Abstract
The majority of 5-methylcytosine in mammalian DNA resides in endogenous transposable elements and is associated with the transcriptional silencing of these parasitic elements. Methylation also plays an important role in the silencing of exogenous retroviruses. One of the difficulties inherent in the study of proviral silencing is that the sites in which proviruses randomly integrate influence the probability of de novo methylation and expression. In order to compare methylated and unmethylated proviruses at the same genomic site, we used a recombinase-based targeting approach to introduce an in vitro methylated or unmethylated Moloney murine leukemia-based provirus in MEL cells. The methylated and unmethylated states are maintained in vivo, with the exception of the initially methylated proviral enhancer, which becomes demethylated in vivo. Although the enhancer is unmethylated and remodeled, the methylated provirus is transcriptionally silent. To further analyze the repressed state, histone acetylation status was determined by chromatin immunoprecipitation (ChIP) analyses, which revealed that localized histone H3 but not histone H4 hyperacetylation is inversely correlated with proviral methylation density. Since members of the methyl-CpG binding domain (MBD) family of proteins recruit histone deacetylase activity, these proteins may play a role in proviral repression. Interestingly, only MBD3 and MeCP2 are expressed in MEL cells. ChIPs with antibodies specific for these proteins revealed that only MeCP2 associates with the provirus in a methylation-dependent manner. Taken together, our results suggest that MeCP2 recruitment to a methylated provirus is sufficient for transcriptional silencing, despite the presence of a remodeled enhancer.
Cytosines in the context of a CpG dinucleotide are frequently methylated in mammalian cells. Such methylation is associated with the transcriptionally repressed state of imprinted genes and endogenous retroelements. Although DNA methylation can repress transcription by directly interfering with the binding of sequence-specific transcription factors (27), the recent discovery and biochemical characterization of the methyl-CpG binding domain (MBD) family of proteins (24) have revealed that an indirect mechanism of methylation-mediated repression also exists. Several MBD proteins, including MBD1 (20), MBD2 (40), MBD3 (47), and the archetypal MeCP2 (38), are thought to play a role in transcriptional repression. The discovery that MeCP2 interacts with a histone deacetylase (HDAC)-containing core complex via recruitment of the Sin3A corepressor (30, 39) has revealed that MeCP2 may function in part by recruiting deacetylase activity to methylated DNA. The recent finding that MBD2 interacts with the Mi-2/NuRD repressor complex, of which MBD3 is an integral subunit (47, 52), and the same HDAC-containing core complex suggests that alteration of the local chromatin structure via recruitment of complexes containing HDACs may be a general mechanism by which MBD proteins mediate transcriptional repression. However, several observations suggest that these proteins serve distinct functions in the cell: murine MBD3 binds weakly (47) or not at all (24, 52) to methylated DNA and, in contrast to MeCP2 and MBD2, does not colocalize with the highly methylated major satellite DNA in murine cells (24). Furthermore, transgenic studies have revealed that while mbd3-null mice die in early embryogenesis and mbd2-null mice are viable and fertile (25), MeCP2-null mice show neurological abnormalities similar to those observed in Rett syndrome (23). These differences suggest that MBD proteins bind distinct loci and may repress a unique complement of genes, yet little is known about the specific roles that these proteins play in vivo.
Retrotransposons have accumulated during the course of vertebrate evolution to the extent that such selfish DNA comprises over 45% of the human genome (33). Long terminal repeat (LTR)-based transposable elements and other repetitive sequences interspersed in the mammalian genome are typically transcriptionally silent and methylated in adult somatic tissues (4). Given this correlation and the fact that the majority of genomic 5-methylcytosine is found in parasitic sequence elements, Bestor proposed that CpG methylation has evolved as a host defense system (4). While this theory remains controversial, evidence has emerged indicating that the transcription of endogenous retroviruses is indeed constrained by methylation (48). Thus, the de novo methylation machinery may preferentially target parasitic elements, perhaps as a result of structural features characteristic of these elements (5). Cytosine methylation also plays an important role in the silencing of exogenous retroviruses in somatic tissues (8). As a result, the propensity for therapeutic retroviral vectors to become methylated and silenced in vivo remains one of the major stumbling blocks to efficacious gene therapy treatment.
Previously, we showed that a Moloney murine leukemia virus (MMuLV)-based retrovirus encoding the green fluorescent protein (GFP) is rapidly de novo methylated and silenced in MEL cells (35). Because retroviruses integrate more or less randomly in the genome, it is not possible to predict a priori the influence of the local chromatin milieu, which may permit or inhibit expression. Thus, to determine the consequences of methylation for proviral expression and chromatin structure, it is desirable to compare unmethylated and methylated proviruses at the same genomic position. Recently, we established that Cre recombinase can be used to target in vitro methylated DNA to defined genomic sites in MEL cells and that the methylation introduced is stably maintained in vivo (45). Here, we use this recombinase mediated-cassette exchange (RMCE) (17) approach to generate either methylated or unmethylated MMuLV provirus in two defined genomic sites in MEL cells. Surprisingly, while these cells have the potential to efficiently methylate proviral DNA (35), the unmethylated provirus remained devoid of CpG methylation with long-term culture, while the methylation state of the in vitro methylated provirus was substantially maintained in vivo. Preservation of these distinct methylation states permitted analysis of the influence of methylation on expression, de novo methylation, chromatin remodeling, histone acetylation state, and MBD protein binding. Using chromatin immunoprecipitation (ChIP), we show that the methylated, silent provirus is associated with deacetylated histones and that MeCP2 is recruited to the provirus in a methylation-dependent manner.
MATERIALS AND METHODS
Generation and in vitro methylation of the L1-MFGhGFP-1L plasmid.
The MMuLV-based retroviral vector MFGhGFP (2, 35) was originally isolated as an EcoRI-HindIII fragment including the complete proviral genome flanked by 396 and 697 bp of mouse genomic sequences 5′ and 3′ of the retroviral genome, respectively (15). To generate a construct for RMCE, MFGhGFP was digested with EcoRI and HindIII and cloned into the L1-1L cloning vector DpBlueKS(+)L1-PL-1L (sequence available upon request) to generate L1-MFGhGFP-1L. In vitro methylation of this construct with SssI methylase (New England Biolabs), which methylates all CpGs, was performed as described elsewhere (http://stke.sciencemag.org/cgi/content/full/OC_sigtrans;2001/83/pl1). To determine that the reaction was carried out to completion, following organic extraction and ethanol precipitation, methylated DNA was digested with the methylation-sensitive enzymes HpaII and HhaI and visualized by electrophoresis on a 0.7% agarose gel as described elsewhere (http://stke.sciencemag.org/cgi/content/full/OC_sigtrans;2001/83/pl1).
Tissue culturing and gene targeting.
MEL 745 cells (16) were maintained in growth medium (Dulbecco modified Eagle medium, 10% bovine calf serum, 100 U of penicillin/ml, 0.05 mM streptomycin, 2 mM glutamine) supplemented with 750 μg of hygromycin (Roche)/ml in log phase for at least 2 weeks prior to transfection to select cells expressing the HYTK (hygromycin B-phosphotransferase-thymidine kinase) fusion gene. Approximately 4 × 106 cells were electroporated in the presence of 15 μg of cytomegalovirus enhancer-Cre expression vector (45), 100 μg of sonicated salmon sperm DNA, and 25 μg of the L1-MFGhGFP-1L plasmid as previously described (45). After 3 days in nonselective medium, the cultures were supplemented with 10 μM ganciclovir and cultured for 7 days to select against HYTK-expressing cells. Ganciclovir-resistant cells were cloned by limiting dilution and screened for Cre-mediated exchange by Southern blotting. Greater than 80% of the clones analyzed contained a cassette integrated in one of the two possible orientations.
