Abstract
Glioblastoma (GBM), which has poor prognosis and low 5-year survival rate, is the most common primary central nervous system malignant tumour in adults. Kinesin family member 18A (KIF18A) plays an important role in multiple tumours and is potential therapeutic target for GBM. Therefore, the present study investigated the role of KIF18A in GBM. The expression level and survival prognosis of KIF18A and protein phosphatase 1 catalytic subunit α (PPP1CA) in GBM patients were analysed using the Chinese Glioma Genome Atlas (CGGA) database. Reverse transcription-quantitative PCR and western blot analysis were applied to measure the expression of KIF18A and PPP1CA in normal and GBM cell lines. KIF18A expression was inhibited through cell transfection with a KIF18A-targeting short hairpin RNA. Cell proliferation was detected with the Cell Counting Kit-8 assay. Flow cytometry was used to detect cell cycle changes. Transwell and wound healing assays were used to measure cell invasion and migration. Western blotting was utilized for the detection of invasion- and migration-related proteins MMP9 and MMP2. Biological General Repository for Interaction Datasets and GeneMANIA databases were used to analyse the interaction between KIF18A and PPP1CA. The correlation between PPP1CA and KIF18A was examined using data from the CGGA database. Immunoprecipitation was used to demonstrate the binding relationship between KIF18A and PPP1CA. PPP1CA was overexpressed using cell transfection technology and its mechanism was further examined. The results demonstrated that KIF18A was upregulated in GBM cells compared with normal microglia HMC3. Compared with that in sh-NC group, silencing of KIF18A reduced cell proliferation, induced G2/M cycle arrest and inhibited the migration and the invasion of A172 GBM cells by interacting with PPP1CA. In conclusion, KIF18A interacted with PPP1CA to promote the proliferation, cycle arrest, migration and invasion of GBM cells.
Keywords: kinesin family member 18A, protein phosphatase 1 catalytic subunit α, glioblastoma, malignant development
Introduction
Glioma is a primary tumour originating from glial cells of the central nervous system, accounting for ~30% of all central nervous system tumours and ~80% of all primary malignant central nervous system tumours worldwide (1). Glioblastoma (GBM) is the most common primary malignant tumour of the central nervous system in adults, which originates from the neuroepithelium and accounts for 40-50% of all intracranial tumours worldwide (2). Even with aggressive multimodal therapy, GBM has poor prognosis, with a 5-year survival rate <5% (3). Therefore, finding novel drug targets and exploring the pathogenesis of the disease is of great significance for the development of new drugs and the treatment of GBM.
Kinesin family member 18A (KIF18A) is a member of the kinesin-8 family, and is closely associated with cell division (4). In recent years, the expression and role of KIF18A in tumours have attracted increasing attention in molecular oncology research (5). It has been shown that chromosomally unstable tumour cells specifically require KIF18A for proliferation (6). KIF18A mutant mice exhibit a stable micronucleus envelope that does not promote the occurrence of tumours (7). KIF18A gene knockdown inhibits the proliferation, migration and invasion of oesophageal cancer cells, while improves the sensitivity of cells to radiotherapy (8). Stangeland et al (9) showed that KIF18A can be a potential therapeutic target for GBM. A relevant clinical study demonstrated that KIF18A expression is higher in human GBM tissues than normal tissues, and its expression is closely associated with the recurrence of GBM in patients (10). However, to the best of our knowledge, the specific mechanism of KIF18A in GBM has not been yet elucidated.
Biological General Repository for Interaction Datasets (BioGRID) and GeneMANIA databases were used to analyse the genes interacting with KIF18A, and a binding relationship between KIF18A and protein phosphatase 1 catalytic subunit α (PPP1CA) was found. PPP1CA is an enzyme that is closely related to cell cycles and it was shown that PPP1CA activates the ERK/MAPK signalling pathway to promote the growth and metastasis of colorectal cancer (11). Inhibition of PPP1CA can significantly inhibit the proliferation and migration of breast cancer cells (12). At the same time, PPP1CA has been found to be a prognostic marker of GBM through a clinical database analysis (13). However, to the best of our knowledge, the regulatory mechanism of KIF18A and PPP1CA in GBM has not been reported.
Therefore, the present study investigated the expression of KIF18A and PPP1CA in GBM cells and further examined the regulatory mechanism of these proteins in malignant GBM cells. The present findings may provide a solid foundation for the clinical treatment of GBM.
Materials and methods
Databases
The Chinese Glioma Genome Atlas (CGGA) database (http://cgga.org.cn/analyse/RNA-data.jsp) was used to analyse the expression levels of PPP1CA and KIF18A, survival prognosis as well as their correlation. Pearson correlation analysis was performed.
BioGRID (version 4.4; https://thebiogrid.org/) and GeneMANIA (https://genemania.org/) databases were used to analyse the interaction between KIF18A and PPP1CA.
