Abstract
Th cells, which orchestrate immune responses to various pathogens, differentiate from naïve CD4 T cells into several subsets that stimulate and regulate immune responses against various types of pathogens, as well as a variety of immune-related diseases. Decades of research have revealed that the fate decision processes are controlled by cytokines, cytokine receptor signaling, and master transcription factors that drive the differentiation programs. Since the Th1 and Th2 paradigm was proposed, many subsets have been added to the list. In this review, I will summarize these events, including the fate decision processes, subset functions, transcriptional regulation, metabolic regulation, and plasticity and heterogeneity. I will also introduce current topics of interest.
Keywords: Helper T cell, T-lymphocyte subset, Cell differentiation, Adaptive immune response, Transcription factors, Cytokines
INTRODUCTION
Pathogens can infect many different tissues and cellular compartments, and the life cycles of these pathogens can vary considerably. To overcome this, the host immune system has developed pathogen-specific strategies. For example, Th cells (effector CD4 T cells) orchestrate immune responses against many different pathogens (1,2,3,4). Naïve CD4 T cells recirculate via the blood and lymphatic systems, gathering information about pathogens by binding to antigens presented by dendritic cells (DCs) in lymphoid tissues. DCs also secrete cytokines when they present these pathogen-specific antigens to naïve CD4 T cells, thereby triggering differentiation pathways (1,2,3,4). Activated CD4 T cells proliferate and differentiate into different immune cell subsets, which determine the type of immune response (1,2,3,4). Research in the field of Th cell differentiation and function began about 30 years ago, and it is still progressing. In this review, I will summarize the basic principles that govern Th cell differentiation (Fig. 1). Other topics in this field, including the microbiota and plasticity/heterogeneity, will be covered by other articles in this issue.
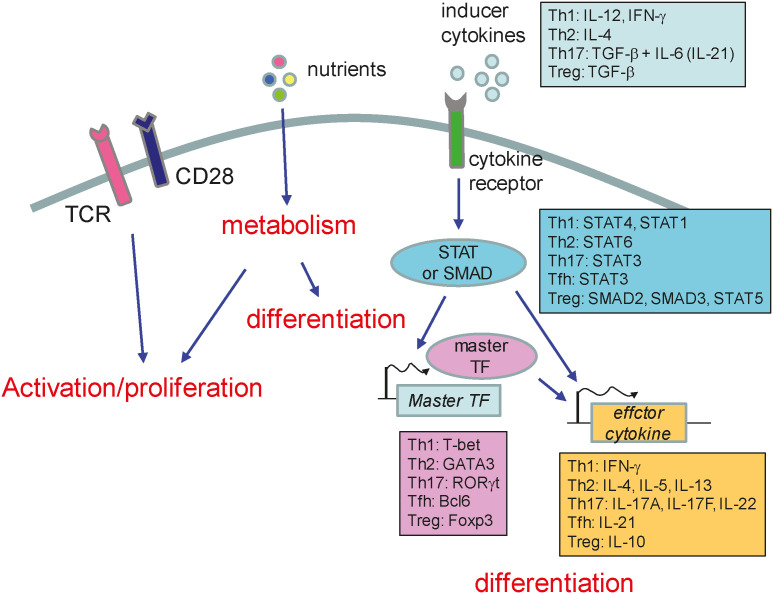
Figure 1. Schematic diagram of Th cell differentiation. Th cell differentiation pathway is determined by inducer cytokines that are present during T cell activation. The cytokine signaling activates STAT or SMAD, which in turn induces master transcription factors. Master transcription factors drive the subset-specific differentiation programs. Th cell proliferation and differentiation are also influenced by nutrient or metabolites.
BRIEF HISTORY OF Th SUBSETS
For a long time, Th cells were regarded as cells that aid activation of B cells and macrophages by producing cytokines. In 1986, Mosmann and colleagues (5) published a seminal work showing that Th cells comprise 2 distinct cell types based on their cytokine profiles, namely, Th1 and Th2. The different cytokines produced by these cell types lead to mutually exclusive functional properties (5). Th1 cells activate macrophages and mediate immune responses against intracellular pathogens, while Th2 cells stimulate B cells (5). Identification of Th cell heterogeneity and differences in functional properties was a major conceptual advance in the field, and triggered further study of the molecular mechanisms that regulate the fate-determining processes and functional properties. These efforts led to identification of cytokines, signaling pathways, and transcription factors as determinants of Th1 and Th2 subset differentiation. In 2005, Th17 cells were identified and proposed as an independent subset (6,7,8). Th17 cells induce inflammation and mediate immune responses against extracellular bacteria and fungi (9,10). Another independent subset, called follicular helper T (Tfh) cells, was also identified (11,12,13). Tfh cells reside in the germinal centers of lymphoid tissue and interact with B cells to trigger maturation. It was thought that B cell stimulation was the role of Th2 cells; however, this is now thought to be the role of Tfh cells. In addition to activator cells, repressor cell types have also been identified; these are termed Treg cells (14,15,16). Many other subsets have also been identified, including IL-9-producing Th9 cells, IL-22-producing Th22 cells, and IL-10 producing type-1 regulatory T (Tr1) cells. Th9, Th22, and Tr1 cells will not be covered in this review because our understanding of their functions and roles is still in its infancy.