Nuclease sensitivity analysis.
DNase I digestion of nuclei was performed as described previously (18). DNase I-digested genomic DNA was purified and digested with BamHI. The GFP probe used for Southern hybridization was generated by digestion of the MFG-hGFP plasmid with NcoI and BamHI, yielding a restriction fragment including the 720-bp hGFP gene.
Northern blot hybridization and RT-PCR analysis.
Northern blot hybridization was conducted by standard procedures with 12 μg of total RNA prepared with Trizol reagent (GibcoBRL) according to the manufacturer's protocol and the GFP probe described above. For reverse transcription (RT)-PCR, total RNA was isolated as described for Northern analysis. SuperScript II (GibcoBRL) reverse transcriptase was used for first-strand cDNA synthesis as described previously (43). Primer pairs specific for MBD1 (plus-strand [+str], CCTGGCTGGAAACGCCGAGAGTCC; minus-strand [−str], GTGAAGCTAGAGCTGTGGCAGTAGG), MBD2 (+str, GATGGAAGAAGGAGGAAGTGATCC; −str, CGTGGTTGTTCATTCATCCGCTGG), MBD3 (+str, GGCGCTCCCGCAGGGCTGGGAAAG; −str, CCTTGGGCAAGTCCATGGTCCTGAC), and MeCP2 (+str, ATGGTAGCTGGGATGTTAGGGCTCAG; −str, CAGTTCCTGGAGCTTTGGGAGATTTG) were used for RT-PCR (32 cycles), yielding products of 346, 366, 466, and 555 bp, respectively.
Bisulfite analysis.
Bisulfite conversion was carried out with minor modifications using the protocol of Clark et al. (11) as described previously (35). Briefly, mixtures containing 5 μl of bisulfite-treated DNA (final volume, 50 μl) were subjected to 25 to 32 amplification cycles using a GeneAmp PCR system 9700 (Perkin-Elmer) with denaturation at 94°C, annealing at 49 to 56°C, and extension at 72°C. Nested or seminested amplification was performed using 2 μl of product from the first round in a 50-μl reaction volume. Primers were designed to favor the amplification of bisulfite-converted DNA. If the template strand included a CpG, degeneracy was incorporated in the primer at the nucleotide position corresponding to the cytosine such that no bias for amplification of the methylated template was introduced. Primers used for the 5′ LTR were as follows: bis+25+ (TAGGTTTGGTAAGTTAGTTTAAGTAAYGTT) with bis+1080− (TAAAAAAATAATAACAAACTAACCCRAAC) in the first round and bis+25+ (TTGTAAGGTATGGAAAAATATATAATTG) with bis+665− (TAAATTACTAACCAACTTACCTCCCRATAA) in the second round. Primers used for the seminested junction reactions were as follows: +bis3LTR (TGATTGGTATAATGGGAAATTGATTTTGAT) with bis1Ldis2 (TTACRATTCCTAACCTTTTACTAACC) and bis1Ldis (ACCRATTCATTAATACAACTAACACRAC) in the first and second rounds, respectively.
Flow cytometry and Western blot analysis.
For fluorescence-activated cell sorting analysis, cells were harvested and resuspended in staining medium (phosphate-buffered saline supplemented with 3% calf serum) supplemented with 1 μg of propidium iodide/ml for live/dead discrimination. Data were collected with a FACSCalibur (Becton Dickinson) equipped with the standard fluorescein filter set. Data for a minimum of 10,000 live cells were collected, and the fluorescence distribution was determined with FlowJo software (Treestar). For Western blot analysis, MEL and HeLa cell nuclear extracts were generated as described by Dignam et al. (14). Mouse brain nuclear extracts (Upstate Biotechnology) were used as a positive control, where appropriate. Western blotting was conducted according to the protocol provided by Santa Cruz Biotechnology with a GFP monoclonal antibody (Clontech) or polyclonal antibodies specific for MBD3 (Santa Cruz Biotechnology) or MeCP2 (Upstate Biotechnology). Chromatin used for Western blotting was generated with and without isopycnic centrifugation (as described below) for MBD3 and MeCP2, respectively, and boiled for 10 min in the presence of electrophoresis sample buffer (Santa Cruz Biotechnology) prior to loading on a denaturing acrylamide gel.
ChIPs.
To generate cross-linked chromatin for ChIPs with antiacetylated histone antibodies, exponentially growing cells (2 × 108) were fixed with 1% formaldehyde at room temperature for 3 min, and chromatin was purified as described previously (44). Briefly, fixed cells were washed once in buffer 1 (10 mM Tris [pH 8], 10 mM EDTA, 0.5 mM EGTA 0.25% Triton X-100), washed twice in buffer 2 (10 mM Tris [pH 8], 1 mM EDTA, 0.5 mM EGTA, 0.2 M NaCl), and resuspended in buffer 3 (10 mM Tris [pH 8], 1 mM EDTA, 0.5 mM EGTA) prior to sonication. All buffers were supplemented immediately prior to use with 10 mM sodium butyrate. After sonication (four times for 30 s each time on ice; Fisher Scientific sonic Dismembrator, highest setting), protein-DNA complexes were purified by isopycnic (CsCl) centrifugation (41). The DNA content of cross-linked chromatin was quantified using a Hoefer Instruments fluorometer. Immunoprecipitations with purified chromatin were conducted as described previously (44) using polyclonal antibodies against all acetylated isoforms of histone H4 (AcH4) or against histone H3 acetylated at lysines 9 and 14 (AcH3) (Upstate Biotechnology). A 1:1 mixture of protein A and G-Sepharose beads (Amersham) was used for all immunoprecipitations. Washes and reversal of the cross-link were conducted as described previously (44). DNA fragments ranged in size from 0.3 to 1.0 kb.
For ChIPs with polyclonal antibodies specific for MeCP2 or MBD3, cells were fixed for 30 min with formaldehyde as described above, washed once in buffer 1, washed twice in buffer 2, resuspended in 2 ml of immunoprecipitation buffer (40 mM Tris [pH 8], 4 mM EDTA, 300 mM NaCl, 1% Triton X-100) supplemented with protease inhibitors [10 μg of aprotonin/ml, 1 μg of pepstatin/ml, 1 μg of leupeptin/ml, and 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF)], and sonicated as described above. Chromatin was centrifuged for 10 min (12,000 × g) at 4οC to remove debris, and 100 μl of supernatant was used for immunoprecipitation or to prepare input DNA. Goat preimmune serum and rabbit immunoglobulin G were used as controls for MBD3 and MeCP2 immunoprecipitations, respectively. Washes and reversal of the cross-link were conducted as described above.