Cell culture
The normal microglia HMC3 (cat. no. CRL-3304), glioblastoma A172 (cat. no. CRL-1620) and LN-18 (cat. no. CRL-2610) and astrocytoma SW1783 (cat. no. HTB-13) cell lines were acquired from the American Type Culture Collection and cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc.) supplemented with 10% foetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin (Gibco; Thermo Fisher Scientific, Inc.) at 37˚C with 5% CO2.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from A172 cells with RNeasy Mini Kit (Qiagen China Co., Ltd). Total RNA was reversely transcribed into cDNA using M-MLV RTase and random primer kit (GeneCopoeia, Inc.) according to the standard protocol. qPCR was subsequently performed on an ABI PRISM 7900HT system (Applied Biosystems; Thermo Fisher Scientific, Inc.) using SYBR Premix Ex Taq™ (Takara Bio, Inc.) or TaqMan probes (cat. no. 450003; Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The thermocycling conditions were as follows: Initial denaturation at 95˚C for 3 min; followed by 40 cycles of denaturation at 95˚C for 30 sec, annealing at 60˚C for 30 sec and extension at 72˚C for 30 sec. GAPDH was used as internal reference and relative mRNA expression levels were calculated using the 2-ΔΔCq method (14). The following primer pairs were used for qPCR: KIF18A forward, 5'-GAGAGGCACATGAAGAGAAGT-3' and reverse, 5'-AAGTCCATGAACGACCACCC-3'; PPP1CA forward, 5'-CAGGGTCCTGACACCCCATT-3' and reverse, 5'-AGGTAAAAGAGACGCCACGG-3'; GAPDH forward, 5'-AATGGGCAGCCGTTAGGAAA-3' and reverse, 5'-GCGCCCAATACGACCAAATC-3'.
Western blotting
Total protein was extracted from A172 cells using RIPA lysis buffer (Beyotime Institute of Biotechnology). Total protein concentration was quantified with a BCA assay kit (Beyotime Institute of Biotechnology) and equal amount of proteins (20 µg per lane) was separated using 10% SDS-PAGE and subsequently transferred onto PVDF membranes (MilliporeSigma) which were blocked with 5% non-fat milk for 2 h at room temperature. The membranes were incubated with primary antibodies targeting KIF18A (1:2,000; cat. no. ab72417; Abcam), CDK1 (1:10,000; cat. no. ab133327; Abcam), cyclin B (1:50,000; cat. no. ab32053; Abcam), MMP9 (1:1,000; cat. no. ab76003; Abcam), MMP2 (1:1,000; cat. no. ab92536; Abcam), PPP1CA (1:20,000; cat. no. ab52619; Abcam), or GAPDH (1:1,000; cat. no. ab9485; Abcam) overnight at 4˚C. Following the rinse with PBS for three times, membranes were incubated with HRP-conjugated secondary antibodies (cat. no. ab6759; 1:5,000; Abcam) for 1.5 h. The protein bands were visualized using ECL detection reagent (MilliporeSigma) and analysed with ImageJ software 1.8.0 (National Institutes of Health). GAPDH was used as internal reference.
Cell transfection
The two GV248-green fluorescent protein (GFP)-short harpin (sh)RNA-KIF18A lentiviral vectors, namely GV248-GFP-sh-KIF18A#1 and GV248-GFP-sh-KIF18A#2, the negative control (NC) GV248-GFP-sh-NC lentiviral vector, PPP1CA-specific pcDNA overexpression vector (Oe-PPP1CA) and pcDNA3.1 empty vector were constructed by Shanghai GeneChem Co., Ltd. A total of 100 nM plasmids were transfected into A172 cells at 37˚C for 24 h using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. After 24 h, the harvested A172 cells were adopted for follow-up experiments. The shRNA targeting sequences were as follows: sh-KIF18A#1, 5'-TTGTTTCAGACTCACATATAA-3'; sh-KIF18A#2, 5'-AGTCCTGAGAGGAAGTCTTAA-3'; and sh-NC, 5'-TTCTCCGAACGTGTCACGT-3'. Cells were then divided into the following groups: Control (incubated in DMEM); sh-NC (transfected with GV248-GFP lentiviral vector); sh-KIF18A (transfected with GV248-GFP-shRNA-KIF18A#2 lentiviral vector); sh-KIF18A + Oe-NC (transfected with GV248-GFP-shRNA-KIF18A#2 lentiviral vector and Oe-NC empty vector); and sh-KIF18A + Oe-PPP1CA (transfected with GV248-GFP-shRNA-KIF18A#2 lentiviral vector and Oe-PPP1CA vector).
Cell Counting Kit-8 (CCK-8) assay
A172 cells were plated in 96-well plates at the density of 2x103 cells/well and incubated for 24, 48 and 72 h. Subsequently, the cells were incubated with CCK-8 solution (MilliporeSigma) at 37˚C for 3 h. The absorbance of each well at 450 nm was measured with a multimode microplate reader (Synergy H1; BioTek Instruments, Inc.).
