Sir,
Polymerase chain reaction (PCR) testing is the gold standard for diagnosis of SARS-CoV-2 infection. However, detection of viral RNA does not necessarily indicate infectious virus [1,2].
Two approaches were established for quarantine practice during the pandemic: first, setting a general timeframe as isolation period; second, aiming to assess infectiousness by measuring viral load [3]. In PCR tests, the cycle threshold (C T)-value, which indicates the PCR cycles needed for virus detection, inversely relates to viral load. Evidence suggests that contagiousness ceases at C T-values >30 [4]. For such matters, repeated PCR tests appear uneconomical and might cause unnecessary burden for laboratories. Point-of-care test (POCT) biomarker combinations previously showed potential for diagnosing COVID-19 in the emergency department [5]. Measurements of tumour necrosis factor-related apoptosis-inducing ligand (TRAIL), interferon-γ-induced protein-10 (IP-10), and C-reactive protein (CRP), initially developed to distinguish bacterial from viral infections when used within a combinatorial score, have been demonstrated to predict COVID-19 severity [[6], [7], [8]].
Here, we performed a sub-analysis to the previously reported DIRECTOR study to detect possible predictors of infectiousness by evaluating the biomarkers' association to SARS-CoV-2 PCR C T-values [7,8]. Adult patients presenting to the Saarland University Hospital in Homburg, Germany, were included. Signed informed consent and a positive SARS-CoV-2 PCR were required. All samples (blood, respiratory samples) were acquired repeatedly during routine care. Ethical approval was granted prior to the study (Ärztekammer des Saarlandes, reference number 019/20). Respiratory samples were tested for SARS-CoV-2 via real-time reverse transcription PCR (RT-qPCR). We operationalized infectiousness as PCR C T-values. For assuming infectiousness, C T of ≤/>30 was used, based on previous literature [4]. TRAIL, IP-10, and CRP were measured on a MeMed Key® platform (MeMed Diagnostics, Tirat Carmel, Israel). Biomarker levels were paired with patients' C T-values when they were measured on the same day or, if not available, the day before PCR testing. Only matched pairs were included in the analyses. Variables were reported as the median with interquartile range (IQR) or as the mean with standard deviation (SD). Correlation analyses were performed using Spearman rank test. Receiver operating characteristics (ROC) curves were generated to assess the performance of the biomarkers' prediction on infectiousness including all possible decision thresholds. In addition, groups were compared by Mann–Whitney U-test. For analyses, R Studio (Version 1.3.1093) was used. Statistical significance was set at P < 0.05.
The adult DIRECTOR study population comprised 132 COVID-19 patients, in whom the host-response biomarkers were measured 899 times (mean of 6.8 times per patient). Patient characteristics, clinical course and biomarker expression were described in detail previously [7]. Overall, 436 C T-values from 123 COVID-19 patients (93.18%) were available (mean of 3.54 per patient). In total, 177 C T-values of 97 patients were matched with host-response biomarkers. The mean of these C T-values was 28.98 (SD: 5.36).
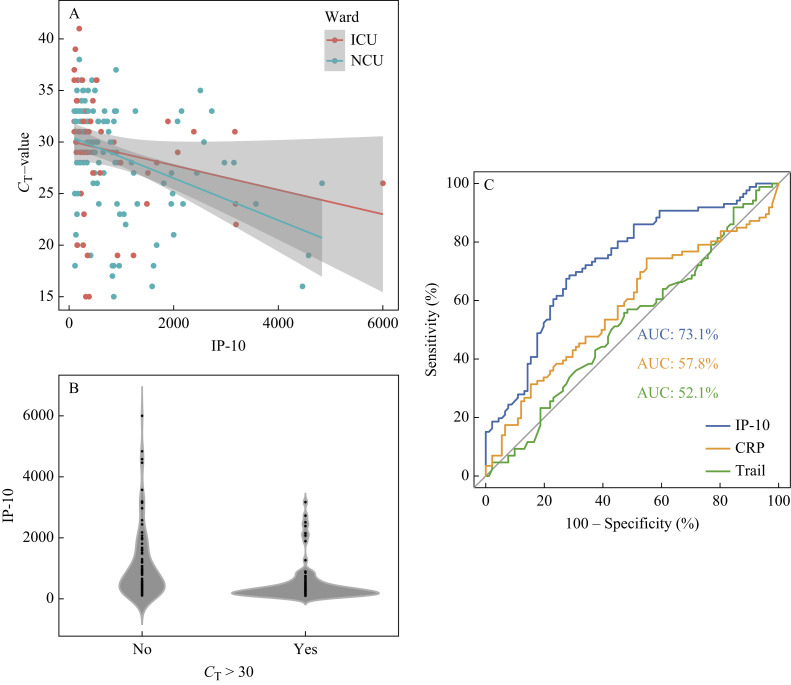
