Abstract
Metabolite biosensors based on metabolite-responsive transcription factors are key synthetic biology components for sensing and precisely controlling cellular metabolism. Biosensors are often designed under laboratory conditions but are deployed in applications where cellular growth rate differs drastically from its initial characterization. Here we asked how growth rate impacts the minimum and maximum biosensor outputs and the dynamic range, which are key metrics of biosensor performance. Using LacI, TetR, and FadR-based biosensors in Escherichia coli as models, we find that the dynamic range of different biosensors have different growth rate dependencies. We developed a kinetic model to explore how tuning biosensor parameters impact the dynamic range growth rate dependence. Our modeling and experimental results revealed that the effects to dynamic range and its growth rate dependence are often coupled, and the metabolite transport mechanisms shape the dynamic range-growth rate response. This work provides systematic understanding on biosensor’s performance under different growth rates, which will be useful for predicting biosensor’s behavior in broad synthetic biology and metabolic engineering applications.
Keywords: metabolite biosensor, dynamic pathway regulation, metabolic engineering, biological circuit, context dependence, transcriptional regulator
Introduction
Metabolite-responsive transcription factor (MRTF)-based biosensors have broad applications in synthetic biology and metabolic engineering, ranging from metabolic detection1–4 screening and selecting for high-metabolite-producing strains5–7, dynamic metabolic control8–13, to strain functional evolution14,15. Nature has evolved various MRTFs that can be harnessed to create biosensors for a wide range of metabolites16–20. Protein engineering and directed evolution of MRTF have further expanded the range of compounds which can be detected21–23. Additionally, promoters regulated by MRTFs can be engineered to tune sensitivity and dynamic range24,25, enabling precise control of biosensor’s performance26,27. These biosensors can be further layered to create complex circuits28,29, which require a well-defined performance for correct operation30.
Most MRTF-based biosensors were designed and tested in well-defined laboratory conditions using rich growth media. These biosensors were often characterized using fluorescent proteins whose expression has little burden to growth and other cellular processes. However, during applications, biosensors are often deployed in different growth environments and used to control burdensome genes that affect cell growth rate. Changes in cell growth rate has been shown to impact several cellular parameters including plasmid copy numbers31, ribosome concentration and mass fraction32, transcription factor abundance and concentration33, gene expression rate34,35, and average cellular volume36,37. Biosensor operation relies on these shared cellular resources, so changes in cell growth rate will unavoidably affect the expression of MRTF and its regulated genes. Additionally, cell growth dilutes all molecular components as the volume of the cell increases (Fig. 1A), so faster growth may lead to a net reduction in the concentration of molecules needed for biosensor operation34,38. Thus, when cell growth rate changes, it may significantly alter a sensor’s behavior, leading to undesirable performance. For example, in two-stage dynamic metabolic control, engineered microbial cells need to shift from a high-growth phase to a low-growth production phase12. Biosensors used in two-stage dynamic metabolic control have to be optimized to perform under both growth conditions39,40.
Figure 1. Molecular components contributing to the operation of a metabolite-responsive transcription factor (MRTF)-based biosensor.
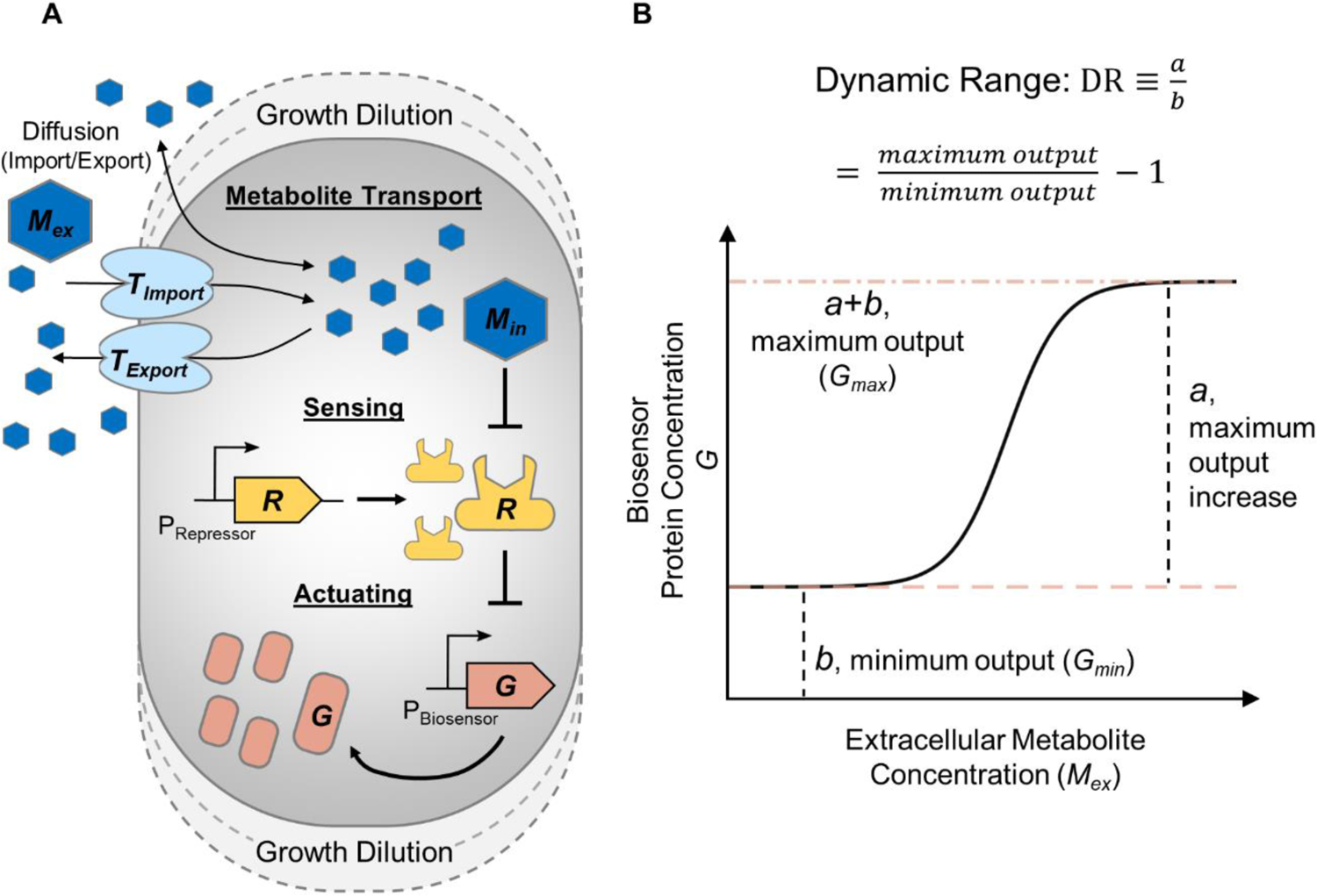
(A) Schematic of general components contributing to overall biosensor response. Extracellular metabolite enters the cell, becoming intracellular metabolite. Intracellular metabolite represses the DNA-binding activity of the MRTF. The MRTF represses the biosensor’s promoter, which controls the expression of a reporter protein. All components are universally affected by growth rate through dilution as the cell volume expands. B) Minimum output, maximum output, and dynamic range (DR) are critical parameters characterizing the dose-response curve of a MRTF-based biosensor.
Many MRTF-based biosensors use a repressed-repressor architecture41,42. In this architecture, the MRTF represses gene expression from its cognate promoter in the absence of its target metabolite, but its expression repression activity is antagonized by the presence of a specific intracellular metabolite (Fig. 1A). The output of a biosensor is the steady-state expression level of the controlled gene at a particular target metabolite concentration (Fig. 1B). A sensor’s behavior can be characterized by its minimum output in the absence of target metabolite, its maximum output under a saturating concentration of extracellular target metabolite, and its dynamic range (DR) which is defined as the ratio of the maximal increase in biosensor output relative to the its minimum output (Fig. 1B)26. In real applications, it is often desirable to keep a low minimum output to prevent unwanted gene expression in the absence of target metabolite43, a high maximum output to reach high signal-to-noise ratios44, and a large DR. Previous studies have shown how TF-repressed gene expression varies under different cell growth rates34, yet, little is known about how growth rate affects a biosensor’s behavior. Experimentally characterizing biosensor’s behavior under a wide range of growth rates, for example by growing the biosensor in medium with different nutrient conditions both with and without the target metabolite, is labor intensive. Thus, it would be beneficial to develop quantitative models to understand how biosensor operation is impacted by growth rate to avoid undesirable sensor performance due to the change of growth rate.
In this work, we use three repressed-repressor type of biosensors to explore the effect of growth rate to sensor behavior. Interestingly, we found that while all sensors displayed both decreasing minimum and maximum outputs, the DR could have either a positive or negative growth rate dependence. By integrating experimental data with kinetic modeling, we provide a mathematical framework to reveal biosensor’s behavior under changing growth conditions. We show how different parameters of the biosensor promoter and TF expression can lead to either a positive or negative dynamic range-growth rate (DR-μ) dependence. Additionally, our results show how growth rate-dependent membrane transport mechanisms of the target metabolite shape the overall DR-μ dependence. Our model and experimental results demonstrate a coupling between DR and sensitivity of DR to changes in growth rate for most biosensor designs, which implies a trade-off between high DR and low sensitivity objectives. Altogether our work provides a framework for tuning or predicting a biosensor’s behavior under varying growth conditions, which will be useful for a wide range of biosensor applications in synthetic biology.
