Abstract
Bioinformatics analysis indicates that lysophosphatidylcholine acyltransferase 1 (LPCAT1) and forkhead box A1 (FOXA1) are highly expressed in breast cancer tissues and their expression levels are correlated. Therefore, the aim of the present study was to investigate their involvement in the malignant progression and drug resistance of breast cancer. The clinical significance of LPCAT1 was analyzed using The Cancer Genome Atlas data. The enrichment of LPCAT1 in breast cancer cells was determined and the effects of LPCAT1 knockdown on cell proliferation, colony formation, migration, invasion and paclitaxel (PTX) resistance were evaluated. The association between LPCAT1 and FOXA1 was verified using luciferase reporter and chromatin immunoprecipitation assays. Thereafter, the ability of FOXA1 overexpression to regulate LPCAT1 regulation was evaluated. The results revealed that a high LPCAT1 level was associated with poor overall survival in patients with breast cancer. Furthermore, LPCAT1 was found to be highly expressed in breast cancer cells, and its knockdown resulted in suppressed proliferation, colony formation, migration and invasion, and weakened PTX resistance. Furthermore, FOXA1 overexpression attenuated the effects of LPCAT1 knockdown on cells, indicating that FOXA1 transcriptionally regulates LPCAT1. In summary, the present study reveals that LPCAT1 is transcriptionally regulated by FOXA1, which influences breast cancer cell proliferation, metastatic potential and PTX resistance.
Keywords: LPCAT1, FOXA1, breast cancer, metastasis, paclitaxel resistance
Introduction
According to cancer statistics released in 2021 (1), breast cancer has surpassed lung cancer as the most frequently occurring malignant tumor. Its high prevalence in women makes it a leading cause of death in women, and its incidence continues to increase. At present, surgical resection, chemotherapy, endocrine therapy, targeted therapy and radiation therapy are the mainstream treatments for breast cancer (2,3). The connections between breast cancer cells are loose and easily broken, and detached cells can migrate via the blood or lymphatic system to develop metastases in other regions of the body (4), which accounts for >90% of breast cancer-associated fatalities (5). Despite the success of emerging immunotherapies in metastatic breast cancer (6), high costs and recurrence rates prevent a large number of patients with advanced cancer from benefiting from them. Paclitaxel (PTX) is a chemotherapeutic medication commonly used in the treatment of breast cancer that blocks microtubule dissociation, hinders cell cycle progression and stops mitosis (7). Resistance to chemotherapeutic drugs, however, is a pressing issue in cancer treatment. Treatment with PTX may induce resistance in patients, resulting in chemotherapy failure (8). Specific heredity, epigenetic aberrations in cancer cells and altered drug transport, where the drug is pumped out of the tumor cells, may all be associated with the induction of drug resistance (9,10).
In a study of glioblastoma, lysophosphatidylcholine acyltransferase 1 (LPCAT1) was suggested to play a role in remodeling the structure of the plasma membrane by modifying the phospholipid composition of the cytoplasmic membrane, increasing the stabilization of epidermal growth factor receptor, and transmitting and amplifying growth signals (11). In addition, LPCAT1 has been reported to promote the advancement of cutaneous squamous cell carcinoma via the protein kinase B and p38MAPK signaling pathways (12). The upregulation of LPCAT1 in hepatocellular carcinoma tissue specimens is associated with a poor prognosis and contributes to progression by encouraging cell growth and metastasis (13). LPCAT1 is also highly expressed in endometrial carcinoma samples, and the silencing of LPCAT1 inhibits endometrial carcinoma cell proliferation (14). To the best of our knowledge, it has not yet been revealed whether LPCAT1 has growth-promoting or pro-metastatic effects in breast cancer. However, analyses performed using UALCAN (15) and Gene Expression Profiling Interactive Analysis (GEPIA) (16) based on data in The Cancer Genome Atlas (TCGA) indicate that LPCAT1 is upregulated in breast cancer tissues and is associated with a poor prognosis.
