Abstract
During the aging process, the reduced osteogenic differentiation of bone marrow mesenchymal stem cells (BM-MSCs) results in decreased bone formation, which contributes to senile osteoporosis. Previous studies have confirmed that interrupted circadian rhythm plays an indispensable role in age-related disease. However, the mechanism underlying the impaired osteogenic differentiation of BM-MSCs during aging and its relationship with interrupted circadian rhythm remains unclear. In this study, we confirmed that the circadian rhythm was interrupted in aging mouse skeletal systems. The level of the core rhythm component BMAL1 but not that of CLOCK in the osteoblast lineage was decreased in senile osteoporotic specimens from both human and mouse. BMAL1 targeted TTK as a circadian-controlled gene to phosphorylate MDM2 and regulate H2Bub1 level, while H2Bub1 in turn regulated the expression of BMAL1. The osteogenic capacity of BM-MSCs was maintained by a positive loop formed by BMAL1-TTK-MDM2-H2Bub1. Furthermore, we demonstrated that using bone-targeting recombinant adeno-associated virus 9 (rAAV9) to enhance Bmal1 or Ttk might have a therapeutic effect on senile osteoporosis and delays bone repair in aging mice. In summary, our study indicated that targeting the BMAL1-TTK-MDM2-H2Bub1 axis via bone-targeting rAAV9 might be a promising strategy for the treatment of senile osteoporosis.
Keywords: MT: Oligonucleotides: Therapies and Applications, senile osteoporosis, mesenchymal stem cells, BMAL1, histone H2B monoubiquitination, rAAV9 therapy
Graphical abstract
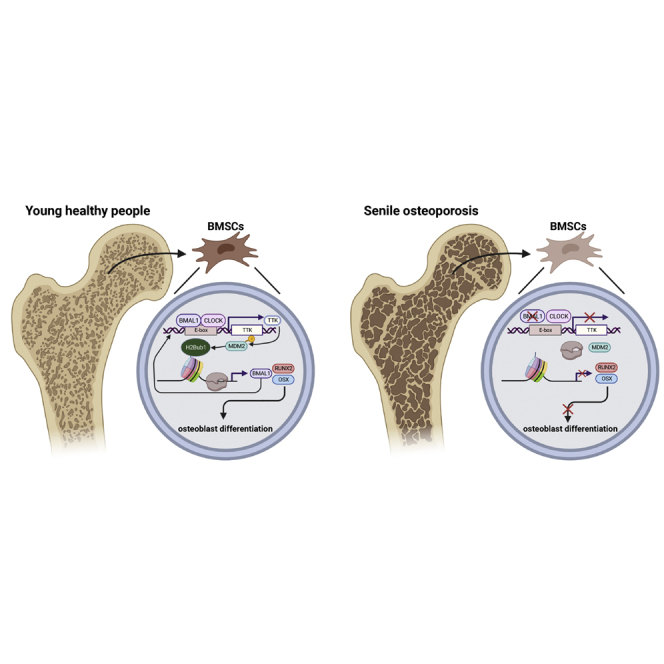
Jinteng and colleagues describe interrupted circadian rhythm and decreased level of BMAL1 in aging skeletal systems and BM-MSCs. They prove that the osteogenic capacity of BM-MSCs is maintained by a BMAL1-TTK-MDM2-H2Bub1 loop, which might be a promising target via bone-targeting rAAV9 for the treatment of senile osteoporosis.
Introduction
Senile osteoporosis is characterized by age-related bone loss and specific biological aging in the skeletal system and generally refers to osteoporosis after the age of 70 years.1 By 2050, the proportion of people aged 60 years or older in the total population will reach 22% worldwide, as estimated by the World Health Organization. With the increase in the aging population, senile osteoporosis and related fractures not only enhance the morbidity and mortality of elderly individuals but also dramatically increase the financial burden on public health.2 Therefore, many attempts have been made to discover novel therapeutic medicines, such as teriparatide, risedronate3 and herbal medicines.4
The “osteogenesis-adipogenesis” balance of bone marrow mesenchymal stem cells (BM-MSCs) is essential for the maintenance of healthy bone homeostasis. However, during the aging process, differentiation of BM-MSCs into osteoblasts decreases and differentiation into adipocytes increases.5,6 This shift in the cell fate of BM-MSCs results in reduced bone formation, which contributes to senile osteoporosis.7 The mechanism underlying this abnormal trend of differentiation during aging is still unclear. Therefore, elucidation of the mechanism of osteogenic impairment of aging BM-MSCs and improving osteogenesis of BM-MSCs in vivo are important for antiaging bone loss.
Circadian rhythms in behavior and physiology are hallmarks of life on earth. The endogenous timekeeping mechanism known as the circadian clock allows organisms to anticipate the periodic fluctuations brought on by the 24 h solar cycle, aligning biological functions with external changes.8 Brain and muscle Arnt-like protein 1 (BMAL1) and circadian locomotor output cycles kaput (CLOCK) heterodimers drive daily changes in the transcription of circadian-controlled genes to control cellular molecular oscillations.9 Previous studies have confirmed that BMAL1/CLOCK heterodimers play an indispensable role in physiological BM-MSC-mediated bone formation both in vitro and in vivo.10,11 Moreover, reduced levels of BMAL1 impair the osteogenic differentiation of BM-MSCs. However, the role of BMAL1 in impaired senile osteoporosis-related bone formation remains unclear and needs further exploration.
Histone modifications and their associated enzymes can affect chromatin compaction, nucleosome dynamics, and gene transcription. Dysregulation of these processes disrupts gene expression homeostasis and is frequently observed during the aging process in different systems, including the skeletal system.12,13,14 Histone H2B is monoubiquitylated on Lys120 in mammals (H2Bub1), and H2Bub1 is primarily associated with the transcribed regions of active genes, where it facilitates transcriptional elongation.15 A stable global level of H2Bub1 is necessary for the osteogenic capacity of BM-MSCs.16 Moreover, multiple histone modifications, including H2Bub1, cooperate with BMAL1/CLOCK heterodimers in regulating gene transcription. During the aging process, abnormal histone modification impairs BMAL1/CLOCK heterodimer function in circadian-controlled gene transcription and leads to morbidity.17 However, as multiple histone modification enzymes or modulators are circadian-controlled genes, BMAL1 deficiency may also lead to dysfunctional histone modification during the aging process.18 As it remains largely unknown whether reduced BMAL1 impairs the osteogenic differentiation of BM-MSCs through changes in histone modification during aging, further investigation is needed.
Bone-targeting therapy remains a worldwide challenge. Drug agents are frequently administered at high doses to reach the effective therapeutic level inside the diseased bone microenvironment, which may lead to severe side effects, including myelosuppression and dose-limiting toxicity to healthy tissues. Thus, a drug delivery system with high bone affinity to reduce off-target side effects is urgently needed. Adeno-associated virus (AAV) is a small (26 nm) nonenveloped parvovirus with a single-stranded genome of ∼4.7 kb.19 Because of its high transduction efficiency, persistent transgene expression, and lack of postinfection immunogenicity and pathogenicity, AAV is an attractive viral vector for use in gene therapy in the clinic.20 Previous studies have already confirmed that the rAAV9 subtype could effectively drive DNA delivery into osteoblast lineage cells in vivo.21 However, the prototype rAAV9 still has other organ tropisms, including that of cardiac muscle, skeletal muscle, lung alveoli, and liver. Thus, the modification of rAAV9 to confer it a strongly osteophilic property and the ability to deliver bone-formation genes into BM-MSCs may be a promising strategy for treating senile osteoporosis.
In this study, we demonstrated that the circadian rhythm was interrupted in the skeletons of aging mice. BMAL1 but not CLOCK was decreased in the osteoblast lineage in both human and mouse senile osteoporotic specimens. BMAL1 targeted TTK as a circadian control gene to regulate the H2Bub1 level, while H2Bub1 in turn regulated the expression of BMAL1 at the transcriptional level. BMAL1-TTK-H2Bub1 formed a positive loop to maintain the osteogenic capacity of BM-MSCs. Finally, we demonstrated that using bone-targeting rAAV9 to enhance Bmal1 or Ttk could have a therapeutic effect on senile osteoporosis and delayed bone repair in aging mice. Taken together, our study indicated that targeting the BMAL1-TTK-H2Bub1 axis via bone-targeting rAAV9 is a promising strategy for treating senile osteoporosis.
Results
Disruption of the circadian rhythm and BMAL1 expression in the skeletal system in senile osteoporosis
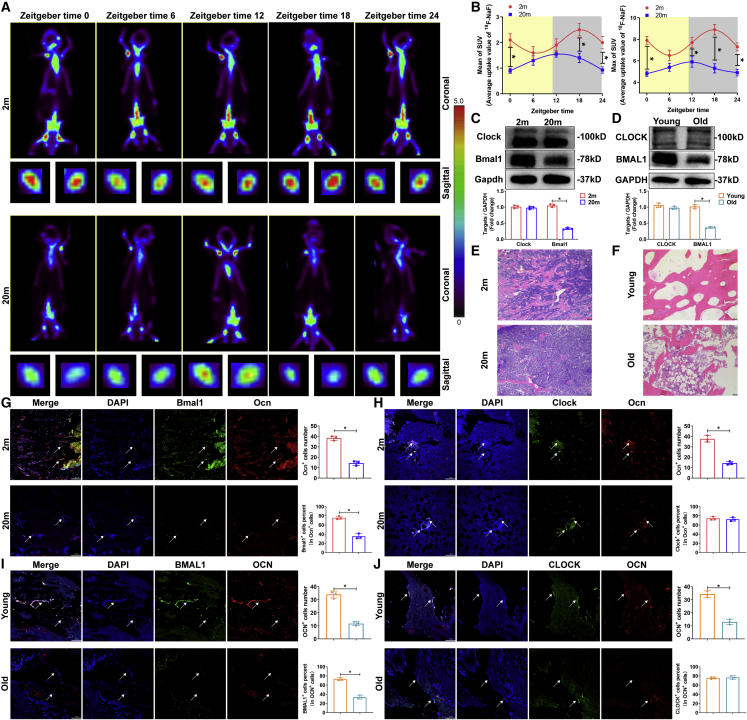
To identify the disruption of the circadian rhythm of bone formation and metabolism in the aging skeletal system, we monitored 18F-labeled sodium fluoride (18F-NaF) uptake in the femur in 2-month-old and 20-month-old C57BL/6 male mice by a micro-positron emission tomography (PET) computed tomographic scanner. During a 24 h dark-light cycle, we found that there was a significant rhythm of 18F-NaF uptake in 2-month-old mice, which reached a peak around zeitgeber time (ZT) 18 and a nadir around ZT6 (Figure 1A). However, in 20-month-old mice, both the maximum and mean rates of 18F-NaF uptake were reduced compared with those in 2-month-old mice, and the physiological variation was disrupted (Figure 1B), suggesting that circadian rhythm disruption occurred in the skeletal system in senile osteoporosis.