Results
Growth Rate Dependence of Biosensor Dynamic Range
To study the relationship between sensor DR and cell growth rate of MRTF-based biosensors, we first tested three TF-promoter systems with a repressed-repressor architecture, TetR-Ptet (Fig. 2A), LacI-PLacUV542 (Fig. 2B), and FadR-PAR (Fig. 2C)45–47 that sense extracellular chemicals anhydrotetracycline (aTc), isopropyl β-D-1-thiogalactopyranoside (IPTG), and fatty acid (FA), respectively. All these three sensors have been commonly used for various applications42,45. For each biosensor, we used a red fluorescent protein (RFP) as a reporter. Each biosensor construct was cloned into plasmids with the stringently regulated SC101 origin of replication to reduce copy number variation due to changes in growth rate31,48. Additionally, for the FA-sensor, native regulations of the fadR and fadD genes were removed, and fadE was knocked-out to prevent FA from being used as a carbon source for growth through β-oxidation47. Thus, all three target metabolites are primarily non-metabolizable activators of the biosensor output gene.
Figure 2. Minimum output, maximum output, and dynamic range of PTet, PLacUV5, and PAR biosensors at different growth rates.
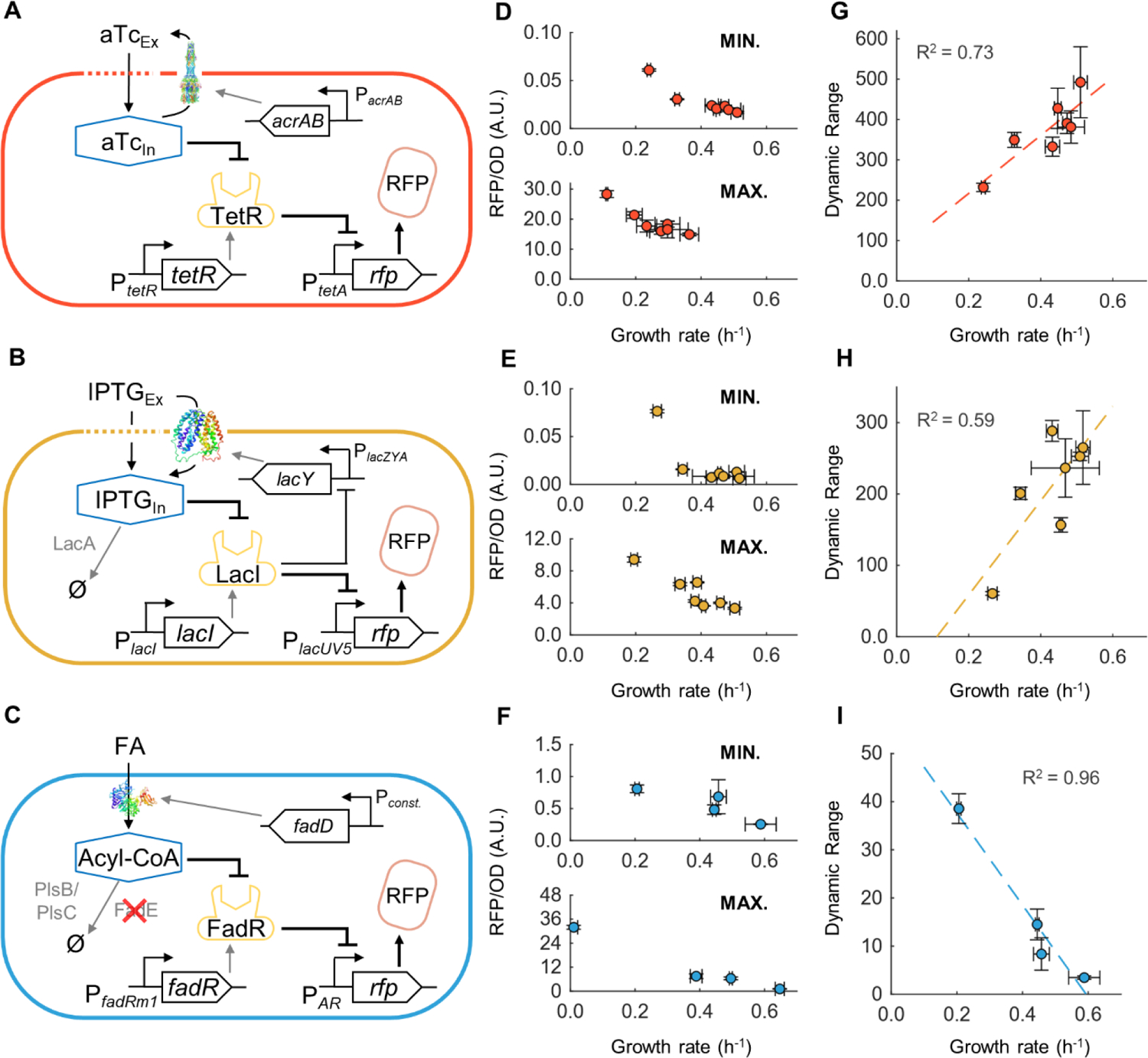
(A-C) Schematic of cellular interactions for the (A) TetR-based biosensor, (B) LacI-based biosensor and (C) FadR-based biosensor. (D-F) Minimum (top) and maximum (bottom) output of the biosensor at different growth rates for (D) TetR, (E) LacI, and (F) FadR-based biosensors. (G-I) Calculated dynamic range of biosensor at different growth rates for (G) TetR, (H) LacI, and (I) FadR-based biosensors. Error bars are S.D. of biological replicates, n=3. Dashed line: Line of best fit.
To manipulate cellular growth rate, biosensor cells were grown in minimal medium supplemented with different commonly-used carbon sources49 including acetate, pyruvate, glycerol, sorbitol, succinate, glycerol with amino acids, and xylose. These differing media conditions supported a range of growth rates from 0.24–0.51 h−1 (see Supplemental Table S3 for a summary of media conditions and growth rates). To quantify the effect of growth rate on DR, sensor minimum and maximum outputs were measured under exponential growth phase under different media conditions either without or with high concentrations of the target metabolites, respectively. The target metabolite concentrations used for aTc (1000 nM)42, IPTG (1 mM)26, and FA (4 mM)45 were previously shown to saturate the output of these biosensors. Our results show that both minimum and maximum outputs decreased with increasing growth rate for all three sensors (Fig. 2D, 2E, 2F). At a saturated target metabolite concentration (i.e. maximum output) a sensor’s promoter is equivalent to a strong constitutive promoter, while in the absence of the target metabolite (i.e. minimum output), a sensor’s promoter is equivalent to a repressor repressed promoter. Previous studies have shown that protein concentration under the control of either a constitutive promoter or a repressor controlled promoter decreases with increasing growth rates34. Thus, our observations are consistent with previous studies. Additionally, we observe that cell growth rates in maximum output are always lower than in minimum output (Fig. 2D, 2E, 2F), suggesting growth burden from activation of these sensors, even when they were only used to express an RFP reporter protein. These observations clearly demonstrated the influence of cell growth rate to sensor output as well as the effect of sensor’s function to cell growth, which cannot be ignored during biotechnology applications50,51.
We next calculated the DR of each biosensor under different growth rates. Interestingly, the aTc and IPTG sensors displayed an increasing DR with increasing growth rate (Fig. 2G, 2H), while the FA sensor showed a decreased DR with increasing growth rate. Our results showed that for aTc and IPTG sensors, although both the minimum and maximum outputs decreased with growth rate, the minimum output decreased more rapidly than the maximum output, thus leading to a relative increase in the ratio of the two, as quantified by the DR. In contrast, the FA strain has a negative DR trend with increasing growth rate (Fig. 2I) because the maximum output decreased more rapidly than the minimum output for this system. One major difference between the aTc/IPTG biosensors, and the FA biosensors is that FA transport requires the protein FadD to enter the intracellular space, whereas aTc and IPTG can diffuse into the cell (Fig. 2A, 2B, 2C). The concentration of these transport proteins is growth-dependent, particularly since FadD is expressed from a constitutive promoter34, which may contribute to the rapid decline in maximum biosensor output at higher growth rates. Thus, these results suggest that although all three biosensors have the same general architecture, their DR-μ dependency are different and may be a tunable property of biosensors.
To gain further insights into the different DR-μ dependencies, we developed a kinetic model to describe the intracellular concentration of the biosensor’s output protein (G) and the intracellular concentrations of TF repressor (R) and target metabolite (MIn) at different growth rates. The volumetric production rate of each species is balanced by dilution due to cellular volume expansion during cell growth. Additionally, transport of the target metabolite across cell membrane via either passive transport by diffusion or enzyme-facilitated active transport were also considered. For active transport, the concentration of protein transporters can also be affected by dilution due to cell growth. For a detailed description of the model, see Supplemental Note 1.1.
