Abstract
The hepatitis B virus X protein (HBx) plays essential roles in viral replication and the generation of hepatocellular carcinoma. In spite of a large number of suggestive cellular targets and functions, a clear picture of its mechanism(s) of action has remained elusive. In this report, we continue to characterize its recently described mitochondrial association and further examine its impact on mitochondrial functions. HBx was previously shown to bind to a voltage-dependent anion channel (VDAC3) and alter the mitochondrial transmembrane potential (ΔΨm). Here we show that, as a consequence of association with mitochondria, HBx constitutively induces activation of transcription factors, which include STAT-3 and NF-κB. This induction of activation was sensitive to the antioxidants N-acetyl l-cysteine and pyrrolidine dithiocarbamate, as well as to overexpression of Mn-superoxide dismutase. These results therefore implicate a potential role of reactive oxygen species (ROS) in a process that ultimately leads to the activation of STAT-3 and NF-κB. Evidence is also presented for the HBx-induced generation of ROS. The ability of HBx to induce the activation of STAT-3 and NF-κB was demonstrated by mobility shift and reporter gene expression assays with lysates from HBx-transfected HepG2 cells. A C-terminal HBx deletion mutant, HBxΔ99, failed to bind VDAC3 and activate STAT-3 and NF-κB. These studies shed new light on the physiological significance of HBx's mitochondrial association and its role in inducing oxidative stress which can contribute to the liver disease pathogenesis associated with the hepatitis B virus infection.
Human hepatitis B virus (HBV) is one of the causative agents of acute chronic hepatitis, cirrhosis, and hepatocellular carcinoma (2). Among the proteins encoded by the HBV genome, the X gene product (HBx) is essential for productive infection of the mammalian HBVs (9, 60) and has drawn considerable attention for its pleiotropic functions. HBx does not directly bind DNA but functions via protein-protein interaction (31). HBx has been shown to transactivate transcription through multiple cis-acting elements, including AP-1, AP-2, ATF/CREB, NF-κB, NF-AT, C/EBP, p53, the DNA-binding repair protein UV-DDB, and Egr-1 (3, 16, 22, 30, 31, 44, 50, 57). While HBx is predominantly localized to the cytoplasm, very little, if any, is probably distributed to the nucleus of transfected cells (15, 42, 58). Based on its staining from cytoplasmic localization, it has been shown to mediate the activation of signal transduction cascades, including the Ras/ mitogen-activated protein kinase-, c-jun N-terminal kinase-, NF-κB-, and Src-dependent pathways (4, 6, 11, 23). HBx-dependent activation of Src has been shown to affect viral replication (24). Stimulation of these signaling pathways can lead to the activation of AP-1- and NF-κB-dependent transcriptional activation (4, 11, 23). Among its other cytoplasmic targets are the components of proteasomes (21). Among components of basal transcriptional machinery, HBx is known to bind to TBP, TFIIB, and TFIIH, including the RPB5 subunit of RNA polymerase (10, 35, 36). HBx also stimulates transcription of promoters regulated by RNA polymerase I and III (1, 27, 51). The latter associations require the presence of HBx in the nucleus.
Using both cell biological fractionation procedures and laser confocal immunofluorescence microscopy, we showed that HBx directly and physically interacts with an outer mitochondrial voltage-dependent anion channel, VDAC3 (37). Mitochondrial targeting of HBx has been described by two other independent reports (20, 48). We further noted that this association led to a decrease in the mitochondrial membrane potential (37), which implicates a key role of HBx in affecting mitochondrial physiology, metabolism and other relevant functions. One of the responses to such stimuli is the generation of reactive oxygen species (ROS) or reactive oxygen intermediates. ROS play an important functional role, both as regulators of transcription factors and inhibitors of protein tyrosine phosphatases (17, 40). In this study, we demonstrate that HBx via its association with mitochondria induces oxidative stress, which in turn leads to activation of a series of transcription factors, including NF-κB and STAT-3. STAT-3 is a member of family of transcription factors that are activated upon tyrosine phosphorylation in response to extracellular signals such as cytokines and growth factors (14, 59). Activated STATs form dimers or multimers through their Src-homology domain II, transported into the nucleus, where they bind to the cognate DNA sequences and activate gene expression. Oxidative stress has been shown to trigger STAT-3 tyrosine phosphorylation and nuclear translocation (8), which correlates with the activation of STAT-3 leading to its DNA binding activity. We further show that HBx constitutively activates STAT-3. This activation requires that HBx is associated with mitochondria. These data collectively demonstrate that STAT-3 is an important component of signaling pathways that become activated by oxidative stress in mitochondria. Our characterizations also included the HBx-mediated activation of NF-κB, which is one of the most widely acknowledged inducer of oxidative stress. NF-κB was one of the first HBx-responsive elements that were identified (44, 49). However, the mechanism by which HBx stimulated transcription via NF-κB motif was not clearly understood. HBx did not directly interact with either subunit of NF-κB and HBx that was predominantly localized to the cytoplasm. The studies described here provide a novel insight into the mechanism of transcription regulation by HBx from its cytoplasmic location and implicate a potential functional role of oxidative stress in the HBV-associated liver disease pathogenesis, including hepatocellular carcinoma.
MATERIALS AND METHODS
Plasmids and oligonucleotides.
Construction of reporter plasmids containing wild-type Footprint V (pFPV3Luc) and its mutant (pFPV4′Luc) have been described previously (25). Plasmids p3x-κB-Luc (luciferase reporter driven by minimal fos promoter with three upstream NF-κB binding sites from major histocompatibility complex class I) and p3x-mut-Luc (in which the NF-κB binding site has been mutated) were generous gifts of J. Martin, University of Colorado, Boulder. The oligonucleotides STAT-3, NF-κB, NF-AT, CRE, AP-1, and NF-1 were purchased from Santa Cruz Biotechnology. The HBx-encoding gene was cloned in pCMV4 with a flag tag cassette at the C terminus of the gene to produce the plasmid pCXF (pCMV4 X-DNA flag). The HBx mutant HBxΔ99, which contains only N-terminal amino acid residues 1 to 99 of HBx, was cloned in pCMV with flag tag. The four conserved cysteine mutants (C7, C61, C69, and C137) of HBx (X1 to X4) individually replaced by threonine residues by site-directed mutagenesis were provided by V. Kumar (ICGEB, New Delhi, India [26]). Manganese-superoxide dismutase (Mn-SOD) expression vector was a gift of S. Flores (University of Colorado Health Sciences Center, Denver).
Preparation of nuclear extracts.
Nuclear lysates were prepared from HepG2 cells transfected with indicated plasmids by lipofectin (Gibco), followed by treatment with the antioxidants PDTC (100 μM) and NAC (30 mM) for 6 h before the cells were harvested at 42 h posttransfection. Mn-SOD expression vector was cotransfected, along with HBx expression plasmid pCXF. Cells were lysed in hypotonic buffer (20 mM HEPES [pH 7.9], 10 mM KCl, 0.1 mM Na3VO4, 1 mM EDTA, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 3 mg of aprotinin/ml, 1 mg of pepstatin/ml, 20 mM NaF, and 1 mM dithiothreitol [DTT] with 0.2% NP-40) on ice for 10 min. After centrifugation at 4°C (13,000 rpm) for 1 min, the nuclear pellet was resuspended in high-salt buffer (hypotonic buffer with 20% glycerol and 420 mM NaCl) at 4°C by rocking for 30 min after centrifugation. The supernatant was collected and stored in aliquots at −80°C.
In vitro binding assay.
