Abstract
Cardiovascular diseases are the leading cause of mortality, morbidity, and hospitalization around the world. Recent technological advances have facilitated analyzing, visualizing, and monitoring cardiovascular diseases using emerging computational fluid dynamics, blood flow imaging, and wearable sensing technologies. Yet, computational cost, limited spatiotemporal resolution, and obstacles for thorough data analysis have hindered the utility of such techniques to curb cardiovascular diseases. We herein discuss how leveraging machine learning techniques, and in particular deep learning methods, could overcome these limitations and offer promise for translation. We discuss the remarkable capacity of recently developed machine learning techniques to accelerate flow modeling, enhance the resolution while reduce the noise and scanning time of current blood flow imaging techniques, and accurate detection of cardiovascular diseases using a plethora of data collected by wearable sensors.
Keywords: Cardiovascular diseases, Computational fluid dynamics, Flow imaging, Wearable sensors, Machine learning
Introduction
Cardiovascular diseases (CVDs) refer to a broad range of pathologies caused by disorders in the heart (i.e., cardiomyopathy, valvular diseases, and arrhythmia) or in the blood vessels (i.e., coronary artery disease, cerebrovascular disease, deep vein thrombosis, and peripheral arterial disease), potentially leading to heart attack, heart failure, and stroke (Nabel 2003). CVDs are the major cause of universal mortality, morbidity, and hospitalization (Virani et al. 2020), claiming the lives of 18.6 million people globally in 2019 (Roth et al. 2020), equivalent to one death in roughly every two seconds. Modern life habits such as obesity, lack of physical activities, unhealthy nutrition, high cholesterol, stress, and smoking as well as genetic disorders are among the major risk factors.
A diverse range of diagnostic means have been employed for early detection and monitoring of CVDs in clinical settings (Celermajer et al. 2012), including regular blood test, blood pressure monitoring, electrocardiogram, echocardiogram, stress test, chest x-ray, cardiac computerized tomography (CT) scan, and cardiac magnetic resonance imaging (MRI) (Hunter 2016).
In recent decades, substantial advancements in computing power, mathematical models, imaging techniques, data acquisition and analysis, microfabrication, and biomaterials have facilitated the introduction of new approaches for curbing CVDs. Notably, computational fluid dynamic (CFD) simulations, recent blood flow imaging techniques such as 4D-flow MRI, and wearable sensors are among the means which can significantly enhance our ability to model, monitor, and predict CVDs. Yet, these pioneer means to explore hemodynamics bear their inherent limitations.
CFD modules leverage advanced computational techniques to solve differential equations governing the balance of mass and momentum in hemodynamics, enabling the prediction of flow patterns and associated shear matrices on the vessel walls and circulating cells (Bazilevs et al. 2009; Rikhtegar et al. 2012). However, excessive computational cost impedes the ability of CFD techniques for real-time analysis of blood flow, especially in complex 3D geometries. 4D-flow MRI allows for visualization and quantification of complex blood flow patterns through the heart chambers/valves and large vessels, enabling the identification of pathophysiological flows caused by various cardiovascular disorders (Markl et al. 2012; Stankovic et al. 2014). This technique is also limited in clinical translation as obtaining high spatial and temporal resolution images over large volumes requires long scan times.
Emerging wearable sensors facilitate the non-invasive and continuous monitoring of vital cardiovascular signals such as heartbeat, heart rhythm, and blood pressure in real-time amassing invaluable wealth of data for research and clinical use (Chen et al. 2021). Yet, in-depth analysis of the collected signals to detect meaningful patterns and facilitate the early detection and monitoring of disease progression using conventional statistical approaches is still an unresolved challenge.
These limitations warrant the development and implementation of new strategies to maximize the utility of CFD, 4D-flow MRI, and wearable sensors to tackle CVDs, wherein machine learning methods have been recently leveraged to help. Early roots of machine learning go back to 1950s, when Arthur Samuel of IBM developed a computer program to play checkers. This program was able to make non-programmed decisions based on scoring an early predecessor machine learning algorithm (Samuel 1959). Machine learning can predict outcomes and make decisions based on a given dataset without explicitly coding each of the input possibilities. This is a major advancement over traditional programming, where the code cannot evolve to solve unseen problems. To achieve this, algorithms based on machine learning build a model from the given training data and can, in many cases, automatically extract the prominent features (patterns) from the training data to make the needed predictions (Sidey-Gibbons and Sidey-Gibbons 2019).
Machine learning is a collection of tens of methods that have been developed over the decades under various names such as statistical signal processing, pattern recognition, and computer vision (Theodoridis 2020). Many of the classical machine learning methods employ the Bayesian approach, which is the use of Bayes’ theorem in inferring the probability of an outcome based on a set of measurements (data) (van de Schoot, et al. 2021). Neural networks, on the other hand, use relatively simple building blocks (neurons) that are typically organized in layers, where the level of abstraction is increased in each of the neural layers. The use of artificial neurons as building blocks is also not new and can be dated back to the 40’s; however, it was until the development of the back propagation algorithms in the 70’s that training neural networks become practical. With the exponential increase in the computational power and the further developments in neural network architectures, the training of such networks become feasible with the ability to handle larger number of layers (i.e., deeper networks); hence, the term deep learning started to be widely adopted (Chollet 2018).
Currently, two major applications are extensively assumed for machine learning in the field of biology. The first is using genome sequencing to take large geometric data sets and create predictive models that transcribe the data and identify diseases. The second is image processing and computer vision to classify cells based on their morphology. Both methods require a large amount of raw data and create a predictive model based on computer detected patterns buried within. Once completely trained, the model can analyze and detect those patterns to either classify or identify the new data (Webb 2018). Continuous learning is also possible, where the model continuously evolves as more data is fed to the system.
In this paper, we discuss recent developments in the application of machine learning methods for improving numerical analysis, visualization, and monitoring of CVDs. We highlight the ability to reduce the computational time of CFD simulations using deep learning methods to make them amenable for clinical applications. We describe methods for enhancing the resolution while reducing the noise and scan time of 4D-flow MRI using deep learning. We also present the ability for accurate detection and classification of CVDs based on data collected by wearable sensors. Finally, we provide some recommendations to facilitate further utilization of machine learning in our fight against CVDs.
Hemodynamics and CVD monitoring
Computational fluid dynamics (CFD)
CFD techniques generally leverage finite difference, finite element, or finite volume methods to solve the partial differential equations governing the balance of mass and momentum (also known as Navier–Stokes equations) in fluidic domains, as given below for incompressible flow of blood (Chung 2010; Versteeg and Malalasekera 2007):
in which is the velocity vector of blood through the blood vessels, and are the density and viscosity of blood, respectively, is time, is pressure, and is the gravitational acceleration.
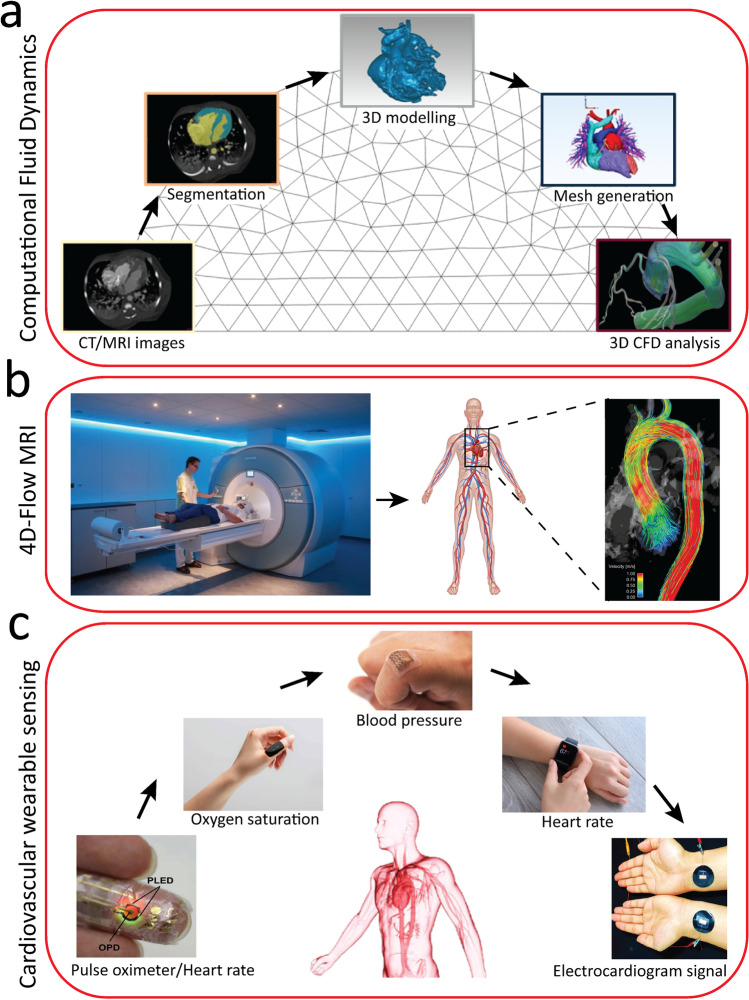
Solving these equations allows for predicting the distribution of velocity and pressure within the blood flow, which can be further processed to obtain the flow streamlines, shear stress, pressure drop, and or solute exchange across the vessels. The process involves creation of geometry, mesh generation, applying boundary conditions, setting thermophysical properties of the blood, and solving differential equations (Fig. 1a). Simulations can be performed in 2D/3D, highly complex/simplified geometries and under steady/transient, and laminar/turbulent conditions. As such, the geometry, transport properties, and boundary conditions of the CFD model can be customized to mimic tailored physiological or pathophysiological conditions (Baratchi et al. 2020; Duenas-Pamplona et al. 2021; Gijsen, et al. 1999; Kim, et al. 2013). In a more complicated setup, simulations can also be conducted in movable structures such as heart valves (Hoeijmakers et al. 2021; Weinberg and Kaazempur Mofrad 2008) as well as soft, deformable structures such as pulsating blood vessels (Chen et al. 2010). In such scenarios, fluidic equations should be coupled to the solid mechanics equations describing the structural dynamics. These so-called fluid–structure-interaction (FSI) models, on top of hemodynamic measures, can also predict the strain in cardiac tissue and vasculature as well as locomotion and constant deformation of red blood cells through the human circulatory system (Cetin and Sahin 2019; Liu and Liu 2006).
