Abstract
Mouse esophagus is lined with a stratified epithelium, which is maintained by the constant renewal of unipotent progenitors. In this study, we profiled mouse esophagus by single-cell RNA sequencing and found taste buds specifically in the cervical segment of the esophagus. These taste buds have the same cellular composition as the ones from the tongue but express fewer taste receptor types. State-of-the-art transcriptional regulatory network analysis allowed the identification of specific transcription factors associated to the differentiation of immature progenitors into the three different taste bud cell types. Lineage tracing experiments revealed that esophageal taste buds arise from squamous bipotent progenitor, thus demonstrating that all esophageal progenitors are not unipotent. Our cell resolution characterization of cervical esophagus epithelium will enable a better understanding of esophageal progenitor potency and insights into the mechanisms involved in the development of taste buds.
Some esophageal progenitors can exit the squamous differentiation program to generate taste buds.
INTRODUCTION
The esophagus is a tube-shaped organ that runs from the throat to the stomach. In rodents and humans, its inner part is lined with a stratified squamous epithelium, which is required to sustain the passing of potentially abrasive food. Mouse esophagus epithelium is described as a uniform tissue, which lacks any glands or other appendages. Proliferation is confined to cells in the basal layer, and once differentiation starts, basal cells exit the cell cycle and then migrate to the tissue surface where they are progressively shed (1). Although esophageal basal keratinocytes are phenotypically heterogeneous (2–5), it has been demonstrated that esophageal squamous epithelium is maintained by a single population of unipotent progenitors under homeostatic conditions (6).
On the tongue and soft palate, lineage tracing experiments revealed that a subset of keratinocytes is bipotent and can either undergo a squamous differentiation program and thus participate in the maintenance of the epithelium or give rise to appendages called taste buds (TBs) (7). TBs are the anatomical structures responsible for gustation. Gustation, or the sense of taste, guides organisms to identify and consume nutrients while avoiding potentially toxic materials. In humans, this means identifying basic tastes, which are usually classified as sweet, umami, sour, salty, and bitter, and transmitting the information to the brain (8). TBs are groups of receptor cells, which transduce chemical stimuli into neural signals to mediate the sense of taste. Each TB is composed of three different cell types (glial-like, receptor, and presynaptic) characterized by the expression of specific markers. However, these structures have not been yet fully characterized at single-cell resolution, and little is known about the existence and the organization of TBs outside the oral cavity in mammals.
Lingual TB cells are continually renewed from mitotically active, keratin 14–positive (Krt14+) basal keratinocytes (9). This Krt14+ population generates postmitotic cells that enter TBs (7, 9–12) and constitute immature progenitors, which are referred to as type IV cells (13). Lineage tracing of these type IV cells has unambiguously demonstrated that they are precursors for all taste cell types within TBs (14). However, the molecular mechanisms and the transcriptional regulatory networks involved in the generation of the three types of TB cells from immature progenitors are still partially unknown.
Here, we identified through single-cell RNA sequencing (scRNA-seq) of mouse esophagus epithelium a population of TBs clustered in the cervical segment of the esophagus. Although some articles mention that TBs can be found in the cervical esophagus (15, 16), these structures have never been characterized and their cellular origin is currently unknown. Through a combination of transcriptomic trajectories, pseudotime analysis, single-cell regulatory network inference, and clustering in mouse esophagus and tongue epithelium, we provide a characterization of the transcriptional regulatory networks involved in the generation of each TB cell type. In addition, using lineage tracing in vivo, we unambiguously demonstrate that esophageal TBs arise from the squamous lineage. These data highlight an alternative fate for a subset of esophageal squamous progenitors in vivo, which participate in the maintenance of a pool of esophagus-specific TBs.
RESULTS
Atlas of mouse esophagus cells highlights the presence of rare epithelial appendages
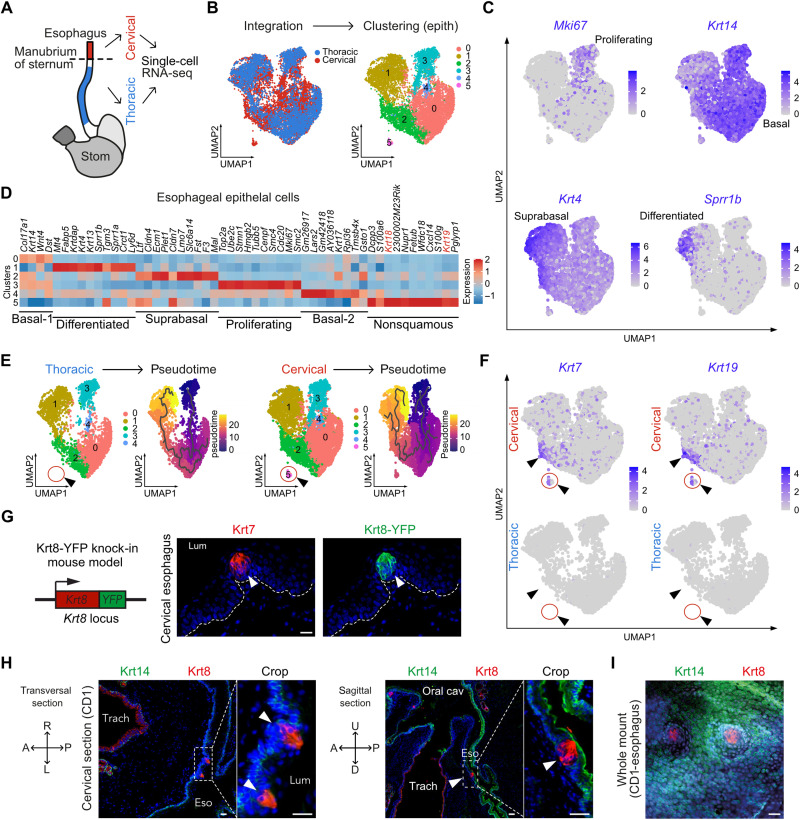
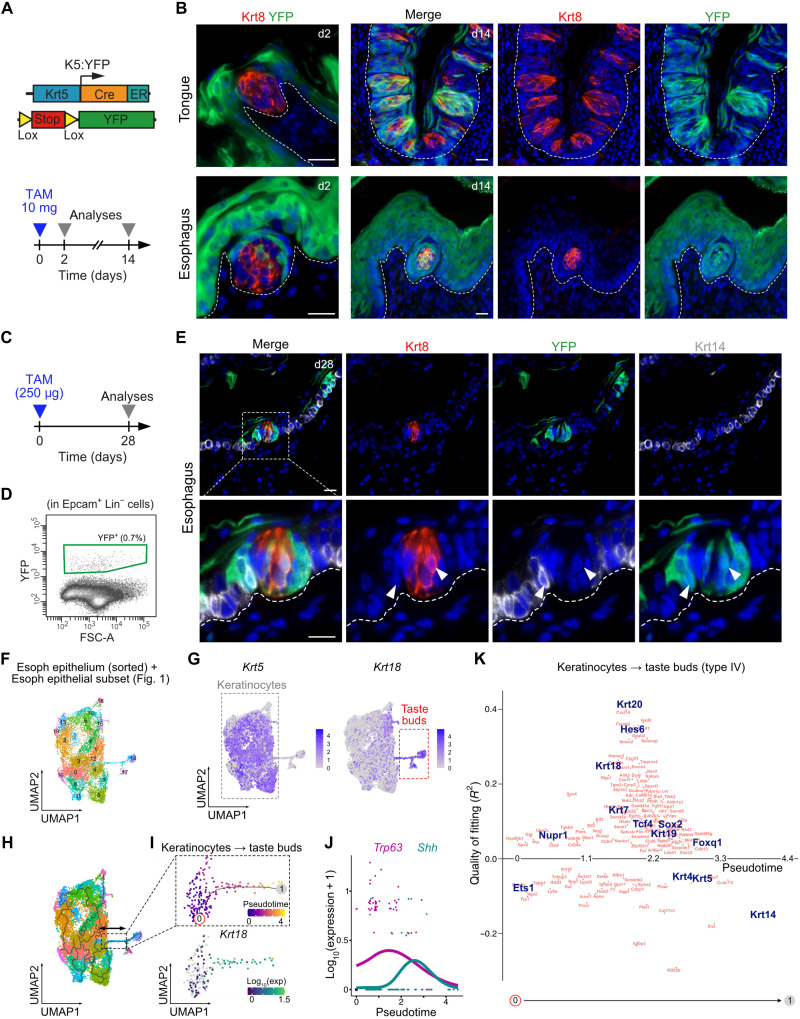
To investigate the heterogeneity of esophagus epithelial cells, we first dissected the cervical and thoracic segments of wild-type mouse esophagus and sequenced about 9000 single living cells from each segment (Fig. 1A and fig. S1A). We subsetted clusters of epithelial cells based on the expression of multiple epithelial (Trp63, Epcam, Krt14, and Krt13) and non-epithelial (Ptprc, Lyve1, Vim, Pdgfra, and Pecam1) markers and then integrated the data (Fig. 1B and fig. S1, A to E). Transcriptomic profiling of epithelial cells allowed the identification of the classical subpopulations associated to the squamous differentiation program: basal proliferating cells (Krt14+ and Mki67+), basal nonproliferating cells (Krt14+ and Mki67−), suprabasal differentiated cells (Krt4+ and Sprr1b−), and terminally differentiated cells (Sprr1b+) (Fig. 1C). Differentially expressed gene analysis highlighted a cell cluster enriched in Krt18 and Krt19, suggesting the presence of nonsquamous epithelial cells (Fig. 1D). By splitting esophageal cells from cervical and thoracic segments, we observed that this cluster is exclusively found in the cervical esophagus epithelium (Fig. 1E). Pseudotime analysis revealed a transcriptomic trajectory compatible with the squamous differentiation program in both segments of the esophagus but did not predict a connection between keratinocytes and the cluster of nonsquamous cells (Fig. 1E). The presence of nonsquamous epithelial cells was unexpected because mouse esophagus is lined with a squamous epithelium devoid of appendages such as submucosal glands. This cluster is composed of cells enriched for markers such as Krt18, Krt19, and Krt7 and expressing low levels of squamous markers Krt14, Krt5, Krt15, Krt13, Krt4, and Trp63 (Fig. 1F and fig. S2, A to C). In the cervical esophagus, some cells outside the nonsquamous cluster express markers such as Krt19 and Krt7 as well, while these markers are completely absent from thoracic esophagus epithelial cells (Fig. 1F). Krt8 and Krt18 are intermediate filament proteins that are typically coexpressed as the primary keratin pair in simple epithelial cells (17). To seek for these nonsquamous cells in situ, we therefore used the Krt8-YFP (yellow fluorescent protein) knock-in mouse model in which all Krt8+ cells are labeled with the fluorescent reporter YFP (18). In these animals, we found clusters of Krt7+/Krt8+ cells in the cervical esophagus (Fig. 1G). In wild-type CD1 mice, we also found small clusters of Krt8+ cells in the most proximal segment of the esophagus on the ventral part, facing the trachea, as shown by transversal and sagittal sections of mouse neck (Fig. 1H). These Krt8+ cells were Krt14−, surrounded by Krt14+/Krt8− squamous cells, and virtually devoid of p63, which is the master regulator of squamous differentiation (Fig. 1H and fig. S2, C and D). Together, these data indicate that the Krt8+/Krt14−/p63− cells observed in the cervical segment of the esophagus are the nonsquamous epithelial cells (Krt7/Krt19 high; Krt14/p63 low) identified by scRNA-seq (fig. S2D). These Krt8+ cells were not scattered but rather clustered in small round structures resembling TBs (Fig. 1I).
Fig. 1. Cervical esophagus epithelium contains a resident population of nonsquamous epithelial cells.
