome; inactive, samples from cells containing an inactive HPRT gene on the inactive human X chromosome; DNA, naked DNA treated with DNase I; cells, DNA from permeabilized cells treated with DNase I; GC boxes, position of a DNase I in vivo footprint over the five GC boxes in the human HPRT promoter; AP-2, position of a DNase I in vivo footprint over a putative consensus AP-2 site in the human HPRT promoter. All position numbers (left and right) are relative to the translation initiation site of the HPRT gene. (A) DNase I in vivo footprint analysis of the upper strand of the HPRT promoter using LMPCR primer set E. Ladder of arrows, apparent 10-bp ladder of DNase I cleavages in permeabilized cells consistent with rotationally positioned nucleosomes on the inactive HPRT promoter. (B) DNase I in vivo footprinting analysis of the lower strand of the HPRT promoter using LMPCR primer set A. All designations and symbols are as described above. This analysis identifies footprints over both a cluster of five GC boxes and a putative AP-2 site in the active HPRT promoter. (C) DNase I in vivo footprinting analysis of the upper strand using LMPCR primer set C. All designations and symbols are as described above. This analysis identifies a DNase in vivo footprint over a putative AP-2 site on the active HPRT promoter.
However, DMS in vivo footprinting (13) also identified a footprint corresponding to a potential initiator element at positions −95 to −86 that overlaps the two major transcription initiation sites of the active promoter and that lies near a critical methylation site at −97 (6). DNase I in vivo footprinting of the 4.12 human-hamster hybrid containing the active X chromosome did not detect an equivalent footprint (Fig. 5, lanes 3 to 5 and 14 to 16), but an in vivo DNase footprint consisting of a DNase I-protected region from positions −106 to −83 was observed in the human male fibrosarcoma cell line, HT1080 (data not shown). This observation suggests that the interactions between the hamster trans-acting factor(s) and the human initiator element are readily detectable by DMS but not DNase I footprinting in 4.12 cells, while the interactions of the human factor(s) with the human sequence in HT1080 cells are possibly more stable and therefore more readily detectable by DNase I in vivo footprinting.
FIG. 5.
DNase I cleavage analysis of the HPRT promoter using LMPCR primer sets CA and E. All designations are the same as for Fig. 4. Asterisks, positions of the two major transcription initiation sites of the HPRT promoter determined by Kim et al. (16); arrows, 10-base ladders suggestive of rotational positioning of nucleosomes on the inactive HPRT allele. (A) DNase I cleavage analysis of the lower strand of the HPRT promoter using LMPCR primer set CA. (B) DNase I cleavage analysis of the upper strand of the HPRT promoter using LMPCR primer set E.
In contrast to what was found for the active allele, DNase I footprint analysis of the inactive promoter did not reveal any DNase I in vivo footprints in permeabilized 8121 cells (Fig. 4, lanes 5 to 7, 15 to 17, and 26 to 28). These results are consistent with previous DMS in vivo footprinting studies that indicate that no transcription factors are bound on the inactive allele (13). The absence of DNase I footprints on the inactive HPRT allele in permeabilized cells is also consistent with the presence of nucleosomes across the entire promoter region and the absence of nuclease-hypersensitive sites on the inactive promoter.
Rotational orientation of nucleosomes at the HPRT promoter.
The high-resolution DNase I cleavage pattern generated by DNase I footprinting can also identify the rotational positioning of nucleosomes in chromatin. DNase I binds within the minor groove of DNA and can cleave DNA wrapped around the histone octamer, but the accessibility of the minor groove to DNase I varies as a function of the helical path of the DNA along the surface of the histone octamer. DNase I is thought to preferentially cleave the DNA where the minor groove faces directly away from the histone octamer and therefore is maximally exposed to the solution. When a nucleosome is rotationally positioned within a population of cells, this differential accessibility of the DNA helix wrapped around the histone octamer results in a pattern of preferential DNase I cleavages at approximately 10-base intervals corresponding to the helical pitch of DNA. This cleavage pattern can be visualized as a ladder of bands with a periodicity and spacing of approximately 10 bases in a DNA sequencing gel (23, 31, 38, 46).
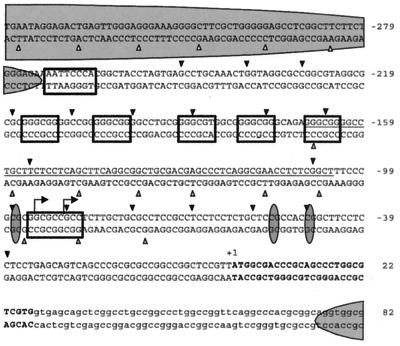
Such a 10-base periodicity was, in fact, observed for DNase I cleavage of the inactive HPRT promoter of permeabilized 8121 cells (Fig. 4A, 5, and 6). This periodic 10-base cleavage pattern was the result of both enhanced DNase I cleavage at 10-base intervals and suppression of cleavage between bands of the 10-base ladder. Figure 5A shows a strong periodic 10-base cleavage ladder which extends from position −75 to −167 on the lower strand of the inactive promoter (lanes 6 to 8) and which covers the region of multiple transcription initiation sites and the potential initiator element. A similar pattern of preferential cleavages at 10-base intervals also occurred on the upper strand, extending upstream, from positions −154 to −249, through the cluster of GC boxes on the inactive allele (Fig. 4A, lanes 5 to 7, and Fig. 6B, lanes 18 to 20), and downstream, from positions −107 to −38 (Fig. 5B, lanes 17 to 19), where the cleavages encompassed the positions of all three critical sites of methylation located at −97, −54, and −48 (6).
