Abstract
Janus kinase/signal transduction and transcription activation (JAK/STAT) pathways were originally thought to be intracellular signaling pathways that mediate cytokine signals in mammals. Existing studies show that the JAK/STAT pathway regulates the downstream signaling of numerous membrane proteins such as such as G-protein-associated receptors, integrins and so on. Mounting evidence shows that the JAK/STAT pathways play an important role in human disease pathology and pharmacological mechanism. The JAK/STAT pathways are related to aspects of all aspects of the immune system function, such as fighting infection, maintaining immune tolerance, strengthening barrier function, and cancer prevention, which are all important factors involved in immune response. In addition, the JAK/STAT pathways play an important role in extracellular mechanistic signaling and might be an important mediator of mechanistic signals that influence disease progression, immune environment. Therefore, it is important to understand the mechanism of the JAK/STAT pathways, which provides ideas for us to design more drugs targeting diseases based on the JAK/STAT pathway. In this review, we discuss the role of the JAK/STAT pathway in mechanistic signaling, disease progression, immune environment, and therapeutic targets.
Keywords: JAK/STAT, disease progression, immune environment, mechanotransduction, therapeutic targets
1 Introduction
Studies have shown that activation of the JAK/STAT pathway promotes the development and progression of various diseases, including various inflammatory diseases, lymphomas, leukemias, various solid tumors, and so on. Their relationship and mechanisms have become crucial for the treatment of various diseases. The JAK/STAT pathway is an important cascade of signal transduction for multiple growth factors and cytokines, which regulates gene expression and cell activation, proliferation, and differentiation (Reddy et al., 2019; Xin et al., 2020; Awasthi et al., 2021).
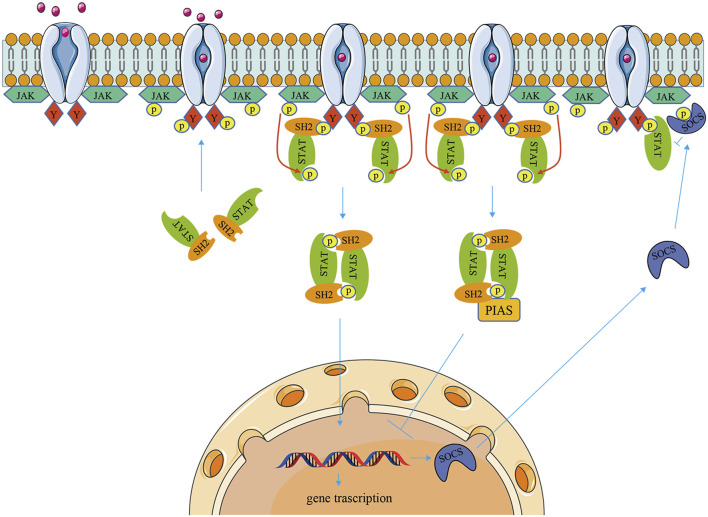
The JAK/STAT pathway has three components: cellular receptors, JAK protein, and STAT protein. The JAK family is a group of non-transmembrane tyrosine kinases, which is mainly composed of four members: JAK1, JAK2, JAK3, and TYK2 with molecular weights ranging from 120 to 140 kDa. JAK1, JAK2, and TYK2 are ubiquitous, while JAK3 is mainly expressed in hematopoietic cells (Villarino et al., 2015; Banerjee et al., 2017). There are seven members of the STAT family in a mammal: STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B, and STAT6 (Banerjee et al., 2017). Each member of the STAT family can be activated by a variety of cytokines and associated JAKs (O'Shea et al., 2015). First, cytokines bind to the corresponding transmembrane receptors and induce dimerization then activating JAK kinases couple to and phosphorylate the receptors. Second, the tyrosine residues on the catalytic domain of the receptor are phosphorylated to form a docking site in which STAT proteins with SH2 domains are recruited to this docking site, and STATs are phosphorylated and form homodimers or heterodimers. Finally, dimerized STATs dissociate from receptors and translocate into the nucleus, where they bind to DNA-binding sites and regulate gene transcription (Murray, 2007; Muller, 2019). Therefore, the activation of the JAK/STAT signaling cascade pathway is necessarily influenced by upstream extracellular cytokines and downstream JAK/STAT family protein types. For example, IFN-α/β activate STAT1, STAT2, and STAT4 via JAK1 and TYK2, whereas IFN-γ actives STAT1 or STAT5 via JAK1 and JAK2. IL-6 and IL-11 activate STAT1, STAT3 via JAK1, JAK2, and TYK2, but IL-12 and IL-23 activate STAT3 and STAT4 via JAK2 and TYK2. At the same time, STAT can be directly activated independently of JAK pathways, such as epidermal growth factor (EGF), platelet-derived growth factor (PDGF), and mitogen-activated protein kinase (MAPK). In addition, the JAK/STAT pathway receives regulation by multiple mechanisms, including that PIAS inhibits gene transcription by directly binding to STAT dimers and thereby blocking STAT binding to DNA. And SOCS protein can negatively regulate the JAK/STAT signaling cascade by inhibiting JAK activity, competing with STAT to bind phosphorylation sites on cytokine receptors, and inducing STAT proteasomal degradation (Boyle et al., 2009; La Manna et al., 2021) (Figure 1).
FIGURE 1.
The signaling mechanism of the JAK/STAT pathway. When cytokines bind to transmembrane receptors, receptor-associated JAKs are activated, which then phosphorylate STAT proteins. Activation of STAT proteins forms homo- or heterodimers that are transferred to the nucleus and regulate gene transcription. The JAK/STAT pathway is negatively regulated through SOCS and PIAS.
2 JAK/STAT pathway in disease progression
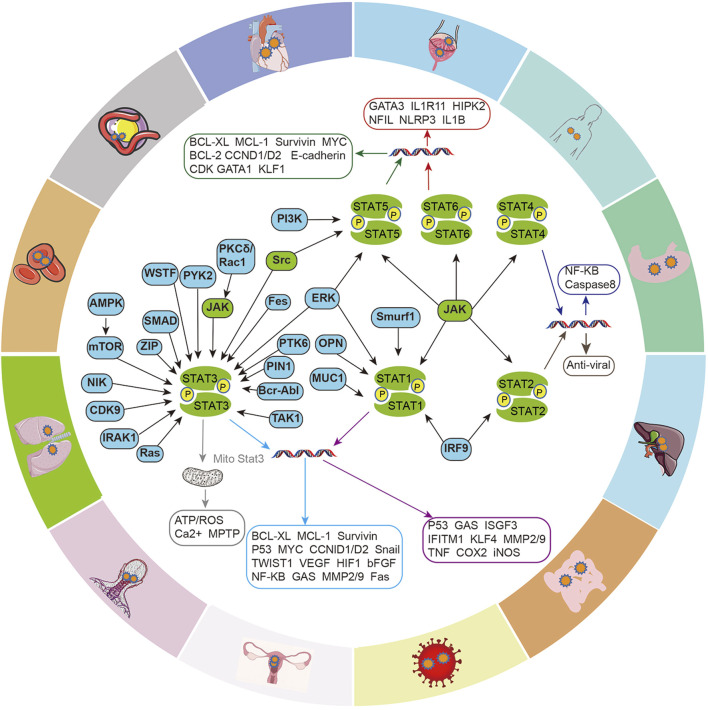
The JAK/STAT signaling axis is a central pathway that mediates the cellular inflammation response, and carcinogenesis, and participates in the transduction of cellular physiological signals, such as renin-angiotensin signaling, insulin-like growth factor (IGF-IR) signaling (Figure 2). STAT promotes the transcriptional activation of target genes in response to specific extracellular stimuli (including cytokines, growth factors, and other agents) through tyrosine phosphorylation-mediated activation, most of which is mediated by JAKs, but with the interaction of multiple intracellular signaling proteins, then affecting key cellular processes, including differentiation, proliferation, survival and functional activation, which in turn are involved in the development of various diseases.
FIGURE 2.
Intracellular signaling crosstalk of the JAK/STAT pathway. Different signaling proteins activate different STATs and induce the transcription and expression of genes for different cellular functions, including cell cycle, apoptosis, cell proliferation, epithelial-mesenchymal transition (EMT), angiogenesis, inflammatory factor production, etc. which in turn are involved in the development of various diseases.
2.1 JAK/STAT pathway in oncopathology
In oncology research, JAK/STAT is attracting more and more attention, increasingly studies show that STAT3 is constitutive activated in tumors and involved in cellular carcinogenesis (Teng et al., 2014). The pathway is involved in a variety of malignant tumors, including leukemia (Dufva et al., 2018), multiple myeloma (Hu and Hu, 2018), lymphoma (von Hoff et al., 2019), head and neck cancer (Sen et al., 2015), colon cancer (Neradugomma et al., 2014), gastric cancer (Mimura et al., 2018), hepatocellular carcinoma (Ren et al., 2019), pancreatic cancer (Chen et al., 2018), breast cancer (Shao et al., 2021), melanoma (Guenterberg et al., 2010), ovarian cancer (Bagratuni et al., 2020), lung cancer (Patel et al., 2019) and prostate cancer (Kroon et al., 2013).
Cancer stem cells (CSCs), a subpopulation of tumor cells with stem cell properties that self-renew and give rise to a variety of more differentiated cells, are a key driver of tumor progression (Dean et al., 2005; Dorritie et al., 2014; Avgustinova and Benitah, 2016). Studies have shown that cancer stem cells have been proposed to explain the development of cancer and resistance to treatment, and activation of the JAK/STAT signalling pathway, or induction of other signals that interact with the JAK/STAT pathway, can promote the production and acquisition of drug resistance by cancer stem cells (Shibue and Weinberg, 2017; Dongre and Weinberg, 2019). Researches show that STAT3 is critical for tumor transformation downstream of oncogenes Src and Ras. Src induces tyrosine phosphorylation and transcriptional activity of STAT3, and Ras phosphorylates STAT3 at Serine 727, which is required for localization to mitochondria. In turn, mitochondrial STAT3 supports Ras oncogenic transformation by supporting a metabolic shift (Avalle et al., 2012). Cao et al. found that STAT3 was consistently activated in Src-transformed cell lines, and the interruption of the STAT3 signal blocked the transformation of mouse fibroblasts by Src oncoprotein (Cao et al., 1996). Furthermore, it has been reported that oncogenes such as Bcr-Abl, v-Eyk, v-Ros, and v-Fps may play similar functions (Mankan and Greten, 2011). JAK/STAT pathways are also involved in various aspects of tumor development, such as invasion and metastasis (Loh et al., 2019). For example, abnormal activation of IL-6-mediated JAK/STAT3 signal transduction frequently occurs in human cancers and is involved in transformation, tumorigenicity, EMT, and metastasis. IL-6/JAK2/STAT3 activation induces EMT by up-regulating EMT-induced transcription factors (EMT-TFs, Snail, Zeb1, JUNB, and Twist-1), and enhances cell motility by activating focal adhesion kinase (FAK), which enhances metastasis (Jin, 2020). Xiao et al. demonstrated that IL-6 can promote EMT in peritoneal mesothelial cells, which is related to the activation of the JAK/STAT pathway (Xiao et al., 2017). Furthermore, reviews have concluded the effects of JAK/STAT3 activation on EMT by multiple intracellular signals protein, including PTK6, Williamʹs syndrome transcription factor (WSTF), Pin1, PYK2, SMAD4, RAC1, and other signals protein (Jin, 2020). NF-κB signaling has been identified as a major pathway to induce inflammation in tumors, where STAT3 directly interact with NF-κB family members to capture it in the nucleus, thereby promoting constitutive activation of NF-κB (Lee et al., 2009), leading to many oncogenic and inflammatory genes activations (Lee et al., 2009; Yu et al., 2009). Ruan et al. found that overexpression of OCT4 (a marker for cancer stem cells in ovarian cancer) increased the activation of the JAK/STAT pathway, especially JAK1 and STAT6, and promoted the translocation of STAT6 from the cytoplasm to nuclear in non-SP cells (CSC-like side population cells), thereby increasing the expression of Cyclin D1, c-Myc, and Bcl-2 (Gough et al., 2013; Ruan et al., 2019). As mentioned earlier, JAK/STAT can be activated by a variety of cytokines, thereby transducing and activating a variety of downstream signaling pathways in cells (Figure 2).
Activated JAKs also induces the activation of other downstream signaling cascades, including the MAPK and PI3K/AKT pathways. Studies demonstrate that ERK signaling regulates MHC II expression in spinal cord microglia through regulation of the STAT1 phosphorylation and promotes bone cancer pain (Song et al., 2017). AMPK inhibits tumor proliferation by suppressing STAT3 activation (Lee et al., 2011). Furthermore, STAT5 was found to form a complex with ERK1/ERK2 in colorectal cancer cells, suggesting a cross-talk between STAT5 and MAPK signaling pathways in the development of human colorectal cancer (Xiong et al., 2009). However, another recent study showed the presence of STAT5 in PI3K immunoprecipitation in leukemic bone marrow cells (Harir et al., 2007), but no STAT5-PI3K complexes were found in CRC cells. The specific cell type and tumor microenvironment may explain this phenomenon. It has been reported that STAT3 down-regulates the expression of important proteins related to apoptosis induction, including P53 (Niu et al., 2005), IFN-β (Wang T. et al., 2004), Fas and its ligands, and BAX (Kunigal et al., 2009; Liang et al., 2011). Abnormal activation of STAT3 also leads to abnormal overexpression of various proteins, including Mcl-1, Bcl-2, Bcl-xl, survivin, Cyclin D1, c-Myc, and VEGF, which leads to tumor development (Gao et al., 2005; Verma et al., 2010). In Barbara’s review, he mentioned that STAT was related to autophagy. PKR-eIF2A pathway is an important inducer of autophagy, and STAT3 inactivates this pathway through binding to PKR and inactivation of eIF2A phosphorylation (Jonchere et al., 2013; Bagca et al., 2016). In addition to STAT3, constitutive activation of STAT1 and STAT5 was also shown in tumor cells and tumor tissues. In chronic myeloid leukemia (CML) and myeloproliferative diseases induced by TEL-JAK2, STAT5 is activated by a variety of hematopoietic and non-hematopoietic cytokines and growth factors to promote the development of these tumors (Lin et al., 2000; Levis et al., 2002; Subramaniam et al., 2020). However, activation of STAT1 usually appears to promote tumor cell apoptosis and anti-proliferative effects. Tumors are more likely to develop in STAT1-deficient mice (Shankaran et al., 2001; Avalle et al., 2012). However, some studies have also shown that STAT1 can induce platinum resistance in breast cancer, which may be independent of the activation of JAK2/3 (Stronach et al., 2011).
In summary, the JAK/STAT pathway is involved in the activation and transduction of various signaling pathways related to tumorigenesis and development, suggesting that the JAK/STAT pathway may be another new target for cancer treatment.
2.2 JAK/STAT pathway in other diseases
Recent studies have shown the involvement of JAK/STAT in multiple diseases and their physiological processes. For example, studies have found that RA phosphorylates JAK2 by binding to AT1, thereby activating JAK and STAT signaling pathways to mediate VCSM growth, migration, and remodeling (Mehta and Griendling, 2007). IGF-IR exerts signaling effects by activating the JAK/STAT pathway. A study unveiled that miR-326 targets MDK to regulate the progression of cardiac hypertrophy by blocking JAK/STAT and MAPK signaling pathways (Zhang J. et al., 2020). Melatonin may have protective and therapeutic effects on hypercholesterolemia by regulating vaspin, STAT-3, DDAH, and ADMA signaling pathways (Sezgin et al., 2020). Increased levels of STAT-1 promote SMC (Smooth muscle cell) de-differentiation, whereas high levels of STAT-3 drive SMC into a more mature phenotype (Kirchmer et al., 2014). Therefore, the study of the JAK/STAT pathway can help us gain a deeper understanding of the pathological and pharmacological mechanisms of multiple diseases.
3 JAK/STAT signaling regulation of the immune environment
The role of the JAK/STAT pathway is critical in immune regulation which has attracted increasing attention. STAT transcription factors were regulated by many cytokines, so as to control the immune response and induce tumor immune escape, promote or inhibit the expansion and activation of various immune cells (Table 1).
TABLE 1.
JAK/STAT pathway mediates the effect of cytokines on immune cells.
| Cytokine | STAT | Effect | PMID |
|---|---|---|---|
| IFN-γ | STAT1 deficiency | reduce suppression by MO-MDSCs | 18272812 |
| IFN-α | STAT1 | induce HSC proliferation and differentiates into CDP | 19212321 |
| IFN-α | STAT1 | regulators of IL-12 production by DCs | 16618773 |
| IFN-α/β | STAT1 | maintain accumulation of proliferative NK cell | 12370359 |
| IL-12/IFN-γ | STAT1 | enhance T-cell infiltration and tumor growth inhibition | 17634555 |
| IL-12 | STAT1 deficiency | increase tumor-specific CTL activity | 16618773 |
| increases the CD8 T-cell density | |||
| IFN-α | STAT2 | antagonize stress-dependent expansions of T cells | 11163195 |
| iNOS/VEGF | STAT3 | increase MDSC suppressive function | 22529296 |
| G-CSF | STAT3 | promotes the development of MDSCs | 25649351 |
| Flt3L | STAT3 | increase MDSC suppressive function | 24639346 |
| GM-CSF | STAT3 | MDSCs expand and suppress antitumor immunity | 27199222 |
| IL-6/IL-10/VEGF | activate STAT3 | enhance the number of MDSC | 25238263 |
| 29100353 | |||
| 22529296 | |||
| IL-6 | STAT3 | increase CD11b+CD14+HLA-DR- myeloid cells | 25238263 |
| IL-6 | STAT3 | suppress DC maturation | 15356132 |
| VEGF/IL-10 | STAT3 | inhibit functional DC maturation | 16288283 |
| 14702630 | |||
| 16371463 | |||
| 14688356 | |||
| 14702634 | |||
| Flt3L | STAT3 | stimulate pDC generation | 20933441 |
| 14670306 | |||
| IL-10/IL-21 | STAT3 | NK-cell activity impaired | 24891320 |
| IL-6/IL-10/VEGF/HGF | STAT3 | regulating the activity of NK cells, toxicity function and interaction with other immune system components | 16288283 |
| 27148255 | |||
| APT2 | STAT3 | promotes Th17 cell differentiation | 33029007 |
| IL-10/TGF-β | STAT3 | induction of the Treg phenotype of the transformed CD4+ T cells | 16766651 |
| IL-6 | STAT3 | promoting naïve CD4+ T cell differentiation into inflammatory Th17 cells | 26912317 |
| IL-6 | STAT3 | inhibit the differentiation of Th9 cells | 26976954 |
| IL-6 | STAT3 | mediated Th17 differentiation | 19564351 |
| 16688182 | |||
| IL-6 | STAT3 | maintains the mitochondrial membrane potential during CD4 cell activation | 25974216 |
| 34809691 | |||
| IL-2 | STAT3 | induce CD4+CD25+ Tregs | 16645171 |
| 14500638 | |||
| 15611254 | |||
| IL-10/TGF-β | STAT3 | tumor-derived CD4+CD25+ regulatory T cells suppress DC maturation | 16612596 |
| IL-17A/IL-6/IL-23 | STAT3 | modulating the balance of Th17 and Treg cells, as well as in promoting CD4 T cell proliferation | 20493732 |
| IL-10 | STAT3 | deactivation of macrophages and neutrophils | 10023769 |
| IL-10 | STAT3 | promote the formation of M2 macrophages | 23169551 |
| IL-6/IL-10 | STAT3 | poor cytotoxicity and anti-tumor immune response | 29222039 |
| IL-12 | STAT4 | promote Th1 cells differentiation | 11086031 |
| T-bet | STAT4 | Tfh cell production of IFN-γ | 29212666 |
| GM-CSF | STAT5 | block pDC development | 18342552 |
| GM-CSF | STAT5 | promotes CD103+ DC development | 23033267 |
| IL-2/IL-15 | STAT5 | accumulation of NK | 28916644 |
| 29105654 | |||
| IL-2 | STAT5 | induce CD4+CD25+ Tregs | 16645171 |
| 14500638 | |||
| 15611254 | |||
| IL-10/TGF-β | STAT5 | tumor-derived CD4+CD25+ regulatory T cells suppress DC maturation | 16612596 |
| IL-2 receptor beta | STAT5 | regulate FoxP3 expression, and promote Treg differentiation | 17182565 |
| GM-CSF | STAT5 | drug resistant to sunitinib | 20406969 |
| IL-4/IL-13 | STAT6 | activation of MDS, increases the suppressive function of MDSCs | 19197294 |
| 19197294 | |||
| IL-4 | STAT6 | promote differentiation of Th2 cells | 11086031 |
| 8624821 | |||
| IL-4 | STAT6 | restrict CD8+ T cell expression | 18566374 |
| IL-4 | pSTAT6 | alternative macrophage polarization | 29343442 |
| STING | STAT6 | antiviral innate immunity | 22000020 |
3.1 MDSC immunosuppressive function
As we know, Myeloid-derived suppressor cells (MDSCs) are immature myeloid cells and have immunosuppressive properties for adaptive immunity and innate immunity. MDSCs are derived from hematopoietic stem cells in bone marrow (Millrud et al., 2017). Signals from tumors and inflammatory tissues stimulate the differentiation of IMC (immature myeloid cells, the progenitors of MDSCs) to MDSC through the STAT pathway and promote their expansion (Salminen et al., 2019). Kim’s review has elucidated that VEGF, G-CSF, GM-CSF, Flt3L, and other anti-inflammatory cytokines (IL-4, IL-6, IL-10) can activate STAT signaling and thus regulate MDSC proliferation and activation (Ko and Kim, 2016). IL-6, IL-10, and VEGF can activate STAT3 on MDSC, which can enhance the number of MDSC (Jayaraman et al., 2012; Chen et al., 2014; Wu et al., 2017). MDSCs immunosuppressive mechanisms may be related to the expression of arginase-I (Vasquez-Dunddel et al., 2013), IDO (Yu et al., 2014), iNOS (Wu et al., 2017), and PD-L1 (Thorn et al., 2016) by JAK-STAT3 signals activation. STAT1 is a major transcription factor for IFN-γ mediated signaling activation and is involved in the upregulation of arginase 1 and iNOS expression by MDSCs (Kusmartsev and Gabrilovich, 2005). For example, research has shown that blocking IFN-γ or disrupting STAT1 partially impaired suppression by MO-MDSCs (Movahedi et al., 2008). Activation of STAT6 by IL-4, and IL-13 leads to activation of MDSC, which causes upregulation of arginase 1, inducible iNOS, and production of transforming growth factor-β (TGFβ), which then increases the suppressive function of MDSCs (Gabrilovich and Nagaraj, 2009).
