Abstract
The mechanism(s) of how bacteria acquire tolerance and then resistance to antibiotics remains poorly understood. Here, we show that glucose abundance decreases progressively as ampicillin-sensitive strains acquire resistance to ampicillin. The mechanism involves that ampicillin initiates this event via targeting pts promoter and pyruvate dehydrogenase (PDH) to promote glucose transport and inhibit glycolysis, respectively. Thus, glucose fluxes into pentose phosphate pathway to generate reactive oxygen species (ROS) causing genetic mutations. Meanwhile, PDH activity is gradually restored due to the competitive binding of accumulated pyruvate and ampicillin, which lowers glucose level, and activates cyclic adenosine monophosphate (cAMP)/cAMP receptor protein (CRP) complex. cAMP/CRP negatively regulates glucose transport and ROS but enhances DNA repair, leading to ampicillin resistance. Glucose and Mn2+ delay the acquisition, providing an effective approach to control the resistance. The same effect is also determined in the intracellular pathogen Edwardsiella tarda. Thus, glucose metabolism represents a promising target to stop/delay the transition of tolerance to resistance.
Ampicillin promotes bacterial antibiotic tolerance switching to resistance via glucose metabolism.
INTRODUCTION
Since the discovery in the late 1920s, the use of antibiotic drugs has led to marked improvements in human health and widespread positive impact in many other human societal areas, including aquaculture, animal husbandry, and veterinary medicine. However, the emergence and spread of antibiotic resistance, in the wake of the widespread use and misuse of antibiotics, now represent a major threat to human health, medical practice, and human society as we know it (1). To efficiently combat the further spread of antibiotic-resistant bacteria, we require a deeper understanding of the mechanisms leading to its emergence and rapid spread. While antibiotic-resistant bacterial cells acquire the capacity to grow in the presence of sublethal concentrations of antibiotics for an indefinite period of time, tolerance is the capacity to survive a transient exposure to low or high concentrations of antibiotics, even concentrations much higher than the minimum inhibitory concentration (MIC) (2, 3). Both tolerance and resistance are important reasons for the failure of antibiotic treatments (4, 5).
Resistance is typically caused by inherited mutations (6, 7). Tolerance has two main forms, “tolerance by slow growth” and “tolerance by lag.” Tolerance by slow growth can be either inherited or noninherited, whereas tolerance by lag is inherited (8–10). Extension of lag time leads rapidly to tolerance after intermittent exposure to a high concentration of ampicillin, being common in clinical scenarios (2, 3, 9). Several genes are frequently mutated in clinical isolates exposed to high drug concentrations, and resistance-conferring mutations can occur in cells already exhibiting tolerance (3, 11). Undoubtedly, these tolerance mutations pave the way for the rapid subsequent evolution of resistance, although this phenomenon is poorly understood (11).
Many previous studies suggest that the metabolic state of a bacterial cell influences its susceptibility to antibiotics (12–14), and the antibiotic-resistant metabolome of some bacterial strains has been characterized (15–17), revealing metabolic features that correlate with and contribute to resistance. It has been suggested that it is possible to “reprogram” the antibiotic-resistant metabolome to an antibiotic-sensitive metabolome and thereby restore sensitivity to antibiotic-induced killing (13, 14, 18). However, information regarding metabolic alteration in antibiotic tolerance and the transition of tolerance to resistance is not available.
Therefore, we propose that crucial metabolic biomarkers are clues to the mechanisms underlying antibiotic tolerance and resistance. Here, we describe the critical role of glucose in dictating the metabolic state of ampicillin-tolerant and ampicillin-resistant Escherichia coli K12 and Edwardsiella tarda. We demonstrate that ampicillin targets pts promoter P1b to promote glucose transport and inhibits pyruvate dehydrogenase (PDH) and thereby glucose fluxes to pentose phosphate pathway (PPP) instead of glycolysis. However, the inhibition of PDH causes pyruvate accumulation to remove the inhibition in reverse. These cause the progressively reduced glucose in tolerance. Tolerance- or resistance-conferring mutations were putatively caused by increased abundance of reactive oxygen species (ROS) and defects in DNA mismatch repair (MMR) in ampicillin-tolerant and ampicillin-resistant bacterial strains. Last, we present evidence that increased expression of the cyclic adenosine monophosphate (cAMP)/cAMP receptor protein (CRP) complex, which is trigged by glucose concentrations, plays a role in the evolution of resistance to ampicillin in E. coli and E. tarda.
RESULTS
Characterization of the antibiotic tolerance– and resistance–associated metabolic profile
In this study, cultures of E. coli K12 BW25113 (K12) were grown in LB medium with or without daily intermittent exposure to ampicillin (100 μg/ml) for 10 days (one cycle per day; 100 μg/ml for 4.5 hours and 0.625 μg/ml for another 16 hours) until resistance was established, as previously reported (11). A slower basal rate of growth than unexposed controls demonstrated that cultures acquired tolerance (T) to ampicillin during the first six cycles. The seventh cycle appeared to be a switch point (S) because cells treated for ≥8 cycles grew at the same rate as control untreated cells in the presence of ampicillin. These cells are designated as resistant (R) (Fig. 1A). MIC was the same in tolerant and untreated cells; however, the MIC gradually increased and resistance gradually increased over time (i.e., with increasing number of cycles in the presence of antibiotic) (Fig. 1B and table S1).
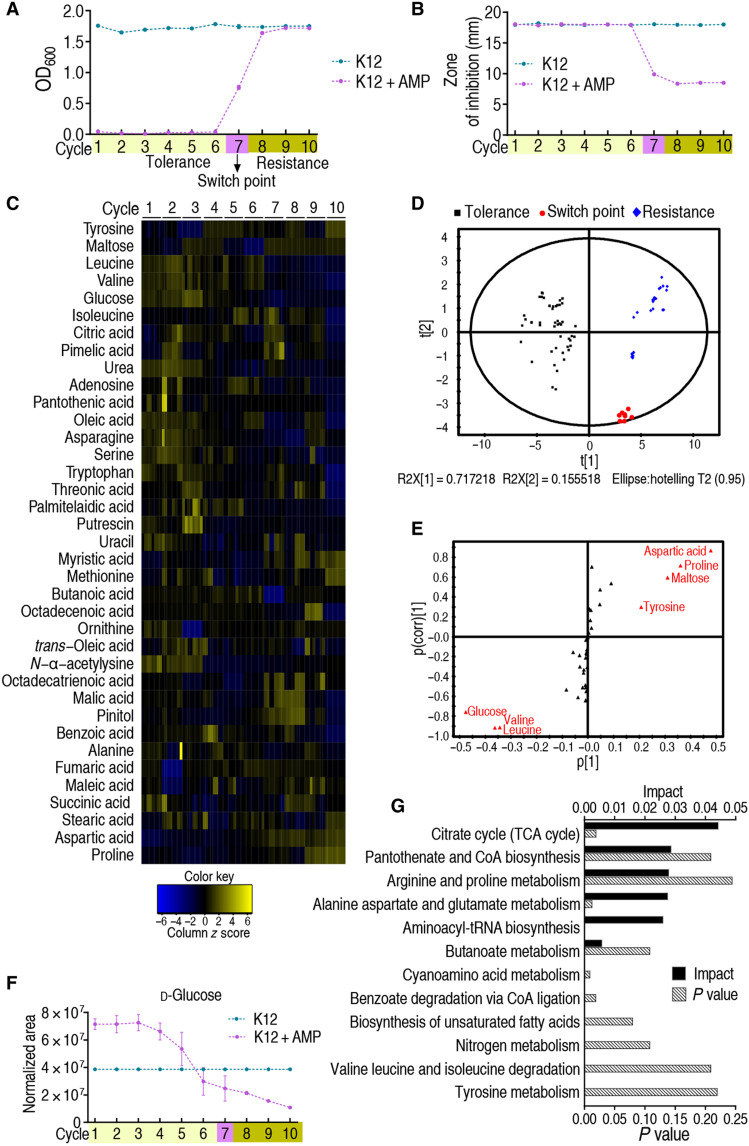
Fig. 1. Metabolomic analyses of E. coli K12 during cycles of intermittent daily exposure to ampicillin.
(A) K12 (n = 4) was cultured in growth medium with or without ampicillin (AMP; 100 μg/ml) for 4.5 hours at 37°C followed by 16 hours in growth medium with or without AMP (0.625 μg/ml), respectively, per day for 10 days/cycles. Cell density/survival after each cycle was measured. Switch point occurs four times and one time in the seventh and sixth cycles, respectively, in five independent experiments. (B) Cells cultured as in (A) were collected at the end of every cycle for MIC by an Oxford cup (n = 4). Results are displayed as means ± SEM. Each experiment was repeated independently at least three times. (C) Cells were also collected for GC-MS analysis. The abundance of 63 metabolites was quantified and presented graphically as a heatmap. Blue and yellow correspond to low to high abundance. See Materials and Methods for details. (D) Principal components analysis of data in (C). Each dot represents one technical replicate. (E) S-plot generated from OPLS-DA. Predictive component p[1] and correlation p(corr)[1] differentiate tolerance from resistance. Each triangle represents a single metabolite. Putative biomarkers (red) based on cutoff of ≥0.05 absolute value of covariance p and ≥0.5 correlation p(corr). (F) Abundance of the putative biomarker glucose as a function of increasing tolerance/resistance to AMP, as shown in (A). (G) Pathway analysis of differential metabolites identified in (C).
Aliquots of bacteria were removed daily to monitor the metabolite profile as the culture became increasingly resistant to ampicillin. For each aliquot/time point, the abundance of 63 metabolites was quantified by gas chromatography–mass spectrometry (GC-MS). Four biological and two technical replicates were performed per sample for a total of 156 metabolite profiles (fig. S1, A to C). Thirty-seven metabolites increased in abundance over three or more successive days/cycles (Fig. 1C). Principal components analysis showed that every day/cycle is distributed in an ordered manner and identified two principal components, t[1] and t[2]: t[1] discriminates between tolerance, switch point, and resistance, and t[2] discriminates by duration of exposure to antibiotic (Fig. 1D). S-plot identified the seven potential biomarkers glucose, valine, leucine, tyrosine, maltose, proline, and aspartic acid (Fig. 1E), where glucose, maltose, and leucine decreased in abundance over time, while aspartic acid, proline, and valine increased in abundance over time; tyrosine did not show a trend with time (Fig. 1F and fig. S2A). The most strongly affected Kyoto Encyclopedia of Genes and Genomes pathway is the tricarboxylic acid cycle (Fig. 1G). Since LB medium formula does not contain additional glucose, we speculate that bacteria may use the residual glucose in the medium. Quantification of glucose in the LB medium showed that approximately 11.8 mg/dl of glucose was present, whose abundance was decreased along culture time (fig. S2B). We tentatively conclude that glucose abundance increases and decreases as E. coli acquires tolerance and resistance to ampicillin, respectively, and postulate that this is linked to a metabolic shift that plays a role in the mechanism of antibiotic resistance.