(A) Scheme of mouse foregut. Cervical and thoracic segments of the esophagus were both profiled by scRNA-seq. (B) UMAP on the fusion between cervical and thoracic esophageal epithelial cells. (C) Feature plots of transcripts associated with keratinocyte differentiation expressed in esophagus epithelium integrated data. (D) Heatmap representing the 10 most up-regulated genes in each cluster compared to the others. (E) Pseudotime analysis on the UMAP from (B) split according to the esophageal segment, thoracic (left), or cervical (right). (F) Feature plots of Krt7 and Krt19 transcripts depicted in thoracic and cervical epithelial esophageal cells. (G) Genetic model (Krt8-YFP). Coimmunostaining for Krt7 and YFP in cervical esophagus of Krt8-YFP knock-in mouse. (H) Coimmunostaining for Krt14 and Krt8 in transversal (left) and sagittal (right) sections of esophagus and trachea in the cervical area of a wild-type CD1 mouse. (I) Whole-mount coimmunostaining for Krt14 and Krt8 in the cervical segment of the esophagus. Scale bar, 20 μm.
Profiling and comparison of esophageal and lingual TBs
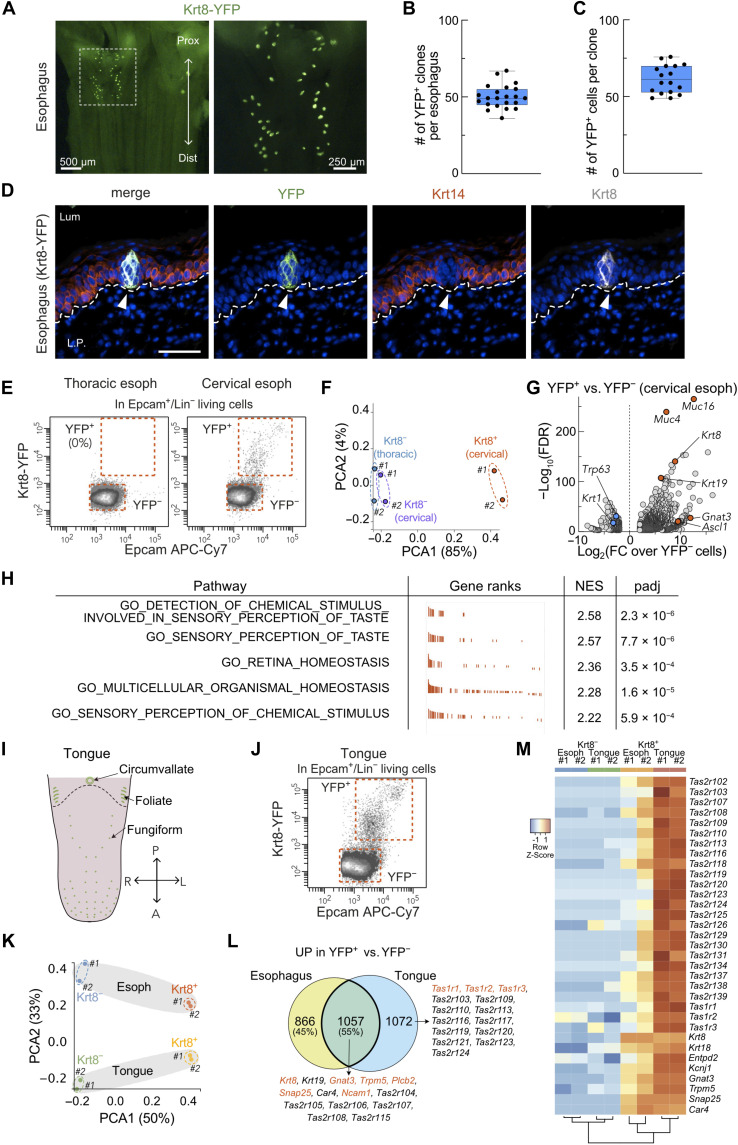
To further characterize the nonsquamous epithelial cells in the esophagus, we used the Krt8-YFP knock-in mouse model that allows the detection of Krt8+ cells in situ. In adult Krt8-YFP mice, clusters of YFP+ cells could be found in the most proximal segment of the esophagus, on the ventral part (Fig. 2A). About 50 clusters of Krt8-YFP+ cells were gathered in adult mouse cervical esophagus (Fig. 2B). Coimmunostaining for YFP, Krt8, and Krt14 showed that these YFP+ histological structures are composed of about 65 elongated Krt8+/Krt14− cells (Fig. 2, C and D). We took advantage of the YFP expression to isolate Krt8+ cells from mouse esophagus using flow cytometry [fluorescence-activated cell sorting (FACS)]. These FACS data confirmed that Krt8+ (YFP+) cells are found in the cervical segment of the esophagus and not in the thoracic one (Fig. 2E). FACS-sorted Krt8+ and Krt8− epithelial cells (YFP+ or YFP− Epcam+/CD45−/CD31−/CD140a−) from the cervical esophagus, as well as Krt8− epithelial cells from the thoracic esophagus, were then profiled by RNA-seq. Principal components analysis (PCA) revealed that Krt8− squamous cells from the cervical and thoracic esophagus were virtually identical, while Krt8+ nonsquamous epithelial cells constituted a clearly separated population (Fig. 2F). Comparison of Krt8+ and Krt8− epithelial cells from the cervical esophagus highlighted strong transcriptomic differences with about 2500 differentially expressed genes between these two populations {abs[log2 fold change (LFC)] > 2; false discovery rate (FDR) < 0.05; Fig. 2G}. Krt8+ cells were characterized by a high Krt19 expression and a low Trp63 expression (Fig. 2G), similar to the cluster of nonsquamous cells identified by scRNA-seq (fig. S2, B and C). We performed unbiased gene set enrichment analysis (GSEA) and found a significant enrichment of transcripts related to “sensory perception of taste,” suggesting that the Krt8+ histological structures found in the cervical segment of the esophagus are TBs (Fig. 2H). Consistent with this notion, many classical markers of TBs and taste receptors (Trpm5, Gnat3, Tas2r104, Tas2r105, Tas2r108, and Tas2r118) are enriched in esophageal Krt8+ cells compared to Krt8− epithelial cells (fig. S3A).
Fig. 2. Presence of a patch of TBs in the cervical esophagus.
(A) Picture of the cervical esophagus mucosa showing endogenous YFP fluorescence in a representative Krt8-YFP mouse. (B) Number of YFP+ clones per esophagus (n = 22). (C) Number of YFP+ cells per clone (n = 17). (D) Coimmunostaining for YFP, Krt14, and Krt8 in cervical esophagus. (E) FACS plot showing Epcam immunostaining and endogenous YFP fluorescence in cells from the thoracic or cervical segment of the esophagus (esoph). (F) PCA on RNA-seq data of FACS-sorted YFP− and YFP+ epithelial cells. (G) Volcano plot showing differentially expressed RNAs between YFP+ and YFP− epithelial cells from cervical esophagus [abs(LFC) > 2; FDR < 0.05]. (H) Table summarizing unbiased GSEA on data shown in (G). Padj < 1 × 10−3 are considered as significant. (I) Scheme representing the papillae on mouse tongue. (J) FACS plot showing Epcam immunostaining and endogenous YFP fluorescence in epithelial cells from the tongue. (K) PCA on RNA-seq data of FACS-sorted YFP− and YFP+ epithelial cells from the cervical esophagus and the tongue. (L) Venn diagram representing the overlap between the genes significantly up-regulated in Krt8-YFP+ cells from cervical esophagus and from tongue compared to Krt8-YFP− cells (LFC > 2; FDR < 0.05). (M) Heatmap representing 25 selected TB markers’ expression in FACS-sorted YFP− and YFP+ epithelial cells from the tongue (Tg) and the cervical esophagus (n = 2 per condition). All data were collected from adult Krt8-YFP mice. Lum, lumen; L.P., lamina propria. Scale bar, 20 μm.
TBs are clustered in three types of papillae on the tongue (Fig. 2I). Foliate and circumvallate papillae are endodermic derivatives, similar to the esophagus mucosa (19). We microdissected the foliate and circumvallate papillae from adult Krt8-YFP mice and profiled FACS-sorted Krt8− squamous epithelial cells and Krt8+ TB cells from the tongue by RNA-seq (Fig. 2J and fig. S3C). PCA analysis of RNA-seq data of Krt8+ and Krt8− cells from the tongue and from the esophagus revealed that the main component segregating cells was associated to Krt8 expression. However, this analysis also showed that both squamous epithelial cells and TBs are different at the transcriptional level, depending on their tissue of origin (Fig. 2K and fig. S3B). Many classical markers of TBs (Entpd1, Trpm5, Gnat3, Plcb2, Snap25, and Car4) were significantly up-regulated in Krt8+ TB cells compared to Krt8− squamous epithelial cells in both the esophagus and the tongue (Fig. 2L). While several taste receptors (Tas2r118, bitter; Pkd2l1, sour; and Sccn1a, salt) were up-regulated (LFC > 1; FDR < 0.05) in Krt8+ cells from both tissues, more receptor types seem to be enriched in the tongue TB cells (Fig. 2, L and M, and fig. S3, A to C). Noticeably, the level of expression of several taste receptors involved in the perception of umami (Tas1r1), sweet (Tas1r2), or bitter (Tas2r120, Tas2r123, Tas2r125) was lower in esophageal TBs compared to their lingual counterpart (Fig. 2M and fig. S3, B and C). In line with this, the expression of the Tas1r2 sweet receptor in esophageal Krt8+ cells appeared to be weaker than in lingual TB cells (fig. S3D). In conclusion, these data suggest that Krt8+ TB cells from the cervical segment of the esophagus share similarities with lingual TBs but might be composed of taste receptor cells with distinct properties, thus raising the question of their cellular composition.
Analysis of the esophageal and lingual TBs at single-cell resolution
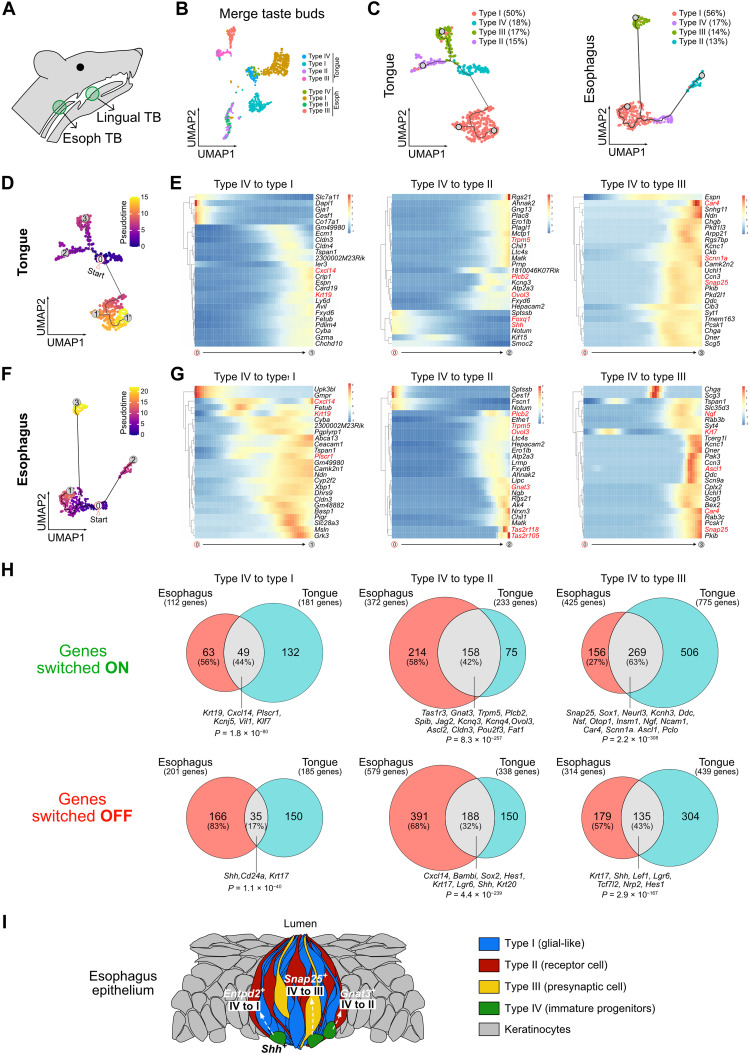
In the oral cavity, each TB is composed of three different cell types: glial-like (type I), receptor cells (type II), and presynaptic (type III). These different TB cell types are characterized by expression of specific markers and arise from immature progenitors referred to as type IV cells (14). To determine whether the differences found between esophageal and lingual TBs may be due to their cellular composition, we analyzed FACS-sorted cervical esophagus and paired tongue epithelial cells by scRNA-seq (Fig. 3A). After normalization of each sample, we merged the datasets, built a clustering, and subsetted the clusters from the TB lineage defined as Krt8+/Krt18+/Krt5−/Krt14− cells (fig. S4, A and B). Then, we built a new clustering on merged TB cells. These scRNA-seq data show different cell clusters with distinct transcriptomic signatures compatible with the TB cell type IV (Shh+), I (Entpd2+), II (Trpm5+), and III (Snap25+) (fig. S4, C and D). Although we observed a good overlap between lingual and esophageal cells from type IV and type II, there was a segregation according to cell origin for type I and type III cell subtypes (Fig. 3B and fig. S4, C and D). Therefore, we split the gustatory cells according to their tissue origin and analyzed them separately (Fig. 3C). The four cell types were in similar proportions in TBs from both origins (Fig. 3C and fig. S4E). In Krt8-YFP mouse, costaining of YFP and Entpd2, Gnat3, or Snap25 confirmed the presence of the three types of TB cells within cervical esophagus (fig. S5, A to C). In addition, receptors for umami (Tas1r2), sweet (Tas1r3), salty (Scnn1a), sour (Pkd2l1), and bitter (Tas2r104, Tas2r105, Tas2r108, Tas2r118, and Tas2r138) tastes were detected in the TB cells from the two origins (fig. S6).