IP-10 showed a moderate correlation with paired C T-values (r = –0.404; P < 0.0001), with a mean level of 804.6 pg/mL (SD: 995.3). TRAIL (r = –0.108; P = 0.153) yielded no correlation, whereas CRP displayed a weak correlation with concurrent CT-values (r = –0.150; P = 0.046). The mean of CRP measurements was 81.44 mg/L (SD: 69.99). When assessing paired values for patients who remained in the normal care unit (NCU) separately (N = 123), we found a stronger correlation for IP-10 (r = –0.412; P < 0.0001; Figure 1 A) and CRP (r = –0.191; P = 0.034). When dividing pairs by C T-values ≤30 (N = 91) and >30 (N = 86), IP-10 levels showed a significant difference in group comparisons (P < 0.0001). In the group assumed to be ‘infectious’ (C T ≤30), IP-10 levels yielded a median of 663 pg/mL (IQR: 1166), whereas IP-10 measurements in the ‘non-infectious’ group (C T >30) showed a median of 274 pg/mL (IQR: 354.6) (Figure 1B). With an area under the ROC curve (AUC) of 73.1%, IP-10 was superior to CRP (AUC: 57.8%) and TRAIL (AUC: 52.1%) in predicting possible SARS-CoV-2 infectiousness (Figure 1C). An IP-10 decision threshold of 410.4 pg/mL IP-10 reached the best combined performance with a sensitivity of 70.9% and specificity of 67%. A sensitivity and specificity of 100% was reached at a threshold of 3177 pg/mL and 109.3 pg/mL, respectively, whereas an IP-10 level of 904.5 pg/mL yielded a sensitivity of 90.7% (specificity 40.7%).
Figure 1.
Correlation of interferon-γ-induced protein-10 (IP-10) (pg/mL) in COVID-19 patients to paired cycle threshold (CT)-values divided by patients in the normal care unit (NCU) and intensive care unit (ICU). Group comparison of COVID-19 patients between paired CT-values ≤30 and >30. Receiver operating characteristics curves indicating areas under the curve (AUC) for the biomarkers' detection of SARS-CoV-2 infectiousness, as assumed at CT-values ≤30. CRP, C-reactive protein; TRAIL, tumour necrosis factor-related apoptosis-inducing ligand.
In this study, we demonstrated a correlation between PCR C T-values and IP-10 levels in COVID-19 patients. With a threshold of >410 pg/mL, IP-10 predicted possible infectiousness with a sensitivity of 70.9%. IP-10's predictive performance on infectiousness increased in mildly ill patients, possibly since clinical deterioration is triggered by hyperinflammation, causing a confounding variable for IP-10 elevation [7]. As limitations of our study, our pairing of C T-values with biomarker measurements may have introduced a certain degree of imprecision, and assessing C T-values for infectiousness is prone to error. Nevertheless, an association of C T-values with growth of SARS-CoV-2 in cell cultures was previously demonstrated [4]. Furthermore, there is no international standard for calibrating C T-values, and they are dependent on, for example, method and reagents used [9].
Nevertheless, our findings indicate that POCT measurements of IP-10 could pose an alternative for costly PCR tests in quarantine practice, especially in the outpatient setting, where mild cases are most prevalent. COVID-19 symptoms should be considered alongside the measurements.
Since IP-10 can predict COVID-19 severity, performing measurements could inform on required clinical management as well, improving patient care next to public health.
In children with respiratory tract infections, TRAIL and IP-10 showed significant correlations with different viral loads [7,10]. This suggests that the biomarkers are not limited to SARS-CoV-2. Future studies are needed to externally validate our findings and to evaluate their clinical impact, as well as to assess a direct relation between biomarkers and virus transmission.
