Abstract
RNA editing of specific residues by adenosine deamination is a nuclear process catalyzed by adenosine deaminases acting on RNA (ADAR). Different promoters in the ADAR1 gene give rise to two forms of the protein: a constitutive promoter expresses a transcript encoding (c)ADAR1, and an interferon-induced promoter expresses a transcript encoding an N-terminally extended form, (i)ADAR1. Here we show that (c)ADAR1 is primarily nuclear whereas (i)ADAR1 encompasses a functional nuclear export signal in the N-terminal part and is a nucleocytoplasmic shuttle protein. Mutation of the nuclear export signal or treatment with the CRM1-specific drug leptomycin B induces nuclear accumulation of (i)ADAR1 fused to the green fluorescent protein and increases the nuclear editing activity. In concurrence, CRM1 and RanGTP interact specifically with the (i)ADAR1 nuclear export signal to form a tripartite export complex in vitro. Furthermore, our data imply that nuclear import of (i)ADAR1 is mediated by at least two nuclear localization sequences. These results suggest that the nuclear editing activity of (i)ADAR1 is modulated by nuclear export.
Adenosine deaminase acting on RNA (ADAR) deaminates adenosines to produce inosines within RNAs that are predominantly double stranded. Since inosine has base-pairing properties similar to those of guanosine, adenosine deamination can change the coding properties of an mRNA and alter the functional role of a gene product, a process known as RNA editing (reviewed in reference 2). The double-stranded RNA (dsRNA) structures of known in vivo ADAR editing substrates are formed through base pairing between an exon and an intron in cis, which has led to the conclusion that editing is a nuclear process preceding splicing (6, 11, 21, 38, 52). Modulation of the properties of neuronal receptors and ion channels by adenosine deaminations at specific sites in the transcripts of, e.g., mammalian glutamate and serotonin receptors has been described (6, 32), Drosophila sodium, calcium, and chloride channels (18, 55, 57), and a squid K+ channel (49).
ADAR proteins belong to a multigene family, and three of the mammalian members, ADAR1, ADAR2, and ADAR3, constitute a structurally related subfamily (reviewed in reference 29). Cell transfection experiments, genetic knockout of the ADAR genes in mice, and in vitro studies indicate that ADAR1 and -2 are the deaminases responsible for the observed editing events (8, 20, 23, 31, 34, 39, 45, 46, 60).
The editing substrates are mostly found in the brain, but the two deaminases are also expressed in other tissues, which suggests that there are still unidentified ADAR substrates (59). In accordance with this, heterozygosity for a null allele of ADAR1 disturbs the hematopoietic erythropoiesis and is embryonically lethal in mice (60).
ADAR1, -2, and -3 share a very similar overall domain structure. Functional domains include two or three repeats of a 70-amino-acid dsRNA binding motif followed by a highly conserved catalytic domain near the C terminus (8, 31, 41). Unique to the ADAR1 gene locus is the presence of both constitutive and interferon-induced promoters that produce transcripts with different first exons (15). The interferon-induced mRNA encodes the form (i)ADAR1, which is extended by 295 amino acids at the N terminus compared to the constitutively expressed form, (c)ADAR1. There is a basal expression of both forms of the protein (31, 48, 60), but treatment with alpha, beta, or gamma interferon increases the level of (i)ADAR1 mRNA two- to threefold (9). The in vitro deamination activities of (c)ADAR1 and (i)ADAR1 are similar. Both enzymes are capable of site-specific editing of pre-mRNA substrates (35, 37) as well as more unspecific deamination of up to half of the adenosines in extended, perfectly double-stranded RNA (36), suggesting that the substrate recognition and catalytic domains are fully retained within (c)ADAR1. The (i)ADAR1 N-terminal extension encompasses a 65-amino-acid domain, Zα, that can bind both DNA and dsRNA in a Z conformation (5, 19). Nuclear magnetic resonance (NMR) and crystal structures of the Zα domain reveal a helix-loop-helix structure similar to that found in many B-DNA binding proteins (53, 54). The functional significance of Zα is still unknown, but the domain has been suggested to direct (i)ADAR1 either to newly synthesized transcripts in the nucleus via binding of Z-DNA formed at transcription or to cytoplasmic viral dsRNA substrates (5, 19).
The nuclear pore complex (NPC) which spans the nuclear membrane generally allows free diffusion of proteins smaller than 40 to 60 kDa, while the translocation of larger proteins or protein complexes is controlled by nuclear localization signals (NLSs) and nuclear export signals (NESs). These elements are recognized by soluble import and export receptors, which can associate with nucleoporins in the NPC, thus leading to transport of cargo across the membrane (reviewed in references 16 and 40).
The classic NLS motif present in most nuclear proteins is composed of one or two clusters of basic residues, and it is recognized by the NLS receptor importin α, which attaches the cargo to the NPC through the import factor importin β. By a largely unknown mechanism, the cargo is translocated through the NPC, and in the nucleus, an interaction between importin β and the small GTPase Ran leads to dissociation of the complex (17, 50, 61). The most common NES motif in proteins is a stretch of characteristically spaced hydrophobic amino acids as first described for the human immunodeficiency virus type 1 Rev and PKI proteins (12, 62). In the nucleus, this motif is recognized by the nuclear export receptor CRM1, which forms a competent nuclear export complex together with RanGTP (13, 14, 47, 58). After translocation through the NPC, cytoplasmic factors, including Ran GTPase activating protein (RanGAP) and Ran binding proteins 1 and 2, promote hydrolysis of Ran-bound GTP and the cargo is released (1, 28).
In accordance with RNA editing preceding splicing, the deamination activity of somatic cells is nuclear (59, 64), and both ADAR2 and (c)ADAR1 have been demonstrated to localize to the nucleus (30, 46, 48). Two putative ADAR1 NLSs have been suggested, one in the Zα domain and one in the N terminus of (c)ADAR1 (22), but despite the suggested NLSs, a fraction of (i)ADAR1 localizes to the cytoplasm (48). These observations prompted us to search for a putative NES in (i)ADAR1, and in this study, we characterized a functional Rev-like NES in the N-terminal part of (i)ADAR1. Via the NES and at least two NLSs, (i)ADAR1 can shuttle between the nucleus and the cytoplasm.
MATERIALS AND METHODS
Plasmid constructs.
