Abstract
There is interest in the application of rapid quantitative polymerase chain reaction (qPCR) methods for recreational freshwater quality monitoring of the fecal indicator bacteria Escherichia coli (E. coli). In this study we determined the performance of 21 laboratories in meeting proposed, standardized data quality acceptance (QA) criteria and the variability of target gene copy estimates from these laboratories in analyses of 18 shared surface water samples by a draft qPCR method developed by the U.S. Environmental Protection Agency (EPA) for E. coli. The participating laboratories ranged from academic and government laboratories with more extensive qPCR experience to “new” water quality and public health laboratories with relatively little previous experience in most cases. Failures to meet QA criteria for the method were observed in 24% of the total 376 test sample analyses. Of these failures, 39% came from two of the “new” laboratories. Likely factors contributing to QA failures included deviations in recommended procedures for the storage and preparation of reference and control materials. A master standard curve calibration model was also found to give lower overall variability in log10 target gene copy estimates than the delta-delta Ct (ΔΔCt) calibration model used in previous EPA qPCR methods. However, differences between the mean estimates from the two models were not significant and variability between laboratories was the greatest contributor to overall method variability in either case. Study findings demonstrate the technical feasibility of multiple laboratories implementing this or other qPCR water quality monitoring methods with similar data quality acceptance criteria but suggest that additional practice and/or assistance may be valuable, even for some more generally experienced qPCR laboratories. Special attention should be placed on providing and following explicit guidance on the preparation, storage and handling of reference and control materials.
Keywords: qPCR, E. coli, Draft Method C, recreational water, QA, variability
1.0. INTRODUCTION
Until recently, the only option for monitoring recreational beach waters for fecal pollution has been by the use of culture-based methods for fecal indicator bacteria (FIB) such as Escherichia coli (E. coli) or Enterococcus spp. These methods typically require 18 or more hours from sample collection to results (Haugland et al., 2005; Noble et al., 2010). This delay limits the ability of beach managers to issue closure or warning notices that are adequate for the protection of public health, particularly on the day of exposure. Advances in molecular techniques now offer attractive alternatives for measuring microbial water quality more rapidly. For example, quantitative polymerase chain reaction (qPCR) is now a well-established technique that can be performed in as little as 3 hours. Based on epidemiologic studies (Wade et al., 2008; 2010), national regulatory criteria and beach action values have been recommended by the EPA for an enterococci qPCR method (U.S. EPA, 2012) and an improved and updated version of this method (U.S. EPA, 2015a) is now being implemented with the EPA criteria for daily beach monitoring in a major metropolitan area (Dorevitch et al., 2017) and in a U.S. National Park (Byappanahalli et al., 2018). In addition, a number of different qPCR methods have also been developed for E. coli FIB (Sivaganesan et al., 2019) and one such method is currently being used for site-specific beach notifications by a Great Lakes beach management authority (Kinzelman et al., 2013).
As discussed in Sivaganesan et al. (2019), qPCR methods that are being considered for widespread use should have standardized data quality acceptance (QA) criteria guidelines. In addition, past evaluations of qPCR-based water quality testing methods have often focused on the performance of these methods by established laboratories with less attention paid to the practical challenges of technology transfer to laboratories with little or no molecular biology-based experience (Griffith & Weisberg, 2011). Therefore, assessments of method performance that account for the ability of personnel from laboratories with varying levels of experience to implement rapid qPCR– based methods are an important consideration for evaluating the general applicability of these methods.
In this study, laboratory performance of a relatively new E. coli qPCR method developed by the EPA (Draft Method C) was investigated based on standardized QA guidelines (Sivaganesan et al., 2019) during the analyses of 18 shared surface water samples, collected from 6 sites in the state of Michigan. A total of 21 laboratories, ranging from academic and government laboratories with more extensive qPCR experience to water quality and public health laboratories with a long history of using culture-based methods but in some cases relatively little previous qPCR experience participated in the study. E. coli 23S rRNA target gene copy concentration estimates were determined for all sample and laboratory analyses meeting the QA guidelines.
In addition, laboratory results were used to characterize the variability in target gene copy concentration estimates using two alternative calibration approaches. The first was the ΔΔCt model used in EPA Methods 1609.1 and 1611.1 for enterococci (U.S. EPA, 2015a; 2015b) and the second was a modified master standard curve calibration approach (Sivaganesan et al., 2010). Results from these analyses should provide informative benchmarks for comparing with variability assessments of other qPCR methods designed for widespread environmental water testing and may offer useful insights for the selection of calibration methods for a final version of Draft Method C as well as for other newer qPCR methods.
2.0. MATERIALS AND METHODS
2.1. Participating laboratories and study design.
Laboratory participants are listed in Table 1 along with categorizations based on prior experience with qPCR technology at the time of the study. “New” laboratories received qPCR equipment in 2015 and a subsequent three-day training course on EPA Draft Method C in 2016 but otherwise had relatively little prior experience. “Experienced” laboratories all had substantial prior knowledge and practice with qPCR technology in general but, in most cases, had not previously performed Draft Method C.
Table 1.
Laboratories participating in the study
| Laboratory | Location | Experience with qPCR a |
|---|---|---|
|
| ||
| Central Michigan District Health Department, Assurance Water Laboratory | Gladwin, MI | New |
| City of Racine Public Health Department | Racine, WI | Experienced |
| Ferris State University, Shimadzu Core Laboratory | Big Rapids, MI | New |
| Georgia Southern University, Department of Environmental Health Sciences | Statesboro, GA | Experienced |
| Grand Valley State University, Annis Water Resources Institute | Muskegon, MI | Experienced |
| Health Department of Northwest Michigan, Northern Michigan Regional Laboratory | Gaylord, MI | New |
| Kalamazoo County Health and Community Services Laboratory | Kalamazoo, MI | New |
| Lake Superior State University, Environmental Analysis Laboratory | Sault Sainte Marie, MI | Experienced |
| Marquette Area Wastewater Facility | Marquette, MI | New |
| Michigan State University, Department of Fisheries and Wildlife | East Lansing, MI | Experienced |
| Northeast Ohio Regional Sewer District Environmental and Maintenance Services Center | Cuyahoga Heights, OH | Experienced |
| Oakland County Health Division Laboratory | Pontiac, MI | New |
| Oakland University, HEART Laboratory | Rochester, MI | Experienced |
| Saginaw County Health Department Laboratory | Saginaw, MI | New |
| Saginaw Valley State University, Department of Chemistry | University Center, MI | Experienced |
| University of Illinois at Chicago, School of Public Health | Chicago, IL | Experienced |
| University of North Carolina at Chapel Hill, Institute of Marine Sciences | Morehead City, NC | Experienced |
| University of Wisconsin Oshkosh, Environmental Research Laboratory | Oshkosh, WI | Experienced |
| US NPS Sleeping Bear Dunes Water Laboratory | Empire, MI | Experienced |
| USEPA National Exposure Research Laboratory | Cincinnati, OH | Experienced |
| USGS Upper Midwest Water Science Center | Lansing, MI | Experienced |
Laboratories categorized as “new” received qPCR equipment in 2015 and subsequent training on using qPCR and Draft Method C in 2016 but otherwise had relatively little prior experience.