In the absence of the target metabolite, an analytical solution can be derived to describe the minimum output (Gmin) at steady state (see Supplemental Note 1.2 for details):
| (1) |
where μ represents cell growth rate. Parameters bG, aG, and KG are intrinsic to the biosensor and describe the basal expression level, the strength of activated expression, and the MRTF-promoter dissociation constant, respectively (Fig. 1B). bR describes the constitutive expression rate of MRTF. While the production rate of the MRTF is constant with growth, the total concentration of MRTF (R = bR / μ) is decreasing due to dilution by cell growth (see Supplemental Note 1.1 and 1.2 for details). Because both the biosensor output protein (G) and its repressor (R) are being diluted by growth, we calculate the dependence of Gmin on μ to understand the combined effect:
| (2) |
The dependence of Gmin on μ is negative for all parameter values. Thus, Gmin always decreases with growth rate, and is consistent with our experimental observation for all three biosensors (Fig. 2D, 2E, 2F). The dependence of maximum output and DR on growth rate depend on sensor parameters and are not monotonic. A biosensor’s DR can be described as:
| (3) |
where KR represents the dissociation constant between the target metabolite and MRTF. Finally, Min describes the intracellular target metabolite concentration which can also be a function of the growth rate, depending on the target metabolite’s transport kinetics. To understand how the model parameters affect the DR-μ dependency, we next calculate the derivative of DR with respect to growth rate μ (dDR/dμ):
| (4) |
Our modeling result suggests that depending on the parameter values of a biosensor and its intracellular target metabolite concentration, dDR/dμ can be either positive (increasing with growth rate) or negative (decreasing with growth rate), depending on the sign of S:
| (5) |
Tuning DR-μ Dependence Through TF Expression
To understand how sensor parameters affect the growth dependence of Gmin, Gmax, and of the biosensor’s DR (i.e. dDR/dμ), we first targeted to the aTc-inducible sensor and tuned the production rate of the MRTF repressor TetR (i.e. bR). According to our model, both Gmin and Gmax should decrease with increasing bR at a fixed growth rate, regardless of other parameters (see Equation (1) and (S12)). Experimentally, we placed TetR under the control of a constitutive promoter and a library of RBS with different strengths to vary the TetR production rate (Fig. 3A), while not changing the promoter or RBS sequence of the biosensor output gene. When growing the library of cells at a fixed growth rate (in glycerol minimal medium), both Gmin and Gmaxdecreased with increasing TetR levels for 3.7- and 1.2-fold change across the library, respectively (Fig. 3B, 3C), which matches our analytical model predictions (Fig. 3E, 3F). Our model also shows that Gmin is sensitive to changes in TetR production rate at low levels of TetR, while it becomes less sensitive at higher TetR levels (Fig. 3E), which matches our observation of high sensitivity in Gmin to TetR expression when TetR is low (Fig. 3B). We further observe that under our experimental parameter regime, Gmin is more sensitive to changes in TetR production rate than Gmax (Fig. 3B, 3C), thus affecting DR mostly by lowering the leaky expression level and leading to an increasing DR (Fig. 3D), consistent with model prediction (Fig. 3G). Additionally, we fit our model to the experimentally measured TetR expression, Gmin and Gmax (see Supplemental Note 2.2 for details). From the fitted model, we calculated the DR and observed a good agreement between the model and the observed DR-TetR expression trend (R2 = 0.49). Overall, these experimental observations are qualitatively consistent with numerical simulations with increasing bR values and with model fitting, thus validating our model.
Figure 3. Tuning biosensor DR through changing TetR expression level at a constant growth rate.
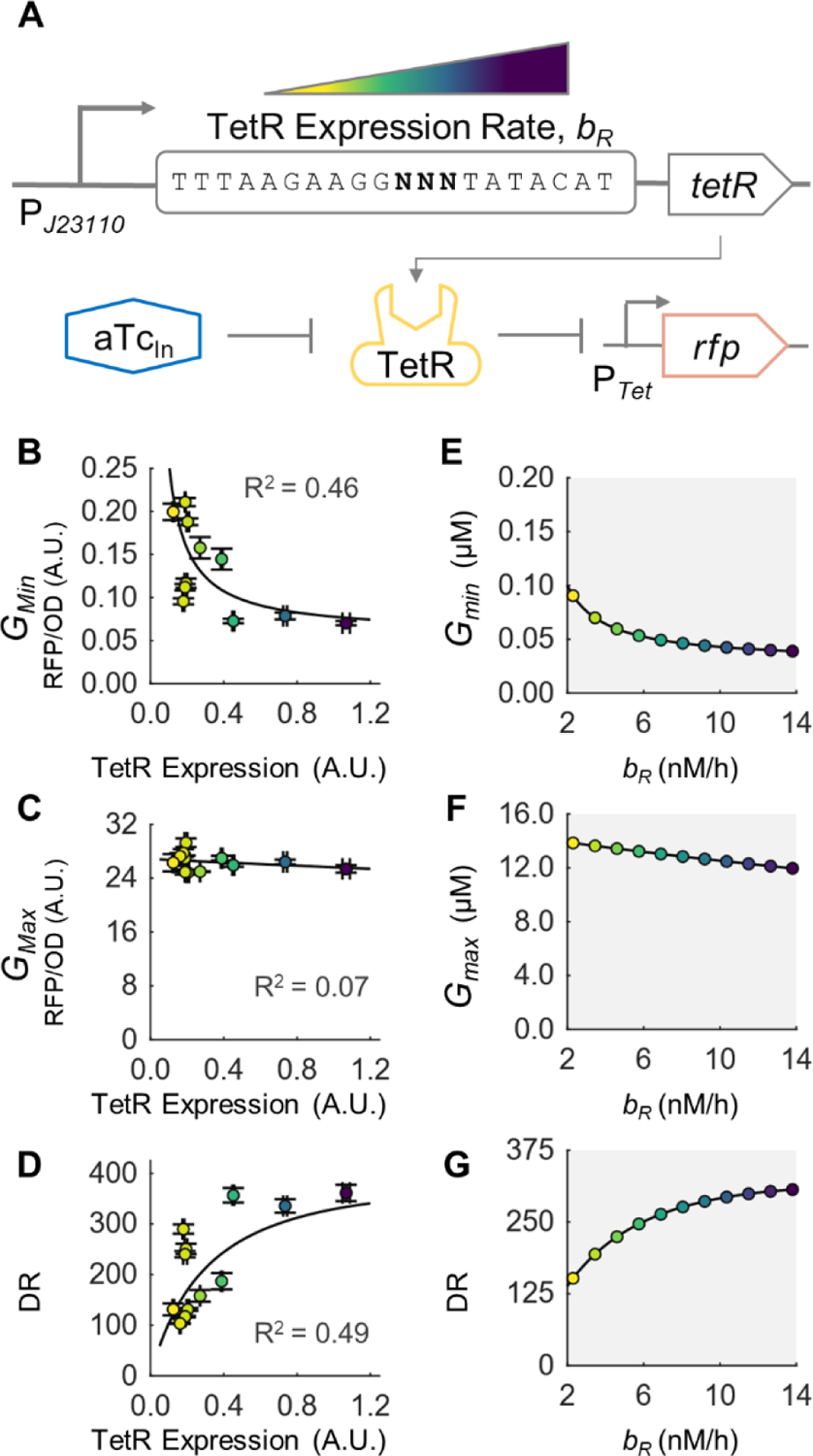
(A) Schematic of the TetR gene used to tune TetR expression level (bR). The TetR level was varied by using a library of TetR genes with different RBS strength. (B-D) Experimentally measured (B) minimum biosensor output, Gmin, (C) maximum biosensor output, Gmax, and (D) dynamic range of TetR RBS library members with varying TetR expression levels. Error bars represent S.D., n=3. RBS sequences and their corresponding TetR expression levels, Gmin, Gmax, and DR are given in Supplemental Table S6. Solid line is fit of model to the experimental data (details in Supplemental Note 2.2) (E-F) Numerical model simulations of (E) minimum output, (F) maximum output, and (G) DR for different values of bR. Parameter values and ranges are given in Supplemental Table S1.
We next measured how Gmin, Gmax, and DR change with growth rate under different TetR production rate (Fig. 4A). According to Equation (2), as bR increases, the dependence of Gmin on μ is expected to be less negative. Experimentally, we indeed observed a less negative slope on the Gmin- μ plot with a higher TetR expression (Fig. 4B). This result indicates that when TetR production is insufficient (low TetR production), leaky expression becomes worse at lower growth rates, while a high TetR production rate can make the sensor less leaky across a wide growth rate range. The dependence of Gmax on growth rate was negative and had similar slope for low and medium TetR production rates and a reduced slope at high TetR production rates (Fig. 4C). This result indicates that at high TetR expression, maximal induction may not be achieved at lower growth rates. Finally, Equation (5) predicts that lowering the TetR production rate bR should reduce the DR-μ dependence, changing it towards a negative regime. Our experimental results indeed showed a less positive dDR/dμ value at a lower TetR production rate (Fig. 4D). Overall, our result showed that for the aTc biosensor, a high TetR production rate provides high DR, however at a cost of higher growth dependence.
