Abstract
Tumors use multiple mechanisms to actively exclude immune cells involved in antitumor immunity. Strategies to overcome these exclusion signals remain limited due to an inability to target therapeutics specifically to the tumor. Synthetic biology enables engineering of cells and microbes for tumor-localized delivery of therapeutic candidates previously unavailable using conventional systemic administration techniques. Here, we engineer bacteria to intratumorally release chemokines to attract adaptive immune cells into the tumor environment. Bacteria expressing an activating mutant of the human chemokine CXCL16 (hCXCL16K42A) offer therapeutic benefit in multiple mouse tumor models, an effect mediated via recruitment of CD8+ T cells. Furthermore, we target the presentation of tumor-derived antigens by dendritic cells, using a second engineered bacterial strain expressing CCL20. This led to type 1 conventional dendritic cell recruitment and synergized with hCXCL16K42A-induced T cell recruitment to provide additional therapeutic benefit. In summary, we engineer bacteria to recruit and activate innate and adaptive antitumor immune responses, offering a new cancer immunotherapy strategy.
Rationally engineered bacteria producing chemokines within the tumor microenvironment promote anti-tumor adaptive immunity.
INTRODUCTION
Surmounting the obstacles tumors use to suppress immune cell infiltration has proven to be an elusive target. Immune infiltration, particularly by CD8+ cytotoxic T lymphocytes and, more specifically, memory CD8+ T cells, within tumors is widely considered a positive prognostic factor (1–4). Immune cells undergo chemotaxis in response to chemokines (5, 6); however, although chemokines may be an attractive therapeutic target in cancer (7), conventional drug delivery methods fail to generate tumor-localized chemokine gradients that sufficiently overcome immune cell exclusion (8, 9). Thus, despite the importance of tumor infiltration by immune cells in treatment outcomes, new approaches are needed to actively attract leukocytes into the tumor microenvironment.
The chemokine CXCL16 was first identified for its ability to recruit activated T cells with extralymphoid homing potential (10, 11). Although some reports have suggested that tumor cell–derived CXCL16 may contribute to tumor invasion and metastasis (12–14), CXCL16 and its receptor, CXCR6 (also known as Bonzo), are positively correlated with T cell infiltration and with increased survival in patients with colon and lung cancer (15, 16). Moreover, recent reports suggest that the CXCL16/CXCR6 axis plays a critical role in generating antitumor immunity and that the manipulation of this axis has therapeutic potential (17–19). However, a method to specifically deliver CXCL16 to the tumor microenvironment to recruit activated T cells has not been available, making the assessment of its potential for directly manipulating antitumor T cell responses in vivo difficult to discern.
Bacteria are increasingly recognized as a component of the tumor microenvironment, with genetic manipulations of bacteria enabling renewed consideration for bacterial cancer therapy (20–22). A previously generated synthetic gene circuit in bacteria, termed the synchronized lysis circuit (SLC), enables repeated delivery of tumor-localized therapeutics following synchronized lysis (23–25). Here, we first use the SLC and engineer bacteria that produce tumor-localized CXCL16 to recruit activated CD8+ T cells and promote antitumor immunity. We then combine the expression of CXCL16 with CCL20 to recruit innate and adaptive immune cells involved in both the priming and response phases of tumor immunity, augmenting the overall antitumor immune response and enhancing therapeutic efficacy, a heretofore unavailable therapeutic approach.
RESULTS
Mutated human CXCL16 variant in probiotic Escherichia coli has bioactivity in vitro
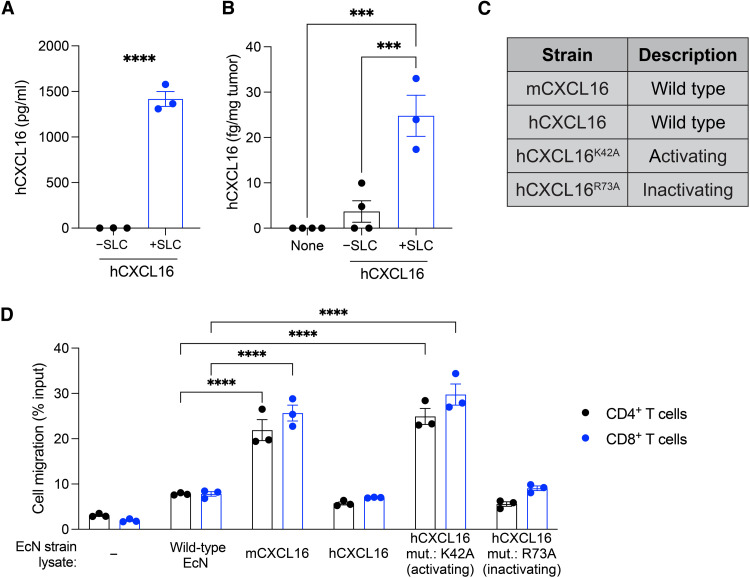
We first hypothesized that SLC-mediated release of CXCL16 in tumors would promote infiltration of activated T cells and support antitumor immunity. Toward this aim, we expressed the bioactive mature region (Asn30-Pro118) of human CXCL16 (hCXCL16) on a previously described (25) high–copy number plasmid in the probiotic E. coli Nissle 1917 (EcN) and confirmed that the release of hCXCL16 was SLC dependent (Fig. 1A). The release of hCXCL16 in tumors in vivo was also SLC dependent, with minimal detection in tumor homogenates from untreated tumors or those treated with EcN expressing hCXCL16 without SLC (−SLC), but with significantly greater detection when SLC was present (+SLC) (Fig. 1B). To identify the optimal variant of CXCL16, we generated mouse CXCL16 (mCXCL16) and previously described (26) activating (hCXCL16K42A) and inactivating (hCXCL16R73A) mutants of human CXCL16 (Fig. 1C). For functional assessment of the probiotic-derived CXCL16 variants, we developed a chemotaxis assay in which activated T cells were assayed for their migration in response to the lysate of wild-type or variant-expressing EcN strains. Compared to wild-type lysate, activated mouse CD4+ and CD8+ T cells significantly migrated in response to lysates of EcN strains expressing mCXCL16 and activating hCXCL16K42A but not wild-type hCXCL16 or inactivating hCXCL16R73A (Fig. 1D). Consistent with previous characterization of the hCXCL16K42A mutation (26), activated human T cells displayed a similar response to the lysate of the hCXCL16K42A-expressing EcN strain (fig. S1). These data suggest that E. coli–derived hCXCL16K42A potently attracts mouse and human activated T cells, potentially easing translational and preclinical studies and warranting an assessment of this strain in vivo using murine tumor models.
Fig. 1. Generation and characterization of CXCL16 variant strains in probiotic E. coli.
