Abstract
Improved copy number variation (CNV) detection remains an area of heavy emphasis for algorithm development; however, both CNV curation and disease association approaches remain in its infancy. The current practice of focusing on candidate CNVs, where researchers study specific CNVs they believe to be pathological while discarding others, refrains from considering the full spectrum of CNVs in a hypothesis-free GWAS. To address this, we present a next-generation approach to CNV association by natively supporting the popular VCF specification for sequencing-derived variants as well as SNP array calls using a PennCNV format. The code is fast and efficient, allowing for the analysis of large (>100,000 sample) cohorts without dividing up the data on a compute cluster. The scripts are condensed into a single tool to promote simplicity and best practices. CNV curation pre and post-association is rigorously supported and emphasized to yield reliable results of highest quality. We benchmarked two large datasets, including the UK Biobank (n > 450,000) and CAG Biobank (n > 350,000) both of which are genotyped at >0.5 M probes, for our input files. ParseCNV has been actively supported and developed since 2008. ParseCNV2 presents a critical addition to formalizing CNV association for inclusion with SNP associations in GWAS Catalog. Clinical CNV prioritization, interactive quality control (QC), and adjustment for covariates are revolutionary new features of ParseCNV2 vs. ParseCNV. The software is freely available at: https://github.com/CAG-CNV/ParseCNV2.
Subject terms: Computational biology and bioinformatics, Pathogenesis, Genetics research
Introduction
PennCNV [1] has emerged as a fast and popular high-sensitivity/specificity tool for CNV detection in single nucleotide polymorphism (SNP) array data. However, no such tool exists as a field consensus tool for whole-genome sequencing data. Compounding this problem is the lack of curation and association tools to apply on CNV calls from these detection algorithms to yield insights into statistical error properties of the calls or disease biology. Plink [2] is an excellent suite of tools for efficient genome-wide association studies on SNP genotypes, but does not support CNV nor structural variation (SV) calls (http://www.cog-genomics.org/plink2#Limitations). ParseCNV has been actively supported and developed since 2008 [3] which led to a Nature paper applying the methods [4] (2011 first posted online, 19 updated releases with new features over 8 years, or almost 2.5 per year) as documented in http://parsecnv.sourceforge.net/#VersionHistory. In contrast, other CNV association tools have stagnated with minimal additional content beyond the initial publication (Table 1).
Table 1.
Comparison of existing CNV association tools.
| CNV Association tool | Reference (First Author) | Reference (Journal) | Year Published | Citations | Citations/Year | Last Updated (Years Ago) | URL |
|---|---|---|---|---|---|---|---|
| CNVRanger | da Silva V | Bioinformatics | 2020 | 7 | 3.50 | 0.25 | https://github.com/waldronlab/CNVRanger |
| CoNVaQ | Larsen SJ | BMC Genomics | 2018 | 15 | 3.75 | 4 | https://github.com/SimonLarsen/convaq |
| Plink1.9 | Chang CC | Gigascience | 2015 | 5632 | 804.57 | 0.33 | https://github.com/chrchang/plink-ng/blob/master/1.9/plink_cnv.c |
| ParseCNV | Glessner JT | Nucleic Acids Res | 2013 | 57 | 6.33 | 3 | http://parsecnv.sourceforge.net/ |
| CNVRuler | Kim JH | Bioinformatics | 2012 | 74 | 7.40 | 10 | www.ircgp.com/CNVRuler/index.html |
| HD-CNV | Butler JL | Bioinformatics | 2012 | 16 | 1.60 | 10 | http://daleylab.org/lab/?page_id=125 |
| CNVassoc | Subirana I | BMC Med Genomics | 2011 | 22 | 2.00 | 3 | https://github.com/isglobal-brge/CNVassoc |
| CONAN | Forer L | BMC Bioinformatics | 2010 | 28 | 2.33 | 3 | http://genepi-conan.i-med.ac.at/https://github.com/lukfor/conan (only readme) |
| R-Gada | Pique-Regi R | BMC Bioinformatics | 2010 | 62 | 5.17 | 3 | https://github.com/isglobal-brge/R-GADA |
| CNVineta | Wittig M | Bioinformatics | 2010 | 9 | 0.75 | 8 | https://www.ikmb.uni-kiel.de/resources/download-tools/software/cnvineta |
| BirdSuite | Korn JM | Nat Genet | 2008 | 903 | 64.50 | 5 | https://github.com/ohsu-comp-bio/birdsuite |
| CNVTools | Barnes C | Nat Genet | 2008 | 209 | 14.93 | 7 | https://sourceforge.net/projects/cnv-tools/ |
| Plink1.07 | Purcell S | Am J Hum Genet | 2007 | 25677 | 1711.80 | 12 | https://zzz.bwh.harvard.edu/plink/download.shtml#download |
The original ParseCNV tool key strengths were: quality tracking information to filter confident associations, uncertainty in CNV calls underlying CNV associations is evaluated to verify significant results, including CNV overlap profiles, genomic context, number of probes supporting the CNV and single-probe intensities. ParseCNV2 started as a code efficiency rewrite from scratch with emphasis on VCF input, the popular consensus format. When optimal quality control parameters are followed using ParseCNV2, 90% of CNVs validate by polymerase chain reaction, an often problematic stage because of inadequate significant association review or low yield of association due to overly strict QC thresholds of input CNV calls.