Fig. 1.
Some common methods for analyzing and monitoring of cardiovascular diseases: (a) computational fluid dynamics (CFD) pipeline (adapted from (Randles et al. 2017)), (b) 4D-flow magnetic resonance imaging (MRI) (adapted from (Allen et al. 2013)), and (c) cardiovascular wearable sensors (adapted from (Chen et al. 2021))
The rapid advancement of computer processing capabilities along with the availability of commercial CFD software such as Fluent (ANSYS Inc.), COMSOL (COMSOL Inc.), Simcenter STAR-CCM + (Siemens) or open-access platforms such as Open Foam (OpenFOAM Foundation Inc.), and Simvascular (a dedicated software for performing cardiovascular CFD simulations) (Updegrove et al. 2017) have facilitated the unprecedented progress of CFD simulations over the past 20 years. Nevertheless, there are some limitations which hinder the capability of CFD techniques in the cardiovascular domain. The generation of geometry and 3D elements might be challenging in complex structures. The simulations can be time-consuming, particularly for transient or FSI simulations, taking from a few hours to days. The computational time is proportional to the number of elements implemented in the model and the number of differential equations that need to be solved. Mesh-free CFD techniques aim to reduce the computational time (Monaghan 2005); however, the problems associated with the stability and accuracy of such techniques can significantly limit their utility in cardiovascular research. Patient-specific CFD simulations can be even more challenging due to the limitations in the reconstruction of the geometry and setting boundary conditions (Tse et al. 2011; Zhong et al. 2018). Additionally, the accuracy of CFD calculations strongly depends on the models used to formulate the complex hemodynamics and mechanobiology of the human vessels (e.g., models to predict the blood viscosity or deformation of red blood cells). The lack of experimental platforms for collecting enough data from the patients to create such models has limited the ability of existing CFD models. The emerging of microfluidic organ-on-chip models of the human cardiovascular system (Nguyen et al. 2021) can potentially address this limitation.
Blood flow imaging
Doppler echocardiography, a non-invasive modality using ultrasound, has been widely used for measuring the direction and velocity of blood flow through the heart chambers and large vessels (Anavekar and Oh 2009) as well as abnormal, regurgitant jets in pathologic heart valves (Nishimura et al. 1985). This technique measures the velocity of moving red blood cells based on the frequency shift between the transmitted and the reflected ultrasound wave. The frequency shift is proportional to the velocity of red blood cells according to Doppler equation (Nishimura et al. 1985):
in which is the frequency shift, is the frequency of the transmitted wave, is the average velocity of blood through the vessel, is the angle between the wave and the vessel axis, and is the velocity of sound in human tissues (~ 1,560 m/s). The measurements are made at arbitrary discrete points, and therefore, this technique has a limited spatiotemporal resolution.
In comparison, phase-contrast magnetic resonance imaging (PC-MRI) enables capturing and encoding of one-directional velocity vectors across imaging planes normal to the vessel. This technique measures the velocity of moving red blood cells based on the net phase shift when exposed to a pair of bipolar magnetic field gradients. The net phase shift is proportional to the velocity of red blood cells along the vessel, as expressed below (Wymer et al. 2020):
wherein is the phase shift, is the gyromagnetic ratio (defined as the ratio of magnetic moment to angular momentum), is the velocity of blood cells through the vessel, and is the change in the magnetic moment. A series of time-resolved images can be taken to capture the dynamics of blood flow during the cardiac cycle at each plane (Pelc et al. 1991).
Advances in PC-MRI have led to capturing and encoding of three-directional flow vectors across a 3D volume throughout the cardiac cycle. This technique, which is known as 4D (3D + time) flow MRI, enables 3D visualization and quantification of blood flow dynamics in the heart and vessels (Markl et al. 2012; Soulat et al. 2020) (Fig. 1b). 4D flow MRI, with its rising interest in clinical research, is used for evaluating various CVDs such as congenital heart disease, cardiac valvular disease, aortic stenosis and aneurysm, and pulmonary hypertension (Azarine et al. 2019).
Despite these advantages, low spatiotemporal resolution, bias noise, velocity aliasing, and eddy current-induced phase offset artifacts (Jiang et al. 2015) along with time-consuming image segmentation, post-processing, and analysis steps (Leiner et al. 2019) can adversely impact the accuracy and utility of 4D flow MRI for the visualization and quantification of blood flow.
Wearable sensors
Commercial wearable sensors such as smartwatches (Apple Inc.), necklaces (toSense, Inc.), and rings (Motiv Inc.) have enabled constant, non-invasive monitoring of useful cardiovascular signals such as heart rate, blood pressure, blood oxygen saturation level, and even blood glucose level without using routine clinical setups and out of clinics (Fig. 1c) (Bayoumy et al. 2021). These wearable sensors are extremely useful for early detection of common cardiovascular conditions such as heart failure, heart valve disease, arrhythmia, hypertension, anoxia, and hypoxia as well as comorbid conditions such as diabetes (Bayoumy et al. 2021). Advances in microfabrication technologies and materials sciences have also facilitated the development of textile-based (also known as e-textile) (Fan et al. 2020) and skin-like (also known as epidermal, e-skin or e-tattoo) (Li et al. 2020) wearable sensors, as comprehensively reviewed in (Khoshmanesh et al. 2021). These sensors leverage a variety of opto-electric, mechano-electric, bio-electric, ultrasonic, and bio-chemical mechanisms for monitoring of vital cardiovascular signals (Chen et al. 2021). This is facilitated by incorporation of miniaturized actuating and sensing elements into a smartwatch, cloths, or skin-like patches. Regardless of the sensing mechanism and the way that they are interfaced with the body, wearable sensors might be equipped with filters, amplifiers, and analog-to-digital converters to facilitate the smooth translation of the signals into meaningful patterns. Wireless transmission components such as Bluetooth modules might be required to transmit the collected signals to a nearby smartphone for further analysis, which might be challenging when dealing with large data.
Artificial intelligence and machine learning
The use of artificial intelligence (AI) has been steadily expanding in many fields including cardiovascular diseases, where the primary aim of such methods is to make automated decisions that can assist or completely replace human operators. This unprecedented progress in AI is mainly due to the rapid advancement of computational power, including graphical processing units and the development of various algorithms for effective training (Russell and Norvig. 2002). The applications in healthcare field cover many aspects including diagnosis, prognosis, and treatment (Graves 2013; Miller and Brown 2018).
Artificial intelligence has different subsets such as machine learning, which specifically aims to train the computer to learn and gain experience through the use of data without being programmed to solve each of the input combinations (Jordan and Mitchell 2015). Broadly, machine learning can be classified into two main categories: supervised and unsupervised machine learning (Tarca et al. 2007). Supervised machine learning relies on labeled training data as a targeted criterion for the algorithm, for example, a description of an object in an image or keywords in a document. It is extensively being used for data classification or diagnostic predictions. Also, supervised machine learning methods are effectively used in solving regression problems, where the output is a numeric value rather than a discrete class. For example, the enhancement of CFD spatiotemporal resolution falls under regression problems category. On the other hand, unsupervised machine learning aims to organize unlabeled data and find hidden patterns or similarities without the necessary prior knowledge. For example, unsupervised machine learning can figure out that patients’ symptoms are clustered into few groups without knowing the underlaying mechanism and processes of the disease. Inherently, the performance of unsupervised learning is much harder to judge, as there is no discernable goal and more susceptible to biases in training data (Jordan and Mitchell 2015; Sidey-Gibbons and Sidey-Gibbons 2019).
Neural networks are a subset of machine learning approaches with basic building blocks called artificial neurons. The fundamental idea of the artificial neuron is a mathematical operation that receives inputs from multiple other neurons, compiles them with different weights, and then fires an output when the input combination reaches a certain level. As such, the output of a simple artificial neuron can be mathematically expressed as follows:
where is the input to the neuron, is the associated weight, is a constant bias, is the total number of inputs, and is a non-linear activation function. The essence of training an artificial neural network (ANN) is to iteratively adjust the weights and biases such that the desired output is obtained. An example of a simple ANN is a single hidden layer feedforward network (fully connected), which consists of (i) an input layer (neurons that each accept a single input), (ii) a hidden layer where each neuron is connected to all input neurons and to all output layer neurons, and (iii) an output layer where each neuron is connected to all hidden layer neurons (Skaria et al. 2019). However, in more complicated ANN networks such as the convolutional neural network (CNN), a sequence of basic mathematical operations is performed at each layer.