Figure 4. The growth rate dependence of DR for TetR-based biosensors with varying TetR production rates.
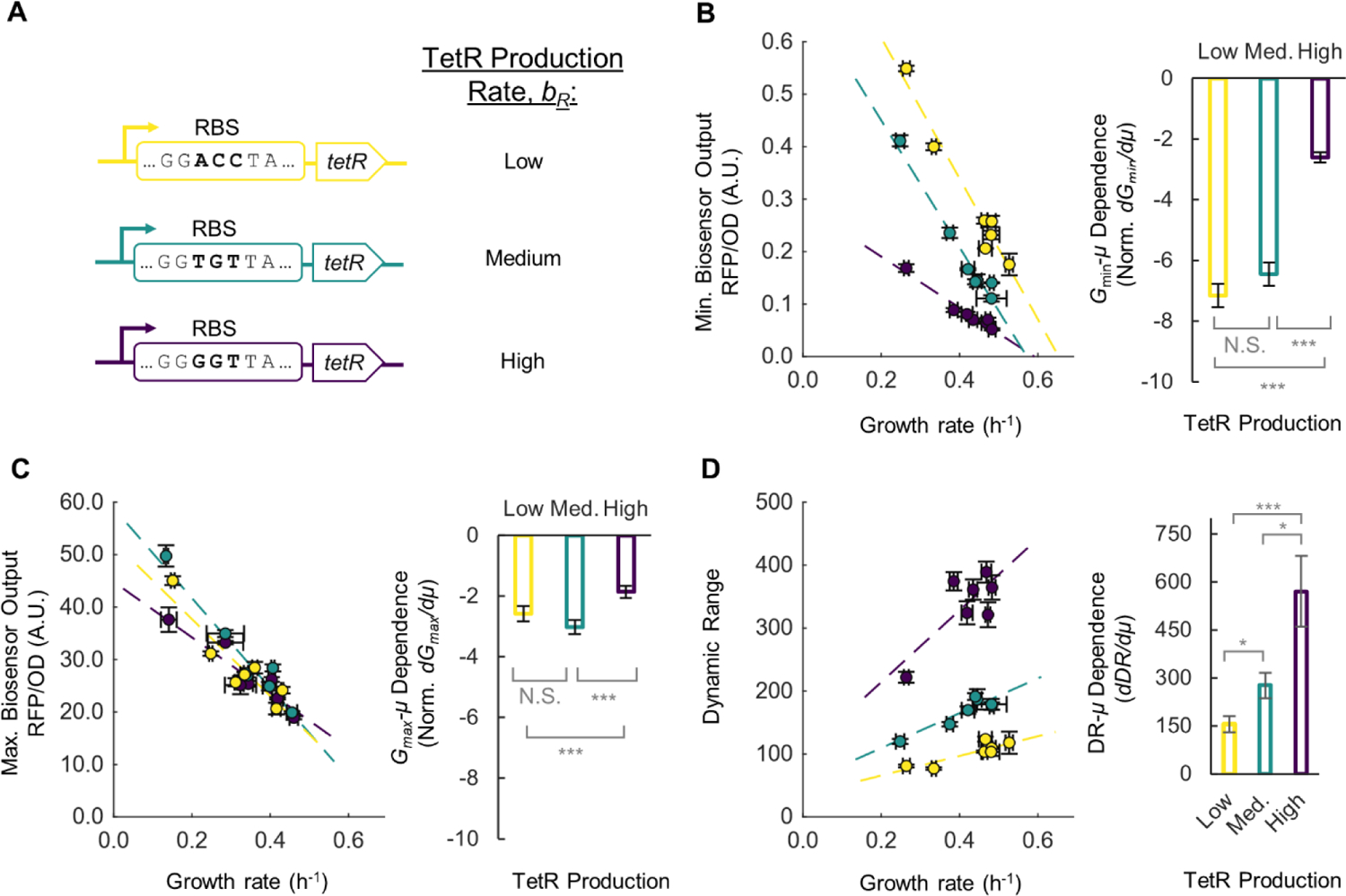
(A) Three tetR genes with different RBS strength to produce TetR at low (yellow), medium (green), and high (purple) levels. (B) Minimum biosensor output at varying growth rate for each library member (left) and the dependence of minimum output on growth rate (right). (C) Maximum biosensor output at varying growth rate for each library member (left) and the dependence of the maximum output on growth rate (right). (D) DR at varying growth rate for each library member (left) and the dependence of DR on growth rate (right). dGmin/dμ and dGmax/dμ are normalized to the average Gmin and Gmax, respectively, from all data. Error bars of individual data points are S.D., n=3. Error bars for Gmin, Gmax, and DR-μ dependence are standard error of the slope. A Student’s t-test was conducted between each pair of growth rate dependence data, stars indicate significant difference (N.S., not significant; * p < 0.05; ** p < 0.01; *** p < 0.001).
Metabolite Transport Affects DR-μ Dependence
Our experimental results on the aTc sensor showed that DR increased with both growth rate and TetR production rate. We next examine which parameter space can simultaneously support these trends and whether such trends can inform us on the parameters of the target metabolite transport mechanism. An extracellular target metabolite can enter or exit a cell via either passive transport (metabolite concentration gradient related) or protein-facilitated active transport (growth rate related), or a combination of both mechanisms. If passive transport is the only mechanism, the intracellular target metabolite concentration is mostly determined by extracellular target metabolite concentration and has little influence from growth rate. In this case, our mathematical analysis indicates that sensor DR cannot increase with both growth rate and MRTF production rate (see Supplemental Note 1.3). On the other hand, DR can increase with growth rate and MRTF production rate if intracellular aTc concentrations are also increasing with growth rate (see Supplemental Note 1.3). One way this could occur is when aTc membrane transport involves proteins whose steady state concentration is regulated by growth rate via cell dilution. Indeed, previous studies demonstrated that aTc can be exported by the AcrAB-TolC multi-drug efflux pump52, and demonstrate that the expression of acrAB is elevated at lower growth rates53. Together, this may lead to reduced aTc concentrations at low growth rates due to elevated efflux. To elucidate this mechanism, we incorporated both passive transport and active export to our model (Fig. 5A) and explored the parameter space that provides positive dependence of DR on both growth rate and TetR production rate. When active export is incorporated in the model, intracellular target metabolite becomes growth-dependent, which causes the gradient-dependent passive export to also be growth-dependent (Fig. S1A). Both passive and active transport rates were varied over 4-orders of magnitude to explore a large parameter space, starting from parameter settings where the target metabolite’s passive and active export rates are equal and considering different target metabolite concentration regimes (Min » KM,ex, Min = KM,ex, or Min « KM,ex, Fig. S1A). Our parameter space covers three modes of target metabolite export, where metabolite export is primarily through the active pathway, the passive pathway, or a mix of both active and passive pathways (Fig 5B). Modeling results show that sensor DR increased as passive transport rate increased and active export decreased (Fig. 5C). This is because intracellular target metabolite concentration is high when the passive transport rate is higher than the active export rate (Fig. S1B). Interestingly, when active export rate is similar to or higher than passive transport rate, DR is more likely to positively correlate with growth (Fig. 5D). And when active export rate is lower than passive transport rate, DR is more likely to positively correlate with TetR production rate (Fig. 5E). As a result, DR can positively correlate with both cell growth and TetR production rate only when active export rate is similar to passive import (Fig. 5F). Our modeling results further show that dMin/dμ strongly increases when active export rate is similar to passive import (Fig. S1C), which is a necessary condition to observe simultaneous positive correlation for DR with cell growth and TetR production rate (see Supplemental Note 1.3). Thus, the aTc biosensor used in our experiments likely fall into this parameter range.
Figure 5. Modeling the impact of passive and active transport mechanism on DR-growth rate dependence for aTc-type biosensors.
