Abstract
Duchenne muscular dystrophy (DMD) is a progressive severe muscle‐wasting disease caused by mutations in DMD, encoding dystrophin, that leads to loss of muscle function with cardiac/respiratory failure and premature death. Since dystrophic muscles are sensed by infiltrating inflammatory cells and gut microbial communities can cause immune dysregulation and metabolic syndrome, we sought to investigate whether intestinal bacteria support the muscle immune response in mdx dystrophic murine model. We highlighted a strong correlation between DMD disease features and the relative abundance of Prevotella. Furthermore, the absence of gut microbes through the generation of mdx germ‐free animal model, as well as modulation of the microbial community structure by antibiotic treatment, influenced muscle immunity and fibrosis. Intestinal colonization of mdx mice with eubiotic microbiota was sufficient to reduce inflammation and improve muscle pathology and function. This work identifies a potential role for the gut microbiota in the pathogenesis of DMD.
Keywords: Duchenne muscular dystrophy, gut microbiota, immunity, skeletal muscle metabolism, T‐lymphocytes
Subject Categories: Digestive System; Microbiology, Virology & Host Pathogen Interaction; Musculoskeletal System
The susceptibility of DMD patients to inflammatory events cannot solely be explained by skeletal muscle genetic defects but rather favors a new paradigm linking the development of chronic inflammation with a strict regulation between epigenetics factors and degenerative environment.

The paper explained.
Problem
In the mucosa of the gastrointestinal tract, immune cells drive innate and adaptive responses, and thus inflammatory events that are common features of Duchenne muscular dystrophy (DMD). Although the gut microbiota likely influences muscle metabolism and physiology, molecular players involved in the gut‐muscle axis remain to be identified. In patients with muscular dystrophies, the lack of dystrophin determines a severe impairment of the intestinal homeostasis. Gastrointestinal disturbances may therefore belong to the clinical symptoms and can appear long before the typical DMD musculoskeletal features occur. Although microbiota modulation could influence DMD inflammatory environment, gastrointestinal dysfunctions in this disease has not been rigorously studied.
Results
We revealed a significant reduction of microbial richness in 3‐month‐old DMD animal model (mdx mice) compared with age‐matched C57Bl mice, with an enrichment of genera Prevotella and different metabolic profiles. Using murine models of DMD mimicking different degrees of disease severity, we highlighted the correlation between gut inflammation and muscular damage. To directly assess the potential contribution of dysbiosis in muscular wasting, we generated germ‐free dystrophic mice (GFmdx). In 3‐month‐old mdx mice, gut microbiota depletion reduced innate immune response while altering muscle metabolism and function. In line with these results, dysbiotic microbiota of mdx negatively affected intestinal, spleen, and muscle inflammation following injection in WT mice, and inversely correlated with muscle function.
Impact
Nutraceutical supplementation and dietary metabolites have been successfully proposed as strategies to co‐adjuvate the treatment of DMD, as well as immune‐modulators. Therefore, the identification of bacteria involved in triggering the inflammation‐driven muscular degeneration could pave the way to manipulate these taxa for therapeutic purposes. Corticosteroids are the current therapeutic standards for treating DMD, prolonging ambulation and muscle function. However, many patients experience serious adverse side effects. The use of nutraceutical with immunomodulating properties would provide DMD patients with an improved quality of life, as well as a reduction of costs associated with recurrent hospitalization that is usually needed to treat corticosteroid side effects.
Introduction
Duchenne muscular dystrophy (DMD) is an X‐linked disease caused by mutations in the DMD gene and loss of the dystrophin protein, leading to myofiber membrane fragility and necrosis with weakness and contractures. Affected DMD boys typically die in their second or third decade of life due to either respiratory failure or cardiomyopathy (Emery, 2002). Although the primary defects rely on skeletal muscle structure, a multitude of secondary defects exist involving deregulated metabolic and inflammatory pathways. Immune cell infiltration into skeletal muscle is, indeed, a typical feature of DMD pathophysiology and is strongly associated with disease severity (Farini et al, 2009). In the dystrophic dystrophin‐deficient mdx murine model, we recently found the presence of activated T lymphocytes and the overexpression of immunoproteasome (IP), an enzymatic complex that cleaves peptides to produce epitopes for antigen presentation to T lymphocytes. We have demonstrated that IP inhibition improved dystrophic muscle functions by reducing the number of both circulating and infiltrating activated T cells, confirming a pathogenic role of immune cells (Farini et al, 2016). Dystrophic muscle features were also improved by the depletion of B‐ and T‐cells in immunodeficient dystrophinopathic (SCID/mdx) and dysferlinopathic (SCID/BlAJ) murine models (Farini et al, 2012). So far, immunosuppressive drugs, such as glucocorticoids, are the only effective therapies to delay the onset and control symptoms (Shan et al, 2017a), ameliorating ambulation and muscle function, but their use in patients is still limited by serious side effects. In this scenario, the individual susceptibility to inflammatory events cannot be simply explained by the genetic defects of skeletal muscles; rather, there is a new emerging paradigm explaining the development of chronic inflammation that comprehend a strict regulation of epigenetics factors, genetic components, and the environment. In particular, environmental intrinsic (as innate and adaptive immunity) and extrinsic (as nutrition) mechanisms are connected to each other in a well‐defined temporal and spatial way, whose dysfunctions are the main causes of chronic inflammatory conditions (Shan et al, 2017b). Tissue‐environment interface is a preferential site critically involved in triggering mechanisms of chronic inflammation, especially in the mucosa of the gastrointestinal tract, where commensal microorganisms forming the so‐called microbiota provide nutrients by digestion of dietary components, modulate the development of the mucosal immune system and protect from pathogens (Yan et al, 2016). Thus, alterations of gut microbial communities (dysbiosis) can cause immune dysregulation and metabolic syndrome, contributing to a multitude of diseases of different aetiologies (Sperduto et al, 2019). The commensal population that constitutes the microbiota is extremely variable among individuals and its composition is dependent on the immune responses that are mediated in the gut and on host genotypes/phenotypes (Spor et al, 2011). Accordingly, the intestinal homeostasis is maintained through the mutualistic interactions between the microbiota and intestinal immune cells: dysfunctions cause serious problem including chronic inflammatory state (Kabat et al, 2016). In some way, gut microbiota modulation can also alter the regulatory molecules secreted by skeletal muscles and adipose tissues—myokines and adipokines—whose function is strictly dependent on the production of short‐chain fatty acids (SCFAs) and branched‐chain amino acids (Suzumura et al, 1986). In muscle tissue, dysbiosis interferes with the proper development of muscle progenitor cells, likely through reactive oxygen species generation and antioxidant genes (Tidball, 2017), and with endothelial cell function (van Bladel et al, 2014). The latter has been already confirmed by the occurrence of vascular development dysfunctions in pathogen‐free mice, possibly dependent on defects in nitric oxide synthase activity and in the expression of vessel inflammatory genes that were reversed by the restoration of normal gut flora (Tidball et al, 2018).
Duchenne muscular dystrophy patients present alterations of gastrointestinal motility and suffer from constipation, pseudo‐obstruction, and acute dilatation. Although no attention was paid to investigate these processes, smooth muscle fibrosis was observed throughout the gastrointestinal tract (Mule et al, 2010). Mdx mice shared impairments in intestinal contractility, linked to important abnormalities of the mucosal epithelial morphology (wider villi, reduced muscular and submucosa layer) normally associated to inflammatory state (Durbeej et al, 2000), and especially to nitric oxide (NO) production (Tomasi et al, 2017a). In addition, it was demonstrated that L‐arginine dietary supplementation improved colonic motility and increased NO signaling, ameliorating the pathological phenotype of mdx mice (Tomasi et al, 2017b). These evidences confirmed the involvement of a muscle‐gut axis‐mediated pathway that contributes to jeopardize the pathophysiology of DMD (Alves et al, 2014). Modulation of microbiota is also responsible for modifications of immunological and inflammatory features in organs distant from the gut. For instance, experiments of gnotobiology have shown that inhibition of microbiota function diminished the development of arthritis and autoimmune encephalomyelitis in murine models, whereas colonization of germ‐free mice with specific bacterial strains modified the expression of circulating monocytes, Th17 T lymphocytes and B lymphocytes (Tonegawa, 1983; Turner & Hilton‐Jones, 2010). Likely, inflammatory molecules derived from microbiota leak from the disrupted gut barrier of the mdx mice, activate inflammatory cells and circulate through blood toward muscles, where they modulate the immune system worsening the dystrophic phenotype.
Thus, it is possible to envision a connection between gut microbiota, immunity, and muscle homeostasis but the molecular details of this cascade of events in muscular dystrophy are still elusive. Here, we demonstrate dysbiosis in mdx mice that is associated with alterations of peripheral and local mdx immune landscape and muscle integrity. Treatment with broad‐spectrum antibiotics depleting the gut microbiota in 3‐month‐old (3m) mdx mice determined reduced muscle inflammation and enhanced fatty acid oxidation with consequent shift in fiber type toward an oxidative phenotype associated with muscle wasting. We also show that germ‐free mdx (GFmdx) mice were impaired in muscle function except for reduced fibrosis and absence of chronic muscle inflammation. Furthermore, intestinal colonization of mdx mice with eubiotic microbiota was sufficient to reduce inflammation improving muscle pathology and function. Our work provides new insights in DMD pathogenesis by highlighting the role of intestinal microbiota in shaping the muscular inflammatory response and conferring a distinct susceptibility to dystrophic muscle disease. From a therapeutic perspective, our results allow the identification of intestinal microorganisms, microbial products, and metabolites as potential targets to tailor innovative therapeutic strategies for the treatment of DMD patients.
Results
Altered gut microbiota composition in mdx mice
Full‐length dystrophin is predominantly expressed by skeletal muscles but dystrophic patients present impairment of gastrointestinal functions, altered motility and histological evidences for smooth muscle fibrosis throughout the gastrointestinal tract (Mule et al, 2010). Mdx mice shared impairments in the intestinal contractility, mainly due to NO dysfunctions (Baccari et al, 2000), increased calcium influx and deregulated tachinergic NK2 receptors (Mule & Serio, 2001). Slow fecal output and transit time of fecal material revealed motor disturbance in mdx mice, highlighting a delay in the propulsion (Mule et al, 2010). We found alterations of histological structures in the colon of 3m mdx mice mainly consisting in epithelial atrophy, shorter villi and thinner muscular and submucosal layers (Fig 1A–C). Total SCFA content in the stool isolated from colon, which contributes to immune regulation and anti‐inflammatory effects (Gul et al, 2014), was found to be similar between 3m C57Bl wild‐type (WT) and 3m mdx mice (Fig 1D). However, the imaging mass spectrometry of lipids in mdx small intestine tissues demonstrated an enrichment of phosphatidylcholines (PC: 34:2; PC 36:2) and PC cleavage by products as lyso‐phosphatidylcholines (LysoPC: 16:0) (Fig 1E). PC and LysoPC activate multiple signaling pathways that are involved in oxidative stress and inflammatory responses triggered through Toll‐like receptors (TLRs) (Vidarsson et al, 2014) leading to increased release of cytokines—i.e. interleukin (IL)‐1β, IL‐6, and tumor necrosis factor‐α (TNFα) (Wang et al, 2019)—and activation of lymphocytes (Wenninger et al, 2018) and pro‐inflammatory M1 macrophages (Yang et al, 2005). Compared with WT, the mdx intestinal tissues showed a trend for increased pro‐inflammatory IL‐6 and TNFα cytokines and TNF receptor‐associated factor 6 (TRAF6) inflammatory mediator (Fig 1F). Accordingly, significant upregulation of mediators of innate immunity, as proteasome subunit beta type‐8 (PMSB8), pentraxin‐3 (PTX‐3) and v‐rel Reticuloendotheliosis viral oncogene homolog B (RelB), was observed in mdx versus WT intestine tissues (Fig 1F). Modifications of neither transforming growth factor (TGF)‐β1 nor TLR4 were found (Fig 1F). Interestingly, we determined that TLR2—which is involved in maintenance of tight junction integrity and regulation of gut chronic inflammation (Wu et al, 2019; Xepapadaki et al, 2019)—was significantly downregulated in mdx intestine (Fig 1F).
Figure 1. Colon characterization of 3m mdx mice.
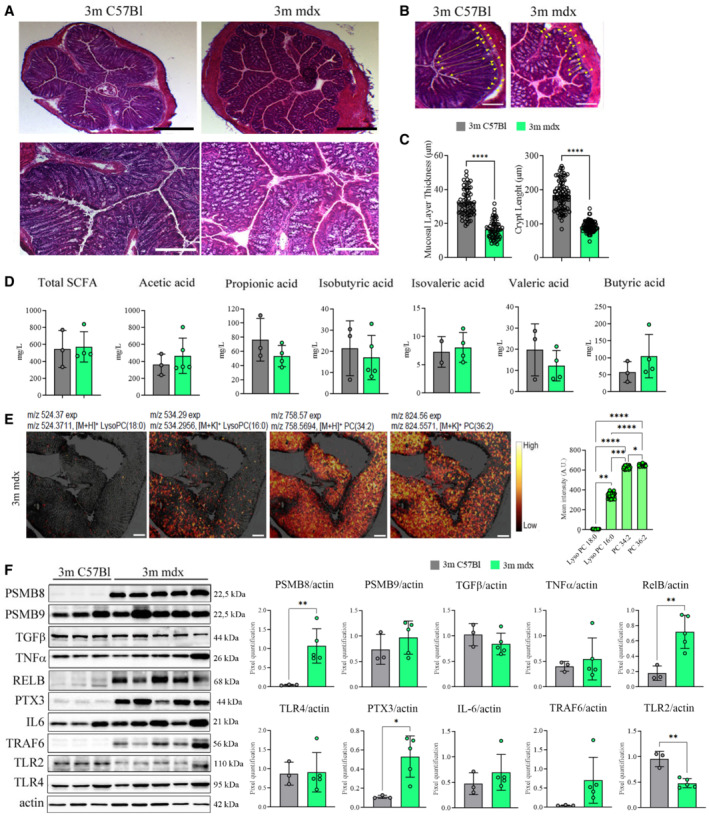
- Representative images of H&E staining of colon from 3m C57Bl (n = 4) and mdx (n = 4) mice. High magnification (scale bar: 20 μm) and low magnification (scale bar: 200 μm).
- Mucus layer, area between yellow dash lines; crypt length, yellow‐headed arrow. Scale bar: 100 μm.
- Mucus layer thickness and crypt length were quantified for n = 4 mice per group (with pooled samples of n = 60 for mucus layer thickness and n = 80 for crypt length).
- Short‐chain fatty acid fecal quantification of 3m C57Bl (n = 3) and mdx (n = 4–5) mice.
- Colon images captured with the iMScope TRIO described altered pattern of expression of different phosphatidylcholines (PC) and lysophosphatidylcholines (LysoPC) (as indicated by m/z values) in 3m mdx mice (n = 3). Scale bar: 50 μm. For each lipid, the mean intensities measured at 12 positions throughout colon images are shown on the right side where bars are mean ± SEM (n = 3).
- Cropped images of representative WB showing the expression of proteins involved in inflammation and fibrosis in colon tissues of 3m C57Bl (n = 3) and 3m mdx (n = 5). Densitometric analyses of protein expression was shown as ratio to actin.
Data information: Data are presented as mean ± SD (*P < 0.05; **P < 0.01, ***P < 0.001, ****P < 0.0001; Student's t‐test).
Source data are available online for this figure.
To verify the intestinal microbial community structure, we performed a metataxonomic analysis. The analysis of gut microbiota alpha‐diversity showed a significant reduction of microbial richness in 3m mdx compared with age matched C57Bl animals (Fig 2A; FDR corrected P < 0.05, pairwise comparisons using Wilcoxon rank‐sum test) suggesting that mdx mice are characterized by a dysbiotic microbiota. The microbial community structures among groups were significantly different as measured by beta‐diversity of Unweighted UniFrac distances and Bray–Curtis dissimilarity (Fig 2B; PERMANOVA P < 0.05). An in‐depth analysis of the gut microbiota (Fig 2C and D) showed the enrichment of different amplicon sequence variants (ASVs) belonging to the genera Alistipes and Prevotella, among others, in 3m mdx compared with C57Bl mice. Notably, LysoPC have been associated with the abundance of the genus Alistipes (Jolles et al, 2014; Heydemann, 2017). By using a Random Forest classifier, we further observed that the gut microbiota was able to classify samples according to status (OOB = 18.2%; P < 0.0001; Accuracy = 0.81; Kappa = 0.725) and that the genus Prevotella was the most important, fully classified, feature to categorize samples according to status (Fig 2E). According to these data, Prevotella was among the taxa whose abundance was significantly higher in 3m mdx (Fig 2F).
Figure 2. Microbiome analysis of 3m mdx mice.
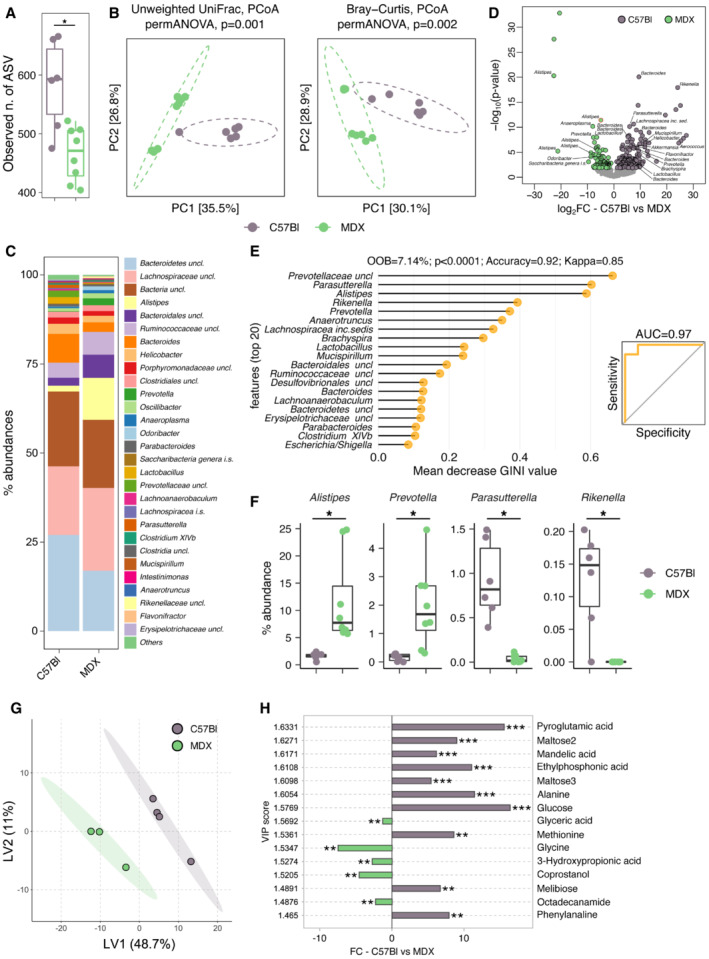
- Observed number of enriched ASVs in 3m C57Bl (n = 6) (maximum: 666; median: 583.67; minimum: 475) and 3m mdx (n = 8) (maximum: 522; median: 465.625; minimum: 404). Data are presented as the exact number of ASVs (*P < 0.05; Student's t‐test).