3.2 DC development
Dendritic cells (DC) are discrete cell populations derived from hematopoietic stem cells (HSCs) and have important functions in immune surveillance (Gardner et al., 2020). DCs are produced by hematopoietic progenitor cells (such as CDP) under the control of exogenous cytokine signals and intrinsic transcriptional regulators (Collin and Ginhoux, 2019). The main cytokines involved in DC development include Flt3L, GM-CSF, and IFN-α, which respectively stimulate STAT3, STAT5, and STAT1, and each STAT has a different role in DC production (Yu et al., 2007). Studies have shown that STAT3 activation is an important factor for Flt3L to regulate DC development, and the absence of STAT3 in hematopoietic cells eliminates the effect of Flt3L on DC (Laouar et al., 2003; Sathaliyawala et al., 2010). Under steady-state conditions, GM-CSF regulates the generation of CD103+DC by inducing STAT5-ld2 signal activation (Li et al., 2012). STAT5 is considered to be the major GM-CSF response signal protein (Gouilleux et al., 1995). Studies have shown that GM-CSF used STAT5 to prevent the development of Flt3L-dependent pDC from the lineage-negative Flt3+ (lin− Flt3+) bone-marrow subset (Esashi et al., 2008). STAT1-mediated IFNα promotes the proliferation of HSCs and then differentiates into CDP (Essers et al., 2009). IL-6 is the main cytokine controlling DCs differentiation in vivo. IL-6 mediates the inhibitory effect of bone marrow-derived DC maturation by regulating STAT3 phosphorylation in DC cells (Park et al., 2004). The inhibitory effect of STAT3 on DC maturation may also be caused by VEGF and IL-10 signaling (Wang T. et al., 2004). The functions of DC, T cells, natural killer (NK) cells, and neutrophils are significantly enhanced in tumor-bearing mice with Stat3−/− hematopoietic cells (Kortylewski et al., 2005).
3.3 NK cell function
Natural killer (NK) cells are an important early effector in the innate immune system to resist multiple viral infections and eliminate tumor cells (Cooper et al., 2001). IL-15 is required for the maturation of NK cells at all stages of development (Becknell and Caligiuri, 2005). In Nguyen’s experiment, they found that in a virus-infected mouse model, the maintenance of IFNα/β on the accumulation of proliferative NK cells is mainly dependent on the production of IL-15 induced by STAT1 (Nguyen et al., 2002). In mice deficient in STAT1, T-bet, or MHC type I molecules, the maturation state of peripheral NK cells is impaired (Robbins et al., 2005), and they are more sensitive to viral and bacterial infections (Sugawara et al., 2004). STAT3 can also indirectly damage the function of NK cells by regulating the expression of NK cell activation receptor ligands and immune checkpoint proteins, such as NKG2D ligand MICA, PD-L1 (Rocca et al., 2013; Zhu et al., 2014; Cacalano, 2016). In a mouse model, STAT3-deficient NK cells enhance tumor immune surveillance and increase DNAM-1 and the lytic enzymes perforin and granzyme B secretion (Gotthardt et al., 2014). STAT5a and STAT5b are important transcription factors for the activation, proliferation, and maturation of NK cells in humans and mice (Vargas-Hernandez and Forbes, 2019; Kee et al., 2020). STAT5b-deficient NK cells have decreasing proliferation and toxicity after stimulation by IL-2 and IL-15 (Imada et al., 1998; Villarino et al., 2017), and circulating NK cells in STAT5b-deficient patients are also significantly reduced, resulting in low cytotoxicity (Bernasconi et al., 2006).
3.4 T cells
T cells are derived from lymphatic stem cells in the thymus. They are the most numerous and complex type of cells in lymphocytes that produce cellular immunity. IL-6 and IL-10 cytokines activate STAT3 on T cells, which is usually related to poor cytotoxicity and anti-tumor immune response (Wang et al., 2018). Studies have shown that IL-6 mediates the differentiation of naive CD4+ T cells into inflammatory Th17 cells through STAT3, while IL-10 targeting IL-10Rα has similar effects (Jones et al., 2016). Furthermore, studies showed that IL-6 maintains the mitochondrial membrane potential during CD4+ cell activation in a STAT3-dependent manner, thereby increasing mitochondrial Ca+ levels and promoting cytokine expression (Yang et al., 2015; Awasthi et al., 2021). Studies found that STAT3 bound to multiple genes involved in Th17 cell differentiation by using chromatin immunoprecipitation and massive parallel sequencing (ChIP-Seq) (Durant et al., 2010). STAT3 and FoxP3 can be used as transcription factors to regulate the biological function of Treg (Woods et al., 2018). IL-12 induces a high intensity of tumor-specific CTL activity in STAT1-deficient mice, increases the CD8+ T-cell density, and induces a T-cell-dependent tumor regression (Torrero et al., 2006). IL-2-induced FOXP3 expression in human Treg cells is mediated by STAT signaling, including both STAT3 and STAT5. STAT5 also can combine with the FoxP3 gene promoter, regulate FoxP3 expression, and promote Treg differentiation (Burchill et al., 2007). The phosphorylation of STAT3 by IL-6 will inhibit the Th9 cell differentiation, which was mediated by the suppression of IL-2 production and STAT5 signaling (Olson et al., 2016). IL-2 selectively up-regulated the expression of FOXP3 in purified CD4+CD25+ T cells which involved the binding of STAT3 and STAT5 proteins (Antov et al., 2003; Anderson et al., 2005; Zorn et al., 2006). And present data demonstrate that CD4+CD25+FoxP3+ regulatory T cells impede dendritic cell function which requires TGF-beta and IL-10 by activating STAT3 (Larmonier et al., 2007). Furthermore, IL-12 is able to transcriptionally regulate STAT4 and thus participated in the development and differentiation of Th1 cells and STAT6-deficient T lymphocytes failed to differentiate into Th2 cells despite under IL-4 stimulation (Ostrand-Rosenberg et al., 2000). Activation of the IFNγ/STAT1/IRF1 axis favors processing and the presentation of tumor antigens, in association with MHC class I or class II molecules (Avalle et al., 2012). Research has shown that endogenously secreted IFNs served to antagonize stress-dependent expansions of T cells through a STAT2-dependent pathway (Park et al., 2000). In addition, STAT1 hyper-phosphorylation can lead to impaired IL-23 signaling, which can in turn result in defective Th17 T cell responses (Smeekens et al., 2011).
Members of the STAT protein family are involved in regulating immune responses in the tumor microenvironment, including pro-tumor or anti-tumor inflammatory responses. On the one hand, abnormal STAT3 expression in tumors is correlated with MDSC and Th17 levels, affecting DCs and thus affecting anti-tumor response (Yu et al., 2009). IL-6 induces STAT3-mediated Th17 differentiation, which maintains inflammatory responses by releasing IL-17 and IL-23 and causes the secretion of VEGF and TGF in fibroblasts and endothelial cells (Langowski et al., 2006; Wang L. et al., 2009). STAT3-mediated IL-23 production also inhibited the proliferation of effector T cells (Kortylewski et al., 2009). STAT3 also regulates the expression of PD-L1 on antigen-presenting cells which affects the drug effect of immunotherapy (Wolfle et al., 2011).
3.5 Indirect effects on immune cells
A study found that STAT3-mediated IL-10 secretion can promote the formation of M2 macrophages, and M2 macrophages regulate the function of breast cancer stem cells through EGFR/STAT3/SOX-2 paracrine signals (Yang et al., 2013). IL-4 induces pSTAT6-mediated inhibition of the activation of multiple genes involved in alternative macrophage polarization, such as NLRP3 and IL-1B, thereby inhibiting inflammasome stimulation and pyroptosis (Czimmerer et al., 2018). In tumor-associated macrophages (TAMs), STAT1 regulates the expression of arginase and NO, which in turn suppresses T cell-mediated immune responses and induces T cell apoptosis (Kusmartsev and Gabrilovich, 2005; Alvaro et al., 2006). On the other hand, inactivated STAT3 in hematopoietic stem cells shows an anti-tumor effect, inhibiting tumor growth and metastasis by affecting the activation of DC, T, and NK cells (Kortylewski et al., 2005). The absence or targeting of STAT3 in myeloid cells can enhance CD8+ T cell responses and activate tumor-associated monocytes and DC cells, leading to anti-tumor responses (Herrmann et al., 2010), and inhibiting the pro-angiogenic factors VEGF, bEGF, MMP9, CXCL2 secretion, thereby inhibiting the formation of vascular-like structures (Kujawski et al., 2008). In addition, STAT1 is involved in the early development of B cells (Najjar et al., 2010).
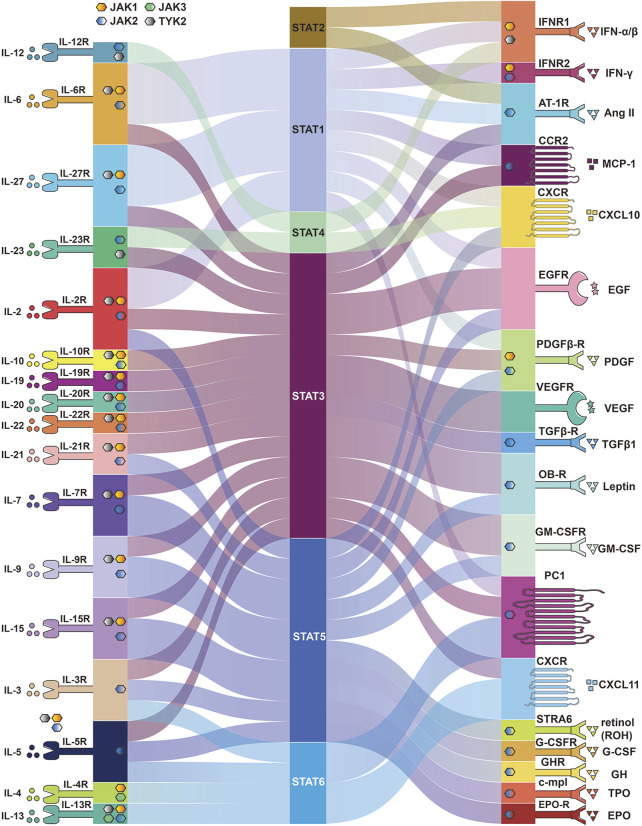
4 Extracellular cytokines affecting the JAK/STAT pathway
Many cytokines activate the JAK/STAT pathway, which transmits signals directly to the nucleus to induce various cellular responses (Figure 3). Below we describe the various cytokines and proteins that affect the JAK/STAT pathway and the various cellular activities to understand how the JAK/STAT pathway is involved in disease development and thus suggest more effective therapeutic approaches.
FIGURE 3.
Membrane proteins and cytokines that activate the JAK/STAT pathway. Different membrane proteins and upstream cytokines phosphorylate different JAK and activate different STATA pathways. Some membrane proteins do not require JAK to activate STAT.
4.1 Interleukin in JAK/STAT pathway
Cytokines of the interleukin family are involved in multiple aspects of cellular life activity functions, including the immune system, physiological functions, inflammatory responses, and cellular metabolism, and all interferons activate members of JAK and STAT. It was shown that IL-6 activated gp130 leading to activation of the JAK/STAT pathway (Rose-John, 2018) and the IL-6/JAK/STAT3 pathway is aberrantly hyperactivated in many types of cancer (Johnson et al., 2018). Studies showed that IL-6 downregulated PTPRO expression leading to enhance PD-L1 secretion in monocytes and macrophages through the JAK2/STAT1 (Zhang W. et al., 2020). IL-4 and IL-13 activate JAK1 by binding to receptors, and JAK1 phosphorylates STAT6, thereby mediating their pulmonary fibrotic effects (Jakubzick et al., 2004), while IL-6 induced fibrosis by activating STAT3 (O'Donoghue et al., 2012). Chemokines trigger receptor dimerization, followed by association and activation of JAK proteins (Soriano et al., 2003). IL-12 is the main driver of STAT4 activation and crucial for IFN-γ production in NK cells (Wang et al., 1999). Studies showed that IL-23 induces IL-17A expression in macrophages through the STAT3 (Hou et al., 2018) and requires STAT4 for IL-17 secretion from memory T helper cells and NKT cells (Glosson-Byers et al., 2014). IL-10 and its subfamily member cytokines IL-19, IL-20, and IL-22 are involved in immune regulation and inflammatory responses by inducing the STAT3 signal transduction pathway (Conti et al., 2003). IL-2 family cytokines including IL-2, IL-4, IL-7, IL-9, IL-15, and IL-21 are involved in NK cell development by activating different JAKs and STATs (Gotthardt et al., 2019). IL-27 mediates signaling predominantly through STAT1 and STAT3 and acts in immune-regulatory functions (Fabbi et al., 2017).
4.2 Other cytokines in JAK/STAT pathway
Cytokines bind to receptor proteins on the cell membrane, activating the downstream JAK/STAT pathway or directly recruiting STAT protein without JAK involvement. Type 1 interferons IFN-α and -β signal bind to their membrane subunits IFNAR1 and IFNAR2 expressed on all cells, then triggering JAK1 and TYK2 phosphorylation and thus recruiting STAT1 and STAT2 monomers for their dimerization activation (Fuertes et al., 2013), thereby facilitating the initiation of dendritic cells (DC) required for T cell activation (Gessani et al., 2014), supporting immune cell migration, stimulation, and differentiation (Hervas-Stubbs et al., 2011), as well as inducing regulation of the PI3K/AKT/mTOR signaling pathway (Bai et al., 2009). Similar to type 1 interferon IFN, IFN-γ signals through two transmembrane receptor subunits, IFNR1 and IFNR2, activating receptor-associated JAK, leading to the selective recruitment of STAT1 (Braumuller et al., 2013), which stimulates the toxic function of CD8+ T cells and NK cells (Bhat et al., 2017; Takeda et al., 2017), as well as promoting macrophage polarization toward the M1 phenotype (Mills et al., 2000). Other studies in human fibroblasts showed that JAK2 has been activated by TGFβ1, then, in turn, phosphorylates STAT3 and leads to its nuclear translocation (Liu et al., 2013). Tumor-derived GM-CSF activated neutrophils and induced neutrophil PD-L1 expression via the JAK-STAT3 pathway (Wang et al., 2017). The effects of EPO, TPO, G-SCF, GH, and Leptin are mainly mediated by JAK2 and mainly STAT5 (Cirillo et al., 2008; Mullen and Gonzalez-Perez, 2016; Muller, 2019). Activation of angiotensin II (AT1) receptors has also been shown to phosphorylate STAT 1, 2, and 3 (Liang et al., 1999; Ram and Iyengar, 2001). Studies have demonstrated that STRA6, a plasma membrane protein, can act as a cell factor receptor that mediates the transport of retinol from serum RBP into cells to activate JAK2/STAT5 signaling (Berry et al., 2012). Studies showed that CCR2 tyrosine phosphorylation is associated with the JAK and STAT1/3 pathway at different stages of rat adjuvant-induced arthritis (AIA), as well as with macrophage and endothelial cell infiltration (Shahrara et al., 2003; Zhu et al., 2021). Studies indicated that JAK1 is a downstream tyrosine kinase in PDGF receptor signaling and is a candidate for activation of STAT1 (Choudhury et al., 1996). Furthermore, the expressions of PDGFRβ, JAK2, and STAT3 can be inhibited by AG490 (Jin et al., 2020). It was also found that in vitro PDGF-induced STAT5 activation was directly mediated by PDGFβ-R and its activation did not require JAK1, JAK2, c-Src, Fyn (Paukku et al., 2000). EGF-R is a transmembrane protein tyrosine kinase and EGF can direct activation of STATs by EGFR binding and indirect activation of STATs through Src-mediated EGFR signaling (Quesnelle et al., 2007). EGF induces activation of STAT1, STAT3, and STAT5 in a variety of EGFR overexpressing cells (Zhang et al., 2003). The study showed no phosphorylation of JAK kinase after the addition of VEGF, suggesting that STAT activation is induced by the intrinsic tyrosine kinase activity of VEGFR (Yahata et al., 2003; Roger et al., 2021). In conclusion, the JAK/STAT pathway is involved in multiple signaling cascades of life activities, and targeting these signaling may provide new ideas for disease treatment.
5 The effect of biophysical forces on the JAK/STAT pathway
\Cells can sense their extracellular environment and respond to chemical (Armstrong et al., 2017), optical (Pail et al., 2011), thermal (McGlynn et al., 2021), and biophysical forces (Pan et al., 1999; Ruwhof and van der Laarse, 2000) signals, which can affect downstream cellular signaling pathways via second messenger cascades (Pan et al., 1999). Ultimately, these changes alter cell behavior and functions (Komuro et al., 1990; Sadoshima and Izumo, 1993; Manokawinchoke et al., 2021). Unlike the effects of drug stimulation or gene editing on cellular activity and pharmacological responses (Bhardwaj et al., 2014), cells are often subjected to continuous and weak mechanical forces from the surroundings (Wang et al., 2010; Wang et al., 2016; Ding et al., 2017). Even various cell-ECM interactions (Guidolin et al., 2018) and 3D cell culture systems have specific biophysical forces (Bouet et al., 2015). These physical interactions continuously transmit extracellular signals to the cell nucleus, which have multiple and profound effects on numerous biological processes including membrane proteins (Dalton and Lemmon, 2021), intracellular organelles (Ma et al., 2019), nuclear transcription and translation processes (Graham and Burridge, 2016; Lee et al., 2021), and intracellular phase separation (Zhang J. Z. et al., 2020).
