PURPOSE
To prospectively examine the association between clonal hematopoiesis (CH) and subsequent risk of lung cancer.
METHODS
Among 200,629 UK Biobank (UKBB) participants with whole-exome sequencing, CH was identified in a nested case-control study of 832 incident lung cancer cases and 3,951 controls (2006-2019) matched on age and year at blood draw, sex, race, and smoking status. A similar nested case-control study (141 cases/652 controls) was conducted among 27,975 participants with whole-exome sequencing in the Mass General Brigham Biobank (MGBB, 2010-2021). In parallel, we compared CH frequency in published data from 5,003 patients with solid tumor (2,279 lung cancer) who had pretreatment blood sequencing performed through Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets.
RESULTS
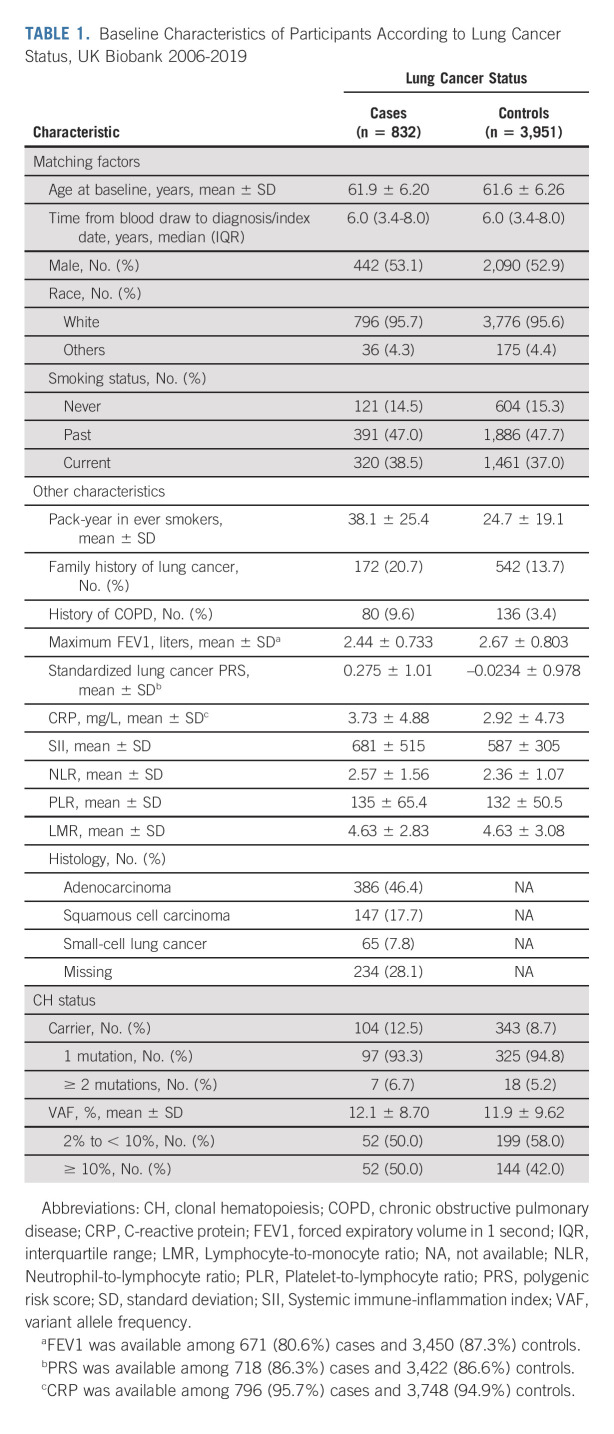
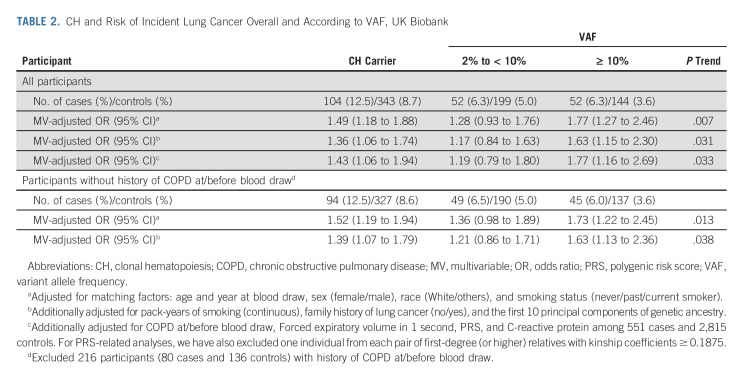
In UKBB, the presence of CH was associated with increased risk of lung cancer (cases: 12.5% v controls: 8.7%; multivariable-adjusted odds ratio [OR], 1.36; 95% CI, 1.06 to 1.74). The association remained robust after excluding participants with chronic obstructive pulmonary disease. No significant interactions with known risk factors, including polygenic risk score and C-reactive protein, were identified. In MGBB, we observed similar enrichment of CH in lung cancer (cases: 15.6% v controls: 12.7%). The meta-analyzed OR (95% CI) of UKBB and MGBB was 1.35 (1.08 to 1.68) for CH overall and 1.61 (1.19 to 2.18) for variant allele frequencies ≥ 10%. In Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets, CH with a variant allele frequency ≥ 10% was enriched in pretreatment lung cancer compared with other tumors after adjusting for age, sex, and smoking (OR for lung v breast cancer: 1.61; 95% CI, 1.03 to 2.53).
CONCLUSION
Independent of known risk factors, CH is associated with increased risk of lung cancer.
INTRODUCTION
Despite steady declines in incidence and mortality, lung cancer remains the leading cause of cancer death worldwide1,2 and accounts for a quarter of all cancer deaths in the United States.3 Given the continuous challenges of low uptake for radiologic screening, with only 14.4% of eligible individuals reporting screening in 2015,4 there is an unmet need to further refine risk assessment to identify high-risk populations.5
CONTEXT
Key Objective
Considering the prominent role of inflammatory elements in the lung microenvironment and emerging evidence positing the lung as a niche of hematopoietic progenitor cells, clonal hematopoiesis (CH) may play a role in lung cancer pathogenesis. This original report prospectively examined the association between CH and subsequent risk of lung cancer, leveraging whole-exome sequencing data in the UK Biobank and Mass General Brigham Biobank. It also compared the frequency of CH in lung cancer with other solid tumors, leveraging Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets.