Fig. 3. The cellular composition of TBs is similar in the tongue and the esophagus.
(A) Scheme of mouse head and neck. TBs from the tongue and the cervical esophagus were dissected on the basis of endogenous YFP expression and profiled by scRNA-seq. (B) UMAP of merge data from TB clusters in lingual and cervical esophageal epithelial cells showing segregation according to the cellular origin. (C) UMAP of gustatory epithelial cells from the tongue (left) and the esophagus (right). Four clusters corresponding to TB cell subtypes are present in both tissues. (D) Pseudotime analysis on the UMAP of lingual epithelial cells (C). The origin is set in the type IV cells and labeled “0.” (E) Smoothened expression heatmap of the 25 switching genes that have the highest pseudo R2 along the path going from type IV to type I (glial-like; “1”), type II (receptor; “2”), or type III (presynaptic; “3”) cells in lingual epithelial cells. (F) Same as in (D) for esophageal epithelial cells. (G) Same as in (E) for esophageal epithelial cells. (H) Venn diagram between genes that are switched on (up) or switched off (down) in the tongue and the esophagus along the paths going from type IV to type I, type II, or type III cells. All the gene lists can be found in table S1. (I) Scheme representing a TB surrounded by squamous epithelial cells in esophageal mucosa.
In the oral cavity, it has already been shown that type IV cells can give rise to the three other cell types (14). Pseudotime analysis of our scRNA-seq data highlighted three trajectories starting from the cluster of progenitors (type IV cells) and leading to each TB cell type, both in tongue and esophagus (Fig. 3, C, D, and F, and fig. S7). Analysis of the transcripts that are turned on and off along these trajectories in both tissues suggests a role for transcriptional regulators such as Creb3l2 and Plscr1 in the differentiation into type I cells (Fig. 3, D to H, and fig. S7, A to D); Pou2f3, Ovol3, and Ascl2 in the differentiation into type II cells (Fig. 3, D to H, and fig. S7, E and F); and Ascl1 in the differentiation into type III cells (Fig. 3, D to H, and fig. S7, G and H). By profiling the transcripts that are significantly modified along each path in the tongue and in the esophagus, we found that 44% of the transcripts up-regulated in the transition from progenitors to type I cells, 42% of the transcripts up-regulated in the transition to type II cells, and 63% of transcripts up-regulated in the transition from progenitors to type III cells in esophagus are also up-regulated in the analogous path in the tongue (Fig. 3H). In the same manner, we observed a significant overlap between the transcripts that are down-regulated along the different trajectories (Fig. 3H). In conclusion, although esophageal TBs and their lingual counterparts show some transcriptional differences, they share the same cellular organization and express common markers, including key transcription factors (Fig. 3I).
Characterization of the transcriptional regulatory networks associated to each esophageal TB cell type
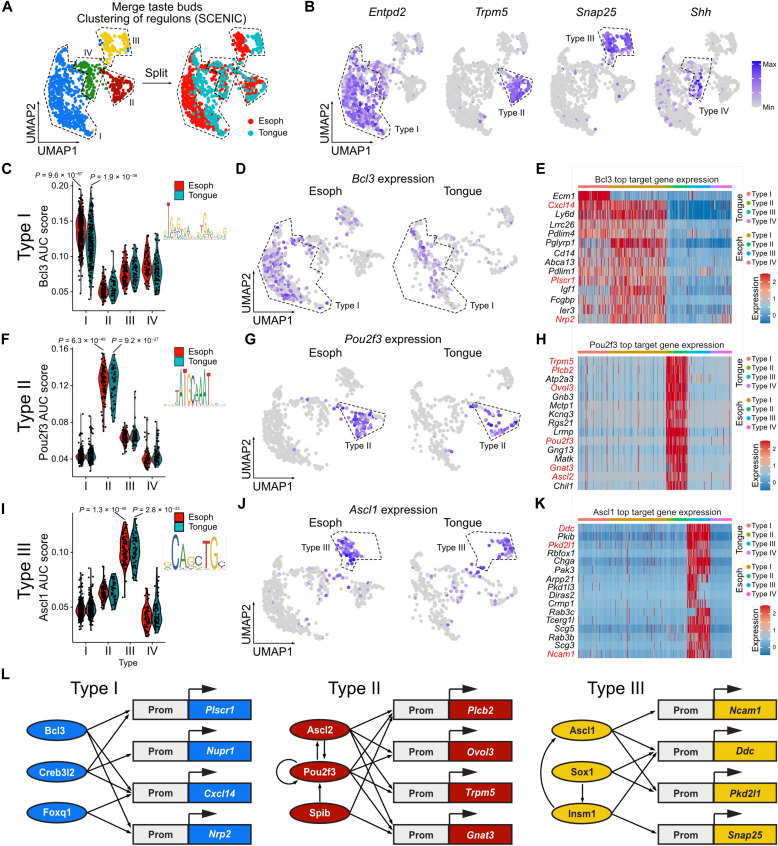
Analysis of the transcripts that are turned on and off along trajectories between immature progenitors and the different TB cell types suggests a role for specific transcription regulators in each TB cell type. To further characterize the different transcriptional regulatory networks associated to each cell fate, we then inferred regulons (i.e., transcription factors and their target genes) and assessed their activity at single-cell level using SCENIC (20, 21). To this end, we analyzed merged scRNA-seq data from paired cervical esophagus and lingual TB cells (Fig. 3, A and B). Clustering of the TB cells based on the regulons highlights four transcriptional profiles that match the pattern of expression of the classical TB cell type markers (Fig. 4, A and B). Although lingual and esophageal cells that belong to type I or type III clusters are not entirely overlaid, they belong to the same clusters, suggesting that they have similar transcriptional profiles. Bcl3 and Creb3l2 regulons are significantly enriched in the type I glial-like cells (Entpd2+) in the tongue and in the esophagus (Fig. 4, C to E, and fig. S8, A and B). These transcription factors have specific motifs and transcriptional networks but share common target genes such as Cxcl14 and Plscr1 (Fig. 4E and fig. S8C). Foxq1 regulon is enriched in the type I cell cluster but is also found in the type IV cells, suggesting that Foxq1 transcriptional network is active in immature progenitors and does not need to be turned off to allow the differentiation into type I cells (fig. S8, A to C). We found Pou2f3 and Ascl2 regulons significantly enriched in the Trpm5+ type II receptor cells (Fig. 4, F and G, and fig. S8, D and E). These transcription factors have different binding motifs but shared common target genes such as Plcb2, Trpm5, Ovol3, Gng13, and Pou2f3, which are known markers of type II cells (Fig. 4H and fig. S8F). We also identified Spib regulon in type II cells that may be important for the differentiation of taste receptor cells by promoting Pou2f3, Plcb2, and Gnat3 expression (fig. S8, D to F).
Fig. 4. Characterization of the transcriptional network associated to TB differentiation in tongue and esophagus.
(A) Clustering based on regulon activity (AUC assay) of gustatory cells from the tongue and the esophagus (merge data). Coloration of the cells according to their origin (right) shows numerous cells from both origins (tongue or esophagus) in all clusters. (B) Feature plots of transcripts associated with TB cell subtypes expressed in gustatory cells from both origins. (C) Violin plot depicting Bcl3 AUC score enriched in gustatory cells from the esophagus (red) and the tongue (turquoise) belonging to type I cluster. (D) Feature plots depicting Bcl3 expression enriched in gustatory cells from the esophagus (left) and the tongue (right) belonging to type I cluster. (E) Heatmap depicting the top target genes of Bcl3 that have the highest expression in lingual cells and esophageal cells belonging to type I cluster. (F) Same as in (C) for Pou2f3 and type II cluster. (G) Same as in (D) for Pou2f3 and type II cluster. (H) Same as in (E) for Pou2f3 and type II cluster. (I) Same as in (C) for Ascl1 and type III cluster. (J) Same as in (D) for Ascl1 and type III cluster. (K) Same as in (E) for Ascl1 and type III cluster. (L) Scheme representing the transcriptional regulatory networks associated to each TB cell subtype. These networks are extrapolated from the regulons identified in the esophagus and tongue scRNA-seq data following SCENIC analysis. Logo of the putative binding motifs for each transcription factor is represented in (C), (F), and (I).
Last, Snap25+ type III presynaptic cells are characterized by Ascl1 and Sox1 associated regulons (Fig. 4, I and J, and fig. S8, G and H). These transcription factors had specific motifs but shared common target genes such as Ddc [l-3,4-dihydroxyphenylalanine (l-DOPA) decarboxylase] and Pkd2l1, which is a candidate receptor for sour taste in type III cells (Fig. 4K and fig. S8I). In addition, we also found Insm1 regulon that is associated with the regulation of Ascl1, Snap25, and Ddc (fig. S8, G to I). Together, these results show that the transcriptional regulatory networks found in the different TB cell types are similar irrespective of their tissue of origin (Fig. 4L).
Challenging the squamous origin of esophageal TBs
Oral TBs originate from bipotent squamous progenitors, while esophageal squamous progenitors are known to be unipotent. To determine whether some esophageal keratinocytes can give rise to TBs, we performed lineage tracing experiments. To this end, we used the Krt5CreER:RosaYFP (K5:YFP) mouse model to map the fate of Krt5+ basal squamous progenitors in esophagus and tongue from adult mice. Taste cells are continually renewed in adult animals, with an average taste cell life span of 10 to 14 days in mouse (22). We therefore analyzed the fate of YFP+ cells 2 and 14 days after tamoxifen (TAM) injection (Fig. 5A). As expected, TBs are initially YFP negative (2 days after TAM injection) because they are devoid of Krt14/Krt5 expression (Fig. 1H and figs. 1D and 9A). However, TBs contain various proportions of YFP+ cells 14 days after TAM administration (Fig. 5B), thus demonstrating that TB cells arise from Krt5+ squamous progenitors both in the tongue and in the esophagus. We then repeated this experiment with a dose of TAM (250ug) that induces the expression of the YFP fluorescent reporter in about 1/150 progenitors and analyzed mouse esophagus 28 days after TAM injection (Fig. 5, C and D). After this delay, it is expected that virtually all the TB cells would have been renewed (22). We observed that at this time point, some YFP+ clones were composed of Krt8+ TB cells and Krt14+ squamous cells like in the tongue (Fig. 5E and fig. S9B). In conclusion, not all esophageal progenitors are unipotent as currently understood (6), and a small pool of these progenitors can give rise to both squamous and TB lineage.
Fig. 5. Esophageal TBs originate from Krt5+ squamous progenitors.
