Abstract
Biogenesis of small nucleolar RNA-protein complexes (snoRNPs) consists of synthesis of the snoRNA and protein components, snoRNP assembly, and localization to the nucleolus. Recently, two nucleoplasmic proteins from mice were observed to bind to a model box C/D snoRNA in vitro, suggesting that they function at an early stage in snoRNP biogenesis. Both proteins have been described in other contexts. The proteins, called p50 and p55 in the snoRNA binding study, are highly conserved and related to each other. Both have Walker A and B motifs characteristic of ATP- and GTP-binding and nucleoside triphosphate-hydrolyzing domains, and the mammalian orthologs have DNA helicase activity in vitro. Here, we report that the Saccharomyces cerevisiae ortholog of p50 (Rvb2, Tih2p, and other names) is required for production of C/D snoRNAs in vivo and, surprisingly, H/ACA snoRNAs as well. Point mutations in the Walker A and B motifs cause temperature-sensitive or lethal growth phenotypes and severe defects in snoRNA accumulation. Notably, depletion of p50 (called Rvb2 in this study) also impairs localization of C/D and H/ACA core snoRNP proteins Nop1p and Gar1p, suggesting a defect(s) in snoRNP assembly or trafficking to the nucleolus. Findings from other studies link Rvb2 orthologs with chromatin remodeling and transcription. Taken together, the present results indicate that Rvb2 is involved in an early stage of snoRNP biogenesis and may play a role in coupling snoRNA synthesis with snoRNP assembly and localization.
Synthesis of small nucleolar RNA protein complexes (snoRNPs) occurs in several stages, which are almost certainly overlapping (for recent summaries, see references 66 and 81). The major phases include (i) synthesis of the snoRNA and protein components, (ii) assembly of the snoRNP, and (iii) movement of the particle to the nucleolar complex, perhaps by way of Cajal bodies (22, 45, 54, 58). In metazoans, most snoRNAs are derived from introns of protein genes, and all of the assembly steps are believed to occur in the nucleoplasm. In Saccharomyces cerevisiae, most snoRNAs are transcribed from independent genes; however, a few are encoded in introns (81). Processing of the pre-snoRNAs in yeast is coupled to mRNA splicing (50, 53), and this situation likely pertains to metazoans as well.
Successful progression of the snoRNA transcript through each biosynthetic stage depends on the box C/D and H/ACA sequences used to classify the two large families of snoRNPs (see, e.g., references 8, 13, 14, 19, 27, 38, 39, 44, 45, 58, 64, and 79). In vivo function studies have shown that the binding of proteins to the motifs defined by the box elements is essential for proper processing of the snoRNA (end formation) and its localization to the nucleolus, presumably as a nascent snoRNP (reviewed in references 35, 66, and 81). Current models propose that protein binding to these motifs may be the first step in snoRNP assembly. Notably, the box elements are also intimately involved in the two types of nucleotide modification reaction mediated by the snoRNPs, i.e., ribose methylation by the C/D snoRNPs and pseudouridine formation by the H/ACA snoRNPs (4–6, 18, 34, 47, 66).
The final list of factors involved in snoRNP synthesis and function will probably be quite extensive. For example snoRNA processing involves several nucleases, some of which also participate in the processing of rRNA, other small RNAs, and pre-mRNA (72; see also references 2, 15, 16, and 71). For the snoRNP particles, four common proteins that are specific for the C/D class and another four that are specific for the H/ACA class have been identified. The core C/D proteins in Saccharomyces cerevisiae include Nop1p, Nop56p, Nop58p, and Snu13p (reviewed in references 35, 52, and 73; see also references 19, 21, 36, 59, 68, 80, and 83). Orthologs of these proteins have been demonstrated to exist in other species as well, including humans (41, 59). Remarkably, several (but not all) archaeal organisms contain C/D-like guide RNAs and snoRNP-like proteins of both classes (20, 49, 77). In addition to the common proteins, some snoRNPs such as U3 have unique proteins (references 17 and 74 and citations therein).
The C/D snoRNP protein Nop1p is a strong candidate for a snoRNP-based methyltransferase, as point mutations in a methylase-like domain disrupt methylation of rRNA globally (67, 76). The Snu13 protein was recently determined to bind specifically to the C/D motif (80). Surprisingly, this protein, known as 15.5 kDa in human cells (48), is also an integral component of the U4/U6.U5-splicing snRNP (65) and binds to a C/D-like motif in U4 (75, 80). It remains to be seen if the presence of Snu13/15.5 kDa in both snoRNPs and snRNPs reflects a mechanistic link between splicing and snoRNP synthesis or simply occurs by coincidence.
Proteins common to the H/ACA family of snoRNAs have been characterized best in S. cerevisiae and include Cbf5p, Gar1p, Nhp2p, and Nop10p (8, 23, 25, 37, 78). As for the C/D core proteins, the H/ACA snoRNP proteins are also evolutionarily conserved (54, 77). Cbf5p is almost certainly the universal H/ACA-associated Ψ synthase, based on the observation that point mutations in known pseudouridine synthase motifs abolish activity in vivo (84). Gar1p has RNA binding activity (6) but is not known to be motif specific.
With the goal of discovering proteins that act early in snoRNP biogenesis, one of our laboratories identified four mouse proteins that associate with a model C/D snoRNA in vitro (46). Two of the proteins are orthologs of yeast Nop56 and Nop58/Nop5p, which were originally identified by genetic strategies (21). The other two proteins, called p50 and p55, are novel and are related to each other at the levels of 42% identity and 68% similarity (46). They are strongly conserved among the Eukarya and many, but not all, Archaea; interestingly, the Archaea contain a single ortholog. Each protein has distinguishing Walker A and B motifs, which suggest ATP/GTP binding and nucleoside triphosphatase (NTPase) functions. Additional sequence relatedness occurs with bacterial DNA helicase RuvB in N- and C-terminal regions that together comprise about 50% of the overall sequence (32). Neither protein has been linked to snoRNAs previously; however, both have been characterized in other contexts. Both proteins have been reported to occur in the nucleoplasm (10, 40, 42, 46), suggesting that they may play a role in an early stage of snoRNP biogenesis.
Studies by others have implicated the novel snoRNA-related proteins in chromatin remodeling and transcription through association with components involved in those processes or stimulation of gene expression (see Discussion). Most relevant to the present study, the p50 and p55 proteins have been shown to associate with each other in yeast and human cells (9, 31, 40, 57, 82) and proteins from rat and human cells exhibit DNA helicase activities of opposite polarity in vitro (28, 31). Historically, the proteins were discovered independently by groups studying different processes in different organisms. Accordingly, they have several names. The mouse protein, called p50 in the previous snoRNA report, had not been described at that time. Subsequent orthologs were identified and given an assortment of names, including Tip49b (TATA-box binding protein [TBP]-interacting protein 49b), ECP-51 (erythrocyte cytosol protein 51), TIP48 (trans-activation domain-interacting protein 48), Reptin52 (repressing pontin 52), TAP54-β (Tip60-associated protein of 54 kDa in humans), P47 (47-kDa protein), scTip49b (S. cerevisiae Tip49b), scRUVBL2 (S. cerevisiae RUVBL2), Rvb2 (RuvB-like protein 2), and Tih2 (Tip49-homology protein 2) (9, 24, 28, 31, 40, 55, 57, 61, 82). The mouse protein called p55 in the snoRNA binding study was first discovered in rats and was called Tip49 (32). Orthologs of this protein have since been reported as well, and various names have been assigned including Tip49 (TBP-interacting protein 49), Pontin52 (Pont meaning bridge and 52 referring to kilodaltons), ECP-54 (erythrocyte cytosol protein 54), NMP238 (nuclear matrix protein 238), RUVBL1 (Ruv B-like protein 1), TIP49 (transactivation domain interacting protein 49), TAP54-alpha (Tip60-associated protein of 54 kDa), p50 (50-kDa protein), Tip49a (TBP-interacting protein 49a), scTip49a (S. cerevisiae Tip49a), scRUVBL1 (S. cerevisiae RuvB-like protein 1), Rvb1 (RuvB-like protein 1), and Tih1 (Tip49 homology protein 1) (9, 10, 26, 28, 31, 33, 40, 55, 57, 61, 82). In this report, which features the yeast p50 variant, we refer to the protein as Rvb2p, as this is the name adopted in the S. cerevisiae Genome Database. The mouse ortholog and other orthologs are referred to simply as p50.
To gain insight into the relationship of p50 with the snoRNAs, we carried out a genetic study of the yeast protein. Depletion and mutation analyses revealed an essential role in the accumulation of the box C/D snoRNAs and, surprisingly, the box H/ACA snoRNAs as well. Depletion also caused core proteins from each snoRNP family to accumulate in the nucleoplasm. Finally, point mutations revealed that snoRNA production requires the phylogenetically conserved ATP/GTP-binding and hydrolysis motifs. We discuss these results in the context of the snoRNP literature and reports that link these proteins with a variety of other processes.
MATERIALS AND METHODS
Gene replacement and tetrad analysis.
The strains and plasmids used are described in Table 1. The RVB2 coding sequence was replaced by transformation of strain MH2-d with a PCR fragment containing the HIS3 gene from plasmid YDp-H (11) flanked by 40 bp of sequence up- and downstream of the RVB2 open reading frame (ORF). The disruption was confirmed by Southern blotting using a 2.2-kb BamHI probe from pTK123 containing the RVB2 ORF and flanking sequence. Yeast transformation, sporulation, and dissection were performed as described previously (1).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description |
|---|---|
| Strains | |
| MH2-d | MATa/α ade2-1/ade2-1 his3Δ200/his3Δ200 leu2-1/leu2-1 ura3-52/ura3-52 trp1Δ101/trp1Δ101 gal2/gal2 |
| MH2-h | MATα ade2-1 his3,200 leu2-1 ura3-52 trp1,101 gal2 |
| YTK84 | MH2-d rvb2::HIS3/RVB2 |
| YTK86 | MH2-h rvb2::HIS3 pTK123 (RVB2 URA3 2 μm) |
| YWD312 | YTK86 Rvb2-GFP (cured of pTK123) |
| YWD313 | YTK86 pGAL-rvb2 (TRP1 CEN6) (cured of pTK123) |
| YWD339 | MH2-h pYES235W (URA3 2 μm) |
| YWD340 | MH2-h pWD296N (URA3 2μm) |
| YTK100 | YTK86 pTK149 (TRP1 CEN6) (cured of pTK123) |
| YTK104 | YTK86 pTK153 (TRP1 CEN6) (cured of pTK123) |
| Plasmids | |
| pTK123 | PCR product of RVB2 with natural flanking sequence inserted into the BamHI site of Yep24 (URA3 2μm) |
| pWD235WGFP | Yeast RVB2 ORF fused in frame to GFP (TRP1 CEN6) |
| GFP/pCDNA3.1 | Super GFP ORF under control of CMV promoter (Invitrogen) |
| pTK135 | RVB2 gene with natural flanking sequences inserted into BamHI site of pRS314 (TRP1 CEN6) |
| pTK147 | PCR-amplified mouse Rvb2 homolog (p50) ORF inserted into MCS of pcDNA 3.1/CT-GFP TOPO (Invitrogen) |
| pTK149 | pTK135 with rvb2 K81A mutation |
| pTK151 | pTK135 with rvb2 G75A mutation |
| pTK153 | pTK135 with rvb2 G80A mutation |
| pTK156 | pTK135 with rvb2 K81R mutation |
| pTK157 | pTK135 with rvb2 T82S mutation |
| pTK159 | pTK135 with rvb2 I295V mutation |
| pTK163 | pTK135 with rvb2 E297D mutation |
| pTK165 | pTK135 with rvb2 V298G mutation |
| pRSY14 | pRS314 with GAL1 promoter and CYC-1 terminator |
| pYES235W | pYES2 with wild-type RVB2 gene |
| pWD296N | Plasmid pYES235W with rvb2 D296N mutation |
| pGAL-rvb2 | RVB2 ORF flanked by GAL1 promoter and CYCI terminator (TRP1 CEN6) |
| pYDp-H | Plasmid carrying HIS3 gene and regulatory sequences (Amp; ori); PCR template for gene replacement |
| Rvb2-GFP | Yeast Rvb2 fused in frame to the N terminus of GFP (TRP1/CEN6) |
Plasmid construction.
