Abstract
In Saccharomyces cerevisiae, AMP biosynthesis genes (ADE genes) are transcriptionally activated in the absence of extracellular purines by the Bas1p and Bas2p (Pho2p) transcription factors. We now show that expression of the ADE genes is low in mutant strains affected in the first seven steps of the pathway, while it is constitutively derepressed in mutant strains affected in later steps. Combined with epistasy studies, these results show that 5′-phosphoribosyl-4-succinocarboxamide-5-aminoimidazole (SAICAR), an intermediate metabolite of the pathway, is needed for optimal activation of the ADE genes. Two-hybrid studies establish that SAICAR is required to promote interaction between Bas1p and Bas2p in vivo, while in vitro experiments suggest that the effect of SAICAR on Bas1p-Bas2p interaction could be indirect. Importantly, feedback inhibition by ATP of Ade4p, catalyzing the first step of the pathway, appears to regulate SAICAR synthesis in response to adenine availability. Consistently, both ADE4 dominant mutations and overexpression of wild-type ADE4 lead to deregulation of ADE gene expression. We conclude that efficient transcription of yeast AMP biosynthesis genes requires interaction between Bas1p and Bas2p which is promoted in the presence of a metabolic intermediate whose synthesis is controlled by feedback inhibition of Ade4p acting as the purine nucleotide sensor within the cell.
In Saccharomyces cerevisiae, biosynthesis pathways are generally negatively regulated by their end product. This regulation usually occurs at two distinct levels, feedback inhibition of an enzyme of the pathway (commonly the first one) and coordinate repression at the transcriptional level of the genes encoding enzymes of the pathway.
Studies on the regulation of the purine biosynthesis pathway in S. cerevisiae revealed that all the genes encoding enzymes required for AMP de novo biosynthesis are repressed at the transcriptional level by the presence of extracellular purines (adenine or hypoxanthine) (6, 7, 10, 23). Two transcription factors, named Bas1p and Bas2p, are required for regulated activation of the ADE genes (6) as well as some histidine biosynthesis genes (2, 7, 35). A LexA-Bas1p fusion can activate a lexAop-lacZ reporter in the presence of Bas2p and in the absence of adenine, suggesting that the regulation process affects the interaction between the two transcription factors (44). A Bas1p subdomain, named BIRD, was identified as being critical for adenine response and Bas1p-Bas2p interaction in vivo (29). However, our understanding of how this domain senses and responds to extracellular adenine is still incomplete.
Our previous work on mutants in which purine biosynthesis genes are no longer repressed by extracellular adenine allowed us to better understand the molecular nature of the signal (13). These mutations, named bra for bypass of repression by adenine, define more than nine complementation groups, several of which have been characterized. BRA7 is FCY2, the gene coding for the purine cytosine permease (Fig. 1) (13). BRA6 is HPT1, the gene encoding hypoxanthine-guanine phosphoribosyltransferase (13), and BRA3 is GUK1, the GMP kinase-encoding gene (21). BRA1 is ADE13 and BRA9 is ADE12, encoding adenylosuccinate lyase and adenylosuccinate synthase, respectively. From these data we proposed that for repression of the ADE genes, adenine has to enter the cell and be metabolized to AMP via formation of hypoxanthine and IMP (Fig. 1). Finally, we have shown that AMP needs to be phosphorylated into ADP to exert its regulatory role (13).
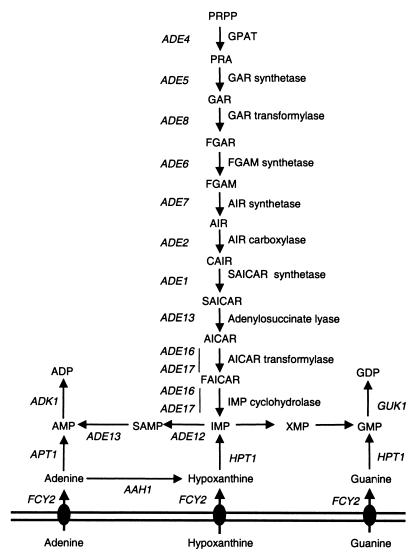
FIG. 1.
Schematic representation of purine metabolism in S. cerevisiae. Abbreviations: AIR, 5′-phosphoribosylaminoimidazole; CAIR, 5′-phosphoribosylaminoimidazole carboxylate; FAICAR, 5′-phosphoribosyl 4-carboxamide-5-formaminoimidazole; FGAM, 5′-phosphoribosyl-N-formylglycinamidine; FGAR, 5′-phosphoribosyl-N-formylglycinamide; GAR, 5′-phosphoribosylglycinamide; IMP, inosine 5′-monophosphate; PRA, 5-phosphoribosylamine; PRPP, 5-phosphoribosyl-1-pyrophosphate; SAMP, adenylosuccinate; XMP, xanthosine 5′-monophosphate. Gene names are italicized, and corresponding enzymatic activities of the IMP biosynthesis pathway are indicated. For simplicity, nucleosides are not represented.
To get a more complete view of AMP biosynthesis regulation in a eukaryotic organism, we have now characterized the remaining three complementation groups (bra2, bra4, and bra5) and six dominant mutations. Additionally, a systematic analysis of the role of each of the de novo pathway genes in their own transcriptional regulation revealed a feedback loop linking the previously characterized ADP signal to a metabolic intermediate in the pathway which activates expression of the ADE genes by affecting the interaction between Bas1p and Bas2p.
MATERIALS AND METHODS
Yeast strains and media.
Yeast strains are listed in Table 1. Strain Y744 is a ura3 segregant from the original prototrophic PUR6 mutant (1) mated to the wild-type PLY122 strain. Strain Y1095 (ade16 ade17) was constructed by mating the Y11583 and Y06561 strains. After sporulation of the resulting diploid, nonparental ditype tetrads containing two spores that were geneticin resistant and auxotrophic for adenine and two spores that were geneticin sensitive and prototrophic for adenine were identified. Results obtained with one such spore, named Y1095, that was both geneticin resistant and an adenine auxotroph are presented in this work, although several other double mutant spores were tested and behaved similarly. Strain Y1124 (ade2) was obtained by sporulating the diploid strain Y22384. Among the resulting spores, ade2 spores were identified by their red color, adenine auxotrophy, and geneticin resistance. One of these spores, named Y1124, isogenic to the wild-type strain BY4742, was used in this work. Strain Y1161 (ade1 homozygous diploid) was constructed by mating strains Y00414 and Y10414. Strain Y1168 (bas1 ade5,7) was constructed by mating strains Y16015 and Y04601. After sporulation of the resulting diploid, nonparental ditype tetrads containing two geneticin-resistant spores and two geneticin-sensitive spores were identified. One such spore, named Y1168, that was both geneticin resistant and auxotrophic for adenine was used in this work. Several other double mutant spores were tested and behaved similarly. Yeast media (YPD, SC, and SD) were prepared according to Sherman et al. (34). SD casa medium is SD supplemented with 0.2% (wt/vol) casamino acids (Difco Laboratories).
TABLE 1.
Yeast strains used in this studya
| Strain | Genotype | Source |
|---|---|---|
| PLY121a | MATα leu2-3,112 ura3-52 lys2-Δ201 his3-Δ200 | P. Ljungdahl |
| 124a | MATα leu2-3,112 ura3-52 lys2-Δ201 his3-Δ200 bra2-2 | Lab collection |
| 127a | MATα leu2-3,112 ura3-52 lys2-Δ201 his3-Δ200 BRA11-1 | Lab collection |
| 128a | MATα leu2-3,112 ura3-52 lys2-Δ201 his3-Δ200 BRA11-2 | Lab collection |
| 131a | MATα leu2-3,112 ura3-52 lys2-Δ201 his3-Δ200 BRA11-3 | Lab collection |
| 133a | MATα leu2-3,112 ura3-52 lys2-Δ201 his3-Δ200 BRA11-4 | Lab collection |
| PLY122a | MATaleu2-3,112 ura3-52 lys2-Δ201 | P. Ljungdahl |
| 206a | MATaleu2-3,112 ura3-52 lys2-Δ201 bra5-2 | Lab collection |
| 211a | MATaleu2-3,112 ura3-52 lys2-Δ201 bra2-3 | Lab collection |
| 213a | MATaleu2-3,112 ura3-52 lys2-Δ201 bra1-2 | Lab collection |
| 215a | MATaleu2-3,112 ura3-52 lys2-Δ201 BRA11-5 | Lab collection |
| 225a | MATaleu2-3,112 ura3-52 lys2-Δ201 BRA11-6 | Lab collection |
| 242a | MATaleu2-3,112 ura3-52 lys2-Δ201 bra4-2 | Lab collection |
| L3852a | MATα ade2 his3 Δ200 leu2-3,112 lys2Δ201 ura3-52 | G. Fink |
| L4233a | MATα leu2-Δ2 ura3-52 gcn4-2 bas1-2 bas2-2 | G. Fink |
| BY4741b | MATahis3-Δ1 leu2-Δ0 met15-Δ ura3-Δ0 | J. Boeke |
| BY4742b | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3Δ0 | J. Boeke |
| Y610a | MATα leu2-3,112 lys2-Δ201 his3-Δ200 ura3-52 ADE13::ADE13-LEU2 | Lab collection |
| Y744 | MATaura3-52 PUR6 | This work |
| Y1095b | MATα his3-Δ1 leu2Δ0 lys2-Δ0 ura3-Δ0 ade16::kanMX4 ade17::kanMX4 | This work |
| Y1124b | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3 Δ0 ade2::kanMX4 | This work |
| Y1161b | MATa/α his3-Δ1/his3-Δ1 leu2-Δ0/leu2-Δ0 lys2-Δ0/LYS2 met15-Δ0/MET15 ura3-Δade1::kanMX4/ade1::kanMX4 | This work |
| Y1168b | MATahis3-Δ1 leu2-Δ0 met15-Δ ura3-Δ 0 bas1::kanMX4 ade5,7::kanMX4 | This work |
| Y1259a | MATα leu2-3 lys2Δ201 ura3-52 his3Δ200 ade13 | This work |
| Y1260a | MATα leu2-3 lys2Δ201 ura3-52 | This work |
| Y1261a | MATaleu2-3 lys2Δ201 ura3-52 his3Δ200 ade2 ade13 | This work |
| Y1262a | MATaleu2-3 lys2Δ201 ura3-52 ade2 | This work |
| Y00414 | MATahis3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 ade1::kanMX4 | Euroscarf |
| Y03803 | MATahis3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 bas2::kanMX4 | Euroscarf |
| Y04601b | MATahis3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 ade5,7::kanMX4 | Euroscarf |
| Y06015b | MATahis3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 bas1::kanMX4 | Euroscarf |
| Y06561b | MATahis3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 ade17::kanMX4 | Euroscarf |
| Y00888b | MATahis3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 ade4::kanMX4 | Euroscarf |
| Y10414b | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 ade1::kanMX4 | Euroscarf |
| Y14244b | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 ade8::kanMX4 | Euroscarf |
| Y14601b | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 ade5,7::kanMX4 | Euroscarf |
| Y14691b | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 ade6::kanMX4 | Euroscarf |
| Y11583b | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 ade16::kanMX4 | Euroscarf |
| Y16015b | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 bas1::kanMX4 | Euroscarf |
| Y10888b | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 ade4::kanMX4 | Euroscarf |
| Y22384b | MATa/α his3-Δ1/his3-Δ1 leu2-Δ0/leu2-Δ0 lys2-Δ0/LYS2 met15-Δ0/MET15 ura3-Δ0 ade2::kanMX4/ADE2 | Euroscarf |
Strains with the same superscript letter are isogenic.