Figure 1.
Disruption of circadian rhythm and BMAL1 expression in the skeletal system in senile osteoporosis
(A) PET scans of 2-month-old and 20-month-old C57BL/6J mice after 18F-NaF tail vein injection at the indicated times. ZT0 is defined as the time the light was turned on (8 a.m.). (B) PET analysis of maximum and mean SUV. (C and D) Western blot analysis of the levels of the core rhythm components BMAL1 and CLOCK in BM-MSCs from 2-month-old and 20-month-old mice, patients with traffic injuries and patients with senile osteoporosis. (E and F) HE staining (scale bar, 100 μm) of bone specimens from 2-month-old and 20-month-old mice, patients with traffic injuries and patients with senile osteoporosis. (G and H) Immunofluorescence staining (scale bar, 100 μm) showed Bmal1 and Clock expression in the Ocn+ osteoblast lineage in 2-month-old and 20-month-old mice (white arrows). (I-J) Immunofluorescence staining (scale bar, 100 μm) showed BMAL1 and CLOCK expression in the OCN+ osteoblast lineage in young patients and patients with senile osteoporosis (white arrows). All data are presented as mean ± SD; n = 3; ∗p < 0.05.
To further establish circadian rhythm disruption, we tested the expression of the circadian rhythm core components Bmal1 and Clock in BM-MSCs from 2-month-old and 20-month-old mice and from young and aged men. Interestingly, western blot results showed that compared with that in the BM-MSCs of young mice or men, the expression of Bmal1 in BM-MSCs of aged mice or men, respectively, was decreased, while the expression of Clock was not (Figures 1C and 1D). Next, to examine whether the expression of BMAL1 and CLOCK in osteoblasts was impaired during senile osteoporosis, we collected bone specimens from 2-month-old and 20-month-old mice, patients with senile osteoporosis and young patients with traffic injuries and analyzed bone sections (Figures 1E and 1F). According to the results of immunofluorescence staining, in the Ocn+ osteoblast lineage, Bmal1 expression was weaker in 20-month-old mice than in 2-month-old mice, whereas Clock expression showed no significant difference between the age groups (Figures 1G and 1H). The results demonstrated that the expression of BMAL1 was also lower in the OCN+ osteoblast lineage in the aged patients than in the young patients, while CLOCK was not altered (Figures 1I and 1J). Moreover, we further explored the expression of BMAL1 and CLOCK in bone samples of ovariectomized (OVX) mice, sham-operated mice, patients with postmenopausal osteoporosis, and age-matched controls, and the results demonstrated that the expression of BMAL1 and CLOCK did not show a significant difference (Figures S1A–S1D) in the OCN+ osteoblast lineage between the OVX mice and the sham-operated mice or between the patients with postmenopausal osteoporosis and the age-matched controls. Taken together, these results indicated that the circadian rhythm and BMAL1 expression were abnormal in the osteoblast lineage in senile osteoporosis.
BMAL1 positively regulated the osteogenic differentiation of MSCs
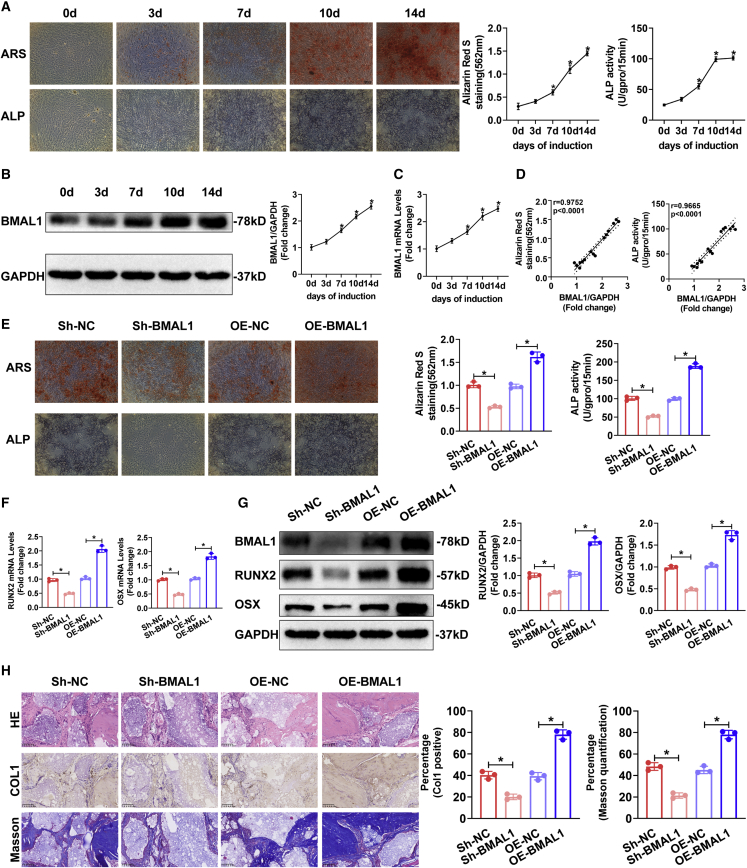
To determine the role of BMAL1 in MSC osteogenic differentiation, we first cultured human BM-MSCs in osteogenic medium (OM) for up to 14 days, and alizarin red S (ARS) staining showed a gradual increase from days 0–14, while alkaline phosphatase (ALP) staining peaked at 10 days (Figure 2A). The results of western blotting and quantitative real-time PCR (qRT-PCR) demonstrated that BMAL1 expression was upregulated gradually during osteogenesis (Figures 2B and 2C). Furthermore, we analyzed the correlation of the BMAL1 expression levels with ARS and ALP staining and found a strong positive relationship in the osteogenic differentiation of MSCs (Figure 2D).
Figure 2.
BMAL1 positively regulated the osteogenic differentiation of human BM-MSCs
(A) ARS, ALP staining, and quantification at different time points during osteogenic differentiation of MSCs. (B and C) BMAL1 protein (B) and mRNA (C) expression in MSCs undergoing osteogenic differentiation as measured at different time points. GAPDH served as the internal control. Bar graphs showing the expression of BMAL1 relative to the GAPDH level. (D) Correlation of expression levels of BMAL1 with ARS quantification and ALP activity during the osteogenic differentiation of MSCs. (E) ARS and ALP staining of the MSCs infected with Sh-NC, Sh-BMAL1, OE-NC, or OE-BMAL1 lentiviruses on the 14th day of osteogenic differentiation. (F and G) Relative mRNA (F) and protein (G) expression of the osteogenesis-associated markers RUNX2 and OSX in the MSCs infected with Sh-NC, Sh-BMAL1, OE-NC, or OE-BMAL1 lentiviruses on the 10th day of osteogenic differentiation. (H) HE and Masson staining and Col I immunohistochemistry of transplanted HA/TCP embedded with the MSCs infected with Sh-NC, Sh-BMAL1, OE-NC, or OE-BMAL1 lentiviruses. All data are presented as mean ± SD; n = 3 (MSCs derived from 3 different donors); ∗p < 0.05.
To further confirm the influence of BMAL1 on MSC osteogenesis, we used lentiviruses to diminish and overexpress BMAL1 in MSCs (Figures S2A and S2B). The osteogenic capacity of MSCs in different groups was evaluated after osteogenic induction of MSCs for 14 days. The results of the ARS staining and ALP assays demonstrated that knocking down BMAL1 expression impaired the osteogenic capacity, whereas the overexpression of BMAL1 promoted osteogenesis (Figure 2E). We also tested the expression of the osteogenesis-associated markers RUNX2, OSX, OPN, and OCN. The results of western blotting and qRT-PCR demonstrated that diminished BMAL1 decreased the expression of RUNX2, OSX, OPN, and OCN in MSCs during osteogenic induction, while overexpressing BMAL1 increased the expression (Figures 2F, 2G, S2A, and S2B). In summary, these results indicated that BMAL1 positively regulated the osteogenic capacity of MSCs in vitro.
MSCs were loaded onto hydroxyapatite/tricalcium phosphate (HA/TCP) scaffolds and implanted into the dorsal subcutaneous space of nude mice. As shown by hematoxylin and eosin (HE) and Masson staining, diminishing BMAL1 led to the reduced bone formation of MSCs on the scaffolds, while overexpression of BMAL1 significantly increased the formation of bone-like tissue. As shown by immunohistochemistry, the expression of collagen I (Col I), a marker of bone formation, was decreased after knocking down BMAL1 and increased by overexpressing BMAL1 (Figure 2H). In summary, BMAL1 positively regulated the osteogenic differentiation of MSCs both in vitro and in vivo.
