Abstract
Kaposi’s sarcoma-associated herpesvirus (KSHV) is the etiological agent of Kaposi’s sarcoma (KS), which primarily affects human immunodeficiency virus (HIV)-infected adults with advanced immunodeficiency. Xinjiang province in China is an endemic area for Kaposi’s sarcoma (KS), however, currently, only limited prevalence data for KSHV infection in HIV-infected individuals living in this endemic area is available. A cross-sectional study of 86 HIV positive participants was conducted in Xinjiang, China from 2014 through 2015. Plasma samples were collected and screened for KSHV and HIV infection. HIV pol gene and KSHV ORF-K1 gene were amplified and sequenced, genotypes were determined by phylogenetic analysis. Over all, prevalence was 48.9% (42/86; 95%CI 38.4%-59.3%) for KSHV. Only CRF07_BC subtype has been identified among all these HIV positive individuals, while the subtype A and C of KSHV were detected in the participants. Meanwhile, we found that those with high CD4 counts (> 500) showed a lower anti-KSHV titer, compare with other groups. Our study indicated a high prevalence of KSHV among HIV positive individuals in Xinjiang, China. Thus, management of HIV/AIDS patients should include KSHV screen and should consider the risk of KSHV associated malignancies.
Keywords: KSHV, HIV, China
INTRODUCTION
Kaposi’s sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8 (HHV8), is the etiological agent of all forms of Kaposi’s sarcoma (KS), primary effusion lymphoma, and multicentric Castleman’s disease. KSHV was first identified in a biopsy tissue from a patient with AIDS-related Kaposi’s sarcoma (AIDS-KS) by representational difference analysis in 1994 by Chang et al. [Chang et al., 1994]. Previous studies have shown that approximately 20% of AIDS patients develop KS in Western countries and AIDS-KS is the major cause of death for about 50% of AIDS patients [Coghill et al., 2015; Engels et al., 2008; Robbins et al., 2015].
In mainland China, there is generally a low incidence of KS. However, a high incidence of classical KS has been reported in the Xinjiang Uygur Autonomous region, indicating that this region is endemic for KS [Dilnur et al., 2001]. Xinjiang Uygur Autonomous region is the largest province in northwestern China, and located on the ancient Silk Road as an important staging post over a thousand years ago. While the ethnic groups in Xinjiang are diverse and distinct, the main ethnic groups are the Uygur (45.7%) and the Han (39.7%), other ethnic minorities include Kazakh, Mongolians, Hui, Kirgiz, Man and Xibo. Previous studies have shown that classic KS are rarely seen in the Han Chinese, but are seen more frequently in the Uygur ethnic group [Dilnur et al., 2001; Ouyang et al., 2014].
Meanwhile, Xinjiang region has been reported with the highest of HIV-1 prevalence in China [China., 2015], which could add the KS burden in this region. Immunosuppression due to HIV-1 also enhances the risk of persistent infection and neoplastic progression of oncogenic viruses. Individuals who are co-infected with KSHV and HIV-1 are at high risk of developing KSHV-associated malignancies [Labo et al.; Yao et al.]. To best interpret these observations, it is necessary to understand the epidemiology of KSHV infection itself within this region.
Although efforts have been made to decipher AIDS-KS in recent years, coinfection with HIV-1 and KSHV still presents a great challenge to the current guidelines on the practice and management of coinfected individuals, especially in this KS endemic region [Olp et al., 2015; Rohner et al., 2016]. Moreover, limited information on KSHV infection is available among HIV-1 infected individuals from this endemic region, which thus hampered the prevention of HIV/AIDS opportunistic infection and cancer. Therefore, especially in endemic settings, identifying those at greatest risk of HIV-associated KS may be useful for the development of both therapeutic and preventative strategies to address this important public health concern.
The aim of the present study was to determine the prevalence of KSHV in a sample of Uygur people living with HIV-1, and further elucidate the KSHV molecular epidemiology among a high risk group from Xinjiang, China.
MATERIAL AND METHOD
Study setting and Participants
This retrospective study was conducted in Xinjiang Uygur Autonomous Region, which locates in the northwest of China, from January 2014 to September 2015. HIV positive patients enrolled in this study were hospitalized in the People’s Hospital of Xinjiang Uygur Autonomous Region (Urumchi, China). Patients were included if they met the following criteria: (1) HIV-positive confirmed by Western blot; (2) 18 years or older; (3) be able to provide a written informed consent; (4) Uygur ethnicity. Demographic data and laboratory values were collected from medical records. The study was approved by the Institutional Review Board of school of public health, Fudan University, Shanghai, China.
Sample collection
Venous blood was collected by experienced nurses using sterilized needles and tubes, and transferred to laboratory within 2 hours after collection while being maintained at 4°C. Plasma specimen were stored at −80°C until being tested serologically. All specimens were coded by means of a unique identification number given to each study participant and were analyzed by two experienced technicians, without knowledge of the personal identity of the study participants.