The sequencing era is advancing rapidly, but the array still holds massive utility for cost-effective screening of SNPs and CNVs. Along with the sequencing era, a number of CNV/SV detection tools and variant call file (VCF) specifications have emerged increasing the complexity of the CNV analysis. In contrast to single nucleotide variants (SNVs), CNVs involve multiple bases and only the chromosome and start positions are given dedicated fields. In some cases, the END position may need to be found by taking the difference in string lengths of reference (REF) and alternate (ALT) alleles. More commonly, the “INFO” field may contain “END”, “SVLEN”, or “SVEND” but cannot be assumed to be located in the same index order of the semi-colon separated INFO list and other fields containing the substring “END” should not be confused, except in the case of “SVEND” or similar. Along with the various CNV detection tools in sequencing are various interpretations of those methods with different CNV specification of the VCFs. While this is not ideal, VCF has remained the popular mainstay of sequencing variant call formats. VCF allows for accessible usage by bioinformaticians and functional biologist alike. VCF was designed chiefly for single nucleotide variants (SNVs) given the singular “POS” field, instead of START and END. A potential solution is the 3rd column ID could be used to contain CHR:POS-END instead of or in addition to (semicolon separated) dbSNP rs ID. Reading the VCF header is needed to get a full description of various abbreviated values, especially QUAL, FILTER, and INFO can be information rich about the variant in the population but have slightly different words used to express the same concept. FORMAT, SAMPLE_ID_1 have genotypes but also sample-specific quality scores of the CNV state predicted by a detection algorithm. Here we present a next-generation approach to CNV association by optimally supporting the popular VCF specification for sequencing-derived variants as well as SNP array PennCNV format.
Methods
ParseCNV2 was coded using Perl primarily with R, Bash, and C modules programming making up the code base as outlined below.
Upfront quality control: SNP microarray data
First, the samples in case and control cohorts must be quality controlled using the SNP genotype call rate, relative intensity standard deviation, intensity waviness, count predicted CNVs, ethnicity principal components, and genotype relatedness as previously described [3] and now expanded in ParseCNV2. CNV calls from passing samples may be further quality controlled using number of probes supporting the CNV signal, genomic length, PennCNV confidence, Mace et al. confidence [5], and DeepCNV probability [6].
Upfront quality control: exome or genome sequencing data
Different yet conceptually similar sample and CNV call QC and confidence metrics are output from each exome or genome sequencing data CNV detection tool for ParseCNV2 -qc usage. For WES, the number of contiguous exons supporting the existence of a CNV is used as the number of probes. The most frequently provided quality control metric across WES and WGS CNV VCF outputs is “Phred-scaled quality score” which should be comparable between different CNV detection tools for WES or WGS. Tool specific QC metrics also exist, for example: CLC Genomics Workbench: Absolute fold change, CNVkit: Mean squared standard error of copy number log2, CNVnator: t-statistic p-value, cn.MOPS: Median informative/non-informative ratio value, CODEX2: Likelihood ratio, ControlFREEC: Wilcoxon rank sum test p-value, DELLY: Genotype quality values, ExomeDepth: Observed/expected read ratio, GATK gCNV: CNQscores (difference between the two best genotype Phred-scaled log posteriors), Lumpy: Number of pieces of evidence supporting the variant across all samples, Manta: CNV quality score. The ParseCNV2 code lends itself to be tested with various QC metric thresholds.
Upfront quality control: modulating metric thresholds
Modulating thresholds of quality metrics such as size, depth of coverage, genotype within CNV regions to increase accuracy while retaining sensitivity can be informed by ParseCNV2 interactive quality control using the -qc option recommended usage: First providing QC inputs and reviewing automatically determined thresholds based on outliers in your dataset. Then you may specify an adjusted threshold for a given quality metric based on your review of the plots. “Field standard” recommended thresholds may vary by array type and refinement, however, there are “essential” CNV QC sample inclusion metrics such as Call Rate >0.98 (similarly BAF_SD < 0.045) and LRR_SD < 0.2 for Illumina SNP arrays.
Input files
The input files allow for PennCNV RAWCNV format, a generic BED format, or a VCF format. The idiosyncrasies of VCF format interpretations from various CNV detection tools in sequencing have been tested to ensure flexibility. Other than the CNV calls file, one must specify the sample IDs of cases for the analysis and the genome build used to map coordinates. The consolidation and support of different CNV calling tools output has been demonstrated to be an important feature [7]. A key new feature is extended interpretation to a further range of CNV calling tools output from sequencing data.
Probe-based CNV statistics
CNVs are mapped to the genomic regions they are predicted to occupy per sample and per copy state. Only observed breakpoints make up the genomic coordinate map for computational efficiency and simplicity of representation. Fisher’s exact test or alternative statistical models, such as logistic regression to take covariates into account, are employed to assess significance of allele frequency differences in cases and controls. The 2 × 2 contingency table with raw counts of observations: cases deletion, cases no deletion, controls deletion, controls no deletion, and similarly for duplications with a separate test statistic. Here we distinguish the Plink options --assoc fisher vs. the --model fisher which provides TEST: GENO, TREND, ALLELIC, DOM, and REC. We use the --model fisher output GENO lines p-values and odd ratios to take forward to be consistent with the previous implementation of ParseCNV which used the perl module Text::NSP::Measures::2D::Fisher::twotailed (or right for case enriched only consideration). The most sophisticated Javascript implementation web-based tool for Fisher’s exact test also is consistent: https://www.langsrud.com/fisher.htm or http://www.liheng.org/fisher.html. We used these other implementations of statistical tests to ensure our computations were consistent. Binary case-control logistic generalized linear model with covariates and quantitative trait linear generalized linear model are powerful options for association testing as well. Continuous trait linear regression is an option in ParseCNV2 beyond binary trait logistic regression or Fisher’s exact test. The Fisher’s exact test was used originally for its precision property in low-count rare variant data. The Plink website states “the statistics computed by --glm are not calibrated well when the minor allele count is very small (<20)”. The -q option is used to provide a quantitative trait value for each sample, instead of the -c option to provide case sample IDs. The -covar option accepts a covariates file (containing sex, age, principal components representing genetic ancestry, residual QC metric values for samples passing initial inclusion QC) The code for conducting logistic regression test with covariates association is in a C++ pre-compiled linux binary executable for efficiency and portability, so the text-based code is not visible. The -stat option specifies the test statistic desired (fisher, logistic, or linear).
