Abstract
Srg3 (SWI3-related gene product) is a mouse homolog of yeast SWI3, Drosophila melanogaster MOIRA (also named MOR/BAP155), and human BAF155 and is known as a core subunit of SWI/SNF complex. This complex is involved in the chromatin remodeling required for the regulation of transcriptional processes associated with development, cellular differentiation, and proliferation. We generated mice with a null mutation in the Srg3 locus to examine its function in vivo. Homozygous mutants develop in the early implantation stage but undergo rapid degeneration thereafter. An in vitro outgrowth study revealed that mutant blastocysts hatch, adhere, and form a layer of trophoblast giant cells, but the inner cell mass degenerates after prolonged culture. Interestingly, about 20% of heterozygous mutant embryos display defects in brain development with abnormal organization of the brain, a condition known as exencephaly. Histological examination suggests that exencephaly is caused by the failure in neural fold elevation, resulting in severe brain malformation. Our findings demonstrate that Srg3 is essential for early embryogenesis and plays an important role in the brain development of mice.
Modification of the nucleosome structure is a fundamental regulatory process during development. Biochemical and genetic studies have isolated and characterized numerous chromatin-remodeling complexes involved in transcription regulation by modifying histones or altering chromatin structure (1, 30, 33, 51, 57). These complexes can be classified into two major groups, which differ in their use of covalent modification to alter chromatin structure. The first class contains the histone acetyltransferase and histone deacetylase complexes. These complexes regulate the transcriptional activity of genes by determining the level of acetylation of amino-terminal domains of nucleosomal histones which are associated with them. Increased acetylation is usually associated with activation of gene expression, whereas decreased acetylation is associated with repression of gene expression (26, 56). The second class consists of ATP-dependent chromatin-remodeling complexes, which use the energy of ATP hydrolysis to locally disrupt or alter the association of histones with DNA. These complexes contain either SWI2/SNF2 or ISWI-related ATPase associated with various subunits and play roles in both gene activation and repression (30, 57).
The yeast SWI/SNF (ySWI/SNF) was originally identified in Saccharomyces cerevisiae. It consists of 11 subunits with a total molecular mass of 2 MDa including SWI2/SNF2 ATPase. Several components have been identified by screening genes involved in the regulation of mating type switching and sucrose-fermenting ability (25, 40). Subsequently, ySWI/SNF genes were shown to be involved in the transcriptional regulation of a wider subset of yeast genes (23). Mutations in both SWI and SNF genes cause pleiotropic phenotypes such as a slow-growth phenotype, defects in mating type switching and sporulation, and inability to utilize sucrose as a carbon source (38, 60). ySWI/SNF has been shown to be highly conserved in all eukaryotes (7, 10, 39, 53). A highly related yeast complex called RSC consists of at least 15 subunits and appears to have a role different from that of SWI/SNF. RSC mutants do not display SWI/SNF transcriptional defects and some, unlike SWI/SNF mutants, are lethal (8).
Homologs of SWI/SNF proteins were identified in Drosophila melanogaster (13, 37). The Drosophila SWI/SNF complex contains eight major proteins, including the ATPase subunit Brahma (BRM), which is essential for oogenesis and embryogenesis. BRM also plays a particularly important role in the maintenance of homeotic gene expression as a member of the trithorax group (4, 55). BRM complex subunits BAP45/SNR1, BAP155/MOIRA, and BAP60 are conserved between yeast and mammals. MOIRA is a homolog of yeast SWI3 (12, 37). This gene was isolated in three independent screenings for loci that undergo dosage-dependent interactions with Polycomb or ectopically expressed Antennapedia (29). Mutations in MOIRA produce many of the genetic and phenotypic characteristics of BRM mutants (5, 15, 16, 54).
The mammalian SWI/SNF complexes consist of 9 to 12 subunits, with those from different tissues showing significant heterogeneity. Subunit diversity of mammalian SWI/SNF suggests that different complexes might have tissue-specific roles during development (58, 59). The complexes fall into two broad classes, depending on whether they contain human BRM (hBRM) or BRG1 as the ATPase. They contain a core set of components, including the DNA-dependent ATPase SWI2/SNF2, SNF5, and SWI3 homologs (30). A minimum-catalytic-core complex of three SWI/SNF components, BRG1 or hBRM, INI1, and BAF155/BAF170, can remodel both mononucleosome and nucleosome arrays (41). In addition, BRG1 or hBRM alone can substitute for the core complex, albeit with less efficiency. Recent studies of targeted mutations of BRM, BRG1, and SNF5/INI1 in the mouse have expanded the understanding of in vivo functions of the mammalian SWI/SNF complex (6, 17, 31, 43, 44). While disruption of mouse BRM (Brm) produced only mild proliferative effects, deficiency of mouse BRG1 (Brg1) or mouse SNF5/INI1 (Snf5/Ini1) resulted in peri-implantation death and predisposition of heterozygotes to exencephaly (Brg1+/−) and tumor formation (Brg1+/− or Snf5/Ini1+/−), particularly in the nervous system.
The gene encoding Srg3, a mouse counterpart of yeast SWI3, Drosophila MOIRA/SWI3D, and human BAF155, was initially isolated as a gene expressed highly in the thymus but at a low level in the periphery by subtractive hybridization (27). Srg3 is a core component of the SWI/SNF complex in mice, as supported by previous studies with its homologs (37, 41). Interestingly, the expression of antisense RNA to Srg3 in a thymoma cell line decreased the apoptosis induced by glucocorticoids (GCs), suggesting that this molecule is involved in the GC-induced apoptosis during T-cell development (27). In the present study, we show that Srg3 is widely expressed during mouse embryogenesis in a spatiotemporal pattern that generally overlaps with that of Brg1. Deficiency in Srg3 expression resulted in early embryonic lethality soon after decidualization by defects in the inner cell mass (ICM) and the primitive endoderm. Similar to BRG1 knockout mice, Srg3 heterozygotes are predisposed to exencephaly, suggesting that the SWI/SNF complex plays an important role in brain development.
MATERIALS AND METHODS
Western blotting.
Western blotting was carried out as previously described (27). Briefly, whole-cell extracts were separated on sodium dodecyl sulfate (SDS)–7.5% polyacrylamide gel and transferred to nitrocellulose. The membrane was then blocked with Tris-buffered saline–Tween-20 containing 5% nonfat dried milk for 1 to 2 h and incubated with antiserum against Srg3 or BRG1, which also has been described previously (27), at room temperature for 2 h. Enhanced chemiluminescence reagents (Amersham Pharmacia) were used for detection.
Targeted disruption of Srg3 and genotyping.