Regulation of ampicillin to glucose metabolism is crucial for the transition from tolerance to resistance
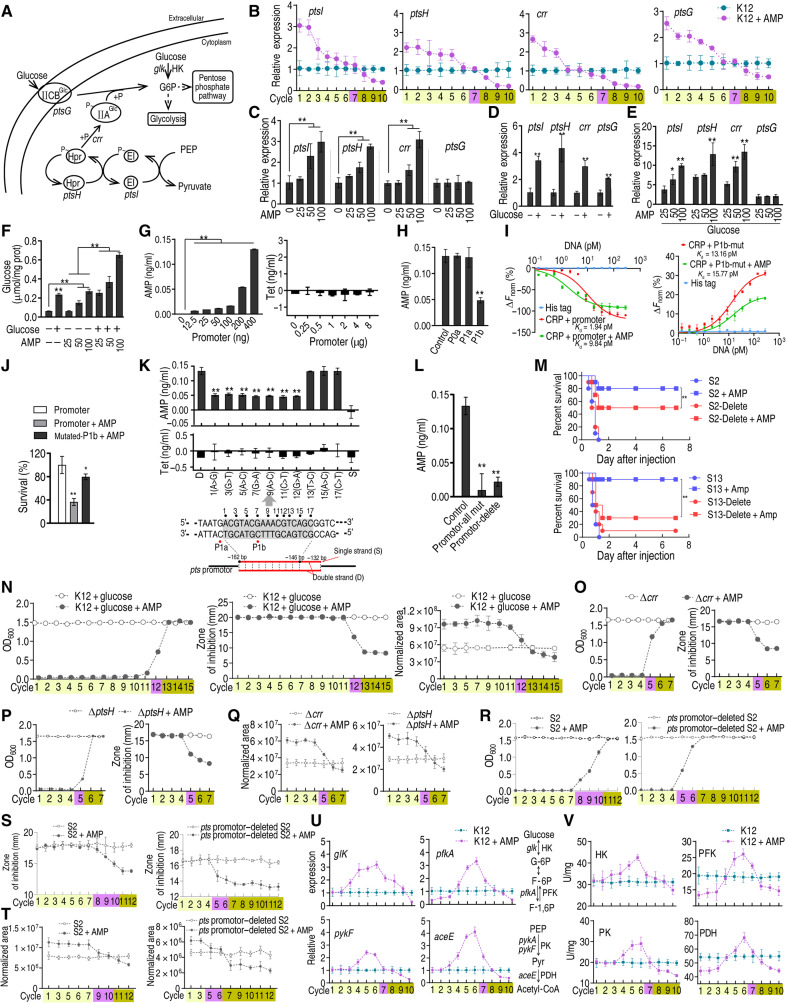
The abovementioned dynamic change of glucose abundance may indicate the involvement of glucose metabolism, including glucose transport and catabolism, on this transition. First, the effect on glucose transport was explored. Glucose uptake and phosphorylation are dependent almost entirely on the IIGlc phosphoenolpyruvate:sugar phosphotransferase system (PTS), containing the genes encoding enzyme I (ptsI), HPr (ptsH), IIAGlc (crr), and IICBGlc (ptsG) (19) (Fig. 2A). In K12 cells, the expression of the four genes was higher in antibiotic-tolerant cells, unchanged at the switch point, and lower in antibiotic-resistant cells during chronic intermittent exposure to ampicillin (Fig. 2B), which is similar to the dynamic change in glucose concentration. The kinetic change of the gene expression might be triggered by ampicillin. Actually, the expression level of ptsH, ptsI, and crr was elevated in an ampicillin dose–dependent manner, but not ptsG (Fig. 2C), which is consistent with ptsH, ptsI, and crr sharing the same promoter (19). It is expected that these four genes were positively regulated by glucose (Fig. 2D), and higher expression was observed when treated with both ampicillin and glucose and increased in an ampicillin-dependent manner (Fig. 2E), which was further confirmed with increased intracellular glucose level (Fig. 2F). To explore the instant effect of ampicillin on the expression of ptsH, ptsI, and crr, we postulated that ampicillin has a direct effect on their expression, e.g., by binding to the promoter site of the pts operon that controls the expression of ptsH, ptsI, and crr as CRP does (20), but tetracycline did not. Thus, we used the promoter DNA sequence to pull down ampicillin or tetracycline that served as a negative control. As expected, the level of ampicillin was increased in a promoter DNA–dependent manner, whereas tetracycline was not detected at any concentration of promoter DNA (Fig. 2G). Ampicillin binding was reduced when P1b was mutated but not P0a and P1a (Fig. 2H). Since P1b is the binding site of CRP (20), competitive binding between ampicillin and CRP with P1b was investigated. Microscale thermophoresis (MST) showed that the binding between CRP and the promoter was inhibited by ampicillin. However, this inhibition was almost recovered when P1b was mutated (Fig. 2I). To further demonstrate the interaction between ampicillin and P1b DNA, the P1b DNA and mutated P1b DNA were separately mixed with ampicillin and then precipitated with isopropanol. The pellets were dissolved in a medium containing ampicillin-sensitive bacteria for plate counting. The P1b DNA group had better killing efficacy than the mutated P1b DNA group (Fig. 2J). Moreover, the motif that binds directly and specifically with ampicillin was explored by constructing a series of DNA fragments with a point mutation around P1b. DNA fragments with a point mutation at 1st, 3rd, 5th, 7th, 9th, 11th, or 12th base pairs (bp) reduced binding to ampicillin by 33 to 50%, whereas others with a point mutation at 13th, 15th, or 17th bp did not. However, no binding of tetracycline with DNA was detected (Fig. 2, H and K). DNA fragments with all mutations or deletion failed to bind to ampicillin (Fig. 2L). These results indicate that the fragment within 1 to 12 bp around P1b is the motif for ampicillin binding. Deletion of pts promoter in clinically isolated antibiotic-sensitive E. coli strains S2 and S13 increased MIC to ampicillin and survival upon ampicillin killing (fig. S3, A and B). Consistently, deletion of pts promoter conferred ampicillin resistance even in the presence of glucose (fig. S3C). In a mouse infection model, strains with deleted pts promoter were more resistant to ampicillin than wild-type strains but had no observable change in their pathogenicity (Fig. 2M). These results demonstrate that the reduced intracellular glucose due to the decrease in glucose phosphorylation promotes bacterial resistance to ampicillin both in vitro and in vivo. These findings indicate that ampicillin has high affinity with P1b and outcompetes CRP to promote the expression of PST, elevating intracellular glucose in tolerance.
Fig. 2. Regulation of glucose and AMP to PTS.
(A) Diagram for glucose transport. (B to E) Gene expression of ptsI, ptsH, crr, and ptsG (n = 3) at indicated cycles (B), AMP concentration (C), 4 mM glucose (D), or both (E). (F) Intracellular glucose concentration (n = 3) exposed to AMP or/and 4 mM glucose. (G and H) Quantification of antibiotics binding to pts promoter (G) or mutated P0a, P1a, and P1b (n = 3) (H). (I) MST for competitive binding of CRP and AMP to pts promoter (left) and mutant (right) (n = 3). (J) K12 survival exposed to AMP preincubated with pts promoter or P1b mutant (n = 3). (K and L) Quantification of antibiotics binding to wild type or point-mutated (K) or multiple mutations or promoter-deleted (L) DNA fragments (n = 3). (M) Survival of mice (n = 10, log-rank test) infected with antibiotic-sensitive E. coli S2, S13, and their promoter-deleted strains. (N) Growth/viability (left), MIC (middle), and glucose (right) at the indicated cycles (n = 4). (O to Q) Growth/viability (O), MIC (P), and glucose (Q) of ∆crr and ∆ptsH at the indicated cycles exposed to AMP (n = 4). (R to T) Growth/viability (R), MIC (S), and glucose (T) of S2 and pts promoter–deleted S2 exposed to AMP (n = 4). (U and V) Gene expression of pfkA, glK, pykF, and aceE (V) and activity of HK, PFK, PK, and PDH (V) during cyclic exposure to AMP (n = 4). Results are displayed as means ± SEM, and statistically significant differences are identified by Kruskal-Wallis followed by Dunn’s multiple comparison post hoc test unless otherwise indicated. *P < 0.05 and **P < 0.01.
The effect of glucose transport on tolerance-to-resistance transition was investigated. Bacteria were cycled as above by supplementing additional glucose (4 mM). Glucose delayed the emergence of resistance, being shifted from cycle 7 to cycle 12. The data were confirmed with MIC, in correlating with increased intracellular glucose concentration (Fig. 2N and table S1). ∆crr and ∆ptsH were cycled the same as their parental strain K12. ∆crr or ∆ptsH advanced the emergence of resistance that occurred at cycle 5 (Fig. 2O and table S1), as well as for their MIC and intracellular glucose concentration (Fig. 2, P and Q, and table S1). As compared to the S2 strain, the promoter-deleted mutant had earlier developed resistance, increased MIC, and decreased glucose (Fig. 2, R to T, and table S1). These data together suggest that the regulation of PTS to glucose in a manner dependent on ampicillin/glucose plays a crucial role in the transition from tolerance to resistance.
Next, the effect of glucose catabolism on tolerance-to-resistance transition was explored. Phosphorylation of glucose is catalyzed by glucokinase (HK), encoded by glK, and then enter into the Embden-Meyerhof-Parnas glycolytic pathway (Fig. 2A) (19). Glycolysis is the process in which glucose is broken down to produce energy, and then the metabolic flux enters the pyruvate cycle (the P cycle), a recently revealed cycle providing respiratory energy in bacteria (21). The expression of glK (HK) and the downstream catabolic genes pfkA [6-phosphofructokinase I (PFK)] in glycolysis and pykF [pyruvate kinase (PK)] and aceE (PDH) in the P cycle was quantified. The expression patterns of glK and HK activity were totally different from those of the PTS system and intracellular glucose concentration. Specifically, glK/HK was progressively increased in a cycle-dependent manner just before the switch point and then decreased afterward and was lower than the nontreated control in the last cycle (Fig. 2, U and V). A similar pattern was obtained for pfkA/PFK, pykF/PK, and ace/PDH but with some characteristic changes. In detail, lower expression of pfkA, pykF, and aceE was detected in the first three and last two cycles when exposed to ampicillin (Fig. 2U). Meanwhile, the activity of PFK and PDH was lower than that of the nontreated control in the first three cycles and last three cycles, as to PK (Fig. 2U). Of particular interest was that PDH activity was 1.78- to 2.62-fold lower in the first cycle than the nontreated group but progressively increased until 1.25-fold at cycle 6 and decreased afterward (Fig. 2V). These data suggest that the gradually increased flux through glycolysis is responsible for the higher abundance than control but progressively reduced abundance of glucose in tolerant cells. The marked change between aceE expression and PDH activity implies its essential role during the transition from tolerance to resistance.
Mechanisms for reduction of PDH activity
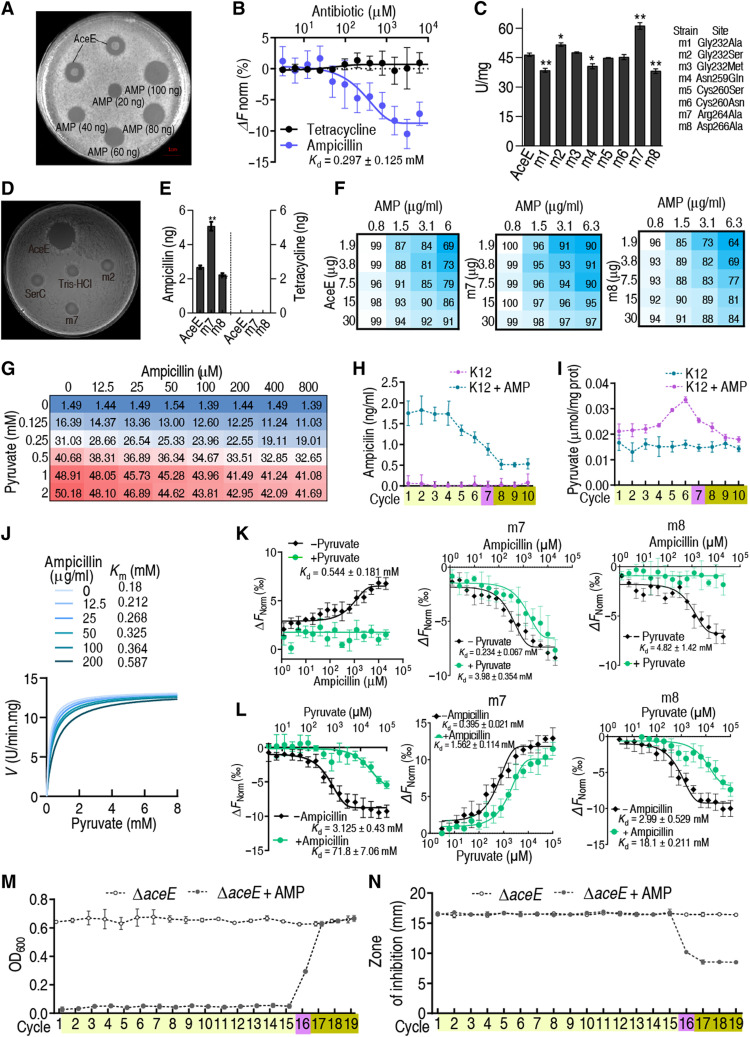
The expression of aceE, a subunit of PDH, was unaffected in the first three cycles, while PDH activity was reduced by 1.78- to 2.62-fold, suggesting that PDH activity was inhibited (Fig. 2, U and V). To test whether PDH interacts with ampicillin, ampicillin binding with recombinant AceE was measured and quantified using the Oxford cup test, and recombinant GLK, PFK1, PFKII, and PK1 in glycolysis were used as controls. The results indicated that AceE bound ampicillin, but other enzymes did not (Fig. 3A and fig. S4). The result was confirmed by MST (Fig. 3B). To probe the amino acid residues that were critical for ampicillin binding, mutations were generated at G232, N259, C260, C264, and D266 as previously reported (22–24). The mutation at the sites should impair the binding of ampicillin to PDH and with altered enzymatic activity. We found that G232 and D266 were the two critical sites whose mutations G232S (m2) and D266A (m7) failed to bind ampicillin and displayed higher PDH activity (Fig. 3, C and D), whereas mutations at other sites, including N259Q (m4), C260S (m5), and C264N (m6), had no effect on ampicillin binding (fig. S5). To further demonstrate the inhibition in vivo, E. coli BL21-pET-28a-AceE, BL21-pET-28a-m7, and BL21-pET-28a-m8 cells were cultured, treated with ampicillin, and collected. Recombinant PDH was isolated by Ni+-nitriloaceate (NTA) super flow resin to measure ampicillin. Higher and lower levels of ampicillin were detected in m7 and m8 than in wild type, respectively. However, no binding was detected if ampicillin was replaced by tetracycline (Fig. 3E). When extracellular ampicillin concentration was at 100 μg/ml, intracellular ampicillin was approximately 1.8 μg/ml (fig. S6). A detailed analysis of the effect of ampicillin on PDH activity was performed by titrating the ampicillin concentration and AceE dose, generating a two-dimensional heatmap. PDH activity was 30.3% lower than in control cells, while higher and lower activity was measured in m7 and m8, respectively (Fig. 3F). In K12 whole-cell lysis, ampicillin inhibited PDH activity in a dose-dependent manner (Fig. 3G). Kinetic intracellular ampicillin concentration was similar to that of glucose (Fig. 3H). High PDH activity and high level of pyruvate were correlated to each other in tolerance (Figs. 3I and 2S), suggesting a competition between pyruvate and ampicillin on binding to AceE. By Michaelis-Menten kinetics, pyruvate outcompetes with ampicillin on the binding of PDH in a dose-dependent manner, as revealed by the Michaelis constant (Km) value, although the maximum reaction rate (Vmax) was similar (Fig. 3J). Furthermore, MST analysis demonstrated that ampicillin had high affinity with AceE, but the binding was entirely and partly inhibited by pyruvate in K12, m8, and m7, respectively (Fig. 3K). Equally, the binding of pyruvate with AceE was interfered by ampicillin, where stronger inhibition was detected in K12 and m8 than m7 (Fig. 3L). Furthermore, the putative ampicillin-binding pocket in AceE was highly conserved among Gram-negative bacteria but was not present in Gram-positive bacteria, indicating a potential conserved regulatory mechanism of PDH by ampicillin in Gram-negative bacteria (fig. S7). In ∆aceE cells, the switch point and MIC from tolerance to resistance were delayed substantially (16 versus 7 days/cycles; Fig. 3, M and N, and table S1) but were recovered in aceE complementation strain (fig. S8). However, the regulation of glucose to the expression of crr and pstH was not affected in the absence of aceE (fig. S9). These results suggest that the ampicillin-dependent inhibition of PDH plays a key role in the glucose metabolic shift, characterized by the reduced level of glycolysis at the beginning and the progressive change from high to low glucose abundance.