Except as noted in section 2.2, all materials used to generate test sample and control filters, including membrane filters, DNA extraction tubes and E. coli cells for spiking, came from central laboratories with the capability to provide these materials to all of the laboratories in order to help ensure the consistency of the samples. E. coli cells were also provided to each of the participant laboratories for the preparation of positive control/calibrator sample filters containing ~1E4 CFU/filter as described in Sivaganesan et al. (2019). With the exception of one laboratory, where the filters from 2 samples were lost, each participant laboratory received a total of 54 test sample filters, prepared from 18 different water samples (three replicate filters/sample) as described in section 2.2, for a total of 1128 filters and 376 samples in the study. A standardized testing procedure including templates for performing the qPCR analyses of the samples in the same order in three instrument runs were provided to each of the laboratories. Laboratories were directed to complete the analyses within approximately one month of receipt of the samples. Three negative control and three positive control/calibrator sample filters were prepared and analyzed by each laboratory for each instrument run as described by Sivaganesan et al. (2019).
2.2. Water sample collection and preparation
Ambient water sample sites (n = 6) and their locations are described in Table 2. Three water samples of approximately 8 L were collected from each site at the same time and location. Water samples were kept at 4° C for no longer than 6 hours between collection and initiation of filtration. With exception of the Metropark Beach, Lake St. Clair (SC) site (Table 2), most probable number (MPN) estimates of the E. coli cultivable cell densities were determined by Colilert™ analysis, as per manufacturer’s procedure (Idexx Laboratories, Westport, ME), for each of the ambient samples. From the first 8 L sample at each site, approximately seventy-five, 100 mL aliquots of unaltered ambient surface water were filtered through 47 mm diameter, 0.4 μm pore size polycarbonate filters (Millipore #HTTP04700). Filters containing retentates from each of the water sample filtrations were transferred to sterile 2 mL, semi-conical microcentrifuge tubes prefilled with 0.3 g (± 0.01 g) of 212–300 μm acid washed glass beads (Sigma-Aldrich). After filtering the ambient water samples, spiked water samples were prepared by inoculating E. coli cell suspensions, originating from known dilutions of MultiShot-10E8 BioBalls™ (BioMerieux, Lombard, IL, catalog #56146, lot B3215, mean colony forming unit (CFU) count: 8.086E7 ± 3.915E6 CFU), into the two remaining 8 L water samples to achieve estimated spike concentrations of 200 CFU per 100 mL and 800 CFU per 100 mL, respectively, and then aliquots were filtered as described above. This spiking protocol was applied to the Meinert Park, Lake Michigan (LMM), Twin Lake Park (ITL) and SC site samples (Table 2). The two remaining samples from the Saginaw river (SR) and Saginaw Bay (SB) sites were spiked with 1 mL and either 10 mL or 20 mL of sewage (Bay County Wastewater Trea™ent Plant) and 50 mL aliquots of the sewage-spiked samples were filtered. E. coli cultivable cell densities were also determined by Colilert™ analysis for these samples (Table 2). Due to the expectation of very high ambient E. coli cell densities in the Memorial Park, Lake St. Clair (ME) site samples (subsequently confirmed by Colilert™ analysis - Table 2), the two remaining samples from this site were diluted 5- and 25-fold with water from another site on Lake St. Clair that was predicted by the sample collector to be uncontaminated in order to obtain an expected corresponding range of E. coli cell densities in the final samples. E. coli cultivable cell densities in these samples were determined by Colilert™ analysis (Table 2) and 100 mL aliquots were filtered as described above for the ambient samples. All spiked and diluted water samples were homogenized by inverting the carboys at least 10 times before filtering. The filter tubes were stored at −80° C for 2–3 months before they were distributed to the participants. Filter tubes were express shipped (< 24 hr) to all participants on dry ice with recommendations to store them at −80° C until they were extracted. With one exception, as detailed in section 2.1, each laboratory received a total of 54 filter tubes labeled with identically coded identifiers: 3 replicates of each of 3 sample types (ambient, low, and high spike/dilution), from each of the 6 sampling locations.
Table 2.
Water samples
| Site ID | Site Name | Site Location | WaterBody Type | GPS | Sample ID | Sample Type | E. coli /100 mL |
|---|---|---|---|---|---|---|---|
| SB | Bay County Pinconning Park (Saginaw Bay) |
Pinconning, MI | Great Lakes | 43.85322,
−83.92283 |
1 | Ambient | 65a |
| 2 | Low Spike | 182 a | |||||
| 3 | High Spike | 1158 a | |||||
| SR | Veterans Memorial Park (Saginaw River) |
Bay City, MI | River | 43.59771,
−83.89447 |
4 | Ambient | 12 a |
| 5 | Low Spike | 50 a | |||||
| 6 | High Spike | 579 a | |||||
| ITL | Twin Lake Park | Twin Lake, MI | Inland Lake | 43.36662,
−86.16682 |
7 | Ambient | 3 a |
| 8 | Low Spike | 203 b | |||||
| 9 | High Spike | 803 b | |||||
| LMM | Meinert Park (Lake Michigan) |
Montague, MI | Great Lakes | 43.45908,
−86.45764 |
10 | Ambient | 56 a |
| 11 | Low Spike | 256 b | |||||
| 12 | High Spike | 856 b | |||||
| ME | Memorial Park (Lake St. Clair) |
St. Clair Shores, MI |
Great Lakes | 42.52727,
−82.87133 |
13 | Ambient | 86596 a |
| 14 | Low Dilution |
20535 a,e | |||||
| 15 | High Dilution |
2371 a,e | |||||
| SC | Metropark Beach (Lake St. Clair) |
Macomb County, MI |
Great Lakes | 42.57106,
−82.79645 |
16 | Ambient | ND c |
| 17 | Low Spike | 200 d | |||||
| 18 | High Spike | 800 d |
Most probable number estimate from Colilert™ method by the laboratory that collected the sample.
Estimated from BioBall™ spike levels plus Colilert™ estimates from corresponding ambient samples.
Not determined, negligible density based on preliminary qPCR analysis by the laboratory that collected the sample.
Estimated from BioBall™ spike levels only with ambient levels assumed to be negligible compared to spike levels.
Samples diluted rather than spiked due to high ambient E. coli densities in the collected water samples.
2.3. DNA extraction and qPCR analyses
DNA extractions were performed as described by Sivaganesan et al. (2019). In brief: 600 μL of SAE extraction buffer, containing 0.2 μg/mL of salmon DNA in Qiagen AE buffer, was added to each filter tube; tubes were sealed, bead milled at 5,000 reciprocations/min for 60 s and centrifuged at 12,000 × g for 1 min; supernatants were transferred to clean, low retention micro-centrifuge tubes and centrifuged for an additional 5 min; and approximately 100 μL of the clarified supernatants were transferred to new microcentrifuge tubes for analysis. All DNA extracts were analyzed immediately after extraction.
Each DNA extract was analyzed in duplicate by the EC23S857 assay for E. coli (Chern et al., 2011) and by the Sketa22 assay for the salmon DNA sample processing control (SPC) (U.S.EPA, 2015a,b) on the same instrument run. Analyses were performed as described by Sivaganesan et al. (2019). In brief: reaction mixtures for both assays contained 12.5 μl of Environmental Master Mix (Thermo Fisher Scientific, Microbiology Division, Lenexa, KS, #4396838), 2.5 μL of 2 mg/mL bovine serum albumin, 3 μL of primer-probe mix (for a final concentration of 1 μM of each primer and 80 nM of TaqMan® probe in the reactions), 2 μL of DNA-free water and 5 μL of the DNA extracts for a total reaction volume of 25 μL. Thermal cycling protocols were 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 60 s at 56°C.
Most of the participants used a StepOnePlus™ real-time PCR sequence detector (Applied Biosystems, Foster City, CA), however, laboratories 10 and 14 used a CFX96 real-time PCR detection system (BioRad, Hercules, CA)..