The glutathione S-transferase (GST) and GST-HVDAC3 fusion proteins were purified as described previously (37). The in vitro synthesis of 35S-labeled HBx and HBxΔ99 was carried out by using the TNT-coupled transcription-translation system (Promega). The precleared proteins were allowed to bind GST and GST fusion proteins immobilized on glutathione-Sepharose beads in buffer B (150 mM KCl, 6 mM MgCl2, 25 mM HEPES [pH 7.9], 10% [vol/vol], glycerol, 0.1% NP-40, 1 mM ATP, 1 mM DTT, 1 mM PMSF, and 10 μg of leupeptin and 9 μg of aprotinin/ml) at 4°C for 2 h. The reaction mixture was washed several times with buffer B, and bound proteins were eluted and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
EMSA.
The oligonucleotides STAT-3, NF-κB, NF-AT, CRE, AP-1, and NF-1 were radiolabeled at their 5′ end with [γ-32P]ATP by T4 polynucleotide kinase. About 20,000 cpm of gel purified probe was incubated with lysates from HepG2 cells not transfected or transfected with HBx expression vector pCXF and a series of HBx mutants (HBxΔ99, X1 to X4) in electrophoretic mobility shift assay (EMSA) buffer [20 mM Tris-HCl (pH 7.9), 10 mM MgCl2, 50 mM KCl, 16.7 μg of poly(dI-dC)/ml, 1 mM EDTA, 1 mM DTT, and 1 μM leupeptin] for 20 min on ice. Competition analyses were carried out in the presence of a 100-fold excess of an unlabeled competitor, and the oligonucleotide was preincubated for 20 min on ice prior to the addition of radiolabeled probe. The DNA-protein complexes were resolved by 5% polyacrylamide gel electrophoresis in 0.5× Tris-borate-EDTA buffer. The gels were dried and subjected to autoradiography.
Immunoprecipitation and Western blot analysis.
Exponentially growing HepG2 cells transfected with the HBx expression vector pCXF and HBxΔ99 were harvested, and cell extracts were prepared by incubation in radioimmunoprecipitation (RIPA) buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM sodium orthovanadate, 1 mM sodium formate, 1 mM PMSF, and 10 μg of aprotinin and 10 μg of leupeptin/ml) for 30 min on ice. Immunoprecipitation was performed with anti-STAT-3 serum. After 4 h of incubation, the immune complexes were captured on protein A-Sepharose, washed three times with RIPA buffer, and boiled for 5 min in SDS-PAGE sample buffer. The samples were subjected to SDS-PAGE. Gels were electroblotted onto polyvinylidene difluoride (PVDF) membrane (Amersham) in 25 mM Tris, 192 mM glycine, and 20% methanol by electrophoresis. Membranes were treated for 1 h in blocking buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.3% polyvinylpyrrolidone, 0.5% Tween 20 [wt/vol]), probed with monoclonal antiphosphotyrosine antibody overnight, and washed twice for 10 min with blocking buffer, followed by incubation with secondary antibody for 45 min. After an additional washing step with blocking buffer, immunoblots were visualized by using the ECL detection system (Amersham).
Luciferase assay.
In transient transfections, HepG2 cells were plated at a density of ∼5 × 105 cells/60-mm dish and maintained in Dulbecco modified Eagle medium containing 10% fetal calf serum and penicillin (75 U/ml)-streptomycin (50 U/ml) at 37 C. Cells (∼50% confluent) were cotransfected with 1 to 2 μg of luciferase reporter plasmid (pFPV3Luc/NF-κB–Luc) with 0.2 μg of pCXF or expression plasmids containing HBx mutants by using Lipofectin reagent (Life Technologies). Mn-SOD expression plasmid was cotransfected along with pCXF. The antioxidants PDTC (100 μM) and NAC (30 mM) were added to the transfected cells for various incubation times before cells were harvested for luciferase activity.
Flow cytometric analysis of cellular ROS levels.
Intracellular ROS levels were measured by using an oxidative-sensitive fluorescent probe, dihydroethidium (DHE; Molecular Probes), as described previously with some modifications (13). Briefly, HepG2 cells (∼2.5 × 105) in a 100-mm plate were transfected with 6 μg of pCMV4, HBx, or HBxΔ99 expression vector by using Lipofectin (Gibco). About 48 h after transfection, the cells were incubated with 4 μM DHE for 45 min at 37C. Cells were harvested, washed with phosphate-buffered saline and analyzed by flow cytometry. The fluorescence from oxidized DHE was detected at a wavelength of 630 nm.
RESULTS
HBx induces tyrosine phosphorylation of STAT-3.
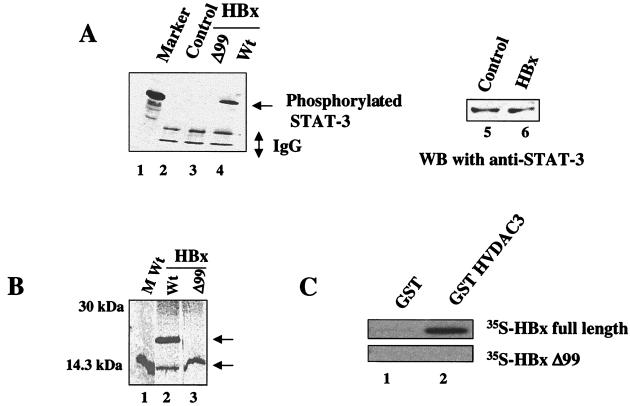
STAT-3 generally remains latent in the cytoplasm in an unphosphorylated state (38). Upon stimulation by cytokines, such as interleukin-6 and epidermal growth factor, STAT-3 is phosphorylated at tyrosine residues and translocated into the nucleus, where it binds cognate DNA sequences as dimers (15, 59). To initiate these studies, we sought to determine whether HBx-transfected HepG2 cells induced tyrosine phosphorylation of STAT-3. Nuclear lysates were immunoprecipitated with a polyclonal anti-STAT-3 serum, fractionated by SDS-PAGE, and electroblotted onto a PVDF membrane. The membrane was then incubated with antiphosphotyrosine monoclonal antibody and analyzed by ECL, a commercial detection kit. The results (Fig. 1A) show that HepG2 cells transfected with wild-type HBx contained phosphorylated STAT-3 protein (lane 4). Neither the HBxΔ99 mutant nor the untransfected HepG2 cells showed activation of STAT-3 protein (lanes 2 and 3). A Western blot analysis of the lysates was carried out to determine the STAT-3 levels in the HepG2 cell. The results (Fig. 1A, lanes 5 and 6) show that the overall STAT-3 level remains unaffected in the control and HBx-transfected cells. The deletion mutant HBxΔ99 contains the N-terminal 99 amino acids of the X gene and has a deletion of the remaining 53 amino acid residues. The expression of wild-type HBx and truncated HBxΔ99 protein was assessed by immunoprecipitation. The results clearly demonstrate that truncated HBxΔ99 protein is stably expressed (Fig. 1B, lane 3). The lower band in the wild-type HBx lane is the product of internal initiation of translation within the X gene which is normally seen (unpublished results). An in vitro 35S-labeled HBxΔ99 protein does not bind GST-VDAC (mitochondrial voltage-dependent anion channel), as shown in Fig. 1, and displayed a diffused pattern of subcellular distribution (K.-W. Huh and A. Siddiqui, unpublished data) in contrast to the punctate pattern observed previously with wild-type HBx (37) and consequently failed to activate STAT-3 (Fig. 1A). These observations clearly demonstrate that HBx is capable of constitutively activating STAT-3 in the absence of any cytokines. While HBx does not possess any kinase activity, the mechanism(s) by which it may influence or trigger activation of STAT-3 via its mitochondrial association remains to be investigated.