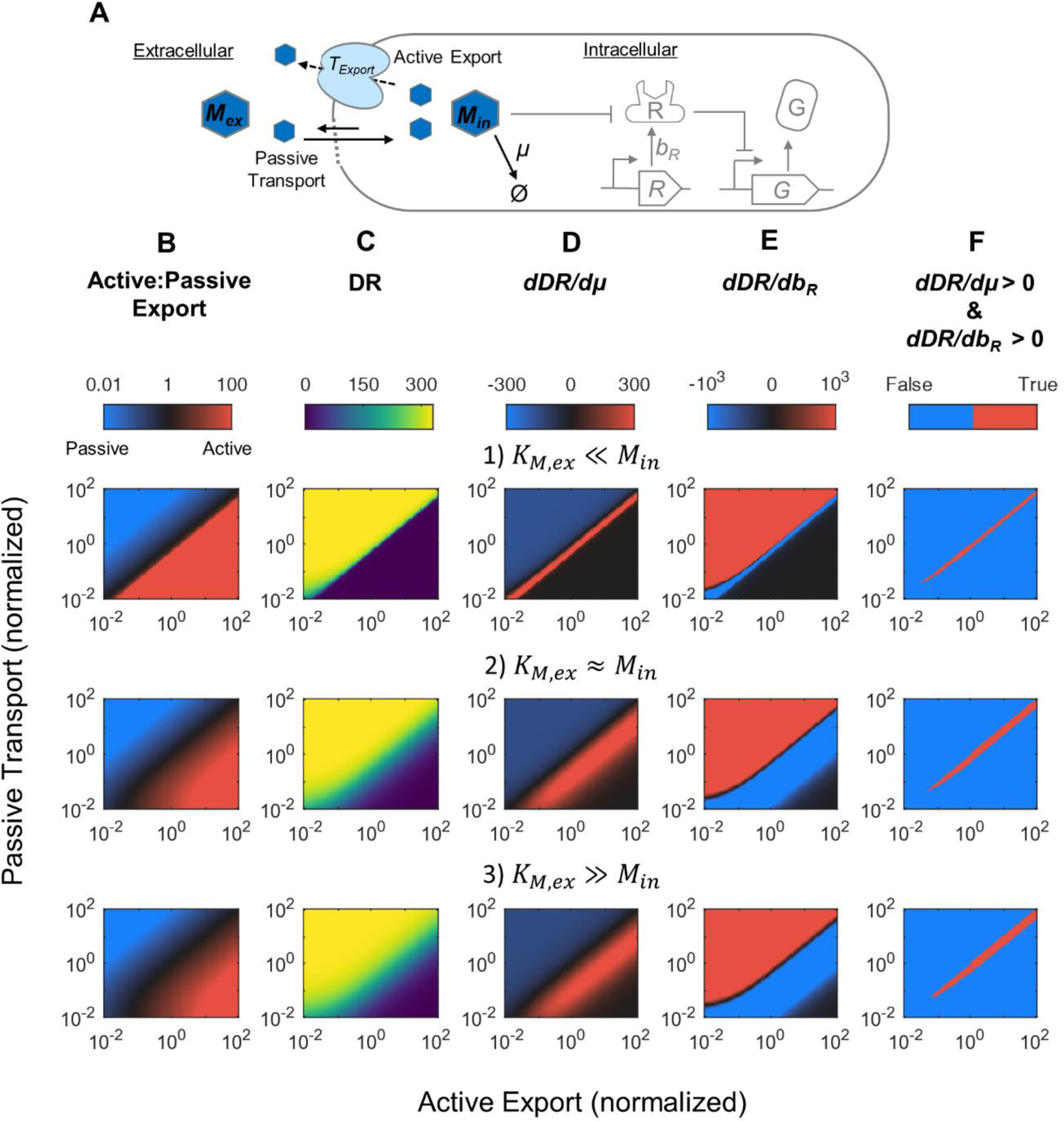
(A) Diagram of transport reactions including passive import of metabolite by diffusion and active export facilitated by a protein transporter. (B) Ratio of active to passive export rates, (C) Calculated DR, (D) dDR/dμ, (E) dDR/dbR, and (F) Parameter region where both dDR/dμ and dDR/dbR are increasing, under different parameter space. The parameters kdiff and kcat,ex were increased to increase passive and active transport rate, respectively. KM,ex was varied to represent three scenarios where its value is either greater, similar, or smaller than the intracellular target metabolite concentration (Min). See Supplemental Fig. S1 and Supplemental Note 2.1 for details. Parameters values are given in Supplemental Table S1.
In contrast to the aTc sensor, many chemicals cannot diffuse across cell membrane passively, but solely rely to protein transporters and enzyme conversion to turn on the sensor. For example, the FA sensor requires a membrane-associated acyl-CoA synthase to convert extracellular FA into intracellular acyl-CoA, which binds to FadR, the MRTF, to turn on reporter expression. Thus, to understand the FA sensor, we modified our model to include both enzyme-based import and export mechanisms without passive transport (Fig. 6A). Similar to the aTc model, DR increases when the active import rate is higher than export rate (Fig. 6B), leading to higher intracellular target metabolite (acyl-CoA) concentration (Fig. 6C). In contrast to aTc-type of sensors, the DR of FA-type of sensors has a negative dependence on cell growth when import rate is higher than export rate, which should occur under most cases since faster target metabolite import is necessary for high biosensor induction (Fig. 6D). Thus, these modeling results are consistent with our experimental results. Additionally, when import is higher than export, the intracellular target metabolite growth dependence (dMin/dμ) is strongly negative (Fig. 6E) indicating a large potential drop in the maximum biosensor output at higher growth rates, which is also consistent with experiments.
Figure 6. Modeling the impact of active transport mechanism on GR-growth rate dependence for FA-type biosensors.
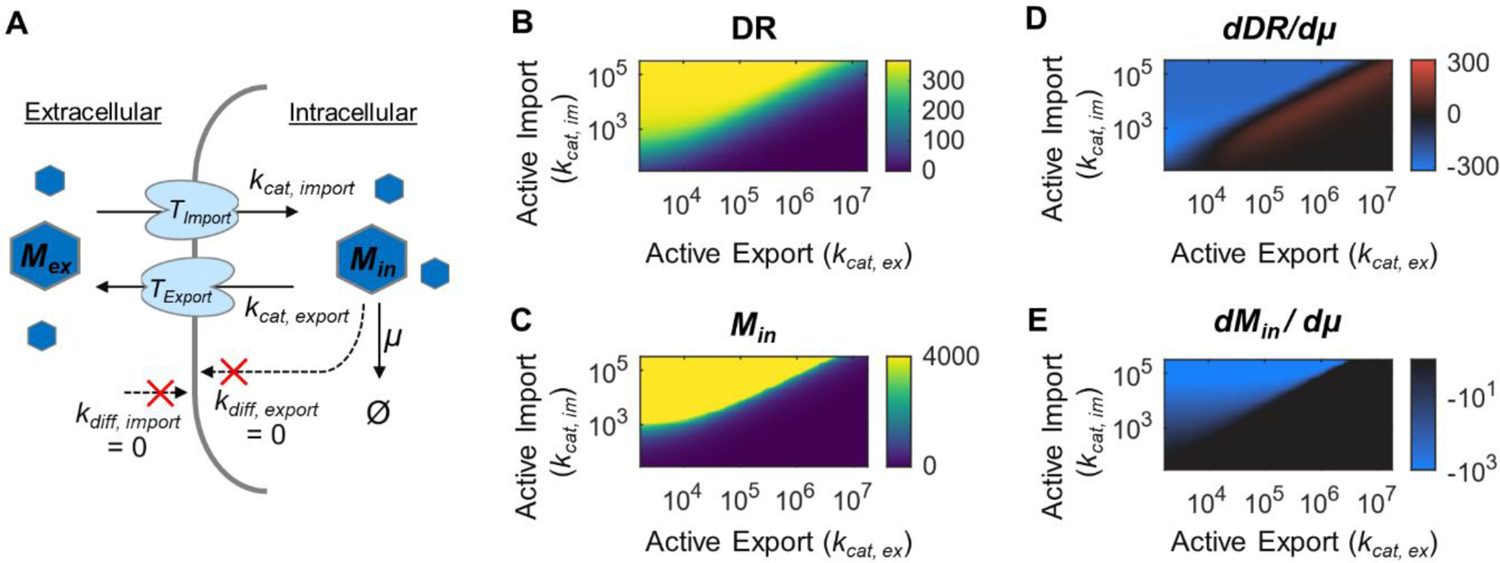
(A) Diagram of transport reactions. Target metabolite transport only occurs by protein-facilitated active mechanisms. (B) Model simulated DR under different import and export rates. All other parameters are held constant and are given in Supplemental Table S1. (C) Intracellular target metabolite concentration (Min), (D) DR-μ dependence, and (E) Intracellular metabolite-μ dependence, under different parameter space.
DR and DR-μ Are Sensitive to Similar Parameters
Thus far, our modeling and experimental results have shown good qualitative agreement and have predicted the effects of bR and transport mechanisms on both DR and DR-μ dependence. In order to gain a deeper understanding of the other parameters effects on DR and DR-μ dependence, we conducted a global parameter sensitivity analysis, for a transport mechanism with both passive and active transport (Supplemental Note 3). Our results show that both the DR (Fig. S2A) and dDR/dμ (Fig. S2B) are most sensitive to parameters intrinsic to the biosensor (aG, bG, and KG), the MRTF expression level, and parameters in MRTF-metabolite interaction (bR, ksr, ksf). The similarity in parameter sets which strongly affect DR and dDR/dμ suggest both are tightly coupled. These results are consistent with our observation of the TetR system where increasing TetR production rate tuned both the DR and the DR-μ dependence in the same direction.
Discussion
Change in cellular growth rate often affects the performance of biosensors and cause problems when using biosensors in a different nutrient or environment54. In this work, we characterized the growth-rate dependent behavior of three commonly-used MRTF-based biosensors through changing the growth medium of the biosensors. Our results show that DR of the aTc-TetR and IPTG-LacI sensors have positive correlations with cell growth rate. In industrial-scale bioreactor settings where growth rates are typically lower than that in rich medium of lab settings, these sensors will have lower DRs and leakier expression before induction (Fig. 2D, 2E)55,56. Thus, these sensors may not be suitable for controlling toxic genes or burdensome pathways whose leaky expression can cause undesirable mutations to deactivate the strain’s function. In contrast, DR of the FA-FadR biosensor has a negative correlation with cell growth rates. A higher DR at lower growth rate can benefit the sensor’s application, although its leaky expression was also high at lower growth rate (Fig. 2F).