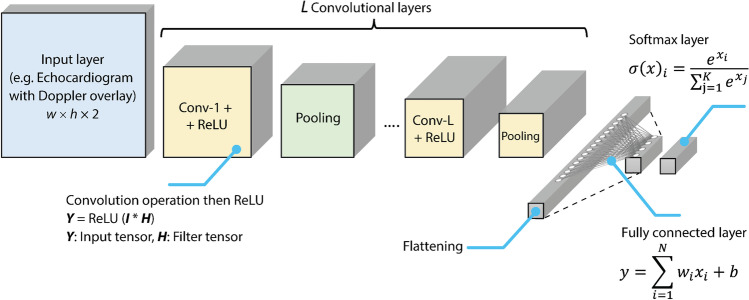
For example, if a CNN network has an input of 2D echocardiogram intensity images of width and a height along with a Doppler overlay having the same dimensions, accordingly, the overall input size in this example is (Fig. 2). A typical CNN network has multiple convolutional layers each composed of a convolution operation, a rectified linear unit (ReLU) operation, and a pooling operation. The output of these convolutional layers is then flatted (converted into a vector) and then passed through a fully connected neural network with a final layer having outputs equal to the number of echocardiogram image classes that needs to be classified. It is worth mentioning that the results from the output are typically normalized by a Softmax function which provides an exponential normalization to the arbitrarily scaled real-value outcomes of the neural network; the resulting values from the Softmax layer indicate the different probability of matching each of the classes. An overview of a typical CNN architecture along with the different mathematical operations involved in each layer is illustrated in Fig. 2.
Fig. 2.
A typical convolutional neural network architecture for classifying multi-dimensional images (e.g., echocardiogram with Doppler overlay) along with the main mathematical operations at each stage
To improve the performance of neural networks, many cascaded layers are added with various intermediary elements to simplify the training process. Such deeper architecture (referred as deep learning (Skaria et al. 2019)) can automate much of the feature extraction phase of machine learning without the need for the designer to explicitly select the prominent measures (features) that needs to be fed to the classifier. For example, deep learning can autonomously identify certain regions in an MRI image that are the main clue for detecting an abnormal function. The drawback of this method is the typical need for large amount of training data to identify the prominent features (LeCun et al. 2015).
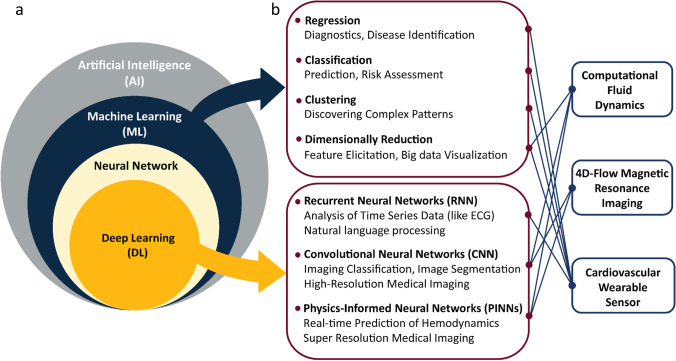
There are several common architectures of deep learning networks, including CNNs, recurrent neural networks (RNNs), and physics informed neural networks (PINNs) (Fig. 3). CNNs use convolutional filters to search for patterns at different abstraction levels, it also uses several polling layers to reduce the spatial dimensionality as patterns progress to the output. CNNs are commonly used for recognizing patterns in 2D and 3D spatial data, including image classification (Gu et al. 2018) and wearable sensors pattern recognition (Hendy et al. 2022; Skaria et al. 2020). RNNs can store, remember, and use the previous data and are commonly used for speech recognition and natural language processing (Graves 2013). PINNs incorporate physical laws described as differential equations into the learning process of the neural network and are extremely useful for solving multi-dimensional problems (Karniadakis et al. 2021; Raissi et al. 2019).
Fig. 3.
Application of artificial intelligence for analysis and monitoring of cardiovascular diseases: (a) relationship between artificial intelligence, machine learning, neural networks, and deep learning and (b) incorporation of machine learning and deep learning into CFD, 4D-flow MRI, and wearable sensors
Artificial intelligence is a powerful tool in the field of cardiovascular imaging and has been used in specialized and time-consuming tasks such as anomaly identification, image segmentation, and increasing image resolution. Moreover, machine learning algorithms can analyze the raw information of patients such as their heart rate, blood pressure, vascular diameter, age, and sex to deduce any hidden or complex patterns for diagnosis of CVDs or risk prediction of surgical procedures (Gupta et al. 2021; Kilic 2020; Podrat et al. 2021). Importantly, machine learning can be used for patient-specific risk stratification of future cardiovascular conditions (death, heart attack, and stroke), diagnosis, prognosis, and treatment of various CVDs (Al'Aref et al. 2019; Arzani et al. 2022; Dey et al. 2019; Krittanawong et al. 2017; Shameer et al. 2018). Machine learning can be incorporated into CFD, 4D-flow MRI, and wearable sensors to facilitate modeling, identification, prediction, and pattern recognition of cardiovascular conditions (Fig. 3).
Incorporation of machine learning for cardiovascular disease monitoring
Computational fluid dynamics (CFD)
A novel approach in cardiovascular medicine is the use of CFD models on a patient-specific basis for diagnostic evaluation (Mousavi et al. 2019; Tajeddini et al. 2020; Williams et al. 2022) and the design of cardiovascular devices such as ventricular assist device (Ghadimi, et al. 2019; Lin et al. 2019), artificial heart valve (Soltany Sadrabadi et al. 2021; Zakerzadeh et al. 2017), and extracorporeal membrane oxygenation (Nezami et al. 2021a, b; Nezami et al. 2021a, b). However, CFD models involving complex structures and dynamic conditions require high computational cost and time. Thus, conventional CFD models do not allow for real-time computations, which might be needed for surgical guidance (real-time analysis of the results during surgery). Therefore, it is essential to devise new strategies to accelerate CFD calculations while maintaining the model accuracy, and machine learning methods and especially deep learning are increasingly being leveraged to this end.
An elegant example is a work by Li et al. (Li et al. 2021), which incorporates deep learning in a CFD model for predicting the flow dynamics in the aorta and coronary artery branches. The training dataset was created by obtaining the geometric details of the aorta and coronary artery branches of 110 patients using computed tomography angiography (CTA) scanning. The dataset was further augmented to 1100 cases and CFD simulations were performed under identical boundary conditions. The simulation results were used to create a neural network model, which could predict with 90% accuracy the hemodynamic state of a new patient almost 600 times faster than a conventional CFD model. A similar approach has been used by Liang et al. (Liang et al. 2020) for modeling the flow dynamics in the thoracic aorta and Wang et al. (Wang et al. 2020) for calculation of myocardial fractional flow reserve.
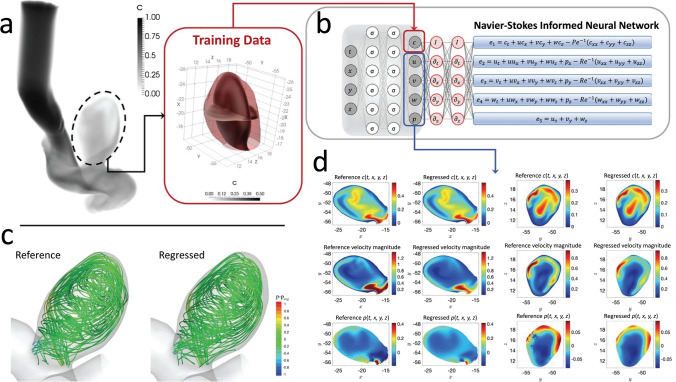
Though promising, the above approach requires a large number of training samples limiting its utility motivating the introduction of a new approach, coined as physics informed neural networks (PINNs), by Raisei et al. (Raissi et al. 2019, 2020) (Fig. 4). These networks are trained by synergic combination of data and mathematical equations, which govern various physical phenomena. In this regard, the incorporation of Navier–Stokes equations into the PINNs model allows for surrogate modeling of the flow field with minimized or total absence of simulated data (Sun et al. 2020), significantly reducing the number of samples to five while achieving an average accuracy of 92%. The PINNs model was later employed by Sun et al. (Sun et al. 2020) for modeling blood flow in stenosed vessel and Yin et al. (Yin et al. 2021) for obtaining of thrombus viscoelastic and permeability properties. The unique features of PINNs model for studying pathophysiological blood flow dynamics have been recently highlighted through calculation of wall shear stress in atherosclerotic vessels or aneurysm by simply measuring the velocity of a few points away from the vessel walls (Arzani et al. 2021). This strategy is particularly important for studying patient-specific blood flow dynamics (Vardhan and Randles 2021), in which access to the boundary conditions is limited due to clinical constraints and ethical concerns.
Fig. 4.
Application of physics informed neural networks (PINNs) for cardiovascular hemodynamics: (a) carotid artery with an aneurysm with the training dataset (pressure and velocity field) being generated by CFD, (b) incorporation of Navier–Stokes equations into the neural network, (c) comparison of flow streamlines obtained by CFD and PINNs, and (d) comparison of velocity and pressure contours obtained by CFD and PINNs (adapted from (Raissi et al. 2020))
Blood flow imaging
4D-flow MRI has been utilized for non-invasive imaging of blood flow over a large volume (Markl et al. 2012; Stankovic et al. 2014). This technique is subjected to multiple limitations, associated with spatiotemporal resolution, signal-to-noise ratio, phase offset errors, velocity aliasing, and velocity encoding (Cibis et al. 2015; Rispoli et al. 2015; Vali, et al. 2017), which further impacts the accuracy of calculating secondary hemodynamic parameters such as wall shear stress (WSS), oscillatory shear index (OSI), and relative residence time (RRT) (Boussel et al. 2009; Fathi et al. 2020). Importantly, long scan time is a major limitation of existing 4D flow MRI. Processing 4D-flow MRI outcome requires roughly half an hour of time, limiting its immediate clinical application, and thus neural networks can play an important role to make clinical use viable by providing predictive interpolation and extrapolation to the measurements.