Th CELL FUNCTIONS
Effector CD4 T cells either stimulate or repress other immune cells. The most important mediators of these functions are cytokines (1,2,3,4). Th1 cells produce IFN-γ, which stimulates macrophages and CTLs. Stimulated macrophages kill intracellular pathogens by activating microbicidal mechanisms, including facilitation of fusion of phagosomes with lysosomes, activation of NADPH oxidase, and generation of reactive oxygen species (17). Activation of CTL requires the help of Th1 cells via secretion of IFN-γ and CD40L signaling, which trigger CTL differentiation. Th2 cells produce IL-4, IL-5, and IL-13, which mobilize or stimulate eosinophils, basophils, and mast cells to combat helminthic parasites (18). IL-13 also stimulates intestinal epithelial cells to increase cell turnover and movement, as well as acting on smooth muscle cells in the intestine to increase contractility, all of which help to remove parasites from the gut. Th17 cells produce IL-17A, IL-17F, and IL-22, which stimulate epithelial cells, fibroblasts, and stromal cells to produce GM-CSF, recruit neutrophils, and induce inflammation, thereby mediating immune responses against extracellular bacteria and fungi (9,10). Tfh cells produce IL-21 and activate B cells in the germinal center to induce maturation via somatic hypermutation and class-switch recombination (19). Treg cells inhibit effector T cell and B cell responses, as well as innate immune responses (20). Treg cells produce IL-10, TGF-β, and IL-35 to inhibit immune responses, although they can use other mechanisms too. Treg cells express high amounts of IL2Rα on their surface, leading to deprivation of IL-2, which is required for growth of effector CD4 T cells. Treg cells express coinhibitory receptors such as Ctla-4 and Lag3, which interfere with the interaction with costimulatory molecules and MHC class II molecules expressed by antigen-presenting cells (APCs), thereby preventing T cell activation (21,22). Treg cells also kill CD4 T cells directly by secreting cytotoxins. They also express high amounts of ectonucleotidases such as CD39 and CD73, leading to production of immunosuppressive adenosine, which promotes T cell anergy (23). Although Treg development in the thymus (tTreg) and Treg in the periphery (pTreg) cells have overlapping functions, tTregs are more specialized at preventing autoimmune responses against self-antigens, whereas pTregs are more specialized in inducing tolerance to foreign antigens.
Each CD4 T cell subset cross regulates other subsets. For example, Th1 cells inhibit Th2 and Th17 cell differentiation by suppressing expression of IL-4 and IL-17, respectively (7,24). Th2 cells repress Th1 differentiation by inhibiting IL-12Rβ2 and IL-17 expression, respectively (7,25). Tfh cells inhibit differentiation of other subsets via Bcl6-mediated repression of Prdma1 (which encodes Blimp-1) expression (26,27).
In addition to the induction of protective immunity against infectious agents, Th cells are responsible for the pathology of many other diseases caused by aberrant immune responses (1,2,3,4). For example, Th1 and Th17 cells plays critical roles in rheumatoid arthritis, type I diabetes, and multiple sclerosis; Th2 cells in allergic diseases such as asthma, rhinitis, and allergic dermatitis; and Treg cells in autoimmune disease, allergy, and cancer. Thus, regulating subset-specific functions is an effective strategy for treating these immune-related diseases. Indeed, based on this knowledge, many therapeutic strategies have been, or are being, developed (28,29).
SIGNAL TRANSDUCTION AND TRANSCRIPTIONAL REGULATION
Naïve CD4 T cells differentiate into various subsets of effector cells when they recognize their cognate antigens presented in conjunction with MHC II molecules on the surface of DCs in lymphoid tissues (1,2,3). These subsets include Th1, Th2, Th17, Tfh, and Treg cells (1,2,3). The fates of these subsets are determined by the cytokines (produced mainly by APCs) present in the environment at the time of activation (1,2,3). The cytokines bind to their cognate receptors on naïve CD4 T cells and activate the STAT or SMAD pathway (1,2,3). Th1 cell differentiation is induced by IL-12 or IFN-γ, which activate Stat4 or Stat1, respectively (1,2,3). Th2 cell differentiation is induced by IL-4, which activates Stat6 (1,2,3). Th17 cell differentiation is induced by TGF-β plus IL-6 (or IL-21), which activate Stat3 (1,2,3). Tfh cell differentiation is induced by IL-21, which also activates Stat3 (1,2,3). Treg cells are rather different in that they can be generated during T cell development in the thymus (tTreg cells) or can differentiate from naïve CD4 pTreg cells (1,2,3). In vitro, Treg cells can be differentiated by TGF-β, which activates Smad2 and Smad3 (1,2,3).
STAT proteins enter the nucleus and induce global epigenetic changes as well as expression of master transcription factors which drive differentiation programs (1,2,3,4). T-bet drives the Th1 program, Gata-3 drives the Th2 program, Rorγt drives the Th17 program, Bcl6 drives the Tfh program, and Foxp3 drives the Treg program. These transcription factors induce expression of a variety of genes required for differentiation, and for the effector functions, of each subset. Division of the roles played by STATs and the master transcription factors is the focus of much interest. Although both factors are important for differentiation, it seems likely that STATs act as pioneering factors that shape global enhancer landscapes and gene expression patterns, whereas master transcription factors regulate a limited set of genes that is required for subset-specific differentiation and function (30,31).
Although master transcription factors drive the differentiation program in Th cells, many other transcription factors play important roles in the process (32,33). In addition to the master transcription factors, Irf1, Runx3, Hlx, Ets-1, Nfatc1, and Bhlhe40 play important roles in Th1 cell differentiation; c-Maf, Notch/CSL, IRF4, Gfi-1, and Yy1 in Th2 differentiation; Ifr4, Batf, and Runx1 in Th17 differentiation; and Runx1, Nr4a2, Foxo family members, Satb1, and Helio in Treg differentiation (32,33). These transcription factors form a coordinated network that drives the differentiation programs (34,35,36), even in the absence of master transcription factors (36). The roles of these transcription factors depend on individual factors. In general, these factors either enhance transcription, induce changes in epigenetic status, or promote organization of the relevant genes or gene loci within the genome (32,33).
EPIGENETIC REGULATION
Along with transcription factors, the genetic loci of characteristic cytokine genes have been investigated extensively with respect to their role in regulating Th cell differentiation (37,38). This is due in part to the fact that the differentiation process is easily controlled in vitro by cytokines, enabling investigation of cell fates within a short period of time. Such studies have reported interesting findings: 1) epigenetic changes accompany gene expression during differentiation; 2) poised state of cytokine loci even before differentiation; 3) coordinate expression of cytokine genes; 4) inter-chromosomal interactions between 2 opposing loci; and 5) insulation of cytokine loci.