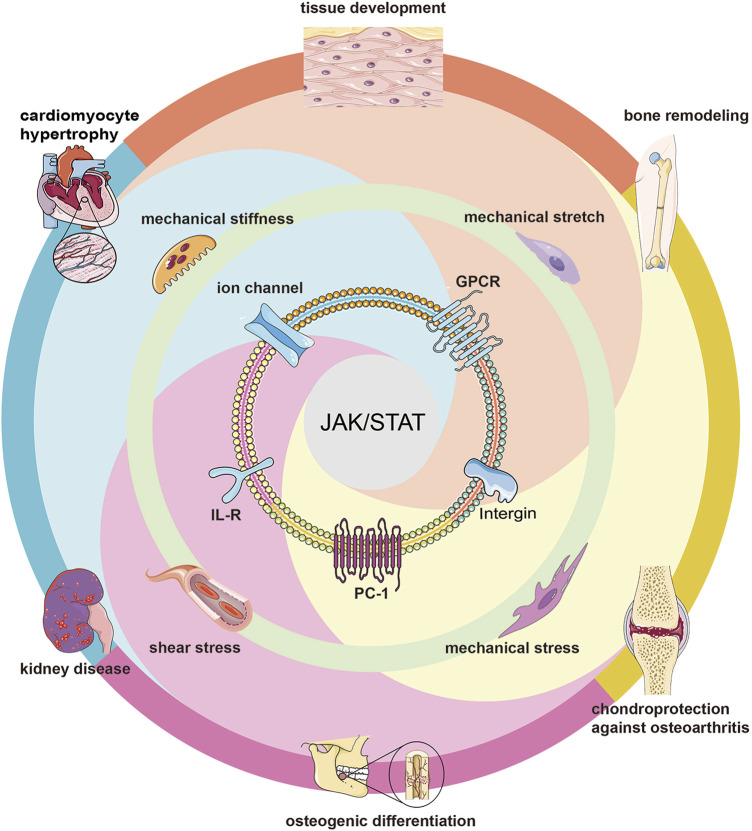
5.1 Biophysical forces in regulating the JAK/STAT pathway
The most common biophysical forces applied to mammalian cells are compression, stretching, shear stress, substrate stiffness, and substrate surface patterns (Wang et al., 2009b; Wang et al., 2009c; Wang et al., 2011; Wang et al., 2012; Wang and Tsai, 2013), which lead to morphological changes, cell membrane deformation (Manokawinchoke et al., 2021), membrane protein conformational changes (Ploscariu et al., 2014; Chen et al., 2017; Martinac and Poole, 2018), and ultimately triggering downstream signalings (Banes et al., 1995; Chen et al., 2017; Schneider et al., 2017). While cytoskeletons, ion channels, integrin receptors, G protein-coupled receptors, a transmembrane protein, and primary cilia are transit points for mechanical signals (Alfieri et al., 2019) and often influence the activation of downstream JAK/STAT pathways (Figure 4).
FIGURE 4.
Mechanotransduction of JAK/STAT pathways. Different biophysical forces activate the JAK/STAT pathway through different membrane proteins, thus affecting many biological processes including tissue development, osteogenic differentiation, cardiomyocyte hypertrophy, etc.
The study of biophysical forces in adult bone differentiation has been of interest, with several studies demonstrating that aged osteoblasts are characterized by impaired mechanosensitivity, and Cui et al. identified changes in the JAK/STAT pathway after transcription of the osteoblast transcriptome by transcriptomics (Cui et al., 2022). For the past few years, Jiliang Li has suggested that the JAK/STAT pathway plays an important role in bone development and metabolism, and that STAT3 has a more profound impact on bone homeostasis compared with other type of STATs (Li, 2013). Similarly, Natalie A Sims also held the same views and believed that the JAK1/STAT3/SOCS3 axis featured in bone development, physiology and pathology (Sims, 2020). Meanwhile, recent studies have shown that mechanical stimulation improves rotator cuff tendon-bone healing by activating IL-4/JAK/STAT signaling pathway mediated macrophage-M2 polarization (Liu et al., 2022). All in all, these studies indicate that JAK/STAT pathway plays an important role in bone development and repair.
Other types of biophysical forces such as extracellular matrix (ECM) stiffness, cell geometry, and shear stress were explored to activate JAK/STAT pathway signaling through activation of associated G proteins as well as rearrangement of the actin cytoskeleton (Erdogmus et al., 2019; Luo and Yu, 2019). Fong et al. found that mechanical shear stress can down-regulate PDGF (Fong et al., 2005) and thus modulate biological responses, a phenomenon that may be related to PDGF stimulation of primary cilia to induce STAT pathway activation. Static mechanical compressive forces lead to IL6 expression, and IL6 may subsequently indirectly activate STATs and translocate them to the nucleus through the JAK (Manokawinchoke et al., 2021). Also, it has been reported that cyclically stretch could induce the expression of MMP-14 and -2 in neonatal rat cardiomyocytes through JAK-STAT1 pathway (Wang T. L. et al., 2004). Braile and Jayaraman et al. found that pulsatile stretching can stimulate VEGF production in cardiomyocytes (CM) and that VEGF receptors can activate STAT phosphorylation, indirectly affecting downstream signaling via the JAK/STAT pathway (Seko et al., 1999; Jayaraman et al., 2012; Braile et al., 2020). In addition, Liang et al. also found that mechanical stretching-induced upregulation of VEGF-A in human mesenchymal cells was also associated with the JAK/STAT pathway, further illustrating the effect of biophysical forces on the JAK/STAT pathway (Liang et al., 2016).
5.2 JAK/STAT pathway mediates the role of biophysical forces
Current studies have shown that JAK/STAT-mediated mechanotransduction often has important effects on cell physiological processes. Matthews et al. found that mechanical stretch could affect the calcium influx by acting on β1 integrin (Matthews et al., 2010) and that Ca2+ plays a key role in stretch-induced activation of STATs (Pan et al., 1999), which has implications for cell and tissue development (Matthews et al., 2010). The research of Xiao et al. claimed that mechanical stretching-induced vascular endothelial growth factor A upregulation was related to the Janus kinase/signal transducer and activator of transcription (JAK/STAT) and Wnt signaling pathway (Liang et al., 2016). Researchers using mechanical stretching of human osteoblasts found that the stretching could upregulate Runx2 gene expression by enhancing the PC1-JAK2/STAT3 signaling axis, which has a decisive effect on bone remodeling (Dalagiorgou et al., 2017). Others showed that mechanical stress induced CCL2 (Zhu et al., 2021) to bind to CCR2 to regulate the production of osteoclasts in pressure grooves (Tsutsumi et al., 2013) and mediated chemotaxis and migration induction through activation of the JAK/STAT pathway in vitro and in vivo (Burysek et al., 2002). He et al. found that mechanical stress in chondrocytes combined with IL-4 to induce CITED2 gene expression in human chondrocytes via JAK/STAT pathway, thereby inhibiting matrix metalloproteinase (MMP13) production and providing chondroprotection against osteoarthritis (OA) (He et al., 2019). In addition, Qin et al. also found that periodontal ligament stem cells (PDLSC) sensitive to mechanical loading may downregulate HHIP-AS1 and promote the osteogenic differentiation potential of PDLSCs under continuous compressive stress, possibly via the JAK/STAT pathway (Qin et al., 2021). The Zyxin/Ajuba family of LIM proteins is a class of proteins that responds to biophysical forces (Jia et al., 2020), and Ajuba can play an important role in cell migration and epithelial morphogenesis by separating JAK1 from the interferon receptor and acting as a bona fide inhibitor of IFN/JAK1/STAT1 function (Sahai and Marshall, 2002; Jia et al., 2020). Machida et al. found that cyclic tensile strain induced the expression of ADAMTS4, ADAMTS5, and MMP13 in human chondrocytes via the underlying JAK/STAT pathway (Machida et al., 2017).
Mechanical stretch studies on rat cardiomyocytes by Pan et al. (Pan et al., 1997; Pan et al., 1999)found that mechanical stretch-induced cardiomyocyte hypertrophy (Kunisada et al., 1996; Kodama et al., 1997; Lammerding et al., 2004; Sims, 2020), which was largely dependent on cytokines of the IL-6 family, with activation of the JAK/STAT (mainly JAK1/STAT1, STAT3, partially binding to JAK2 and TYK2 (Sims, 2020)) pathway mediated by its receptor gp130 (Ruwhof and van der Laarse, 2000). Additionally, studies have shown that renal epithelial cells are subjected to flow-induced shear stress within the nephron and that kidney disease is affected by activation of the JAK/STAT pathway, as found by RNA sequencing (Kunnen et al., 2018). Honsho et al. found that pressure-mediated hypertrophy and mechanical stretch produced a low-level expression of IL-1β (subinflammatory), whereas JAK/STAT pathway-mediated production of IGF-1 could maintain its adaptive compensation for hypertrophy and inhibition of interstitial fibrosis (Honsho et al., 2009). Otherwise, in the neurodevelopmental process, Ciliary neurotrophic factor (CNTF) could directly stimulate JAK-STAT and RAS-MAPK cascaded reactions, and STAT3 signaling was considered as a potential component of neural response to stress stimuli (Peterson et al., 2000). More interestingly, it has been demonstrated that mechanical stress stimulates cellular immune response through JAK/STAT signaling pathway in Drosophila larvae (Tokusumi et al., 2018). In addition, other immunological studies have illustrated that the FTO/SOCS1/YTHDF1 regulatory axis was vital to the stiffness-controlled macrophage inflammatory response, including the culture environment of hydrogel with higher hardness could inhibit the expression of FTO gene through JAK-STAT and NF-κB signals (Hu et al., 2022).
In conclusion, biophysical forces occupy an important role in the induction of downstream signaling in the JAK/STAT pathway, playing a crucial role in the development of individual tissues, including bone, liver, heart, brain, nerves and immune regulation.
6 Clinical status of JAK/STAT pathway inhibitors
Based on the critical role of JAK/STAT in disease pathology and pharmacology, it is not surprising that inhibitors targeting JAK/STAT have been proposed to treat these diseases. Many inhibitors based on the JAK/STAT pathway have entered preclinical studies and clinical trials in a variety of diseases to evaluate their safety and clinical efficacy.
6.1 JAK/STAT inhibitors
At present, a variety of inhibitors targeting the JAK/STAT pathway have been used clinically, mainly for the treatment of rheumatoid arthritis, canine dermatitis, psoriasis, ulcerative colitis, myelofibrosis, polycythemia vera, and Primary thrombocytosis (Table 2). Ruxolitinib, a JAK inhibitor, has been identified by the FDA as a clinical treatment for Polycythemia, myelofibrosis, chronic graft-versus-host disease (cGVHD), and Atopic dermatitis by targeting JAK1 and JAK2. A number of clinical trial studies are also validating its efficacy and safety for the treatment of other diseases such as Chronic myelomonocytic leukemia (CMML) (Hunter et al., 2021), peripheral T-cell lymphoma (PTCL) (Moskowitz et al., 2021), lichen planus (Brumfiel et al., 2022), and COVID-2019 (Cao et al., 2020). The JAK inhibitor Tofacitinib is approved by the FDA as a treatment for rheumatoid arthritis, and it has also been used to study its effectiveness in Psoriasis (Abe et al., 2017), ulcerative colitis (Mukherjee et al., 2022), juvenile idiopathic arthritis (JIA) (Ruperto et al., 2021), transplant rejection (Vincenti et al., 2012), systemic sclerosis (SSc) (Khanna et al., 2022), Sarcoidosis (Damsky et al., 2022), systemic lupus erythematosus (SLE) (Hasni et al., 2021), and ankylosing spondylitis (AS) (Deodhar et al., 2021). Investigators are enrolling patients in a Phase IV clinical study of baricitinib for the treatment of Rheumatoid Arthritis, which is expected to become the standard of care for RA (NCT05238896). Oclacitinib is another inhibitor that targets JAK1 and is used to treat Canine allergic dermatitis (Little et al., 2015). Preclinical studies have shown that specific JAK2 inhibitors can inhibit the growth of tumors in vivo, including pancreatic cancer, colorectal cancer, gastric cancer, liver cancer, lung cancer, ovarian cancer, and breast cancer (Hedvat et al., 2009; Sun et al., 2011). Mohrherr et al. found that the JAK inhibitor Ruxolitinib reduced the proliferation of human K-ras-mutated A549 cells transplanted into immunodeficient mice, and decreased the expression of tumor cell-derived pro-cancer factors IL-1β and IL-6 (Mohrherr et al., 2019). And studies have shown that JAK2 inhibitor TG101209 can inhibit T cell acute lymphoblastic leukemia (T-ALL) proliferation by inhibiting JAK/STAT pathway activation and regulating the interaction between apoptosis and autophagy (Cheng et al., 2017).
TABLE 2.
Clinical study of JAK/STAT inhibitors.
| Agent | Target(s) | Disease(s) | Phase | Status* | ClinicalTrials.gov identifier(s) |
|---|---|---|---|---|---|
| Baricitinib | JAK1, JAK2 | Rheumatoid Arthritis (RA) | Phase 4 | Recruiting | NCT05238896 |
| Atopic Dermatitis | Phase 3 | Completed | NCT03559270 NCT03435081 | ||
| Diabetic Nephropathy | Phase 2 | Completed | NCT01683409 | ||
| Psoriasis | Phase 2 | Completed | NCT01490632 | ||
| Alopecia Areata | Phase 2/3 | Active, not recruiting | NCT03570749 | ||
| Systemic Lupus Erythematosus (SLE) | Phase 3 | Completed | NCT03616964 | ||
| Pyoderma Gangrenosum | Phase 2 | Recruiting | NCT04901325 | ||
| COVID-19 | Phase 2/3 | Completed | NCT04358614 | ||
| Human Immunodeficiency Virus | phase 2 | Not yet recruiting | NCT05452564 | ||
| Dermatomyositis | Phase 3 | Recruiting | NCT04972760 | ||
| Amyotrophic Lateral Sclerosis | Phase 1/2 | Recruiting | NCT05189106 | ||
| Graft-versus-host-disease | Phase 1/2 | Active, not recruiting | NCT04131738 | ||
| Systemic Sclerosis | Phase 4 | Recruiting | NCT05300932 | ||
| Immune Thrombocytopenia | Phase 2 | Recruiting | NCT05446831 | ||
| Juvenile Idiopathic Arthritis | Phase 3 | Completed | NCT03773978 | ||
| Aicardi Goutieres Syndrome | Phase 2 | Active, not recruiting | NCT03921554 | ||
| Liver Diseases | Phase 1/2 | Completed | NCT01870388 | ||
| Arteritis | Phase 2 | Completed | NCT03026504 | ||
| Sjogren’s Syndrome | Phase 2 | Recruiting | NCT05016297 | ||
| Allergic Contact Dermatitis | Early Phase 1 | Recruiting | NCT03945760 | ||
| Vitiligo | Phase 2 | Active, not recruiting | NCT04822584 | ||
| Cutaneous Lichen Planus | Phase 2 | Recruiting | NCT05188521 | ||
| Polymyalgia Rheumatic (PMR) | Phase 2 | Recruiting | NCT04027101 | ||
| Idiopathic Inflammatory Myopathies | Phase 2 | Recruiting | NCT04208464, NCT05400889 | ||
| Chronic Kidney Diseases | Phase 2 | Recruiting | NCT05237388 | ||
| Ruxolitinib | JAK1, JAK2 | Polycythemia, Myelofibrosis, chronic Graft-versus-host disease (cGVHD), Atopic Dermatitis | FDA approved | ||
| Chronic Myelomonocytic Leukemia (CMML) | phase 2 | Recruiting | NCT03722407 | ||
| Chronic Lymphocytic Leukemia | Phase 1/2 | Completed | NCT02015208 | ||
| Lymphoma | Phase 2 | Recruiting | NCT02974647, NCT01965119 | ||
| Lichen Planus | phase 2 | Not yet recruiting | NCT05593432, NCT05593445 | ||
| Bronchiolitis Obliterans Syndrome | Phase 2 | Recruiting | NCT05413356 | ||
| Vitiligo | Phase 3 | Active, not recruiting | NCT04530344 | ||
| Chronic Hand Eczema (CHE) | Phase 3 | Recruiting | NCT05233410 | ||
| COVID-19 | Phase 2 | Unknown | NCT04414098 | ||
| COVID-19 Induced Lung Injury ARDS | Phase 2 | Completed | NCT04359290 | ||
| COVID-19 Associated Cytokine Storm | Phase 3 | Completed | NCT04362137 | ||
| Thrombocythemia and Polycythemia Vera | Phase 2 | Recruiting | NCT04644211 | ||
| Hemophagocytic Syndrome (HPS) | Phase 2 | Completed | NCT02400463 | ||
| Solid Organ Transplant Recipients with Advanced Cutaneous Squamous Cell Carcinoma | Phase 2 | Recruiting | NCT04807777 | ||
| Head and Neck Squamous Cell Carcinoma | Phase 2 | Recruiting | NCT03153982 | ||
| Premalignant Breast Disease | Phase 2 | Recruiting | NCT02928978 | ||
| Non-small Cell Lung Cancer cachexia | Early Phase 1 | Recruiting | NCT04906746 | ||
| Tofacitinib | JAK3, JAK1, JAK2 | Rheumatoid Arthritis (RA) | FDA approved | ||
| Psoriasis | phase 2 | Completed | NCT01831466 | ||
| Kidney Transplantation | Phase 2 | Completed | NCT00263328 | ||
| Systemic Sclerosis (SSc) | phase 1/2 | Completed | NCT03274076 | ||
| Sarcoidosis | phase 1 | Completed | NCT03910543, NCT03793439 | ||
| Systemic Lupus Erythematosus (SLE) | Phase 2 | Recruiting | NCT03288324 | ||
| Ankylosing Spondylitis (AS) | Phase 3 | Completed | NCT03502616 | ||
| Ulcerative Colitis | Phase 3 | Recruiting | NCT04624230 | ||
| Juvenile Idiopathic Arthritis (JIA) | phase 3 | Completed | NCT02592434 | ||
| Alopecia Areata | Phase 4 | Completed | NCT03800979 | ||