Association statistic options
Fisher’s exact test is optimized for rare (low population frequency) variant association where other statistics tend to be inflated. RvTests [8] implements many of the latest developed statistical models motivated by the influx of sequencing data and rare variants to increase statistical power (https://github.com/zhanxw/rvtests#models). RvTests is used as a module that generates p-values and direction of effect that are direct inputs to ParseCNV2_Insert.pl if done by the user separately or internally to the ParseCNV2.pl main script by command line option. The -stat option to ParseCNV2 supports classical fisher, logistic, or linear test options or RvTests including Single variant (score, wald, exact, dominantExact, famLRT, famScore, famGrammarGamma, firth), Burden (cmc, zeggini, mb, fp, exactCMC, cmcWald, rarecover, cmat, famcmc, famzeggini), Variable threshold (price, analytic, famAnalytic), Kernel (skat, skato, kbac, famSkat), and Meta-Analysis (score, dominant, recessive, cov, bolt, boltCovA).
Merging probe-based statistics into Copy Number Variable Regions (CNVRs)
CNV breakpoint coordinates with similar p-values (1 power of 10 default) and not exceeding the maximum distance parameter (1 MB default) are collapsed together into CNVRs to reduce redundancy of reporting similar significance nearby regions. The tag SNP of the CNVR is deemed the representative probe-based CNV statistic to characterize the CNVR in a discrete manner. The CNV detection tool uses SNP/Exon small region data points to call individual-level CNV segments which are larger genomic spans. ParseCNV2 takes these individual-level CNV segments and converts them to population-level CNV resolved to SNP/Exon small region data points granularity for association testing. Once ParseCNV2 has the p-values and direction of effects (OddsRatio/BETA), then these SNP/Exon small region data points are re-segmented into population level CNV segments based on minimal variation in p-value, consistent direction of effect, and minimal distance of neighboring SNP/Exon small region data points. Overall, SNP/Exon small region data points are assessed by one or more CNV Detection Tools resulting in CNV segments (Compose), then ParseCNV2 Association creates SNP/Exon small region data points p-values and direction of effects (Decompose), lastly, ParseCNV2 CNVR Calling outputs the Association CNVRs (Compose). If CNVs in a genomic region are very few the CNVR boundaries will indeed be less confident. Also, since these CNVs are rarely observed, SNP arrays or WES capture kits likely have not tested such variation in developing their content. This is also why we re-calculate CNVR boundary definition based solely on the input CNVs provided from a specific platform, rather than having a static list of CNVR boundaries provided up-front and used for all platforms and studies. While a static list of CNVR boundaries provides consistency and comparability, it does not model rare CNVs well into CNVRs.
Review of association signals by quality tracking
Various genomic features are annotated for each CNVR to further characterize the genomic context of significant signals from the genome-wide CNV analysis. This includes CNV disease and healthy control databases such as Decipher and ClinVar as well as DGV [9] and gnomadSV [10]. Sophisticated CNV call characterization has been done in 1000 Genome Project [11] and Autism Families [12]. Quality tracking remains a paramount concern and is addressed by “red flags” which are annotated on CNVRs to establish confidence in the overall association signal. Predetermined significance criteria examples of best practices: Red Flags Average CNV Length <1000 bp, Database of Genomic Variants Overlapping Entries >10, Maximum p-value in the CNVR >0.5 (as opposed to the minimum p-value which is reported), Population Frequency >0.01, Segmental Duplications (regions >1 kb with 90% matching) Overlapping Entries >10, Known Recurrent False Positive Regions, Inflated Samples Frequency >0.5, Average Confidence <10; p < 0.0005 and direction = “case” (green flag), exon overlap (green flag). Finally, to determine overall CNVR Pass or Fail: RF > 2 Red Flag Pass Fail = “FAIL” otherwise Red Flag Pass Fail = “PASS”. Standard Filter for high significant and confident results: Sort Red Flags <= 3, deletion p-value < 5 × 10^−4 and Odds Ratio deletion>1 or duplication p-value <5 × 10^−4 and Odds Ratio Duplication>1 (on exon).
Multiple testing correction
P-value threshold of 5 × 10−4 is a conservative bar for CNV genome-wide significance surviving multiple testing correction based on analysis of Illumina and Affymetrix genome-wide SNP arrays, including a count of less than 100 CNVs per sample (corrected P value of <0.05). The typical bar of 5 × 10−8 used in GWAS is not appropriate for CNV association considering:
The number of probes with a nominal frequency of CNV occurrence (only probes with some CNV detected are informative) in a high-quality SNP-array sample amounts to fewer than 100 CNVs.
The number of probes with enrichment in cases vs. controls and vice versa (evidence of more case-enriched loci than control-enriched loci)
We are principally interested in probes with less than 1% population frequency of CNV (optionally).
The number of CNVRs (multiple probes are needed to detect a single CNV and should not count separately for multiple testing correction) is well below 100 per sample, rendering P value of 5 × 10−4 being appropriate for multiple testing correction.
2 × 10−5 multiple testing correction threshold according to permutation studies was observed.
In an independent recent study [13], Bonferroni correction was used for multiple testing adjustment to control the family-wise error rate (FWER) of α = 0.05. The P-value threshold for genome-wide significance was 0.05/23 = 2.2 × 10–3, where 23 is the total number of CNVRs tested.
CNV validation by quantitative Polymerase Chain Reaction (QPCR)
Orthogonal validation by an independent method is still a crucial step to verify CNVs and address if a given CNV is enriched in cases and not an artifact of technology issues of a given assay. qPCR is the primary workhorse for such verification. Droplet Digital PCR (ddPCR) is a more expensive yet more sensitive method, especially for non-integer copy states. In the case of detection of CNVs based primarily on sequencing, microarrays run on the same samples can serve as independent validation as well. We ran 393 across various loci on various sample sources with a qPCR validation success rate of 0.8.
Clinical
Prioritization of CNVs based on the predicted pathogenicity, dosage sensitivity scores, and concordance with disease CNV entries in Decipher and ClinVar in individual patient-by-patient basis is another horizon for ParseCNV2 [14, 15]. Deleteriousness of the CNV on the genomic span of bases is integrated. SG-ADVISER [16] and AnnotSV [17] are comparable tools in this approach. Clinical CNV prioritization integrates Annotations: OMIM, DGV, ClinGen, Known Syndromic, Internal Controls (with matched genomic assay platform), Gene exon, pLI, and HGMD CNVs. Quality control filtering of CNV calls for clinical utility typically requires more strict thresholds than research-based GWAS to optimize specificity, even at the cost of lowering sensitivity.