For example, Hammernik et al. (Hammernik et al. 2018) have proposed a novel method for fast MRI reconstruction with high-quality using variational network, followed up by Vishnevskiy et al. (Vishnevskiy et al. 2020), which uses a deep neural network. The network can rapidly reconstruct and transform unsampled data into MRI data. A deep variational neural networks approach enabled researchers to use undersampled data with their network. Researchers used images from 11 healthy subjects to train their model initially and, by using an iterative image reconstruction architecture, were able to reconstruct 4D-flow MRI data in less than one minute. In a similar study, Gong et al. (Gong et al. 2019) used a deep learning method to reconstruct tomographic images which allowed for imaging within the reconstruction providing high resolution images to easily detect diseases such as Alzheimer's.
Alternatively, Ferdian et al. (Ferdian et al. 2020) leveraged CFD simulations to generate synthetic 4D-flow MRI data in thoracic aorta. These synthetic images were further downsampled and injected with white Gaussian noise for using them as the test dataset. A deep learning network, solely trained by such synthetic images, was then able to denoise the synthetic low-resolution dataset to produce low noise 4D flow MRI output images with higher resolution than the input ones. A similar strategy was implemented by Rutkowski et al. (Rutkowski et al. 2021) for denoising velocity contours in the cerebral blood vessels.
To eliminate the need for CFD analysis, Fathi et al. (Fathi et al. 2020) utilized a PINNs model to enhance the resolution and denoise 4D flow MRI results (Fig. 5a). Complex Cartesian images containing the magnitude and direction of velocity vectors are generated using low-resolution MRI data, following which regularization terms are defined to enforce flow physics (Navier–Stokes and continuity equations) within the flow domain. A loss function was introduced to produce high resolution and noise-free images, and the developed deep neural network increased the algorithm’s efficiency. The model yielded a 100-fold improvement in spatial resolution, a fivefold increase in temporal resolution, and a 50% reduction in velocity normalized root mean square error and ultimately were verified against particle image velocimetry results (Fig. 5b and c).
Fig. 5.
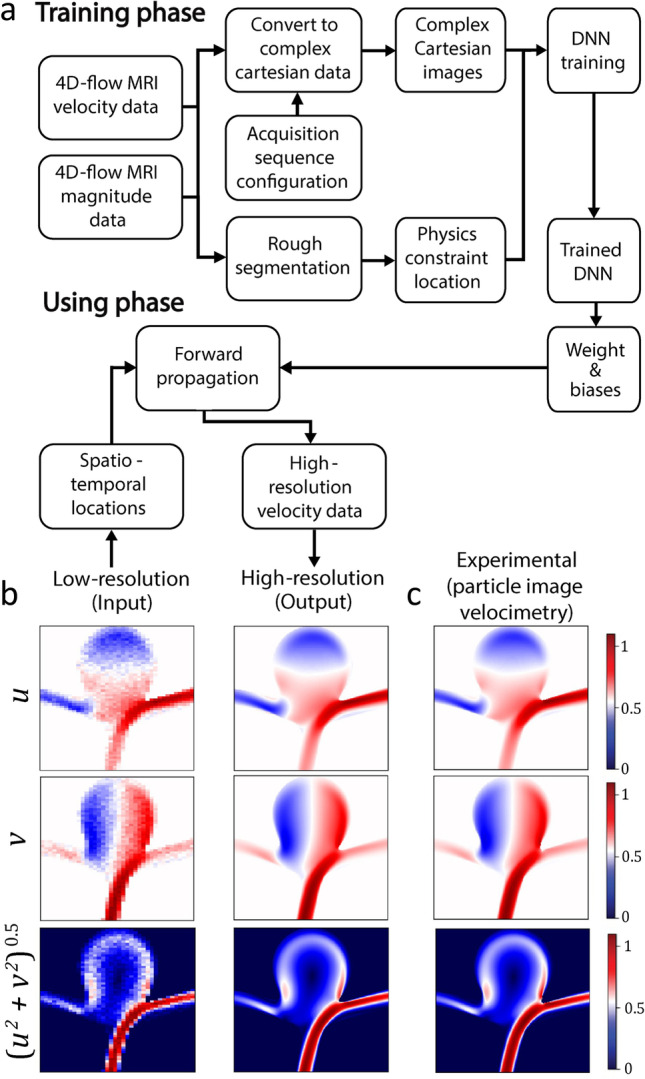
Application of physics-informed neural networks (PINNs) for enhancing the resolution of 4D-flow MRI results: (a) flow chart of the algorithm composed of training and using phases, (b) representative images showing the conversion of low-resolution velocity contours obtained by 4D-flow MRI to high-resolution images using PINNs, and (c) experimental results verified against particle image velocimetry (adapted from (Fathi et al. 2020))
Wearable sensors
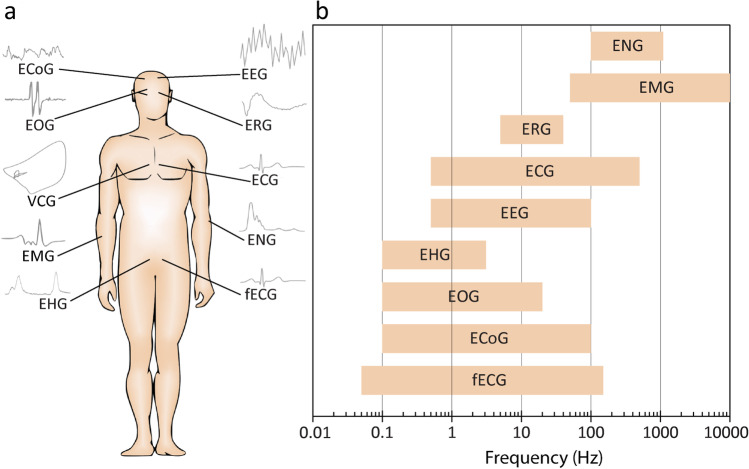
Wearable technologies have facilitated continuous monitoring and collection of cardiovascular bio-signals (Chen et al. 2021). However, analysis and classification of the collected data using standard statistical methods remain challenging (Ghosh et al. 2020). For example, cardiac disorders such as atrial fibrillation, atrial flutter, ventricular tachycardia, ventricular fibrillation, and myocardial infarction all lead to irregular heartbeats (arrhythmia) and abnormal electrocardiogram signals (Bayoumy et al. 2021; Krittanawong et al. 2021). Although an experienced cardiologist can distinguish the above cardiac disorders, the automated classification of these cardiac diseases, especially in the presence of noise, is challenging and prone to inaccuracy using statistical methods (Austin et al. 2013; Luo et al. 2016; Park et al. 2018). This can be even more challenging when dealing with multiple bio-signals such as heart rate, blood pressure, skin temperature, and electrocardiogram signals (Ge et al. 2021; Martin et al. 2021; Tadesse et al. 2021) or mental stress (Patlar Akbulut et al. 2020). Additionally, the variety and complexity of bio-signals (Fig. 6) along with the diverse frequency and amplitude range thereof (Table 1) make it challenging for conventional analysis methods to discover any meaningful patterns within these signals, which correspond to specific medical conditions (Martinek et al. 2021).
Fig. 6.
Human bio-signals: (a) schematics showing the diversity and complexity of bio-signals and (b) the frequency range of various bio-signals (adapted from (Martinek et al. 2021))
Table 1.