The open reading frame of full-length human ADAR1 (splice version a) cloned into the XhoI and XbaI restriction sites of pBluescript II KS(+) (Stratagene) was a kind gift from André Gerber and Walter Keller (46). The open reading frame was subcloned into pcDNA3 (Invitrogen) using the same restriction sites. Insertion of different fragments of this plasmid into the pEGFP C1 vector (Clontech) yielded four constructs with enhanced green fluorescent protein (EGFP) fused in frame to the N terminus of the ADAR1 fragments. The EcoRV-ApaI fragment of (i)ADAR1 inserted into the Ecl136II and ApaI sites of the vector generates a fusion protein with the full-length (i)ADAR1 sequence [EGFP-(i)ADAR1]. The EcoRV-BamHI and the BamHI-ApaI fragments of (i)ADAR1 inserted into the Ecl136II-BamHI and BglII-ApaI sites of the vector, respectively, generate proteins with EGFP fused to the 269 N-terminal amino acids (EGFP–N-term) or the 958 C-terminal amino acids of (i)ADAR1 (EGFP–C-term), respectively. The Escherichia coli expression plasmids were generated by digesting the PCR product from primers 5′ ACT GGA TCC ATG AAT CCG CGG CAG GG 3′ and 5′ ACT GAA TTC CTA GTC TAA AAA CTC AAG AGG 3′ on full-length (i)ADAR1 with BamHI and EcoRI and inserting the fragment into the equivalent sites in the pGEX-GTH vector, yielding the (i)ADAR1 1–295 sequence with an N-terminal glutathione S-transferase (GST) tag [pGEX-GTH-(i)ADAR1(1–295)]. Mutagenesis of the NES (L133A/I135A double mutation, referred to as NESmut herein) was achieved by a modified version of the megaprimer method (27). The N-terminal fragment was also subcloned into pET vectors for in vitro translation. An3-NESmut, encoding An3 with mutations in the NES, has been described elsewhere (1). The editing construct R2L, kindly provided by Trine E. Larsen, contains the sequence from exon 13 to exon 16 of rat GluR2 inserted into pcDNA3 (T. E. Larsen and J. Egebjerg, unpublished results).
Cell cultures.
The mouse neuroblastoma cell line N2A and the human fibrosarcoma cell line HT1080 were cultured at 37°C and 7.5% CO2 in Dulbecco's modified Eagle medium containing 5 and 10% fetal calf serum, respectively. Lipofectamine (Life Technologies) was used for transient transfections according to the manufacturer's instructions. Leptomycin B (LMB), a generous gift from Minoru Yoshida, was added to the medium to a final concentration of 2 nM.
Localization of endogenous (i)ADAR1.
HT1080 cells were seeded in 100-mm dishes, LMB was added 24 h later, and after another 6 h, the cells were harvested with 2 mM EDTA in phosphate-buffered saline (PBS) and washed. Cells were lysed for 5 min on ice in buffer L (0.5% NP-40 in PBS with protease inhibitors). Nuclei were sedimented at 4,500 × g through a sucrose cushion (24% sucrose–1% NP-40 in PBS) in a swing-out bucket rotor and resuspended in buffer L. The nuclei were extracted with a glass homogenizer (Douncer; Wheaton) and incubated with 200 U of Benzoase (Roche) at 0°C overnight. Nuclear and cytoplasmic fractions were resolved on a sodium dodecyl sulfate (SDS)–6% polyacrylamide gel, and Western blotting was performed with a primary polyclonal antibody against human ADAR1. Bands were visualized by standard techniques using a secondary antibody and enhanced chemiluminescence (Pharmacia Biotech). Gels run in parallel were stained with Coomassie blue to ensure equal loading, and to verify the separation of nuclear and cytoplasmic fractions, Western blots were stripped and reprobed with antibodies against topoisomerase type I (Genosys Biotechnologies Inc.) and the eukaryotic release factor 3a, which are constitutively nuclear and cytoplasmic proteins, respectively.
Oocyte injections.
Microinjection of proteins into oocytes, incubations, and extractions were performed as described previously (1). In vitro translations were done with the TNT coupled reticulocyte lysate system (Promega) according to the manufacturer's instructions. Proteins were dialyzed into PBS with 8.7% glycerol.
Protein expression.
pGEX-GTH-(i)ADAR1(1-295) and pGEX-GTH- (i)ADAR1(1-295)NESmut were expressed in the E. coli strain BL21(DE3), yielding the proteins GST-(i)ADAR1(1-295) and GST-(i)ADAR1(1-295)NESmut. Bacterial cultures were induced in early log phase with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h at 37°C. Bacteria recovered by centrifugation were resuspended in buffer A (300 mM NaCl, 50 mM Tris-HCl [pH 7.5], 5 mM 2-mercaptoethanol, protease inhibitors), lysed by a French press, and cleared by centrifugation. The supernatant was incubated with glutathione agarose (Pharmacia Biotech) for 1 h at 4°C, followed by washing with buffer A. The bound proteins were eluted with SG buffer (120 mM NaCl, 100 mM Tris-HCl [pH 8.0], 20 mM glutathione) for 30 min at 4°C. Top fractions were diluted into 50 mM phosphate (pH 6.6) and purified by fast protein liquid chromatography on an SP-Sepharose column using a 0 to 1,200 mM NaCl gradient. Finally, the proteins were dialyzed into storage buffer (150 mM NaCl, 50 mM Tris-HCl [pH 7.5], 1 mM dithiothreitol [DTT], 10% glycerol) and stored at −80°C. The purification of His-tagged CRM1, Ran, and Rna1p was performed as previously described (44).
Hydrolysis assay.
Four nanomolar Ran [γ-32P]GTP (5,000 Ci/mmol; Pharmacia Biotech) was incubated with 200 nM GST-(i)ADAR1(1–295) or GST- (i)ADAR1(1–295)NESmut and 0 to 400 nM CRM1 in reaction buffer (40 mM Tris-HCl [pH 8.0], 8 mM MgCl2, 1 mM DTT, 2 mM GTP, 1 mg of BSA per ml). Complex formation on ice was allowed for 15 min before addition of RanGAP to a final concentration of 5 nM, followed by incubation for 2 min at 25°C. Reactions were stopped by adding 1 ml of charcoal suspension (7% [wt/vol] charcoal, 10% [vol/vol] ethanol, 0.1 M HCl, 10 mM KH2PO4), and the released γ-32P was measured by counting on 0.7 ml of the supernatant.
Coprecipitation assay.
GST-(i)ADAR1(1–295) or GST-(i)ADAR1(1–295)NESmut (1 μM each) was incubated with 200 nM CRM1 in either the presence or absence of 4 μM RanGTP in TB buffer (20 mM HEPES-KOH [pH 7.3], 110 mM potassium acetate [KOAc], 5 mM NaOAc, 2 mM MgOAc, 1 mM DTT). After 30 min of incubation on ice, 20 μl of glutathione beads (Pharmacia Biotech) was added and incubation was continued for 60 min. Unbound protein was removed, beads were washed with 20 column volumes, and bound proteins eluted with SDS loading buffer were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE).
In vivo editing.