Comparison to other methods
We compared existing CNV association tools by benchmarking different: public CNV call data (1KG, UKBiobank, CAG CNV Map), CNV Calling genomic platform, and association type (Case-Control or Quantitative Trait) monitoring: CPU use, runtime, memory use, and results nominally significant p < 0.05 CNV loci split into (where available): deletion/duplication, direction of effect (OR for case-control, BETA for quantitative trait), and Pass/Fail CNV overlap Red Flag association QC (Table 2).
Table 2.
Benchmark results of ParseCNV2 on various Genomic Data.
| RUN | Cases | Controls | CPU | TIME (h:mm:ss) | MEM (kbytes) | Del Case Enrich (Pass + Fail) | Del Control Enrich (Pass + Fail) | Dup Case Enrich (Pass + Fail) | Dup Control Enrich (Pass + Fail) |
|---|---|---|---|---|---|---|---|---|---|
| ParseCNV2 Array | 5 | 5 | 85% | 0:00:11 | 280,888 | 0 + 0 | 0 + 0 | 0 + 0 | 0 + 0 |
| ParseCNV2 Array (550k) | 785 | 1110 | 94% | 0:01:29 | 280,888 | 49 + 11 | 31 + 5 | 33 + 2 | 37 + 4 |
| ParseCNV2 Array (2.5 M) | 700 | 797 | 96% | 0:06:35 | 280,716 | 143 + 1 | 190 + 4 | 124 + 13 | 73 + 4 |
| ParseCNV2 WES | 758 | 1344 | 97% | 0:06:29 | 280,896 | 42 + 2 | 2 + 0 | 154 + 25 | 49 + 3 |
| ParseCNV2 WGS | 205 | 689 | 96% | 0:56:43 | 758,776 | 556 + 17 | 455 + 9 | 293 + 13 | 353 + 14 |
| ParseCNV(original) Array | 5 | 5 | 80% | 0:0:16 | 280,616 | 0 + 0 | 0 + 0 | 0 + 0 | 0 + 0 |
| ParseCNV(original) Array (550k) | 785 | 1110 | 93% | 0:0:32 | 280,620 | 17 + 39 | 3 + 27 | 17 + 13 | 9 + 31 |
| ParseCNV(original) Array (2.5 M) | 700 | 797 | 97% | 0:1:49 | 789,468 | 66 + 56 | 18 + 148 | 41 + 86 | 24 + 42 |
| ParseCNV(original) WES | 758 | 1344 | 98% | 0:5:19 | 991,372 | 0 + 45 | 0 + 17 | 0 + 179 | 0 + 49 |
| ParseCNV(original) WGS | 205 | 689 | 98% | 1:06:11 | 14,440,980 | 0 + 458 | 0 + 335 | 0 + 145 | 0 + 115 |
| Plink2 UKB AguirreEtAl | 6564 | 15,480 | 99% | 0:34:21 | 104,528 | 0 | 55 | 0 | 2 |
| ParseCNV2 UKB AguirreEtAl | 6564 | 15,480 | 99% | 2:32:20 | 973,120 | 17 + 8 | 83 + 8 | 8 + 4 | 22 + 2 |
| Plink2 CNVMAP2 CAG INTERNAL | 20,637 | 48,314 | 99% | 3:51:17 | 298,496 | 4567 | 980 | 2707 | 570 |
| ParseCNV2 CNVMAP2 CAG INTERNAL | 20,637 | 48,314 | 99% | 33:51:57 | 14,907,708 | 777 + 2 | 871 + 1 | 801 + 0 | 680 + 3 |
| CNVRanger 1KG qt | 2504 values | 98% | 0:46:36 | 2,629,344 | 901 | ||||
| ParseCNV2 1KG cc | 767 | 1,737 | 98% | 0:23:59 | 871,864 | 241 + 81 | 170 + 88 | 256 + 28 | 174 + 16 |
| ParseCNV2 1KG qt | 2504 values | 97% | 0:27:18 | 872,832 | 324 + 96 | 390 + 112 | 276 + 81 | 256 + 83 | |
| ParseCNV(original) 1KG cc | 767 | 1,737 | 99% | 0:14:08 | 3,923,728 | 239 + 70 | 32 + 232 | 193 + 59 | 123 + 83 |
| ParseCNV(original) 1KG qt | 2504 values | 91% | 0:46:15 | 4,409,288 | 398 | 470 | 342 | 313 | |
| Plink2 1KG cc | 767 | 1737 | 99% | 0:00:22 | 705,608 | 140 | 55 | 155 | 36 |
Whole Exome Sequencing (WES) data for validation
The WES input to ParseCNV2 was downloaded from the database of genotypes and phenotypes (dbGaP) released by the Pediatric Cardiac Genomics Consortium (PCGC) [18] (accession phs001194.v2.p2 and phs001194.v2.p2.c1) including 2103 individuals with exome capture Nimblegen SeqCap Exome V2 and sequencing on Illumina HiSeq 2000 platform. Sequence reads were aligned to the human reference genome hg19 using Burrows-Wheeler Aligner (BWA −0.7.17 r1188) and duplicates were marked with Picard. Insertion deletion (Indel) realignment and Base Quality Score Recalibration was done with GATK. To generate potential CNV calls and quality metrics, we used the XHMM pipeline [19] consisting of 6 steps: (1) depth of coverage calculated for all targets. (2) Filter out target regions with extreme GC content (<10% or >90%) and complexity regions. (3) PCA normalization of read depth (also included in recently released GATK gCNV). (4) Remove samples with extreme variability in normalized read depth. (5) Per-Sample CNV Detection with a hidden Markov model (HMM). (6) Quality Metrics assigned to discovered CNVs. CNV association was performed using the ParseCNV2 pipeline presented here, based on VCF files from XHMM.