Knowledge Generated
Independent of known risk factors, CH was associated with an increased risk of incident lung cancer.
Relevance
CH may serve as a novel biomarker for lung cancer prevention and early detection. Future studies are warranted to elucidate the underlying mechanisms of this positive association.
Clonal hematopoiesis (CH) defines the age-related expansion of hematopoietic clones harboring somatic mutations in the absence of overt hematologic malignancies or abnormalities. These mutations, either single-nucleotide variants or short insertions/deletions, often involve genes implicated in hematologic malignancies such as DNMT3A, TET2, and ASXL1.6-11 In addition to its potential as a premalignant state in hematologic malignancy, CH can induce an aberrant inflammatory state,12,13 linked with subsequent risk of a variety of health outcomes.14,15
Although CH has been associated with increased mortality in patients with solid cancer,16 the relationship between CH and solid tumor carcinogenesis remains largely unexplored. The presence of CH in solid tumor samples has been detected by multiple studies, confirming infiltration of the solid tumor microenvironment by CH-mutated hematopoietic cells.17,18 Considering the prominent role of inflammatory elements in the lung microenvironment19 and emerging evidence positing the lung as a niche of hematopoietic progenitor cells,20 CH may play a role in lung cancer pathogenesis. Indeed, a recent study aimed at improving early lung cancer detection using cell-free DNA screening techniques discovered CH mutations in 13.5% of 104 patients with non–small-cell lung cancer (NSCLC), but only 7.1% of 56 matched controls.21 In line with these findings, several clinical sequencing studies also reported more frequent CH mutations in blood samples of patients with lung cancer (15%-45%) compared with patients with other solid tumors.16,22-24 The emerging role of CH in lung cancer etiology is further supported by a recent animal study demonstrating that infiltrated myeloid cells harboring CH mutation in TET2 exacerbate lung cancer progression by promoting tumor angiogenesis.25
Although promising, prospective studies on the associations between CH and risk of incident lung cancer are lacking. We leveraged whole-exome sequencing (WES) data from participants in the UK Biobank (UKBB) and the Mass General Brigham Biobank (MGBB) to conduct a nested case-control study to examine the association between CH and subsequent risk of lung cancer. We also compared the frequency of CH in lung cancer with that of other solid tumors, leveraging pretreatment blood sequencing performed through Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT).
METHODS
CH and Subsequent Risk of Lung Cancer, UKBB
Study population.
The UKBB is a population-based prospective study of more than half a million participants age 37-73 years between 2006 and 2010.26 At the baseline visit, participants went through a physical examination; answered a touchscreen questionnaire about sociodemographic, lifestyle, and health information; and provided blood samples. Among 200,629 participants with WES data (released between 2019 and 2020), we conducted a nested case-control analysis of incident lung cancer.
Ascertainment of cases and controls.
Incident lung cancer cases after baseline blood draw were ascertained using cancer registries and death records provided by the National Health Service Information Centre and the National Health Service Central Register (July 31, 2019), National Records of Scotland (October 31, 2015), on the basis of International Classification of Diseases, Tenth Revision, Clinical Modification code C34. Using risk set/incidence density sampling,27 at the time of diagnosis for each case, we selected up to five controls from the rest of the cohort who were free of cancer, matching on age at blood draw (± 1 year), year at blood draw, sex (female/male), race (White/others), and smoking status (never/past/current smoker). We excluded cases and controls with (1) prior malignant neoplasm, except nonmelanoma skin cancer and (2) prior hematologic disorders.
Whole-exome sequencing.
DNA samples collected from whole blood were sequenced by the Regeneron Genetics Center28 on the Illumina NovaSeq 6000 platform with a mean depth of 55× using 75-base-pair paired-end reads with two 10-base-pair index reads (Data Supplement, online only). Sequencing reads were aligned to the human genome (hg 38) using Burrows-Wheeler alignment tool-maximal exact matches. Samples showing low coverage (< 80% of targeted bases reaching 20× coverage), discordance with microarray genotyping data or reported sex, high rates of heterozygosity/contamination (FREEMIX score > 5%), or duplication were removed (Data Supplement).28,29 Polymerase chain reaction duplicate reads were filtered using Genome Analysis Toolkit (GATK) MarkDuplicates (v2.21.2).30
CH variant calling and annotation.
CH calling and annotation were performed blinded to case-control status by an independent group (B.W. and K.L.B.) from the team involved in the statistical analysis (R.T., X.Z., and Y.C.). After base quality score recalibration (GATK BQSRPipelineSpark, v4.2.0.0), single-nucleotide variants and insertions/deletions were called using Mutect2 (v4.2.1.0) and VarDictJava (v1.6.0). Variants were retained if passed by both callers with a variant allele frequency (VAF) of 2% or more and with at least three supporting reads including at least one from both the forward and reverse strands. Mutations were annotated with the Ensembl Variant Effect Predictor31 using the canonical transcript. We applied a series of additional postprocessing filters to further remove sequencing artifacts and germline polymorphisms.32 Putative driver mutations were annotated according to prior evidence for functional relevance in cancer as previously described (Data Supplement).32
Assessment of covariates.
At study baseline, participants self-reported age, sex, race, smoking status and intensity (pack-years), and family history of lung cancer. Personal history of chronic obstructive pulmonary disease (COPD) was identified from self-report, hospital admission, and death records.33 Forced expiratory volume in 1 second (FEV1) at baseline was obtained from the best measure of three technically satisfactory blows using handheld pneumotachograph spirometers (Vitalograph Escort). We also retrieved information on the first 10 principal components of genetic ancestry.34
Assessment of genetic predisposition and circulating markers of inflammation.
Imputed genotype data were used to derive a validated polygenic risk score (PRS) on the basis of 128 single-nucleotide polymorphisms,35-37 identified from (1) known susceptibility loci of lung cancer and conditions related to lung cancer and (2) single-nucleotide polymorphisms with P < 5 × 10−6 in the National Human Genome Research Institute-European Bioinformatics Institute lung cancer genome-wide association studies.35 Baseline circulating levels of a list of inflammation markers (Data Supplement) including C-reactive protein (CRP, mg/L) were extracted.