The core circadian component BMAL1 regulated histone H2B monoubiquitination levels
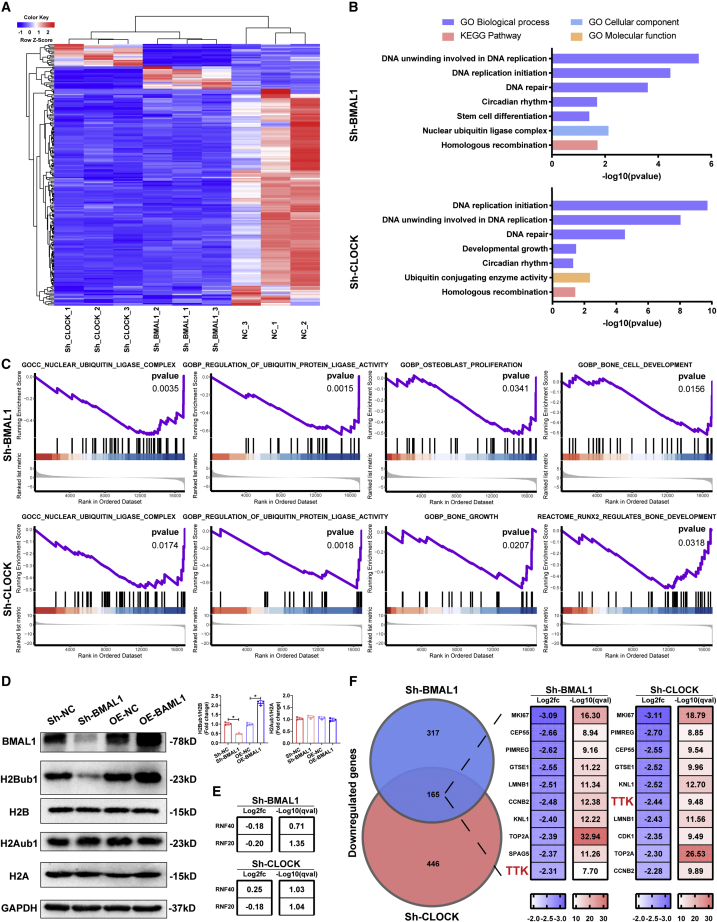
To determine how BMAL1 regulates the osteogenic capacity of MSCs as a part of the core circadian components, we performed RNA sequencing (RNA-seq) to study the impact of diminishing the core component BMAL1 or CLOCK in MSCs. A heatmap showed distinct expression patterns before and after knocking down each component (Figure 3 A). In Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses, the differentially expressed genes in either the Sh-BMAL1 or Sh-CLOCK group were enriched in terms regulated by histone modification, including DNA replication, DNA unwinding, DNA repair, and homologous recombination, and terms related to ubiquitination (Figure 3B). Furthermore, gene set enrichment analysis (GSEA) of the RNA-seq data showed obvious enrichment of genes related to histone ubiquitination-related and osteogenesis-related terms, including nuclear ubiquitin ligase complex, regulation of ubiquitin protein ligase, osteoblast proliferation, bone growth, and bone cell development. (Figure 3C). As reported previously, histone H2B monoubiquitination positively regulates the osteoblastic differentiation of MSCs.16,22 Thus, we investigated whether the core components BMAL1 and CLOCK affect H2Bub1 levels. As shown by western blot analysis, the level of H2Bub1 was decreased after BMAL1 or CLOCK expression was diminished and increased following overexpression of BMAL1 or CLOCK, while the level of H2Aub1 showed no significant difference between control expression and over- or underexpression of either component (Figures 3D and S3A). The results of RNA-seq showed that following knockdown of BMAL1 or CLOCK, the expression of RNF20 and RNF40, which act as classical factors monoubiquitinating histone H2B,15 was not significantly altered (Figure 3E). The top 10 intersecting genes with the most similar expression between the two groups are listed according to log2 fold change (FC) and −log10(q value) values, and TTK, which can regulate H2Bub1 levels according to the literature,23,24 is highlighted (Figure 3F) and shown in a volcano plot (Figure S4A). Together, these data suggested that the core circadian component BMAL1 regulated histone H2B monoubiquitination by managing H2Bub1 level-regulated gene TTK expression.
Figure 3.
The core circadian component BMAL1 regulated histone H2B monoubiquitination levels
(A) RNA-seq heatmap comparing the MSCs infected with Sh-NC, Sh-BMAL1, or Sh-CLOCK lentiviruses on the 7th day of osteogenic differentiation. (B) GO analysis of the RNA-seq data between the Sh-NC and Sh-BMAL1 groups and the Sh-NC and Sh-CLOCK groups. Bar graph showing the p values of the enriched terms. (C) GSEA of the RNA-seq data between the Sh-NC and Sh-BMAL1, Sh-NC, and Sh-CLOCK groups. (D) H2Bub1 and H2Aub1 levels in the MSCs infected with Sh-NC, Sh-BMAL1, or Sh-CLOCK lentiviruses on the 10th day of osteogenic differentiation. H2B and H2A served as the internal controls. Bar graphs showing the relative levels. Data are presented as mean ± SD; n = 3; ∗p < 0.05. (E) log2 FC and −log10(q value) of differential RNF20/40 expression between the Sh-NC and Sh-BMAL1 groups and the Sh-NC and Sh-CLOCK groups as obtained from the RNA-seq data. (F) Circos plot showing the terms with enriched genes and log2 FC and −log10(q value). TTK, the regulator of histone H2B monoubiquitination, is highlighted (G).
BMAL1 targeted the circadian-controlled gene TTK to phosphorylate MDM2 and regulate H2Bub1 levels to affect the osteogenic capacity of MSCs
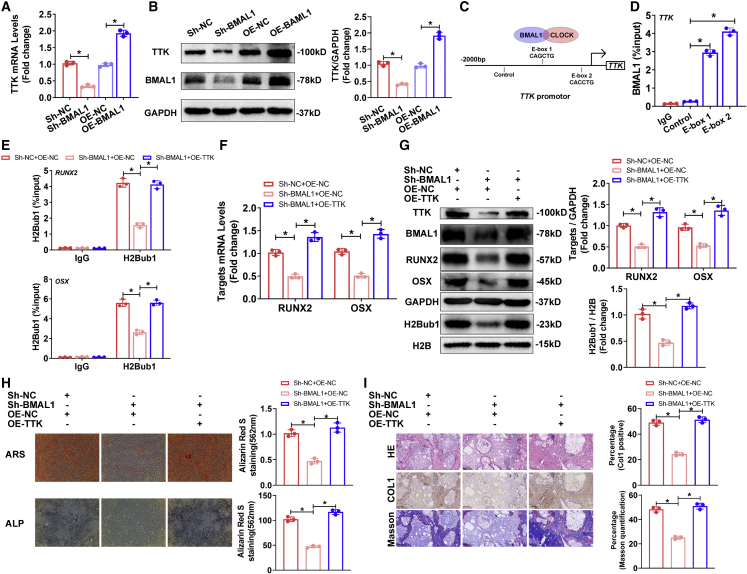
As mentioned above, the RNA-seq results suggested that diminishing BMAL1 impaired TTK expression, which affected H2Bub1 levels. We then evaluated whether and how BMAL1 regulated TTK expression, and the results of qRT-PCR and western blotting demonstrated that diminished BMAL1 decreased the expression of TTK in MSCs, while overexpressing BMAL1 increased the expression (Figures 4A and 4B). BMAL1 regulation of gene expression relies on heterodimers consisting of the circadian rhythm core components BMAL1 and CLOCK, which bind to the E-box elements (5′-CANNTG-3′) located within the promoters of circadian-controlled genes to activate their transcription.25 Through bioinformatic analysis, we identified candidate binding sites: two putative E-box motifs within the TTK promoter region (Figure 4C). Cleavage under targets and tagmentation (CUT&Tag) assays using primers targeting the E-box 1 or E-box 2 sites showed that BMAL1 bound to these regions in the TTK promoter (Figure 4D). The results above demonstrated that BMAL1 regulated the circadian-controlled gene TTK by BMAL1/CLOCK dimers.
Figure 4.
BMAL1 targeted the circadian-controlled gene TTK to regulate H2Bub1 levels to affect the osteogenic capacity of MSCs
(A and B) Relative mRNA (A) and protein (B) expression of TTK in the MSCs infected with Sh-NC, Sh-BMAL1, OE-NC, or OE-BMAL1 lentiviruses on the 10th day of osteogenic differentiation. (C) The putative E-boxes in the TTK promoter region. (D) CUT&Tag-qPCR showed the percentage of BMAL1 occupancy on the TTK promoter. Data are shown as the proportion of input level and normalized to the IgG control. (E) CUT&Tag-qPCR analysis showing the H2Bub1 occupancy on RUNX2 and OSX in the MSCs infected with Sh-NC and OE-NC, Sh-BMAL1 and OE-NC or Sh-BMAL1 and OE-TTK lentiviruses that were undergoing osteogenic differentiation. (F and G) Relative mRNA expression (F) and protein expression (G) of RUNX2 and OSX in MSCs infected with Sh-NC and OE-NC, Sh-BMAL1 and OE-NC, or Sh-BMAL1 and OE-TTK lentiviruses that were undergoing osteogenic differentiation. Bar graphs showing the relative expression. (H) ARS and ALP staining of the MSCs infected with Sh-NC and OE-NC, Sh-BMAL1 and OE-NC, or Sh-BMAL1 and OE-TTK lentiviruses on the 14th day of osteogenic differentiation. (I) HE and Masson staining and Col I immunohistochemistry of transplanted HA/TCP embedded with the MSCs infected with Sh-NC and OE-NC, Sh-BMAL1 and OE-NC, or Sh-BMAL1 and OE-TTK lentiviruses. All data are presented as mean ± SD; n = 3; ∗p < 0.05.
To further verify the effect of BMAL1 on MSC osteogenesis, which mediated H2Bub1 levels by regulating TTK expression, we used lentiviruses to knock down and overexpress TTK in MSCs. The results of qRT-PCR, western blotting, CUT&Tag-qPCR, ARS staining, and ALP assays demonstrated that TTK positively regulated osteogenic capacity by affecting H2Bub1 levels (Figures S5A–S5D). Furthermore, we diminished BMAL1 while overexpressing TTK in MSCs, and the results of CUT&Tag-qPCR showed that overexpressing TTK rescued the decreased recruitment of H2Bub1 to RUNX2 and OSX promoters by knocking down BMAL1 (Figure 4E). As expected, qRT-PCR, western blotting, ARS staining, ALP assays, HE staining, Masson staining, and immunohistochemistry demonstrated that diminishing BMAL1 impaired the osteogenic differentiation of MSCs, while overexpression of TTK significantly rescued the osteogenic capacity of MSCs (Figures 4F–4I).
According to the literature, TTK regulates H2Bub1 levels by phosphorylating MDM2,23,24 while MDM2 acts as a regulatory factor in differentiating osteoblasts.26 To confirm the role of MDM2 in MSC osteogenesis and the BMAL1-TTK-H2Bub1 axis, we knocked down MDM2 in MSCs. The results of western blotting, ARS staining, and ALP assays showed that diminished MDM2 expression impaired the osteogenic differentiation of MSCs (Figures S6A and S6B). We conducted co-immunoprecipitation (IP)/western blot assays to confirm that BAML1 can regulate TTK expression to modulate MDM2 phosphorylation and ultimately regulate H2Bub1 levels (Figure S6C). Collectively, these results indicated that BMAL1 acts as a critical regulator of MSC osteogenesis by regulating TTK expression to phosphorylate MDM2 and affect H2Bub1 levels.