Laboratory Methods
HIV serology
All plasma samples were screened for HIV antibody using an enzyme-linked immunosorbent assay (ELISA; Vironostika HIV Uni-Form II plus O ELISA Kit, Biomerieux Shanghai Company Ltd., Shanghai, China), according to the manufacturer’s instructions. All positive samples were further confirmed by Western blot assay (Genelabs Diagnostic, Singapore).
KSHV testing
Plasma samples were tested by immunofluoresence assay, as reported previously [Minhas et al., 2008]. Briefly, BC-3 cells (HHV8 positive and Epstein-Barr virus negative B cell line, American Type Culture Collection, Manassas, VA), stimulated by tetradecanoyl phorbol acetate (TPA) were fixed and permeabilized and used for monoclonal enhanced immunefluorescence assay. A sample was considered KSHV seropositive only if it was positive at a standard serum dilution of 1:40. Each slide was read independently by two experienced laboratory workers.
To determine geometric mean titer (GMT) of KSHV antibody, KSHV seropositive subjects were further tested by IFA on serially diluted samples ranging from 1:40 to 1:10240.
All above serological tests were performed by the same two experienced technicians according to the manufacturers’ standard protocols. Duplicate negative, positive and blank controls were always analyzed in parallel.
Viral DNA/RNA extraction and amplification
Plasma was separated from the whole blood within two hours after collection and stored at −80 °C until use. HIV Viral DNA/RNA was extracted from 200 µL plasma samples using High Pure Viral Nucleic Acid Kit (Roche Inc., Mannheim, Germany) according to the manufacturer’s instructions.
The HIV-1 pol gene (protease 1-99 amino acids and part of reverse transcriptase 1-300 amino acids) was amplified by using a nested reverse transcription polymerase chain reaction (RT-PCR) method. The target sequence was amplified with TaKaRa One-Step RNA PCR kit (TaKaRa Biotechnology, Dalian, China) using primers MAW26 (5’-TTG GAA ATG TGG AAA GGA AGG AC-3’) and RT21 (5’-CTG TAT TTC TGC TAT TAA GTC TTT TGA TGG G-3’) in a 25 µL reaction. Samples were submitted to the following temperature cycles: 50°C for 30 min, 94°C for 5 min in first-round RT-PCR, followed by 35 cycles at 94°C for 30s, 55°C for 30s, 72°C for 2 min, and an extension at 72°C for 10 min. The nested PCR was performed using Takara Ex Taq PCR kit (TaKaRa Biotechnology, Dalian, China), using primers PRO-1 (5’-CAG AGC CAA CAG CCC CAC CA-3’) and RT20 (5’-CTG CCA GTT CTA GCT CTG CTT C-3’) in a 50 ul reaction and the cycling conditions were 94°C for 5 min in first-round RT-PCR, followed by 30 cycles at 94°C for 30 s, 65°C for 30 s, 72°C 3 min, and an extension at 72°C for 10 min in a thermocycler (Eppendorf Mastercycler 1, Eppendorf, Hamburg, Germany).
KSHV ORF K1 was amplified in a nested PCR with the primer pairs described previously [Ouyang et al., 2014]. Briefly, PCR reactions were performed in a 50-μl reaction mixture containing 100-200 ng of DNA template, 1.5 mM MgCl2, 50 μM of each dNTP, 20 pmol of each primer and 2 U of HotStartTaq DNA polymerase (Qiagen, GmbH, Hildelberg, Germany). The PCR thermal conditions for both first and second rounds were included as follows: an initial 15-min denaturation at 95°C, followed by 35 cycles of 95°C for 30 s, 55°C for 45 s, and 72°C for 50 s, with a final extension at 72°C for 5 min. PCR products were directly loaded onto a 1.5% agarose gel, stained with DNA Safe stain, and visualized by UV Transilluminator.
The PCR products were subjected to DNA sequencing after TA cloning using an automated ABI 3730 DNA sequencer (Applied Biosystems Inc., USA). All the nucleotide sequences obtained were first screened using the BLAST program (National Center for Biotechnology Information, USA) to search for sequence similarities to previously reported sequences in the databases and to eliminate potential laboratory errors.