CH and Subsequent Risk of Lung Cancer, MGBB
MGBB is a hospital-based biobank with linkage to the electronic health record (EHR) at the Mass General Brigham hospital system with 132,660 participants (Data Supplement). MGBB maintains banked samples (plasma, serum, DNA, and buffy coats) collected from patients who consented to broad-based research. WES was performed in 27,975 individuals (2010-2021) with a mean sequencing depth of 85× (Data Supplement). Similar to UKBB, using risk set sampling, incident lung cancer after blood draw was matched with up to five controls on age, sex, year at blood draw, and smoking status (never/past/current smoker). CH calling and annotation were performed (M.M.U.) independent of statistical analyses (B.T.). As described previously,14,38-40 exome variant calling was performed using GATK Mutect2. We excluded common sequencing artifacts and germline mutations frequently observed in the population (> 1%). CH carriers were defined as individuals containing one or more of a prespecified list of CH variants with a VAF > 2%. A total of 141 cases and 652 controls were included.
CH in Lung Cancer Versus Other Solid Cancers, MSK-IMPACT
To compare the frequency of CH across major solid tumors, we leveraged published and publicly accessible data from patients with solid tumors treated at the MSK Cancer Center (2013-2019).41 In this analysis, we restricted to a cohort of patients with the five most prevalent primary tumor sites and who had not received cytotoxic therapy or radiation therapy before blood draw. Blood samples were sequenced using MSK-IMPACT, a validated targeted panel capturing common CH genes with a mean sequencing depth of 497× (Data Supplement). Data from a total of 5,003 patients, including 2,279 lung cancer cases, were retrieved.
Statistical Analysis
UKBB.
As primary analyses, we examined the association between CH carrier status and risk of incident lung cancer in the UKBB. As the planned stratified analysis disassembles the matching, unconditional instead of conditional logistic regression was used to estimate odds ratio (ORs) and 95% CIs to retain samples and allow for direct comparison of effect estimates throughout the analyses. In addition to matching factors (age and year at blood draw, sex, race, and smoking status), we adjusted for smoking pack-years and the first 10 principal components of ancestry. Considering the role of lung function in lung cancer risks,42 we additionally adjusted for COPD and FEV1. We also examined the dose-response relationship on the basis of the size of the CH clone using VAF (2% to < 10%, ≥ 10%) and the number of CH mutations (0, 1, and ≥ 2) and tested for linear trends. As secondary analyses, we also conducted gene-specific analyses for most frequently mutated CH genes (DNMT3A, ASXL1, and TET2).
In stratified analyses, we examined whether the association between CH and risk of lung cancer differed according to sex, family history of lung cancer, smoking status, age at blood draw, lung cancer PRS, CRP, and other chronic inflammation markers. P for interaction was estimated using the Wald test. We further evaluated whether the association differs by lung cancer age at diagnosis and histology. P for heterogeneity was calculated using polytomous regression.
We conducted multiple sensitivity analyses. First, we excluded participants with a history of COPD at/before baseline to assess whether the association was independent of COPD. Second, we excluded participants with ASXL1 mutations, which are strongly associated with smoking.32,43 Third, we excluded cases and controls within 2 years of blood draw to minimize the impact of reverse causation. Fourth, we excluded one individual from each pair of first-degree (or higher) relatives to reduce bias from lung cancer risk carried within families.
MGBB.
Similar to analyses in UKBB, logistic regressions were conducted to examine the association between CH and risk of lung cancer adjusted for the same set of covariates.
Meta-analyses of UKBB and MGBB.
We further meta-analyzed results from UKBB and MGBB using fixed-effects models44 for overall CH and lung cancer risk, according to VAF. P for heterogeneity was estimated using Cochran's Q test. To rule out the possibility of novel rare germline variants, we performed a sensitivity analysis removing individuals with VAF ≥ 35%.
MSK-IMPACT.
Using MSK-IMPACT data, we conducted binary and multinomial logistic regressions to evaluate how CH (yes/no) or CH (VAF ≥ 10%/VAF 2% to < 10%/no) might differ in patients with colorectal, pancreatic, prostate, and lung cancers compared with breast carcinoma, the most common type of cancer other than lung cancer in the MSK-IMPACT. The models were adjusted for age at diagnosis/blood draw, sex, and smoking status (never/ever). P heterogeneity was calculated for each cancer type, which reflected how the association between CH and a particular cancer type (eg, lung cancer) differed from the association between CH and breast carcinoma. All statistical analyses were performed using R (version 4.0.5) and considered significant with two-sided P < .05.
RESULTS
UKBB
In UKBB, a total of 832 incident lung cancer cases and 3,951 controls (median follow-up: 6.0 years; interquartile range, 3.4-8.0 years) matched on age and year at blood draw, sex, race, and smoking status were included (Table 1). The frequency of CH at baseline increased with age and was higher in lung cancer cases (12.5%) compared with controls (8.7%), with differences observed across all age groups (Fig 1A). CH at baseline was associated with an increased risk of subsequent lung cancer after multivariable adjustment. Compared with CH-negative individuals, CH-positive individuals had a 36% higher risk of lung cancer (OR, 1.36; 95% CI, 1.06 to 1.74; Table 2). The association remained robust after further adjusting for history of COPD, FEV1, PRS, and CRP or using conditional logistic regression (Data Supplement). The association was also similar in the sensitivity analyses, especially when restricted to individuals without COPD or ASXL1 mutations (Data Supplement).
TABLE 1.
Baseline Characteristics of Participants According to Lung Cancer Status, UK Biobank 2006-2019
FIG 1.

Frequency of CH mutations among incident lung cancer cases and controls, UKBB. (A) Frequency of overall CH mutations (general additive modeling with spline smoothing) by case-control status and age at blood draw. (B) CH mutations with VAF 2% to < 10%. (C) CH mutations with VAF ≥ 10%. (D) The top 10 most frequent mutated CH genes according to case-control status. CH, clonal hematopoiesis; UKBB, UK Biobank; VAF, variant allele frequency.
TABLE 2.