A brief overview of bio-signals along with their frequency and amplitude range
| Bio-signal | Descriptions | Frequency (Hz) | Voltage amplitude (V) |
|---|---|---|---|
| Electroencephalogram (EEG) (Jurcak et al. 2007) | Electrical signal/activity of the brain | 0.5–100 | 0.005–10 |
| Electrocardiogram (ECG) (Clifford, et al. 2006) | Electrical signal/activity of the heart | 0.05–250 | 0.01–5 |
| Fetal electrocardiogram (fECG) (Sameni and Clifford 2010) | Electrical signal/activity of the fetal heart | 0.05–150 | 0.01–0.02 |
| Vectorcardiogram (VCG) (Vozda and Cerny 2015) | Spatial and temporal cardiac electrical activities | - | - |
| Electromyogram (EMG) (Reaz et al. 2006) | Electrical signal/activity in muscles in response to neural stimulation | 50–10,000 | 0.05–0.3 |
| Electrooculogram (EOG) (López et al. 2020) | Electrical signal corresponding to the movement of the eye | 0.1–20 | 0.05–3.5 |
| Electroretinogram (ERG) (Heckenlively and Arden 2006) | Electrical signal/activity of the retina in response to light stimulation | 5–35 | 0.005–1 |
| Electrohysterogram (EHG) (Rabotti et al. 2008) | Uterine electrical activity | 0.1–3 | 0.1–1 |
| Electrocorticogram (ECoG) (Nakasatp et al. 1994) | Electrical signal/activity of the cerebral cortex | 0.1–100 | 0.005–10 |
| Electroneurogram (ENG) (Cogan 2008) | Electrical signal/activity of axons in peripheral nerves | 100–1000 | 0.005–10 |
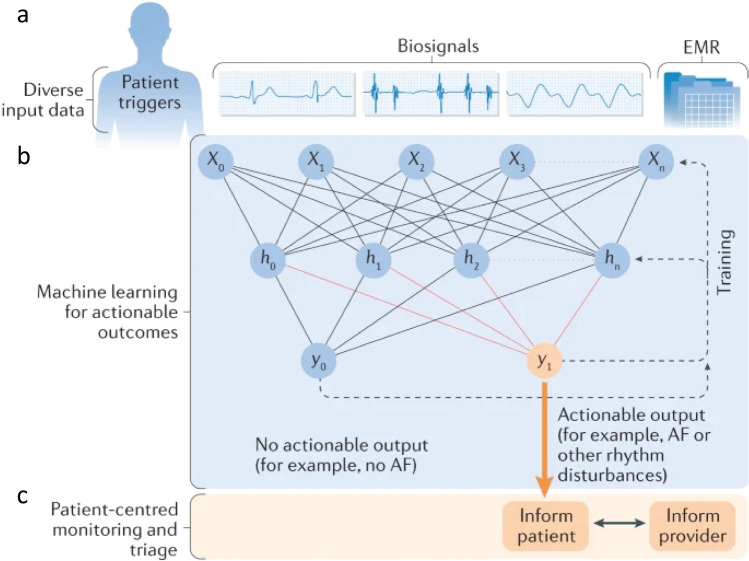
To address this limitation, the cardiovascular bio-signals collected by wearable sensors could be wirelessly transferred to a more powerful analytical platform to be processed using various machine learning algorithms. Any meaningful patterns in the data, which might correspond to the diagnosis, risk prediction or prognosis of cardiovascular diseases, can be shared with the patient or their healthcare provider (Fig. 7). The data collected from thousands of individuals can also be collated to make data-driven decisions (Krittanawong et al. 2021; Quer et al. 2021).
Fig. 7.
Application of machine learning for the analysis of cardiovascular bio-signals collected by wearable sensors: (a) in situ collection of patient’s bio-signals and transfer to a data center, (b) analysis using deep learning algorithms to predict and diagnose potential cardiovascular diseases, and (c) communication of the results with the patients and/or healthcare providers (adapted from (Krittanawong et al. 2021))
Recent published literature has demonstrated the viability of accurate detection and classification of cardiovascular diseases using commercial wearable technologies. For instance, Tison et al. (Tison et al. 2018) developed a deep neural network model to detect atrial fibrillation (irregular heart rhythm) based on photoplethysmography signals obtained by wrist-worn smartwatches (Apple Inc.). Comparing the results against standard 12-lead electrocardiography revealed a 98% sensitivity and a 90% specificity for a population of 51 patients. Likewise, Green et al. (Green et al. 2019) developed a machine learning based classifier for detection of hypertrophic cardiomyopathy (a genetic disorder characterized by thickening of the cardiac muscle) based on photoplethysmography signals obtained by a wrist-worn sensor (Wavelet Health). More recently, Stehlik et al. (Stehlik et al. 2020) developed a similarity-based machine learning model to predict the risk of heart failure based on heart rate, electrocardiography waveforms, and temperature obtained by a commercial chest-worn sensor (Vital Connect). Analysis of results in 100 patients aged between 58 and 78 over a three-month period enabled the prediction of the risk of imminent heart failure hospitalization with an 88% sensitivity and 85% specificity.
Conclusions and future steps
In this paper, we briefly discussed the incorporation of machine learning in hemodynamic analysis and monitoring, showcasing its application in studies with computational fluid dynamics, blood flow imaging, and wearable sensors for enhanced diagnosis, prognosis, and therapy. Highlighting the potential of machine learning for reducing computational cost and time, we presented its promise in improving the resolution and quality of images while keeping long scan time at bay. We as well depicted the promising application of machine learning for early detection and risk stratification of cardiovascular diseases to motivate the promise that machine learning with its accelerated rate of development can significantly improve cardiovascular monitoring, care, and management.
We ultimately outline the following suggestions to facilitate more effective translation of machine learning models for cardiovascular research:
Further interaction: given the multidisciplinary nature of this field, a closer collaboration between the stakeholders, including experts in the areas of computational fluid dynamics, blood flow imaging, cardiology, wearable sensors, and machine learning experts, is required to close the remaining gaps. This can be facilitated by organizing special issues in cardiovascular journals, organizing dedicated conferences, symposiums, and meetings as well as dedicating research grants and designing post-graduate courses to maximize the interaction between engineers, computer scientists, cardiologists, clinicians, and vascular biologists.
User-friendly platforms: machine learning algorithms are advancing rapidly. However, the lack of user-friendly platforms makes it challenging for users with limited knowledge and expertise in computer and data sciences to employ such algorithms. Development of user-friendly machine learning platforms with comprehensive libraries facilitates the adoption of such models by the experts working in the areas of computational fluid dynamics, blood flow imaging, and wearable sensors. A successful role model in this regard is the open-source ImageJ software, supported by the National Institutes of Health funding, which provides a user-friendly platform for processing of microscopic images to calculate cells’ density, morphology, and viability by cellular biologists who are not necessarily expert in image processing. We believe that the adoption of similar machine learning platforms can significantly improve the diagnosis, prognosis, and risk prediction of CVDs.
Dedicated physic-informed neural networks models for cardiovascular diseases: the PINN models have proven their capability for solving complex flow fields. We envisage that the development of dedicated hemodynamics informed neural network models is necessary for better understanding of the complex hemodynamics of human vessels under various physiological and pathophysiological conditions. Such models can be further improved by incorporating fluid–structure-interaction models to predict the deformation of blood vessels under pulsatile flows, movement of blood cells inside the vessels, as well as the complex dynamics of blood cells at the vicinity of heart valves.
Author contribution
H.M. wrote the manuscript and prepared the figures. A.H., G.C., F.K., F.R.N., and S.N. wrote and reviewed the manuscript. S.B. and K.K. conceptualized and led the work, wrote, and reviewed the manuscript.
Funding
S.N., S.B., and K.K. acknowledge the Australian Research Council (ARC) for the Linkage grant (LP190100728).
Declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Al'Aref SJ, et al. Clinical applications of machine learning in cardiovascular disease and its relevance to cardiac imaging. Eur Heart J. 2019;40:1975–1986. doi: 10.1093/eurheartj/ehy404. [DOI] [PubMed] [Google Scholar]
- Allen BD, Barker AJ, Kansal P, Collins JD, Carr JC, Malaisrie SC, Markl M. Impact of aneurysm repair on thoracic aorta hemodynamics. Circulation. 2013;128:e341–e343. doi: 10.1161/CIRCULATIONAHA.112.000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anavekar NS, Oh JK. Doppler echocardiography: a contemporary review. J Cardiol. 2009;54:347–358. doi: 10.1016/j.jjcc.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Arzani A, Wang J-X, D'Souza RM. Uncovering near-wall blood flow from sparse data with physics-informed neural networks. Phys Fluids. 2021;33:071905. doi: 10.1063/5.0055600. [DOI] [Google Scholar]
- Arzani A, Wang J-X, Sacks MS, Shadden SC. Machine learning for cardiovascular biomechanics modeling: challenges and beyond. Ann Biomed Eng. 2022;50:615–627. doi: 10.1007/s10439-022-02967-4. [DOI] [PubMed] [Google Scholar]
- ATmega32-avr. https://atmega32-avr.com/. Accessed Oct 2021