The editing construct R2L and various EGFP-(i)ADAR1-expressing constructs were cotransfected into N2A cells. Cytoplasmic RNA was purified 2 days after transfection with the RNeasy kit (Qiagen), and Superscript II reverse transcriptase (RT) (Life Technologies) was used for reverse transcription of the RNA with a GluR2 exon 16-specific primer (Ex16; 5′ CCG GGA CTT GTA GCA GAA CTC 3′) at 42°C for 40 min followed by 10 min at 45°C. The cDNA was amplified by PCR using Ex16 and a T7 primer. A PCR product with a length of 590 bp corresponding to spliced GluR2 RNA was purified after agarose gel electrophoresis and used to estimate the editing at the R/G site by extension of primer 5′ [32P]ACA CCT AAA GGA TCC TCA TT 3′ (radiolabeled using T4 polynucleotide kinase (New England BioLabs) with [γ-32P]ATP (3,000 Ci/mmol; Pharmacia Biotech) in the presence of 5 mM ddGTP and 0.5 mM dATP, dCTP, and dTTP or 5 mM ddATP and 0.5 mM dGTP, dCTP, and dTTP. The reactions were analyzed by gel electrophoresis on an 18% polyacrylamide gel, and the intensities of the bands were quantified by PhosphorImager. Extensions with ddATP showed that the A 5′ to the R/G site was deaminated only if the R/G site was edited; thus, the editing level at the R/G site was determined as the sum of the signals obtained from extension of the primer by one nucleotide (both A's deaminated) and two nucleotides (only the R/G site edited) relative to the sum of the signals from primers extended by one, two, and three nucleotides (the latter corresponding to unedited RNA).
The transfection efficiencies were estimated as the fraction of green fluorescing cells 48 h after transfection by visual inspection. Cells were lysed, and the amount of total protein loaded was estimated with the Bio-Rad protein assay (Bio-Rad Laboratories). The lysates were separated on SDS–6% polyacrylamide gels, and Western blotting with the ADAR1-specific antibody was performed as described above to estimate the expression levels of the constructs.
RESULTS
(i)ADAR1 is exported to the cytoplasm by an LMB-sensitive pathway.
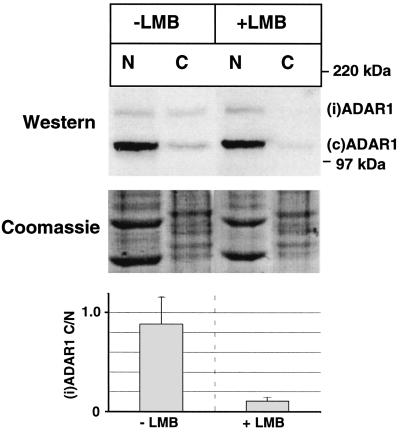
To investigate the localization of endogenous ADAR1, we performed SDS-PAGE and Western blotting of nuclear and cytoplasmic fractions of the human fibrosarcoma cell line HT1080 using an antibody specific for human ADAR1. It confirmed that (c)ADAR1 is a nuclear protein with only a minor fraction present in the cytoplasm, while (i)ADAR1 is equally distributed between the nucleus and the cytoplasm (Fig. 1, −LMB). To determine whether the observed cytoplasmic localization of (i)ADAR1 is a consequence of CRM1-mediated nuclear export, we treated the cells with the fungicide LMB, which binds CRM1 covalently and inhibits its function (13, 33). The LMB treatment reduced the cytoplasmic fraction of (i)ADAR1 to less than 10%, whereas (c)ADAR1 resided unaffected in the nucleus independent of LMB treatment (Fig. 1, +LMB). CRM1 is therefore a likely candidate as the nuclear export receptor for (i)ADAR1.
FIG. 1.
The localization of endogenous (i)ADAR1 is changed by LMB treatment. HT1080 cells were incubated for 6 h with or without LMB as indicated and separated into nuclear (N) and cytoplasmic (C) fractions. Equivalent amounts of total protein were subjected to SDS-PAGE and Western blotting with an antibody specific for human ADAR1 and visualized using a secondary antibody, enhanced chemiluminescence, and fluorography (top panel). Bands corresponding to (c)ADAR1 and (i)ADAR1 and a size marker are indicated on the right. Part of the samples were subjected to SDS-PAGE and stained with Coomassie blue to ensure equal loading (middle panel). The ratio of cytoplasmic (i)ADAR1 to nuclear (i)ADAR1 (C/N) was determined in six parallel experiments, and the average values and error thresholds are presented in the lower panel. Nuclear and cytoplasmic fractions were also probed with antibodies against topoisomerase type I and eukaryotic release factor 3a, representing constitutively nuclear and cytoplasmic proteins, respectively. This showed that there was no detectable leakage between the two compartments during the separation procedure and confirmed equal loading (data not shown).
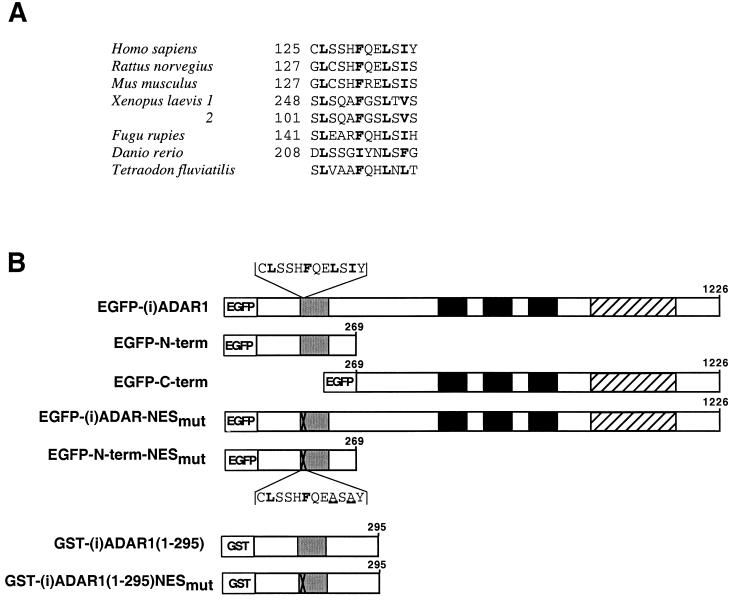
A phylogenetically conserved NES-like sequence in (i)ADAR1.
The difference in distribution of (c)ADAR1 and (i)ADAR1 is consistent with the existence of a NES in the (i)ADAR1 N terminus. The RNA-binding and catalytic domains of ADAR1 are highly conserved among mammals, Xenopus laevis, and fish, while the N-terminal regions are far more variable (56). Nonetheless, an examination of the N termini revealed that all ADAR1 species encompass the putative NES motif LXXXΦXXLXΦ (where X denotes any amino acid and Φ denotes one of the hydrophobic residues I, V, F or L) (Fig. 2A) that conforms to the consensus leucine-rich NES found in several other nucleocytoplasmic shuttling proteins that utilize CRM1 as their transport receptor (Fig. 2A). An N-terminal NES may therefore be phylogenetically conserved in (i)ADAR1.
FIG. 2.
(A) Alignment of putative ADAR1 NESs from different species. The conserved hydrophobic motif, characteristic of a NES protein exported by the CRM1 pathway, is highlighted in bold. The amino acid positions are indicated on the left; the 5′ end of the Tetraodon fluviatilis ADAR1 open reading frame has not been identified. The corresponding GenBank accession numbers are (from the top): NM 001111.2, U18942.2, AF052506.2, U88065.1, U88066.1, AF124048.1, AF124332.1, and AF124334.1. (B) Schematic representation of the functional domains in the EGFP-(i)ADAR1 proteins expressed in transient transfections and the GST-(i)ADAR1 fusion proteins used for in vitro binding studies. Numbers denote the amino acid position relative to the N terminus of human (i)ADAR1 (splice version a) (46), and the boxes indicate functionally characterized domains: grey box, Z-DNA binding domain Zα; black boxes, dsRNA binding domains; hatched box, catalytic domain. The sequence of the wild-type NES is shown above the EGFP-(i)ADAR1 construct, and the mutated NES sequence is denoted by a cross and is shown below the EGFP–N-term–NESmut construct. The mutated amino acids are underlined.