Cell cycle analysis
Flow cytometry was used for cell cycle analysis. The transfected A172 cells at a density of 4x105 cells per well were fixed with 75% ethanol at 4˚C for 24 h, followed by the staining with PI solution [200 µg/ml RNase A, 50 µg/ml PI and 0.1% (v/v) Triton X-100 in PBS] for 30 min at room temperature. Finally, the cell cycle was analysed using CellQuest software v.2.0 (BD Biosciences) and ModFit LT v.2.0 (Verity Software House, Inc.).
Transwell assay
Transwell assays were performed using Transwell inserts (8-µm pore size; Costar; Corning, Inc.) precoated with 50 µl Matrigel (BD Biosciences). The inserts were put into 24-well plates. A172 cells that injected into the serum-free medium at a density of 2x104 cells were seeded into the upper chambers while the lower chambers were filled with DMEM supplemented with 10% FBS. Following incubation at 37˚C for 24 h, the residual cells on upper chambers were removed, and the cells under the surface were stained with 0.5% crystal violet for 10 min at room temperature and then fixed with 70% ethanol at 4˚C. The number of cells in five randomly selected fields were counted with a light microscope (Olympus Corporation) and were analyzed using ImageJ 1.8.0 software (National Institutes of Health).
Wound healing assay
A172 cells were seeded in 6-well plates at the density of 5x105 cells/well. After 95% cell confluence was achieved, artificial wounds were gently made using a 200-µl sterile pipette tip and confluent monolayers for wounding were yielded. The serum-starved culture medium was replaced with DMEM supplemented with 10 mg/ml mitomycin C (Sigma-Aldrich; Merck KGaA). Cells in the scratched area were imaged at 0 and 24 h using a light microscope (Thermo Fisher Scientific, Inc.). The relative migration rate (%)=(wound width at 0 h-wound width at 24 h)/wound width at 0 h x100.
Immunoprecipitation
A172 cell lysates were prepared in RIPA buffer. The supernatant was collected after centrifugation at 13,000 x g for 10 min at 4˚C. Subsequently, 0.2 mg protein A agarose beads (cat. no. 20366; Thermo Fisher Scientific, Inc.) washed with 100 µl PBS buffer were added to 500 µg lysis buffer and incubated with 2 µg IgG antibody (1:1,000; cat. no. ab172730; Abcam) or PPP1CA antibody (1:500; cat. no. GTX105255; GeneTex, Inc.) or KIF18A antibody (1:500; cat. no. ab72417; Abcam) overnight at 4˚C with slow shaking. Following the IP reaction and centrifugation at 1,000 x g at 4˚C for 2 min, the agarose beads were washed with lysis buffer and boiled for 5 min at 100˚C by adding 15 µl 2X SDS loading dye before being subjected to western blot analysis as aforementioned.
Statistical analysis
The data from three independent experiments are presented as the mean ± standard deviation and were evaluated using SPSS 19 software (IBM Corp.). One-way ANOVA followed by Tukey's post hoc test was used for statistical analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
KIF18A is upregulated in GBM
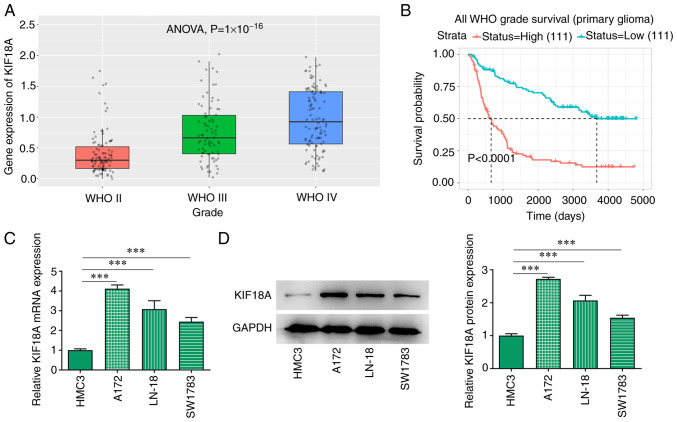
The CGGA database analysis showed that the expression of KIF18A was markedly increased in WHO IV group compared with the WHO II group (Fig. 1A). KIF18A expression was closely associated with the prognosis of patients with GBM and the higher the KIF18A expression, the lower the survival probability of patients with GBM (Fig. 1B). RT-qPCR and western blotting results showed that the mRNA and protein expression of KIF18A in GBM cell lines was significantly increased compared with the HMC3 group (Fig. 1C and D). The expression level of KIF18A in A172 cells was higher than that in LN-18 and SW1783 cells, and consequently A172 cells were selected for subsequent experiments.
Figure 1.
KIF18A is upregulated in glioblastoma cells. (A) KIF18A expression level and (B) associated survival probability were analysed using the Chinese Glioma Genome Atlas database. The expression level of KIF18A was detected using (C) reverse transcription-quantitative PCR and (D) western blotting. ***P<0.001. KIF18A, kinesin family member 18A; WHO, World Health Organization.