- PCA of beta‐diversity of 3m C57Bl (n = 6) and 3m mdx (n = 7) as measured by Unweighted UniFrac distance and Bray–Curtis dissimilarity.
- Mean relative abundance at genus level among groups. All genera with relative abundance < 0.1% are reported together and labeled as “others.”
- Volcano plots of 3m C57Bl (n = 6) and 3m mdx (n = 7) showing the significantly enriched bacterial amplicon sequence variants (ASVs) (with P < 0.05) by the DEseq2 analysis. The names of the significantly enriched bacterial ASVs classified to the genus level and P < 0.005 are reported. All P‐values were false discovery rate–corrected.
- Random forest analysis. The top 20 bacterial genera with the highest discriminatory power sorted by mean decrease GINI value are showed.
- Relative abundance of different genus in 3m C57Bl (n = 6) and 3m mdx (n = 7). Prevotella: 3m C57Bl: maximum: 0.28438734; median: 0.162312199; minimum: 0. 3m mdx: maximum: 4.650449086; median: 1.928565583; minimum: 0.308938765. Alistipes: 3m C57Bl: maximum: 2.381488226; median: 1.622780995; minimum: 0.52990159. 3m mdx: maximum: 24.79693926; median: 11.74146327; minimum: 5.71434417. Parasutterella: 3m C57Bl: maximum: 1.491499069; median: 0.923747366; minimum: 0.390776848. 3m mdx: maximum: 0.114573317; median: 0.036571394; minimum: 0.002045952. Rikenella: 3m C57Bl: maximum: 0.202549256; median: 0.124121093; minimum: 0.067516419. 3m mdx: maximum: 0; median: 0; minimum: 0. *P < 0.05; Student's t‐test.
- Multidimensional scaling analysis of small intestinal metabolomic profiles from of 3m C57Bl (n = 4) and 3m mdx (n = 3) mice calculated by samples' distance similarities (Bray‐Curtis) with the most discriminatory metabolites (top variable importance in projection score) identified.
- Concentration of the significantly different metabolites isolated from the small intestinal content of 3m C57Bl (n = 4) and 3m mdx (n = 3) mice. *P < 0.05, **P < 0.01 and ***P < 0.001; Wilcoxon rank‐sum test.
Source data are available online for this figure.
Multidimensional scaling (MDS) analysis evidenced significantly different metabolic profiles among groups (Fig 2G). Accordingly, the predicted functional potential of the mdx‐associated microbiota showed alterations of metabolic pathways related to carbohydrate and amino acid metabolism (Metabolic Maps C57Bl and 3m mdx; Dataset EV1 and Fig 3A). Indeed, 3m mdx mice showed a significant reduction in the predicted gene content of the key SCFA biosynthetic enzymes propionyl‐CoA:succinate CoA transferase (scpC), propionate CoA‐transferase (pct), butyryl‐CoA:acetate CoA‐transferase (but) and butyrate kinase (buk) while no differences where observed for propionate (tdcD) and acetate (ack) kinases (Fig 3B). We observed a significant reduction in the concentration of different amino acids, namely alanine, aspartic acid, methionine, and phenylalanine, in 3m mdx mice compared with C57Bl animals (Fig 2H). Of note, tartarate may act as a muscle toxin by inhibiting the production of malic acid (Junghans et al, 2001).
Figure 3. Metabolic maps of 3m mdx mice.
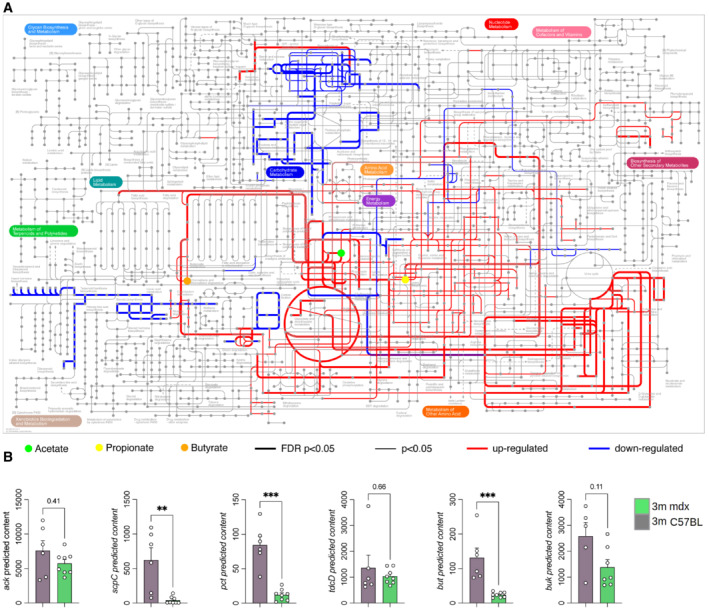
- The iPath3.0 representation of KEGG metabolic pathways inferred from Piphillin analysis significantly upregulated (in red) or downregulated (in blue) in 3m C57Bl (n = 6) versus 3m mdx (n = 8) mice. Nodes in the map colored in green, yellow, and orange correspond to acetate, propionate, and butyrate, respectively. Line thickness represents the level of statistical significance for the inferred pathways; thick lines with FDR‐corrected P‐value < 0.05, thin lines with nominal P‐value < 0.05.
- Predicted metagenomic gene content of the key enzymes catalyzing the final steps for the production of microbiota‐derived SCFAs in GI of 3m C57Bl (n = 5/6) and 3m mdx (n = 7/8) mice. Data are presented as mean ± SD (**P < 0.01, ***P < 0.001; Kruskal–Wallis test).
Source data are available online for this figure.
To evaluate the extent of gut microbiota effects on systemic and muscle immunity, as well as inflammation, we correlated metataxonomic and immunophenotyping data (Dataset EV2 and Fig EV1). Specifically, we investigated the correlations among FACS analysis of different subsets of T cells (naïve, central memory, effector T cells and Tregs) and CD11b+ myeloid subset of spleen and muscle tissues in 3m mdx and C57Bl mice and the most representative microbiota genera (Dataset EV2 and Fig EV1). We observed that Prevotella significantly correlated with the frequency of splenic CD44+CD4+/CD8+ T cells and Tregs as well as with muscle effector/memory CD44+CD8+ T cells and central memory CD4+ T cells (Fig EV1).
Figure EV1. Correlation between bacterial genera and immunity.
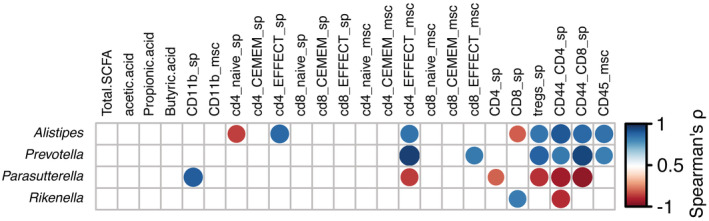
Heatmap of Spearman's rho correlations between the relative abundance of the most represented bacterial genera (with relative abundance > 0.1%) in the gut microbiota of 3m mdx animals (n = 3–8) with the indicated metabolites and immunological parameters. The significant correlations with FDR‐corrected P‐value < 0.1 are indicated with bubbles. Spearman correlation plots for the significant correlations between Prevotella and the indicated immunological parameters are also shown. Abbreviations used in the Figure: sp, spleen‐derived; msc, muscle‐derived; CEMEM, central memory cells; EFFECT, effector cells.Source data are available online for this figure.
Gut microbiota depletion reduces innate immune response but alters muscle metabolism and function in 3‐month‐old mdx mice
Since mdx mice harbor alterations in intestinal microbiome, we investigate whether early secondary effects of muscular dystrophy could be affected by the absence of gut microbiota in GFmdx dystrophic model or by long‐term depletion of intestinal bacteria of mdx by oral treatment with a cocktail of ampicillin, metronidazole, and vancomycin (ABX) (Krebs et al, 2016; Xie et al, 2017). ABX can affect luminal secondary metabolites and gut signaling (Zschuntzsch et al, 2016). Metabolomic analysis of the small intestine in 3m mdx+ABX demonstrated significant alterations of the metabolic profiles and inferred pathways (FRD‐corrected P < 0.05, ANOVA) suggesting an important role of the gut microbiota in affecting host metabolome (Fig 4A and B). Given that combined output of host‐microbes interactions influencing host energy metabolism, development, and function of the immune system (Juliao et al, 2017) is determined by the metabolome, we performed a metabolomic analysis of the small intestinal content from mdx and mdx+ABX mice. Metabolic pathways reconstruction based on the metabolites with the highest discriminatory power (Dataset EV1 and Fig 4C and D) revealed that mdx over‐expressed glycine, coprostanol, threonine, phosphate and galacticol related to age‐matched C57Bl while leucine, methionine, aspartic acid and alanine were downregulated. Interestingly, 3m mdx+ABX mice showed metabolites' expression similar to WT mice, except for maltose and mandelic acid, whose amount was lower in dystrophic mice related to 3m C57Bl (Fig 4C and D). Hence, ABX changed the SCFA pool of mdx, most notably by decreasing butyric, propionic, isopropionic, and valeric acids to undetectable levels, while acetic acid was significantly decreased compared with untreated mdx mice (Fig 4E). We further highlighted the influence of gut microbiota depletion on immune response of 3m mdx. The amount of splenic CD45+CD11b+CD4−CD8− myeloid cells was not modified by ABX treatment (Fig 5A) neither the effector CD4+ T cells which remained upregulated in both mdx and mdx+ABX compared with C57Bl (Fig 5B). In muscle, we did not find differences in the CD45+ myeloid cells (Fig 5C) since ABX only slightly modified the effector CD4+ T cells in mdx compared with untreated age‐matched C57Bl (Fig 5D). According to these evidences and to literature describing modulation of immune system and microbiota in GF mice (Kapur et al, 2015), we investigated the consequences of gut microbiota depletion on the amounts of lymphocytes in 3m GFmdx. We observed similar amount of CD4+ and CD8+ T cells between 3m mdx and 3m GFmdx (Fig 5E), whereas a significant reduction of splenic Treg cells and effector/memory CD44+ T cells (Fig 5F) was shown in 3m GFmdx compared with age‐matched mdx mice. The amount of inflammatory CD45+ cells was also diminished in skeletal muscle of 3m GFmdx related to 3m mdx mice as demonstrated by FACS analysis (Fig 5G) and specific‐muscle staining (Fig 5H). Similarly, the number of CD3+ inflammatory cells was higher in 3m mdx muscles as assessed by quantification of immunofluorescence staining (Fig 5I). Compared with 3m mdx, ABX‐treated mdx and GFmdx muscles showed significant decrease in IL‐6 but not in TNFα cytokines, as well as a significant reduction of Nuclear Factor kappa‐light‐chain‐enhancer of activated B cells (NF‐kB) and RelB inflammatory mediators toward the levels of WT (Fig EV2A), suggesting a reduced muscle innate immune response (Ticinesi et al, 2017). There was no change in the expression of TLR4 and osteopontin (OPN), but mdx, mdx+ABX and GFmdx muscles showed an increase in matrix metallopeptidase (MMP) 9 relative to WT (Fig EV2A). To evaluate the influence of the gut microbiota on the dystrophic skeletal muscle architecture and function, we performed RNAseq of tibialis anterior (TA) muscles from mdx, ABX‐treated mdx and GFmdx mice. Principal component analysis (PCA) of RNAseq datasets showed treatment‐dependent clustering of samples, separating mdx+ABX and GFmdx from mdx (Dataset EV3 and Fig 6A). As additional quality control, mdx RNA datasets clustered separately from the ones of age‐matched, background‐matched C57Bl muscles (Fig 6A). Comparing ABX‐treated and GFmdx with age‐matched mdx muscle, we found 1,381 genes convergently upregulated and 2,722 genes convergently downregulated (Fig 6B). Gene abundance cutoff was set at 10CPM for these initial analyses to focus our comparisons on genes of mid‐to‐high expression in muscle. We performed gene ontology (GO) analysis on both groups of convergent genes. GO analysis of the convergent upregulated genes showed enrichment for pathways of oxidative metabolism (Lpin1, Ppard, Ppargc1a, Ppara) and nutrient uptake/processing (Pfkm, Pck1, Pfkfb3, Pcx, Slc2a3, Slc2a5, Tkt, Pygl, Plin1, Lipe, Acer2) (Dataset EV4 and Fig 6B and C). Conversely, ABX and GF treatments converged on downregulating genes involved in inflammation and fibrosis (Timp1, Mmp15, several members of the Adamts extracellular proteases and Collagen gene families) (Dataset EV4 and Fig 6B and C). Furthermore, gene set enrichment analysis (GSEA) revealed alterations in inflammatory response, epithelial‐to‐mesenchymal transition, complement activity, angiogenesis and interferon‐γ response in 3m mdx versus age‐matched C57Bl muscles (Fig EV3). Moreover, genes involved in G2M checkpoint transition, interferon‐α and ‐γ response, E2F transcriptional activity and myogenesis were reduced in 3m mdx+ABX versus age‐matched mdx muscles (Fig EV3). Muscle genes involved in adipogenesis, fatty acid metabolism and cholesterol homeostasis were upregulated in 3m GFmdx versus age‐matched mdx mice; conversely, muscle genes involved in interferon‐αresponse, E2F transcriptional activity and inflammatory response were downregulated in the 3m GFmdx versus age‐matched mdx mice (Fig EV3).
Figure 4. Characterization of gut tissue metabolome in 3m mdx mice and following antibiotics treatment.
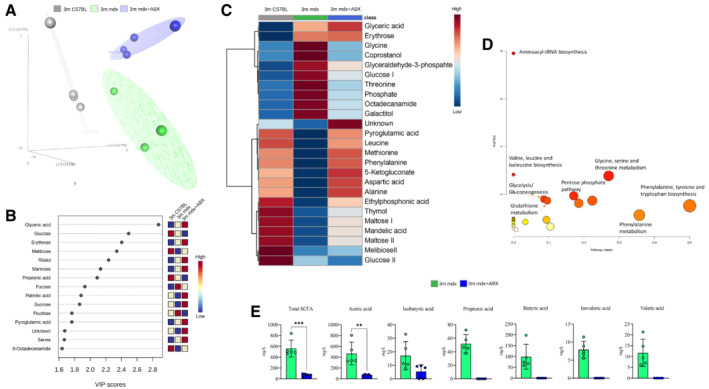
- Partial least square discriminant analysis (PLS‐DA) models score plot used to evaluate the differences among 3m C57B1 (in gray), 3m mdx mice (in green) and 3m mdx+ABX (dark purple), with n = 4 each.
- Relevant metabolites (top variable importance in projection score) in the corresponding PLS‐DA separation, in blue metabolites with a negative fold change and in red metabolites with a positive fold change.
- Heatmap showing all the relevant metabolites concentration change among the groups. Both metabolites and classes were clusterized according to the Wald method. In blue metabolites' concentration with a negative fold change and in red metabolites' concentration with a positive fold change.
- Metabolic pathways involving the relevant metabolites obtained using the MetPa algorithm. The color and size of each circle are based on the P‐value and pathway impact value, respectively. The x‐axis represents the pathway impact, and the y‐axis represents the −log of P values from the pathway enrichment analysis for the key differential metabolites of 3m mdx and 3m mdx+ABX mice.
- Fecal content quantification of SCFAs in 3m mdx and 3m mdx+ABX mice (n = 5 per group). Data equal to 0. Data are presented as mean ± SD (**P < 0.01, ***P < 0.001; Student's t‐test).
Source data are available online for this figure.
Figure 5. Microbiota depletion induces modulation of immune cells.
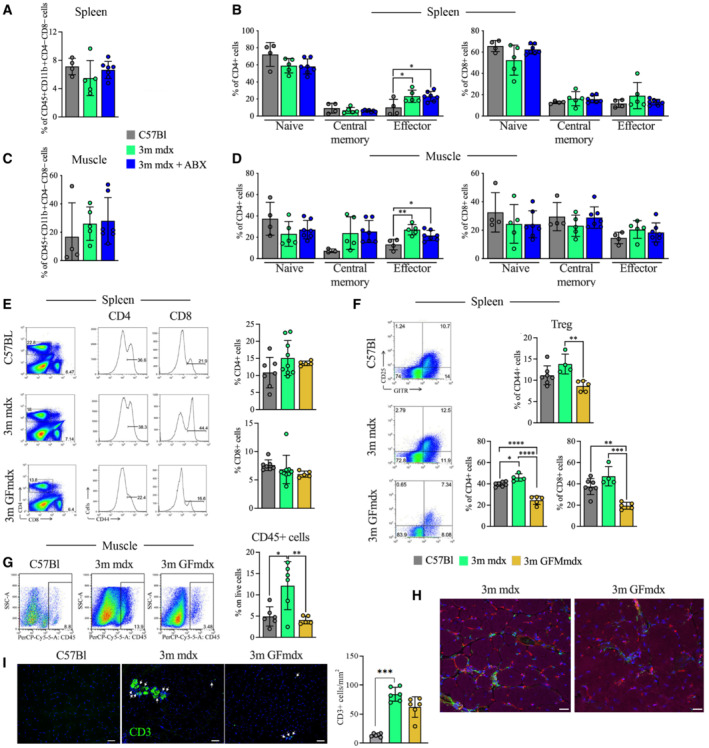
-
A–DFACS analysis of spleen and muscle homogenates from 3m C57Bl (n = 4), mdx (n = 5) and mdx+ABX (n = 7) mice demonstrates no significant alteration of CD45+CD11b+CD4−CD8− myeloid cells (A and C) and few differences in CD4+ or CD8+ naïve (CD62L+ CD44−), central memory (CD62L+ CD44+) and effector (CD62L− CD44+) T cells (B and D).