Epigenetic changes during Th cell differentiation
Marked changes in gene expression occur during CD4 T cell activation and differentiation. Many of the genes expressed by activated T cells are required for cell division. Since naïve T cells are in a quiescent state, transition to actively dividing cells requires systemic changes in gene expression. Yet more genes are required for subset-specific effector functions; for example, genes encoding cytokines that mediate the effector functions of a particular subset. Not surprisingly, naïve CD4 T cells undergo epigenetic changes such as increased accessibility by DNase I, histone acetylation, and DNA methylation (37,38). Ifng and the Th2 cytokine locus (the Il4-Il13-Il5 locus) show subset-specific epigenetic changes. During Th1 differentiation, the Ifng locus opens, but the Th2 cytokine locus remains closed. Likewise, the Th2 cytokine locus opens during Th2 differentiation, but the Ifng locus remains closed. This subset-specific chromatin modification is influenced by master transcription factors such as T-bet and Gata-3. Thus, one of the roles of these transcription factors is to open the chromatin at the loci of subset-specific genes to make it “competent” for expression of these genes.
Poised state of cytokine loci before differentiation
Certain genetic loci, such as the Ifng locus, are demethylated even before Th1 differentiation. This hypomethylation pattern seems to be established early during T cell development, including that of naïve CD4 T cells, suggesting that these loci are predisposed to expression (37,38). During Th1 differentiation, this demethylation pattern at the Ifng locus is preserved; however, the locus becomes methylated during Th2 differentiation. Genome-wide studies of the epigenetic status of chromatin and gene expression in CD4 T cells revealed that the histone modification status of key cytokine and transcription factor loci correlates strongly with gene expression patterns (39). Interestingly, this study also found that Tbx21 and Gata3 loci contain both active (H3K4me3) and suppressive (H3K27me3) histone marks (39). This bivalent status with respect to histone marks also predispose gene expression, suggesting that cells are preparing for rapid changes in gene expression that occur during activation and differentiation. The poised status of multiple key genetic loci facilitates easy conversion from one differentiated state to another.
Coordinate expression of cytokine genes
The Ifng locus and the Th2 cytokine locus are regulated by promoters, enhancers, and silencers in the same manner as other genetic loci (37,38). Since the Il4, Il13, and Il5 genes are clustered, it is of interest to see whether they are coordinately regulated in Th2 cells. One of the regulatory elements that exerts coordinate expression of clustered genes is the locus control region (LCR), best exemplified by the β-globin LCR (40). The LCR is defined as a sequence that enables copy number-dependent expression in transgenic mice (40). The Th2 cytokine locus contains an LCR, called the Th2 LCR, which is interspersed in the intron regions of the Rad50 gene located between the Il13 and Il5 genes (41). The Th2 LCR undergoes epigenetic changes during Th2 differentiation by recruiting GATA3 and other transcription factors such as Yy1, Stat6, Satb1, and Irf4 (42,43,44). This process is essential for Th2 cytokine expression since deletion of the Th2 LCR or its components abolishes Th2 cytokine expression (42,45,46). The Th2 LCR induces chromosomal looping to facilitate interactions with the promoters of Th2 cytokine genes, thereby forming a basis for coordinated regulation (47).
Inter-chromosomal interactions between 2 opposing loci
Chromosomal interactions occur not only intra-chromosomally, but also inter-chromosomally. Chromosomal interaction between 2 different chromosomes has been suggested for long time, but it was proved experimentally in the study of Ifng and Th2 loci, shedding light on a novel mechanism of gene regulation (48). Although they are located on different chromosomes, Ifng and RHS6, a component of the Th2 LCR, in naïve CD4 T cells interact with each other. This physical interaction seems to maintain a poised state for expression of each gene until the cell is activated, since this interaction is replaced with intrachromosomal interactions upon Th2 cell differentiation (48). In naïve CD4 T cells, RHS6 also interacts with the Il17 promoter through Oct-1 and CTCF, suggesting occurrence of inter-chromosomal interactions among multiple chromosomes (49). When Oct-1, CTCF, or the interacting genetic loci are deficient, the interaction does not occur and expression of Il17 increases (49), supporting its role in cytokine gene regulation.
Insulation of cytokine loci
The Ifng locus is located about 40 kb downstream of the IL22 gene in humans and 245 kb downstream of the Il22 gene in mice. Since Il22 is a Th17-specific gene, expression of Ifng and Il22 should be regulated separately. Indeed, an insulator sequence between these gene loci was identified by genome-wide screening of CTCF binding sites and cohesion binding sites (50,51). The Ifng locus is insulated by −70 and +66 boundary elements, which contain CTCF and the cohesion component Rad21 (50,51). Although recruitment of CTCF to the −70 site is conserved in naïve, Th1, and Th2 cells, CTCF recruitment to the Ifng intronic region and +66 site is specific to Th1 cells (50,51), suggesting that this insulator sequence, and its binding to CTCF, may play a role in forming the intra-chromosomal loops needed for the Ifng gene transcription.
PLASTICITY AND HETEROGENEITY
I will provide a very brief overview of this topic as it is covered in depth by another article in this issue. Initially, the status of Th1 and Th2 cells was thought to be stable and terminally differentiated; this is because reversal between subsets cannot be induced after long-term culture (52), and production of specific cytokines seems to be stably maintained (53). However, later studies showed that Th subsets are not permanently differentiated; rather they can differentiate into another subset under certain circumstances. This phenomenon is called plasticity (54,55). Early in vitro studies suggested that forced expression of master regulators by already differentiated cells induces alternative differentiation pathways (55). For example, Th1 cells produce IL-4 upon transfection with Gata-3, and Th2 cells can express IFN-γ upon transfection of T-bet. A fate-mapping study using a mouse model of experimental autoimmune encephalomyelitis (EAE) revealed that most cells producing IFN-γ previously expressed IL-17A (56). Studies in animal models of diabetes and cancer show that Th17 cells acquire Th1-like properties (57,58).