(A) Genetic strategy and experimental design. (B) Coimmunostaining for Krt8 and YFP in the tongue and in the cervical esophagus from K5:YFP mice 2 and 14 days after TAM administration. Scale bar, 20 μm. (C) Experimental design. (D) FACS plot showing the YFP fluorescence in esophageal cells of a K5:YFP mouse 2 days after TAM administration of 250 μg. (E) Coimmunostaining for Krt8, YFP, and Krt14 in the cervical esophagus from K5:YFP mice 28 days after TAM administration of 250μg. Scale bar, 20 μm (F) UMAP on cervical esophageal epithelial cells. Detailed strategy to generate this object is shown in fig. S9. (G) Feature plots of Krt5 and Krt18 transcripts depicted in cervical esophageal epithelial cells. (H) Transcriptomic trajectory analysis of the UMAP. (I) Analysis of the continuum between keratinocytes and the TB lineage, showing the calculation of a pseudotime and the expression of the TB marker Krt18. (J) Trp63 and Shh expression smoothened along the continuum between keratinocytes and TB lineage. (K) Plot of the genes whose expression changes along the continuum between keratinocytes and TB lineage. Switched-on and -off genes are respectively plotted above or below the line. Selected genes (transcriptional regulators and classical markers) have been highlighted in blue.
Clustering of scRNA-seq data from cervical esophagus epithelial cells combined with mapping squamous marker Krt5 and TB marker Krt18 expression clearly shows that these two lineages are segregated on the basis of their transcriptomic profiles (Fig. 5, F and G, and fig. S9, C to H). To identify the molecular mechanisms regulating the differentiation of squamous progenitors into gustatory epithelium in the esophageal mucosa, we then used transcriptomic trajectories (Fig. 5H) and pseudotime analysis (Fig. 5, I and K, and fig. S9J). These data highlight a putative transcriptional trajectory associated with the repression of the squamous differentiation program and the acquisition of the TB markers Krt19, Krt18, and Krt7 (Fig. 5I and fig. S9, I to K). This process is characterized by progressive loss of Trp63 and Krt14 associated with a transitory up-regulation of Shh (Fig. 5J and fig. S9, K and L). The analysis of the transcripts that are turned on and off along this trajectory highlights the up-regulation of several transcription regulators such as Nupr1, Tcf4, Sox2, and Foxq1 (Fig. 5K and fig. S9L). Tcf4 is a transcription factor downstream of the Wnt pathway; hence, we looked at other regulators of this pathway and also observed a strong up-regulation of Lef1 along this trajectory (fig. S9L).
Characterization of the transcriptional regulatory networks associated to the squamous-to-TB transition
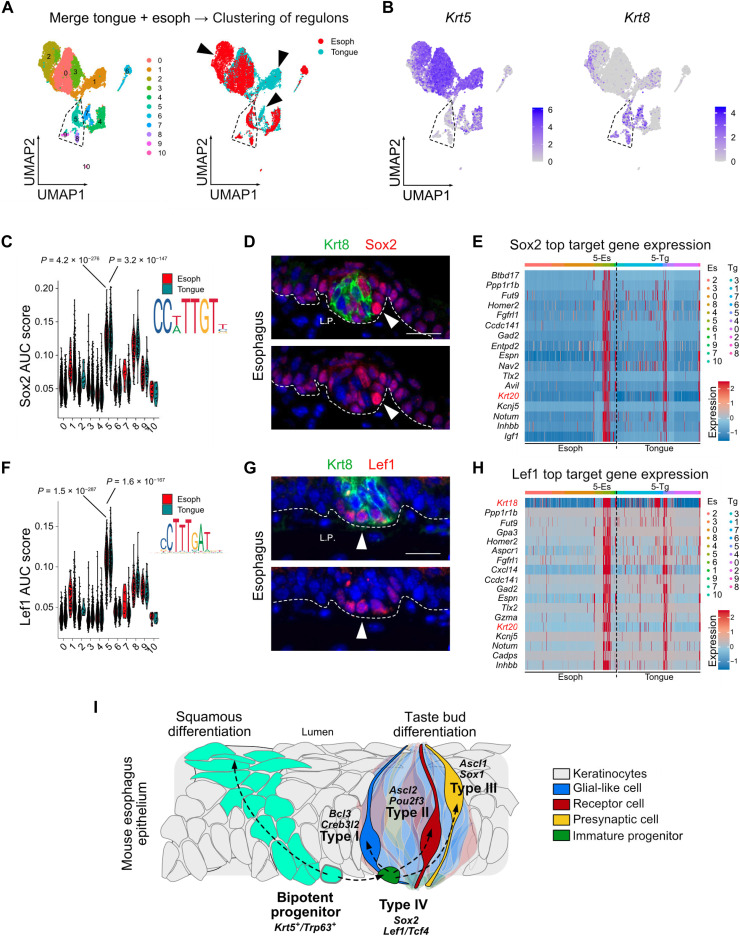
Analysis of the transcripts that are turned on and off along trajectories between keratinocytes and type IV immature progenitors suggests a role for transcription regulators such as Sox2 and Tcf4 (Fig. 5K). To compare the transcriptional regulatory networks associated with TB generation in the esophagus and the tongue, we inferred regulons using SCENIC (20, 21) on merged scRNA-seq data from cervical esophagus and lingual epithelium (Fig. 6A and fig. S10A). Clustering of the cells based on the regulon activity highlighted that while some clusters are tissue specific (#1, #2, and #7), most of the clusters share a similar transcriptional network profile in the tongue and the esophagus (Fig. 6A and fig. S10A). We used the pattern of expression of Krt5, Krt8, and Krt18 to identify TB clusters (Fig. 6B and fig. S10B). Notably, the cluster#7 was enriched in Krt18 and Krt8 but found only in the tongue. This cluster was composed of submucosal glandular cells expressing Muc5b and did not express TB markers; it was therefore not included in our further analyses (fig. S10B). In the cluster#5, which contained a mix of type I and type IV TB cells (fig. S10C), Sox2, Lef1, and Tcf4 regulons are significantly enriched compared to squamous epithelial cells (Fig. 6, C and F, and fig. S10, D to G). These three transcription factors are also specifically and highly expressed in this cluster (Fig. 6, D and G, and fig. S10, E and G). Numerous target genes of these transcription factors are TB markers such as Krt18, Krt20, or Krt19 (Fig. 6, E and H, and fig. S10H), suggesting that these transcription factors could play a role in the transition between squamous and nonsquamous lineage. Notably, while Sox2 and Lef1 regulons are specific of the cluster#5 (Fig. 6, C and F; composed of type IV and type I cells; see fig. S10C), Tcf4 regulon is also enriched in some clusters of squamous cells from the tongue (fig. S10F), indicating that tongue and esophagus squamous epithelia may partially differ, as already suggested by our bulk RNA-seq data (Fig. 2K). Because Lef1, Tcf4, and Tcf7l2 regulons are associated to the type IV immature progenitors, the process of differentiation into TB cells may require the activation of the Wnt signaling pathway. To determine what may promote the activation of this pathway in the gustatory epithelium, we analyzed the non-epithelial cells from the cervical and the thoracic segments of the esophagus by scRNA-seq (fig. S11, A and B). We observed that a subset of Pdgfra+ fibroblasts from the cervical segment of the esophagus was characterized by a low expression of the Wnt inhibitors Sfrp1 and Dkk2, as well as a high expression of Wnt5a compared to the same subset of fibroblasts in the thoracic segment of the esophagus (fig. S11, C to I). Fibroblasts from the thoracic esophagus may therefore create a microenvironment repressing the Wnt pathway. Such a repressive environment may explain at least partially why TBs are restricted to the most proximal part of the esophagus. Together, our results suggest that some esophageal progenitors can either undergo a classical squamous differentiation program or escape their squamous lineage to give rise to TBs in vivo (Fig. 6I).
Fig. 6. Characterization of the transcriptional networks associated to the transition from keratinocytes to the TB lineage.
(A) Clustering based on regulon activity of all lingual epithelial cells merged with all esophageal epithelial cells. Coloration of the cells according to their cellular origin (right) shows numerous clusters composed of cells from both origins (tongue or esophagus). Dashed pentagon surrounds the clusters of TBs. (B) Feature plots of Krt5 and Krt8 lingual epithelial cells merged with esophageal epithelial cells. (C) Feature plot depicting Sox2 AUC scores calculated using SCENIC in epithelial cells from the tongue and the cervical esophagus. (D) Coimmunostaining for Krt8 and Sox2 in a TB from the cervical esophagus. Scale bar, 20 μm. (E) Heatmap depicting the top target genes of Sox2 that have the highest expression in lingual cells and esophageal cells belonging to cluster#5. (F) Same as in (C) for Lef1. (G) Same as in (D) for Lef1. (H) Same as in (E) for Lef1. (I) Model depicting our observations.
DISCUSSION
In the present study, we report the existence of about 50 TBs clustered in a 1-mm2 area on the ventral part of the cervical segment from the mouse esophagus. To the best of our knowledge, although some articles mention that TBs can be found in the cervical esophagus of rodents (15, 16) and humans (23), these histological structures have never been characterized in mammals. Our data show that esophageal TBs share similarities with the TBs from the tongue at the cellular and molecular level. The different TB cell types are characterized by the preferential expression of classical markers such as Entpd2 (type I), Trpm5, Plcb2, Gnat3 (type II), and Snap25 or Car4 (type III). Our scRNA-seq data show that the relative frequency of each subtype is comparable in the tongue and in the esophagus, with type I cells representing about 50% of the population, whereas type II and type III represent about 15%. Several receptors to the different types of tastes such as sweet, umami, salt, sour, and bitter are expressed in type II or type III cells from the esophagus, suggesting that the spectrum of taste detected in the oral cavity and in the esophagus may be similar. However, the low expression of some of these receptors in esophageal TBs suggests that these structures may be less sensitive than their lingual counterparts. Determining the precise sensitivity to the different tastes (sweet, umami, sour, salty, and bitter) will require a complete in vitro pharmacological characterization of FACS-sorted type II and III cells from the tongue and the esophagus.
Lineage tracing in the soft palate and circumvallate and fungiform papillae from the tongue showed that Sonic hedgehog–positive (Shh+) progenitors give rise to all three TB lineages (14). Our pseudotime analysis confirms the existence of a common progenitor in the tongue and shows the same cellular hierarchy in the esophagus. In addition, our data point to the role of specific transcription regulators in the fate of each TB cell type. Type I cells are characterized by the expression of the transcription regulators Creb3l2, Foxq1, and Bcl3, whose functions are currently unknown in TBs. Our data show that the transcription factors Pou2f3, Ascl2, and Spib may be important for the generation and maintenance of type II cells. Consistent with this notion, it has been reported that Pou2f3 is preferentially expressed by type II cells and its genetic invalidation leads to the loss of type II cells and an increase of type III cell number with no significant impact on type I cells (24). Our data suggest that Pou2f3 and Ascl2 could directly regulate the transcription factor Ovol3, whose expression is very specific to type II cells in the tongue and in the esophagus. Ovol3 has already been reported to label type II cells in TB organoids ex vivo (25), yet its role in these cells is unknown. Last, we showed that Ascl1, Sox1, and Insm1 expression may be important for type III cells and could directly regulate the expression of neuronal markers. Notably, the question of Pou2f3 and Ascl1 coexpression in TB cells was recently raised (26). Our scRNA-seq data suggest that they are not coexpressed and that Ascl1 and Ascl2 might be mutually exclusive. Our data thus provide a repertoire of the transcription factors that regulate the differentiation of basal progenitors into the three cell types constituting TBs. In future studies, it will be important to modulate the expression of each of these transcription factors in vivo to better understand their interplay and their role in TB lineage specification. Our characterization of the regulons (transcription factors and their target genes) reveals similarities between the transcriptional networks associated with the different TB cell types in the tongue and in the esophagus.