CH and Risk of Incident Lung Cancer Overall and According to VAF, UK Biobank
The differences in CH frequency between cases and controls were largely driven by expanded CH mutations with VAF ≥ 10% (OR, 1.63; 95% CI, 1.15 to 2.30), but not VAF 2% to < 10% (OR, 1.17; 95% CI, 0.84 to 1.63; P trend = .031; Table 2; Figs 1B and 1C). Few participants with CH had very high VAF (≥ 35%), not driving the differences between cases and controls (Data Supplement). When the number of mutations was examined, a positive linear trend was documented (Data Supplement). Gene-specific analysis on the top three most frequently mutated CH genes (DNMT3A, ASXL1, and TET2; Fig 1D) suggested that each of these genes and other CH mutations all contributed to the association between CH and risk of lung cancer (Data Supplement).
The association between CH and lung cancer risk did not differ significantly by lung cancer risk factors, including PRS and CRP (all P interaction > .05; Fig 2A and Data Supplement). Positive association was observed for NSCLC including both adenocarcinoma (OR, 1.68; 95% CI, 1.23 to 2.29) and squamous cell carcinoma (OR, 1.59; 95% CI, 1.51 to 1.68), but not for small-cell lung cancer (OR, 0.82; 95% CI, 0.82 to 0.83; Fig 2B).
FIG 2.

CH and risk of incident lung cancer according to (A) lung cancer risk factors and (B) lung cancer characteristics, UKBB. All models were adjusted for age and year at blood draw, sex (female/male), race (White/others), smoking status (never/past/current smoker), pack-years of smoking (continuous), family history of lung cancer (no/yes), and the first 10 principal components of genetic ancestry. Stratified analyses by PRS were additionally adjusted for genotype array (BiLEVE/Axiom) and standardized lung cancer PRS (continuous). Stratified analyses by CRP were additionally adjusted for CRP (continuous). P for interaction was estimated using the Wald test. P for heterogeneity was estimated using polytomous logistic regression. CH, clonal hematopoiesis; CRP, C-reactive protein; OR, odds ratio; PRS, polygenic risk score; UKBB, UK Biobank.
MGBB
In MGBB, 141 incident lung cancer cases and 652 matched controls were included (median follow-up: 5.1 years; interquartile range, 3.4-7.2 years; Data Supplement). We observed similar enrichment of CH in lung cancer (cases: 15.6% v controls: 12.7%; OR, 1.33; 95% CI, 0.82 to 2.14; Fig 3).
FIG 3.
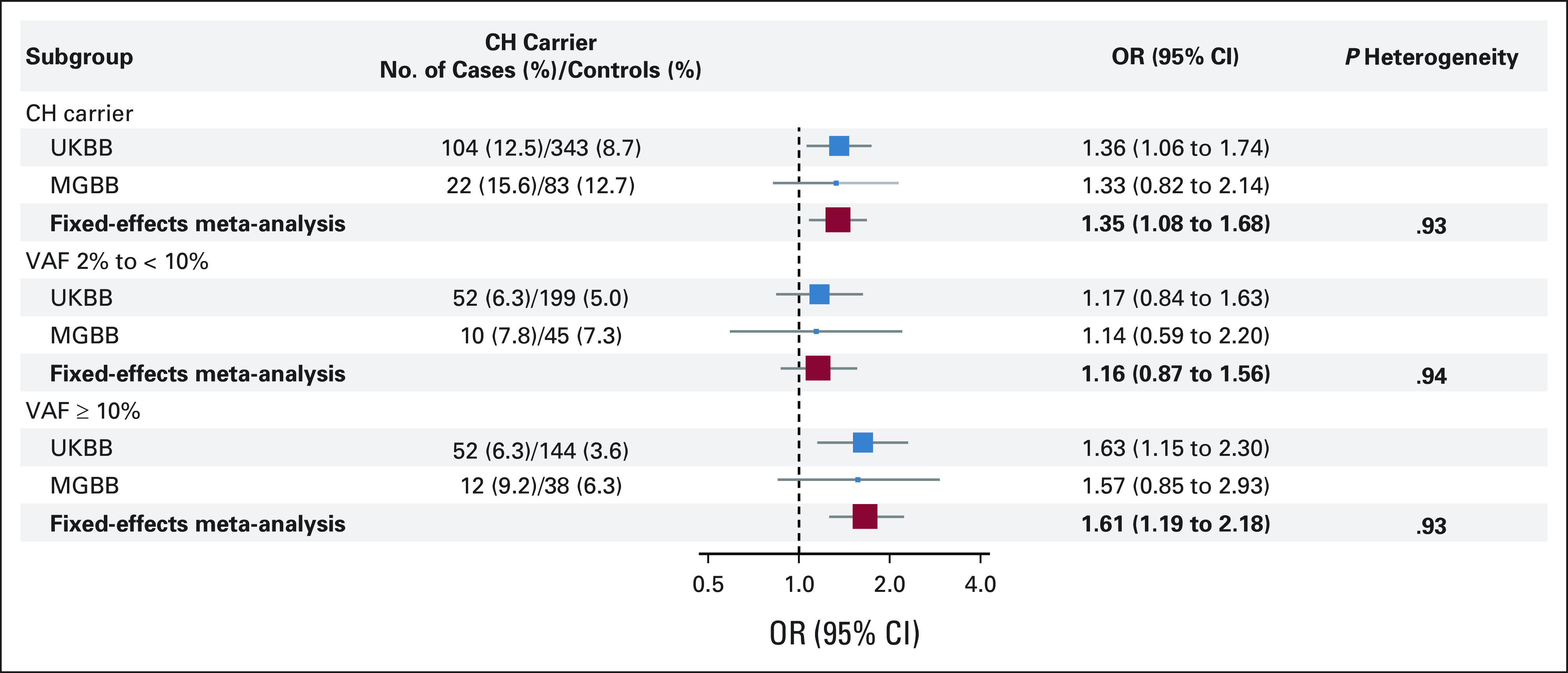
Meta-analyzed association between CH and risk of incident lung cancer, UKBB and MGBB. Meta-analyzed ORs were bolded. CH-negative individuals were considered as the referent group in all models. All models were adjusted for age and year at blood draw, sex (female/male), race (White/others), smoking status (never/past/current smoker), pack-years of smoking (continuous), family history of lung cancer (no/yes), and the first 10 principal components of genetic ancestry. P for heterogeneity was estimated using Cochran's Q test. CH, clonal hematopoiesis; MGBB, Mass General Brigham Biobank; OR, odds ratio; UKBB, UK Biobank; VAF, variant allele frequency.