| Primary Sjögren’s Syndrome | Phase 2 | Recruiting | NCT05087589 | ||
| Dermatomyositis | phase 1 | Completed | NCT03002649 | ||
| Glioblastoma | Phase 2 | Recruiting | NCT05326464 | ||
| Myasthenia Gravis | Early Phase 1 | Recruiting | NCT04431895 | ||
| Psoriatic Arthritis | phase 3 | Completed | NCT03736161, NCT03486457 | ||
| COVID-19 | phase 2 | Completed | NCT04750317 | ||
| Takayasu Arteritis | Phase 4 | Recruiting | NCT05102448 | ||
| Upadacitinib (ABT494) | JAK1 | Rheumatoid Arthritis | Phase 3 | Completed | NCT02955212 |
| Psoriatic Arthritis | Phase 3 | Active, not recruiting | NCT03104374, NCT03104400 | ||
| Atopic Dermatitis | Phase 3 | Active, not recruiting | NCT04195698, NCT03569293 | ||
| Hidradenitis Suppurativa (HS) | Phase 2 | Completed | NCT04430855 | ||
| Spondyloarthritis | Phase 3 | Active, not recruiting | NCT04169373 | ||
| Juvenile Idiopathic Arthritis (JIA) | Phase 1 | Recruiting | NCT03725007 | ||
| Ulcerative Colitis (UC) | Phase 3 | Completed | NCT03653026 | ||
| Crohn’s Dsease | Phase 3 | Completed | NCT03345836, NCT03345849 | ||
| Takayasu Arteritis (TAK) | Phase 3 | Recruiting | NCT04161898 | ||
| Ankylosing Spondylitis (AS) | Phase 2 | Completed | NCT03178487 | ||
| Non-Segmental Vitiligo | Phase 2 | Active, not recruiting | NCT04927975 | ||
| Giant Cell Arteritis (GCA) | Phase 3 | Recruiting | NCT03725202 | ||
| Itacitinib (INCB039110) | JAK1, JAK2 | Plaque Psoriasis | Phase 2 | Completed | NCT01634087 |
| Myelofibrosis | Phase 2 | Completed | NCT01633372 | ||
| Non-Severe Hemophagocytosis Lymphohistiocytosis | Phase 2 | Recruiting | NCT05063110 | ||
| Advanced Hepatocellular Carcinoma | Phase 1 | Recruiting | NCT04358185 | ||
| Graft-versus-host-disease | Phase 2 | Completed | NCT03846479 | ||
| Bronchiolitis Obliterans Syndrome | Phase 1/2 | Active, not recruiting | NCT03978637 | ||
| Systemic Sclerosis | phase 2 | Not yet recruiting | NCT04789850 | ||
| Metastatic Synovial Sarcoma | Phase 1 | Recruiting | NCT03670069 | ||
| Rheumatoid Arthritis | Phase 2 | Completed | NCT01626573 | ||
| Cytokine Release Syndrome | Phase 2 | Recruiting | NCT04071366 | ||
| Filgotinib | JAK1 | Rheumatoid Arthritis (RA) | Phase 3 | Active, not recruiting | NCT03025308 |
| Cutaneous lupus erythematosus (CLE) | Phase 2 | Completed | NCT03134222 | ||
| Fistulizing Crohn’s Disease | Phase 2 | Completed | NCT03077412 | ||
| Ulcerative Colitis | Phase 3 | Completed | NCT02914522 | ||
| Ankylosing Spondylitis | Phase 2 | Completed | NCT03117270 | ||
| Lupus Membranous Nephropathy (LMN) | Phase 2 | Completed | NCT03285711 | ||
| Small Bowel Crohn’s Disease | Phase 2 | Completed | NCT03046056 | ||
| Psoriatic Arthritis | Phase 2 | Completed | NCT03101670 | ||
| Sjogren’s Syndrome | Phase 2 | Completed | NCT03100942 | ||
| Deucravacitinib | Tyk2 | Psoriasis | Phase 3 | Recruiting | NCT05478499, NCT04036435 |
| Psoriatic Arthriti | Phase 3 | Recruiting | NCT04908202, NCT04908189 | ||
| Nail Psoriasis | Early Phase 1 | Not yet recruiting | NCT05124080 | ||
| Plaque Psoriasis | Phase 3 | Recruiting | NCT04772079 | ||
| Alopecia Areata | Phase 2 | Not yet recruiting | NCT05556265 | ||
| Crohn Disease | Phase 2 | Recruiting | NCT04877990 | ||
| Ulcerative Colitis | Phase 2 | Active, not recruiting | NCT03934216 | ||
| Subacute Cutaneous Lupus Erythematosus (SCLE) | Phase 2 | Recruiting | NCT04857034 | ||
| Lestaurtinib (CEP-701) | JAK2 | Myelofibrosis | Phase 2 | Completed | NCT00494585 |
| Acute Myeloid Leukemia | Phase 2 | Completed | NCT00079482 | ||
| Neuroblastoma | Phase 1 | Completed | NCT00084422 | ||
| Polycythemia Vera | Phase 2 | Completed | NCT00586651 | ||
| Chronic Beryllium Disease (CBD) | Phase 2 | Completed | NCT00586651 | ||
| Psoriasis | Phase 2 | Completed | NCT00236119 | ||
| Prostate Cancer | Phase 2 | Completed | NCT00081601 | ||
| Ritlecitinib | JAK3 | Rheumatoid Arthritis (RA) | Phase 2 | Completed | NCT04413617 |
| Alopecia Areata | Phase 2/3 | Completed | NCT03732807 | ||
| Non-segmental Vitiligo | phase 3 | Not yet recruiting | NCT05583526 | ||
| Cicatricial Alopecia | phase 2 | Not yet recruiting | NCT05549934 | ||
| Abrocitinib | JAK1 | Atopic Dermatitis | Phase 3 | Completed | NCT04345367 |
| Prurigo Nodularis | Phase 2 | Completed | NCT05038982 | ||
| Food Allergy | Phase 1 | Recruiting | NCT05069831 | ||
| Brepocitinib | JAK1, Tyk2 | Cicatricial Alopecia | Phase 2 | Recruiting | NCT05076006 |
| Active Non-Infectious Non-Anterior Uveitis | Phase 2 | Recruiting | NCT05523765 | ||
| Dermatomyositis | Phase 3 | Recruiting | NCT05437263 | ||
| Delgocitinib | JAK1, JAK2, JAK3, Tyk2 | Atopic Dermatitis | Phase 2 | Completed | NCT03725722 |
| Frontal Fibrosing Alopecia | Phase 2 | Recruiting | NCT05332366 | ||
| Chronic Hand Eczema | Phase 3 | Recruiting | NCT05355818 | ||
| Danvatirsen (AZD9150) | STAT3 | Advanced Colorectal Carcinoma | Phase 2 | Active, not recruiting | NCT02983578 |
| Advanced Lung Non-Small Cell Carcinoma | Phase 2 | Active, not recruiting | NCT03819465 | ||
| Metastatic Squamous Cell Carcinoma of the Head and Neck | Phase 1/2 | Active, not recruiting | NCT02499328 | ||
| Fedratinib | JAK2 | Myeloproliferative Neoplasm | Phase 2 | Recruiting | NCT05177211 |
| Myelofibrosis | Phase 3 | Recruiting | NCT03952039 | ||
| OPB-31121 | STAT3 | Advanced Solid Tumors | Phase 1 | Completed | NCT00955812 |
| Hepatocellular Carcinoma | Phase 1/2 | Completed | NCT01406574 | ||
| OPB-51602 | STAT3 | Malignant Solid Tumour | Phase 1 | Completed | NCT01423903, NCT01184807 |
| Oclacitinib | JAK1 | Canine Allergic Dermatitis | FDA approved | ||
| Momelitinib | JAK1, JAK2 | Anemic Myelofibrosis | Phase 3 | Active, not recruiting | NCT04173494 |
| Peficitinib | JAK1, JAK3 | Rheumatoid Arthritis (RA) | Phase 3 | Completed | NCT03660059, NCT02305849 |
| Decernotinib (VX509) | JAK3 | Rheumatoid Arthritis (RA) | Phase 2/3 | Completed | NCT01830985 |
| AZD1480 | JAK1, JAK2 | Myelofibrosis | Phase 1 | Completed | NCT00910728 |
| Gandotinib (LY2784544) | JAK2V617F | Myeloproliferative Neoplasms | Phase 2 | Active, not recruiting | NCT01594723 |
Inhibitors targeting STAT3, Danvatirsen (Reilley et al., 2018; Nishina et al., 2022), OPB-31121 (Bendell et al., 2014; Oh et al., 2015), OPB-51602 (Ogura et al., 2015; Wong et al., 2015), are in clinical trials in a variety of solid tumors, including colorectal cancer, non-small cell lung cancer, liver cancer, head and neck cancer, etc. Although still in Phase 1 trials, their feasibility for treating tumors is being further demonstrated. In addition, several inhibitors targeting the JAK/STAT pathway are being tested in multi-stage clinical trials in a variety of diseases, such as Momelitinib (Mesa et al., 2022), Peficitinib (Tanaka et al., 2021), itacitinib (Naing et al., 2022), AZD1480 (Plimack et al., 2013), Fedratinib (Harrison et al., 2022b), Gandotinib (Berdeja et al., 2018), Filgotinib (Meng et al., 2022), Upadacitinib (Deodhar et al., 2022), Decernotinib (Genovese et al., 2016), lestatitinib (Brown et al., 2021), Decernotinib (Fleischmann et al., 2015), lestaurtinib (Mascarenhas et al., 2019), abrocitinib (Reich et al., 2022), ritlecitinib (Winnette et al., 2022), brepocitinib (Peeva et al., 2022), deucravacitinib (Mease et al., 2022), delgocitinib (Worm et al., 2022). The JAK/STAT pathway is activated in a variety of common solid tumors contributing to an aggressive phenotype (Roxburgh and McMillan, 2016). Studies also have shown that Ruxolitinib can regulate the expression of phosphorylated STAT1 in patients with STAT1 Gain-of-function mutations, thereby restoring the toxicity function of NK cells (Vargas-Hernandez et al., 2018). Silibinin is a direct STAT3 targeting agent, which can not only reduce therapy-associated nephrotoxicity, neurotoxicity and cardiotoxicity in preclinical models but also has the potential to reverse cancer cell drug resistance (Bosch-Barrera et al., 2017). STAT3 inhibitor WP1066 also inhibited Treg and increased T cell toxicity in patients with melanoma brain metastasis (Kong et al., 2009).
6.2 Combination therapy with JAK/STAT inhibitors
As we know, the clinical drug tolerance of some tumors is gradually emerging, so combined JAK/STAT inhibitor therapy may be a new treatment strategy (Table 3). JAK inhibitor (AZD1480) combined with EGFR inhibitor (cediranib) reduces tumor volume and microvascular density by reducing hypoxia and macrophage infiltration (de Groot et al., 2012). Sun’s results showed that TG101209 increased radiosensitivity by inducing apoptosis and decreasing cell proliferation and vascular density in lung cancer (Zhang et al., 2015). But in our experiments, we found that the JAK2 inhibitor, WP1066, combined with radiotherapy failed to reduce cell viability in gastric cell lines, which may be related to the cancer specificity. Matthew conducted a phase I/II trial to study the safety and efficacy of combining trastuzumab with ruxolitinib in patients with trastuzumab-resistant metastatic HER2+ breast cancer. However, the results did not observe an improvement in patient PFS (Kearney et al., 2021). And Sukhmani found that momelotinib in combination with erlotinib did not appear to enhance the benefit of patients with EGFR-mutated NSCLC (Padda et al., 2022). Robert evaluated the JAKA1 inhibitor itacitinib in combination with corticosteroids or placebo for the treatment of acute GVHD, and the observed improvement in ORR at day 28 in the combination group did not reach the prespecified significance level (Zeiser et al., 2022). Filgotinib was found to improve signs and symptoms of rheumatoid arthritis, improve physical function, inhibit radiographic progression, and be well tolerated by RA patients with an inadequate response to methotrexate (MTX) (Combe et al., 2021). Studies have shown that after combined treatment with the STAT3/5 inhibitor Static, the Oncolytic Adenovirus XVir-N-31 has increased viral replication and increased virus-induced death of bladder cancer cells (Hindupur et al., 2020). Furthermore, some clinical trials are ongoing. A phase II clinical trial investigated the efficacy and safety of adding the BCL-XL/BCL-2 inhibitor navitoclax in patients with myelofibrosis who progressed on ruxolitinib therapy or responded suboptimally to ruxolitinib monotherapy. The results demonstrated that patients achieved durable SVR35 (≥35% spleen volume reduction) and improved TSS50 (≥50% reduction in total symptom score) (Harrison et al., 2022a).
TABLE 3.
JAK/STAT inhibitors are used in combination with other drugs.
| Agent | Disease(s) | Phase | Status* | ClinicalTrials.gov identifier(s) |
|---|---|---|---|---|
| Ruxolitinib + Chidamide | Peripheral Blood Stem Cell Transplantation | Phase 2 | Recruiting | NCT05088226 |
| NCT04582604 | ||||
| Ruxolitinib + Radiation and Temozolomide | Glioma | Phase 1 | Active, not recruiting | NCT03514069 |
| Ruxolitinib + Trastuzumab | Metastatic HER2 Positive Breast Cancer | Phase 1/2 | Completed | NCT02066532 |
| Itacitinib + Everolimus | Classical Hodgkin Lymphoma | Phase 1/2 | Recruiting | NCT03697408 |
| Itacitinib + Low-Dose Ruxolitinib | Myeloproliferative Neoplasms (MPN) | Phase 2 | Completed | NCT03144687 |
| Itacitinib + Osimertinib | Non-Small Cell Lung Cancer | Phase 1/2 | Active, not recruiting | NCT02917993 |
| Itacitinib + Alemtuzumab | T-Cell Prolymphocytic Leukemia | Phase 1 | Recruiting | NCT03989466 |
| Itacitinib + Ibrutinib | Diffuse Large B-Cell Lymphoma | Phase 1/2 | Completed | NCT02760485 |
| Itacitinib + Corticosteroids | Acute Graft-versus-host disease | Phase 3 | Completed | NCT03139604 |
| Itacitinib + Gemcitabine and Nab-Paclitaxel | Pancreatic Cancer | Phase 1/2 | Completed | NCT01858883 |
| Itacitinib + Dabrafenib and Trametinib | Melanoma | Phase 1 | Active, not recruiting | NCT03272464 |
| Itacitinib + Pembrolizumab | Colorectal Cancer | Phase 1 | Completed | NCT02646748 |
| Fedratinib + Decitabine | Myeloproliferative Neoplasms (MPN) | Phase 1 | Recruiting | NCT05524857 |
| Fedratinib + Nivolumab | Myelofibrosis | Phase 2 | Recruiting | NCT05393674 |
| Decitabine + Ruxolitinib or Fedratinib | Accelerated/Blast Phase Myeloproliferative Neoplasms | Phase 2 | Recruiting | NCT04282187 |
| Filgotinib + Methotrexate | Rheumatoid Arthritis | Phase 3 | Completed | NCT02886728 |
| NCT02889796 | ||||
| Upadacitinib + Methotrexate | Rheumatoid Arthritis | Phase 3 | Recruiting | NCT05121298 |
| Upadacitinib + Corticosteroids | Atopic Dermatitis | Phase 3 | Completed | NCT03661138 |
| Upadacitinib + Elsubrutinib | Systemic Lupus Erythematosus (SLE) | Phase 2 | Completed | NCT03978520 |
| Lestaurtinib + Chemotherapy | Acute Lymphoblastic Leukemia | Phase 3 | Active, not recruiting | NCT00557193 |
| Upadacitinib + Methotrexate | Rheumatoid Arthritis | Phase 2 | Completed | NCT01960855 |
| Danvatirsen + Tremelimumab | Diffuse large B-cell lymphoma | Phase 1 | Completed | NCT02549651 |
Moreover, potential natural products such as plant-derived cucurbitacin I and curcumin analogue ASC-J9 could be used as JAK/STAT inhibitors to treat related diseases (Yin et al., 2023), and the small molecule drugs, which screened from the databases (Dogra et al., 2023), targeting JAK/STAT could also be regarded as treatment strategies for related diseases (various organ fibrosis, etc.) (Liu et al., 2023).
7 Conclusion
The JAK/STAT pathway, a key pathway for protein signaling on the membrane, is crucial in human cells. The dysregulation of this pathway has been considered one of the causes leading to disease progression and tumor growth. In tumors, JAK/STAT acts as a regulatory hub for transduction signals, which affects the activation of various inflammatory factors, growth factors, and angiogenic factors in the tumor microenvironment (TME), and participates in regulating the maturation, proliferation, and differentiation of various immune cells. In addition, JAK/STAT pathway is also affected by many extracellular mechanical signals and consequently mediates numerous downstream biological processes. Therefore, inhibition of this pathway has attracted widespread attention as a potential therapeutic strategy. Based on the underlying molecular and genomic mechanisms of JAK/STAT, the internal and external factors affecting JAK/STAT activity, epigenetic and transcription factors, and genetic causes of dysregulated JAK/STAT signaling, these provide good ideas for the development and implementation of targeted drugs. In order to properly incorporate JAK/STAT targeted drugs into multimodality therapies, including combinations with chemotherapy, radiotherapy, immunotherapy, and physiotherapy, we also need to find predictive biomarkers, not just the overactivation of pathways. In view of the different sensitivity of individuals to drugs, it will be a hot research direction for us to learn more about the changes in individual tumor genomes and help us make therapeutic regimens for different gene mutations in the future.
Funding Statement
PW thanks the support from the Ministry of Science and Technology of China (2019YFE0113000); the National Natural and Science Foundation of China (31870988).