-
EFACS analysis of spleen of 3m C57Bl (n = 7), mdx (n = 9) and GFmdx (n = 5) mice revealed similar proportions of CD4+ and CD8+ T cells but reduced activated CD44+ T cells in GFmdx mice. Representative plots are depicted.
-
FGraphs show cumulative frequencies of CD4+ and CD8+ T cells on live cells of 3m C57Bl (n = 7), mdx (n = 4) and GFmdx (n = 5) mice. Representative dot plots and cumulative frequencies of splenic CD4+GITR+CD25+ Treg. Frequencies of effector CD44+ T cells were significantly decreased in spleen of GFmdx mice.
-
GRepresentative dot‐plots showing the proportion of muscle‐infiltrating CD45+ cells of 3m C57Bl (n = 6), mdx (n = 6) and GFmdx (n = 5) mice. Cumulative frequencies of muscle‐infiltrating CD45+ cells are shown.
-
HRepresentative images of TA muscles from 3m mdx and GFmdx mice stained for CD45 (in green), isolectin (in red), and phalloidin (in purple). Nuclei were counterstained with DAPI (in blue). Scale bar: 10 μm.
-
IAbsolute number of CD3+ inflammatory cells (white arrows) were quantified in n = 12 images of TA of 3m C57Bl, 3m mdx, and 3m GFmdx mice (n = 6 each). CD3 staining is shown in green and DAPI in blue. Scale bars: 50 μm.
Data information: Data are presented as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 ordinary one‐way ANOVA, Tukey's multiple‐comparison test. The comparisons among the averages of CD3+ cells were evaluated using unpaired t‐test.
Source data are available online for this figure.
Figure EV2. Gene and protein expression in muscles from 3m mdx, mdx+ABX, and GFmdx.
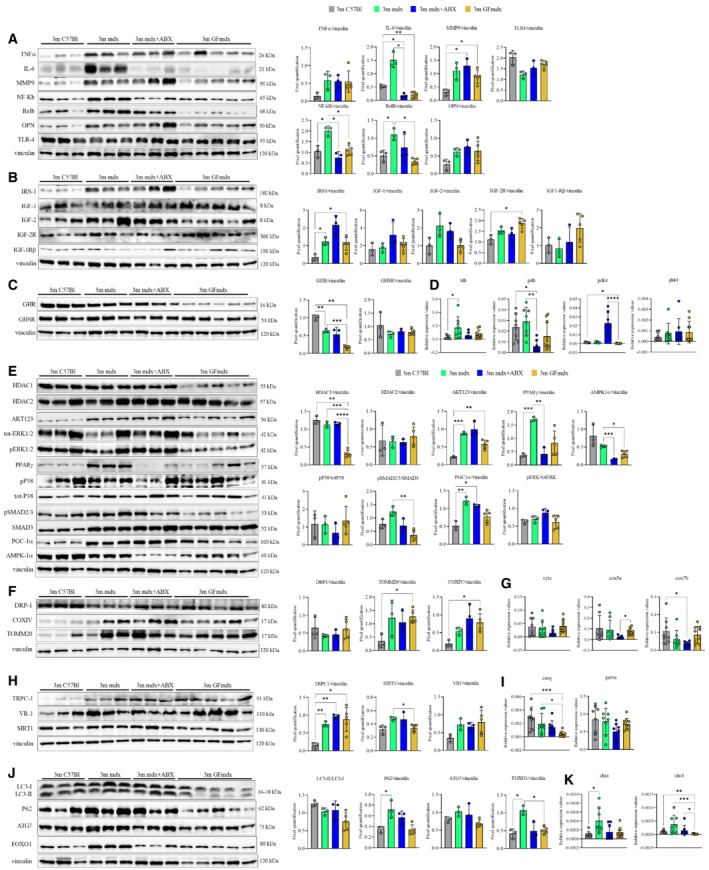
-
A–KCropped images of representative WB and RT‐qPCR analysis of TA muscle of 3m mdx (n = 3/4), 3m mdx+ABX (n = 3/4), and 3m GFmdx (n = 5) showing the expression of the proteins specifically involved in inflammation/fibrosis (A), skeletal muscle metabolism (B–E), mitochondrial biogenesis (F and G), calcium conducting channels (H and I), autophagy (J), and nicotinic acetylcholine receptors (K). Densitometric data were normalized on vinculin and expressed as mean ± SD. Data are presented as mean ± SD (*P < 0.05, **P < 0.01, ***P < 0.001; ****P < 0.0001, ordinary one‐way ANOVA, Tukey's multiple‐comparison test for WB and non‐parametric test followed by Kruskal–Wallis test for RT‐qPCR).
Source data are available online for this figure.
Figure 6. Muscle homeostasis of 3m mdx mice is influenced by microbiota.
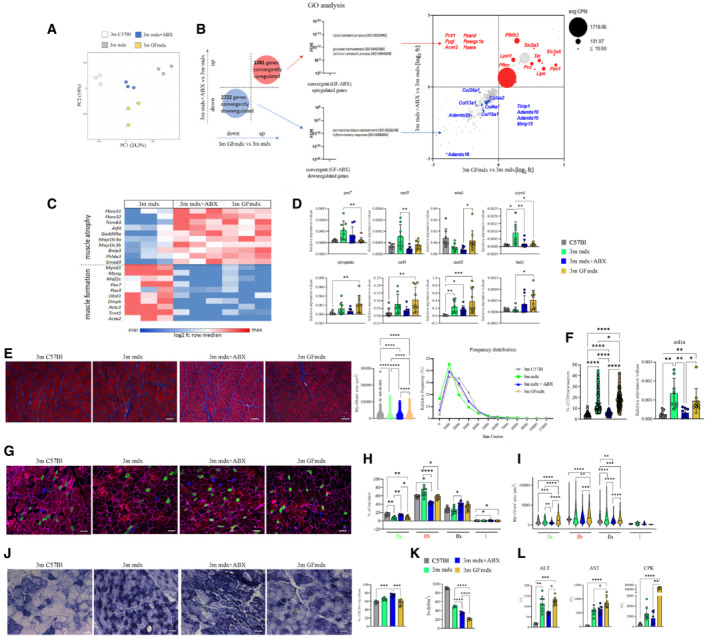
-
A, BRNA datasets clustering (A) and convergently up‐ and downregulated genes (B) of muscles of 3m mdx (n = 3), mdx+ABX (n = 3) and GFmdx (n = 3) mice.
-
CGene ontology (GO) analysis on both groups of convergent genes.
-
DRT‐qPCR analysis of TA muscles of two independent experiments with 3m mdx (n = 4), mdx+ABX (n = 4), and GFmdx (n = 5) mice determined the expression of myogenic markers.
-
ERepresentative Gomori‐modified staining and quantification of myofiber area and relative frequency of the myofiber cross‐sectional area (CSA) expressed as the frequency distribution of the TA muscles of 3m C57Bl (n = 4), 3m mdx (n = 4), mdx+ABX (n = 4) and GFmdx mice (n = 5). Pooled samples for each group with n = 6,240 for 3m C57Bl; n = 6,001 for 3m mdx; n = 10,556 for 3m mdx+ABX; n = 23,059 for GFmdx. For morphometric analysis, images were quantified with Image J software for each mouse. Scale bars: 50 μm.
-
FQuantification of the fibrotic area from Gomori stained images (pooled samples for each group with n = 223 for 3m C57Bl, 3m mdx and 3m mdx+ABX; n = 215 for GFmdx) and RT‐qPCR analysis of Col1a (two independent experiments with n = 4 animals each group).
-
GRepresentative images of skeletal muscle showed the distribution and composition of the myosin heavy chain (MyHC) isoforms (Type IIa, Type IIx, and Type IIb).
-
H, IGraph portrays (H) the percentage of myofibers expressing different MyHC isoforms and (I) myofibers area per type of MyHC in TAs of 3m mdx (n = 4), mdx+ABX (n = 4), and GFmdx (n = 5) mice (n = 12 images per animal).
-
JRepresentative SDH staining and quantification of percentage of SDH+ myofibers of TAs from 3m mdx (n = 4), mdx+ABX (n = 4), and GFmdx mice (n = 5) (n = 12 images per animal). Scale bars: 50 μm.
-
KTetanic force of TA muscle of 3m C57Bl (n = 4), mdx (n = 4), mdx+ABX (n = 4), and GFmdx (n = 5) mice.
-
LALT, AST, and CPK serum levels were measured in 3m mdx (n = 4), mdx+ABX (n = 4), and GFmdx mice (n = 5) (two independent experiments).
Data information: Data are presented as mean ± SD (*P < 0.05, **P < 0.01, ***P < 0.001; ****P < 0.0001, ordinary one‐way ANOVA, Tukey's multiple‐comparison test for WB and non‐parametric test followed by Kruskal–Wallis test for RT‐qPCR).
Source data are available online for this figure.
Figure EV3. Gene set enrichment analysis of 3m C57bl, mdx, mdx+ABX, and GFmdx mice RNA sequencing data.
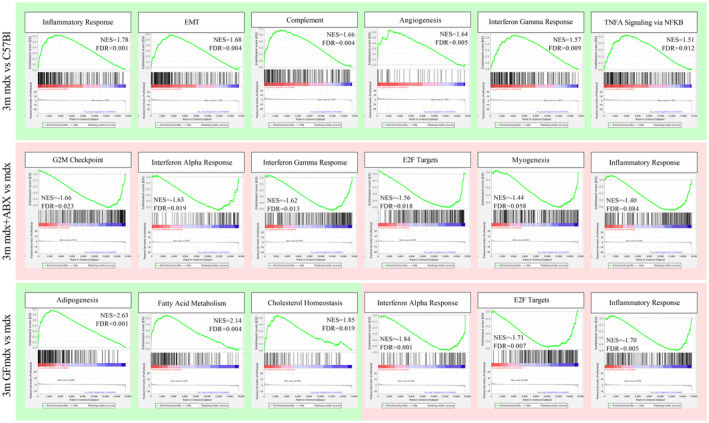
The annotated dataset “Hallmark” collection by the Molecular Signatures Database (MSigDB) was used as a reference. A green background refers to positive Normalized Enrichment Score (NES) (enrichment in positive phenotype, or upregulation); a red background refers to negative NES (enrichment in negative phenotype, or downregulation). FDR, false discovery rate. Genes involved in inflammatory response, epithelial‐to‐mesenchymal transition, complement activity, angiogenesis, and interferon‐γ response are upregulated in 3m mdx (n = 3) versus age‐matched C57Bl (n = 3) mice (top lane). Genes involved in G2M checkpoint transition, interferon‐α and ‐γ response, E2F transcriptional activity and myogenesis are downregulated in 3m mdx+ABX (n = 3) versus age‐matched mdx (n = 3) mice (mid lane). Genes involved in adipogenesis, fatty acid metabolism, and cholesterol homeostasis are upregulated in 3m GFmdx (n = 3) versus age‐matched mdx (n = 3) mice; conversely, genes involved in interferon‐alpha response, E2F transcriptional activity, and inflammatory response are downregulated in the 3m GFmdx versus age‐matched mdx mice (bottom lane).Source data are available online for this figure.
We further validated muscle RNAseq analysis for genes involved in myogenesis. Compared with mdx, mdx+ABX and GFmdx muscles showed decreases in genes involved in early myogenesis as MyoD, Pax7 and Myf5 with similar levels of later genes of myogenesis as myogenin and MRF4 (Fig 6D). Relative to WT controls, mdx+ABX and GFmdx muscles showed no difference in expression for the majority of these genes except for myogenin and MRF4, where GFmdx muscles showed significant increases (Fig 6D). Transcript expression of the E3 ubiquitin ligase gene, MuRF‐1, which promotes protein degradation and muscle catabolism (Sasson et al, 2019), was similarly and significantly increased in mdx, mdx+ABX and GFmdx muscles compared with WT (Fig 6D). These results suggest that microbiota depletion may likewise alter the regulation of genes related to muscle growth and differentiation in mdx.
To uncover the effects of changes seen in microbiota‐depleted dystrophin‐deficient mice, we examined the muscle cross‐sectional areas (CSAs) of mdx+ABX and GFmdx compared with age‐matched mdx and WT mice. The ABX‐treated mdx and GFmdx displayed increased CSAs relative to mdx, with the former presenting the highest area of myofibers (mean fiber area for TA ± SEM: 3m mdx 1,625.01 ± 19.59 μm2; 3m mdx+ABX 1,855.572 ± 11.903 μm2; 3m GFmdx: 1,754.22 ± 7.937 μm2; 3m C57Bl 1,998.21 ± 16.79 μm2), and the latter a lower size variability (Fig 6E). In particular, the values of frequency distribution confirmed the smaller area of myofibers in 3m mdx mice (25% Percentile: 3m mdx: 661.68; 3m mdx+ABX: 1,003.75; 3m GFmdx: 838.447; 3m C57Bl: 1,131.217. 75% Percentile: 3m mdx: 2,061.384; 3m mdx+ABX: 2,443.35; 3m GFmdx: 2,398.803; 3m C57Bl: 2,621.3125) (Fig 6E). Reduced immune response correlated with reduced fibrosis deposition in mdx+ABX and GFmdx (% of fibrosis per muscular section: 3m mdx 20.62%; 3m mdx+ABX 5.016%; 3m GFmdx 17.58%) (Fig 6F). As an additional confirmation of the RNAseq data, RT‐qPCR analysis for fibrotic genes confirmed the downregulation of col1a in mdx+ABX compared with age‐matched mdx and GFmdx (Fig 6F).
To determine whether these features were associated with fiber switch, the immunoreactivity for adult myosin heavy chain isoforms (MyHC) were detected and quantified by immunofluorescence (Fig 6G). Compared with mdx, the percentage of oxidative MyHC‐I and oxidative/glycolytic MyHC‐IIa/IIx fibers remained unchanged in GFmdx, whereas the percentage of type IIa and IIx fibers significantly increased in mdx+ABX (Fig 6H). Interestingly, mdx+ABX and GFmdx showed a significant increase of type I fibers relative to WT and significant reduction of glycolytic MyHC‐IIb fibers compared with mdx (Fig 6H). Consistent with these findings, there was no change in the expression of α‐actinin‐3 (ACTN3)—a sarcomeric protein mainly expressed in fast type‐IIb myofibers‐ except for significant differences between mdx+ABX and GFmdx which correspond to their relative proportions of type IIb fibers (Fig 6D). Moreover, the oxidative switching of myofibers in mdx+ABX and GFmdx was confirmed by a trend for increased expression of slow skeletal muscle troponin T‐1 (TNNT1) compared with mdx and WT (Fig 6D). Interestingly, compared with mdx, the myofiber areas of type IIb and IIx fibers were decreased in both 3m mdx+ABX and 3m GFmdx mice, whereas type IIa area was increased in 3m GFmdx and decreased in 3m mdx+ABX (Fig 6I). We further evaluated the metabolism of microbiota‐depleted mdx muscle by staining for succinate dehydrogenase (SDH) activity, indicative of oxidative metabolism (Bejaoui et al, 1995; Bansal et al, 2003). The ABX‐treated mdx muscles showed higher percentage of SDH‐positive fibers, in favor of higher oxidative metabolism than 3m GFmdx and WT (Fig 6J). These data supported a shift in fiber type toward an oxidative phenotype in microbiota‐depleted mdx exacerbating the hallmark of muscular dystrophy, through a fiber‐type switch from damage‐sensitive glycolytic type IIb fibers toward damage‐resistant glycolytic‐oxidative type IIx and IIa fibers. In addition, ABX and GF conditions differentially affect change toward a slower MyHC profile: unlike germ‐free conditions, under which microbiota depletion is maintained throughout life allowing gradual plastic remodeling in response to metabolic changes, ABX‐treated muscles might undergo a substantial increase of muscle catabolism as suggested by the reduced size of the more abundant glycolytic‐oxidative fibers.
The decrease of tetanic force is a hallmark of muscular dystrophies. We noticed a significant reduction of tetanic force in TA muscle of both mdx+ABX and GFmdx compared with mdx and WT. However, TA tetanic force of GFmdx was significantly lower than the force exerted by mdx+ABX (Fig 6K). To better understand muscular weakness observed in ABX‐treated mdx and GFmdx mice, we investigated serum concentration of alanine aminotransferase (ALT) and aspartate aminotransferase (AST): both ALT and AST were upregulated in GFmdx (Fig 6L). Of note, we found an increase of creatine phosphokinase (CPK) in 3m GFmdx versus mdx+ABX and WT, suggesting an important damage of skeletal muscle membranes (Fig 6L).
Muscle metabolism modifications induced by depletion of mdx microbiota prompted us to analyze the insulin‐like signaling and the orexigenic gut‐peptide hormone ghrelin (GHR), which is known to affect whole body energy metabolism. Among insulin‐like growth factor (IGF) pathways, we detected a significant upregulation of the insulin receptor substrates 1 (IRS‐1), a key modulator of insulin resistance (Confalonieri et al, 2003), in muscles of GFmdx and mdx relative to WT (Fig EV2B). Moreover, we showed that ghrelin was downregulated in muscles of GFmdx compared with mdx and WT mice (Fig EV2C). We thus measured the expression of the mitochondrial pyruvate dehydrogenase lipoamide kinase isozyme 4 (pdk4), whose activity is regulated by insulin and is necessary to decrease glycolytic metabolism and conserve glucose (de Morree et al, 2013). The pdk4 was upregulated in mdx+ABX related to mdx and GFmdx, whereas no differences were found in the expression of glucose transporter glut4 (Fig EV2D), which is involved in the uptake of lactic acid in oxidative fibers for oxidation (Liu et al, 1998). Consistent with this, we observed a trend for decreased expression of lactate dehydrogenase (Ldh) and pyruvate dehydrogenase (Pdh) in mdx+ABX and GFmdx compared with mdx (Fig EV2D). Notably, inhibition of Pdh activity by Pdk4 reduces the conversion of glycolytically derived pyruvate into acetyl‐CoA, thereby diverting glucose flux to lactate and away from oxidation in the TCA cycle (Mammen et al, 2011). All these data suggest that microbiota depletion in mdx induces alterations in cellular glucose metabolism recognized as aerobic glycolysis (Mohassel et al, 2019).