The aim of the present study was to investigate the involvement of LPCAT1 in breast cancer and to explore its mechanism. The Human Transcription factor Database (HumanTFDB) (17) indicates that the transcription factor forkhead box A1 (FOXA1) can bind to the LPCAT1 promoter and regulate its transcriptional regulation. As a corollary, it was hypothesized that the FOXA1-mediated transcriptional upregulation of LPCAT1 promotes the malignant progression and drug resistance of breast cancer cells.
Materials and methods
Bioinformatics
The UALCAN database (ualcan.path.uab.edu) was used to compare LPCAT1 and FOXA1 expression levels between tumor and normal tissues using TCGA data. GEPIA (gepia.cancer-pku.cn) was used to compare overall and disease-free survival between patients with low and high LPCAT1 expression using TCGA data. The HumanTFDB database (bioinfo.life.hust.edu.cn) predicted the binding sites for FOXA1 and the LPCAT1 promoter.
Cell culture
MCF-10A mammary epithelial cells and MDA-MB-231, BT-549, HCC1937, SK-BR-3 and MCF-7 breast cancer cell lines were obtained from Shanghai EK-Bioscience Biotechnology Co., Ltd. PTX-resistant MDA-MB-231 (MDA-MB-231/PTX) cells were generated by 3 months of continuous exposure to a stepwise steadily increasing concentration of PTX (0–100 nM; MedChemExpress) at 37°C as previously described (18). MDA-MB-231 cells were cultured in Leibovitz's L-15 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS and 1% penicillin/streptomycin (P/S; all Gibco; Thermo Fisher Scientific, Inc.) without CO2 at 37°C. The other cell lines were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS and 1% P/S at 37°C in a 5% CO2 atmosphere.
Cell transfection
In order to reduce the expression of LPCAT1 and overexpress FOXA1, MDA-MB-231 cells were transfected with short hairpin (sh)RNAs targeting LPCAT1 (sh-LPCAT1-1 and −2) and FOXA1-overexpression plasmids (oe-FOXA1), respectively. Cells transfected with non-targeting shRNA and empty plasmid served as the negative controls (sh-NC and oe-NC, respectively). These pLVX-shRNAs and pcDNA3.1 plasmids were constructed by VectorBuilder, Inc. Briefly, cells (1×104/well) were seeded in 96-well plates 1 day before transfection, and transfection with a final concentration of 50 nM shRNA and/or 15 nM overexpression plasmids was then performed for 48 h at 37°C using FuGENE® transfection reagents (Promega Corporation). The interval between transfection and subsequent experiments was 48 h. The target sequences were as follows: sh-LPCAT1-1, 5′-GGAACTCTGATCCAGTATATA-3′; sh-LPCAT1-2, 5′-GGGAACTCTGATCCAGTATAT-3′; and sh-NC, 5′-GCACTACCAGAGCTAACTCAG-3′.
Reverse transcription-quantitative PCR (RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was added to the cells, mixed and allowed to stand at room temperature for 5 min. Chloroform was then added, the lysate was centrifuged at 12,000 × g for 12 min at 4°C, the upper aqueous phase was collected and the RNA was precipitated with isopropanol. The isolated RNA was reverse transcribed to generate cDNA using a PrimeScript™ RT Reagent Kit (Takara Bio, Inc.). The reaction conditions for reverse transcription were as follows: 30°C for 10 min, 42°C for 30 min and 70°C for 15 min. A QuantiTect SYBR Green PCR kit (Qiagen, Inc.) was used for qPCR according to the manufacturer's protocol. The qPCR was performed in a 20-µl reaction system containing 10 µl Master Mix, 10 ng DNA template and 500 nM specific forward and reverse primers. The thermocycling reaction conditions were as follows: Predenaturation at 95°C for 15 min and 40 cycles of denaturation at 94°C for 30 sec, annealing at 60°C for 30 sec and extension at 68°C for 30 sec. The relative mRNA levels were measured using the 2−∆∆Cq method (19) following normalization against GAPDH. The primer sequences were as follows (5′-3′): LPCAT1, forward, ATGAGGCTGCGGGGATG and reverse, GATGGCCTTCAGCAGGAAGT; FOXA1, forward, CCCTCTGGCGCCTCTAAC and reverse, TGGAGAACGGGTGGTTGAAG; GAPDH, forward, GACTCATGACCACAGTCCATGC and reverse, AGAGGCAGGGATGATGTTCTG.