In this work we used a simplified kinetic model for understanding the DR-μ dependency. The core modeling approach expands upon previous phenomenological models which were successful in capturing design constraints between various tunable molecular interactions and biosensor dose-response parameters26. We extended these phenological models with kinetic interactions between TF and intracellular target metabolite, as well as metabolite transport, which have been shown to capture key dynamic features of bacterial response to external metabolites47. Many of the model parameters are related to key biological processes that can be tuned experimentally to optimize a sensor’s performance, such as the TF-operator interactions57,58 and the presence of exporters59,60. Our model captures new design constraints for tuning DR across growth rates. For example, while the DR can be increased by reducing the minimum output through a stronger binding of the MRTF to the biosensor promoter, our model predicts this can result in a reduction of the maximum output at some growth rates, because there is a limit to how much the sensor promoter can be activated by the intracellular target metabolite. Thus, once model parameters are characterized at a single growth rate, the model can be used to predict the change in DR under different medium compositions by measuring the growth rate, allowing engineers to check for a potential loss of sensor performance without the need for lengthy experimental re-characterization of the biosensor at different growth rates or under different growth mediums. Additionally, while our results focus on biosensor’s with a repressed-repressor architecture, previous phenomenological models26 can be similarly extended following our procedures to identify growth-dependent design constraints for other biosensor architectures, such as the commonly used activated-activator architecture.
Importantly, our model also highlights some fundamental couplings in biosensor’s performance across varying growth rates, which are important when engineering a MRTF-based sensor. For both types of sensors explored in this study, strategies in increasing the DR always led to stronger growth rate dependence of DR (regardless of whether the dependence is positive or negative), thus making the DR more sensitive to changes in growth (Fig. 4D, Fig. 5, Fig. 6). Therefore, there is a trade-off between high DR and low DR-μ sensitivity, which is important in applications when the value of the DR itself is critical. As an example, MRTF-based biosensors used for high-throughput screening usually require high DRs to reduce the identification of false-positive and false-negative strains25,61. However, a sensor with too high of a DR also has high DR-μ sensitivity, which will lead to a high false-hit rate when screening under different conditions. Thus, reducing DR in exchange for a less sensitive DR-μ dependence may present a better strategy in sensor design.
Additionally, our model suggests that transport of the target metabolite plays important roles in the growth rate dependence of DR. Different biosensors with similar underlying parameters can show different DR-μ dependencies. For biosensors where the target metabolite’s transport is controlled by passive diffusion and active export (e.g. aTc), there is a narrow parameter region where increasing TF expression can improve DR while also maintaining a positive DR-μ dependence (Fig. 5E). In contrast, for biosensors where intracellular target metabolite levels are controlled solely by enzymes (e.g. FA), the metabolite-μ dependence is negative over a large parameters space, so DR cannot be improved by increasing TF levels without leading to a more negative DR-μ dependence under these conditions. Thus, our model highlights metabolite transport or production as an important consideration for understanding the impact of growth on a sensor’s DR.
MRTF-based biosensors are key components of synthetic biology systems and enable a diverse range of sensing applications. These applications require sensors with robust and predictable operation under a wide range of nutrient conditions and growth environments. Our work has uncovered new design considerations and trade-offs for metabolite biosensors under changing growth conditions, which will be found useful for a wide range of metabolic engineering and synthetic biology applications.
Methods
Materials.
Phusion polymerase, restriction enzymes, and T4 ligase were purchased from Thermo Fisher Scientific (Waltham, MA, U.S.A.). DNA primers were synthesized by Integrated DNA Technologies (Coralville, IA, U.S.A.). DNA plasmid miniprep kits were purchased from iNtRON Biotechnology (Burlington, MA, U.S.A). All other reagents were purchased from Sigma-Aldrich (St. Louis, MO, U.S.A.)
Plasmids and Strains.
E. coli MDS42pdu62 was used to clone and isolate plasmids. E. coli MG1655 was used to host biosensor plasmids. All plasmids were constructed through PCR amplification and standard Golden-Gate DNA assembly techniques. Plasmids were transformed into electro-competent strains by electroporation and selected on LB agar plates with corresponding antibiotics (ampicillin, 100 mg/L; kanamycin, 50 mg/L; streptomycin 100 mg/L). Plasmids (Table S4), strains (Table S5), and sequences of constructed plasmids are given in the supplementary information (Supplemental Note 5).
FA biosensor construction.
The FA-FadR biosensor was created in E. coli strain MG1655 by modifying a previous FA sensor47. These modifications include the deletion of fadE, replacement of fadD’s native promoter with a constitutive promoter Ppro4 to deactivate the positive feedback loop in FA transport47, and deletion of the negative autoregulation of FadR63. These genome modifications were performed using CRISPR-Cas9 following previous methods64,65 using pTargetF plasmids as listed in Table S4. The sequence of Ppro4 was taken from a library of insulated constitutive promoters66 and constructed from PCR of overlapping primers. Deletion of the negative autoregulation of FadR was done by replacing fadR’s native promoter with the constitutive PfadRm1 (Supplemental Note 5.1). Additionally, the constitutively controlled fadR was cloned to the pA6a BglBrick plasmid, resulting in plasmid pAfadRm1a-fadR (Supplemental Note 5.1 for full sequence).
TetR RBS library construction.
A library of plasmids with varying TetR RBS strength (namely pSk-PTet-rfp-PJ23110-RBSLibrary-tetR) was constructed by introducing a terminator, followed by the constitutive Bba_J23110 promoter, followed by an RBS 3’ of pTetR promoter and 5’ of the TetR coding sequence on a pS2k-rfp plasmid. Terminator and promoter were constructed from overlapping primers. The RBS library was then introduced by primers with degenerate nucleotide sequences. To evaluate the RBS strength, we constructed another plasmid library, namely pSk-PJ23110-RBSLibrary-rfp, by replacing the tetR gene of pSk-PJ23110-RBSLibrary-tetR with rfp. This library was constructed by individually amplifying the promoter, RBS sequence and first 30 codons of the TetR coding sequence from aTc-biosensor-TetR-RBS-library members which had characterized minimum and maximum outputs. These sequences were then individually cloned 5’ of rfp and introduced to a BglBrick backbone with SC101 origin and kanamycin resistance. The final constructed sequences are shown in Supplemental Note 5.1. A list of the tested RBS sequences is given in Supplemental Table S6.
Cell Growth and Induction Conditions.
Cells were cultivated in different growth media following previous protocols67,68. Specifically, single colonies from an overnight Luria-Bertani (LB) plate were cultivated for 3–5 hours in LB medium (225 rpm, 37 °C) supplemented with appropriate antibiotic. Cells were then washed twice by centrifugation (4500 rcf, 3 minutes) into M9 minimal medium supplemented with 1% (w/v) glycerol and diluted to OD600 ~0.04 and grown overnight. Overnight cultures were again washed in M9 media without supplemented carbon. To achieve different growth rates, washed cells were transferred to M9 media supplemented with the following nutrient sources: 75 mM sodium acetate, 20 mM sodium pyruvate, 1% Glycerol, 20 mM sorbitol, 15 mM sodium succinate, 1% glycerol + 7 amino acids (0.8 mM Glycine, 0.2 mM Histidine, 0.4 mM isoleucine, 0.8 mM leucine, 0.4 mM lysine, 0.2 mM methionine, 0.4 mM phenylalanine), 20 mM xylose. For the FadR-based biosensor only, media was supplemented with 0.5% Tergitol NP-40 and the following nutrient sources were used: 75 mM sodium acetate, 1% glycerol, 1% glycerol+7 amino acids, and 0.4% glucose. A summary of growth rates achieved for each media condition is given in Supplemental Table S3. To induce biosensor activity, 1000 nM anhydrotetracycline (aTc), 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), and 4 mM of sodium oleate, were added to the media for the aTc, IPTG, and FA sensors, respectively.
Biosensor Growth Rate, Minimum, Maximum, and Dynamic Range Assays.
Cells growing in different carbon sources, with or without the target metabolite were diluted to OD600 at ~0.0007. Exponentially growing cells were transferred to a Falcon 96-Well Imaging Microplate (Corning, NY, U.S.A.). An Infinite F200PRO plate reader (TECAN, Männedorf, Switzerland) was used to take automated OD600 and red fluorescence measurements (Excitation: 584±9 nm, Emission: 620±20 nm) every 15 minutes with constant shaking at 37°C. Following previously established procedures for calculating growth rate67,68 and biosensor output26, growth rate was calculated as the slope of natural log(OD600) from OD600 0.1 to 0.4. RFP values were normalized by the OD600, and the minimum and maximum RFP/OD values were calculated as the average of RFP/OD measurements over the same OD600 range (0.1–0.4). The dynamic range was calculated from the minimum and maximum outputs measured from the same biological replicate (colony).