H2Bub1 positively modulated BMAL1 expression at the transcript level
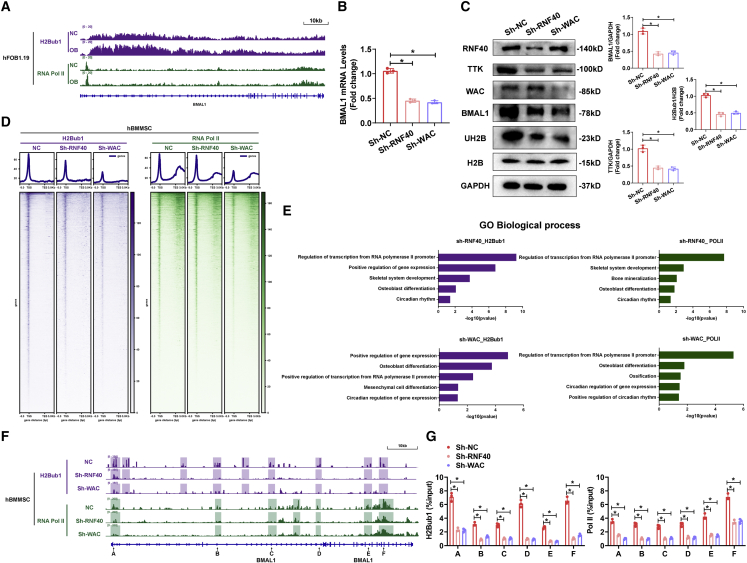
The role of H2Bub1 in regulating part of the clock component has been proposed,17 but the influence of H2Bub1 levels on BMAL1 expression has never been reported. To answer this question, we performed integrative analyses to compare the Pol II and H2Bub1 chromatin immunoprecipitation sequencing (ChIP-seq) data of human fetal osteoblasts (hFOB) 1.19 cells on days 0 and 10 of osteogenic differentiation. The results revealed that Pol II and H2Bub1 occupancy on BMAL1 was increased after osteogenic differentiation (Figure 5A). To confirm whether H2Bub1 level regulates the expression of BMAL1, we used lentiviruses to diminish the levels of RNF40 and WAC, which are parts of the obligate complex and monoubiquitinated histone H2B,27 in MSCs (Figures S7A and S7B). The results of qRT-PCR and western blotting demonstrated that diminished levels of RNF40 or WAC decreased BMAL1 expression and H2Bub1 levels (Figures 5B and 5C). Furthermore, to explore the impact of H2Bub1 on BMAL1, Pol II, and H2Bub1, we used CUT&Tag-seq data from MSCs with decreased RNF40 or WAC. Heatmaps showing the general CUT&Tag-seq data signals revealed that loss of RNF40 or WAC resulted in a significant reduction in Pol II and H2Bub1 occupancy in the gene body (Figure 5D). GO biological process analysis revealed that the differentially expressed H2Bub1- or Pol II-occupied genes in both the Sh-RNF40 and Sh-WAC groups were enriched in terms related to RNA transcription, osteoblast differentiation, and circadian rhythm (Figure 5E). As expected, the occupancy of H2Bub1 and Pol II on BMAL1, which was examined by CUT&Tag-seq and CUT&Tag-qPCR, was decreased in both the Sh-RNF40 and Sh-WAC groups, especially in colored region and at sites A–F (Figures 5F and 5G). Taken together, these observations suggest that the expression of BMAL1 was positively modulated by H2Bub1 at the transcript level.
Figure 5.
H2Bub1 positively modulated the expression of BMAL1 at the transcript level
(A) Signal traces of ChIP-seq data showing H2Bub1 and Pol II occupancy on BMAL1 in hFOB1.19 cells on day 0 or 7 of osteogenic differentiation. (B and C) Relative mRNA expression (B) and protein expression (C) of BMAL1 in the MSCs infected with Sh-NC, Sh-RNF40, or Sh-WAC lentiviruses. Bar graphs showing the relative expression. Data are presented as mean ± SD; n = 3; ∗p < 0.05. (D) CUT&Tag-seq average binding profiles and heatmaps depicting occupancy of H2Bub1 and Pol II in the MSCs infected with Sh-NC, Sh-RNF40, or Sh-WAC lentiviruses. (E) GO biological process analyses of the CUT&Tag-seq data comparing between the Sh-NC and Sh-RNF40 groups and the Sh-NC and Sh-WAC groups. Bar graph showing the p values of the enriched terms. (F) Signal traces of CUT&Tag-seq data showing H2Bub1 and Pol II occupancy on BMAL1 in the MSCs infected with Sh-NC, Sh-RNF40, or Sh-WAC lentiviruses. The colorful shadows showing regions with difference (G) CUT&Tag-qPCR analysis showing the H2Bub1 and Pol II occupancy on BMAL1 sites A–F in the MSCs infected with Sh-NC, Sh-RNF40 or Sh-WAC lentiviruses. Data are presented as mean ± SD; n = 3; ∗p < 0.05.
Bone-targeted BMAL1 or TTK rescue-treated senile osteoporosis
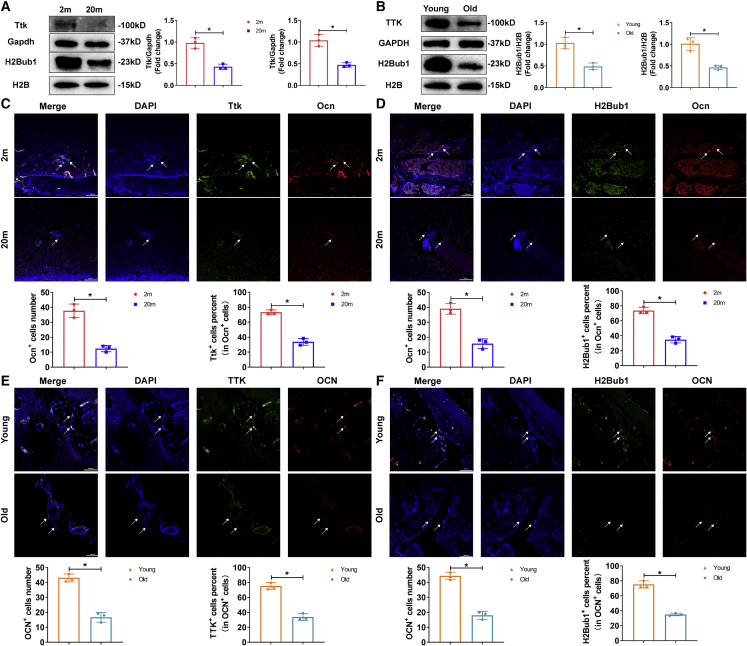
To establish the role of TTK and H2Bub1 in the impaired age-related osteogenic capacity, we examined TTK expression and H2Bub1 levels in MSCs during senile osteoporosis using western blotting and immunofluorescence staining. As expected, in BM-MSCs, the expression of Ttk and the level of H2Bub1 were lower in 20-month-old mice than in 2-month-old mice and lower in aged patients than in young patients (Figures 6A–6F). To exclude the possible effect of RNF20/40 on H2Bub1 level, we also examined RNF20/40 expression by western blotting and immunofluorescence staining. The results showed that RNF20/40 expression did not significantly differ between younger stages and senile osteoporosis (Figures S8A–S8F).
Figure 6.
TTK expression and H2Bub1 levels were decreased in BM-MSCs in senile osteoporosis
(A and B) Western blot analysis of the levels of TTK and H2Bub1 in BM-MSCs from 2-month-old and 20-month-old mice, patients with traffic injuries and patients with senile osteoporosis. (C and D) Immunofluorescence staining (scale bar, 100 μm) showed Ttk expression and H2Bub1 levels in the Ocn+ osteoblast lineage in 2-month-old and 20-month-old mice (white arrows). (E and F) Immunofluorescence staining (scale bar, 100 μm) showed TTK expression and H2Bub1 levels in the OCN+ osteoblast lineage in young patients with traffic injuries and patients with senile osteoporosis (white arrows). All data are presented as mean ± SD; n = 3; ∗p < 0.05.
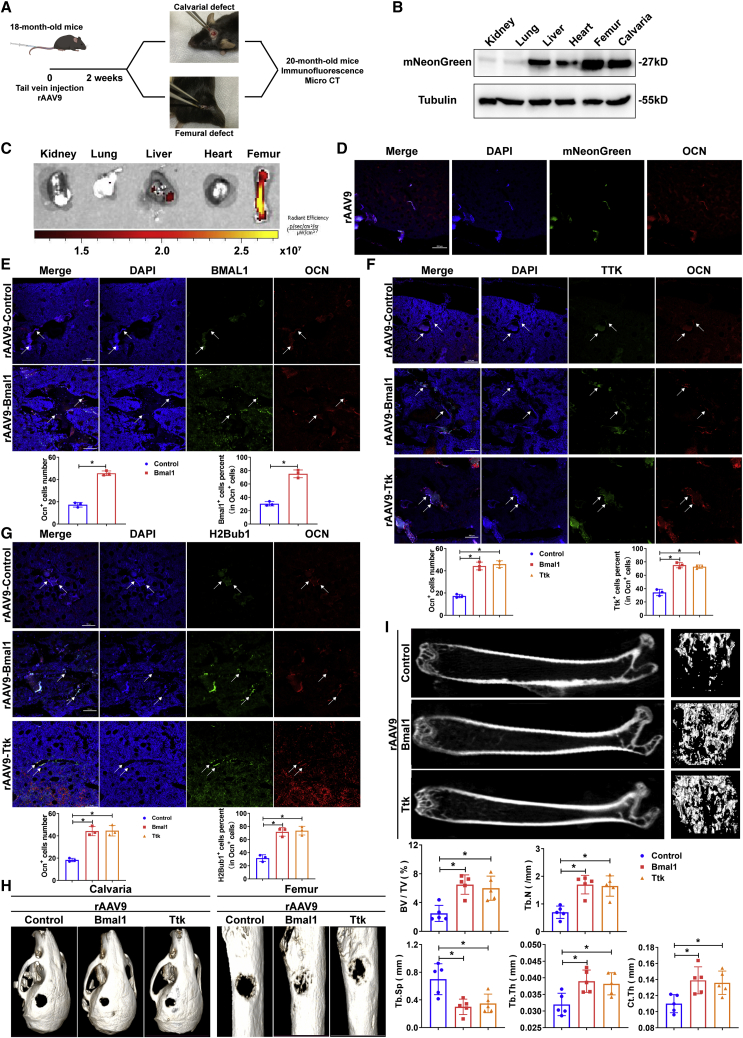
To evaluate whether BMAL1 and TTK can be used as therapeutic targets for osteoporosis, we constructed bone-targeting recombinant adeno-associated virus 9 (rAAV9) as previously described21 to overexpress Bmal1 or Ttk in vivo. To verify the therapeutic effect of Bmal1 and Ttk, we intravenously injected rAAV9-control/Bmal1/Ttk into senile osteoporotic (18-month-old) mice, which were subjected to procedures creating defects on the calvaria or femur and were sacrificed for immunofluorescence staining and microcomputed tomography (micro-CT) analysis (Figure 7A). The tissue distribution of rAAV9 was assessed by mNeonGreen expression, which appeared in calvarias and femurs, showing that rAAV9 specifically infected these skeletal systems (Figure 7B). Additionally, fluorescence imaging of different organs verified the effective bone-targeting delivery of rAAV9 (Figure 7C). As shown by fluorescence microscopy, mNeonGreen was expressed primarily in osteoblasts in cortical and trabecular bones (Figure 7D). These results demonstrated that the rAAV9 vector targets osteoblast lineage cells residing in the endosteal bone. As shown by the results of immunofluorescence staining and western blotting, the expression of Bmal1 and Ttk and the levels of H2Bub1 in the rAAV9-Bmal1 group as well as Ttk expression and H2Bub1 levels in the rAAV9-Ttk group were increased in bone sections (Figures 7E–7G, S9A and S9B). Compared with those in the rAAV9-control group, the mice in either the rAAV9-Bmal1 or rAAV9-Ttk group showed accelerated calvarial- and femoral-defect healing as well as increased bone density and improvements in other femur parameters, including BV/TV, Tb.Th, Tb.N, Tb.Sp, and Ct.Th, after infection with rAAV9 (Figures 7H and 7I). Thus, rAAV9-Bmal1 and rAAV9-Ttk may be potential drugs for the treatment of senile osteoporosis as potent bone anabolic agents.