Characterization of an N-terminal NES in (i)ADAR1.
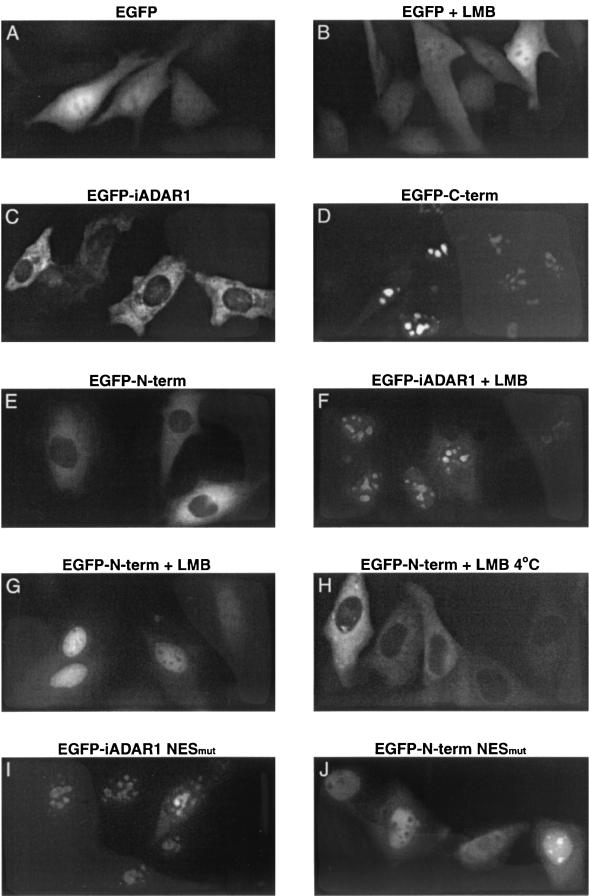
To characterize the ADAR1 localization signals further, we transiently transfected the mouse neuroblastoma cell line N2A with an expression plasmid encoding human ADAR1 fused to EGFP at the N terminus [EGFP-(i)ADAR1] (Fig. 2B) and examined the localization of the fusion protein by fluorescence microscopy (Fig. 3). The distribution of nuclear and cytoplasmic EGFP signals is quantitated in Table 1. EGFP-(i)ADAR1 localizes almost exclusively to the cytoplasm (Fig. 3C; Table 1), whereas EGFP alone localizes diffusely to both the cytoplasm and to the nucleus, except for the nucleoli (Fig. 3A).
FIG. 3.
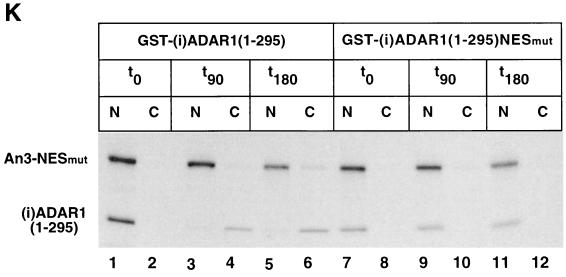
Subcellular localization of ADAR1 proteins. (A to J) Fluorescence microscope images (Olympus IX70) of living N2A cells were taken 24 h after transfections with the indicated EGFP-(i)ADAR1 constructs (Fig. 2B). (B, F, G, and H) Cells were treated with 2 nM LMB for 4 h. (H) Cells were transferred from 37 to 4°C immediately after the addition of LMB. The nucleocytoplasmic distribution is quantitated for panels C to J in Table 1. (K) Nuclear export of GST-(i)ADAR1(1–295) in X. laevis oocytes. X. laevis oocyte nuclei were injected with in vitro-translated GST-(i)ADAR1(1–295) or GST-(i)ADAR1(1–295)NESMut together with An3 NESmut protein as a control. Oocytes were dissected into nuclear (N) and cytoplasmic (C) fractions immediately (lanes 1, 2, 7, and 8) or after incubation for 90 min (lanes 3, 4, 9, and 10) or 180 min (lanes 5, 6, 11, and 12) and labeled proteins were visualized by SDS-PAGE and fluorography.
TABLE 1.
Localization of EGFP fusion proteins
| Constructa | % of cells with signals localized inb:
|
||
|---|---|---|---|
| Nucleus | Nucleus and cytoplasm | Cytoplasm | |
| EGFP-(i)ADAR1 (86) | 1 | 6 | 93 |
| EGFP–C-term (112) | 68 | 32 | 0 |
| EGFP–N-term (147) | 0 | 2 | 98 |
| EGFP-(i)ADAR1 + LMB (152) | 92 | 8 | 0 |
| EGFP–N-term + LMB (138) | 60 | 40 | 0 |
| EGFP–N-term + LMB, 4°C (220) | 0 | 6 | 94 |
| EGFP-(i)ADAR1-NESmut (140) | 54 | 44 | 2 |
| EGFP–N-term–NESmut (158) | 35 | 65 | 0 |
The protein expression plasmids and conditions correspond to Fig. 3C to J. The total numbers of counted cells expressing the proteins are in parentheses.
Nucleus, only a nuclear signal was clearly visible; nucleus and cytoplasm, a signal was visible in both nucleus and cytoplasm; cytoplasm, only a cytoplasmic signal was clearly visible.
Like EGFP-(i)ADAR1, EGFP fused to the N-terminal 269 amino acids (EGFP–N-term) (Fig. 2B) localizes almost exclusively to the cytoplasm (Fig. 3E; Table 1), while EGFP fused to the C-terminal 958 amino acids (EGFP–C-term)(Fig. 2B) is primarily nuclear (Fig. 3D; Table 1). In agreement with the observations made with endogenous ADAR1, these results indicate that (c)ADAR1 harbors a functional NLS and that (i)ADAR1 has an N-terminal NES counteracting the NLS.
To confirm that the cytoplasmic localization is mediated by the predicted NES, we mutated two of its hydrophobic residues, L133 and I135, to alanines in the constructs expressing full-length (i)ADAR1 [EGFP-(i)ADAR1-NESmut] (Fig. 2B) and the N-terminal fragment (EGFP-N-term-NESmut) (Fig. 2B). For both constructs, nuclear localization was evident in nearly all of the cells (Fig. 3I and J; Table 1), while nuclear localization was observed in <10% of cells transfected with the wild-type constructs (Fig. 3C and E; Table 1). Since mutations in the proposed NES change the localization of EGFP fusion proteins from cytoplasmic to nuclear, the sequence is likely to direct the nuclear export of (i)ADAR1.