It has been demonstrated that lingual TBs arise from squamous progenitors in the tongue (7, 9). This process relies on the migration of squamous basal cells into TBs and their exit from the cell cycle to generate ovoid immature type IV TB cells (13). Type IV taste precursor cells then give rise to the three types of TB cells (type I, glial-like cells; type II, taste receptor cells; and type III, presynaptic) (14). This flux of cells is required to replace TBs, which are lost over time. TB average life span is about 10 days (9), although TB cell individual life span can vary from 2 to 24 days (22). Lineage tracing has demonstrated that some Krt14+ basal progenitors from the tongue can participate in the renewal of the TBs and the maintenance of squamous epithelium (7). In the esophagus, Krt5+ basal progenitors have been described as a homogenous population with two equiprobable fates, either dividing to generate two progenitors or undergoing asymmetrical cell division to give rise to differentiated cells (6). This process is balanced under homoeostatic conditions but can be unbalanced upon wound healing (6) or oncogenic mutations (27). Here, we show that some Krt5+ basal progenitors in the cervical segment of the esophagus can have an alternative fate and give rise to TBs. Lineage tracing of single Krt5+ progenitors using low concentration of TAM highlights the existence of clones composed of Krt14+/Krt8− squamous cells and Krt14−/Krt8+ gustatory cells. This observation suggests that some squamous progenitors from the esophagus are bipotent. In addition, because esophageal TBs are composed of both YFP− and YFP+ cells 28 days after induction, it also suggests that TBs are maintained by multiple squamous progenitors as recently shown in the gustatory epithelium from the oral cavity (28). The transition from the squamous lineage to gustatory epithelium in the esophagus is associated with the up-regulation of transcription factors such as Sox2, Lef1, Tcf4, and Tcf7l2, which is in line with what has been previously described in oral TBs (26). Analysis of the regulons suggests that some of these transcription factors may be important for the generation of immature progenitors and still play a role in some differentiated cells.
TB cell renewal is regulated by several signaling pathways including Wnt/β-catenin and Shh (29, 30), which are both required for the generation of the three types of TB cells (31). Our data show that differentiation of squamous progenitors into TBs is associated with the up-regulation of several transcription factors downstream of the Wnt/β-catenin pathway such as Lef1 or Tcf4, both in the esophagus and in the tongue. Our data revealed that the stromal cells from the thoracic and the cervical segment of the esophagus are different. Some subpopulations of fibroblasts from the cervical esophagus are characterized by a lower expression of Wnt inhibitors (Sfrp1 and Dkk2) and a higher expression of Wnt5a compared to their thoracic counterpart. Microenvironment may therefore facilitate the activation of the Wnt pathway and subsequently the development and/or the maintenance of TBs in the cervical segment of the esophagus. Characterization of the esophageal TB population following inhibition of the Wnt/β-catenin pathway in vivo will be required to determine the precise role of this pathway in the gustatory epithelium physiology. Moreover, other cellular components of the microenvironment and other pathways may modulate the maintenance of TBs in vivo. Although we cannot rule out differences between progenitors depending on their tissue of origin, we think that the microenvironment may constitute a major difference between cervical and thoracic esophagus. Hence, a thorough characterization of the non-epithelial populations in the esophagus will be important to identify all the cues regulating the differentiation of squamous progenitors into TBs.
The Hedgehog pathway activator Shh is expressed by type IV immature progenitors, which will give rise to the three other cell types (14). Ectopic activation of the Shh pathway in tongue epithelium induces development of ectopic TBs (29). Our group recently showed that esophageal keratinocytes have the potential to transdifferentiate in vivo. We found that the Shh pathway is activated in the subset of squamous keratinocytes that have the competence to give rise to intestinal-like metaplasia in vivo (32). In addition, activation of the Shh pathway in esophageal epithelium induces a squamous-to-columnar transition in a subset of basal progenitors in vivo. This process relies on a step of dedifferentiation and then a Sox9-dependent differentiation program leading to appearance of intestinal markers (32). It will be important to determine whether basal progenitors are intrinsically different in the thoracic and the cervical segment of the esophagus, if their differences depend on their localization in distinct microenvironments, or both. Although preliminary, our data do not show clear differences between progenitors depending on their localization but rather a different microenvironment, which raises questions on the composition of the mesenchyme and its influence on cell plasticity and differentiation programs. It will be also interesting to compare the mechanisms involved in the squamous-to-columnar transition induced by Shh in the thoracic esophagus with the transition leading to the development of TBs in the cervical esophagus.
Under physiological conditions, activation of the Shh pathway in TBs is stimulated by afferent nerve fibers, which secrete the Shh ligand (33, 34). This central role of the Shh pathway is consistent with the loss of taste sensation in patients under chemotherapy targeting the Hedgehog pathway (34, 35). Innervation is tightly correlated with the presence of all differentiated taste cell types, as well as of Shh+ type IV cells (33). Gustatory innervation is required for TB maintenance and presumably restricts where TBs form in the tongue surface (36). These results show that a local production of Shh creates a permissive niche for TB development. It will be important to determine whether esophageal TBs also dwell in permissive areas created by afferent fibers and whether the production of Shh by these neurons would be required for their maintenance.
TBs in the oral cavity are innervated by three different cranial nerves: VII (facial), IX (glossopharyngeal), and X (vagus). These three nerves can transmit information to the gustatory cortex (37). The vagal afferent innervation is very dense in the cervical segment of the esophagus (38). Physiological studies showed that the vagal esophageal motor reflexes including swallowing, reflex regulation of upper esophageal sphincter, and vagal respiratory reflexes are modulated by the acid in the esophagus (39). This modulation is at least partially explained by the presence of vagal afferent nerve fibers in the esophageal mucosa (40). We use taste to select nutritious foods, which are typically sweet, salty, and/or savory. On the opposite, we reject bitter compounds, which are often identified as toxic (41). Recent studies have shed light on taste receptors outside the oral cavity, notably in the gastrointestinal tract (42) and the laryngeal epithelium (43), suggesting that these receptors would not be limited to the detection of each type of taste and their projection to the brain. Because of their specific location, the TBs from the esophagus are exposed to taste molecules once they have already been ingested. TBs from the esophagus thus probably play a distinct role from their oral counterpart. These TBs may stimulate vagal esophageal motor reflexes including swallowing or induce nausea. While mice and other rodents are incapable of vomiting, some stimuli that evoke nausea and vomiting in humans cause behavioral aversion to sensory cues (44). Bitter taste is innately aversive, and many poisons taste bitter to humans. Esophageal TBs may therefore constitute gateway receptors, which would inform that a potentially toxic substance has entered the body. Addressing whether esophageal TBs play a role in esophageal motor reflexes or whether they transmit information to the gustatory cortex will require very specific functional assays such as calcium imaging and physiological measurements in vivo as illustrated in a recent study (43). Investigating whether their function is conserved in humans will be critical as well.
In conclusion, our work reveals the presence of a patch of TBs in murine esophagus and provides elements to understand how esophageal squamous progenitors give rise to these receptors. In addition, our results highlight multiple gene regulatory networks that are associated to each TB cell type in the tongue and in the esophagus. Future directions should focus on the physiological role of these structures.
MATERIALS AND METHODS
Experimental model and subject details
All the experiments were approved by the ethical committee from the university and conform with regulatory standards (LA1230406, project 666N). Mice were bred and maintained under pathogen-free conditions in a certified animal facility in accordance with the European guidelines. Adult (8-week-old) wild-type CD1 mice or Krt8-YFP knock-in mice (18) were used for sequencing and histology. Krt5-CreERT2 knock-in mice (the Jackson Laboratory, stock no. 029155; RRID:IMSR_JAX:029155) were crossed with R26R-EYFP (the Jackson Laboratory, stock no. 006148; RRID:IMSR_JAX:006148) to generate Krt5-CreERT2:R26R-EYFP (K5:RYFP) mice. For lineage tracing experiments, 10 or 0.25 mg of TAM resuspended in sunflower oil was injected intraperitoneally to 8-week-old K5:RYFP mice, and the expression of the YFP was analyzed in esophagi up to 4 weeks after TAM administration. In all the experiments, littermates of the same sex were randomly assigned to experimental groups.
Method details
For all experiments presented in this study, sample size was large enough to measure the effect size. No randomization and no blinding were performed in this study.
Histology and immunostaining
For the staining on frozen sections, tissues were harvested, directly embedded in O.C.T. (Tissue Tek) and flash-frozen for cryopreservation. For the following staining: Krt14, Krt8-YFP, Gnat3, Lef1, and Sox2, tissues were prefixed in 4% formaldehyde for 2 hours at room temperature, washed in phosphate-buffered saline (PBS), incubated overnight in 30% sucrose in PBS at 4°C, embedded in O.C.T. (Tissue Tek), and flash-frozen for cryopreservation. Samples were sectioned at 6μm using a M1860 cryostat (Leica Microsystems GmbH). Nonspecific antibody binding was blocked with 5% horse serum, 1% bovine serum albumin (BSA), and 0.2% Triton X-100 for 1 hour at room temperature. Primary antibodies were incubated overnight at 4°C in the same blocking buffer. Sections were rinsed three times in PBS and incubated with secondary antibodies for 1 hour at room temperature. Nuclei were stained with Hoechst (4 mM). Slides were mounted using Glycergel (Dako) supplemented with 2.5% DABCO (Sigma-Aldrich).
Cervical esophageal whole mount was prepared by cutting the cervical part of the esophagus, which is delimited by the cricoid cartilage proximally and the manubrium of the sternum distally. Tissue was incubated for 1 hour in 5 mM EDTA at 37°C on a rocking plate. Esophageal epithelium was separated from underlying tissue using forceps and then opened longitudinally with scissors. Tissue was fixed in 4% formaldehyde for 30 min at room temperature then rinsed in PBS. For staining, whole mount was blocked for 1 hour with 5% horse serum, 1% BSA, and 0.5% Triton X-100 in a 12-well plate. Whole mount was then stained with primary antibodies in blocking buffer (0.2% Triton X-100) overnight at 4°C. The following dilution has been used for the primary antibodies: anti-Krt14 (polyclonal chicken; 1:1000; BioLegend), anti-Krt8 (rat monoclonal; 1:1000; DSHB, RRID:AB_531826). Then, whole mount was incubated with secondary antibodies and Hoechst (4 mM) in blocking buffer (0.2% Triton X-100) overnight at 4°C using classical dilution of the secondary antibodies described below. Following each antibody incubation, whole mount was washed four times with 0.2% Tween 20 in PBS for 2 hours in total. Slides were mounted using Glycergel (Dako) supplemented with 2.5% Dabco (Sigma-Aldrich).
Antibodies
The following primary antibodies were used: anti-Krt14 (polyclonal chicken; 1:10,000; BioLegend), anti-GFP (green fluorescent protein; polyclonal rabbit; 1:1000; Abcam, RRID:AB_305564), anti-GFP (polyclonal goat; 1:1000; Abcam, RRID AB_305643), anti-GFP (polyclonal chicken; 1:500; Invitrogen, RRID:AB_2534023), anti-p63 (rabbit monoclonal; 1:1000; Abcam, RRID:AB_10971840), anti-Krt8 (rat monoclonal; 1:250; DSHB, RRID:AB_531826), anti-Entpd2 (rabbit; 1:4000; http://ectonucleotidases-ab.com, mN2-36Li6), anti-Gnat3 (goat polyclonal; 1:500; Novus Biologicals, NBP1-20926), anti-Snap25 (rabbit polyclonal; 1:500; Sigma-Aldrich, S9684), anti-Tas1r2 (rabbit; 1:500; Invitrogen, PA5-99935), anti-Lef1 (rabbit monoclonal; 1:100; Thermo Fisher Scientific, MA5-14966), anti-Sox2 (rabbit monoclonal; 1:200; Abcam ab92494).
The following secondary antibodies were used: anti-rabbit, anti-rat, anti-chicken, anti-goat conjugated to Alexa Fluor 488 (1:500; Jackson ImmunoResearch), to rhodamine Red-X (1:500; Jackson ImmunoResearch), or to Cy5 (1:1000; Jackson ImmunoResearch).
Image acquisition
Imaging was performed on a Zeiss Axio Imager M2 fluorescence microscope with a Zeiss Axiocam 503 mono camera and a Zeiss microscope Axio Zoom V16 with Axiocam MRN camera for immunofluorescence microscopy using Zen Blue (Zeiss) software. Brightness, contrast, and picture size were adjusted using Zen Blue (Zeiss). For Gnat3 staining (fig. S5), Z-stack combined with maximum intensity projection was used.