Fig. 3. AMP-binding capability of AceE and effect.
(A) Oxford cup test for the AMP-binding capability of AceE. Purified recombinant AceE was mixed with AMP, and then AceE was precipitated by acetone. The precipitated protein was diluted and used for the test. (B) MST for binding of AceE with AMP. Tetracycline was used as a control (n = 3). (C) Activity of PDH with point mutation on AceE (n = 3). (D) Oxford cup test for AMP binding to AceE with point mutation. (E) Quantification for AMP or tetracycline binding to recombinant proteins (n = 3). (F) Effect of AMP on the activity of AceE, m7, and m8. (G) Activity of PDH in the presence of AMP. (H) Intracellular AMP concentration during cyclic daily intermittent exposure to AMP (0.625 μg/ml) (see Materials and Methods for details) (n = 4). (I) Pyruvate level in E. coli K12 during cyclic daily intermittent exposure to AMP. (J) Michaelis-Menten kinetics of PDH in AMP (0 to 200 μg). (K and L) MST for competition between pyruvate and AMP with AceE, m7, and m8 (n = 4). (M) Growth/viability of the E. coli K12 ∆aceE after the indicated number of cycles exposed to AMP (n = 4). (N) MIC of (M) (n = 4). Experiments were performed the same as described in Fig. 1 (A, B, and F). Results are displayed as means ± SEM, and statistically significant differences are identified by Kruskal-Wallis followed by Dunn’s multiple comparison post hoc test unless otherwise indicated. *P < 0.05 and **P < 0.01.
Glucose metabolism shift correlates with higher abundance of ROS
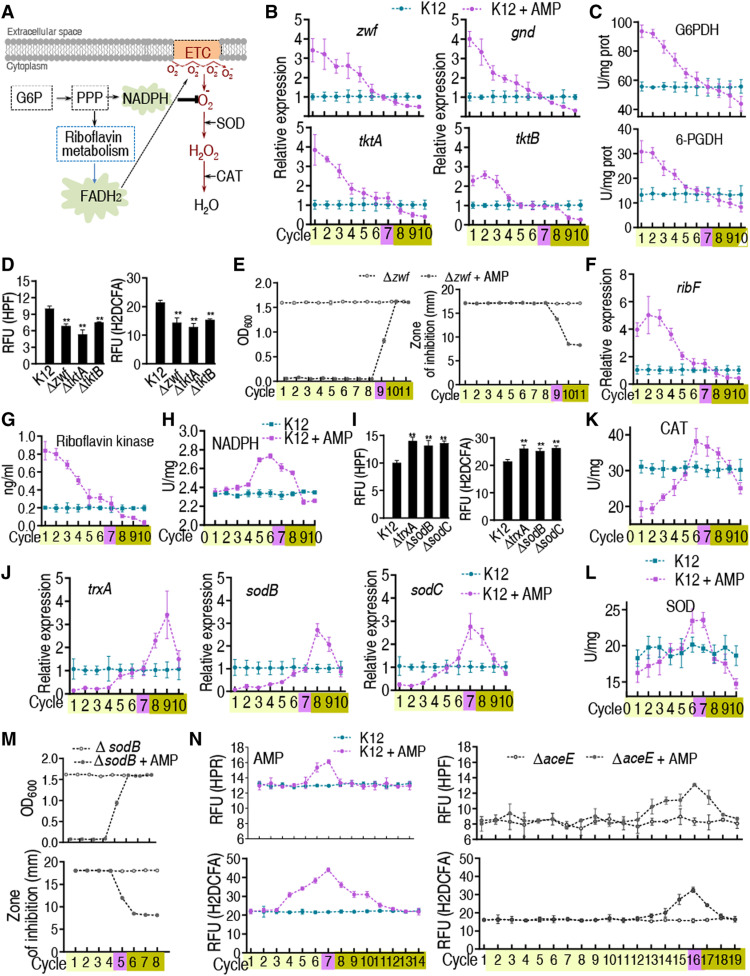
Glucose-6-phosphate (G6P) fluxes the glycolysis and the PPP. Therefore, investigation on the PPP is especially important. It is well known that PPP, riboflavin metabolism, and respiratory chain oxidative phosphorylation are linked to FADH2-dependent release of and increased abundance of ROS (Fig. 4A) (25–27). Oxidative stress and high abundance of ROS cause mutations (25, 28–30). Genetic mutations are frequently associated with antibiotic tolerance and resistance (11). The disrupted glycolysis suggests that the glucose metabolic flow may drive the PPP, possibly generating more ROS. Therefore, the effect of the disrupted glycolysis on these pathways is investigated. Genes zwf, gnd, tktA, and tktB play a role in the PPP. Exposure to ampicillin correlates with the expression of the four genes and the activity of glucose-6-phosphate dehydrogenase (G6PDH) and 6-phosphogluconate dehydrogenase (6-PGDH) with glucose abundance and increases and decreases their expression before tolerance and resistance, respectively (Fig. 4, B and C), thereby increasing the risk of oxidative stress. To test this possibility, ∆zwf, ∆tktA, or ∆tktB had lower ROS (Fig. 4D and fig. S10), and ∆zwf delayed the switch point and MIC from tolerance to resistance (9 versus 7 days/cycles; Fig. 4E and table S1). When ∆zwf was complemented by full-length zwf, the delay was recovered (fig. S11). These data indicate that activation of the PPP promotes ROS production.
Fig. 4. Gene expression of PPP and ROS abundance at cycles exposed to AMP in bacteria.
K12, ∆aceE, or other mutant cells were cultured and collected as in Fig. 1. (A) ROS generation during oxidative phosphorylation via PPP. (B) Gene expression of zwf, gnd, tktA, and tktB of PPP in cycles (n = 4). (C) Activity of G6PDH and 6-PGDH in the evolutionary cycle (n = 4). (D) ROS in the indicated mutants (n = 3). (E) Growth/viability and MIC of the E. coli K12 ∆zwf after the indicated number of cycles exposed to AMP (n = 4). (F) Gene expression of ribF in the evolutional cycle (n = 4). (G) Activity of riboflavin kinase in the cycles (n = 4). (H) NADPH content in the cycles (n = 4). (I) ROS in the indicated mutants (n = 3). (J) Gene expression of antioxidant reactive pathways in the cycles (n = 4). (K and L) Activity of CAT (K) and SOD (L) of K12 in the cycles (n = 4). (M) Growth/viability and MIC of the E. coli K12 ∆sodB were estimated from OD600 after the indicated number of cycles of exposure to AMP (n = 4). (N) ROS of K12 and ∆aceE in the cycles (n = 4). Results are displayed as means ± SEM, and statistically significant differences are identified by Kruskal-Wallis followed by Dunn’s multiple comparison post hoc test unless otherwise indicated. **P < 0.01.
ribF is an essential gene in riboflavin metabolism, which exhibits a similar expression pattern as glucose abundance did during exposure to ampicillin (Fig. 4F). Consistently, the activity of riboflavin kinase was reduced along with the cycles (Fig. 4G). The relationships between ROS abundance, the reduced form of nicotinamide adenine dinucleotide phosphate (NADPH), and the expression and activity of antioxidants were also investigated. NADPH provides reducing power required for protection against the toxicity of ROS (27). After chronic intermittent exposure to ampicillin, NADPH was slightly elevated (i.e., five to eight cycles of growth for NADPH and in the presence/absence of ampicillin) (Fig. 4H). Loss of trxA (encoding thioredoxin), sodB, and sodC [encoding superoxide dismutase (SOD)] up-regulated ROS (Fig. 4I). In antibiotic-tolerant strains of E. coli K-12, exposure to ampicillin decreased the expression of trxA, sodB, and sodC. However, in antibiotic-resistant strains, the expression of trxA, sodB, and sodC was gradually increased and then became normal (Fig. 4J). Activity of catalase (CAT) was lower in tolerant cells but slightly increased in these cycles around the switch point and then decreased with additional cycles of exposure to ampicillin (Fig. 4K). Similar results were obtained for the expression of SOD (Fig. 4L). Consistently, ∆sodB advanced the switch point and MIC from tolerance to resistance (5 versus 7 days/cycles; Fig. 4M and table S1). We interpret these data to indicate that exposure to ampicillin generates ROS and oxidative stress due to glucose metabolism shift.
The delayed resistance in ∆aceE can be related to ROS. ROS abundance was measured in E. coli and ∆aceE cells after exposure to ampicillin for 19 days/cycles. Higher ROS was detected in E. coli K12 (13.21 ± 0.26 for HPF and 21.89 ± 0.24 for H2DCFA) than in ∆aceE cells (9.59 ± 0.56 for HPF and 18.48 ± 0.18 for H2DCFA) without exposure to ampicillin, and ROS abundance increased in E. coli K12 and ∆aceE exposed to ampicillin. In E. coli K12 exposed to ampicillin, ROS level was approximately 2.03-fold (H2DCFA) and 1.35-fold (HPF) higher than in control cells (no ampicillin) in 7 days/cycles, respectively. In contrast, in ∆aceE cells exposed to ampicillin for 19 days/cycles, ROS abundance was reaching the peak, being consistent with the delayed occurrence of resistance. Meanwhile, ROS abundance was approximately 1.96-fold (H2DCFA) and 1.58-fold (HPF) higher than in control cells in 16 days/cycles (Fig. 4N). This supports the idea that glucose metabolism shift induces a cellular stress response that stimulates ROS production related to PDH.
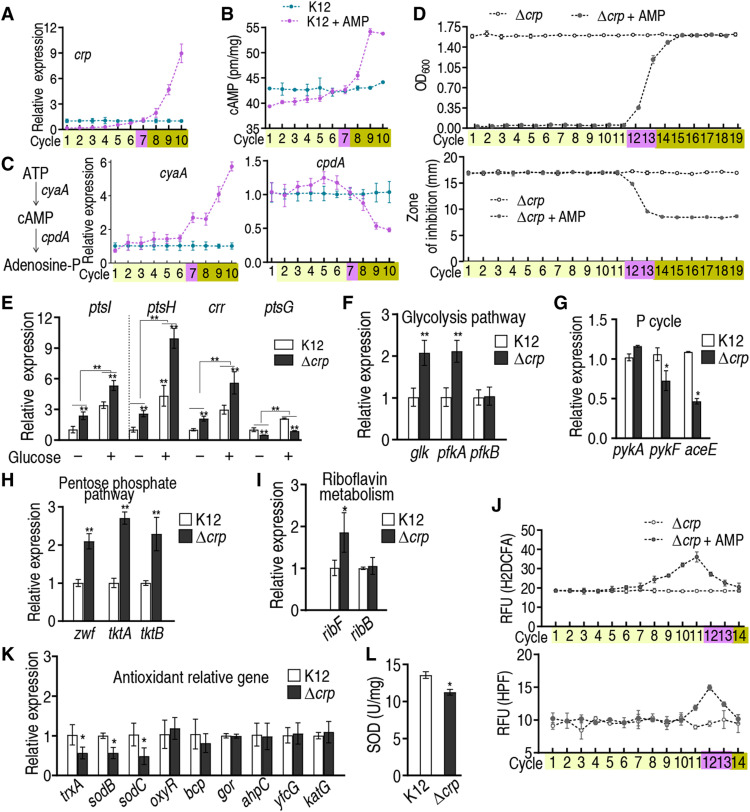
Reduced glucose activates cAMP/CRP complex expression and activity for resistance
Low level of glucose was frequently observed in antibiotic resistance and in our study (14, 15), but the mechanism is unknown. Glucose availability negatively regulates the expression and activity of cAMP/CRP complex (31). Our above results demonstrated that glucose transport and catabolism were decreased along with tolerance-to-resistance transition. Thus, cAMP/CRP expression can be activated by the decreased level of glucose, contributing to the evolution of resistance. Here, we demonstrated that the expression of crp mRNA was increased with increasing resistance to ampicillin and decreasing glucose concentration (Figs. 5A and 1F); cAMP level showed a similar change (Fig. 5B), as did the transcription of cyaA, encoding adenylate cyclase. CyaA regulates cAMP biosynthesis. In contrast, the expression of cpdA, encoding cAMP phosphodiesterase, which degrades cAMP, decreased over time with increasing antibiotic resistance (Fig. 5C). These results suggest that the cAMP/CRP complex plays a role in the evolution of bacterial resistance to ampicillin.
Fig. 5. Expression of cAMP/CRP during cycles of intermittent daily exposure to AMP.
In (A) to (D) and (J), E. coli K12 was cultured in LB medium with or without AMP (100 μg/ml) for 4.5 hours at 37°C followed by 16 hours in LB medium with or without AMP (0.625 μg/ml) per day, respectively, for the indicated number of cycles/days. (A) Expression of crp with increasing number of cycles exposed to AMP (n = 4). (B) Quantitation of cAMP with increasing number of cycles exposed to AMP (n = 4). (D) Growth/viability and MIC of the indicated ∆crp cells after the indicated number of cycles exposed to AMP (n = 4). (E) Expression of ptsL, ptsH, crr, and ptsG with and without 4 mM glucose in wild type and ∆crp (n = 3). (F to I) qRT-PCR for the expression of the indicated genes in wild type and ∆crp E. coli K12 (n = 3). (J) ROS of ∆crp in the presence or absence of AMP (n = 4). (K) Expression of the indicated genes in wild type and ∆crp (n = 3). (L) SOD activity in wild type and ∆crp (n = 3). Results are displayed as means ± SEM, and statistically significant differences are identified by Kruskal-Wallis followed by Dunn’s multiple comparison post hoc test for all the tests unless otherwise indicated. *P < 0.05 and **P < 0.01.