To determine the nuclear export pathway of the transiently expressed EGFP-(i)ADAR1, we examined the effect of treating the transfected cells with LMB. After 4 h of LMB-treatment, nuclear accumulation was evident in nearly all cells transfected with a construct encoding either EGFP-(i)ADAR1 or EGFP–N-term (Fig. 3F and G; Table 1), whereas LMB had no effect on EGFP localization (Fig. 3B). This implies that the observed cytoplasmic localization of (i)ADAR1 is dependent on functional CRM1 and indicates that (i)ADAR1 can shuttle between the nucleus and the cytoplasm.
The ability of the characterized NES to directly mediate export from the nucleus was addressed by injecting in vitro-translated GST-(i)ADAR1(1–295) or GST-(i)ADAR1(1– 295)NESmut into the nuclei of X. laevis oocytes and dissecting the oocytes into nuclear and cytoplasmic fractions at different incubation times (Fig. 3K). After 90 min, GST-(i)ADAR1(1–295) had accumulated in the cytoplasm (Fig. 3K, lanes 1 to 4), while GST-(i)ADAR1(1–295)NESmut remained exclusively in the nucleus (Fig. 3K, lanes 7 to 10). Even after prolonged incubation, GST-(i)ADAR1(1–295)NESmut did not show any export activity (Fig. 3K, lanes 11 and 12). The An3 protein (1) with a mutated NES served as a negative control.
The N terminus of (i)ADAR1 is thus able to directly access the CRM1 export pathway.
In vitro interaction between (i)ADAR1 and CRM1.
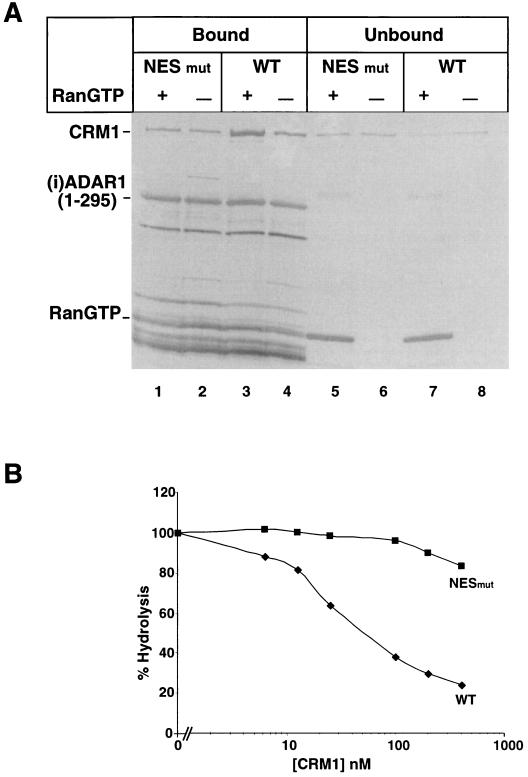
To demonstrate a direct interaction between (i)ADAR1 and CRM1, we tested if the N-terminal fragment of (i)ADAR1 could form a complex with CRM1 and RanGTP in vitro. Protein fragments with the N-terminal 295 amino acids of (i)ADAR1, carrying either wild-type NES or a NES with the L133A and I135A mutations, were expressed as GST fusion proteins [GST-(i)ADAR1(1–295) and GST-(i)ADAR1(1–295)NESmut, respectively] (Fig. 2B) in E. coli, and the purified proteins were incubated with recombinant CRM1 in either the presence or absence of RanGTP. ADAR1 and proteins associated with ADAR1 were precipitated from solution by the addition of glutathione agarose beads, and after washing, bound proteins were eluted and analyzed by SDS-PAGE (Fig. 4A). In the absence of RanGTP, only small amounts of CRM1 were retained, but the addition of RanGTP allowed CRM1 to bind ADAR1 (Fig. 4A, lanes 3 and 4). This interaction was sensitive to mutations in the ADAR1 NES, as this mutant only bound residual amounts of CRM1 (Fig. 4A, lanes 1 and 2).
FIG. 4.
In vitro interaction between CRM1 and the (i)ADAR1 NES. (A) Coprecipitation of CRM1 by the N-terminal 295 amino acids of (i)ADAR1. 1 GST-(i)ADAR1(1–295) or GST-(i)ADAR1(1–295) NESmut (1 μM) was incubated with 200 nM CRM1 in either the presence or absence of 4 μM RanGTP. GST-(i)ADAR1 and bound proteins were precipited by addition of glutathione beads, and, after washing, bound proteins were resolved by SDS-PAGE. The identities of the bands are indicated on the left. (B) Binding of a NES to CRM1 occurs in a tripartite complex with RanGTP. The GTPase activity of Ran is blocked when it is complexed to NES-CRM1 and can therefore be used as a measure of NES binding (1, 44). This assay was used to assess the capacity of GST-(i)ADAR1(1–295) (diamonds) and GST-(i)ADAR1(1–295)NESmut (squares) proteins to form a tripartite complex. The degree of hydrolysis was measured as the fraction of released γ32P 2 min after the addition of 5 nM RanGAP to 200 nM of the (i)ADAR1 fragments preincubated with 0 to 400 nM CRM1.
To estimate the Kd of complex formation, we used an assay based on the observation that RanGAP-induced GTP hydrolysis by Ran is blocked when RanGTP is in complex with CRM1 and a NES substrate (1, 4). Ran charged with [γ32P]GTP was incubated with GST-(i)ADAR1(1–295) or GST-(i)ADAR1(1–295)NESmut and increasing concentrations of CRM1. The amount of complexed RanGTP was measured as the release of γ-32P upon addition of RanGAP, and the level of complex formation was determined from this (Fig. 4B). From the 50% hydrolysis, we estimate the Kd between CRM1 and wild-type (i)ADAR1 NES to be 45 nM, while mutations in the NES lower the binding substantially (Fig. 4B). The above results support the idea that CRM1 binds specifically to the suggested NES in (i)ADAR1.
NLSs in (i)ADAR1.
The nuclear accumulations of EGFP–N-term in LMB-treated cells and EGFP–N-term–NESmut (Fig. 3J and G) suggest the presence of an NLS in the N-terminal fragment. Alternatively, the expected molecular mass of about 60 kDa (including the EGFP) may permit diffusion through the NPC, and the protein might then be retained in the nucleus, e.g., via the Zα domain. To distinguish between the models, EGFP–N-term was transiently expressed in N2A cells and transferred to 4°C immediately after the addition of LMB (Fig. 3H). Receptor-mediated nuclear import and export are energy-dependent processes that will be blocked by this shift in temperature, while diffusion is unaffected (42). At the low temperature, LMB treatment had no effect, and EGFP–N-term remained cytoplasmic (compare Fig. 3G and H; Table 1), but when the temperature was raised to 37°C after the 4°C treatment, EGFP–N-term became nuclear (data not shown). This strongly suggests that an NLS within the N-terminal 269 amino acids directs active import of the protein. Moreover, the nuclear localization of the C-terminal ADAR1 fragment (Fig. 3D), which is too large (>130 kDa) to diffuse across the nuclear membrane, implies that an additional NLS is located in this part of the protein.