The RVB2 gene was cloned by PCR amplification from yeast genomic DNA using Vent polymerase (New England Biolabs) under conditions recommended by the manufacturer. The primers were designed to amplify a 2.2-kb genomic region containing 500 bp of upstream and 300 bp of downstream sequence flanking the RVB2 ORF. The oligonucleotide sequences were as follows: upstream, GCGCGGATCCTTATCCCTAGTCAGTCGC; downstream, GCGCGGATCCCTAGAGGCTCTGAATGAG. The amplified RVB2 ORF was inserted into Yep24 and pRS314 to generate pTK123 and pTK135, respectively. Subcloning the mouse p50 cDNA into the CT-GFP/pCDNA 3.1 TOPO vector was performed as follows. The mouse p50 ORF was amplified from cDNA by PCR using Taq polymerase according to conditions recommended by Invitrogen. The primers were as follows: upstream, CCATCGATTGCATCATGGCAACCGTGG; downstream, GGGAGGTGTCCATTGTTTCG. The PCR product was gel purified and inserted into the TOPO vector as described in the green fluorescent protein (GFP) fusion cloning manual (Invitrogen).
Yeast Rvb2-GFP was generated in two steps. First pGFPCYC was constructed by insertion of a 988-bp DNA fragment containing the GFP-UV ORF (Clontech, Palo Alto, Calif.) and CYC-1 terminator into the PstI and NotI sites of PRS314. Then, a PCR fragment containing the RVB2 gene and 509 bp of upstream sequence was digested with KpnI and PstI and inserted into the KpnI/PstI sites of pGFPCYC immediately upstream and in-frame with the GFP-UV ORF.
pGal-rvb2 was generated by cloning a BamHI/PstI-digested PCR fragment containing the RVB2 gene into the BamHI and PstI sites of pRSY14. The PCR product was amplified with primers with sequences ATTGGATCCATGTCGATTCAAACTAGTGATCCAAATG and GGTTCTGCAGTGCTGGCCAACTGCTGCC using yeast genomic DNA as the template. pRSY14 was created by cloning a SalI/AvrII-digested PCR product containing the GAL1 promoter, cloning sites, and terminator from Invitrogen plasmid pYES2 into XhoI/SpeI-digested pRS314 (63).
Fluorescence microscopy.
Nop1p and Gar1p were localized in yeast as follows. For Nop1p, wild-type (MH2-h) and GAL::rvb2 (YWD313) cells were grown to early log phase in histidine-free or minimal complete medium, respectively, each containing 2% raffinose and 2% galactose. After being washed in yeast nitrogen base and incubated in minimal histidine-free medium containing 4% glucose for 10 h, cells were fixed and processed for immunofluoresence as previously described (12). Fluorescein isothiocyanate-conjugated anti-mouse Nop1p antibodies were diluted 1/200 in phosphate-buffered saline containing 1% bovine serum albumin (3). For Gar1p, the experimental and wild-type cells were transformed with a plasmid expressing a Gar1-GFP fusion protein (62). Transformants were processed as described above, except that growth was in medium lacking histidine and uracil. In situ hybridization against U3 was performed as previously described (58) with a Cy3-labeled oligonucleotide.
For localization of p50 in monkeys, COS-7 cells were transformed with a mouse p50-GFP fusion under the control of a cytomegalovirus (CMV) promoter (pTK147). Living cells were observed with fluorescence microscopy to visualize GFP and with differential interphase contrast (DIC) to observe cell morphology.
RNA procedures.
Steady-state levels of snoRNA were estimated by Northern blotting or 5′-[32P]pCp labeling as described previously (7). For Northern blotting, gel loading was adjusted to yield approximately equal quantities of tRNA in each lane. RNAs were fractionated in agarose-formaldehyde or 8% acrylamide–7 M urea–Tris-borate EDTA gels. For pCp labeling, 1 μg of total RNA was 3′ end labeled with 5′-[32P]pCp as described elsewhere (7). Blots and dried [32P]pCp gels were exposed to a Molecular Dynamics PhosphorImager screen for 14 h. Oligonucleotides used for detecting snoRNAs were as follows: U3, CATAGGATGGGTCAAGATCATCGCGCC; U14, CGATGGGTTCGTACTCCTACCGTGG; U18, GTCAGATACTGTGATAGTC; snR37, GATAGTATTAACCACTACTG; snR8, CACTCGCGCAGCTACCGATCTGGGCCAATGGGAGAC; snR46, CTTCCCTTTGGAAATCGGAAATTCAATGATATGCCCTATGCCC; 5S RNA, AGGCTCTTACCAGCTTAACT; MRP RNA, AATAGAGGTACCAGGTCAAGAAGC; snR35, GAACAAAATGATGATCTCTCCGATGGACTTGACGC; snR41, GGGTTGTCGACATGTAGTTAAACCACTATTCAGTCGG; U24, GGTATGTCTCATTCGGAACTCAAAGTTCCATCTGAAGTAGC; snR38, GAGAGGTTACCTATTATTACCCATTCAGACAGGGATAACTG; snR48, GGAGAGTACTTAAACTTCACATCCTAACATTAGAGATGCCAG; U5 (s,l), AAGTTCCAAAAAATATGGCAA; U6, AAAACGAAATAAATCTCTTTGTAAAAC.
Antisense RNA probes for hybridization analyses were prepared by transcription with T7 RNA polymerase in the presence of [∝-32P]CTP. Probes were constructed for RNase P, snR3, snR13, snR10, snR11, snR30, EF-1β, ASC1, TEF4, RPS5, RPL32, and RPS26B RNAs. Hybridization was in a solution containing 30% formamide, 5× SSPE (1× SSPE is 0.15 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.4]) buffer, 50 μg of heparin/ml, 0.2% sodium dodecyl sulfate (SDS) at 42°C for 16 h. Posthybridization washes were performed in 7× SSPE for 10 min at 25°C with agitation for oligonucleotide probes and in 0.2× SSPE–1% SDS for 1 h at 65°C for antisense probes.
Primer extension analyses were carried out as follows. Ten micrograms of total RNA was hybridized to 1 pmol of 32P-labeled oligonucleotides in a volume of 6 μl by heating to 95°C followed by cooling to 25°C over 10 min. To this mixture was added 2 μl of 5× reverse transcription buffer, and 4 U of avian myeloblastosis virus reverse transcriptase (U.S. Biochemicals [USB]). The mixture was incubated for 30 min at 37°C, and the reaction was stopped by the addition of 5 μl of USB stop solution. Extension products were fractionated and detected as described for [32P]pCp labeling. Placement of the oligonucleotides used for the extension reactions is shown in Fig. 4A. The sequences are as follows: oligo 1 (35S end), AGCGACTCTCTCCACCG; oligo 3 (site A1), CATGGCCTTAATCTTTGAGAC; oligo 5 (site A2), GGCCCCGATTGCTCGAATGCCCAAAG.
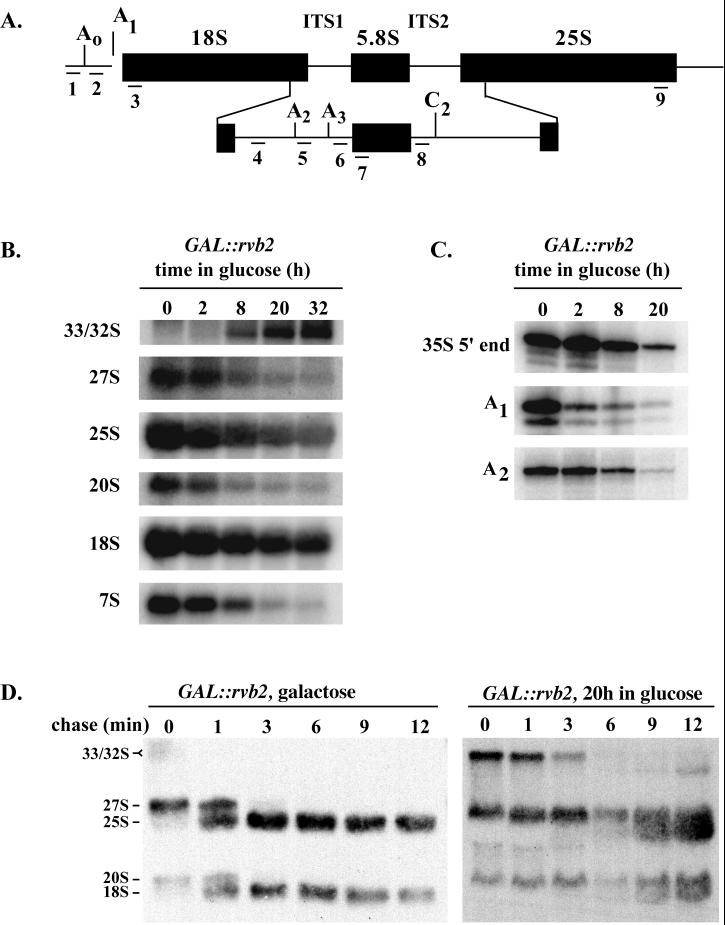
FIG. 4.
Rvb2p is required for pre-rRNA processing. (A) Structure of the 35S pre-rRNA, showing the positions of cleavage sites and regions complementary to oligonucleotide probes 1 to 8. ITS, internal transcribed spacer. (B) Steady-state levels of pre-rRNA species in cells depleted of Rvb2 mRNA for 0 to 32 h. Total RNA was extracted from cells at the times indicated following shift from galactose to glucose and subjected to Northern analysis. Loading was adjusted to obtain approximately equal quantities of tRNA. The oligonucleotides used to identify the various rRNA species are as follows: probes 3 and 9, mature 18S and 25S rRNAs, respectively; probe 4, 20S and 35S precursors; probe 8, 27S and 7S precursors. (C) Mapping of the 5′ ends of the 35S precursor and products of the A1 and A2 cleavage reactions. The 5′ ends were identified by primer extension analysis using as the template total RNA from cells depleted of Rvb2 mRNA as described for panel B. Oligonucleotides 1, 3, and 5 were extended to the 5′ end of 35S rRNA and the A1 and A2 cleavage sites, respectively. (D) Pulse-chase analysis of pre-rRNA processing in GAL::rvb2 cells incubated in galactose (left) or glucose medium for 20 h (right). Cells were labeled with [3H]methionine for 2.5 min and chased with cold methionine. Samples were removed at the times indicated, and total RNA was extracted and fractionated on an agarose-formaldehyde gel.