FIG. 6.
DNase I cleavage analysis of the HPRT minimal promoter using LMPCR primer sets A and C. All designations are the same as for Fig. 4 and 5. Dashed arrows, 10-base cleavage ladder suggestive of a rotationally positioned nucleosome on the active HPRT promoter. (A) DNase I cleavage analysis of the lower strand immediately upstream of the HPRT promoter using LMPCR primer set A. (B) DNase I cleavage analysis of the upper strand immediately upstream of the HPRT promoter using LMPCR primer set C.
Together, the patterns of 10-base periodic cleavages on the upper and lower strands of the HPRT promoter in permeabilized 8121 cells suggest rotational nucleosomal positioning over a region of at least 210 bp, starting from approximately 110 bp downstream of the GC boxes and extending to just downstream of the AP-2 site (Fig. 7); no other region analyzed in the inactive promoter exhibited this 10-base periodicity of DNase I cleavages on either strand. While there was some overlap between the periodic cleavages on the upper and lower strands, the 10-base cleavage ladder (and rotational positioning of nucleosomes) was evident on only one strand at a time. This pattern of strand-specific rotational positioning has also been described for the X-linked PGK-1 gene by Pfeifer and Riggs (38). The two- to four-base shift between the positions of preferential cleavage on the upper and lower strands reflects differences in the positions of maximal exposure of two the strands in the minor groove. The presence of rotationally positioned nucleosomes over the minimal promoter of the HPRT gene, and in particular the strong rotational positioning over the region of multiple transcription initiation sites identified by Patel et al. (32) as well as the two major transcription initiation sites identified by Kim et al. (16), suggest that rotational positioning of nucleosomes in the promoter may be involved in transcriptional repression.
FIG. 7.
Summary of the 10-base DNase I cleavage ladders of chromatin from the active and inactive HPRT promoters. Boldface letters, protein-coding region of the first exon; lowercase letters, nucleotides within the first intron; partial ovals, approximate positions of the translationally positioned nucleosomes on the active HPRT promoter as determined by MNase cleavage; open boxes, positions of transcription factor (TF) binding sites. From top to bottom, left to right, the TF binding sites are a putative AP-1 site (−271 to −264), five GC boxes (centered at −213, −201, −187, −177, and −166), and a putative initiator element (−94 to −86). Bent arrows, positions of the two major transcription initiation sites identified by Kim et al. (16); line between the nucleotide sequence of the upper and lower strands, region of multiple transcription initiation sites described by Patel et al. (32); black triangles above the sequence, positions of DNase I cleavage sites on the upper strand comprising the 10-bp ladder suggestive of rotationally positioned nucleosomes in the inactive promoter; gray triangles below the sequence, positions of DNase I cleavages on the lower strand comprising the 10-bp ladder suggestive of rotationally positioned nucleosomes in the inactive promoter; white triangles, positions of DNase I cleavages on the lower strand making up the 10-bp ladder, suggestive of rotational positioning of a nucleosome on the active promoter region in permeabilized cells; vertical ovals, positions of three CpG dinucleotides whose methylation is strongly correlated with transcriptional repression of the HPRT gene on the inactive allele (6).
In contrast, this same 210-bp region exhibited no apparent evidence of rotational positioning on the active promoter (Fig. 4A, lanes 2 to 4, Fig. 5, lanes 3 to 5 and 14 to 16, and Fig. 6B, lanes 15 to 17). In fact, in permeabilized 4.12 cells, with the exception of the DNase I footprints (Fig. 4), the DNase I cleavage pattern of this region of the active promoter was very similar to that of naked DNA (Fig. 4 to 6). This similarity to naked DNA, when coupled with the increased accessibility to MNase and the hypersensitivity to DNase I observed in the active promoter, strongly suggests that the functional promoter on the active X chromosome is devoid of nucleosomes.
However, evidence for rotational positioning on the active promoter was detected immediately upstream of the AP-2 site in the form of a series of 10-base periodic cleavages from positions −284 to −387 on the lower strand of the active promoter (Fig. 6A, lanes 4 to 6). This rotationally positioned region occurs within a translationally positioned nucleosome identified by MNase analysis of the active promoter (from positions −260 to −460), suggesting that this nucleosome is both translationally and rotationally positioned. However, a similar cleavage pattern was also observed in the naked DNA from 4.12 cells (Fig. 6A, lanes 1 to 3), making the interpretation of rotational positioning in this region somewhat uncertain. The pattern of DNase I cleavage sites indicative of rotational positioning on the inactive HPRT promoter is summarized in Fig. 7.