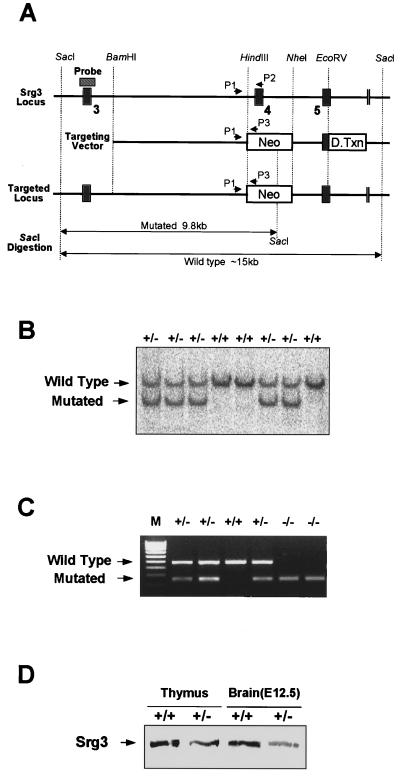
The Srg3 genomic DNA clone was obtained from a mouse 129/Sv genomic library (Stratagene) using a 5′ 0.6-kb XhoI-HindIII fragment of cDNA as a probe. The targeting vector was constructed by cloning a 5.4-kb BamHI-HindIII genomic DNA fragment encompassing sequences upstream of exon 4 as the long arm and a 1.4-kb NheI-EcoRV fragment including portions of intron 4 (intron between exons 4 and 5) and exon 5 as the short arm into a neo expression cassette (PGKneolox2DTA) (49). This vector contains the diphtheria toxin gene driven by the Pgk promoter for negative selection. The construct was linearized by XhoI digestion and electroporated into 129/Sv-derived AK7 embryonic stem (ES) cells (49). ES cell colonies were selected with G418, and correctly targeted clones were identified by PCR using primers corresponding to the Neo gene (5Neo; 5′-TCGCAGCGCATCGCCTTCTA-3′) and a genomic sequence outside of the targeting construct (3XB; 5′-ATCGTGTCTATTACCCTGATGC-3′). Seventeen PCR-positive clones were obtained from screening 750 G418-resistant clones. Clones identified as homologous recombinants were confirmed by Southern blot analysis using a 5′ external genomic probe (see Fig. 3A and B). Eight out of 17 positive clones were used for microinjection into the C57BL/6J host blastocysts, and all clones gave rise to chimeric mice. To establish heterozygous lines, chimeric males were mated with C57BL/6 females. Germ line transmission of the mutated allele was verified by Southern blotting and three-primer PCR analysis of tail DNA from agouti coat colored F1 offspring using common 5′ primer P1 (5′-ACAACGAAATCTGTGGAGTAGC-3′), in combination with Srg3-specific 3′ primer P2 (5′-GGGATGGGTTCTGAAGATCA-3′) and Neo-specific 3′ primer P3 (5′-CTAAAGCGCATGCTCCAGAC-3′) (see Fig. 3A and C). Primers P1 and P2 amplify a wild-type 450-bp fragment, whereas P1 and P3 amplify a 250-bp fragment specific for the mutant allele. For PCR-based genotyping, embryonic day 3.5 (E3.5) to E8.5 embryos and small pieces of E9.5 to E18.5 embryos underwent lysis by boiling in 10 μl of lysis buffer containing 0.035 N NaOH and 0.05% SDS.
FIG. 3.
Targeting of the Srg3 gene. (A) Schematic diagram of the Srg3 locus and targeting vector and predicted structure of targeted Srg3 allele. Restriction enzyme sites and three exons (3 to 5; black boxes) are shown. The locations of the 5′ flanking Southern probe (hatched box) and PCR primers (P1 to P3) used in panels B and C are indicated. The targeting construct was designed to replace an exon (exon 4) with a PGK-Neo gene. Neo, neomycin resistance gene; D.Txn, diphtheria toxin gene. (B) Southern blot analysis showing correct targeting of the Srg3 locus. Genomic DNA samples were prepared from tails of the progeny derived from Srg3 heterozygous intercrosses, digested with SacI, and probed as indicated. The resulting 15- and 9.8-kb bands correspond to the wild-type and mutated genotypes, respectively. (C) Three-primer PCR analysis of blastocysts from Srg3 heterozygous intercrosses showing wild-type (450-bp) and mutated (250-bp) alleles. M, 100-bp marker. (D) Western blot analysis of Srg3 and Brg1 expression in the thymus (4 to 6 weeks old) and brain at E12.5 from wild-type littermate and Srg3 heterozygous mice.
Blastocyst culture and confocal microscope analysis.
E3.5 blastocysts generated from Srg3 heterozygous intercrosses were collected by uterine flush and individually cultured in Dulbecco's modified Eagle medium supplemented with 15% fetal bovine serum (FBS) in 5% CO2 at 37°C. For confocal microscope analysis, freshly isolated blastocysts and blastocysts after 3 and 5 days of culture were stained with goat anti-BAF155 (Santa Cruz Biotechnology; sc-9747) and rabbit anti-BRG1 (previously described [27]) antibodies. Fluorescein isothiocyanate-conjugated anti-goat immunoglobulin (IgG) and tetramethyl rhodamine isocyanate-conjugated anti-rabbit IgG secondary antibodies were purchased from Jackson Laboratory. Embryos were then collected and genotyped by three-primer PCR as described above.
Histological analysis and in situ hybridization.
For paraffin sections, embryos were isolated and fixed in Bouin's solution (Sigma) for 2 to 24 h at room temperature, dehydrated in an ethanol series, cleared in xylene, and embedded in paraffin. Sections were cut 6 to 7 μm thick and processed for immunohistochemistry or BF-1 in situ hybridization. In situ hybridization was carried out as previously described (19, 63). In brief, deparaffinated and rehydrated sections were hybridized at 72°C in a moisture chamber with a digoxigenin-labeled BF-1-specific riboprobe. Following hybridization, a high-stringency wash was carried out in 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate)–50% formamide at 72°C for 1 h. BF-1 expression was detected with alkaline phosphatase-coupled antidigoxigenin antibodies (Boehringer Mannheim) followed by a reaction with nitroblue tetrazolium and BCIP (5-bromo-4-chloro-3-indolylphosphate). Frozen sections were used for the detection of Srg3 and Brg1 expression in mouse embryogenesis. To avoid possible redundancy with the mouse BAF170 transcript, which is related to Srg3/BAF155 but encoded by a different gene (59), 0.5-kb XbaI/PstI cDNA (nucleotides 2826 to 3309), showing specificity in a BLASTn search, was used as an Srg3 antisense probe. The Brg1 probe was prepared as previously described (42).
BrdU labeling and immunohistochemistry.
Bromodeoxyuridine (BrdU) (50 μg/g of body weight) was injected intraperitoneally into pregnant females at E13.5. The mice were sacrificed 2 h after injection, and, for paraffin sections, dissected embryos were fixed in Bouin's solution and processed as described above. The detection of BrdU incorporation was performed in accordance with the manufacturer's instructions (cell proliferation kit; Boehringer Mannheim). For microtubule-associated protein 2 (MAP2) staining, a mouse anti-MAP2 antibody (clone HM-2; Sigma) was used, and the detection was carried out using the LSAB kit (DAKO).
RESULTS
Expression of Srg3 during mouse development.