For pulse-chase labeling of RNA with methionine, GAL::rvb2 cells were grown in minimal medium containing 2% (each) raffinose, sucrose, and galactose but lacking methionine. When the optical density at 600 nm (OD600) reached 1.0, 3 ml of cells was incubated with 200 μl of l-[3H]methionine (85 Ci/mmol) for 2.5 min. To this sample was added 250 μl of unlabeled l-methionine (200 mM), and 500 μl of the mixture was removed at the time points indicated (see Fig. 4D) and snap-frozen in liquid nitrogen. RNA extraction, fractionation, and autoradiography were carried out as described elsewhere (83). The remaining cells were washed with prewarmed minimal medium containing 3% glucose and incubated at 30°C for up to 20 h. These cells were harvested and resuspended at 25 OD600 units/ml. To 6 ml of this suspension was added 800 μl of prewarmed l-[3H]methionine; all other steps were performed as described above except that the sample was chased with 1 ml of unlabeled l-methionine, and 1-ml aliquots were harvested at each time point. For electrophoretic analysis, 30,000 cpm of labeled RNA was loaded per lane.
Protein analysis.
Protein detection in Western blots was with rabbit anti-Rvb2p polyclonal immunoglobulin G (IgG) and a goat anti-rabbit IgG-peroxidase conjugate (Calbiochem). Protein extraction, SDS-polyacrylamide gel electrophoresis (PAGE), blotting, and chemiluminescence detection were carried out as described previously (84).
Construction of rvb2 mutant strains.
Site-directed mutations were created in Walker A and B motifs as follows. For all Walker B mutations except D296N, a 220-bp megaprimer was generated by PCR using oligonucleotides complementary to plasmid pTK123; the upstream primer contained one or two mismatches to the template to create the desired amino acid change, and the downstream primer was ATTGTCTTAATCTCCTGCTC. The megaprimer was used in conjunction with primer GACCGCTCTTGCCATGGGTG and template pTK123 in a second round of PCR to generate an 840-bp product. This fragment was digested with PflMI and ligated into pTK135 from which the corresponding wild-type PflMI fragment had been removed. For Walker A mutations, a 270-bp megaprimer was generated by PCR using primers complementary to plasmid pTK135; the upstream primer was GTAAAGCGGCCGCATGTCGGATTCAAACTAGTG, and the downstream primer was complementary to the Walker A region, except for mismatches introduced to create selected point mutations. The megaprimer was used with primer ACTGCTGTAACGCAAACTAG in a second round of PCR to amplify a 1-kb fragment from pTK135. This fragment was digested with SpeI and ligated into the SpeI site of pTK135.
Plasmids were introduced into yeast strain YTK86 by lithium acetate transformation, and transformants were selected on yeast nitrogen base with dextrose lacking tryptophan and uracil. Transformants were then cured of pTK123 (wild-type p50) by two rounds of streaking on 0.1% 5-fluoroorotic acid (5-FOA) at 30°C. Isolates no longer able to grow on YND lacking uracil were restreaked onto YPD and incubated at 16, 30, or 37°C. For strain and plasmid descriptions and nomenclature, see Table 1.
The rvb2 Walker B D296N allele was generated as follows. A region of the RVB2 ORF was amplified from pGAL-rvb2 using primers CCTTCAACTGGTAAGACCGCTC and CATTCTATATCCAACATGTGGACTTCGTTGATAAATAATACACCAGGAAC. The PCR product was digested with PflMI and ligated into PflMI-digested pGAl-rvb2, generating pGAL-ATP235. A BamHI/XhoI fragment carrying the mutant rvb2 DNA was liberated from pGAL-ATP235 and inserted into BamHI/XhoI-digested pYES2 (Invitrogen), generating pWD296N. This plasmid was transformed into strain MH2-h; the resulting strain is called YWD340.
RESULTS
Rvb2p is required for accumulation of C/D and H/ACA snoRNAs.
The essentiality of the RVB2 gene was examined by transforming diploid cells with a DNA fragment composed of HIS3::rvb2. Tetrad analysis of HIS3::rvb2/+ heterozygotes yielded a 2:2 segregation pattern, and surviving spores were HIS−, demonstrating that the RVB2 gene is required for growth. Evidence that gene replacement occurred at the RVB2 locus came from transforming HIS3::rvb2/+ cells with a plasmid (pTK123) harboring the RVB2 gene and natural flanking sequences. Tetrads from URA+ diploids contained four viable spores, confirming that gene replacement occurred at the RVB2 locus and that plasmid pTK123 carries a functional copy of RVB2 (data not shown). High-copy-number expression of the related p55 gene (RVB1) was not able to rescue the effects of RVB2 deletion (results not shown). While this work was in progress, another group also showed that the RVB2 gene is essential (31).
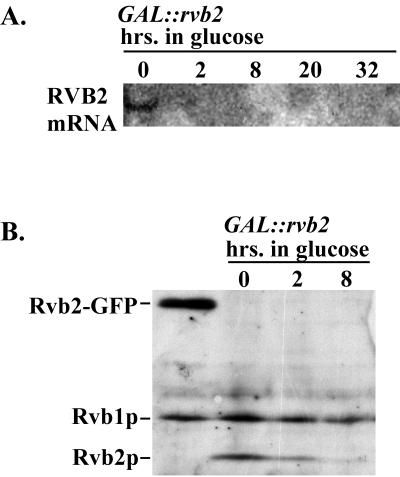
The possibility that Rvb2p is involved in the production of box C/D snoRNAs was examined by first creating a galactose-dependent test strain in which synthesis of RVB2 mRNA can be repressed by incubating cells in glucose medium and monitoring snoRNA levels after the onset of repression. Characterization of the test strain (YWD313; Table 1) showed that the levels of RVB2 mRNA and protein drop soon after shifting from galactose to glucose medium (Fig. 1). The mRNA was undetectable 2 h after the medium shift (Fig. 1A), and Western blotting showed that the protein level was substantially reduced after 2 h and effectively depleted by 8 h (the next time point; Fig. 1B).
FIG. 1.
Kinetics of Rvb2 depletion in galactose-dependent test cells. Test cells dependent on a GAL::rvb2 allele (strain YWD313) were grown in medium containing 2% galactose, washed, and shifted to medium containing 2% glucose. Cell samples were collected before and at various intervals after the medium shift. (A) Northern analysis of RVB2 mRNA accumulation. Total RNA was extracted from cells at the time points indicated and fractionated on an agarose-formaldehyde gel, and a blot was probed with radiolabeled DNA that recognizes RVB2 mRNA. (B) Western analysis of Rvb2p protein accumulation. Total protein was extracted at the time points indicated, fractionated by SDS-polyacrylamide gel electrophoresis, and electroblotted to a nitrocellulose membrane. Proteins were detected with rabbit polyclonal IgG (raised against recombinant Rvb2p) and a goat anti-rabbit–peroxidase conjugate. Left lane, total protein from a strain (YWD312) carrying HIS3::rvb2 and Rvb2-GFP expressed from a plasmid; the other lanes show total proteins from GAL::rvb2 cells incubated in glucose for 0, 2, and 8 h, as indicated.
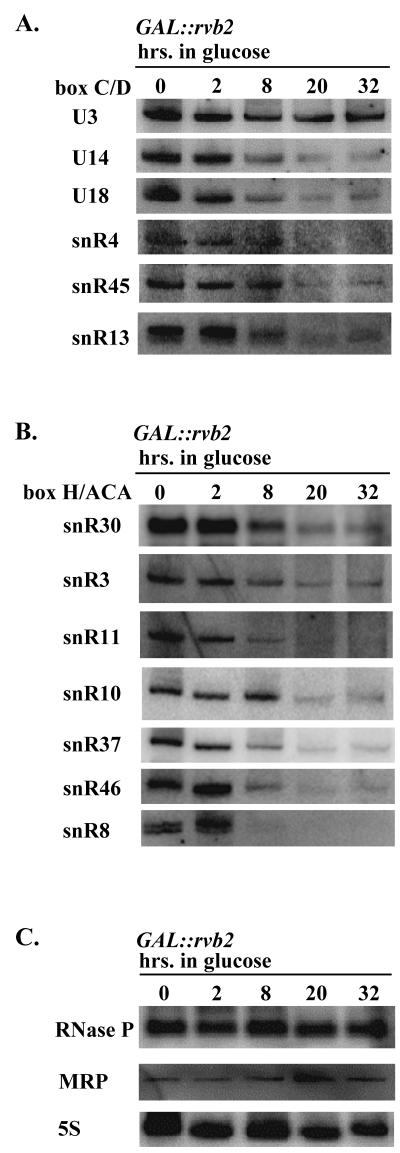
The importance of Rvb2p for snoRNA accumulation was evaluated by Northern analyses of total RNA prepared from cells after 0 to 32 h in glucose. In addition to examining the C/D snoRNAs, we also assessed potential effects on the H/ACA snoRNAs, the MRP snoRNA, 5S rRNA, and the RNase P RNA. Loss of Rvb2p strongly impaired accumulation of the C/D snoRNAs and, to our surprise, the H/ACA snoRNAs as well. The steady-state levels of the five box C/D and seven H/ACA snoRNAs probed decreased markedly over the course of Rvb2p depletion (Fig. 2A and B). In contrast, little or no effect was seen for the other small RNAs examined (Fig. 2C). Interestingly, the level of the U3 box C/D snoRNA appeared to be nearly constant over the same period (Fig. 2A, top, and data not shown).
FIG. 2.
Rvb2p is required for accumulation of box C/D and H/ACA snoRNAs in yeast. Test cells dependent on a GAL::rvb2 allele (strain YWD313) were grown in galactose medium and shifted to glucose medium. Total RNA was extracted from cells harvested at various intervals following the shift, and snoRNAs were examined by Northern blot analysis. RNA loading was adjusted to obtain approximately equal amounts of tRNA per lane. The gel patterns show the effects of Rvb2p depletion on the accumulation of box C/D snoRNAs (A), box H/ACA snoRNAs (B), and RNase P, MRP, and 5S RNAs (C).
These blotting results were extended by examining the electrophoretic pattern of [32P]pCp-labeled total RNA using the same samples (data not shown). In addition to verifying the snoRNA effects, the patterns showed that accumulation of some other small RNAs including 5S and 5.8S was somewhat impaired at later time points, after 20 to 32 h in glucose. Interestingly, at late time points (20 to 32 h), two novel small RNA species in the size range of 240 to 245 nucleotides were observed to accumulate. The identities of these RNAs are not known. The time course shows that Rvb2p loss has a rapid effect on snoRNA or snoRNP production and, eventually, less-immediate effects on other RNAs. Taken together the results indicate that loss of snoRNAs occurs soon after repression of Rvb2p expression and that other effects also occur, but much later.