Whole Genome Sequencing (WGS) data for validation
WGS input to ParseCNV2 was generated by the Center for Applied Genomics (CAG) at The Children’s Hospital of Philadelphia (CHOP), leveraging CAG’s Biobank including comprehensive electronic medical records (EMR). We reviewed and analyzed 205 Attention Deficit Hyperactivity Disorder (ADHD) cases and 670 controls of European and African Ancestry. All subjects were thoroughly phenotyped [20]. The structural variations (SVs), including deletions, duplications, insertions, and inversions, were detected by MANTA which leverages read-pair information content. We included SVs that passed MANTA’s default filters. VCF files from MANTA were input to ParseCNV2.
Results
We outlined the workflow structure for the variables incorporated into ParseCNV2 runs for both array and sequencing data inputs (Fig. 1). We also provide a graphical CNV analysis workflow which includes the steps: Quality assessment, ParseCNV2 CNV association, Red Flag review, and Raw signal (BAF/LRR) Review (Fig. 2). ParseCNV2 starts with a comprehensive quality control (QC) stringency of both samples and individual CNV calls. The software performs genome-wide CNV association which can be accomplished in less than 10 min for 2000 samples run with 32 GB memory on a x86_64 GNU/Linux system with Intel(R) Xeon(R) Platinum 8176 CPU @ 2.10 GHz processor (Table 2). The data used for computational benchmarking were: AGRE Autism samples genotyped on Illumina 550 K SNP microarray: 5 cases vs. 5 controls and 785 cases vs. 1110 controls. Neurodevelopmental disorder samples genotyped on Illumina Omni2-5-8v1-3 SNP microarray: 700 cases vs. 797 controls. Congenital heart defect samples genotyped on Nimblegen SeqCap Exome V2 WES: 758 cases vs. 1344 controls. Neurodevelopmental disorder samples genotyped on Illumina WGS: 205 cases vs. 689 controls. Publicly available CNV call sets were used including: 1000 Genomes Population control samples genotyped on Illumina WGS including the widely studied gold standard sample NA12878: 767 cases vs. 1737 controls (randomly assigned), UKBiobank CNV calls [21], and CAG CNV Map [22] (Table 2). CNV association tools were run with publicly available CNV call sets listed in Table 2 and resulting nominally significant genomic regions compared (Table 3 and Fig. 3).
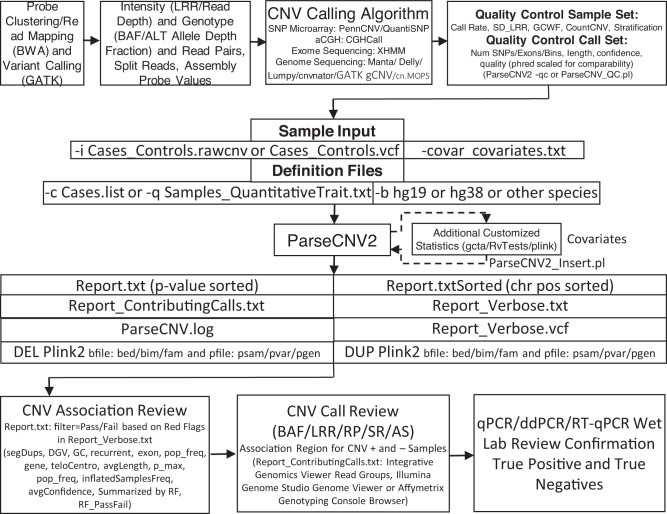
Fig. 1. ParseCNV2 process flow.
Mirroring ParseCNV Nucleic Acids Research Fig. 1 [3] to show advances made at each step in ParseCNV2. Data processing, information content extraction and assessment, CNV calling based on genomic regions of aberration, Quality control metric assessment for samples and calls (in passing samples), CNV calls are provided for all samples in PennCNV rawcnv or VCF formats, covariates (age, sex, race/top principal components), case sample IDs list or quantitative trait value for all sample IDs, and genome build are the main ParseCNV2 command line options. Optionally, ParseCNV2 output files can be used by other statistical methods and imported back to ParseCNV2 reporting using ParseCNV2_Insert. The main output is Report.txt (p-value sorted) listing genomic segments and ParseCNV2 Red Flag filter for CNV Association Review (details in Report_Verbose output), Report_ContributingCalls.txt contains the input calls underlying significant associations for review. Lastly, samples with predicted CNV and samples without predicted CNV for each remaining significant CNV region are confirmed by lab PCR method.
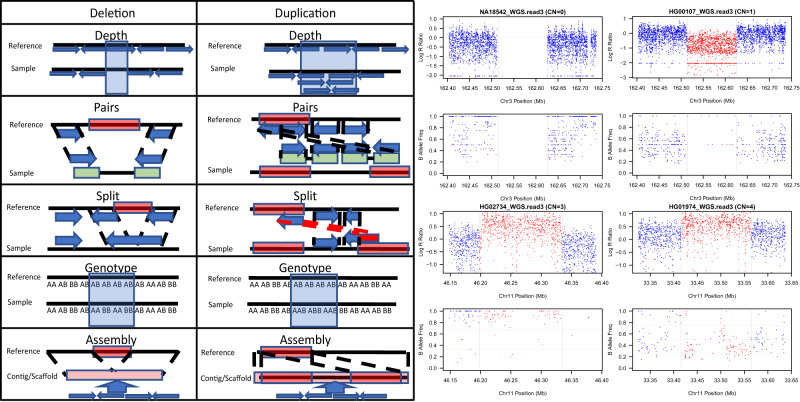
Fig. 2. ParseCNV2 process flow graphical representation.
Image representation mirroring top row boxes and bottom row boxes of ParseCNV2 process flow text-based Fig. 1.
Table 3.