Silencing KIF18A reduces cell proliferation and induces G2/M cycle arrest in A172 cells
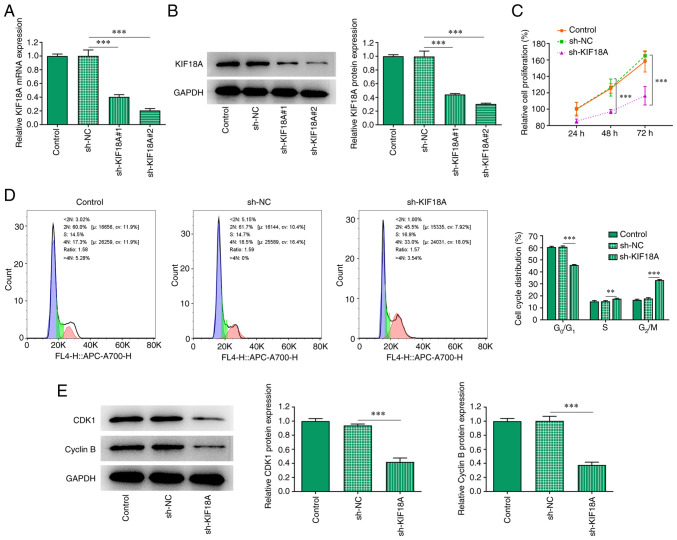
After silencing KIF18A expression, RT-qPCR and western blotting were used to estimate the knockdown efficiency (Fig. 2A and B). Compared with the sh-KIF18A#1 group, KIF18A exhibited lower mRNA and protein expression in the sh-KIF18A#2 group, likely indicating an improved transfection efficacy for sh-KIF18A#2. Therefore, sh-KIF18A#2 was chosen for subsequent experiments. CCK-8 was utilized to measure the cell proliferation rate and the results showed that the proliferation of the sh-KIF18A group was significantly decreased compared with sh-NC (Fig. 2C). The cell cycle was analysed using flow cytometry, and the results showed that compared with sh-NC, the G0/G1 proportion in the sh-KIF18A group was significantly decreased and the G2/M proportion was significantly increased (Fig. 2D). Western blot analysis was used to examine the expression of the cycle-related proteins cyclin-dependent kinase 1 (CDK1) and cyclin B. The results showed that KIF18A silencing could significantly inhibit the expression of CDK1 and cyclin B (P<0.001; Fig. 2E).
Figure 2.
Silencing of KIF18A reduces proliferation and induces G2/M cycle arrest in A172 cells. Expression level of KIF18A was detected by (A) reverse transcription-quantitative PCR and (B) western blotting after transfection of sh-KIF18A. (C) Cell Counting Kit-8 assay. (D) Flow cytometry cell cycle assay. (E) Western blot analysis of the expression of CDK1 and cyclin B. **P<0.01 and ***P<0.001. KIF18A, kinesin family member 18A; CDK1, cyclin-dependent kinase 1; NC, negative control; sh, short hairpin.
Silencing of KIF18A inhibits the migration and invasion of A172 cells
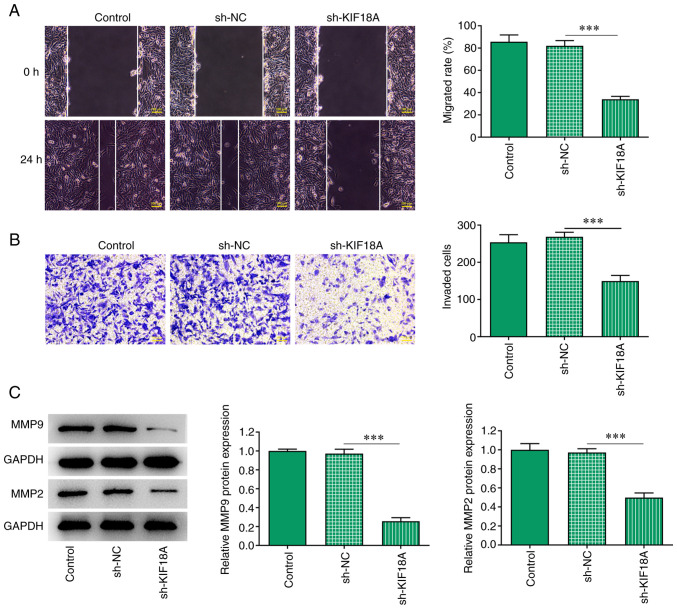
Transwell and wound healing assays were performed to assess cell invasion and migration, and the results showed that the migration and invasion of A172 cells were significantly decreased after KIF18A knockdown (P<0.001 vs. sh-NC; Fig. 3A and B). Western blotting results showed that the expression of the migration-related proteins MMP2 and MMP9 were decreased significantly after KIF18A silencing (P<0.001 vs. sh-NC; Fig. 3C).
Figure 3.
Silencing of KIF18A inhibits the migration and invasion of A172 cells. (A) Wound healing cell migration assay (magnification, x100). (B) Transwell cell invasion assay (magnification, x100). (C) Western blot analysis of the expression of MMP9 and MMP2. ***P<0.001. KIF18A, kinesin family member 18A; NC, negative control; sh, short hairpin.