Meta-Analysis of UKBB and MGBB
No heterogeneity between UKBB/MGBB was identified (all P heterogeneity > .05). The meta-analyzed OR (95% CI) of overall CH and lung cancer was 1.35 (95% CI, 1.08 to 1.68) and 1.61 (95% CI, 1.19 to 2.18) for VAF ≥ 10% (Fig 3) and remains robust after excluding individuals with CH VAF ≥ 35% (Data Supplement). We also observed a potential slightly stronger association for CH with VAF ≥ 10% and lung cancer among never smokers than among past/current smokers (Data Supplement).
MSK-IMPACT
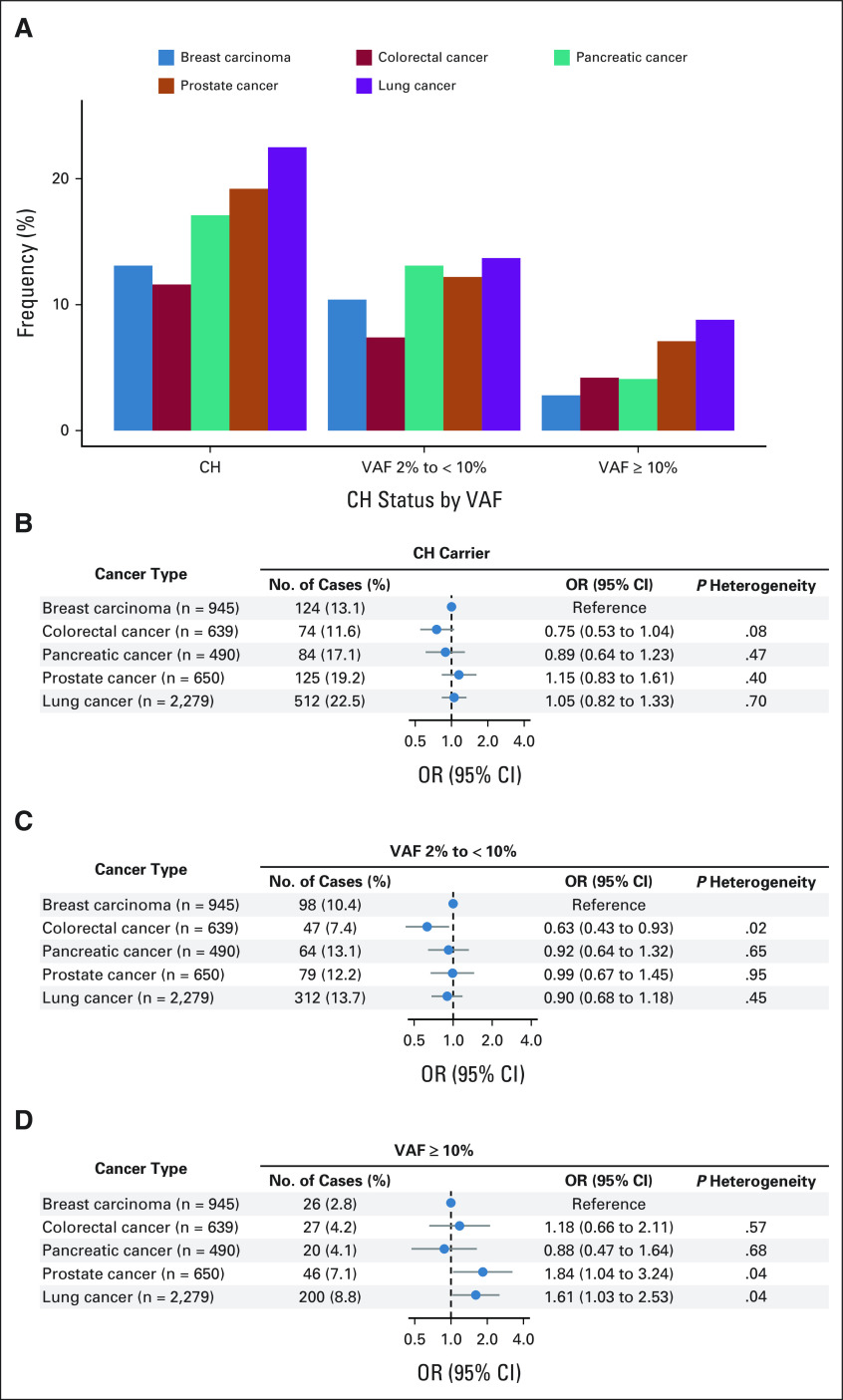
In MSK-IMPACT, the frequency of CH was the highest in lung cancer (22.5%, n = 2,279) compared with other common cancer types. Using breast cancer as the referent (13.1%), the crude OR (95% CI) was 1.92 (95% CI, 1.55 to 2.37), which was reduced to 1.15 (95% CI, 0.92 to 1.45) after adjusting for age at diagnosis (Fig 4A and Data Supplement). After further adjustment for sex and smoking, patients with lung cancer were not significantly different from patients with breast carcinoma for associations with overall CH (OR, 1.05; 95% CI, 0.82 to 1.33) or CH with VAF 2% to < 10% (OR, 0.90; 95% CI, 0.68 to 1.18; Figs 4B and 4C). However, patients with lung cancer were 61% more likely than those with breast carcinoma to harbor CH with VAF ≥ 10% (OR, 1.61; 95% CI, 1.03 to 2.53; Fig 4D).
FIG 4.
Comparison of CH frequency overall and according to VAF using pretreatment blood samples from patients with major solid tumors, MSK-IMPACT. (A) Frequency of CH mutations in the five most common cancer subtypes. (B) Differential associations between CH and the cancer type, using breast carcinoma as the referent cancer type. (C) Differential associations between CH and the cancer type for VAF 2% to < 10%, using breast cancer as the referent cancer type. (D) Differential associations between CH and the cancer type for VAF ≥ 10%, using breast carcinoma as the referent cancer type. All models were adjusted for age at diagnosis/blood draw, sex (female/male), and smoking status (never/ever). CH, clonal hematopoiesis; MSK‐IMPACT, Memorial Sloan Kettering‐Integrated Mutation Profiling of Actionable Cancer Targets; OR, odds ratio; VAF, variant allele frequency.