Author contributions
JM and QH conceived the structure of the manuscript. QH, DR, and JS made the figures and tables. QB and JZ completed the literature collection. JM, QH, LW, HH, and PW revised the manuscript. All authors approved the final manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2023.1110765/full#supplementary-material
References
- Abe M., Nishigori C., Torii H., Ihn H., Ito K., Nagaoka M., et al. (2017). Tofacitinib for the treatment of moderate to severe chronic plaque psoriasis in Japanese patients: Subgroup analyses from a randomized, placebo-controlled phase 3 trial. J. Dermatol 44 (11), 1228–1237. 10.1111/1346-8138.13956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfieri R., Vassalli M., Viti F. (2019). Flow-induced mechanotransduction in skeletal cells. Biophys. Rev. 11 (5), 729–743. 10.1007/s12551-019-00596-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvaro T., Lejeune M., Camacho F. I., Salvado M. T., Sanchez L., Garcia J. F., et al. (2006). The presence of STAT1-positive tumor-associated macrophages and their relation to outcome in patients with follicular lymphoma. Haematologica 91 (12), 1605–1612. [PubMed] [Google Scholar]
- Anderson P. O., Sundstedt A., Yazici Z., Minaee S., O'Neill E. J., Woolf R., et al. (2005). IL-2 overcomes the unresponsiveness but fails to reverse the regulatory function of antigen-induced T regulatory cells. J. Immunol. 174 (1), 310–319. 10.4049/jimmunol.174.1.310 [DOI] [PubMed] [Google Scholar]
- Antov A., Yang L., Vig M., Baltimore D., Van Parijs L. (2003). Essential role for STAT5 signaling in CD25+CD4+ regulatory T cell homeostasis and the maintenance of self-tolerance. J. Immunol. 171 (7), 3435–3441. 10.4049/jimmunol.171.7.3435 [DOI] [PubMed] [Google Scholar]
- Armstrong J. P., Holme M. N., Stevens M. M. (2017). Re-engineering extracellular vesicles as smart nanoscale therapeutics. ACS Nano 11 (1), 69–83. 10.1021/acsnano.6b07607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avalle L., Pensa S., Regis G., Novelli F., Poli V. (2012). STAT1 and STAT3 in tumorigenesis: A matter of balance. JAKSTAT 1 (2), 65–72. 10.4161/jkst.20045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avgustinova A., Benitah S. A. (2016). Epigenetic control of adult stem cell function. Nat. Rev. Mol. Cell Biol. 17 (10), 643–658. 10.1038/nrm.2016.76 [DOI] [PubMed] [Google Scholar]
- Awasthi N., Liongue C., Ward A. C. (2021). STAT proteins: A kaleidoscope of canonical and non-canonical functions in immunity and cancer. J. Hematol. Oncol. 14 (1), 198. 10.1186/s13045-021-01214-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagca B. G., Ozalp O., Kurt C. C., Mutlu Z., Saydam G., Gunduz C., et al. (2016). Ruxolitinib induces autophagy in chronic myeloid leukemia cells. Tumour Biol. 37 (2), 1573–1579. 10.1007/s13277-015-3947-4 [DOI] [PubMed] [Google Scholar]
- Bagratuni T., Mavrianou N., Gavalas N. G., Tzannis K., Arapinis C., Liontos M., et al. (2020). JQ1 inhibits tumour growth in combination with cisplatin and suppresses JAK/STAT signalling pathway in ovarian cancer. Eur. J. Cancer 126, 125–135. 10.1016/j.ejca.2019.11.017 [DOI] [PubMed] [Google Scholar]
- Bai D., Ueno L., Vogt P. K. (2009). Akt-mediated regulation of NFκB and the essentialness of NFκB for the oncogenicity of PI3K and Akt. Int. J. Cancer 125 (12), 2863–2870. 10.1002/ijc.24748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S., Biehl A., Gadina M., Hasni S., Schwartz D. M. (2017). JAK-STAT signaling as a target for inflammatory and autoimmune diseases: Current and future prospects. Drugs 77 (5), 521–546. 10.1007/s40265-017-0701-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banes A. J., Tsuzaki M., Yamamoto J., Fischer T., Brigman B., Brown T., et al. (1995). Mechanoreception at the cellular level: The detection, interpretation, and diversity of responses to mechanical signals. Biochem. Cell Biol. 73 (7-8), 349–365. 10.1139/o95-043 [DOI] [PubMed] [Google Scholar]
- Becknell B., Caligiuri M. A. (2005). Interleukin-2, interleukin-15, and their roles in human natural killer cells. Adv. Immunol. 86, 209–239. 10.1016/S0065-2776(04)86006-1 [DOI] [PubMed] [Google Scholar]
- Bendell J. C., Hong D. S., Burris H. A., 3rd, Naing A., Jones S. F., Falchook G., et al. (2014). Phase 1, open-label, dose-escalation, and pharmacokinetic study of STAT3 inhibitor OPB-31121 in subjects with advanced solid tumors. Cancer Chemother. Pharmacol. 74 (1), 125–130. 10.1007/s00280-014-2480-2 [DOI] [PubMed] [Google Scholar]
- Berdeja J., Palandri F., Baer M. R., Quick D., Kiladjian J. J., Martinelli G., et al. (2018). Phase 2 study of gandotinib (LY2784544) in patients with myeloproliferative neoplasms. Leuk. Res. 71, 82–88. 10.1016/j.leukres.2018.06.014 [DOI] [PubMed] [Google Scholar]
- Bernasconi A., Marino R., Ribas A., Rossi J., Ciaccio M., Oleastro M., et al. (2006). Characterization of immunodeficiency in a patient with growth hormone insensitivity secondary to a novel STAT5b gene mutation. Pediatrics 118 (5), e1584–e1592. 10.1542/peds.2005-2882 [DOI] [PubMed] [Google Scholar]
- Berry D. C., O'Byrne S. M., Vreeland A. C., Blaner W. S., Noy N. (2012). Cross talk between signaling and vitamin A transport by the retinol-binding protein receptor STRA6. Mol. Cell Biol. 32 (15), 3164–3175. 10.1128/MCB.00505-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj N., Katyal P., Sharma A. K. (2014). Suppression of inflammatory and allergic responses by pharmacologically potent fungus Ganoderma lucidum. Recent Pat. Inflamm. Allergy Drug Discov. 8 (2), 104–117. 10.2174/1872213x08666140619110657 [DOI] [PubMed] [Google Scholar]
- Bhat P., Leggatt G., Waterhouse N., Frazer I. H. (2017). Interferon-gamma derived from cytotoxic lymphocytes directly enhances their motility and cytotoxicity. Cell Death Dis. 8 (6), e2836. 10.1038/cddis.2017.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch-Barrera J., Queralt B., Menendez J. A. (2017). Targeting STAT3 with silibinin to improve cancer therapeutics. Cancer Treat. Rev. 58, 61–69. 10.1016/j.ctrv.2017.06.003 [DOI] [PubMed] [Google Scholar]
- Bouet G., Cruel M., Laurent C., Vico L., Malaval L., Marchat D. (2015). Validation of an in vitro 3D bone culture model with perfused and mechanically stressed ceramic scaffold. Eur. Cell Mater 29, 250–267. discussion 266-257. 10.22203/ecm.v029a19 [DOI] [PubMed] [Google Scholar]
- Boyle K., Zhang J. G., Nicholson S. E., Trounson E., Babon J. J., McManus E. J., et al. (2009). Deletion of the SOCS box of suppressor of cytokine signaling 3 (SOCS3) in embryonic stem cells reveals SOCS box-dependent regulation of JAK but not STAT phosphorylation. Cell Signal 21 (3), 394–404. 10.1016/j.cellsig.2008.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braile M., Marcella S., Cristinziano L., Galdiero M. R., Modestino L., Ferrara A. L., et al. (2020). VEGF-A in cardiomyocytes and heart diseases. Int. J. Mol. Sci. 21 (15), 5294. 10.3390/ijms21155294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braumuller H., Wieder T., Brenner E., Assmann S., Hahn M., Alkhaled M., et al. (2013). T-helper-1-cell cytokines drive cancer into senescence. Nature 494 (7437), 361–365. 10.1038/nature11824 [DOI] [PubMed] [Google Scholar]
- Brown P. A., Kairalla J. A., Hilden J. M., Dreyer Z. E., Carroll A. J., Heerema N. A., et al. (2021). FLT3 inhibitor lestaurtinib plus chemotherapy for newly diagnosed KMT2A-rearranged infant acute lymphoblastic leukemia: Children's Oncology Group trial AALL0631. Leukemia 35 (5), 1279–1290. 10.1038/s41375-021-01177-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumfiel C. M., Patel M. H., Severson K. J., Zhang N., Li X., Quillen J. K., et al. (2022). Ruxolitinib cream in the treatment of cutaneous lichen planus: A prospective, open-label study. J. Invest. Dermatol 142 (8), 2109–2116.e4. 10.1016/j.jid.2022.01.015 [DOI] [PubMed] [Google Scholar]
- Burchill M. A., Yang J., Vogtenhuber C., Blazar B. R., Farrar M. A. (2007). IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J. Immunol. 178 (1), 280–290. 10.4049/jimmunol.178.1.280 [DOI] [PubMed] [Google Scholar]
- Burysek L., Syrovets T., Simmet T. (2002). The serine protease plasmin triggers expression of MCP-1 and CD40 in human primary monocytes via activation of p38 MAPK and janus kinase (JAK)/STAT signaling pathways. J. Biol. Chem. 277 (36), 33509–33517. 10.1074/jbc.M201941200 [DOI] [PubMed] [Google Scholar]
- Cacalano N. A. (2016). Regulation of natural killer cell function by STAT3. Front. Immunol. 7, 128. 10.3389/fimmu.2016.00128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Tay A., Guy G. R., Tan Y. H. (1996). Activation and association of Stat3 with Src in v-Src-transformed cell lines. Mol. Cell Biol. 16 (4), 1595–1603. 10.1128/MCB.16.4.1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Wei J., Zou L., Jiang T., Wang G., Chen L., et al. (2020). Ruxolitinib in treatment of severe coronavirus disease 2019 (COVID-19): A multicenter, single-blind, randomized controlled trial. J. Allergy Clin. Immunol. 146 (1), 137–146.e3. 10.1016/j.jaci.2020.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Zhou D., Liu Z., Huang X., Liu Q., Kang Y., et al. (2018). Combination of gemcitabine and erlotinib inhibits recurrent pancreatic cancer growth in mice via the JAK-STAT pathway. Oncol. Rep. 39 (3), 1081–1089. 10.3892/or.2018.6198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. F., Kuan F. C., Yen T. C., Lu M. S., Lin P. Y., Chung Y. H., et al. (2014). IL-6-stimulated CD11b+ CD14+ HLA-DR- myeloid-derived suppressor cells, are associated with progression and poor prognosis in squamous cell carcinoma of the esophagus. Oncotarget 5 (18), 8716–8728. 10.18632/oncotarget.2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Ju L., Rushdi M., Ge C., Zhu C. (2017). Receptor-mediated cell mechanosensing. Mol. Biol. Cell 28 (23), 3134–3155. 10.1091/mbc.E17-04-0228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Yi Y., Xie S., Yu H., Peng H., Zhang G. (2017). The effect of the JAK2 inhibitor TG101209 against T cell acute lymphoblastic leukemia (T-ALL) is mediated by inhibition of JAK-STAT signaling and activation of the crosstalk between apoptosis and autophagy signaling. Oncotarget 8 (63), 106753–106763. 10.18632/oncotarget.22053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury G. G., Marra F., Kiyomoto H., Abboud H. E. (1996). PDGF stimulates tyrosine phosphorylation of JAK 1 protein tyrosine kinase in human mesangial cells. Kidney Int. 49 (1), 19–25. 10.1038/ki.1996.3 [DOI] [PubMed] [Google Scholar]
- Cirillo D., Rachiglio A. M., la Montagna R., Giordano A., Normanno N. (2008). Leptin signaling in breast cancer: An overview. J. Cell Biochem. 105 (4), 956–964. 10.1002/jcb.21911 [DOI] [PubMed] [Google Scholar]
- Collin M., Ginhoux F. (2019). Human dendritic cells. Semin. Cell Dev. Biol. 86, 1–2. 10.1016/j.semcdb.2018.04.015 [DOI] [PubMed] [Google Scholar]
- Combe B., Kivitz A., Tanaka Y., van der Heijde D., Simon J. A., Baraf H. S. B., et al. (2021). Filgotinib versus placebo or adalimumab in patients with rheumatoid arthritis and inadequate response to methotrexate: A phase III randomised clinical trial. Ann. Rheum. Dis. 80 (7), 848–858. 10.1136/annrheumdis-2020-219214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti P., Kempuraj D., Frydas S., Kandere K., Boucher W., Letourneau R., et al. (2003). IL-10 subfamily members: IL-19, IL-20, IL-22, IL-24 and IL-26. Immunol. Lett. 88 (3), 171–174. 10.1016/s0165-2478(03)00087-7 [DOI] [PubMed] [Google Scholar]
- Cooper M. A., Fehniger T. A., Caligiuri M. A. (2001). The biology of human natural killer-cell subsets. Trends Immunol. 22 (11), 633–640. 10.1016/s1471-4906(01)02060-9 [DOI] [PubMed] [Google Scholar]
- Cui J., Shibata Y., Zhu T., Zhou J., Zhang J. (2022). Osteocytes in bone aging: Advances, challenges, and future perspectives. Ageing Res. Rev. 77, 101608. 10.1016/j.arr.2022.101608 [DOI] [PubMed] [Google Scholar]
- Czimmerer Z., Daniel B., Horvath A., Ruckerl D., Nagy G., Kiss M., et al. (2018). The transcription factor STAT6 mediates direct repression of inflammatory enhancers and limits activation of alternatively polarized macrophages. Immunity 48 (1), 75–90.e6. 10.1016/j.immuni.2017.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalagiorgou G., Piperi C., Adamopoulos C., Georgopoulou U., Gargalionis A. N., Spyropoulou A., et al. (2017). Mechanosensor polycystin-1 potentiates differentiation of human osteoblastic cells by upregulating Runx2 expression via induction of JAK2/STAT3 signaling axis. Cell Mol. Life Sci. 74 (5), 921–936. 10.1007/s00018-016-2394-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton C. J., Lemmon C. A. (2021). Fibronectin: Molecular structure, fibrillar structure and mechanochemical signaling. Cells 10 (9), 2443. 10.3390/cells10092443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damsky W., Wang A., Kim D. J., Young B. D., Singh K., Murphy M. J., et al. (2022). Inhibition of type 1 immunity with tofacitinib is associated with marked improvement in longstanding sarcoidosis. Nat. Commun. 13 (1), 3140. 10.1038/s41467-022-30615-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot J., Liang J., Kong L. Y., Wei J., Piao Y., Fuller G., et al. (2012). Modulating antiangiogenic resistance by inhibiting the signal transducer and activator of transcription 3 pathway in glioblastoma. Oncotarget 3 (9), 1036–1048. 10.18632/oncotarget.663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M., Fojo T., Bates S. (2005). Tumour stem cells and drug resistance. Nat. Rev. Cancer 5 (4), 275–284. 10.1038/nrc1590 [DOI] [PubMed] [Google Scholar]
- Deodhar A., Sliwinska-Stanczyk P., Xu H., Baraliakos X., Gensler L. S., Fleishaker D., et al. (2021). Tofacitinib for the treatment of ankylosing spondylitis: A phase III, randomised, double-blind, placebo-controlled study. Ann. Rheum. Dis. 80 (8), 1004–1013. 10.1136/annrheumdis-2020-219601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deodhar A., Van den Bosch F., Poddubnyy D., Maksymowych W. P., van der Heijde D., Kim T. H., et al. (2022). Upadacitinib for the treatment of active non-radiographic axial spondyloarthritis (SELECT-AXIS 2): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 400 (10349), 369–379. 10.1016/S0140-6736(22)01212-0 [DOI] [PubMed] [Google Scholar]
- Ding S., Kingshott P., Thissen H., Pera M., Wang P. Y. (2017). Modulation of human mesenchymal and pluripotent stem cell behavior using biophysical and biochemical cues: A review. Biotechnol. Bioeng. 114 (2), 260–280. 10.1002/bit.26075 [DOI] [PubMed] [Google Scholar]
- Dogra N., Jakhmola-Mani R., Potshangbam A. M., Buch S., Pande Katare D. (2023). CXCR4 as possible druggable target linking inflammatory bowel disease and Parkinson's disease. Metab. Brain Dis. 10.1007/s11011-022-01155-6 [DOI] [PubMed] [Google Scholar]
- Dongre A., Weinberg R. A. (2019). New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 20 (2), 69–84. 10.1038/s41580-018-0080-4 [DOI] [PubMed] [Google Scholar]
- Dorritie K. A., Redner R. L., Johnson D. E. (2014). STAT transcription factors in normal and cancer stem cells. Adv. Biol. Regul. 56, 30–44. 10.1016/j.jbior.2014.05.004 [DOI] [PubMed] [Google Scholar]
- Dufva O., Kankainen M., Kelkka T., Sekiguchi N., Awad S. A., Eldfors S., et al. (2018). Aggressive natural killer-cell leukemia mutational landscape and drug profiling highlight JAK-STAT signaling as therapeutic target. Nat. Commun. 9 (1), 1567. 10.1038/s41467-018-03987-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant L., Watford W. T., Ramos H. L., Laurence A., Vahedi G., Wei L., et al. (2010). Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity 32 (5), 605–615. 10.1016/j.immuni.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdogmus S., Storch U., Danner L., Becker J., Winter M., Ziegler N., et al. (2019). Helix 8 is the essential structural motif of mechanosensitive GPCRs. Nat. Commun. 10 (1), 5784. 10.1038/s41467-019-13722-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esashi E., Wang Y. H., Perng O., Qin X. F., Liu Y. J., Watowich S. S. (2008). The signal transducer STAT5 inhibits plasmacytoid dendritic cell development by suppressing transcription factor IRF8. Immunity 28 (4), 509–520. 10.1016/j.immuni.2008.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers M. A., Offner S., Blanco-Bose W. E., Waibler Z., Kalinke U., Duchosal M. A., et al. (2009). IFNα activates dormant haematopoietic stem cells in vivo . Nature 458 (7240), 904–908. 10.1038/nature07815 [DOI] [PubMed] [Google Scholar]
- Fabbi M., Carbotti G., Ferrini S. (2017). Dual roles of IL-27 in cancer biology and immunotherapy. Mediat. Inflamm. 2017, 1–14. 10.1155/2017/3958069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann R. M., Damjanov N. S., Kivitz A. J., Legedza A., Hoock T., Kinnman N. (2015). A randomized, double-blind, placebo-controlled, twelve-week, dose-ranging study of decernotinib, an oral selective JAK-3 inhibitor, as monotherapy in patients with active rheumatoid arthritis. Arthritis Rheumatol. 67 (2), 334–343. 10.1002/art.38949 [DOI] [PubMed] [Google Scholar]
- Fong K. D., Trindade M. C., Wang Z., Nacamuli R. P., Pham H., Fang T. D., et al. (2005). Microarray analysis of mechanical shear effects on flexor tendon cells. Plast. Reconstr. Surg. 116 (5), 1393–1404. ; discussion 1405-1396. 10.1097/01.prs.0000182345.86453.4f [DOI] [PubMed] [Google Scholar]
- Fuertes M. B., Woo S. R., Burnett B., Fu Y. X., Gajewski T. F. (2013). Type I interferon response and innate immune sensing of cancer. Trends Immunol. 34 (2), 67–73. 10.1016/j.it.2012.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrilovich D. I., Nagaraj S. (2009). Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 9 (3), 162–174. 10.1038/nri2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L. F., Xu D. Q., Wen L. J., Zhang X. Y., Shao Y. T., Zhao X. J. (2005). Inhibition of STAT3 expression by siRNA suppresses growth and induces apoptosis in laryngeal cancer cells. Acta Pharmacol. Sin. 26 (3), 377–383. 10.1111/j.1745-7254.2005.00053.x [DOI] [PubMed] [Google Scholar]
- Gardner A., de Mingo Pulido A., Ruffell B. (2020). Dendritic cells and their role in immunotherapy. Front. Immunol. 11, 924. 10.3389/fimmu.2020.00924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese M. C., Yang F., Ostergaard M., Kinnman N. (2016). Efficacy of VX-509 (decernotinib) in combination with a disease-modifying antirheumatic drug in patients with rheumatoid arthritis: Clinical and MRI findings. Ann. Rheum. Dis. 75 (11), 1979–1983. 10.1136/annrheumdis-2015-208901 [DOI] [PubMed] [Google Scholar]