FIG. 1.
(A) HBx constitutively activates STAT-3 tyrosine phosphorylation. Activated STAT-3 protein in cell extracts transfected with wild-type HBx and HBxΔ99 expression vectors were immunoprecipitated with anti-STAT-3 polyclonal antibody, and the immunoprecipitates were captured on protein A-Sepharose, subjected to SDS-PAGE, and then immunoblotted with antiphosphotyrosine monoclonal antibody. The antibody binding was detected by using an ECL detection system. Lane 1, prestained low-molecular-mass standard (range, 20 to 113 kDa); lane 2, untransfected lysates; lane 3, lysates from cells transfected with HBxΔ99; lane 4, lysates from cells transfected with wild-type (Wt) HBx expression vector; lanes 5 and 6, Western blot analysis of HepG2 cell lysates (lane 5) and lysates transfected with wild-type HBx (lane 6) resolved by SDS-PAGE and probed with anti-STAT-3 antibody. (B) Expression of wild-type and truncated HBxΔ99 protein. HepG2 cells were transfected with HBx and HBxΔ99 expression vectors. Cells were rediolabeled with [35S]methionine, and lysates were prepared and immunoprecipitated with flag antibody. Lane 1, molecular weight (M Wt) standard; lanes 2 and 3, expressed HBx and HBxΔ99 proteins, respectively. (C) HBx interacts with VDAC3 in vitro. HBx and HBxΔ99 were translated in vitro with the TNT system (Promega) in the presence of [35S]methionine. Precleared 35S-labeled HBx and HBxΔ99 were allowed to bind with GST and GST-VDAC3 fusion protein. Bound proteins were washed with binding buffer (see Materials and Methods) eluted, resolved by SDS-PAGE, and exposed to X-ray film. Lane 1, binding of 35S-labeled HBx and HBxΔ99 with GST; lane 2, binding with GST-VDAC3.
HBx activates STAT-3 and NF-κB.
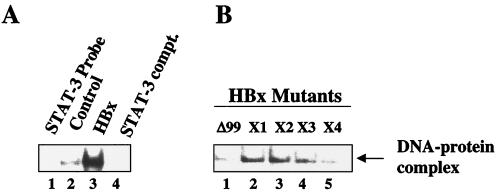
To examine whether HBx was able to induce the formation of DNA-STAT-3 protein complexes, EMSAs were carried out by using γ-32P-labeled STAT-3 cognate oligonucleotide as a probe in the presence of nuclear lysates from HepG2 cells transfected with vectors expressing wild-type HBx and a series of HBx mutants (HBxΔ99, X1 to X4). The HBx cysteine mutants (X1 to X4) have been described previously (26). They contain point mutations of cysteine residues (C7, C61, C69, and C137) within the HBx protein. The results (Fig. 2A) show that the STAT-3 binding activity was substantially enhanced by ca. six- to eightfold in the presence of lysates from HepG2 cells transfected with wild-type HBx (lane 3) compared to lysates from untransfected HepG2 cells (lane 2). The DNA-protein complex observed in lane 3 was specifically competed for by a 100-fold excess of unlabeled STAT-3 oligonucleotides (lane 4). Lysates prepared from various HBx mutants showed various degrees of binding with the STAT-3 probe. HBxΔ99- and the cysteine mutant X4 (C137)-transfected lysates were found to display reduced STAT-3 activation (Fig. 2B, lanes 1 and 5), whereas other cysteine mutants formed a STAT-3–DNA complex with slightly lower efficiency than that of the wild type (compare Fig. 2A, lane 3, with Fig. 2B, lanes 2 to 4).
FIG. 2.
HBx activates STAT-3 transcription factor via its mitochondrial association. (A) EMSA was carried out in the presence of γ-32P-labeled STAT-3 cognate nucleotide probe and the nuclear lysates prepared from cells transfected with wild-type HBx and a series of HBx mutants. Lane 1, probe alone; lanes 2 and 3, equal amounts of untransfected and HBx-transfected nuclear lysates; lane 4, a 100-fold excess of STAT-3 cognate oligonucleotide as the competitor. (B) Equal amounts of a series of HBx mutant-tranfected nuclear lysates were incubated with γ-32P-labeled STAT-3 cognate oligonucleotide probe. Lane 1, probe incubated with lysates from HBxΔ99-transfected cells; lanes 2, 3, 4, and 5, STAT-3 probe incubated with HBx cysteine mutants designated X1(C6), X2 (C61), X3 (C69), and X4 (C137), respectively.
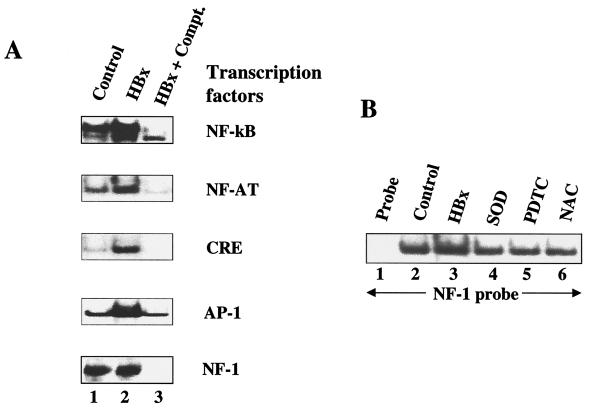
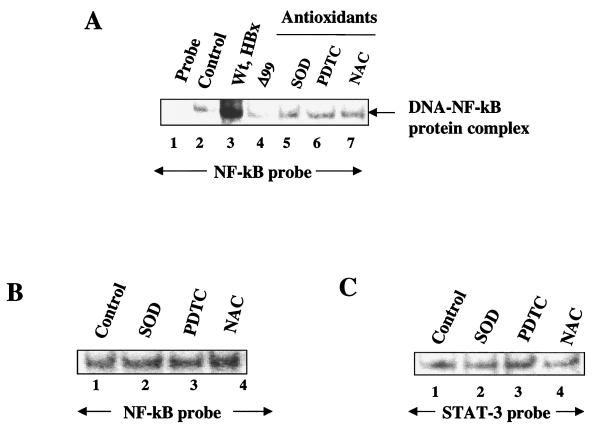
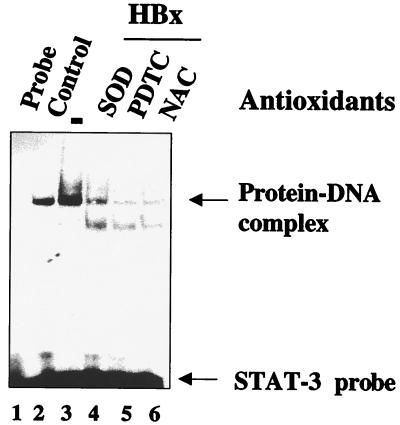
Next, we examined a whole host of transcription factors for similar stimulatory responses to HBx. The nuclear lysates from untransfected HepG2 cells (control) and transfected with HBx were incubated with γ-32P-labeled oligonucleotide probes representing cognate sequences for NF-κB, NF-AT, AP-1, CRE, and NF-1. EMSA revealed that an appreciable degree of activation of several of these factors, including NF-κB, NF-AT, CRE, and AP-1, occurred in the presence of HBx (Fig. 3A, lanes 2), but the DNA binding activity of NF-1 remained unchanged (Fig. 3A). The specificity of DNA-protein complexes were confirmed by competition with a 100-fold excess of respective unlabeled cognate oligonucleotides (Fig. 3A, lanes 3). We next pursued NF-κB activation by using wild-type HBx and the deletion mutant HBxΔ99. NF-κB has been previously shown to activate NF-κB-driven gene expression (43, 48). Here we revisited this earlier observation with a different emphasis. In the present study, the ability of HBx to activate NF-κB is being examined in the context of its association with mitochondria. Nuclear lysates from HepG2 cells transfected with wild-type HBx were initially used and compared with untransfected cells (Fig. 4A, lanes 2 and 3). As can be seen, there is a significant level of stimulation of NF-κB activity in the presence of wild-type HBx. In the next step, lysates from HBxΔ99 were utilized. The carboxy-terminal deletion mutant HBxΔ99 is unable to produce the NF-κB–DNA complex. Several conditions can lead to the activation of NF-κB. Since HBx is localized to mitochondria, the most logical scenario is the elevation of ROS, which