TetR RBS library characterization.
Colonies of freshly transformed aTc-biosensor-TetR-RBS-library members were picked and grown following the cell growth and induction methods in M9 media supplemented with 1% glycerol. The minimum, maximum, and dynamic range of several colonies were measured, and colonies with different dynamic range were chosen for further characterization. The RBS sequence was determined by Sanger sequencing to confirm uniqueness of tested library members and duplicate members were combined. The minimum, maximum, and DR of the sequence verified library members were then re-measured with biological triplicates in 1% glycerol medium. To get the TetR expression level, TetR expression library members were individually constructed, and then also grown in M9 with 1% glycerol, and the growth rate and steady state RFP/OD were measured by plate reader. A summary of the RBS sequences characterized, and their TetR expression level, minimum and maximum outputs is provided in Supplemental Table S6.
Dynamic Range Modeling and Simulations.
Details of the model development and parameterizations are given in Supplemental Note 2.1. Kinetic model simulations were performed using MATLAB 2020b ode15s (The Mathworks, Natick, MA, U.S.A.) from initial conditions where the concentration each species is zero. Simulations were run for 109 seconds to reach steady state, and the end point of each simulation was used as the model output. To obtain the model minimum and maximum outputs, the model was run with either 0 μM or 4000 μM external target metabolite (Mex), respectively. dDR/dbR was calculated directly from the simulations using Equation (S21), dDR/dμ was calculated numerically using a 5-point central difference formula by running 5 simulations at nearby μ (equally spaced on the range μ ± 0.0039).
Supplementary Material
Acknowledgments
This work is supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R35GM133797.
Abbreviations
- aTc
anhydrotetracycline
- DR
dynamic range
- DR-μ
Dynamic range-growth rate
- FA
fatty acid
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- MRTF
metabolite-responsive transcription factor
Footnotes
- Supplemental Notes 1–4: Details of model construction, derived results, model parameterization, model fitting, and parameter sensitivity analysis, Supplemental Figures S1–S3, Supplemental Tables S1–S2. Supplemental Note 5: Supplemental Tables S3–S6 of growth conditions, strains, plasmids, and DNA sequences of selected plasmids.
The authors declare no competing financial interest.
References
- (1).Xu T, Close DM, Sayler GS, and Ripp S (2013) Genetically modified whole-cell bioreporters for environmental assessment. Ecol. Indic 28, 125–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Layton AC, Muccini M, Ghosh MM, and Sayler GS (1998) Construction of a bioluminescent reporter strain to detect polychlorinated biphenyls. Appl. Environ. Microbiol 64, 5023–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Rogers JK, and Church GM (2016) Genetically encoded sensors enable real-time observation of metabolite production. Proc. Natl. Acad. Sci. U. S. A 113, 2388–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Xiao Y, Jiang W, and Zhang F (2017) Developing a genetically encoded, cross-species biosensor for detecting ammonium and regulating biosynthesis of cyanophycin. ACS Synth. Biol 6, 1807–1815. [DOI] [PubMed] [Google Scholar]
- (5).Raman S, Rogers JK, Taylor ND, and Church GM (2014) Evolution-guided optimization of biosynthetic pathways. Proc. Natl. Acad. Sci 111, 17803–17808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Cheng F, Tang X-L, and Kardashliev T (2018) Transcription factor-based biosensors in high-throughput screening: Advances and applications. Biotechnol. J 13, 1700648. [DOI] [PubMed] [Google Scholar]
- (7).Bentley GJ, Narayanan N, Jha RK, Salvachúa D, Elmore JR, Peabody GL, Black BA, Ramirez K, De Capite A, Michener WE, Werner AZ, Klingeman DM, Schindel HS, Nelson R, Foust L, Guss AM, Dale T, Johnson CW, and Beckham GT (2020) Engineering glucose metabolism for enhanced muconic acid production in Pseudomonas putida KT2440. Metab. Eng 59, 64–75. [DOI] [PubMed] [Google Scholar]
- (8).Liu D, Xiao Y, Evans BS, and Zhang F (2015) Negative feedback regulation of fatty acid production based on a malonyl-CoA sensor–actuator. ACS Synth. Biol 4, 132–140. [DOI] [PubMed] [Google Scholar]
- (9).Schmitz ACAC, Hartline CJCJ, and Zhang F (2017) Engineering microbial metabolite dynamics and heterogeneity. Biotechnol. J 12, 1700422. [DOI] [PubMed] [Google Scholar]
- (10).Liang C, Zhang X, Wu J, Mu S, Wu Z, Jin J-M, and Tang S-Y (2020) Dynamic control of toxic natural product biosynthesis by an artificial regulatory circuit. Metab. Eng 57, 239–246. [DOI] [PubMed] [Google Scholar]
- (11).Wu Y, Chen T, Liu Y, Tian R, Lv X, Li J, Du G, Chen J, Ledesma-Amaro R, and Liu L (2020) Design of a programmable biosensor-CRISPRi genetic circuits for dynamic and autonomous dual-control of metabolic flux in Bacillus subtilis. Nucleic Acids Res. 48, 996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Hartline CJ, Schmitz AC, Han Y, and Zhang F (2021) Dynamic control in metabolic engineering: Theories, tools, and applications. Metab. Eng 63, 126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Verma BK, Mannan AA, Zhang F, and Oyarzún DA (2022) Trade-offs in biosensor optimization for dynamic pathway engineering. ACS Synth. Biol 11, 228–240. [DOI] [PubMed] [Google Scholar]
- (14).Chou HH, and Keasling JD (2013) Programming adaptive control to evolve increased metabolite production. Nat. Commun 4, 1–8. [DOI] [PubMed] [Google Scholar]
- (15).Xiao Y, Bowen CH, Liu D, and Zhang F (2016) Exploiting nongenetic cell-to-cell variation for enhanced biosynthesis. Nat. Chem. Biol 12, 339–344. [DOI] [PubMed] [Google Scholar]
- (16).Li S, Si T, Wang M, and Zhao H (2015) Development of a synthetic malonyl-CoA sensor in Saccharomyces cerevisiae for intracellular metabolite monitoring and genetic screening. ACS Synth. Biol 4, 1308–1315. [DOI] [PubMed] [Google Scholar]
- (17).Liang C, Xiong D, Zhang Y, Mu S, and Tang SY (2015) Development of a novel uric-acid-responsive regulatory system in Escherichia coli. Appl. Microbiol. Biotechnol 99, 2267–2275. [DOI] [PubMed] [Google Scholar]
- (18).Liu D, Evans T, and Zhang F (2015) Applications and advances of metabolite biosensors for metabolic engineering. Metab. Eng 31, 35–43. [DOI] [PubMed] [Google Scholar]
- (19).Thompson MG, Costello Z, Hummel NFC, Cruz-Morales P, Blake-Hedges JM, Krishna RN, Skyrud W, Pearson AN, Incha MR, Shih PM, Garcia-Martin H, and Keasling JD (2019) Robust characterization of two distinct glutarate sensing transcription factors of Pseudomonas putida L-lysine metabolism. ACS Synth. Biol 8, 2385–2396. [DOI] [PubMed] [Google Scholar]
- (20).Hanko EKR, Paiva AC, Jonczyk M, Abbott M, Minton NP, and Malys N (2020) A genome-wide approach for identification and characterisation of metabolite-inducible systems. Nat. Commun 11, 1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Taylor ND, Garruss AS, Moretti R, Chan S, Arbing MA, Cascio D, Rogers JK, Isaacs FJ, Kosuri S, Baker D, Fields S, Church GM, and Raman S (2016) Engineering an allosteric transcription factor to respond to new ligands. Nat. Methods 13, 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Koch M, Pandi A, Borkowski O, Cardoso Batista A, and Faulon JL (2019) Custom-made transcriptional biosensors for metabolic engineering. Curr. Opin. Biotechnol 59, 78–84. [DOI] [PubMed] [Google Scholar]
- (23).Flachbart LK, Gertzen CGW, Gohlke H, and Marienhagen J (2021) Development of a biosensor platform for phenolic compounds using a transition ligand strategy. ACS Synth. Biol 10, 2002–2014. [DOI] [PubMed] [Google Scholar]
- (24).Chen Y, Ho JML, Shis DL, Gupta C, Long J, Wagner DS, Ott W, Josić K, and Bennett MR (2018) Tuning the dynamic range of bacterial promoters regulated by ligand-inducible transcription factors. Nat. Commun 9, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Dabirian Y, Li X, Chen Y, David F, Nielsen J, and Siewers V (2019) Expanding the dynamic range of a transcription factor-based biosensor in Saccharomyces cerevisiae. ACS Synth. Biol 8, 1968–1975. [DOI] [PubMed] [Google Scholar]
- (26).Mannan AA, Liu D, Zhang F, and Oyarzún DA (2017) Fundamental design principles for transcription-factor-based metabolite biosensors. ACS Synth. Biol 6, 1851–1859. [DOI] [PubMed] [Google Scholar]
- (27).Liu D, Mannan AA, Han Y, Oyarzún DA, and Zhang F (2018) Dynamic metabolic control: towards precision engineering of metabolism. J. Ind. Microbiol. Biotechnol 45, 535–543. [DOI] [PubMed] [Google Scholar]