DISCUSSION
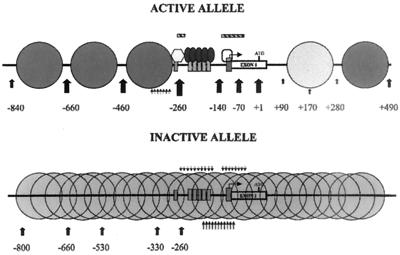
Analysis of both translational and rotational nucleosomal positioning at the HPRT promoter indicates that the nucleosomal organization of the promoter differs dramatically between the active and inactive X chromosomes. A schematic summary of these differences is shown in Fig. 8. The active promoter is assembled into an ordered array of six upstream and at least two downstream translationally positioned nucleosomes which flank a 350-bp nuclease-accessible region that includes the minimal promoter (42) and the region of multiple transcription initiation sites (16, 32) in the HPRT gene. Consistent with previous DMS in vivo footprinting studies (13), DNase I footprinting analysis of permeabilized 4.12 cells (i.e., the active promoter) shows that the potential AP-2 site and the cluster of GC boxes that reside in this region are occupied on the active promoter; no new footprints were identified by DNase I footprinting on either the active or inactive HPRT promoter. This 350-bp region on the active allele appears to be devoid of nucleosomes since it is hypersensitive to DNase I (Fig. 2B, lanes 19 and 20), is accessible to MNase cleavage (Fig. 2A, lanes 14 to 18, and B, lanes 13 to 16), and resembles naked DNA in its high-resolution DNase I cleavage pattern (Fig. 4 to 6).
FIG. 8.
Summary of the nucleosomal organization of the active and inactive HPRT promoter regions. Large dark gray circles on the active allele, positions of translationally positioned nucleosomes; light gray circle on the active allele, first nucleosome downstream of the promoter, which may be modified, shifted, or absent in a subpopulation of cells since an intranucleosomal MNase cleavage occurs at position +170 (see text); large gray overlapping circles on the inactive allele, nucleosomes with random translational positioning on the inactive promoter; hexagon and vertical ovals, bound transcription factors identified by DNase I and DMS in vivo footprinting (13) (vertical rectangles, their binding sites); bent arrow, position of the two major transcription initiation sites on the HPRT promoter; white box, first exon of the HPRT gene; ATG, position of the translation initiation site; thick vertical arrows, approximate positions and relative intensities of the major MNase cleavage sites in the HPRT promoter; clusters of thin triangular dashed arrows and barbed arrows, positions of the high-resolution DNase I cleavage ladders suggestive of rotationally positioned nucleosomes on the active and inactive HPRT promoters, respectively, in permeabilized cells (the slightly longer arrows on the lower strand in the inactive allele indicate that this ladder was unusually prominent); hatched bars, approximate locations of the DNase I-hypersensitive sites on the active HPRT promoter in permeabilized cells; All position numbers are relative to the translation initiation site.
In contrast, no translational positioning of nucleosomes on the inactive allele was observed by in vivo MNase analysis (Fig. 2A, lanes 5 to 9, and B, lanes 5 to 8). This absence of translational nucleosomal positioning on the inactive promoter is evident both in the lack of a 200-bp periodic MNase cleavage pattern in vivo (Fig. 2A, lanes 5 to 9, and B, lanes 5 to 8) and the similarity between the patterns of MNase cleavage of chromatin of the inactive allele (Fig. 2A, lanes 5 to 9, and B, lanes 5 to 8) and naked DNA (Fig. 2A, lanes 1 to 4, and B, lanes 1 to 4). Unlike the active allele, the inactive allele exhibits no DNase I hypersensitivity (26), and MNase accessibility is greatly reduced, as indicated by the significant reduction or absence of MNase cleavages at positions −260, −140, −70, and +1 (Fig. 2A, lanes 5 to 9, and B, lanes 5 to 8). Consistent with this finding, the AP-2 site and the cluster of GC boxes on the inactive promoter appear to be unoccupied by sequence-specific DNA binding proteins, as determined by both DNase I footprinting and previous DMS in vivo footprinting analysis (13). However, DNase I cleavage analysis of permeabilized cells did identify rotational positioning of nucleosomes over a region of at least 210 bp, from immediately downstream of the potential AP-2 binding site through the cluster of GC boxes and the region of multiple transcription initiations site to about 40 bp upstream of the translation start site (Fig. 7 and 8). This region of rotational positioning on the inactive promoter is particularly strong over the region of multiple transcription initiation sites and falls completely within the 350-bp nuclease-accessible region of the active allele. The location of this rotational positioning and its unique association with the inactive promoter argue that it may be involved in maintaining the transcriptional repression of the inactive HPRT allele.