In addition to plasticity, CD4 T cells show functional heterogeneity. Cells co-expressing T-bet and RORγt are present in the gut and inflamed brain tissue (56,59). These cells express both IFN-γ and IL-17, and are thought to play an important role in immune responses to Mycobacterium tuberculosis (60). Treg cells often co-express T-bet, Gata3, or Rorγt. T-bet-expressing Treg cells inhibit type I immune responses (61). Gata3 is frequently co-expressed with Foxp3 by Treg cells and is thought to help control Th2-type immune response (62). Treg cells co-expressing Irf4 or Stat3 control Th2- and Th17-mediated immune responses, respectively (63,64). Thus, co-expression of transcription factors by other subsets seems to endow Treg cells with the ability to control specific effector functions.
The plasticity and heterogeneity of Th subsets modifies and expands the characteristics of each subset. Plasticity is thought to be an adaptation to a changing environment or to pathogens. Since some pathogens change their habitats (e.g., by infecting different cells/organs) and their virulence strategy during their life cycle, the immune system needs to change its defensive strategies to combat them.
METABOLIC REGULATION
When naïve CD4 T cells are activated and differentiate into effector cells, they undergo drastic changes in their metabolic programs (1,65,66,67). During this process, differentiating and proliferating cells require energy, as well as the building blocks to synthesize DNA, RNA, proteins, and lipids. Effector subsets of Th cells, namely, Th1, Th2, and Th17 cells, rely more heavily on glycolysis than Treg cells (1,34,65,66). Most cells generate ATP via oxidative phosphorylation (OXPHOS), which occurs in the mitochondria. However, rapidly dividing cells such as cancer cells use aerobic glycolysis instead of OXPHOS to generate ATP. Although it generates less ATP than OXPHOS, aerobic glycolysis can provide many of the materials required for biosynthesis in cells proliferating under hypoxic conditions. This phenomenon, called the “Warburg effect,” also occurs in effector T cells, which are rapidly dividing cells. Tfh cells also show enhanced glycolysis, but cell-autonomous IL-2 inhibits the differentiation and metabolic programs of Tfh cells. Th17 cells also use glutaminolysis and fatty acid synthesis programs, whereas fatty acid synthesis antagonizes differentiation of Treg cells. By contrast, Treg cells rely on mitochondrial activity for their differentiation, homeostasis, and function.
Since Th cell differentiation requires metabolic reprogramming, factors that regulate this process affect Th cell differentiation (65). These factors include 1) nutrients/metabolites, 2) transcription factors, and 3) signaling molecules.
Nutrients/metabolites
Glucose is a readily available nutrient that provides energy to activated and proliferating T cells, as well as building blocks for nucleic acids, proteins, lipids, and carbohydrates. Glucose-6-phosphate, the first intermediate of glycolysis, can be converted to fructose-6-phosphate to yield pyruvate, which is used to synthesize glycogen, or shuttled to synthesize purines and pyrimidines via the pentose phosphate pathway. TCR and/or costimulatory signaling through CD28 triggers rapid uptake of glucose by upregulating the glucose transporter Glut1 and by increasing glycolytic flux (1,65,66,67). Thus, activated CD4 T cells are highly dependent on glucose uptake. Indeed, Th1 and Th17 cells have an increased glycolytic rate. Chemical or genetic inhibition of specific steps within the glycolytic pathway leads to a marked reduction in effector T cell proliferation and cytokine production. Deprivation of glucose impairs T cell activation and survival, highlighting the importance of glucose for the T cell activation and differentiation processes (1,34,65,66). Glucose deprivation also inhibits IFN-γ production, as well as reducing production of phosphoenolpyruvate, which supports Ca2+/NFAT signaling required for glycolytic reprogramming in T cells (1,65,66,67). Unlike effector T cell subsets, Treg cells are not affected by Glut1 deficiency (1,34,65,66). Blocking glycolysis with 2-DG, an inhibitor of hexokinase (an enzyme that regulates the rate limiting step of glycolysis), inhibits differentiation of CD4 and CD8 T cells into effector cells (1,65,66,67). Deficiency of lactate dehydrogenase A, an enzyme that converts lactate to pyruvate, inhibits ATP generation and PI3K/Akt activation, thereby repressing T cell activation (68). High levels of glycolytic metabolism inhibit induction of Foxp3, instead inducing Th17 cell differentiation (69).
Fatty acid metabolism is a critical part of T cell differentiation. Fatty acid oxidation is essential for Treg cell generation and function, and for making membrane components by proliferating effector T cells, including Th1, Th2, and Th17 cells (70). Proliferating activated T cells show increased fatty acid synthesis, with a concomitant reduction in fatty acid oxidation. Fatty acid metabolism is important for the balance between Th17 and Treg cells: fatty acids aid the generation and function of Treg cells, while controlling the pathogenicity of Th17 cells (70).
Transcription factors
Myc, induced by TCR stimulation, is essential for metabolic reprogramming of T cell activation and function. Myc (71), which plays an essential role in glycolysis and glutaminolysis in activated T cells, induces Glut1 and several glycolytic enzymes (71). When Myc is ablated, glycolysis is impaired, resulting in decreased T cell activation and growth (72). Myc also stimulates glutaminolysis by increasing expression of glutaminase and glutamine transporters (73).
HIF-1α plays an important role in Th cell differentiation and function (74) by enhancing Th17 cell differentiation via stimulation of Il17 gene transcription while reducing Treg cell generation via Foxp3 degradation (75). Conditional deletion of HIF-1α causes resistance to EAE, with reduced infiltration of the CNS by Th17 cells (69). In addition, HIF-1α induces glycolysis under hypoxic conditions. The oxygen-sensing function is regulated by 2 proteins; the von Hippel-Lindau (Vhl) complex supports and prolyl-4-hydroxylase domain (Phd) proteins oppose the function. Upon activation, Vhl-deficient T cells show augmented glycolysis but reduced mitochondrial oxidative metabolism, leading to enhanced effector programming, effector/memory cell differentiation, and cell death (76,77). Phd-deficient CD4 T cells, however, show increased effector cell differentiation but decreased Treg cell differentiation (78).