Western blotting
Protein was isolated from cells following treatment with RIPA lysis buffer (Life-iLab Bio) and quantified using a Nano 300 protein detector (YPH-Bio). Protein separation (30 µg per lane) was achieved using 10% SDS-polyacrylamide gel electrophoresis and the separated proteins were transferred to PVDF membranes (Roche Diagnostics). The membranes were incubated with 5% skimmed milk for 2 h at room temperature, with primary antibodies overnight at 4°C and HRP-conjugated secondary antibodies for 2 h at room temperature in sequence. The primary antibodies against LPCAT1 (cat. no. ab214034; 1:2,000), FOXA1 (cat. no. ab170933; 1:1,000), Ki67 (cat. no. ab92742; 1:5,000), proliferating cell nuclear antigen (PCNA; cat. no. ab92552; 1:1,000), MMP2 (cat. no. ab92536; 1:1,000), MMP9 (cat. no. ab76003; 1:1,000), Bcl-2 (cat. no. ab32124; 1:2,000), Bax (cat. no. ab32503; 1:1,000), cleaved caspase 3 (cat. no. ab32042; 1:500) and GAPDH (cat. no. ab9485; 1:2,500), and the secondary antibodies (cat. no. ab6721; 1:4,000) were all from Abcam. Blots were visualized after treatment with Immobilon ECL Ultra Western HRP (Merck KGaA) and gray values were analyzed with ImageJ software (v1.8.0; National Institutes of Health).
Cell counting kit-8 (CCK-8) assay
CCK-8 assay was used to evaluate the proliferation of transfected cells and the viability of resistant cells. In brief, transfected cells (3×103/well) were seeded in 96-well plates and cultured for 24, 48 and 72 h. The resistant cells were treated with PTX (0–100 nM) for 72 h at 37°C. CCK-8 solution (Absin Bioscience, Inc.) was added to each well and incubation was continued for another 2 h. The optical density (OD) was then determined at 450 nm using a microplate reader (Thermo Fisher Scientific, Inc.).
Colony formation
Control and transfected MDA-MB-231 cells were seeded into culture dishes at a density of 500 cells/dish. They were cultured for 2 weeks and the medium was changed every 3 days. Thereafter, cells were washed twice with PBS, fixed with 4% paraformaldehyde (Merck KGaA) for 20 min at room temperature and stained with 0.5% crystal violet (Shanghai Gefan Biotechnology Co., Ltd.) for 20 min at room temperature. The colonies were counted manually. A cluster of >50 cells was considered a colony.
Wound healing and Transwell
The migration and invasion of the control and transfected MDA-MB-231 cells were separately assessed using wound healing and Transwell assays, respectively. In the wound healing assay, cells were cultured until a confluent monolayer formed and a sterile pipette tip was used to generate a wound in the middle of the cells that were cultured in serum-free Leibovitz's L-15 medium. Images were captured at 0 and 24 h. In the Transwell assay, cells (1×104 cells/well) were cultivated in serum-free Leibovitz's L-15 medium in the upper chamber, which was pre-coated with Matrigel (Corning, Inc.) at 37°C for 1 h. Leibovitz's L-15 containing 20% FBS was loaded into the lower chamber. Following 24 h of incubation at 37°C, the invasive cells were fixed and stained with 0.1% crystal violet solution at room temperature for 15 min. Results for both assays were observed under a light microscope (magnification ×100; Olympus Corporation).