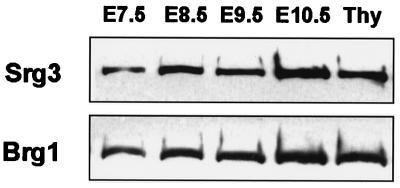
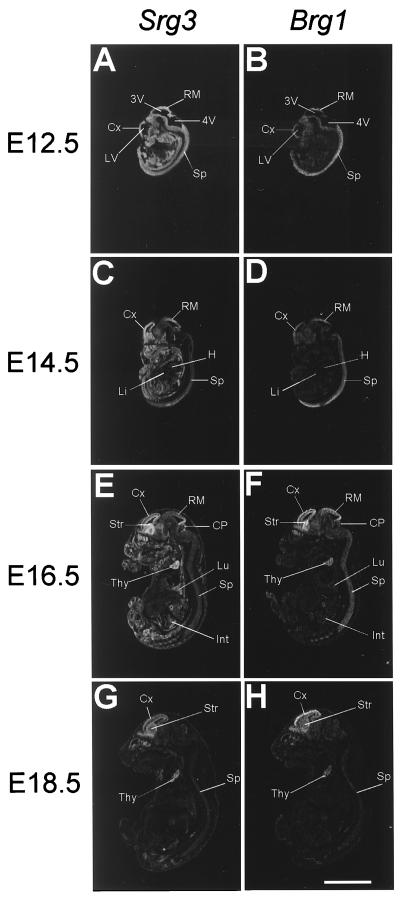
To examine the expression pattern of Srg3 during mouse development, Western blotting and in situ hybridization experiments were performed. In the embryonic stages, Brg1 is known to be dominantly expressed about 20- to 30-fold more than Brm (43). Thus, we also examined the expression of Brg1 to address where the SWI/SNF complex is expressed during development. Embryos from E7.5 to E10.5 developmental stages were dissected at the proper times, and whole embryonic extracts were prepared and subjected to Western blotting. Thymocyte extract was used as a quantitative control simply because the Srg3 protein is most highly expressed in the thymus in adult mice (27). As shown in Fig. 1, Srg3 is expressed constitutively at a high level in embryonic stages from E7.5 to E10.5, similar to Brg1 (43). To determine the spatiotemporal pattern of Srg3 expression and to compare the relative localization of expression with Brg1 expression in mid- and late-embryonic stages from E12.5 to E18.5, in situ hybridization was performed. Frozen sagittal sections of E12.5, E14.5, E16.5, and E18.5 mouse embryos were probed with Srg3 or Brg1 cDNA fragments. The expression pattern of Srg3 overlaps with that of Brg1, which is highly expressed in the spinal cord, brain, and thymus as previously reported (42), and Srg3 appears more ubiquitously than Brg1 (Fig. 2). At E12.5 and E14.5 (Fig. 2A and C), high expression of Srg3 is apparent in almost all developing organs except the heart and liver. At E16.5, high expression of Srg3 is evident in the lung and intestine as well as the central nervous system (CNS) and thymus (Fig. 2E), where Brg1 is also highly expressed (Fig. 2F). This overall expression pattern of Srg3 is shown to be altered as the embryos grow to near birth in such a way that the level of expression gradually diminishes. At E18.5, Srg3 expression appears to be restricted mostly to the CNS and thymus, where Brg1 is also expressed (Fig. 2G and H, respectively).
FIG. 1.
Expression of Srg3 and Brg1 proteins during mouse early embryogenesis. Embryos were dissected at the indicated times (E7.5 to E10.5). To estimate the amount of Srg3 and Brg1, thymocyte extract (Thy) was used as a control. Thirty micrograms of total extract was subjected to SDS-polyacrylamide gel electrophoresis and analyzed by Western blotting. Antisera against Srg3 and BRG1 were used as described in Materials and Methods.
FIG. 2.
In situ hybridization analysis of Srg3 (A, C, E, and G) and Brg1 (B, D, F, and H) expression at E12.5 (A and B), E14.5 (C and D), E16.5 (E and F), and E18.5 (G and H). Sagittal sections of indicated embryos were hybridized with Srg3- or Brg1-specific probes. To avoid possible redundant signals with the mouse BAF170 transcript, the Srg3 antisense probe was made of the nonredundant region (see Materials and Methods). A sense strand probe was also made, and no signal was detected (data not shown). Cx, cerebral cortex; LV, lateral ventricle; RM, roof of midbrain; 3V, third ventricle; 4V, fourth ventricle; Sp, spinal cord; H, heart; Li, liver; Lu, lung; Int, intestine; CP, cerebellar primordium; Str, striatum; Thy, thymus. Scale bar, 5 mm.
Targeted disruption of Srg3.
To investigate the role of the Srg3 protein in vivo, we created an Srg3 null mutation via homologous recombination in mouse ES cells. A targeting vector that replaces a 0.9-kb HindIII-NheI fragment including exon 4, which encodes amino acid residues 131 to 158, with the neomycin resistance gene (Neo) was used (Fig. 3A). Correct homologous recombination in ES cells was confirmed by Southern blot analysis (Fig. 3B). Since remaining exons 3 and 5 are out of frame, this deletion is expected to result in a null allele. Eight different Srg3+/− ES cell clones were independently injected into blastocysts and gave rise to germ line-transmitting chimeric mice that were crossed into a C57BL/6 background. The resulting Srg3+/− mice appeared normal and fertile. Genotyping progeny from heterozygous intercrosses was performed by Southern blotting; however, we could not find any homozygous mutants in postnatal progeny. Homozygous embryos were found at the blastocyst stage by three-primer PCR analysis (Fig. 3C). Immunohistochemical analysis of blastocysts confirmed the absence of protein in homozygous mutants (Fig. 4A). The expression of the Srg3 protein in heterozygous mice was reduced to about one-half the level in control mice in the thymus and developing brain (Fig. 3D), where the Srg3 protein is highly expressed (27). Thymocytes expressing reduced levels of Srg3 displayed reduced sensitivity to GC-induced apoptosis (data not shown).
FIG. 4.
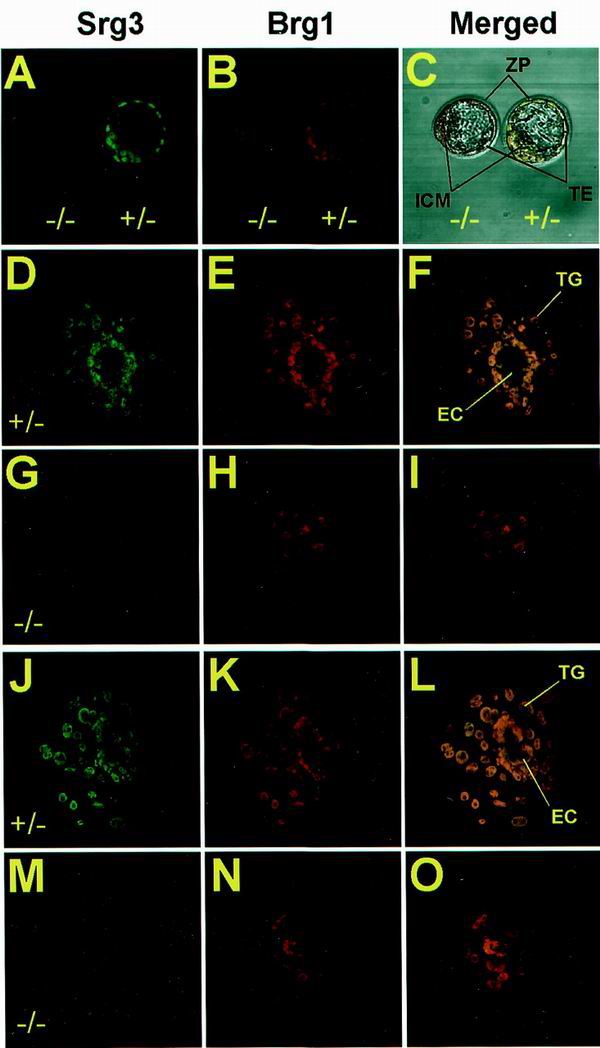
In vitro outgrowth defects of Srg3−/− blastocysts. Freshly isolated blastocysts at E3.5 from Srg3 heterozygous intercross were directly stained (A to C) or cultured for 3 (D to I) and 5 (J to O) days in Dulbecco's modified Eagle medium supplemented with 15% FBS followed by staining with anti-Srg3 and anti-BRG1 antibodies. Confocal images of each stage show Srg3 (A, D, G, J, and M) and Brg1 (B, E, H, K, and N) expression patterns. Merged images are also provided (C, F, I, L, and O). In the blastocyst stage, Srg3 null mutants appeared morphologically normal (C); however, after culture for 3 (D to I) and 5 (J to O) days, they failed to form the three-dimensional egg cylinder structure enclosed by the primitive endoderm (I and O). ZP, zona pellucida; TE, trophoectoderm; TG, trophoblast giant cells; EC, egg cylinder.