Signaling molecules
The mTOR is a serine/threonine kinase that transmits signals from the PI3K-Akt pathway. mTOR forms 2 functionally distinct complexes, mTORC1 and mTORC2, and is essential for regulating glucose metabolism and upregulating glycolysis. It is also required for effector T cell differentiation; indeed, mTOR deficiency causes failure of effector T cell differentiation and leads to Treg cell generation instead (79). mTORC1 and mTORC2 play distinct roles during differentiation of Th subsets. CD4 T cells lacking mTORC1 activity due to the absence of one of its components (Rheb) failed to generate Th1 and Th17 cells (80), whereas those lacking mTORC2 activity due to the absence of Rictor failed to generate Th2 cells (80,81). Excessive mTOR activity caused by Pten-deficiency reduces the stability of Treg cells, leading to uncontrolled Th1 and Tfh cell responses (82,83).
AMP-activated protein kinase (AMPK) is a glucose-sensitive metabolic sensor that regulates mRNA translation and glutamine-dependent mitochondrial metabolism (84). AMPK promotes catabolic processes but inhibits anabolic processes. It also promotes OXPHOS, mitochondrial biogenesis, fatty acid oxidation, and autophagy, but inhibits glycolysis, glutaminolysis, and glycogen and fatty acid synthesis (84). AMPK promotes energy conservation by T cells, as well as maintaining T cell bioenergetics and viability (84). It also negatively regulates effector Th cell differentiation by inhibiting mTORC1 activity. Ampka1-deficient T cells show reduced metabolic plasticity and mitochondrial bioenergetic when glucose is limited (84). Under glucose-limiting conditions, reduced expression of IFN-γ mRNA and protein affects Th1 differentiation. By contrast, AMPK induces differentiation of naïve CD4 T into Treg cells (85).
CHALLENGES TO THE Th SUBSET PARADIGM
The paradigm of Th subsets has contributed greatly to our understanding of the immune system. The paradigm divides immune responses into categories, and provides a simple framework for understanding otherwise complex phenomena. Based on this paradigm, many immune-related diseases have been categorized as type I, type II, and type III diseases. This categorization is also applied to innate immune cells, such as innate lymphoid cells and macrophages. The paradigm was originally proposed, and mainly studied, in the context of in vitro-differentiated CD4 T cells, generation of which requires excessive amounts of cytokines and neutralizing antibodies.
The plasticity and heterogeneity of Th cells modifies their functions rather elegantly without necessarily challenging the classical view of Th cell subsets, which states that these subsets are terminally differentiated and stable. Recent technological advances such as single cell RNA sequencing (scRNA-seq) and CyTOF have made it possible to analyze gene expression at the single cell level. Studies using these technologies show that effector Th cells are much more heterogeneous than the paradigm suggests.
scRNA-seq and computational analysis show that Th17 cells isolated from animal models of autoimmune diseases are heterogeneous and show a spectrum of cellular states (86,87). Tortola et al. (88) used CyTOF to analyze expression of subset-specific marker proteins expressed by in vitro-differentiated Th cells and by Th cells in animal disease models. They found that effector Th cells are far more heterogeneous than expected, even under controlled stimulation conditions. Kiner et al. (89) analyzed gut CD4 T cells using scRNA-seq and ATAC-seq. Surprisingly, they found that CD4 T cells do not follow the classic subset-specific gene expression pattern. IFN-γ and IL-17A are expressed by most effector cells, many of which co-express both genes (89). Transcriptional and epigenetic patterns are determined by infectious agents rather than by cytokine and master transcription factor signatures (89). scRNA-seq followed by trajectory analysis revealed that Th1 cells and Tfh cells bifurcate during their differentiation in malaria infection model (90). These results challenge the Th subset paradigm, and suggest that Th cells exist in a heterogeneous state of differentiation, which may change according to the environment. Thus, more studies in this area are needed.
CONCLUSIONS AND PERSPECTIVES
Th cells play essential roles in coordinating immune responses to different pathogens. They are also responsible for many immune pathologies. Thus, research on Th cells (either the fundamental mechanisms underlying differentiation and function, or therapeutic applications based on the results of such studies) is ongoing. New technological advances will speed up progress. For example, scRNA-seq can reveal detailed gene expression patterns in individual cells within a large population, providing information about possible differentiation pathways. CRISPR/Cas9 technology enables functional study of genes of interest on a large scale, either in vitro or in vivo. Introduction of the concept of super-enhancers, which are defined as regulatory regions that are highly enriched in transcription factors and activating/suppressive epigenetic marks, will facilitate more rapid study of regulatory regions, since they can be identified by genome wide approaches and bioinformatics analysis. These new technologies or concepts will enable comprehensive analysis of the molecular mechanisms underlying these processes, while challenging the current paradigm. I anticipate and hope that more and more new and interesting findings about the underlying molecular mechanisms will come to light, and that we will make progress in the treatment of difficult-to-cure diseases.
ACKNOWLEDGEMENTS
This work was supported by the National Research Foundation of Korea (NRF) grants funded by the Korean government (2021K2A9A2A06048161, 2022R1A2B5B03001840, 2022R1A4A5032688 to GRL).