in turn can activate NF-κB. To address this issue, we incubated the HBx-transfected HepG2 cells with the antioxidants PDTC and NAC. In the presence of these reagents, NF-κB activation is abrogated (Fig. 4A, lanes 6 and 7), indicating that NAC and PDTC counteract the effect of ROS. Similarly, when Mn-SOD expression vector was cotransfected along with HBx, leading to the overexpression of Mn-SOD, the ability of HBx to induce NF-κB was also reduced (lane 5). Mn-SOD converts superoxide anion radicals into water and molecular oxygen, thereby preventing the effects of ROS in the cellular environment. Overexpression of Mn-SOD has been previously shown to abolish NF-κB activation induced by oxidants (32). We carried out similar analysis by using STAT-3 and NF-1 oligonucleotide probes. The results shown in Fig. 5 demonstrate that the HBx-expressing HepG2 cells display activated STAT-3 binding (lane 3), which is abolished in the presence of PDTC, NAC, and overexpression of Mn-SOD (lanes 4 to 6), whereas the NF-1 protein-DNA complex was not affected by antioxidants (Fig. 3B). These data support the results shown in Fig. 4 and implicate ROS as second messengers in the activation of STAT-3 by HBx. Treatment of untransfected HepG2 cellular lysates with antioxidants did not appreciably diminish the intensity of the protein-DNA complexes formed by STAT-3 or NF-κB proteins (Fig. 4B and C), a finding consistent with similar earlier observation (33). Collectively, these data point to the involvement of ROS in the activation of transcription factors STAT-3 and NF-κB by HBx.
FIG. 3.
(A) HBx-induced DNA binding activities of several transcription factors. EMSA was carried out in the presence of γ-32P-labeled oligonucleotides representing cognate sequences for the transcription factors NF-κB, NF-AT, AP-1, CRE, and NF-1 cognate sequences and the nuclear lysates prepared from untransfected and cells transfected with wild-type HBx expression vector. Lanes 1 and 2, equal amounts of untransfected and HBx-transfected nuclear lysates; lane 3, a 100-fold excess of respective unlabeled oligonucleotides as competitors. (B) EMSA with NF-1 probe with untransfected HepG2 lysates. Lane 1, probe alone; lane 2, equal amount of untransfected nuclear lysates; lanes 3 to 6, HBx-transfected nuclear lysates; lane 4, cotransfected with Mn-SOD expression vector; lanes 5 and 6, treated with PDTC and NAC.
FIG. 4.
Mitochondrial association of HBx activates NF-κB. (A) EMSA was carried out in the presence of γ-32P-labeled NF-κB cognate sequences and nuclear lysates prepared from cells transfected with wild-type HBx and a series of HBx mutants. Lane 1, probe alone; lanes 2 and 3, equal amounts of untransfected and HBx-transfected nuclear lysates; lane 4, NF-κB probe incubated with HBxΔ99. The binding of probe with lysates from cells overexpressed with Mn-SOD (lane 5) or treated with 100 μM PDTC (lane 6) and 30 mM NAC (lane 7) is shown. (B and C) Lack of STAT-3 and NF-κB activation in HepG2 cells. Untreated HepG2 cell nuclear lysate (lane 1) or HepG2 cell nuclear lysate treated with antioxidants (lanes 2, 3, and 4) were incubated with NF-κB and STAT-3 probes.
FIG. 5.
Inhibition of HBx-mediated STAT-3 activation by various antioxidants. Nuclear lysates were prepared from cells transfected with wild-type HBx expression vector and treated with NAC (30 mM) and PDTC (100 μM) for 6 h. Mn-SOD and HBx expression vectors were cotransfected. The STAT-3 binding activity was assayed by EMSA by using equal amounts of nuclear lysate and γ-32P-labeled STAT-3 cognate sequences. Lane 1, probe alone; lanes 2 and 3, equal amounts of untransfected and HBx-transfected nuclear lysates; lanes 4, 5, and 6, lysates from cells cotransfected with Mn-SOD (lane 4) or treated with PDTC (lane 5) or NAC (lane 6). The fast-migrating band in lanes 4, 5, and 6 is a nonspecific band.
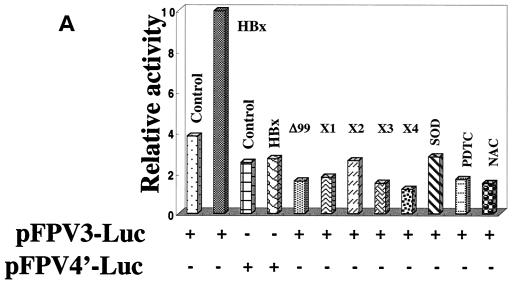
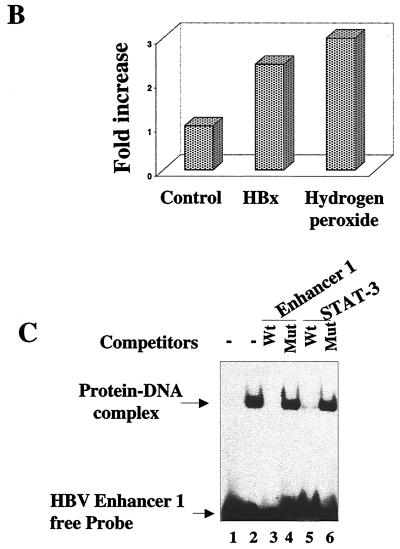
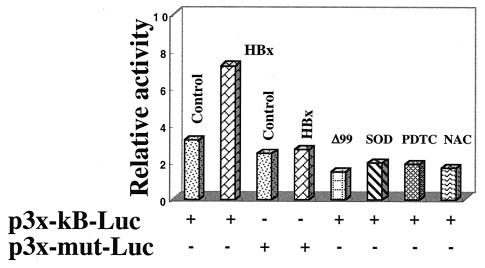
We next used the luciferase reporter expression vectors in support of EMSA data. A STAT-3 binding site exists in the core domain of HBV enhancer 1, as evidenced by EMSA (Fig. 6C). The plasmids with the wild type and a mutated STAT-3 binding site linked to the luciferase reporter are termed pFPV3-Luc and pFPV4′-Luc, respectively (25; Waris and Siddiqui, unpublished). HepG2 cells were cotransfected with the indicated plasmids as shown in Fig. 6A. HBx was able to induce luciferase activity via the STAT-3 motif, whereas the mutated vector did not show activation (Fig. 6A). HBx mutants (both deletion and cysteine point mutants) did not stimulate luciferase expression. Wild-type HBx transfected HepG2 cells, when treated with the antioxidants NAC and PDTC or cotransfected with Mn-SOD expression vector, failed to stimulate STAT-3 activity, as evidenced by the reduced luciferase activities (Fig. 6A). Hydrogen peroxide is an oxidant which has been shown to activate NF-κB and STAT-3. We have included this control here to show that hydrogen peroxide indeed activates STAT-3 (Fig. 6B). H2O2 treatment, as well as HBx expression, stimulated luciferase activity which is under the control of STAT-3-binding nucleotide sequences. We carried out the same experiment with NF-κB luciferase vector (p3x-κB-Luc) and a mutated NF-κB vector (p3x-mut-Luc). The results illustrate similar patterns of NF-κB-regulated stimulation with wild-type HBx and reduction of that activity in the presence of antioxidants (NAC and PDTC) and the overexpresssion of Mn-SOD (Fig. 7). The reporter plasmid with mutated NF-κB binding sites was unable to respond to HBx.