RNA editing by ADAR1 is regulated by localization.
Since site-specific editing by adenosine deamination in all studied cases precedes splicing, it must occur in the nucleus. One well-studied example is the most 3′ adenosine of the GluR2 exon 13 (the R/G site), which is differentially edited in the mammalian brain, and in vitro as well as knockout studies suggest that it is a substrate for both ADAR1 and ADAR2 (34, 38, 60). To determine the nuclear editing activity of the ADAR1 fusion proteins, we expressed a splicing- and editing-competent RNA that contained the sequence of GluR2 pre-mRNA from exon 13 to exon 16 (R2L). N2A cells were transiently cotransfected with R2L and various EGFP-(i)ADAR1 constructs, and after 48 h cytoplasmic RNA was purified and amplified by RT-PCR using R2L-specific primers. The PCR product corresponding to spliced RNA was gel purified, and the editing level at the R/G site was determined by primer extension (Fig. 5).
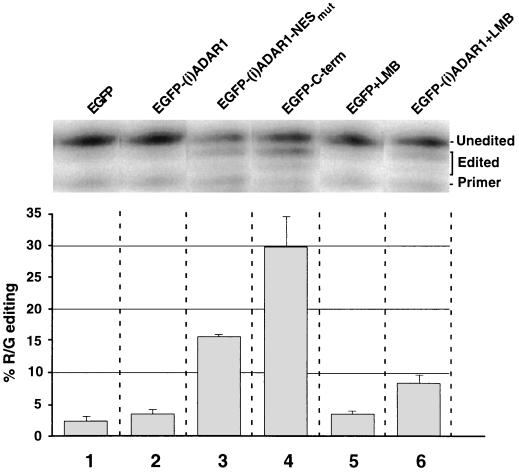
FIG. 5.
Nuclear RNA editing by ADAR1. N2A cells were cotransfected with a minimal editing and splicing construct, R2L, encompassing the R/G site of the rat GluR2 pre-mRNA and expression plasmids, encoding EGFP-(i)ADAR1 fusion constructs. +LMB, LMB was added to 2 nM 18 h posttransfection (lanes 5 and 6). At 48 h after transfection, cytoplasmic RNA was purified and subjected to RT-PCR, and the editing level at the R/G site was determined by primer extension. The editing levels were calculated from the sum of the intensities of the two bands (marked “edited”) corresponding to deamination of one or two adenosines at the R/G site relative to the signal from the band representing unedited RNA (marked “unedited”) (see Materials and Methods). The signals were quantitated by phosphorimaging, and the averages and standard deviations of the result from three parallel experiments are shown.
The GluR2 construct was edited <5% in cells cotransfected with EGFP alone or with EGFP fused to (i)ADAR1 (Fig. 5, lanes 1 and 2), while cotransfection with EGFP-(i)ADAR1-NESmut raised the level to 16% (Fig. 5, lane 3). This indicates that the fusion proteins are catalytically active but that the cytoplasmic localization of EGFP-(i)ADAR1 abolishes nuclear editing. Removal of the N terminus, including the NES, in EGFP–C-term further raised the editing level to 30% (Fig. 5, lane 4). Western blot analysis of transfected cells showed that the concentrations of the transiently expressed ADAR1 proteins were more than 100-fold higher than the concentration of endogenous ADAR1, explaining why endogenous (c)ADAR1 edits the GluR2 construct to only a very low degree (data not shown).
To ensure that the NES mutation was not directly responsible for the change in catalytic activity, we added LMB 18 h after transfection with EGFP-(i)ADAR1. The nuclear editing level was raised to 8% (Fig. 5, lane 6), while the drug had no effect on the level in EGFP-transfected cells (Fig. 5, lane 5). This confirms that EGFP-(i)ADAR1 is catalytically active if retained in the nucleus but exhibits a reduced nuclear editing level because of its primarily cytoplasmic localization.
DISCUSSION
All known examples of site-specific RNA editing by adenosine deamination depend on RNA structures formed between exon and intron segments in the pre-mRNA, and the process has therefore been regarded as strictly nuclear. However, in this study we have demonstrated that the interferon-induced form of human ADAR1 is exported from the nucleus via the NES sequence LSSHFQELSI (conserved hydrophobic amino acids are in boldface) in the N terminus. The sequence conforms to the consensus leucine-rich-type NES whose export receptor is CRM1. Four lines of evidence suggest that the NES we have characterized in (i)ADAR1 is functional and that CRM1 is its cognate export receptor. (i) Treatment of cells with LMB, which specifically targets CRM1, induces nuclear accumulation of (i)ADAR1 proteins. This was demonstrated for both endogenous (i)ADAR1 and transiently expressed EGFP-(i)ADAR1. (ii) Mutations in the NES also result in nuclear accumulation. (iii) In vitro CRM1 binds specifically to a fragment of (i)ADAR1 containing the wild-type NES in the presence of RanGTP, and mutations in the NES lower the binding substantially. (iv) The N terminus of (i)ADAR1 is rapidly exported to the cytoplasm when injected into the nuclei of X. laevis oocytes, while mutations in the NES abolish this export.
The number and the spacing of hydrophobic residues in the human (i)ADAR1 NES described here are conserved in ADAR1 from different organisms, and a study of the ADAR1 localization in X. laevis supports the existence of a functional N-terminal NES. Full-length ADAR1 in X. laevis localizes mainly to the cytoplasm, but when approximately 30 kDa of the N terminus, including the putative NES, are proteolytically removed, the protein becomes nuclear (10).
The NES that we have characterized overlaps with the N-terminal part of the Z-DNA binding domain, Zα, which has been studied by NMR and X-ray crystallography, and in the NMR structure, the entire NES region is included (53, 54). Caution must be taken when the structures of protein termini in NMR studies are assigned, but the suggested (i)ADAR1 NES folds into a α-helix-like structure, which is similar to what has been described for other putative NESs, including p53, IκB, the 14-3-3 peptide, Stat 1, and β-actin (3, 24–26, 43, 51). As previously noted by Rittinger et al. for the 14-3-3 peptide (51), the first three hydrophobic residues can cluster to one side of an aliphatic helix and provide a potential hydrophobic binding platform for CRM1, whereas the last C-terminal residue is located at the opposite side of the helix.
Given the size of (c)ADAR1, its nuclear localization must depend on active import and our transfection experiments with the N-terminal part of (i)ADAR1 demonstrate an NLS sequence in this region. This is based on the observation that inhibition of CRM1 leads to nuclear accumulation of EGFP–N-term at 37°C, but if active import is also inhibited by lowering the temperature, EGFP–N-term remains in the cytoplasm. We therefore conclude that (i)ADAR1 is likely to shuttle between the nucleus and the cytoplasm via a NES and at least two NLSs.