Is the snoRNA effect direct or indirect?
The role of Rvb2p in snoRNA accumulation could be direct or indirect. Direct effects could reflect a mechanistic role in processes such as snoRNA transcription or maturation, assembly of snoRNP particles, and localization of nascent snoRNPs to the nucleolus. Similar effects can be imagined to occur if Rvb2p loss disrupts processes that overlap or are otherwise coupled to these events. Plausible examples include impaired expression of protein genes that encode snoRNAs within intron elements and interference with expression of ribosomal proteins or rRNA. Loss of intron-encoded snoRNAs might lead to loss of all snoRNAs including those expressed from independent snoRNA genes (via regulation). Defects in the synthesis of r- proteins or rRNA would interfere with ribosome biogenesis, of course, and this could lead to reduced levels of snoRNAs and snoRNPs. For example, defective rRNP intermediates that are associated with snoRNPs might form. These complexes might turn over and lead to a deficiency in snoRNAs. Combinations of both direct and indirect effects on snoRNA levels can also be envisioned.
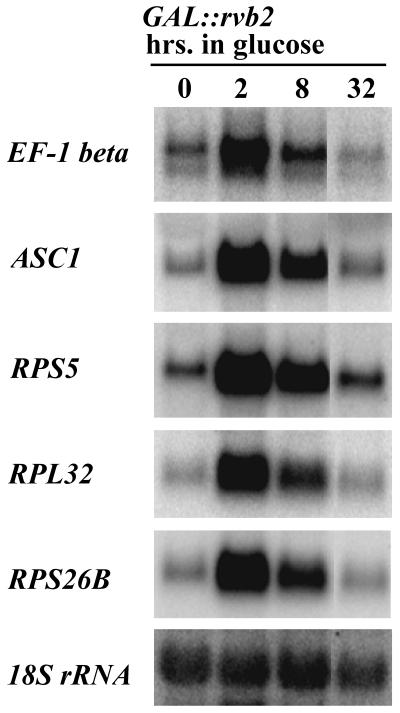
To test the possibility that reduced expression of snoRNA host genes is a contributing factor, we analyzed the effects of Rvb2p loss on one such gene, EF-1β. This gene encodes the U18 snoRNA in addition to the EF-1β protein. Our initial analysis showed that U18 is affected by Rvb2p loss (Fig. 2A). We probed total RNA for EF-1β mRNA at three time points after repressing Rvb2p expression. The results revealed that mRNA transcription increased dramatically soon after the shift to glucose (i.e., 2 and 8 h) and then dropped. After 32 h in glucose, the mRNA level had declined to about the same as that detected before the shift (Fig. 3). Eight hours after the shift to glucose, the level of U18 was dramatically reduced while the level of EF-1β was greatly increased compared with the levels in cells grown in galactose. We conclude that loss of the U18 intronic snoRNA that occurs during Rvb2p depletion is not caused by reduced expression of its host gene. An additional snoRNA host gene, ASC1, was analyzed in the same way and gave a similar pattern (Fig. 3). This gene encodes the intronic snoRNA U24, which we show below also requires Rvb2p for accumulation.
FIG. 3.
Accumulation of selected mRNAs in Rvb2p-depleted cells. GAL::rvb2 test cells were grown in medium containing 2% galactose, washed, and shifted to medium containing 2% glucose. Total RNA was extracted from cells collected before (left lane) and after (other lanes) the medium shift, fractionated on a 1.5% agarose-formaldehyde gel, and incubated with appropriate radiolabeled antisense probes. Incubation times in glucose are indicated above each lane. 18S rRNA served as a loading control.
While this work was in progress, another group reported that inactivation of a temperature-sensitive variant of Rvb2p causes a substantial decrease in the levels of mRNAs for ribosomal proteins (40). We tested the possibility that snoRNA loss might be linked to disruption of r-protein expression by examining the steady-state levels of three r-protein mRNAs during Rvb2p depletion. The transcripts tested are from the RPS5, RPL32, and RPS26B genes. As observed for the snoRNA host genes above, the level of r-protein mRNAs increased immediately after the shift to glucose and then dropped off to a level approximately equal to that found in cells grown in galactose (Fig. 3). While snoRNA levels are reduced after 8 h in glucose, the level of ribosomal protein mRNAs is greatly elevated. Furthermore, after 32 h in glucose, snoRNAs are nearly undetectable while r-protein mRNAs are present at about the same level as in galactose medium. We conclude that the loss of snoRNAs that occurs during Rvb2p depletion is not a consequence of reduced accumulation of r-protein mRNAs.
To determine if the loss of snoRNAs in Rvb2p-depleted cells could be caused by a defect in rRNA transcription, we carried out a pulse-labeling experiment in which wild-type and GAL::rvb2 cells were labeled with [3H]methionine or [3H]uracil before and at various time points after a shift from galactose to glucose medium. Total RNA was extracted and fractionated, and newly synthesized rRNA was visualized by autoradiography. The results indicated that after 8 h of incubation in glucose, the amount of rRNA made in GAL::rvb2 cells was approximately the same as that made in wild-type cells. After 24 h in glucose, the accumulation of new rRNA was somewhat impaired in GAL::rvb2 cells (data not shown); we do not know if this reduction reflects impaired synthesis or accelerated degradation of rRNA. Thus, following lengthy repression of Rvb2p synthesis, rRNA accumulation is impaired, and this occurs after the onset of snoRNA loss. We conclude that the snoRNA accumulation defect associated with Rvb2p depletion is probably not due to impaired rRNA transcription.
Rvb2p is required for pre-rRNA processing.
The requirement for Rvb2p in snoRNA accumulation predicts that it also affects pre-rRNA processing, since a few C/D and H/ACA snoRNAs are needed for cleavage reactions. To test this possibility, the steady-state levels of various pre-rRNAs were determined (for YWD313 cells) following depletion of Rvb2p in glucose medium. Northern analysis was carried out on total RNA, and processing intermediates and the mature rRNAs were detected using oligonucleotide probes specific for internally and externally transcribed spacer regions and mature 18S or 25S rRNA (Fig. 4A). As anticipated, loss of Rvb2p impaired rRNA processing. All major intermediates leading to the production of 25S, 18S, and 5.8S rRNAs decreased in abundance, while the 32S precursor transcript accumulated. The intermediates affected include the 27S and 7S species, which are precursors of 25S and 5.8S rRNAs, and the 20S species, which is the direct precursor of 18S rRNA. The levels of the mature rRNA species were correspondingly low compared with those for wild-type cells, and each decreased at about the same rate (Fig. 4B). The data demonstrate that Rvb2p is required for the processing steps that generate the 25S, 18S, and 5.8S rRNA species.
Consistent with this situation, results from a pulse-chase labeling analysis showed that both 18S and 25S RNAs are produced at a considerably slower rate in Rvb2p-depleted cells (Fig. 4D). The effect of Rvb2p on rRNA production is unusual, as genes involved in rRNA processing typically affect production of either large- or small-subunit rRNAs. To our knowledge, this is the first example of a gene that affects the majority of snoRNAs from both major snoRNA families. The unusual pattern of rRNA processing could reflect loss of an unknown snoRNA(s) that affects processing of 25S rRNA or negative effects on other factors important for rRNA maturation.
To determine which pre-rRNA cleavages are affected, primer extension analyses were performed on total RNA from cells depleted of Rvb2p. The primers used are suitable for detecting the 5′ ends of the primary transcript, and the products that result from cleavages at the A1 and A2 sites (primers 1, 3, and 5 in Fig. 4A). The levels of the intermediates generated by the two cleavages decreased approximately 80- and 30-fold, respectively, after 20 h in glucose (Fig. 4C), while the relative abundance of the 35S transcript decreased 10-fold. The data indicate that Rvb2p loss leads to impairment of the A1 and A2 cleavages and a decrease in the accumulation of the 35S precursor. The A1 and A2 cleavages depend on C/D snoRNA U14 and H/ACA snoRNA snR30, which accumulate poorly in Rvb2p-depleted cells. Thus, loss of these snoRNAs seems a likely basis for these defects. The relative decrease in 35S pre-rRNA is more difficult to explain; one possibility is degradation of aberrant rRNP intermediates. We note that enhanced turnover could also contribute to loss of products generated by cleavages at A1 and A2.
The yeast and mouse homologs of Rvb2p occur in the nucleoplasm.
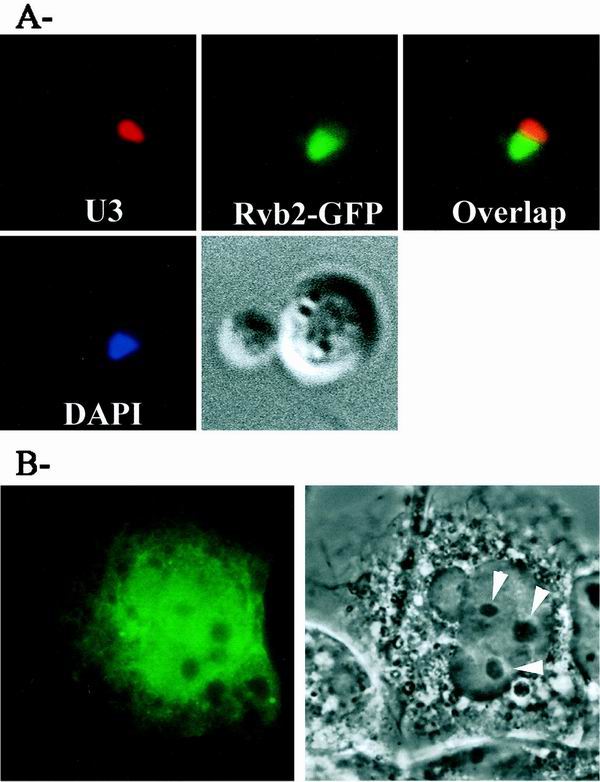
The Rvb2p protein and its orthologs have been reported to be present in the nucleoplasm of yeast and vertebrate cells (40, 46). The observation that Rvb2p has strong effects on accumulation of snoRNAs and rRNA prompted us to carry out an independent evaluation of its nuclear location using different strategies. The analyses were done with both yeast Rvb2p and its mouse counterpart (p50), fused in each case to GFP and expressed in yeast or monkey COS-7 cells. For the yeast analysis, a strain harboring plasmid-borne Rvb2-GFP (YWD312; rvb2::HIS3 pWD235WGFP) was grown to log phase in selective medium. The cells were incubated with a Cy3-labeled oligonucleotide probe that recognizes U3, and the subnuclear localizations of Rvb2-GFP and U3 were examined by fluorescence microscopy (see Materials and Methods). The results show that Rvb2p occurs throughout the nucleoplasm and appears to be absent from the nucleolus (Fig. 5A).
FIG. 5.