(A) ELISA quantification of hCXCL16 concentration in culture supernatants of EcN expressing hCXCL16 with (“+SLC”) or without (“−SLC”) SLC. Data are representative of three independent experiments (****P < 0.0001, two-sided unpaired Student’s t test). (B) Quantification of hCXCL16 concentration in homogenates of A20 tumors intratumorally injected with EcN expressing hCXCL16 with (+SLC) or without (−SLC) SLC by ELISA. Data are representative of two independent experiments (***P < 0.001, one-way ANOVA with Holm-Sidak post hoc test). (C) Table of CXCL16 variants expressed in EcN and used throughout the current study. (D) Flow cytometric quantification of the migration of activated mouse T cells in response to each of the indicated EcN strain lysates. Media without any bacteria was used as a control (“−”). Data are representative of three independent experiments (****P < 0.0001, two-way ANOVA with Holm-Sidak post hoc test). All data are displayed as means ± SEM.
Probiotic E. coli–derived CXCL16 promotes tumor regression
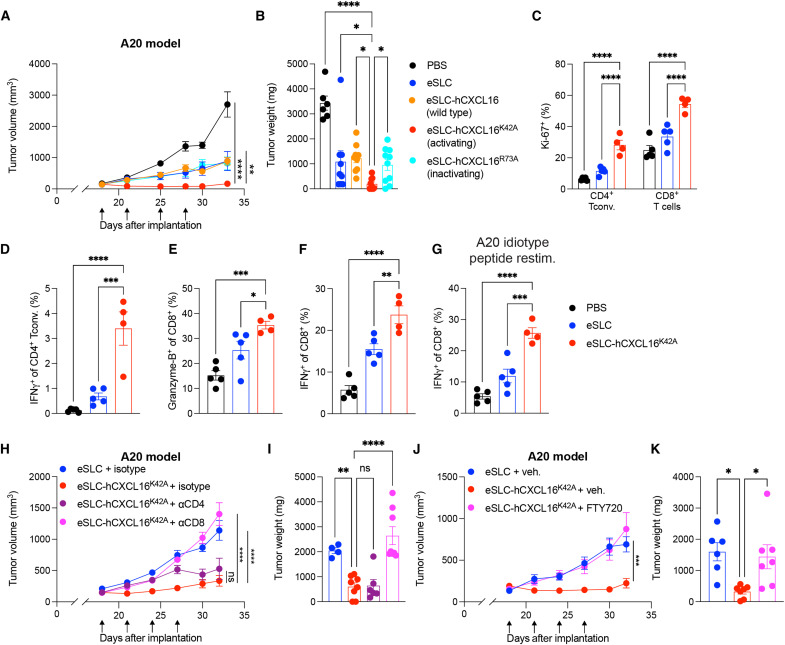
We examined the efficacy of EcN-derived hCXCL16K42A in vivo by treating subcutaneously growing murine tumors after they were established and palpable (~100 mm3). We first tested variants of hCXCL16 in the A20 B cell lymphoma model, performing intratumoral injections of bacteria every 3 to 4 days for a total of four treatments, with multiple injections performed to minimize the likelihood of plasmid loss. Significant reductions in tumor growth were observed in mice treated with SLC EcN strains expressing activating hCXCL16K42A (eSLC-hCXCL16K42A) as compared to those treated with phosphate-buffered saline (PBS) or EcN expressing SLC alone (eSLC) (Fig. 2, A and B, and fig. S2A), confirming the therapeutic activity of EcN-mediated intratumoral delivery of hCXCL16K42A in vivo. Furthermore, wild-type hCXCL16 (eSLC-hCXCL16)– and inactivating mutant hCXCL16R73A (eSLC-hCXCL16R73A)–expressing SLC EcN strains offered similar responses to eSLC alone (Fig. 2, A and B, and fig. S2A), suggesting a lack of additional therapeutic benefit from these strains in vivo. Most notably, the hCXCL16K42A strain induced complete regression of three of eight treated A20 tumors. Phenotyping of tumor-infiltrating lymphocytes revealed that the hCXCL16K42A strain induced an increase in activated and proliferating CD4+ conventional T cells, as assessed by Ki-67 expression and cytokine production (Fig. 2, C and D, and fig. S2B). Treatment with the hCXCL16K42A strain also led to increased CD8+ T cell activation in A20 tumors, as assessed by Ki-67 (Fig. 2C), Granzyme-B expression (Fig. 2E), and their production of the cytokines interferon-γ (IFN-γ) and tumor necrosis factor–α (TNFα) following ex vivo restimulation with phorbol 12-myristate 13-acetate (PMA) and ionomycin (Fig. 2F and fig. S2C). Moreover, upon ex vivo restimulation with a major histocompatibility complex class I (MHC-I) (H-2Kd)–restricted A20 idiotype peptide, CD8+ T cells from A20 tumors treated with the eSLC-hCXCL16K42A strain demonstrated increased production of IFN-γ and TNFα (Fig. 2G and fig. S2D), suggesting increased effector function of tumor antigen–specific T cells following hCXCL16K42A treatment. CXCR6 expression was increased among intratumoral CD4+ T conventional cells following hCXCL16K42A treatment (fig. S2E), consistent with a role for CXCR6 in mediating activated CD4+ T cell recruitment. Levels of endogenous mCXCL16 were unchanged in tumor homogenate (fig. S2F), highlighting the specific role for hCXCL16K42A delivered by our engineered bacteria to mediate therapeutic efficacy and indicating that exogenously delivered CXCL16 does not activate pathways that result in the induction of endogenous (mouse-derived) CXCL16. Overall, these data suggest that the eSLC-hCXCL16K42A strain promotes A20 tumor regression via an expansion of activated T cells, specifically tumor antigen–specific T cells.
Fig. 2. Activating hCXCL16K42A strain promotes tumor regression in a subcutaneous murine B cell lymphoma model.
(A and B) Subcutaneous A20 tumor growth curves and weights from BALB/c mice (n = 5 per group) that received intratumoral injections (black arrows) with the indicated treatment. Representative of three independent experiments [*P < 0.05, **P < 0.01, and ****P < 0.0001, (A) two-way ANOVA with Holm-Sidak post hoc test; (B) one-way ANOVA with Holm-Sidak post hoc test]. (C to G) Flow cytometric analysis of tumor-infiltrating lymphocytes from subcutaneous A20 tumors (n ≥ 4 per group) following the indicated treatment. (C) Frequencies of Ki-67+CD4+Foxp3− and Ki-67+CD8+ T cells. (D) Frequencies of IFN-γ+CD4+Foxp3− T cells following PMA/ionomycin ex vivo restimulation. (E) Frequencies of Granzyme-B+CD8+ T cells. (F and G) Frequencies of IFN-γ+CD8+ T cells following ex vivo restimulation with (F) PMA/ionomycin and (G) MHC-I–restricted A20 idiotype peptide. Data are representative of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, (C) two-way ANOVA with Holm-Sidak post hoc test and (D to G) one-way ANOVA with Holm-Sidak post hoc test. (H and I) A20 tumor growth curves and weights (n = 5 mice per group) with indicated intratumoral bacterial injections and intraperitoneal injections of the indicated antibody. **P < 0.01 and ****P < 0.0001, (H) two-way ANOVA with Holm-Sidak post hoc test; (I) ****P < 0.0001, one-way ANOVA with Holm-Sidak post hoc test. (J and K) A20 tumor growth curves and weights (n = 3 to 4 mice per group) that received intratumoral treatment and daily administration of vehicle or FTY720, as indicated. (J) ***P < 0.001, two-way ANOVA with Holm-Sidak post hoc test; (K) *P < 0.01, one-way ANOVA with Holm-Sidak post hoc test. Data are displayed as means ± SEM. Tconv, conventional T cells; ns, not significant.