Plasmids.
P79 is a URA3 centromeric plasmid carrying the BAS1 gene in the Ycp50 backbone (30). B273 is a URA3 centromeric plasmid carrying the BAS1-BAS2 fusion (29). Plasmids used in the two-hybrid experiments have already been described. pSH18-34 is a 2μm URA3 plasmid carrying the lexAop-lacZ reporter (14). pEG202 is a 2μm HIS3 plasmid carrying lexA (12). p2099 is a 2μm HIS3 plasmid carrying a lexA-BAS1 fusion (44), and pSH17-4 is a 2μm HIS3 plasmid carrying a lexA-GAL4 fusion (15). B354 is a centromeric LEU2 plasmid carrying a BAS2-VP16 fusion (29).
Plasmids used for overexpression of ADE genes are derivatives of YEp13 (4). YEp13:(ADE1)1 (5), pYeADE5,7(5.2) (16), pPM13 (23), and P1199 (laboratory collection) are LEU2 2μm plasmids carrying ADE1, ADE5,7, ADE4, and ADE17, respectively. P1933, the plasmid carrying the Tet-ADE4 fusion, was constructed as follows. A 1,536-bp fragment carrying the ADE4 coding sequence was amplified by PCR from yeast genomic DNA using synthetic oligonucleotides 429 (5′-AAACTGCAGTCAATAATCTGCACAATTATATAATC-3′) and 48 (5′-CGCGGATCCAAATGTGTGGTATTTTAG-3′). The PCR product was cut with BamHI and PstI and introduced into pCM189 (9) that had been linearized with BamHI and PstI.
Complementation of bra2-2 mutation.
An ADE17-TPK2 fusion was constructed by successive cloning of TPK2 and ADE17 in pSK (Stratagene). A PCR fragment carrying the TPK2 coding sequence was amplified with oligonucleotides 184 (5′-CGCGGATCCATGGAATTCGTTGCAGAA-3′) and 185 (5′-GCTCTAGATGAATCTCTAAGATCTA-3′). The PCR fragment was then restricted with EcoRI and XbaI and inserted in pSK linearized with the same enzymes. The resulting plasmid (named P1540) was linearized with EcoRI and KpnI and used to insert the ADE17 promoter region amplified from P753 (ADE17-lacZ-carrying plasmid [7]) with oligonucleotides 102 and 310 and cut with EcoRI and KpnI. In the resulting plasmid (P1548), the TPK2 coding region is placed under control of ADE17 transcription signals. The sequences of oligonucleotides 102 and 310 were 5′-GCAGCGAGTCAGTGAGCG-3′ and 5′-GGAATTCCATATTTGATGGTGATATG-3′, respectively. Finally, an XbaI-KpnI restriction fragment carrying the ADE17-TPK2 fusion was cloned in the YEplac181 LEU2 2μm vector (11). The resulting plasmid, named P1721, was used to clone the bra2 mutants.
Since overexpression of TPK2 is lethal (26), the ADE17-TPK2 plasmid is toxic. Toxicity was only observed in the absence of adenine when expression of TPK2 was high, while ADE17-TPK2-transformed cells were able to grow in the presence of adenine (repression conditions). In the bra mutants, ADE17-TPK2 was toxic both in the absence and in the presence of adenine. The bra2-2 mutant was cotransformed with the ADE17-TPK2 plasmid and with a genomic library constructed in the pFL38 vector (kind gift from F. Lacroute). Cotransformants able to grow in the presence of adenine were tested further, and plamid DNA was extracted from those unable to grow in the absence of adenine, i.e., behaving like the wild type. Indeed, one plasmid that was able to complement the bra2-2 mutation was isolated (P1813). This plasmid carries a 3,074-bp insert containing 726 bp upstream and 897 bp downstream of ADE13, which is the only complete open reading frame in this insert.
Linkage analyses.
Linkage between bra2 and ADE13 was established using a wild-type strain carrying the LEU2 marker inserted at the ADE13 locus (Y610). This strain was crossed to the bra2-2 mutant carrying an ADE1-lacZ plasmid. The resulting strain was sporulated, and 21 tetrads were analyzed. In all tetrads, the Bra phenotype, monitored by the lacZ fusion, segregated 2:2 and all adenine-derepressed spores were Leu−. bra2 and ADE13 are therefore tightly linked. Similar linkage analysis was done with bra4-2 and bra5-2. In both cases, the Bra phenotype was found to segregate 2:2 in the cross in 20 and 21 tetrads, respectively. As in the case of bra2, constitutive expression of the ADE1-lacZ fusion always cosegregated with the Leu− phenotype, demonstrating the tight linkage between bra4, bra5, and ADE13.
Three dominant purine-excreting mutants (named 128, 131, and 133) were crossed to the ade4 knockout strain, and the meiotic progeny were analyzed. In all tetrads (15, 13, and 16, respectively) the two geneticin-sensitive spores were excreting purines in the medium, demonstrating that these mutations are tightly linked to ade4. The BRA11-1 mutant was crossed to the two remaining dominant purine-excreting mutants, named 215 and 225 (13). In both cases, all spores in the meiotic progeny were excreting purines into the medium (16 and 17 tetrads tested, respectively). Therefore, the mutations in all six dominant BRA mutants excreting purines are tightly linked to ADE4.
lacZ fusions and βGal assays.
The lacZ fusions used in this study have been described previously (6, 13). P115 is a plasmid carrying an ADE1-lacZ fusion in the 2μm URA3 vector YEp356R (25). P473 is a plasmid carrying an ADE1-lacZ fusion in the 2μm LEU2 vector YEp367 (25). β-Galactosidase (βGal) assays were performed as described by Kippert (18) on cells grown for 6 h in the presence or absence of adenine. βGal units are defined as [(optical density at 420 nm [OD420] × 1,000)/(OD600 × minutes × milliliters)]. In each experiment, at least two independent βGal assays were performed, and each assay was done on three independent transformants. The variation between assays in each experiment was <20%.
HPLC analysis of excreted purine compounds of BRA11-1 mutant.
Wild-type and BRA11-1 strains were grown in adenine- and uracil-free SC medium. Cells were then harvested, and the medium was filtered. Separation of purine compounds of the medium was achieved by high-pressure liquid chromatography (HPLC) using a Supelcosil LC-18 5-μm reversed-phase column with a step gradient set up with buffer 1 (0.01 M KH2PO4) and buffer 2 (20% buffer 1, 80% methanol). The following proportions of buffer 1/buffer 2 were used at the indicated run times: 0 min (97/3), 13 min (89/11), 17 min (75/25), 19 min (30/70), and 27 min (97/3), and the flow rate was 1 ml/min. Excreted purine compound separation was monitored by following absorbance at 260 nm at the column end, and the different peaks obtained were identified by comparison with the retention times of known standards. The hypoxanthine and inosine peaks in the BRA11-1 mutant strain growth medium were confirmed by treating the growth medium for 60 min at 37°C with either purine nucleoside phosphorylase (Sigma; 0.01 U/ml) or xanthine oxidase (Sigma; 0.01 U/ml), which metabolize inosine to hypoxanthine and hypoxanthine to uric acid, respectively.