2.4. Raw data reporting and analysis
Instrument-generated Excel export files containing raw, sequence detector-determined quantitative cycle threshold (Cq) measurement data (referred to as Ct measurements, values or data in this article) for each sample and analysis were sent by each of the participants to a central laboratory for compilation and data analysis by a qualified statistician. The compiled Ct data for EC23S857 and Sketa22 assay analyses were identified by assigned lab number, instrument run number, sample type (positive control/calibrator, negative control or unknown test sample) and sample identification number for unknown test samples (see Table 2). Data analyses were performed using SAS (Version 9.2; Cary, NC) and WinBugs (http://www.mrc-bsu.cam.ac.uk/bugs), as detailed in the following sections.
2.5. Data QA analysis and trimming
Except as noted below, data collected by each laboratory was subject to the QA criteria described in Sivaganesan et al. (2019). The hierarchy of parameters for QA evaluation was as follows: 1) standard curve (slope acceptance range: −3.23 to −3.74 and intercept Ct acceptance range: 36.66 to 39.25); 2) positive control/calibrator samples (EC23S857 assay Ct acceptance range: 26.48 to 29.63; Sketa22 assay Ct acceptance range: 18.58 to 22.01); 3) negative control samples (EC23S857 assay Ct acceptance range: > lower limit of quantification Ct value described below); 4) matrix interference by test samples (Sketa22 assay Ct acceptance range for test samples: within 3 units of mean from corresponding positive control samples); 5) variability of test sample EC23S857 assay Ct measurements (acceptance range for standard deviation from duplicate measurements: <1.44); and 6) lower limit of quantification (LLOQ) for EC23S857 assay Ct measurements of test samples (Ct acceptance range: > global LLOQ Ct value of 35.17). Standard curve results from Sivaganesan et al. (2019) were used to determine acceptability of the master standard curves for each laboratory. In addition, test samples were deemed eligible for EC23S857 target copy estimations if at least 2 of 3 replicate filters and at least 4 out of 6 corresponding positive and negative control samples in each instrument run met the QA criteria described above. For Sketa22 analyses, only one replicate test sample filter was required to meet the matrix interference QA criterion. For the EC23S857 assay analyses, only one of the duplicate analyses of a test sample filter was required to give a Ct measurement within 40 cycles (the total number of thermal cycles performed for each analysis) for that filter to be used for further analyses.
2.6. Delta-Delta Ct calibration method
The ΔΔCt model used in this study for estimating the number of E. coli target gene copies (X0 ) from an unknown water sample is given by:
| (1) |
where ΔCt0 is the difference between the mean EC23S857 assay and mean Sketa22 assay Ct measurements of each unknown sample filter, and are the master standard curve intercept and slope, previously defined for each laboratory (Sivaganesan et al., 2019), Ctc and Ctcs are the mean EC23S857 assay and mean Sketa22 assay Ct values, respectively, for corresponding instrument run calibrator/positive control sample analyses passing QA, denoted by . is the mean of all initial calibrator sample EC23S857 assay Ct values previously defined for each laboratory (Sivaganesan et al., 2019). , and were assumed to have the following known normal distributions:
| (2) |
where, and are the estimated mean and standard deviation of the ΔCt0 values for the three replicate sample filters of each unknown water sample, and Δmc and Δsc are the estimated mean and standard deviation of the corresponding calibrator/positive control samples ΔCtc values from the same instrument run. Filter variability was accounted for via a one-way analysis of variance (ANOVA) model in estimating Δs0 and Δsc. Moreover, the overall mean and standard deviation estimates of Ct values of the initial calibrators were used to estimate mean (mic) and standard deviation (𝑠𝑖𝑐) of . Instrument run to run variability, as well as filter within run variability were incorporated in estimating 𝑠𝑖𝑐 via a nested ANOVA model. WinBugs codes for performing these analyses are provided as supplemental material.
2.7. Master standard curve calibration method
When using the master standard curve model to estimate the number of E. coli target gene copies (X0) per analysis in the unknown water sample extracts, equation (1) was modified to:
| (3) |
which does not include the initial or instrument run specific calibrator Ct measurements for the EC23S857 assay, but does continue to include the mean of the Sketa22 assay Ct measurements (Ctcs) from the corresponding calibrator/positive control samples in the respective instrument run (again assumed to have a known normal distribution with mean ct1 and standard deviation s1). As in the case of ΔΔCt model, filter variability was incorporated in s1:
| (4) |
Thus the ΔΔCt and master curve models agree, in theory, if the mean EC23S857 assay Ct values of the run specific calibrator/positive control samples and the initial calibrator samples are the same, i.e. . WinBugs codes for performing these analyses are provided as supplemental material.
2.8. Method variability analysis
For a given sample, between-laboratory and total variabilities were estimated from all results passing the QA criteria described above via the following model:
| (5) |
Where m is the number of labs for a given sample, and are estimated mean and standard deviation of log10 copies for ith lab and are respectively the overall mean log10 copies per analysis and standard deviation of between lab variability. is the estimated standard deviation of overall within lab variability, which is the average of all ’s (I = 1, 2..,m). The standard deviation for the total variability, including both within and between laboratories is denoted by . Non-informative normal and gamma prior distributions are assumed for and , respectively. For each sample, the posterior distribution of a normal random variable with mean ɤ̅ and standard deviation σ𝑇 was used to estimate the mean and 95% Bayesian credible interval (BCI) for the overall mean log10 gene copies per PCR analysis.
As sometimes different or unequal numbers of samples passed all QA criteria for each laboratory, a subset of samples and labs were selected to generate a balanced data set to estimate between-lab, within-lab and total variabilities. Data from 11 labs and 13 samples were selected for this analysis as described above with m = 11. Codes for performing these analyses are provided as supplemental material as indicated above.