Early embryonic lethality of homozygous mutant.
Heterozygous mice were intercrossed in an effort to generate homozygous mutant progeny. However, no homozygous mutants were identified in the resulting progeny at times ranging from E7.5 to the postnatal stage, and about 26 to 27% (95 of 359) of the decidua from E7.5 to E18.5 were empty or resorbed (Table 1). Histological analysis of E5.5 and E6.5 decidua from heterozygous intercrosses showed that about 28% (11 of 39) of them were empty or seemed to contain traces of degenerated embryos mixed with maternal blood cells (data not shown). These empty decidua are presumed to be Srg3 homozygous mutant embryos if one assumes a normal Mendelian distribution of genotypes in the F1 generation. To examine this assumption, the homozygous mutants recovered at the blastocyst stage (E3.5) were cultured in vitro. Very recently, it was reported that Brg1 null mutant dies during the peri-implantation stage, and in vitro blastocyst outgrowth studies revealed that neither the ICM nor the trophoectoderm survives (6). Because Srg3 is a core component of the SWI/SNF complex, it is presumed that Srg3 null mutants die from the lack of the activity of the complex. Freshly isolated blastocysts were stained by anti-BAF155, which is also reactive to Srg3, and anti-BRG1 antibodies. As shown in Fig. 4A to C, the Srg3-deficient blastocyst appears morphologically normal. The Brg1 protein in the homozygous mutant was stained almost at the same level as that of the heterozygous control, indicating that the lack of Srg3 does not greatly affect the Brg1 protein level. To pursue the developmental defect(s) of the Srg3 homozygous mutant, we cultured blastocysts from heterozygous intercrosses in individual microdrops of medium containing 15% FBS. After 3 to 5 days of culture, wild-type and heterozygous mutant blastocysts hatched from the zona, adhered, and developed into a giant trophoblast layer around the incipient egg cylinder, which consists of the developing ICM and primitive endoderm cells (Fig. 4D to F and J to L). However, the Srg3-deficient embryo failed to form the egg cylinder structure and had a few dispersed ICMs and no discernible visceral endoderm layer (Fig. 4G to I and M to O). Stable Brg1 expression was also detected in cultured embryos (Fig. 4H and N). Compared to the lethality of the Brg1 null mutant, it is notable that Srg3-deficient embryos undergo hatching and partial outgrowth with normal trophoblast giant cell differentiation ex vivo.
TABLE 1.
Summary of genotypes resulting from Srg3+/− intercrosses
| Stage | No. of mice with genotype:
|
Total | |||
|---|---|---|---|---|---|
| +/+ | +/− | −/− | NDa | ||
| Postnatal | 131 | 185 | 0 | 316 | |
| E9.5–E18.5 | 61 | 126 (24b) | 0 | 71c | 258 |
| E7.5–E8.5 | 26 | 51 | 0 | 24d | 101 |
| E3.5 | 22 | 45 | 20 | 3e | 90 |
ND, not determined.
Number of embryos with neural tube defects resulting in exencephaly.
Resorption.
Empty decidua.
Genotyping failure.
Exencephaly in a portion of heterozygous mutants.
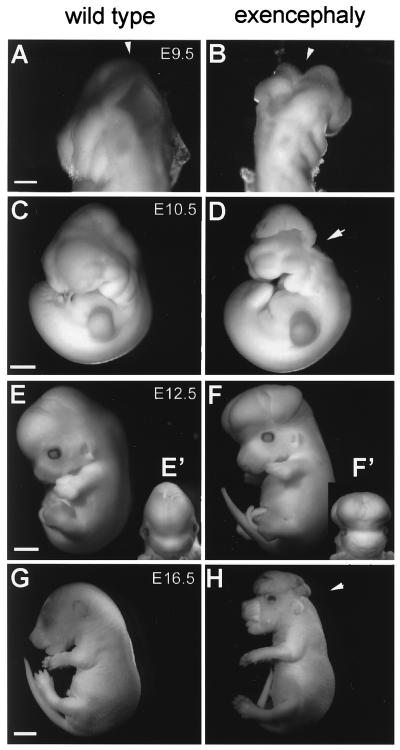
Interestingly, about 20% (24 of 126; Table 1) of Srg3 heterozygous embryos exhibit exencephaly. It is well known that almost all exencephaly of genetic origin is caused by neural tube defects (NTDs), causing failure in neural fold elevation followed by outward expansion of neural tissue via the eversion of the neural plate (18). Normally, the neural tube begins to close at E8.5 and completes closure by E9.5 in mice. Gross morphological analysis at E9.5 revealed that some heterozygous embryos display the failure of neural fold elevation (Fig. 5A and B). Subsequently, a severe abnormal brain structure with a failure of neural plate closing in the mid-hind cephalic region is apparent at E10.5 (Fig. 5C and D). Further analyses of the brain structure at E12.5 and E16.5 revealed severe gross perturbations of the whole cephalic structure including extensive malformation of the forebrain (Fig. 5E to H).
FIG. 5.
Gross appearance of wild-type littermates (A, C, E, and G) and exencephalic Srg3+/− (B, D, F, and H) mice. (A and B) Dorsal view of E9.5 embryos. In this stage, wild-type embryos show normally closed neural tubes (A, arrowhead). In contrast, the exencephalic embryo shows an open neural tube (B, arrowhead). (C and D) Lateral view of E10.5 embryos. Note the defects in the midbrain-hindbrain junction region (D, arrowhead). (E and F) E12.5, lateral view. (E′ and F′) Frontal view. Exencephalic embryos show laterally swollen brains (F′). (G and H) E16.5, lateral view. Note the typical exencephaly without the skull (H, arrowhead). Scale bars, 0.2 (A and B); 0.4 (C and D), 1 (E and F), and 2 mm (G and H).