Western blot analyses.
Total yeast protein extracts were made as described previously (19). Proteins were separated by 10% Tris–glycine sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and Western blot analysis was done as described (29), using anti-Bas2p (diluted 1:25,000; kind gift from O. S. Gabrielsen) or purified anti-Bas1p (diluted 1:1,000 [29]) as primary antibodies and peroxidase-conjugated anti-rabbit immunoglobulin G (IgG; diluted 1:10,000, Pierce) as the secondary antibody.
Glutamine PRPP amidotransferase (Ade4p) activity determination.
Yeast strain Y00888 (ade4) was transformed with either plasmid p1933 or pCM189, containing and not containing the ADE4 gene, respectively. Cells (400 ml) were grown to an OD600 of 1 in SD casa medium (containing tryptophan and adenine), harvested by centrifugation, rinsed with 2 ml of buffer A (24), and finally resuspended in 2 ml of the same buffer. Glass beads (3 g) were added, and cells were disrupted by four vigorous vortexing cycles for 1 min at room temperature, followed by 2 to 3 min of incubation on ice. Supernatant was transferred in a 2-ml Eppendorf tube. Beads were rinsed with 2 ml of buffer A, and the second supernatant was combined with the first one. Samples were centrifuged for 10 min at 20,000 × g and 4°C, pellets were discarded, and supernatants were dialyzed for 1 h against buffer A (4 liters). Protein concentration was measured after dialysis using the Bio-Rad protein assay kit. Extracts usually contained about 8 mg of proteins/ml.
Glutamine 5-phosphoribosyl-1-pyrophosphate (PRPP) amidotransferase activity was then determined by measuring the initial rate of glutamate formation as described (24) in either the presence or absence of nucleotides. For each determination, a control reaction was done in the same conditions but in the absence of PRPP. Measurements were done with 500 to 600 μg of total yeast protein extract per assay for 10 min at 37°C. The reaction was stopped by 2 min of incubation at 100°C. The glutamate formed was measured by the glutamate dehydrogenase method. Aliquots of the first reaction (100 μl) were transferred in a spectrophotometer cuvette containing 400 μl of buffer B (KH2PO4/K2HPO4, 0.12 M [pH 8] containing NADP, 1.1 mg/ml, and glutamate dehydrogenase, 2.6 U/ml). Absorbance was recorded for 10 min at room temperature. In these conditions, the reaction is linear up to consumption of 200 nmol of glutamate. The PRPP-dependent Ade4p activity measured in the absence of nucleotide was 52.3 ± 5.6 nmol of glutamate/min/mg of protein. No measurable activity was obtained for the ade4 yeast strain (Y00888 transformed by the pCM189 plasmid) not containing the ADE4 gene.
Synthesis of SAICAR.
5′-Phosphoribosyl-4-succinocarboxamide-5-aminoimidazole (SAICAR) was prepared from 5′-phosphoribosyl-4-carboxamide-5-aminoimidazole (AICAR) by an enzymatic method. The reaction mixture containing 0.5 M fumarate, 23 mM AICAR (Sigma), and 20 mg of adenylosuccinase (Sigma) was incubated for 4 h at room temperature and passed through a QAE Sephadex A column. SAICAR was eluted from the column with a 20 to 800 mM gradient of NH4HCO3.
Electrophoresis mobility shift assays.
The 81-bp probe containing Bas1p and Bas2p DNA-binding sites was obtained by PCR on yeast genomic DNA in the presence of 5 μl of [α-32P]dATP (400 Ci/mmol) with oligonucleotides 125 (5′-CGCCCCGTCGGTAG-3′) and 126 (5′-AGTTCAAGCCCATCGC-3′). The 22-bp oligonucleotide probe was obtained by labeling oligonucleotides 463 (5′-GTGCCGACTGACTCGTGTCCTG-3′) and 464 (5′-CAGGACACGAGTCAGTCGGCAC-3′) with T4 polynucleotide kinase (Promega) in the presence of 10 μl of [γ-32P]ATP. Protein purification and electrophoresis mobility shift assays were performed as described previously (30, 40).
RESULTS
Mutations of bra2, bra4, and bra5 complementation groups are complementing alleles of ADE13
A plasmid complementing the bra2-2 mutation was isolated by rescuing the toxicity of an ADE17-TPK2 fusion in the presence of adenine (see Materials and Methods for details). Sequencing of both ends of the plasmid insert revealed that it carries a 3-kb insert containing the ADE13 gene. This plasmid fully complemented the derepressed phenotype of the bra2-2 mutation and also complemented other bra2 alleles (see, for example, bra2-3, in Fig. 2). ADE13 was previously shown to complement the bra1 and bra8 mutations (13). Linkage between bra2 and ADE13 was then established (see Materials and Methods for details). We conclude that bra2 mutants define a new case of intragenic complementation at the ADE13 locus.
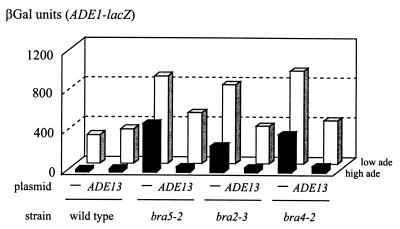
FIG. 2.
Bra phenotype of bra5-2, bra2-3, and bra4-2 mutant strains is complemented by ADE13. Yeast strains 206 (bra5-2), 211 (bra2-3), 242 (bra4-2), and the isogenic wild-type PLY122 were cotransformed with a plasmid carrying the ADE1-lacZ fusion (P473) and a plasmid carrying the ADE13 gene (P1813) or the corresponding empty centromeric vector (pFL38, lanes −). βGal assays were performed after growth for 6 h in SC medium in the absence (low ade) or presence (high ade) of adenine (0.3 mM), as described in Materials and Methods.
Since bra4 and bra5 mutants share several phenotypes with bra2 mutants (13) we suspected that they could also be alleles of ADE13. Indeed, we found that bra4-2 and bra5-2 were fully complemented by an ADE13 centromeric plasmid (Fig. 2). Linkage analysis was done as detailed in Materials and Methods and demonstrated tight linkage between bra4, bra5, and ADE13. Finally, adenylosuccinate lyase activity was found to be strongly impaired in the bra2-3, bra4-2, and bra5-2 mutants (data not shown). Therefore the three previously uncharacterized bra complementation groups (bra2, bra4, and bra5) correspond to complementing alleles of the ADE13 locus.
These data complete our study of the recessive mutations of the bra1 to bra9 complementation groups. These mutations affect five different genes: ADE13 (bra1, bra2, bra4, bra5, and bra8), GUK1 (bra3), HPT1 (bra6), FCY2 (bra7), and ADE12 (bra9) (Fig. 1) (13, 21). Since several mutants were found in most of these complementation groups, we believe that the screen was reasonably exhaustive. An intriguing observation emerging from the whole data set was that most of the ade13 mutants in the bra1, bra2, bra4, bra5, and bra8 complementation groups belong to a specific class of mutants previously named class 3 mutants (13). In these mutants, expression of the ADE genes is higher than in the wild-type strain under both repression and derepression conditions (presence and absence of adenine, respectively; Fig. 2).
Why should expression of the ADE genes increase in the ade13 mutants? We previously proposed (see discussion in reference 13) that this specific behavior of ade13 mutants could reflect the fact that Ade13p catalyzes two steps in AMP synthesis, the eighth step of IMP synthesis and the last step of AMP synthesis (Fig. 1) and thus participates in both de novo purine synthesis and salvage pathways. We therefore tested whether an interruption of the de novo pathway upstream from ade13 would result in a similar phenotype.
Synthesis of metabolic intermediate SAICAR is critical for ADE gene expression.
Expression of the ADE1-lacZ fusion was monitored in a set of isogenic strains each carrying a deletion of a specific ADE gene encoding one of the first seven steps of IMP biosynthesis (ade4 to ade1; Fig. 1). Since these mutants are auxotrophic for adenine, expression of the fusion could not be monitored in the absence of adenine. We therefore used a low adenine concentration (0.025 mM), which sustained growth of the auxotrophic strains during the 6-h shift but was not high enough to induce repression in the wild-type strain (Fig. 3A). Expression of the ADE1-lacZ fusion was very low in the ade mutants with mutations affecting the first seven enzymes of the pathway and was not efficiently induced at low adenine concentrations (Fig. 3A). This result suggested that, in these mutants, something required for activation of the ADE genes by Bas1p and Bas2p was missing even under adenine starvation conditions.
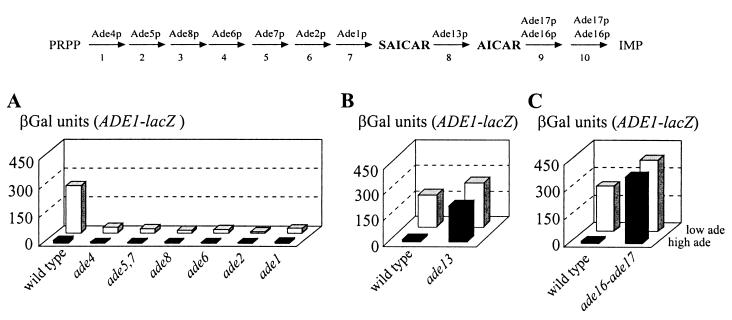
FIG. 3.