KIF18A interacts with PPP1CA in GBM
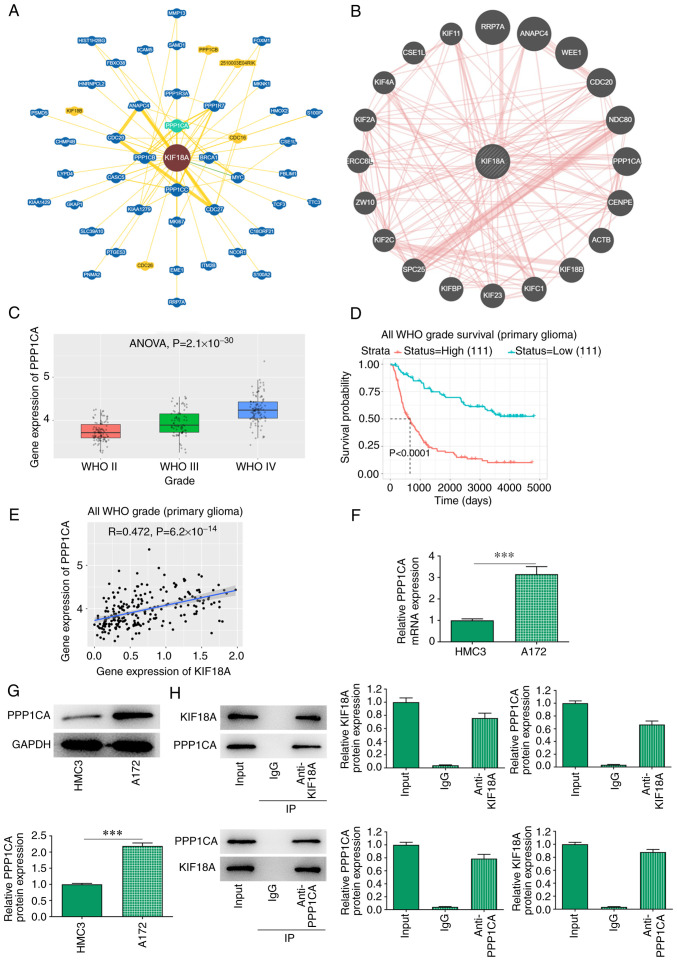
BioGRID and GeneMANIA database analysis revealed a potential interaction between KIF18A and PPP1CA (Fig. 4A and B). Through the CGGA database, it was found that the expression of PPP1CA was significantly increased in WHO IV group compared with the WHO II group (Fig. 4C). The expression of PPP1CA was closely related to the prognosis of GBM and the higher the KIF18A expression, the lower the survival probability of patients with GBM (Fig. 4D). In addition, CGGA database also revealed a highly positive correlation between KIF18A and PPP1CA in GBM (R=0.472, P=6.2x10-14; Fig. 4E). RT-qPCR and western blot analysis showed that PPP1CA mRNA and protein expression were significantly increased in A172 cells compared with that in HMC3 cells (P<0.001; Fig. 4F and G). Co-immunoprecipitation assay also verified the targeted binding relationship between KIF18A and PPP1CA (Fig. 4H).
Figure 4.
KIF18A interacts with PPP1CA in glioblastoma cells. (A) Biological General Repository for Interaction Datasets and (B) GeneMANIA databases were utilized to analyse the interaction between KIF18A and PPP1CA. (C) Expression level of PPP1CA, (D) associated survival probability and (E) correlation with KIF18A were analysed utilizing the Chinese Glioma Genome Atlas database. The expression level of PPP1CA was measured via (F) reverse transcription-quantitative PCR and (G) western blot analysis. (H) IP was utilized to investigate the targeted binding relationship between KIF18A and PPP1CA. ***P<0.001. KIF18A, kinesin family member 18A; PPP1CA protein phosphatase 1 catalytic subunit α; WHO, World Health Organization; IP, immunoprecipitation.