To further unravel a potential role of microbiota depletion in altering muscle glucose uptake and fatty acid oxidation of mdx, we investigated downstream signaling of AMP‐activated protein kinase (AMPK). Histone deacetylase (HDAC) activity is partly modulated through activation of AMPK (Merrill et al, 1997). Downregulation of HDAC1 and similar amounts of HDAC2 were observed in GFmdx mice (Fig EV2E). Compared with mdx, both ABX‐treated mdx and GFmdx mice exhibited reduced muscle 5′‐AMP‐activated protein kinase catalytic subunit alpha‐1 (AMPK‐1α) and downregulation of the peroxisome proliferator activated receptor gamma (PPARγ) and Small mother against decapentaplegic 2/3 (SMAD2/3) that acts as afatty acid sensors to control adipogenesis (Fig EV2E). The reduction of AMPK‐1α observed in muscles of mdx+ABX and GFmdx was associated to unmodified levels of the insulin‐dependent downstream pathways that control energy homeostasis, including serine–threonine protein kinase 1‐2‐3 isoform (AKT 1‐2‐3) and extracellular signal‐regulated kinase (ERK) (Fig EV2E). Among downstream targets of AMPK, we found significant increase of peroxisome proliferator–activated receptor gamma coactivator 1α (PGC1α) without modifications of p38 mitogen‐activated protein kinases (p38 MAPKs) in mdx, mdx+ABX and GFmdx relative to WT muscles (Fig EV2E). In accordance with the increase of PGC1α, which is a master regulator of mitochondrial biogenesis and function (Allegra et al, 1997), we found a trend for increased mitochondrial mass revealed by translocase of outer mitochondrial membrane 20 (TOMM‐20) and for increased mitochondrial activity identified by cytochrome c oxidase (COX) IV in mdx, mdx+ABX and GFmdx relative to WT muscles (Fig EV2F). GTPase dynamin‐related protein 1 (DRP1), which is critical for mitochondrial fission machinery and mitochondrial dynamics, was not affected (Fig EV2F). However, ABX‐treated mdx muscle displayed downregulation of mitochondrial genes as CoxVa, CoxVIIb, but not cytc, related to 3m GFmdx (Fig EV2G).
Since muscle calcium dysfunctions are common in DMD (Walther et al, 2000; Sparks et al, 2007; Maguire et al, 2019), we investigated the muscle amount of calcium channel proteins as transient receptor potential canonical 1 (TRPC1) and vanilloid receptor 1 (VR‐1). Compared with WT, the TRPC1 expression was similarly upregulated in mdx, mdx+ABX and GFmdx, whereas VR‐1 was comparable among animal groups. Interestingly, sirtuin 1 (SIRT1) was found to be downregulated in GFmdx versus mdx (Fig EV2H). We further evaluated the expression of calcium ion binding genes as parvalbumin (Pvalb) and calsequestrin 1 (Casq1), whose activities mediate calcium contraction and release in the lumen of sarcoplasmic reticulum (SR) of muscle fibers: only Casq1 was significantly downregulated in 3m GFmdx compared with mdx and WT (Fig EV2I).
AMP‐activated protein kinase activation is also involved in autophagy activation signaling (Perez et al, 2017). In response to nutrient deficiency and exercise, AMPK increases autophagy activity by activating Forkhead box (FOX) O1 (Dejardin, 2006). The FOXO transcription factors, including FOXO1, FOXO3, and FOXO4, have recently been implicated as key regulators of gene expression during skeletal muscle atrophy (Sun, 2017) and FOXO1 mRNA in particular is upregulated during fasting and dexametasone treatment (Porter et al, 2002). Consistent with decrease of AMPK determined by microbiota depletion, we found a trend for reduced ratio of autophagy marker light chain 3‐I and II (LC3‐I/LC3‐II) in GFmdx with no modifications of autophagy receptor (P62) and autophagy related 7 (ATG7) in all groups. However, FOXO1 was diminished in both ABX‐treated mdx and GFmdx compared with age matched mdx (Fig EV2J). As microbiota depletion was found to regulate neuromuscular junction (NMJ), we studied AChR genes. As reported previously by others (Ieronimakis et al, 2016), we found down regulation of fast‐channel acetylcholine receptor subunits in microbiota‐depleted mdx (Fig EV2K).
All these data highlighted that AMPK‐related pathways could be the mediator of microbiota depletion in exacerbating the dysmetabolic hallmarks of DMD such as deregulation of muscle glucose uptake and enhanced fatty acid oxidation with consequent shift in fiber type toward an oxidative phenotype.
Dysbiotic microbiota of mdx affects intestinal, spleen and muscle inflammation and inversely correlates with muscle function
An important characteristic of the gut microbiota is its ability to modulate host immune responses (Geva‐Zatorsky et al, 2017). To determine whether restoration of microbial dysbiosis of mdx normalizes inflammatory responses, ABX‐treated 3m mdx mice were colonized with eubiotic microbiota of age‐matched C57Bl (ABX‐mdxFMT_C57Bl) via fecal microbiota transplant (FMT). The lamina propria (LP) immune cell compositions from the entire colon of ABX‐mdxFMT_C57Bl mice were analyzed using flow cytometry.
Colon LP T cell repertoire characterization revealed a significant decrease of CD3+ T cells in ABX‐mdxFMT_C57Bl (Fig 7A). Specifically, colon LP CD8+ T cells, but not CD4+, were reduced in ABX‐mdxFMT_C57Bl (Figs 7A and EV4A). Likewise, reductions were seen in IFNγ (Th1) or IL17 (Th17) production of colon LP pro‐inflammatory CD8+ T cells in ABX‐mdxFMT_C57Bl (Fig 7A). Otherwise, central memory CD62+ and activated CD69+ colon LP CD8+ T cells were similar between mdx and ABX‐mdxFMT_C57Bl (Fig EV4A). Furthermore, we measured reduced activated (CD69+CD4+ and CD69+Ki67+CD4+) and pro‐inflammatory (IFNγ+CD4+ and IL10+CD4+) CD4+ T cells in the colon LP of ABX‐mdxFMT_C57Bl mice, whereas there were no differences in central memory CD4+CD62+ and CD4+IL10+ Th17 cells in the colon LP between mdx and ABX‐mdxFMT_C57Bl mice (Figs 7A and EV4A). Altogether, these data strongly support the correction of innate immune activation of CD4+/CD8+ T cells in colon LP of mdx via eubiotic FMT.
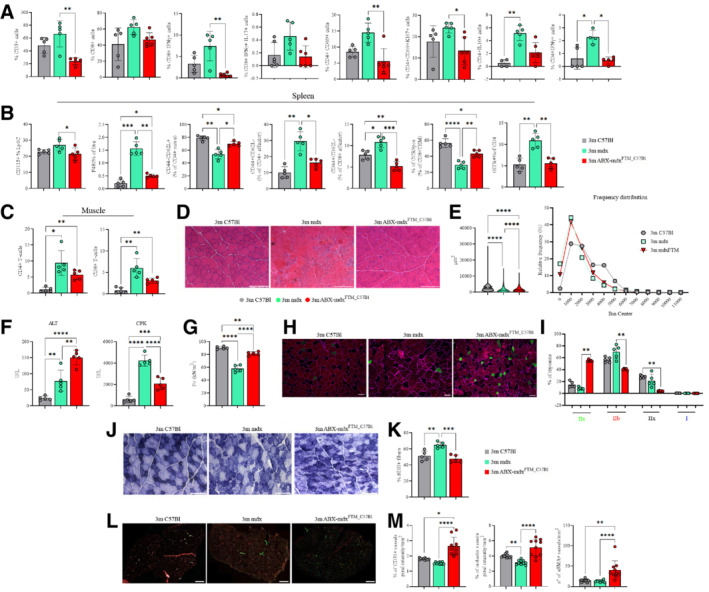
Figure 7. Effects of dysbiotic microbiota of mdx on intestine, spleen and muscle inflammation and muscle function.
-
AFACS analysis of T cell subsets from lamina propria in 3m C57Bl (n = 5), mdx (n = 5), and ABX‐mdxFMT_C57Bl (n = 5/6) showing decrease in CD3+ T cells in ABX‐mdxFMT_C57Bl. Infiltrating CD3+CD4+, and regulatory CD69+ subsets of CD4+ and CD8+ were decreased in ABX‐mdxFMT_C57Bl. Eubiotic FMT in mdx modulates T helper response, with reductions in the cumulative frequencies of CD4+ IFNγ+ (Th1) and CD4+ IL‐10+ cells in ABX‐mdxFMT_C57Bl. Data are presented as mean ± SD (*P < 0.05; **P < 0.01 ordinary one‐way ANOVA, Tukey's multiple‐comparison test).
-
BFACS analysis of spleen and muscle homogenates from 3m C57Bl (n = 5), mdx (n = 5), and ABX‐mdxFMT_C57Bl (n = 5/6). Analysis of the spleen revealed downregulation of Ly6C+ inflammatory monocytes and F4/80+ macrophages in ABX‐mdxFMT_C57Bl. Eubiotic FMT in mdx mice determined a decrease of CD4+/CD8+ CD44+CD62L effector and GITR+CD4+ T‐cells in ABX‐mdxFMT_C57Bl. Gut‐derived CCR9+CD8+TEM+ cells were increased in ABX‐mdxFMT_C57Bl. Data are presented as mean ± SD (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, ordinary one‐way ANOVA, Tukey's multiple‐comparison test).
-
CGraphs showing cumulative frequencies of infiltrating CD45+CD4+ and CD45+CD8+ cells in muscles of 3m C57Bl (n = 5), mdx (n = 5) and ABX‐mdxFMT_C57Bl (n = 5/6) were decreased in ABX‐mdxFMT_C57Bl related to mdx mice. Data are presented as mean ± SD (*P < 0.05; **P < 0.01, ordinary one‐way ANOVA, Tukey's multiple‐comparison test).
-
D, ERepresentative H&E staining (D) and quantification of myofiber area with Image J software (E) of TA muscles from 3m C57Bl (n = 5), mdx (n = 5) and ABX‐mdxFMT_C57Bl (n = 6). Scale bars for H&E: 200 μm.
-
FMeasurement of ALT, and CPK in the serum of 3m C57Bl (n = 5), mdx (n = 5), and ABX‐mdxFMT_C57Bl (n = 6).
-
GTetanic force of TA muscles from mdx (n = 5) and ABX‐mdxFMT_C57Bl (n = 6).
-
HRepresentative images of skeletal muscle showed the distribution and composition of MyHC isoforms (Type IIa, Type IIx, and Type IIb). Scale bar: 50 μm.
-
IGraph portrays the percentage of myofibers expressing different MyHC isoforms. n = 12 images were analyzed for each mouse.
-
J, KRepresentative SDH staining and quantification of percentage of SDH+ myofibers of TA muscles from mdx (n = 5) and ABX‐mdxFMT_C57Bl (n = 6) (n = 12 images per mouse). Scale bar: 200 μm.
-
L, MRepresentative image of CD31 (in cyan), α‐SMA (in green), and isolectin (in red) staining and their quantification in TA muscles from mdx (n = 5) and ABX‐mdxFMT_C57Bl (n = 6) mice. Scale bar: 500 μm.
Data information: Data for tetanic force, ALT and CPK concentration, and staining quantification are presented as mean ± SD (*P < 0.05, **P < 0.01, ***P < 0.001; ****P < 0.0001, one‐way ANOVA, Tukey's multiple‐comparison test).
Source data are available online for this figure.
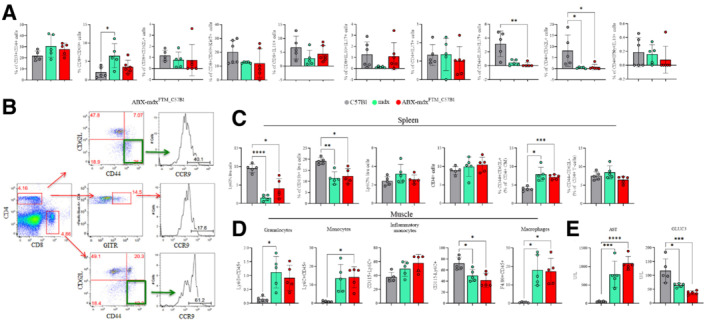
Figure EV4. Effects of dysbiotic microbiota of mdx on intestine, spleen and muscle inflammation.
-
AFACS analysis of colon lamina propria of mdx (n = 5) and ABX‐mdxFMT_C57Bl (n = 5/6) for quantification of T cell subsets.
-
BRepresentative plots of FACS analysis for the expression of CCR9 in ABX‐mdxFMT_C57Bl and ABX‐C57BlFMT_mdx are depicted. The numbers within the panels indicate the percentage of each population of live cells. Each analysis included at least 5–10 × 104 events for each gate.
-
C, DFACS analysis of T cells of the spleen (C) and granulocyte, monocyte and macrophage of muscle (D) tissues from mdx (n = 5) and ABX‐mdxFMT_C57Bl (n = 5/6).
-
ESerum levels of AST and GLUC3 in 3m C57Bl (n = 5), mdx (n = 5) and ABX‐mdxFMT_C57Bl (n = 6).
Data information: Data are presented as mean ± SD (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 ordinary one‐way ANOVA, Tukey's multiple‐comparison test).
Source data are available online for this figure.
As a specialized immune organ, the spleen immune system plays a significant role in innate and adaptive immunity. Analysis of splenic tissue revealed that inflammatory F4/80+ macrophages as well as naïve CD4+ T cells and effector memory CD4+/CD8+ T cells (TEM) were significantly reduced in ABX‐mdxFMT_C57Bl compared with mdx mice (Fig 7B). In agreement with the normalization of the effector T cell compartment CD4+GITR+ regulatory T cells were significantly less abundant in the ABX‐mdxFMT_C57Bl mice than in mdx mice. The notable exception was the gut‐derived subset of CCR9+CD8+TEM+ which was significantly increased in ABX‐mdxFMT_C57Bl compared with mdx mice (Figs 7B and EV4B). Since eubiotic FMT may have profound effects on inflammatory responses of dystrophic muscle, we further characterized muscles of ABX‐mdxFMT_C57Bl mice. No differences in granulocytes, monocytes and macrophages percentages were observed (Fig EV4D); however, muscle CD45+CD4+ and CD45+CD8+ cells were decreased in ABX‐mdxFMT_C57Bl mice compared with mdx mice (Fig 7C), suggesting the amelioration of inflammation in dystrophic muscle tissues.
Morphometric analysis of TAs of ABX‐mdxFMT_C57Bl mice showed increased CSAs of the myofibers (mean fiber area ± SEM: 3m mdx 1,644.77 ± 18.86 μm2, 3m C57Bl 1,757.41 ± 9.87 μm2, ABX‐mdxFMT_C57Bl 1,741.80 ± 8.33 μm2) and reduced fibrotic infiltrate suggesting an amelioration of the dystrophic phenotype (Fig 7D). The values of frequency distribution confirmed the higher area of myofibers in ABX‐mdxFMT_C57Bl mice related to untreated age‐matched mice (25% Percentile: 3m mdx: 648.221; 3m C57Bl: 911.385; 3m ABX‐mdxFMT_C57Bl: 837.8725; 75% Percentile: 3m mdx: 2,168.295; 3m C57Bl: 2,328.3305; 3m ABX‐mdxFMT_C57Bl: 2,293.675) (Fig 7E).
Furthermore, ALT serum level was higher in ABX‐mdxFMT_C57Bl mice than those in mdx mice suggesting increased lipid metabolism after eubiotic FMT (Fig 7F), whereas similar levels of AST were found in both groups of mice (Fig EV4E). Of note, CPK values were significantly reduced in ABX‐mdxFMT_C57Bl (Fig 7F). Strength evaluation demonstrated a significant increase of tetanic force of TA muscle in ABX‐mdxFMT_C57Bl mice compared with untreated mdx mice (Fig 7G).
Consistent with the increase of myofibers area, we observed rescue of the number of oxidative/glycolytic MyHC type IIa myofibers in TAs of ABX‐mdxFMT_C57Bl to levels observed in healthy C57Bl (Fig 7H and I). To further support this evidence, we analyzed SDH levels, and observed a significantly decrease in oxidative SDH+ fibers in ABX‐mdxFMT_C57Bl compared with mdx mice (Fig 7J and K). These data support the hypothesis that correction of dysbiotic Prevotella‐enriched microbiota in ABX‐mdxFMT_C57Bl promotes amelioration of skeletal muscle carbohydrates uptake and metabolism.
By analyzing all the measured variables in combination, we observed that ABX‐mdxFMT_C57Bl clustered apart from mdx animals and inversely correlated with gut IFNγ‐producing pro‐inflammatory, proliferating and activated CD4+ T cells as well as splenic and muscle infiltrating T cells (Fig EV5), suggesting that eubiotic FMT modulates immune response of mdx.
Figure EV5. Principal component analysis (PCA) biplot of samples and analyzed variables from spleen, gut, and muscle.
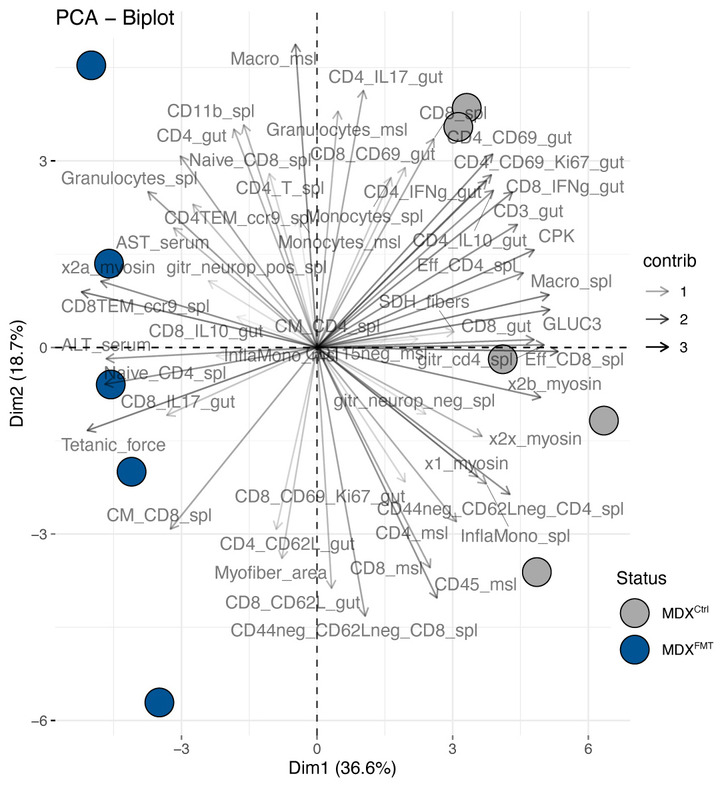
The biplot shows the PCA scores of the explanatory variables as vectors and samples colored according to treatment and genetic backgrounds. The color intensity of the vectors (lines) shows the strength of their contribution to each PC. Vectors pointing in similar directions indicate positively correlated variables, and vectors pointing in opposite directions indicate negatively correlated variables.