Effect of mutations in IMP biosynthesis genes on expression of ADE1-lacZ. Yeast strains transformed with a plasmid carrying the ADE1-lacZ fusion (P115) were grown for 6 h in SD casa medium containing adenine at either a low (0.025 mM, low ade) or high concentration (0.3 mM, high ade), and βGal activity was then measured as described in Materials and Methods. The following strains were used: (A) BY4742 (wild type), Y10888 (ade4), Y14601 (ade5,7), Y14244 (ade8), Y14691 (ade6), Y1124 (ade2), and Y10414 (ade1); (B) PLY121 (wild type) and 124 (ade13); (C) BY4742 (wild type) and Y1095 (ade16 ade17). Strains used in each panel are isogenic. The 10 steps of IMP biosynthesis are schematically represented at the top of the figure.
To get a complete picture of the role of the IMP synthesis pathway on ADE gene regulation, we then investigated mutations further downstream in the pathway. Mutations affecting the three final steps of IMP synthesis were studied (Fig. 1). Since deletion of ADE13 strongly affects growth, expression of ADE1-lacZ was measured in the bra2-2 mutant and isogenic wild-type strain (Fig. 3B). In this mutant and in all ade13 mutants tested, expression of ADE1-lacZ was derepressed, suggesting that these mutants accumulate something required for activation of the ADE genes. Finally, the effect of mutations affecting the two final steps was studied. These steps are catalyzed by bifunctional enzymes encoded by the ADE16 and ADE17 genes, and knockout of both genes is required to obtain an adenine auxotrophic mutant (39). Expression of ADE1-lacZ measured in the double knockout is presented in Fig. 3C. As in the ade13 mutants, expression was fully derepressed, suggesting that the double mutant accumulates something required for activation of the ADE genes. Furthermore, the high level of ADE1-lacZ expression in the ade16 ade17 auxotrophic mutant clearly demonstrated that the low expression of the fusion in the other ade mutants was not due to their adenine auxotrophy.
Altogether, these data strongly suggest that blocking the synthesis of SAICAR by invalidating one of the first seven steps of IMP synthesis results in a lack of activation of the ADE1-lacZ fusion while impairing the last three steps by mutations in either ADE13 or ADE16 and ADE17 results in constitutively derepressed expression of the fusion. These results point to SAICAR (and possibly AICAR; see Discussion section) as an important molecule in the signal transduction process (Fig. 1). A prediction from this hypothesis is that mutations in the first part of the pathway should be epistatic to mutations in the second part.
An ade2 ade13 (bra1-2) double mutant was constructed by mating isogenic ade2 and ade13 mutant strains L3852 and 213 and sporulating the resulting diploid. Ten tetrads were obtained, and expression of an ADE1-lacZ fusion was measured in the four spores of each tetrad. In each tetrad, the two ade2 spores were identified by their typical red color and auxotrophy for adenine. In all 10 tetrads, the two ade2 spores showed constitutively repressed expression of ADE1-lacZ. Results obtained for a tetratype are presented in Table 2. As expected, the ade2 ade13 spores have the same repressed phenotype as the ade2 ADE13 spores, and this phenotype is abolished by transformation with an ADE2 CEN plasmid (pASZ11 [37]).
TABLE 2.
Expression of an ADE1-lacZ fusion in a tetratype obtained by mating ade2 and ade13 (bra1-2) mutant strains
| Spore | Relevant genotype | βGal activity (U)
|
|||||
|---|---|---|---|---|---|---|---|
| No plasmid
|
ADE2 plasmid
|
||||||
| Adenine concn (mM)
|
DR/Ra | Adenine concn (mM)
|
DR/R | ||||
| 0.025 | 0.3 | 0 | 0.3 | ||||
| Y1259 | ade13 | 541 | 198 | 2.7 | —b | — | — |
| Y1260 | Wild type | 252 | 21 | 12.0 | — | — | — |
| Y1261 | ade2 ade13 | 37 | 22 | 1.7 | 512 | 165 | 3.1 |
| Y1262 | ade2 | 26 | 11 | 2.4 | 192 | 20 | 9.6 |
DR/R, ratio of the activities measured under derepressing conditions (0 or 0.025 mM adenine, as indicated) versus repressing conditions (0.3 mM adenine).
—, not done.
These results clearly show that ade2 is epistatic to ade13. Therefore, the deregulation observed in the ade13 mutants, presumably due to accumulation of SAICAR, is prevented when synthesis of SAICAR through the de novo pathway is abolished by the upstream ade2 mutation.
Synthesis of SAICAR is correlated to Bas1p Bas2p interaction in vivo.
The molecular mechanism of adenine repression of the ADE genes is thought to involve an alteration of Bas1p-Bas2p interactions (29, 44). Our data presented above now suggest that SAICAR is required for expression of ADE1-lacZ, possibly by favoring Bas1p-Bas2p interaction. The link between production of this metabolic intermediate and interaction of Bas1p and Bas2p was investigated. First we took advantage of a Bas1p-Bas2p fusion protein previously shown to activate transcription of ADE1-lacZ independently of adenine (29). The Bas1p-Bas2p fusion protein or the Bas1p protein alone (control) was expressed in a bas1 ade5,7 double mutant and in a bas1 ADE5,7 isogenic control strain. Expression of the Bas1p-Bas2p fusion protein in both strains resulted in constitutively derepressed expression of the ADE1-lacZ fusion (Fig. 4A), showing that the absence of SAICAR synthesis in the ade5,7 mutant does not affect activation by the fusion. On the contrary, expression of Bas1p alone in the bas1 ade5,7 mutant resulted in a constitutively repressed phenotype (Fig. 4A). Finally, expression of Bas1p in the bas1 ADE5,7 strain led to adenine-regulated expression of the ADE1-lacZ fusion (Fig. 4A). Thus, a covalent interaction between Bas1p and Bas2p can bypass the requirement for the metabolic intermediate.
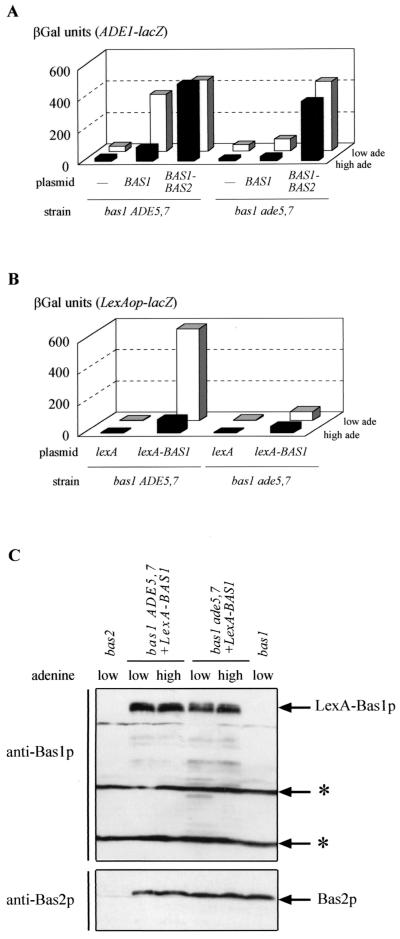
FIG. 4.
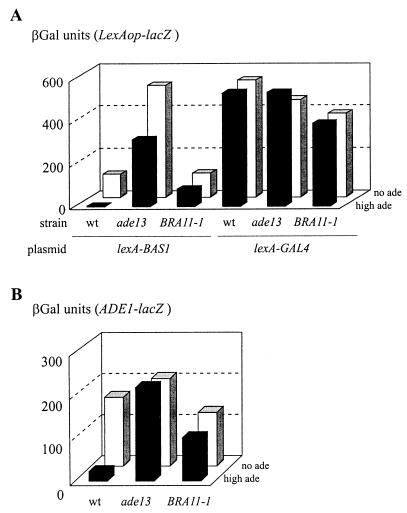
Effect of an ade5,7 mutation on transcriptional activation by a Bas1p-Bas2p fusion and on interaction between LexA-Bas1p and Bas2p. Yeast strains were cultured on SC medium supplemented with adenine at either a low (0.025 mM, low ade) or high adenine concentration (0.3 mM, high ade). βGal assays were performed as described in Materials and Methods. (A) Strains Y06015 (bas1 ADE5,7) and Y1168 (bas1 ade5,7) were cotransformed with a plasmid carrying the ADE1-lacZ fusion (P473) and either a control plasmid (YCp50) or the same plasmid carrying BAS1 (P79) or a BAS1-BAS2 fusion (B273). (B) Strains Y06015 (bas1 ADE5,7) and Y1168 (bas1 ade5,7) were cotrans- formed with a plasmid carrying the lexAop-lacZ fusion (pSH18-34) and a plasmid carrying either a lexA-BAS1 fusion (p2099) or unfused lexA (pEG202). (C) Strains Y06015 (bas1 ADE5,7), Y1168 (bas1 ade5,7), and Y03803 (bas2), transformed or not with a plasmid carrying lexA-BAS1 (p2099) as indicated, were cultured in the presence of a low or high concentration of adenine. Total yeast protein extracts were prepared and subjected to Western blot analyses with antibodies against Bas1p or Bas2p, as described in Materials and Methods. The cross-reacting bands are marked by asterisks.