3.0. RESULTS
3.1. Laboratory QA performance
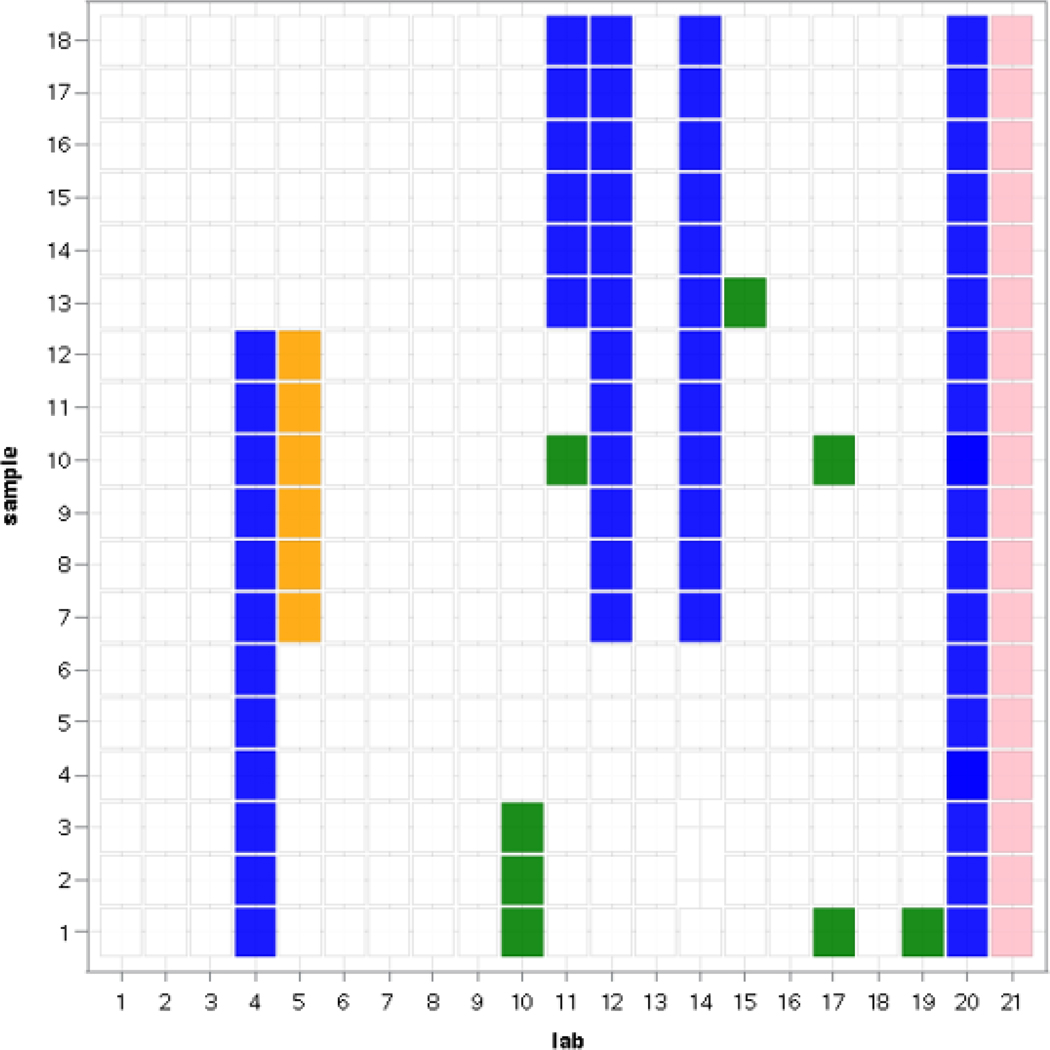
One of the major objectives of this study was to investigate how consistently a diverse group of laboratories with varying levels of prior qPCR experience could perform Draft Method C, based on their success in meeting of standardized QA guidelines. Fig. 1 shows the laboratory and sample combinations that had to be excluded due to QA failures as indicated in section 2.5. The first 5 QA categories were considered to specifically relate to laboratory performance of the method. Failures to meet the sixth category: EC23S857 assay Ct measurements > global LLOQ, were considered indicative of unusable data but not necessarily to reflect laboratory performance. All of these QA categories are equally applicable to the ΔΔCt and standard curve calibration methods. As indicated in section 2.5, data from Laboratory 21 was excluded due to the failure to meet QA criteria for their master standard curve. A total of 60 out of the remaining total of 358 sample analyses (excluding Laboratory 21 and 2 missing sample analyses from Laboratory 14) failed to meet the QA criteria for the positive control/calibrator samples EC23S857 and Sketa22 assay Ct values. More informatively, since each failure to meet these criteria affected 6 sample analyses in an instrument run, 10 out of the total of 60 instrument runs (17%, excluding Laboratory 21) failed these criteria. Among the 10 failed runs, 2 were caused by EC23S857 assay failures, 5 by Sketa22 assay failures and 3 by failures of both assays (data not shown). Only 6 sample analyses (1 of the 60 instrument runs) failed the negative control samples EC23S857 assay criterion. Only 8 individual sample analyses, originating from 5 laboratories and 5 samples, failed the Sketa22 assay criterion for assessing matrix interference and no eligible water sample analyses failed the criterion for excessive variability of duplicate EC23S857 assay Ct measurements.
Fig. 1.
Diagram showing laboratory/sample analysis combinations that failed to meet any one of a hierarchy of five data quality acceptance categories under the conventions of this study as described in section 2.5. Color codes for QA categories: pink = standard curve (slope and intercept); blue = positive control/calibrator samples EC23S587 and Sketa22 assay Ct values; orange = negative control samples EC23S587 assay Ct values; green = matrix interference by test samples as assessed by Sketa22 assay Ct values.
3.2. Variability of water sample analysis results
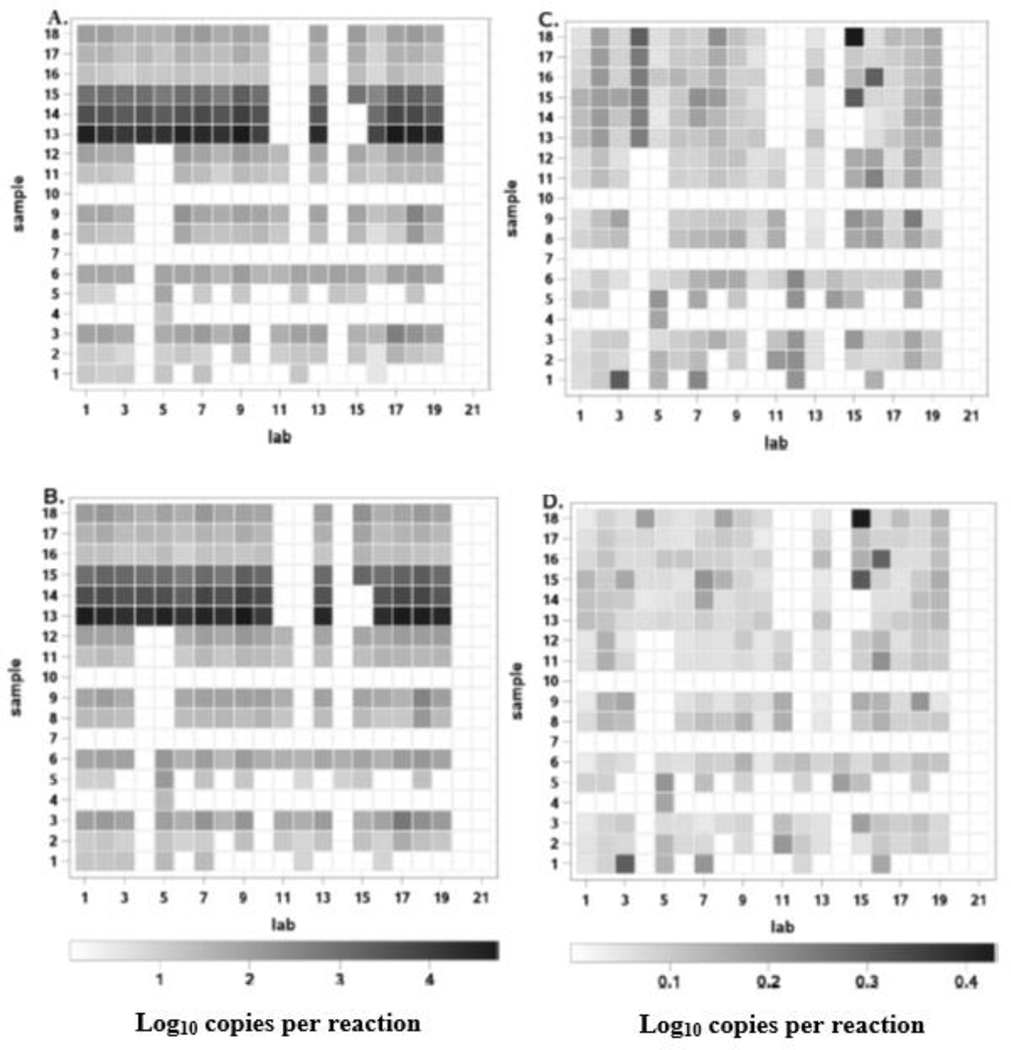
A second major objective of the study was to comprehensively evaluate the variability of target gene copy estimates obtained by the laboratories from analyses of a variety of ambient water matrices when all QA criteria were met. As indicated in the introduction, another component of this analysis was to compare the variability of target gene copy estimates obtained by the two alternative calibration methods. Fig. 2 shows heat maps representing either the posterior means of log10 EC23S857 assay target gene copies per reaction for each of the 18 samples from each lab’s data that passed the QA criteria, as estimated by the ΔΔCt (Fig. 2A) and standard curve (Fig. 2B) methods, respectively. Corresponding standard deviations are represented in Fig. 2C and Fig. 2D. The quantitative estimates ranged from log10 0.5 to 4.8 for means and from log10 0.03 to 0.43 for standard deviations. Samples analyses that failed any of the first 5 QA criteria listed in section 2.5 (see Fig. 1) or where mean Ct measurements by the EC23S857 assay exceeded the global LLOQ are shown in white in all panels. Corresponding numeric mean and standard deviation values and QA failure codes for each sample analysis are provided in Table S1.