FIG. 6.
(A) HBx stimulates the STAT-3-dependent transcriptional activation in HepG2 cells. The reporter plasmid pFPV3Luc, which contains the STAT-3 binding site, was cotransfected with wild-type HBx and with HBxΔ99 X1 to X4 expression vectors. HBx and Mn-SOD cotransfected cells or cells treated with PDTC and NAC for 6 h before luciferase assay. (B) HBx and hydrogen peroxide activates STAT-3 in vivo. HepG2 cells were transfected with HBx expression vector or treated with hydrogen peroxide (2 mM) for 30 min. The cells were harvested to determine the luciferase activity. (C) Interaction between cellular STAT-3 protein and HBV enhancer 1. EMSA was carried out in the presence of γ-32P-labeled Enhancer 1 probe and STAT-3 protein synthesized in bacteria. Lane 1, probe alone; lane 2, 1 μg of STAT-3 protein; lanes 3 and 5, a 100-fold excess of wild-type (Wt) unlabeled HBV Enhancer 1 and STAT-3 oligonucleotides; lanes 4 and 6, a 100-fold excess of unlabeled mutants (Mut), Enhancer 1, and STAT-3 competitor oligonucleotides.
FIG. 7.
HBx transactivates NF-κB-dependent transcriptional activation. The reporter plasmid p3x-κB-Luc with an NF-κB binding site was cotransfected with HBx and HBxΔ99. The results obtained with HBx- and Mn-SOD-cotransfected cells or cells treated with PDTC and NAC for 6 h before luciferase assay are shown.
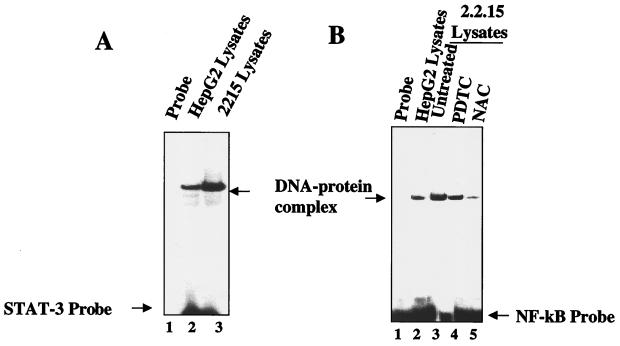
Constitutive activation of transcription factor STAT-3 and NF-κB in 2.2.15 cells.
Next, we wished to determine if oxidative stress is induced by HBx in the context of the rest of viral genes and ongoing replication cycle. We employed stable cell line 2.2.15 developed by Sells et al. (42). The 2.2.15 cell line is derived from the HepG2 cell line, which was transfected with a dimer molecule of the HBV genome by using a selectable neomycin resistance marker. This cell line expresses all of the HBV genes, including HBx, and has a modest HBV replicative cycle (42). Nuclear lysates were prepared from HepG2 (negative control) and HBV-positive 2.2.15 cells and subjected to EMSA by using γ-32P-labeled STAT-3 and NF-κB cognate oligonucleotides as probes. In each case, the 2.2.15 nuclear lysates displayed an elevated level of STAT-3 (Fig. 8A, lane 3) and NF-κB (Fig. 8B, lane 3) but not the HepG2 cell lysates (control). The DNA–NF-κB complexes were markedly reduced in 2.2.15 cell lysates when treated with the antioxidant NAC but slightly reduced in PDTC-treated lysates (Fig. 8B, lanes 4 and 5). In the HBV-expressing 2.2.15 cell line, the HBx gene is under the control of its native promoter-enhancer and is expressed at a much lower level than in the vector utilized in previous studies presented here. These results clearly indicate that HBx expressed under the control of the native promoter is still capable of inducing both NF-κB and STAT-3 transcription factors. This allays the concern that overexpression of HBx from the cytomegalovirus promoter-enhancer is responsible for the observed effects of HBx.
FIG. 8.
Antioxidants inhibit the constitutive activation of STAT-3 and NF-κB in the 2.2.15 cell line. EMSA was carried out in the presence of γ-32P-labeled STAT-3 (A) and NF-κB (B) cognate sequences and nuclear lysates prepared from HepG2 cells (control) and the HepG2 2.2.15 cell line. (A) Lane 1, probe alone; lanes 2 and 3, equal amounts of lysates from HepG2 and 2.2.15 cells, respectively. (B) Lane 1, probe alone; lanes 2 and 3, equal amounts of lysates from HepG2 and 2.2.15 cells, respectively; lanes 4 and 5, lysates from 2.2.15 cells treated with the antioxidants PDTC and NAC, respectively.
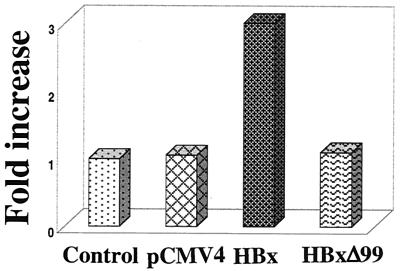
To examine whether HBx expression increases ROS levels, HepG2 cells were transfected with HBx, HBxΔ99 expression vector, or pCMV4 (as a control). Transfected cells were stained with DHE, an oxidation-sensitive fluorescent probe. The fluorescence intensities of the cells were analyzed by fluorescence-activated cell sorting. DHE is oxidized to ethidium in the presence of cellular ROS (13). HBx cotransfected cells show higher DHE fluorescence intensities compared to those of HBxΔ99- and pCMV4-transfected cells (Fig. 9). These experiments, which were repeated at least four times, produced similar results. An overnight treatment of control cells with antimycin A (100 μM), which is an inhibitor of complex III electron flow, exhibited a fold increase similar to that observed in HBx-expressing cells (data not shown). These data suggest that HBx expression leads to the generation of higher levels of ROS in the cells. Similarly, 2.2.15 cells also displayed higher levels of ROS than did the parental HepG2 cells (Huh and Siddiqui, unpublished).
FIG. 9.
ROS levels of untranfected or HepG2 cells transfected with pCMV4, HBx, or HBxΔ99 expression vector were determined by flow cytometry as described in Materials and Methods. The bars show the fold increase in oxidized DHE fluorescence. The experiment was repeated at least four times.
In summary, the results described above collectively demonstrate that HBx induces the generation of oxidants, which by a mechanism(s) that is not yet clearly understood can induce the activation of transcription factors STAT-3 and NF-κB.