Quantitative PCR was performed with a Perkin-Elmer 9700 thermocycler and 0.5 to 1.5 ng of reverse-cross-linked DNA from input and antibody-bound chromatin. Conditions for linear amplification (see Fig. 2 and reference 44) were achieved for all reactions using 27 to 29 cycles of amplification and a 60οC annealing temperature. Each 25-μl reaction mixture was supplemented with 1 μCi of [α-32P]dCTP (NEN). Primer pairs for the proviral LTR (−9LTR+ST, CATGTGAAAGACCCCACCTGTAG; 5LTR329−, AATAAGGCACAGGGTCATTTCAGG) and the GFP gene (GFP1, ACATGAAGCAGCACGACTTC; GFP2, TGCTCAGGTAGTGGTTGTC) are specific for the introduced cassette and give product sizes of 364 and 377 bp, respectively. Control primer pairs for the mouse amylase gene (amy4 and amy6) and the mouse β-major promoter (mubmp1 and mubmp2) of the B-globin locus (44) give products of 400 and 320 bp, respectively, permitting duplex PCR with the transgene primer sets. One-third of the reaction product was loaded on a 5% nondenaturing polyacrylamide gel and subjected to electrophoresis. Products were quantified with a PhosphorImager and ImageQuant software (Molecular Dynamics). To determine the level of protein enrichment at a given region in the provirus, the ratio of the two PCR products was calculated for the antibody-bound fraction and normalized to the ratio obtained for the input material.
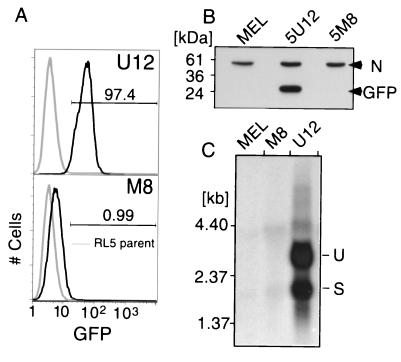
FIG. 2.
Methylation of the MFGhGFP provirus is necessary and sufficient for proviral silencing. (A) Representative in vitro methylated (M8) and unmethylated (U12) L1-MFGhGFP-1L clones (black histograms) in the same genomic orientation and control RL5 cells (gray histograms) were analyzed by flow cytometry at day 46 posttransfection. Greater than 97% of clone U12 cells show GFP expression, while greater than 99% of M8 cells show low to undetectable levels of expression. (B) The lack of GFP expression was confirmed by Western blot analysis using an anti-GFP antibody. This reagent also recognizes a nonspecific protein (N), which serves as an internal control for protein loading. (C) To confirm that repression acted at the level of transcription, Northern blotting was performed using equal amounts of total RNA isolated 74 days after electroporation. As expected, the unmethylated clone expresses both spliced (S) and unspliced (U) isoforms, while the methylated clone and RL5 MEL parent cell line show no detectable signal.
RESULTS
Targeting of methylated and unmethylated proviral DNA to defined genomic sites.
In order to study the mechanism of methylation-mediated proviral silencing, we used a Cre-loxP-based system, which allows for the introduction of DNA constructs flanked by inverted loxP sites at specific sites in the genome (Fig. 1A). This method, RMCE (17), involves selection against thymidine kinase (TK) expression from the preexisting cassette rather than for expression from the introduced cassette. Thus, potentially nonexpressing, methylated constructs can be introduced and clones can be efficiently isolated for further analysis (45).
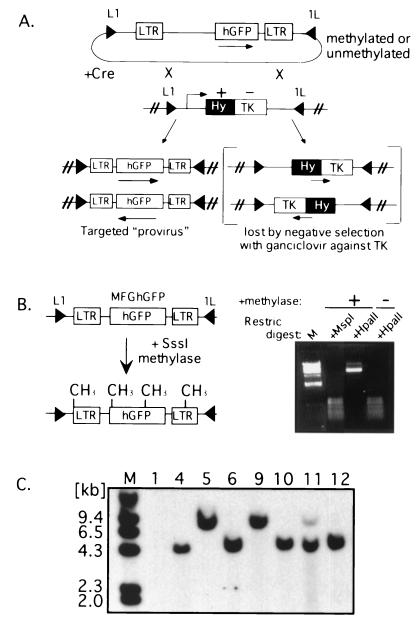
FIG. 1.
Principle and application of Cre-mediated targeting of methylated and unmethylated proviruses at a defined genomic site. (A) A cell line containing a stably integrated L1-HYTK-1L gene flanked by inverted loxP sites (black triangles) is transfected with the proviral construct L1-MFGhGFP-1L (also flanked by inverted loxP sites) together with a Cre recombinase expression plasmid. Recombination between the loxP sites in the two constructs results in exchange of the cassettes and loss of the TK-negative selectable marker. Alternatively, recombination between the inverted loxP sites on the same DNA molecule occurs, resulting in the inversion of the intervening DNA. Cells that have undergone the latter recombination event still express the HYTK gene and thus can be selected against with ganciclovir, allowing the isolation of cells that have undergone the targeting reaction. Note that selection is not dependent on the expression of the introduced cassette. hGFP, humanized GFP. (B) The L1-MFGhGFP-1L construct, containing the MFGhGFP retroviral vector, was methylated with SssI methylase and digested with the methylation-sensitive restriction (Restric) enzyme HpaII and its insensitive isoschizomer MspI to establish that the reaction was carried to completion. Methylated DNA was introduced into RL5 or RL6 L1-HYTK-1L MEL cells, and ganciclovir-resistant clones were generated by limiting dilution. Clones with an unmethylated L1-MFGhGFP-1L cassette were also generated. (C) Genomic DNA was isolated from RL5 clones after ganciclovir selection and digested with BamHI, which cuts only at the 3′ end of the GFP gene and thus generates junction fragments of different sizes depending on the orientation of the insertion. Southern blot analysis using a GFP probe to confirm the fidelity of targeting reveals that the majority of clones contain the provirus preferentially integrated in one of the two possible orientations. Marker lanes (M) are included on each gel.
L1-MFGhGFP-1L, a construct containing the retroviral vector MFGhGFP (2) flanked by inverted loxP sites, was methylated in vitro with the bacterial methyltransferase SssI, yielding a cassette methylated at all CpGs (Fig. 1B). The previously characterized MEL cell lines RL5 and RL6 (16), which harbor a stably integrated HYTK cassette flanked by inverted loxP sites, were transfected with methylated or control, unmethylated L1-MFGhGFP-1L DNA in combination with a Cre recombinase expression plasmid. Cre-mediated recombination of this construct, which includes the complete proviral genome and flanking mouse genomic DNA, yields a single integrated provirus which appears topologically as it would following conventional proviral integration. After ganciclovir selection, clones were isolated and analyzed by Southern blot hybridization for the presence and genomic orientation of the L1-MFGhGFP-1L cassette. As expected, the majority of TK-resistant RL5 and RL6 clones contained the provirus integrated in one of the two possible genomic orientations (Fig. 1C).
The proviral transcription state depends upon the initial density of methylation.