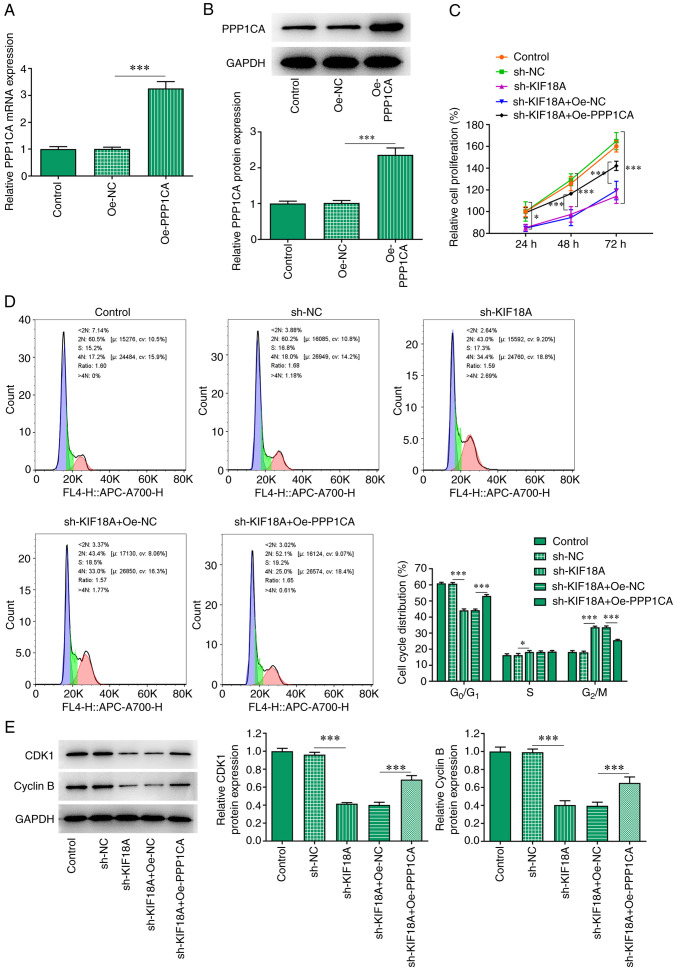
Overexpression of PPP1CA reduces the effect of KIF18A knockdown on the proliferation and G2/M cycle arrest of A172 cells
The Oe-PPP1CA vector was constructed and the transfection efficiency was examined using RT-qPCR and western blot analysis (Fig. 5A and B). Cells were divided into control, sh-NC, sh-KIF18A, sh-KIF18A + Oe-NC and sh-KIF18A + Oe-PPP1CA groups. CCK-8 results showed that KIF18A knockdown reduced the proliferation of A172 cells compared with the sh-NC group. Nevertheless, the reduced proliferation in KIF18A-depleted A172 cells was significantly increased by PPP1CA overexpression compared with that of sh-KIF18A + Oe-NC group (Fig. 5C). As presented in Fig. 5D, KIF18A interference decreased the number of cells at G0/G1 and S phase but increased that of cells at G2/M phase relative with the sh-NC, whereas PPP1CA overexpression exhibited the opposite impacts on them, evidenced by an increased number of cells at G0/G1 and S phase and decreased number of cells at G2/M phase relative with the sh-KIF18A + Oe-NC group (Fig. 5D). Moreover, results obtained from western blot analysis demonstrated the declined contents of CDK1 and cyclin B in A172 cells resulted from KIF18A knockdown were enhanced after overexpressing PPP1CA expression in contrast with those in sh-KIF18A + Oe-NC group (Fig. 5E).
Figure 5.
Overexpression of PPP1CA reduces the effect of KIF18A silencing on the proliferation and G2/M cycle arrest of A172 cells. The mRNA and protein expression of PPP1CA were measured via (A) reverse transcription-quantitative PCR and (B) western blotting. (C) Cell Counting Kit-8 assay. (D) Cell cycle was detected by flow cytometry. (E) Western blotting was used to detect the expression of CDK1 and cyclin B. *P<0.05 and ***P<0.001. PPP1CA protein phosphatase 1 catalytic subunit α; KIF18A, kinesin family member 18A; CDK1, cyclin-dependent kinase 1; NC, negative control; sh, short hairpin; Oe, overexpression.
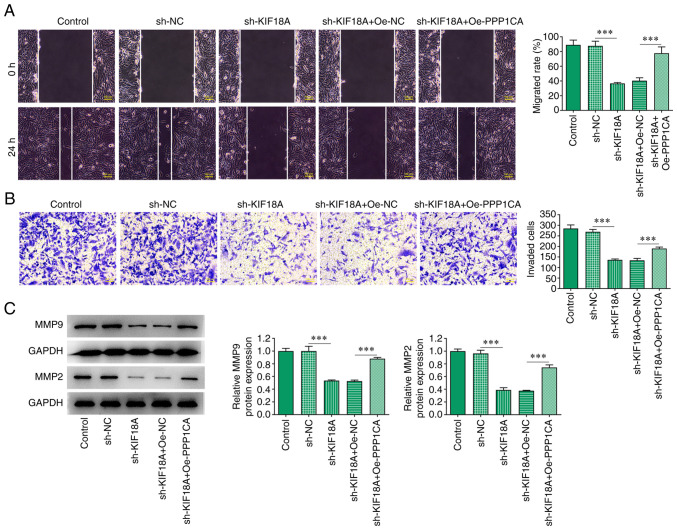
Overexpression of PPP1CA reduces the effect of KIF18A knockdown on the migration and invasion of A172 cells
Wound healing and Transwell assays showed that compared with the sh-KIF18A + Oe-Nc group, cell migration and invasion were significantly increased in the sh-KIF18A + Oe-PPP1CA group (Fig. 6A and B). Western blotting results showed that MMP9 and MMP2 expression was significantly increased in the sh-KIF18A + Oe-PPP1CA group compared with that in the sh-KIF18A + Oe-NC group (P<0.001; Fig. 6C).
Figure 6.