Second, using a two-hybrid approach, interaction between LexA-Bas1p and Bas2p was assayed in various genetic backgrounds by monitoring expression of a lexAop-driven reporter gene in vivo. As shown in Fig. 4B, in the bas1 strain, expression of lexAop-lacZ relies on the presence of LexA-Bas1p and was much more efficient at a low adenine concentration. In the bas1 ade5,7 strain, lexAop-lacZ expression was low at both high and low adenine concentrations, while the amount of LexA-Bas1p and Bas2p was not affected (Fig. 4C). Therefore, the ade5,7 mutation leading to low expression of ADE1-lacZ (Fig. 3A) impaired the LexA-Bas1p/Bas2p interaction in vivo (Fig. 4B). In these experiments, activation by LexA-Bas1p was strictly dependent on the presence of Bas2p (data not shown).
A similar two-hybrid approach was used to evaluate Bas1p-Bas2p interaction in constitutively derepressed mutants. Since these mutants carry the wild-type BAS1 gene, we coexpressed a BAS2-VP16 fusion to avoid competition for Bas2p between endogenous Bas1p and LexA-Bas1p. Indeed, such competition was observed in our strains (data not shown) and was previously reported by Zhang et al. (44). As expected, in the wild-type strain interaction between LexA-Bas1p and Bas2p-VP16 was strong in the absence of adenine and low in the presence of adenine (Fig. 5A). The same protein interaction monitored in a bra2-2 (ade13) or BRA11-1 strain (see next section) was high in both the presence and absence of adenine (Fig. 5A), showing that the LexA-Bas1p/Bas2p-VP16 in vivo interaction tightly reflected ADE gene expression (Fig. 5B).
FIG. 5.
Effect of mutations in ADE13 and BRA11 on interaction between Bas1p and Bas2p in a two-hybrid assay. (A) Strains PLY121 (wild type), 124 (ade13), and 127 (BRA11-1) were cotransformed with a plasmid carrying the lexAop-lacZ fusion (pSH18-34), a plasmid carrying the BAS2-VP16 fusion (P2013), and a plasmid carrying either a lexA-BAS1 (p2099) or a lexA-GAL4 (pSH17-4) fusion. Cells were grown in SC medium lacking histidine, uracil, and leucine and in the absence (no ade) or presence (high ade) of 0.3 mM adenine. βGal assays were performed as described in Materials and Methods. (B) Strains PLY121 (wild type), 124 (ade13), and 127 (BRA11-1) were transformed with a plasmid carrying the ADE1-lacZ fusion (P115). βGal assays were done as described in Materials and Methods after growth for 6 h in SD casa medium lacking uracil, in the absence (no ade) or presence (high ade) of 0.3 mM adenine.
In the same experiment, expression of lexAop-lacZ driven by LexA-Gal4p was totally insensitive to adenine and mutations in the ADE pathway (Fig. 5A). Therefore, these two-hybrid studies further substantiate the role of SAICAR in promoting interaction between Bas1p and Bas2p in vivo. In this hypothesis, the regulation by adenine of ADE gene transcription would operate by modulating the amount of SAICAR, and we therefore intended to find out how SAICAR synthesis is regulated in yeast cells. We reasoned that a mutation that makes the cell unable to sense the signal should lead to dominant derepression of SAICAR synthesis, and thus the study of dominant BRA mutations was undertaken.
Dominant purine-excreting BRA11 mutations are allelic to ade4
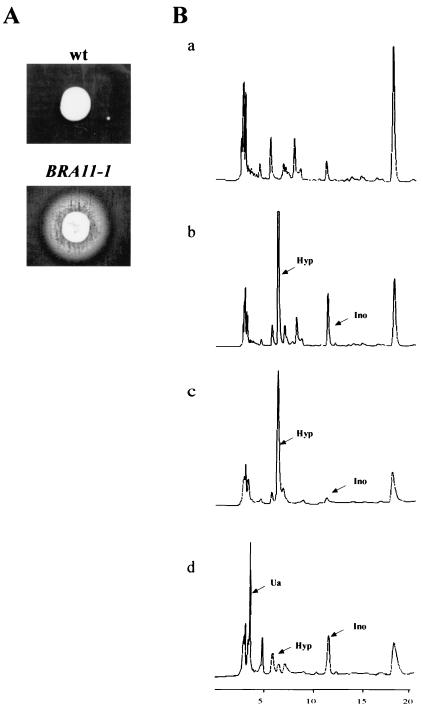
In the original screen, 15 BRA mutations were found to be fully or partially dominant (13), and 6 of these mutants excreted purines into the medium, as determined by cross-feeding experiments (Fig. 6A). The excreted compounds were analyzed by chromatography. The excretion profiles showed two major additional peaks compared to the isogenic wild-type strain profile (Fig. 6B). These peaks were identified as hypoxanthine and inosine by their specific retention times before and after treatment with purine nucleoside phosphorylase or xanthine oxidase, which specifically metabolize inosine to hypoxanthine and hypoxanthine to uric acid, respectively (Fig. 6B).
FIG. 6.
BRA11-1 excretion analysis. (A) The wild-type (wt) strain PLY121 and the BRA11-1 strain 127 were spotted on a lawn of strain Y1161 (ade1) plated on purine-free SC medium. Purine excretion was monitored after 3 days at 30°C. (B) HPLC analysis of excreted purine compounds. Wild-type and BRA11-1 strains were grown in adenine- and uracil-free SC medium. Cells were then harvested, and the medium was filtered. Separation of purine compounds was achieved by HPLC and monitored by following absorbance at 260 nm at the column end. Wild-type (a) and BRA11-1 (b) growth medium was analyzed. BRA11-1-specific peaks were identified as hypoxanthine and inosine by their retention times and after treatment with purine nucleoside phosphorylase (c), which metabolizes inosine to hypoxanthine, and xanthine oxidase (d), which metabolize hypoxanthine to uric acid. Arrows indicate the identified peaks. Abbreviations: Hyp, hypoxanthine; Ino, inosine; Ua, uric acid.
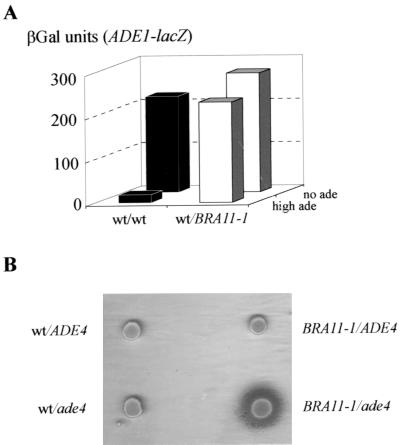
One of the dominant mutations, named BRA11-1 (mutant 127), was studied further. The mutant showed derepressed expression of an ADE1-lacZ fusion, and this phenotype was found to be dominant (Fig. 7A), while the purine excretion phenotype was recessive (Fig. 7B). The BRA11-1 mutant was crossed to the BY4741 wild-type strain, and both derepression and excretion phenotypes were monitored in the meiotic progeny (18 tetrads). As expected, the derepression phenotype segregated 2:2 in the cross and cosegregated with the purine excretion phenotype (data not shown).
FIG. 7.
BRA11-1 derepression and excretion phenotype. (A) Diploid strains (PLY121 × PLY122 [+/+] and 127 × PLY122 [+/BRA11-1]) were transformed with an ADE1-lacZ fusion (P115). Expression of the ADE1-lacZ fusion was monitored by measuring βGal activity in the two diploid strains after growth for 6 h in the presence (high ade) or absence (no ade) of adenine. (B) Purine excretion phenotype in various diploid backgrounds. The BRA11-1 mutant and isogenic wild-type (wt) strain PLY121 were crossed to isogenic strains either wild-type (BY4741, ADE4) or disrupted for ADE4 (Y00888, ade4). The resulting diploid strains were assayed for cross-feeding of a lawn of ade1-homozygous diploid cells (Y1161). Purine excretion, resulting in growth of ade1 red-pigmented colonies, was monitored after 3 days at 30°C.
Interestingly, one such dominant purine-excreting mutant had been described previously as carrying a PUR6 mutation which was shown to be allelic to ade4 (1, 22). We therefore tested whether BRA11-1 could be allelic to ade4. The BRA11-1 mutant was crossed to the Y00888 strain, in which ADE4 is disrupted by a geneticin resistance cassette (ade4::kanMX4) and to the isogenic wild-type control BY4741 (ADE4). Purine excretion was observed only in the BRA11-1/ade4 diploid (Fig. 7B). Linkage between BRA11-1 and ade4 was then estimated by sporulating the BRA11-1/ade4::kanMX4 diploid. Among 35 tetrads, the 2 geneticin-sensitive spores were excreting purines into the medium, demonstrating that BRA11-1 is tightly linked to ade4. Finally, expression of the ADE1-lacZ fusion was also tested in the original PUR6 mutant (1) and found to be deregulated (data not shown). Therefore, PUR6 and BRA11-1 behaved very similarly. Finally, the five other dominant purine excretion mutations were shown to be alleles of ADE4 (see Materials and Methods for details).