Quantification
For the number of YFP+ (TBs) per esophagus, native YFP fluorescence was used from Krt8-YFP knock-in mice to spot the YFP+ clone in the cervical part of 22 esophagi. For the number of cells per YFP clone (TB), coimmunostaining for YFP and Krt14 or Krt8 were used and 17 clones from three different animals were counted.
Tissue digestion
Esophagi were dissected, minced, and digested in collagenase I (2 mg/ml; A&E Scientific) for 1 hour and 30 min. Collagenase I activity was blocked by the addition of EDTA (5 mM). After 20 min of incubation, trypsin (0.125%) was added for 15 min, and then, the cells were rinsed in PBS supplemented with 2% fetal bovine serum (FBS). All incubations have been done on a rocking plate at 37°C.
Tongues from K8-YFP mice were dissected, and the circumvallate and foliate papillae were microdissected under a fluorescence binocular microscope using endogenous YFP fluorescence. Tissues were then washed in Hanks’ balanced salt solution with 10 mM Hepes and digested in collagenase I (2 mg/ml; A&E Scientific) with 20 mM Hepes for 1 hour and 30 min. Collagenase I activity was blocked by the addition of EDTA (5 mM). After 15 min of incubation, trypsin (0.125%) was added for 15 min, and then, the cells were rinsed in PBS supplemented with 2% FBS. All incubations have been done on a rocking plate at 37°C.
TBs from the esophagus of K8-YFP mice were dissected and dissociated using the same protocol. For all the tissues, after rinsing in PBS supplemented with 2% FBS, cells were filtered through 70-μm cell strainers (BD) to ensure the elimination of undigested tissue.
FACS isolation
Immunostaining was performed on single-cell suspension using phycoerythrin (PE)–conjugated anti-CD45 (1:500; BioLegend), PE-conjugated anti CD31 (1:500; BioLegend), PE-conjugated anti-CD140a (1:500, BioLegend), and Allophycocyanin-Cyanine7 (APC-Cy7)–conjugated anti-EpCam (clone G8.8; 1:250; BioLegend) for 45 min at 4°C on a rocking plate. Living epithelial cells were selected by forward scatter, side scatter, doublets discrimination, and Hoechst dye exclusion. EpCam+/Lin− cells were selected on the basis of the expression of EpCam and the exclusion of CD45, CD31, and CD140a (Lin−). For Krt8-YFP mice, YFP+ and YFP− cells were selected within the EpCam+/Lin− population. FACS analysis was performed using FACSAria III and FACSDiva software (BD Biosciences).
RNA extraction
Sorted cells were collected into TRK lysis buffer (Omega Bio-tek), and RNA was extracted using E.Z.N.A Total RNA Kit (Omega Bio-tek) according to the manufacturer’s recommendations with deoxyribonuclease I digestion protocol on column (Omega Bio-tek).
RNA-seq and analysis of bulk samples
RNA quality was checked using a Bioanalyzer 2100 (Agilent Technologies). Indexed cDNA libraries were obtained using the Ovation SoLo RNA-Seq System (NuGEN) following the manufacturer’s recommendations. The multiplexed libraries were loaded on a NovaSeq 6000 (Illumina) using an S2 flow cell, and sequences were produced using a 200-cycle kit. Paired-end reads were mapped against the mouse reference genome GRCm38 using STAR software (version 2.5.3a) to generate read alignments for each sample. Annotations Mus_musculus.GRCm38.90.gtf were obtained from ftp.Ensembl.org. After transcript assembling, gene level counts were obtained using HTSeq (45). Total raw counts were loaded on degust 4.1.1 (46). All analyses were performed using EdgeR, TMM normalization, and “Min gene read count” set at 10.
We sequenced Krt8-YFP− and Krt8-YFP+ cells from cervical esophagus and tongue from Krt8-YFP knock-in mice, as well as Krt8-YFP− cells from thoracic esophagus. The Krt8-YFP cells were first compared to Krt8-YFP− cells from the same cervical esophagi (n = 2). Then, Krt8-YFP cells from the cervical esophagus and the tongue were compared to each other.
Heatmap was generated using “heatmap.2” function and represent values in logCPM scaled by row for selected genes typically found in lingual TBs, for thoracic and cervical esophagus YFP− cells and thoracic and cervical esophagus YFP+ cells. All volcano plots represent results of RNA-seq as the statistical significance versus the magnitude of fold change (FC) and were generated using the package “EnhancedVolcano” (47) from Bioconductor in R version 3.6.3. Multidimensional scaling (MDS) plots have been generated using degust 4.1.1 (46).
GSEA analysis
GSEA was performed using the fgsea package (Korotkevich and Sukhov) in R version 3.6.3. Unbiased gene ontology analyze was performed with the C5 collection adapted for mouse that contains gene sets annotated by Gene Ontology terms that has been downloaded on http://bioinf.wehi.edu.au/software/MSigDB/. The function “fgseaMultilevel” has been used with ranked fold change values corresponding to Krt8-YFP+ cells over Krt8-YFP- cells from cervical esophagus [abs(LFC) > 2; FDR < 0.05].
scRNA-seq and analysis
Single-cell sequencing preparation was carried out as follows: Sorted cells were collected in 1 ml of PBS + 0.04% BSA at 4°C. Samples were loaded on a chromium chip targeting a recovery between 2500 and 7000 cells. Samples were then processed using the Chromium Single Cell 3′ Reagent Kits v3 (10x Genomics) following the manufacturer’s recommendations. The multiplexed libraries were loaded on a NovaSeq 6000 (Illumina) using a S1 flow cell, and paired-end sequences were produced using a 100-cycle kit (Read1, 28 cycles; i7 Index, 8 cycles; i5 Index, 0 cycles; and Read2, 87 cycles). Cell Ranger version 3.1.0 pipeline with the RNA-seq aligner STAR were used to generate output files aligned on mm10 reference genome. Package Seurat v4.2 from Bioconductor (48) was used in R to perform all the analyses. The analyses followed recommendations from Satija Lab (https://satijalab.org).
The datasets were converted into Seurat objects. For each sample, thresholds for “nCount_RNA” were defined to remove potential empty or low-quality droplets and multiplets. Between 9 and 13% of the cells have been removed for each sample. The data were normalized using “SCTransform” that outperforms traditional global scaling normalization methods derived from bulk RNA-seq (49). To avoid the influence of mitochondrial reads in the analyses, we used “vars.to.regress=percent.mt” with “percent.mt” calculated using “PercentageFeaturesSet.”
For each Seurat object, the following steps have been run: PCAs were calculated using “RunPCA” (with npcs = 50), and Uniform Manifold Approximation and Projection (UMAP) analyses have been done using “RunUMAP” (with dims = 1:20). Distances between cells were defined using “FindNeighbors” (with dims = 1:20). The cells were grouped using “FindClusters” with resolution depending on the datasets. These steps were repeated for the clustering of each Seurat object newly generated (after subset, merge, or integration). Last, for visualization and differential expression (DE) analysis, RNA counts were normalized using “NormalizeData.”
In this study, five samples were sequenced at single-cell level:
1) Sample#1 (Tho Tot esophagus). For all living cells of thoracic esophagus, 300,000 living cells were sorted from one wild-type mouse. A total of 10,672 of these cells were sequenced. The thresholds were defined as follows: nCount_RNA > 1000 and nCount_RNA < 50,000. Ten percent of the cells were eliminated, and a total of 9707 cells were used for the analysis. This sample has been used in Fig. 1 and related supplementary figures and fig. S11.
2) Sample#2 (Up Tot esophagus). For all living cells of cervical esophagus, 100,000 living cells were sorted from the cervical esophagus of 15 wild-type mice. A total of 9102 of these cells were sequenced. The thresholds were defined as follows: nCount_RNA > 750 and nCount_RNA < 15,000. Twelve percent of the cells were eliminated, and a total of 8000 cells were used for the analysis. This sample has been used in Figs. 1 and 5 and related supplementary figures and fig. S11.
3) Sample#3 (Up Epith esophagus). For epithelial cells of cervical esophagus, 20,000 EpCam+ cells were sorted from the cervical esophagus of the same 15 wild-type mice used for sample#2 (Up Tot), and 5163 of these cells were sequenced on the same chip. The thresholds were defined as follows: nCount_RNA > 750 and nCount_RNA < 30,000. Thirteen percent of the cells were eliminated, and a total of 4491 cells were used for the analysis. As some non-epithelial cells were detected despite the cell sorting, a filter to eliminate vimentin-positive cells has been used (Vim < 0.0001). This sample has been used in Fig. 5 and related supplementary figures.
4) Sample#4 (Up Epith esophagus paired with tongue). A total of 35,000 EpCam+ cells enriched with 15,000 YFP+ cells were sorted from cervical esophagus of 20 Krt8-YFP mice, and 6767 of these cells were sequenced. The thresholds were defined as follows: nCount_RNA > 750 and nCount_RNA < 25,000. Eleven percent of the cells were eliminated, and a total of 6030 cells were used for the analysis. This sample has been used in Figs. 3, 4, and 6 and related supplementary figures.
5) Sample#5 (Tg Epith paired with esophagus). A total of 35,000 EpCam+ cells enriched with 15,000 YFP+ cells were sorted from foliate and circumvallate papillae of 20 tongues from the same Krt8-YFP mice that have been used for sample#4 (Up Epith esophagus paired with tongue), and 5952 of these cells were sequenced on the same chip. The thresholds were defined as follows: nCount_RNA > 750 and nCount_RNA < 30,000. Nine percent of the cells were eliminated, and a total of 5455 cells were used for the analysis. This sample has been used in Figs. 3, 4, and 6 and related supplementary figures.
To merge the data, after SCTransform normalization, samples were merged using “merge” function (with parameter merge.data = TRUE). Variable features were reattributed using the command “SelectIntegrationFeatures” on the original objects.
To integrate the data, SCTransform normalization was applied to a list containing the Seurat objects that we wanted to integrate. Features were defined using “SelectIntegrationFeatures” on the same list; then, anchors were defined with “FindIntegrationAnchors” before integration of the datasets using the function “IntegrateData.”
Differentially expressed markers for each group comparing to remaining ones were identified with Wilcoxon test using “FindMarkers” and “adjusted P value <0.05”. Gene expressions were illustrated using heatmaps with the function “DoHeatmap” combined or not with “AverageExpression” function, feature plots with the function “FeaturePlot,” or violin plots with the function “VlnPlot.” In the violin plots, Wilcoxon tests were used to calculate adjusted P values (Padj) for clusters of interest.
To perform pseudotime analysis, Seurat objects were manually converted to be compatible with Monocle3 (50). Starting points were chosen on the basis of biology (proliferative squamous progenitors for the esophagus or type IV immature progenitors for the TBs). Following Monocle3 analysis, cells belonging to a specific trajectory were selected using “choose_graph_segments” and reordered.
Analysis of the genes switching on and off along the pseudotime was performed using the GeneSwitches package (51). The GeneSwitches package first binarizes the gene expression data into 1 (on) or 0 (off) state. To do so, for each trajectory, a fast binarization threshold is defined. To identify the global threshold, we plot a histogram of expression of all the genes in all cells and look for a break between the zero and expressed distributions. Last, a logistic regression is applied to model the binary states (on or off) of gene expression. A McFadden’s pseudo R2 is calculated, and we can therefore identify the best fitted genes that have a high McFadden’s pseudo R2. The switching pseudotime point is determined by the time at which the fitted line crosses the probability threshold 0.5. Random downsampling of zero expressions (downsample = TRUE) is used to rescue the prediction of switching time for genes with high zero inflation. We can therefore highlight genes up-regulated or down-regulated along a trajectory. This specific package allows the comparison of genes switched on and off between two chosen trajectories from different Seurat objects. For each GeneSwitches analysis, we defined thresholds to represent the transcripts with the highest variation along pseudotime. Moreover, all the results with default threshold FDR < 0.05 and pseudo R2 > 0.03 can be found in the table S1. To illustrate gene expression dynamics along the pseudotime, we used the package tradeseq (52). To use tradeseq downstream of Monocle3, we extracted pseudotimes and cell weights and used them as input of the “fitGAM” function. We generated heatmaps after smoothing the expression of the genes along the time axis using the function “predictSmooth” with “nPoints” set to 50 from the tradeseq package and “pheatmap” from the pheatmap package. We represented the expression of specific chosen genes along the selected trajectory using “plotSmoothers” function from tradeseq and “plot_cells” from Monocle3.