Venn diagram (upset plot) quadrant counts for upset plot input.
| 1KG association results File 1 | 1KG association results File 2 | Intersect |
|---|---|---|
| CNVRanger_1KG_qt | ParseCNV2_1KG_cc | 1518 |
| CNVRanger_1KG_qt | ParseCNV2_1KG_qt | 2322 |
| CNVRanger_1KG_qt | ParseCNVOriginal_1KG_cc | 1286 |
| CNVRanger_1KG_qt | ParseCNVOriginal_1KG_qt | 429 |
| CNVRanger_1KG_qt | Plink2_1KG_cc | 15,529 |
| ParseCNV2_1KG_cc | ParseCNV2_1KG_qt | 194 |
| ParseCNV2_1KG_cc | ParseCNVOriginal_1KG_cc | 828 |
| ParseCNV2_1KG_cc | ParseCNVOriginal_1KG_qt | 39 |
| ParseCNV2_1KG_cc | Plink2_1KG_cc | 2090 |
| ParseCNV2_1KG_qt | ParseCNVOriginal_1KG_cc | 158 |
| ParseCNV2_1KG_qt | ParseCNVOriginal_1KG_qt | 219 |
| ParseCNV2_1KG_qt | Plink2_1KG_cc | 3851 |
| ParseCNVOriginal_1KG_cc | ParseCNVOriginal_1KG_qt | 32 |
| ParseCNVOriginal_1KG_cc | Plink2_1KG_cc | 1939 |
| ParseCNVOriginal_1KG_qt | Plink2_1KG_cc | 854 |
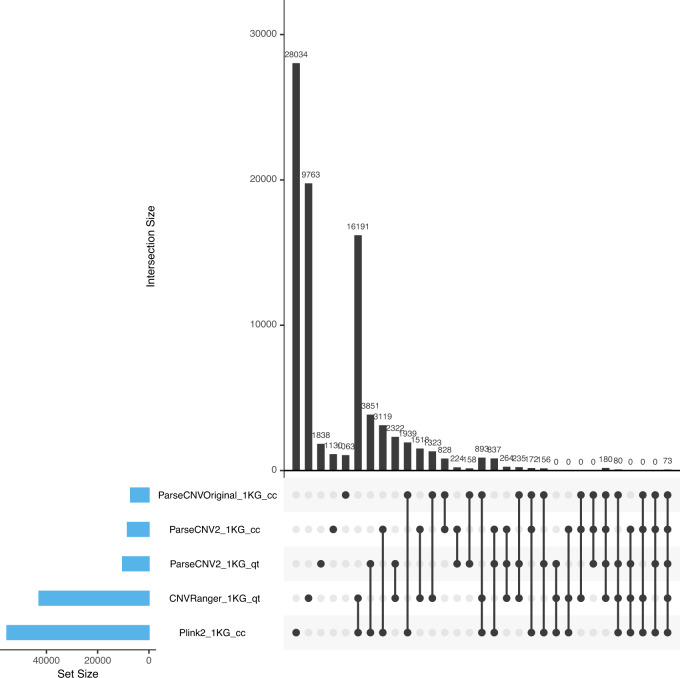
Fig. 3. Upset Plot Comparing Nominally Significant CNVR Loci.
1000 Genomes CNV callset phase 3 from Sudmant et al. [11] with samples split randomly into cases and controls or assigned quantitative trait values to conduct CNV association testing with different methods. The Upset Plot shows 73 nominally significant CNVR loci are detected by all tested CNV association methods. The Upset Plot was generated by R package UpSetR [25].
Chromosome, start and end (base pair position based on the genome build used), p-value, odds ratio (OR), cases (count), controls (count), and filters used are statistical association fields in the output file generated. Direction, type, count cases, count controls, caseIDs, controlIDs are annotated to track the kind of association signals produced. Segmental duplications, DGV, Guanine/Cytosine base content, cytoband, recurrent events, exon impact, gene(s) impacted and telomere/centromere involvement are provided as genomic feature annotations. Collectively, this approach provides high quality association results with robust confidence. The output is also provided in a brief and succinct format to be readable in a Linux terminal and Microsoft Excel format. Significant CNVRs are then QC reviewed for further curation and bias screening and either kept or dropped based on predetermined significance criteria.
Development of a unified CNV VCF parser for diverse applications
While VCF parsers exist, few if any support the variety of VCF presentations and interpretations of CNV genotypes. Therefore, we implemented a flexible VCF to rawcnv/bed format conversion tool. Key challenges include the variety of ways to represent the end genomic position and the alternative alleles. The allele coding for the genotypes may be phased “|” or unphased “/” and can use any combination and order of alternative allele copy number states such as ALT = 0,2 and GT = 0/0 meaning CN = 2 or GT = 0/1 meaning CN = 1 or GT = 2/2 meaning cn = 4. This lack of strict convention for encoding CNVs into VCF creates a strong challenge to make a unified parser that will accept and correctly interpret diverse sources of VCFs.
Validation based on real NGS data
Figure 4 shows WES and WGS CNVR association results from ParseCNV2 results based on sequencing inputs. CNV detection algorithm concordance and potential unique features such as type of information content assessed can produce a CNV callset balancing sensitivity and specificity.
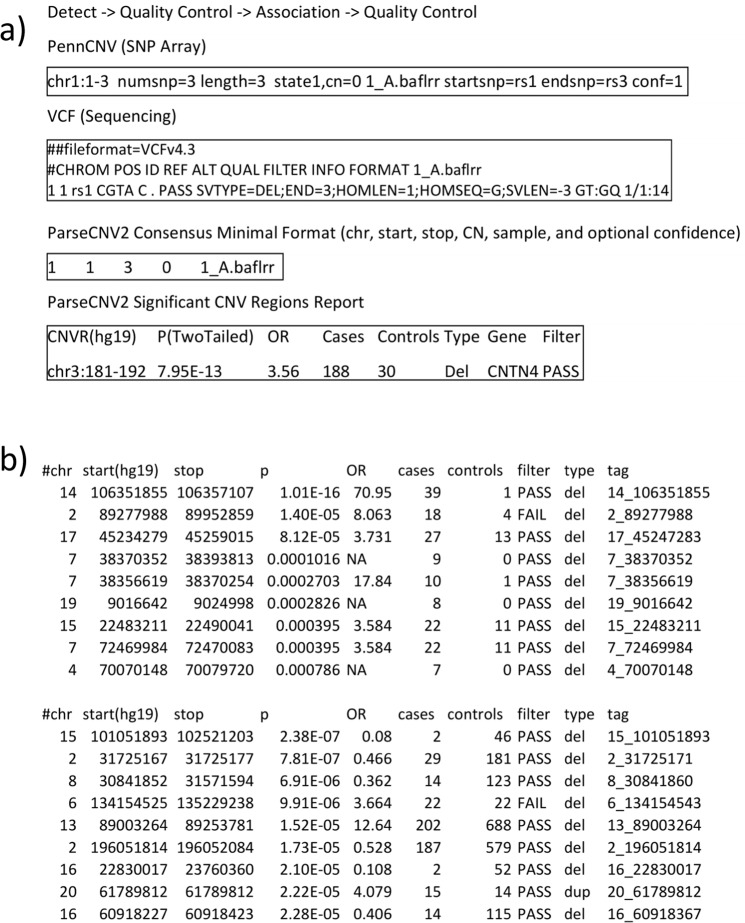
Fig. 4. ParseCNV2 Input and Output Formats.