We further sought to investigate whether amelioration of sustained immunity in ABX‐mdxFMT_C57Bl mice muscles might be in part driven by rescue of altered vessels. Previous examination of the mdx muscle microvasculature showed defective endothelial layer and reduced vessels (Bella et al, 2020). Accumulating evidence has shown that gut microbiome can influence the balance of vascular homeostasis (Kiouptsi & Reinhardt, 2018; Battson et al, 2019; Brunt et al, 2019; Maiuolo et al, 2022). Staining of whole TA muscle sections of ABX‐mdxFMT_C57Bl mice demonstrated that eubiotic FMT efficiently increased the number of CD31+ capillaries and small arterioles expressing α‐smooth muscle actin (αSMA) (Fig 7L and M). Importantly, we found a significant increase of total isolectin positive vessels of ABX‐mdxFMT_C57Bl mice (Fig 7L and M).
All these data suggested that restoration of gut microbiota composition improves muscle pathology and function in dystrophic mdx.
Discussion
The gut microbiota is central player in shaping and modulating immune system responses, with gut microbial dysbiosis linked to several autoimmune and immune‐mediated diseases (Kosiewicz et al, 2011). The commensal population that constitutes the microbiota is extremely variable among individuals but functionally stable (Kapur et al, 2014) and its composition is also dependent on the immune responses that are mediated in the gut and on host genotypes/phenotypes (Spor et al, 2011). The interactions between the microbiota and intestinal immune cells are so strictly and hierarchical regulated that their dysfunctions cause serious problem as chronic inflammatory state, contributing to alter homeostasis even at distant sites including skeletal muscles (Kabat et al, 2016). Since immune system dysregulation was recently shown as a fundamental component of DMD pathogenesis we investigated the gut microbiota composition of dystrophic mice and its effects on muscle metabolism and function.
In DMD patients, the lack of dystrophin and dystrophin glycoprotein complex proteins is likely to cause dysfunctions in the structural integrity of the intestinal barrier. Altered bowel permeability can represent a source of molecules that might produce muscle wasting by entering the circulation, via a disrupted gut barrier, and traveling toward muscles. In addition, this condition could be responsible for the uncontrolled passage of components of the microbiota toward the immune cells of the LP, where they induce the pathological activation of these cells. Recent studies showed that chronic inflammation is partially dependent on the tissue‐environment interface; in the mucosa of gastrointestinal tract, antigen‐presenting cells and lymphocytes drive innate and adaptive responses whose deregulation determine the rising of inflammatory events that are the common features of muscular dystrophies. Dietary metabolites function as immune‐modulators: accordingly, the identification of bacteria involved in triggering the inflammation‐driven muscular degeneration could open the door to manipulate these taxa for therapeutic purposes.
For the first time, we demonstrated that microbiota composition in 3m mdx is different from age‐matched C57Bl and, more importantly, we found a reduction in diversity and a selection of few bacterial species. Of note, family‐level Prevotellaceae, genus‐level Prevotella were highly associated with dystrophic mdx mice. Prevotella has been associated with either good or bad metabolic outcomes and, probably, both depend on the presence of metabolic and inflammatory alterations (Maulucci et al, 2019). Prevotella was more abundant in patients suffering from untreated rheumatoid arthritis than in healthy individuals with a reduction in the abundance of beneficial microbes. In a mouse model of gut inflammation, animals colonized with Prevotella had more severe disease than controls, consistent with a pro‐inflammatory function of this organism (Mielke et al, 2017). Interestingly, Prevotella‐dominated microbiome is a community shift away from Bacteroides, previously reported to be associated with an anti‐inflammatory state and Treg production (Muller et al, 1996). This could account, in part, for the observed differences in susceptibility to inflammation in Prevotella‐dominated microbiome of elderly individuals (Ticinesi et al, 2017).
Following these evidences, we addressed whether modulation of the microbiota composition through antibiotics (Krebs et al, 2016; Xie et al, 2017) and germ‐free mdx dystrophic model could foster DMD pathophysiology. GF condition enables to study DMD progression from birth, whereas antibiotics treatment in adult mdx mice allows for study of the role of microbiota in modulating muscle functionality and signaling pathways at specific times of disease. We showed that microbiota depletion in mdx is associated with a shift in fiber type toward an oxidative phenotype. Interestingly, ABX and GF conditions differentially affect change toward a slower MyHC profile mirroring differences in the muscle adaptive responses to metabolic changes. This could be supported by findings in differential activation of AMPK‐related pathways that mediate deregulation of muscle glucose uptake and enhanced fatty acid oxidation in mdx+ABX and GFmdx mice.
As such, the ABX‐treated mdx and GFmdx mice displayed increased oxidative metabolism with reduced inflammation and fibrosis. The severe increase in the values of CPK in GFmdx revealed a more complex muscle‐gut microbiota interaction. Our observations are in line with recent data describing the fundamental role of gut microbiota in degradation of CPK (Nugraha et al, 2019), suggesting the hypothesis that GFmdx could suffer from colonic dysfunctions as CPK modifications are associated to these pathologies (Zuppinger, 2019; Farini et al, 2020).
We thus verified whether correction of dysbiotic Prevotella‐enriched microbiota of mdx can induce substantial modifications in skeletal muscle physiology and function. Intestinal Prevotella colonization results in metabolic changes in the microbiota determining intestinal inflammation and systemic autoimmunity (Iljazovic et al, 2021). Since gnotobiotic colonization of GF mice with Prevotella species was shown to be ineffective (Geva‐Zatorsky et al, 2017), we tested the effect of eubiotic microbiota in mdx. Our main finding was that eubiotic fecal transplantation in mdx mice was able to reduce gut‐distal muscle immune responses with a parallel recovery of dystrophic muscle features. The increase of muscle carbohydrate metabolism and hepatic fatty acid oxidation could explain, at least in part, the recovered muscle metabolism and function of mdx mice. Other potentially interesting immunomodulatory activities were not reported previously, such as the augmentation of gut‐derived regulatory CCR9+CD4+GITR+ subset in mdx mice receiving eubiotic microbiota. It will be interesting to see whether gut‐derived regulatory CCR9+CD4+GITR+ will perform additional activities in skeletal muscles.
Multiple lines of investigation have revealed that DMD is characterized by primary and secondary features caused by dystrophin absence that occurs in sequential phases. Notably, adaptive immunity is one of the secondary features of DMD, which proposes that an environmental factor triggers chronic muscle inflammation in the context of pre‐existent innate immunity activation. Although a role for the gut microbiota has been clearly established in muscle homeostasis (Sasson et al, 2019), it is not known if dysbiosis influences DMD. Our data suggest that residing gut microbial communities could be implicated in DMD progression influencing clinical and phenotypic variability of dystrophic patients by modulating metabolic and immune response. All considering, the balance of microbiota composition is crucial to maintain correct muscle function and the GF murine models retain both beneficial and deleterious effects (Guo et al, 2020). Despite the great research efforts, an effective cure for DMD is still missing. However, since patients are living longer, multidisciplinary management of DMD became fundamental to prevent complications and alleviate disease progression. This study demonstrates that modulation of gut microbiota strain displacement presents interesting therapeutic opportunities to ameliorate DMD symptoms in patients. In addition, dietary interventions are the most effective and less‐expensive strategies and represent a valuable co‐adjuvant for DMD treatment by counter‐acting the damaging effects of chronic inflammation. Further characterization of microbiota changes in dystrophic patients should provide deeper insight into whether dysbiosis contributes to the progression of DMD.
Materials and Methods
List of abbreviations in alphabetical order is indicated in Table 1.
Table 1.
Abbreviations used in the text in alphabetical order.
| AKT 1–2‐3 (Serine–threonine protein kinase 1–2‐3 isoform) |
| AMPK‐1α (5′‐AMP‐activated protein kinase catalytic subunit alpha‐1) |
| ATG7 (Autophagy Related 7) |
| COX IV (Cytochrome c oxidase IV) |
| DRP‐1 (Dynamin‐related protein 1) |
| FKHR‐FOXO1 (Forkhead box protein O1) |
| FKHRL1‐FOXO3 (Forkhead box protein O3) |
| GHR (Ghrelin) |
| GHSR (Growth Hormone Secretagogue Receptor) |
| HDAC (Histone Deacetylase inhibitors 1) |
| IGF (Insulin‐like growth) |
| IL‐33 (Interleukin 33) |
| IL‐6 (Interleukin 6) |
| IRS‐1 (Insulin receptor substrate 1) |
| LC3 (Autophagy marker Light Chain 3) |
| MMP9 (Matrix Metallopeptidase 9) |
| MyD88 (MyD88 Innate Immune Signal Transduction Adaptor) |
| NF‐kB (Nuclear Factor kappa‐light‐chain‐enhancer of activated B cells) |
| OPN (Osteopontin) |
| P38 (Mitogen‐activated protein kinases MAPK P38) |
| P62 (Autophagy receptor) |
| PGC‐1α (Peroxisome proliferator‐activated receptor gamma coactivator 1‐alpha) |
| PPARγ (Peroxisome proliferator‐activated receptor γ) |
| PSMB5 (Proteasome 20S Subunit Beta 5) |
| PSMB8 (Proteasome 20S Subunit Beta 8) |
| PSMB9 (Proteasome 20S Subunit Beta 9) |
| PTX3 (Pentraxin 3) |
| RELB (v‐rel Reticuloendotheliosis viral oncogene homolog B) |
| SIRT‐1 (Sirtuin 1) |
| SMAD2 (Small mother against decapentaplegic 2) |
| SMAD3 (Small mother against decapentaplegic 3) |
| TGF‐β (Transforming growth factor Beta) |
| TLR2 (Toll‐like receptor 2) |
| TLR4 (Toll‐like receptor 4) |
| TNF‐α (Tumor necrosis factor Alfa) |
| TOMM20 (Translocase of Outer Mitochondrial Membrane 20) |
| TRAF‐6 (Tumor necrosis factor Receptor Associated Factor 6) |
| TRPC‐1 (Transient Receptor Potential Cation Channel Subfamily C Member 1) |
| VDAC1 (Voltage‐Dependent Anion‐selective Channel 1) |
| VR‐1 (Capsaicin Receptor) |
Animal ethics statement
Procedures involving living animals were conformed to Italian law (D.L.vo 116/92) and approved by local ethics committees. This work was authorized by the Ministry of Health and Local University of Milan Committee, authorization number 859/2017‐PR (5247B.35, 10/07/2017). Two‐month‐old C57BL/10 and mdx male mice from Charles River were maintained at the Policlinico Hospital animal facility. All animals were housed in ventilated cages in a 12 h light/dark cycle, with free access to water and standard autoclaved chow. Upon arrival, mice were allowed to acclimate to the animal facility environment for at least 4 weeks and analysis performed on 3‐month‐old animals. Food intake was measured. No food intake differences were observed between C57BL/10 and mdx mice.
Individual mice were placed in empty autoclaved cages and monitored for defecation: stool samples were collected with autoclaved toothpicks. For the microbiota depletion experiments, 3‐month‐old mice were orally gavaged with a mix of antibiotics (ABX) containing vancomycin (1.25 mg), ampicillin (2.5 mg) and metronidazole (1.25 mg) in 200 μl of water for 4 weeks. For the Fecal Microbiota Transplantation (FMT) experiments, 3‐month‐old mdx mice were pre‐treated with the ABX cocktail in 200 μl water per mouse by oral gavage for 7 days to promote a more efficient bacterial colonization (Ji et al, 2017) and then transplanted with feces from C57Bl/10 (ABX‐mdxFMT_C57Bl), as previously described (Burrello et al, 2018; Strati et al, 2021). Feces collected from donor mice were diluted in PBS (1:10 w/v), briefly centrifuged to remove large debris and 200 μl of this fecal slurry was given to recipients daily for 5 days by oral gavage.
Randomization within blocks was performed to allocate the animals to different experimental procedures. To avoid that the effects of our treatments on mice had been overestimated, thus diminishing the reliability of our results, the laboratory members that analyzed the mice were blinded regarding the treatment(s) that animals received, during all the experimental procedures. Animals that eventually suffered from clinical complications during each treatment (enhancement of stress, motor impairments) were excluded from the experimental plan.
GFmdx derivation
We participated at the EC Horizon 2020 funded INFRAFRONTIER2020 project (2017–2020), to obtain mouse axenic service implemented as a Trans‐national Access activity. INFRAFRONTIER is the European Research Infrastructure for phenotyping, archiving and distribution of model mammalian genomes by the European Mouse Mutant Archive (EMMA), providing access to tools and data for biomedical research (www.infrafrontier.eu). Through the “INFRAFRONTIER2020 project and microbiome research”, in collaboration with the Gnoto/Axenic Facility of the Instituto Gulbenkian de Ciência—partner and founding member of the ECGnoto network http://www.ecgnoto.eu—we generated the GFmdx. Detailed procedure is described below.
Standard operative procedure for generating germ‐free (axenic) mice using cesarean section rederivation
Equipment
Sterile isolator and set up for rearing germ‐free mice.
Transfer chamber compatible with the isolator.
Autoclaved water inside the transfer chamber.
Surgical equipment
Medroxiprogesterone acetate (150 mg/ml, Pfizer).
VirkonS, 1% solution at room temperature (RT) (Antec Int. Ltd.).
Method
Ensure availability of isolator reared, germ‐free surrogate mother with newborn pups (< 5 days old) at day 19 of procedure (see below).
Day 2—Set up the relevant mating of foster strain inside the recipient isolator (usually on Sundays).
Day 1—Check for mating plugs inside the isolator, and identify the foster females.
Day 0—Check for mating plugs inside the isolator and identify the foster females. If more than two plugs between day 1 and 0, set up the relevant mating of mouse strain to be converted to germ‐free status (usually on Wednesdays).
Day 1—Check for mating plugs inside and outside the isolator. Identify the foster and donor female(s) for the experiment.
Day 2—Check for mating plugs inside and outside the isolator. Identify the foster and donor female(s) for the experiment. Separate females from males (if some remain without plug) inside the isolator.
Day 3—Check for mating plugs outside the isolator. Identify the donor female(s) for the experiment. Separate females from males (if some remain without plug), from the strain to be converted to germ‐free.
Day 18—Check pregnancies inside and outside the isolators. Give pregnant donor female(s) from day 1, at 17.5 days post coitus (dpc), a subcutaneous injection of medroxiprogesterone acetate (5 mg/0.1 ml).
Day 19—Carefully following the SOP for isolator entry procedures, transfer the sterile instruments and supplies required for surgery into the isolator in which the surrogate female(s) are housed. Prepare the hysterectomy suite/surgical transfer chamber: fill up the reservoir with 1% VirkonS, sterilize the surgical compartment and ventilate it overnight. Give pregnant donor female(s) from day 2, at 17.5 dpc, a subcutaneous injection of medroxiprogesterone acetate (5 mg/0.1 ml).
Day 20—Give pregnant donor female(s) from day 3, at 17.5 dpc, a subcutaneous injection of medroxiprogesterone acetate (5 mg/0.1 ml). Transfer water, paper towels and surgical instruments from the isolator to the sterilized compartment of the transfer chamber. Working in the non‐sterile compartment of the surgical transfer chamber or the place where the animals are allocated, sacrifice the donor female by cervical dislocation and submerge the whole animal in the 1% VirkonS solution for 1 min. Use sterile scissors to open the abdomen. Clamp the top of each uterine horn and the base of the uterus close to the cervix, with mosquito scissors. Cut out the “uterine package,” and place it in the transfer chamber reservoir filled with 1% VirkonS for 1 min. This procedure can be performed for a maximum of two females at the same time. Inside the sterile compartment of the transfer chamber rinse the “uterine package” with sterile water to remove the VirkonS (200 ml of minimal volume of water). On top of a heating pad at 37°C, open the “uterine package” with scissors and take out the pups, taking care do not cut the umbilical cord. After removing the pup from the placenta gently pull the umbilical cord with your forceps. Stimulate breathing of the pups while cleaning them with dry paper towel. When pups are breathing normally and have gained a healthy skin color, transfer them to the isolator housing the foster mother. Gently rub the pups with bedding material from the foster mother's cage. Leave them mixed with the bedding 1 or 2 min. Remove some of the original pups so that the foster mother has the same number of pups to feed. If some pups from the foster mother remain in the cage, mix the adopted ones with them (clean the bedding). Check for adoption not earlier than 24 h after transfer. Monitor a microbiological status of the isolator and the animals it houses 3 weeks after transfer.
Day 21/22—repeat step 20 for pregnant donor females of days 2 and 3, if necessary.
FACS analysis
Murine lamina propria mononuclear cells (LPMC) were isolated as described in (Weigmann et al, 2007). Briefly, colonic LPMCs were isolated via incubation with 5 mM EDTA at 37°C for 30 min, followed by mechanical disruption with GentleMACS (Miltenyi Biotec). After filtration with 100 μm and 70 μm nylon strainers (BD), the LPMC were counted and stained for immunophenotyping.