The observation that both EGFP-(i)ADAR1-NESmut and EGFP–C-term are capable of editing the R/G site raises the possibility that the N terminus mainly serves to target (i)ADAR1 to different substrates than those of (c)ADAR1 and ADAR2. One scenario is that nuclear export of (i)ADAR1 is used as a means of modulating the nuclear deaminase activity. Regulating the activity of a protein by controlled compartmentalization is a fast mechanism utilized in, e.g., signal transduction and cell cycle regulation (3, 63). Alternatively, (i)ADAR1 may have specific cytoplasmic targets, such as dsRNA viruses (reviewed in reference 7). Although further experiments are required to test for such an effect, this would clearly explain why (i)ADAR1 contains an NES.
ACKNOWLEDGMENTS
H.P. and J.N. contributed equally to this work.
We thank Marie Öhman, Brenda L. Bass, and Herbert L. Ley III for the ADAR1-specific antibody, Just Justesen for the eRF3a-specific antibody, André Gerber and Walter Keller for the ADAR1 plasmid, Iain W. Mattaj for the CRM1 plasmid, Dirk Görlich for the Ran and Rna1p plasmids, Trine E. Larsen for the R2L R/G editing construct, and Minoru Yoshida for a generous supply of LMB. We are grateful to Ray Brown for critical reading of the manuscript. Finally, we are indebted to Rita Rosendahl for excellent technical assistance.
The work was supported in part by grants from the Danish National Science and Medical Research Councils, The Carlsberg Foundation, Novo Nordisk Foundation, and Karen Elise Jensen Foundation. H.P., J.N., and C.K.D. were supported by the University of Aarhus.
REFERENCES
- 1.Askjaer P, Bachi A, Wilm M, Bischoff F R, Weeks D L, Ogniewski V, Ohno M, Niehrs C, Kjems J, Mattaj I W, Fornerod M. RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol Cell Biol. 1999;19:6276–6285. doi: 10.1128/mcb.19.9.6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bass B L. RNA editing and hypermutation by adenosine deamination. Trends Biochem Sci. 1997;22:157–162. doi: 10.1016/s0968-0004(97)01035-9. [DOI] [PubMed] [Google Scholar]
- 3.Begitt A, Meyer T, van Rossum M, Vinkemeier U. Nucleocytoplasmic translocation of Stat1 is regulated by a leucine-rich export signal in the coiled-coil domain. Proc Natl Acad Sci USA. 2000;97:10418–10423. doi: 10.1073/pnas.190318397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bischoff F R, Görlich D. RanBP1 is crucial for the release of RanGTP from importin beta-related nuclear transport factors. FEBS Lett. 1997;419:249–254. doi: 10.1016/s0014-5793(97)01467-1. [DOI] [PubMed] [Google Scholar]
- 5.Brown B A, Lowenhaupt K, Wilbert C M, Hanlon E B, Rich A. The Zα domain of the editing enzyme dsRNA adenosine deaminase binds left-handed Z-RNA as well as Z-DNA. Proc Natl Acad Sci USA. 2000;97:13532–13536. doi: 10.1073/pnas.240464097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns C M, Chu H, Rueter S M, Hutchinson L K, Canton H, Sanders-Bush E, Emeson R B. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 7.Cattaneo R. Biased (A→I) hypermutation of animal RNA virus genomes. Curr Opin Genet Dev. 1994;4:895–900. doi: 10.1016/0959-437x(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 8.Dabiri G A, Lai F, Drakas R A, Nishikura K. Editing of the GluR-B ion channel RNA in vitro by recombinant double-stranded RNA adenosine deaminase. EMBO J. 1996;15:34–45. [PMC free article] [PubMed] [Google Scholar]
- 9.Der S D, Zhou A, Williams B R, Silverman R H. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckmann C R, Jantsch M F. The RNA-editing enzyme ADAR1 is localized to the nascent ribonucleoprotein matrix on Xenopus lampbrush chromosomes but specifically associates with an atypical loop. J Cell Biol. 1999;144:603–615. doi: 10.1083/jcb.144.4.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egebjerg J, Kukekov V, Heinemann S F. Intron sequence directs RNA editing of the glutamate receptor subunit GluR2 coding sequence. Proc Natl Acad Sci USA. 1994;91:10270–10274. doi: 10.1073/pnas.91.22.10270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 13.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 14.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 15.George C X, Samuel C E. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc Natl Acad Sci USA. 1999;96:4621–4626. doi: 10.1073/pnas.96.8.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 17.Görlich D, Pante N, Kutay U, Aebi U, Bischoff F R. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 18.Hanrahan C J, Palladino M J, Ganetzky B, Reenan R A. RNA editing of the Drosophila para Na(+) channel transcript. Evolutionary conservation and developmental regulation. Genetics. 2000;155:1149–1160. doi: 10.1093/genetics/155.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herbert A, Alfken J, Kim Y G, Mian I S, Nishikura K, Rich A. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc Natl Acad Sci USA. 1997;94:8421–8426. doi: 10.1073/pnas.94.16.8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higuchi M, Maas S, Single F N, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg P H. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 21.Higuchi M, Single F N, Kohler M, Sommer B, Sprengel R, Seeburg P H. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 22.Hough R F, Bass B L. Analysis of Xenopus dsRNA adenosine deaminase cDNAs reveals similarities to DNA methyltransferases. RNA. 1997;3:356–370. [PMC free article] [PubMed] [Google Scholar]
- 23.Hurst S R, Hough R F, Aruscavage P J, Bass B L. Deamination of mammalian glutamate receptor RNA by Xenopus dsRNA adenosine deaminase: similarities to in vivo RNA editing. RNA. 1995;1:1051–1060. [PMC free article] [PubMed] [Google Scholar]
- 24.Huxford T, Huang D B, Malek S, Ghosh G. The crystal structure of the IκBα/NF-κB complex reveals mechanisms of NF-κB inactivation. Cell. 1998;95:759–770. doi: 10.1016/s0092-8674(00)81699-2. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs M D, Harrison S C. Structure of an IκBα/NF-κB complex. Cell. 1998;95:749–758. doi: 10.1016/s0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- 26.Jeffrey P D, Gorina S, Pavletich N P. Crystal structure of the tetramerization domain of the p53 tumor suppressor at 1.7 angstroms. Science. 1995;267:1498–1502. doi: 10.1126/science.7878469. [DOI] [PubMed] [Google Scholar]
- 27.Ke S H, Madison E L. Rapid and efficient site-directed mutagenesis by single-tube ‘megaprimer’ PCR method. Nucleic Acids Res. 1997;25:3371–3372. doi: 10.1093/nar/25.16.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kehlenbach R H, Dickmanns A, Kehlenbach A, Guan T, Gerace L. A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J Cell Biol. 1999;145:645–657. doi: 10.1083/jcb.145.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keller W, Wolf J, Gerber A. Editing of messenger RNA precursors and of tRNAs by adenosine to inosine conversion. FEBS Lett. 1999;452:71–76. doi: 10.1016/s0014-5793(99)00590-6. [DOI] [PubMed] [Google Scholar]