To characterize this brain abnormality at the histological level, comparable coronal sections through the forebrains of wild-type littermate and exencephalic embryos at E12.5 were stained with hematoxylin and eosin (Fig. 6A and B). In the exencephalic embryo, the third ventricle was clearly open (Fig. 6B) and the diencephalon had burst out to the upper side of the head (Fig. 6A and B), accompanied by the lateral ventricles turned inside out (Fig. 6B). Furthermore, the stenosis of the lateral ventricles in the exencephalic mutant was evident, as well as prominent expansion of the corpus striatum mediale (Fig. 6B and D). To clarify the identity of the telencephalic region in exencephaly, BF-1 (brain factor-1), in situ hybridization was performed. BF-1 is a molecular marker gene whose expression is restricted within the developing basal telencephalon and cerebral cortex (19, 24). In situ hybridization with the BF-1 gene of exencephalic embryos showed that the telencephalic neuroepithelia were located below the thalamus (Fig. 6D). Additionally, the ganglionic eminence of the exencephalic embryo exhibited a rotation toward the midline axis of the brain, so that the eminence region faces outward from rather than inward to the brain. These observations suggest the possibility that Srg3 plays an important role during brain development, probably in a dosage-dependent manner. High expression of the Srg3 protein during E8.5 to E9.5 (Fig. 1), when the neural tube closes, and prolonged constitutively high expression in the CNS region thereafter (Fig. 2) are compatible with this possibility. Recent studies revealed that BRG1 and BAF155 interact with the retinoblastoma suppressor gene product (Rb) and cyclin E, indicating that they participate in the control of the cell cycle (47, 50, 64). Therefore, we investigated the proliferation of the neuroepithelial cells in exencephalic embryos at E13.5 by BrdU labeling. At this stage, developing neuroepithelial cells proliferate within the ventricular zones and subsequently migrate out of this zone to become fully differentiated mature neurons expressing neuronal marker MAP2 (11). Control littermate embryos showed normal neuronal proliferation and differentiation (Fig. 7A, C, E, and F). However, in exencephalic embryos, expression of MAP2 was shown to be relatively reduced in the telencephalic striatum region (Fig. 7B and D). In addition, ectopic proliferating cells were found in the upper posterior region (Fig. 7B, D, G, and H). These cells appear to be neuronal epithelial cells producing an excessive cell mass in the diencephalic region. It was also found that the mesenchymal tissues of exencephalic embryos were obviously expanded (Fig. 7D). Our observations suggest that the haploinsufficiency of Srg3 results in a susceptibility to brain malformation accompanied by inappropriate cellular proliferation and differentiation.
FIG. 6.
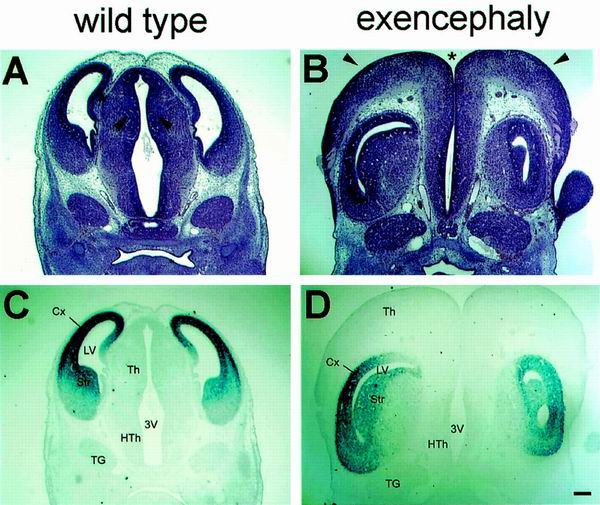
Histological analysis and BF-1 in situ hybridization of exencephalic Srg3+/− embryos at E12.5. (A and B) Hematoxylin- and eosin-stained coronal brain sections of wild-type littermate and exencephalic embryos. Note the open third ventricle (asterisk), expansion of the thalamic cells (arrowheads) covering the upper side of the head, and shrunken lateral ventricles in the exencephalic embryo (B). (C and D) BF-1 in situ hybridization of sections adjacent to those shown in panels A and B, respectively. Note that the cerebral cortex is turned upside down. The cerebral cortex appears to be pushed by the enlarged thalamus bursting out to the upper side of the head, with an open third ventricle in the exencephalic embryo. Cx, cerebral cortex; LV, lateral ventricle; Th, thalamus; HTh, hypothalamus; 3V, third ventricle; Str, striatum; TG, trigeminal ganglion. Scale bar, 0.2 mm.
FIG. 7.
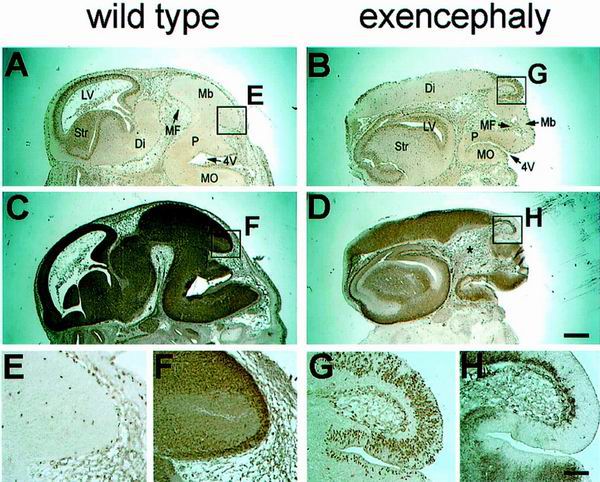
Neuronal cell proliferation and differentiation of wild-type littermate and exencephalic embryos at E13.5. Sagittal sections are shown. (A and B) BrdU incorporation of the littermate control and exencephalic embryo, respectively, after a 2-h pulse. (C and D) MAP2 staining of sections adjacent to those shown in panels A and B, respectively, showing differentiation of neuronal cells. Note the ectopic proliferating cells in the upper posterior region (box G in panel B). These cells are thought to be the diencephalon-producing neuroblasts. Note also the irregular position of midbrain flexure (MF), open fourth ventricle (4V), and enlarged head mesenchyme (D, asterisk) in the exencephalic embryo (B and D). (E and F) High-power views of the upper posterior regions of panels A and C, respectively, showing the posterior part of midbrain in the control embryo. (G and H) High-power views of the upper posterior regions of panels B and D, respectively, showing the ectopic proliferating cells in the exencephalic embryo. LV, lateral ventricle; Str, striatum; Di, diencephalon; MF, midbrain flexure; Mb, midbrain; P, pons; 4V, fourth ventricle; MO, medulla oblongata. Scale bars, 0.5 (A and D) and 0.1 mm (E and H).
DISCUSSION
Here we report the first study addressing the in vivo role of Srg3, a mouse counterpart of yeast SWI3, Drosophila MOIRA, and human BAF155. Srg3 is expressed ubiquitously in postimplantation embryos with particularly high levels of expression in the CNS and thymus. Srg3 deficiency results in peri-implantation lethality, resulting from a defective development of the ICM and primitive endoderm, as suggested by blastocyst outgrowth studies. Similar to Brg1-deficient mice, heterozygotes are predisposed to exencephaly (6). Histological analysis of brains of exencephalic embryos reveals defects in proliferation and differentiation of neural cells, implying that the CNS is sensitive to Srg3 dosage. Previous studies reported that both Brg1 and Snf5/Ini1 heterozygous mice were susceptible to spontaneous neoplasia (6, 17, 31, 44). However, we have not yet looked for such a phenotype although it might be expected in light of the Brg1 and Snf5/Ini1 knockout results.
The similar expression patterns of Srg3 and Brg1 and the resemblance of the phenotypes of the knockout mice strongly suggest that Srg3, as previously shown for Brg1 and Snf5/Ini1, is required for full activity of mammalian SWI/SNF complexes in vivo (6, 17, 31, 44). Further, the present study confirms the essential function of Srg3/SWI/SNF in peri-implantation development and control of neural cell differentiation and proliferation.