From both the RL5 and the RL6 cell lines, at least two clones with unmethylated or methylated cassettes in each orientation were expanded for further analysis. The expression state of these clones was determined by flow cytometry, and representative orientation-matched RL5 clones M8 (M, methylated) and U12 (U, unmethylated) are shown in Fig. 2A. All MEL clones with an unmethylated cassette show stable expression in greater than 95% of cells (data not shown). In contrast, all SssI-methylated clones show fluorescence intensity comparable to that seen in control RL5 MEL cells. The absence of GFP in the methylated clone was confirmed by Western blotting with an antibody raised against GFP (Fig. 2B), and Northern hybridization analysis revealed that expression is blocked at the level of transcription (Fig. 2C). The active and repressed transcription states of the unmethylated and methylated clones, respectively, suggest that the methylation state generated in vitro is maintained in vivo upon genomic integration of the provirus and that de novo methylation of the unmethylated cassette does not occur. Comparable results were found at the RL6 integration site (data not shown).
The methylation state is maintained in vivo, with the exception of the 5′ LTR enhancer, which is preferentially demethylated.
Preliminary analysis by Southern hybridization of genomic DNA isolated from several L1-MFGhGFP-1L MEL clones 21 days after electroporation revealed that the initially unmethylated provirus is not methylated de novo, while the initially methylated cassette becomes demethylated, specifically in the 5′ LTR enhancer (data not shown). As similar results were found for both integration sites, we focused on the methylated and unmethylated RL5 clones, M8 and U12, respectively.
The PCR-based bisulfite sequencing method (11) was used to determine the methylation state of all CpGs within several regions of the provirus, including the plasmid-L1-proviral junction upstream of the 5′ LTR, the 5′ LTR itself, the GAG region (data not shown), and the GFP gene (Fig. 3A). Consistent with the Southern hybridization data, unmethylated clone U12 remains virtually devoid of methylation across the introduced cassette (Fig. 3B to D). Methylated clone M8, in contrast, remains methylated throughout the provirus, with the exception of the CpGs in the enhancer region, which are consistently demethylated (Fig. 3C), and several CpGs in the promoter and GFP gene, which are sporadically demethylated (Fig. 3C and D). Interestingly, regardless of the methylation status of the introduced provirus, no methylation was detected in the region upstream of the introduced cassette (Fig. 3B), suggesting that spreading of methylation does not occur at this integration site. The stability of two distinct methylation states of a provirus integrated at the same genomic site in the same orientation allowed us to study the properties of proviral methylation in the absence of position effects.
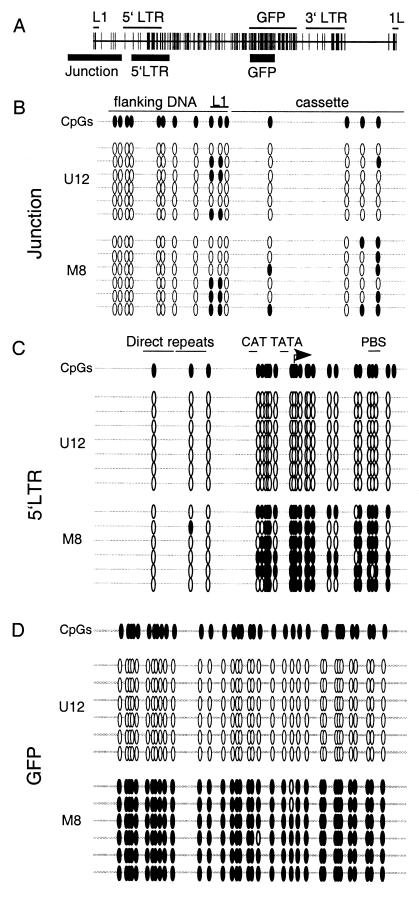
FIG. 3.
Detailed methylation mapping of methylated and unmethylated proviral clones. Genomic DNA isolated 56 days after electroporation was bisulfite converted, and the regions of interest were PCR amplified, subcloned, and sequenced (see Materials and Methods). (A) The regions amplified, including the junction region, the 5′ LTR, and the GFP gene (thick black lines), are shown relative to the L1-MFGhGFP-1L map. (B to D) Open and filled ovals correspond to CpGs and methylated CpGs, respectively. For each amplified region, at least six molecules from clones M8 and U12 were sequenced. Bisulfite sequencing of the junction region (B), the 5′ LTR (C), and the GFP gene (D) reveals the absence of methylation in the initially unmethylated clone U12 and the maintenance of methylation, with the exception of the enhancer region, in the methylated clone M8. PBS, primer binding site.
Remodeling of the LTR enhancer is not influenced by the proviral methylation state.
Having shown that demethylation of the enhancer region occurs independently of the transcription state, we next sought to study the chromatin structure of the proviral 5′ LTR by assaying for the formation of DNase I-hypersensitive sites (HSs) previously described for this region (46). Nuclei isolated from M8 and U12 cells were incubated with increasing amounts of DNase I, and purified genomic DNA was analyzed by Southern blot hybridization after digestion with BamHI (Fig. 4). While the promoter HS forms only in the unmethylated clone, the enhancer HS forms regardless of the methylation state of flanking DNA, indicating that recruitment of nuclear factors to the enhancer occurs independently of transcription or methylation state at the promoter and coding region.
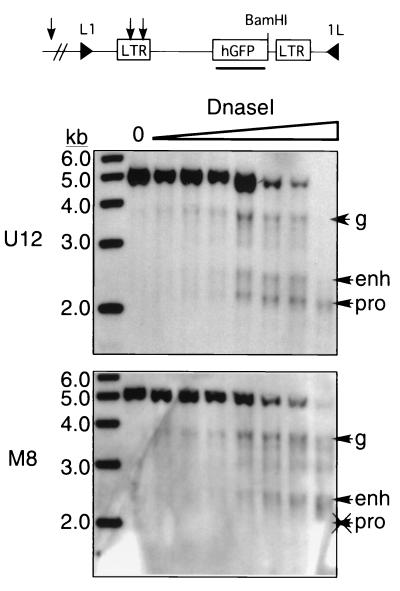
FIG. 4.
Dependence of LTR enhancer and promoter remodeling on methylation state. Nuclei were isolated from RL5 clones M8 and U12 and digested with increasing concentrations of DNase I. Subsequently, genomic DNA was isolated, digested with BamHI, and subjected to Southern hybridization with a GFP probe. Predicted HSs at the proviral enhancer (enh) and promoter (pro), as well as a genomic site (g) upstream of the introduced cassette, are labeled with arrows. A sample with no DNase I added (lane 0) is also shown. Note the absence of the promoter HS in the methylated clone. hGFP, humanized GFP.
Proviral methylation correlates with histone H3 deacetylation.
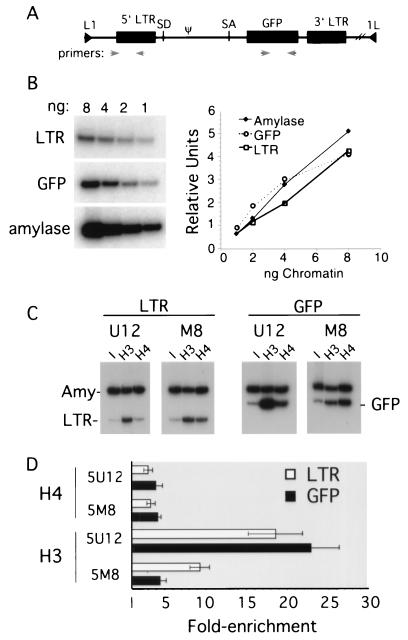
We used ChIPs to determine if histones associated with the methylated provirus are hypoacetylated relative to the unmethylated provirus. Primer pairs specific for the proviral 5′ LTR (which includes the direct repeat enhancer) and GFP gene (Fig. 5A), in addition to the endogenous amylase 2.1y gene, were generated, and PCR conditions were established to ensure linear amplification (Fig. 5B). Formaldehyde-cross-linked chromatin from clones M8 and U12 was immunoprecipitated with antisera specific for AcH3 or AcH4, and antibody-bound DNA was eluted and analyzed by duplex PCR using either of the proviral primer pairs in combination with the amylase primer pair. The latter serves as a control in MEL cells, as this gene is in a closed chromatin conformation characterized by relative hypoacetylation for histones H3 and H4 (44). The ratio of the two PCR products was determined for the antibody-bound fraction and normalized to the ratio obtained from the input chromatin.