Pooled muscle from the leg (vastus medialis, vastus lateralis, rectus femoris, biceps femoris, adductors and gastrocnemius) and spleen of ABX‐treated mdx, GFmdx, ABX‐mdxFMT_C57Bl and age‐matched untreated mdx or C57Bl mice were minced slightly to removed blood trapped vessels and dispersed with scissors to increase total surface area, to enhance the efficiency of digestion while shortening the time required for this procedure. Tissues were washed in PBS, and then digested at 37°C with 0.2 mg/ml Liberase in DMEM culture medium. Undigested tissues were mashed with a plunger through the filters and washed with DMEM with serum. Then, they were filtered through a 70 μm filter, placed on Histopaque 1077 gradient and centrifuged at 400 g for 45 min. We harvested the cells at the interface, washed two times with PBS and then used for flow cytometry analysis. Cells obtained from the muscle, spleen and colon LP tissues from the same mice were evaluated for the expression of different immunological subpopulations. Cells were multiple‐labeled with different groups of antibodies to recognize specific sub‐populations (for muscle: CD45 PerCp, CD4 Pe‐Cy7 and Pacific Blue, CD8 efluor 450, CD44 FITC, CD62L PE, CD25 APC, B220 APC‐Cy7, GITR Pe‐Cy7, CD3 FITC. For spleen: CD4 Pe‐Cy7 and Pacific Blue, CD8 efluor 450, CD44 FITC, CD62L PE, Foxp3 Alexa fluor 488, CD25 APC, F4/80 Pe‐Cy7, CD11b PE, CD11c FITC, IL‐17 PE, IFN‐γ APC. For GI: CD45 PerCp, CD11b PE, Lys). All the antibodies were purchased from eBioscience (San Diego, USA), except for CD45 PerCp, CD44 FITC obtained from BD (New Jersey, USA) and CD4 Pacific Blue from BioLegend (San Diego, USA). For FACS characterization, data were acquired with the BD Canto II machine and analyzed with FlowJo 9 software. Each analysis included at least 5–10 × 104 events for each gate.
Serum analysis
CPK, ALT, AST, and GLUC3 analysis were performed on serum samples of ABX‐treated mdx, GFmdx, ABX‐mdxFMT_C57Bl, and untreated mdx mice with CPK/ALT/AST/GLUC3 kit (Cobas), according to manufacturer's instructions.
Analysis of tetanic force
Tetanic force of TA muscle was determined as described in (Farini et al, 2016), normalized to muscle cross section area and expressed as kN/m2.
Histological analysis
Colon tissues were collected from 3m C57Bl/10 and mdx, frozen in liquid‐nitrogen cooled isopentane and cut on a cryostat into 10 μm slices. H&E staining was performed as in Farini et al (2021). TA muscle tissues were collected from ABX‐treated mdx, GFmdx, ABX‐mdxFMT_C57Bl, and untreated mdx mice, frozen in liquid‐nitrogen cooled isopentane and cut on a cryostat into 10 μm. Gömöri trichrome staining was performed to evaluate the morphology and the percentage of fibrosis. Adjacent sections were stained with H&E. Frozen sections were brought to RT and placed in preheated Bouin's fluid (BF) at 56°C for 15 min. Equal volumes of Hematoxylin Weigert's Iron Part A and B (Bio‐Optica, Milan S.p.A. Italy) were applied to tissue sections for 5 min. Then, acid alcohol solution (0.5%) was applied to sections for 10 s, to stain cytoplasm, followed by Acid Fuchsin solution (Bio‐Optica, Milan S.p.A. Italy) diluted 1:2 in deionized water for 5 min. Tissue sections were incubated with phosphomolybdic acid (Bio‐Optica, Milan S.p.A. Italy) for 5 min to block the staining of all tissue components other than connective tissue fibers. Then, slides were incubated with Aniline Blue solution (Bio‐Optica, Milan S.p.A., Italy) for 5 min to stain collagen fibers. Finally, slides were washed in deionized water combined with 1% glacial acetic acid (Carlo Erba, Milan, Italy) and incubated for 30 s in 100% ethanol solution, for dehydration. 100% Xylene (Sigma‐Aldrich, USA) for 1 min before mounting with DPX reagent (VWR International, USA) and coverslips. Frozen sections were characterized by immunofluorescence staining. Slides were fixed with 4% paraformaldehyde for 10 min, permeabilized with 0.3% Triton X‐100 for 15 min and incubated with 10% donkey serum to block non‐specific binding for 1 h and then incubated with the primary antibodies (overnight at 4°C) diluted in blocking solution. Fluorochrome‐conjugated secondary antibodies were diluted in PBS and added for 1 h at RT. Primary antibodies were used at the following dilutions: CD3 1:50 (AB135372, Abcam, UK); fibers type I 1:50 (BA‐D5, Developmental Studies Hybridoma Bank, Douglas Houston); fiber type IIA 1:50 (sc‐71, Developmental Studies Hybridoma Bank); fiber type IIB 1:50 (BF‐F3, Developmental Studies Hybridoma Bank). Slides were then mounted with Prolong Gold® Antifade Reagent with DAPI (Thermo Fisher, Carlsbad, CA). Leica Dmi8 fluorescence microscope was used for acquiring images. Histological identification of slow/type I, fast fatigue resistant/type Iia, and fast fatigable/type Iib fibers was performed by staining for either myosin ATPases or oxidative enzyme capacity (succinate dehydrogenase, SDH). Enzymatic activity of SDH was assayed by placing the slides in SDH incubating solution, containing sodium succinate as a substrate and nitro‐blue tetrazolium (NBT) for visualization of reaction for 1 h at 37°C. At first, slides were incubated for 10 s in 30–60–90–60–30% acetone solution and, then, for 30 s in 80–90–100% ethanol solution for dehydration. Finally, 100% Xylene (Sigma‐Aldrich, USA) for 1 min before mounting with DPX reagent (VWR International, USA) and coverslips. For Gömöri trichrome and SDH staining, images were captured by Leica microdissector (CTR6000).
Imaging mass spectrometry
Colon tissues of 3‐month‐old mdx mice were frozen for preparation of cryosections (thickness of 10 μm) with the use of a cryostat (CM 1900; Leica Microsystems, Wetzlar, Germany). For imaging mass spectrometry, the sections were thaw‐mounted on indium–tin oxide (ITO) slides (Bruker Daltonik, Bremen, Germany), dried in silica gel–containing plastic tubes, and then sprayed with 9‐aminoacridine (5 mg in 4 ml of 80% ethanol) with the use of a 0.2‐mm nozzle caliber airbrush (Procon Boy FWA Platinum; Mr Hobby, Tokyo, Japan) for matrix‐assisted laser desorption‐ionization (MALDI) imaging mass spectrometry in positive‐ion mode. Adjacent sections were stained with H&E. Imaging mass spectrometry was performed with iMScope TRIO Mass Microscope (Shimadzu, Kyoto, Japan). MALDI mass spectra were acquired with a laser diameter of 50 μm, 200 shots/spot, scanning pitch of 20 μm, and scanning m/z range of 615–931. Regions of tissue samples exposed to the laser radiation were determined by light and fluorescence microscopic observations. For each lipid, the mean intensity was measured by ImageJ Software at 12 positions (sample area of 100 × 100 μm2) throughout the colon images.
Qualitative (RT‐qPCR) experiments
Total RNA was extracted from TA muscle of ABX‐treated, GF and age‐matched untreated mdx or C57Bl mice and cDNA generated using the Reverse Transcriptase Kit (ThermoFisher Scientific, California, USA). We quantified the expression of genes through SYBR‐Green method. All the samples were tested in duplicate and the threshold cycles (Ct) of target genes were normalized against the housekeeping gene, β‐actin. Relative transcript levels were calculated from the Ct values as X = 2−ΔΔct where X is the fold difference in amount of target gene versus β‐actin and ΔCt = Cttarget − Ctβ‐actin. The efficiency of primers used was calculated between 95.2 and 98.9%. The sequence of primers used is listed in Table 2.
Table 2.
List of primers (5′→3′) for RT‐qPCR.
| Actn3 f AATCGCCAACGTTAACAAGG |
| Actn3 r AGTGTTCAGGTTTCCGATGG |
| Chrnd f TCGTCGCAAACCGCTCTT |
| Chrnd r GATGGCCAGCGAGGTGAT |
| Col1a f CCTCAGGGTATTGCTGGACAAC |
| Col1a r CAGAAGGACCTTGTTTGCCAGG |
| Col3a‐f CCTTAACATGTGTCTTTAAAGCCC |
| Col3a‐r AAATGCTTTTAAAGGTGCTTCTCT |
| CoxVa‐f TTGATGCCTGGGAATTGCGTAAAG |
| CoxVa‐r AACAACCTCCAAGATGCGAACAG |
| CoxVIIb‐f TTTCAGGACGCTTTGCAAGG |
| CoxVIIb‐r TGCTTCGAACTTGGAGACGG |
| CytC f CATCTCAACGGCTTATTATGACTTT |
| CytC r GCTAACCACCAGGAGGCAACTGT |
| Ldh f TATCTTAATGAAGGACTTGGCGGATGAG |
| Ldh r GGAGTTCGCAGTTACACAGTAGTC |
| matp2a1‐f TGTTTGTCCTATTTCGGGGTG |
| matp2a1‐r AATCCGCACAAGCAGGTCTTC |
| Mcad f TAC GGC ACA AAA GAA CAG ATC G |
| Mcad r CAG GCT CTG TCA TGG CTA TGG |
| mFoxP3‐f TCAAGTACCACAATATGCGA |
| mFoxP3‐r GATTTCATTGAGTGTCCTCTG |
| mGPx1‐f AGTTCGGACATCAGGAGAATGGCA |
| mGPx1‐r TCACCATTCACCTCGCACTTCTCA |
| miNOS‐f CTCACTGGGACAGCACAGAA |
| miNOS‐r GGCCTTGTGGTGAAGAGTGT |
| mMurF1‐f CAGAGGCAGTTGGATCGTCTATG |
| mMurF1‐r TGAGGCAGAGTCTCTCTATGT |
| mMYHCs12‐r TTCACCTGGGACTCAGCAATG |
| mMYHCsl2‐f AAGCTGAGGAGGCTGAGGAAC |
| mNRF1‐f GGCACTGTCTCACTTATCCAGGTT |
| mNRF1‐r CAGCCACGGCAGAATAATTCA |
| mp62‐f AGGCGCACTACCGCGAT |
| mp62‐r CGTCACTGGAAAAGGCAACC |
| mpdk4‐f GTCTCAATAGTGTCACCTGTGTAA |
| mpdk4‐r CCTGGGCATTTAGCATCTATCT |
| mPGC1α‐f GCTAAACGACTCCGAGAACAA |
| mPGC1α‐r ACTGACCCAAACATCATACCC |
| mPPARα‐f TGATTGGTTCCAGGCAATTAGA |
| mPPARα‐r CACTCGTACAGTCAGTTCAGTC |
| Mrf4 f GCACGCAGTGCTTCTTC |
| Mrf4 r CATGCTGCTGTCTGAAGGTC |
| mRORγt‐f GACTGACAATCAGCAGGGATAA |
| mRORγt‐r GGGAAATACAATGAGGTATTGAAAGG |
| mTbet‐f GATCATCACTAAGCAAGGAC |
| mTbet‐r ACATCCACAAACATCCTGTA |
| Myf5 f CTGCTCTGAGCCACCAG |
| Myf5 r GACAGGGCTGTTACATTCAGG |
| MyHC‐IIb f CAAGAGACAAGCTGAAGAGGCT |
| MyHC‐IIb r GATATACAGGACAGTGACAAAGAACT |
| MyoD f AGCACTACAGTGGCGACTA |
| MyoD r GGCCGCTGTAATCCATCA |
| Myogenin f CCTTGCTCAGCTCCCTCA |
| Myogenin r TGGGAGTTGCATTCACTGG |
| Myogl f GAGGGAGCTGGTGTCAACAG |
| Myogl r CTTGCAAAACCACACTGCTC |
| Pax7 f AAAAAACCCTTTCCCTTCCTACA |
| Pax7 r AGCATGGGTAGATGGCACACT |
| Tnnt1 f AAGGGGAGCGTGTGGATTTTG |
| Tnnt1 r TCCTCCTTTTTCCGCTGTTCA |
| β‐actin f GGCTGTATTCCCCTCCATCG |
| β‐actin r CCAGTTGGTAACAATGCCATGT |
WB analysis
Tibialis anterior skeletal muscles and colonic tissues were isolated from ABX‐treated mdx, GFmdx, ABX‐mdxFMT_C57Bl and age‐matched untreated mdx or C57Bl mice and total proteins were obtained as in (Parolini et al, 2009). Samples were resolved on polyacrylamide gels (ranging from 6 to 14%) and transferred to nitrocellulose membranes (Bio‐Rad Laboratories, California, USA). Filters were incubated overnight with following antibodies: PSMB5 (1:500, AB3330, Abcam); PSMB8 (1:500, AB3329, Abcam); PSMB9 (1:500, AB42987, Abcam); β‐actin (1:500, a2066, Sigma‐Aldrich); LC3B (1:500, L7543, Sigma‐Aldrich); TGFβ (1:500, e‐ab‐33090, Elabscience); TNFα (1:500, e‐ab‐40015, Elabscience); NF‐kB (1:500, sc‐514451, Santa Cruz Biotechnology – SCB); TRAF‐6 (1:500, sc‐8409, SCB); RELb (1:500, sc‐48366, SCB); PTX3 (1:500, AB90806, Abcam); IL‐6 (1:500, sc‐57315, SCB); VDAC1/Porin (1:500, sc‐390996, SCB); PGC1α (1:500, sc‐518038, SCB); FKHR‐FOXO1 (1:500, sc‐374427, SCB); FKHRL1‐FOXO3 (1:500, sc‐48348, SCB); IGF1 (1:500, sc‐9013, SCB); IGF2 (1:500, sc‐5622, SCB); TRPC‐1 (1:500, sc‐20110, SCB); MTCO‐1 (1:500, sc‐58347, SCB); IKK‐I (1:500, sc‐10760, SCB); AMPK‐1α (1:500, sc‐74461, SCB); GSK‐3 αβ (1:500, sc‐81496, SCB); TLR2 (1:500, orb229137, Biorbyt): TLR4 (1:500, sc‐293072, SCB); vinculin (1:500, MA5‐11690, Invitrogen); FGF21 (1:500, sc‐292879, SCB); MMP9 (1:500, ab38898, Abcam); SIRT‐1 (1:500, PA5‐17074, Invitrogen); ATG7 (1:500, sab4200304, Sigma‐Aldrich); GHSR (1:500, eab12471, Elabscience); GHRELIN (1:500, pa1‐1070, Invitrogen); TOMM20 (1:500, AB186735, Abcam); DRP1 (1:500, AB184247, Abcam); SMAD3 (1:500, e‐ab‐32921, Elabscience); SMAD2 (1:500, e‐ab‐32916, Elabscience); P38 (1:500, E‐AB‐32460, Elabscience); P62 (1:500, P0067, Sigma‐Aldrich); phosphoERK1‐2 (1:500, E‐AB‐20868, Elabscience); phosphoP38 (1:500, E‐AB‐20949, Elabscience); phosphoSMAD2‐3 (1:500, E‐AB‐21‐040, Elabscience); COX IV (1:500, AB16056, Abcam): IRS‐1 (1:500, ab131487, Abcam); AKT 1‐2‐3 (1:500, ab179463, Abcam); PPARγ (1:500, AB59256, Abcam); IL‐33 (1:500, af3626, R&D); ERK 1‐2 (1:500, ab54230, Abcam); OPN (1:500, AF808, R&D); MyD88 Innate Immune Signal Transduction Adaptor (MyD88) (1:500, 23230‐I‐AP, Proteintech); HDAC1 (1:500, MA5‐1807, Invitrogen); HDAC2 (1:500, 51‐5100, Invitrogen); VR‐1 (1:500, sc‐12503, SCB); IGF1Rβ (1:500, sc‐9038, SCB); and IGF2R (1:500, sc‐14413, SCB). Filters were detected with peroxidase conjugated secondary antibodies (Agilent Technologies, California, USA) and developed by ECL (Amersham Biosciences, UK).
Microbiota analysis
DNA extraction, 16S rRNA gene amplification, purification, library preparation and pair‐end sequencing on the Illumina MiSeq platform were performed as described in (Nunes et al, 2017a). Reads were pre‐processed using the MICCA pipeline (v.1.7.0; http://www.micca.org; Nunes et al, 2017b). Forward and reverse primers trimming and quality filtering were performed using micca trim and micca filter, respectively. Filtered sequences were denoised using the UNOISE algorithm implemented in micca I to determine true biological sequences at the single nucleotide resolution by generating ASVs. Bacterial ASVs were taxonomically classified using micca classify and the Ribosomal Database Project (RDP) Classifier v2.11 (Nunes & Resende, 2017). Multiple sequence alignment of 16S sequences was performed using the Nearest Alignment Space Termination (NAST) algorithm (Nunes et al, 2017c) implemented micca msa with the template alignment clustered at 97% similarity of the Greengenes database (Nunes et al, 2017d) (release 13_08). Phylogenetic trees were inferred using micca tree (Nunes et al, 2017e). Sampling heterogeneity was reduced rarefying samples at the depth of the less abundant sample using micca tablerare. Alpha (within‐sample richness) and beta‐diversity (between‐sample dissimilarity) estimates were computed using the phyloseq R package (Nunes et al, 2017f). Permutational multivariate analysis of variance (PERMANOVA) test was performed using the adonis function in the R package vegan with 999 permutations. Differential abundance testing was carried out using the R package DESeq2 (Nunes et al, 2017g) using the non‐rarefied data (Nunes et al, 2017h). P‐values were false discovery rate corrected using the Benjamini–Hochberg procedure implemented in DESeq2. Random Forest (Yang et al, 2020) analyses of 16S rRNA gene sequencing data were performed using the randomForest R package; permutation tests with 1,000 permutations were performed to assess model significance (Nunes et al, 2017i). Spearman's correlation tests were computed using the psych R package. Prediction of functional metagenomic content was inferred by using Piphillin (Parchem et al, 2019) with the reference curated databases BioCyc (Pezzilli & Mauloni, 2019) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (Pinciotti et al, 2019). Metabolic pathway maps were visualized using iPATH 3 (https://pathways.embl.de/; Rodrigues et al, 2017).