Nuclear location of Rvb2p and mouse p50. (A) Distribution of Rvb2-GFP in yeast. Yeast cells (YWD312) each harboring a plasmid-borne copy of Rvb2-GFP (pWD235WGFP) were grown to log phase in selective medium. The cells were incubated with a Cy3-labeled oligonucleotide probe specific for U3, and the subnuclear localizations of p50-GFP and U3 were examined by fluorescence microscopy (top row). Nuclear DNA was visualized with DAPI (bottom left), and cell morphology was visualized with DIC (bottom right). (B) Localization of the mouse homolog of Rvb2 in COS-7 cells. Cells were transformed by the calcium-phosphate procedure (58) with a plasmid (pTK147) carrying mouse Rvb2 fused to GFP. Localization of Rvb2-GFP was observed by fluorescence microscopy (left), and nucleoli were visualized with phase contrast (right). Each field is 30 by 30 μm; arrows, nucleoli.
For the analysis of monkey cells, expression of the p50-GFP coding sequence was from a CMV promoter. The fusion construct was transformed into COS-7 cells, and the location of the fusion protein was examined by fluorescence microscopy; morphology was visualized by DIC. The GFP fluorescence (Fig. 5B, left) was very evident in the nucleoplasm in all cells examined and here too appears to be confined to this compartment. As in the yeast analysis, little or no p50 signal was observed in the nucleolus; however, it is too soon to rule out a presence in this location. No fluorescence is apparent in the cytoplasm. The GFP sequence is not believed to affect the localization of p50, as GFP alone accumulates in the cytosol and the yeast p50-GFP fusion protein is functional in yeast (Fig. 5A). These patterns corroborate earlier biochemical fractionation and microscopy data from other laboratories (40, 46). In addition, the results include a nucleolar reference molecule (U3) for yeast and represent the first high-resolution analysis for p50 in a mammalian cell.
Depletion of Rvb2p causes delocalization of C/D and H/ACA core snoRNP proteins.
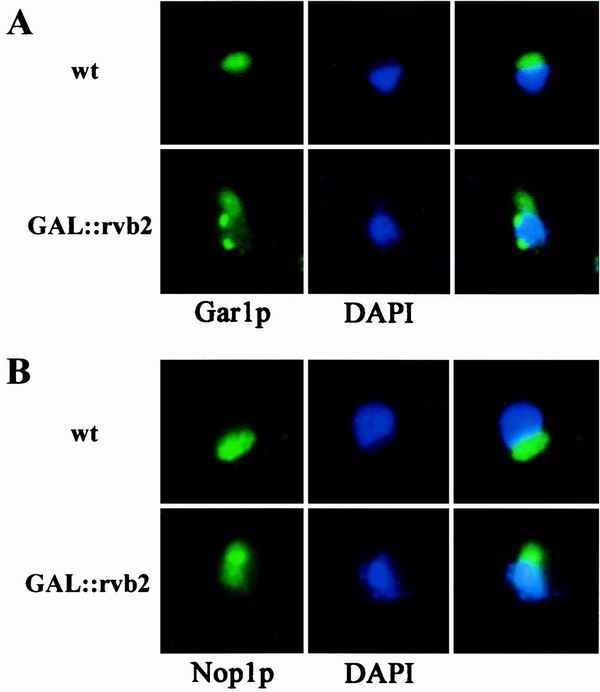
In addition to the depletion effects described above, the fact that Rvb2p is a nucleoplasmic protein also suggests that Rvb2p influences snoRNA synthesis at an early stage, perhaps at the level of snoRNP assembly or trafficking. Interference with snoRNP formation is known to lead to loss of snoRNAs, presumably because free snoRNAs and defective RNP intermediates are degraded. In principle, a defect in snoRNP localization might also lead to snoRNA loss, although this has not been established. More directly, of course, the loss of snoRNAs could reflect a defect in snoRNA gene expression. To further assess the role of Rvb2p, we examined the effect of blocking its production on the distribution of two core snoRNP proteins in the nucleus, one from each major snoRNP family. The proteins were Nop1p (C/D family) and Gar1p (H/ACA family). The locations of the snoRNP proteins were analyzed by immunofluorescence microscopy (Nop1p) or by direct fluorescence microscopy (Gar1-GFP) 10 h after shifting the test cells from galactose to glucose medium.
In wild-type cells, both core proteins were localized in a crescent-shaped structure characteristic of the nucleolus, and no signal was apparent in the nucleoplasm (visualized with DAPI [4′,6′-diamidino-2-phenylindole]). After Rvb2p depletion, the Nop1p signal was no longer restricted to the nucleolus but was dispersed throughout the nucleoplasm (Fig. 6B) and, in some cells, Nop1p was also evident in the cytoplasm. The signal for Gar1p was also delocalized, and in many cases punctate structures were seen throughout the nucleoplasm (Fig. 6A). These results indicate that Rvb2p is required for localization of the snoRNP proteins and are consistent with a defect in the formation or localization of snoRNP complexes. It should be noted that a similar mislocalization of Nop1p has been observed following depletion of Nop58p or Sen1p; both conditions induce a loss of box C/D snoRNAs (69, 83). For Rvb2p, however, both classes of snoRNPs are affected. The basis of these effects remains to be determined.
FIG. 6.
Loss of Rvb2p causes delocalization of C/D and H/ACA snoRNP proteins. GAL::rvb2 test strains containing chromosomally encoded Nop1p (YWD313) or a plasmid encoding a Gar1p-GFP fusion were grown in minimal selective galactose medium and shifted to glucose medium for 10 h to halt production of RVB2 mRNA (see Materials and Methods). Nop1p was detected with a primary monoclonal antibody, followed by a fluorescein isothiocyanate-coupled secondary antibody and observed by immunofluorescence microscopy. Gar1p-GFP was observed by direct fluorescence microscopy. (A) Localization of Gar1p in wild-type cells (upper left) and cells depleted of Rvb2p (lower left). (B) Localization of Nop1p in wild-type cells (upper left) and Rvb2p-depleted cells (lower left). The nucleus was visualized with a DNA stain (DAPI; middle). A merged view is also shown (right).
The Walker A and B motifs are essential for snoRNA production.
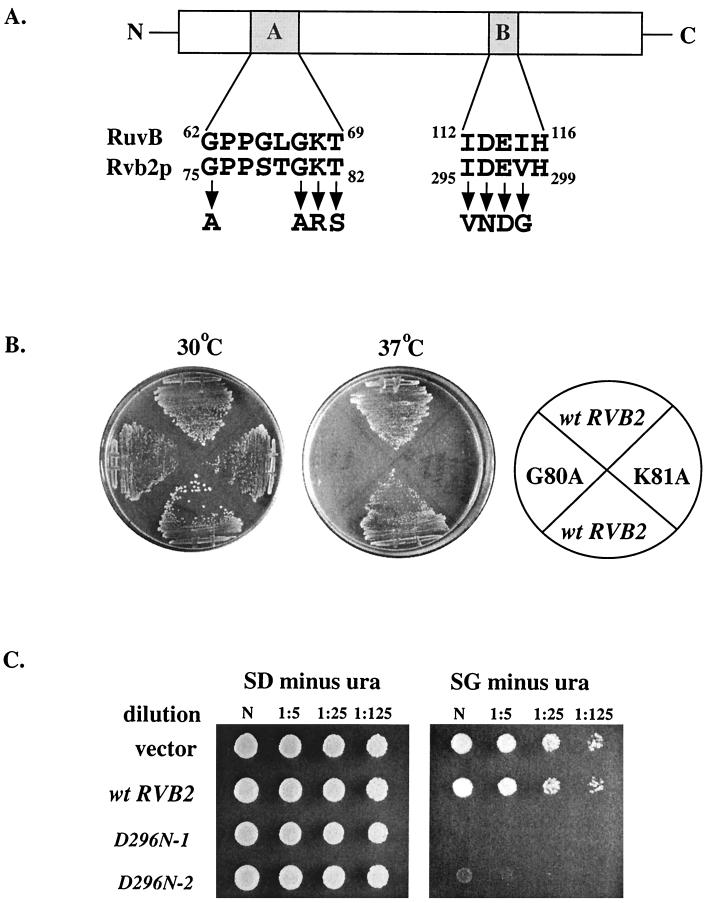
To assess the functional importance of the Walker A and B domains, we substituted conserved amino acids in each motif. The substitutions made were similar or identical to those that impair the function of bacterial DNA helicase RuvB and mammalian translation factor eIF4a; both are known ATPases (43, 51). Plasmids harboring the mutant alleles were introduced into yeast test strain YTK86, and the plasmid containing the wild-type allele was evicted by counter-selection on medium containing 5-FOA.
Of nine mutations constructed, two were lethal (G75A and D296N), three caused temperature sensitivity (G80A, K81R, and K81A), and the remainder caused little or no growth inhibition (Table 2). The rvb2 K81R mutation was reported earlier to be lethal (40), but in our test cells a conditional temperature sensitivity (ts) phenotype is observed. The cause of this difference is not known, but variation in genetic background could be a factor. The ts growth phenotypes caused by the G80A and K81A alleles are shown in Fig 7B. Interestingly, high-copy-number expression of the rvb2 D296N allele in wild-type MH2-h cells severely impaired growth (Fig. 7C). These results demonstrate that the putative nucleotide binding and ATPase domains are required in vivo. The phenotypes do not appear to be due to instability of the mutant proteins, at least for two ts variants. Fluorescence imaging analysis of the mutant K81A and G80A proteins showed that each occurs in the nucleus at levels comparable to that of wild-type Rvb2p (data not shown).
TABLE 2.
Growth phenotypes caused by point mutations in the Walker motifs of Rvb2pa
| Motif | Mutation | Phenotype at:
|
||
|---|---|---|---|---|
| 16°C | 30°C | 37°C | ||
| Walker A | G75A | − | − | − |
| G80A | + | + | − | |
| K81R | − | +++ | − | |
| K81A | + | + | − | |
| T82S | + | + | + | |
| Walker B | I295V | +++ | +++ | +++ |
| E297D | ++ | +++ | ++ | |
| V298G | +++ | +++ | +++ | |
| D296N | − | − | − | |
Mutations were made in the A and B elements by site-directed mutagenesis. The mutant alleles were inserted individually into a TRP/CEN vector (Table 1) and then introduced into a strain with an inactive rvb2 chromosomal gene (YTK86, with an rvb2::HIS3 allele) and a plasmid-borne copy of wild-type RVB2 (PTK123). The latter plasmid was evicted by curing on 5-FOA. Growth phenotypes were assessed on YPD at 3 days (30°C) or 7 days (16 and 37°C). +++, wild-type growth rate; ++, slow growth; +, very slow growth; −, lethal.
FIG. 7.
Point mutations in the Walker A and B domains disrupt growth. (A) Selected residues in Walker domains A and B of Rvb2p were substituted by site-directed mutagenesis. (B) Point mutations G80A and K81A (Walker A motif) cause temperature-sensitive growth defects. Left plate, permissive temperature (30°C); right plate, restrictive temperature (37°C). (C) The D296N allele is lethal and also toxic when overexpressed in a wild-type background. Growth of a wild-type strain (MH2-h) expressing a high-copy-number GAL1::rvb2 D296N allele is severely impaired in galactose medium (right, bottom two rows) but unaffected when expression is repressed by incubation in glucose (left, bottom two rows). No effects occur with the empty vector (left and right, top rows) or a vector containing a wild-type RVB2 allele under the control of the GAL1 promoter (left and right, second rows from top). The dilution series is indicated above each panel. N, no dilution.