- Austin PC, Tu JV, Ho JE, Levy D, Lee DS. Using methods from the data-mining and machine-learning literature for disease classification and prediction: a case study examining classification of heart failure subtypes. J Clin Epidemiol. 2013;66:398–407. doi: 10.1016/j.jclinepi.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azarine A, Garcon P, Stansal A, Canepa N, Angelopoulos G, Silvera S, Sidi D, Marteau V, Zins M. Four-dimensional flow MRI: principles and cardiovascular applications. Radiographics. 2019;39:632–648. doi: 10.1148/rg.2019180091. [DOI] [PubMed] [Google Scholar]
- Baratchi S, et al. Transcatheter aortic valve implantation represents an anti-inflammatory therapy via reduction of shear stress–induced, Piezo-1–mediated monocyte activation. Circulation. 2020;142:1092–1105. doi: 10.1161/CIRCULATIONAHA.120.045536. [DOI] [PubMed] [Google Scholar]
- Bayoumy K, et al. Smart wearable devices in cardiovascular care: where we are and how to move forward. Nat Rev Cardiol. 2021;18:581–599. doi: 10.1038/s41569-021-00522-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazilevs Y, Hsu MC, Benson DJ, Sankaran S, Marsden AL. Computational fluid–structure interaction: methods and application to a total cavopulmonary connection. Comput Mech. 2009;45:77–89. doi: 10.1007/s00466-009-0419-y. [DOI] [Google Scholar]
- Boussel L, Rayz V, Martin A, Acevedo-Bolton G, Lawton MT, Higashida R, Smith WS, Young WL, Saloner D. Phase-contrast magnetic resonance imaging measurements in intracranial aneurysms in vivo of flow patterns, velocity fields, and wall shear stress: comparison with computational fluid dynamics. Magn Reson Med. 2009;61:409–417. doi: 10.1002/mrm.21861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celermajer DS, Chow CK, Marijon E, Anstey NM, Woo KS. Cardiovascular disease in the developing world: prevalences, patterns, and the potential of early disease detection. J Am Coll Cardiol. 2012;60:1207–1216. doi: 10.1016/j.jacc.2012.03.074. [DOI] [PubMed] [Google Scholar]
- Cetin A, Sahin M. A monolithic fluid-structure interaction framework applied to red blood cells. Int J Numer Method Biomed Eng. 2019;35:e3171. doi: 10.1002/cnm.3171. [DOI] [PubMed] [Google Scholar]
- Chen HY, Zhu L, Huo Y, Liu Y, Kassab GS. Fluid–structure interaction (FSI) modeling in the cardiovascular system. In: Guccione JM, Kassab GS, Ratcliffe MB, editors. Computational cardiovascular mechanics: modeling and applications in heart failure. Boston, MA: Springer US; 2010. pp. 141–157. [Google Scholar]
- Chen S, Qi J, Fan S, Qiao Z, Yeo JC, Lim CT. Flexible wearable sensors for cardiovascular health monitoring. Adv Healthc Mater. 2021;10:e2100116. doi: 10.1002/adhm.202100116. [DOI] [PubMed] [Google Scholar]
- Chollet F. Manning Publications Co. (in English) 2018. Deep learning with Python. [Google Scholar]
- Chung TJ. Computational fluid dynamics. Cambridge University Press; 2010. [Google Scholar]
- Cibis M, Jarvis K, Markl M, Rose M, Rigsby C, Barker AJ, Wentzel JJ. The effect of resolution on viscous dissipation measured with 4D flow MRI in patients with Fontan circulation: evaluation using computational fluid dynamics. J Biomech. 2015;48:2984–2989. doi: 10.1016/j.jbiomech.2015.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford, G. D., Azuaje, F., and McSharry, P., Advanced methods and tools for ECG data analysis. Artech house Boston, 2006. N/A
- Cogan SF. Neural stimulation and recording electrodes. Annu Rev Biomed Eng. 2008;10:275–309. doi: 10.1146/annurev.bioeng.10.061807.160518. [DOI] [PubMed] [Google Scholar]
- Dey D, Slomka PJ, Leeson P, Comaniciu D, Shrestha S, Sengupta PP, Marwick TH. Artificial intelligence in cardiovascular imaging: JACC state-of-the-art review. J Am Coll Cardiol. 2019;73:1317–1335. doi: 10.1016/j.jacc.2018.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duenas-Pamplona J, Garcia JG, Sierra-Pallares J, Ferrera C, Agujetas R, Lopez-Minguez JR. A comprehensive comparison of various patient-specific CFD models of the left atrium for atrial fibrillation patients. Comput Biol Med. 2021;133:104423. doi: 10.1016/j.compbiomed.2021.104423. [DOI] [PubMed] [Google Scholar]
- Elkworks. https://elkeworks.com/product/_1500946.html. Accessed Oct 2021)
- Fan W, He Q, Meng K, Tan X, Zhou Z, Zhang G, Yang J, Wang ZL. Machine-knitted washable sensor array textile for precise epidermal physiological signal monitoring. Sci Adv. 2020;6:eaay2840. doi: 10.1126/sciadv.aay2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathi MF, Perez-Raya I, Baghaie A, Berg P, Janiga G, Arzani A, D'Souza RM. Super-resolution and denoising of 4D-flow MRI using physics-informed deep neural nets. Comput Methods Prog Biomed. 2020;197:105729. doi: 10.1016/j.cmpb.2020.105729. [DOI] [PubMed] [Google Scholar]
- Ferdian E, Suinesiaputra A, Dubowitz DJ, Zhao D, Wang A, Cowan B, Young AA (2020) 4DFlowNet: super-resolution 4D flow MRI using deep learning and computational fluid dynamics. Front Phys 8. 10.3389/fphy.2020.00138
- Ge R, Shen T, Zhou Y, Liu C, Zhang L, Yang B, Yan Y, Coatrieux JL, Chen Y (2021) Convolutional squeeze-and-excitation network for ECG arrhythmia detection. Artif Intell Med 121. 10.1016/j.artmed.2021.102181 [DOI] [PubMed]
- Ghadimi B, Nejat A, Nourbakhsh SA, Naderi N. Shape optimization of a centrifugal blood pump by coupling CFD with metamodel-assisted genetic algorithm. J Artif Organs. 2019;22:29–36. doi: 10.1007/s10047-018-1072-z. [DOI] [PubMed] [Google Scholar]
- Ghosh SK, Tripathy RK, Paternina MRA, Arrieta JJ, Zamora-Mendez A, Naik GR. Detection of atrial fibrillation from single Lead ECG signal using multirate cosine filter Bank and deep neural network. J Med Syst. 2020;44:114. doi: 10.1007/s10916-020-01565-y. [DOI] [PubMed] [Google Scholar]
- Gijsen FJH, van de Vosse FN, Janssen JD. The influence of the non-Newtonian properties of blood on the flow in large arteries: steady flow in a carotid bifurcation model. J Biomech. 1999;32:601–608. doi: 10.1016/s0021-9290(99)00015-9. [DOI] [PubMed] [Google Scholar]
- Gong K, Catana C, Qi J, Li Q. PET image reconstruction using deep image prior. IEEE Trans Med Imaging. 2019;38:1655–1665. doi: 10.1109/TMI.2018.2888491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves A (2013) Generating sequences with recurrent neural networks. arXiv:1308.0850v5. 10.48550/arXiv.1308.0850
- Green EM, van Mourik R, Wolfus C, Heitner SB, Dur O, Semigran MJ. Machine learning detection of obstructive hypertrophic cardiomyopathy using a wearable biosensor. NPJ Digit Med. 2019;2:57. doi: 10.1038/s41746-019-0130-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, et al. Recent advances in convolutional neural networks. Pattern Recogn. 2018;77:354–377. doi: 10.1016/j.patcog.2017.10.013. [DOI] [Google Scholar]
- Gupta T, Joseph DT, Goel SS, Kleiman NS. Predicting and measuring mortality risk after transcatheter aortic valve replacement. Expert Rev Cardiovasc Ther. 2021;19:247–260. doi: 10.1080/14779072.2021.1888715. [DOI] [PubMed] [Google Scholar]
- Hammernik K, Klatzer T, Kobler E, Recht MP, Sodickson DK, Pock T, Knoll F. Learning a variational network for reconstruction of accelerated MRI data. Magn Reson Med. 2018;79:3055–3071. doi: 10.1002/mrm.26977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckenlively JR, Arden GB (2006) Principles and practice of clinical electrophysiology of vision. MIT Press. 10.7551/mitpress/5557.001.0001
- Hendy N, Fayek HM, Al-Hourani A (2022) Deep learning approaches for air-writing using single UWB radar. IEEE Sensors J:1. 10.1109/JSEN.2022.3172727
- Hoeijmakers M, Huberts W, Rutten MCM, van de Vosse FN. The impact of shape uncertainty on aortic-valve pressure-drop computations. Int J Numer Method Biomed Eng. 2021;37:e3518. doi: 10.1002/cnm.3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter J (2016) Cardiovascular diseases: pathophysiology, diagnosis and treatment, 1st edn, Foster Academics N/A
- Jiang J, Kokeny P, Ying W, Magnano C, Zivadinov R, Mark Haacke E. Quantifying errors in flow measurement using phase contrast magnetic resonance imaging: comparison of several boundary detection methods. Magn Reson Imaging. 2015;33:185–193. doi: 10.1016/j.mri.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan MI, Mitchell TM. Machine learning: trends, perspectives, and prospects. Science. 2015;349:255–260. doi: 10.1126/science.aaa8415. [DOI] [PubMed] [Google Scholar]
- Jurcak V, Tsuzuki D, Dan I. 10/20, 10/10, and 10/5 systems revisited: their validity as relative head-surface-based positioning systems. Neuroimage. 2007;34:1600–1611. doi: 10.1016/j.neuroimage.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Karniadakis GE, Kevrekidis IG, Lu L, Perdikaris P, Wang S, Yang L. Physics-informed machine learning. Nat Rev Phys. 2021;3:422–440. doi: 10.1038/s42254-021-00314-5. [DOI] [Google Scholar]
- Khoshmanesh F, Thurgood P, Pirogova E, Nahavandi S, Baratchi S. Wearable sensors: at the frontier of personalised health monitoring, smart prosthetics and assistive technologies. Biosens Bioelectron. 2021;176:112946. doi: 10.1016/j.bios.2020.112946. [DOI] [PubMed] [Google Scholar]