FIG. 5.
Histone acetylation is dependent on proviral methylation status. (A) A map of the L1-MFGhGFP-1L provirus is shown with the locations of the primer pairs used to amplify either the 5′ LTR or the GFP gene. SA, splice acceptor; SD, splice donor. (B) Amplification of titrated input DNA using these primer pairs and a control primer pair specific for the endogenous mouse amylase 2.1y gene is linear under the conditions used for duplex PCR (see Materials and Methods). Formaldehyde-cross-linked chromatin was purified by isopycnic centrifugation and immunoprecipitated with antibodies recognizing AcH4 or AcH3. After reversal of the cross-link, duplex PCR was performed for the input and antibody-bound chromatin fractions (equivalent to approximately 1 to 2 ng of DNA) with the amylase 2.1y primer pair in combination with either the 5′ LTR or the GFP gene primer pair in the presence of radiolabeled deoxycytidine. (C) The PCR products from the input (I) and antibody-bound DNA (H3 and H4) were resolved by electrophoresis on a nondenaturing acrylamide gel. Amy, amylase 2.1y. (D) To determine the enrichment of proviral sequences relative to the nonexpressed, hypoacetylated amylase gene, products from three independent duplex amplifications were quantified by PhosphorImager analysis. The ratio of the two PCR products was determined for the antibody-bound fraction and normalized to the ratio obtained from the input material prior to immunoprecipitation. The mean and standard error of the mean are plotted. The x axis is set at 1, which reflects no enrichment relative to the amylase 2.1y gene.
A representative set of duplex PCRs is shown in Fig. 5C, and a summary of three independent amplifications is shown in Fig. 5D. The acetylation state of histone H4 was not influenced by the presence of methylation. In contrast, unmethylated clone U12 shows an 18-fold enrichment of AcH3 relative to amylase within the LTR, versus only 9-fold for methylated clone M8, indicating that H3 is relatively hypoacetylated in the methylated clone in this region. Similarly, U12 shows a 23-fold enrichment of AcH3 within the GFP gene, while M8 shows only a 4-fold enrichment in this region. Thus, relative to the methylated clone, the unmethylated clone shows two- and fivefold greater levels of enrichment of AcH3 in the LTR and GFP regions, respectively, demonstrating that the methylation state of the provirus strongly influences the acetylation state of histone H3.
Expression of MBD proteins in MEL cells.
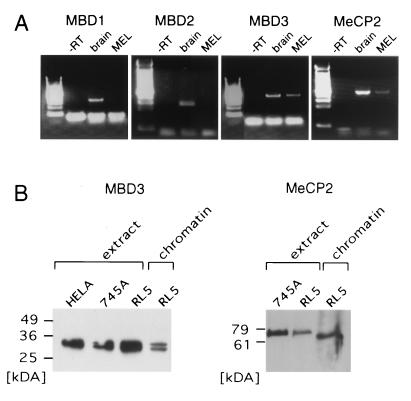
The association of hypoacetylated histone H3 with the silent proviral reporter suggests that HDAC activity plays a role in methylation-mediated repression. Given that MeCP2, MBD2, and MBD3 all interact with complexes containing HDAC1 and/or HDAC2, this result is not informative with respect to which of these proteins, if any, are bound to the methylated provirus. Although MBD proteins are ubiquitously expressed in somatic tissues (24), RT-PCR with primer pairs specific for MBD1, MBD2, MBD3, and MeCP2 revealed that only MBD3 and MeCP2 are expressed in MEL cells (Fig. 6A). These results were confirmed by Western blotting using antisera specific for each MBD protein (Fig. 6B and data not shown). Simultaneous analysis of reverse-cross-linked chromatin preparations revealed that MeCP2 and MBD3 are detectable in formaldehyde-cross-linked chromatin.
FIG. 6.
MeCP2 and MBD3 are expressed in MEL cells and are detectable in chromatin preparations. (A) mRNAs from MEL cells and mouse brain as a positive control were reverse transcribed, and the cDNA generated was used as a template for PCR with primers specific for MBD1, MBD2, MBD3, and MeCP2. Only MBD3 and MeCP2 are expressed in MEL cells. −RT, without reverse transcriptase. (B) Western blot analyses were conducted with antibodies specific for MeCP2 and MBD3. Nuclear extracts were prepared from HeLa, MEL 745A, and MEL RL5 cells, and 10 to 20 μg of protein was loaded per lane. Crude (for MeCP2) or purified (for MBD3) chromatin preparations (see Materials and Methods) were also generated and subjected to electrophoresis in parallel with the nuclear extracts.
MeCP2 is associated with the methylated provirus.
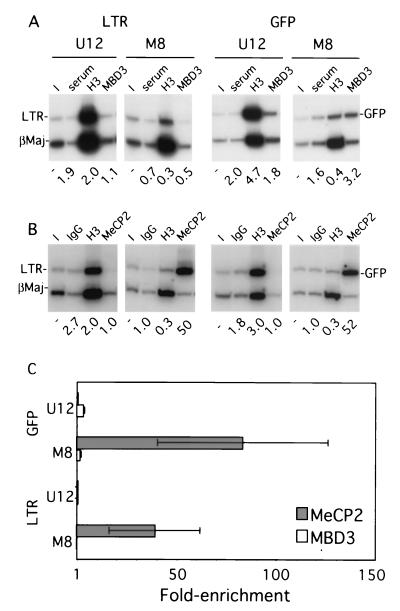
A series of ChIP experiments using MBD3 or MeCP2 antisera were carried out to establish which, if either, of these proteins is recruited to the methylated provirus in vivo. Chromatin from clones M8 and U12 was generated without isopycnic centrifugation (see Materials and Methods), and the immunoprecipitated material was subjected to duplex PCR. The transcriptionally active (44) and presumably unmethylated endogenous β-major globin gene promoter was used, rather than the silent amylase gene, as it is less likely to be associated with MBD proteins. The level of enrichment of MBD3 was low to undetectable, regardless of the methylation state (Fig. 7). Similar results were found when chromatin purified by isopycnic centrifugation was used (data not shown). The absence of enrichment is not likely to be due to insufficient cross-linking, as MBD3 can be detected in purified cross-linked chromatin (Fig. 6B). In contrast, while clone U12 shows no enrichment for MeCP2 relative to the β-major promoter, clone M8 is significantly enriched for MeCP2 in both the proviral 5′ LTR and the GFP gene (Fig. 7B), suggesting that methylation is necessary and sufficient for the recruitment of MeCP2 in vivo. Repetition of the MeCP2 and MBD3 immunoprecipitations with independently generated chromatin confirmed these results (Fig. 7C). Thus, MeCP2 is apparently the only known MBD protein targeted to the methylated provirus in MEL cells.