Phylogenetic Tree Analysis
The nucleotide sequences obtained were aligned with Clustal W in Bioedit (version 7.0.0), and were compared to previously reported sequences of different genotypes and subgenotypes obtained from GenBank. The phylogenetic trees were constructed using the neighbour-joining method in MEGA 5.0. Bootstrap re-sampling and reconstruction were performed with 1000 replicates to confirm the reliability of the phylogenic trees. HIV/KSHV genotypes and subgenotypes were determined by phylogenetic analysis
Statistical analysis
Both medical recorder and laboratory testing results were double entered in EpiData 3.1, and then the database was transformed to a SAS database for further analyses. Demographic characteristics clinical data were analyzed using descriptive statistics, i.e., mean, median and interquartile range (IQR) for continuous variables, and proportions for categorical variables. KSHV seroprevalence was computed using the normal approximation to a binomial distribution. The Kruskal-Wallis nonparametric was used to evaluate overall difference of geometric mean titers (GMTs) of anti-KSHV IgG, and paired comparison were analyzed by post hoc test of Dunnett. All statistical analyses were performed using the SAS system for windows version 9.13 (SAS, Cary, NC, USA) and GraphPad Prism 5.0 (GraphPad, La, Jolla, CA, USA). A two-sided P-value of 0.05 or less was considered statistically significant for all analysis.
RESULTS
Patients’ characteristics
A total of 86 HIV positive individuals were recruited to participate in the present study. Table 1 summarizes the characteristics of the study participants. All the participants were Uygur. Majority (83.7%) of them was male and the mean age of the participants was 36.67±5.35 yrs. (Range 24-47). Among the study subjects, the median CD4+T lymphocyte count was 398 cell/cm3, with a wide range (50-1628 cell/m3), the percentage of CD4+ T cells was 0.2 (IQR: 0.1-0.3), and the median CD4/CD8 ratio was 0.35, (IQR: 0.22-0.58). Among the participants, the overall prevalence for KSHV was 48.8% (42/86; 95%CI 38.4%-59.3%). And the prevalence was 46.2%, 46.9% and 54.2% for the those who < 30 years old, 30~40 years old and ≥41 years old, respectively.
Table 1.
Characteristics of the HIV-infected Uygurs in the study
| Characteristics | Total (N=86) |
|---|---|
| Age (Mean±SD) | 36.76±5.34 |
| Gender No (%) | |
| Male | 72 (83.7%) |
| Female | 14 (16.3%) |
| CD4+ percentage (range) | 0.2 (0.0-0.5) |
| CD4+ cell count (cells/mm3) | |
| ≤200 | 10 (11.4%) |
| 201-500 | 48 (54.5%) |
| ≥501 | 30 (34.1%) |
| CD4/CD8 ratio, median (IQR) | 0.35 (0.22-0.59) |
The molecular characteristics of HIV and KSHV
Of 86 HIV-1 positive participants, 56 (63.6%) were successfully genotyped for the HIV-1 pol gene. Phylogenetic tree of these HIV subtypes is presented in Figure 1A. Only CRF07_BC subtype has been identified among all these HIV positive individuals.
Figure 1A.
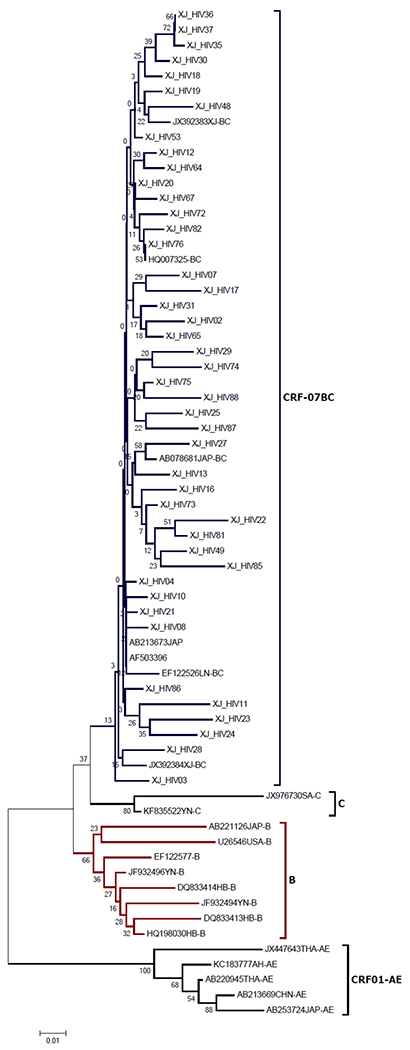
Phylogenetic tree of HIV strains isolated from Uygurs in Xinjiang.
The phylogenetic trees were constructed using the neighbor-joining method in MEGA 5.0. Bootstrap re-sampling and reconstruction were performed with 1000 replicates to confirm the reliability of the phylogenic trees. HIV genotypes and subgenotypes were determined by phylogenetic analysis.
While only 4 KSHV positive samples were successfully amplified with primer pairs specific for the K1 locus and PCR products subjected to direct sequencing analysis. Phylogenetic tree of K1 sequences was achieved comparing with K1 reference sequences of common KSHV subtypes available in GenBank. Based on the MEGA phylogenetic trees, the 4 KSHV positive cases were categorized into subtypes A (A1, A2 and A3) and C (Figure 1B).
Figure 1B.