We next performed in vivo functional analyses of the therapeutic response to eSLC-hCXCL16K42A treatment. To functionally assess the cell types involved, we performed antibody depletion experiments in mice bearing A20 tumors. Depletion of CD8+ cells, but not CD4+ cells, in eSLC-hCXCL16K42A–treated mice resulted in similar tumor growth kinetics as observed in mice treated with control eSLC bacteria alone (Fig. 2, H and I), consistent with CD8+ T cells, but not CD4+ T cells, being responsible for the efficacy of the eSLC-hCXCL16K42A strain. We next asked whether T cell migration was required for the therapeutic benefit afforded by treatment with the eSLC-hCXCL16K42A strain. Blockade of T cell trafficking by injection of the sphingosine 1-phosphate receptor agonist FTY720 restored tumor growth in mice treated with the eSLC-hCXCL16K42A strain to a similar level to those treated with the eSLC control strain (Fig. 2, J and K). Together, these data suggest that the eSLC-hCXCL16K42A strain offers therapeutic benefit via recruitment of activated CD8+ T cells.
Probiotic CXCL16 generates a systemic antitumor response
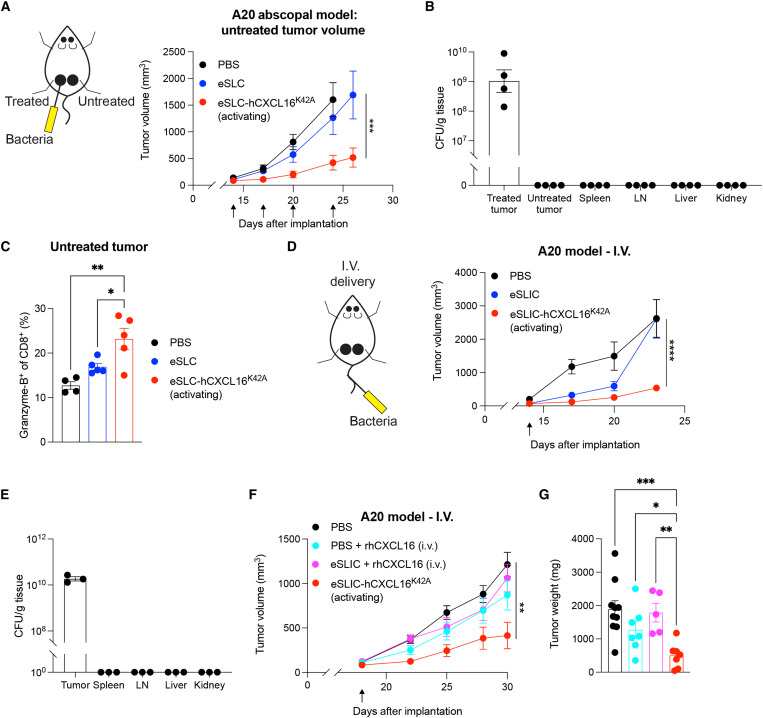
To increase the translational relevance, we next aimed to broaden the applicability of this approach beyond accessible subcutaneous tumors. We first assessed the ability of treatment of a single tumor to generate a systemic response to the slow growth of untreated distant tumors, termed an “abscopal effect.” Using the A20 B cell lymphoma model, we observed that treatment with the hCXCL16K42A strain in one tumor led to slowed tumor growth in distant untreated tumors, compared to eSLC alone or PBS (Fig. 3A and fig. S3A). Consistent with prior reports (24, 25), following intratumoral injection, colonies of eSLC-hCXCL16K42A were found only in the treated tumor and were not found in the lymph nodes, spleen, liver, kidney, or untreated tumor tissue (Fig. 3B), suggesting that bacteria trafficking and colonization of distant tumors were not responsible for the therapeutic benefit observed in the untreated tumor. Rather, we observed an increased frequency of Granzyme-B+ CD8+ T cells in the untreated tumor following the treatment of contralateral tumors with the eSLC-hCXCL16K42A strain (Fig. 3C), consistent with the hypothesis that CD8+ T cells, locally activated within treated tumors, circulate systemically to provide therapeutic benefit within untreated tumors sharing a similar antigenic profile. To examine the efficacy of our therapeutics in tumors that cannot be easily accessed for intratumoral injection, we explored intravenous injection as an alternative delivery approach. To this end, we performed a single intravenous injection of bacteria, an approach that specifically colonizes tumors (24, 25), using an EcN strain in which the SLC has been integrated into the genome (eSLIC) (25). Treatment with eSLIC expressing hCXCL16K42A (eSLIC-hCXCL16K42A) significantly reduced the growth of A20 tumors following a single intravenous injection, as compared to treatment with PBS or eSLIC (Fig. 3D and fig. S3B). Consistent with prior reports (24, 25), colonies of the eSLIC-hCXCL16K42A strain were only detected in the tumor tissue following intravenous delivery and were not detected in the spleen, lymph nodes, liver, or kidney (Fig. 3E). Furthermore, we compared the necessity of each component of the engineered bacteria delivery system. Intravenous injection of recombinant CXCL16 protein, with or without coinjection of control eSLIC, did not mediate a significant effect when compared with intravenous injection of the eSLIC-hCXCL16K42A strain (Fig. 3, F and G), suggesting a requirement for bacteria expressing hCXCL16K42A to actively grow and lyse within the tumor microenvironment in order for sufficient, therapeutically beneficial levels of chemokine to be locally established. Together, these data demonstrate that hCXCL16K42A-expressing EcN strains promote tumor regression, including in distant tumors left untreated and those treated via intravenous injection, and an expansion of activated CD8+ T cells.
Fig. 3. Treatment with eSLC-hCXCL16K42A provides systemic therapeutic benefit.
(A) Tumor growth curve of untreated tumor from BALB/c mice (n ≥ 5 per group) implanted subcutaneously with A20 cells on both hind flanks that received intratumoral injections (black arrows) of the indicated strain into a single tumor. Data are representative of two independent experiments (***P < 0.001, two-way ANOVA with Holm-Sidak post hoc test). (B) Colony-forming unit (CFU) analysis in BALB/c mice (n = 4 per group) treated as in (A). Data are representative of two independent experiments. (C) Flow cytometric analysis of tumor-infiltrating lymphocytes isolated from untreated subcutaneous A20 tumors (n ≥ 4 mice per group) on day 8 following treatment with the indicated strain. Frequencies of intratumoral Granzyme-B+CD8+ T cells. Data are representative of two independent experiments. *P < 0.05 and **P < 0.01, one-way ANOVA with Holm-Sidak post hoc test. (D) Tumor growth curves of BALB/c mice (n ≥ 5 per group) injected with A20 cells into both hind flanks that received a single intravenous (I.V.) injection (black arrow) of the indicated strain. Data are representative of two independent experiments (****P < 0.0001, two-way ANOVA with Holm-Sidak post hoc test). (E) CFU analysis in BALB/c mice (n = 3 per group) treated as in (D). (F and G) Tumor growth curves and weights of BALB/c mice (n ≥ 4 per group) that were injected with A20 cells into both hind flanks and received a single intravenous injection (black arrow) of the indicated treatment. (F) **P < 0.01, two-way ANOVA with Holm-Sidak post hoc test; (G) *P < 0.05, **P < 0.01, and ***P < 0.001, one-way ANOVA with Holm-Sidak post hoc test. Data are displayed as means ± SEM. LN, lymph node.