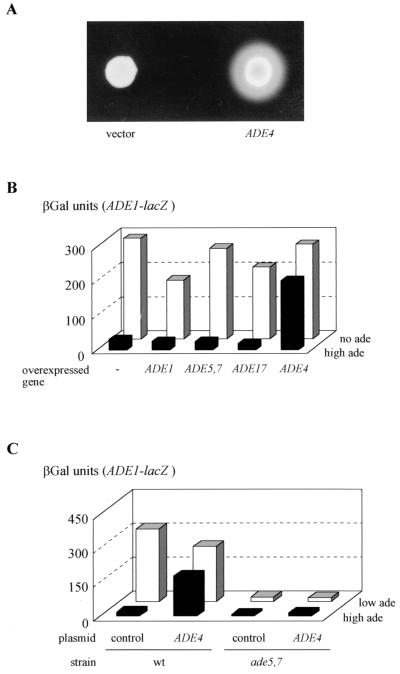
We also observed that overexpression of wild-type ADE4 resulted in both purine excretion (Fig. 8A) and derepression of the ADE1-lacZ fusion, while overexpression of other ADE genes had no effect (Fig. 8B). Therefore, either dominant mutations or overexpression of the ADE4 gene results in purine excretion and derepression of the ADE genes. These results suggested an important role for ADE4 in regulation of the ADE genes and was interpreted as a consequence of increased metabolic flow through the pathway, resulting in increased synthesis of both SAICAR and IMP: the former leading to constitutive transcriptional activation by Bas1p and Bas2p, and the latter being degraded to inosine and hypoxanthine and ultimately excreted into the medium. The dominance effects observed in the heterozygous BRA11-1/wild-type diploid (Fig. 7) are interpreted as follows: synthesis of SAICAR in this strain is efficient enough to result in dominant deregulation of ADE1-lacZ, while IMP synthesis is not sufficient to lead to detectable purine excretion.
FIG. 8.
Effect of ADE4 overexpression. (A) Purine excretion phenotype of strain overexpressing the ADE4 gene. The wild-type (wt) strain BY4742 transformed either with a control plasmid (pCM189) or with a plasmid expressing the tet-ADE4 fusion (P1933) was spotted on a lawn of ade1-homozygous diploid cells (Y1161) plated on purine-free SD casa medium. Purine excretion was monitored after 7 days at 30°C. (B) Effect of overexpression of several ADE genes on activation of ADE1-lacZ. The wild-type strain BY4742 was cotransformed with a plasmid carrying the ADE1-lacZ fusion (P115) and a 2μm plasmid carrying one of the following genes: ADE1 [YEp13:(ADE1)1], ADE5,7 [pYeADE5,7(5.2)], ADE17 (P1199), ADE4 (pPM13), or the corresponding empty vector (YEp13). βGal assays were performed on the transformed strains grown for 6 h in the absence (no ade) or presence (high ade) of 0.3 mM adenine. (C) Effect of overexpression of ADE4 on activation of the ADE1-lacZ fusion in the ade5,7 mutant strain. Yeast strains BY4741 (wild type) and Y04601 (ade5,7) were cotransformed with a plasmid carrying the ADE1-lacZ fusion (P115) and a 2μm plasmid carrying the ADE4 gene (pPM13) or the corresponding empty vector (YEp13). βGal assays were performed after 6 h of growth in SC medium in the presence of adenine at either a low (0.025 mM, low ade) or high concentration (0.3 mM, high ade), as described in Materials and Methods.
Ade4p is the sensor of purine nucleotides.
Since ADE4 encodes the first committed step of the pathway, it was tempting to assume that it could play a critical role in ADE gene transcriptional regulation by modulating the synthesis of the coactivator SAICAR. Should this hypothesis be correct, dominant ADE4 mutations or overexpression of ADE4 should be hypostatic to mutations blocking the synthesis of SAICAR. This prediction was assayed by overexpressing ADE4 in wild-type and ade5,7 strains. We observed that overexpression of ADE4 had no effect on the low expression of ADE1-lacZ in the ade5,7 strain (Fig. 8C).
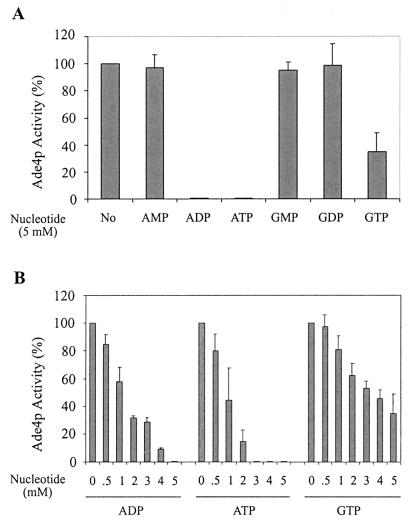
An important additional prediction would be that Ade4p enzymatic activity should be inhibited by ADP (or a derivative), previously shown to be the effector for adenine repression (13). This was investigated by in vitro measurement of Ade4p activity. This activity was hardly detectable in a wild-type strain, and the enzyme could not be efficiently purified in an active form from Escherichia coli. We therefore measured it in an ade4 knockout mutant strain (Y00888) carrying either a control plasmid (pCM189) or an ADE4 overexpression plasmid (P1933). In the absence of added nucleotides, Ade4p activity was not detectable in the ade4 strain transformed with the control plasmid and was 52.3 ± 5.6 U (nanomoles of glutamate formed per minute per milligram of protein) in the ADE4-overexpressing strain. As shown in Fig. 9A, only ADP, ATP, and, to a lesser extent, GTP had an effect on Ade4p activity. AMP, GMP, and GDP had no inhibitory effect even at high concentration (5 mM). For ADP, ATP, and GTP, a greater range of concentrations were tested, confirming that ADP and ATP are better inhibitors of Ade4p than GTP (Fig. 9B). From these data, we conclude that ADP and ATP produced from extracellular adenine regulate synthesis of the coactivator SAICAR through inhibition of the first enzymatic step of the pathway.
FIG. 9.
Inhibitory effect of purine nucleotides on glutamine PRPP amidotransferase activity. Yeast strain Y00888 (ade4) was transformed with the P1933 plasmid constitutively overexpressing Ade4p. Yeast protein extracts were prepared as described in Materials and Methods. Glutamine PRPP amidotransferase (Ade4p) activity was measured for 10 min at 37°C in either the absence (No) or the presence of different nucleotides at 5 mM (A) or at increasing concentrations of various nucleotides (B). The rate of glutamate formation was then determined by the glutamate dehydrogenase method as described in the text. The PRPP-dependent Ade4p activity measured in the absence of nucleotide is 52.3 ± 5.6 nmol of glutamate/min/mg of protein (100% activity). No activity was measurable when the ade4-disrupted yeast strain was transformed with the pCM189 empty vector. Results are averages of three to nine independent experiments.
Does SAICAR directly affect Bas1p-Bas2p interaction?
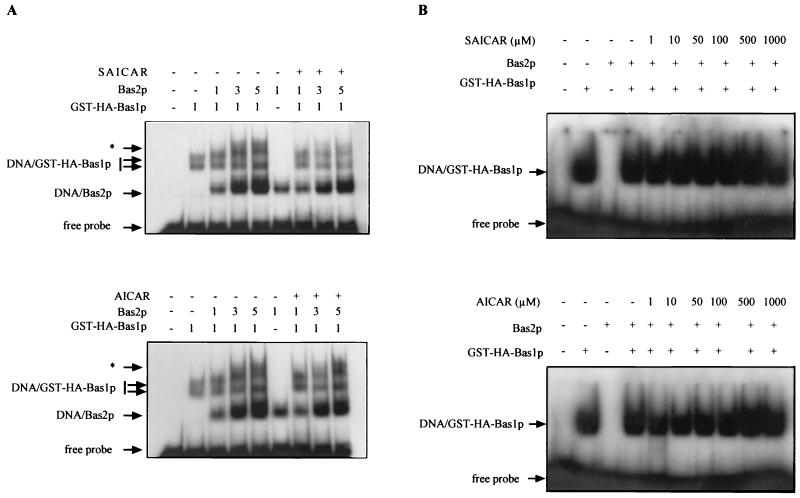
The major remaining question is how SAICAR affects the interaction between Bas1p and Bas2p. This question was addressed using purified Bas1p and Bas2p from E. coli. We first assayed whether SAICAR could enhance cooperative binding of the two proteins to the ADE5,7 promoter (32). No such cooperative binding could be found in previous studies on the HIS4 promoter (40) or ADE1 and ADE17 promoters (our unpublished data). Clearly, the addition of SAICAR or AICAR did not result in any cooperative binding (Fig. 10A). We then used a fragment of the ADE5,7 promoter allowing binding of Bas1p but not of Bas2p (32) to test whether, when Bas1p can bind to the probe, SAICAR (or AICAR) could promote the formation of a Bas1p-Bas2p complex in vitro. As shown in Fig. 10B, no supershift could be detected that would suggest the formation of such a complex. We therefore conclude that SAICAR (or AICAR) is not likely to directly promote the interaction between the two transcription factors.
FIG. 10.
Effect of SAICAR and AICAR on in vitro binding of glutathione S-transferase (GST)-hemagglutinin (HA)-Bas1p and Bas2p to DNA. Electrophoresis mobility shift assays were done as described in Materials and Methods. GST-HA-Bas1p and Bas2p purified from E. coli were incubated for 15 min at room temperature with 100 μM SAICAR or AICAR. Samples were then incubated for 15 min with the ADE5,7 promoter-radiolabeled probe and separated on a 4% nondenaturing gel. The gel was dried, and radioactivity was revealed by autoradiography. (A) Effect of SAICAR and AICAR on cooperative in vitro binding of Bas1p and Bas2p to the ADE5,7 promoter. The probe was an 81-bp fragment spanning the region between nucleotides −212 and −131 in the ADE5,7 promoter, containing both Bas1p and Bas2p DNA-binding sites (32). The complexes marked by an asterisk were only observed in the presence of both Bas2p and GST-HA-Bas1p and may therefore correspond to the binding of both proteins to the probe. Amounts of proteins added are indicated in arbitrary units. (B) Effect of SAICAR and AICAR on in vitro binding of Bas2p to Bas1p on the ADE5,7 promoter. The probe was a 22-bp oligonucleotide spanning the region between oligonucleotides −191 and −169 in the ADE5,7 promoter, containing only the Bas1p DNA-binding site (32).