Flow cytometry
The apoptosis of MDA-MB-231 and MDA-MB-231/PTX cells with or without transfection was analyzed using Annexin V-FITC Apoptosis Detection Kit (Beyotime Institute of Biotechnology) and flow cytometry. Briefly, cells (1×105) were washed twice with precooled PBS and suspended in 1 ml binding buffer. A 100-µl sample of the cell suspension was transferred in a culture tube and incubated with Annexin V-FITC and propidium iodide at room temperature in the absence of light for 15 min. Results were obtained using flow cytometry using a BD FACSCanto™ instrument (BD Biosciences) and FlowJo version 10 software (GlowJo LLC).
Luciferase reporter
The promoter site of LPCAT1 and a mutated form (CGCCCAGGC) of this site were cloned into a dual-luciferase reporter vector (Promega Corporation). The reporter vector was co-transfected along with oe-FOXA1 or oe-NC into MDA-MB-231 cells using FuGENE® transfection reagents (Promega Corporation). At 48 h post-transfection, the luciferase activity was assessed using the Dual-luciferase Reporter Assay System (Promega Corporation), according to the manufacturer's protocol, and normalized to Renilla luciferase activity.
Chromatin immunoprecipitation (ChIP)
The association between LPCAT1 and FOXA1 was evaluated using a ChIP Detection Kit (cat. no. 17–295; EZ-ChIP; MilliporeSigma). Briefly, the MDA-MB-231 cells were treated with 1% formaldehyde, followed by lysis buffer and then sonicated. The cells were subsequently incubated with an anti-FOXA1 (cat. no. ab170933; 1:50; Abcam) or anti-IgG antibody (cat. no. ab172730; 1:50; Abcam) overnight at 4°C. Following the incubation, 60 µl protein A agarose beads was added to harvest the protein-DNA complex. The complex was washed in low-salt and high-salt washing buffers at 4°C, for 5 min each time, 4 times in total. The liquid was removed by centrifugation at 1,000 × g for 1 min at 4°C, and 5 mmol/l NaCl was added to retrieve the DNA. The enrichment of LPCAT1 was determined using RT-qPCR.
Statistical analysis
For statistical analysis, GraphPad Prism 8.0 (GraphPad Software, Inc.) was utilized. All data are presented as the mean ± SD and all experiments were performed ≥3 times independently. To compare differences between two and multiple groups, the unpaired Student's t-test and one-way ANOVA followed by Tukey's post hoc test were used, respectively. P<0.05 was considered to indicate a statistically significant difference.
Results
Role of LPCAT1 in cell proliferation and metastatic potential
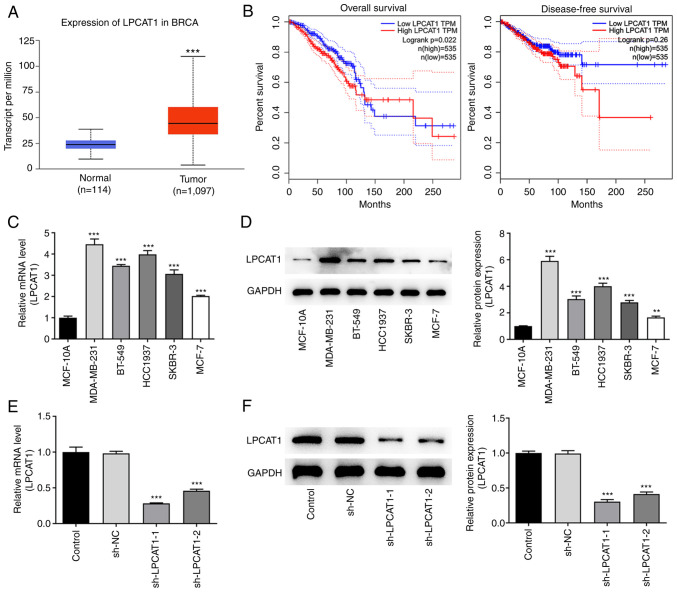
Analysis performed using the UALCAN database revealed that LPCAT1 is expressed at significantly higher levels in breast cancer tissues compared with normal tissues (Fig. 1A). Furthermore, survival analysis performed using GEPIA indicated that patients with high LPCAT1 expression were more likely than those with low LPCAT1 expression to have a poor prognosis in terms of overall survival and disease-free survival within 10 years; LPCAT1 is significantly associated with the poor overall survival of patients (Fig. 1B). Thereafter, the expression levels of LPCAT1 in various cell lines were determined using RT-qPCR (Fig. 1C) and western blotting (Fig. 1D). The results revealed that LPCAT1 expression was significantly elevated in breast cancer cell lines compared with MCF-10A cells. To highlight the potential role of LPCAT1 in breast cancer, the MDA-MB-231 cell line was selected for further analysis. The expression of LPCAT1 in the transfected MDA-MB-231 cell line was markedly reduced by transfection with sh-LPCAT1 as shown by the results of RT-qPCR (Fig. 1E) and western blotting (Fig. 1F). Since the level of knockdown was superior in cells in the sh-LPCAT1-1 group, the proliferation of cells transfected with sh-LPCAT1-1 (henceforth referred to as sh-LPCAT1) was further examined.