Probiotic CXCL16 has therapeutic efficacy in murine colorectal and breast cancer models
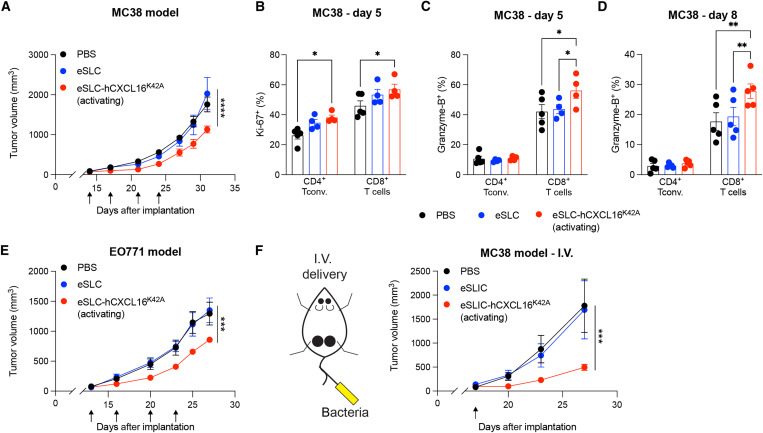
To assess the broader applicability of this approach, we next examined the therapeutic efficacy of the hCXCL16K42A strain in more aggressive murine cancer models. Treatment of established MC38 colorectal tumors with eSLC-hCXCL16K42A slowed tumor growth, compared to PBS and eSLC alone (Fig. 4A and fig. S4A). In this colorectal cancer model, we observed an expansion of proliferating conventional CD4+ and CD8+ T cells following treatment with the hCXCL16K42A strain at day 5 after treatment (Fig. 4B). Furthermore, treatment with the eSLC-hCXCL16K42A led to an expansion of Granzyme-B+ CD8+ T cells at days 5 and 8 after treatment (Fig. 4, C and D). CXCL16 has been reported to contribute to antitumor immunity in mouse models of triple-negative breast cancer (TNBC) (27). Consistent with this observation, intratumoral injection of eSLC-hCXCL16K42A significantly slowed tumor growth in the TNBC EO771 model, compared to PBS or eSLC (Fig. 4E and fig. S4B). To examine more translational approaches, we again explored the therapeutic efficacy of intravenous delivery. A single intravenous dose of eSLIC-hCXCL16K42A significantly reduced the growth of MC38 tumors compared to injection of PBS or eSLIC (Fig. 4F and fig. S4C). These data show that hCXCL16K42A-expressing strains are therapeutically effective, including with a single intravenous treatment, and lead to increased activation of tumor-infiltrating T cells in multiple different murine cancer models, with a more modest benefit in MC38 colorectal and EO771 breast cancer models than in the A20 B cell lymphoma model.
Fig. 4. Activating hCXCL16K42A strain offers therapeutic benefit in colorectal and breast cancers.
(A) Tumor growth curves from C57BL/6 mice (n = 5 per group) subcutaneously implanted with MC38 colorectal tumor cells on both hind flanks that received intratumoral injections (indicated by black arrows) with the indicated strain. Data are representative of four independent experiments (****P < 0.0001, two-way ANOVA with Holm-Sidak post hoc test at the final time point). (B to D) Flow cytometric analysis of tumor-infiltrating lymphocytes from subcutaneous MC38 tumors (n ≥ 4 mice per group) on (B and C) day 5 or (D) day 8 following intratumoral injections [as in (A)]. (B) Frequencies of intratumoral Ki-67+ CD4+Foxp3− and CD8+ T cells. (C and D) Frequencies of intratumoral Granzyme-B+ CD4+Foxp3− and Granzyme-B+ CD8+ T cells on (C) day 5 after injection and (D) day 8 after injection. For (B) to (D), data are representative of three independent experiments (*P < 0.05 and **P < 0.01, two-way ANOVA with Holm-Sidak post hoc test). (E) Tumor growth curves from C57BL/6 mice (n = 5 per group) subcutaneously implanted with EO771 tumor cells on both hind flanks that received intratumoral injections (indicated by black arrows) with the indicated strain. Data are representative of two independent experiments (***P < 0.001, two-way ANOVA with Holm-Sidak post hoc test at the final time point). (F) C57BL/6 mice (n ≥ 5 per group) subcutaneously implanted with MC38 colorectal tumor cells and received a single intravenous injection (black arrow) of the indicated strain. Data are representative of two independent experiments (***P < 0.001, two-way ANOVA with Holm-Sidak post hoc test at the final time point). Data are displayed as means ± SEM.
Recruitment of dendritic cells synergizes with activated T cell recruitment
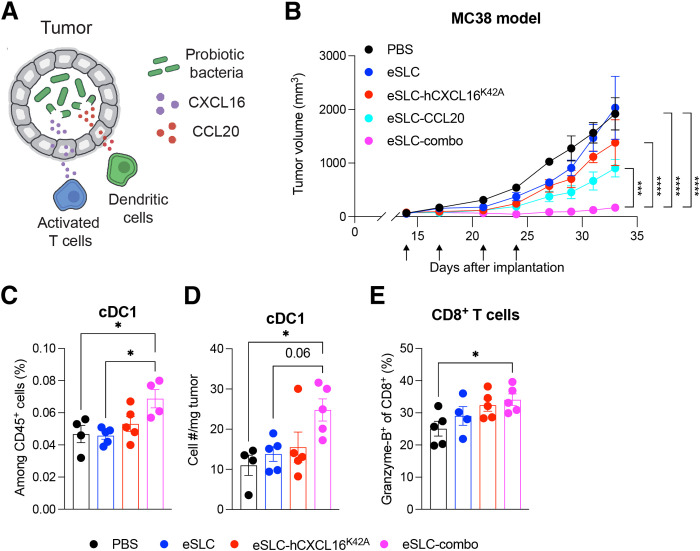
As the therapeutic benefit of hCXCL16K42A was more modest in models of colorectal and breast cancer than in B cell lymphoma, we explored additional approaches to augment the immune response. T cells are primed by dendritic cells (DCs); in particular, type 1 conventional DCs (cDC1) cross-present antigens to activate CD8+ T cells. We therefore considered how to recruit DCs to increase T cell priming and thus the availability of activated T cells that could be recruited by treatment with eSLC-hCXCL16K42A. Such an approach leverages the unique components of our system, namely, (i) the ability to generate tumor-localized chemokine gradients and (ii) potentiating DC maturation, particularly to cDC1s, as enforced by the recognition of bacterial components detected within the SLC-generated bacterial lysate. The chemokine CCL20 recruits pre-DCs (28, 29) and, when expressed by tumor cells, demonstrates therapeutic potential and DC recruitment in vivo (30, 31). We therefore hypothesized that probiotic-derived CCL20 and CXCL16 would synergize to promote antitumor immunity (Fig. 5A). The combination of hCXCL16K42A and CCL20 strains (“eSLC-combo”) demonstrated a synergistic therapeutic effect in MC38 tumors, slowing tumor growth significantly compared to PBS and eSLC treatment, as well as compared to treatment with either eSLC-hCXCL16K42A or eSLC-CCL20 strains individually (Fig. 5B and fig. S5). Moreover, treatment with the combination of eSLC-hCXCL16K42A and eSLC-CCL20 strains promoted the expansion, both by frequency and by number, of cDC1s (Fig. 5, C and D). Last, this combination also led to the expansion of tumor-infiltrating, Granzyme-B–expressing CD8+ T cells (Fig. 5E), consistent with increased activation of CD8+ T cells by cDC1s.