- (28).Lo TM, Chng SH, Teo WS, Cho HS, and Chang MW (2016) A two-layer gene circuit for decoupling cell growth from metabolite production. Cell Syst. 3, 133–143. [DOI] [PubMed] [Google Scholar]
- (29).Zhou S, Yuan S-F, Nair PH, Alper HS, Deng Y, and Zhou J (2021) Development of a growth coupled and multi-layered dynamic regulation network balancing malonyl-CoA node to enhance (2S)-naringenin biosynthesis in Escherichia coli. Metab. Eng 67, 41–52. [DOI] [PubMed] [Google Scholar]
- (30).Brophy JAN, and Voigt CA (2014) Principles of genetic circuit design. Nat. Methods 11, 508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Klumpp S (2011) Growth-rate dependence reveals design principles of plasmid copy number control. PLoS One 6, e20403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Scott M, Klumpp S, Mateescu EM, and Hwa T (2014) Emergence of robust growth laws from optimal regulation of ribosome synthesis. Mol. Syst. Biol 10, 747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Schmidt A, Kochanowski K, Vedelaar S, Ahrné E, Volkmer B, Callipo L, Knoops K, Bauer M, Aebersold R, and Heinemann M (2016) The quantitative and condition-dependent Escherichia coli proteome. Nat. Biotechnol 34, 104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Klumpp S, Zhang Z, and Hwa T (2009) Growth rate-dependent global effects on gene expression in bacteria. Cell 139, 1366–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Klumpp S, and Hwa T (2014) Bacterial growth: Global effects on gene expression, growth feedback and proteome partition. Curr. Opin. Biotechnol 28, 96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Volkmer B, and Heinemann M (2011) Condition-dependent cell volume and concentration of Escherichia coli to facilitate data conversion for systems biology modeling. PLoS One (Langowski J, Ed.) 6, e23126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Taheri-Araghi S, Bradde S, Sauls JT, Hill NS, Levin PA, Paulsson J, Vergassola M, and Jun S (2015) Cell-size control and homeostasis in bacteria. Curr. Biol 25, 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Hintsche M, and Klumpp S (2013) Dilution and the theoretical description of growth-rate dependent gene expression. J. Biol. Eng 7, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Qian Y, Huang H-H, Jiménez JI, and Del Vecchio D (2017) Resource competition shapes the response of genetic circuits. ACS Synth. Biol 6, 1263–1272. [DOI] [PubMed] [Google Scholar]
- (40).Moreb EA, Ye Z, Efromson JP, Hennigan JN, Menacho-Melgar R, and Lynch MD (2020) Media robustness and scalability of phosphate regulated promoters useful for two-stage autoinduction in E. coli . ACS Synth. Biol 9, 1483–1486. [DOI] [PubMed] [Google Scholar]
- (41).Canton B, Labno A, and Endy D (2008) Refinement and standardization of synthetic biological parts and devices. Nat. Biotechnol 26, 787–793. [DOI] [PubMed] [Google Scholar]
- (42).Lee TS, Krupa RA, Zhang F, Hajimorad M, Holtz WJ, Prasad N, Lee SK, and Keasling JD (2011) BglBrick vectors and datasheets: A synthetic biology platform for gene expression. J. Biol. Eng 5, 15–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Anthony LC, Suzuki H, and Filutowicz M (2004) Tightly regulated vectors for the cloning and expression of toxic genes. J. Microbiol. Methods 58, 243–250. [DOI] [PubMed] [Google Scholar]
- (44).Nevoigt E, Fischer C, Mucha O, Matthäus F, Stahl U, and Stephanopoulos G (2007) Engineering promoter regulation. Biotechnol. Bioeng 96, 550–558. [DOI] [PubMed] [Google Scholar]
- (45).Zhang F, Carothers JM, and Keasling JD (2012) Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat. Biotechnol 30, 354–359. [DOI] [PubMed] [Google Scholar]
- (46).Liu D, and Zhang F (2018) Metabolic feedback circuits provide rapid control of metabolite dynamics. ACS Synth. Biol 7, 347–356. [DOI] [PubMed] [Google Scholar]
- (47).Hartline CJ, Mannan AA, Liu D, Zhang F, and Oyarzún DA (2020) Metabolite sequestration enables rapid recovery from fatty acid depletion in Escherichia coli. MBio 11, e03112–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Jahn M, Vorpahl C, Hübschmann T, Harms H, and Müller S (2016) Copy number variability of expression plasmids determined by cell sorting and Droplet Digital PCR. Microb. Cell Fact 15, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).You C, Okano H, Hui S, Zhang Z, Kim M, Gunderson CW, Wang YP, Lenz P, Yan D, and Hwa T (2013) Coordination of bacterial proteome with metabolism by cyclic AMP signalling. Nature 500, 301–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Han Y, and Zhang F (2020) Control strategies to manage trade-offs during microbial production. Curr. Opin. Biotechnol 66, 158–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Han Y, and Zhang F (2020) Heterogeneity coordinates bacterial multi-gene expression in single cells. PLoS Comput. Biol 16, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Le TT, Emonet T, Harlepp S, Guet CC, and Cluzel P (2006) Dynamical determinants of drug-inducible gene expression in a single bacterium. Biophys. J 90, 3315–3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Rand JD, Danby SG, Greenway DLA, and England RR (2002) Increased expression of the multidrug efflux genes acrAB occurs during slow growth of Escherichia coli. FEMS Microbiol. Lett 207, 91–95. [DOI] [PubMed] [Google Scholar]
- (54).Cardinale S, and Arkin AP (2012) Contextualizing context for synthetic biology - Identifying causes of failure of synthetic biological systems. Biotechnol. J 7, 856–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Hollinshead W, He L, and Tang YJ (2014) Biofuel production: An odyssey from metabolic engineering to fermentation scale-up. Front. Microbiol 5, 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Nadal-Rey G, McClure DD, Kavanagh JM, Cornelissen S, Fletcher DF, and Gernaey KV (2021) Understanding gradients in industrial bioreactors. Biotechnol. Adv 46, 107660. [DOI] [PubMed] [Google Scholar]
- (57).Hao N, Krishna S, Ahlgren-Berg A, Cutts EE, Shearwin KE, and Dodd IB (2014) Road rules for traffic on DNA-systematic analysis of transcriptional roadblocking in vivo. Nucleic Acids Res. 42, 8861–8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Chen S-Y, Wei W, Yin B-C, Tong Y, Lu J, and Ye B-C (2019) Development of a highly sensitive whole-cell biosensor for arsenite detection through engineered promoter modifications. ACS Synth. Biol 8, 2295–2302. [DOI] [PubMed] [Google Scholar]
- (59).Diao J, Charlebois DA, Nevozhay D, Bódi Z, Pál C, and Balázsi G (2016) Efflux pump control alters synthetic gene circuit function. ACS Synth. Biol 5, 619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).De Paepe B, Peters G, Coussement P, Maertens J, and De Mey M (2017) Tailor-made transcriptional biosensors for optimizing microbial cell factories. J. Ind. Microbiol. Biotechnol 44, 623–645. [DOI] [PubMed] [Google Scholar]
- (61).Lin J-L, Wagner JM, and Alper HS (2017) Enabling tools for high-throughput detection of metabolites: Metabolic engineering and directed evolution applications. Biotechnol. Adv 35, 950–970. [DOI] [PubMed] [Google Scholar]
- (62).Csörgő B, Fehér T, Tímár E, Blattner FR, and Pósfai G (2012) Low-mutation-rate, reduced-genome Escherichia coli: an improved host for faithful maintenance of engineered genetic constructs. Microb. Cell Fact 11, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Zhang F, Ouellet M, Batth TS, Adams PD, Petzold CJ, Mukhopadhyay A, and Keasling JD (2012) Enhancing fatty acid production by the expression of the regulatory transcription factor FadR. Metab. Eng 14, 653–660. [DOI] [PubMed] [Google Scholar]
- (64).Jiang Y, Chen B, Duan C, Sun B, Yang J, and Yang S (2015) Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol 81, 2506–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Jiang W, Qiao JB, Bentley GJ, Liu D, and Zhang F (2017) Modular pathway engineering for the microbial production of branched-chain fatty alcohols. Biotechnol. Biofuels 10, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Davis JH, Rubin AJ, and Sauer RT (2011) Design, construction and characterization of a set of insulated bacterial promoters. Nucleic Acids Res. 39, 1131–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Basan M, Honda T, Christodoulou D, Hörl M, Chang YF, Leoncini E, Mukherjee A, Okano H, Taylor BR, Silverman JM, Sanchez C, Williamson JR, Paulsson J, Hwa T, and Sauer U (2020) A universal trade-off between growth and lag in fluctuating environments. Nature 584, 470–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Hartline CJ, Zhang R, and Zhang F (2022) Transient antibiotic tolerance triggered by nutrient shifts from gluconeogenic carbon sources to fatty acid. Front. Microbiol 13, 854272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