The idea that CRP plays an important role in the mechanism of bacterial antibiotic resistance was tested in the following experiments. ∆crp cells were subjected to daily intermittent exposure to ampicillin or untreated, and both were evaluated for the ability to acquire ampicillin resistance over time. The results show that acquisition of ampicillin resistance is delayed by approximately 5 to 6 days/cycles in ∆crp cells exposed to ampicillin (∆crp switch point at cycles 12 and 13 versus switch point at cycle 7 in control E. coli K12 cells with ampicillin). Consistently, loss of crp increased MIC at the switch point from cycles 7 to 12 and 13 (Fig. 5D and table S1). The finding was confirmed by crp complementation (fig. S12). These results support the idea that the cAMP/CRP complex plays a crucial role in the mechanism of resistance to ampicillin, in the context of the experimental system described here.
Loss of crp caused higher expression of the glucose transport genes ptsH, ptsI, crr, and ptsG, and this promotion was enlarged by glucose (Fig. 5E), supporting crp-mediated negative regulation to glucose transport. Expression of glk and pfkA of glycolysis was higher, and expression of pykF and aceE of the P cycle was lower in ∆crp cells (Fig. 5, F and G). In ∆crp cells, expression of zwf, tktA, and tktB of PPP and ribF of riboflavin metabolism was higher than in control cells (Fig. 5, H and I). These data suggest that cAMP/CRP down-regulates glucose transport, glycolysis, and PPP and up-regulates the P cycle. Thus, cAMP/CRP leads to the acquisition of resistance.
Therefore, up-regulation of cAMP/CRP may reduce the abundance of ROS, decreasing the risk of oxidative stress–induced mutations. To test this possibility, ROS abundance was measured in ∆crp cells after exposure to ampicillin for 14 days/cycles. Lower ROS abundance was detected in ∆crp cells (18.48 ± 0.18 for H2DCFA and 9.59 ± 0.56 for HPF) than in E. coli K12 (21.89 ± 0.24 for H2DCFA and 13.21 ± 0.26 for HPF) without exposure to ampicillin, and ROS abundance increased in E. coli K12 and ∆aceE exposed to ampicillin. In E. coli K12 exposed to ampicillin, ROS level was approximately 2.03-fold (H2DCFA) and 1.35-fold (HPF) higher than in control cells (no ampicillin) in 7 days/cycles, respectively (Fig. 4N). In contrast, in ∆crp cells exposed to ampicillin for 14 days/cycles, ROS abundance was approximately 1.96-fold (H2DCFA) and 1.58-fold (HPF) higher than in control cells in 11 and 12 days/cycles, respectively (Fig. 5J). When crp was complemented, the period for the increased ROS was in advance (fig. S13). cAMP/CRP also negatively regulated the expression of ribF (Fig. 5I) and positively regulated the expression of trxA, sodB, and sodC but not oxyR, bcp, gor, ahpC, yfcG, and katG (Fig. 5K). Loss of crp decreased the activity of SOD (encoded by sodB and sodC) (Fig. 5L). We interpret these data to indicate that cAMP/CRP is activated to reduce ROS and oxidative stress generated by ampicillin.
ROS abundance contributes to the occurrence of tolerance and switch to resistance
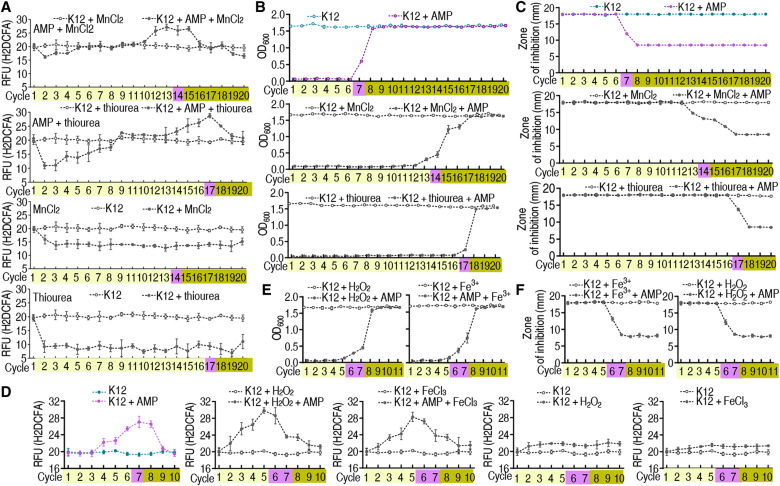
Mn2+ and thiourea are potent antioxidants capable of scavenging ROS. Exogenous Mn2+ and thiourea reduced ROS by approximately one-quarter and one-half at all times, respectively. The synergistic use of Mn2+ or thiourea and ampicillin postponed ROS elevation at the top by 14 and 17 cycles (versus 7 cycles in K12 exposure to ampicillin) (Fig. 6A and fig. S14A). Consistently, the synergy delayed the switch and MIC change from antibiotic tolerance to antibiotic resistance from 7 cycles/days to 14 to 17 cycles/days (Fig. 6, B and C, and table S1). On the contrary, H2O2 and Fe3+ are potent oxidants capable of producing ROS. H2O2 and Fe3+ complements caused ROS elevation in advance and thereby promoted the switch from antibiotic tolerance to antibiotic resistance from 7 to 5 cycles/days (Fig. 6D and fig. S14B). A similar switch was detected in optical density (OD) and MIC (Fig. 6, E and F, and table S1). These results support the conclusion that ROS plays a crucial role in the switch from antibiotic sensitive to antibiotic tolerant and then to antibiotic resistant.
Fig. 6. ROS abundance, growth, and MIC in the synergistic use of AMP and MnCl2, thiourea, FeCl3, and H2O2 in E. coli K12 or ∆aceE with increasing number of cycles of exposure to AMP.
(A) ROS of E. coli K12 is quantified in the presence of MnCl2 or thiourea (n = 4). (B) Growth of E. coli K12 in the presence of MnCl2 or thiourea (n = 4). (C) MIC of E. coli K12 in the presence of MnCl2 or thiourea (n = 4). (D) ROS of E. coli K12 is quantified in the presence of FeCl3 or H2O2 (n = 4). (E) Growth of E. coli K12 in the presence of FeCl3 or H2O2 (n = 4). (F) MIC of E. coli K12 in the presence of FeCl3 or H2O2 (n = 4). Results are displayed as means ± SEM.
cAMP/CRP positively regulates DNA repair and promotes acquisition of resistance to ampicillin
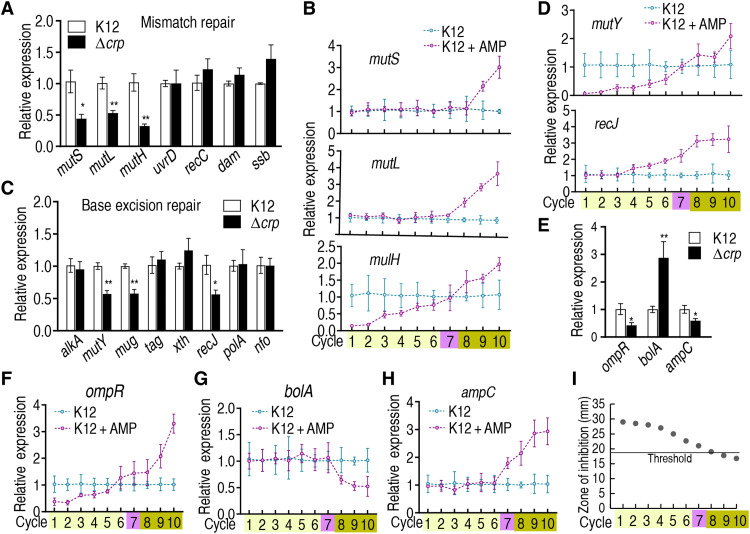
Inefficient or inadequate capacity for DNA MMR, homologous recombination, base excision repair, or nucleotide excision repair increases the probability that DNA lesions will lead to mutations that confer resistance to antibiotics (as well as other phenotypic changes). Gene mutations in the occurrence of tolerance and ampC resistance are mostly classified to MMR, and others belong to base excision repair (one is nucleotide excision repair) (3, 11). With respect to DNA MMR, cAMP/CRP positively regulates the expression of mutS, mutL, and mutH (Fig. 7A). Expression of these MMR genes was lower or constant in antibiotic-tolerant cells but higher in antibiotic-resistant cells during chronic intermittent exposure to ampicillin (Fig. 7B). Similar results were detected in the expression of mutY, mug, and recJ, which work for base excision repair (Fig. 7, C and D). Thus, cAMP/CRP plays a role in inactivation of DNA repair.
Fig. 7. Expression of DNA repair genes and ampC in wild type and ∆crp E. coli K12 and during cyclic intermittent exposure to AMP.
(A, C, and E) qRT-PCR for the expression of the indicated genes in wild type and ∆crp. (A) MMR, (C) base excision repair system, and (E) ampC and its related genes (n = 3). (B, D, and F to H) E. coli K12 cells were cultured as described in the legend of Fig. 1 (n = 4). qRT-PCR was used to quantify the expression of the indicated genes during cyclic exposure to AMP, as indicated. (B) MMR, (D) base excision repair system, and (F to H) ompR, bola, and ampC, respectively. (I) Activity of AmpC in the evolutionary cycle was measured using susceptibility test disks. Threshold value = 18 (n = 4). Results are displayed as means ± SEM, and statistically significant differences are identified by Kruskal-Wallis followed by Dunn’s multiple comparison post hoc test for all the tests unless otherwise indicated. *P < 0.05 and **P < 0.01.
We further investigate how antibiotic-tolerant cells acquire antibiotic resistance. Previous reports indicate that cAMP/CRP positively and negatively regulates ompR (32), while ompR negatively regulates bolA (33), and bolA positively regulates ampC (34). Thus, ampC is indirectly regulated by cAMP/CRP. Our results showed that defect/loss of crp decreases the expression of ompR and ampC but increases the expression of bolA (Fig. 7E), and exposure to ampicillin decreases and increases the expression of ompR before and after the switch point, respectively, and decreases the expression of bolA after the switch point (Fig. 7, F and G). Last, expression of ampC increases after the switch point during chronic intermittent exposure to ampicillin (Fig. 7H). To confirm that the increased ampC expression was associated with resistance, AmpC activity was measured. Inhibition zone was decreased along with the evolutionary cycle. A value close to and lower than the positive threshold emerged at cycle 8 and the following cycles, respectively, supporting the crucial role played by AmpC (Fig. 7I and fig. S15). These results suggest that cAMP/CRP induces ompR expression, inhibits bolA expression, and induces ampC, promoting evolution from antibiotic tolerance to antibiotic resistance. Since reports have indicated that an RpoS-DinB mechanism contributes to MMR in antibiotic-induced mutagenesis (25, 35), we further demonstrated that elevated expression of rpoS and dinB was similar to that of ROS in the evolutionary cycle (fig. S16), suggesting that RpoS-DinB also contributes to the transition. Last, whole-genome sequencing of the first to ninth cycled bacteria identified 27 mutations from 23 genes, where mutated genes associated with antibiotic resistance included envZ (cycles 7 to 9), cyoE (cycle 9), frlA (cycle 9), cpxA (cycle 8), and ompR and wecC (cycle 7) (fig. S17). These data support the conclusion that the switch point at cycle 7 is crucial for the development of mutations for antibiotic resistance and rationalize the higher level of ROS in this cycle than in other cycles.