DISCUSSION
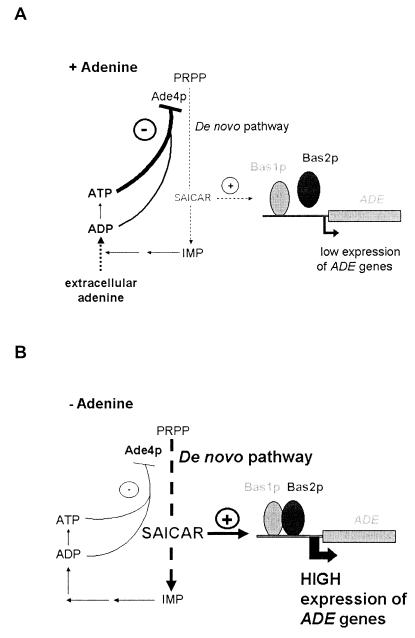
In this paper we show that transcriptional regulation of the AMP biosynthesis genes is intimately connected to feedback inhibition of the first step of the pathway. A model of this regulation is presented in Fig. 11. In adenine-replete cells (Fig. 11A), ADP and ATP synthesis from adenine leads to feedback inhibition of Ade4p, which is the controlling enzyme of the pathway. Lower Ade4p activity results in decreased synthesis of SAICAR and reduced interaction between Bas1p and Bas2p. Consequently, transcriptional activation of the ADE genes, including ADE4, is less efficient, and the amount of Ade4p decreases, further diminishing SAICAR synthesis. However, SAICAR synthesis will never be turned down totally because part of ADE gene transcription is Bas1p and Bas2p independent (6). This residual synthesis of the pathway enzymes in adenine-replete cells will allow reactivation of the pathway when adenine in the growth medium becomes limiting. Under such conditions (Fig. 11B), less ADP and ATP will be available, and inhibition of Ade4p activity will be less efficient, leading to increased synthesis of SAICAR and transcriptional activation of the ADE genes. This activation will in turn increase the enzyme level and the flow through the pathway and finally result in increased ADP and ATP synthesis until the system has reached a new equilibrium, leading to high expression of the ADE genes in the absence of adenine. This expression level is not the highest possible one, since in the ade13 mutants, in which SAICAR is poorly metabolized, expression of the ADE genes is higher than in the wild-type strain. Therefore, in wild-type cells, SAICAR appears to be limiting for activation by Bas1p and Bas2p.
FIG. 11.
Model for regulation of AMP biosynthesis genes (see Discussion for details). The presence or absence of extracellular adenine is indicated by + Adenine (A) and − Adenine, respectively (B).
How does SAICAR affect interaction between Bas1p and Bas2p? The in vivo SAICAR-dependent Bas1p-Bas2p interaction, observed in the two-hybrid assay, suggests that DNA binding of these factors is not required for the interaction. We found that SAICAR did not promote cooperative DNA binding of the two factors and was not sufficient to complex Bas2p to DNA-bound Bas1p. We therefore believe that SAICAR itself might not interact directly with the transcription factors but requires a protein intermediate that has not yet been identified or would need to be further metabolized. Indeed, AICAR was found in its di- and triphosphate forms in Salmonella enterica serovar Typhimurium and as such was proposed to play an important signaling role in folate metabolism (3). Yeast ade16 ade17 double mutants cannot metabolize AICAR and presumably accumulate it. Such double mutants were shown to deregulate ADE gene transcription (Fig. 3C), suggesting that AICAR could either play a signaling role similar to that of SAICAR or that AICAR accumulation could feedback inhibit adenylosuccinate lyase and result in SAICAR accumulation, thus phenocopying an ade13 mutation. Indeed, we found that AICAR can feedback inhibit Ade13p activity, and the 50% inhibitory concentration was estimated to be 65 μM (data not shown).
Other cases of metabolic intermediates activating yeast transcription factors have been reported. For example, α-isopropyl malate was shown to be required for transcription activation by Leu3p (38) and either orotate or dihydrorotate activates Ppr1p-dependent transcription (8). Therefore, utilization of small molecules, which are metabolic intermediates, as coactivators appears to be an efficient regulatory strategy developed during evolution. Indeed, such a strategy combines high specificity and sensitivity, since the coactivator is involved in a single metabolic pathway and its synthesis can be tightly regulated by other means.
In yeast, regulation of the AMP biosynthesis pathway coactivator depends on feedback inhibition of Ade4p catalyzing the first step of the pathway. Ade4p is regulated by both ADP and ATP, which were previously found to be important signal molecules in the adenine response process (13). A previous study on partially purified yeast glutamine PRPP amidotransferase has shown that this enzyme is feedback inhibited by AMP, ADP, and ATP. In this study, AMP was the most effective inhibitor (33), whereas we found (this work) that AMP did not have any major inhibitory effect (Fig 9A). The difference between these two results could be due to the partial purification of the enzyme in the study by Satyanarayana et al. (33), while our study was done with total yeast protein extract.
Another report on Ade4p activity measured in total yeast protein extract by Nieto and Woods (27) revealed that AMP has a significant inhibitory effect only at very high concentrations (88% inhibition at 20 mM AMP), which are far from physiological concentrations. Indeed, in yeast, the ATP concentration is in the millimolar range (17, 20), and it is more abundant than ADP and AMP: the ATP/ADP ratio is about 5, and the ATP/AMP ratio is >20 (17, 20, 42). Therefore, under physiological conditions, AMP is not likely to play a critical role in Ade4p regulation, and we believe that ATP is most probably the important molecule responsible for regulation of Ade4p activity. This assumption is in good agreement with genetic studies showing that ADP, or a derivative of ADP, is the important molecule for regulation by adenine (13).
This work constitutes the first description of AMP biosynthesis gene regulation in a eukaryotic organism. In prokaryotes, repression of pur operons by extracellular purine is achieved by very different processes. In E. coli, a specific repressor named PurR binds to its target site and represses transcription only when the corepressor hypoxanthine or guanine is present; therefore, in this bacterium, the purine bases are directly regulating transcription (31). In Bacillus subtilis, the regulation is less direct. The repressor interaction with its binding site is inhibited by PRPP. Since the synthesis of PRPP is itself regulated at the enzymatic level by ADP, it therefore responds to the presence of extracellular adenine, which modulates ADP levels (41). It is striking that this highly conserved metabolic pathway is tightly regulated by extracellular bases in bacteria and yeasts, although through very different mechanisms.
Could the mechanism described in S. cerevisiae and involving SAICAR apply to mammals? Interestingly, accumulation of SAICAR has been reported in adenylosuccinate lyase-deficient patients and was associated with autism (36). Strikingly, purine overproduction and uric acid excretion were found for about 20% of autistic patients (28) and could indeed be a consequence of AMP biosynthesis deregulation. Moreover, retroinhibition of human amidophosphoribosyltransferase by purine nucleotides was shown to play an important role in regulation of the de novo pathway and cellular proliferation (43). The mechanism of AMP biosynthesis regulation described in yeast could therefore give important clues to its mammalian counterpart and elucidate its possible implication in pathologies.
ACKNOWLEDGMENTS
We are grateful to G. Fink, O. S. Gabrielsen, S. Henikoff, E. Herrero, D. B. Kaback, F. Lacroute, I. Lascu, R. Rolfes, R. Woods, and H. Zalkin for providing biological materials. We thank C. Napias for helpful discussions concerning enzymatic assay and I. Belloc for technical assistance with HPLC.
This work was supported by grants from Fondation pour la Recherche Médicale, Conseil Régional d'Aquitaine, and CNRS (UMR5095). C.D. was supported by a Conseil Régional d'Aquitaine postdoctoral fellowship. K.R. was supported by a Ministère de la Recherche training fellowship.