Overexpression of PPP1CA reduces the effect of KIF18A silencing on the migration and invasion of A172 cells. (A) Wound healing cell migration assay (magnification, x100). (B) Transwell cell invasion assay (magnification, x100). (C) Western blot analysis of the expression of MMP9 and MMP2. ***P<0.001. PPP1CA protein phosphatase 1 catalytic subunit α; KIF18A, kinesin family member 18A; NC, negative control; sh, short hairpin; Oe, overexpression.
Discussion
The incidence of GBM has been increasing annually lately, whereas the effective treatment methods for GBM were not improved, with radiotherapy and chemotherapy still being the standard treatment for patients with GBM after surgical resection (15). Moreover, the use of chemotherapy drugs only increased the median survival time of patients with GBM by 2-3 months, while it did not significantly prolong the survival time of patients (16). Therefore, it is of great urgency to find more effective and sensitive therapeutic targets to improve the treatment of GBM.
Through CGGA database analysis, the present study found that the expression of KIF18A in patients with GBM is significantly increased in the WHO IV group compared with the WHO II group and is closely related to the prognosis of GBM. The present study revealed that the expression of KIF18A is also significantly increased in GBM cell lines when compared with the normal microglia HMC3. Previously, it has been shown that KIF18A can be used as a potential therapeutic target for GBM (9). Therefore, it is reasonable to hypothesize that the abnormal expression of KIF18A is closely related to the occurrence and development of GBM. A previous study has shown that knockdown of KIF18A could suppress the proliferation, migration and invasion of oesophageal cancer cells (8). In hepatocellular carcinoma cells, KIF18A may promote cell proliferation, invasion and metastasis by promoting the cell cycle and Akt and MMP7/MMP9 related signalling pathways (17). In prostate cancer, KIF18A silencing in PC-3 prostate cells has been shown to significantly inhibit cell proliferation and metastasis (18). These results indicated that KIF18A may have an oncogenic role in multiple tumours. The present results showed that KIF18A knockdown decreased the proliferation, invasion and migration of GBM A172 cells but induced the cell cycle. These results indicated that KIF18A may have an oncogenic role in GBM.
The current study further explored the mechanism of KIF18A in regulating GBM. Firstly, the interaction between KIF18A and PPP1CA was investigated through BioGRID and GeneMANIA database analysis. Moreover, CGGA database analysis showed a high correlation between PPP1CA and KIF18A in GBM. Furthermore, the interaction between PPP1CA and KIF18A was experimentally investigated and demonstrated. A previous study has shown that the transcriptional level of PPP1CA in pancreatic cancer is higher than that in normal pancreas and the high transcriptional level of PPP1CA is associated with the poor survival rate of patients with pancreatic cancer (19). In the current study, PPP1CA was also found to be associated with GBM survival prognosis, which is consistent with the study by Shastry et al (13). It has been shown that PPP1CA deficiency may significantly inhibit the proliferation and migration of breast cancer cells (12). PPP1CA is upregulated in clinical specimens of maxillary sinus squamous cell carcinoma. Silencing PPP1CA gene can significantly inhibit the proliferation and invasion of cancer cells (20). Therefore, PPP1CA shows oncogenic characteristics. However, it has also been reported that PPP1CA downregulation could significantly promote the proliferation, migration and invasion of gastric cancer cells (21). Therefore, PPP1CA can also act as a tumour suppressor gene. This raises questions about the role of PPP1CA in GBM. In this context, the present study found that PPP1CA expression is significantly elevated in A172 cells compared with that in HMC3 cells. Overexpression of PPP1CA countervailed the inhibitory effects of KIF18A knockdown on the proliferation, cell cycle arrest, migration and invasion of GBM cells, suggesting that PPP1CA may act as an oncogenic gene in GBM.
The present study only detected the expression of PPP1CA in GMB A172 cells and investigated its regulatory mechanism. In the future, the mechanism of PPP1CA and KIF18A in different stages of GBM should be further examined.