Glucose metabolism regulates the evolution from tolerance to resistance in E. tarda
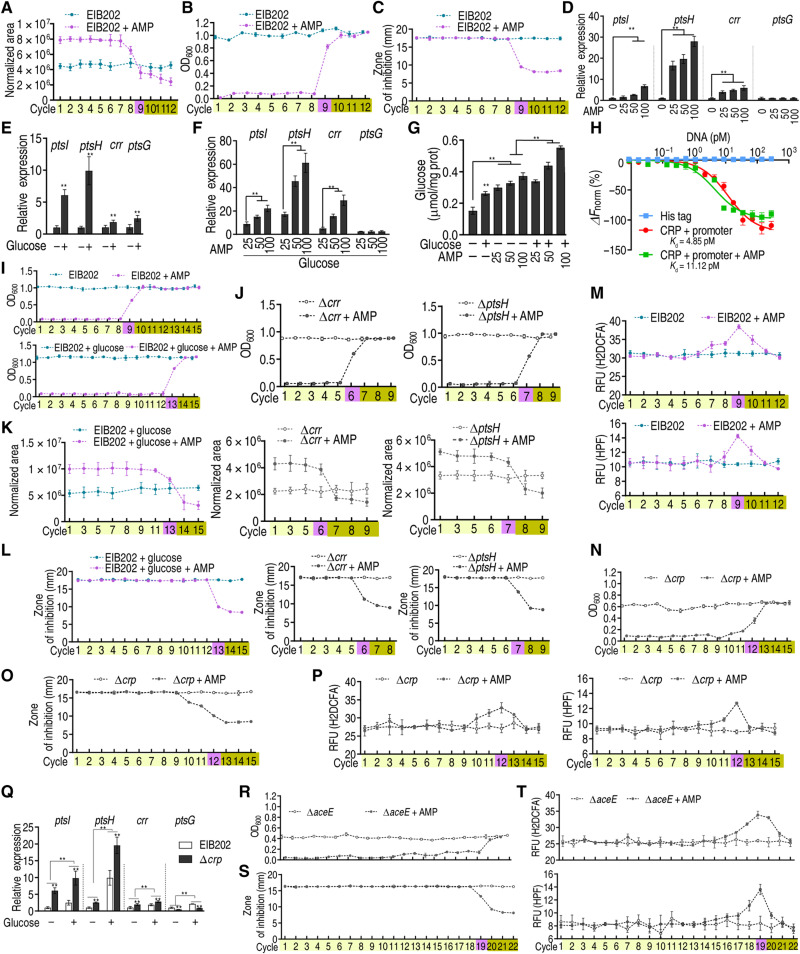
To investigate whether the regulation of glucose metabolism is a general pathway that facilitates the transition from tolerance to resistance, we test this model in an intracellular organism, E. tarda, whose multidrug-resistant isolates were frequently found in the environment (36). The abundance of glucose was decreased in a cycle-dependent manner, similar to that in E. coli (Fig. 8A). The switch point occurred, and resistance emerged since cycle 9 (Fig. 8B). Resistance was confirmed by the Oxford cup showing that the bacteria of cycle 9 were more resistant to ampicillin than the bacteria of cycles 1 to 8 (Fig. 8C and table S1). Expression of ptsI, ptsH, and crr was elevated in an ampicillin dose–dependent manner, and that of the three genes and ptsG was promoted by glucose (Fig. 8, D and E). The expression of ptsI, ptsH, and crr was markedly increased by the synergistic use of ampicillin and glucose (Fig. 8F). Consistently, intracellular glucose was elevated by the synergy (Fig. 8G). The binding of CRP with the promoter was inhibited by ampicillin (Fig. 8H). The resistance was delayed and advanced in the presence of 4 mM glucose and the absence of either crr or ptsH, respectively (Fig. 8, I and J), when glucose was elevated and decreased (Fig. 8K). Consistently, MIC was increased later if glucose was supplemented, but earlier if crr or ptsH was absent, when compared to wildtype cells(Fig. 8L and table S1). The abundance of ROS was substantially boosted in cycle 9, supporting our hypothesis that ROS plays critical roles in mutation generation during the switch point (Fig. 8M and fig. S18). Resistance and ROS elevation did not occur until cycle 12 in crp deletion mutant, which was confirmed by the Oxford cup experiment and ROS abundance (Fig. 8, N to P, and table S1). Expression of ptsI, ptsH, and crr was positively regulated by crp (ptsG was negatively regulated), which was promoted by exogenous glucose (Fig. 8Q). Similarly, the aceE deletion mutant has a more profound effect in delaying the occurrence of resistance and ROS increase (Fig. 8, R to T, and table S1). However, one aspect should be noticed that unlike the sudden shift from tolerance to resistance, the two deletion mutants undergo an extended time of the transition. These data together suggest that glucose metabolism plays a key role in mediating tolerance-to-resistance transition in E. tarda.
Fig. 8. AMP-PTS/PDH-glucose-cAMP/CRP-ROS regulatory cascade in E. tarda.
EIB202, EIB202 ∆crp, or EIB202 ∆aceE cells were treated as in Fig. 1. (A to C) Glucose (A), cell density/survival (B), and MIC (C) of EIB202 (n = 4) were measured after each cycle. (D to F) Gene expression of ptsI, ptsH, crr, and ptsG at the indicated AMP concentration (D), 4 mM glucose (E), or 4 mM glucose plus AMP (F) (n = 3). (G) Intracellular glucose level in the presence of 4 mM glucose or/and indicated AMP concentrations (n = 3). (H) MST for the binding of CRP with promoter in the presence of AMP (n = 3). (I and J) Growth/viability of EIB202 (I) and ∆crr and ∆ptsH (J) was measured after the indicated number of cycles exposed to AMP (n = 4). (K and L) Glucose concentration (K) and MIC (L) on data (I and K) (n = 4). (M) ROS abundance in EIB202 with AMP (n = 4). (N to P) Cell density/survival (N), MIC (O), and ROS abundance (P) of ∆crp after each cycle (n = 4). (Q) Gene expression of ptsI, ptsH, crr, and ptsG exposed to 4 mM glucose in EIB202 and ∆crp (n = 3). (R to T) Cell density/survival (R), MIC (S), and ROS abundance (T) of ∆aceE after each cycle (n = 4). Results are displayed as means ± SEM, and statistically significant differences are identified by Kruskal-Wallis followed by Dunn’s multiple comparison post hoc test for all the tests unless otherwise indicated. **P < 0.01.
DISCUSSION
Acquired antibiotic resistance is a rapidly growing problem worldwide. Several lines of evidence suggest that bacteria acquire tolerance before acquiring resistance to antibiotics (37, 38). Furthermore, unlike the phenotype of antibiotic resistance, the phenotype of antibiotic tolerance is not linked to specific genetic mutations (3, 11). Unfortunately, the mechanisms underlying tolerance and the transition from tolerance to resistance are largely unexplored and poorly understood.
Cellular metabolic state influences bacterial susceptibility to antibiotics, and antibiotic resistance is associated with a metabolic shift (13–15, 18, 21, 39). However, how metabolic state determines bacterial tolerance and the acquisition of resistance is largely unexplored. Here, we provide evidence that intracellular glucose concentration is reduced with increasing cycles of daily intermittent exposure to ampicillin. Specifically, high, unchanged, and low levels of glucose are detected in tolerance, switch point, and resistance, respectively. Ampicillin and cAMP/CRP consecutively manipulate tolerance and resistance. In tolerance, ampicillin binds with pts promoter P1b to promote PTS and with E1 (encoded by aceE) to inhibit PDH activity, driving glucose to PPP. However, the accumulated pyruvate resulting from the inhibition of PDH competes with ampicillin to activate PDH, promoting glucose consumption. Meanwhile, the phosphoenolpyruvate/pyruvate ratio is critical for glucose phosphorylation. Higher pyruvate reduces the ratio, making the utilization of glucose less effective (40). These cause a progressive decrease in glucose and elevation of ROS, which reach the plateau at tolerance. When glucose is progressively lower than normal, cAMP/CRP is activated to trigger the resistance. The activated cAMP/CRP has a strong ability to down-regulate PTS, which is consistent with reports on the repression of ptsG and ptsH in crp+ mutant (41, 42), thereby countering the up-regulation caused by ampicillin and leading to progressively reduced glucose. Furthermore, cAMP/CRP promotes ROS reduction, DNA repair, and ampC expression (Fig. 9). Therefore, the ampicillin-PTS/PDH-glucose-cAMP/CRP-ROS/AmpC regulatory cascade manipulates the evolution from tolerance to resistance via controlling glucose metabolism in bacteria, where PTS/PDH and cAMP/CRP play a crucial role in the manipulation of tolerance and resistance, respectively. Specifically, a high glucose level supports the tolerance, which is regulated by ampicillin through PTS and PDH, while a low glucose level promotes the resistance, which is regulated by cAMP/CRP. The conclusion is further supported by the data that the external glucose, aceE, or crp absence and pts promoter, crr, or pstH loss delay and promote the resistance, respectively. In addition, RpoS-DinB also contributes to MMR for antibiotic-induced mutagenesis.
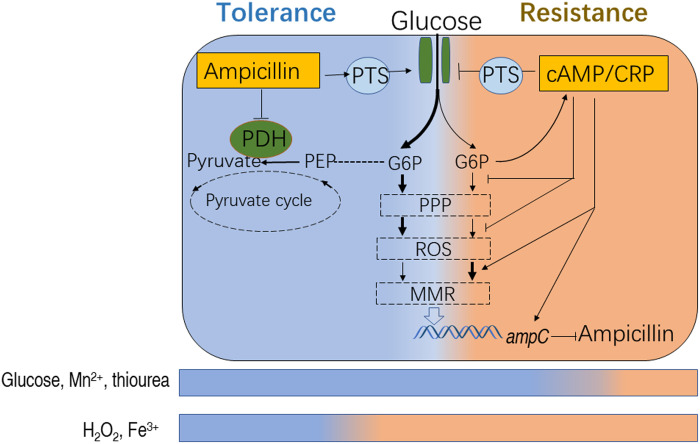
Fig. 9. Proposed model.
In tolerance, AMP enters bacterial cells, promotes pts, and inhibits PDH, which drive glucose fluxes into PPP and promote ROS production. In resistance, the reduced glucose gradually activates cAMP/CRP to promote DNA MMR and up-regulate the expression of ampC. The elevated AmpC confers AMP resistance. Reagents that scavenge ROS delay the occurrence of resistance, while those that promote ROS prime antibiotic resistance.
The present study has three core findings. First, it is ampicillin that targets the promoter P1b of PTS and the E1 of PDH to trigger the following events, which highlights the way in exploring antibiotic tolerance and resistance mechanisms based on the trigger antibiotic. Bacterial evolution to antibiotic is dependent on regulatory mechanisms that alter cellular physiology or genetic changes that lead to the degradation or sequestration of the antibiotic, prevent its uptake, pump it out of the cell, or prevent it from binding to the target molecule (43). However, which steps are triggered by antibiotics is not defined. The present study reveals that ampicillin binds with the pts promoter P1b and enzyme E1 to activate PTS and inhibit PDH, respectively, for regulating glucose abundance and flux and triggering the tolerance and following the evolution from tolerance to resistance. Information regarding antibiotics binding to DNA is not available. Here, one unexpected finding is that ampicillin directly targets DNA and modulates the gene expression to manipulate tolerance maintenance and the evolution from tolerance to resistance. We speculate that the interaction of antibiotic with DNA will emerge as an unrecognized mechanism for the development of antibiotic resistance.
Then, it is ampicillin-controlled pts and E1 that regulate glucose metabolism to support tolerance. The ampicillin-activated PTS promotes glucose content in the form of G6P, which is not out of the cell. However, glucose does not flux to glycolysis in the first three cycles. This is because ampicillin binds to and inhibits PDH, a core enzyme that links glycolysis to the P cycle (21); instead, it fluxes to PPP. The inhibition of PDH increases intracellular pyruvate to compete with ampicillin to E1, and thereby, PDH activity is recovered and even higher than in control in the fifth to sixth cycles of tolerance. The elevated PDH activity consumes glucose and is responsible for the progressively reduced glucose in these cycles of tolerance. All genes (glk, pfkA, and pykF) and enzymes (HK, PFK, PK, and PDH) in the glycolysis and P cycle display a progressive elevation in contrast to all genes (ptsI, ptsH, crr, ptsG, zwf, gnd, tktA, and tktB) and enzymes (G6PDH and 6-GPDH) in PTS and PPP that exhibit a progressive reduction in tolerance. Repression of glucose is reported in antibiotic-resistant bacterial cells and persisters (14, 15, 39, 44), but underlying mechanisms are not known. The findings not only reveal that the reduced glucose is responsible for the transition to resistance but also explain why ampicillin stress causes the progressive reduction of intracellular glucose, which is attributed to the balance between pts and E1 regulation.
Third, the present study establishes a prominent role of cAMP/CRP system in the occurrence of antibiotic resistance. cAMP and CRP act as a nutrient availability and energy sufficiency sensor and a transcription activator, respectively. Expression and activity of cAMP/CRP complex are negatively regulated by glucose availability (31). The system plays a key role in regulating the expression of many regulons and operons that encode enzymes and transporters involved in the catabolism of different nutrient sources (45). Reports have indicated that the role of cAMP/CRP system in aminoglycoside and mecillinam resistance is related to the regulation of l-alanine metabolism and the natural lipopolysaccharide defectiveness, respectively (46, 47). However, information regarding the decisive role of cAMP/CRP system in the occurrence of antibiotic resistance is unknown. The present study shows that intracellular glucose concentration correlates with the induction of cAMP/CRP system, which is consistent with the report (31). cAMP/CRP system is activated and plays a role in resistance when glucose is lower than in untreated control. The activated cAMP/CRP system inhibits glucose transport, reduces ROS and oxidative stress, and promotes DNA repair, especially DNA MMR. These cause decreased glucose, oxidative stress, and gene mutation rate, which promote the transition to an antibiotic-resistant state. These findings are consistent with the reports on lower total ROS and OH− levels in ampicillin-treated E. coli persisters and antibiotic-resistant bacteria than in control (14, 15, 48). However, the underlying mechanism is not known. The present study reveals that cAMP/CRP regulation of glucose transport and glucose catabolism, together with ampC overexpression and mutations on EnvZ and CpxA that control antibiotic uptake (13), plays crucial roles in developing antibiotic resistance.
Moreover, control measures are established to delay the transition since the development of strategies able to eliminate tolerant cells is a potential approach to preclude the evolution of resistance (11, 49). Increased survival and mutation rates in persisters jointly affect the likelihood of evolving clinical resistance, and anti-persister strategies may slow down resistance development (38). Inhibition of the AcrAB-TolC efflux pump improved treatment outcome by decreasing mutation rates (50). The present study shows that loss of crp or aceE and complement of glucose, Mn2+, and thiourea delay the transition to an antibiotic-resistant state and extend the period of the transition by approximately twofold. Thus, the CRP of cAMP/CRP system and AceE of the P cycle can be used as two targets and glucose and Mn2+ can be used as drugs to delay the transition. The two proteins and glucose, Mn2+, and thiourea work for cellular metabolism. These findings highlight the way to battle against antibiotic resistance via metabolic modulation to incorporate anti-tolerance therapies. Comparatively, the delays rank as thiourea > aceE loss > glucose > Mn2+ > crp loss. This is because thiourea has a stronger ability to quench ROS than Mn2+, and PDH/aceE is regulated by both cAMP/CRP and ampicillin, but CRP is regulated only by glucose.