Fig. 2.
Heat map representations of posterior means and standard deviations of log10 copies per reaction for each laboratory/sample analysis combination generated by different calibration models. Panel A: posterior means from ΔΔCt model. Panel B: posterior means from Master Standard Curve model. Panel C: standard deviations from ΔΔCt model. Panel D: standard deviations from Master Standard Curve model. For all panels: white cells indicate <LLOQ, QA failure (see Fig.1) or missing data.
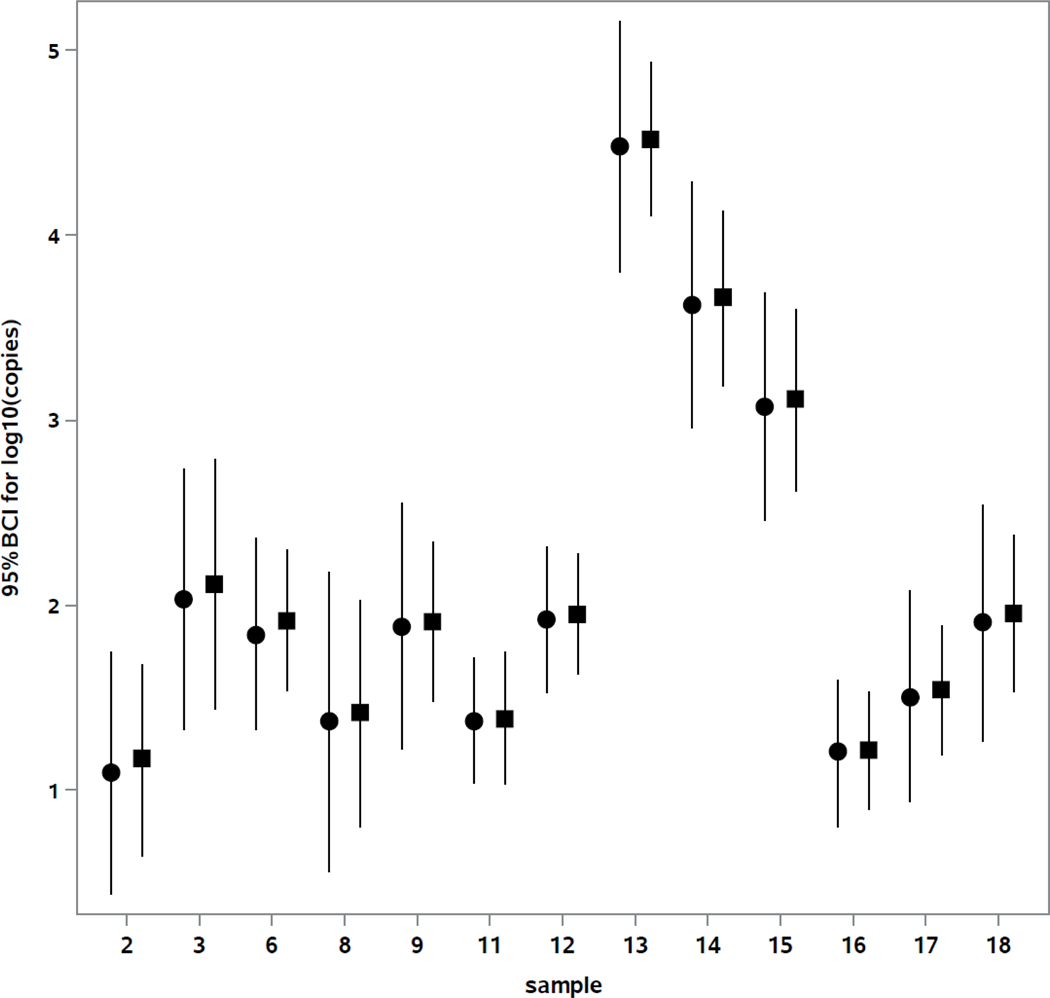
Table 3 shows the means and standard deviations based on total lab variabilities of log10 EC23S857 assay target gene copies per reaction, as determined using both the ΔΔCt and standard curve methods, for each of 13 water samples where analyses by > 50% of the laboratories produced data that met all 6 QA categories described in section 2.5. Four of the 5 excluded samples where most of the laboratories obtained mean EC23S857 assay Ct measurements exceeding the global LLOQ Ct value were un-spiked water samples with low ambient densities of culturable E. coli (Table 2). As indicated in Table 3, the total number of laboratories contributing acceptable data for each these 13 water samples ranged from 15 to 18 and there was not complete consistency from sample to sample in the laboratories that were excluded due to QA failures. To obtain a more statistically balanced assessment of method variability, a second analysis using results from only 11 of the labs that produced acceptable data for all 13 of these samples was therefore conducted. Results from these analyses, including estimates of between-lab (), within-lab () as well as total variabilities, are also shown in Table 3. Estimates of means and total variability, expressed as 95% BCI’s, for each of the 13 samples from this subset of data are also graphically portrayed in Fig. 3.
Table 3.
Estimated mean log10 target gene copies/reaction and variability by sample
| All Lab Data | Balanced Data (N = 11 labs)a | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method: | DDb | SCc | DD | SC | DD | SC | DD | SC | DD | SC | DD | SC | |
| Sample | Labs (N) | Mean()d | Total ()e | Mean() d | Total ()e | Between ()f | Within ()g | ||||||
| 2 | 15 | 1.103 | 1.164 | 0.289 | 0.239 | 1.096 | 1.169 | 0.307 | 0.242 | 0.293 | 0.230 | 0.090 | 0.074 |
| 3 | 16 | 1.950 | 2.000 | 0.327 | 0.342 | 2.035 | 2.110 | 0.328 | 0.314 | 0.314 | 0.304 | 0.095 | 0.077 |
| 6 | 18 | 1.802 | 1.850 | 0.235 | 0.227 | 1.842 | 1.915 | 0.241 | 0.177 | 0.216 | 0.151 | 0.103 | 0.089 |
| 8 | 15 | 1.361 | 1.427 | 0.337 | 0.262 | 1.373 | 1.417 | 0.381 | 0.287 | 0.361 | 0.265 | 0.118 | 0.105 |
| 9 | 15 | 1.856 | 1.907 | 0.276 | 0.179 | 1.882 | 1.909 | 0.306 | 0.198 | 0.284 | 0.170 | 0.110 | 0.096 |
| 11 | 15 | 1.324 | 1.360 | 0.188 | 0.164 | 1.374 | 1.386 | 0.161 | 0.170 | 0.117 | 0.141 | 0.106 | 0.091 |
| 12 | 15 | 1.856 | 1.906 | 0.242 | 0.175 | 1.926 | 1.950 | 0.179 | 0.154 | 0.148 | 0.130 | 0.096 | 0.080 |
| 13 | 15 | 4.420 | 4.474 | 0.318 | 0.202 | 4.481 | 4.518 | 0.314 | 0.196 | 0.290 | 0.170 | 0.117 | 0.095 |
| 14 | 15 | 3.608 | 3.668 | 0.281 | 0.192 | 3.625 | 3.663 | 0.307 | 0.224 | 0.283 | 0.201 | 0.116 | 0.095 |
| 15 | 16 | 3.023 | 3.084 | 0.284 | 0.217 | 3.072 | 3.111 | 0.289 | 0.230 | 0.257 | 0.203 | 0.128 | 0.105 |
| 16 | 16 | 1.213 | 1.245 | 0.179 | 0.151 | 1.209 | 1.215 | 0.189 | 0.150 | 0.133 | 0.102 | 0.130 | 0.106 |
| 17 | 16 | 1.479 | 1.539 | 0.259 | 0.160 | 1.505 | 1.541 | 0.266 | 0.163 | 0.242 | 0.138 | 0.109 | 0.084 |
| 18 | 16 | 1.867 | 1.934 | 0.297 | 0.205 | 1.907 | 1.954 | 0.304 | 0.198 | 0.285 | 0.182 | 0.103 | 0.078 |
All analyses contributing to the balanced dataset were generated on StepOnePlus™ instruments
DD = ΔΔCt model
SC = Master standard curve model.