- Kilic A. Artificial intelligence and machine learning in cardiovascular health care. Ann Thorac Surg. 2020;109:1323–1329. doi: 10.1016/j.athoracsur.2019.09.042. [DOI] [PubMed] [Google Scholar]
- Kim SK, Na Y, Kim JI, Chung SK. Patient specific CFD models of nasal airflow: overview of methods and challenges. J Biomech. 2013;46:299–306. doi: 10.1016/j.jbiomech.2012.11.022. [DOI] [PubMed] [Google Scholar]
- Krittanawong C, Rogers AJ, Johnson KW, Wang Z, Turakhia MP, Halperin JL, Narayan SM. Integration of novel monitoring devices with machine learning technology for scalable cardiovascular management. Nat Rev Cardiol. 2021;18:75–91. doi: 10.1038/s41569-020-00445-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krittanawong C, Zhang H, Wang Z, Aydar M, Kitai T. Artificial Intelligence in Precision Cardiovascular Medicine. J Am Coll Cardiol. 2017;69:2657–2664. doi: 10.1016/j.jacc.2017.03.571. [DOI] [PubMed] [Google Scholar]
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Leiner T, Rueckert D, Suinesiaputra A, Baessler B, Nezafat R, Isgum I, Young AA. Machine learning in cardiovascular magnetic resonance: basic concepts and applications. J Cardiovasc Magn Reson. 2019;21:61. doi: 10.1186/s12968-019-0575-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Wang H, Zhang M, Tupin S, Qiao A, Liu Y, Ohta M, Anzai H. Prediction of 3D cardiovascular hemodynamics before and after coronary artery bypass surgery via deep learning. Commun Biol. 2021;4:99. doi: 10.1038/s42003-020-01638-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. Wearable skin-like optoelectronic systems with suppression of motion artifacts for cuff-less continuous blood pressure monitor. Natl Sci Rev. 2020;7:849–862. doi: 10.1093/nsr/nwaa022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Mao W, Sun W. A feasibility study of deep learning for predicting hemodynamics of human thoracic aorta. J Biomech. 2020;99:109544. doi: 10.1016/j.jbiomech.2019.109544. [DOI] [PubMed] [Google Scholar]
- Lin WCP, Doyle MG, Roche SL, Honjo O, Forbes TL, Amon CH. Computational fluid dynamic simulations of a cavopulmonary assist device for failing Fontan circulation. J Thorac Cardiovasc Surg. 2019;158:1424–1433 e5. doi: 10.1016/j.jtcvs.2019.03.008. [DOI] [PubMed] [Google Scholar]
- Liu Y, Liu WK. Rheology of red blood cell aggregation by computer simulation. J Comput Phys. 2006;220:139–154. doi: 10.1016/j.jcp.2006.05.010. [DOI] [Google Scholar]
- López A, Ferrero F, Villar JR, Postolache O. High-performance analog front-end (AFE) for EOG systems. Electronics. 2020;9:970. doi: 10.3390/electronics9060970. [DOI] [Google Scholar]
- Luo J, Wu M, Gopukumar D, Zhao Y. Big data application in biomedical research and health care: a literature review. Biomed Inform Insights. 2016;8:1–10. doi: 10.4137/BII.S31559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markl M, Frydrychowicz A, Kozerke S, Hope M, Wieben O. 4D flow MRI. J Magn Reson Imaging. 2012;36:1015–1036. doi: 10.1002/jmri.23632. [DOI] [PubMed] [Google Scholar]
- Martin H, Morar U, Izquierdo W, Cabrerizo M, Cabrera A, Adjouadi M (2021) Real-time frequency-independent single-Lead and single-beat myocardial infarction detection. Artif Intell Med 121. 10.1016/j.artmed.2021.102179 [DOI] [PubMed]
- Martinek R, Ladrova M, Sidikova M, Jaros R, Behbehani K, Kahankova R, Kawala-Sterniuk A. Advanced bioelectrical signal processing methods: past, present and future approach—part I: cardiac signals. Sensors. 2021;21:5186. doi: 10.3390/s21155186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DD, Brown EW. Artificial intelligence in medical practice: the question to the answer? Am J Med. 2018;131:129–133. doi: 10.1016/j.amjmed.2017.10.035. [DOI] [PubMed] [Google Scholar]
- Monaghan JJ. Smoothed particle hydrodynamics. Rep Prog Phys. 2005;68:1703–1759. doi: 10.1088/0034-4885/68/8/r01. [DOI] [Google Scholar]
- Mousavi SJ, Farzaneh S, Avril S. Patient-specific predictions of aneurysm growth and remodeling in the ascending thoracic aorta using the homogenized constrained mixture model. Biomech Model Mechanobiol. 2019;18:1895–1913. doi: 10.1007/s10237-019-01184-8. [DOI] [PubMed] [Google Scholar]
- Nabel EG. Cardiovascular disease. N Engl J Med. 2003;349:60–72. doi: 10.1056/NEJMra035098. [DOI] [PubMed] [Google Scholar]
- Nakasatp N, Levesque MF, Barth DS, Baumgartner C, Rogers RL, Sutherling WW. Comparisons of MEG, EEG, and ECoG source localization in neocortical partial epilepsy in humans. Electroencephalogr Clin Neurophysiol. 1994;91:171–178. doi: 10.1016/0013-4694(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Nezami FR, Khodaee F, Edelman ER, Keller SP. A computational fluid dynamics study of the extracorporeal membrane oxygenation-failing heart circulation. ASAIO J. 2021;67:276–283. doi: 10.1097/MAT.0000000000001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezami FR, Ramezanpour M, Khodaee F, Goffer E, Edelman ER, Keller SP (2021b) Simulation of fluid-structure interaction in extracorporeal membrane oxygenation circulatory support systems. J Cardiovasc Transl Res. 10.1007/s12265-021-10143-7 [DOI] [PubMed]
- Nguyen N, Thurgood P, Sekar NC, Chen S, Pirogova E, Peter K, Baratchi S, Khoshmanesh K (2021) Microfluidic models of the human circulatory system: versatile platforms for exploring mechanobiology and disease modeling. Biophys Rev. 10.1007/s12551-021-00815-8 [DOI] [PMC free article] [PubMed]
- Nishimura RA, Miller FA, Callahan MJ, Benassi RC, Seward JB, Tajik AJ. Doppler echocardiography: theory, instrumentation, technique, and application. Mayo Clin Proc. 1985;60:321–343. doi: 10.1016/s0025-6196(12)60540-0. [DOI] [PubMed] [Google Scholar]
- Park C, Took CC, Seong JK. Machine learning in biomedical engineering. Biomed Eng Lett. 2018;8:1–3. doi: 10.1007/s13534-018-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patlar Akbulut F, Ikitimur B, Akan A (2020) Wearable sensor-based evaluation of psychosocial stress in patients with metabolic syndrome. Artif Intell Med 104. 10.1016/j.artmed.2020.101824 [DOI] [PubMed]
- Pelc NJ, Herfkens RJ, Shimakawa A, Enzmann DR. Phase contrast cine magnetic resonance imaging. Magn Reson Q. 1991;7:229–254. [PubMed] [Google Scholar]
- Podrat JL, Del Val FR, Pei KY. Evolution of risk calculators and the Dawn of artificial intelligence in predicting patient complications. Surg Clin N Am. 2021;101:97–107. doi: 10.1016/j.suc.2020.08.012. [DOI] [PubMed] [Google Scholar]
- Quer G, Arnaout R, Henne M, Arnaout R. Machine learning and the future of cardiovascular care. J Am Coll Cardiol. 2021;77:300–313. doi: 10.1016/j.jacc.2020.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabotti C, Mischi M, van Laar JO, Oei GS, Bergmans JW. Estimation of internal uterine pressure by joint amplitude and frequency analysis of electrohysterographic signals. Physiol Meas. 2008;29:829. doi: 10.1088/0967-3334/29/7/011. [DOI] [PubMed] [Google Scholar]
- Raissi M, Perdikaris P, Karniadakis GE. Physics-informed neural networks: a deep learning framework for solving forward and inverse problems involving nonlinear partial differential equations. J Comput Phys. 2019;378:686–707. doi: 10.1016/j.jcp.2018.10.045. [DOI] [Google Scholar]
- Raissi M, Yazdani A, Karniadakis GE. Hidden fluid mechanics: learning velocity and pressure fields from flow visualizations. Science. 2020;367:1026–1030. doi: 10.1126/science.aaw4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randles A, Frakes DH, Leopold JA. Computational fluid dynamics and additive manufacturing to diagnose and treat cardiovascular disease. Trends Biotechnol. 2017;35:1049–1061. doi: 10.1016/j.tibtech.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaz MBI, Hussain MS, Mohd-Yasin F. Techniques of EMG signal analysis: detection, processing, classification and applications. Biol Proced Online. 2006;8:11–35. doi: 10.1251/bpo115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikhtegar F, Knight JA, Olgac U, Saur SC, Poulikakos D, Marshall W, Jr, Cattin PC, Alkadhi H, Kurtcuoglu V. Choosing the optimal wall shear parameter for the prediction of plaque location-a patient-specific computational study in human left coronary arteries. Atherosclerosis. 2012;221:432–437. doi: 10.1016/j.atherosclerosis.2012.01.018. [DOI] [PubMed] [Google Scholar]
- Rispoli VC, Nielsen JF, Nayak KS, Carvalho JL. Computational fluid dynamics simulations of blood flow regularized by 3D phase contrast MRI. Biomed Eng Online. 2015;14:110. doi: 10.1186/s12938-015-0104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth GA, et al. Global burden of cardiovascular diseases and risk factors, 1990–2019. J Am Coll Cardiol. 2020;76:2982–3021. doi: 10.1016/j.jacc.2020.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell S, Norvig P. Artificial intelligence: a modern approach. Prentice Hall; 2002. [Google Scholar]