An important result of the present study is that specific metabolic signatures are associated with bacterial states of tolerance and resistance to ampicillin, a likely strategy by which bacteria cope with antibiotic-induced cellular stress (18, 21, 39, 51). Although genetic factors such as isocitrate lyase that mediate broad antibiotic tolerance in Mycobacterium tuberculosis are described (52), metabolic factors are still less explored. The tolerance metabolome observed in the present study is characterized by seven putative biomarkers: glucose, maltose, leucine, aspartic acid, proline, valine, and tyrosine, where the abundance of glucose, maltose, and leucine is differentially lower, and that of aspartic acid, proline, and valine is differentially higher. This study looks in detail only at glucose metabolism, and the roles of other metabolites in the acquisition of antibiotic tolerance and/or resistance has and/or will be addressed separately.
E. coli is an extracellular bacterium, while E. tarda is an intracellular bacterium. To demonstrate that the ampicillin-PTS/PDH-glucose-cAMP/CRP-ROS regulatory cascade is general in mediating the transition from tolerance to resistance in bacteria, the present study continues to use E. tarda as a model to investigate the transition. Similar results are obtained in E. tarda, including the following: Ampicillin positively regulates PTS, glucose abundance decreases progressively as ampicillin-sensitive strains acquire increasing resistance to ampicillin, ROS elevation around the switch point, the MIC gradually increased and resistance gradually increased over time, and the absence of crr or pstH promotes and the loss of crp or aceE and exogenous glucose delays the transition. These results indicate that extracellular and intracellular bacteria contain the same ampicillin-PTS/PDH-glucose-cAMP/CRP-ROS regulatory cascade in mediating the transition from tolerance to resistance. Recently, Zeng et al. (53) used phenol to enrich tolerant mutants of E. coli and identified ptsI with a point mutation as a consequence of phenol tolerance. ΔptsI conferred a pan-tolerance phenotype to both disinfectants and antimicrobials, including β-lactams, quinolones, and aminoglycosides. They thus reveal a PtsI-CyaA-Crp–mediated death process that is dependent on ROS (53). Therefore, PTS/PDH-glucose-cAMP/CRP-ROS regulatory cascade can be activated by diverse stresses.
In summary, here we describe metabolic profiles specifically associated with ampicillin-tolerant and ampicillin-resistant E. coli and E. tarda. These findings provide insight into the mechanism(s) underlying antibiotic tolerance and resistance in E. coli and E. tarda and could facilitate the development of effective tools for combating antimicrobial tolerance and resistance.
MATERIALS AND METHODS
Bacterial strains
E. coli K12 and its knockout strains were obtained from the Keio collection. Each mutant strain was validated using polymerase chain reaction (PCR). E. tarda EIB202 (a gift from Y. Zhang, East China University of Science and Technology, China) and its knockout strains, clinically isolated E. coli S2 and S13, and aceE-, crp-, and zwf-complemented strains were from the collection of our laboratory.
The cyclic evolution of E. coli
The cyclic evolution was carried out as previously described (11). In detail, a single colony of E. coli K12, ∆crr E. coli K12, ∆ptsH E. coli K12, ∆crp E. coli K12, ∆crp-pACYC184, ∆crp-pACYC184-crp, ∆aceE E. coli K12, ∆aceE-pACYC184, ∆aceE-pACYC184-aceE, ∆zwf E. coli K12, ∆zwf-pACYC184, ∆zwf-pACYC184-zwf, E. coli S2, ∆pst promoter E. coli S2 and E. tarda, ∆crr E. tarda, ∆ptsH E. tarda, ∆crp E. tarda, and ∆aceE E. tarda was inoculated in LB medium (10 g of bacteriological peptones, 5 g of yeast powder, and 10 g of NaCl/L; from Guangdong Huankai Microbial Science and Technology Co. Ltd., China) and grown at 37°C for 16 hours. Aliquots of 4 mM glucose, 2.5 mM MnCl2, 40 mM thiourea, 320 μM C6H10FeNO8, and 1 mM H2O2 were added if desired. The cultures were collected by centrifugation, washed three times, and diluted with LB medium to 2 × 109 colony-forming units (CFU)/ml. Then, 500 μl of diluted cultures was resuspended in 50 ml of LB medium with ampicillin (100 μg/ml) and incubated at 37°C and 200 rpm for 4.5 hours. After incubation, antibiotic was washed out. The cultures were resuspended in fresh LB medium with ampicillin (0.625 μg/ml) and grown at 37°C for 16 hours, where nearly all of the surviving cells were kept from cycle to cycle. At the end of every cycle, some regrown cultures were diluted with LB medium for the next cycle, and the remainder was frozen in −80°C for further analysis. No ampicillin was used as a control. Instead, 28°C was used for E. tarda.
Determination of glucose in LB medium
Overnight cultures were subcultured at a 1:100 dilution in 50 ml of LB medium for 0 (without bacteria), 2, 4, and 6 hours. Supernatant was collected by centrifugation for glucose content using the CheKine Micro Glucose Assay Kit (KTB1300, Abbkine Scientific Co. Ltd., China). Briefly, 50 μl of supernatant was added to 1000 μl of o-toluidine reagent and vortexed. The mixture was heated in a boiling water bath for 8 min and cooled to room temperature in a cool water bath for 4 min. Then, 200 μl of each mixture was added to a 96-well plate, and absorbance at 630 nm was measured.
GC-MS analysis of cyclic evolution of bacteria
GC-MS analysis was carried out as described previously (13, 15). Briefly, 10 ml of OD600 = 1.0 cells [survived in medium with ampicillin (0.625 μg/ml) at the end of every cycle] was quenched with 1 ml of precooled methanol (Sigma-Aldrich) and then by ultrasonication. Metabolites were prepared by centrifugation at 12,000 rpm for 10 min, and ribitol (0.1 mg/ml; Sigma-Aldrich) was used as an internal standard. Supernatant (500 μl) was transferred into a 1.5-ml microtube and dried by a vacuum centrifugation device (LABCONCO). GC-MS analysis was carried out on the two-stage technique. The mass fragmentation spectrum was analyzed using XCalibur software (Thermo Fisher Scientific, version 2.1) to identify compounds by matching the data with the National Institute of Standards and Technology (NIST) library and NIST MS search 2.0 program. Peak areas of all identified metabolites were normalized by ribitol. Each sample had four biological repeats with two technical replicas.
Point mutation of pts promoter
Point mutation of pts promoter for P0a, P1a, and P1b positions was performed by two-step PCR method using Q5 DNA polymerase (New England Biolabs). Briefly, primers F1R1 and F2R2 were used for the first round of PCR. After the first step of PCR, quantity and quality of PCR products were checked in 2% agarose gel. The PCR products were digested with Dpn I (to remove methylated DNA to prevent its interference in the second PCR step). Fragments were purified by QIAGEN MinElute spin column. Equal concentration of each purified fragment from the first PCR amplification was taken as templates for the next round of reaction with primer F1R2. The purified gene product was cloned into the T vector. Mutation was confirmed from the selected colonies by Sanger sequencing. For the point mutation of fragment, the mutated single chains were synthesized by Guangzhou IGE Biotechnology Company and were annealed to form a double chain. All primers are listed in tables S2 and S3.
Measurement of ampicillin and tetracycline binding with DNA
PCR products of pts promoter fragments were purified by a PCR purification kit (Sangon Biotech, Shanghai, China), and their concentration was quantified. Various concentrations of DNA, ranging from 12.5 to 400 ng, were mixed with tris-HCl buffer (100 μg/ml, pH 8.0) at 37°C for 3 hours. Twenty tubes were pooled and precipitated by 1:1 isopropanol for 10 min and centrifuged at 14,000g for 10 min. The precipitation was washed three times with 75% ethanol and dissolved with 10 μl of tris-HCl buffer (pH 8.0) for measurement of ampicillin (Shenzhen Technology, China) or tetracycline (Shanghai Enzyme-linked Biotechnology Co. Ltd., China) with enzyme-linked immunosorbent assay (ELISA) kits and bactericidal assay.
Antibiotic bactericidal assay by plate counting
Antibiotic bactericidal assay was performed as described previously (13). A single colony was propagated in 50 ml of LB medium in a 250-ml flask for 14 hours at 37°C in a shaker. The cultures were collected by centrifugation at 8000 rpm for 5 min. The samples were washed three times with saline solution; resuspended in M9 minimal medium (M9) supplemented with 10 mM acetate, 2 mM MgSO4, and 100 mM CaCl2; and then diluted to an OD600 of 0.6. Glucose or ampicillin (2 μg/ml) was added and incubated at 37°C and 200 rpm for 6 hours. To determine bacterial counts at specified time points, 100-μl portions of samples were removed and serially diluted. An aliquot of 10 μl of each dilution was plated onto LB agar plates and cultured at 37°C for 12 hours to determine CFU. Only dilutions that yielded 20 to 200 colonies were available. Percent survival was determined by dividing the CFU obtained from a treated sample by the CFU obtained from a control sample.
Bactericidal assay using ampicillin binding with pts promoter
Overnight E. coli K12 cultures were collected and washed with saline solution. The cultures were adjusted to OD600 of 0.6 with M9 medium and diluted into 1:100,000. Then, 50 μl of bacteria was incubated with or without pst promoter precipitated by isopropanol as described above at 37°C with shaking at 200 rpm for 6 hours. CFU per milliliter was determined as described above.
Survival capability
Survival capability test was performed as previously described (54). Frozen stocks were grown overnight on fresh LB at 37°C. The saturated bacteria were diluted (1:1000) into fresh LB medium, and ampicillin (2 μg/ml) or an indicated concentration of glucose was added. After shaking at 37°C for 6 hours, the OD600 was measured, survival rate was calculated in each group, and without antibiotics was used as a control.
Construction for pts promoter–deleted mutants
Gene pts promoter–deleted mutants were constructed to resemble those in Keio knockout collection. KanR cassette was amplified by using pKD13 as a template with sequences homologous to the 5′ and 3′ ends of the target genes in the chromosome. Primer pairs used for gene deletion are listed in table S2. Amplified DNA was electroporated into clinically isolated antibiotic-sensitive E. coli S2 and S13 harboring pSIM5 that expresses the lambda red genes from a high-temperature–inducible promoter (pSIM5). Recombinants were selected on LB agar plate supplemented with kanamycin (50 μg/ml). The KanR mutants were transformed with pCP20, and chloramphenicol-resistant transformants were selected at 30°C, after which a few were colony-purified once nonselective at 43°C and then tested for loss of kanamycin and chloramphenicol resistances. The majority lost the FRT (FLP recognition target) flanked resistance gene and the FLP helper plasmid simultaneously.
Bacterial infection by pts promoter–deleted mutants and treated with ampicillin in a mouse model
The experiment was carried out as described previously (13). Male mice (BALB/c, pathogen-free), weighing 20 ± 2 g from the same litter, were obtained from the Animal Center of Sun Yat-sen University. Mice were reared in cages and fed with sterile water and dry pellet diets. Mice were intraperitoneally challenged with E. coli S2, S13, or their corresponding pst promoter mutants (4 × 106 CFU per mouse; n = 20). One hour later, they were treated with ampicillin (20 mg/kg) or the same volume of sterile saline (n = 10). Survivals were monitored for 7 days.
Construction of point mutations in aceE gene
Point mutations in aceE gene were constructed using double-enzyme digestion and enzyme ligation. Briefly, PCR amplification was performed according to aceE-F and aceE-R primers in (table S4) to obtain the full-length sequence of aceE gene and ligated between Sac I and Xho I sites in pET-28a vector. Nine pairs of primers were designed and amplified by PCR to separately introduce nine mutation sites. The fragments containing the mutation sites and the unmutated pET-28a-aceE plasmid were digested by Eco RI and Hind III and then ligated by T4 ligase to obtain an expression vector containing the mutation site. Last, the unmutated or mutated vector was transferred to BL21(DE3) competent cells for protein expression.
Expression and purification of AceE and its mutants
For AceE and eight mutants’ protein expression and purification, a colony of DE3 containing recombinant plasmids pET-28a-aceE or each of the eight pET-28a-mutant genes was cultured in 5 ml of LB medium with ampicillin (100 μg/ml) at 37°C with shaking for 16 hours. The cultures were diluted in 1:100 to 500 ml of LB medium supplemented with ampicillin and incubated at 37°C until the OD600 reached 0.5. Then, 0.1 mM isopropyl-β-d-thiogalactoside (IPTG) was added to induce the recombinant protein at 16°C for 12 hours. Bacteria were harvested by centrifugation at 8000g and resuspended in 0.01 mM (pH 7.4) phosphate-buffered saline (1 × PBS). The resulting cells were disrupted by high pressure (1.586 × 10+8 Pa) using a cell disruptor (Constant Systems Ltd., Daventry, UK), and supernatant was obtained by centrifugation at 8000g for 30 min at 4°C. The supernatant was applied onto a nickel-affinity column and incubated at 4°C for 2 hours. After protein binding, the column was washed with 10 volumes of washing buffer (1× PBS buffer with 5 mM imidazole) to remove unbound protein. Recombinant proteins were eluted with elution buffer (1× PBS buffer with 100 mM imidazole), respectively. Purity and concentration of recombinant proteins were analyzed and quantified by SDS–polyacrylamide gel electrophoresis and a bicinchoninic acid protein concentration determination kit (Beyotime, P0009).