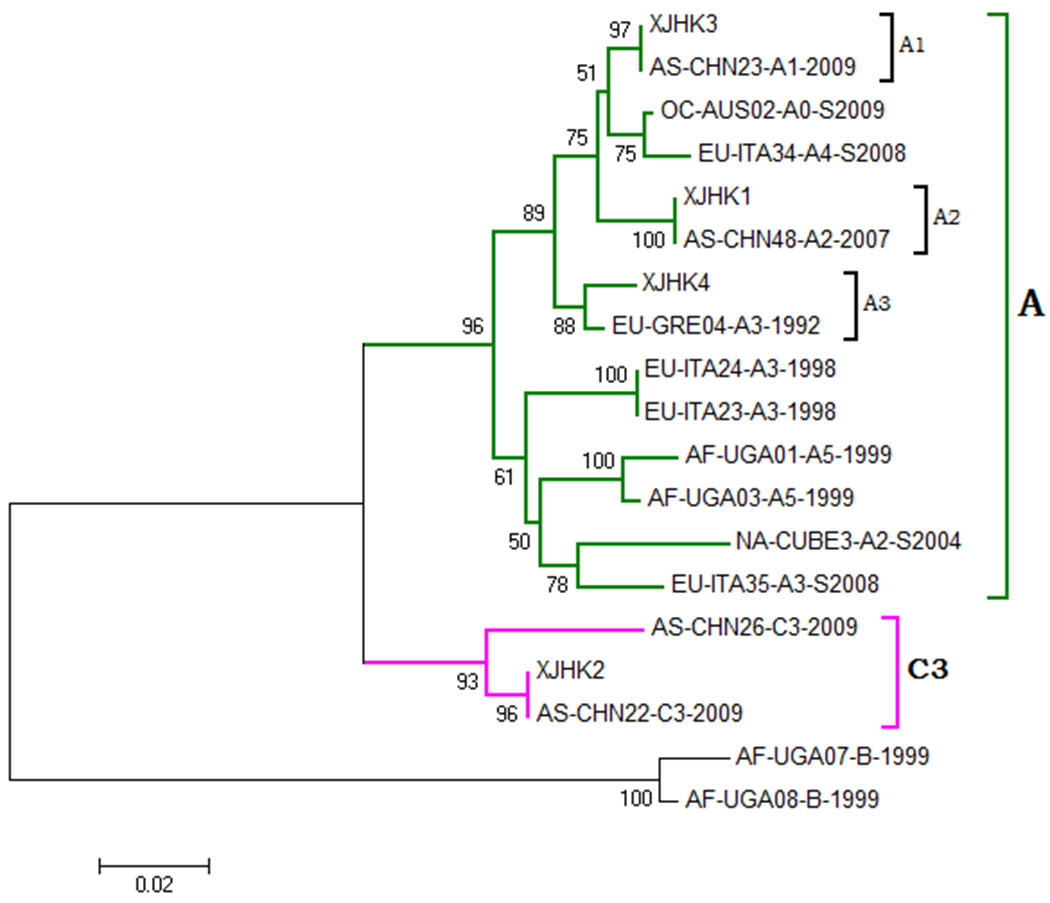
Phylogenetic tree of KSHV strains isolated from Uygurs in Xinjiang.
The phylogenetic trees were constructed using the neighbor-joining method in MEGA 5.0. Bootstrap re-sampling and reconstruction were performed with 1000 replicates to confirm the reliability of the phylogenic trees. KSHV genotypes and subgenotypes were determined by phylogenetic analysis.
KSHV seroprevalence and antibody titers
In order to clarify the influence of concomitant HIV-1 infection on the KSHV antibodies titer, we compared the KSHV antibodies titer distribution across the CD4+ counts subgroups. The geometric mean titer (GMT) of KSHV antibodies was 1289 (IQR: 640-2560), 960 (IQR: 640-1280) and 320 (IQR: 200-640) for CD4+ counts <200 cells/mm3, CD4+ counts 201-500 cells/mm3 and CD4+ counts >500 cells/mm3 group, respectively. A significant difference in the GMT of KSHV antibodies across 3 groups was detected (Kruskal-Wallis H-test, P=0.001). Further pairwise comparison showed that participants who had higher level of CD4+ counts were more likely to be with low KSHV antibodies titers, when compared with CD4+ counts>500 cells/mm3 group (Figure 2).
Figure 2.