DISCUSSION
The HBx protein remains an unsolved puzzle in the HBV field. While a large number of cellular functions have been attributed to HBx, such as the activation of signal transduction pathways, the sensitization of cells to apoptosis, the loss of cell cycle control checkpoints (5), and possible direct interactions with several components of the transcription apparatus (reviewed in reference 56), the exact mechanism of its action has remained elusive. It is clear, however, that HBx is a multifunctional protein, one that is indispensable for viral replication and production (9, 60). Circumstantial evidence suggests that it may play an indirect role in the genesis of hepatocellular carcinoma. HBx is frequently studied as a transactivator of transcription, which has led to the identification of a wide variety of transcriptional elements and factors as its possible target(s) (19, 30, 31, 35, 36, 54, 55, 56). Despite this wealth of information, there has been considerable difficulty in establishing the exact mechanism(s) through which HBx activates transcription. The in vitro interaction of HBx with components of the transcriptional machinery has led to the idea that HBx functions directly in the nucleus (10, 35, 36, 54). While most of the available evidence points to its predominantly cytoplasmic distribution, very little, if any, HBx has been found in the nucleus (15, 19). We recently demonstrated direct physical interaction between HBx and mitochondrial VDAC3 and showed that this association altered the mitochondrial transmembrane potential, ΔΨ (37). We further noted that HBx's association did not lead to cytochrome c release from mitochondria. These studies imply a key functional role of HBx in mitochondrial functions. Mitochondria are key organelles that generate cellular energy and control apoptosis by releasing death-promoting proteins into the cytoplasm (18). It is also the principal organelle in which ROS are generated in response to stress induced by a variety of conditions, including viral infection (41). In the present study, we explored this principle and provide evidence in support of this notion. HBx expression led to generation of ROS and ultimately to the activation of a whole host of transcription factors. Here, we have focused on two transcriptional factors, STAT-3 and NF-κB, and provide evidence that mitochondrial association of HBx is necessary for the activation of these factors. Using EMSA and schemes of reporter gene expression, we show that the ability of HBx to activate STAT-3 and NF-κB is affected by antioxidants, thereby implicating a key role of ROS in triggering pathways that translocate the latent transcription factors to the nucleus. This scenario suggests that HBx from its cytoplasmic (mitochondrial) residence can induce activation of gene expression via a number of transcription factors that are known to respond to oxidative stress. These include NF-κB, AP-1, NF-AT, and others, all of which have been previously shown to respond to HBx's ability to transactivate (28, 33, 44, 47, 49). Meyer et al. (33) have shown that MHBs, a hepatitis B surface antigen derivative, as well as HBx transactivated NF-κB and that these activities were sensitive to NAC and PDTC, implicating the involvement of ROS.
Oxidative stress is associated with nearly all pathological states, especially those involving inflammatory processes. The role of ROS in viral pathogenesis has been documented for influenza virus and human immunodeficiency virus (34). High doses of ROS are produced during chronic and acute inflammatory diseases or as a result of environmental stress (41). An overwhelming number of studies support the role of free radicals in the initiation and progression of multistage carcinogenesis (7, 45). Consistent with this idea, free-radical scavengers and antioxidant enzymes are downregulated in tumor cells (12). For instance, glutathione levels were found to be low in hepatocullular carcinoma tumors (12). The production of hepatocellular carcinoma by HBV probably involves a combination of indirect mechanisms. Chronic liver injury leads to necrosis, inflammation, and liver regeneration, which over a period of time can contribute to cirrhosis, thus paving a way for events preceding the development of hepatocellular carcinoma.
It is well established that the activation of STAT-3 requires tyrosine phosphorylation, which occurs upon cytokine signaling, such as by epidermal growth factor or interleukin-6 (53). The mechanism by which ROS generated by oxidative stress in mitochondria directly activate STAT-3 is not clearly understood. One possibility is that the alteration of their redox status could directly alter their conformation in such a way that their interaction with cytosolic proteins responsible for nuclear targeting is triggered. The other likely explanation is the ability of oxidants to act as inhibitors of tyrosine phosphatases, thereby inducing STAT-3 nuclear translocation by enhanced tyrosine phosphorylation. Lee and Yun (29) showed that HBx binds to JAK1 and may be directly responsible for the activation of STAT-3. In contrast, our studies present a view in which HBx was able to constitutively activate STAT-3 via ROS, as evidenced by the failure to activate STAT-3 in the presence of NAC and PDTC. HBx mutants, which differ in their association from that of the wild-type HBx, failed to activate STAT-3 and NF-κB. In this context, we observed that HBx was able to constitutively activate tyrosine phosphorylation of STAT-3 in HepG2 cells transfected with wild-type HBx expression vector (Fig. 1, lane 4). The HBx mutant HBxΔ99, which does not target mitochondria (Huh and Siddiqui, unpublished), failed to activate STAT-3 (Fig. 1A, lane 3). According to this model, HBx induces the generation of ROS, which then activate STAT-3. No direct physical interaction between HBx and STAT-3 was observed (data not shown).
NF-κB was one of the first HBx-responsive motifs to be identified (15, 30, 44, 49). However, the mechanism by which HBx promoted activation of NF-κB transcription through this motif was not understood. For instance, HBx neither physically interacted with the NF-κB subunit nor altered the binding affinity of NF-κB in mobility shift assays when it was exogenously added, suggesting a lack of protein-protein interactions between HBx and NF-κB. Recently, direct binding of HBx with the IκB subunit of the NF-κB complex has been described which permits its entry into the nucleus (52). The physiological relevance of this observation at present is not clear. NF-κB is a multiprotein complex which is found in the cytoplasm in an inactive state. Phosphorylation of IκB triggers its disassembly, which ultimately leads to translocation of two subunits, p50 and p65, into the nucleus, where they directly interact with their cognate sequences and regulate gene expression. Our findings here show that HBx, without migrating to the nucleus, has the ability to induce NF-κB activation via generation of ROS. H2O2 has been implicated in the positive modulation of the activity of a number of protein tyrosine kinases whose function is critical for lymphocyte function. The oxidative stress-induced activation of NF-κB has recently been shown to be modulated by the tyrosine phosphorylation (Y42) and the PEST sequences of IκB (39). It was further demonstrated that such IκB molecules are degraded by calpains rather than by the proteosome pathway (39). Our future work will focus on whether HBx induces NF-κB via these motifs of IκB. Su and Schneider (46) have shown the stimulation of IκB phopshorylation in cells infected with an adenovirus vector containing HBx. Whether this stimulation involved tyrosine residue 42 and the PEST sequences of IκB remains to be determined.
The direct consequence of activation of these transcription factors is the induction of genes whose functions can be protective and antiapoptotic in the final analysis. STAT-3 and NF-κB motifs are found in a wide variety of cellular genes whose functions range from growth promotion, to proliferation, to DNA replication and repair, and to functions involved in cell death and cancer programs.
In summary, the results of our study collectively suggest a novel mechanism of HBx's action in inducing the activation of transcription factors. These transcription factors include those which respond to oxidative stress. Interestingly, several of these have been previously shown by investigators to respond to HBx (4, 11, 28, 51). We propose here a model in which HBx's ability to transactivate gene expression via a large number of transcription factors can be explained by a common mechanism. This model does not require nuclear localization of HBx. From its mitochondrial residence, HBx induces oxidative stress in mitochondria. Rising levels of ROS then, by a mechanism not clearly understood, induce the activation of whole host of transcription factors, which ultimately translocate to the nucleus and activate gene expression. This notion is further strengthened by the observations of Doria et al. (15), who showed that nuclear targeting of HBx by the addition of the nuclear localization signal abrogated its ability to transactivate AP-1 and NF-κB.
ACKNOWLEDGMENTS
This work was supported by NIH grants CA64415 and CA92187 to A.S.