FIG. 7.
MeCP2 is recruited to the methylated provirus. Formaldehyde-cross-linked chromatin was prepared from MEL clones M8 and U12 without isopycnic centrifugation (as described in Materials and Methods). (A) Chromatin was immunoprecipitated with goat preimmune serum or antibodies specific for AcH3 (H3) or MBD3. Duplex PCR was conducted with the 5′ LTR or GFP gene primers described in Fig. 5, in combination with a control primer pair specific for the actively transcribed β-major promoter region (βMaj) of the endogenous β-globin locus. A representative acrylamide gel is shown, along with the levels of enrichment, as measured by PhosphorImager analysis, relative to the input (I) sample. The goat-derived MBD3 antibody shows no significant enrichment relative to the control goat serum. In contrast, concurrent immunoprecipitation with the AcH3 antibody shows a 7- to 10-fold difference in abundance between the unmethylated and the methylated proviruses, confirming the fidelity of the chromatin preparation. (B) Immunoprecipitation with control rabbit immunoglobulin G or antibodies specific for AcH3 (H3) or MeCP2 reveals that MeCP2 is significantly enriched in the 5′ LTR and the GFP gene of the methylated clone. (C) Quantification of duplex PCR products from two chromatin preparations (three independent MeCP2 or MBD3 immunoprecipitations) confirms these results. The mean and standard error of the mean for the enrichment are plotted (see the text). The x axis is set at 1, which reflects no enrichment.
DISCUSSION
Genomic targeting of methylated provirus.
Random integration in the host genome is an integral component of the retroviral life cycle. Analysis of the mechanism(s) of methylation-mediated proviral silencing has been complicated by the fact that the chromatin structure of the integration site influences the level and stability of expression, the propensity for de novo methylation, and the complement of associated DNA binding factors (13). To avoid the inherent complications associated with such position effects, we modified the Cre recombinase-based targeting system, RMCE (17), to target in vitro methylated DNA into the genome (45; http://stke.sciencemag.org/cgi /content/full/OC_sigtrans;2001/83/pl1). Here, we introduced an MMuLV-based provirus, unmethylated or methylated in vitro, into two defined genomic sites in MEL cells. With the striking exception of the proviral enhancer, CpG methylation is stably maintained in vivo, while at the same integration site, an initially unmethylated provirus remains devoid of methylation for at least 2 months in culture. Given that 100% of the cells in clone U12 express GFP for at least 6 months in cultures (data not shown), we infer that the virus will remain unmethylated indefinitely at this site. In contrast, we found previously that this retroviral vector is particularly prone to de novo methylation when introduced by infection in MEL 745 cells, with the majority of initially GFP-positive clones being silenced and presumably methylated after 1 month in culture (35). The absence of de novo methylation of the provirus integrated at RL5 suggests that the integration site itself is somehow protected from de novo methyltransferase activity. Alternatively, the difference in susceptibility to de novo methylation may be due to the fact that the structures of the integration intermediates differ between viral integrase-mediated and Cre-mediated rearrangements (5). However, a comparison of single-copy proviruses introduced by transfection versus infection did not reveal a difference in the rate of silencing (3). Regardless, the generation of stable, complementary proviral methylation states at a defined genomic site permitted us to study the influence of preexisting methylation on de novo methylation, chromatin structure, and MBD protein binding.
Dense CpG methylation is not sufficient to promote methylation spreading.
While the methylated provirus introduced at RL5 remained methylated and transcriptionally inactive after long-term culture, CpGs in the region flanking the introduced cassette were never methylated, indicating that a methylated provirus does not necessarily act as a focus for the initiation of methylation spreading, as has been previously reported (29, 32). The absence of methylation spreading is not a peculiarity of proviral constructs, as methylation spreading from a methylated β-globin reporter construct integrated at this site was not observed either (45). Nor is the absence of methylation spreading due to the presence of the LTR enhancer element, since no methylation was detected in the flanking DNA of a methylated construct from which the 5′ LTR enhancer region was deleted (data not shown). Taken together, these results suggest that the integration site, rather than features of the heterologous element itself, may be the dominant factor in determining the probability of de novo methylation.
The LTR enhancer is demethylated and remodeled regardless of the transcription state.
In the majority of clones analyzed, the two initially premethylated CpGs in the proviral enhancer are demethylated, results consistent with those previously reported for germ line-transmitted retroviral genomes (28) and for MEL cell clones infected with the MFGhGFP vector (35). These CpG sites overlap with a putative NF-1 binding site present in each of the direct repeats (21), raising the possibility that the enhancer may contain a binding site(s) for a complex with intrinsic demethylating activity. Consistent with this hypothesis, the DNase I-HS in the 5′ LTR enhancer region (46) still forms. Interestingly, Zhu et al. (53) recently showed that the hormone receptor RXRα interacts with a G/T-mismatched 5-methylcytosine DNA glycosylase which demethylates CpGs around the receptor DNA binding site in the absence of a ligand and regardless of the transcription state. The demethylated CpGs within the proviral tandem repeat enhancer are located within 20 bp of a glucocorticoid-responsive element site, raising the possibility that in MEL cells, the enhancer may be bound by endogenous hormone receptor complexes which demethylate adjacent CpGs.
Alternatively, transcription factor binding may itself be sufficient to trigger the demethylation of nearby methylated CpGs (26). In support of the latter model, demethylation of the HS2 enhancer element from the β-globin locus in another methylated construct introduced in RL5 was also observed (45). Nevertheless, the transcriptional activators bound to the HS2 or LTR enhancers are insufficient to overcome methylation-mediated repression, suggesting that dense methylation of the promoter and downstream regions in some way neutralizes enhancer function.
The methylated provirus is hypoacetylated for histone H3.
The modification of histone tails by acetylation is strongly associated with transcriptional competence (10). Conversely, histone deacetylation is associated with transcriptional silencing, and a number of repressor proteins have recently been shown to interact with HDAC complexes (42). In vitro and in vivo experiments have revealed that several MBD proteins, including MeCP2 (30, 39), MBD2 (52), and MBD3 (47, 52), are associated with repressor complexes that include HDACs, implicating a role for local histone deacetylation in methylation-mediated silencing (6). The ChIP experiments presented here revealed that histone H3 associated with the GFP gene in particular and the LTR to a lesser extent is hypoacetylated in the methylated provirus relative to the unmethylated provirus. In contrast, no difference in acetylation was observed for histone H4. Considering that the LTR is demethylated in vivo, the H3 acetylation state correlates closely with the location of methylated CpGs in the provirus. These results are consistent with a previous analysis of a β-globin reporter construct in MEL cells (44). However, consistent with previous results obtained with the MFGhGFP retroviral vector introduced by infection (35), treatment with the HDAC inhibitor trichostatin A (TSA) failed to induce GFP expression from the methylated provirus in RL5 (data not shown).