Fig. 5. Combination of CXCL16 and CCL20 synergizes to promote antitumor immunity.
(A) Schematic overview of the approach to recruit DCs to tumors via CCL20 production in probiotic bacteria to complement activated T cell recruitment via CXCL16 (as in Fig. 1A). (B) Tumor growth curves from C57BL/6 mice (n = 5 per group) subcutaneously implanted with 5 × 105 MC38 colorectal tumor cells on both hind flanks. When tumor volumes were 50 to 150 mm3, mice received intratumoral injections (indicated by black arrows) every 3 to 4 days with PBS, eSLC, eSLC-hCXCL16K42A, eSLC-CCL20, or eSLC-combo (1:1 mixture of eSLC-hCXCL16K42A and eSLC-CCL20). Data are representative of four independent experiments (***P < 0.001 and ****P < 0.0001, two-way ANOVA with Holm-Sidak post hoc test at the final measurement time point). (C to E) Flow cytometric analysis of tumor-infiltrating immune cells isolated from subcutaneously growing MC38 tumors (n ≥ 4 mice per group) following intratumoral injections [performed as in (B)] with PBS, eSLC, eSLC-hCXCL16K42A, or eSLC-combo [as in (A)]. (C) Frequencies and (D) numbers of cDC1s on day 5 after treatment. (E) Frequencies of intratumoral Granzyme-B+ CD8+ T cells. Data are representative of three independent experiments (*P < 0.05, one-way ANOVA with Holm-Sidak post hoc test). All data are displayed as means ± SEM.
DISCUSSION
Here, we demonstrate the utility of recruiting cells responsible for the adaptive immune response into tumors. We find that expression of hCXCL16K42A recruits activated mouse T cells, offers therapeutic benefit in vivo, and leads to the expansion of effector T cells in the tumor. Furthermore, we observe synergy between the recruitment of DCs and activated T cells in slowing tumor growth. We believe that this system offers a new set of therapeutic strategies to elicit immune cell recruitment and may complement existing immunotherapies that target the activation of immunity systemically but are unable to act locally to increase the intratumoral infiltration of cells.
Bacteria-derived CXCL16 alone has substantial therapeutic efficacy, and the use of an activating mutation of human CXCL16 that is bioactive on mouse and human T cells in our report may reduce translational studies required to show therapeutic preclinical benefits. CXCL16 alone does not promote tumor regression in all of the murine tumor models assessed, with one possible explanation being a paucity of activated T cells. In this regard, the combination of CXCL16 with CCL20 shows promise for addressing the possible lack of activated T cells in certain tumor types. Our study focuses on the chemokines CXCL16 and CCL20, but other chemokines, or combinations of chemokines, may offer additional therapeutic benefit. Our approach also has advantages over engineering tumor cells to express chemokines using oncolytic viruses and other gene delivery approaches because of the simplicity of bacterial growth and delivery to the tumor microenvironment, either by intratumoral or by intravenous injection, with repeated release of therapeutic payloads occurring through multiple growth and lysis cycles.
Although checkpoint blockade offers durable survival in a previously untreatable disease (32), it only achieves disease regression in a small subset of patients, with immunologically “hot” tumors offering greater responsiveness compared to “cold” tumors (2–4). The recruitment of peripheral tumor-specific T cells may be one mechanism to enhance the therapeutic response of Programmed Cell Death Protein-1/Programmed Death-L1 blockade (33, 34). Furthermore, the effectiveness of tumor vaccines and cell therapies, such as ex vivo expanded tumor-infiltrating lymphocytes and chimeric antigen receptor T cells, is hindered by the inability of primed T cells to infiltrate tumors (35, 36). Our approach may therefore offer a means to overcome challenges in other immunotherapies, broadening their efficacy and applicability.
While the current study is limited by the use of preclinical subcutaneous mouse models, future work will assess the efficacy of bacterially produced CXCL16 in orthotopic mouse models, as well as its potential combination with checkpoint blockade, which may broaden its potential use. Moreover, in this study, we focused on using the bacterial delivery system to explore potential combinations of chemokines, whereas CXCL16 or CCL20 may better combine with therapeutics targeting other immune molecules, such as “do not eat me” signals or immune-activating or immune-polarizing cytokines. Future work will explore additional potential combinations of therapeutics using the SLC delivery system.
In summary, we show the effectiveness of engineered bacteria to recruit activated T cells and DCs to tumors, which results in slowed tumor growth and increased effector T cell and DC infiltration. This approach offers a new range of therapeutic strategies in the quest for novel anticancer therapies.
MATERIALS AND METHODS
Experimental design
This study was designed to assess the use of bacteria engineered to express and release chemokines within the tumor environment as a potential therapeutic for antitumor immunity. The experiments were designed to first assess whether the bacteria could produce a functional protein and then assess their efficacy in vivo using a variety of different preclinical mouse models and delivery approaches. Studies in mice were also carried out to address the mechanisms through which these therapies function.
Bacterial strains
The mature bioactive regions of murine (Asn27-Pro114, UniProt accession number Q8BSU2) and human (Asn30-Pro118, UniProt accession number Q9H2A7) CXCL16 and murine CCL20 (Ala27-Met96, UniProt accession number O89093) were cloned into plasmid p246 (25) via Gibson Assembly, (co)transformed with or without the SLC plasmid (p15a) into electrically competent EcN, and maintained as previously described (25). Briefly, strains with only a therapeutic plasmid (p246) were grown in LB broth with kanamycin (50 μg/ml). Strains with the therapeutic and SLC plasmids were grown in LB broth with kanamycin (50 μg/ml) and spectinomycin (100 μg/ml) with 0.2% glucose. Mutant human CXCL16K42A and CXCL16R73A plasmids were generated from the wild-type hCXCL16 p246 plasmid using the New England BioLabs Q5 Site-Directed Mutagenesis Kit, as per the manufacturer’s instructions.