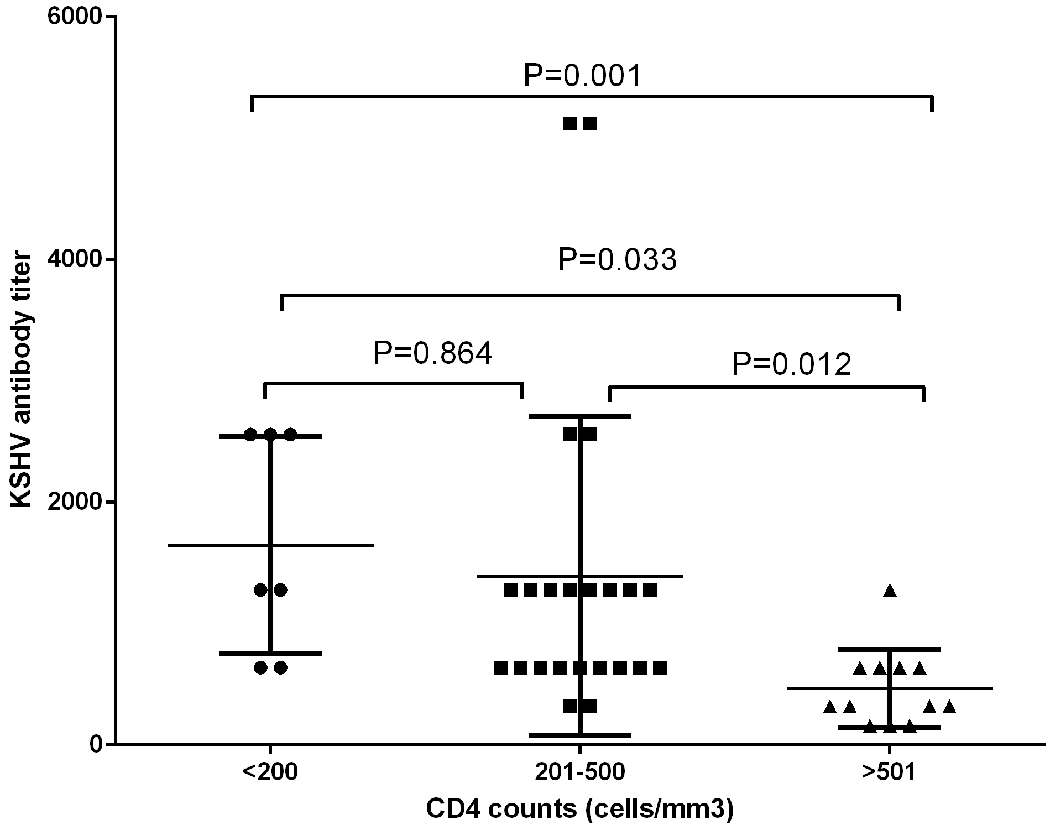
GMT of KSHV antibodies among patients with different CD4+ cell count level
DISCUSSION
The present study investigates the epidemiology of KSHV among HIV positive Uygur individuals in Xinjiang, China, where is highly prevalent for both HIV and KS. To the best of our knowledge, it is the first time to analyze HIV/KSHV co-infected patients characterising both KSHV and HIV parameters in this endemic region. The rate of increase for HIV/AIDS in Xinjiang is currently one of the highest in China[China., 2015]. According to official estimates, there are more than 10,000 HIV-positive persons living in Xinjiang [Zhang et al., 2013]. Previous data have shown that KSHV infection is endemic in the area and there is a high incidence of KS, particularly among Uygur ethnicity [Fu et al., 2009; Wang et al., 2011; Wu et al., 2014; Zhang et al., 2008]. KS is one of the most frequently occurring malignant tumor in individuals infected with HIV-1 in western and African countries[Plummer et al., 2016]. Previous studies from China have shown a high seroprevalence of KSHV in the range of 25.5-46.6% in Xinjiang, but limited data is available for KSHV infection among HIV-1 positive individuals, even among Uygur ethnicity[Zhang et al., 2012]. As expected, a high KSHV prevalence was witnessed among the HIV-1 positive Uygur patients in the present study, and was increased with the age group. It is reasonable to speculate that Uygur people living with HIV are at a considerable risk of developing KS. Whereas we previously reported that a relatively lower prevalence of KSHV infection among HIV positive individuals in central China [Zhang et al., 2011]. Together, these results suggest that Xinjiang is a unique region where the KSHV seroprevalence is significantly higher than other parts of China, and Uygur may carry a high risk for KSHV infection. This high seroprevalence of KSHV may mirror the high diseases burden of KS in this region, particularly among HIV-1 positive Uygur individuals. Such notable findings may contribute to our understanding of HIV/KSHV coinfected patients in order to focus further prevention and therapy.