The failure of TSA-mediated activation of densely methylated genes may be the rule rather than the exception (7, 36), suggesting that an HDAC1- or HDAC2-independent mechanism of transcriptional repression plays a role in maintaining the silent state of methylated genes. Deacetylated histone H3 associated with methylated DNA may be marked by additional covalent modifications (10). For example, methylation of deacetylated lysine 9 in the H3 histone tail might render it refractory to histone acetyltransferase activity, thus consolidating the silent state (37). Interestingly, treatment of the repressed provirus with TSA revealed no change in the histone H3 acetylation state of the LTR or the GFP gene (data not shown), results consistent with those of Coffee et al. (12) and supportive of the hypothesis that the inhibition of HDAC activity is insufficient for the acetylation of histones in densely methylated DNA.
CpG methylation is necessary and sufficient for the recruitment of MeCP2 to the MMuLV provirus.
While the repressor complexes with which several of the MBD proteins interact have been characterized, little is known about the sequences to which these proteins are recruited in vivo. Recently, Magdinier and Wolffe (36) showed that MBD2 is recruited in a methylation-dependent manner to the p14/p16 locus in human neoplastic cells. Using a model system for proviral methylation, we showed that MeCP2 is recruited in vivo to a proviral construct in a methylation-dependent manner. In contrast, we did not detect binding of MBD3 to the provirus. The latter result is not surprising, given that purified murine MBD3 binds weakly (47) or not at all (24, 52) to methylated oligonucleotides in gel shift analyses. In fact, methylation-dependent recruitment of the NuRD complex, of which MBD3 is an integral component, depends upon the presence of MBD2 (52). As MBD2 is not expressed in MEL cells, recruitment of the Mi-2/NuRD complex to DNA is presumably dependent upon its interaction with other DNA binding proteins (42). Since MBD1 is also not expressed in MEL cells, MeCP2 seems to be the only known MBD protein associated with the silent, methylated provirus. While we observed hypoacetylation of histone H3 but not histone H4 associated with the methylated provirus, a lack of functional MeCP2 was recently shown to result in hyperacetylation of H4, as measured in a bulk assay for histone acetylation (49). In contrast, Gregory et al. reported that MeCP2 is associated exclusively with the methylated maternal allele of the imprinted gene U2af1-rs1 (22), which is deacetylated at histone H3 exclusively, results entirely consistent with our own. Thus, the influence of MeCP2 on the acetylation of specific histones may depend on the locus at which the MeCP2 protein is bound.
It has been hypothesized that the activation of endogenous retroelements may disrupt normal patterns of tissue-specific gene expression (50). Given that Rett syndrome is linked to mutations in the MeCP2 gene (1), it is tempting to speculate that the loss of MeCP2 function results in the activation of endogenous retroelements, which in turn disrupt the transcription of neuronal genes. The recent generation of MeCP2-null mice (9, 23) should allow for the detection of aberrant expression of both retroelements and endogenous genes in murine tissues.
What role might MeCP2 play in repressing proviral transcription, given that the 5′ LTR enhancer is demethylated? In vitro experiments with MeCP2-Gal4 fusions show that this MBD protein is capable of repressing transcription when positioned over 400 bp from the transcriptional start site (38). Although the mechanism of long-range repression remains to be determined, Kaludov and Wolffe observed a direct interaction between MeCP2 and TFIIB, a component of the basal transcription machinery, suggesting that MeCP2 may directly prevent components of RNA polymerase from functioning during assembly of the preinitiation complex (31). This alternative mechanism of repression may also explain why the inhibition of HDAC activity fails to induce proviral expression. In support of this theory, using MeCP2-Gal4 fusions and a Gal4-simian virus 40 reporter construct to mimic methylation-mediated recruitment of the MBD protein, Yu et al. recently showed that MeCP2-mediated repression is refractory to TSA induction (51). Taken together, these results suggest that MeCP2 is capable of repressing transcription at a distance and independent of the HDAC activity associated with the Sin3A complex.
MeCP2 is associated with pericentromeric heterochromatin in MEL cells (data not shown), as has been observed previously in other cell types (34). While the predominantly centromeric staining of MeCP2 does not exclude its presence in other parts of the interphase nucleus, it is possible that the densely methylated provirus colocalizes in an MeCP2-dependent manner with pericentromeric heterochromatin. Recruitment to this nuclear compartment has been shown to correlate with transcriptional repression (19) and may represent a general mechanism by which silencing of retroelements is stably maintained. The targeting system described here may be useful in determining the influence of DNA methylation on nuclear localization and in further defining the biochemical characteristics that distinguish the methylated and unmethylated states of the provirus.
ACKNOWLEDGMENTS
This work was supported by NIH fellowship GM 19767/01to M.C.L., a fellowship from the Rett Syndrome Research Foundation to D.S., and NIH grants DK44746 and HL57620 to M.G.
We thank M. Bender for mouse brain cDNA; Eric Bouhassira and the members of the Groudine laboratory for suggestions; Claire Francastel and Tomoyuki Sawado for comments on the manuscript; and Joan Hamilton, David Scalzo, Jennifer Stout, and Urszula Maliszewski for technical assistance.
REFERENCES
- 1.Amir R E, Van den Veyver I B, Wan M, Tran C Q, Francke U, Zoghbi H Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 2.Anderson M T, Tjioe I M, Lorincz M C, Parks D R, Herzenberg L A, Nolan G P, Herzenberg L A. Simultaneous fluorescence-activated cell sorter analysis of two distinct transcriptional elements within a single cell using engineered green fluorescent proteins. Proc Natl Acad Sci USA. 1996;93:8508–8511. doi: 10.1073/pnas.93.16.8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baer A, Schübeler D, Bode J. Transcriptional properties of genomic transgene integration sites marked by electroporation or retroviral infection. Biochemistry. 2000;39:7041–7049. doi: 10.1021/bi992957o. [DOI] [PubMed] [Google Scholar]
- 4.Bestor T H. The host defence function of genomic methylation patterns. Novartis Found Symp. 1998;214:187–195. doi: 10.1002/9780470515501.ch11. [DOI] [PubMed] [Google Scholar]
- 5.Bestor T H, Tycko B. Creation of genomic methylation patterns. Nat Genet. 1996;12:363–367. doi: 10.1038/ng0496-363. [DOI] [PubMed] [Google Scholar]
- 6.Bird A P, Wolffe A P. Methylation-induced repression—belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 7.Cameron E E, Bachman K E, Myohanen S, Herman J G, Baylin S B. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 8.Challita P M, Kohn D B. Lack of expression from a retroviral vector after transduction of murine hematopoietic stem cells is associated with methylation in vivo. Proc Natl Acad Sci USA. 1994;91:2567–2571. doi: 10.1073/pnas.91.7.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen R Z, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- 10.Cheung P, Allis C D, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- 11.Clark S J, Harrison J, Paul C L, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coffee B, Zhang F, Warren S T, Reines D. Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat Genet. 1999;22:98–101. doi: 10.1038/8807. [DOI] [PubMed] [Google Scholar]