Chemotaxis assay
Activated T cells were generated as previously described (10). Briefly, T cells were isolated from wild-type adult C57BL/6 mouse spleen and lymph nodes using the Dynabeads FlowComp Mouse Pan T (CD90.2) Kit (Thermo Fisher Scientific) as per the manufacturer’s protocol. Isolated T cells were cultured with anti-CD3/CD28 beads (Dynabeads, catalog number 11452D, Thermo Fisher Scientific) in a 1:1 ratio in 10% complete RPMI [RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), penicillin/streptomycin (Pen/Strep), nonessential amino acids, GlutaMAX, Hepes, sodium pyruvate, and 2-mercaptoethanol]. After 5 days, the beads were removed by magnetic separation, and T cells were replated at 106 cells/ml for 4 days in 10% complete RPMI supplemented with recombinant human IL-2 (100 IU/ml). T cells were then washed and resuspended at 5.9 × 106 cells/ml in serum-free complete RPMI in preparation for the chemotaxis assay. Human T cells from anonymous donors sourced by STEMCELL Technologies were isolated by negative selection for CD3+ populations as per the manufacturer’s instructions (EasySep, STEMCELL Technologies) and cultured and prepared identically to murine T cells using corresponding reagents for human cells.
Overnight cultures of each bacterial strain (without SLC) were grown in LB with appropriate antibiotics and then subcultured at a 1:100 dilution in a shaking incubator for 90 min at 37°C. Bacteria were washed twice in serum-free complete RPMI, matched at optical density at 600 nm (OD600), and lysed via sonication in serum-free complete RPMI. Lysates were centrifuged to remove debris (20,817g for 10 min at 4°C), and 235 μl of the supernatant was entered into the lower chamber of a Corning HTS transwell plate (well area = 0.143 cm2; pore size = 5 μm). Activated T cells (75 μl of the preparation described above) were added to the upper chamber, and the plate was incubated for 3 hours in a humidified 37°C 5% CO2 incubator. The bottom chamber was then harvested, washed, and stained with anti-mouse CD3 violetFluor 450 (Tonbo Biosciences, clone 17A2), CD4 allophycocyanin (Tonbo Biosciences, clone RM4-5), and CD8 phycoerythrin (Tonbo Biosciences, clone 53-6.7). Human T cells were stained with anti-human CD3 BUV395 (BD Biosciences, clone SK7), CD4 BV785 (BioLegend, clone RPA-T4), and CD8 BV510 (BD Biosciences, clone RPA-T8). Samples were then acquired on a BD Fortessa for 60 s. Cell counts were normalized to the number of cells entered into the assay.
CXCL16 ELISA
For in vitro characterization, relevant strains were grown overnight as described above and then subcultured (1:100 dilution) for 3 hours in a shaking incubator at 37°C. Cultures were then centrifuged (20,817g for 10 min at 4°C), and supernatants were entered into an hCXCL16 enzyme-linked immunosorbent assay (ELISA) (R&D Human CXCL16 DuoSet, catalog number DY1164), performed as per the manufacturer’s protocol. For quantification of mouse (R&D mouse CXCL16 DuoSet) or human CXCL16 concentrations in tumor homogenates, 3 days after the final intratumoral injection of bacteria, tumors were harvested, weighed, and homogenized in tissue lysis buffer [100 mM tris (pH 7.4), 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1% Triton X-100, and 0.5% sodium deoxycholate in water] with protease inhibitor and EDTA. Homogenates were then centrifuged (5000g for 10 min at 4°C) to remove debris, and supernatants were used for ELISA, performed as per the manufacturer’s protocol.
Animal models
All animal experiments were approved by the Institutional Animal Care and Use Committee of Columbia University (protocols AC-AAAQ8474, AC-AABD8554, and AC-AAAZ4470). The protocol requires animals to be euthanized when the tumor burden reaches 2 cm in diameter or upon recommendation by veterinary staff. A20 cells were maintained in RPMI supplemented with 10% FBS, Pen/Strep, and 2-mercaptoethanol. MC38 and EO771 cells were maintained in Dulbecco’s Modified Eagle Medium supplemented with 10% FBS, Pen/Strep, nonessential amino acids, GlutaMAX, Hepes, sodium pyruvate, and 2-mercaptoethanol. Cultures were maintained in a humidified 37°C, 5% CO2 incubator. Before injection, A20 cells were resuspended in RPMI without phenol red at 5 × 107 cells/ml. A20 cells were subcutaneously implanted at 100 μl (5 × 106 cells) per hind flank. MC38 and EO771 cells were washed in PBS and resuspended at 5 × 106 and 10 × 106 cells/ml in PBS, respectively, and 100 μl of cell suspension was then injected subcutaneously into both hind flanks. Seven- to 8-week-old female BALB/c (for A20 tumors) or C57BL/6N (EO771 and MC38 tumors) mice were purchased from Taconic Biosciences or the Jackson Laboratory, allowed to acclimate for a week, and then injected with tumor cells. A20 tumor volume was determined by caliper measurements (length × width2 × 0.5), and mice were randomly assigned to treatment groups after tumors reached a volume of 100 to 150 mm3. MC38 and EO771 tumor volume was calculated as length × width × height, and mice were randomly assigned to treatment groups after tumors reached a volume of 50 to 150 mm3. For treatment with bacteria, bacteria were cultured in a 37°C shaking incubator for up to 12 hours to reach the stationary phase of growth in LB broth with appropriate antibiotics and 0.2% glucose. Bacteria were then subcultured (1:100 dilution) until a maximum OD600 of 0.15 was reached, again in LB broth with appropriate antibiotics and 0.2% glucose, then washed three times, and resuspended in ice-cold PBS. A total of 40 μl of each suspension containing 5 × 106 (for A20 tumors) or 1 × 106 bacteria (for MC38 and EO771 tumors) was injected intratumorally. Intratumoral injections were performed every 3 to 4 days for a total of three to four treatments (for tumor growth assays) or one to two treatments (for immunophenotyping assays), as indicated. For intravenous treatment experiments, bacteria were prepared as above and injected via the tail vein at a concentration of 5 × 107/ml in PBS, with a total volume of 100 μl injected per mouse. In some experiments, recombinant human CXCL16 (1 μg per mouse; R&D Systems) was injected intravenously in PBS or in combination with 5 × 106 bacteria diluted in PBS. In some experiments, antibody depletion was performed. In these experiments, isotype (clone 2A3, Bio X Cell), αCD4 (clone GK1.5, Bio X Cell), or αCD8 (clone 53-6.7, Bio X Cell) antibody was injected intraperitoneally (30 mg/kg) at the time of bacteria treatment. In some experiments, mice were treated with vehicle (ethanol) or FTY720 (1.25 mg/kg; Cayman Chemical) via intraperitoneal injection each day for the duration of the experiment once bacterial therapy was initiated.