Interestingly, the genotype of HIV was homogeneous among the study participants. After B/C recombinants (CRF07_BC and CRF08_BC) was first being identified in Xinjiang, now it is still the predominate strain in this high prevalent region [Li et al., 2016]. Given the fact that CRF07_BC was highly prevalent in Xinjiang and the subjects were mainly intravenous drug users (IDUs), it is possible to detect the CRF_07BC strains among the study participants. With regard to the KSHV subtypes, we did not observe any specific geographical or ethnical subtype or subgroup segregation. Only A and C were successfully detected, which is in consistent with previous studies in Xinjiang region [Ouyang et al., 2014; Wang et al., 2012; Zhang et al., 2008]. Since genetic recombination may happen during the evolution, we first envisioned that KSHV may recombination. However, we found no evidence for such a genetic event by Simplot analysis either. KSHV has different subtypes, these subtypes have distinct geographic distributions, and appear to migrate with the human populations [Hayward and Zong, 2007; Meng et al., 1999; Zong et al., 2007; Zong et al., 1999; Zong et al., 1997]. KSHV subtypes A and C are predominant in the Mediterranean, Middle Eastern and Asian regions [Davidovici et al., 2001; Elyamany et al., 2015; Jalilvand et al., 2012; Otvos et al., 2014]. In geographic terms, Xinjiang is located in the center of Asia connected to Europe and was part of the ancient Great Silk Route through these regions. Therefore, it is likely that KSHV migrated to the Xinjiang region with certain ethnic groups in ancient times and persisted in the population. Technically, Xinjiang is a good candidate for further phylogenetic analysis of KSHV subtype distribution and polymorphism as it is inhabited by a multitude of ethnic groups, which could better the understanding of the pathogenies of KSHV within ethnically diverse group.
Regarding the KSHV antibody titer among the study participants, it is notable that KSHV antibody was significantly higher in those who with lower CD4 cell counts, which correlates well with the previous studies [Sullivan et al., 2010]. Generally, KSHV antibody titer distribution may have a prognostic value in predicting the development and progression of KS [Guadalupe et al., 2011; Labo et al., 2015; Olp et al., 2015]. A possible reason is the compromised immune system caused by HIV making the host susceptible to KSHV infection. Besides, the HIV Tat activates lytic cycle replication of HHV-8 via JAK/STAT signaling or HHV-8 RTA induction, and the RTA is the ORF 50 gene product that controls the transition from latency to lytic replication [Zeng et al., 2007]. The present study indicated that the low CD4 count means advanced immunodeficiency, which predisposed a person at high risk of developing KS.
There are several limitations to this study. First, a relatively small sample (86 cases) was included in the present study, yet, despite a small sample size, our results were consistent with previous reports. Second, only 4 KSHV ORFK1 genes were successfully amplified, caution is therefore warranted when interpreting the data. Third, due to the anonymous of the study of HIV subjects, it is hard to access the accurate socio-demographic information.
In conclusion, we identified a high prevalence of KSHV infection in the HIV positive patients from a high endemic region of China. The unremittingly high prevalence of KSHV in HIV-1-infected persons indicates the importance of KSHV co-infection in this population. This is of particular concern, given the accumulating evidence on the continuing occurrence of Kaposi’s sarcoma in HIV-1-infected individuals and the associated decrease in life expectancy.
ACKNOWLEDGEMENTS
We would like to thank all the study subjects for their participation in the study. This study was supported by Natural Science Foundation of Shanghai (17ZR1401400), Doctoral Fund of Ministry of Education of China (Grant No. 20120071120050), the Shanghai Municipal Bureau of Health (grant No. 08GWQ058), The Fourth Round of Three-Year Public Health Action Plan of Shanghai, China (15GWZK0101), and the United States National Institutes of Health Fogarty International Center (grant No. D43 TW001492, RO1 CA75903 and P30 GM103509 to CW).
Footnotes
TRANSPARENCY DECLARATION
The authors declare that there are no conflicts of interests.