This pattern of nucleosomal organization on the active and inactive HPRT alleles is unlike those of two other endogenous promoters and one enhancer that have been examined for both rotational and translational positioning in vivo, specifically, the mouse albumin enhancer, the human pS2 promoter, and the mung bean phaseolin promoter (23, 30, 46). These promoters and the enhancer exhibit translational positioning of nucleosomes on the active allele, but only the albumin enhancer and the HPRT promoter contain a region that appears to be free of nucleosomes. On the inactive allele, the pS2, phaseolin, and HPRT promoters all exhibit rotational positioning of nucleosomes while the albumin enhancer does not. However, the inactive pS2 and phaseolin promoters also exhibit translational positioning of the rotationally positioned nucleosomes, whereas the inactive HPRT promoter does not. These differences in the nucleosomal organization of various promoters and the enhancer suggest that different nucleosomal architectures can be utilized to facilitate both transcriptional activation and transcriptional silencing.
The inactive HPRT promoter appears to be the first promoter described to demonstrate rotational positioning of nucleosomes in the absence of translational positioning (either on the active or inactive allele), indicating that translational and rotational positioning of nucleosomes are not only conceptually distinct but also physically distinct and separable modes of nucleosomal organization.
Translational positioning of nucleosomes and the active HPRT promoter.
While a causal relationship between nucleosomal organization and transcription state cannot be established by these studies, it is likely that the different nucleosomal architectures of the active and inactive HPRT promoters play a significant role in promoter function. The strict translational positioning of nucleosomes over the HPRT promoter may act to preferentially expose specific transcription factor binding sites and the multiple transcription initiation sites to facilitate transcriptional activation. This hypothesis is consistent with several studies that indicate that the promoters and enhancers of expressed genes are assembled into translationally positioned nucleosomal arrays in which the nucleosomes over the transcription initiation site are absent or modified (2, 33, 46, 51). In contrast, the randomly positioned nucleosomes on the inactive promoter may serve to obstruct access to critical cis-acting elements by their cognate transcriptional activators and interfere with transcription initiation since transcriptional initiation (but not elongation) is strongly inhibited by nucleosomal assembly (28).
The mechanisms responsible for establishing and maintaining the translational positioning of nucleosomes in vivo are not well understood, but both nucleosomal positioning sequences and boundary proteins have been implicated (25). Unlike a number of promoters (which are predominantly inducible promoters) (29, 39, 40, 45, 46), the HPRT promoter region does not appear to harbor a nucleosome-positioning sequence. In vitro reconstitution of nucleosomes on the promoter region using a Drosophila chromatin assembly extract (1) does not generate a uniform translationally positioned nucleosomal array (Fig. 3). In addition, the inactive HPRT promoter region is not organized into translationally positioned nucleosomes in vivo (Fig. 2A, lanes 5 to 9, and B, lanes 5 to 8), in contrast to promoters that contain a translational positioning sequence and invariably show translational positioning of nucleosomes on both the active and inactive alleles (29, 39, 40, 45, 46).
The role of boundary proteins in the translational nucleosomal positioning of the active HPRT promoter is less clear. Numerous DNA-binding proteins, when bound to their cognate site, have been shown to translationally position nucleosomes both in vitro (8, 19, 34, 36, 37, 47, 55) and in vivo (2, 30, 47). The position of the potential AP-2 binding site (from positions −86 to −94) in the HPRT promoter immediately adjacent to the first upstream translationally positioned nucleosome in the active promoter (between positions −260 and −460; Fig. 8) suggests that the protein bound to the AP-2 site may act as a boundary element to maintain the positioning of the translationally positioned nucleosome array upstream of the AP-2 site. However, Litt et al. (27) have shown that, during the reactivation of the HPRT gene by 5aCdr, chromatin remodeling from a nuclease-resistant to a nuclease-accessible conformation occurs prior to the detectable binding of transcription factors to the promoter region (see below). Even if the protein bound to the AP-2 site does not mediate the initial remodeling and opening of chromatin during 5aCdr-induced reactivation, it may nevertheless be involved in positioning the first nucleosome of the upstream array and preventing nucleosomes from assembling on or sliding into the highly nuclease-accessible 350-bp region in the active allele.
Because differential DNA methylation of the active and inactive HPRT promoters is a prominent feature of the HPRT locus and because demethylation of the promoter by 5aCdr is closely associated with the remodeling of chromatin in the promoter region (27, 44), it was conceivable that DNA methylation could play a role in establishing and/or maintaining the differential nucleosomal architecture of the active and inactive HPRT promoter regions. However, our analysis of in vitro chromatin assembly on methylated and unmethylated HPRT promoter templates (Fig. 3) suggests that DNA methylation has little, if any, role in facilitating or preventing the translational positioning of nucleosomes.
Results of in vivo analysis of the chromatin structure of the X-linked PGK-1 promoter on the active and inactive X chromosomes appear to be somewhat inconsistent with our findings for the HPRT promoter. Pfeifer and Riggs (38) reported that the human PGK-1 promoter region on the inactive X chromosome is organized into nucleosomal arrays that are translationally positioned. This conclusion was based on the observation that DNase I generated 10-base cleavage ladders, similar to those found in the HPRT promoter region, on the inactive allele in permeabilized cells. While these ladders may be indicative of rotational positioning of nucleosomes, they do not necessarily indicate translational positioning. No MNase analysis or other direct examination of translational nucleosomal positioning in the PGK-1 promoter was performed, so it is conceivable that nucleosomes are rotationally positioned but not translationally positioned in the inactive PGK-1 promoter (as we find on the inactive HPRT promoter).