Measurement for ampicillin binding with PDH in vivo
E. coli BL21-pET-28a-AceE, BL21-pET-28a-m7, and BL21-pET-28a-m8 were cultured in LB medium overnight at 37°C, with shaking at 200 rpm, and then diluted into 1:100 using fresh LB medium growing to an OD600 of 0.5. Protein expressions were induced with 0.01 mM IPTG after optimization of the expression conditions. Ampicillin (100 μg/ml) or tetracycline (100 μg/ml) was added and cultured for 4.5 hours. Bacteria were collected by centrifugation, washed, resuspended with 1 × PBS, and disrupted by intermittent sonic oscillation in an ice bath, followed by centrifugation. Cell debris was removed, and clarified supernatant was loaded onto a column packed with Ni+-NTA super flow resin for purification of recombinant proteins. Concentrations of the recombinant proteins were determined using the Bradford method. Aliquot of a 200-μg purified protein was taken for antibiotic detection by using commercially available ELISA kits (Lusiyuan Biotech. Corp., China, for ampicillin detection; Fantai Biotech. Corp., China, for tetracycline detection).
Quantitative reverse transcription PCR
Quantitative reverse transcription (qRT)–PCR was carried out as described previously (55). Briefly, total RNA was isolated with TRIzol (Invitrogen, USA) as templates. qRT-PCR was performed by a PrimeScript RT reagent kit with genomic DNA eraser (Takara, Japan) in 384-well plates. Each well contained a total volume of 10 μl of liquid, including 5 μl of 2× SYBR Premix Ex Taq, 2.6 μl of PCR-grade water, 2 μl of complementary DNA template, and 0.2 μl of each pair of primers (10 mM) (table S3). All samples were performed on a LightCycle 480 system (Roche, Germany), and four independent samples were assayed for both control and test groups. The cycling parameters are listed as follows: 95°C for 30 s to activate the polymerase; 45 cycles of 95°C for 10 s; and 57°C for 30 s. Fluorescence measurements were performed at 70°C for 1 s during each cycle. Cycling was terminated at 95°C with a calefactive velocity of 5°C/s, and a melting curve was obtained. 16S ribosomal RNA was used as an internal control to normalize the relative expression level of the target gene. We converted the data to percentages relative to the value of the no-treatment group. At least triplicate repeats were carried out.
Antimicrobial susceptibility test to quantify ampicillin bound with recombinant proteins
Antimicrobial susceptibility test was performed by the Oxford cup method as described previously (11). Briefly, cells were resuspended and diluted to 107 CFU/ml using LB medium. Aliquots of 100 μl of cells were spread on LB agar plates, and 40 μl of ampicillin (0.125 mg/ml) was poured into Oxford cups on the LB plates. Inhibition zone was measured overnight. For quantification of ampicillin bound with recombinant AceE, GLK, PFK1, PFKII, and PK1, these recombinant proteins were purified by a nickel-affinity column. Aliquots of 800 μg of proteins were mixed with 2000 μg of ampicillin at 37°C for 4 hours, and then we added 10 times the volume of acetone precooled at −20°C. Following precipitation at −20°C for 2 hours, proteins were harvested by centrifugation at 8000g at 4°C for 5 min. The acetone precipitation was repeated. Residual acetone was removed by spin drying. The resulting proteins were suspended with 50 mM tris-HCl and quantified. Aliquots of 100 μg of proteins were added to the Oxford cup, and inhibition zone was measured overnight. Ampicillin of 20, 40, 60, 80, and 100 ng was used as a reference standard. All plates were incubated at 37°C for 16 hours. The ampicillin bound with AceE was quantified using the reference standard curve.
Measurement of enzyme activity
Measurement of enzyme activity was performed as described previously (56). Cells (10 ml, OD600 = 1.0) were resuspended in 500 μl of PBS and broken down by sonication for 2 min at a 200-W power setting on ice and then centrifuged at 14,000g for 10 min to remove insoluble material. Supernatants containing 400 μg of total proteins were transferred to PDH reaction mix (0.5 mM MTT, 1 mM MgCl2, 6.5 mM PMS, 0.2 mM TPP, 2 mM sodium pyruvate, and 50 mM PBS) to a final volume of 200 μl in a 96-well plate. The plate was incubated at 37°C for 5 min and then measured at 566 nm for colorimetric readings. The plate was protected from light during the incubation. Activity of SOD, CAT, HK, PFK, PK, and citrate synthase was measured using commercial kits (Nanjing Jiancheng, A007-1). The samples were incubated with reagents given in the kit and analyzed by a microplate reader (Varioskan LUX, Thermo Fisher Scientific, USA) at 405 nm. Light was avoided during the test.
Microscale thermophoresis
MST assay was carried out as previously described (57). Briefly, recombinant CRP and AceE were purified with amylose resin (New England BioLabs). The purified CRP (10 μM) was incubated with 30 μM labeling dye for 30 min, followed by 16 serial twofold dilutions into buffer containing DNA. After 5 min of incubation, 4 μl of each reaction was enclosed in premium-coated glass capillaries and subjected to MST on a Monolith NT.115 NanoTemper instrument, at 40% MST power and 40% light-emitting diode power. Data were analyzed by MST software (MO.Affinity, Munich, Germany). The purified AceE and N-hydroxysuccinimide (NHS) ester fluorescent dye solutions were mixed at a ratio of 1:1 and incubated in the dark at room temperature for 30 min. Serial dilutions of ampicillin or pyruvate with PBS buffer were mixed with 20 μM NHS fluorescent-labeled AceE and then incubated at room temperature for 5 min. The sample was loaded into MST-glass capillaries, and a NanoTemper Monolith NT.115T instrument was used for MST analysis. By plotting the concentration of antibiotic or metabolite as the per mil change of normalized fluorescence [∆Fnorm (‰) = (Antibiotic or metabolite fluorescence − Control fluorescence) / Control fluorescence], curve fitting was performed using GraphPad Prism software 8.0, and dissociation constant (Kd) values were determined. Three independent samples were carried out.
cAMP assay
Cells were resuspended in 0.1 N HCl, and total lysate was extracted. The concentration of cAMP was measured with a cAMP ELISA kit (Enzo Life Sciences, Farmingdale, NY, USA) according to the manufacturer’s protocol.
Quantification of ROS production
ROS production was quantified by fluorescence as described previously (58). Cells were resuspended in PBS and incubated with 10 mM carboxy-H2DCFDA (Sigma-Aldrich) or 5 μM hydroxyphenyl fluorescein solution (Sigma-Aldrich) at 37°C for 30 min in the dark. The cells were washed twice with PBS and analyzed by a microplate reader (Varioskan LUX, Thermo Fisher Scientific, USA) at an excitation and emission wavelength of 495 and 525 nm, respectively.
Determination of NADPH
Intracellular NADPH concentrations were measured using the Enzychrom NADP/NADPH assay kit (BioAssay Systems) according to the manufacturer’s protocol.
Measurement of intracellular ampicillin concentration
Measurement of intracellular ampicillin was carried out with an ampicillin ELISA rapid diagnostic kit (ShangHai QiYi Biological Technology Group, QY142). Briefly, E. coli K12 was incubated with ampicillin (100 μg/ml) in LB medium at 37°C for 4.5 hours. The cells were washed three times with sterile saline and centrifuged at 14,000g for 10 min. Liquid was removed, and precipitation was made as dry as possible for analysis. To measure intracellular ampicillin concentration, we calculated the intracellular water volume. To do this, dry weight and fresh weight of the incubated cells were measured, and the ratio between the dry weight and fresh weight was calculated. Volume of intracellular water is calculated using the equation: fresh weight × (1 − 0.23 − dry weight/fresh weight). Intracellular ampicillin was measured with an ampicillin ELISA rapid diagnostic kit, and ampicillin concentration is determined by the mass of intracellular ampicillin/volume of intracellular water.
Measurement of AmpC activity
The assay was performed using susceptibility test disks (Oxoid), according to 2010 CLSI guidelines. Bacteria cultured in ampicillin (0.625 μg/ml) at 37°C for 16 hours were adjusted to a 0.5 McFarland standard. These bacteria were uniformly inoculated with Mueller-Hinton agar, and then, a 30-μg cefoxitin disk was placed at the center of the disk. After overnight incubation at 37°C, zone of inhibition was detected. Zone of inhibition 18 was defined as a positive threshold. Klebsiella pneumoniae ATCC700603 and Enterobacter cloacae ATCC13047 were used as negative and positive controls, respectively.
Whole-genome sequencing and bioinformatic analysis
E. coli K12 cultured with and without ampicillin (0.625 μg/ml) at 37°C for 16 hours was collected, 20 clones with ampicillin and 3 clones without ampicillin for each cycle, yielding 206 datasets (one absent in the third cycle with ampicillin). Fragmented DNA was purified using sample purification beads (Illumina), and whole-genome resequencing was performed by Biomarker Technologies (Beijing, China) with Illumina platforms. According to standard Illumina protocol, each DNA library was sequenced with the Illumina HiSeq X. The high-quality filtered data were compared to the reference genome (E. coli BW25113) using the bwa-0.7.15 program according to the default parameters. SAMtools (v. 0.1.19-44428cd) was used to convert the SAM format file from the comparison to the bam file format. We adopted the GATK package (v4.02) (https://broadinstitute.org/gatk/download/) to sort the BAM file and remove duplicates. Subsequently, the Unified Genotype module of GATK software was used to detect single-nucleotide polymorphism (SNP). To guarantee the reliability of SNP sites, the obtained SNP sites were filtered according to the filter parameter “QD<2.0 | | MQ<40.0 | | FS>60.0 | | SOR>3.0 | | QUAL<30 | | DP<5”. SnpEff software was used to obtain SNP sites (https://pcingola.github.io/SnpEff/).
Acknowledgments
We thank S.-r. Liu and X. Ma for technical assistance.
Funding: This work was supported by the International Cooperation and Exchanges NSFC 32061133007 (to B.P.); National Natural Science Foundation of China 31822058 (to B.P.), 42276125 (to H.L.), 32102829 (to M.J.), and 31902414 (to J.-z.Y.); and Innovation Group Project of Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai) no. 311020006 (to X.-x.P.). Ethics statement: This study was conducted in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and maintained according to the standard protocols. All experiments were approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University (SYSU-IACUC-2020-B1267).
Author contributions: Conceptualization: B.P. Methodology: M.J., Y.-b.S., J.-z.Y., H.L., S.-f.K., and J.-h.W. Investigation: M.J., Y.-b.S., J.-z.Y., H.L., S.-f.K., J.-h.W., and S.-h.L. Visualization: M.J., Y.-b.S., J.-z.Y., and H.L. Funding acquisition: B.P., H.L., M.J., J.-z.Y., and X.-x.P. Project administration: B.P. and X.-x.P. Supervision: B.P. and X.-x.P. Writing—original draft: B.P. Writing—review and editing: B.P., X.-x.P., H.L., and M.J.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The whole-genome sequencing data are deposited to the National Library of Medicine with accession no PRJNA909844 (https://dataview.ncbi.nlm.nih.gov/object/PRJNA909844?reviewer=uqsab7ht9jjl44lcsphtmfp9).