a Input CNV Data Formatting and Harmonization by ParseCNV2 from array and sequencing data and b CNVR Association Results for WES and WGS.
To illustrate advantages of ParseCNV2, we applied ParseCNV2 association to the XHMM and exomeDepth detection algorithm outputs from the PCGC data set. Based on annotations and filters in ParseCNV2, the number of CNV candidates in the PCGC WES dataset was reduced to 242.
Various datatypes and sizes were tested and benchmarked for speed of computation. Results are listed as the number of nominally significant (p < 0.05 unadjusted for multiple testing) CNVRs enriched in cases/enriched in controls. For ParseCNV comparison, pre-parsed VCFs to RAWCNVs were required and run with the splitByChr option to avoid memory overflow.
Discussion
In this study, we present a next-generation approach to CNV association by supporting VCF specification for sequencing-derived variants and SNP array data. The code is fast and efficient, allowing for the analysis of large cohorts without dividing up the data on a compute cluster. The scripts are condensed into a single tool to promote simplicity and best practices with association CNV curation that is rigorously supported to yield reliable results and of higher quality than existing tools. In this study, we present a formalization of CNV association. CNV association curation is rigorously supported to yield reliable results of higher quality than existing tools. The UK Biobank samples were CNV called using previously described methods [21]. We would like to provide sensitivity and specificity data in the benchmarking table but establishing a “truth set” or “gold standard” for CNV associations genome-wide remains challenging. The closest thing is probably 1KG P3 curated CNVs release [11] and in particular sample NA12878. The 1KG VCF including 2500 samples is the best gold standard for CNV detection which is also freely accessible for raw read mapping data and CNV call data [11].
1KGP4 released 9 samples (3 trios: Han Chinese, Puerto Rican, and Yoruban) sequenced and arrayed by many platform types and algorithms [23]. Namely Pacific Biosciences, Oxford Nanopore, Illumina short insert, Illumina liWGS, Illumina 7 kb JMP, 10X Chromium, Bionano Genomics, Tru-Seq SLR, Strand-seq, and Hi-C.
Genome in a Bottle (GIAB) has independently undertaken a similar effort [24] typing 7 samples on diverse platforms.
Limitations to address in future releases include: extending functionality to support the full range of SV types, including SVA (SINE, VNTR and Alu), ALU, LINE1, CNV, INVersion, INSertion, STR (Short Tandem Repeat), ROH (Run of Homozygosity), MOS (Mosaic CNV). Need more Gold Standards for CNV and SV association and Reference Map Definitions in sizeable cohorts. Modeling Detection Error Profile of each SV type is needed to inform metrics and thresholds for QC filtering. The Weight of each SV in Association could be further delineated. Formalizing CNV and SV association and reporting is needed for inclusion of significant CNV and SV associations in a widely used reference platform like the GWAS Catalog. QC filtering of samples for CNV detection and passing samples CNV calls could provide the passing sample and call with the actual value of all the QC metrics as a covariate for association to further adjust the association testing.
Conclusion
Being able to interpret CNVs effectively is severely hampered by lack of confidence of current CNV detection tools as well as lack of effective association tools. The same phase existed prior to PennCNV being released where CNV variant calls from array data were completely unreliable. Today, we are in a similar phase with sequencing data CNV variant calls. Fast and easy curation and association is a must have tool as implemented in ParseCNV2. ParseCNV2 is an efficient algorithm for both QC and statistical disease phenotype associations, a feature lacking in current tools, and supports many species genomic CNV analysis. With respect to key attributes of the ParseCNV2 we emphasize the fast and easy curation and association of CNVs in both population and family-based disease association settings.
Acknowledgements
We thank the study participants who allowed for the use of genotyping, sequencing, and disease phenotype data for this study, and to testers of the codes used in this study. Funding This work was supported in part by CHOP’s Endowed Chair in Genomic Research (Hakonarson), by 5U01HG011175-03 and U01-HG006830 (NHGRI-sponsored eMERGE Network), by a sponsored research agreement from Aevi Genomic Medicine Inc. (HH), Intellectual and Developmental Disabilities Research Center (IDDRC), Kids First Gabriella Miller Pediatric Research Program, and by an Institutional Development Award from Children’s Hospital of Philadelphia (HH).
Author contributions
JTG conceived, designed, and implemented the code and wrote the paper. JL provided strategic guidance and ran other CNV association tools in benchmarking. YL provided and ran WES and WGS data CNV calls for validation of the ParseCNV2 algorithm and wrote those sections. MK compared ParseCNV2 with ParseCNV original version outputs to delineate reproducibility vs. new associations based on feature improvement. XC designed experiments and helped write the manuscript. PMAS contributed to data extraction. HH provided feedback on the report.