To analyze the gene regulatory networks associated with TB development, we used SCENIC tool (20, 21). We first merged sample#4 (Up Epith paired) and sample#5 (Tg Epith paired) (fig. S4) and extracted the expression matrix in “.tsv.” PySCENIC has been installed with Conda and Python 3.7. The sets of genes coexpressed with transcription factors have been identified using “grn” on the extracted expression matrix and the list of transcription factors “mm_mgi_tfs.txt” downloaded from https://github.com/aertslab/pySCENIC/tree/master/resources. The selection of potential direct-binding targets (regulons) based on DNA-motif analysis have been done using “ctx” with the two cistarget databases available for mouse genome mm10 version [500 base pairs (bp) up and 100 bp down transcription start sites (TSS), as well as 10 kb up and down TSS] and the transcription factor annotation motifs v9 collection from mouse, downloaded from https://resources.aertslab.org/cistarget/. The network activity in each individual cell has been calculated using “aucell.” Because it is a prediction based on the coexpression, the results are quite stochastic; therefore, each step has been repeated 11 times. The 12 Area Under the Curve (AUC) matrix obtained have been fused and averaged using Python. Regulon files have been concatenated and then fused using Python. Then, AUC values have been reallocated to each cell and stored in AUC assay of the original merged Seurat object in R. Consequently, clustering can be calculated on the basis of AUC assay.
DE analysis using AUC assay allowed us to highlight specific regulons for chosen clusters. For each highlighted regulon, the expression of the transcription factor has been assessed with the generation of violin and feature plots with the functions described previously; we added the parameter “split.by” to show the values in the cells from the tongue and esophagus separately. Adjusted P values were calculated for one cluster compared to all the others from the same origin (esophagus or tongue) using Wilcoxon tests. The best regulons for each cluster have been selected on the basis of the DE results, the expression of the transcription factor, and the AUC scores. All target genes of each regulon have been retrieved from SCENIC analysis. We used heatmaps to depict the expression of the 10 target genes that had the highest expression in the esophageal cluster of interest and the 10 target genes that had the highest expression in lingual cluster of interest. Notably, the function “DoHeatmap” automatically eliminates duplicates; therefore, sometimes, less than 20 genes are depicted in the heatmaps. Logo motifs have been retrieved from the JASPAR database, and Bcl3 logo was retrieved from the ISMARA database. By default, clustering was calculated on the basis of normalized transcript expression (SCT assay or integrated assay). When clustering was calculated on the basis of regulon activity (AUC assay), it is clearly indicated in the legend of the figure.
In Fig. 1 and related supplementary figures, we compared epithelial cells from thoracic and cervical esophagus. We used the two samples in which all living cells have been sequenced. After normalization of sample#1 (Tho Tot) and sample#2 (Up Tot), we defined the resolution of “FindCluster” to 0.45 and selected epithelial cells using classical markers of epithelial and non-epithelial cells (fig. S1). For sample#1, our DE analysis showed that the 10 most expressed genes in cluster#2 compared to others were mitochondrial genes. This could reflect a high fraction of apoptotic or lysing cells. Therefore, this cluster was excluded from further analysis. We merged epithelial cells selected from sample#1 and sample#2 and observed a major batch effect due to coverage differences (fig. S1, B and C). Moreover, these samples were not sequenced on the same chip; therefore, it is recommended to use integration to compare the datasets. After integration, we removed one cluster that was enriched for fibroblast markers in both samples. We set the resolution of “FindClusters” to 0.2 and observed that the quality of the cells found in cluster#5 was similar to the quality of cells from other clusters in the same sample (fig. S1C).
In Fig. 3 and related supplementary figures, we compared lingual TB cells to esophageal TB cells. These cells were isolated from the same mice and sequenced on the same chip. After normalization, we merged sample#4 (Up Epith paired) and sample#5 (Tg Epith paired). Although we observed some differences associated to the tissue of origin, TB cells from the tongue and the esophagus were clustered together (fig. S4, A and B). TB clusters were identified on the basis of the expression of squamous markers (Krt5 and Krt14) and TB markers (Krt8 and Krt18) (fig. S4B). After the clustering of the subset, we eliminated a cluster enriched in keratinocyte markers (such as Trp63, Krt14, and Krt13) and set the resolution of “FindClusters” to 0.2. As we noticed segregation associated to cell origin for some clusters (Fig. 3B), we pursued the analysis separately for the tongue and the esophagus. After the split according to the tissue of origin, we set the resolution of “FindClusters” to 0.3 for both tissues and identified the different TB cell subtypes in the tongue and the esophagus. Trajectory analysis with monocle3 highlighted different paths ending in the three different TB cell subtypes in the tongue and the esophagus (Fig. 3C and fig. S4E). We defined the starts of the pseudotimes for the tongue and the esophagus in type IV (Shh+) cells (Fig. 3, D and F) because these cells are known to be the common progenitors of the different TB cell types (14). Using the GeneSwitches package, we identified the 25 best-fitted genes (with the highest McFadden’s pseudo R2) for each path leading to the different cell subtypes. We represented their expression smoothened along the time axis with tradeseq and pheatmap (Fig. 3, E and G). We used the GeneSwitches package to compare the switched genes along the different paths in the tongue and the esophagus. We represented the 60 common switched genes that had the best pseudo R2 along the scaled pseudotimes of the tongue (turquoise) and the esophagus (red) (fig. S7). In the tongue, two predicted paths from type IV to type I cells were identified (Fig. 3D). Around 350 genes were switched along the path going to end 1, while more than 1000 genes were switched along the path going to end 1′. Because around 300 genes were switched along the path going to end 1 (type I) in the esophagus, we decided to show the path going to end 1 in the main figure (similar number of genes modified). We show the transcripts modified along the alternative path (end 1′) in fig. S5. Notably, the 25 switched genes that had the best pseudo R2 along the scaled pseudotimes were the same in the two predicted paths. We compared the switching genes between the tongue and the esophagus along the different paths and represent them as Venn diagrams. For better visualization, we used order = TRUE to generate Tas receptor feature plots in fig. S6.
In Fig. 4 and related supplementary figures, we compared gene regulatory networks associated with TB development in esophagus and tongue using SCENIC. We calculated a new clustering based on AUC assay on the Seurat object of Fig. 3B (merge of TB cell subtypes from the esophagus and the tongue). Testing different resolutions for “FindClusters,” we observed that the cells were not segregated depending on their origin. Therefore, we set the resolution of “FindClusters” to 0.2 and analyzed the merged object. We performed DE analysis using AUC assay to highlight specific regulons for each TB cell subtypes and followed the method described before.
In Fig. 5 and related supplementary figures, to highlight a link in silico between keratinocytes and TBs, we studied the trajectory between the keratinocyte lineage and the TB lineage at single-cell level. To increase the resolution of our analysis and enrich for rare populations, we combined sample#2 (Up Tot esophagus) and sample#3 (Up Epith esophagus). The merge of the two samples showed a major segregation according to the sample (fig. S9G). Because these epithelial cells were isolated from the same mice, it is expected that cells should be similar in the two samples. Segregation according to the sample was therefore consistent with a batch effect. Hence, we integrated the two samples and eliminated non-epithelial cells based on the expression of selected markers from epithelial and non-epithelial lineages. The clustering of epithelial cells enriched in TB cells revealed a continuum between keratinocytes and the gustatory epithelium. This continuum was expected because our lineage tracing experiments (Fig. 5F) clearly demonstrated that TB cells arise from Krt5+ keratinocytes. We tested different resolutions. In this analysis, we focused on the continuum and set the resolution of “FindClusters” to 1. Although a resolution of 1 may not be optimal for the squamous lineage (overclustering), this resolution was required for the analysis of the lineages that goes from keratinocytes to gustatory epithelium, as it allows the identification of TB clusters (fig. S9H). We defined the start of the trajectory in the keratinocyte cluster at the beginning of the continuum and the end of the trajectory just before the bifurcation for the differentiation of TB cell subtypes, as it fits our lineage tracing experiments (Fig. 5H). Using the GeneSwitches package, we displayed the switching genes along this path using default parameters (pseudo R2 > 0.03 and FDR < 0.05; Fig. 5K). We showed the expression of specific gene smoothened along the time axis using tradeseq and monocle3 (Fig. 5, I to J, and fig. S9, K and L). To generate Trp63 feature plot, we used order = TRUE for better visualization (fig. S9I).
In Fig. 6 and related supplementary figures, we compared gene regulatory networks associated with TB emergence from keratinocytes in esophagus and tongue using SCENIC. We first merged sample#4 (Up Epith esophagus paired) and sample#5 (Tg Epith paired) (like in fig. S4A). Then, we calculated a clustering based on AUC assay. For cluster#7, our DE analysis showed that genes associated with glandular fate were enriched, confirming that this cluster was not composed of TB cells. To sustain our results, we represent Muc5b expression as feature plot with the option order = T (fig. S10B). This new clustering showed the presence of cells from the tongue and the esophagus in the cluster#5, which contains cells of interest for the analysis, type IV (Shh+) and type I (Entpd2+) cells (fig. S10). Therefore, we set the resolution of “FindClusters” to 0.3 and analyzed the merged object. We performed DE analysis using AUC assay to highlight specific regulons for the cluster#5 and followed the method described before.
In fig. S11, to understand why TBs emerge specifically in the proximal part of the esophagus, we compared the microenvironment of thoracic and cervical esophagus. We used the two samples in which all living cells have been sequenced. After normalization of sample#1 (Tho Tot) and sample#2 (Up Tot), we defined the resolution of “FindCluster” to 0.45 and selected all the clusters excluded from the analysis in Fig. 1, which are composed of non-epithelial cells from thoracic and cervical esophagus. For the reasons described in Fig. 1, we integrated the two datasets. Then, for each cluster, we performed DE analysis according to the origin of the cells (thoracic or cervical esophagus). We displayed several genes associated with the Wnt pathway, differentially expressed in the fibroblasts of the cervical esophagus compared to the fibroblast of the thoracic esophagus.
Quantification and statistical analysis
Statistical and graphical data analyses were performed using Prism 8 (GraphPad) and R software. All experiments shown were replicated at least twice. All data in histograms represent means. Statistical significance was calculated by Mann-Whitney or Fisher exact test using the GraphPad Prism and R software, considering P < 0.05 as statistically significant. Venn diagrams were generated online using Venny2.1 (53). Hypergeometric probability was used to calculate the significance of gene set overlap, setting the total number of RNA as 30,000. All tests are two-sided.
Acknowledgments
We thank the FACS core facility and the animal house facility of the ULB for the help. Sequencing was performed at the Brussels Interuniversity Genomics High Throughput Core (www.brightcore.be). The Krt8-YFP knock-in mice have been provided by R. Leube. B.B. is an investigator of Walloon Excellence in Lifesciences and Biotechnology (WELBIO) and FNRS at the ULB.
Funding: This work is supported by WELBIO (FRFS-WELBIO-CR-2019S-01R; recipient: B.B), Fondation contre le cancer (FCC/ULB 2018-067; recipient: B.B.), the region Wallonie-Bruxelles (ARC consolidator 2016-2021; recipient: B.B), and Fonds Paul Genicot (recipient: B.B.). A.V.D. is supported by a fellowship from the Fondation Contre le Cancer and from the FNRS.
Author contributions: Conceptualization: A.V.D. and B.B. Methodology: A.V.D. Investigation: A.V.D. Funding acquisition: B.B. Supervision: B.B. Writing (original draft): B.B. Writing (review and editing): A.V.D. and B.B.
Competing interests: The authors declare they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. scRNA-seq and RNA-seq data reported here are deposited in the Gene Expression Omnibus database under accession no. GSE220117, GSE220065, and GSE220064.