Figure 7.
Bone-targeted Bmal1 or Ttk rescue-treated senile osteoporosis
(A) Diagram showing the workflow of rAAV9 injection in 18-month-old mice with calvarial and femoral defects and bone section analysis. (B) Immunoblot analysis showing mNeonGreen expression in different organs of the mice injected with rAAV9. (C) Fluorescence images of different organs of mice injected with rAAV9. (D) Immunofluorescence staining (scale bar, 100 μm) showing mNeonGreen-expressing osteoblasts in the femurs of the mice injected with rAAV9. (E) Immunofluorescence staining (scale bar, 100 μm) showing Bmal1 expression in the Ocn+ osteoblast lineage in the mice injected with rAAV9-control or rAAV9-Bmal1 (white arrows). (F) Immunofluorescence staining (scale bar, 100 μm) showing Ttk expression in the Ocn+ osteoblast lineage in the mice injected with rAAV9-control, rAAV9-Bmal1, or rAAV9-Ttk (white arrows). (G) Immunofluorescence staining (scale bar, 100 μm) showing H2Bub1 levels in the Ocn+ osteoblast lineage in the mice injected with rAAV9-control, rAAV9-Bmal1, or rAAV9-Ttk (white arrows). Data are presented as mean ± SD; n = 3; ∗p < 0.05. (H) Micro-CT analysis comparing the healing rates of calvarial and femoral defects in the mice injected with rAAV9-control, rAAV9-Bmal1, or rAAV9-Ttk. (I) Representative micro-CT images showing the trabecular bone of mice with senile osteoporosis injected with rAAV9-control, rAAV9-Bmal1, or rAAV9-Ttk. Bone morphometric analysis, including the analysis of BV/TV, Tb.Th, Tb.N, Tb.Sp, and Ct.Th., was performed. Data are presented as mean ± SD; n = 5; ∗p < 0.05.
Discussion
Senile osteoporosis is the most common age-related bone metabolic disorder worldwide. According to the recent report of the National Osteoporosis Foundation (NOF), every second woman and fourth man worldwide older than 50 years will encounter bone fracture due to senile osteoporosis in their remaining lives.28 Senile osteoporosis and related fractures not only enhance the morbidity and mortality of elderly individuals but also dramatically increase the financial burden on public health. Thus, further study to explore the mechanism of the aging process in the skeletal system and conduct corresponding clinical translation research will not only decrease the morbidity and mortality from senile osteoporosis-related fracture but will also reduce the economic burden on public health.
In contrast to postmenopausal osteoporosis, bone formation impairment instead of osteoclastic hyperfunction is the major cause of senile osteoporosis. However, there are far fewer approaches to effectively promote BM-MSC-mediated bone formation than inhibit osteoclastic activity in the clinic for treating osteoporosis.29,30 Only two anabolic agents, parathyroid hormone and parathyroid hormone-related protein, can promote osteoblast function to treat patients with osteoporosis. However, these agents are costly, require frequent injections and can promote the development of osteosarcomas. Recently developed agents, including an anti-clerostin antibody and a small-molecule inhibitor of cathepsin K, can increase bone mass and reduce fracture risk in osteoporosis. However, these drugs showed severe adverse cardiovascular events in clinical trials.29 Thus, a therapeutic strategy that can effectively promote BM-MSC-mediated bone formation without adverse effects is still an unmet need.
BMAL1/CLOCK heterodimers drive daily changes in the transcription of circadian-controlled genes to control physiological system development and homeostasis.31 However, multiple studies have shown that during the aging process, the expression of BMAL1/CLOCK heterodimers is reduced, which impairs the physiological function of various tissues and organs32,33,34,35; for example, BMAL1 deficiency has been found to contribute to the decline in visual function during aging, and circadian rhythm disruption has been shown to play a role in osteoarthritis-like changes in chondrocyte activity.36 Consistent with these studies, our research demonstrated that BMAL1 but not CLOCK was specifically decreased in osteoblast lineage cells of senile osteoporotic specimens from both human and mouse but not postmenopausal osteoporotic specimens. Thus, we conclude that similar to that in multiple other organs, BMAL1/CLOCK heterodimer dysfunction is indispensable for the aging-related osteogenic impairment in senile osteoporosis. Further study to reveal the mechanism of BMAL1 deficiency-mediated BM-MSC osteogenic impairment is necessary to elucidate the mechanism of senile osteoporosis.
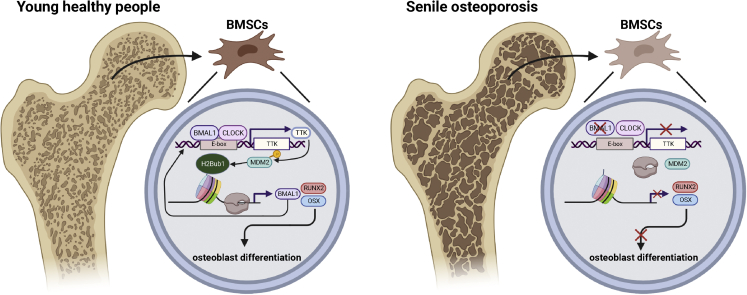
BMAL1/CLOCK heterodimers conservatively recognize the E-box element (CANNTG) in the promoter to initiate circadian-controlled gene transcription in mammals.25 The transcription of circadian-controlled genes by BMAL1/CLOCK heterodimers also requires multiple types of histone modifications. During aging, abnormal histone modification impairs BMAL1/CLOCK heterodimer function in circadian-controlled gene transcription, thereby leading to morbidity.32 However, as multiple histone modification enzymes or modulators are circadian-controlled genes,37,38 BMAL1 deficiency may also lead to dysfunctional histone modification during the aging process. H2Bub1 in mammals is a type of histone modification that is essential for the early steps in transcriptional initiation and elongation. A stable level of H2Bub1 not only facilitates the transcriptional function of BMAL1/CLOCK heterodimers but also is indispensable for the osteogenic capacity of BM-MSCs. H2Bub1 deficiency leads to gene transcription dysfunction and contributes to multiple diseases.39,40 However, whether H2Bub1 dysfunction plays a role in aging status, including senile osteoporosis, has never been explored. In this study, we first discovered that BMAL1 can positively regulate H2Bub1 level by targeting the H2Bub1 modulator TTK as a circadian-controlled gene. Knocking down BMAL1 significantly decreased the expression of TTK and further reduced the level of H2Bub1 in BM-MSCs. Overexpression of TTK rescued the H2Bub1 level as well as the osteogenic capacity of BM-MSCs under BMAL1 deficiency. Accordingly, TTK-H2Bub1 deficiency was also observed in osteoblast lineage cells in senile osteoporotic specimens from both human and mouse. Interestingly, we also confirmed that H2Bub1 can positively regulate the transcription of BMAL1. Inhibiting H2Bub1 significantly decreased the expression of BMAL1. The results described above lead to the intriguing possibility that BMAL1-TTK-H2Bub1 may form a positive cycle to maintain the osteogenic capacity of BM-MSCs. However, during the aging process, BMAL1 deficiency decreases H2Bub1, which in turn further aggravates BMAL1 deficiency. BMAL1-TTK-H2Bub1 cycle impairment significantly reduces the osteogenic capacity of BM-MSCs during the aging process and ultimately leads to senile osteoporosis (Figure 8).
Figure 8.
Model showing that the disruption of the BMAL1-TTK-MDM2-H2Bub1 positive loop led to the impaired osteogenic capacity of BM-MSCs in senile osteoporosis
AAV vectors have a long track record of both safety due to their lack of pathogenicity and efficacy in treating multiple diseases in the clinic.41 Moreover, unlike nonviral vectors, such as nanoparticles and liposomes, which can be rapidly degraded and cleared in circulation, AAVs are capable of persisting in postmitotic cells of host tissues for several years.42 As senile osteoporosis is a chronic disease throughout aging life, AAV is a relatively suitable delivery vehicle for treating senile osteoporosis. Previous studies have confirmed that the AAV9 subtype could target osteoblast lineage cells in the bone to drive RNA delivery in vivo. However, there are still other organs, including cardiac muscle, skeletal muscle, lung alveoli, and liver, with a high affinity for rAAV9. Thus, delivery of DNA via rAAV9 to treat senile osteoporosis will still have multiple off-target effects. Yang et al.21 previously demonstrated that grafting the bone-targeting peptide motif (Asp-Ser-Ser)6 onto the AAV9-VP2 capsid protein could improve the affinity of AAV9 to osteoblast lineage cells while substantially reducing tropism to cardiac muscle, skeletal muscle, lung alveoli, and liver. We also used this rational strategy to modify AAV9 to systemically deliver Bmal1 or Ttk to treat senile osteoporosis in aging mice. We obtained satisfactory results in which only a single systemic injection of this modified AAV9 carrying Bmal1 or Ttk plasmids could sufficiently improve the H2Bub1 level in osteoblast lineage cells in aging mice and further significantly promote bone mass. These results indicated that targeting the BMAL-TTK-H2Bub1 axis via bone-targeting AAV9 might have potential in treating senile osteoporosis in the clinic.
To our knowledge, our study is the first to demonstrate that BMAL1 deficiency contributes to circadian rhythm disruption and impaired BM-MSC-mediated bone formation in senile osteoporosis. BMAL1 positively regulated the osteogenic capacity of BM-MSCs by maintaining H2Bub1 levels by targeting the H2Bub1 modulator TTK as a circadian-controlled gene. Furthermore, we are the first to discover that H2Bub1 in turn regulates BMAL1 expression at the transcriptional level. Thus, BMAL1-TTK-H2Bub1 forms a positive cycle to maintain the osteogenic capacity of BM-MSCs. However, BMAL1 deficiency deteriorated this cycle, which significantly impaired the osteogenic capacity of BM-MSCs during the aging process and finally led to senile osteoporosis. Finally, we demonstrated that only a single injection to supplement BMAL1 or TTK in osteoblast lineage cells via bone-targeting rAAV9 could significantly promote bone mass and bone repair speed in senile mice. However, our study has some limitations. First, we did not clarify the mechanism of BMAL1 deficiency in osteoblast lineage cells in senile osteoporosis. Moreover, current bone-targeting rAAV9 still has some affinity for other organs. To translate our research into the clinic, we will further explore the mechanism of BMAL1 deficiency in senile osteoporosis and further modify this bone-targeting rAAV9 delivery system to improve its bone affinity as well as reduce its off-target effects in the future.