Figure 1.
LPCAT1 levels and their clinical significance. (A) Data from the UALCAN database based on The Cancer Genome Atlas data suggest that LPCAT1 is upregulated in breast cancer tissues. ***P<0.001 vs. normal. (B) Gene Expression Profiling Interactive Analysis database analysis implicates an association of LPCAT1 expression with overall and disease-free survival in breast cancer. Expression levels of LPCAT1 in various cell lines were determined using (C) RT-qPCR and (D) western blotting. **P<0.01, ***P<0.001 vs. MCF-10A. Expression levels of LPCAT1 in transfected MDA-MB-231 cells were determined using (E) RT-qPCR and (F) western blotting. ***P<0.001 vs. sh-NC. LPCAT1, lysophosphatidylcholine acyltransferase 1; RT-qPCR, reverse transcription-quantitative PCR; sh, short hairpin; NC, negative control; BRCA, breast invasive carcinoma.
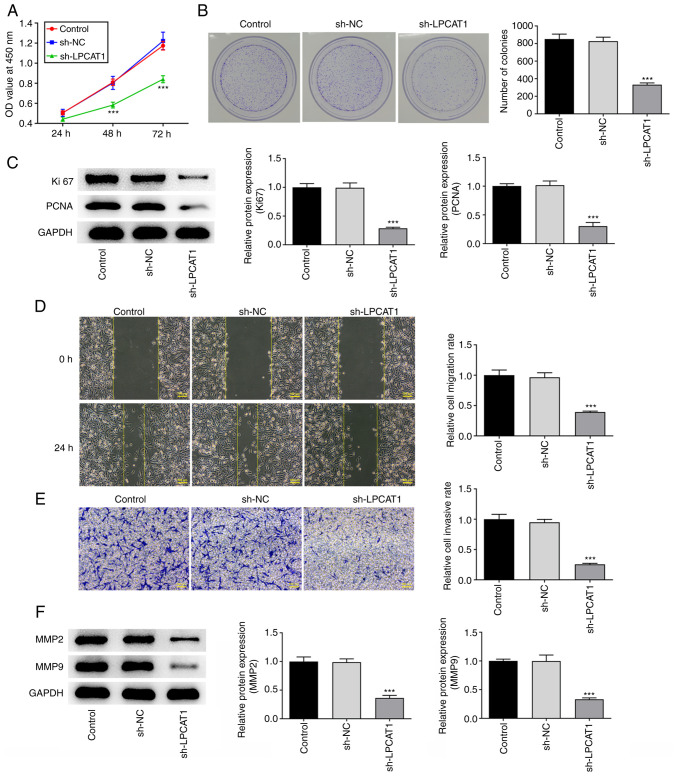
CCK-8 (Fig. 2A) and colony formation (Fig. 2B) assay results showed that LPCAT1 knockdown significantly suppressed the proliferation and colony formation of the cells. The expression levels of Ki67 and PCNA were also significantly decreased due to the reduction in LPCAT1 expression (Fig. 2C). In addition, the knockdown of LPCAT1 inhibited the migration and invasion of the cells (Fig. 2D and E), which was supported by a reduction in MMP2 and MMP9 expression levels (Fig. 2F).