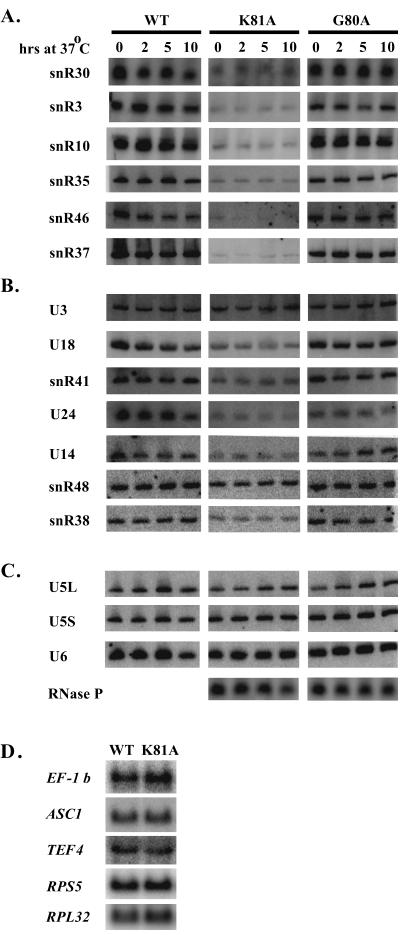
To determine if the Walker A domain is required for snoRNA production, levels of snoRNA were examined in cells dependent on either of two alleles conferring temperature sensitivity. Cells with Walker A (P-loop) G80A or K81A mutations were grown to mid-log phase at the permissive temperature of 30°C and shifted to 37°C. Total RNA was extracted from cells sampled before and at various intervals after the shift and examined by Northern blot analysis. The patterns revealed that the K81A mutation caused a noticeable defect in the accumulation of six box H/ACA and six box C/D snoRNAs examined. While the defect was evident for the majority of snoRNAs tested, the magnitude of the effects varied (compare, for example, snR48 with snR37; Fig. 8). As with the depletion analysis, the variation appears to be unrelated to the genetic origins of the RNAs, i.e., whether the snoRNAs are intronic or independently transcribed, and the accumulation of U3 was not affected. Accumulation patterns were also analyzed for three other small nuclear RNAs: the U5 and U6 splicesomal RNAs and RNase P RNA. These species were not affected (Fig. 8C).
FIG. 8.
Accumulation of snoRNA is impaired in cells harboring rvb2 ts mutations. Results of Northern blot analysis are shown for box H/ACA snoRNAs (A), box C/D snoRNAs (B), splicing snRNAs and RNase P (C), and EF-1β, ASC1, TEF4, RPS5, and RPL32 mRNAs (D). Panels A to C include total RNA from wild-type cells (WT) and cells with the rvb2 K81A allele (K81A) or G80A allele (G80A). Times of incubation at 37°C are indicated above each lane. Time zero identifies samples harvested prior to the temperature shift. Northern blots in panel D are of RNA extracted from cells grown at 30°C.
The accumulation patterns of mRNAs for genes EF-1β, ASC1, TEF4, RPS5, and RPL32 were also analyzed. The patterns were not altered by the point mutation (Fig. 8D; see below). Three of these genes encode snoRNAs affected by the K81A mutation (EF-1β, ASC1, and TEF4), and two specify ribosomal proteins (RPS5 and RPL32). Taken together, the results indicate that the Walker A domain is specifically required for production of snoRNAs and that the effects of Rvb2p depletion and inactivation are essentially the same.
Analysis of pCp-labeled RNA from one point mutant (G80A) revealed that the two aberrant small RNA species observed in the protein depletion analysis were also evident here, after incubation at the nonpermissive temperature for 2 to 10 h (data not shown).
Interestingly, the defect in snoRNA production is allele specific. Cells harboring a K81R allele, which preserves the charge at position 81, or a G80A substitution accumulate snoRNAs to wild-type levels (Fig. 8, right, and data not shown). In several ATPases of known three-dimensional structure, the ɛ amino group of the lysine corresponding to K81 forms a salt bridge with the terminal phosphate(s) of nucleoside diphosphates and nucleoside triphosphates (NTPs) (29). This situation could explain why loss of charge in the K81A protein has such a strong effect. The defect was observed at the permissive temperature and was not exacerbated by prolonged incubation at 37°C, indicating that no correlation exists between the thermal instability of mutant Rvb2p and its ability to function in snoRNA production. These results argue that Rvb2p has NTP binding and/or hydrolysis activities and that these activities are required for snoRNA production.
DISCUSSION
The results show that Rvb2p has an essential role in snoRNP biogenesis and that this function most likely occurs at an early stage and in the nucleoplasm. Depletion or inactivation of the protein results in loss of both major classes of snoRNAs and impaired localization of core snoRNP proteins. Processing of rRNA is impaired as well, and these effects could also reflect loss of snoRNAs, since a few snoRNAs in each major family are needed for rRNA processing (35, 73). Two factors point to a nucleoplasmic role for Rvb2p. First, it is clear that the vast majority and possibly all of the protein is in the nucleoplasm (Fig. 5) (40, 46). Second, results from a variety of other studies unrelated to snoRNAs have linked Rvb2p and its vertebrate counterparts with nucleoplasmic components (see below). On this basis, it seems most likely that Rvb2p functions in the nucleoplasm, although the possibility that it has a nucleolar phase is still formally open.
Several clues about the role of Rvb2p in snoRNA production are provided by studies of Rvb1p and Rvb2p orthologs (p50 and p55) in other organisms and in other contexts. The most definitive information available is that both proteins have been demonstrated to have DNA helicase activity in vitro (see below). Based on interactions with proteins of known function, the p50 and p55 proteins have been suggested to participate, variously, in transcription, chromatin remodeling, DNA recombination, signal transduction, and the cell cycle (9, 10, 28, 31, 32, 40, 55, 61, 82). The potential for overlap among these processes is high, of course. It will be interesting to learn if some or all of the effects observed, including those described here, are linked to a single biochemical process or rather reflect similar DNA- or RNA-based roles in multiple processes. The key findings from these studies are discussed in the following sections, with a view to identifying common links and clues about snoRNA accumulation.
The earliest ties to transcription are based on observations that rat p55 was in immunoprecipitates of TBP prepared from nuclear extracts (32) and, in a separate study, copurified with the RNA polymerase II (Pol II) holoenzyme through several chromatographic steps and density gradient centrifugation (55). The interaction with the Pol II complex appears to be of high affinity. However, association of p55 with TBP and Pol II has not been determined to occur in the cell, and these effects may not extend to p50.
More recently, p50 and p55 have been implicated in transcription on other grounds. In a yeast study, Rvb2p (p50) was shown to be required for accumulation of mRNAs that specify ribosomal proteins (40); in the present study we show that Rvb2p depletion affects snoRNAs before r-protein production. In rat cells, p50 and p55 have been determined to influence oncogenic transformation in response to transcription factors c-Myc and H-Ras (82). In the context of the present study, c-Myc may have limited relevance, since p50 is highly conserved and Myc-related factors are not. In other studies, human p50 and p55 were shown to interact biochemically with β-catenin and TBP and to serve as antagonistic regulators for activation of the Wnt signaling pathway (9, 10). All of these effects could be manifest at the level of transcription, but effects on downstream processes are also possible.
Intriguing new results appear to place the p50 and p55 proteins in position to influence transcription by remodeling chromatin. In one study Rvb2p and Rvb1p were both found to be present in a megadalton complex affinity purified from yeast using an epitope-tagged variant of chromatin-remodeling factor INO80 (61). The complex contained 3′→5′ helicase activity and stimulated transcription from chromatin. In another study, human p50 and p55 were both identified in a complex isolated through binding to histone acetylase TIP60 (28). The TIP60 complex acetylates nucleosomes in vitro and has ATPase and DNA helicase activities as well. Interestingly, the yeast and human complexes contain several common protein homologs (including p50 and p55) and are probably functionally equivalent in the two species. Notably, both Rvb2p and Rvb1p are essential for growth in yeast while INO80 is not.
More recently, a different affinity enrichment procedure was used to isolate a yeast complex containing Rvb1p and Rvb2p, and chromatin-remodeling activity was once again demonstrated (30). In that study, it was shown that the two proteins are responsible for activation or repression of a subset of yeast genes that comprise up to 5% of the genome (30). It would be interesting to learn the basis of these effects and the physiological significance.
It has been proposed that the p50 and p55 orthologs are involved in other aspects of DNA function as well. Based on modest sequence similarity with the bacterial RuvB protein, which participates in homologous recombination and repair of double-stranded breaks, it was suggested that p55 has a similar function(s) in eukaryotic cells (55). That case is somewhat weakened, however, by the fact that the RuvB-like Walker A and B domains in p50 and p55 are much further apart than in RuvB. A role in recombination was also suggested by a positive two-hybrid interaction of human p55 with a subunit of human replication protein A (hsRPA3), but there is no other experimental evidence supporting this suggestion.
Results from two other studies are also interesting and difficult to interpret. In one, p55 was reported to occur in nuclear matrix preparations from human and rat cells. Specifically, p55 was enriched in an insoluble pellet following treatment of nuclei with detergent, buffer of high ionic strength, and DNase I (26). A different study, with human erythrocytes, found that p50 and p55 (named ECP-51 and ECP-54) bind a fragment of the membrane protein stomatin in vitro (57). These results could reflect important subcellular associations for p50- and p55-containing complexes, although the erythrocyte situation may not be relevant to snoRNP synthesis, since these cells do not have a nucleus.
In light of the complexity and number of processes in which Rvb2p is involved, it is possible that the effects on snoRNA accumulation and snoRNP protein localization could arise from alterations in more than one nuclear process. An important goal for the future will be to carry out systematic studies on the order and kinetics of the various snoRNP-related events that occur during Rvb2p depletion and inactivation. These results should yield valuable insights into the precise cause-and-effect relationships between Rvb2p function and the biogenesis of snoRNPs and ribosomes.
The results from our point mutation studies show that the Walker A and B domains are essential for cell growth (see also reference 40) and for accumulation of snoRNAs. The growth phenotypes (Table 2 and Fig. 7) would be predicted in some cases, based on effects of mutations in known ATPases such as eIF-4A and RuvB (43, 51). For example, G75 and K81 in yeast Rvb2p (Walker A) are vital for function and correspond to residues required for ATP binding and/or ATPase activity in eIF-4A (51). Similarly, the D296N mutation blocks function. Overexpression of the latter protein causes a dominant lethal growth phenotype in cells with a wild-type gene (Fig. 7C). This situation is analogous to that observed for Escherichia coli RuvB, where the equivalent mutation (D113N) causes a loss of ATPase activity and overexpression of the allele in vivo results in a dominant-negative UV-sensitive phenotype (43). These results strongly suggest that Rvb2p has ATPase activity in vivo and are consistent with reports showing that p50 has ATPase and ATP-dependent DNA helicase activities in vitro (28, 31). DNA helicase activity can be imagined to be important in gene transcription, for example, in remodeling chromatin structure or influencing the binding of transcription factors. Our present results argue that Rvb2p does not influence snoRNA accumulation by altering transcription of host mRNA genes. Thus, for these and perhaps the other snoRNAs, the effect of Rvb2p is apparently downstream of transcription.