Materials and methods
Animal models
Wild-type C57BL/6 mice at 2, 18, and 20 months of age and BALB/c-nu/nu mice at 2 months of age were purchased from the Laboratory Animal Center of Sun Yat-sen University and housed in cages under standard animal housing conditions with 12 h light and 12 h dark cycles. Mice in poor health were excluded from the study. All mouse experiments were approved by the animal ethics committee of Sun Yat-sen University (SYSU-IACUC-2021-000811).
In vivo osteogenic induction
For in vivo osteogenic induction, MSCs in the fourth passage infected with lentiviruses (Sh-NC, Sh-BMAL1, Sh-TTK, OE-NC, OE-BMAL1, and OE-TTK) were cultured in OM before use in the in vivo study. After 7 days of osteogenic induction, MSCs (5 × 105) were trypsinized and seeded onto HA/TCP (Zimmer) for 24 h. HA/TCP loaded with MSCs was implanted into the subcutaneous dorsal space of 8-week-old BALB/c nu/nu mice. Specimens were harvested 8 weeks after implantation, and the animals were euthanized via cervical dislocation. The grafts were retrieved for subsequent HE and Masson trichrome staining and histochemical analysis.
Calvarial and femoral bone defects in mice
Calvarial bone defects were created with an electric bone drill applied to the parietal bones of 18-month-old mice. Briefly, after anesthetization, disinfection, and skin incision, the subcutaneous tissue was carefully peeled away to expose the parietal bones. A 2.5 mm calvarial defect was created with an electric bone drill 2 weeks after ejection, and the incision was sutured with a thin thread. A 1.0 mm femoral bone defect was created following the same method used to create calvarial defects 8 weeks after ejection. Ten weeks after ejection, the whole skulls and femurs were collected for micro-CT and subsequent analysis.
OVX mice
For the generation of OVX mice, 2-month-old female wild-type C57BL/6 mice underwent bilateral ovariectomy or sham operation. Mice were allowed 2 months to recover from the ovariectomy surgery and then were sacrificed and used in various assays.
Bone-targeting Bmal1/Ttk overexpression in vivo
Bone-targeting rAAV9 overexpressing Bmal1/Ttk was designed and constructed as previously described.21 In summary, the DNA sequence encoding the bone-specific peptide motif DSS (Asp-Ser-Ser)6 was codon-optimized and inserted into the AAV9 capsid protein VP2 to construct bone-targeting rAAV9 vectors that overexpressed Bmal1/Ttk in vivo.
PET imaging
PET imaging of mice was designed as previously described. In summary, PET imaging was performed using an Inveon MM system (Siemens). A total of 120 μCi sodium fluoride labeled with fluorine-18 was intraperitoneally injected into the animals 40 min before imaging for all four different time points. Quantification of bone metabolism was obtained using the maximum standardized uptake value (SUVmax) and the mean standardized uptake value (SUVmean), which were normalized to body weight. SUV = CT/DInj/Ws. CT is the maximum/mean 18F-NaF radioactivity per unit volume in the femur head, DInj is the injection dose, and WS is the mouse weight.
Isolation and culture of MSCs
MSC isolation
Bone marrow was extracted from the posterior superior iliac spine of healthy volunteers under sterile conditions. Then, MSCs were isolated and purified by density gradient centrifugation as described in our previously reported methods.43 Briefly, MSCs were isolated and purified from bone marrow using density gradient centrifugation at 600 × g for 30 min. The isolated MSCs were resuspended in a medium composed of 90% Dulbecco’s modified Eagle’s medium (DMEM; Gibco) and 10% fetal bovine serum (FBS; Hangzhou Sijiqing Biological Engineering Material Company) seeded in flasks and cultured in incubators at 37°C in a 5% CO2 atmosphere, and the culture medium was replaced every third day. When the cells reached 80%–90% confluence, the MSCs were digested using 0.25% trypsin containing 0.53 mM EDTA and reseeded in new flasks or plates, and cells in the second through fifth passages were used for all experiments.
Osteogenic induction
MSCs (0.5 × 105 cells/well) were seeded in 12-well plates and cultured in DMEM (with 1,000 mg/L glucose) with 10% FBS. After 24 h, the culture medium was replaced with OM, consisting of DMEM (1,000 mg/L glucose) with 10% FBS, 100 IU/mL penicillin, 100 IU/mL streptomycin, 0.1 μM dexamethasone, 10 mM β-glycerol phosphate, and 50 μM ascorbic acid (Sigma-Aldrich). The OM was changed every 3 days.
Lentivirus construction and infection
Protocols were reported previously.44 Lentiviruses encoding short hairpin RNA for BMAL1, TTK, RNF40, WAC, and MDM2 and negative control lentivirus were generated by IGE (Guangzhou, China). BMAL1 and TTK overexpression lentiviruses and their vector controls were also purchased from IGE.
Detailed information on the reagents is shown in Table S1.
ARS and ALP staining
Before staining, MSCs were first fixed with 4% paraformaldehyde (PFA) for 30 min.
ARS staining
For visualization of calcium deposition, MSCs were incubated with 1% ARS (pH 4.3) (catalog no. G8550; Solarbio) for 15 min at room temperature. The MSCs were rinsed three times with PBS to remove the nonspecific stain, and images were subsequently captured directly with a microscope. For ARS quantification, 10% cetylpyridinium chloride monohydrate (Sigma-Aldrich) was used to destain the cells for 1 h at room temperature. Then, 200 μL of the extraction mixture was transferred to a 96-well plate, and the spectrophotometric absorbance was measured at 562 nm.
ALP staining
MSCs were stained using a BCIP/NBT alkaline phosphatase kit (catalog no. C3206; Beyotime Institute of Biotechnology) following the manufacturer’s instructions. Stained cells were photographed. For the ALP activity assay, ALP activity kits (catalog no. A059-2; Nanjing Jiancheng Biotech, Nanjing, China) were used according to the manufacturer’s instructions. In summary, MSCs were lysed in ice-cold radioimmunoprecipitation assay (RIPA) buffer containing protease and phosphatase inhibitors (Beyotime Institute of Biotechnology). After centrifugation at 14,000 rpm and 4°C for 30 min, the supernatants were incubated with reaction buffer at 37°C for 15 min. After addition of the stop solution, the absorbance was measured at 405 nm. A Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific) was used to detect the total protein concentration. ALP activity was normalized to the total protein content at the end of the experiment and reported as units per gram of protein per 15 min (U/gpro/15 min).
Protein extraction and western blot analysis
The western blot protocols were reported previously. Briefly, the cells were harvested and lysed in RIPA buffer containing protease and phosphatase inhibitors for 30 min on ice. Lysates were obtained via centrifugation at 14,000 rpm at 4°C for 10 min. Equal amounts of each sample diluted in 5× sodium dodecyl sulfate (SDS) loading buffer were subjected to SDS-PAGE and were subsequently transferred to polyvinylidene fluoride membranes (Millipore). The membranes were blocked in 5% nonfat dry milk dissolved in TBST (150 mM NaCl, 50 mM Tris-HCl [pH 7.5], and 0.05% Tween 20) at room temperature for 1 h and then incubated overnight with primary antibodies at 4°C. After incubation with horseradish peroxidase (HRP)-conjugated anti-mouse antibody (catalog no. 7076; Cell Signaling Technology) or HRP-conjugated anti-rabbit antibody (catalog no. 7074; Cell Signaling Technology) for 1 h at room temperature, the protein levels were detected using chemiluminescent reagents (catalog no. WBKLS0500; Millipore) according to the instructions provided by the manufacturer. The following primary antibodies were used: anti-BMAL1 antibody (catalog no. 14020; Cell Signaling Technology), anti-TTK antibody (catalog no. ab11108; Abcam), anti-CLOCK antibody (catalog no. ab3517; Abcam), anti-OCN antibody (catalog no. 29560; Sab), anti-GAPDH (catalog no. 5174S; Cell Signaling Technology), anti-RUNX2 (catalog no. 12556S; Cell Signaling Technology), anti-OSX (catalog no. ab209484; Abcam), anti-β-tubulin (catalog no. 2128; Cell Signaling Technology), anti-OPN antibody (catalog no. 42036; Sab), anti-RNF20 antibody (catalog no. ab181104; Abcam), anti-RNF40 antibody (catalog no. ab191309; Abcam), anti-WAC antibody (catalog no. ab109486; Abcam), anti-H2B antibody (catalog no. 12364; Cell Signaling Technology), anti-H2Bub1 antibody (catalog no. 5546; Cell Signaling Technology), anti-H2A antibody (catalog no. 12349; Cell Signaling Technology) anti-H2Aub1 antibody (catalog no. 8240; Cell Signaling Technology), anti-MDM2 antibody (catalog no. ab226939; Abcam), and anti-pan phosphoserine/threonine antibody (catalog no. AP1067; Abclonal).
RNA isolation and qRT-PCR analysis
Total RNA was extracted from MSCs using RNAiso Plus (catalog no. 9109; TaKaRa), and the isolated RNA was reverse-transcribed into cDNA using a PrimeScript RT reagent kit (catalog no. RR036A; TaKaRa) according to the manufacturer’s recommendations. qRT-PCR was then performed on a LightCycler R480 PCR system (Roche) using SYBR Premix Ex Taq (catalog no. RR420A; TaKaRa). The results were analyzed using the 2-ΔΔCt method, and the reference gene GAPDH was used to normalize the amplification data obtained for the target genes. Each qRT-PCR analysis was performed in triplicate. The primer sequences are listed in Table S1.
Histology
The HA/TCP grafts loaded with MSCs and bone tissues were collected, fixed in 4% PFA, and then decalcified for 10 days in 10% EDTA (pH 7.4). After decalcification, the specimens were dehydrated and subsequently embedded in paraffin. Sections (5 mm thickness) were stained with HE stain (catalog no. AR1180; Boster) and Masson stain (catalog no. G1340-100; Solarbio) according to the manufacturers’ instructions. The slides were subjected to immunohistochemistry with anti-collagen I (catalog no. ab34710; Abcam).
Spine section biopsies
We recruited three male patients with compression fractures of the vertebra who had been diagnosed with senile osteoporosis and three male patients with fractures of the spine who had experienced traffic accidents as a control group. All of the patients needed treatment for percutaneous vertebroplasty or subtotal vertebrectomy. The characteristics of the study subjects are presented in Table S2. The other bone sections biopsied were obtained as described.44 The specimen was obtained during the surgery.