DISCUSSION
In a nested case-control study within the UKBB, CH was associated with 36% increased risk of incident lung cancer after taking into account major risk factors. This positive association remained similar in sensitivity analyses restricted to individuals without COPD or ASXL1 mutations. Similar findings were observed in MGBB, a hospital-based biobank with EHR linkage. Within patients with treatment-naïve solid tumor in the MSK-IMPACT, we also identified an enrichment of CH in patients with lung cancer compared with other cancer types. Using breast cancer as the referent and after adjusting for covariates, the enrichment was not significant for overall CH or CH with 2% to < 10% (driven by age adjustment) but remained for CH with VAF ≥ 10%. These findings suggest that the association between CH, especially CH with VAF ≥ 10%, and cancer risk may be specific to certain tumor sites such as lung cancer.
These findings provide robust evidence that CH may be an independent risk factor for lung cancer. Potential shared risk factors between lung cancer and CH, predominantly smoking,45 have been thoroughly considered in our analyses through matching and additional adjustment for pack-years. Of note, our observation of similar findings among individuals without ASXL1 CH mutations and a larger effect size for individuals with NSCLC compared with small-cell lung cancer (mainly caused by exposure to tobacco)46 together supports that the association between CH and lung cancer risk may be independent of smoking. Similarly, we did not find evidence of COPD confounding the association between CH and lung cancer risk.
The biologic mechanisms underlying the observed association remain to be investigated. Hematopoietic cells play key roles in the tumor microenvironment, either suppressing or supporting tumor growth. The potential of CH to promote dysregulation of immune- and inflammatory-related pathways has been supported by a growing body of evidence both clinical and in vivo.47,48 It has been reported that deletion of TET2 switched immunosuppressive tumor–associated macrophages to proinflammatory ones in a mouse model of melanoma. Furthermore, inhibition of a key inflammatory mediator interleukin-1β has shown potential in reducing incidence and mortality of lung cancer.49 Taken together, one plausible mechanism through which CH might play a causal role in promoting lung cancer pathogenesis is through CH-induced modification of the tumor microenvironment. Targeting of hematopoietic cells through enhancement of the immune response has led to major advances in treatment of many cancers. If CH plays a role in modifying the tumor-immune response, this would have therapeutic implications.50 Future studies clarifying the casual basis of this association are needed including experimental models and Mendelian randomization studies for CH as the genetic basis of CH continues to be clarified.38 Multiomics characterization of the impact of CH on normal lung parenchyma and the lung cancer tumor microenvironment will be important to clarify these mechanisms. Alternatively, it is known that CH mutations reflect positive selection within the complex and dynamic interplays between genetic predisposition, epigenetic configuration, and environmental carcinogens.51 CH may serve as an indicator of not yet identified shared risk factors rather than playing a causal role although less likely given that CH was preferentially enriched in patients with lung cancer. Even in this situation, CH as a biomarker of lung cancer risk could be clinically relevant.
Our study has notable strengths. The use of a large populational cohort with linked genomic and phenotypic data allowed us to conduct prospective investigation of CH and lung cancer with comprehensive adjustment for known confounders. Moreover, directionally consistent results from an EHR-based cohort and a hospital-based case-only study support the validity and generalizability of our findings. Our study also has limitations. First, although the positive association between CH and lung cancer was primarily observed for large clones with VAF ≥ 10%, it is worth noting that because of low sequencing coverage in WES, misclassification of CH, especially for VAF below 10%, is likely. Deeper and/or more targeted sequencing of CH variants is warranted. This would be particularly important in further characterizing the relationship between lower VAF CH and risk of lung cancer. Future studies with serial sampling using higher-sensitivity CH assays will help to quantitate the impact of low VAF CH and elucidate the role of CH trajectory over time in enhancing lung cancer risk.12 Finally, we have limited power to test the association between CH and lung cancer risk in specific genes and racial/ethnic minority groups among whom inherited genetic factors might mediate the association.38 Additional studies with a larger sample size are warranted.
In conclusion, independent of known risk factors, especially smoking, CH was associated with an increased risk of incident lung cancer. These findings were also supported in EHR-based biobank and patient-only data. CH may serve as a novel biomarker/target for lung cancer prevention/early detection.
ACKNOWLEDGMENT
This research has been conducted using the UK Biobank Resource under Application Number 55288.
Semanti Mukherjee
Stock and Other Ownership Interests: Regeneron
Varun Puri
Stock and Other Ownership Interests: Intuitive Surgical (I)
Honoraria: Precisca
Benjamin L. Ebert
Stock and Other Ownership Interests: Skyhawk Therapeutics, Exo Therapeutics, Neomorph, TenSixteen Bio
Consulting or Advisory Role: Exo Therapeutics, Skyhawk Therapeutics, Neomorph, TenSixteen Bio
Research Funding: Celgene (Inst), Deerfield Management (Inst), Novartis Institutes for BioMedical Research (Inst), Calico (Inst)
Patents, Royalties, Other Intellectual Property: Patents related to the prediction of risk of cardiovascular disease (Inst)
Ramaswamy Govindan
Consulting or Advisory Role: Merck, Inivata
Pradeep Natarajan
Employment: Vertex (I)
Stock and Other Ownership Interests: Vertex (I), TenSixteen Bio
Consulting or Advisory Role: TenSixteen Bio, Blackstone, Apple, AstraZeneca, Foresite Capital, Novartis, Genentech/Roche, Allelica
Research Funding: AstraZeneca, Novartis
Kelly L. Bolton
Consulting or Advisory Role: GoodCell
Research Funding: GRAIL, Bristol Myers Squibb Foundation, Servier
Yin Cao
Consulting or Advisory Role: Geneoscopy
No other potential conflicts of interest were reported.