In principle, it is possible that Rvb2p can alter helices in RNA as well as DNA. Several proteins with DNA helicase activity can also unwind RNA:RNA and RNA:DNA helices in vitro (reference 60 and citations therein). One protein of this class, yeast Sen1p, is particularly relevant to the present study, as it also affects snoRNP biogenesis, with similar consequences. Sen1p from Schizosaccharomyces pombe has helicase activity with both RNA and DNA in vitro, and the S. cerevisiae ortholog affects the processing of snoRNAs and localization of the snoRNP proteins Nop1 (C/D class) and Sbp1 (H/ACA class) (56, 69, 70). RNA helicases can be imagined to function at different stages of snoRNP synthesis, including dissociation of snoRNA transcripts from DNA, remodeling of RNPs during pre-snoRNA processing or snoRNP assembly, and perhaps interactions with the nucleolar trafficking machinery.
The fact that p50 and p55 were affinity selected in vitro with a model C/D snoRNA is consistent with a role for one or both proteins at the RNA or RNP level. Specificity is suggested by the finding that complex formation required an intact C/D motif, as mutations in box D blocked assembly (46). The two C/D snoRNP core proteins, Nop56p and Nop58p, were present in the complex in nearly stoichiometric amounts, as seen in natural complexes from yeast (21). The p50 and p55 proteins occurred at similar levels (21). Attempts to immunoprecipitate snoRNAs from mouse extracts with rabbit antibodies to mouse p50 were negative (46), and in the present study we were unable to detect snoRNAs in immunoprecipitates of protein A-tagged yeast Rvb2p (data not shown). These results suggest that p50 may not be present in mature snoRNP particles. To date, neither p50 nor p55 has been detected in preparations of natural C/D (and H/ACA) snoRNPs enriched from yeast or human extracts by affinity enrichment schemes. However, the complexes analyzed thus far were incomplete. Because p50 and p55 exhibited stable, stoichiometry-like binding in the initial isolation strategy, it seems likely that their association with snoRNAs is relevant and that this association probably occurs in vivo as well, in the nucleoplasm.
The p50 and p55 proteins could bind directly to snoRNA or indirectly through protein-protein interactions. The observation that loss or mutation of Rvb2p impairs the accumulation of H/ACA snoRNAs implies that the association may not be limited to C/D snoRNAs. In the context of snoRNA binding, this result has several possible interpretations. One possibility is that RNA binding is direct and occurs through RNA structure elements common to both H/ACA and C/D snoRNAs. Simple sequence elements are not likely to be involved here, as the only conserved sequences known are the family-specific box elements. Alternatively, direct binding to RNA could involve secondary- or tertiary-structure domains common to both major classes of snoRNAs. Arguing against this possibility is the belief that the C/D snoRNAs do not have conserved secondary structures beyond the C/D motif.
More likely is the possibility that p50 and p55 associate with both major classes of snoRNAs through common elements that occur in the snoRNP proteins. The signals could be present in a protein common to all C/D and H/ACA snoRNAs, although none has yet been identified. Alternatively, p50 could bind to different family-specific proteins with common motifs. The latter possibility is especially attractive at present, as the two sets of unique core proteins each include one member with striking sequence relatedness to the other. One of these corresponds to the Snu13p/15.5 kDa protein, which binds directly to the C/D motif and which is present in natural C/D snoRNPs (80). The related H/ACA protein is Nhp2p in yeast (hNHP2 in humans) (54, 65). Yeast Snu13p and Nhp2p exhibit an overall relatedness of 33% identity and 44% similarity. The related sequences are concentrated in the middle of the protein, where a stretch of 22 amino acids is 55% identical and 82% similar. These core proteins provide a common link between the two major classes of snoRNAs and are excellent candidates for the binding of p50 and p55. We note that Snu13p was not observed in the affinity isolation procedure that identified p50 and p55; however, it might have been overlooked in the gel analysis because of its small size (46).
The fact that Snu13p is also present in the U4 snRNP raises the interesting possibility that mRNA splicing and snoRNA synthesis may be linked. It is possible that Snu13p has separate, unrelated functions in splicing and snoRNP production. However, the processes are clearly coupled, and Snu13p is a common link. Similarly, the ties between p50 and chromatin remodeling and gene expression suggest that p50 (and p55) might participate in coupling the processes that coordinate ribosome and protein synthesis. Relevant to this, the Snu13p and Nhp2p sequences have some similarity to ribosomal protein L30 (RPL30) in S. cerevisiae, and corresponding proteins in a wide range of organisms from the Archaea to humans (80). The sequence relatedness, 24% identity and 33% similarity, between Snu13p or Nhp2 and RPL30 is modest; however, these similarities raise the possibility of yet other links between splicing, snoRNP production, and ribosomes. Sorting out these relationships promises to be both interesting and challenging.
ACKNOWLEDGMENTS
We gratefully acknowledge the expert help of Celine Verheggen with the protein localization analyses and Heidi Galonek for help with a yeast Rvb2-GFP plasmid construction.
This study was supported by a fellowship from the Foundation Pour la Recherche Medicale to E.B. and NIH grant GM19351 to M.J.F.
REFERENCES
- 1.Adams A. Methods in yeast genetics, 1997 ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- 2.Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aris J P, Blobel G. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J Cell Biol. 1988;107:17–31. doi: 10.1083/jcb.107.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachellerie J P, Cavaille J. Guiding ribose methylation of rRNA. Trends Biochem Sci. 1997;22:257–261. doi: 10.1016/s0968-0004(97)01057-8. [DOI] [PubMed] [Google Scholar]
- 5.Bachellerie J P, Cavaillé J. Small nucleolar RNAs guide the ribose methylations of eukaryotic rRNAs. In: Grosjean H, Benne R, editors. Modification and editing of RNA. Washington, D.C.: ASM Press; 1998. pp. 255–272. [Google Scholar]
- 6.Bagni C, Lapeyre B. Gar1p binds to the small nucleolar RNAs snR10 and snR30 in vitro through a nontypical RNA binding element. J Biol Chem. 1998;273:10868–10873. doi: 10.1074/jbc.273.18.10868. [DOI] [PubMed] [Google Scholar]
- 7.Balakin A G, Lempicki R A, Huang G M, Fournier M J. Saccharomyces cerevisiae U14 small nuclear RNA has little secondary structure and appears to be produced by post-transcriptional processing. J Biol Chem. 1994;269:739–746. [PubMed] [Google Scholar]
- 8.Balakin A G, Smith L, Fournier M J. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 9.Bauer A, Chauvet S, Huber O, Usseglio F, Rothbacher U, Aragnol D, Kemler R, Pradel J. Pontin52 and reptin52 function as antagonistic regulators of beta-catenin signalling activity. EMBO J. 2000;19:6121–6130. doi: 10.1093/emboj/19.22.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bauer A, Huber O, Kemler R. Pontin52, an interaction partner of beta-catenin, binds to the TATA box binding protein. Proc Natl Acad Sci USA. 1998;95:14787–14792. doi: 10.1073/pnas.95.25.14787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berben G, Dumont J, Gilliquet V, Bolle P A, Hilger F. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- 12.Bertrand E, Chartrand P, Schaefer M, Shenoy S M, Singer R H, Long R M. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 13.Bortolin M L, Ganot P, Kiss T. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. EMBO J. 1999;18:457–469. doi: 10.1093/emboj/18.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caffarelli E, Fatica A, Prislei S, De Gregorio E, Fragapane P, Bozzoni I. Processing of the intron-encoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J. 1996;15:1121–1131. [PMC free article] [PubMed] [Google Scholar]
- 15.Chanfreau G, Legrain P, Jacquier A. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J Mol Biol. 1998;284:975–988. doi: 10.1006/jmbi.1998.2237. [DOI] [PubMed] [Google Scholar]
- 16.Chanfreau G, Rotondo G, Legrain P, Jacquier A. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 1998;17:3726–3737. doi: 10.1093/emboj/17.13.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colley A, Beggs J D, Tollervey D, Lafontaine D L. Dhr1p, a putative DEAH-box RNA helicase, is associated with the box C+D snoRNP U3. Mol Cell Biol. 2000;20:7238–7246. doi: 10.1128/mcb.20.19.7238-7246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganot P, Bortolin M L, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 19.Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- 20.Gaspin C, Cavaille J, Erauso G, Bachellerie J P. Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: lessons from the Pyrococcus genomes. J Mol Biol. 2000;297:895–906. doi: 10.1006/jmbi.2000.3593. [DOI] [PubMed] [Google Scholar]
- 21.Gautier T, Berges T, Tollervey D, Hurt E. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol Cell Biol. 1997;17:7088–7098. doi: 10.1128/mcb.17.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerbi S A, Borovjagin A. U3 snoRNA may recycle through different compartments of the nucleolus. Chromosoma. 1997;105:401–406. doi: 10.1007/BF02510476. [DOI] [PubMed] [Google Scholar]
- 23.Girard J P, Lehtonen H, Caizergues-Ferrer M, Amalric F, Tollervey D, Lapeyre B. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 1992;11:673–682. doi: 10.1002/j.1460-2075.1992.tb05099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gohshi T, Shimada M, Kawahire S, Imai N, Ichimura T, Omata S, Horigome T. Molecular cloning of mouse p47, a second group mammalian RuvB DNA helicase-like protein: homology with those from human and Saccharomyces cerevisiae. J Biochem (Tokyo) 1999;125:939–946. doi: 10.1093/oxfordjournals.jbchem.a022372. [DOI] [PubMed] [Google Scholar]
- 25.Henras A, Henry Y, Bousquet-Antonelli C, Noaillac-Depeyre J, Gelugne J P, Caizergues-Ferrer M. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 1998;17:7078–7090. doi: 10.1093/emboj/17.23.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holzmann K, Gerner C, Korosec T, Poltl A, Grimm R, Sauermann G. Identification and characterization of the ubiquitously occurring nuclear matrix protein NMP 238. Biochem Biophys Res Commun. 1998;252:39–45. doi: 10.1006/bbrc.1998.9604. [DOI] [PubMed] [Google Scholar]
- 27.Huang G M, Jarmolowski A, Struck J C, Fournier M J. Accumulation of U14 small nuclear RNA in Saccharomyces cerevisiae requires box C, box D, and a 5′, 3′ terminal stem. Mol Cell Biol. 1992;12:4456–4463. doi: 10.1128/mcb.12.10.4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikura T, Ogryzko V V, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 29.Johnson E R, McKay D B. Crystallographic structure of the amino terminal domain of yeast initiation factor 4A, a representative DEAD-box RNA helicase. RNA. 1999;5:1526–1534. doi: 10.1017/s1355838299991410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jonsson Z O, Dhar S K, Narlikar G J, Auty R, Wagle N, Pellman D, Pratt R E, Kingston R, Dutta A. Rvb1p and Rvb2p are essential components of a chromatin remodeling complex that regulates transcription of over 5% of yeast genes. J Biol Chem. 2001;276:16279–16288. doi: 10.1074/jbc.M011523200. [DOI] [PubMed] [Google Scholar]