RNAseq analysis
Library Preparation and DNA Sequencing: 150–300 ng of total RNA determined by InvitrogenTM QubitTM high‐sensitivity spectrofluorometric measurement was poly‐A selected and reverse transcribed using Illumina's TruSeq stranded mRNA library preparation kit. Each sample was fitted with one of 96 adapters containing a different 8‐base molecular barcode for high‐level multiplexing. After 15 cycles of PCR amplification, completed libraries were sequenced on an Illumina NovaSeqTM 6000, generating 20 million or more high‐quality, 100‐base, long‐paired end reads per sample. RNA‐Seq Analysis: A quality control check on the fastq files was performed using FastQC. Upon passing basic quality metrics, the reads were trimmed to remove adapters and low‐quality reads using default parameters in Trimmomatic1 (Populo et al, 2017). Alignment, Transcript Abundance and Differential Gene Expression Analysis: The trimmed reads were then mapped to a reference genome using default parameters with strandness (R for single‐end and RF for paired‐end) option in Hisat22 (Prukop et al, 2020). In the next step, transcript/gene abundance was determined using kallisto3 (Recke et al, 2014). We first created a transcriptome index in kallisto using Ensembl cDNA sequences for the reference genome. This index was then used to quantify transcript abundance in raw counts and transcript per million. Fold‐changes between groups were calculated using EdgeR from the Bioconductor package (Cao et al, 2020). PCA on differentially expressed genes was performed using ClustVis (Richner et al, 2017). Gene ontology (GO) analysis was conducted submitting gene lists to the PANTHER Enrichment Test (release 16.0), built‐in analytical tool in the AmiGO2 software suite by the GO consortium (Rispens et al, 2014). GO analyses were conducted on the GO database (version 2021‐05‐01), using all genes in the Mus musculus database as reference list and the GO Biological Process Complete as annotation dataset. Significantly enriched GO terms were identified by adjusted P‐value < 0.05. GSEA was performed via dedicated software (release 4.2.3) by Molecular Signatures Database (MSigDB). The “Hallmark” annotated gene set collection was used for analysis of ranked gene lists.
Metabolome analysis
To extract the metabolome from the GI tissues of ABX‐treated and age‐matched untreated mdx or C57Bl mice we used the the MetaboPrep kit (Theoreo, Montecorvino Pugliano, SA) as in (Lee et al, 2020; Long et al, 2020) according to manufacturers' protocol. Analysis was conducted in gas‐chromatography coupled with mass spectrometry (GC‐2010 Plus gas chromatograph and 2010 Plus single quadrupole mass spectrometer; Shimadzu Corp., Kyoto, Japan). Chromatographic separation was achieved as previously reported using a 30 m 0.25 mm CP‐Sil 8 CB fused silica capillary GC column with 1.00 μm film thickness from Agilent (J&W Scientific, Folsom, CA, USA), with helium as carrier gas. Untargeted metabolites were identified by comparing the mass spectrum of each peak with the NIST library collection (NIST, Gaithersburg, MD, USA). To identify metabolites, the linear index difference max tolerance was set at 50, while the minimum matching for the NIST library search was set at 85%. According to MSI level 1 standard (Roquette et al, 2017), the relevant putative metabolites was further confirmed using an independent analytical standard analysis. The normalization procedures consisted of data transformation and scaling. Statistical analyses were conducted on transformed (og transformation) and autoscaled (mean‐centered and divided by the standard deviation of each variable) data. Partial least square discriminant analysis (PLS‐DA) was performed on internal standard peak area normalized chromatogram using R. Classification and cross‐validation were performed using the wrapper function included in the caret package. A permutation test was performed to assess the significance of class discrimination. Variable importance in projection (VIP) scores were calculated for each component. For each relevant metabolite, the Mouse Metabolome Database ID number was determined. Metabolic pathways associated with these metabolites were analyzed using the MetScape application (Santos et al, 2017a). Metabolic pathways involvement was also evaluated using the MetPa tool (Santos et al, 2017b).
Image quantification
Histological images were captured by Leica microdissector, fluorescent microscope and confocal microscopy. Quantitative analyses were performed by ImageJ Software (NIH). Threshold color Plug in of ImageJ Software was used to quantify the Gömöri trichrome staining as percentage of area over a fixed grid area. For IF quantification, confocal acquisition of n = 12 muscle cross‐sections for distinct TA muscles were obtained from each experimental animal used for each protocol. Data were analyzed by GraphPad Prism and expressed as means ± SD.
Statistics
To determine the significance of the variation of cellular concentration throughout the time, we used the linear regression for repeated measures. To compare multiple‐group means, one‐way ANOVA followed by Tukey's multiple‐comparison test or non‐parametric test followed by Kruskal–Wallis test were used to determine significance (*P < 0.05, **P < 0.01, ***P < 0.001; ****P < 0.0001). To compare two groups, Student's t‐test was applied assuming equal variances (*P < 0.05, **P < 0.01, ***P < 0.001; ****P < 0.0001). Sample size was determined by using a sample‐size calculator freely available on internet. All the samples that did not rich quality control standards due to the presence of contaminants for RNA or to problems in freezing procedures for histological analysis were excluded. The analysis of Alpha diversity index to evaluate microbiota richness was based on Wilcoxon rank‐sum test on row data. The exact P‐values of the manuscript are summarized in the Appendix Table S1.
Author contributions
Andrea Farini: Conceptualization; data curation; formal analysis; investigation; writing – original draft; writing – review and editing. Luana Tripodi: Formal analysis; investigation. Chiara Villa: Formal analysis; validation; writing – original draft. Francesco Strati: Investigation; methodology. Amanda Facoetti: Investigation. Guido Baselli: Validation; methodology. Jacopo Troisi: Resources; funding acquisition. Annamaria Landolfi: Software; visualization. Caterina Lonati: Methodology. Davide Molinaro: Data curation; software. Michelle Wintzinger: Methodology. Stefano Gatti: Supervision; validation. Barbara Cassani: Conceptualization; data curation; investigation. Flavio Caprioli: Supervision; writing – review and editing. Federica Facciotti: Supervision; writing – original draft. Mattia Quattrocelli: Data curation; validation; investigation; writing – original draft. Yvan Torrente: Conceptualization; funding acquisition; methodology; writing – original draft; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Dataset EV3
Dataset EV4
Source Data for Expanded View
PDF+
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Acknowledgements
We thank Luca Valenti and Daniele Prati of Department of Transfusion Medicine and Hematology, Fondazione IRCCS Ca′ Granda Ospedale Maggiore Policlinico (Milan, Italy) for helpful critical discussion. We thank Ann‐Christin Niehoff from Shimadzu for the outstanding assistance in the acquisition and interpretation of Imaging mass spectrometry data. We thank Francesco Fortunato (IRCCS Foundation Ca′ Granda Ospedale Maggiore Policlinico, Dino Ferrari Center, Neuroscience Section, Department of Pathophysiology and Transplantation, University of Milan, Milan 20122, Italy) and Filomena Napolitano (Laboratorio di Chimica Clinica e Microbiologia, Fondazione IRCCS Cà Granda Ospedale Maggiore Policlinico, Milano, Italy) for technical assistance. This work was supported by INFRAFRONTIER2020 Project to derive the germ‐free mdx mice and support microbiome research and Associazione Amici Centro Dino Ferrari. AF is the recipient of Fondazione IRCCS Cà Granda, Ospedale Maggiore Policlinico 5x1000 Research Award; YT is the recipient of Fondo Europeo di Sviluppo Regionale 2014‐2020, POR FESR 2014‐2020, Ricerca Innovazione and Gruppo familiari beta‐sarcoglicanopatie, PR‐0394, GFB‐ONLUS; FF and FC are the recipients of Premio Ricerca Biomedica Fondazione CARIPLO; MQ is the recipient of DK121875 (NIH), Start‐up funds (CCHMC), Trustee Award (CCHMC) and Heart Institute Translational Funds (CCHMC) MQ. CV is the recipient of Piano nazionale di ripresa e resilienza (PNRR) Missione 4 Componente 2 (National Center for Gene Therapy and Drugs based on RNA Technology, Spoke 1), NextGenerationEU, MUR.
EMBO Mol Med (2023) 15: e16244
See also: A Jayaraman & S Pettersson (March 2023)
Data availability
The RNAseq data have been deposited to GEO Database (accession number: GSE218370) and are available at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE218370.
References
- Allegra A, Corsonello A, Buemi M, D'Angelo R, di Benedetto A, Bonanzinga S, Cucinotta D, Ientile R, Corica F (1997) Plasma, erythrocyte and platelet magnesium levels in type 1 diabetic patients with microalbuminuria and clinical proteinuria. J Trace Elem Med Biol 11: 154–157 [DOI] [PubMed] [Google Scholar]
- Alves GA, Silva LR, Rosa EF, Aboulafia J, Freymuller‐Haapalainen E, Souccar C, Nouailhetas VL (2014) Intestine of dystrophic mice presents enhanced contractile resistance to stretching despite morphological impairment. Am J Physiol Gastrointest Liver Physiol 306: G191–G199 [DOI] [PubMed] [Google Scholar]
- Baccari MC, Romagnani P, Calamai F (2000) Impaired nitrergic relaxations in the gastric fundus of dystrophic (mdx) mice. Neurosci Lett 282: 105–108 [DOI] [PubMed] [Google Scholar]
- Bansal D, Miyake K, Vogel SS, Groh S, Chen CC, Williamson R, McNeil PL, Campbell KP (2003) Defective membrane repair in dysferlin‐deficient muscular dystrophy. Nature 423: 168–172 [DOI] [PubMed] [Google Scholar]
- Battson ML, Lee DM, Li Puma LC, Ecton KE, Thomas KN, Febvre HP, Chicco AJ, Weir TL, Gentile CL (2019) Gut microbiota regulates cardiac ischemic tolerance and aortic stiffness in obesity. Am J Physiol Heart Circ Physiol 317: H1210–H1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejaoui K, Hirabayashi K, Hentati F, Haines JL, Ben Hamida C, Belal S, Miller RG, McKenna‐Yasek D, Weissenbach J, Rowland LP et al (1995) Linkage of Miyoshi myopathy (distal autosomal recessive muscular dystrophy) locus to chromosome 2p12‐14. Neurology 45: 768–772 [DOI] [PubMed] [Google Scholar]
- Bella P, Farini A, Banfi S, Parolini D, Tonna N, Meregalli M, Belicchi M, Erratico S, D'Ursi P, Bianco F et al (2020) Blockade of IGF2R improves muscle regeneration and ameliorates Duchenne muscular dystrophy. EMBO Mol Med 12: e11019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunt VE, Gioscia‐Ryan RA, Richey JJ, Zigler MC, Cuevas LM, Gonzalez A, Vazquez‐Baeza Y, Battson ML, Smithson AT, Gilley AD et al (2019) Suppression of the gut microbiome ameliorates age‐related arterial dysfunction and oxidative stress in mice. J Physiol 597: 2361–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrello C, Garavaglia F, Cribiu FM, Ercoli G, Lopez G, Troisi J, Colucci A, Guglietta S, Carloni S, Guglielmetti S et al (2018) Therapeutic faecal microbiota transplantation controls intestinal inflammation through IL10 secretion by immune cells. Nat Commun 9: 5184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Tu WJ, Cheng W, Yu L, Liu YK, Hu X, Liu Q (2020) Clinical features and short‐term outcomes of 102 patients with coronavirus disease 2019 in Wuhan, China. Clin Infect Dis 71: 748–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Confalonieri P, Oliva L, Andreetta F, Lorenzoni R, Dassi P, Mariani E, Morandi L, Mora M, Cornelio F, Mantegazza R (2003) Muscle inflammation and MHC class I up‐regulation in muscular dystrophy with lack of dysferlin: an immunopathological study. J Neuroimmunol 142: 130–136 [DOI] [PubMed] [Google Scholar]
- de Morree A, Flix B, Bagaric I, Wang J, van den Boogaard M, Grand Moursel L, Frants RR, Illa I, Gallardo E, Toes R et al (2013) Dysferlin regulates cell adhesion in human monocytes. J Biol Chem 288: 14147–14157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejardin E (2006) The alternative NF‐kappaB pathway from biochemistry to biology: pitfalls and promises for future drug development. Biochem Pharmacol 72: 1161–1179 [DOI] [PubMed] [Google Scholar]
- Durbeej M, Cohn RD, Hrstka RF, Moore SA, Allamand V, Davidson BL, Williamson RA, Campbell KP (2000) Disruption of the beta‐sarcoglycan gene reveals pathogenetic complexity of limb‐girdle muscular dystrophy type 2E. Mol Cell 5: 141–151 [DOI] [PubMed] [Google Scholar]
- Emery AE (2002) The muscular dystrophies. Lancet 359: 687–695 [DOI] [PubMed] [Google Scholar]
- Farini A, Razini P, Erratico S, Torrente Y, Meregalli M (2009) Cell based therapy for Duchenne muscular dystrophy. J Cell Physiol 221: 526–534 [DOI] [PubMed] [Google Scholar]
- Farini A, Sitzia C, Navarro C, D'Antona G, Belicchi M, Parolini D, Del Fraro G, Razini P, Bottinelli R, Meregalli M et al (2012) Absence of T and B lymphocytes modulates dystrophic features in dysferlin deficient animal model. Exp Cell Res 318: 1160–1174 [DOI] [PubMed] [Google Scholar]
- Farini A, Sitzia C, Cassani B, Cassinelli L, Rigoni R, Colleoni F, Fusco N, Gatti S, Bella P, Villa C et al (2016) Therapeutic potential of immunoproteasome inhibition in Duchenne muscular dystrophy. Mol Ther 24: 1898–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farini A, Villa C, Di Silvestre D, Bella P, Tripodi L, Rossi R, Sitzia C, Gatti S, Mauri P, Torrente Y (2020) PTX3 predicts myocardial damage and fibrosis in Duchenne muscular dystrophy. Front Physiol 11: 403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farini A, Sitzia C, Villa C, Cassani B, Tripodi L, Legato M, Belicchi M, Bella P, Lonati C, Gatti S et al (2021) Defective dystrophic thymus determines degenerative changes in skeletal muscle. Nat Commun 12: 2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geva‐Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz‐Lopez A, Yanortsang TB, Yang L, Jupp R, Mathis D et al (2017) Mining the human gut microbiota for immunomodulatory organisms. Cell 168: 928–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gul N, Babes L, Siegmund K, Korthouwer R, Bogels M, Braster R, Vidarsson G, ten Hagen TL, Kubes P, van Egmond M (2014) Macrophages eliminate circulating tumor cells after monoclonal antibody therapy. J Clin Invest 124: 812–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Qiu X, Deis J, Lin TY, Chen X (2020) Pentraxin 3 deficiency exacerbates lipopolysaccharide‐induced inflammation in adipose tissue. Int J Obes (Lond) 44: 525–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydemann A (2017) Severe murine limb‐girdle muscular dystrophy type 2C pathology is diminished by FTY720 treatment. Muscle Nerve 56: 486–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieronimakis N, Hays A, Prasad A, Janebodin K, Duffield JS, Reyes M (2016) PDGFRalpha signalling promotes fibrogenic responses in collagen‐producing cells in Duchenne muscular dystrophy. J Pathol 240: 410–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iljazovic A, Roy U, Galvez EJC, Lesker TR, Zhao B, Gronow A, Amend L, Will SE, Hofmann JD, Pils MC et al (2021) Perturbation of the gut microbiome by Prevotella spp. enhances host susceptibility to mucosal inflammation. Mucosal Immunol 14: 113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji SK, Yan H, Jiang T, Guo CY, Liu JJ, Dong SZ, Yang KL, Wang YJ, Cao ZJ, Li SL (2017) Preparing the gut with antibiotics enhances gut microbiota reprogramming efficiency by promoting xenomicrobiota colonization. Front Microbiol 8: 1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolles S, Jordan SC, Orange JS, van Schaik IN (2014) Immunoglobulins: current understanding and future directions. Clin Exp Immunol 178: 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliao M, Oliveira F, Nunes B, Carneiro AV, Barbosa A (2017) Effect of dignity therapy on end‐of‐life psychological distress in terminally ill Portuguese patients: a randomized controlled trial. Palliat Support Care 15: 628–637 [DOI] [PubMed] [Google Scholar]
- Junghans RP, Ebralidze A, Tiwari B (2001) Does (CUG)n repeat in DMPK mRNA ‘paint’ chromosome 19 to suppress distant genes to create the diverse phenotype of myotonic dystrophy?: a new hypothesis of long‐range cis autosomal inactivation. Neurogenetics 3: 59–67 [DOI] [PubMed] [Google Scholar]
- Kabat AM, Pott J, Maloy KJ (2016) The mucosal immune system and its regulation by autophagy. Front Immunol 7: 240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur R, Einarsdottir HK, Vidarsson G (2014) IgG‐effector functions: “the good, the bad and the ugly”. Immunol Lett 160: 139–144 [DOI] [PubMed] [Google Scholar]
- Kapur R, Della Valle L, Verhagen OJ, Hipgrave Ederveen A, Ligthart P, de Haas M, Kumpel B, Wuhrer M, van der Schoot CE, Vidarsson G (2015) Prophylactic anti‐D preparations display variable decreases in Fc‐fucosylation of anti‐D. Transfusion 55: 553–562 [DOI] [PubMed] [Google Scholar]