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
Legends for tables S1 to S9
Other Supplementary Material for this : manuscript includes the following:
Tables S1 to S9
REFERENCES AND NOTES
- 1.J. P. Marques-Pereira, C. P. Leblond, Mitosis and differentiation in the stratified squamous epithelium of the rat esophagus. Am. J. Anat. 117, 73–87 (1965). [DOI] [PubMed] [Google Scholar]
- 2.D. Croagh, W. A. Phillips, R. Redvers, R. J. S. Thomas, P. Kaur, Identification of candidate murine esophageal stem cells using a combination of cell kinetic studies and cell surface markers. Stem Cells 25, 313–318 (2007). [DOI] [PubMed] [Google Scholar]
- 3.J. Kalabis, K. Oyama, T. Okawa, H. Nakagawa, C. Z. Michaylira, D. B. Stairs, J.-L. Figueiredo, U. Mahmood, J. A. Diehl, M. Herlyn, A. K. Rustgi, A subpopulation of mouse esophageal basal cells has properties of stem cells with the capacity for self-renewal and lineage specification. J. Clin. Invest. 118, 3860–3869 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.A. D. DeWard, J. Cramer, E. Lagasse, Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population. Cell Rep. 9, 701–711 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.V. Giroux, A. A. Lento, M. Islam, J. R. Pitarresi, A. Kharbanda, K. E. Hamilton, K. A. Whelan, A. Long, B. Rhoades, Q. Tang, H. Nakagawa, C. J. Lengner, A. J. Bass, E. P. Wileyto, A. J. Klein-Szanto, T. C. Wang, A. K. Rustgi, Long-lived keratin 15+ esophageal progenitor cells contribute to homeostasis and regeneration. J. Clin. Invest. 127, 2378–2391 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D. P. Doupé, M. P. Alcolea, A. Roshan, G. Zhang, A. M. Klein, B. D. Simons, P. H. Jones, A single progenitor population switches behavior to maintain and repair esophageal epithelium. Science 337, 1091–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.T. Okubo, C. Clark, B. L. M. Hogan, Cell lineage mapping of taste bud cells and keratinocytes in the mouse tongue and soft palate. Stem Cells 27, 442–450 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.N. Chaudhari, S. D. Roper, The cell biology of taste. J. Cell Biol. 190, 285–296 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.L. M. Beidler, R. L. Smallman, Renewal of cells within taste buds. J. Cell Biol. 27, 263–272 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.A. I. Farbman, Renewal of taste bud cells in rat circumvallate papillae. Cell Tissue Kinet. 13, 349–357 (1980). [DOI] [PubMed] [Google Scholar]
- 11.H. Miura, L. A. Barlow, Taste bud regeneration and the search for taste progenitor cells. Arch. Ital. Biol. 148, 107–118 (2010). [PMC free article] [PubMed] [Google Scholar]
- 12.J. M. Sullivan, A. A. Borecki, S. Oleskevich, Stem and progenitor cell compartments within adult mouse taste buds. Eur. J. Neurosci. 31, 1549–1560 (2010). [DOI] [PubMed] [Google Scholar]
- 13.H. Miura, Y. Kusakabe, S. Harada, Cell lineage and differentiation in taste buds. Arch. Histol. Cytol. 69, 209–225 (2006). [DOI] [PubMed] [Google Scholar]
- 14.H. Miura, J. K. Scott, S. Harada, L. A. Barlow, Sonic hedgehog-expressing basal cells are general post-mitotic precursors of functional taste receptor cells. Dev. Dyn. 243, 1286–1297 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.I. J. Miller, Jr., D. V. Smith, Quantitative taste bud distribution in the hamster. Physiol. Behav. 32, 275–285 (1984). [DOI] [PubMed] [Google Scholar]
- 16.S. P. Travers, K. Nicklas, Taste bud distribution in the rat pharynx and larynx. Anat. Rec. 227, 373–379 (1990). [DOI] [PubMed] [Google Scholar]
- 17.M. B. Omary, Intermediate filament proteins of digestive organs: physiology and pathophysiology. Am. J. Physiol. Gastrointest. Liver Physiol. 312, G628–G634 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.N. Schwarz, R. Windoffer, T. M. Magin, R. E. Leube, Dissection of keratin network formation, turnover and reorganization in living murine embryos. Sci. Rep. 5, 9007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.M. Rothova, H. Thompson, H. Lickert, A. S. Tucker, Lineage tracing of the endoderm during oral development. Dev. Dyn. 241, 1183–1191 (2012). [DOI] [PubMed] [Google Scholar]
- 20.S. Aibar, C. B. González-Blas, T. Moerman, V. A. Huynh-Thu, H. Imrichova, G. Hulselmans, F. Rambow, J.-C. Marine, P. Geurts, J. Aerts, J. van den Oord, Z. K. Atak, J. Wouters, S. Aerts, SCENIC: Single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.B. Van de Sande, C. Flerin, K. Davie, M. De Waegeneer, G. Hulselmans, S. Aibar, R. Seurinck, W. Saelens, R. Cannoodt, Q. Rouchon, T. Verbeiren, D. De Maeyer, J. Reumers, Y. Saeys, S. Aerts, A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat. Protoc. 15, 2247–2276 (2020). [DOI] [PubMed] [Google Scholar]
- 22.R. Hamamichi, M. Asano-Miyoshi, Y. Emori, Taste bud contains both short-lived and long-lived cell populations. Neuroscience 141, 2129–2138 (2006). [DOI] [PubMed] [Google Scholar]
- 23.D. E. Haines, G. A. Mihailoff, Fundamental Neuroscience for Basic and Clinical Applications (Elsevier, 2018).
- 24.I. Matsumoto, M. Ohmoto, M. Narukawa, Y. Yoshihara, K. Abe, Skn-1a (Pou2f3) specifies taste receptor cell lineage. Nat. Neurosci. 14, 685–687 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.W. Ren, E. Aihara, W. Lei, N. Gheewala, H. Uchiyama, R. F. Margolskee, K. Iwatsuki, P. Jiang, Transcriptome analyses of taste organoids reveal multiple pathways involved in taste cell generation. Sci. Rep. 7, 4004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.T. E. Finger, L. A. Barlow, Cellular diversity and regeneration in taste buds. Curr. Opin. Physiol. 20, 146–153 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.M. P. Alcolea, P. Greulich, A. Wabik, J. Frede, B. D. Simons, P. H. Jones, Differentiation imbalance in single oesophageal progenitor cells causes clonal immortalization and field change. Nat. Cell Biol. 16, 615–622 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.M. Ohmoto, S. Nakamura, H. Wang, P. Jiang, J. Hirota, I. Matsumoto, Maintenance and turnover of Sox2+ adult stem cells in the gustatory epithelium. PLOS ONE 17, e0267683 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D. Castillo, K. Seidel, E. Salcedo, C. Ahn, F. J. de Sauvage, O. D. Klein, L. A. Barlow, Induction of ectopic taste buds by SHH reveals the competency and plasticity of adult lingual epithelium. Development 141, 2993–3002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D. Gaillard, L. A. Barlow, Taste bud cells of adult mice are responsive to Wnt/β-catenin signaling: Implications for the renewal of mature taste cells. Genesis 49, 295–306 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D. Gaillard, S. G. Bowles, E. Salcedo, M. Xu, S. E. Millar, L. A. Barlow, β-Catenin is required for taste bud cell renewal and behavioral taste perception in adult mice. PLoS Genet. 13, e1006990 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.A. Vercauteren Drubbel, S. Pirard, S. Kin, B. Dassy, A. Lefort, F. Libert, S. Nomura, B. Beck, Reactivation of the Hedgehog pathway in esophageal progenitors turns on an embryonic-like program to initiate columnar metaplasia. Cell Stem Cell 28, 1411–1427.e7 (2021). [DOI] [PubMed] [Google Scholar]
- 33.H. Miura, H. Kato, Y. Kusakabe, M. Tagami, J. Miura-Ohnuma, Y. Ninomiya, A. Hino, A strong nerve dependence of sonic hedgehog expression in basal cells in mouse taste bud and an autonomous transcriptional control of genes in differentiated taste cells. Chem. Senses 29, 823–831 (2004). [DOI] [PubMed] [Google Scholar]
- 34.W.-J. Lu, R. K. Mann, A. Nguyen, T. Bi, M. Silverstein, J. Y. Tang, X. Chen, P. A. Beachy, Neuronal delivery of Hedgehog directs spatial patterning of taste organ regeneration. Proc. Natl. Acad. Sci. U.S.A. 115, E200–E209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.H. Yang, W.-N. Cong, J. S. Yoon, J. M. Egan, Vismodegib, an antagonist of hedgehog signaling, directly alters taste molecular signaling in taste buds. Cancer Med. 4, 245–252 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.M. Cheal, B. Oakley, Regeneration of fungiform taste buds: Temporal and spatial characteristics. J. Comp. Neurol. 172, 609–625 (1977). [DOI] [PubMed] [Google Scholar]
- 37.R. Vincis, A. Fontanini, Central taste anatomy and physiology. Handb. Clin. Neurol. 164, 187–204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.W. L. Neuhuber, M. Raab, H.-R. Berthoud, J. Wörl, Innervation of the mammalian esophagus. Adv. Anat. Embryol. Cell. Biol. 185, 1–73 (2006). [PubMed] [Google Scholar]
- 39.M. Hennel, M. Brozmanova, M. Kollarik, Cough reflex sensitization from esophagus and nose. Pulm. Pharmacol. Ther. 35, 117–121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.J. Harsanyiova, F. Ru, T. Zatko, M. Kollarik, M. Hennel, Vagus nerves provide a robust afferent innervation of the mucosa throughout the body of the esophagus in the mouse. Dysphagia 35, 471–478 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.T. Reynolds, Hemlock alkaloids from Socrates to poison aloes. Phytochemistry 66, 1399–1406 (2005). [DOI] [PubMed] [Google Scholar]
- 42.K. Iwatsuki, H. Uneyama, Sense of taste in the gastrointestinal tract. J. Pharmacol. Sci. 118, 123–128 (2012). [DOI] [PubMed] [Google Scholar]
- 43.S. L. Prescott, B. D. Umans, E. K. Williams, R. D. Brust, S. D. Liberles, An airway protection program revealed by sweeping genetic control of vagal afferents. Cell 181, 574–589.e14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.P. L. Andrews, Physiology of nausea and vomiting. Br. J. Anaesth. 69, 2S–19S (1992). [DOI] [PubMed] [Google Scholar]
- 45.S. Anders, D. J. McCarthy, Y. Chen, M. Okoniewski, G. K. Smyth, W. Huber, M. D. Robinson, Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protocols 8, 1765–1786 (2013). [DOI] [PubMed] [Google Scholar]
- 46.D. Powell, M. Milton, A. Perry, K. Santos, drpowell/degust 4.1.1 (Zenodo, 2019).
- 47.K. Blighe, kevinblighe/EnhancedVolcano (2020).
- 48.T. Stuart, A. Butler, P. Hoffman, C. Hafemeister, E. Papalexi, W. M. Mauck, M. Stoeckius, P. Smibert, R. Satija, Comprehensive integration of single cell data. bioRxiv , 460147 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.C. A. Vallejos, D. Risso, A. Scialdone, S. Dudoit, J. C. Marioni, Normalizing single-cell RNA sequencing data: Challenges and opportunities. Nat. Methods 14, 565–571 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.C. Trapnell, D. Cacchiarelli, J. Grimsby, P. Pokharel, S. Li, M. Morse, N. J. Lennon, K. J. Livak, T. S. Mikkelsen, J. L. Rinn, The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.E. Y. Cao, J. F. Ouyang, O. J. L. Rackham, GeneSwitches: Ordering gene expression and functional events in single-cell experiments. Bioinformatics 36, 3273–3275 (2020). [DOI] [PubMed] [Google Scholar]
- 52.K. Van den Berge, H. Roux de Bézieux, K. Street, W. Saelens, R. Cannoodt, Y. Saeys, S. Dudoit, L. Clement, Trajectory-based differential expression analysis for single-cell sequencing data. Nat. Commun. 11, 1201 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.J. C. Oliveros, VENNY. An Interactive Tool for Comparing Lists With Venn’s Diagrams. (2007); https://bioinfogp.cnb.csic.es/tools/venny/index.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Legends for tables S1 to S9
Tables S1 to S9