- 31.Kanemaki M, Kurokawa Y, Matsu-ura T, Makino Y, Masani A, Okazaki K, Morishita T, Tamura T A. TIP49b, a new RuvB-like DNA helicase, is included in a complex together with another RuvB-like DNA helicase, TIP49a. J Biol Chem. 1999;274:22437–22444. doi: 10.1074/jbc.274.32.22437. [DOI] [PubMed] [Google Scholar]
- 32.Kanemaki M, Makino Y, Yoshida T, Kishimoto T, Koga A, Yamamoto K, Yamamoto M, Moncollin V, Egly J M, Muramatsu M, Tamura T. Molecular cloning of a rat 49-kDa TBP-interacting protein (TIP49) that is highly homologous to the bacterial RuvB. Biochem Biophys Res Commun. 1997;235:64–68. doi: 10.1006/bbrc.1997.6729. [DOI] [PubMed] [Google Scholar]
- 33.Kikuchi N, Gohshi T, Kawahire S, Tachibana T, Yoneda Y, Isobe T, Lim C R, Kohno K, Ichimura T, Omata S, Horigome T. Molecular shape and ATP binding activity of rat p50, a putative mammalian homologue of RuvB DNA helicase. J Biochem (Tokyo) 1999;125:487–494. doi: 10.1093/oxfordjournals.jbchem.a022312. [DOI] [PubMed] [Google Scholar]
- 34.Kiss-Laszlo Z, Henry Y, Bachellerie J P, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 35.Kressler D, Linder P, de La Cruz J. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7897–7912. doi: 10.1128/mcb.19.12.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lafontaine D L, Tollervey D. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA. 1999;5:455–467. doi: 10.1017/s135583829998192x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lafontaine D L J, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. Genes Dev. 527–537. 1998. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lange T S, Borovjagin A, Maxwell E S, Gerbi S A. Conserved boxes C and D are essential nucleolar localization elements of U14 and U8 snoRNAs. EMBO J. 1998;17:3176–3187. doi: 10.1093/emboj/17.11.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lange T S, Ezrokhi M, Borovjagin A V, Rivera-Leon R, North M T, Gerbi S A. Nucleolar localization elements of Xenopus laevis U3 small nucleolar RNA. Mol Biol Cell. 1998;9:2973–2985. doi: 10.1091/mbc.9.10.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim C R, Kimata Y, Ohdate H, Kokubo T, Kikuchi N, Horigome T, Kohno K. The Saccharomyces cerevisiae RuvB-like protein, Tih2p, is required for cell cycle progression and RNA polymerase II-directed transcription. J Biol Chem. 2000;275:22409–22417. doi: 10.1074/jbc.M001031200. [DOI] [PubMed] [Google Scholar]
- 41.Lyman S K, Gerace L, Baserga S J. Human Nop5/Nop58 is a component common to the box C/D small nucleolar ribonucleoproteins. RNA. 1999;5:1597–1604. doi: 10.1017/s1355838299991288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makino Y, Mimori T, Koike C, Kanemaki M, Kurokawa Y, Inoue S, Kishimoto T, Tamura T. TIP49, homologous to the bacterial DNA helicase RuvB, acts as an autoantigen in human. Biochem Biophys Res Commun. 1998;245:819–823. doi: 10.1006/bbrc.1998.8504. [DOI] [PubMed] [Google Scholar]
- 43.Mezard C, Davies A A, Stasiak A, West S C. Biochemical properties of RuvBD113N: a mutation in helicase motif II of the RuvB hexamer affects DNA binding and ATPase activities. J Mol Biol. 1997;271:704–717. doi: 10.1006/jmbi.1997.1225. [DOI] [PubMed] [Google Scholar]
- 44.Narayanan A, Lukowiak A, Jady B E, Dragon F, Kiss T, Terns R M, Terns M P. Nucleolar localization signals of box H/ACA small nucleolar RNAs. EMBO J. 1999;18:5120–5130. doi: 10.1093/emboj/18.18.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narayanan A, Speckmann W, Terns R, Terns M P. Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol Biol Cell. 1999;10:2131–2147. doi: 10.1091/mbc.10.7.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newman D R, Kuhn J F, Shanab G M, Maxwell E S. Box C/D snoRNA-associated proteins: two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA. 2000;6:861–879. doi: 10.1017/s1355838200992446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ni J, Tien A L, Fournier M J. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 48.Nottrott S, Hartmuth K, Fabrizio P, Urlaub H, Vidovic I, Ficner R, Luhrmann R. Functional interaction of a novel 15.5kD [U4/U6.U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. EMBO J. 1999;18:6119–6133. doi: 10.1093/emboj/18.21.6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Omer A D, Lowe T M, Russell A G, Ebhardt H, Eddy S R, Dennis P P. Homologs of small nucleolar RNAs in Archaea. Science. 2000;288:517–522. doi: 10.1126/science.288.5465.517. [DOI] [PubMed] [Google Scholar]
- 50.Ooi S L, Samarsky D A, Fournier M J, Boeke J D. Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: intron length effects and activity of a precursor snoRNA. RNA. 1998;4:1096–1110. doi: 10.1017/s1355838298980785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peculis B A. RNA-binding proteins: if it looks like a sn(o)RNA. Curr Biol. 2000;10:R916–R918. doi: 10.1016/s0960-9822(00)00851-4. [DOI] [PubMed] [Google Scholar]
- 53.Petfalski E, Dandekar T, Henry Y, Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol Cell Biol. 1998;18:1181–1189. doi: 10.1128/mcb.18.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pogacic V, Dragon F, Filipowicz W. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol Cell Biol. 2000;20:9028–9040. doi: 10.1128/mcb.20.23.9028-9040.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qiu X B, Lin Y L, Thome K C, Pian P, Schlegel B P, Weremowicz S, Parvin J D, Dutta A. A eukaryotic RuvB-like protein (RUVBL1) essential for growth. J Biol Chem. 1998;273:27786–27793. doi: 10.1074/jbc.273.43.27786. [DOI] [PubMed] [Google Scholar]
- 56.Rasmussen T P, Culbertson M R. The putative nucleic acid helicase Sen1p is required for formation and stability of termini and for maximal rates of synthesis and levels of accumulation of small nucleolar RNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:6885–6896. doi: 10.1128/mcb.18.12.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salzer U, Kubicek M, Prohaska R. Isolation, molecular characterization, and tissue-specific expression of ECP-51 and ECP-54 (TIP49), two homologous, interacting erythroid cytosolic proteins. Biochim Biophys Acta. 1999;1446:365–370. doi: 10.1016/s0167-4781(99)00104-9. [DOI] [PubMed] [Google Scholar]
- 58.Samarsky D A, Fournier M J, Singer R H, Bertrand E. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 1998;17:3747–3757. doi: 10.1093/emboj/17.13.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schimmang T, Tollervey D, Kern H, Frank R, Hurt E C. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 1989;8:4015–4024. doi: 10.1002/j.1460-2075.1989.tb08584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seybert A, Hegyi A, Siddell S G, Ziebuhr J. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA. 2000;6:1056–1068. doi: 10.1017/s1355838200000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 62.Sicard H, Faubladier M, Noaillac-Depeyre J, Leger-Silvestre I, Gas N, Caizergues-Ferrer M. The role of the Schizosaccharomyces pombe gar2 protein in nucleolar structure and function depends on the concerted action of its highly charged N terminus and its RNA-binding domains. Mol Biol Cell. 1998;9:2011–2023. doi: 10.1091/mbc.9.8.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Speckmann W A, Terns R M, Terns M P. The box C/D motif directs snoRNA 5′-cap hypermethylation. Nucleic Acids Res. 2000;28:4467–4473. doi: 10.1093/nar/28.22.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stevens S W, Abelson J. Purification of the yeast U4/U6.U5 small nuclear ribonucleoprotein particle and identification of its proteins. Proc Natl Acad Sci USA. 1999;96:7226–7231. doi: 10.1073/pnas.96.13.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 67.Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt E C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- 68.Tyc K, Steitz J A. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989;8:3113–3119. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ursic D, DeMarini D J, Culbertson M R. Inactivation of the yeast Sen1 protein affects the localization of nucleolar proteins. Mol Gen Genet. 1995;249:571–584. doi: 10.1007/BF00418026. [DOI] [PubMed] [Google Scholar]
- 70.Ursic D, Himmel K L, Gurley K A, Webb F, Culbertson M R. The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic Acids Res. 1997;25:4778–4785. doi: 10.1093/nar/25.23.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol. 2000;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Hoof A, Parker R. The exosome: a proteasome for RNA? Cell. 1999;99:347–350. doi: 10.1016/s0092-8674(00)81520-2. [DOI] [PubMed] [Google Scholar]
- 73.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 74.Venema J, Vos H R, Faber A W, van Venrooij W J, Raue H A. Yeast Rrp9p is an evolutionarily conserved U3 snoRNP protein essential for early pre-rRNA processing cleavages and requires box C for its association. RNA. 2000;6:1660–1671. doi: 10.1017/s1355838200001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vidovic I, Nottrott S, Hartmuth K, Luhrmann R, Ficner R. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol Cell. 2000;6:1331–1342. doi: 10.1016/s1097-2765(00)00131-3. [DOI] [PubMed] [Google Scholar]
- 76.Wang H, Boisvert D, Kim K K, Kim R, Kim S H. Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 A resolution. EMBO J. 2000;19:317–323. doi: 10.1093/emboj/19.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Watanabe Y, Gray M W. Evolutionary appearance of genes encoding proteins associated with box H/ACA snoRNAs: cbf5p in Euglena gracilis, an early diverging eukaryote, and candidate Gar1p and Nop10p homologs in archaebacteria. Nucleic Acids Res. 2000;28:2342–2352. doi: 10.1093/nar/28.12.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Watkins N J, Gottschalk A, Neubauer G, Kastner B, Fabrizio P, Mann M, Luhrmann R. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA. 1998;4:1549–1568. doi: 10.1017/s1355838298980761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Watkins N J, Leverette R D, Xia L, Andrews M T, Maxwell E S. Elements essential for processing intronic U14 snoRNA are located at the termini of the mature snoRNA sequence and include conserved nucleotide boxes C and D. RNA. 1996;2:118–133. [PMC free article] [PubMed] [Google Scholar]
- 80.Watkins N J, Segault V, Charpentier B, Nottrott S, Fabrizio P, Bachi A, Wilm M, Rosbash M, Branlant C, Luhrmann R. A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–466. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- 81.Weinstein L B, Steitz J A. Guided tours: from precursor snoRNA to functional snoRNP. Curr Opin Cell Biol. 1999;11:378–384. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- 82.Wood M A, McMahon S B, Cole M D. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol Cell. 2000;5:321–330. doi: 10.1016/s1097-2765(00)80427-x. [DOI] [PubMed] [Google Scholar]
- 83.Wu P, Brockenbrough J S, Metcalfe A C, Chen S, Aris J P. Nop5p is a small nucleolar ribonucleoprotein component required for pre-18 S rRNA processing in yeast. J Biol Chem. 1998;273:16453–16463. doi: 10.1074/jbc.273.26.16453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zebarjadian Y, King T, Fournier M J, Clarke L, Carbon J. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol Cell Biol. 1999;19:7461–7472. doi: 10.1128/mcb.19.11.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]