REFERENCES
- 1.Aufiero B, Schneider R J. The hepatitis B virus X-gene product transactivates both RNA polymerase II and III. EMBO J. 1990;9:497–504. doi: 10.1002/j.1460-2075.1990.tb08136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beasley R P, Liu C C, Huang L Y, Chien C S. Hepatocellular carcinoma and hepatitis B virus. A prospective study of 22707 men in Taiwan. Lancet. 1981;ii:1129–1133. doi: 10.1016/s0140-6736(81)90585-7. [DOI] [PubMed] [Google Scholar]
- 3.Becker S A, Lee T H, Butel J S, Slagle B L. Hepatitis B virus X protein interferes with cellular DNA repair. J Virol. 1998;72:266–272. doi: 10.1128/jvi.72.1.266-272.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benn J, Schneider R J. Hepatitis B virus HBx protein activates Ras-GTP complex formation and establishes a Ras, Raf, MAP kinase signaling cascade. Proc Natl Acad Sci USA. 1994;91:10350–10354. doi: 10.1073/pnas.91.22.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benn J, Schneider R J. Hepatitis B virus HBx protein deregulates cell cycle check point control. Proc Natl Acad Sci USA. 1995;92:11215–11219. doi: 10.1073/pnas.92.24.11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benn J, Su F, Doria M, Schneider R J. Hepatitis B virus HBx protein induces transcription factor AP-1 by activation of extracellular signal-related and c-Jun N-terminal mitogen activated protein kinases. J Virol. 1996;70:4978–4985. doi: 10.1128/jvi.70.8.4978-4985.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butel J. Viral carcinogenesis: revelation of molecular mechanisms and etiology of human disease. Carcinogenesis. 2000;21:405–426. doi: 10.1093/carcin/21.3.405. [DOI] [PubMed] [Google Scholar]
- 8.Carballo M, Conde M, El Bekay R, Martin-Nieto J, Camacho M J, Monteseiris J, Conde J, Bedoya F J, Sobrino F. Oxidative stress triggers STAT-3 tyrosine phosphorylation and nuclear translocation in human lymphocytes. J Biol Chem. 1999;274:17580–17586. doi: 10.1074/jbc.274.25.17580. [DOI] [PubMed] [Google Scholar]
- 9.Chen H S, Kaneko R, Girones R W, Anderson W E, Hornbuckle B C, Tennant P J, Cote J L, Gerin R H, Purcell R H, Miller R H. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J Virol. 1993;67:1218–1226. doi: 10.1128/jvi.67.3.1218-1226.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheong J-H, Yi M-K, Lin Y, Murakami S. Human RPB5, a subunit shared by eukaryotic nuclear polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. EMBO J. 1995;14:143–150. doi: 10.1002/j.1460-2075.1995.tb06984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chirillo P, Faleco M, Puri P L, Artini M, Balsano C, Levrero M, Natoli G. Hepatitis B virus pX activates NF-κB-dependent transcription through a Raf-independent pathway. J Virol. 1996;70:641–646. doi: 10.1128/jvi.70.1.641-646.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corrocher R, Casaril M, Bellisola G, Gabrielli G B, Nicoli N, Guidi G C, De Sandre G. Severe impairment of anti-oxidants system in human hepatoma. Cancer. 1986;58:1658–1662. doi: 10.1002/1097-0142(19861015)58:8<1658::aid-cncr2820580814>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 13.Costa-Pereira A, Cotter T. Metabolic alterations associated with apoptosis. In: Studzinski G P, editor. Apoptosis: a practical approach. Oxford, England: Oxford University Press; 1999. pp. 141–156. [Google Scholar]
- 14.Darnell J E., Jr STATs and gene regulation. Science. 1997;277:630–635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 15.Doria M, Klein N, Lucito R, Schneider R J. The hepatitis B virus HBx protein is a dual specificity cytoplasmic activator of Ras and nuclear activator of transcription factors. EMBO J. 1995;14:4747–4757. doi: 10.1002/j.1460-2075.1995.tb00156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faktor O, Shaul Y. The identification of hepatitis B virus X gene responsive elements reveals functional similarity of X and HTLV-I tax. Oncogene. 1990;5:867–872. [PubMed] [Google Scholar]
- 17.Fialkow L, Chan C K, Rotis D, Grinstein S, Downey G P. Activation of the mitogen-activated protein kinase signaling pathway in neutrophils. Role of oxidants. J Biol Chem. 1994;269:31234–31242. [PubMed] [Google Scholar]
- 18.Green D R, Reed J C. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 19.Haviv I, Shamay M, Doitsh G, Shaul Y. Hepatitis B virus pX target TFII B in transcription coactivator. Mol Cell Biol. 1998;18:1562–1569. doi: 10.1128/mcb.18.3.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henkler F, Hoare J, Waseem N, Goldin R, McGarvey M, Koshy R, King I. Intracellular localization of the hepatitis B virus HBx protein. J Gen Virol. 2001;82:871–882. doi: 10.1099/0022-1317-82-4-871. [DOI] [PubMed] [Google Scholar]
- 21.Hu Z, Zhang Z, Doo E, Coux O, Goldberg A L, Liang T J. Hepatitis B virus X protein is both a substrate and a potential inhibitor of the proteasome complex. J Virol. 1999;73:7231–7240. doi: 10.1128/jvi.73.9.7231-7240.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kekule A, Lauer U, Weiss L, Luber B, Hofschneider P. Hepatitis B virus transactivator HBx uses a tumor promoter signaling pathway. Nature. 1993;361:742–745. doi: 10.1038/361742a0. [DOI] [PubMed] [Google Scholar]
- 23.Klein N P, Schneider R J. Activation of Src family of kinase by HBV HBx protein, and coupled signaling to Ras. Mol Cell Biol. 1997;17:6427–6436. doi: 10.1128/mcb.17.11.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein N, Bouchard M, Wang L-H, Kobarg C, Schneider R J. Src kinases involved in hepatitis B virus replication. EMBO J. 1999;18:5019–5020. doi: 10.1093/emboj/18.18.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kosovsky M J, Huan B, Siddiqui A. Purification and properties of rat liver nuclear proteins that interact with the hepatitis B virus enhancer 1. J Biol Chem. 1996;271:21859–21869. doi: 10.1074/jbc.271.36.21859. [DOI] [PubMed] [Google Scholar]
- 26.Kumar V, Jayasuryan N, Kumar R. A truncated mutant (residues 58 to 40) of the hepatitis B virus X protein retains transactivation function. Proc Natl Acad Sci USA. 1996;93:5647–5652. doi: 10.1073/pnas.93.11.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwee L, Lucito R, Aufiero B, Schneider R J. Alternate translation initiation on hepatitis B virus X mRNA produces multiple polypeptides that differentially transactivates class II and class III promoters. J Virol. 1992;66:4382–4389. doi: 10.1128/jvi.66.7.4382-4389.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lara-Pezzi E, Armesilla A L, Majano P L, Redondo J M, Lopez-Cabrera M. The hepatitis B virus X protein activates nuclear factor of activated T cells (NF-ATc) by a cyclosporin A-sensitive pathway. EMBO J. 1998;17:7066–7077. doi: 10.1093/emboj/17.23.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee Y H, Yun Y. HBx protein of hepatitis B virus activates JAK1-STAT signaling. J Biol Chem. 1998;273:25510–25515. doi: 10.1074/jbc.273.39.25510. [DOI] [PubMed] [Google Scholar]
- 30.Lucito R, Schneider R J. Hepatitis B virus X protein activates transcription factor NF-κB without a requirement for protein kinase C. J Virol. 1992;66:983–991. doi: 10.1128/jvi.66.2.983-991.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maguire H F, Hoeffler J P, Siddiqui A. HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science. 1991;252:842–844. doi: 10.1126/science.1827531. [DOI] [PubMed] [Google Scholar]