Data availability
• Project name: ParseCNV2
• Project (source code) home page: https://github.com/CAG-CNV/ParseCNV2
• Operating systems: Linux (32/64-bit), OS X (64-bit Intel), Windows (32/64-bit)
• Programming language: Perl, R, Bash
• Other requirements (when recompiling): none
• License: GNU General Public License version 3.0 (GPLv3)
• Any restrictions to use by non-academics: none
Ethical approval
All subjects were recruited through IRB-approved protocols. Participants enrolled in various studies and completed a broad informed consent, including consent for prospective analyses of EHRs. Confidentiality is guarded to address issues of privacy and insurability. Each subject is assigned a study number upon recruitment, using complex algorithms to remove personal identification. Encrypted patient data is integrated into the lab’s custom phenotype browser, where it can be coupled with genotyping and sequencing data.
Competing interests
The authors declare no competing interests. Unrelated to this manuscript, we disclose that HH and CHOP own stock in Aevi Genomic Medicine.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–74. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glessner JT, Li J, Hakonarson H. ParseCNV integrative copy number variation association software with quality tracking. Nucleic Acids Res. 2013;41:e64. doi: 10.1093/nar/gks1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–73. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mace A, Tuke MA, Beckmann JS, Lin L, Jacquemont S, Weedon MN, et al. New quality measure for SNP array based CNV detection. Bioinformatics. 2016;32:3298–305.. doi: 10.1093/bioinformatics/btw477. [DOI] [PubMed] [Google Scholar]
- 6.Glessner JT, Hou X, Zhong C, Zhang J, Khan M, Brand F, et al. DeepCNV: a deep learning approach for authenticating copy number variations. Brief Bioinform. 2021. [DOI] [PMC free article] [PubMed]
- 7.Kim JH, Hu HJ, Yim SH, Bae JS, Kim SY, Chung YJ. CNVRuler: a copy number variation-based case-control association analysis tool. Bioinformatics. 2012;28:1790–2. doi: 10.1093/bioinformatics/bts239. [DOI] [PubMed] [Google Scholar]
- 8.Zhan X, Hu Y, Li B, Abecasis GR, Liu DJ. RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics. 2016;32:1423–6. doi: 10.1093/bioinformatics/btw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacDonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:D986–92. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins RL, Brand H, Karczewski KJ, Zhao X, Alfoldi J, Francioli LC, et al. A structural variation reference for medical and population genetics. Nature. 2020;581:444–51. doi: 10.1038/s41586-020-2287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Werling DM, Brand H, An JY, Stone MR, Zhu L, Glessner JT, et al. An analytical framework for whole-genome sequence association studies and its implications for autism spectrum disorder. Nat Genet. 2018;50:727–36.. doi: 10.1038/s41588-018-0107-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhan X, Girirajan S, Zhao N, Wu MC, Ghosh D. A novel copy number variants kernel association test with application to autism spectrum disorders studies. Bioinformatics. 2016;32:3603–10.. doi: 10.1093/bioinformatics/btw500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexander-Bloch A, Huguet G, Schultz LM, Huffnagle N, Jacquemont S, Seidlitz J, et al. Copy Number Variant Risk Scores Associated With Cognition, Psychopathology, and Brain Structure in Youths in the Philadelphia Neurodevelopmental Cohort. JAMA Psychiatry. 2022;79:699–709. doi: 10.1001/jamapsychiatry.2022.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins RL, Glessner JT, Porcu E, Lepamets M, Brandon R, Lauricella C, et al. A cross-disorder dosage sensitivity map of the human genome. Cell. 2022;185:3041–55.e25. doi: 10.1016/j.cell.2022.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erikson GA, Deshpande N, Kesavan BG, Torkamani A. SG-ADVISER CNV: copy-number variant annotation and interpretation. Genet Med. 2015;17:714–8. doi: 10.1038/gim.2014.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geoffroy V, Herenger Y, Kress A, Stoetzel C, Piton A, Dollfus H, et al. AnnotSV: an integrated tool for structural variations annotation. Bioinformatics. 2018;34:3572–4. doi: 10.1093/bioinformatics/bty304. [DOI] [PubMed] [Google Scholar]
- 18.Glessner JT, Bick AG, Ito K, Homsy J, Rodriguez-Murillo L, Fromer M, et al. Increased frequency of de novo copy number variants in congenital heart disease by integrative analysis of single nucleotide polymorphism array and exome sequence data. Circ Res. 2014;115:884–96. doi: 10.1161/CIRCRESAHA.115.304458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fromer M, Purcell SM. Using XHMM software to detect copy number variation in whole-exome sequencing data. Curr Protoc Hum Genet. 2014;81:7 23 1–1. doi: 10.1002/0471142905.hg0723s81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elia J, Glessner JT, Wang K, Takahashi N, Shtir CJ, Hadley D, et al. Genome-wide copy number variation study associates metabotropic glutamate receptor gene networks with attention deficit hyperactivity disorder. Nat Genet. 2011;44:78–84. doi: 10.1038/ng.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aguirre M, Rivas MA, Priest J. Phenome-wide burden of copy-number variation in the UK Biobank. Am J Hum Genet. 2019;105:373–83. doi: 10.1016/j.ajhg.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li YR, Glessner JT, Coe BP, Li J, Mohebnasab M, Chang X, et al. Rare copy number variants in over 100,000 European ancestry subjects reveal multiple disease associations. Nat Commun. 2020;11:1–9. doi: 10.1038/s41467-019-13624-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaisson MJP, Sanders AD, Zhao X, Malhotra A, Porubsky D, Rausch T, et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat Commun. 2019;10:1784. doi: 10.1038/s41467-018-08148-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenside P, Zook J, Salit M, Cule M, Poplin R, DePristo M. CrowdVariant: a crowdsourcing approach to classify copy number variants. Pac Symp Biocomput. 2019;24:224–35.. [PubMed] [Google Scholar]
- 25.Conway JR, Lex A, Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33:2938–40.. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
• Project name: ParseCNV2
• Project (source code) home page: https://github.com/CAG-CNV/ParseCNV2
• Operating systems: Linux (32/64-bit), OS X (64-bit Intel), Windows (32/64-bit)
• Programming language: Perl, R, Bash
• Other requirements (when recompiling): none
• License: GNU General Public License version 3.0 (GPLv3)
• Any restrictions to use by non-academics: none