Rotational positioning of nucleosomes and the inactive HPRT promoter.
To date, the promoters of very few genes have been examined for rotational positioning of nucleosomes. Thus, the prevalence of rotational positioning and its significance for the regulation of transcription in vivo are neither well understood nor well documented. Of the four other genes examined, the inactive human PGK-1 (38), mung bean phaseolin gene (23), and human pS2 (46) promoters show rotationally positioned nucleosomes, while the inactive mouse albumin enhancer does not. However, rotational positioning is modified, but not abolished, in the active pS2 and phaseolin genes. Therefore, specific patterns of rotationally positioned nucleosomes are strongly associated with transcriptionally repressed promoters, though it is not entirely clear in each case if rotational positioning contributes to transcriptional silencing or is the result of silencing.
If rotational positioning of nucleosomes contributes to transcriptional repression, it might modulate the accessibility of transcription factors to their cognate sites in DNA since the binding of certain transcription factors appears to be sensitive to the rotational orientation of their binding sites relative to the histone octamer surface (14, 24, 59). Thus, rotational positioning of nucleosomes in the HPRT promoter may inhibit transcription factor binding to one or more of the GC boxes (particularly those at −212 and −198, which are located predominantly between periodic DNase I cleavage sites; Fig. 7) or to a potential transcriptional initiator element associated with the two major transcription initiation sites (13, 16). It is also possible that the rotational orientation of specific sequences inward toward the surface of the histone octamer hinders the binding of transiently bound factors (including chromatin-remodeling factors) that are not detectable by in vivo footprinting.
Alternatively, the rotational positioning of nucleosomes in the HPRT promoter may insure that a nucleosomal core, rather than a linker region, is preferentially associated with the transcription initiation sites of the promoter. For the HPRT promoter, the region that exhibits the most distinct 10-base cleavage periodicity (from positions −167 to −75) also coincides almost perfectly with the multiple sites of transcriptional initiation for the HPRT promoter reported by Patel et al. (32) and Kim et al. (16), extending from positions −89 to −168. If a nucleosomal core (rather than a linker region) is preferentially situated over the region of transcriptional initiation, it would likely inhibit transcription initiation (28).
The basis for rotational nucleosomal positioning is not well understood and has not been closely examined but may involve the underlying DNA sequence (23, 46, 50) and/or epigenetic modifications such as DNA methylation (7, 50). For the HPRT promoter, the underlying sequence alone is unlikely to influence rotational positioning since the active promoter does not exhibit rotational positioning. However, since the active and inactive HPRT promoters are differentially methylated in vivo (11), DNA methylation may be involved in allele-specific rotational nucleosomal positioning (though it does not appear to affect translational positioning; Fig. 3).
5-Azadeoxycytidine-induced chromatin remodeling of the HPRT promoter.
Litt et al. (27) have shown that, during the course of HPRT gene reactivation on the inactive X chromosome by 5aCdr, chromatin remodeling of the promoter region from a nuclease-resistant to a nuclease-accessible conformation occurs prior to both transcription factor binding and the appearance of mRNA. Presumably, the 5aCdr-induced remodeling of the promoter region consists of converting the nucleosomal organization on the inactive promoter, where nucleosomes are not translationally positioned across the promoter region but rather are rotationally positioned over key regulatory regions, to a transcriptionally competent structure that contains translationally positioned nucleosomal arrays flanking a nucleosome-free region. The findings of Litt et al. suggest that initiation of this remodeling of the nucleosomal architecture in response to 5aCdr treatment does not require the stable binding of sequence-specific transcription factors and that the remodeling may in fact facilitate the subsequent transcription factor binding by exposing transcription factor binding sites in the promoter. Thus, factors responsible for remodeling the nucleosomal architecture of the HPRT promoter region during 5aCdr-induced reactivation may bind only transiently and are subsequently replaced by stably bound sequence-specific DNA-binding transcription factors that initiate HPRT gene transcription. This scenario is supported by recent findings of Kontaraki et al. (17) that indicate that changes in chromatin structure in the lysozyme promoter region during development are initiated at developmental stages well before end stage sequence-specific transcriptional activators bind to the promoter and activate transcription of the gene. Conversely, one allele of the HPRT gene also must undergo the reverse remodeling of nucleosomal architecture in the promoter region, from the active nucleosomal organization to an inactive organization, during the process of X chromosome inactivation in female embryogenesis. Little is known about the mechanisms or factors that mediate these changes in nucleosomal organization in vivo, but repression of the HPRT promoter (and, presumably, maintenance of a repressive chromatin conformation) appears to involve methylation of specific critical CpG sites in the promoter region (6).
Overall, these data suggest that the nucleosomal organization of the promoter may play an important role in activation or repression of transcription at the HPRT locus. On the active allele, the translational positioning of nucleosomes preferentially exposes the functional promoter, potentially allowing efficient transcription, whereas, on the inactive allele, rotationally positioned nucleosomes may sterically hinder transcription factor binding and formation of the preinitiation complex. The role of DNA methylation and transcription factor binding in establishing and maintaining this differential nucleosomal organization of the HPRT promoter on the active and inactive X chromosomes is currently under investigation.