REFERENCES
- 1.Armitt S, Woods R A. Purine-excreting mutants of Saccharomyces cerevisiae. I. Isolation and genetic analysis. Genet Res. 1970;15:7–17. doi: 10.1017/s0016672300001324. [DOI] [PubMed] [Google Scholar]
- 2.Arndt K T, Styles C, Fink G R. Multiple global regulators control HIS4 transcription in yeast. Science. 1987;237:874–880. doi: 10.1126/science.3303332. [DOI] [PubMed] [Google Scholar]
- 3.Bochner B R, Ames B N. ZTP (5-amino-4-imidazole carboxamide riboside 5′-triphosphate): a proposed alarmone for 10-formyl-tetrahydrofolate deficiency. Cell. 1982;29:929–937. doi: 10.1016/0092-8674(82)90455-x. [DOI] [PubMed] [Google Scholar]
- 4.Broach J R, Strathern J N, Hicks J B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979;8:121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- 5.Crowley J C, Kaback D B. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: isolation of the ADE1 gene. J Bacteriol. 1984;159:413–417. doi: 10.1128/jb.159.1.413-417.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daignan-Fornier B, Fink G R. Coregulation of purine and histidine biosynthesis by the transcriptional activators BAS1 and BAS2. Proc Natl Acad Sci USA. 1992;89:6746–6750. doi: 10.1073/pnas.89.15.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denis V, Boucherie H, Monribot C, Daignan-Fornier B. Role of the Myb-like protein Bas1p in Saccharomyces cerevisiae: a proteome analysis. Mol Microbiol. 1998;30:557–566. doi: 10.1046/j.1365-2958.1998.01087.x. [DOI] [PubMed] [Google Scholar]
- 8.Flynn P J, Reece R J. Activation of transcription by metabolic intermediates of the pyrimidine biosynthetic pathway. Mol Cell Biol. 1999;19:882–888. doi: 10.1128/mcb.19.1.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gari E, Piedrafita L, Aldea M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 10.Giani S, Manoni M, Breviario D. Cloning and transcriptional analysis of the ADE6 gene of Saccharomyces cerevisiae. Gene. 1991;107:149–154. doi: 10.1016/0378-1119(91)90309-y. [DOI] [PubMed] [Google Scholar]
- 11.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 12.Golemis E A, Brent R. Fused protein domains inhibit DNA binding by LexA. Mol Cell Biol. 1992;12:3006–3014. doi: 10.1128/mcb.12.7.3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guetsova M L, Lecoq K, Daignan-Fornier B. The isolation and characterization of Saccharomyces cerevisiae mutants that constitutively express purine biosynthetic genes. Genetics. 1997;147:383–397. doi: 10.1093/genetics/147.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 15.Hanes S D, Brent R. DNA specificity of the bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell. 1989;57:1275–1283. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- 16.Henikoff S. The Saccharomyces cerevisiae ADE5,7 protein is homologous to overlapping Drosophila melanogaster Gart polypeptides. J Mol Biol. 1986;190:519–528. doi: 10.1016/0022-2836(86)90238-x. [DOI] [PubMed] [Google Scholar]
- 17.Hinze H, Holzer H. Analysis of the energy metabolism after incubation of Saccharomyces cerevisiae with sulfite or nitrite. Arch Microbiol. 1986;145:27–31. doi: 10.1007/BF00413023. [DOI] [PubMed] [Google Scholar]
- 18.Kippert F. A rapid permeabilization procedure for accurate quantitative determination of beta-galactosidase activity in yeast cells. FEMS Microbiol Lett. 1995;128:201–206. doi: 10.1111/j.1574-6968.1995.tb07523.x. [DOI] [PubMed] [Google Scholar]
- 19.Kushnirov V V. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 20.Larsson C, Nilsson A, Blomberg A, Gustafsson L. Glycolytic flux is conditionally correlated with ATP concentration in Saccharomyces cerevisiae: a chemostat study under carbon- or nitrogen-limiting conditions. J Bacteriol. 1997;179:7243–7250. doi: 10.1128/jb.179.23.7243-7250.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lecoq K, Konrad M, Daignan-Fornier B. Yeast GMP kinase mutants constitutively express AMP biosynthesis genes by phenocopying a hypoxanthine-guanine phosphoribosyltransferase defect. Genetics. 2000;156:953–961. doi: 10.1093/genetics/156.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lomax C A, Woods R A. A complex genetic locus controlling purine nucleotide biosynthesis in yeast. Mol Gen Genet. 1973;120:139–149. doi: 10.1007/BF00267242. [DOI] [PubMed] [Google Scholar]
- 23.Mantsala P, Zalkin H. Glutamine nucleotide sequence of Saccharomyces cerevisiae ADE4 encoding phosphoribosylpyrophosphate amidotransferase. J Biol Chem. 1984;259:8478–8484. [PubMed] [Google Scholar]
- 24.Messenger L J, Zalkin H. Glutamine phosphoribosylpyrophosphate amidotransferase from Escherichia coli: purification and properties. J Biol Chem. 1979;254:3382–3392. [PubMed] [Google Scholar]
- 25.Myers A M, Tzagoloff A, Kinney D M, Lusty C J. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene. 1986;45:299–310. doi: 10.1016/0378-1119(86)90028-4. [DOI] [PubMed] [Google Scholar]
- 26.Nehlin J O, Ronne H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J. 1990;9:2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nieto D J, Woods R A. Studies on mutants affecting amidophosphoribosyltransferase activity in Saccharomyces cerevisiae. Can J Microbiol. 1983;29:681–688. doi: 10.1139/m83-111. [DOI] [PubMed] [Google Scholar]
- 28.Page T, Coleman M. Purine metabolism abnormalities in a hyperuricosuric subclass of autism. Biochim Biophys Acta. 2000;1500:291–296. doi: 10.1016/s0925-4439(99)00113-1. [DOI] [PubMed] [Google Scholar]
- 29.Pinson B, Kongsrud T L, Ording E, Johansen L, Daignan-Fornier B, Gabrielsen O S. Signaling through regulated transcription factor interaction: mapping of a regulatory interaction domain in the Myb-related Bas1p. Nucleic Acids Res. 2000;28:4665–4673. doi: 10.1093/nar/28.23.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinson B, Sagot I, Borne F, Gabrielsen O S, Daignan-Fornier B. Mutations in the yeast Myb-like protein Bas1p resulting in discrimination between promoters in vivo but not in vitro. Nucleic Acids Res. 1998;26:3977–3985. doi: 10.1093/nar/26.17.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rolfes R J, Zalkin H. Purification of the Escherichia coli purine regulon repressor and identification of corepressors. J Bacteriol. 1990;172:5637–5642. doi: 10.1128/jb.172.10.5637-5642.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rolfes R J, Zhang F, Hinnebusch A G. The transcriptional activators BAS1, BAS2, and ABF1 bind positive regulatory sites as the critical elements for adenine regulation of ADE5,7. J Biol Chem. 1997;272:13343–13354. doi: 10.1074/jbc.272.20.13343. [DOI] [PubMed] [Google Scholar]
- 33.Satyanarayana T, Kaplan J G. Regulation of the purine pathway in bakers yeast: activity and feedback inhibition of phosphoribosyl-pyrophosphate amidotransferase. Arch Biochem Biophys. 1971;142:40–47. doi: 10.1016/0003-9861(71)90257-8. [DOI] [PubMed] [Google Scholar]
- 34.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 35.Springer C, Kunzler M, Balmelli T, Braus G H. Amino acid and adenine cross-pathway regulation act through the same 5′-TGACTC-3′ motif in the yeast HIS7 promoter. J Biol Chem. 1996;271:29637–29643. doi: 10.1074/jbc.271.47.29637. [DOI] [PubMed] [Google Scholar]
- 36.Stone R L, Aimi J, Barshop B A, Jaeken J, Van den Berghe G, Zalkin H, Dixon J E. A mutation in adenylosuccinate lyase associated with mental retardation and autistic features. Nat Genet. 1992;1:59–63. doi: 10.1038/ng0492-59. [DOI] [PubMed] [Google Scholar]
- 37.Stotz A, Linder P. The ADE2 gene from Saccharomyces cerevisiae: sequence and new vectors. Gene. 1990;95:91–98. doi: 10.1016/0378-1119(90)90418-q. [DOI] [PubMed] [Google Scholar]
- 38.Sze J Y, Woontner M, Jaehning J A, Kohlhaw G B. In vitro transcriptional activation by a metabolic intermediate: activation by Leu3 depends on alpha-isopropylmalate. Science. 1992;258:1143–1145. doi: 10.1126/science.1439822. [DOI] [PubMed] [Google Scholar]
- 39.Tibbetts A S, Appling D R. Saccharomyces cerevisiae expresses two genes encoding isozymes of 5-aminoimidazole-4-carboxamide ribonucleotide transformylase. Arch Biochem Biophys. 1997;340:195–200. doi: 10.1006/abbi.1997.9919. [DOI] [PubMed] [Google Scholar]
- 40.Tice-Baldwin K, Fink G R, Arndt K T. BAS1 has a Myb motif and activates HIS4 transcription only in combination with BAS2. Science. 1989;246:931–935. doi: 10.1126/science.2683089. [DOI] [PubMed] [Google Scholar]
- 41.Weng M, Nagy P L, Zalkin H. Identification of the Bacillus subtilis pur operon repressor. Proc Natl Acad Sci USA. 1995;92:7455–7459. doi: 10.1073/pnas.92.16.7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson W A, Hawley S A, Hardie D G. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr Biol. 1996;6:1426–1434. doi: 10.1016/s0960-9822(96)00747-6. [DOI] [PubMed] [Google Scholar]
- 43.Yamaoka T, Yano M, Kondo M, Sasaki H, Hino S, Katashima R, Moritani M, Itakura M. Feedback inhibition of amidophosphoribosyltransferase regulates the rate of cell growth via purine nucleotide, DNA, and protein syntheses. J Biol Chem. 2001;276:21285–21291. doi: 10.1074/jbc.M011103200. [DOI] [PubMed] [Google Scholar]
- 44.Zhang F, Kirouac M, Zhu N, Hinnebusch A G, Rolfes R J. Evidence that complex formation by Bas1p and Bas2p (Pho2p) unmasks the activation function of Bas1p in an adenine-repressible step of ADE gene transcription. Mol Cell Biol. 1997;17:3272–3283. doi: 10.1128/mcb.17.6.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]