- Kiouptsi K, Reinhardt C (2018) Contribution of the commensal microbiota to atherosclerosis and arterial thrombosis. Br J Pharmacol 175: 4439–4449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosiewicz MM, Zirnheld AL, Alard P (2011) Gut microbiota, immunity, and disease: a complex relationship. Front Microbiol 2: 180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs CF, Paust HJ, Krohn S, Koyro T, Brix SR, Riedel JH, Bartsch P, Wiech T, Meyer‐Schwesinger C, Huang J et al (2016) Autoimmune renal disease is exacerbated by S1P‐receptor‐1‐dependent intestinal Th17 cell migration to the kidney. Immunity 45: 1078–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Lin RTP, Renia L, Ng LFP (2020) Serological approaches for COVID‐19: epidemiologic perspective on surveillance and control. Front Immunol 11: 879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Aoki M, Illa I, Wu C, Fardeau M, Angelini C, Serrano C, Urtizberea JA, Hentati F, Hamida MB et al (1998) Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nat Genet 20: 31–36 [DOI] [PubMed] [Google Scholar]
- Long QX, Liu BZ, Deng HJ, Wu GC, Deng K, Chen YK, Liao P, Qiu JF, Lin Y, Cai XF et al (2020) Antibody responses to SARS‐CoV‐2 in patients with COVID‐19. Nat Med 26: 845–848 [DOI] [PubMed] [Google Scholar]
- Maguire D, Talwar D, Burns A, Catchpole A, Stefanowicz F, Robson G, Ross DP, Young D, Ireland A, Forrest E et al (2019) A prospective evaluation of thiamine and magnesium status in relation to clinicopathological characteristics and 1‐year mortality in patients with alcohol withdrawal syndrome. J Transl Med 17: 384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuolo J, Carresi C, Gliozzi M, Mollace R, Scarano F, Scicchitano M, Macri R, Nucera S, Bosco F, Oppedisano F et al (2022) The contribution of gut microbiota and endothelial dysfunction in the development of arterial hypertension in animal models and in humans. Int J Mol Sci 23: 3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammen AL, Chung T, Christopher‐Stine L, Rosen P, Rosen A, Doering KR, Casciola‐Rosen LA (2011) Autoantibodies against 3‐hydroxy‐3‐methylglutaryl‐coenzyme A reductase in patients with statin‐associated autoimmune myopathy. Arthritis Rheum 63: 713–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maulucci G, Cohen O, Daniel B, Ferreri C, Sasson S (2019) The combination of whole cell lipidomics analysis and single cell confocal imaging of fluidity and micropolarity provides insight into stress‐induced lipid turnover in subcellular organelles of pancreatic beta cells. Molecules 24: 3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill GF, Kurth EJ, Hardie DG, Winder WW (1997) AICA riboside increases AMP‐activated protein kinase, fatty acid oxidation, and glucose uptake in rat muscle. Am J Physiol 273: E1107–E1112 [DOI] [PubMed] [Google Scholar]
- Mielke GI, Brown WJ, Nunes BP, Silva ICM, Hallal PC (2017) Socioeconomic correlates of sedentary behavior in adolescents: systematic review and meta‐analysis. Sports Med 47: 61–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohassel P, Landon‐Cardinal O, Foley AR, Donkervoort S, Pak KS, Wahl C, Shebert RT, Harper A, Fequiere P, Meriggioli M et al (2019) Anti‐HMGCR myopathy may resemble limb‐girdle muscular dystrophy. Neurol Neuroimmunol Neuroinflamm 6: e523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mule F, Serio R (2001) Increased calcium influx is responsible for the sustained mechanical tone in colon from dystrophic (mdx) mice. Gastroenterology 120: 1430–1437 [DOI] [PubMed] [Google Scholar]
- Mule F, Amato A, Serio R (2010) Gastric emptying, small intestinal transit and fecal output in dystrophic (mdx) mice. J Physiol Sci 60: 75–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HE, Brenner DJ, Fanning GR, Grimont PA, Kampfer P (1996) Emended description of Buttiauxella agrestis with recognition of six new species of Buttiauxella and two new species of Kluyvera: Buttiauxella ferragutiae sp. nov., Buttiauxella gaviniae sp. nov., Buttiauxella brennerae sp. nov., Buttiauxella izardii sp. nov., Buttiauxella noackiae sp. nov., Buttiauxella warmboldiae sp. nov., Kluyvera cochleae sp. nov., and Kluyvera georgiana sp. nov. Int J Syst Bacteriol 46: 50–63 [DOI] [PubMed] [Google Scholar]
- Nugraha B, Buono MF, von Boehmer L, Hoerstrup SP, Emmert MY (2019) Human cardiac organoids for disease modeling. Clin Pharmacol Ther 105: 79–85 [DOI] [PubMed] [Google Scholar]
- Nunes B, Resende ST (2017) Cholinesterase characterization of two autochthonous species of Ria de Aveiro (Diopatra neapolitana and Solen marginatus) and comparison of sensitivities towards a series of common contaminants. Environ Sci Pollut Res Int 24: 12155–12167 [DOI] [PubMed] [Google Scholar]
- Nunes B, Nunes J, Soares A, Figueira E, Freitas R (2017a) Toxicological effects of paracetamol on the clam Ruditapes philippinarum: exposure vs recovery. Aquat Toxicol 192: 198–206 [DOI] [PubMed] [Google Scholar]
- Nunes B, Pinho C, Sousa C, Melo AR, Bandarra N, Silva MC (2017b) Relevance of omega‐3 and omega‐6/omega‐3 ratio in preventing cognitive impairment. Acta Med Port 30: 213–223 [DOI] [PubMed] [Google Scholar]
- Nunes B, Serdoura F, Vital L, Seara M, Pinto R (2017c) Posterior interosseous nerve syndrome after pneumatic hammer use: an uncommon condition. Am J Ind Med 60: 670–672 [DOI] [PubMed] [Google Scholar]
- Nunes B, Silva V, Campos I, Pereira JL, Pereira P, Keizer JJ, Goncalves F, Abrantes N (2017d) Off‐site impacts of wildfires on aquatic systems—biomarker responses of the mosquitofish Gambusia holbrooki . Sci Total Environ 581–582: 305–313 [DOI] [PubMed] [Google Scholar]
- Nunes BG, Loures FV, Bueno HMS, Cangussu EB, Goulart E, Coatti GC, Caldini EG, Condino‐Neto A, Zatz M (2017e) Immunoglobulin therapy ameliorates the phenotype and increases lifespan in the severely affected dystrophin‐utrophin double knockout mice. Eur J Hum Genet 25: 1388–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes BM, Xavier TC, Martins RR (2017f) Antimicrobial drug‐related problems in a neonatal intensive care unit. Rev Bras Ter Intensiva 29: 331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes BP, Chiavegatto Filho ADP, Pati S, Cruz Teixeira DS, Flores TR, Camargo‐Figuera FA, Munhoz TN, Thume E, Facchini LA, Rodrigues Batista SR (2017g) Contextual and individual inequalities of multimorbidity in Brazilian adults: a cross‐sectional national‐based study. BMJ Open 7: e015885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes BP, Soares MU, Wachs LS, Volz PM, Saes MO, Duro SMS, Thume E, Facchini LA (2017h) Hospitalization in older adults: association with multimorbidity, primary health care and private health plan. Rev Saude Publica 51: 43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes JD, Saes MO, Nunes BP, Siqueira FCV, Soares DC, Fassa MEG, Thume E, Facchini LA (2017i) Functional disability indicators and associated factors in the elderly: a population‐based study in Bage, Rio Grande do Sul, Brazil. Epidemiol Serv Saude 26: 295–304 [DOI] [PubMed] [Google Scholar]
- Parchem K, Sasson S, Ferreri C, Bartoszek A (2019) Qualitative analysis of phospholipids and their oxidised derivatives—used techniques and examples of their applications related to lipidomic research and food analysis. Free Radic Res 53: 1068–1100 [DOI] [PubMed] [Google Scholar]
- Parolini D, Meregalli M, Belicchi M, Razini P, Lopa R, Del Carlo B, Farini A, Maciotta S, Bresolin N, Porretti L et al (2009) CD20‐related signaling pathway is differently activated in normal and dystrophic circulating CD133+ stem cells. Cell Mol Life Sci 66: 697–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez EE, Orange JS, Bonilla F, Chinen J, Chinn IK, Dorsey M, El‐Gamal Y, Harville TO, Hossny E, Mazer B et al (2017) Update on the use of immunoglobulin in human disease: a review of evidence. J Allergy Clin Immunol 139: S1–S46 [DOI] [PubMed] [Google Scholar]
- Pezzilli R, Mauloni PA (2019) Acute pancreatitis associated with myotonic dystrophy type I. Pancreas 48: e63–e64 [DOI] [PubMed] [Google Scholar]
- Pinciotti CM, Allen CE, Milliken JM, Orcutt HK, Sasson S (2019) Reliability and findings from an instrument examining sexual assault disclosure content and context: the sexual assault inventory of disclosure. Violence Vict 34: 260–295 [DOI] [PubMed] [Google Scholar]
- Populo H, Nunes B, Sampaio C, Batista R, Pinto MT, Gaspar TB, Miranda‐Alves L, Cai RZ, Zhang XY, Schally AV et al (2017) Inhibitory effects of antagonists of growth hormone‐releasing hormone (GHRH) in thyroid cancer. Horm Cancer 8: 314–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JD, Khanna S, Kaminski HJ, Rao JS, Merriam AP, Richmonds CR, Leahy P, Li J, Guo W, Andrade FH (2002) A chronic inflammatory response dominates the skeletal muscle molecular signature in dystrophin‐deficient mdx mice. Hum Mol Genet 11: 263–272 [DOI] [PubMed] [Google Scholar]
- Prukop T, Wernick S, Boussicault L, Ewers D, Jager K, Adam J, Winter L, Quintes S, Linhoff L, Barrantes‐Freer A et al (2020) Synergistic PXT3003 therapy uncouples neuromuscular function from dysmyelination in male Charcot‐Marie‐Tooth disease type 1A (CMT1A) rats. J Neurosci Res 98: 1933–1952 [DOI] [PubMed] [Google Scholar]
- Recke A, Trog LM, Pas HH, Vorobyev A, Abadpour A, Jonkman MF, van Zandbergen G, Kauderer C, Zillikens D, Vidarsson G et al (2014) Recombinant human IgA1 and IgA2 autoantibodies to type VII collagen induce subepidermal blistering ex vivo . J Immunol 193: 1600–1608 [DOI] [PubMed] [Google Scholar]
- Richner JM, Jagger BW, Shan C, Fontes CR, Dowd KA, Cao B, Himansu S, Caine EA, Nunes BTD, Medeiros DBA et al (2017) Vaccine mediated protection against Zika virus‐induced congenital disease. Cell 170: 273–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rispens T, Davies AM, Ooijevaar‐de Heer P, Absalah S, Bende O, Sutton BJ, Vidarsson G, Aalberse RC (2014) Dynamics of inter‐heavy chain interactions in human immunoglobulin G (IgG) subclasses studied by kinetic fab arm exchange. J Biol Chem 289: 6098–6109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues S, Antunes SC, Nunes B, Correia AT (2017) Histological alterations in gills and liver of rainbow trout (Oncorhynchus mykiss) after exposure to the antibiotic oxytetracycline. Environ Toxicol Pharmacol 53: 164–176 [DOI] [PubMed] [Google Scholar]
- Roquette R, Painho M, Nunes B (2017) Spatial epidemiology of cancer: a review of data sources, methods and risk factors. Geospat Health 12: 504 [DOI] [PubMed] [Google Scholar]
- Santos AJ, Kislaya I, Machado A, Nunes B (2017a) Beliefs and attitudes towards the influenza vaccine in high‐risk individuals. Epidemiol Infect 145: 1786–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AJ, Nunes B, Kislaya I, Gil AP, Ribeiro O (2017b) Exploring the correlates to depression in elder abuse victims: abusive experience or individual characteristics? J Interpers Violence 36: NP115–NP134 [DOI] [PubMed] [Google Scholar]
- Sasson SC, Corbett A, McLachlan AJ, Chen R, Adelstein SA, Riminton S, Limaye S (2019) Enhanced serum immunoglobulin G clearance in myotonic dystrophy‐associated hypogammaglobulinemia: a case series and review of the literature. J Med Case Reports 13: 338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan C, Muruato AE, Jagger BW, Richner J, Nunes BTD, Medeiros DBA, Xie X, Nunes JGC, Morabito KM, Kong WP et al (2017a) A single‐dose live‐attenuated vaccine prevents Zika virus pregnancy transmission and testis damage. Nat Commun 8: 676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan C, Muruato AE, Nunes BTD, Luo H, Xie X, Medeiros DBA, Wakamiya M, Tesh RB, Barrett AD, Wang T et al (2017b) A live‐attenuated Zika virus vaccine candidate induces sterilizing immunity in mouse models. Nat Med 23: 763–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks S, Rakocevic G, Joe G, Manoli I, Shrader J, Harris‐Love M, Sonies B, Ciccone C, Dorward H, Krasnewich D et al (2007) Intravenous immune globulin in hereditary inclusion body myopathy: a pilot study. BMC Neurol 7: 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperduto PW, Fang P, Li J, Breen W, Brown PD, Cagney D, Aizer A, Yu J, Chiang V, Jain S et al (2019) Survival and prognostic factors in patients with gastrointestinal cancers and brain metastases: have we made progress? Transl Res 208: 63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spor A, Koren O, Ley R (2011) Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 9: 279–290 [DOI] [PubMed] [Google Scholar]
- Strati F, Pujolassos M, Burrello C, Giuffre MR, Lattanzi G, Caprioli F, Troisi J, Facciotti F (2021) Antibiotic‐associated dysbiosis affects the ability of the gut microbiota to control intestinal inflammation upon fecal microbiota transplantation in experimental colitis models. Microbiome 9: 39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC (2017) The non‐canonical NF‐kappaB pathway in immunity and inflammation. Nat Rev Immunol 17: 545–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzumura A, Yamada H, Matsuoka Y, Sobue I (1986) Immunoglobulin abnormalities in patients with myotonic dystrophy. Acta Neurol Scand 74: 132–139 [DOI] [PubMed] [Google Scholar]
- Ticinesi A, Milani C, Lauretani F, Nouvenne A, Mancabelli L, Lugli GA, Turroni F, Duranti S, Mangifesta M, Viappiani A et al (2017) Gut microbiota composition is associated with polypharmacy in elderly hospitalized patients. Sci Rep 7: 11102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidball JG (2017) Regulation of muscle growth and regeneration by the immune system. Nat Rev Immunol 17: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidball JG, Welc SS, Wehling‐Henricks M (2018) Immunobiology of inherited muscular dystrophies. Compr Physiol 8: 1313–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi E, Cesar MA, Neves RG, Schmidt PR, Thume E, da Silveira DS, Siqueira FC, Nunes BP, Fassa AG, Saes MO et al (2017a) Diabetes care in Brazil: program to improve primary care access and quality‐PMAQ. J Ambul Care Manage 40: S12–S23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi E, Fernandes PA, Fischer T, Siqueira FC, Silveira DS, Thume E, Duro SM, Saes MO, Nunes BP, Fassa AG et al (2017b) Quality of prenatal services in primary healthcare in Brazil: indicators and social inequalities. Cad Saude Publica 33: e00195815 [DOI] [PubMed] [Google Scholar]
- Tonegawa S (1983) Somatic generation of antibody diversity. Nature 302: 575–581 [DOI] [PubMed] [Google Scholar]
- Turner C, Hilton‐Jones D (2010) The myotonic dystrophies: diagnosis and management. J Neurol Neurosurg Psychiatry 81: 358–367 [DOI] [PubMed] [Google Scholar]
- van Bladel ER, Laarhoven AG, van der Heijden LB, Heitink‐Polle KM, Porcelijn L, van der Schoot CE, de Haas M, Roest M, Vidarsson G, de Groot PG et al (2014) Functional platelet defects in children with severe chronic ITP as tested with 2 novel assays applicable for low platelet counts. Blood 123: 1556–1563 [DOI] [PubMed] [Google Scholar]
- Vidarsson G, Dekkers G, Rispens T (2014) IgG subclasses and allotypes: from structure to effector functions. Front Immunol 5: 520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther LE, Winnefeld K, Solch O (2000) Determination of iron, copper, zinc, magnesium and selenium in plasma and erythrocytes in neurosurgical patients. J Trace Elem Med Biol 14: 92–95 [DOI] [PubMed] [Google Scholar]
- Wang T, Zhu H, Hou Y, Gu W, Wu H, Luan Y, Xiao C, Zhou C (2019) Galantamine reversed early postoperative cognitive deficit via alleviating inflammation and enhancing synaptic transmission in mouse hippocampus. Eur J Pharmacol 846: 63–72 [DOI] [PubMed] [Google Scholar]
- Weigmann B, Tubbe I, Seidel D, Nicolaev A, Becker C, Neurath MF (2007) Isolation and subsequent analysis of murine lamina propria mononuclear cells from colonic tissue. Nat Protoc 2: 2307–2311 [DOI] [PubMed] [Google Scholar]
- Wenninger S, Montagnese F, Schoser B (2018) Core clinical phenotypes in myotonic dystrophies. Front Neurol 9: 303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YF, Huang WH, Gu MH, Lin SR, Chu SC, Wang TF, Li CC (2019) Higher CD56+ or CD2+ lymphocyte percentage predicts poor steroid response in patients with immune thrombocytopenia. Thromb Res 183: 63–68 [DOI] [PubMed] [Google Scholar]
- Xepapadaki E, Maulucci G, Constantinou C, Karavia EA, Zvintzou E, Daniel B, Sasson S, Kypreos KE (2019) Impact of apolipoprotein A1‐ or lecithin:cholesterol acyltransferase‐deficiency on white adipose tissue metabolic activity and glucose homeostasis in mice. Biochim Biophys Acta Mol Basis Dis 1865: 1351–1360 [DOI] [PubMed] [Google Scholar]
- Xie X, Yang Y, Muruato AE, Zou J, Shan C, Nunes BT, Medeiros DB, Vasconcelos PF, Weaver SC, Rossi SL et al (2017) Understanding Zika virus stability and developing a chimeric vaccine through functional analysis. MBio 8: e02134‐16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Diao H, Xiao Y, Li W, Yu B, He J, Yu J, Zheng P, Mao X, Luo Y et al (2016) Gut microbiota can transfer fiber characteristics and lipid metabolic profiles of skeletal muscle from pigs to germ‐free mice. Sci Rep 6: 31786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LV, Radu CG, Wang L, Riedinger M, Witte ON (2005) Gi‐independent macrophage chemotaxis to lysophosphatidylcholine via the immunoregulatory GPCR G2A. Blood 105: 1127–1134 [DOI] [PubMed] [Google Scholar]
- Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, Wu Y, Zhang L, Yu Z, Fang M et al (2020) Clinical course and outcomes of critically ill patients with SARS‐CoV‐2 pneumonia in Wuhan, China: a single‐centered, retrospective, observational study. Lancet Respir Med 8: 475–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zschuntzsch J, Zhang Y, Klinker F, Makosch G, Klinge L, Malzahn D, Brinkmeier H, Liebetanz D, Schmidt J (2016) Treatment with human immunoglobulin G improves the early disease course in a mouse model of Duchenne muscular dystrophy. J Neurochem 136: 351–362 [DOI] [PubMed] [Google Scholar]
- Zuppinger C (2019) 3D cardiac cell culture: a critical review of current technologies and applications. Front Cardiovasc Med 6: 87 [DOI] [PMC free article] [PubMed] [Google Scholar]