Overall mean log10 copies per analysis reaction.
Standard deviation for total variability, including both within and between labs
Standard deviation of overall between lab variability
Standard deviation of overall within lab variability
Fig. 3.
Overall mean log10 copies plus 95% BCI per reaction for balanced data from 11 laboratories. ●: ΔΔCt calibration model, ■: Master Standard Curve calibration model.
4.0. DISCUSSION
4.1. Data QA criteria and laboratory performance
The goal of utilizing QA criteria in Draft Method C is to ensure, to the greatest extent possible, that potentially extreme and erroneous test sample quantitative results will be identified in future implementations of the method. A total of 92 of the 376 test sample analyses conducted (24%) failed at least 1 of the 5 QA performance related categories (excluding the global LLOQ metric). This seemingly high percentage of failures raises the question of whether the imposed QA metrics are overly restrictive or whether they are truly detecting and excluding what would otherwise be erroneous estimates of analyte (E. coli target gene copy) concentrations in the samples due to suboptimal laboratory performance. The availability of reference variability estimations (standard deviations and 95% BCI values) of E. coli target gene copy estimates from the laboratories that met the QA criteria for each of the 13 samples that also gave acceptable quantitative data (>LLOQ) in this study allowed for an assessment of this question.
As described in Sivaganesan, et al. (2019) and shown in Fig. 1, results of all sample analyses by Laboratory 21 were excluded due to a high standard curve intercept value. Results shown in Table S1 also reveal that this laboratory failed the QA criteria for positive control/calibrator samples associated with samples 13–18. It was observed that after excluding these samples, the mean E. coli target gene copy estimates for 6 of the 7 quantifiable (>LLOQ) samples remaining from this laboratory were outside of the 95% BCI bounds established by the laboratories that passed all QA criteria when using the master standard curve calibration model. Conversely, none of the estimates for these samples were outside of the 95% BCI bounds established by the ΔΔCt model. These results illustrate the primary difference between the two calibration models wherein the Ct values from the EC23S857 assay for the calibrator samples from each instrument run are essentially used to adjust the intercept of the master standard curve for that run in the ΔΔCt model. In this instance, the high master standard curve intercept value obtained by Laboratory 21 was offset by comparably high calibrator sample Ct values in the ΔΔCt model which resulted in comparable E. coli target gene copy estimates to those of the other laboratories. Nevertheless, results of this nature also may be indicative of subpar sensitivity of the method in this laboratory.
The QA category responsible for the largest number of failures by far was the positive control/calibrator samples EC23S857 and/or Sketa22 assay results with 65% of all failures being associated with this category as illustrated in Fig. 1. Within this category, failures to meet the Sketa22 assay acceptance criterion were more common with 80% of the failed sample analyses being associated with this assay as opposed to 50% for the EC23S857 assay (30% failed the criteria for both assays). Possible reasons for these failures were identified in some instances and are yet to be confirmed in others but could include storage temperatures and numbers of freeze/thaw cycles of the salmon DNA and cellular E. coli control materials, holding time between preparation and extraction of positive control/calibrator samples and variability in preparing new working stock solutions of SAE extraction buffer. Several laboratories reported an inability to store DNA standards and cellular E. coli control materials at the recommended temperature of −80° C due to unavailability of an appropriate freezer. It is again noteworthy that failures to meet the acceptance criteria established for this QA category often did not equate to extreme E. coli target sequence copy estimates from the quantification models in relation to the results of the other laboratories that passed all QA categories. For example, among the 46 otherwise quantifiable (>LLOQ) sample analyses that failed this QA category, E. coli target gene copy estimates were outside of the 95% BCI bounds established by the laboratories that passed all QA categories for only 11 (24%) and 8 (17%) of these analyses using the master standard curve and ΔΔCt calibration models, respectively (data not shown). Most of these instances (9 and 6, respectively) occurred in a total of just 3 instrument runs by 2 laboratories suggesting that these runs were compromised. Thus, the multi-laboratory data available from this study indicated that the proposed QA criteria metrics for positive control/calibrator sample results can exclude valid sample analysis results but also showed that these criteria were able to identify compromised instrument runs. While further refinements to the proposed QA criteria for positive controls in Draft Method C should be explored, the availability of such criteria are particularly important when, as normally would be the case, no other option is readily available for objectively assessing day-to-day laboratory performance of this method.
The next most common source of failed analyses was presumptive matrix interference based on a previously established acceptance criterion for Sketa22 assay results (U.S. EPA, 2015a; 2015b; Sivaganesan, et al., 2019). This criterion is designed to assess the suitability of a DNA extract for qPCR amplification and is typically not considered an indicator of laboratory performance. The highly sporadic and rare occurrence of analysis failures across the laboratories in this QA category (Fig. 1) suggests, however, that none of the samples truly harbored substances that interfered with qPCR testing. Instead, these failures more likely suggest suboptimal performance of specific analyses, possibly caused by operator error in setting up these reactions. Previous studies have suggested a possible inverse relationship between frequencies of sample matrix interference and the assay amplification efficiency [10(−1/master slope) −1] (Haugland et al., 2016) but no such relationship was evident in this study. The broader question of how diverse ambient surface waters from more widely different geographic locations in Michigan and across the entire U.S. affect the performance of this method has been examined (U.S. EPA, unpublished data) and will be addressed in subsequent reports.
The last two QA categories considered to be related to laboratory performance: negative control samples EC23S857 assay Ct values and excessive variability of duplicate EC23S857 assay Ct measurements in individual test sample filters, were the source of only six failed sample analyses in the entire study (all coming from unacceptably low negative control Ct values in just one instrument run). The conventions described in section 2.5 for applying these criteria in this study: at least two of the three replicate filters of each test sample and at least four out of the six analyses of the corresponding negative control samples in each instrument run having to meet the established criteria, would not be applicable in other studies where replicate filters of test samples are not analyzed. Results from this study therefore may be underestimating the potential impact of these two QA categories on frequencies of failed sample analyses in other such studies.
Close inspection of the results shown in Fig. 2 reveals several rare instances of sample analysis results by individual laboratories that passed all five of the QA categories but still showed atypical mean target sequence copy estimates in relation to the other laboratories. The most striking example of this occurred in the analysis results for sample 14 by Laboratory 15 where most of the laboratories obtained mean log10 target sequence copy estimates of ~3.5 whereas the estimate from this laboratory was < global LLOQ. Examination of Ct measurements from this laboratory revealed that while the six Sketa22 assay results for this sample were generally consistent and acceptable following the conventions of this study, the EC23S857 assay results were highly variable (Ct values ranging from 25.87 to non-detect or “undetermined”, as indicated by the instrument outputs) suggesting operator error in setting up the EC23S857 reactions for this sample. Such errors could be detected by the variability test for duplicate EC23S857 assay Ct measurements if Ct values of 40 were assigned to “undetermined” reactions and also likely would be detected by the duplex internal amplification control (IAC) assay that is available as an option for Draft Method C but was not employed in this study (Sivaganesan et al., 2019).