Abbreviations
- AMPK
AMP-activated protein kinase
- APC
antigen-presenting cell
- DC
dendritic cell
- EAE
experimental autoimmune encephalomyelitis
- LCR
locus control region
- OXPHOS
oxidative phosphorylation
- Phd
prolyl-4-hydroxylase domain
- pTreg
regulatory T cell in the periphery
- scRNA-seq
single cell RNA sequencing
- Tfh
follicular helper T
- Tr1
type-1 regulatory T
- tTreg
regulatory T cell development in the thymus
- Vhl
von Hippel-Lindau
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
References
- 1.Saravia J, Chapman NM, Chi H. Helper T cell differentiation. Cell Mol Immunol. 2019;16:634–643. doi: 10.1038/s41423-019-0220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy KM, Ouyang W, Farrar JD, Yang J, Ranganath S, Asnagli H, Afkarian M, Murphy TL. Signaling and transcription in T helper development. Annu Rev Immunol. 2000;18:451–494. doi: 10.1146/annurev.immunol.18.1.451. [DOI] [PubMed] [Google Scholar]
- 4.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136:2348–2357. [PubMed] [Google Scholar]
- 6.Bettelli E, Kuchroo VK. IL-12- and IL-23-induced T helper cell subsets: birds of the same feather flock together. J Exp Med. 2005;201:169–171. doi: 10.1084/jem.20042279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 8.Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel DD, Kuchroo VK. Th17 cell pathway in human immunity: lessons from genetics and therapeutic interventions. Immunity. 2015;43:1040–1051. doi: 10.1016/j.immuni.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 10.Ouyang W, Kolls JK, Zheng Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity. 2008;28:454–467. doi: 10.1016/j.immuni.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chtanova T, Tangye SG, Newton R, Frank N, Hodge MR, Rolph MS, Mackay CR. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J Immunol. 2004;173:68–78. doi: 10.4049/jimmunol.173.1.68. [DOI] [PubMed] [Google Scholar]
- 12.Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, Ma L, Wang YH, Watowich SS, Jetten AM, Tian Q, et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29:138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim CH, Lim HW, Kim JR, Rott L, Hillsamer P, Butcher EC. Unique gene expression program of human germinal center T helper cells. Blood. 2004;104:1952–1960. doi: 10.1182/blood-2004-03-1206. [DOI] [PubMed] [Google Scholar]
- 14.Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol. 1995;155:1151–1164. [PubMed] [Google Scholar]
- 15.Veldhoen M, Uyttenhove C, van Snick J, Helmby H, Westendorf A, Buer J, Martin B, Wilhelm C, Stockinger B. Transforming growth factor-beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat Immunol. 2008;9:1341–1346. doi: 10.1038/ni.1659. [DOI] [PubMed] [Google Scholar]
- 16.Dardalhon V, Awasthi A, Kwon H, Galileos G, Gao W, Sobel RA, Mitsdoerffer M, Strom TB, Elyaman W, Ho IC, et al. IL-4 inhibits TGF-β-induced Foxp3+ T cells and, together with TGF-β, generates IL-9+ IL-10+ Foxp3- effector T cells. Nat Immunol. 2008;9:1347–1355. doi: 10.1038/ni.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maizels RM, Pearce EJ, Artis D, Yazdanbakhsh M, Wynn TA. Regulation of pathogenesis and immunity in helminth infections. J Exp Med. 2009;206:2059–2066. doi: 10.1084/jem.20091903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shulman Z, Gitlin AD, Weinstein JS, Lainez B, Esplugues E, Flavell RA, Craft JE, Nussenzweig MC. Dynamic signaling by T follicular helper cells during germinal center B cell selection. Science. 2014;345:1058–1062. doi: 10.1126/science.1257861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitagawa Y, Sakaguchi S. Molecular control of regulatory T cell development and function. Curr Opin Immunol. 2017;49:64–70. doi: 10.1016/j.coi.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Huang CT, Workman CJ, Flies D, Pan X, Marson AL, Zhou G, Hipkiss EL, Ravi S, Kowalski J, Levitsky HI, et al. Role of LAG-3 in regulatory T cells. Immunity. 2004;21:503–513. doi: 10.1016/j.immuni.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 22.Paust S, Lu L, McCarty N, Cantor H. Engagement of B7 on effector T cells by regulatory T cells prevents autoimmune disease. Proc Natl Acad Sci U S A. 2004;101:10398–10403. doi: 10.1073/pnas.0403342101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deaglio S, Dwyer KM, Gao W, Friedman D, Usheva A, Erat A, Chen JF, Enjyoji K, Linden J, Oukka M, et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J Exp Med. 2007;204:1257–1265. doi: 10.1084/jem.20062512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Djuretic IM, Levanon D, Negreanu V, Groner Y, Rao A, Ansel KM. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat Immunol. 2007;8:145–153. doi: 10.1038/ni1424. [DOI] [PubMed] [Google Scholar]
- 25.Szabo SJ, Dighe AS, Gubler U, Murphy KM. Regulation of the interleukin (IL)-12R beta 2 subunit expression in developing T helper 1 (Th1) and Th2 cells. J Exp Med. 1997;185:817–824. doi: 10.1084/jem.185.5.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, Dong C. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JB, Kim HR, Ha SJ. Immune checkpoint inhibitors in 10 years: contribution of basic research and clinical application in cancer immunotherapy. Immune Netw. 2022;22:e2. doi: 10.4110/in.2022.22.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung SM, Kim WU. Targeted immunotherapy for autoimmune disease. Immune Netw. 2022;22:e9. doi: 10.4110/in.2022.22.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vahedi G, Takahashi H, Nakayamada S, Sun HW, Sartorelli V, Kanno Y, O’Shea JJ. STATs shape the active enhancer landscape of T cell populations. Cell. 2012;151:981–993. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samstein RM, Arvey A, Josefowicz SZ, Peng X, Reynolds A, Sandstrom R, Neph S, Sabo P, Kim JM, Liao W, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151:153–166. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chalmin F, Humblin E, Ghiringhelli F, Végran F. Transcriptional programs underlying Cd4 T cell differentiation and functions. Int Rev Cell Mol Biol. 2018;341:1–61. doi: 10.1016/bs.ircmb.