- Rutkowski DR, Roldan-Alzate A, Johnson KM. Enhancement of cerebrovascular 4D flow MRI velocity fields using machine learning and computational fluid dynamics simulation data. Sci Rep. 2021;11:10240. doi: 10.1038/s41598-021-89636-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sameni R, Clifford GD. A review of fetal ECG signal processing; issues and promising directions. The Open Pacing, Electrophysiology & Therapy Journal. 2010;3:4. doi: 10.2174/1876536X01003010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel AL. Some studies in machine learning using the game of checkers. IBM J Res Dev. 1959;3:210–229. doi: 10.1147/rd.33.0210. [DOI] [Google Scholar]
- Shameer K, Johnson KW, Glicksberg BS, Dudley JT, Sengupta PP. Machine learning in cardiovascular medicine: are we there yet? Heart. 2018;104:1156–1164. doi: 10.1136/heartjnl-2017-311198. [DOI] [PubMed] [Google Scholar]
- Sidey-Gibbons JAM, Sidey-Gibbons CJ. Machine learning in medicine: a practical introduction. BMC Med Res Methodol. 2019;19:64. doi: 10.1186/s12874-019-0681-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simvascular software. https://simvascular.github.io/. Accessed Oct 2021
- Skaria S, Al-Hourani A, Evans RJ. Deep-learning methods for hand-gesture recognition using ultra-wideband radar. IEEE Access. 2020;8:203580–203590. doi: 10.1109/ACCESS.2020.3037062. [DOI] [Google Scholar]
- Skaria S, Al-Hourani A, Lech M, Evans RJ. Hand-gesture recognition using two-antenna Doppler radar with deep convolutional neural networks. IEEE Sensors J. 2019;19:3041–3048. doi: 10.1109/JSEN.2019.2892073. [DOI] [Google Scholar]
- Soltany Sadrabadi M, Hedayat M, Borazjani I, Arzani A. Fluid-structure coupled biotransport processes in aortic valve disease. J Biomech. 2021;117:110239. doi: 10.1016/j.jbiomech.2021.110239. [DOI] [PubMed] [Google Scholar]
- Soulat G, McCarthy P, Markl M. 4D flow with MRI. Annu Rev Biomed Eng. 2020;22:103–126. doi: 10.1146/annurev-bioeng-100219-110055. [DOI] [PubMed] [Google Scholar]
- Stankovic Z, Allen BD, Garcia J, Jarvis KB, Markl M. 4D flow imaging with MRI. Cardiovasc Diagn Ther. 2014;4:173–192. doi: 10.3978/j.issn.2223-3652.2014.01.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehlik J, et al. Continuous wearable monitoring analytics predict heart failure hospitalization: the LINK-HF multicenter study, circulation. Heart Failure. 2020;13:e006513. doi: 10.1161/CIRCHEARTFAILURE.119.006513. [DOI] [PubMed] [Google Scholar]
- Sun L, Gao H, Pan S, Wang J-X (2020) Surrogate modeling for fluid flows based on physics-constrained deep learning without simulation data. Comput Methods Appl Mech Eng 361. 10.1016/j.cma.2019.112732
- Tadesse GA, Javed H, Weldemariam K, Liu Y, Liu J, Chen J, Zhu T (2021) DeepMI: deep multi-lead ECG fusion for identifying myocardial infarction and its occurrence-time. Artif Intell Med 121. 10.1016/j.artmed.2021.102192 [DOI] [PubMed]
- Tajeddini F, Nikmaneshi MR, Firoozabadi B, Pakravan HA, Ahmadi Tafti SH, Afshin H. High precision invasive FFR, low-cost invasive iFR, or non-invasive CFR?: optimum assessment of coronary artery stenosis based on the patient-specific computational models. Int J Numer Methods Biomed Eng. 2020;36:e3382. doi: 10.1002/cnm.3382. [DOI] [PubMed] [Google Scholar]
- Tarca AL, Carey VJ, Chen X-w, Romero R, Drăghici S. Machine learning and its applications to biology. PLoS Comput Biol. 2007;3:e116. doi: 10.1371/journal.pcbi.0030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoridis S. Chapter 1 - introduction. In: Theodoridis S, editor. Machine learning. Second Edition. Academic Press; 2020. pp. 1–17. [Google Scholar]
- Tison GH, et al. Passive detection of atrial fibrillation using a commercially available smartwatch. JAMA Cardiol. 2018;3:409–416. doi: 10.1001/jamacardio.2018.0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse KM, Chiu P, Lee HP, Ho P. Investigation of hemodynamics in the development of dissecting aneurysm within patient-specific dissecting aneurismal aortas using computational fluid dynamics (CFD) simulations. J Biomech. 2011;44:827–836. doi: 10.1016/j.jbiomech.2010.12.014. [DOI] [PubMed] [Google Scholar]
- Updegrove A, Wilson NM, Merkow J, Lan H, Marsden AL, Shadden SC. SimVascular: An Open Source Pipeline for Cardiovascular Simulation. Ann Biomed Eng. 2017;45:525–541. doi: 10.1007/s10439-016-1762-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vali A, Abla AA, Lawton MT, Saloner D, Rayz VL. Computational fluid dynamics modeling of contrast transport in basilar aneurysms following flow-altering surgeries. J Biomech. 2017;50:195–201. doi: 10.1016/j.jbiomech.2016.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Schoot R, et al. Bayesian statistics and modelling. Nat Rev Methods Primers. 2021;1:1. doi: 10.1038/s43586-020-00001-2. [DOI] [Google Scholar]
- Vardhan M, Randles A. Application of physics-based flow models in cardiovascular medicine: current practices and challenges. Biophys Rev. 2021;2:011302. doi: 10.1063/5.0040315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg HK, Malalasekera W. An introduction to computational fluid dynamics, 2 ed. Pearson; 2007. [Google Scholar]
- Virani SS, et al. Heart disease and stroke Statistics-2020 update: a report from the American Heart Association. Circulation. 2020;141:e139–e596. doi: 10.1161/CIR.0000000000000757. [DOI] [PubMed] [Google Scholar]
- Vishnevskiy V, Walheim J, Kozerke S. Deep variational network for rapid 4D flow MRI reconstruction. Nat Mach Intell. 2020;2:228–235. doi: 10.1038/s42256-020-0165-6. [DOI] [Google Scholar]
- Vozda M, Cerny M. Methods for derivation of orthogonal leads from 12-lead electrocardiogram: a review. Biomedical signal processing and control. 2015;19:23–34. doi: 10.1016/j.bspc.2015.03.001. [DOI] [Google Scholar]
- Wageningen University & Research. https://www.wur.nl/en/show/3t-magnetic-resonance-imaging-3t-mri.htm. Accessed Oct 2021
- Wang G, Ye JC, De Man B. Deep learning for tomographic image reconstruction. Nat Mach Intell. 2020;2:737–748. doi: 10.1038/s42256-020-00273-z. [DOI] [Google Scholar]
- Webb S. Deep learning for biology. Nature. 2018;554:555–557. doi: 10.1038/d41586-018-02174-z. [DOI] [PubMed] [Google Scholar]
- Weinberg EJ, Kaazempur Mofrad MR. A multiscale computational comparison of the bicuspid and tricuspid aortic valves in relation to calcific aortic stenosis. J Biomech. 2008;41:3482–3487. doi: 10.1016/j.jbiomech.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Williams JG et al (2022) Aortic dissection is determined by specific shape and hemodynamic interactions. Ann Biomed Eng. 10.1007/s10439-022-02979-0 [DOI] [PMC free article] [PubMed]
- World Health Organization: Cardiovascular diseases. https://www.who.int/health-topics/cardiovascular-diseases#tab=tab_1. Accessed Oct 2021
- Wymer DT, Patel KP, III, Burke WF, Bhatia VK. Phase-contrast MRI: physics, techniques, and clinical applications. RadioGraphics. 2020;40:122–140. doi: 10.1148/rg.2020190039. [DOI] [PubMed] [Google Scholar]
- Yin M, Zheng X, Humphrey JD, Em Karniadakis G (2021) Non-invasive inference of Thrombus material properties with physics-informed neural networks. Comput Methods Appl Mech Eng 375. 10.1016/j.cma.2020.113603 [DOI] [PMC free article] [PubMed]
- Zakerzadeh R, Hsu MC, Sacks MS. Computational methods for the aortic heart valve and its replacements. Expert Rev Med Devices. 2017;14:849–866. doi: 10.1080/17434440.2017.1389274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Zhang J-M, Su B, Tan RS, Allen JC, Kassab GS (2018) Application of patient-specific computational fluid dynamics in coronary and intra-cardiac flow simulations: challenges and opportunities. Front Physiol 9. 10.3389/fphys.2018.00742 [DOI] [PMC free article] [PubMed]
- "https://www.docwirenews.com/docwire-pick/future-of-medicine-picks/researchers-use-ai-and-wrist-worn-sensor-to-detect-heart-disease/." Accessed Oct 2021
Further reading
- "ATmega32-avr." https://atmega32-avr.com/ (accessed October 2021)
- "Elkworks." https://elkeworks.com/product/_1500946.html (accessed October 2021)
- "https://www.docwirenews.com/docwire-pick/future-of-medicine-picks/researchers-use-ai-and-wrist-worn-sensor-to-detect-heart-disease/." (accessed October 2021)
- "Simvascular software." https://simvascular.github.io/ (accessed October 2021)
- "Wageningen University & Research." https://www.wur.nl/en/show/3t-magnetic-resonance-imaging-3t-mri.htm (accessed October 2021)
- "World Health Organization: Cardiovascular diseases." https://www.who.int/health-topics/cardiovascular-diseases#tab=tab_1 (accessed October 2021)