Comparison of the Srg3 expression pattern with that of Brg1 in embryogenesis.
The expression pattern of Srg3 largely overlaps with that of Brg1 but appeared to be more ubiquitous. From E7.5 to E10.5, both Srg3 and Brg1 were expressed at constitutively high levels. During this period, embryos undergo gastrulation, neurulation, and early organogenesis, in which exclusive cell proliferation and differentiation occur. High Srg3 and Brg1 expression is consistent with the presumptive need for the SWI/SNF complex to regulate the expression of various genes by remodeling the chromatin structure. In situ hybridization studies using middle- and late-stage embryos (Fig. 2) revealed widespread expression of Srg3 mRNA in developing embryonic tissues. The expression patterns of Srg3 and Brg1 appeared to be similar but somewhat different during mouse embryogenesis. Whereas Brg1 is highly expressed in some restricted organs such as the brain, spinal cord, and thymus, Srg3 shows a pattern of high expression additionally in the ventral parts including the lungs and intestines of the developing embryos (Fig. 2). Interestingly, this overall high expression pattern gradually weakened as the embryo neared birth. These observations suggest that there may be an unknown function of Srg3 independent of Brg1 in mouse development. Previously, it was reported that Brm is expressed at very low levels at all stages of development in the embryonic tissue (35, 43). On the basis of these findings, it is possible that some Srg3 proteins may not be components of either Brg1- or Brm-containing SWI/SNF complexes in some tissues. It is also worthwhile to note that certain human cell lines containing very little or no BRG1 and hBRM express BAF155 (58). In addition, MOIRA, a Drosophila counterpart of Srg3, also exhibited an expression pattern which overlaps with those of BRM and SNR1 but which is more widespread (12). These findings support the possibility that Srg3 may have an alternative novel function(s) besides being a subunit of the SWI/SNF complex.
An essential role for Srg3 in early embryogenesis.
A previous study of yeast demonstrated that a mutation in SWI3 and SWI2 resulted in identical phenotypes (38). In Drosophila, the SWI3 homolog MOIRA appears to be essential for BRM function in vivo, and mutation of the corresponding genes resulted in identical phenotypes, i.e., homozygous mutants die at the unhatched-larva stage (12, 37). These observations suggest that Srg3 knockout represents inactivation of the SWI/SNF complex. We demonstrated here that Srg3 is required for peri-implantation development. In vitro culture of Srg3-deficient embryos showed that the trophoectoderm developed into trophoblast giant cells, but no visceral endoderm and egg cylinder formation was found. Recently, it was reported that Brg1 null mutant died during the peri-implantation stage (6). Brg1-deficient embryos did not undergo even hatching processes, and neither the ICM nor the trophoectoderm survived in vitro. Although Srg3 and Brg1 mutants showed similar phenotypes of very early embryonic lethality, there appears to be a time gap in the death of the two mutants. The mutants showed differences in blastocyst hatching and differentiation of the trophoectoderm into trophoblast giant cells. As shown in Fig. 4B, the Brg1 protein seems to be stable in Srg3-deficient embryos. Therefore, it is likely that a basal chromatin-remodeling activity due to BRG1 alone (41) is present in Srg3 null mutants. This may have caused the phenotypic difference between the Brg1-deficient and Srg3-deficient mutants. The robust activity of the SWI/SNF complex may be required for the development of the ICM and primitive endoderm into the egg cylinder. However, Snf5/Ini1 null embryos display defects in the hatching process similar to those displayed by Brg1 null mutants (6, 17, 31). It is not clear at present what causes the time gap in death between Srg3 null and Snf5/Ini1 null embryos. It is possible that Snf5/Ini1 is more critical for the activity of Brg1 in vivo at the peri-implantation stage. This possibility needs to be further investigated. In addition, it should be noted that Brg1-deficient embryos have Brm-containing SWI/SNF complexes. Although Brm has been shown to be dispensable for mouse development (43), one cannot exclude the effect of the Brm-containing SWI/SNF complex on normal embryogenesis. In fact, there seems to be a correlation between cellular differentiation and expression of BRM (36). Therefore, it is possible that the failure of Brg1-deficient blastocysts to develop further may be due to the remaining Brm complex, which does not favor the cellular proliferation required for embryonic development.
Predisposal to exencephaly in the heterozygous mutant.
Another conspicuous phenotype of the Srg3 mutant is exencephaly in a portion of heterozygous embryos. This brain anomaly seems to be due to NTDs, reflecting failure in neural fold elevation. In fact, it is well known that most exencephaly or spina bifida aperta of genetic origin is caused by failure in neural fold elevation (18). However, for most mouse NTDs, it is not clear how the genes involved in NTDs contribute to neural fold elevation (28).
The SWI/SNF complex has been implicated in the regulation of cellular growth and proliferation (36, 57). For example, the BRG1-containing SWI/SNF complex is inactivated by phosphorylation of BAF155 and BRG1 during the G2/M phase of the cell cycle (48), whereas Brm knockout mice show increased cell proliferation (43). Several studies carried out with human cell lines have shown that BRG1 and hBRM physically interact with Rb. These studies showed that the SWI/SNF complex induced the growth arrest of cells in an Rb-dependent manner (14, 50). In addition, BRG1 and BAF155 have been shown to coimmunoprecipitate with cyclin E. It was revealed that the cdk2-cyclin E complex can phosphorylate both proteins (47). These features of the SWI/SNF complex suggest that the predisposal to NTDs in Srg3 heterozygous embryos may be due to a failure in cell cycle regulation caused by haploinsufficiency of Srg3. Exencephalic embryos showed excessive increases in the number of diencephalic neuronal cells flowing over to the cranial region (Fig. 6). BrdU incorporation and MAP2 staining studies showed that some actively proliferating cells in the upper posterior region seemed to produce an excessive cell mass of the diencephalon and relatively poorly differentiated telencephalic neuronal cells in the striatum (Fig. 7). It is not clear whether these are causes or results of NTDs. However, there seems to be a correlation between abnormal cell proliferation and/or differentiation and NTDs. Studies with some NTD mutants suggest that genes with a basic mitotic function also have a function specific to neural fold elevation (2, 20, 22, 46).
Recently, it was reported that Brg1 is highly expressed during embryogenesis, including the neurulation stage, and that high-level accumulation of its transcript is observed in the spinal cord and brain after neural tube closure (42, 43). Furthermore, Brg1 heterozygotes also show susceptibility to exencephaly with almost the same penetrance (15 to 30% of heterozygous mutants) (6) as that of Srg3 heterozygotes. Thus, it is likely that reduced Srg3 protein expression might result in down-regulation of Brg1-containing SWI/SNF complexes, causing cell cycle perturbation and resulting in NTDs.
Another possible cause of NTDs in Srg3 heterozygous mutants could be based on the fact that the SWI/SNF complex contains actin itself and an actin-related protein such as BAF53 (59). Some genes in a growing list of mouse NTD mutants have been shown to play roles in the organization of actin molecules in the cytoskeleton (3, 9, 21, 32, 34, 52, 61, 62). It was suggested that part of the force required to change the shape of the neural folds from flat or convex to concave or elevated is a “pulse-string” rearrangement of actin at the luminal-apical surface, changing the neuroepithelial cells from columnar to wedge shaped (45). Thus, it is possible that the SWI/SNF complex may participate in neural fold elevation by its actin-related property or by regulation of some genes organizing actin rearrangement. Whether it is cell cycle regulation, an actin-related property, or another unknown mechanism, the fact that the haploinsufficiency of Srg3, as well as of Brg1, confers susceptibility to NTDs indicates the quantitative importance of this molecule for brain development.