ACKNOWLEDGMENTS
We thank Jorg Bungert and Peter Becker for providing Drosophila chromatin assembly extracts and Jorg Bungert and Kelly Leach for helpful advice in the nucleosome reconstitution experiments.
This work was supported by NIH grant RO1 GM44286 to T.P.Y.
REFERENCES
- 1.Becker P B, Tsukiyama T, Wu C. Chromatin assembly extracts from Drosophila embryos. Methods Cell Biol. 1994;44:207–223. doi: 10.1016/s0091-679x(08)60915-2. [DOI] [PubMed] [Google Scholar]
- 2.Belikov S, Gelius B, Almouzni G, Wrange O. Hormone activation induces nucleosome positioning in vivo. EMBO J. 2000;19:1023–1033. doi: 10.1093/emboj/19.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benezra R, Cantor C R, Axel R. Nucleosomes are phased along the mouse beta-major globin gene in erythroid and nonerythroid cells. Cell. 1986;44:697–704. doi: 10.1016/0092-8674(86)90835-4. [DOI] [PubMed] [Google Scholar]
- 4.Blomquist P, Belikov S, Wrange O. Increased nuclear factor 1 binding to its nucleosomal site mediated by sequence-dependent DNA structure. Nucleic Acids Res. 1999;27:517–525. doi: 10.1093/nar/27.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckle R, Balmer M, Yenidunya A, Allan J. The promoter and enhancer of the inactive chicken beta-globin gene contains precisely positioned nucleosomes. Nucleic Acids Res. 1991;19:1219–1226. doi: 10.1093/nar/19.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C, Yang M C, Yang T P. Evidence that silencing of the HPRT promoter by DNA methylation is mediated by critical CpG sites. J Biol Chem. 2001;276:320–328. doi: 10.1074/jbc.M007096200. [DOI] [PubMed] [Google Scholar]
- 7.Davey C, Pennings S, Allan J. CpG methylation remodels chromatin structure in vitro. J Mol Biol. 1997;267:276–288. doi: 10.1006/jmbi.1997.0899. [DOI] [PubMed] [Google Scholar]
- 8.Fedor M J, Lue N F, Kornberg R D. Statistical positioning of nucleosomes by specific protein-binding to an upstream activating sequence in yeast. J Mol Biol. 1988;204:109–127. doi: 10.1016/0022-2836(88)90603-1. [DOI] [PubMed] [Google Scholar]
- 9.Gartler S M, Dyer K A, Goldman M A, editors. Mammalian X chromosome inactivation. Vol. 2. New York, N.Y: Academic Press; 1992. [DOI] [PubMed] [Google Scholar]
- 10.Gross D S, Garrard W T. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 11.Hornstra I K, Yang T P. High-resolution methylation analysis of the human hypoxanthine phosphoribosyltransferase gene 5′ region on the active and inactive X chromosomes: correlation with binding sites for transcription factors. Mol Cell Biol. 1994;14:1419–1430. doi: 10.1128/mcb.14.2.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hornstra I K, Yang T P. In vivo footprinting and genomic sequencing by ligation-mediated PCR. Anal Biochem. 1993;213:179–193. doi: 10.1006/abio.1993.1407. [DOI] [PubMed] [Google Scholar]
- 13.Hornstra I K, Yang T P. Multiple in vivo footprints are specific to the active allele of the X-linked human hypoxanthine phosphoribosyltransferase gene 5′ region: implications for X chromosome inactivation. Mol Cell Biol. 1992;12:5345–5354. doi: 10.1128/mcb.12.12.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imbalzano A N, Kwon H, Green M R, Kingston R E. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 15.Kadonaga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 16.Kim S H, Moores J C, David D, Respess J G, Jolly D J, Friedmann T. The organization of the human HPRT gene. Nucleic Acids Res. 1986;14:3103–3118. doi: 10.1093/nar/14.7.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kontaraki J, Chen H, Riggs A D, Bonifer C. Chromatin fine structure profiles for a developmentally regulated gene: reorganization of the lysozyme locus before trans-activator binding and gene expression. Genes Dev. 2000;14:2106–2122. [PMC free article] [PubMed] [Google Scholar]
- 18.Kornberg R D, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 19.Kornberg R D, Stryer L. Statistical distributions of nucleosomes: nonrandom locations by a stochastic mechanism. Nucleic Acids Res. 1988;16:6677–6690. doi: 10.1093/nar/16.14.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krebs J E, Peterson C L. Understanding “active” chromatin: a historical perspective of chromatin remodeling. Crit Rev Eukaryot Gene Expr. 2000;10:1–12. [PubMed] [Google Scholar]