Immunofluorescence
For antigen retrieval, bone tissue sections were deparaffinized and rehydrated, immersed in 10 mM citrate buffer (pH 6.0), and microwaved for 15 min. Bone sections were permeabilized with 0.5% Triton X-100 for 20 min and blocked with 10% fetal bovine serum in PBS for 1 h. An anti-BMAL1 antibody (catalog no. ab230822; Abcam), an anti-TTK antibody (catalog no. ab11108; Abcam), an anti-CLOCK antibody (catalog no. ab3517; Abcam), an anti-OCN antibody (catalog no. 29560; Sab), an anti-OCN antibody (catalog no. MAB1419; Novus), or an anti-H2Bub1 antibody (catalog no. 5546; Cell Signaling Technology) was incubated with the samples overnight at 4°C. The secondary antibodies used were anti-rabbit Alexa 488 (catalog no. 2975; Cell Signaling Technology), anti-mouse Alexa 488 (catalog no. 4408; Cell Signaling Technology), anti-rabbit Alexa 555 (catalog no. 4413; Cell Signaling Technology), and anti-mouse Alexa 555 (catalog no. 4409; Cell Signaling Technology). Antifade mounting medium with DAPI (catalog no. P0131; Beyotime) was used for mounting. The samples were viewed under a laser scanning confocal microscope at wavelengths of 488 nm (green, BMAL1, TTK, CLOCK, H2Bub1), 555 nm (red, OCN), and 405 nm (blue, DAPI). Images were captured using a Nikon Eclipse Ni-E confocal microscope.
RNA-seq library preparation and sequencing
Total RNA was extracted using RNAiso Plus (catalog no. 9109; TaKaRa) according to the manufacturer’s instructions. Oligo(dT)-attached magnetic beads were used to purify mRNA. Purified mRNA was fragmented into small pieces with fragment buffer at the appropriate temperature. Then, first-strand cDNA was generated using random hexamer-primed reverse transcription, and second-strand cDNA was synthesized. Next, an A-Tailing Mix and RNA Index Adaptors were added and incubated for end repair. The cDNA fragments obtained in the previous step were amplified by PCR, and the products were purified with AMPure XP Beads and then dissolved in EB solution. The product was validated with an Agilent Technologies 2100 bioanalyzer for quality control. The double-stranded PCR products obtained in the previous step were heated, denatured, and circularized with a splint oligo sequence to generate the final library. Single-strand circle DNA (ssCirDNA) was formatted in the final library. The final library was amplified with phi29 to generate DNA nanoballs (DNBs) with more than 300 copies of a molecule. The DNBs were loaded into a patterned nanoarray, and single-end 50 base reads were generated with a BGISeq500 platform (BGI-Shenzhen).
RNA-seq data analysis
The sequencing data were filtered with SOAPnuke (version 1.5.2)45 by removing reads containing sequencing adaptors, a low-quality base rate (base quality less than or equal to 5) greater than 20%, or an unknown base (“N” base) rate greater than 5%. Then, clean reads were obtained and stored in FASTQ format. The clean reads were mapped to the hg38 reference genome for coverage calculation and quality control using HISAT2 (version 2.0.4).46 Bowtie2 (version 2.5.1)47 was used to align the clean reads to the reference coding gene set, and the gene expression levels were calculated with RSEM software (version 1.2.12).48 A heatmap was drawn using pheatmap (version 1.0.8) (https://CRAN.R-project.org/package=pheatmap) according to the gene expression in different samples. Differential expression analysis was performed using DESeq2 (version 1.4.5)49 with a cutoff Q value of ≤0.05.
KEGG (https://www.kegg.jp/) and GO (http://www.geneontology.org/) enrichment analyses of annotated differentially expressed genes were performed using the R package clusterProfiler (version 3.11). The significance levels of the terms and pathways were corrected according to the Q value on the basis of a rigorous threshold (Q value ≤ 0.05) through the Bonferroni method. GSEA was carried out using OmicStudio tools (https://www.omicstudio.cn/tool) according to the instructions.
CUT&Tag assay, qPCR, and sequencing
CUT&Tag assay
The CUT&Tag assay was performed using the NovoNGS CUT&Tag 3.0 High-Sensitivity Kit (catalog no. N259-YH01; NovoProtein). MSCs (1.0 × 105) were washed twice with 0.5 mL of wash buffer, mixed with ConA beads and incubated with an anti-BMAL1 antibody (catalog no. 14020; Cell Signaling Technology), an anti-H2Bub1 antibody (catalog no. 5546; Cell Signaling Technology), or an anti-RNA polymerase II CTD antibody (catalog no. ab26721; Abcam) overnight at 4°C. Goat anti-rabbit IgG antibody (NovoProtein) was diluted at 1:500 and added to the sample; then, the sample was incubated for 1 h at room temperature (RT). The cells were washed and incubated with pAG-Tn5 for 1 h at RT. Then, MgCl2 was added to activate tagmentation for 1 h at 37°C. Next, DNA was isolated by NovoNGS DNA Extract Beads and dissolved in TE buffer. DNA was amplified with N5 and N7 primers, which can be enriched by PCR to form sequencing-ready libraries. After PCR, libraries were purified with NovoNGS DNA Clean Beads, and library quality was assessed using the Agilent Bioanalyzer 2100 system.
CUT&Tag-qPCR
Part of the purified DNA from the CUT&Tag assay was diluted 10 times and used for CUT&Tag-qPCR analysis. Primer sequences are listed in Table S1.
CUT&Tag sequencing
The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s instructions. The library preparations were sequenced on the Illumina NovaSeq platform at Novogene Science and Technology (Beijing, China), and 150 bp paired-end reads were generated.
CUT&Tag/ChIP-seq data analysis
CUT&Tag/Chip-seq data analysis was performed as previously described with slight modification.50 In summary, raw reads were filtered and trimmed to remove adaptor sequences and low-quality reads using TrimGalore (version 0.6.6) with the parameters -q 20 --phred33 --stringency 3. Clean reads were mapped to the human genome version hg38 using Bowtie2 (version 2.5.1)51 with the parameters -p 6 -q. Only the uniquely mapped reads were retained. All peak calling was performed with MACS2 using ‘macs2 -q 0.05 --call-summits --nomodel --shift −100 --extsize 200 --keep-dup all’. For simulations of peaks called per input read, aligned and deduplicated BAM files were used without any additional filtering. Normalization (in reads per kilobase per million [RPKM]) of the CUT&Tag-seq data was performed using the bamCoverage command, and heatmaps were plotted using the computeMatrix and plotHeatmap commands in deepTools (version 2.3.6.0).52 The normalized CUT&Tag-seq data were visualized using the Integrative Genomics Viewer (IGV).
The raw data obtained from hFOB1.19 H2Bub1 and RNAPII ChIP-seq data from the GSE82295 dataset were downloaded. We generated hBM-MSC H2Bub1 and RNAPII CUT&Tag-seq data, and datasets have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database under accession number GSE210426.
Micro-CT
For evaluation of the mass and microarchitecture of the bone between different groups, micro-CT was performed using an Inveon MM system (Siemens). Images were acquired at each of the 360 rotational steps with a pixel size of 8.82 lm, a voltage of 80 kV, a current of 500 lA, and an exposure time of 1,500 ms. The parameters BV/TV, Tb.N, Tb.Th, Tb.Sp, and Ct.Th in the trabecular region (1–2 mm distal to the proximal epiphysis) were calculated using an Inveon Research Workplace (Siemens) according to the guidelines set by the American Society for Bone and Mineral Research. In addition, two-dimensional and three-dimensional bone structure image slices were reconstructed.
Statistical analysis
The data in this study were analyzed using SPSS 26.0. The results are presented as the mean ± SD. Student’s t test and one-way analysis of variance followed by the Bonferroni test and the Pearson correlation test were performed for statistical analyses. p values < 0.05 were considered to indicate statistical significance.
Data availability
All data generated or analyzed during this study are included in this published article (and its supplemental information files). The data from RNA-seq and CUT&Tag-seq have been deposited in NCBI’s Gene Expression Omnibus database under GEO accession number GSE210426.
Acknowledgments
This study was supported by the Shenzhen Key Medical Discipline Construction Fund (ZDSYS20190902092851024), the National Natural Science Foundation of China (82172385, 82172349, and 82002267), and the Shenzhen Science and Technology Program (RCBS20200714114909007 and JCYJ20210324115007019).
Author contributions
S.H. designed the study. L.J.T., X.P., Y.W., Y.Z., Y.G., Y.F., X.X., S.Z., L.J.J., C.Y., Z.Z., Z.Y., and L.Z. performed experiments and analyzed the data. L.J.T., X.P., and Y.W. wrote the manuscript. F.P., C.Q., and L.D. provided technical assistance. W.Y. and X.Z. provided valuable comments.
Declaration of interests
The authors declare no conflict of interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2023.02.014.
Contributor Information
Xie Zhongyu, Email: xiezhy23@mail.sysu.edu.cn.
Wu Yanfeng, Email: wuyf@mail.sysu.edu.cn.
Shen Huiyong, Email: shenhuiy@mail.sysu.edu.cn.