The lack of extensive prior laboratory experience with qPCR technology appeared to contribute to the overall frequencies of failed analyses based on the five QA categories with the seven laboratories categorized as “new” in Table 1 contributing 48% of the total failed analyses. Of these seven laboratories, however, only two were responsible for 39% of the total failures. Both of these laboratories also exhibited high run to run variability in Sketa22 assay Ct measurements during preliminary analyses (Sivaganesan et al., 2019) suggesting within-lab variability in the preparation of SAE buffer working stocks. These laboratories in particular will require further assistance before implementing the method. The inability of these laboratories to maintain the DNA standards and cellular control materials at the recommended −80° C storage temperature also may have contributed to their failed analyses. Thus, the availability of a −80° C freezer, ideally with a backup power system, may be necessary for laboratories that intend to use qPCR analyses for beach monitoring. Conversely, the QA failure rate among the other five “new” laboratories was only 9% compared to 19% for the 14 “experienced” laboratories. These results suggest that the training received by these five laboratories on Draft Method C was beneficial and that additional practice or assistance, specifically with Method C, also may be beneficial for some otherwise more experienced laboratories. The current effort by the state of Michigan to establish a network of multiple laboratories and personnel with demonstrated proficiency in Draft Method C gives an example of one approach for providing support to newer laboratories that may be initially struggling with the implementation of a new qPCR method.
4.2. Calibration models and method variability
The design of this study provided an opportunity to directly compare the variability of target gene copy estimates generated by the ΔΔCt calibration model used in current EPA qPCR methods for enterococci (U.S. EPA, 2015a,b) and by a master standard curve model similar to those used in many other qPCR studies (Sivaganesan et al., 2010). Since Draft Method C has not yet been published as an EPA method, it is conceivable that its final version could incorporate either of these calibration models based on relative performance in minimizing variability and other considerations. A demonstration that these models produce similar mean target sequence copy estimates also could be useful for future comparisons of results from this method with those of other EPA or non-EPA qPCR methods that may use different models.
The results in Table 3 and Fig. 3 show that the total standard deviations and 95% BCIs of E. coli target gene copy estimates were lower for most sample analyses using the master standard curve model. This observation is expected from the standpoint that the repeated EC23S857 assay measurements of the preliminary and run-specific calibrator samples introduce additional sources of random and sample preparation-related variability into the ΔΔCt calibration model that are not included in the master standard curve model. Day-to-day or laboratory-specific differences in the measurements of run-specific calibrator samples by this assay must reflect corresponding differences in the measurements of test samples for the ΔΔCt calibration model to overcome its greater inherent potential for variability. Specific examples where this compensation may have occurred can be seen, such as in the Laboratory 21 results from this study referred to in section 4.1. However, the overall results of this study suggest that this was not normally the case.
The results in Table 3 and Fig. 3 also show that the mean estimates of E. coli target gene copies were higher from the master standard curve model than from ΔΔCt calibration model for all sample analyses. The mean, median and maximum differences among the samples were only 0.054, 0.054 and 0.069 log10 copies per reaction, respectively, based on all accepted laboratory analyses. These differences were not significant in relation to the 95% BCIs for each of the sample estimates as shown in Fig. 3. As mentioned in section 2.7, the ΔΔCt and standard curve models should agree in theory if the mean EC23S857 assay Ct values of the run specific calibrator/positive control samples and the initial calibrator samples are the same. Additional studies using different calibrator cell preparations would be needed to conclude whether the slight differences in target gene copy estimates observed in this study reflect any consistent, systematic bias in using one or the other of the two models in this method.
4.3. Comparison of method variability with other qPCR methods and studies
Several previous studies have employed similar designs to this study in evaluating the inter-laboratory variability of other qPCR methods for genetic markers from general or source-specific FIB in surface waters (Griffith and Weisberg, 2006; Ebentier et al., 2013; Kinzelman et al., 2013; Shanks et al., 2012; Shanks, et al., 2016). The differences in the quantitative models, data analysis methods and/or the reporting of results from those studies largely preclude direct comparisons with the results of this study. For example, many of the earlier studies modeled and reported FIB density estimates as cell equivalents rather than target sequence copy estimates. Also, the scaling of target sequence copy estimates to be expressed on a per water sample volume rather than on a per reaction basis and conversions from original log10 unit estimates from standard curves to arithmetic values are common practices. Such practices often may be necessary for comparing the results of qPCR methods with those of other methods such as culture or with existing recreational water quality criteria values, or when different water sample or DNA extract volumes are analyzed. However, arithmetic qPCR results should be reported as median copies and assessments of the variability of qPCR methods should be reported in the log10 scale on a per reaction basis with information provided on any deviations from the standard sample extraction and analysis volumes of the method.
The data reporting from this study followed this approach and employed traditional and Bayesian statistics to take into account most sources of variability in the quantification models, including: replicate standards curves from replicate analyses; replicate whole cell calibrator samples and analyses (when applicable for the ΔΔCt calibration model); replicate test sample filters and analyses within laboratories; and finally variability across laboratories, for data analysis of water samples from diverse sites that contained a wide range of E. coli cell densities (Table 2). One common observation from this (Table 3) and other studies (Ebentier et al., 2013) is that variability between laboratories was consistently the greatest contributor to overall method variability. Despite this, the overall 95% BCIs for many of the samples in this study, particularly from the master standard curve model, were nearly the same as previously reported values of ~0.6 from sample analyses by a single laboratory using EPA Methods 1609.1 and 1611.1 (Sivaganesan et al., 2014). This value of 0.6 log10 copies (equating to a ± two-fold target sequence recovery range from the expected values for spiked samples) has been used as a benchmark in subsequent studies to evaluate the performance of Methods 1609.1 and 1611.1 by multiple laboratories (Haugland et al., 2016). The use of similar study designs and data analysis approaches to those of the present study is recommended for future studies that wish to assess the quantitative variability of qPCR methods for FIB in water. The variability ranges of log10 gene copy/reaction estimates obtained from samples with varying densities of target organisms determined in this study may be useful for evaluating the results of such similarly designed studies of Draft Method C or other comparable methods.
5.0. CONCLUSIONS
Previously established standardized data quality acceptance criteria for five QA categories were used to evaluate the results of 21 laboratories with varying levels of prior experience with qPCR technology in performing Draft Method C for qPCR analysis of E. coli in diverse freshwater recreational waters. Failures to meet these criteria occurred in 24% of all laboratory/sample analyses with the major contributing category being unacceptable E. coli and salmon DNA SPC assay Ct values from calibrator/positive control samples.
Failures to meet acceptable E. coli and salmon DNA SPC assay Ct values for calibrator/positive control samples were often not associated with extreme or erroneous E. coli target sequence copy estimates in the samples. Instances of such associations occurred in approximately 20% of the analyses that failed this QA category and primarily in just three instrument runs in the entire study. These observations point to the need for these controls and criteria in evaluating the day-to-day performance of the method by individual laboratories but also suggest that the development of further refinements to the proposed QA criteria for positive controls in Draft Method C should be explored.
Variations in the percentages of sample analyses meeting all QA criteria among both “new” and more generally experienced laboratories suggest that further assistance or practice with Draft Method C may be beneficial for some laboratories in both groups.
Variations in the storage and preparation of reference and control materials, particularly by two of the laboratories, were suggested to be potentially important factors in contributing to QA failures.