Immune phenotyping by flow cytometry
Tumors were treated as above and harvested at the indicated time points. For isolation of tumor-infiltrating lymphocytes, tumors were excised, then minced, and digested in wash media (RPMI 1640 supplemented with 5% fetal calf serum, Hepes, GlutaMAX, and Pen/Strep) with collagenase A (1 mg/ml) and deoxyribonuclease I (0.5 μg/ml) in a shaking incubator for up to 45 min at 37°C to achieve a single-cell suspension. Once a single-cell suspension was achieved, samples were washed and then either restimulated or stained for flow cytometry analysis. For cytokine staining and ex vivo restimulation with PMA and ionomycin, aliquots of tumor homogenates were incubated for 3 hours at 37°C in 10% complete RPMI (as above) with PMA (50 ng/ml), ionomycin (500 ng/ml), and brefeldin A (1 μg/ml) before flow cytometry staining. For cytokine staining and ex vivo restimulation with A20 idiotype peptide, aliquots of tumor homogenates were incubated for 5 hours at 37°C in 10% complete RPMI (as above) with the A20 idiotype peptide (DYWGQGTEL; 1 μg/ml) and brefeldin A (1 μg/ml) before staining for flow cytometry. Live/dead staining was performed via Ghost Dye Red 780 labeling (Tonbo Biosciences), as per the manufacturer’s protocol. Cells were then stained for flow cytometry, with intracellular staining performed using the Tonbo Foxp3/Transcription Factor Staining Buffer Kit per the manufacturer’s instructions. Antibodies used were anti-CD45 (clone 30-F11, BioLegend), NK1.1 (clone PD136, BD Biosciences), CD3e (clone 145-2C11, Tonbo Biosciences), TCRβ (clone H57-597, BD Biosciences), CD4 (clone RM4-5, BD Biosciences), CD8 (clone 53-6.7, Tonbo Biosciences), Foxp3 (clone FJK-16s, Thermo Fisher Scientific), CXCR6 (clone SA051D1, BioLegend), Granzyme-B (clone QA16A02, BioLegend), Ki-67 (clone SolA15, Thermo Fisher Scientific), IFN-γ (clone XMG1.2, Tonbo Biosciences), B220 (clone RA3-6B2, BD Biosciences), CD11c (clone N418, Tonbo Biosciences), Ly6G (clone 1A8, Tonbo Biosciences), CD11b (clone M1/70, Tonbo Biosciences), MHC-II (clone M5/114.15.2, Tonbo Biosciences), and CD103 (clone 2E7, BioLegend).
Statistical analysis
Statistical analysis was performed in GraphPad Prism 9, using Student’s t test or one- or two-way analysis of variance (ANOVA) with post hoc testing as indicated. A P value of <0.05 was considered statistically significant. The specific statistical test, sample sizes, and P values for each figure are indicated in the figure legends.
Acknowledgments
We thank the members of the Arpaia and Danino laboratories for helpful discussions and members of the S.H. Sternberg laboratory for technical advice. Research reported in this publication was performed in the Columbia University Department of Microbiology and Immunology Flow Cytometry Core facility. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Funding: This work was supported by the following: National Institutes of Health/National Cancer Institute grant R01CA249160 (to N.A. and T.D.), National Institutes of Health/National Cancer Institute grant R01CA259634 (to N.A.), Searle Scholars Program grant SSP-2017-2179 (to N.A.), Roy and Diana Vagelos Precision Medicine Pilot Grant (to N.A. and T.D.), and National Institutes of Health/National Institute of General Medical Sciences grant T32GM007367 (to T.M.S.).
Author contributions: T.M.S. and N.A. conceived and designed the project with input from T.D.; T.M.S. performed experiments and analyzed data jointly with N.A.; R.L.V. assisted with transwell assays; S.S.R., A.A., F.L., and K.d.l.S.-A. assisted with tumor immunophenotyping; L.H.H., K.P., and C.C. assisted with a portion of the animal studies. T.M.S. and N.A. wrote the manuscript.
Competing interests: T.M.S., R.L.V., T.D., and N.A. are inventors on a patent application describing the use of chemokine-expressing engineered bacteria for cancer immunotherapy (International Application No. PCT/US2022/016775). N.A. and T.D. have a financial interest in GenCirq Inc. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The bacterial strains and plasmids can be provided by N.A. pending scientific review and a completed material transfer agreement with Columbia University. Requests for the bacterial strains and plasmids should be submitted to N.A.
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
REFERENCES AND NOTES
- 1.Fridman W. H., Pages F., Sautes-Fridman C., Galon J., The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 12, 298–306 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Tumeh P. C., Harview C. L., Yearley J. H., Shintaku I. P., Taylor E. J. M., Robert L., Chmielowski B., Spasic M., Henry G., Ciobanu V., West A. N., Carmona M., Kivork C., Seja E., Cherry G., Gutierrez A. J., Grogan T. R., Mateus C., Tomasic G., Glaspy J. A., Emerson R. O., Robins H., Pierce R. H., Elashoff D. A., Robert C., Ribas A., PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auslander N., Zhang G., Lee J. S., Frederick D. T., Miao B., Moll T., Tian T., Wei Z., Madan S., Sullivan R. J., Boland G., Flaherty K., Herlyn M., Ruppin E., Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat. Med. 24, 1545–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu D., Schilling B., Liu D., Sucker A., Livingstone E., Jerby-Arnon L., Zimmer L., Gutzmer R., Satzger I., Loquai C., Grabbe S., Vokes N., Margolis C. A., Conway J., He M. X., Elmarakeby H., Dietlein F., Miao D., Tracy A., Gogas H., Goldinger S. M., Utikal J., Blank C. U., Rauschenberg R., von Bubnoff D., Krackhardt A., Weide B., Haferkamp S., Kiecker F., Izar B., Garraway L., Regev A., Flaherty K., Paschen A., van Allen E., Schadendorf D., Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 25, 1916–1927 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hughes C. E., Nibbs R. J. B., A guide to chemokines and their receptors. FEBS J. 285, 2944–2971 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffith J. W., Sokol C. L., Luster A. D., Chemokines and chemokine receptors: Positioning cells for host defense and immunity. Annu. Rev. Immunol. 32, 659–702 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Nagarsheth N., Wicha M. S., Zou W., Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 17, 559–572 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckert E. C., Nace R. A., Tonne J. M., Evgin L., Vile R. G., Russell S. J., Generation of a tumor-specific chemokine gradient using oncolytic vesicular stomatitis virus encoding CXCL9. Mol. Ther. Oncolytics 16, 63–74 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J., O'Malley M., Urban J., Sampath P., Guo Z. S., Kalinski P., Thorne S. H., Bartlett D. L., Chemokine expression from oncolytic vaccinia virus enhances vaccine therapies of cancer. Mol. Ther. 19, 650–657 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matloubian M., David A., Engel S., Ryan J. E., Cyster J. G., A transmembrane CXC chemokine is a ligand for HIV-coreceptor Bonzo. Nat. Immunol. 1, 298–304 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Kim C. H., Kunkel E. J., Boisvert J., Johnston B., Campbell J. J., Genovese M. C., Greenberg H. B., Butcher E. C., Bonzo/CXCR6 expression defines type 1-polarized T-cell subsets with extralymphoid tissue homing potential. J. Clin. Invest. 