- 21.Lee H, Archer T K. Nucleosome-mediated disruption of transcription factor-chromatin initiation complexes at the mouse mammary tumor virus long terminal repeat in vivo. Mol Cell Biol. 1994;14:32–41. doi: 10.1128/mcb.14.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee M S, Garrard W T. Transcription-induced nucleosome 'splitting': an underlying structure for DNase I sensitive chromatin. EMBO J. 1991;10:607–615. doi: 10.1002/j.1460-2075.1991.tb07988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li G, Chandler S P, Wolffe A P, Hall T. Architectural specificity in chromatin structure at the TATA box in vivo: nucleosome displacement upon β-phaseolin gene activation. Proc Natl Acad Sci USA. 1998;95:4772–4777. doi: 10.1073/pnas.95.8.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q, Wrange O. Accessibility of a glucocorticoid response element in a nucleosome depends on its rotational positioning. Mol Cell Biol. 1995;15:4375–4384. doi: 10.1128/mcb.15.8.4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Q, Wrange O, Eriksson P. The role of chromatin in transcriptional regulation. Int J Biochem Cell Biol. 1997;29:731–742. doi: 10.1016/s1357-2725(97)00016-2. [DOI] [PubMed] [Google Scholar]
- 26.Lin D, Chinault A C. Comparative study of DNase I sensitivity at the X-linked human HPRT locus. Somat Cell Mol Genet. 1988;14:261–272. doi: 10.1007/BF01534587. [DOI] [PubMed] [Google Scholar]
- 27.Litt M D, Hansen R S, Hornstra I K, Gartler S M, Yang T P. 5-Azadeoxycytidine-induced chromatin remodeling of the inactive X-linked HPRT gene promoter occurs prior to transcription factor binding and gene reactivation. J Biol Chem. 1997;272:14921–14926. doi: 10.1074/jbc.272.23.14921. [DOI] [PubMed] [Google Scholar]
- 28.Lorch Y, LaPointe J W, Kornberg R D. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987;49:203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- 29.Lu Q, Wallrath L L, Elgin S C. The role of a positioned nucleosome at the Drosophila melanogaster hsp26 promoter. EMBO J. 1995;14:4738–4746. doi: 10.1002/j.1460-2075.1995.tb00155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McPherson C E, Shim E Y, Friedman D S, Zaret K S. An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell. 1993;75:387–398. doi: 10.1016/0092-8674(93)80079-t. [DOI] [PubMed] [Google Scholar]
- 31.Noll M. Internal structure of the chromatin subunit. Nucleic Acids Res. 1974;1:1573–1578. doi: 10.1093/nar/1.11.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel P I, Framson P E, Caskey C T, Chinault A C. Fine structure of the human hypoxanthine phosphoribosyltransferase gene. Mol Cell Biol. 1986;6:393–403. doi: 10.1128/mcb.6.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel S A, Graunke D M, Pieper R O. Aberrant silencing of the CpG island-containing human O6-methylguanine DNA methyltransferase gene is associated with the loss of nucleosome-like positioning. Mol Cell Biol. 1997;17:5813–5822. doi: 10.1128/mcb.17.10.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pazin M J, Bhargava P, Geiduschek E P, Kadonaga J T. Nucleosome mobility and the maintenance of nucleosome positioning. Science. 1997;276:809–812. doi: 10.1126/science.276.5313.809. [DOI] [PubMed] [Google Scholar]
- 35.Pazin M J, Hermann J W, Kadonaga J T. Promoter structure and transcriptional activation with chromatin templates assembled in vitro. A single Gal4-VP16 dimer binds to chromatin or to DNA with comparable affinity. J Biol Chem. 1998;273:34653–34660. doi: 10.1074/jbc.273.51.34653. [DOI] [PubMed] [Google Scholar]
- 36.Pazin M J, Kamakaka R T, Kadonaga J T. ATP-dependent nucleosome reconfiguration and transcriptional activation from preassembled chromatin templates. Science. 1994;266:2007–2011. doi: 10.1126/science.7801129. [DOI] [PubMed] [Google Scholar]
- 37.Pazin M J, Sheridan P L, Cannon K, Cao Z, Keck J G, Kadonaga J T, Jones K A. NF-kappa B-mediated chromatin reconfiguration and transcriptional activation of the HIV-1 enhancer in vitro. Genes Dev. 1996;10:37–49. doi: 10.1101/gad.10.1.37. [DOI] [PubMed] [Google Scholar]
- 38.Pfeifer G P, Riggs A D. Chromatin differences between active and inactive X chromosomes revealed by genomic footprinting of permeabilized cells using DNase I and ligation-mediated PCR. Genes Dev. 1991;5:1102–1113. doi: 10.1101/gad.5.6.1102. [DOI] [PubMed] [Google Scholar]
- 39.Pina B, Barettino D, Truss M, Beato M. Structural features of a regulatory nucleosome. J Mol Biol. 1990;216:975–990. doi: 10.1016/S0022-2836(99)80015-1. [DOI] [PubMed] [Google Scholar]
- 40.Pina B, Bruggemeier U, Beato M. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell. 1990;60:719–731. doi: 10.1016/0092-8674(90)90087-u. [DOI] [PubMed] [Google Scholar]