Supplemental information
References
- 1.Compston J.E., McClung M.R., Leslie W.D. Osteoporosis. Lancet. 2019;393:364–376. doi: 10.1016/S0140-6736(18)32112-3. [DOI] [PubMed] [Google Scholar]
- 2.Von Bank H., Kirsh C., Simcox J. Aging adipose: depot location dictates age-associated expansion and dysfunction. Ageing Res. Rev. 2021;67:101259. doi: 10.1016/j.arr.2021.101259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kendler D.L., Marin F., Zerbini C.A.F., Russo L.A., Greenspan S.L., Zikan V., Bagur A., Malouf-Sierra J., Lakatos P., Fahrleitner-Pammer A., et al. Effects of teriparatide and risedronate on new fractures in post-menopausal women with severe osteoporosis (VERO): a multicentre, double-blind, double-dummy, randomised controlled trial. Lancet (London, England) 2018;391:230–240. doi: 10.1016/S0140-6736(17)32137-2. [DOI] [PubMed] [Google Scholar]
- 4.Lee J.H., Wei Y.J., Zhou Z.Y., Hou Y.M., Wang C.L., Wang L.B., Wu H.J., Zhang Y., Dai W.W. Efficacy of the herbal pair, Radix Achyranthis bidentatae and eucommiae cortex, in preventing glucocorticoid-induced osteoporosis in the zebrafish model. J. Integr. Med. 2022;20:83–90. doi: 10.1016/J.JOIM.2021.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Chen Q., Shou P., Zheng C., Jiang M., Cao G., Yang Q., Cao J., Xie N., Velletri T., Zhang X., et al. Fate decision of mesenchymal stem cells: adipocytes or osteoblasts? Cell Death Differ. 2016;23:1128–1139. doi: 10.1038/cdd.2015.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demontiero O., Vidal C., Duque G. Aging and bone loss: new insights for the clinician. Ther. Adv. Musculoskelet. Dis. 2012;4:61–76. doi: 10.1177/1759720X11430858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang T.L., Shen H., Liu A., Dong S.S., Zhang L., Deng F.Y., Zhao Q., Deng H.W. A road map for understanding molecular and genetic determinants of osteoporosis. Nat. Rev. Endocrinol. 2020;16:91–103. doi: 10.1038/s41574-019-0282-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allada R., Bass J. Circadian mechanisms in medicine. N. Engl. J. Med. 2021;384:550–561. doi: 10.1056/nejmra1802337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patke A., Young M.W., Axelrod S. Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol. 2020;21:67–84. doi: 10.1038/s41580-019-0179-2. [DOI] [PubMed] [Google Scholar]
- 10.Samsa W.E., Vasanji A., Midura R.J., Kondratov R.V. Deficiency of circadian clock protein BMAL1 in mice results in a low bone mass phenotype. Bone. 2016;84:194–203. doi: 10.1016/j.bone.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan G., Hua B., Yang Y., Xu L., Cai T., Sun N., Yan Z., Lu C., Qian R. The circadian gene clock regulates bone formation via PDIA3. J. Bone Miner. Res. 2017;32:861–871. doi: 10.1002/jbmr.3046. [DOI] [PubMed] [Google Scholar]
- 12.Berson A., Nativio R., Berger S.L., Bonini N.M. Epigenetic regulation in neurodegenerative diseases. Trends Neurosci. 2018;41:587–598. doi: 10.1016/j.tins.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang W., Song M., Qu J., Liu G.H. Epigenetic modifications in cardiovascular aging and diseases. Circ. Res. 2018;123:773–786. doi: 10.1161/CIRCRESAHA.118.312497. [DOI] [PubMed] [Google Scholar]
- 14.Li G., Ma L., He S., Luo R., Wang B., Zhang W., Song Y., Liao Z., Ke W., Xiang Q., et al. WTAP-mediated m 6 A modification of lncRNA NORAD promotes intervertebral disc degeneration. Nat. Commun. 2022;13:1469. doi: 10.1038/S41467-022-28990-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pavri R., Zhu B., Li G., Trojer P., Mandal S., Shilatifard A., Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 16.Karpiuk O., Najafova Z., Kramer F., Hennion M., Galonska C., König A., Snaidero N., Vogel T., Shchebet A., Begus-Nahrmann Y., et al. The histone H2B monoubiquitination regulatory pathway is required for differentiation of multipotent stem cells. Mol. Cell. 2012;46:705–713. doi: 10.1016/j.molcel.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 17.Tamayo A.G., Duong H.A., Robles M.S., Mann M., Weitz C.J. Histone monoubiquitination by Clock-Bmal1 complex marks Per1 and Per2 genes for circadian feedback. Nat. Struct. Mol. Biol. 2015;22:759–766. doi: 10.1038/nsmb.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samoilova E.M., Belopasov V.V., Ekusheva E.V., Zhang C., Troitskiy A.V., Baklaushev V.P. Epigenetic clock and circadian rhythms in stem cell aging and rejuvenation. J. Pers. Med. 2021;11:1050. doi: 10.3390/jpm11111050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Q., Wang T., Zhu X., Tian X., Zhong C., Chang G., Ran G., Xie Y., Zhao B., Zhu L., Ling C. Low endotoxin E. coli strain-derived plasmids reduce rAAV vector-mediated immune responses both in vitro and in vivo. Mol. Ther. Methods Clin. Dev. 2021;22:293–303. doi: 10.1016/J.OMTM.2021.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang D., Tai P.W.L., Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019;18:358–378. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Y.S., Xie J., Wang D., Kim J.M., Tai P.W.L., Gravallese E., Gao G., Shim J.H. Bone-targeting AAV-mediated silencing of Schnurri-3 prevents bone loss in osteoporosis. Nat. Commun. 2019;10:2958. doi: 10.1038/s41467-019-10809-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Najafova Z., Liu P., Wegwitz F., Ahmad M., Tamon L., Kosinsky R.L., Xie W., Johnsen S.A., Tuckermann J. RNF40 exerts stage-dependent functions in differentiating osteoblasts and is essential for bone cell crosstalk. Cell Death Differ. 2021;28:700–714. doi: 10.1038/s41418-020-00614-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu Z.C., Huang Y.F., Shieh S.Y. Requirement for human mps1/TTK in oxidative DNA damage repair and cell survival through MDM2 phosphorylation. Nucleic Acids Res. 2016;44:1133–1150. doi: 10.1093/nar/gkv1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang Q., Chen X.L., Zhang L., Li Y.B., Sun T.Z., Yang C.X., Chang J.F., Yang X.M., Sun F. The essential roles of Mps1 in spermatogenesis and fertility in mice. Cell Death Dis. 2021;12:531. doi: 10.1038/s41419-021-03815-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017;18:164–179. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olivos D.J., Perrien D.S., Hooker A., Cheng Y.H., Fuchs R.K., Hong J.M., Bruzzaniti A., Chun K., Eischen C.M., Kacena M.A., Mayo L.D. The proto-oncogene function of Mdm2 in bone. J. Cell. Biochem. 2018;119:8830–8840. doi: 10.1002/jcb.27133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang F., Yu X. WAC, a functional partner of RNF20/40, regulates histone H2B ubiquitination and gene transcription. Mol. Cell. 2011;41:384–397. doi: 10.1016/j.molcel.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reid I.R. A broader strategy for osteoporosis interventions. Nat. Rev. Endocrinol. 2020;16:333–339. doi: 10.1038/s41574-020-0339-7. [DOI] [PubMed] [Google Scholar]
- 29.Khosla S., Hofbauer L.C. Osteoporosis treatment: recent developments and ongoing challenges. Lancet Diabetes Endocrinol. 2017;5:898–907. doi: 10.1016/S2213-8587(17)30188-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reid I.R., Billington E.O. Drug therapy for osteoporosis in older adults. Lancet. 2022;399:1080–1092. doi: 10.1016/S0140-6736(21)02646-5. [DOI] [PubMed] [Google Scholar]
- 31.Sancar A., Van Gelder R.N. Clocks, cancer, and chronochemotherapy. Science. 2021;371:eabb0738. doi: 10.1126/science.abb0738. [DOI] [PubMed] [Google Scholar]
- 32.Longo V.D., Panda S. Fasting, circadian rhythms, and time-restricted feeding in healthy lifespan. Cell Metab. 2016;23:1048–1059. doi: 10.1016/j.cmet.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobi D., Liu S., Burkewitz K., Kory N., Knudsen N.H., Alexander R.K., Unluturk U., Li X., Kong X., Hyde A.L., et al. Hepatic Bmal1 regulates rhythmic mitochondrial dynamics and promotes metabolic fitness. Cell Metab. 2015;22:709–720. doi: 10.1016/j.cmet.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ulgherait M., Midoun A.M., Park S.J., Gatto J.A., Tener S.J., Siewert J., Klickstein N., Canman J.C., Ja W.W., Shirasu-Hiza M. Circadian autophagy drives iTRF-mediated longevity. Nature. 2021;598:353–358. doi: 10.1038/s41586-021-03934-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato S., Solanas G., Peixoto F.O., Bee L., Symeonidi A., Schmidt M.S., Brenner C., Masri S., Benitah S.A., Sassone-Corsi P. Circadian reprogramming in the liver identifies metabolic pathways of aging. Cell. 2017;170:664–677.e11. doi: 10.1016/j.cell.2017.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dudek M., Gossan N., Yang N., Im H.J., Ruckshanthi J.P.D., Yoshitane H., Li X., Jin D., Wang P., Boudiffa M., et al. The chondrocyte clock gene Bmal1 controls cartilage homeostasis and integrity. J. Clin. Invest. 2016;126:365–376. doi: 10.1172/JCI82755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hung F.Y., Chen F.F., Li C., Chen C., Lai Y.C., Chen J.H., Cui Y., Wu K. The Arabidopsis LDL1/2-HDA6 histone modification complex is functionally associated with CCA1/LHY in regulation of circadian clock genes. Nucleic Acids Res. 2018;46:10669–10681. doi: 10.1093/nar/gky749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song Q., Huang T.Y., Yu H.H., Ando A., Mas P., Ha M., Chen Z.J. Diurnal regulation of SDG2 and JMJ14 by circadian clock oscillators orchestrates histone modification rhythms in Arabidopsis. Genome Biol. 2019;20:170. doi: 10.1186/s13059-019-1777-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarcic O., Pateras I.S., Cooks T., Shema E., Kanterman J., Ashkenazi H., Boocholez H., Hubert A., Rotkopf R., Baniyash M., et al. RNF20 links histone H2B ubiquitylation with inflammation and inflammation-associated cancer. Cell Rep. 2016;14:1462–1476. doi: 10.1016/j.celrep.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao Z., Su Z., Liang P., Liu D., Yang S., Wu Y., Ma L., Feng J., Zhang X., Wu C., et al. USP38 couples histone ubiquitination and methylation via KDM5B to resolve inflammation. Adv. Sci. 2020;7:2002680. doi: 10.1002/advs.202002680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner H.J., Weber W., Fussenegger M. Synthetic biology: emerging concepts to design and advance adeno-associated viral vectors for gene therapy. Adv. Sci. 2021;8:2004018. doi: 10.1002/advs.202004018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li C., Samulski R.J. Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 2020;21:255–272. doi: 10.1038/s41576-019-0205-4. [DOI] [PubMed] [Google Scholar]
- 43.Colter D.C., Class R., DiGirolamo C.M., Prockop D.J. Rapid expansion of recycling stem cells in cultures of plastic-adherent cells from human bone marrow. Proc. Natl. Acad. Sci. USA. 2000;97:3213–3218. doi: 10.1073/pnas.97.7.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J., Wang P., Xie Z., Wang S., Cen S., Li M., Liu W., Tang S., Ye G., Zheng G., et al. TRAF4 positively regulates the osteogenic differentiation of mesenchymal stem cells by acting as an E3 ubiquitin ligase to degrade Smurf2. Cell Death Differ. 2019;26:2652–2666. doi: 10.1038/s41418-019-0328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li R., Li Y., Kristiansen K., Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 46.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramírez F., Dündar F., Diehl S., Grüning B.A., Manke T. DeepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its supplemental information files). The data from RNA-seq and CUT&Tag-seq have been deposited in NCBI’s Gene Expression Omnibus database under GEO accession number GSE210426.