Supplementary Materials
This PDF file includes:
Figs. S1 to S18
Tables S1 to S4
REFERENCES AND NOTES
- 1.M. L. Nadimpalli, S. J. Marks, M. C. Montealegre, R. H. Gilman, M. J. Pajuelo, M. Saito, P. Tsukayama, S. M. Njenga, J. Kiiru, J. Swarthout, M. A. Islam, T. R. Julian, A. J. Pickering, Urban informal settlements as hotspots of antimicrobial resistance and the need to curb environmental transmission. Nat. Microbiol. 5, 787–795 (2020). [DOI] [PubMed] [Google Scholar]
- 2.A. Brauner, O. Fridman, O. Gefen, N. Q. Balaban, Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 14, 320–330 (2016). [DOI] [PubMed] [Google Scholar]
- 3.O. Fridman, A. Goldberg, I. Ronin, N. Shoresh, N. Q. Balaban, Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 513, 418–421 (2014). [DOI] [PubMed] [Google Scholar]
- 4.C. W. Hall, T. Mah, Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 41, 276–301 (2017). [DOI] [PubMed] [Google Scholar]
- 5.D. Martins, G. McKay, G. Sampathkumar, M. Khakimova, A. M. English, D. Nguyen, Superoxide dismutase activity confers (p)ppGpp-mediated antibiotic tolerance to stationary-phase Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 115, 9797–9802 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.I. El Meouche, M. J. Dunlop, Heterogeneity in efflux pump expression predisposes antibiotic-resistant cells to mutation. Science 362, 686–690 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.R. Guérillot, L. Li, S. Baines, B. Howden, M. B. Schultz, T. Seemann, I. Monk, S. J. Pidot, W. Gao, S. Giulieri, A. Gonçalves da Silva, A. D'Agata, T. Tomita, A. Y. Peleg, T. P. Stinear, B. P. Howden, Comprehensive antibiotic-linked mutation assessment by resistance mutation sequencing (RM-seq). Genome Med. 10, 63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O. Gefen, C. Gabay, M. Mumcuoglu, G. Engel, N. Q. Balaban, Single-cell protein induction dynamics reveals a period of vulnerability to antibiotics in persister bacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 6145–6149 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.K. Lewis, Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5, 48–56 (2007). [DOI] [PubMed] [Google Scholar]
- 10.D. Madar, E. Dekel, A. Bren, A. Zimmer, Z. Porat, U. Alon, Promoter activity dynamics in the lag phase of Escherichia coli. BMC Syst. Biol. 7, 136 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.I. Levin-Reisman, I. Ronin, O. Gefen, I. Braniss, N. Shoresh, N. Q. Balaban, Antibiotic tolerance facilitates the evolution of resistance. Science 355, 826–830 (2017). [DOI] [PubMed] [Google Scholar]
- 12.A. Gutierrez, S. Jain, P. Bhargava, M. Hamblin, M. A. Lobritz, J. J. Collins, Understanding and sensitizing density-dependent persistence to quinolone antibiotics. Mol. Cell 68, 1147–1154.e3 (2017). [DOI] [PubMed] [Google Scholar]
- 13.X.-L. Zhao, Z.-G. Chen, T.-C. Yang, M. Jiang, J. Wang, Z.-X. Cheng, M.-J. Yang, J.-X. Zhu, T.-T. Zhang, H. Li, B. Peng, X.-X. Peng, Glutamine promotes antibiotic uptake to kill multidrug-resistant uropathogenic bacteria. Sci. Transl. Med. 13, eabj0716 (2021). [DOI] [PubMed] [Google Scholar]
- 14.B. Peng, Y. B. Su, H. Li, Y. Han, C. Guo, Y. M. Tian, X. X. Peng, Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metab. 21, 249–262 (2015). [DOI] [PubMed] [Google Scholar]
- 15.S. Zhang, J. Wang, M. Jiang, D. Xu, B. Peng, X. X. Peng, H. Li, Reduced redox-dependent mechanism and glucose-mediated reversal in gentamicin-resistant Vibrio alginolyticus. Environ. Microbiol. 21, 4724–4739 (2019). [DOI] [PubMed] [Google Scholar]
- 16.S.-F. Kuang, D.-Y. Feng, Z.-G. Chen, Z.-Z. Liang, J.-J. Xiang, H. Li, X.-X. Peng, T. Zhang, Inactivation of nitrite-dependent nitric oxide biosynthesis is responsible for overlapped antibiotic resistance between naturally and artificially evolved Pseudomonas aeruginosa. mSystems 6, e0073221 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Y.-B. Su, S.-F. Kuang, J.-Z. Ye, J.-J. Tao, H. Li, X.-X. Peng, B. Peng, Enhanced biosynthesis of fatty acids is associated with the acquisition of ciprofloxacin resistance in Edwardsiella tarda. mSystems 6, e0069421 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.B. Peng, H. Li, X. X. Peng, Functional metabolomics: From biomarker discovery to metabolome reprogramming. Protein Cell 6, 628–637 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.G. Gosset, Improvement of Escherichia coli production strains by modification of the phosphoenolpyruvate:sugar phosphotransferase system. Microb. Cell Fact. 4, 14 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.S. Ryu, S. Garges, Promoter switch in the Escherichia coli pts operon. J. Biol. Chem. 269, 4767–4772 (1994). [PubMed] [Google Scholar]
- 21.Y.-B. Su, B. Peng, H. Li, Z.-X. Cheng, T.-T. Zhang, J.-X. Zhu, D. Li, M.-Y. Li, J.-Z. Ye, C.-C. Du, S. Zhang, X.-L. Zhao, M.-J. Yang, X.-X. Peng, Pyruvate cycle increases aminoglycoside efficacy and provides respiratory energy in bacteria. Proc. Natl. Acad. Sci. U.S.A. 115, E1578–E1587 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.P. Arjunan, N. Nemeria, A. Brunskill, K. Chandrasekhar, M. Sax, Y. Yan, F. Jordan, J. R. Guest, W. Furey, Structure of the pyruvate dehydrogenase multienzyme complex E1 component from Escherichia coli at 1.85 Å resolution. Biochemistry 41, 5213–5221 (2002). [DOI] [PubMed] [Google Scholar]
- 23.M. Fries, H. J. Chauhan, G. J. Domingo, H.-I. Jung, R. N. Perham, Site-directed mutagenesis of a loop at the active site of E1 (α2β2) of the pyruvate dehydrogenase complex. A possible common sequence motif. Eur. J. Biochem. 270, 861–870 (2003). [DOI] [PubMed] [Google Scholar]
- 24.J. Yi, N. Nemeria, A. McNally, F. Jordan, R. S. Machado, J. R. Guest, Effect of substitutions in the thiamin diphosphate-magnesium fold on the activation of the pyruvate dehydrogenase complex from Escherichia coli by cofactors and substrate. J. Biol. Chem. 271, 33192–33200 (1996). [DOI] [PubMed] [Google Scholar]
- 25.J. Blázquez, J. Rodríguez-Beltrán, I. Matic, Antibiotic-induced genetic variation: How it arises and how it can be prevented. Annu. Rev. Microbiol. 72, 209–230 (2018). [DOI] [PubMed] [Google Scholar]
- 26.J.-Y. Liang, J.-M. P. Yuann, C.-W. Cheng, H.-L. Jian, C.-C. Lin, L.-Y. Chen, Blue light induced free radicals from riboflavin on E. coli DNA damage. J. Photochem. Photobiol. B. 119, 60–64 (2013). [DOI] [PubMed] [Google Scholar]
- 27.J.-Z. Ye, Y.-b. Su, X.-m. Lin, S.-s. Lai, W.-x. Li, F. Ali, J. Zheng, B. Peng, Alanine enhances aminoglycosides-induced ROS production as revealed by proteomic analysis. Front. Microbiol. 9, 29 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.M. N. Ahmed, A. Porse, A. Abdelsamad, M. Sommer, N. Høiby, O. Ciofu, Lack of the major multifunctional catalase KatA in Pseudomonas aeruginosa accelerates evolution of antibiotic resistance in ciprofloxacin-treated biofilms. Antimicrob. Agents Chemother. 63, e00766-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.P. Belenky, J. J. Collins, Antioxidant strategies to tolerate antibiotics. Science 334, 915–916 (2011). [DOI] [PubMed] [Google Scholar]
- 30.C. C. Gruber, V. M. P. Babu, K. Livingston, H. Joisher, G. C. Walker, Degradation of the Escherichia coli essential proteins DapB and Dxr results in oxidative stress, which contributes to lethality through incomplete base excision repair. mBio 13, e0375621 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.T. Shimada, N. Fujita, K. Yamamoto, A. Ishihama, Novel roles of cAMP receptor protein (CRP) in regulation of transport and metabolism of carbon sources. PLOS ONE 6, e20081 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.L. Huang, P. Tsui, M. Freundlich, Positive and negative control of ompB transcription in Escherichia coli by cyclic AMP and the cyclic AMP receptor protein. J. Bacteriol. 174, 664–670 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.K. Yamamoto, R. Nagura, H. Tanabe, N. Fujita, A. Ishihama, R. Utsumi, Negative regulation of the bolA1p of Escherichia coli K-12 by the transcription factor OmpR for osmolarity response genes. FEMS Microbiol. Lett. 186, 257–262 (2000). [DOI] [PubMed] [Google Scholar]
- 34.J. M. Santos, M. Lobo, A. P. A. Matos, M. A. D. Pedro, C. M. Arraiano, The gene bolA regulates dacA (PBP5), dacC (PBP6) and ampC (AmpC), promoting normal morphology in Escherichia coli. Mol. Microbiol. 45, 1729–1740 (2002). [DOI] [PubMed] [Google Scholar]
- 35.R. S. Galhardo, R. Do, M. Yamada, E. C. Friedberg, P. J. Hastings, T. Nohmi, S. M. Rosenberg, DinB upregulation is the sole role of the SOS response in stress-induced mutagenesis in Escherichia coli. Genetics 182, 55–68 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Q. Wang, M. Yang, J. Xiao, H. Wu, X. Wang, Y. Lv, L. Xu, H. Zheng, S. Wang, G. Zhao, Q. Liu, Y. Zhang, Genome sequence of the versatile fish pathogen Edwardsiella tarda provides insights into its adaptation to broad host ranges and intracellular niches. PLOS ONE 4, e7646 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.J. Liu, O. Gefen, I. Ronin, M. Bar-Meir, N. Q. Balaban, Effect of tolerance on the evolution of antibiotic resistance under drug combinations. Science 367, 200–204 (2020). [DOI] [PubMed] [Google Scholar]
- 38.E. M. Windels, J. E. Michiels, M. Fauvart, T. Wenseleers, B. Van den Bergh, J. Michiels, Bacterial persistence promotes the evolution of antibiotic resistance by increasing survival and mutation rates. ISME J. 13, 1239–1251 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.A. J. Lopatkin, S. C. Bening, A. L. Manson, J. M. Stokes, M. A. Kohanski, A. H. Badran, A. M. Earl, N. J. Cheney, J. H. Yang, J. J. Collins, Clinically relevant mutations in core metabolic genes confer antibiotic resistance. Science 371, eaba0862 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.B. M. Hogema, J. C. Arents, R. Bader, K. Eijkemans, H. Yoshida, H. Takahashi, H. Aiba, P. W. Postma, Inducer exclusion in Escherichia coli by non-PTS substrates: The role of the PEP to pyruvate ratio in determining the phosphorylation state of enzyme IIAGlc. Mol. Microbiol. 30, 487–498 (1998). [DOI] [PubMed] [Google Scholar]
- 41.R. Yao, Y. Hirose, D. Sarkar, K. Nakahigashi, Q. Ye, K. Shimizu, Catabolic regulation analysis of Escherichia coli and its crp, mlc, mgsA, pgi and ptsG mutants. Microb. Cell Fact. 10, 67 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.S. Adhya, Escherichia coli and Salmonella typhimurium; Cellular and Molecular Biology (American Society for Microbiology, 1987), pp. 1503–1512. [Google Scholar]
- 43.L. Sandegren, D. I. Andersson, Bacterial gene amplification: Implications for the evolution of antibiotic resistance. Nat. Rev. Microbiol. 7, 578–588 (2009). [DOI] [PubMed] [Google Scholar]
- 44.X.-k. Tang, Y.-b. Su, H.-q. Ye, Z.-y. Dai, H. Yi, K.-x. Yang, T.-t. Zhang, Z.-g. Chen, Glucose-potentiated amikacin killing of cefoperazone/sulbactam resistant Pseudomonas aeruginosa. Front. Microbiol. 12, 800442 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.G. Almagro, A. M. Viale, M. Montero, F. J. Muñoz, E. Baroja-Fernández, H. Mori, J. Pozueta-Romero, A cAMP/CRP-controlled mechanism for the incorporation of extracellular ADP-glucose in Escherichia coli involving NupC and NupG nucleoside transporters. Sci. Rep. 8, 15509 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.D. N. Antón, Resistance to mecillinam produced by the co-operative action of mutations affecting lipopolysaccharide, spoT, and cya or crp genes of Salmonella typhimurium. Mol. Microbiol. 16, 587–595 (1995). [DOI] [PubMed] [Google Scholar]
- 47.M. Jiang, S.-f. Kuang, S.-s. Lai, S. Zhang, J. Yang, B. Peng, X.-x. Peng, Z.-g. Chen, H. Li, Na+-NQR confers aminoglycoside resistance via the regulation of l-alanine metabolism. mBio 11, e02086-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.R. C. Molina-Quiroz, C. Silva-Valenzuela, J. Brewster, E. Castro-Nallar, S. B. Levy, A. Camilli, Cyclic AMP regulates bacterial persistence through repression of the oxidative stress response and SOS-dependent DNA repair in uropathogenic Escherichia coli. mBio 9, e02144-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.E. M. Windels, J. E. Michiels, B. Van den Bergh, M. Fauvart, J. Michiels, Antibiotics: Combatting tolerance to stop resistance. mBio 10, e02095-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.J. Frimodt-Møller, A. Løbner-Olesen, Efflux-pump upregulation: From tolerance to high-level antibiotic resistance? Trends Microbiol. 27, 291–293 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Z.-x. Cheng, M.-J. Yang, B. Peng, X.-x. Peng, X.-m. Lin, H. Li, The depressed central carbon and energy metabolisms is associated to the acquisition of levofloxacin resistance in Vibrio alginolyticus. J. Proteomics 181, 83–91 (2018). [DOI] [PubMed] [Google Scholar]
- 52.M. Nandakumar, C. Nathan, K. Y. Rhee, Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nat. Commun. 5, 4306 (2014). [DOI] [PubMed] [Google Scholar]
- 53.J. Zeng, Y. Hong, N. Zhao, Q. Liu, W. Zhu, L. Xiao, W. Wang, M. Chen, S. Hong, L. Wu, Y. Xue, D. Wang, J. Niu, K. Drlica, X. Zhao, A broadly applicable, stress-mediated bacterial death pathway regulated by the phosphotransferase system (PTS) and the cAMP-Crp cascade. Proc. Natl. Acad. Sci. U.S.A. 119, e2118566119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.H. Li, X.-m. Lin, S.-y. Wang, X.-x. Peng, Identification and antibody-therapeutic targeting of chloramphenicol-resistant outer membrane proteins in Escherichia coli. J. Proteome Res. 6, 3628–3636 (2007). [DOI] [PubMed] [Google Scholar]
- 55.W. Li, Z. Yao, L. Sun, W. Hu, J. Cao, W. Lin, X. Lin, Proteomics analysis reveals a potential antibiotic cocktail therapy strategy for Aeromonas hydrophila infection in biofilm. J. Proteome Res. 15, 1810–1820 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Z.-x. Cheng, Q.-y. Gong, Z. Wang, Z.-g. Chen, J.-z. Ye, J. Li, J. Wang, M.-j. Yang, X.-p. Ling, B. Peng, Edwardsiella tarda tunes tricarboxylic acid cycle to evade complement-mediated killing. Front. Immunol. 8, 1706 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.M. Jiang, Z.-g. Chen, H. Li, T.-t. Zhang, M.-j. Yang, X.-x. Peng, B. Peng, Succinate and inosine coordinate innate immune response to bacterial infection. PLOS Pathog. 18, e1010796 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.T. S. Kou, J. H. Wu, X. W. Chen, Z. G. Chen, J. Zheng, B. Peng, Exogenous glycine promotes oxidation of glutathione and restores sensitivity of bacterial pathogens to serum-induced cell death. Redox Biol. 58, 102512 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S18
Tables S1 to S4