- Gessani S., Conti L., Del Corno M., Belardelli F. (2014). Type I interferons as regulators of human antigen presenting cell functions. Toxins (Basel) 6 (6), 1696–1723. 10.3390/toxins6061696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glosson-Byers N. L., Sehra S., Kaplan M. H. (2014). STAT4 is required for IL-23 responsiveness in Th17 memory cells and NKT cells. JAKSTAT 3 (3), e955393. 10.4161/21623988.2014.955393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotthardt D., Putz E. M., Straka E., Kudweis P., Biaggio M., Poli V., et al. (2014). Loss of STAT3 in murine NK cells enhances NK cell-dependent tumor surveillance. Blood 124 (15), 2370–2379. 10.1182/blood-2014-03-564450 [DOI] [PubMed] [Google Scholar]
- Gotthardt D., Trifinopoulos J., Sexl V., Putz E. M. (2019). JAK/STAT cytokine signaling at the crossroad of NK cell development and maturation. Front. Immunol. 10, 2590. 10.3389/fimmu.2019.02590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough D. J., Koetz L., Levy D. E. (2013). The MEK-ERK pathway is necessary for serine phosphorylation of mitochondrial STAT3 and Ras-mediated transformation. PLoS One 8 (11), e83395. 10.1371/journal.pone.0083395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouilleux F., Pallard C., Dusanter-Fourt I., Wakao H., Haldosen L. A., Norstedt G., et al. (1995). Prolactin, growth hormone, erythropoietin and granulocyte-macrophage colony stimulating factor induce MGF-Stat5 DNA binding activity. EMBO J. 14 (9), 2005–2013. 10.1002/j.1460-2075.1995.tb07192.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham D. M., Burridge K. (2016). Mechanotransduction and nuclear function. Curr. Opin. Cell Biol. 40, 98–105. 10.1016/j.ceb.2016.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenterberg K. D., Grignol V. P., Raig E. T., Zimmerer J. M., Chan A. N., Blaskovits F. M., et al. (2010). Interleukin-29 binds to melanoma cells inducing Jak-STAT signal transduction and apoptosis. Mol. Cancer Ther. 9 (2), 510–520. 10.1158/1535-7163.MCT-09-0461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidolin D., Marcoli M., Tortorella C., Maura G., Agnati L. F. (2018). G protein-coupled receptor-receptor interactions give integrative dynamics to intercellular communication. Rev. Neurosci. 29 (7), 703–726. 10.1515/revneuro-2017-0087 [DOI] [PubMed] [Google Scholar]
- Harir N., Pecquet C., Kerenyi M., Sonneck K., Kovacic B., Nyga R., et al. (2007). Constitutive activation of Stat5 promotes its cytoplasmic localization and association with PI3-kinase in myeloid leukemias. Blood 109 (4), 1678–1686. 10.1182/blood-2006-01-029918 [DOI] [PubMed] [Google Scholar]
- Harrison C. N., Garcia J. S., Somervaille T. C. P., Foran J. M., Verstovsek S., Jamieson C., et al. (2022a). Addition of navitoclax to ongoing ruxolitinib therapy for patients with myelofibrosis with progression or suboptimal response: Phase II safety and efficacy. J. Clin. Oncol. 40 (15), 1671–1680. 10.1200/JCO.21.02188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison C. N., Schaap N., Vannucchi A. M., Kiladjian J. J., Passamonti F., Zweegman S., et al. (2022b). Safety and efficacy of fedratinib, a selective oral inhibitor of Janus kinase-2 (JAK2), in patients with myelofibrosis and low pretreatment platelet counts. Br. J. Haematol. 198 (2), 317–327. 10.1111/bjh.18207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasni S. A., Gupta S., Davis M., Poncio E., Temesgen-Oyelakin Y., Carlucci P. M., et al. (2021). Phase 1 double-blind randomized safety trial of the Janus kinase inhibitor tofacitinib in systemic lupus erythematosus. Nat. Commun. 12 (1), 3391. 10.1038/s41467-021-23361-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., Leong D. J., Xu L., Hardin J. A., Majeska R. J., Schaffler M. B., et al. (2019). CITED2 mediates the cross-talk between mechanical loading and IL-4 to promote chondroprotection. Ann. N. Y. Acad. Sci. 1442 (1), 128–137. 10.1111/nyas.14021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedvat M., Huszar D., Herrmann A., Gozgit J. M., Schroeder A., Sheehy A., et al. (2009). The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer Cell 16 (6), 487–497. 10.1016/j.ccr.2009.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann A., Kortylewski M., Kujawski M., Zhang C., Reckamp K., Armstrong B., et al. (2010). Targeting Stat3 in the myeloid compartment drastically improves the in vivo antitumor functions of adoptively transferred T cells. Cancer Res. 70 (19), 7455–7464. 10.1158/0008-5472.CAN-10-0736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervas-Stubbs S., Perez-Gracia J. L., Rouzaut A., Sanmamed M. F., Le Bon A., Melero I. (2011). Direct effects of type I interferons on cells of the immune system. Clin. Cancer Res. 17 (9), 2619–2627. 10.1158/1078-0432.CCR-10-1114 [DOI] [PubMed] [Google Scholar]
- Hindupur S. V., Schmid S. C., Koch J. A., Youssef A., Baur E. M., Wang D., et al. (2020). STAT3/5 inhibitors suppress proliferation in bladder cancer and enhance oncolytic Adenovirus therapy. Int. J. Mol. Sci. 21 (3), 1106. 10.3390/ijms21031106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honsho S., Nishikawa S., Amano K., Zen K., Adachi Y., Kishita E., et al. (2009). Pressure-mediated hypertrophy and mechanical stretch induces IL-1 release and subsequent IGF-1 generation to maintain compensative hypertrophy by affecting Akt and JNK pathways. Circ. Res. 105 (11), 1149–1158. 10.1161/CIRCRESAHA.109.208199 [DOI] [PubMed] [Google Scholar]
- Hou Y., Zhu L., Tian H., Sun H. X., Wang R., Zhang L., et al. (2018). IL-23-induced macrophage polarization and its pathological roles in mice with imiquimod-induced psoriasis. Protein Cell 9 (12), 1027–1038. 10.1007/s13238-018-0505-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Hu W. X. (2018). Targeting signaling pathways in multiple myeloma: Pathogenesis and implication for treatments. Cancer Lett. 414, 214–221. 10.1016/j.canlet.2017.11.020 [DOI] [PubMed] [Google Scholar]
- Hu Z., Li Y., Yuan W., Jin L., Leung W. K., Zhang C., et al. (2022). N6-methyladenosine of Socs1 modulates macrophage inflammatory response in different stiffness environments. Int. J. Biol. Sci. 18 (15), 5753–5769. 10.7150/ijbs.74196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter A. M., Newman H., Dezern A. E., Steensma D. P., Niyongere S., Roboz G. J., et al. (2021). Integrated human and murine clinical study establishes clinical efficacy of ruxolitinib in chronic myelomonocytic leukemia. Clin. Cancer Res. 27 (22), 6095–6105. 10.1158/1078-0432.CCR-21-0935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imada K., Bloom E. T., Nakajima H., Horvath-Arcidiacono J. A., Udy G. B., Davey H. W., et al. (1998). Stat5b is essential for natural killer cell-mediated proliferation and cytolytic activity. J. Exp. Med. 188 (11), 2067–2074. 10.1084/jem.188.11.2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubzick C., Choi E. S., Carpenter K. J., Kunkel S. L., Evanoff H., Martinez F. J., et al. (2004). Human pulmonary fibroblasts exhibit altered interleukin-4 and interleukin-13 receptor subunit expression in idiopathic interstitial pneumonia. Am. J. Pathol. 164 (6), 1989–2001. 10.1016/S0002-9440(10)63759-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman P., Parikh F., Lopez-Rivera E., Hailemichael Y., Clark A., Ma G., et al. (2012). Tumor-expressed inducible nitric oxide synthase controls induction of functional myeloid-derived suppressor cells through modulation of vascular endothelial growth factor release. J. Immunol. 188 (11), 5365–5376. 10.4049/jimmunol.1103553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Peng H., Hou Z. (2020). Ajuba: An emerging signal transducer in oncogenesis. Pharmacol. Res. 151, 104546. 10.1016/j.phrs.2019.104546 [DOI] [PubMed] [Google Scholar]
- Jin W. (2020). Role of JAK/STAT3 signaling in the regulation of metastasis, the transition of cancer stem cells, and chemoresistance of cancer by epithelial-mesenchymal transition. Cells 9 (1), 217. 10.3390/cells9010217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Ding L., Ding Z., Fu Y., Song Y., Jing Y., et al. (2020). Tensile force-induced PDGF-BB/PDGFRβ signals in periodontal ligament fibroblasts activate JAK2/STAT3 for orthodontic tooth movement. Sci. Rep. 10 (1), 11269. 10.1038/s41598-020-68068-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. E., O'Keefe R. A., Grandis J. R. (2018). Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 15 (4), 234–248. 10.1038/nrclinonc.2018.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonchere B., Belanger A., Guette C., Barre B., Coqueret O. (2013). STAT3 as a new autophagy regulator. JAKSTAT 2 (3), e24353. 10.4161/jkst.24353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L. L., Alli R., Li B., Geiger T. L. (2016). Differential T cell cytokine receptivity and not signal quality distinguishes IL-6 and IL-10 signaling during Th17 differentiation. J. Immunol. 196 (7), 2973–2985. 10.4049/jimmunol.1402953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney M., Franks L., Lee S., Tiersten A., Makower D. F., Cigler T., et al. (2021). Phase I/II trial of ruxolitinib in combination with trastuzumab in metastatic HER2 positive breast cancer. Breast Cancer Res. Treat. 189 (1), 177–185. 10.1007/s10549-021-06306-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee B. L., Morman R. E., Sun M. (2020). Transcriptional regulation of natural killer cell development and maturation. Adv. Immunol. 146, 1–28. 10.1016/bs.ai.2020.01.001 [DOI] [PubMed] [Google Scholar]
- Khanna D., Padilla C., Tsoi L. C., Nagaraja V., Khanna P. P., Tabib T., et al. (2022). Tofacitinib blocks IFN-regulated biomarker genes in skin fibroblasts and keratinocytes in a systemic sclerosis trial. JCI Insight 7 (17), e159566. 10.1172/jci.insight.159566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchmer M. N., Franco A., Albasanz-Puig A., Murray J., Yagi M., Gao L., et al. (2014). Modulation of vascular smooth muscle cell phenotype by STAT-1 and STAT-3. Atherosclerosis 234 (1), 169–175. 10.1016/j.atherosclerosis.2014.02.029 [DOI] [PubMed] [Google Scholar]
- Ko H. J., Kim Y. J. (2016). Signal transducer and activator of transcription proteins: Regulators of myeloid-derived suppressor cell-mediated immunosuppression in cancer. Arch. Pharm. Res. 39 (11), 1597–1608. 10.1007/s12272-016-0822-9 [DOI] [PubMed] [Google Scholar]
- Kodama H., Fukuda K., Pan J., Makino S., Baba A., Hori S., et al. (1997). Leukemia inhibitory factor, a potent cardiac hypertrophic cytokine, activates the JAK/STAT pathway in rat cardiomyocytes. Circ. Res. 81 (5), 656–663. 10.1161/01.res.81.5.656 [DOI] [PubMed] [Google Scholar]
- Komuro I., Kaida T., Shibazaki Y., Kurabayashi M., Katoh Y., Hoh E., et al. (1990). Stretching cardiac myocytes stimulates protooncogene expression. J. Biol. Chem. 265 (7), 3595–3598. 10.1016/s0021-9258(19)39631-0 [DOI] [PubMed] [Google Scholar]
- Kong L. Y., Wei J., Sharma A. K., Barr J., Abou-Ghazal M. K., Fokt I., et al. (2009). A novel phosphorylated STAT3 inhibitor enhances T cell cytotoxicity against melanoma through inhibition of regulatory T cells. Cancer Immunol. Immunother. 58 (7), 1023–1032. 10.1007/s00262-008-0618-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortylewski M., Kujawski M., Wang T., Wei S., Zhang S., Pilon-Thomas S., et al. (2005). Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat. Med. 11 (12), 1314–1321. 10.1038/nm1325 [DOI] [PubMed] [Google Scholar]
- Kortylewski M., Xin H., Kujawski M., Lee H., Liu Y., Harris T., et al. (2009). Regulation of the IL-23 and IL-12 balance by Stat3 signaling in the tumor microenvironment. Cancer Cell 15 (2), 114–123. 10.1016/j.ccr.2008.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon P., Berry P. A., Stower M. J., Rodrigues G., Mann V. M., Simms M., et al. (2013). JAK-STAT blockade inhibits tumor initiation and clonogenic recovery of prostate cancer stem-like cells. Cancer Res. 73 (16), 5288–5298. 10.1158/0008-5472.CAN-13-0874 [DOI] [PubMed] [Google Scholar]
- Kujawski M., Kortylewski M., Lee H., Herrmann A., Kay H., Yu H. (2008). Stat3 mediates myeloid cell-dependent tumor angiogenesis in mice. J. Clin. Invest. 118 (10), 3367–3377. 10.1172/JCI35213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunigal S., Lakka S. S., Sodadasu P. K., Estes N., Rao J. S. (2009). Stat3-siRNA induces Fas-mediated apoptosis in vitro and in vivo in breast cancer. Int. J. Oncol. 34 (5), 1209–1220. 10.3892/ijo_00000249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisada K., Hirota H., Fujio Y., Matsui H., Tani Y., Yamauchi-Takihara K., et al. (1996). Activation of JAK-STAT and MAP kinases by leukemia inhibitory factor through gp130 in cardiac myocytes. Circulation 94 (10), 2626–2632. 10.1161/01.cir.94.10.2626 [DOI] [PubMed] [Google Scholar]
- Kunnen S. J., Malas T. B., Semeins C. M., Bakker A. D., Peters D. J. M. (2018). Comprehensive transcriptome analysis of fluid shear stress altered gene expression in renal epithelial cells. J. Cell Physiol. 233 (4), 3615–3628. 10.1002/jcp.26222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusmartsev S., Gabrilovich D. I. (2005). STAT1 signaling regulates tumor-associated macrophage-mediated T cell deletion. J. Immunol. 174 (8), 4880–4891. 10.4049/jimmunol.174.8.4880 [DOI] [PubMed] [Google Scholar]
- La Manna S., De Benedictis I., Marasco D. (2021). Proteomimetics of natural regulators of JAK-STAT pathway: Novel therapeutic perspectives. Front. Mol. Biosci. 8, 792546. 10.3389/fmolb.2021.792546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerding J., Kamm R. D., Lee R. T. (2004). Mechanotransduction in cardiac myocytes. Ann. N. Y. Acad. Sci. 1015, 53–70. 10.1196/annals.1302.005 [DOI] [PubMed] [Google Scholar]
- Langowski J. L., Zhang X., Wu L., Mattson J. D., Chen T., Smith K., et al. (2006). IL-23 promotes tumour incidence and growth. Nature 442 (7101), 461–465. 10.1038/nature04808 [DOI] [PubMed] [Google Scholar]
- Laouar Y., Welte T., Fu X. Y., Flavell R. A. (2003). STAT3 is required for Flt3L-dependent dendritic cell differentiation. Immunity 19 (6), 903–912. 10.1016/s1074-7613(03)00332-7 [DOI] [PubMed] [Google Scholar]
- Larmonier N., Marron M., Zeng Y., Cantrell J., Romanoski A., Sepassi M., et al. (2007). Tumor-derived CD4(+)CD25(+) regulatory T cell suppression of dendritic cell function involves TGF-beta and IL-10. Cancer Immunol. Immunother. 56 (1), 48–59. 10.1007/s00262-006-0160-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B., Lee S., Lee Y., Park Y., Shim J. (2021). Emerin represses STAT3 signaling through nuclear membrane-based spatial control. Int. J. Mol. Sci. 22 (13), 6669. 10.3390/ijms22136669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Herrmann A., Deng J. H., Kujawski M., Niu G., Li Z., et al. (2009). Persistently activated Stat3 maintains constitutive NF-κB activity in tumors. Cancer Cell 15 (4), 283–293. 10.1016/j.ccr.2009.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. H., Hsu E. C., Guh J. H., Yang H. C., Wang D., Kulp S. K., et al. (2011). Targeting energy metabolic and oncogenic signaling pathways in triple-negative breast cancer by a novel adenosine monophosphate-activated protein kinase (AMPK) activator. J. Biol. Chem. 286 (45), 39247–39258. 10.1074/jbc.M111.264598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis M., Allebach J., Tse K. F., Zheng R., Baldwin B. R., Smith B. D., et al. (2002). A FLT3-targeted tyrosine kinase inhibitor is cytotoxic to leukemia cells in vitro and in vivo . Blood 99 (11), 3885–3891. 10.1182/blood.v99.11.3885 [DOI] [PubMed] [Google Scholar]
- Li H. S., Yang C. Y., Nallaparaju K. C., Zhang H., Liu Y. J., Goldrath A. W., et al. (2012). The signal transducers STAT5 and STAT3 control expression of Id2 and E2-2 during dendritic cell development. Blood 120 (22), 4363–4373. 10.1182/blood-2012-07-441311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. (2013). JAK-STAT and bone metabolism. JAKSTAT 2 (3), e23930. 10.4161/jkst.23930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Venema V. J., Wang X., Ju H., Venema R. C., Marrero M. B. (1999). Regulation of angiotensin II-induced phosphorylation of STAT3 in vascular smooth muscle cells. J. Biol. Chem. 274 (28), 19846–19851. 10.1074/jbc.274.28.19846 [DOI] [PubMed] [Google Scholar]
- Liang X., Huang X., Zhou Y., Jin R., Li Q. (2016). Mechanical stretching promotes skin tissue regeneration via enhancing mesenchymal stem cell homing and transdifferentiation. Stem Cells Transl. Med. 5 (7), 960–969. 10.5966/sctm.2015-0274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z. W., Guo B. F., Li Y., Li X. J., Li X., Zhao L. J., et al. (2011). Plasmid-based Stat3 siRNA delivered by hydroxyapatite nanoparticles suppresses mouse prostate tumour growth in vivo . Asian J. Androl. 13 (3), 481–486. 10.1038/aja.2010.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T. S., Mahajan S., Frank D. A. (2000). STAT signaling in the pathogenesis and treatment of leukemias. Oncogene 19 (21), 2496–2504. 10.1038/sj.onc.1203486 [DOI] [PubMed] [Google Scholar]
- Little P. R., King V. L., Davis K. R., Cosgrove S. B., Stegemann M. R. (2015). A blinded, randomized clinical trial comparing the efficacy and safety of oclacitinib and ciclosporin for the control of atopic dermatitis in client-owned dogs. Vet. Dermatol 26 (1), 23–30. 10.1111/vde.12186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Wang F., Luo F. (2023). The role of JAK/STAT pathway in fibrotic diseases: Molecular and cellular mechanisms. Biomolecules 13 (1), 119. 10.3390/biom13010119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu H., Meyer C., Li J., Nadalin S., Konigsrainer A., et al. (2013). Transforming growth factor-beta (TGF-beta)-mediated connective tissue growth factor (CTGF) expression in hepatic stellate cells requires Stat3 signaling activation. J. Biol. Chem. 288 (42), 30708–30719. 10.1074/jbc.M113.478685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Wang L., Li S., Zhang T., Chen C., Hu J., et al. (2022). Mechanical stimulation improves rotator cuff tendon-bone healing via activating IL-4/JAK/STAT signaling pathway mediated macrophage M2 polarization. J. Orthop. Transl. 37, 78–88. 10.1016/j.jot.2022.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh C. Y., Arya A., Naema A. F., Wong W. F., Sethi G., Looi C. Y. (2019). Signal transducer and activator of transcription (STATs) proteins in cancer and inflammation: Functions and therapeutic implication. Front. Oncol. 9, 48. 10.3389/fonc.2019.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Yu F. X. (2019). GPCR-hippo signaling in cancer. Cells 8 (5), 426. 10.3390/cells8050426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Meng Z., Chen R., Guan K. L. (2019). The hippo pathway: Biology and pathophysiology. Annu. Rev. Biochem. 88, 577–604. 10.1146/annurev-biochem-013118-111829 [DOI] [PubMed] [Google Scholar]