- 41.Rincon Limas D E, Amaya Manzanares F, Nino Rosales M L, Yu Y, Yang T P, Patel P I. Ubiquitous and neuronal DNA-binding proteins interact with a negative regulatory element of the human hypoxanthine phosphoribosyltransferase gene. Mol Cell Biol. 1995;15:6561–6571. doi: 10.1128/mcb.15.12.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rincon Limas D E, Krueger D A, Patel P I. Functional characterization of the human hypoxanthine phosphoribosyltransferase gene promoter: evidence for a negative regulatory element. Mol Cell Biol. 1991;11:4157–4164. doi: 10.1128/mcb.11.8.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Sasaki T, Hansen R S, Gartler S M. Hemimethylation and hypersensitivity are early events in transcriptional reactivation of human inactive X-linked genes in a hamster × human somatic cell hybrid. Mol Cell Biol. 1992;12:3819–3826. doi: 10.1128/mcb.12.9.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schild C, Claret F X, Wahli W, Wolffe A P. A nucleosome-dependent static loop potentiates estrogen-regulated transcription from the Xenopus vitellogenin B1 promoter in vitro. EMBO J. 1993;12:423–433. doi: 10.1002/j.1460-2075.1993.tb05674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sewack G F, Hansen U. Nucleosome positioning and transcription-associated chromatin alterations on the human estrogen-responsive pS2 promoter. J Biol Chem. 1997;272:31118–31129. doi: 10.1074/jbc.272.49.31118. [DOI] [PubMed] [Google Scholar]
- 47.Shim E Y, Woodcock C, Zaret K S. Nucleosome positioning by the winged helix transcription factor HNF3. Genes Dev. 1998;12:5–10. doi: 10.1101/gad.12.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor I C, Workman J L, Schuetz T J, Kingston R E. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev. 1991;5:1285–1298. doi: 10.1101/gad.5.7.1285. [DOI] [PubMed] [Google Scholar]
- 49.Tormanen V T, Swiderski P M, Kaplan B E, Pfeifer G P, Riggs A D. Extension product capture improves genomic sequencing and DNase I footprinting by ligation-mediated PCR. Nucleic Acids Res. 1992;20:5487–5488. doi: 10.1093/nar/20.20.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Travers A, Drew H. DNA recognition and nucleosome organization. Biopolymers. 1997;44:423–433. doi: 10.1002/(SICI)1097-0282(1997)44:4<423::AID-BIP6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 51.Truss M, Bartsch J, Schelbert A, Hache R J, Beato M. Hormone induces binding of receptors and transcription factors to a rearranged nucleosome on the MMTV promoter in vivo. EMBO J. 1995;14:1737–1751. doi: 10.1002/j.1460-2075.1995.tb07163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Truss M, Candau R, Chavez S, Beato M. Transcriptional control by steroid hormones: the role of chromatin. Ciba Found Symp. 1995;191:7–17. doi: 10.1002/9780470514757.ch2. [DOI] [PubMed] [Google Scholar]
- 53.Venter U, Svaren J, Schmitz J, Schmid A, Horz W. A nucleosome precludes binding of the transcription factor Pho4 in vivo to a critical target site in the PHO5 promoter. EMBO J. 1994;13:4848–4855. doi: 10.1002/j.1460-2075.1994.tb06811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vettese-Dadey M, Walter P, Chen H, Juan L J, Workman J L. Role of the histone amino termini in facilitated binding of a transcription factor, GAL4-AH, to nucleosome cores. Mol Cell Biol. 1994;14:970–981. doi: 10.1128/mcb.14.2.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Widlak P, Gaynor R B, Garrard W T. In vitro chromatin assembly of the HIV-1 promoter. ATP-dependent polar repositioning of nucleosomes by Sp1 and NFkappaB. J Biol Chem. 1997;272:17654–17661. doi: 10.1074/jbc.272.28.17654. [DOI] [PubMed] [Google Scholar]
- 56.Wolf S F, Migeon B R. Clusters of CpG dinucleotides implicated by nuclease hypersensitivity as control elements of housekeeping genes. Nature. 1985;314:467–469. doi: 10.1038/314467a0. [DOI] [PubMed] [Google Scholar]
- 57.Wolffe A P. New insights into chromatin function in transcriptional control. FASEB J. 1992;6:3354–3361. doi: 10.1096/fasebj.6.15.1464369. [DOI] [PubMed] [Google Scholar]
- 58.Wolffe A P. Transcription: in tune with the histones. Cell. 1994;77:13–16. doi: 10.1016/0092-8674(94)90229-1. [DOI] [PubMed] [Google Scholar]
- 59.Wong J, Li Q, Levi B Z, Shi Y B, Wolffe A P. Structural and functional features of a specific nucleosome containing a recognition element for the thyroid hormone receptor. EMBO J. 1997;16:7130–7145. doi: 10.1093/emboj/16.23.7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 61.Yang T P, Caskey C T. Nuclease sensitivity of the mouse HPRT gene promoter region: differential sensitivity on the active and inactive X chromosomes. Mol Cell Biol. 1987;7:2994–2998. doi: 10.1128/mcb.7.8.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yaniv M, Cereghini S. Structure of transcriptionally active chromatin. CRC Crit Rev Biochem. 1986;21:1–26. doi: 10.3109/10409238609113607. [DOI] [PubMed] [Google Scholar]