The master standard curve calibration model produced lower overall variability, expressed as standard deviations and 95% BCIs, in log10 target gene copy estimates than the ΔΔCt calibration model in most sample analyses. However, mean log10 target sequence copy concentration estimates in the sample analyses were not significantly different using these two calibration models and variability between the laboratories was the greatest contributor to the overall variability of the method in either case.
To the extent to which comparisons were possible, the variability of Draft Method C quantitative estimates in this study appears to be consistent with those from previous studies of other qPCR methods for analysis of FIB genetic markers in ambient water samples.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank each of the organizations and laboratories listed in Table 1 that participated in this study as well as Brian Scull (Grand Valley State University), Dr. David Szlag and Hamzah Ansari (Oakland University) and Matt Flood (Michigan State University) for providing water samples and logistical support. Also thanked are Drs. Orin Shanks and Eric Villegas (EPA, Office of Research and Development), Drs. Lemuel Walker and John Ravenscroft (EPA, Office of Water), Dr. Susan Spencer (USDA) and Dr. Christina Kellogg (USGS) for their critical review of this manuscript. The views expressed in this article are those of the authors and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
REFERENCES
- Byappanahalli MN, Nevers MB, Shively DA, Spoljaric A, Otto C, 2018. Real-Time Water Quality Monitoring at a Great Lakes National Park. J. Environ. Qual. Spec. Sec. Microb. Water Qual. Monit. Model 47(5), 1086–1093. [DOI] [PubMed] [Google Scholar]
- Chern EC, Siefring SC, Paar J, Doolittle M, Haugland RA, 2011. Comparison of quantitative PCR assays for Escherichia coli targeting ribosomal RNA and single copy genes. Lett. Appl. Microbiol 52, 298–306. [DOI] [PubMed] [Google Scholar]
- Dorevitch S, Shrestha A, DeFlorio-Barker S, Breitenbach C, Heimler I, 2017. Monitoring urban beaches with qPCR vs. culture measures of fecal indicator bacteria: Implications for public notification. Environ. Health 16(1), 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebentier DL, Hanley KT, Cao Y, Badgley BD, Boehm AB, Ervin JS, Goodwin KD, Gourmelon M, Griffith JF, Holden PA, Kelty CA, Lozach S, McGee C, Peed LA, Raith M, Ryu H, Sadowsky MJ, Scott EA, Domingo JS, Schriewer A, Sinigalliano CD, Shanks OC, Van De Werfhorst LC, Wang D, Wuertz S, Jay JA, 2013. Evaluation of the repeatability and reproducibility of a suite of qPCR-based microbial source tracking methods. Water Res. 47, 6839–6848. [DOI] [PubMed] [Google Scholar]
- Griffith JF, Weisberg SB, 2006. Evaluation of rapid microbiological methods for measuring recreational water quality. Southern California Coastal Water Research Project, Costa Mesa, CA. Technical report 485. [Google Scholar]
- Griffith JF, Weisberg SB, 2011. Challenges in implementing new technology for beach water quality monitoring: Lessons from a California demonstration project. Mar. Technol. Soc. J 45(2), 65–71. [Google Scholar]
- Haugland RA, Siefring SC, Wymer LJ, Brenner KP, Dufour AP, 2005. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res. 39 (4), 559–568. [DOI] [PubMed] [Google Scholar]
- Haugland RA, Siefring S, Varma M, Oshima KH, Sivaganesan M, Cao Y, Raith M, Griffith J, Weisberg SB, Noble RT, Blackwood AD, Kinzelman J, Anan’eva T, Bushon RN, Harwood VJ, Gordon KV, Sinigalliano C, 2016. Multi-laboratory survey of qPCR enterococci analysis method performance in U.S. coastal and inland surface waters. J. Microbiol, Methods 123, 114–125. [DOI] [PubMed] [Google Scholar]
- Kinzelman J, Anan’eva T, Mudd D, 2013. Evaluation of rapid bacteriological analytical methods for use as fecal indicators of beach contamination. Report prepared for: U.S. Environmental Protection Agency, Office of Science and Technology, Region 5 Water Division, EPA Contract #EP-11–5-000072. [Google Scholar]
- Noble RT, Blackwood AD, Griffith JF, McGee CD, & Weisberg SB, 2010. Comparison of rapid QPCR-based and conventional culture-based methods for enumeration of Enterococcus sp. and Escherichia coli in recreational waters. Applied and Environ. Microbiol. 76(22), 7437–7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks O, Sivaganesan M, Peed L, Kelty C, Blackwood A, Greene M, Noble R, Bushon R, Stelzer E, Kinzelman J, Anna’eva T, Sinigalliano C, Wanless D Griffith J, Cao Y, Weisberg S, Harwood VJ, Staley C, Oshima K, Varma M, Haugland R, 2012. Inter-laboratory comparison of real-time PCR methods for quantification of general fecal indicator bacteria. Environ. Sci. Technol 46, 945–953. [DOI] [PubMed] [Google Scholar]
- Shanks O, Kelty C, Oshiro R, Haugland R, Madi T, Brooks L, Field K, Sivaganesan M, 2016. Data Acceptance Criteria for Standardized Human-Associated Fecal Source Identification Quantitative Real-Time PCR Methods. Appl. Environ. Microbiol 82, 2773–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaganesan M, Haugland RA, Chern EC, Shanks OC, 2010. Improved strategies and optimization of calibration models for real-time PCR absolute quantification. Water Res. 44, 4726–35. [DOI] [PubMed] [Google Scholar]
- Sivaganesan M, Siefring S, Varma M, Haugland RA, 2014. Comparison of Enterococcus quantitative polymerase chain reaction analysis results from midwestern U.S. river samples using EPA Method 1611 and Method 1609 PCR reagents. J. Microbiol. Methods 101, 9–17, Corrigendum 115, 166. [DOI] [PubMed] [Google Scholar]
- Sivaganesan M, Aw TG, Briggs S, Dreelin E, Aslan A, Dorevitch S, Shrestha A, Isaacs N, Kinzelman J, Kleinheinz G, Noble R, Rediske R, Scull B, Rosenberg L, Weberman B, Sivy T, Southwell B, Siefring S, Oshima K, Haugland R, 2019. Standardized data quality acceptance criteria for a rapid Escherichia coli qPCR method (Draft Method C) for water quality monitoring at recreational beaches. Water Res. 10.1016/j.watres.2019.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. EPA, 2012. Recreational Water Quality Criteria. United States Environmental Protection Agency, Office of Water, Washington, D.C. EPA-820-F-12–058. [Google Scholar]
- U.S. EPA, 2015a. Method 1609.1: Enterococci in water by TaqMan® quantitative polymerase chain reaction (qPCR) assay with internal amplification control (IAC) assay. [Google Scholar]
- United States Environmental Protection Agency, Office of Water, Washington, D.C. EPA-820-R-15–009. [Google Scholar]
- U.S. EPA, 2015b. Method 1611.1: Enterococci in water by TaqMan® quantitative polymerase chain reaction (qPCR) assay. United States Environmental Protection Agency, Office of Water, Washington, D.C. EPA-820-R-12–008. [Google Scholar]
- Wade TJ, Calderon RL, Brenner KP, Sams E, Beach M, Haugland R, Wymer L, Dufour AP, 2008. High sensitivity of children to swimming-associated gastrointestinal illness: results using a rapid assay of recreational water quality. Epidemiology 19(3), 375–383. [DOI] [PubMed] [Google Scholar]
- Wade TJ, Sams E, Brenner KP, Haugland R, Chern E, Beach M, Wymer L, Rankin CC, Love D, Li Q, Noble R, Dufour AP, 2010. Rapidly measured indicators of recreational water quality and swimming-associated illness at marine beaches: a prospective cohort study. Environ. Health 9, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.