107, 595–601 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu W., Zhen X., Xiong B., Wang B., Zhang W., Zhou W., CXCR6 is expressed in human prostate cancer in vivo and is involved in the in vitro invasion of PC3 and LNCap cells. Cancer Sci. 99, 1362–1369 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu W., Liu Y., Zhou W., Si L., Ren L., CXCL16 and CXCR6 are coexpressed in human lung cancer in vivo and mediate the invasion of lung cancer cell lines in vitro. PLOS ONE 9, e99056 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mir H., Kaur G., Kapur N., Bae S., Lillard J. W. Jr., Singh S., Higher CXCL16 exodomain is associated with aggressive ovarian cancer and promotes the disease by CXCR6 activation and MMP modulation. Sci. Rep. 9, 2527 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hald S. M., Kiselev Y., al-Saad S., Richardsen E., Johannessen C., Eilertsen M., Kilvaer T. K., al-Shibli K., Andersen S., Busund L. T., Bremnes R. M., Donnem T., Prognostic impact of CXCL16 and CXCR6 in non-small cell lung cancer: Combined high CXCL16 expression in tumor stroma and cancer cells yields improved survival. BMC Cancer 15, 441 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hojo S., Koizumi K., Tsuneyama K., Arita Y., Cui Z., Shinohara K., Minami T., Hashimoto I., Nakayama T., Sakurai H., Takano Y., Yoshie O., Tsukada K., Saiki I., High-level expression of chemokine CXCL16 by tumor cells correlates with a good prognosis and increased tumor-infiltrating lymphocytes in colorectal cancer. Cancer Res. 67, 4725–4731 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Di Pilato M., Kfuri-Rubens R., Pruessmann J. N., Ozga A. J., Messemaker M., Cadilha B. L., Sivakumar R., Cianciaruso C., Warner R. D., Marangoni F., Carrizosa E., Lesch S., Billingsley J., Perez-Ramos D., Zavala F., Rheinbay E., Luster A. D., Gerner M. Y., Kobold S., Pittet M. J., Mempel T. R., CXCR6 positions cytotoxic T cells to receive critical survival signals in the tumor microenvironment. Cell 184, 4512–4530.e22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma C., Han M., Heinrich B., Fu Q., Zhang Q., Sandhu M., Agdashian D., Terabe M., Berzofsky J. A., Fako V., Ritz T., Longerich T., Theriot C. M., McCulloch J. A., Roy S., Yuan W., Thovarai V., Sen S. K., Ruchirawat M., Korangy F., Wang X. W., Trinchieri G., Greten T. F., Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 360, eaan5931 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lesch S., Blumenberg V., Stoiber S., Gottschlich A., Ogonek J., Cadilha B. L., Dantes Z., Rataj F., Dorman K., Lutz J., Karches C. H., Heise C., Kurzay M., Larimer B. M., Grassmann S., Rapp M., Nottebrock A., Kruger S., Tokarew N., Metzger P., Hoerth C., Benmebarek M.-R., Dhoqina D., Grünmeier R., Seifert M., Oener A., Umut Ö., Joaquina S., Vimeux L., Tran T., Hank T., Baba T., Huynh D., Megens R. T. A., Janssen K.-P., Jastroch M., Lamp D., Ruehland S., di Pilato M., Pruessmann J. N., Thomas M., Marr C., Ormanns S., Reischer A., Hristov M., Tartour E., Donnadieu E., Rothenfusser S., Duewell P., König L. M., Schnurr M., Subklewe M., Liss A. S., Halama N., Reichert M., Mempel T. R., Endres S., Kobold S., T cells armed with C-X-C chemokine receptor type 6 enhance adoptive cell therapy for pancreatic tumours. Nat. Biomed. Eng. 5, 1246–1260 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forbes N. S., Engineering the perfect (bacterial) cancer therapy. Nat. Rev. Cancer 10, 785–794 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chien T., Doshi A., Danino T., Advances in bacterial cancer therapies using synthetic biology. Curr. Opin. Syst. Biol. 5, 1–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou S., Gravekamp C., Bermudes D., Liu K., Tumour-targeting bacteria engineered to fight cancer. Nat. Rev. Cancer 18, 727–743 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Din M. O., Danino T., Prindle A., Skalak M., Selimkhanov J., Allen K., Julio E., Atolia E., Tsimring L. S., Bhatia S. N., Hasty J., Synchronized cycles of bacterial lysis for in vivo delivery. Nature 536, 81–85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chowdhury S., Castro S., Coker C., Hinchliffe T. E., Arpaia N., Danino T., Programmable bacteria induce durable tumor regression and systemic antitumor immunity. Nat. Med. 25, 1057–1063 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gurbatri C. R., Lia I., Vincent R., Coker C., Castro S., Treuting P. M., Hinchliffe T. E., Arpaia N., Danino T., Engineered probiotics for local tumor delivery of checkpoint blockade nanobodies. Sci. Transl. Med. 12, eaax0876 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimaoka T., Nakayama T., Hieshima K., Kume N., Fukumoto N., Minami M., Hayashida K., Kita T., Yoshie O., Yonehara S., Chemokines generally exhibit scavenger receptor activity through their receptor-binding domain. J. Biol. Chem. 279, 26807–26810 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Matsumura S., Wang B., Kawashima N., Braunstein S., Badura M., Cameron T. O., Babb J. S., Schneider R. J., Formenti S. C., Dustin M. L., Demaria S., Radiation-induced CXCL16 release by breast cancer cells attracts effector T cells. J. Immunol. 181, 3099–3107 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dieu M. C., Vanbervliet B., Vicari A., Bridon J. M., Oldham E., Aït-Yahia S., Brière F., Zlotnik A., Lebecque S., Caux C., Selective recruitment of immature and mature dendritic cells by distinct chemokines expressed in different anatomic sites. J. Exp. Med. 188, 373–386 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bell D., Chomarat P., Broyles D., Netto G., Harb G. M., Lebecque S., Valladeau J., Davoust J., Palucka K. A., Banchereau J., In breast carcinoma tissue, immature dendritic cells reside within the tumor, whereas mature dendritic cells are located in peritumoral areas. J. Exp. Med. 190, 1417–1426 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furumoto K., Soares L., Engleman E. G., Merad M., Induction of potent antitumor immunity by in situ targeting of intratumoral DCs. J. Clin. Invest. 113, 774–783 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fushimi T., Kojima A., Moore M. A., Crystal R. G., Macrophage inflammatory protein 3α transgene attracts dendritic cells to established murine tumors and suppresses tumor growth. J. Clin. Invest. 105, 1383–1393 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pardoll D. M., The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yost K. E., Satpathy A. T., Wells D. K., Qi Y., Wang C., Kageyama R., McNamara K. L., Granja J. M., Sarin K. Y., Brown R. A., Gupta R. K., Curtis C., Bucktrout S. L., Davis M. M., Chang A. L. S., Chang H. Y., Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat. Med. 25, 1251–1259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yost K. E., Chang H. Y., Satpathy A. T., Recruiting T cells in cancer immunotherapy. Science 372, 130–131 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Sterner R. C., Sterner R. M., CAR-T cell therapy: Current limitations and potential strategies. Blood Cancer J. 11, 69 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirtane K., Elmariah H., Chung C. H., Abate-Daga D., Adoptive cellular therapy in solid tumor malignancies: Review of the literature and challenges ahead. J. Immunother. Cancer 9, e002723 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5