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 33.Christie D, Zhu J. Transcriptional regulatory networks for CD4 T cell differentiation. Curr Top Microbiol Immunol. 2014;381:125–172. doi: 10.1007/82_2014_372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkhurst CN, Muratet M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A, Wu C, Karwacz K, Xiao S, Jorgolli M, et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 2013;496:461–468. doi: 10.1038/nature11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu W, Ergun A, Lu T, Hill JA, Haxhinasto S, Fassett MS, Gazit R, Adoro S, Glimcher L, Chan S, et al. A multiply redundant genetic switch ‘locks in’ the transcriptional signature of regulatory T cells. Nat Immunol. 2012;13:972–980. doi: 10.1038/ni.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 38.Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24:369–379. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 39.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levings PP, Bungert J. The human β-globin locus control region. Eur J Biochem. 2002;269:1589–1599. doi: 10.1046/j.1432-1327.2002.02797.x. [DOI] [PubMed] [Google Scholar]
- 41.Lee GR, Fields PE, Griffin TJ, 4th, Flavell RA. Regulation of the Th2 cytokine locus by a locus control region. Immunity. 2003;19:145–153. doi: 10.1016/s1074-7613(03)00179-1. [DOI] [PubMed] [Google Scholar]
- 42.Koh BH, Hwang SS, Kim JY, Lee W, Kang MJ, Lee CG, Park JW, Flavell RA, Lee GR. Th2 LCR is essential for regulation of Th2 cytokine genes and for pathogenesis of allergic asthma. Proc Natl Acad Sci U S A. 2010;107:10614–10619. doi: 10.1073/pnas.1005383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hwang SS, Kim YU, Lee S, Jang SW, Kim MK, Koh BH, Lee W, Kim J, Souabni A, Busslinger M, et al. Transcription factor YY1 is essential for regulation of the Th2 cytokine locus and for Th2 cell differentiation. Proc Natl Acad Sci U S A. 2013;110:276–281. doi: 10.1073/pnas.1214682110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hwang SS, Jang SW, Lee KO, Kim HS, Lee GR. RHS6 coordinately regulates the Th2 cytokine genes by recruiting GATA3, SATB1, and IRF4. Allergy. 2017;72:772–782. doi: 10.1111/all.13078. [DOI] [PubMed] [Google Scholar]
- 45.Lee GR, Spilianakis CG, Flavell RA. Hypersensitive site 7 of the TH2 locus control region is essential for expressing TH2 cytokine genes and for long-range intrachromosomal interactions. Nat Immunol. 2005;6:42–48. doi: 10.1038/ni1148. [DOI] [PubMed] [Google Scholar]
- 46.Williams A, Lee GR, Spilianakis CG, Hwang SS, Eisenbarth SC, Flavell RA. Hypersensitive site 6 of the Th2 locus control region is essential for Th2 cytokine expression. Proc Natl Acad Sci U S A. 2013;110:6955–6960. doi: 10.1073/pnas.1304720110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- 48.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 49.Kim LK, Esplugues E, Zorca CE, Parisi F, Kluger Y, Kim TH, Galjart NJ, Flavell RA. Oct-1 regulates IL-17 expression by directing interchromosomal associations in conjunction with CTCF in T cells. Mol Cell. 2014;54:56–66. doi: 10.1016/j.molcel.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sekimata M, Pérez-Melgosa M, Miller SA, Weinmann AS, Sabo PJ, Sandstrom R, Dorschner MO, Stamatoyannopoulos JA, Wilson CB. CCCTC-binding factor and the transcription factor T-bet orchestrate T helper 1 cell-specific structure and function at the interferon-gamma locus. Immunity. 2009;31:551–564. doi: 10.1016/j.immuni.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murphy E, Shibuya K, Hosken N, Openshaw P, Maino V, Davis K, Murphy K, O’Garra A. Reversibility of T helper 1 and 2 populations is lost after long-term stimulation. J Exp Med. 1996;183:901–913. doi: 10.1084/jem.183.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grogan JL, Mohrs M, Harmon B, Lacy DA, Sedat JW, Locksley RM. Early transcription and silencing of cytokine genes underlie polarization of T helper cell subsets. Immunity. 2001;14:205–215. doi: 10.1016/s1074-7613(01)00103-0. [DOI] [PubMed] [Google Scholar]
- 54.Ballesteros-Tato A, Randall TD, Lund FE, Spolski R, Leonard WJ, León B. T follicular helper cell plasticity shapes pathogenic T helper 2 cell-mediated immunity to inhaled house dust mite. Immunity. 2016;44:259–273. doi: 10.1016/j.immuni.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 56.Hirota K, Duarte JH, Veldhoen M, Hornsby E, Li Y, Cua DJ, Ahlfors H, Wilhelm C, Tolaini M, Menzel U, et al. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat Immunol. 2011;12:255–263. doi: 10.1038/ni.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bending D, De la Peña H, Veldhoen M, Phillips JM, Uyttenhove C, Stockinger B, Cooke A. Highly purified Th17 cells from BDC2.5NOD mice convert into Th1-like cells in NOD/SCID recipient mice. J Clin Invest. 2009;119:565–572. doi: 10.1172/JCI37865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muranski P, Borman ZA, Kerkar SP, Klebanoff CA, Ji Y, Sanchez-Perez L, Sukumar M, Reger RN, Yu Z, Kern SJ, et al. Th17 cells are long lived and retain a stem cell-like molecular signature. Immunity. 2011;35:972–985. doi: 10.1016/j.immuni.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 60.Okada S, Markle JG, Deenick EK, Mele F, Averbuch D, Lagos M, Alzahrani M, Al-Muhsen S, Halwani R, Ma CS, et al. Impairment of immunity to Candida and Mycobacterium in humans with bi-allelic RORC mutations. Science. 2015;349:606–613. doi: 10.1126/science.aaa4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wan YY, Flavell RA. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature. 2007;445:766–770. doi: 10.1038/nature05479. [DOI] [PubMed] [Google Scholar]