- 30.Kim U, Garner T L, Sanford T, Speicher D, Murray J M, Nishikura K. Purification and characterization of double-stranded RNA adenosine deaminase from bovine nuclear extracts. J Biol Chem. 1994;269:13480–13489. [PubMed] [Google Scholar]
- 31.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc Natl Acad Sci USA. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohler M, Burnashev N, Sakmann B, Seeburg P H. Determinants of Ca2+ permeability in both TM1 and TM2 of high affinity kainate receptor channels: diversity by RNA editing. Neuron. 1993;10:491–500. doi: 10.1016/0896-6273(93)90336-p. [DOI] [PubMed] [Google Scholar]
- 33.Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner E P, Wolff B, Yoshida M, Horinouchi S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai F, Chen C X, Carter K C, Nishikura K. Editing of glutamate receptor B subunit ion channel RNAs by four alternatively spliced DRADA2 double-stranded RNA adenosine deaminases. Mol Cell Biol. 1997;17:2413–2424. doi: 10.1128/mcb.17.5.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Emeson R B, Samuel C E. Serotonin-2C receptor pre-mRNA editing in rat brain and in vitro by splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase ADAR1. J Biol Chem. 1999;274:18351–18358. doi: 10.1074/jbc.274.26.18351. [DOI] [PubMed] [Google Scholar]
- 36.Liu Y, George C X, Patterson J B, Samuel C E. Functionally distinct double-stranded RNA-binding domains associated with alternative splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase. J Biol Chem. 1997;272:4419–4428. doi: 10.1074/jbc.272.7.4419. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y, Samuel C E. Editing of glutamate receptor subunit B pre-mRNA by splice-site variants of interferon-inducible double-stranded RNA-specific adenosine deaminase ADAR1. J Biol Chem. 1999;274:5070–5077. doi: 10.1074/jbc.274.8.5070. [DOI] [PubMed] [Google Scholar]
- 38.Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger J R, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg P H. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 39.Maas S, Melcher T, Herb A, Seeburg P H, Keller W, Krause S, Higuchi M, O'Connell M A. Structural requirements for RNA editing in glutamate receptor pre-mRNAs by recombinant double-stranded RNA adenosine deaminase. J Biol Chem. 1996;271:12221–12226. doi: 10.1074/jbc.271.21.12221. [DOI] [PubMed] [Google Scholar]
- 40.Mattaj I W, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 41.Melcher T, Maas S, Herb A, Sprengel R, Seeburg P H, Higuchi M. A mammalian RNA editing enzyme. Nature. 1996;379:460–464. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- 42.Michael W M, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 43.Mittl P R, Chene P, Grutter M G. Crystallization and structure solution of p53 (residues 326-356) by molecular replacement using an NMR model as template. Acta Crystallogr D. 1998;54(Pt. 1):86–89. doi: 10.1107/s0907444997006550. [DOI] [PubMed] [Google Scholar]
- 44.Nilsson J, Askjaer P, Kjems J. A role for the basic patch and the C terminus of RanGTP in regulating the dynamic interactions with importin beta, CRM1 and RanBP1. J Mol Biol. 2001;305:231–243. doi: 10.1006/jmbi.2000.4313. [DOI] [PubMed] [Google Scholar]
- 45.O'Connell M A, Gerber A, Keller W. Purification of human double-stranded RNA-specific editase 1 (hRED1) involved in editing of brain glutamate receptor B pre-mRNA. J Biol Chem. 1997;272:473–478. doi: 10.1074/jbc.272.1.473. [DOI] [PubMed] [Google Scholar]
- 46.O'Connell M A, Krause S, Higuchi M, Hsuan J J, Totty N F, Jenny A, Keller W. Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase. Mol Cell Biol. 1995;15:1389–1397. doi: 10.1128/mcb.15.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 48.Patterson J B, Samuel C E. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol Cell Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patton D E, Silva T, Bezanilla F. RNA editing generates a diverse array of transcripts encoding squid Kv2 K+ channels with altered functional properties. Neuron. 1997;19:711–722. doi: 10.1016/s0896-6273(00)80383-9. [DOI] [PubMed] [Google Scholar]
- 50.Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 51.Rittinger K, Budman J, Xu J, Volinia S, Cantley L C, Smerdon S J, Gamblin S J, Yaffe M B. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol Cell. 1999;4:153–166. doi: 10.1016/s1097-2765(00)80363-9. [DOI] [PubMed] [Google Scholar]
- 52.Rueter S M, Dawson T R, Emeson R B. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- 53.Schade M, Turner C J, Lowenhaupt K, Rich A, Herbert A. Structure-function analysis of the Z-DNA-binding domain Z-α of dsRNA adenosine deaminase type I reveals similarity to the (alpha + beta) family of helix-turn-helix proteins. EMBO J. 1999;18:470–479. doi: 10.1093/emboj/18.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwartz T, Rould M A, Lowenhaupt K, Herbert A, Rich A. Crystal structure of the Z-α domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science. 1999;284:1841–1845. doi: 10.1126/science.284.5421.1841. [DOI] [PubMed] [Google Scholar]
- 55.Semenov E P, Pak W L. Diversification of Drosophila chloride channel gene by multiple posttranscriptional mRNA modifications. J Neurochem. 1999;72:66–72. doi: 10.1046/j.1471-4159.1999.0720066.x. [DOI] [PubMed] [Google Scholar]
- 56.Slavov D, Crnogorac-Jurcevic T, Clark M, Gardiner K. Comparative analysis of the DRADA A-to-I RNA editing gene from mammals, pufferfish and zebrafish. Gene. 2000;250:53–60. doi: 10.1016/s0378-1119(00)00175-x. [DOI] [PubMed] [Google Scholar]
- 57.Smith L A, Peixoto A A, Hall J C. RNA editing in the Drosophila DMCA1A calcium-channel alpha 1 subunit transcript. J Neurogenet. 1998;12:227–240. doi: 10.3109/01677069809108560. [DOI] [PubMed] [Google Scholar]
- 58.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 59.Wagner R W, Yoo C, Wrabetz L, Kamholz J, Buchhalter J, Hassan N F, Khalili K, Kim S U, Perussia B, McMorris F A, Nishikura K. Double-stranded RNA unwinding and modifying activity is detected ubiquitously in primary tissues and cell lines. Mol Cell Biol. 1990;10:5586–5590. doi: 10.1128/mcb.10.10.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Q, Khillan J, Gadue P, Nishikura K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- 61.Weis K, Dingwall C, Lamond A I. Characterization of the nuclear protein import mechanism using Ran mutants with altered nucleotide binding specificities. EMBO J. 1996;15:7120–7128. [PMC free article] [PubMed] [Google Scholar]
- 62.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 63.Yang J, Bardes E S, Moore J D, Brennan J, Powers M A, Kornbluth S. Control of cyclin B1 localization through regulated binding of the nuclear export factor CRM1. Genes Dev. 1998;12:2131–2143. doi: 10.1101/gad.12.14.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang J H, Sklar P, Axel R, Maniatis T. Purification and characterization of a human RNA adenosine deaminase for glutamate receptor B pre-mRNA editing. Proc Natl Acad Sci USA. 1997;94:4354–4359. doi: 10.1073/pnas.94.9.4354. [DOI] [PMC free article] [PubMed] [Google Scholar]