SUPPORT
Supported by grants from the National Institutes of Health (NIH), including P30CA091842 and K08CA241318 (to K.L.B.) from the National Cancer Institute and R01HL151283, R01HL148050, and R01HL148565 (to P.N.) from the National Heart, Lung, and Blood Institute.
R.T., B.W., J.L., X.Z., and B.T. contributed equally to this work. R.G., P.N., K.L.B., and Y.C. contributed equally to this work.
DATA SHARING STATEMENT
The UK Biobank (UKBB) data can be accessed through the UKBB portal (web link: https://www.ukbiobank.ac.uk/, e-mail: ukbiobank@ukbiobank.ac.uk). The genome‐wide association studies summary statistics used for calculating PRS are available at https://www.ebi.ac.uk/gwas/, with contact e-mail at gwas-info@ebi.ac.uk. The weights used for the PRS are available at the PGS Catalog (https://www.pgscatalog.org/; pgs-info@ebi.ac.uk), at https://www.pgscatalog.org/publication/PGP000148/. The remaining data are available within the article, Data Supplement, or available from the authors upon request.
AUTHOR CONTRIBUTIONS
Conception and design: Ramaswamy Govindan, Pradeep Natarajan, Kelly L. Bolton, Yin Cao
Financial support: Pradeep Natarajan, Kelly L. Bolton, Yin Cao
Administrative support: Yin Cao
Provision of study materials or patients: Pradeep Natarajan, Kelly L. Bolton, Yin Cao
Collection and assembly of data: Brian Wiley, Xiaoyu Zong, Buu Truong, Benjamin L. Ebert, Pradeep Natarajan, Kelly L. Bolton, Yin Cao
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Clonal Hematopoiesis and Risk of Incident Lung Cancer
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Semanti Mukherjee
Stock and Other Ownership Interests: Regeneron
Varun Puri
Stock and Other Ownership Interests: Intuitive Surgical (I)
Honoraria: Precisca
Benjamin L. Ebert
Stock and Other Ownership Interests: Skyhawk Therapeutics, Exo Therapeutics, Neomorph, TenSixteen Bio
Consulting or Advisory Role: Exo Therapeutics, Skyhawk Therapeutics, Neomorph, TenSixteen Bio
Research Funding: Celgene (Inst), Deerfield Management (Inst), Novartis Institutes for BioMedical Research (Inst), Calico (Inst)
Patents, Royalties, Other Intellectual Property: Patents related to the prediction of risk of cardiovascular disease (Inst)
Ramaswamy Govindan
Consulting or Advisory Role: Merck, Inivata
Pradeep Natarajan
Employment: Vertex (I)
Stock and Other Ownership Interests: Vertex (I), TenSixteen Bio
Consulting or Advisory Role: TenSixteen Bio, Blackstone, Apple, AstraZeneca, Foresite Capital, Novartis, Genentech/Roche, Allelica
Research Funding: AstraZeneca, Novartis
Kelly L. Bolton
Consulting or Advisory Role: GoodCell
Research Funding: GRAIL, Bristol Myers Squibb Foundation, Servier
Yin Cao
Consulting or Advisory Role: Geneoscopy
No other potential conflicts of interest were reported.
REFERENCES
- 1.Barta JA, Powell CA, Wisnivesky JP: Global epidemiology of lung cancer. Ann Glob Health 85:8, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, et al. : Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68:394-424, 2018 [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Fuchs HE, et al. : Cancer statistics, 2021. CA Cancer J Clin 71:7-33, 2021 [DOI] [PubMed] [Google Scholar]
- 4.Zahnd WE, Eberth JM: Lung cancer screening utilization: A behavioral risk factor surveillance system Analysis. Am J Prev Med 57:250-255, 2019 [DOI] [PubMed] [Google Scholar]
- 5.Chu GCW, Lazare K, Sullivan F: Serum and blood based biomarkers for lung cancer screening: A systematic review. BMC Cancer 18:181, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network, Ley TJ, Miller C, et al. : Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368:2059-2074, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bejar R, Stevenson K, Abdel-Wahab O, et al. : Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med 364:2496-2506, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papaemmanuil E, Gerstung M, Malcovati L, et al. : Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 122:3616-3627, 2013; quiz 3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie M, Lu C, Wang J, et al. : Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 20:1472-1478, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaiswal S, Fontanillas P, Flannick J, et al. : Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 371:2488-2498, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genovese G, Kahler AK, Handsaker RE, et al. : Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 371:2477-2487, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaiswal S: Clonal hematopoiesis and nonhematologic disorders. Blood 136:1606-1614, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arends CM, Galan-Sousa J, Hoyer K, et al. : Hematopoietic lineage distribution and evolutionary dynamics of clonal hematopoiesis. Leukemia 32:1908-1919, 2018 [DOI] [PubMed] [Google Scholar]
- 14.Bick AG, Pirruccello JP, Griffin GK, et al. : Genetic interleukin 6 signaling deficiency attenuates cardiovascular risk in clonal hematopoiesis. Circulation 141:124-131, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Libby P, Sidlow R, Lin AE, et al. : Clonal hematopoiesis: Crossroads of aging, cardiovascular disease, and cancer: JACC review topic of the week. J Am Coll Cardiol 74:567-577, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coombs CC, Zehir A, Devlin SM, et al. : Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell 21:374-382 e4, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coombs CC, Gillis NK, Tan X, et al. : Identification of clonal hematopoiesis mutations in solid tumor patients undergoing unpaired next-generation sequencing assays. Clin Cancer Res 24:5918-5924, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Severson EA, Riedlinger GM, Connelly CF, et al. : Detection of clonal hematopoiesis of indeterminate potential in clinical sequencing of solid tumor specimens. Blood 131:2501-2505, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altorki NK, Markowitz GJ, Gao D, et al. : The lung microenvironment: An important regulator of tumour growth and metastasis. Nat Rev Cancer 19:9-31, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lefrancais E, Ortiz-Munoz G, Caudrillier A, et al. : The lung is a site of platelet biogenesis and a reservoir for haematopoietic progenitors. Nature 544:105-109, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chabon JJ, Hamilton EG, Kurtz DM, et al. : Integrating genomic features for non-invasive early lung cancer detection. Nature 580:245-251, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conces M, Ni Y, Bazeley P, et al. : Clonal hematopoiesis of indeterminate potential (CHIP) mutations in solid tumor malignancies. J Clin Oncol 37, 2019. (abstr 1507) [Google Scholar]