- 63.Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, Chu TT, Corcoran L, Treuting P, Klein U, Rudensky AY. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control TH2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, Rudensky AY. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roy S, Rizvi ZA, Awasthi A. Metabolic checkpoints in differentiation of helper T cells in tissue inflammation. Front Immunol. 2019;9:3036. doi: 10.3389/fimmu.2018.03036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Almeida L, Dhillon-LaBrooy A, Carriche G, Berod L, Sparwasser T. CD4+ T-cell differentiation and function: unifying glycolysis, fatty acid oxidation, polyamines NAD mitochondria. J Allergy Clin Immunol. 2021;148:16–32. doi: 10.1016/j.jaci.2021.03.033. [DOI] [PubMed] [Google Scholar]
- 67.Yang W, Yu T, Cong Y. CD4+ T cell metabolism, gut microbiota, and autoimmune diseases: implication in precision medicine of autoimmune diseases. Precis Clin Med. 2022;5:pbac018. doi: 10.1093/pcmedi/pbac018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu K, Yin N, Peng M, Stamatiades EG, Chhangawala S, Shyu A, Li P, Zhang X, Do MH, Capistrano KJ, et al. Glycolytic ATP fuels phosphoinositide 3-kinase signaling to support effector T helper 17 cell responses. Immunity. 2021;54:976–987.e7. doi: 10.1016/j.immuni.2021.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, Chi H. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, Sullivan SA, Nichols AG, Rathmell JC. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Preston GC, Sinclair LV, Kaskar A, Hukelmann JL, Navarro MN, Ferrero I, MacDonald HR, Cowling VH, Cantrell DA. Single cell tuning of Myc expression by antigen receptor signal strength and interleukin-2 in T lymphocytes. EMBO J. 2015;34:2008–2024. doi: 10.15252/embj.201490252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Man K, Kallies A. Synchronizing transcriptional control of T cell metabolism and function. Nat Rev Immunol. 2015;15:574–584. doi: 10.1038/nri3874. [DOI] [PubMed] [Google Scholar]
- 74.McNamee EN, Korns Johnson D, Homann D, Clambey ET. Hypoxia and hypoxia-inducible factors as regulators of T cell development, differentiation, and function. Immunol Res. 2013;55:58–70. doi: 10.1007/s12026-012-8349-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dang EV, Barbi J, Yang HY, Jinasena D, Yu H, Zheng Y, Bordman Z, Fu J, Kim Y, Yen HR, et al. Control of TH17/Treg balance by hypoxia-inducible factor 1. Cell. 2011;146:772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Doedens AL, Phan AT, Stradner MH, Fujimoto JK, Nguyen JV, Yang E, Johnson RS, Goldrath AW. Hypoxia-inducible factors enhance the effector responses of CD8+ T cells to persistent antigen. Nat Immunol. 2013;14:1173–1182. doi: 10.1038/ni.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Phan AT, Doedens AL, Palazon A, Tyrakis PA, Cheung KP, Johnson RS, Goldrath AW. Constitutive glycolytic metabolism supports CD8+ T cell effector memory differentiation during viral infection. Immunity. 2016;45:1024–1037. doi: 10.1016/j.immuni.2016.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Clever D, Roychoudhuri R, Constantinides MG, Askenase MH, Sukumar M, Klebanoff CA, Eil RL, Hickman HD, Yu Z, Pan JH, et al. Oxygen sensing by T cells establishes an immunologically tolerant metastatic niche. Cell. 2016;166:1117–1131.e14. doi: 10.1016/j.cell.2016.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, Worley PF, Kozma SC, Powell JD. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, Xiao B, Worley PF, Powell JD. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee K, Gudapati P, Dragovic S, Spencer C, Joyce S, Killeen N, Magnuson MA, Boothby M. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity. 2010;32:743–753. doi: 10.1016/j.immuni.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shrestha S, Yang K, Guy C, Vogel P, Neale G, Chi H. Treg cells require the phosphatase PTEN to restrain TH1 and TFH cell responses. Nat Immunol. 2015;16:178–187. doi: 10.1038/ni.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huynh A, DuPage M, Priyadharshini B, Sage PT, Quiros J, Borges CM, Townamchai N, Gerriets VA, Rathmell JC, Sharpe AH, et al. Control of PI(3) kinase in Treg cells maintains homeostasis and lineage stability. Nat Immunol. 2015;16:188–196. doi: 10.1038/ni.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hardie DG. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 2011;25:1895–1908. doi: 10.1101/gad.17420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Meares GP, Qin H, Liu Y, Holdbrooks AT, Benveniste EN. AMP-activated protein kinase restricts IFN-γ signaling. J Immunol. 2013;190:372–380. doi: 10.4049/jimmunol.1202390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gaublomme JT, Yosef N, Lee Y, Gertner RS, Yang LV, Wu C, Pandolfi PP, Mak T, Satija R, Shalek AK, et al. Single-cell genomics unveils critical regulators of Th17 cell pathogenicity. Cell. 2015;163:1400–1412. doi: 10.1016/j.cell.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Karmaus PW, Chen X, Lim SA, Herrada AA, Nguyen TM, Xu B, Dhungana Y, Rankin S, Chen W, Rosencrance C, et al. Metabolic heterogeneity underlies reciprocal fates of TH17 cell stemness and plasticity. Nature. 2019;565:101–105. doi: 10.1038/s41586-018-0806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tortola L, Jacobs A, Pohlmeier L, Obermair FJ, Ampenberger F, Bodenmiller B, Kopf M. High-dimensional T helper cell profiling reveals a broad diversity of stably committed effector states and uncovers interlineage relationships. Immunity. 2020;53:597–613.e6. doi: 10.1016/j.immuni.2020.07.001. [DOI] [PubMed] [Google Scholar]
- 89.Kiner E, Willie E, Vijaykumar B, Chowdhary K, Schmutz H, Chandler J, Schnell A, Thakore PI, LeGros G, Mostafavi S, et al. Gut CD4+ T cell phenotypes are a continuum molded by microbes, not by TH archetypes. Nat Immunol. 2021;22:216–228. doi: 10.1038/s41590-020-00836-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lönnberg T, Svensson V, James KR, Fernandez-Ruiz D, Sebina I, Montandon R, Soon MS, Fogg LG, Nair AS, Liligeto U, et al. Single-cell RNA-seq and computational analysis using temporal mixture modelling resolves Th1/Tfh fate bifurcation in malaria. Sci Immunol. 2017;2:eaal2192. doi: 10.1126/sciimmunol.aal2192. [DOI] [PMC free article] [PubMed] [Google Scholar]