In conclusion, biomarkers such as IP-10 represent possible tools in estimating infectiousness of SARS-CoV-2 infection and could overcome the economic limitations of exhaustive PCR testing in quarantine practice.
Acknowledgements
We thank all patients and their families who participated in this study. We also wish to express our gratitude to all staff members involved in the conduct of the study, as well as in patient care.
Author contributions
S.T. and C.P. conceived the study and its design, had full access to the data, and take responsibility for the integrity of the data and accuracy of the analysis. Funding acquisition was made by C.P.; S.T., J.E., and C.P. organized and entered data. S.T. and C.P. performed data analyses. All authors contributed to data interpretation. S.T. and C.P. wrote the main draft of the manuscript. All authors contributed to the final drafting of the manuscript.
Conflict of interest statement
None declared.
Funding sources
This study has been funded by a 2021 CAREer Grant from the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) to C.P. Additionally, it was supported by a grant awarded to MeMed from the European Commission, Executive Agency for Small and Medium-sized Enterprises H2020-EIC-SMEInst-2018-2020-2 [grant number 88124]. The funding sources played no role in study design, data collection, data analysis, data interpretation, writing of the manuscript, or in the decision to submit the manuscript for publication.
Ethics approval statement
This study was approved by the ethics committee of the Ärztekammer des Saarlandes (reference number 019/20).
References
- 1.Cevik M., Tate M., Lloyd O., Maraolo A.E., Schafers J., Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2021;2:e13–e22. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puhach O., Meyer B., Eckerle I. SARS-CoV-2 viral load and shedding kinetics. Nat Rev Microbiol. 2023;21:147–161. doi: 10.1038/s41579-022-00822-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeong Y.D., Ejima K., Kim K.S., Iwanami S., Bento A.I., Fujita Y., et al. Revisiting the guidelines for ending isolation for COVID-19 patients. eLife. 2021;10 doi: 10.7554/eLife.69340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Onakpoya I.J., Heneghan C.J., Spencer E.A., Brassey J., Rosca E.C., Maltoni S., et al. Viral cultures for assessing fomite transmission of SARS-CoV-2: a systematic review and meta-analysis. J Hosp Infect. 2022;130:63–94. doi: 10.1016/j.jhin.2022.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark T.W., Brendish N.J., Poole S., Naidu V.V., Mansbridge C., Norton N., et al. Diagnostic accuracy of the FebriDx host response point-of-care test in patients hospitalised with suspected COVID-19. J Infect. 2020;81:607–613. doi: 10.1016/j.jinf.2020.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papan C., Argentiero A., Porwoll M., Hakim U., Farinelli E., Testa I., et al. A host signature based on TRAIL, IP-10, and CRP for reducing antibiotic overuse in children by differentiating bacterial from viral infections: a prospective, multicentre cohort study. Clin Microbiol Infect. 2022;28:723–730. doi: 10.1016/j.cmi.2021.10.019. [DOI] [PubMed] [Google Scholar]
- 7.Tegethoff S.A., Danziger G., Kühn D., Kimmer C., Adams T., Heintz L., et al. TNF-related apoptosis-inducing ligand, interferon gamma-induced protein 10, and C-reactive protein in predicting the progression of SARS-CoV-2 infection: a prospective cohort study. Int J Infect Dis. 2022;122:178–187. doi: 10.1016/j.ijid.2022.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fröhlich F., Gronwald B., Bay J., Simon A., Poryo M., Geisel J., et al. Expression of TRAIL, IP-10, and CRP in children with suspected COVID-19 and real-life impact of a computational signature on clinical decision-making: a prospective cohort study. Infection. 2023:1–8. doi: 10.1007/s15010-023-01993-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Safiabadi Tali S.H., LeBlanc J.J., Sadiq Z., Oyewunmi O.D., Camargo C., Nikpour B., et al. Tools and techniques for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)/COVID-19 detection. Clin Microbiol Rev. 2021;34:e00228–e320. doi: 10.1128/CMR.00228-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papan C., Argentiero A., Adams O., Porwoll M., Hakim U., Farinelli E., et al. Association of viral load with TRAIL, IP-10, CRP biomarker signature and disease severity in children with respiratory tract infection or fever without source: a prospective, multicentre cohort study. J Med Virol. 2023;95 doi: 10.1002/jmv.28113. [DOI] [PubMed] [Google Scholar]