ACKNOWLEDGMENTS
We are grateful to Kyoung-Li Kim and Young-Ho Ahn for their expert technical assistance.
This work was supported in part by the Molecular Medicine Research Group Program (98-MM-01-01-A-02) and International Cooperative Research Program (1-04-006) from the Korean Ministry of Science and Technology and in part by grants from the Korea Science and Engineering Foundation, through the Protein Network Research Center. R.H.S. is a Hae-Eun LSRI investigator. H. Choi, C. Lee, and D. Shin were supported by the BK21 Research Fellowship from the Korea Ministry of Education.
REFERENCES
- 1.Armstrong J A, Emerson B M. Transcription of chromatin; these are complex times. Curr Opin Genet Dev. 1998;8:165–172. doi: 10.1016/s0959-437x(98)80137-8. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong J F, Kaufman M H, Harrison D J, Clarke A R. High-frequency developmental abnormalities in p53-deficient mice. Curr Biol. 1995;5:931–936. doi: 10.1016/s0960-9822(95)00183-7. [DOI] [PubMed] [Google Scholar]
- 3.Blackshear P J, Lai W S, Tuttle J S, Stumpo D J, Kennington E, Nairn A C, Sulik K K. Developmental expression of MARCKS and protein kinase C in mice in relation to the exencephaly resulting from MARCKS deficiency. Dev Brain Res. 1996;96:62–75. doi: 10.1016/0165-3806(96)00097-1. [DOI] [PubMed] [Google Scholar]
- 4.Brizuela B J, Elfring L, Billiard J, Tamkun J W, Kennison J A. Genetic analysis of the brahma gene of Drosophila melanogaster and polytene chromosome subdivisions 72AB. Genetics. 1994;137:803–813. doi: 10.1093/genetics/137.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brizuela B J, Kennison J A. The Drosophila homeotic gene moira regulates expression of engrailed and HOM genes in imaginal tissues. Mech Dev. 1997;65:209–220. doi: 10.1016/s0925-4773(97)00081-6. [DOI] [PubMed] [Google Scholar]
- 6.Bultman S, Gebuhr T, Yee D, Mantia C L, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 7.Cairns B R, Kim Y-J, Sayre M H, Laurent B C, Kornberg R D. A multisubuit complex containing the SWI1/ADR6, SW12/SNF2, SW13, SNF5, and SNF6 gene products isolated from yeast. Proc Natl Acad Sci USA. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Chang S, Duncan S A, Okano H J, Fishell G, Aderem A. Disruption of the MacMARCKS gene prevents cranial neural tube closure and results in anencephaly. Proc Natl Acad Sci USA. 1996;93:6275–6279. doi: 10.1073/pnas.93.13.6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cote J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 11.Crandall J E, Jacobson M, Kosik K S. Ontogenesis of microtubule-associated protein 2 (MAP2) in embryonic mouse cortex. Brain Res. 1986;393:127–133. doi: 10.1016/0165-3806(86)90072-6. [DOI] [PubMed] [Google Scholar]
- 12.Crosby M A, Miller C, Alon T, Watson K L, Verrijzer C P, Goldman-Lebi R, Zak N B. The trithorax group gene moira encodes a brahma-associated putative chromatin-remodeling factor in Drosophila melanogaster. Mol Cell Biol. 1999;19:1159–1170. doi: 10.1128/mcb.19.2.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dingwall A K, Beek S J, McCallum C M, Tamkun J W, Kalpana G V, Goff S P, Scott M P. The Drosophila snr and brm proteins are related to yeast SWI/SNF proteins and are components of a large protein complex. Mol Biol Cell. 1995;6:777–791. doi: 10.1091/mbc.6.7.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunaief J L, Strober B E, Guha S, Khavari P A, Alin K, Luban J, Begemann M, Crabtree G R, Goff S P. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 15.Elfring L K, Deuring R, McCallum C M, Peterson C L, Tamkun J W. Identification and characterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol Cell Biol. 1994;14:2225–2234. doi: 10.1128/mcb.14.4.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elfring L K, Daniel C, Papoulas O, Deuring R, Sarte M, Moseley S, Beek S J, Waldrip W R, Daubresse G, DePace A, Kennison J A, Tamkun J W. Genetic analysis of brahma: the Drosophila homolog of the yeast chromatin remodeling factor SWI2/SNF2. Genetics. 1998;148:251–265. doi: 10.1093/genetics/148.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guidi C J, Sands A T, Zambrowicz B P, Turner T K, Demers D A, Webster W, Smith T W, Imbalzano A N, Jones S N. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol Cell Biol. 2001;21:3598–3603. doi: 10.1128/MCB.21.10.3598-3603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris M J, Juriloff D M. Toward understanding mechanisms of genetic neural tube defects in mice. Teratology. 1999;60:292–305. doi: 10.1002/(SICI)1096-9926(199911)60:5<292::AID-TERA10>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 19.Hatini V, Tao W, Lai E. Expression of winged helix genes, BF-1 and BF-2, define adjacent domains within the developing forebrain and retina. J Neurobiol. 1994;25:1293–1309. doi: 10.1002/neu.480251010. [DOI] [PubMed] [Google Scholar]
- 20.Herrera E, Samper E, Blasco M A. Telomere shortening in mTR−/− embryos is associated with failure to close the neural tube. EMBO J. 1999;18:1172–1181. doi: 10.1093/emboj/18.5.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hildebrand J D, Soriano P. Shroom, a PDZ domain-containing actin-binding protein, is required for neural tube morphogenesis in mice. Cell. 1999;99:485–497. doi: 10.1016/s0092-8674(00)81537-8. [DOI] [PubMed] [Google Scholar]
- 22.Hollander M C, Shaikh M S, Bulavin D V, Lundgren K, Augeri-Henmueller L, Shehee R, Molinaro T A, Kim K E, Tolosa E, Ashwell J D. Genomic instability in Gadd45a-deficient mice. Nat Genet. 1999;23:176–184. doi: 10.1038/13802. [DOI] [PubMed] [Google Scholar]
- 23.Holstege F C P, Jennings E G, Wyrick J J, Lee T I, Hengarner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 24.Huh S, Hatini V, Marcus R C, Li S C, Lai E. Dorsal-ventral patterning defects in the eye of BF-1 deficient mice associated with a restricted loss of shh expression. Dev Biol. 1999;211:53–63. doi: 10.1006/dbio.1999.9303. [DOI] [PubMed] [Google Scholar]
- 25.Imbalzano A N. Energy-dependent chromatin remodelers: complex complexes and their components. Crit Rev Eukaryot Gene Expr. 1998;8:225–255. doi: 10.1615/critreveukargeneexpr.v8.i3-4.10. [DOI] [PubMed] [Google Scholar]
- 26.Imhof A, Wolffe A P. Transcription: gene control by targeted histone acetylation. Curr Biol. 1998;8:R422–R424. doi: 10.1016/s0960-9822(98)70268-4. [DOI] [PubMed] [Google Scholar]
- 27.Jeon S H, Kang M G, Kim Y H, Jin Y H, Lee C, Chung H Y, Kwon H, Park S D, Seong R H. A new mouse gene, Srg3, related to the SWI3 of Saccharomyces cerevisiae, is required for apoptosis induced by glucocorticoids in a thymoma cell line. J Exp Med. 1997;185:1827–1836. doi: 10.1084/jem.185.10.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Juriloff D M, Harris M J. Mouse model for neural tube closure defects. Hum Mol Genet. 2000;9:993–1000. doi: 10.1093/hmg/9.6.993. [DOI] [PubMed] [Google Scholar]