REFERENCES
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science (New York, NY) 266(5192):1865–1869. [DOI] [PubMed] [Google Scholar]
- China. NHaFPCotPsRo. 2015. 2015 China AIDS Response Progress Report. [Google Scholar]
- Coghill AE, Shiels MS, Suneja G, Engels EA. 2015. Elevated Cancer-Specific Mortality Among HIV-Infected Patients in the United States. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovici B, Karakis I, Bourboulia D, Ariad S, Zong J, Benharroch D, Dupin N, Weiss R, Hayward G, Sarov B, Boshoff C. 2001. Seroepidemiology and molecular epidemiology of Kaposi’s sarcoma-associated herpesvirus among Jewish population groups in Israel. J Natl Cancer Inst 93(3):194–202. [DOI] [PubMed] [Google Scholar]
- Dilnur P, Katano H, Wang ZH, Osakabe Y, Kudo M, Sata T, Ebihara Y. 2001. Classic type of Kaposi’s sarcoma and human herpesvirus 8 infection in Xinjiang, China. Pathology international 51(11):845–852. [DOI] [PubMed] [Google Scholar]
- Elyamany G, Alzahrani AM, Aljuboury M, Mogadem N, Rehan N, Alsuhaibani O, Alabdulaaly A, Al-Mussaed E, Elhag I, AlFiaar A. 2015. Clinicopathologic features of plasmablastic lymphoma: Single-center series of 8 cases from Saudi Arabia. Diagnostic pathology 10:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels EA, Biggar RJ, Hall HI, Cross H, Crutchfield A, Finch JL, Grigg R, Hylton T, Pawlish KS, McNeel TS, Goedert JJ. 2008. Cancer risk in people infected with human immunodeficiency virus in the United States. International journal of cancer Journal international du cancer 123(1):187–194. [DOI] [PubMed] [Google Scholar]
- Fu B, Sun F, Li B, Yang L, Zeng Y, Sun X, Xu F, Rayner S, Guadalupe M, Gao SJ, Wang L. 2009. Seroprevalence of Kaposi’s sarcoma-associated herpesvirus and risk factors in Xinjiang, China. Journal of medical virology 81(8):1422–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guadalupe M, Pollock BH, Westbrook S, Redding S, Bullock D, Anstead G, Agan BK, Marconi VC, Barbieri S, Sankar V, Rebeles J, Flahive Y, Schoolfield J, Wang L, Lei X, Dow D, Yeh CK, Dang H, Infante AJ, Gao SJ. 2011. Risk factors influencing antibody responses to Kaposi’s sarcoma-associated herpesvirus latent and lytic antigens in patients under antiretroviral therapy. Journal of acquired immune deficiency syndromes (1999) 56(1):83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward GS, Zong JC. 2007. Modern evolutionary history of the human KSHV genome. Current topics in microbiology and immunology 312:1–42. [DOI] [PubMed] [Google Scholar]
- Jalilvand S, Tornesello ML, Buonaguro FM, Buonaguro L, Naraghi ZS, Shoja Z, Ziaee AA, Hamkar R, Shahmahmoodi S, Nategh R, Mokhtari-Azad T. 2012. Molecular epidemiology of human herpesvirus 8 variants in Kaposi’s sarcoma from Iranian patients. Virus research 163(2):644–649. [DOI] [PubMed] [Google Scholar]
- Labo N, Miley W, Benson CA, Campbell TB, Whitby D. Epidemiology of Kaposi’s sarcoma-associated herpesvirus in HIV-1-infected US persons in the era of combination antiretroviral therapy. AIDS (London, England) 29(10):1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labo N, Miley W, Benson CA, Campbell TB, Whitby D. 2015. Epidemiology of Kaposi’s sarcoma-associated herpesvirus in HIV-1-infected US persons in the era of combination antiretroviral therapy. AIDS (London, England) 29(10):1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li W, Zhong P, Fang K, Zhu K, Musa TH, Song Y, Du G, Gao R, Guo Y, Yan W, Xuan Y, Wei P. 2016. Nationwide Trends in Molecular Epidemiology of HIV-1 in China. AIDS research and human retroviruses 32(9):851–859. [DOI] [PubMed] [Google Scholar]
- Meng YX, Spira TJ, Bhat GJ, Birch CJ, Druce JD, Edlin BR, Edwards R, Gunthel C, Newton R, Stamey FR, Wood C, Pellett PE. 1999. Individuals from North America, Australasia, and Africa are infected with four different genotypes of human herpesvirus 8. Virology 261(1):106–119. [DOI] [PubMed] [Google Scholar]
- Minhas V, Crosby LN, Crabtree KL, Phiri S, M’Soka TJ, Kankasa C, Harrington WJ, Mitchell CD, Wood C. 2008. Development of an immunofluorescence assay using recombinant proteins expressed in insect cells to screen and confirm presence of human herpesvirus 8-specific antibodies. Clin Vaccine Immunol 15(8):1259–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olp LN, Minhas V, Gondwe C, Kankasa C, Wojcicki J, Mitchell C, West JT, Wood C. 2015. Effects of Antiretroviral Therapy on Kaposi’s Sarcoma-Associated Herpesvirus (KSHV) Transmission Among HIV-Infected Zambian Children. Journal of the National Cancer Institute 107(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otvos R, Juhasz A, Szalai E, Ujvari D, Otvos K, Szabo K, Remenyik E, Szekely L, Gergely L, Konya J. 2014. Molecular typing of human herpesvirus 8 isolates from patients with Kaposi’s sarcoma in Hungary. Anticancer research 34(2):893–898. [PubMed] [Google Scholar]