- 23.Ptashkin RN, Mandelker DL, Coombs CC, et al. : Prevalence of clonal hematopoiesis mutations in tumor-only clinical genomic profiling of solid tumors. JAMA Oncol 4:1589-1593, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zink F, Stacey SN, Norddahl GL, et al. : Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood 130:742-752, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen YTM, Fujisawa M, Nguyen TB, et al. : Tet2 deficiency in immune cells exacerbates tumor progression by increasing angiogenesis in a lung cancer model. Cancer Sci 112:4931-4943, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sudlow C, Gallacher J, Allen N, et al. : UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 12:e1001779, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vandenbroucke JP, Pearce N: Case-control studies: Basic concepts. Int J Epidemiol 41:1480-1489, 2012 [DOI] [PubMed] [Google Scholar]
- 28.Backman JD, Li AH, Marcketta A, et al. : Exome sequencing and analysis of 454,787 UK Biobank participants. Nature 599:628-634, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szustakowski JD, Balasubramanian S, Kvikstad E, et al. : Advancing human genetics research and drug discovery through exome sequencing of the UK Biobank. Nat Genet 53:942-948, 2021 [DOI] [PubMed] [Google Scholar]
- 30.Ebbert MT, Wadsworth ME, Staley LA, et al. : Evaluating the necessity of PCR duplicate removal from next-generation sequencing data and a comparison of approaches. BMC Bioinformatics 17:239, 2016. (suppl 7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLaren W, Gil L, Hunt SE, et al. : The Ensembl variant effect predictor. Genome Biol 17:122, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bolton KL, Ptashkin RN, Gao T, et al. : Cancer therapy shapes the fitness landscape of clonal hematopoiesis. Nat Genet 52:1219-1226, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Q, Bush K, Nolan J, et al. : Definitions of Chronic Obstructive Pulmonary Disease for UK Biobank Phase 1 Outcomes Adjudication. UK Biobank Outcome Adjudication Group, version 1.0, 2018 [Google Scholar]
- 34.Data-Field 22009, 2022, https://biobank.ctsu.ox.ac.uk/crystal/field.cgi?id=22009 [Google Scholar]
- 35.Hung RJ, Warkentin MT, Brhane Y, et al. : Assessing lung cancer absolute risk trajectory based on a polygenic risk model. Cancer Res 81:1607-1615, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi SW, Mak TS, O'Reilly PF: Tutorial: A guide to performing polygenic risk score analyses. Nat Protoc 15:2759-2772, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi SW, O'Reilly PF: PRSice-2: Polygenic Risk Score software for biobank-scale data. Gigascience 8:giz082, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bick AG, Weinstock JS, Nandakumar SK, et al. : Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature 586:763-768, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu B, Roberts MB, Raffield LM, et al. : Supplemental association of clonal hematopoiesis with incident heart failure. J Am Coll Cardiol 78:42-52, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhattacharya R, Zekavat SM, Haessler J, et al. : Clonal hematopoiesis is associated with higher risk of stroke. Stroke 53:788-797, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolton KL, Koh Y, Foote MB, et al. : Clonal hematopoiesis is associated with risk of severe Covid-19. Nat Commun 12:5975, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muller DC, Johansson M, Brennan P: Lung cancer risk prediction model incorporating lung function: Development and validation in the UK biobank prospective cohort study. J Clin Oncol 35:861-869, 2017 [DOI] [PubMed] [Google Scholar]
- 43.Dawoud AAZ, Tapper WJ, Cross NCP: Clonal myelopoiesis in the UK Biobank cohort: ASXL1 mutations are strongly associated with smoking. Leukemia 34:2660-2672, 2020 [DOI] [PubMed] [Google Scholar]
- 44.Schwarzer G, Carpenter JR, Rücker G: Fixed Effect and Random Effects Meta-Analysis, Meta-Analysis with R. Use R! Cham, Switzerland, Springer International Publishing, 2015, pp 21-53 [Google Scholar]
- 45.Hong W, Li A, Liu Y, et al. : Clonal hematopoiesis mutations in patients with lung cancer are associated with lung cancer risk factors. Cancer Res 82:199-209, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.George J, Lim JS, Jang SJ, et al. : Comprehensive genomic profiles of small cell lung cancer. Nature 524:47-53, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan W, Zhu S, Qu K, et al. : The DNA methylcytosine dioxygenase Tet2 sustains immunosuppressive function of tumor-infiltrating myeloid cells to promote melanoma progression. Immunity 47:284-297 e5, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh AS, Mencia-Trinchant N, Griffiths EA, et al. : DNMT3A and TET2 mutant clonal hematopoiesis may drive a proinflammatory state and predict enhanced response to immune checkpoint inhibitors. Blood 138:4295, 2021 [Google Scholar]
- 49.Ridker PM, MacFadyen JG, Thuren T, et al. : Effect of interleukin-1beta inhibition with canakinumab on incident lung cancer in patients with atherosclerosis: Exploratory results from a randomised, double-blind, placebo-controlled trial. Lancet 390:1833-1842, 2017 [DOI] [PubMed] [Google Scholar]
- 50.Li K, Shi H, Zhang B, et al. : Myeloid-derived suppressor cells as immunosuppressive regulators and therapeutic targets in cancer. Signal Transduct Target Ther 6:362, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kakiuchi N, Ogawa S: Clonal expansion in non-cancer tissues. Nat Rev Cancer 21:239-256, 2021 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The UK Biobank (UKBB) data can be accessed through the UKBB portal (web link: https://www.ukbiobank.ac.uk/, e-mail: ukbiobank@ukbiobank.ac.uk). The genome‐wide association studies summary statistics used for calculating PRS are available at https://www.ebi.ac.uk/gwas/, with contact e-mail at gwas-info@ebi.ac.uk. The weights used for the PRS are available at the PGS Catalog (https://www.pgscatalog.org/; pgs-info@ebi.ac.uk), at https://www.pgscatalog.org/publication/PGP000148/. The remaining data are available within the article, Data Supplement, or available from the authors upon request.