- 13.Conklin K F, Groudine M. Varied interactions between proviruses and adjacent host chromatin. Mol Cell Biol. 1986;6:3999–4007. doi: 10.1128/mcb.6.11.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dranoff G, Jaffee E, Lazenby A, Golumbek P, Levitsky H, Brose K, Jackson V, Hamada H, Pardoll D, Mulligan R C. Vaccination with irradiated tumor cells engineered to secrete murine granulocyte-macrophage colony-stimulating factor stimulates potent, specific, and long-lasting anti-tumor immunity. Proc Natl Acad Sci USA. 1993;90:3539–3543. doi: 10.1073/pnas.90.8.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng Y Q, Lorincz M C, Fiering S, Greally J M, Bouhassira E E. Position effects are influenced by the orientation of a transgene with respect to flanking chromatin. Mol Cell Biol. 2001;21:298–309. doi: 10.1128/MCB.21.1.298-309.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng Y Q, Seibler J, Alami R, Eisen A, Westerman K A, Leboulch P, Fiering S, Bouhassira E E. Site-specific chromosomal integration in mammalian cells: highly efficient CRE recombinase-mediated cassette exchange. J Mol Biol. 1999;292:779–785. doi: 10.1006/jmbi.1999.3113. [DOI] [PubMed] [Google Scholar]
- 18.Forrester W C, Epner E, Driscoll M C, Enver T, Brice M, Papayannopoulou T, Groudine M. A deletion of the human beta-globin locus activation region causes a major alteration in chromatin structure and replication across the entire beta-globin locus. Genes Dev. 1990;4:1637–1649. doi: 10.1101/gad.4.10.1637. [DOI] [PubMed] [Google Scholar]
- 19.Francastel C, Schubeler D, Martin D I, Groudine M. Nuclear compartmentalization and gene activity. Nat Rev Mol Cell Biol. 2000;1:137–143. doi: 10.1038/35040083. [DOI] [PubMed] [Google Scholar]
- 20.Fujita N, Takebayashi S, Okumura K, Kudo S, Chiba T, Saya H, Nakao M. Methylation-mediated transcriptional silencing in euchromatin by methyl-CpG binding protein MBD1 isoforms. Mol Cell Biol. 1999;19:6415–6426. doi: 10.1128/mcb.19.9.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Granger S W, Fan H. In vivo footprinting of the enhancer sequences in the upstream long terminal repeat of Moloney murine leukemia virus: differential binding of nuclear factors in different cell types. J Virol. 1998;72:8961–8970. doi: 10.1128/jvi.72.11.8961-8970.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gregory R I, Randall T E, Johnson C A, Khosla S, Hatada I, O'Neill L P, Turner B M, Feil R. DNA methylation is linked to deacetylation of histone h3, but not h4, on the imprinted genes snrpn and u2af1-rs1. Mol Cell Biol. 2001;21:5426–5436. doi: 10.1128/MCB.21.16.5426-5436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guy J, Hendrich B, Holmes M, Martin J E, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- 24.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendrich B, Guy J, Ramsahoye B, Wilson V A, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15:710–723. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsieh C L. Dynamics of DNA methylation pattern. Curr Opin Genet Dev. 2000;10:224–228. doi: 10.1016/s0959-437x(00)00064-2. [DOI] [PubMed] [Google Scholar]
- 27.Iguchi-Ariga S M, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- 28.Jahner D, Jaenisch R. Chromosomal position and specific demethylation in enhancer sequences of germ line-transmitted retroviral genomes during mouse development. Mol Cell Biol. 1985;5:2212–2220. doi: 10.1128/mcb.5.9.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jahner D, Jaenisch R. Retrovirus-induced de novo methylation of flanking host sequences correlates with gene inactivity. Nature. 1985;315:594–597. doi: 10.1038/315594a0. [DOI] [PubMed] [Google Scholar]
- 30.Jones P L, Veenstra G J, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 31.Kaludov N K, Wolffe A P. MeCP2 driven transcriptional repression in vitro: selectivity for methylated DNA, action at a distance and contacts with the basal transcription machinery. Nucleic Acids Res. 2000;28:1921–1928. doi: 10.1093/nar/28.9.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kass S U, Goddard J P, Adams R L. Inactive chromatin spreads from a focus of methylation. Mol Cell Biol. 1993;13:7372–7379. doi: 10.1128/mcb.13.12.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lander E S, Linton L M, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 34.Lewis J D, Meehan R R, Henzel W J, Maurer F I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 35.Lorincz M C, Schübeler D, Goeke S C, Walters M, Groudine M, Martin D I. Dynamic analysis of proviral induction and de novo methylation: implications for a histone deacetylase-independent, methylation density-dependent mechanism of transcriptional repression. Mol Cell Biol. 2000;20:842–850. doi: 10.1128/mcb.20.3.842-850.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magdinier F, Wolffe A P. Selective association of the methyl-CpG binding protein MBD2 with the silent p14/p16 locus in human neoplasia. Proc Natl Acad Sci USA. 2001;98:4990–4995. doi: 10.1073/pnas.101617298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakayama J, Rice J C, Strahl B D, Allis C D, Grewal S I. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 38.Nan X, Campoy F J, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 39.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 40.Ng H H, Zhang Y, Hendrich B, Johnson C A, Turner B M, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 41.Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 42.Pazin M J, Kadonaga J T. What's up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 43.Reik A, Telling A, Zitnik G, Cimbora D, Epner E, Groudine M. The locus control region is necessary for gene expression in the human beta-globin locus but not the maintenance of an open chromatin structure in erythroid cells. Mol Cell Biol. 1998;18:5992–6000. doi: 10.1128/mcb.18.10.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schübeler D, Francastel C, Cimbora D M, Reik A, Martin D I, Groudine M. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes Dev. 2000;14:940–950. [PMC free article] [PubMed] [Google Scholar]
- 45.Schübeler D, Lorincz M C, Cimbora D M, Telling A, Feng Y Q, Bouhassira E E, Groudine M. Genomic targeting of methylated DNA: influence of methylation on transcription, replication, chromatin structure, and histone acetylation. Mol Cell Biol. 2000;20:9103–9112. doi: 10.1128/mcb.20.24.9103-9112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson T, Fan H. Mapping of DNase I-hypersensitive sites in the 5′ and 3′ long terminal repeats of integrated Moloney murine leukemia virus proviral DNA. Mol Cell Biol. 1985;5:601–609. doi: 10.1128/mcb.5.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wade P A, Gegonne A, Jones P L, Ballestar E, Aubry F, Wolffe A P. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 48.Walsh C P, Chaillet J R, Bestor T H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 49.Wan M, Zhao K, Lee S S, Francke U. MECP2 truncating mutations cause histone H4 hyperacetylation in Rett syndrome. Hum Mol Genet. 2001;10:1085–1092. doi: 10.1093/hmg/10.10.1085. [DOI] [PubMed] [Google Scholar]
- 50.Whitelaw E, Martin D I. Retrotransposons as epigenetic mediators of phenotypic variation in mammals. Nat Genet. 2001;27:361–365. doi: 10.1038/86850. [DOI] [PubMed] [Google Scholar]
- 51.Yu F, Thiesen J, Stratling W H. Histone deacetylase-independent transcriptional repression by methyl-CpG-binding protein 2. Nucleic Acids Res. 2000;28:2201–2206. doi: 10.1093/nar/28.10.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, Ng H H, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu B, Benjamin D, Zheng Y, Angliker H, Thiry S, Siegmann M, Jost J P. Overexpression of 5-methylcytosine DNA glycosylase in human embryonic kidney cells EcR293 demethylates the promoter of a hormone-regulated reporter gene. Proc Natl Acad Sci USA. 2001;98:5031–5036. doi: 10.1073/pnas.091097298. [DOI] [PMC free article] [PubMed] [Google Scholar]