- Ouyang X, Zeng Y, Fu B, Wang X, Chen W, Fang Y, Luo M, Wang L. 2014. Genotypic analysis of Kaposi’s sarcoma-associated herpesvirus from patients with Kaposi’s sarcoma in Xinjiang, China. Viruses 6(12):4800–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer M, de Martel C, Vignat J, Ferlay J, Bray F, Franceschi S. 2016. Global burden of cancers attributable to infections in 2012: a synthetic analysis. The Lancet Global health 4(9):e609–616. [DOI] [PubMed] [Google Scholar]
- Robbins HA, Pfeiffer RM, Shiels MS, Li J, Hall HI, Engels EA. 2015. Excess cancers among HIV-infected people in the United States. Journal of the National Cancer Institute 107(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohner E, Wyss N, Heg Z, Faralli Z, Mbulaiteye SM, Novak U, Zwahlen M, Egger M, Bohlius J. 2016. HIV and human herpesvirus 8 co-infection across the globe: Systematic review and meta-analysis. International journal of cancer Journal international du cancer 138(1):45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan SG, Hirsch HH, Franceschi S, Steffen I, Amari EB, Mueller NJ, Magkouras I, Biggar RJ, Rickenbach M, Clifford GM. 2010. Kaposi sarcoma herpes virus antibody response and viremia following highly active antiretroviral therapy in the Swiss HIV Cohort study. AIDS (London, England) 24(14):2245–2252. [DOI] [PubMed] [Google Scholar]
- Wang H, Liu J, Dilimulati, Li L, Ren Z, Wen H, Wang X. 2011. Seroprevalence and risk factors of Kaposi’s sarcoma-associated herpesvirus infection among the general Uygur population from south and north region of Xinjiang, China. Virology journal 8:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang H, He B, Hui Y, Lv G, Li L, Wen H. 2012. Virological and molecular characterization of Kaposi’s sarcoma-associated herpesvirus strains from Xinjiang, China. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology 31(1):53–59. [DOI] [PubMed] [Google Scholar]
- Wu XJ, Pu XM, Kang XJ, Halifu Y, An CX, Zhang DZ, Yakeya B, Mijit J. 2014. One hundred and five Kaposi sarcoma patients: a clinical study in Xinjiang, Northwest of China. Journal of the European Academy of Dermatology and Venereology : JEADV 28(11):1545–1552. [DOI] [PubMed] [Google Scholar]
- Yao S, Hu M, Hao T, Li W, Xue X, Xue M, Zhu X, Zhou F, Qin D, Yan Q, Zhu J, Gao SJ, Lu C. MiRNA-891a-5p mediates HIV-1 Tat and KSHV Orf-K1 synergistic induction of angiogenesis by activating NF-kappaB signaling. Nucleic Acids Res 43(19):9362–9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Zhang X, Huang Z, Cheng L, Yao S, Qin D, Chen X, Tang Q, Lv Z, Zhang L, Lu C. 2007. Intracellular Tat of human immunodeficiency virus type 1 activates lytic cycle replication of Kaposi’s sarcoma-associated herpesvirus: role of JAK/STAT signaling. Journal of virology 81(5):2401–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Pu X, Wu W, Jin Y, Juhear M, Wu X. 2008. Genotypic analysis on the ORF-K1 gene of human herpesvirus 8 from patients with Kaposi’s sarcoma in Xinjiang, China. Journal of genetics and genomics = Yi chuan xue bao 35(11):657–663. [DOI] [PubMed] [Google Scholar]
- Zhang L, Chow EP, Jing J, Zhuang X, Li X, He M, Sun H, Li X, Gorgens M, Wilson D, Wang L, Guo W, Li D, Cui Y, Wang L, Wang N, Wu Z, Wilson DP. 2013. HIV prevalence in China: integration of surveillance data and a systematic review. The Lancet Infectious diseases 13(11):955–963. [DOI] [PubMed] [Google Scholar]
- Zhang T, He N, Ding Y, Crabtree K, Minhas V, Wood C. 2011. Prevalence of human herpesvirus 8 and hepatitis C virus in a rural community with a high risk for blood-borne infections in central China. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases 17(3):395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Shao X, Chen Y, Zhang T, Minhas V, Wood C, He N. 2012. Human herpesvirus 8 seroprevalence, China. Emerging infectious diseases 18(1):150–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong JC, Arav-Boger R, Alcendor DJ, Hayward GS. 2007. Reflections on the interpretation of heterogeneity and strain differences based on very limited PCR sequence data from Kaposi’s sarcoma-associated herpesvirus genomes. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology 40(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong JC, Ciufo DM, Alcendor DJ, Wan X, Nicholas J, Browning PJ, Rady PL, Tyring SK, Orenstein JM, Rabkin CS, Su IJ, Powell KF, Croxson M, Foreman KE, Nickoloff BJ, Alkan S, Hayward GS. 1999. High-level variability in the ORF-K1 membrane protein gene at the left end of the Kaposi’s sarcoma-associated herpesvirus genome defines four major virus subtypes and multiple variants or clades in different human populations. Journal of virology 73(5):4156–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong JC, Metroka C, Reitz MS, Nicholas J, Hayward GS. 1997. Strain variability among Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genomes: evidence that a large cohort of United States AIDS patients may have been infected by a single common isolate. Journal of virology 71(3):2505–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]