- 29.Kennison J A, Tamkun J W. Dosage-dependent modifiers of Polycomb and Antennapedia mutations in Drosophila. Proc Natl Acad Sci USA. 1988;85:8136–8140. doi: 10.1073/pnas.85.21.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kingston R E, Narlikar G J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 31.Klochendler-Yeivin A, Fiette L, Barra J, Muchardt C, Babinet C, Yaniv M. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 2000;1:500–506. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koleske A J, Gifford A M, Scott M L, Nee M, Bronson R T. Essential roles for the Abl and Arg tyrosine kinases in neurulation. Neuron. 1998;21:1259–1272. doi: 10.1016/s0896-6273(00)80646-7. [DOI] [PubMed] [Google Scholar]
- 33.Kornberg R D, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 34.Lanier L M, Gates M A, Witke W, Menzies A S, Wehman A M. Mena is required for neurulation and commissure formation. Neuron. 1999;22:313–325. doi: 10.1016/s0896-6273(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 35.Legouy E, Thompson E M, Muchardt C, Renald J P. Differential preimplantation regulation of two mouse homologs of the yeast SWI2 protein. Dev Dyn. 1998;212:38–48. doi: 10.1002/(SICI)1097-0177(199805)212:1<38::AID-AJA4>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 36.Muchardt C, Yaniv M. The mammalian SWI/SNF complex and the control of cell growth. Semin Cell Dev Biol. 1999;10:189–195. doi: 10.1006/scdb.1999.0300. [DOI] [PubMed] [Google Scholar]
- 37.Papoulas O, Beek S J, Moseley S L, McCallum C M, Sarte M, Shearn A, Tamkun J K. The Drosophila trithorax group proteins BRM, ASH, and ASH are subunits of distinct protein complexes. Development. 1998;125:3955–3966. doi: 10.1242/dev.125.20.3955. [DOI] [PubMed] [Google Scholar]
- 38.Peterson C L, Herskowitz I. Characterization of the yeast SWI1, SWI2, SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 39.Peterson C L, Dingwall A, Scott M P. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc Natl Acad Sci USA. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peterson C L. Multiple SWItches to turn on chromatin? Curr Opin Genet Dev. 1996;6:171–175. doi: 10.1016/s0959-437x(96)80047-5. [DOI] [PubMed] [Google Scholar]
- 41.Phelan M L, Sif S, Narlikar G J, Kingston R E. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunit. Mol Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 42.Randazzo F M, Khavari M, Crabtree G, Tamkun J, Rossant J. brg1: a putative murine homologue of the Drosophila brahma gene, a homeotic gene regulator. Dev Biol. 1994;161:229–242. doi: 10.1006/dbio.1994.1023. [DOI] [PubMed] [Google Scholar]
- 43.Reyes J C, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α) EMBO J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts C W, Galusha S A, McMenamin M E, Fletcher C D, Orkin S H. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc Natl Acad Sci USA. 2000;97:13796–13800. doi: 10.1073/pnas.250492697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sadler T W, Greenberg D, Coughlin P, Lessard J L. Actin distribution patterns in the mouse neural tube during neurulation. Science. 1982;215:172–174. doi: 10.1126/science.7031898. [DOI] [PubMed] [Google Scholar]
- 46.Sah V P, Attardi L D, Mulligan G J, Williams B O, Bronson R T, Jacks T. A subset of p53-deficient embryos exhibit exencephaly. Nat Genet. 1995;10:175–180. doi: 10.1038/ng0695-175. [DOI] [PubMed] [Google Scholar]
- 47.Shanahan F, Seghezzi W, Parry D, Mahony D, Lees E. Cyclin E associates with BAF155 and BRG1, components of the mammalian SWI-SNF complex, and alters the ability of BRG1 to induce growth arrest. Mol Cell Biol. 1999;19:1460–1469. doi: 10.1128/mcb.19.2.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sif S, Stukenberg P T, Kirschner M W, Kingston R E. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 1998;12:2842–2851. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soriano P. The PDGF receptor is required for neural crest cell development and for normal patterning of the somites. Development. 1997;124:2691–2700. doi: 10.1242/dev.124.14.2691. [DOI] [PubMed] [Google Scholar]
- 50.Strober B E, Dunaief J L, Guha S, Goff S P. Functional interactions between the hBRM/hBRG1 transcriptional activators and the pRB family of proteins. Mol Cell Biol. 1996;16:1576–1583. doi: 10.1128/mcb.16.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Struhl K. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell. 1999;98:1–4. doi: 10.1016/S0092-8674(00)80599-1. [DOI] [PubMed] [Google Scholar]
- 52.Stumpo D J, Bock C B, Tuttle J S, Blackshear P J. MARCKS deficiency in mice leads to abnormal brain development and perinatal death. Proc Natl Acad Sci USA. 1995;92:944–948. doi: 10.1073/pnas.92.4.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sudarsanam P, Winston F. The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet. 2000;16:345–351. doi: 10.1016/s0168-9525(00)02060-6. [DOI] [PubMed] [Google Scholar]
- 54.Tamkun J W, Deuring R, Scott M P, Kissinger M, Pattatucci A M, Kaufman T C, Kennison J A. Brahma: a regulator of Drosophila homeotic genes and structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 55.Tamkun J W. The role of brahma and related proteins in transcription and development. Curr Opin Genet Dev. 1995;5:473–477. doi: 10.1016/0959-437x(95)90051-h. [DOI] [PubMed] [Google Scholar]
- 56.Varga-Weisz P D, Becker P B. Chromatin-remodeling factors: machines that regulate? Curr Opin Cell Biol. 1998;10:346–353. doi: 10.1016/s0955-0674(98)80010-0. [DOI] [PubMed] [Google Scholar]
- 57.Vignali M, Hassan A H, Neely K E, Workman J L. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang W, Cote J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Goff S P, Yaniv M, Workman J L, Crabtree G R. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 59.Wang W, Xue Y, Zhou S, Kuo A, Cairns B R, Crabtree G R. Diversity and specialization of mammalian SWI/SNF complex. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 60.Winston F, Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 61.Wu M, Chen D F, Sasaoka T, Tonegawa S. Neural tube defects and abnormal brain development in F52-deficient mice. Proc Natl Acad Sci USA. 1996;93:2110–2115. doi: 10.1073/pnas.93.5.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu W, Baribault H, Adamson E D. Vinculin knockout results in heart and brain defects during embryonic development. Development. 1998;125:327–337. doi: 10.1242/dev.125.2.327. [DOI] [PubMed] [Google Scholar]
- 63.Xuan S, Baptista C, Balas G, Tao W, Soares V, Lai E. Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron. 1995;14:1141–1152. doi: 10.1016/0896-6273(95)90262-7. [DOI] [PubMed] [Google Scholar]
- 64.Zhang S H, Gavin M, Dahiya A, Postigo A A, Ma D, Luo R X, Harbour J W, Dean D C. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]