- Machida T., Nishida K., Nasu Y., Nakahara R., Ozawa M., Harada R., et al. (2017). Inhibitory effect of JAK inhibitor on mechanical stress-induced protease expression by human articular chondrocytes. Inflamm. Res. 66 (11), 999–1009. 10.1007/s00011-017-1083-x [DOI] [PubMed] [Google Scholar]
- Mankan A. K., Greten F. R. (2011). Inhibiting signal transducer and activator of transcription 3: Rationality and rationale design of inhibitors. Expert Opin. Investig. Drugs 20 (9), 1263–1275. 10.1517/13543784.2011.601739 [DOI] [PubMed] [Google Scholar]
- Manokawinchoke J., Pavasant P., Limjeerajarus C. N., Limjeerajarus N., Osathanon T., Egusa H. (2021). Mechanical loading and the control of stem cell behavior. Arch. Oral Biol. 125, 105092. 10.1016/j.archoralbio.2021.105092 [DOI] [PubMed] [Google Scholar]
- Martinac B., Poole K. (2018). Mechanically activated ion channels. Int. J. Biochem. Cell Biol. 97, 104–107. 10.1016/j.biocel.2018.02.011 [DOI] [PubMed] [Google Scholar]
- Mascarenhas J., Baer M. R., Kessler C., Hexner E., Tremblay D., Price L., et al. (2019). Phase II trial of Lestaurtinib, a JAK2 inhibitor, in patients with myelofibrosis. Leuk. Lymphoma 60 (5), 1343–1345. 10.1080/10428194.2018.1532509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B. D., Thodeti C. K., Tytell J. D., Mammoto A., Overby D. R., Ingber D. E. (2010). Ultra-rapid activation of TRPV4 ion channels by mechanical forces applied to cell surface β1 integrins. Integr. Biol. (Camb) 2 (9), 435–442. 10.1039/c0ib00034e [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn M. L., Schnitzler H., Shute R., Ruby B., Slivka D. (2021). The acute effects of exercise and temperature on regional mtDNA. Int. J. Environ. Res. Public Health 18 (12), 6382. 10.3390/ijerph18126382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mease P. J., Deodhar A. A., van der Heijde D., Behrens F., Kivitz A. J., Neal J., et al. (2022). Efficacy and safety of selective TYK2 inhibitor, deucravacitinib, in a phase II trial in psoriatic arthritis. Ann. Rheum. Dis. 81 (6), 815–822. 10.1136/annrheumdis-2021-221664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P. K., Griendling K. K. (2007). Angiotensin II cell signaling: Physiological and pathological effects in the cardiovascular system. Am. J. Physiol. Cell Physiol. 292 (1), C82–C97. 10.1152/ajpcell.00287.2006 [DOI] [PubMed] [Google Scholar]
- Meng A., Anderson K., Nelson C., Ni L., Chuang S. M., Bellanti F., et al. (2022). Exposure-response relationships for the efficacy and safety of filgotinib and its metabolite GS-829845 in subjects with rheumatoid arthritis based on phase 2 and phase 3 studies. Br. J. Clin. Pharmacol. 88 (7), 3211–3221. 10.1111/bcp.15239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa R., Harrison C., Oh S. T., Gerds A. T., Gupta V., Catalano J., et al. (2022). Overall survival in the SIMPLIFY-1 and SIMPLIFY-2 phase 3 trials of momelotinib in patients with myelofibrosis. Leukemia 36 (9), 2261–2268. 10.1038/s41375-022-01637-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millrud C. R., Bergenfelz C., Leandersson K. (2017). On the origin of myeloid-derived suppressor cells. Oncotarget 8 (2), 3649–3665. 10.18632/oncotarget.12278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills C. D., Kincaid K., Alt J. M., Heilman M. J., Hill A. M. (2000). M-1/M-2 macrophages and the Th1/Th2 paradigm. J. Immunol. 164 (12), 6166–6173. 10.4049/jimmunol.164.12.6166 [DOI] [PubMed] [Google Scholar]
- Mimura K., Teh J. L., Okayama H., Shiraishi K., Kua L. F., Koh V., et al. (2018). PD-L1 expression is mainly regulated by interferon gamma associated with JAK-STAT pathway in gastric cancer. Cancer Sci. 109 (1), 43–53. 10.1111/cas.13424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrherr J., Haber M., Breitenecker K., Aigner P., Moritsch S., Voronin V., et al. (2019). JAK-STAT inhibition impairs K-RAS-driven lung adenocarcinoma progression. Int. J. Cancer 145 (12), 3376–3388. 10.1002/ijc.32624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz A. J., Ghione P., Jacobsen E., Ruan J., Schatz J. H., Noor S., et al. (2021). A phase 2 biomarker-driven study of ruxolitinib demonstrates effectiveness of JAK/STAT targeting in T-cell lymphomas. Blood 138 (26), 2828–2837. 10.1182/blood.2021013379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movahedi K., Guilliams M., Van den Bossche J., Van den Bergh R., Gysemans C., Beschin A., et al. (2008). Identification of discrete tumor-induced myeloid-derived suppressor cell subpopulations with distinct T cell-suppressive activity. Blood 111 (8), 4233–4244. 10.1182/blood-2007-07-099226 [DOI] [PubMed] [Google Scholar]
- Mukherjee A., Tsuchiwata S., Nicholas T., Cook J. A., Modesto I., Su C., et al. (2022). Exposure-response characterization of tofacitinib efficacy in moderate to severe ulcerative colitis: Results from phase II and phase III induction and maintenance studies. Clin. Pharmacol. Ther. 112 (1), 90–100. 10.1002/cpt.2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen M., Gonzalez-Perez R. R. (2016). Leptin-induced JAK/STAT signaling and cancer growth. Vaccines (Basel) 4 (3), 26. 10.3390/vaccines4030026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller R. (2019). JAK inhibitors in 2019, synthetic review in 10 points. Eur. J. Intern Med. 66, 9–17. 10.1016/j.ejim.2019.05.022 [DOI] [PubMed] [Google Scholar]
- Murray P. J. (2007). The JAK-STAT signaling pathway: Input and output integration. J. Immunol. 178 (5), 2623–2629. 10.4049/jimmunol.178.5.2623 [DOI] [PubMed] [Google Scholar]
- Naing A., Powderly J. D., Nemunaitis J. J., Luke J. J., Mansfield A. S., Messersmith W. A., et al. (2022). Exploring the safety, effect on the tumor microenvironment, and efficacy of itacitinib in combination with epacadostat or parsaclisib in advanced solid tumors: a phase I study. J. Immunother. Cancer 10 (3), e004223. 10.1136/jitc-2021-004223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najjar I., Deglesne P. A., Schischmanoff P. O., Fabre E. E., Boisson-Dupuis S., Nimmerjahn F., et al. (2010). STAT1-dependent IgG cell-surface expression in a human B cell line derived from a STAT1-deficient patient. J. Leukoc. Biol. 87 (6), 1145–1152. 10.1189/jlb.1109714 [DOI] [PubMed] [Google Scholar]
- Neradugomma N. K., Subramaniam D., Tawfik O. W., Goffin V., Kumar T. R., Jensen R. A., et al. (2014). Prolactin signaling enhances colon cancer stemness by modulating Notch signaling in a Jak2-STAT3/ERK manner. Carcinogenesis 35 (4), 795–806. 10.1093/carcin/bgt379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen K. B., Salazar-Mather T. P., Dalod M. Y., Van Deusen J. B., Wei X. Q., Liew F. Y., et al. (2002). Coordinated and distinct roles for IFN-αβ, IL-12, and IL-15 regulation of NK cell responses to viral infection. J. Immunol. 169 (8), 4279–4287. 10.4049/jimmunol.169.8.4279 [DOI] [PubMed] [Google Scholar]
- Nishina T., Fujita T., Yoshizuka N., Sugibayashi K., Murayama K., Kuboki Y. (2022). Safety, tolerability, pharmacokinetics and preliminary antitumour activity of an antisense oligonucleotide targeting STAT3 (danvatirsen) as monotherapy and in combination with durvalumab in Japanese patients with advanced solid malignancies: A phase 1 study. BMJ Open 12 (10), e055718. 10.1136/bmjopen-2021-055718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu G., Wright K. L., Ma Y., Wright G. M., Huang M., Irby R., et al. (2005). Role of Stat3 in regulating p53 expression and function. Mol. Cell Biol. 25 (17), 7432–7440. 10.1128/MCB.25.17.7432-7440.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donoghue R. J., Knight D. A., Richards C. D., Prele C. M., Lau H. L., Jarnicki A. G., et al. (2012). Genetic partitioning of interleukin-6 signalling in mice dissociates Stat3 from Smad3-mediated lung fibrosis. EMBO Mol. Med. 4 (9), 939–951. 10.1002/emmm.201100604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea J. J., Schwartz D. M., Villarino A. V., Gadina M., McInnes I. B., Laurence A. (2015). The JAK-STAT pathway: Impact on human disease and therapeutic intervention. Annu. Rev. Med. 66, 311–328. 10.1146/annurev-med-051113-024537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura M., Uchida T., Terui Y., Hayakawa F., Kobayashi Y., Taniwaki M., et al. (2015). Phase I study of OPB-51602, an oral inhibitor of signal transducer and activator of transcription 3, in patients with relapsed/refractory hematological malignancies. Cancer Sci. 106 (7), 896–901. 10.1111/cas.12683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D. Y., Lee S. H., Han S. W., Kim M. J., Kim T. M., Kim T. Y., et al. (2015). Phase I study of OPB-31121, an oral STAT3 inhibitor, in patients with advanced solid tumors. Cancer Res. Treat. 47 (4), 607–615. 10.4143/crt.2014.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M. R., Verdan F. F., Hufford M. M., Dent A. L., Kaplan M. H. (2016). STAT3 impairs STAT5 activation in the development of IL-9-secreting T cells. J. Immunol. 196 (8), 3297–3304. 10.4049/jimmunol.1501801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrand-Rosenberg S., Grusby M. J., Clements V. K. (2000). Cutting edge: STAT6-deficient mice have enhanced tumor immunity to primary and metastatic mammary carcinoma. J. Immunol. 165 (11), 6015–6019. 10.4049/jimmunol.165.11.6015 [DOI] [PubMed] [Google Scholar]
- Padda S. K., Reckamp K. L., Koczywas M., Neal J. W., Kawashima J., Kong S., et al. (2022). A phase 1b study of erlotinib and momelotinib for the treatment of EGFR-mutated, tyrosine kinase inhibitor-naive metastatic non-small cell lung cancer. Cancer Chemother. Pharmacol. 89 (1), 105–115. 10.1007/s00280-021-04369-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pail G., Huf W., Pjrek E., Winkler D., Willeit M., Praschak-Rieder N., et al. (2011). Bright-light therapy in the treatment of mood disorders. Neuropsychobiology 64 (3), 152–162. 10.1159/000328950 [DOI] [PubMed] [Google Scholar]
- Pan J., Fukuda K., Kodama H., Makino S., Takahashi T., Sano M., et al. (1997). Role of angiotensin II in activation of the JAK/STAT pathway induced by acute pressure overload in the rat heart. Circ. Res. 81 (4), 611–617. 10.1161/01.res.81.4.611 [DOI] [PubMed] [Google Scholar]
- Pan J., Fukuda K., Saito M., Matsuzaki J., Kodama H., Sano M., et al. (1999). Mechanical stretch activates the JAK/STAT pathway in rat cardiomyocytes. Circ. Res. 84 (10), 1127–1136. 10.1161/01.res.84.10.1127 [DOI] [PubMed] [Google Scholar]
- Park C., Li S., Cha E., Schindler C. (2000). Immune response in Stat2 knockout mice. Immunity 13 (6), 795–804. 10.1016/s1074-7613(00)00077-7 [DOI] [PubMed] [Google Scholar]
- Park S. J., Nakagawa T., Kitamura H., Atsumi T., Kamon H., Sawa S., et al. (2004). IL-6 regulates in vivo dendritic cell differentiation through STAT3 activation. J. Immunol. 173 (6), 3844–3854. 10.4049/jimmunol.173.6.3844 [DOI] [PubMed] [Google Scholar]
- Patel M. R., Dash A., Jacobson B. A., Ji Y., Baumann D., Ismail K., et al. (2019). JAK/STAT inhibition with ruxolitinib enhances oncolytic virotherapy in non-small cell lung cancer models. Cancer Gene Ther. 26 (11-12), 411–418. 10.1038/s41417-018-0074-6 [DOI] [PubMed] [Google Scholar]
- Paukku K., Valgeirsdottir S., Saharinen P., Bergman M., Heldin C. H., Silvennoinen O. (2000). Platelet-derived growth factor (PDGF)-induced activation of signal transducer and activator of transcription (Stat) 5 is mediated by PDGF beta-receptor and is not dependent on c-src, fyn, jak1 or jak2 kinases. Biochem. J. 345 Pt 3, 759–766. 10.1042/bj3450759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeva E., Guttman-Yassky E., Banerjee A., Sinclair R., Cox L. A., Zhu L., et al. (2022). Maintenance, withdrawal, and re-treatment with ritlecitinib and brepocitinib in patients with alopecia areata in a single-blind extension of a phase 2a randomized clinical trial. J. Am. Acad. Dermatol 87 (2), 390–393. 10.1016/j.jaad.2021.12.008 [DOI] [PubMed] [Google Scholar]
- Peterson W. M., Wang Q., Tzekova R., Wiegand S. J. (2000). Ciliary neurotrophic factor and stress stimuli activate the Jak-STAT pathway in retinal neurons and glia. J. Neurosci. 20 (11), 4081–4090. 10.1523/JNEUROSCI.20-11-04081.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plimack E. R., Lorusso P. M., McCoon P., Tang W., Krebs A. D., Curt G., et al. (2013). AZD1480: a phase I study of a novel JAK2 inhibitor in solid tumors. Oncologist 18 (7), 819–820. 10.1634/theoncologist.2013-0198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploscariu N., Kuczera K., Malek K. E., Wawrzyniuk M., Dey A., Szoszkiewicz R. (2014). Single molecule studies of force-induced S2 site exposure in the mammalian Notch negative regulatory domain. J. Phys. Chem. B 118 (18), 4761–4770. 10.1021/jp5004825 [DOI] [PubMed] [Google Scholar]
- Qin Q., Yang H., Zhang C., Han X., Guo J., Fan Z., et al. (2021). lncRNA HHIP-AS1 promotes the osteogenic differentiation potential and inhibits the migration ability of periodontal ligament stem cells. Stem Cells Int. 2021, 1–12. 10.1155/2021/5595580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesnelle K. M., Boehm A. L., Grandis J. R. (2007). STAT-mediated EGFR signaling in cancer. J. Cell Biochem. 102 (2), 311–319. 10.1002/jcb.21475 [DOI] [PubMed] [Google Scholar]
- Ram P. T., Iyengar R. (2001). G protein coupled receptor signaling through the src and Stat3 pathway: Role in proliferation and transformation. Oncogene 20 (13), 1601–1606. 10.1038/sj.onc.1204186 [DOI] [PubMed] [Google Scholar]
- Reddy D., Kumavath R., Ghosh P., Barh D. (2019). Lanatoside C induces G2/M cell cycle arrest and suppresses cancer cell growth by attenuating MAPK, Wnt, JAK-STAT, and PI3K/AKT/mTOR signaling pathways. Biomolecules 9 (12), 792. 10.3390/biom9120792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich K., Thyssen J. P., Blauvelt A., Eyerich K., Soong W., Rice Z. P., et al. (2022). Efficacy and safety of abrocitinib versus dupilumab in adults with moderate-to-severe atopic dermatitis: A randomised, double-blind, multicentre phase 3 trial. Lancet 400 (10348), 273–282. 10.1016/S0140-6736(22)01199-0 [DOI] [PubMed] [Google Scholar]
- Reilley M. J., McCoon P., Cook C., Lyne P., Kurzrock R., Kim Y., et al. (2018). STAT3 antisense oligonucleotide AZD9150 in a subset of patients with heavily pretreated lymphoma: Results of a phase 1b trial. J. Immunother. Cancer 6 (1), 119. 10.1186/s40425-018-0436-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W., Wu S., Wu Y., Liu T., Zhao X., Li Y. (2019). MicroRNA-196a/-196b regulate the progression of hepatocellular carcinoma through modulating the JAK/STAT pathway via targeting SOCS2. Cell Death Dis. 10 (5), 333. 10.1038/s41419-019-1530-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins S. H., Tessmer M. S., Van Kaer L., Brossay L. (2005). Direct effects of T-bet and MHC class I expression, but not STAT1, on peripheral NK cell maturation. Eur. J. Immunol. 35 (3), 757–765. 10.1002/eji.200425797 [DOI] [PubMed] [Google Scholar]
- Rocca Y. S., Roberti M. P., Arriaga J. M., Amat M., Bruno L., Pampena M. B., et al. (2013). Altered phenotype in peripheral blood and tumor-associated NK cells from colorectal cancer patients. Innate Immun. 19 (1), 76–85. 10.1177/1753425912453187 [DOI] [PubMed] [Google Scholar]
- Roger I., Milara J., Montero P., Cortijo J. (2021). The role of JAK/STAT molecular pathway in vascular remodeling associated with pulmonary hypertension. Int. J. Mol. Sci. 22 (9), 4980. 10.3390/ijms22094980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose-John S. (2018). Interleukin-6 family cytokines. Cold Spring Harb. Perspect. Biol. 10 (2), a028415. 10.1101/cshperspect.a028415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roxburgh C. S., McMillan D. C. (2016). Therapeutics targeting innate immune/inflammatory responses through the interleukin-6/JAK/STAT signal transduction pathway in patients with cancer. Transl. Res. 167 (1), 61–66. 10.1016/j.trsl.2015.08.013 [DOI] [PubMed] [Google Scholar]
- Ruan Z., Yang X., Cheng W. (2019). OCT4 accelerates tumorigenesis through activating JAK/STAT signaling in ovarian cancer side population cells. Cancer Manag. Res. 11, 389–399. 10.2147/CMAR.S180418 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ruperto N., Brunner H. I., Synoverska O., Ting T. V., Mendoza C. A., Spindler A., et al. (2021). Tofacitinib in juvenile idiopathic arthritis: A double-blind, placebo-controlled, withdrawal phase 3 randomised trial. Lancet 398 (10315), 1984–1996. 10.1016/S0140-6736(21)01255-1 [DOI] [PubMed] [Google Scholar]
- Ruwhof C., van der Laarse A. (2000). Mechanical stress-induced cardiac hypertrophy: Mechanisms and signal transduction pathways. Cardiovasc Res. 47 (1), 23–37. 10.1016/s0008-6363(00)00076-6 [DOI] [PubMed] [Google Scholar]
- Sadoshima J., Izumo S. (1993). Mechanical stretch rapidly activates multiple signal transduction pathways in cardiac myocytes: Potential involvement of an autocrine/paracrine mechanism. EMBO J. 12 (4), 1681–1692. 10.1002/j.1460-2075.1993.tb05813.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E., Marshall C. J. (2002). ROCK and Dia have opposing effects on adherens junctions downstream of Rho. Nat. Cell Biol. 4 (6), 408–415. 10.1038/ncb796 [DOI] [PubMed] [Google Scholar]
- Salminen A., Kauppinen A., Kaarniranta K. (2019). AMPK activation inhibits the functions of myeloid-derived suppressor cells (MDSC): Impact on cancer and aging. J. Mol. Med. Berl. 97 (8), 1049–1064. 10.1007/s00109-019-01795-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathaliyawala T., O'Gorman W. E., Greter M., Bogunovic M., Konjufca V., Hou Z. E., et al. (2010). Mammalian target of rapamycin controls dendritic cell development downstream of Flt3 ligand signaling. Immunity 33 (4), 597–606. 10.1016/j.immuni.2010.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A. K., Cama G., Ghuman M., Hughes F. J., Gharibi B. (2017). Sprouty 2, an early response gene regulator of FosB and mesenchymal stem cell proliferation during mechanical loading and osteogenic differentiation. J. Cell Biochem. 118 (9), 2606–2614. 10.1002/jcb.26035 [DOI] [PubMed] [Google Scholar]
- Seko Y., Seko Y., Takahashi N., Shibuya M., Yazaki Y. (1999). Pulsatile stretch stimulates vascular endothelial growth factor (VEGF) secretion by cultured rat cardiac myocytes. Biochem. Biophys. Res. Commun. 254 (2), 462–465. 10.1006/bbrc.1998.9969 [DOI] [PubMed] [Google Scholar]