In conclusion, the present study results demonstrated that KIF18A interacts with PPP1CA to promote the proliferation, cell cycle arrest, migration and invasion of GBM A172 cells.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JY and XL conceived and designed the study. QZ, ZY, JS, LZ, YY and XW performed the experiments. JY and XL analysed the experimental data and wrote and revised the manuscript. JY and XL confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Davis ME. Epidemiology and overview of gliomas. Semin Oncol Nurs. 2018;34:420–429. doi: 10.1016/j.soncn.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Preusser M, Lim M, Hafler DA, Reardon DA, Sampson JH. Prospects of immune checkpoint modulators in the treatment of glioblastoma. Nat Rev Neurol. 2015;11:504–514. doi: 10.1038/nrneurol.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen R, Smith-Cohn M, Cohen AL, Colman H. Glioma subclassifications and their clinical significance. Neurotherapeutics. 2017;14:284–297. doi: 10.1007/s13311-017-0519-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malaby HL, Lessard DV, Berger CL, Stumpff J. KIF18A's neck linker permits navigation of microtubule-bound obstacles within the mitotic spindle. Life Sci Alliance. 2019;2(e201800169) doi: 10.26508/lsa.201800169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tao BY, Liu YY, Liu HY, Zhang ZH, Guan YQ, Wang H, Shi Y, Zhang J. Prognostic biomarker KIF18A and its correlations with immune infiltrates and mitosis in glioma. Front Genet. 2022;13(852049) doi: 10.3389/fgene.2022.852049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marquis C, Fonseca CL, Queen KA, Wood L, Vandal SE, Malaby HLH, Clayton JE, Stumpff J. Chromosomally unstable tumour cells specifically require KIF18A for proliferation. Nat Commun. 2021;12(1213) doi: 10.1038/s41467-021-21447-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sepaniac LA, Martin W, Dionne LA, Stearns TM, Reinholdt LG, Stumpff J. Micronuclei in Kif18a mutant mice form stable micronuclear envelopes and do not promote tumorigenesis. J Cell Biol. 2021;220(e202101165) doi: 10.1083/jcb.202101165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian LX, Cao X, Du MY, Ma CX, Zhu HM, Peng Y, Hu XY, He X, Yin L. KIF18A knockdown reduces proliferation, migration, invasion and enhances radiosensitivity of oesophageal cancer. Biochem Biophys Res Commun. 2021;557:192–198. doi: 10.1016/j.bbrc.2021.04.020. [DOI] [PubMed] [Google Scholar]
- 9.Stangeland B, Mughal AA, Grieg Z, Sandberg CJ, Joel M, Nygard S, Meling T, Murrell W, Vik Mo EO, Langmoen IA. Combined expressional analysis, bioinformatics and targeted proteomics identify new potential therapeutic targets in glioblastoma stem cells. Oncotarget. 2015;6:26192–26215. doi: 10.18632/oncotarget.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang LB, Zhang XB, Liu J, Liu QJ. The proliferation of glioblastoma is contributed to kinesin family member 18A and medical data analysis of GBM. Front Genet. 2022;13(858882) doi: 10.3389/fgene.2022.858882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun H, Ou B, Zhao S, Liu X, Song L, Liu X, Wang R, Peng Z. USP11 promotes growth and metastasis of colorectal cancer via PPP1CA-mediated activation of ERK/MAPK signalling pathway. EBioMedicine. 2019;48:236–247. doi: 10.1016/j.ebiom.2019.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie W, Sun Y, Zeng Y, Hu L, Zhi J, Ling H, Zheng X, Ruan X, Gao M. Comprehensive analysis of PPPCs family reveals the clinical significance of PPP1CA and PPP4C in breast cancer. Bioengineered. 2022;13:190–205. doi: 10.1080/21655979.2021.2012316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shastry AH, Thota B, Srividya MR, Arivazhagan A, Santosh V. Nuclear Protein Phosphatase 1 α (PP1A) expression is associated with poor prognosis in p53 expressing glioblastomas. Pathol Oncol Res. 2016;22:287–292. doi: 10.1007/s12253-015-9928-5. [DOI] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Hombach-Klonisch S, Mehrpour M, Shojaei S, Harlos C, Pitz M, Hamai A, Siemianowicz K, Likus W, Wiechec E, Toyota BD, et al. Glioblastoma and chemoresistance to alkylating agents: Involvement of apoptosis, autophagy, and unfolded protein response. Pharmacol Ther. 2018;184:13–41. doi: 10.1016/j.pharmthera.2017.10.017. [DOI] [PubMed] [Google Scholar]
- 16.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 17.Luo W, Liao M, Liao Y, Chen X, Huang C, Fan J, Liao W. The role of kinesin KIF18A in the invasion and metastasis of hepatocellular carcinoma. World J Surg Oncol. 2018;16(36) doi: 10.1186/s12957-018-1342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H, Shen T, Zhang Z, Li Y, Pan Z. Expression of KIF18A is associated with increased tumor stage and cell proliferation in prostate cancer. Med Sci Monit. 2019;25:6418–6428. doi: 10.12659/MSM.917352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hang J, Lau SY, Yin R, Zhu L, Zhou S, Yuan X, Wu L. The role of phosphoprotein phosphatases catalytic subunit genes in pancreatic cancer. Biosci Rep. 2021;41(BSR20203282) doi: 10.1042/BSR20203282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nohata N, Hanazawa T, Kikkawa N, Sakurai D, Fujimura L, Chiyomaru T, Kawakami K, Yoshino H, Enokida H, Nakagawa M, et al. Tumour suppressive microRNA-874 regulates novel cancer networks in maxillary sinus squamous cell carcinoma. Br J Cancer. 2011;105:833–841. doi: 10.1038/bjc.2011.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verdugo-Sivianes EM, Navas L, Molina-Pinelo S, Ferrer I, Quintanal-Villalonga A, Peinado J, Garcia-Heredia JM, Felipe-Abrio B, Muñoz-Galvan S, Marin JJ, et al. Coordinated downregulation of Spinophilin and the catalytic subunits of PP1, PPP1CA/B/C, contributes to a worse prognosis in lung cancer. Oncotarget. 2017;8:105196–105210. doi: 10.18632/oncotarget.22111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.