- 32.Manna S K, Zhang H J, Tao Y, Oberley L W, Aggarwal B B. Overexpression of manganese superoxide dismutase suppress tumor necrosis factor-induced apoptosis and activation of nuclear transcription factor-κB and activated protein-1. J Biol Chem. 1998;273:13245–13254. doi: 10.1074/jbc.273.21.13245. [DOI] [PubMed] [Google Scholar]
- 33.Meyer M, Caselmann W H, Schluter V, Schreck R, Hofschneider P H, Baeuerle P A. Hepatitis B virus transactivator MHBs: activation of NF-κB, selective inhibition by antioxidants and integral membrane localization. EMBO J. 1992;11:2991–3001. doi: 10.1002/j.1460-2075.1992.tb05369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muller F. Reactive oxygen intermediates and human immunodeficiency virus (HIV) infection. Free Rad Biol Med. 1992;13:651–657. doi: 10.1016/0891-5849(92)90039-j. [DOI] [PubMed] [Google Scholar]
- 35.Qadri I, Maguire H F, Siddiqui A. Hepatitis B virus transactivator protein X interacts with the TATA binding protein. Proc Natl Acad Sci USA. 1995;92:1003–1007. doi: 10.1073/pnas.92.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qadri I, Conaway J W, Conaway R C, Schaack J, Siddiqui A. Hepatitis B virus transactivator protein, HBx, associates with the components of TFIIH and stimulates the DNA helicase activity of TFIIH. Proc Natl Acad Sci USA. 1996;93:10578–10583. doi: 10.1073/pnas.93.20.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rahmani Z, Huh K-W, Lasher R L, Siddiqui A. Hepatitis B virus X protein colocalize to mitochondria with a human voltage-dependent anion channel, hVDAC3 and alters its transmemebrane potential. J Virol. 2000;74:2840–2846. doi: 10.1128/jvi.74.6.2840-2846.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schindler C, Shuai K, Prezioso V R, Darnell J E., Jr Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science. 1992;257:809–813. doi: 10.1126/science.1496401. [DOI] [PubMed] [Google Scholar]
- 39.Schoonbroodt S, Ferreira V, Best-Belpomme M, Boelaert J, Legrand-Poels S, Korner M, Peitte J. Crucial role of the amino-terminal tyrosine residue 42 and the carboxyl-terminal PEST domain of IκBα in NF-κB activation by an oxidative stress. J Immunol. 2000;164:4292–4300. doi: 10.4049/jimmunol.164.8.4292. [DOI] [PubMed] [Google Scholar]
- 40.Schreck R, Rieber P, Baeurele P A. Reactive oxygen intermediates as apparently widely used messengers in the activation of NF-κB transcription factor and HIV. EMBO J. 1991;10:2247–2258. doi: 10.1002/j.1460-2075.1991.tb07761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwarz K B. Oxidative stress during viral infection: a review. Free Rad Biol Med. 1996;21:641–649. doi: 10.1016/0891-5849(96)00131-1. [DOI] [PubMed] [Google Scholar]
- 42.Sells M A, Chen M-L, Acs G. Production of hepatitis B virus particles in HepG2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siddiqui A, Jameel S, Mapoles J. Expression of the hepatitis B virus X gene in mammalian cells. Proc Natl Acad Sci USA. 1987;84:2513–2517. doi: 10.1073/pnas.84.8.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siddiqui A, Gayner R, Srinivasan A, Mapoles J, Farr R W. Trans-activation of viral enhancers inducing the long terminal repeat of the human immunodeficiency virus by the hepatitis B virus X protein. Virology. 1989;169:479–484. doi: 10.1016/0042-6822(89)90177-3. [DOI] [PubMed] [Google Scholar]
- 45.Sun Y. Free radicals, anti-oxidant enzymes, and carcinogenesis. Free Rad Biol Med. 1990;8:583–599. doi: 10.1016/0891-5849(90)90156-d. [DOI] [PubMed] [Google Scholar]
- 46.Su F, Schneider R J. Hepatitis B virus HBx protein sensitize cells to apoptotic killing by TNF- Proc Natl Acad Sci USA. 1997;94:8744–8749. doi: 10.1073/pnas.94.16.8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Su F, Theodosis C N, Schneider R J. Role of NF-κB and Myc proteins in apoptosis induced by hepatitis B virus HBx protein. J Virol. 2001;75:215–225. doi: 10.1128/JVI.75.1.215-225.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takada S, Shriakat Y, Kaneniwa N, Koike K. Association of HBV X protein with mitochondria causes mitochondrial aggregation at the nuclear periphery, leading to cell death. Oncogene. 1999;18:6965–6973. doi: 10.1038/sj.onc.1203188. [DOI] [PubMed] [Google Scholar]
- 49.Twu J-S, Chu K, Robinson W S. Hepatitis B virus X gene can transactivate heterologous viral sequence. Proc Natl Acad Sci USA. 1989;86:2046–2050. doi: 10.1073/pnas.86.6.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X W, Forrester K, Yeh H, Feitelson M A, Gu J R, Harris C C. Hepatitis B virus X protein inhibits p53 sequence specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc Natl Acad Sci USA. 1994;91:2230–2234. doi: 10.1073/pnas.91.6.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang H O, Trivedi A, Johnson D L. Regulation of RNA polymerase 1-dependent promoters by the hepatitis B virus X protein via activated Ras and TATA-binding protein. Mol Cell Biol. 1998;18:7086–7094. doi: 10.1128/mcb.18.12.7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weil R, Sirma H, Giannini C, Kremsdorf D, Bessia C, Dargemont C, Brechot C, Israel A. Direct association and nuclear import of the hepatitis B virus X protein with the NF-κB inhibitor IκBα. Mol Cell Biol. 1999;19:6345–6354. doi: 10.1128/mcb.19.9.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wen Z, Zhong Z, Darnell J E., Jr Maximal activation of transcription by STAT-1 and STAT-3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 54.Williams J S, Andrisani O M. The hepatitis B virus X protein targets the basic region-leucine zipper domain of CREB. Proc Natl Acad Sci USA. 1995;92:3819–3823. doi: 10.1073/pnas.92.9.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wollersheim M, Debelka U, Hofschneider P H. A transactivating function encoded in the hepatitis B virus X gene is conserved in the integrated state. Oncogene. 1988;3:545–552. [PubMed] [Google Scholar]
- 56.Yen T B S. Hepadnaviral X protein: review of recent progress. J Biomed Sci. 1996;3:20–30. doi: 10.1007/BF02253575. [DOI] [PubMed] [Google Scholar]
- 57.Yoo Y D, Ueda H, Park K, Flanders K C, Lee Y I, Jay G, Kim S J. Regulation of transformimg growth factor-β1 expression by the hepatitis B virus (HBV) X transactivator. J Clin Investig. 1996;97:388–395. doi: 10.1172/JCI118427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zentgraf H G, Herrmann R, Klein P, Schranz I, Loncarenic D, Herrmann K, Hubner H, Schroder C. Mouse monoclonal antibody directed against hepatitis B virus X protein synthesized in Escherichia coli: detection of reactive antigen in liver cell carcinoma and chronic hepatitis. Oncology. 1990;47:143–148. doi: 10.1159/000226807. [DOI] [PubMed] [Google Scholar]
- 59.Zhong Z, Wen W, Darnell J E., Jr Stat-3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 60.Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis B virus X gene is important for viral infection in vivo. J Virol. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]