- Sen M., Pollock N. I., Black J., DeGrave K. A., Wheeler S., Freilino M. L., et al. (2015). JAK kinase inhibition abrogates STAT3 activation and head and neck squamous cell carcinoma tumor growth. Neoplasia 17 (3), 256–264. 10.1016/j.neo.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezgin D., Aslan G., Sahin K., Tuzcu M., Ilhan N., Sahna E. (2020). The effects of melatonin against atherosclerosis-induced endothelial dysfunction and inflammation in hypercholesterolemic rats. Arch. Physiol. Biochem., 1–8. 10.1080/13813455.2020.1838550 [DOI] [PubMed] [Google Scholar]
- Shahrara S., Amin M. A., Woods J. M., Haines G. K., Koch A. E. (2003). Chemokine receptor expression and in vivo signaling pathways in the joints of rats with adjuvant-induced arthritis. Arthritis Rheum. 48 (12), 3568–3583. 10.1002/art.11344 [DOI] [PubMed] [Google Scholar]
- Shankaran V., Ikeda H., Bruce A. T., White J. M., Swanson P. E., Old L. J., et al. (2001). IFNγ and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature 410 (6832), 1107–1111. 10.1038/35074122 [DOI] [PubMed] [Google Scholar]
- Shao F., Pang X., Baeg G. H. (2021). Targeting the JAK/STAT signaling pathway for breast cancer. Curr. Med. Chem. 28 (25), 5137–5151. 10.2174/0929867328666201207202012 [DOI] [PubMed] [Google Scholar]
- Shibue T., Weinberg R. A. (2017). EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 14 (10), 611–629. 10.1038/nrclinonc.2017.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims N. A. (2020). The JAK1/STAT3/SOCS3 axis in bone development, physiology, and pathology. Exp. Mol. Med. 52 (8), 1185–1197. 10.1038/s12276-020-0445-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S. P., Plantinga T. S., van de Veerdonk F. L., Heinhuis B., Hoischen A., Joosten L. A., et al. (2011). STAT1 hyperphosphorylation and defective IL12R/IL23R signaling underlie defective immunity in autosomal dominant chronic mucocutaneous candidiasis. PLoS One 6 (12), e29248. 10.1371/journal.pone.0029248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Xiong B., Zheng H., Manyande A., Guan X., Cao F., et al. (2017). STAT1 as a downstream mediator of ERK signaling contributes to bone cancer pain by regulating MHC II expression in spinal microglia. Brain Behav. Immun. 60, 161–173. 10.1016/j.bbi.2016.10.009 [DOI] [PubMed] [Google Scholar]
- Soriano S. F., Serrano A., Hernanz-Falcon P., Martin de Ana A., Monterrubio M., Martinez C., et al. (2003). Chemokines integrate JAK/STAT and G-protein pathways during chemotaxis and calcium flux responses. Eur. J. Immunol. 33 (5), 1328–1333. 10.1002/eji.200323897 [DOI] [PubMed] [Google Scholar]
- Stronach E. A., Alfraidi A., Rama N., Datler C., Studd J. B., Agarwal R., et al. (2011). HDAC4-regulated STAT1 activation mediates platinum resistance in ovarian cancer. Cancer Res. 71 (13), 4412–4422. 10.1158/0008-5472.CAN-10-4111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam D., Angulo P., Ponnurangam S., Dandawate P., Ramamoorthy P., Srinivasan P., et al. (2020). Suppressing STAT5 signaling affects osteosarcoma growth and stemness. Cell Death Dis. 11 (2), 149. 10.1038/s41419-020-2335-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara I., Yamada H., Mizuno S. (2004). STAT1 knockout mice are highly susceptible to pulmonary mycobacterial infection. Tohoku J. Exp. Med. 202 (1), 41–50. 10.1620/tjem.202.41 [DOI] [PubMed] [Google Scholar]
- Sun Y., Moretti L., Giacalone N. J., Schleicher S., Speirs C. K., Carbone D. P., et al. (2011). Inhibition of JAK2 signaling by TG101209 enhances radiotherapy in lung cancer models. J. Thorac. Oncol. 6 (4), 699–706. 10.1097/JTO.0b013e31820d9d11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K., Nakayama M., Hayakawa Y., Kojima Y., Ikeda H., Imai N., et al. (2017). IFN-gamma is required for cytotoxic T cell-dependent cancer genome immunoediting. Nat. Commun. 8, 14607. 10.1038/ncomms14607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Takeuchi T., Izutsu H., Kaneko Y., Kato D., Fukuda M., et al. (2021). Patient- and physician-reported outcomes from two phase 3 randomized studies (RAJ3 and RAJ4) of peficitinib (ASP015K) in Asian patients with rheumatoid arthritis. Arthritis Res. Ther. 23 (1), 221. 10.1186/s13075-021-02590-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y., Ross J. L., Cowell J. K. (2014). The involvement of JAK-STAT3 in cell motility, invasion, and metastasis. JAKSTAT 3 (1), e28086. 10.4161/jkst.28086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorn M., Guha P., Cunetta M., Espat N. J., Miller G., Junghans R. P., et al. (2016). Tumor-associated GM-CSF overexpression induces immunoinhibitory molecules via STAT3 in myeloid-suppressor cells infiltrating liver metastases. Cancer Gene Ther. 23 (6), 188–198. 10.1038/cgt.2016.19 [DOI] [PubMed] [Google Scholar]
- Tokusumi Y., Tokusumi T., Schulz R. A. (2018). Mechanical stress to Drosophila larvae stimulates a cellular immune response through the JAK/STAT signaling pathway. Biochem. Biophys. Res. Commun. 502 (3), 415–421. 10.1016/j.bbrc.2018.05.192 [DOI] [PubMed] [Google Scholar]
- Torrero M. N., Xia X., Henk W., Yu S., Li S. (2006). Stat1 deficiency in the host enhances interleukin-12-mediated tumor regression. Cancer Res. 66 (8), 4461–4467. 10.1158/0008-5472.CAN-05-3554 [DOI] [PubMed] [Google Scholar]
- Tsutsumi T., Kajiya H., Goto K. T., Takahashi Y., Okabe K. (2013). Hyperocclusion up-regulates CCL3 expression in CCL2-and CCR2-deficient mice. J. Dent. Res. 92 (1), 65–70. 10.1177/0022034512467803 [DOI] [PubMed] [Google Scholar]
- Vargas-Hernandez A., Forbes L. R. (2019). JAK/STAT proteins and their biological impact on NK cell development and function. Mol. Immunol. 115, 21–30. 10.1016/j.molimm.2018.12.005 [DOI] [PubMed] [Google Scholar]
- Vargas-Hernandez A., Mace E. M., Zimmerman O., Zerbe C. S., Freeman A. F., Rosenzweig S., et al. (2018). Ruxolitinib partially reverses functional natural killer cell deficiency in patients with signal transducer and activator of transcription 1 (STAT1) gain-of-function mutations. J. Allergy Clin. Immunol. 141 (6), 2142–2155.e5. 10.1016/j.jaci.2017.08.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez-Dunddel D., Pan F., Zeng Q., Gorbounov M., Albesiano E., Fu J., et al. (2013). STAT3 regulates arginase-I in myeloid-derived suppressor cells from cancer patients. J. Clin. Invest. 123 (4), 1580–1589. 10.1172/JCI60083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma N. K., Davies A. M., Long A., Kelleher D., Volkov Y. (2010). STAT3 knockdown by siRNA induces apoptosis in human cutaneous T-cell lymphoma line Hut78 via downregulation of Bcl-xL. Cell Mol. Biol. Lett. 15 (2), 342–355. 10.2478/s11658-010-0008-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarino A. V., Kanno Y., Ferdinand J. R., O'Shea J. J. (2015). Mechanisms of Jak/STAT signaling in immunity and disease. J. Immunol. 194 (1), 21–27. 10.4049/jimmunol.1401867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarino A. V., Sciume G., Davis F. P., Iwata S., Zitti B., Robinson G. W., et al. (2017). Subset- and tissue-defined STAT5 thresholds control homeostasis and function of innate lymphoid cells. J. Exp. Med. 214 (10), 2999–3014. 10.1084/jem.20150907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincenti F., Tedesco Silva H., Busque S., O'Connell P., Friedewald J., Cibrik D., et al. (2012). Randomized phase 2b trial of tofacitinib (CP-690,550) in de novo kidney transplant patients: Efficacy, renal function and safety at 1 year. Am. J. Transpl. 12 (9), 2446–2456. 10.1111/j.1600-6143.2012.04127.x [DOI] [PubMed] [Google Scholar]
- von Hoff L., Kargel E., Franke V., McShane E., Schulz-Beiss K. W., Patone G., et al. (2019). Autocrine LTA signaling drives NF-κB and JAK-STAT activity and myeloid gene expression in Hodgkin lymphoma. Blood 133 (13), 1489–1494. 10.1182/blood-2018-08-871293 [DOI] [PubMed] [Google Scholar]
- Wang K. S., Ritz J., Frank D. A. (1999). IL-2 induces STAT4 activation in primary NK cells and NK cell lines, but not in T cells. J. Immunol. 162 (1), 299–304. 10.4049/jimmunol.162.1.299 [DOI] [PubMed] [Google Scholar]
- Wang L., Yi T., Kortylewski M., Pardoll D. M., Zeng D., Yu H. (2009a). IL-17 can promote tumor growth through an IL-6-Stat3 signaling pathway. J. Exp. Med. 206 (7), 1457–1464. 10.1084/jem.20090207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P. Y., Chow H. H., Lai J. Y., Liu H. L., Tsai W. B. (2009b). Dynamic compression modulates chondrocyte proliferation and matrix biosynthesis in chitosan/gelatin scaffolds. J. Biomed. Mater Res. B Appl. Biomater. 91 (1), 143–152. 10.1002/jbm.b.31384 [DOI] [PubMed] [Google Scholar]
- Wang P. Y., Chow H. H., Tsai W. B., Fang H. W. (2009c). Modulation of gene expression of rabbit chondrocytes by dynamic compression in polyurethane scaffolds with collagen gel encapsulation. J. Biomater. Appl. 23 (4), 347–366. 10.1177/0885328208093684 [DOI] [PubMed] [Google Scholar]
- Wang P. Y., Thissen H., Kingshott P. (2016). Modulation of human multipotent and pluripotent stem cells using surface nanotopographies and surface-immobilised bioactive signals: A review. Acta Biomater. 45, 31–59. 10.1016/j.actbio.2016.08.054 [DOI] [PubMed] [Google Scholar]
- Wang P. Y., Tsai W. B. (2013). Modulation of the proliferation and matrix synthesis of chondrocytes by dynamic compression on genipin-crosslinked chitosan/collagen scaffolds. J. Biomater. Sci. Polym. Ed. 24 (5), 507–519. 10.1080/09205063.2012.696310 [DOI] [PubMed] [Google Scholar]
- Wang P. Y., Tsai W. B., Voelcker N. H. (2012). Screening of rat mesenchymal stem cell behaviour on polydimethylsiloxane stiffness gradients. Acta Biomater. 8 (2), 519–530. 10.1016/j.actbio.2011.09.030 [DOI] [PubMed] [Google Scholar]
- Wang P. Y., Yu H. T., Tsai W. B. (2010). Modulation of alignment and differentiation of skeletal myoblasts by submicron ridges/grooves surface structure. Biotechnol. Bioeng. 106 (2), 285–294. 10.1002/bit.22697 [DOI] [PubMed] [Google Scholar]
- Wang P. Y., Yu J., Lin J. H., Tsai W. B. (2011). Modulation of alignment, elongation and contraction of cardiomyocytes through a combination of nanotopography and rigidity of substrates. Acta Biomater. 7 (9), 3285–3293. 10.1016/j.actbio.2011.05.021 [DOI] [PubMed] [Google Scholar]
- Wang T. L., Yang Y. H., Chang H., Hung C. R. (2004b). Angiotensin II signals mechanical stretch-induced cardiac matrix metalloproteinase expression via JAK-STAT pathway. J. Mol. Cell Cardiol. 37 (3), 785–794. 10.1016/j.yjmcc.2004.06.016 [DOI] [PubMed] [Google Scholar]
- Wang T., Niu G., Kortylewski M., Burdelya L., Shain K., Zhang S., et al. (2004a). Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat. Med. 10 (1), 48–54. 10.1038/nm976 [DOI] [PubMed] [Google Scholar]
- Wang T. T., Zhao Y. L., Peng L. S., Chen N., Chen W., Lv Y. P., et al. (2017). Tumour-activated neutrophils in gastric cancer foster immune suppression and disease progression through GM-CSF-PD-L1 pathway. Gut 66 (11), 1900–1911. 10.1136/gutjnl-2016-313075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Shen Y., Wang S., Shen Q., Zhou X. (2018). The role of STAT3 in leading the crosstalk between human cancers and the immune system. Cancer Lett. 415, 117–128. 10.1016/j.canlet.2017.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnette R., Banerjee A., Sikirica V., Peeva E., Wyrwich K. (2022). Characterizing the relationships between patient-reported outcomes and clinician assessments of alopecia areata in a phase 2a randomized trial of ritlecitinib and brepocitinib. J. Eur. Acad. Dermatol Venereol. 36 (4), 602–609. 10.1111/jdv.17909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfle S. J., Strebovsky J., Bartz H., Sahr A., Arnold C., Kaiser C., et al. (2011). PD-L1 expression on tolerogenic APCs is controlled by STAT-3. Eur. J. Immunol. 41 (2), 413–424. 10.1002/eji.201040979 [DOI] [PubMed] [Google Scholar]
- Wong A. L., Soo R. A., Tan D. S., Lee S. C., Lim J. S., Marban P. C., et al. (2015). Phase I and biomarker study of OPB-51602, a novel signal transducer and activator of transcription (STAT) 3 inhibitor, in patients with refractory solid malignancies. Ann. Oncol. 26 (5), 998–1005. 10.1093/annonc/mdv026 [DOI] [PubMed] [Google Scholar]
- Woods D. M., Ramakrishnan R., Laino A. S., Berglund A., Walton K., Betts B. C., et al. (2018). Decreased suppression and increased phosphorylated STAT3 in regulatory T cells are associated with benefit from adjuvant PD-1 blockade in resected metastatic melanoma. Clin. Cancer Res. 24 (24), 6236–6247. 10.1158/1078-0432.CCR-18-1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worm M., Thyssen J. P., Schliemann S., Bauer A., Shi V. Y., Ehst B., et al. (2022). The pan-JAK inhibitor delgocitinib in a cream formulation demonstrates dose response in chronic hand eczema in a 16-week randomized phase IIb trial. Br. J. Dermatol 187 (1), 42–51. 10.1111/bjd.21037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Deng Z., Peng Y., Han L., Liu J., Wang L., et al. (2017). Ascites-derived IL-6 and IL-10 synergistically expand CD14(+)HLA-DR(-/low) myeloid-derived suppressor cells in ovarian cancer patients. Oncotarget 8 (44), 76843–76856. 10.18632/oncotarget.20164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Gong Y., Chen Y., Yu D., Wang X., Zhang X., et al. (2017). IL-6 promotes epithelial-to-mesenchymal transition of human peritoneal mesothelial cells possibly through the JAK2/STAT3 signaling pathway. Am. J. Physiol. Ren. Physiol. 313 (2), F310–F318. 10.1152/ajprenal.00428.2016 [DOI] [PubMed] [Google Scholar]
- Xin P., Xu X., Deng C., Liu S., Wang Y., Zhou X., et al. (2020). The role of JAK/STAT signaling pathway and its inhibitors in diseases. Int. Immunopharmacol. 80, 106210. 10.1016/j.intimp.2020.106210 [DOI] [PubMed] [Google Scholar]
- Xiong H., Su W. Y., Liang Q. C., Zhang Z. G., Chen H. M., Du W., et al. (2009). Inhibition of STAT5 induces G1 cell cycle arrest and reduces tumor cell invasion in human colorectal cancer cells. Lab. Invest. 89 (6), 717–725. 10.1038/labinvest.2009.11 [DOI] [PubMed] [Google Scholar]
- Yahata Y., Shirakata Y., Tokumaru S., Yamasaki K., Sayama K., Hanakawa Y., et al. (2003). Nuclear translocation of phosphorylated STAT3 is essential for vascular endothelial growth factor-induced human dermal microvascular endothelial cell migration and tube formation. J. Biol. Chem. 278 (41), 40026–40031. 10.1074/jbc.M301866200 [DOI] [PubMed] [Google Scholar]
- Yang J., Liao D., Chen C., Liu Y., Chuang T. H., Xiang R., et al. (2013). Tumor-associated macrophages regulate murine breast cancer stem cells through a novel paracrine EGFR/Stat3/Sox-2 signaling pathway. Stem Cells 31 (2), 248–258. 10.1002/stem.1281 [DOI] [PubMed] [Google Scholar]
- Yang R., Lirussi D., Thornton T. M., Jelley-Gibbs D. M., Diehl S. A., Case L. K., et al. (2015). Mitochondrial Ca²⁺ and membrane potential, an alternative pathway for Interleukin 6 to regulate CD4 cell effector function. Elife 4, e06376. 10.7554/eLife.06376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q., Wolkerstorfer A., Lapid O., Niessen F. B., Van Zuijlen P. P. M., Gibbs S. (2023). The JAK-STAT pathway in keloid pathogenesis A systematic review with qualitative synthesis. Exp. Dermatol. 10.1111/exd.14747 [DOI] [PubMed] [Google Scholar]
- Yu H., Kortylewski M., Pardoll D. (2007). Crosstalk between cancer and immune cells: Role of STAT3 in the tumour microenvironment. Nat. Rev. Immunol. 7 (1), 41–51. 10.1038/nri1995 [DOI] [PubMed] [Google Scholar]
- Yu H., Pardoll D., Jove R. (2009). STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 9 (11), 798–809. 10.1038/nrc2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Wang Y., Yan F., Zhang P., Li H., Zhao H., et al. (2014). Noncanonical NF-κB activation mediates STAT3-stimulated Ido upregulation in myeloid-derived suppressor cells in breast cancer. J. Immunol. 193 (5), 2574–2586. 10.4049/jimmunol.1400833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiser R., Socie G., Schroeder M. A., Abhyankar S., Vaz C. P., Kwon M., et al. (2022). Efficacy and safety of itacitinib versus placebo in combination with corticosteroids for initial treatment of acute graft-versus-host disease (GRAVITAS-301): A randomised, multicentre, double-blind, phase 3 trial. Lancet Haematol. 9 (1), e14–e25. 10.1016/S2352-3026(21)00367-7 [DOI] [PubMed] [Google Scholar]
- Zhang J., Wei X., Zhang W., Wang F., Li Q. (2020a). MiR-326 targets MDK to regulate the progression of cardiac hypertrophy through blocking JAK/STAT and MAPK signaling pathways. Eur. J. Pharmacol. 872, 172941. 10.1016/j.ejphar.2020.172941 [DOI] [PubMed] [Google Scholar]
- Zhang J. Z., Lu T. W., Stolerman L. M., Tenner B., Yang J. R., Zhang J. F., et al. (2020b). Phase separation of a PKA regulatory subunit controls cAMP compartmentation and oncogenic signaling. Cell 182 (6), 1531–1544.e15. 10.1016/j.cell.2020.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Zhang C., He J., Guo Q., Hu D., Yang X., et al. (2015). STAT3 inhibitor stattic enhances radiosensitivity in esophageal squamous cell carcinoma. Tumour Biol. 36 (3), 2135–2142. 10.1007/s13277-014-2823-y [DOI] [PubMed] [Google Scholar]
- Zhang T., Ma J., Cao X. (2003). Grb2 regulates Stat3 activation negatively in epidermal growth factor signalling. Biochem. J. 376 (2), 457–464. 10.1042/BJ20030668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Liu Y., Yan Z., Yang H., Sun W., Yao Y., et al. (2020c). IL-6 promotes PD-L1 expression in monocytes and macrophages by decreasing protein tyrosine phosphatase receptor type O expression in human hepatocellular carcinoma. J. Immunother. Cancer 8 (1), e000285. 10.1136/jitc-2019-000285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Liu M., Bennett S., Wang Z., Pfleger K. D. G., Xu J. (2021). The molecular structure and role of CCL2 (MCP-1) and C-C chemokine receptor CCR2 in skeletal biology and diseases. J. Cell Physiol. 236 (10), 7211–7222. 10.1002/jcp.30375 [DOI] [PubMed] [Google Scholar]
- Zhu S., Phatarpekar P. V., Denman C. J., Senyukov V. V., Somanchi S. S., Nguyen-Jackson H. T., et al. (2014). Transcription of the activating receptor NKG2D in natural killer cells is regulated by STAT3 tyrosine phosphorylation. Blood 124 (3), 403–411. 10.1182/blood-2013-05-499707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn E., Nelson E. A., Mohseni M., Porcheray F., Kim H., Litsa D., et al. (2006). IL-2 regulates FOXP3 expression in human CD4+CD25+ regulatory T cells through a STAT-dependent mechanism and induces the expansion of these cells in vivo . Blood 108 (5), 1571–1579. 10.1182/blood-2006-02-004747 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.