Figure 2.
Role of LPCAT1 in cell proliferation and metastasis. (A) Cell proliferation was examined using a Cell Counting Kit-8 assay. (B) Colony formation was examined using a colony formation assay. (C) Expression levels of Ki67 and PCNA were determined using western blotting. (D) Cell migration and (E) invasion potential were assessed using wound healing and Transwell assays, respectively. Scale bar, 100 µm. (F) Expression levels of MMP2 and MMP9 were determined using western blotting. ***P<0.001 vs. sh-NC. LPCAT1, lysophosphatidylcholine acyltransferase 1; PCNA, proliferating cell nuclear antigen; sh, short hairpin; NC, negative control; OD, optical density.
Role of LPCAT1 in PTX resistance
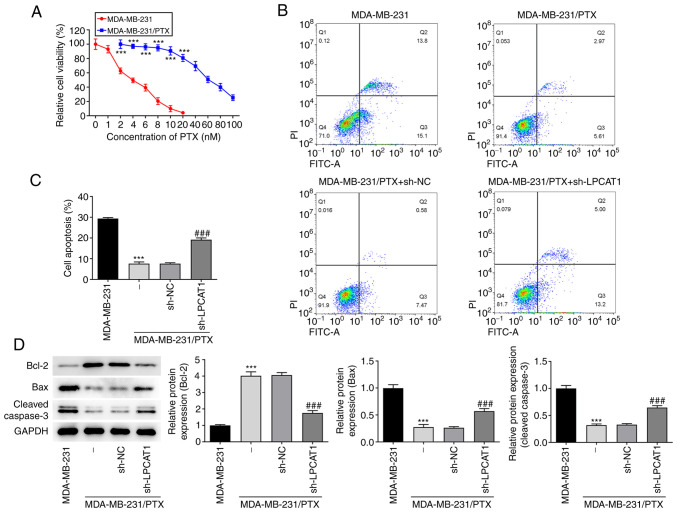
The viability of MDA-MB-231 and MDA-MB-231/PTX cells following treatment with PTX (0–100 nM) was detected by a CCK-8 assay. The results indicated that MDA-MB-231 cells were significantly more sensitive to PTX than were the MDA-MB-231/PTX cells (Fig. 3A). The level of apoptosis following treatment with 4 nM PTX was assessed by flow cytometry. The apoptosis rate of the MDA-MB-231/PTX cells was significantly lower than that of the MDA-MB-231 cells, and LPCAT1 knockdown increased the apoptosis rate of the MDA-MB-231/PTX cells compared with that of the MDA-MB-231/PTX cells transfected with sh-NC (Fig. 3B and C). The levels of apoptosis-associated proteins in the cells treated with PTX were also determined. Western blot results revealed that Bcl-2 was more abundant in MDA-MB-231/PTX cells compared with MDA-MB-231 cells, whereas Bax and cleaved caspase 3 levels were lower. Furthermore, LPCAT1 knockdown reduced the expression of Bcl-2 and increased Bax and cleaved caspase 3 levels in MDA-MB-231/PTX cells (Fig. 3D).
Figure 3.
Role of LPCAT1 in PTX resistance. (A) Viability of MDA-MB-231 and MDA-MB-231/PTX cells after PTX intervention was detected by a Cell Counting Kit-8 assay. (B and C) Proportion of apoptotic cells following treatment with 4 nM PTX was assessed by flow cytometry. (B) Representative plots and (C) quantified results are shown. (D) Enrichment of apoptosis-associated proteins in the cells was determined using western blotting. ***P<0.001 vs. MDA-MB-231; ###P<0.001 vs. MDA-MB-231/PTX + sh-NC. LPCAT1, lysophosphatidylcholine acyltransferase 1; PTX, paclitaxel; sh, short hairpin; NC, negative control; PI, propidium iodide; FITC-A, fluorescein isothiocyanate-Annexin V.
Association between FOXA1 and LPCAT1
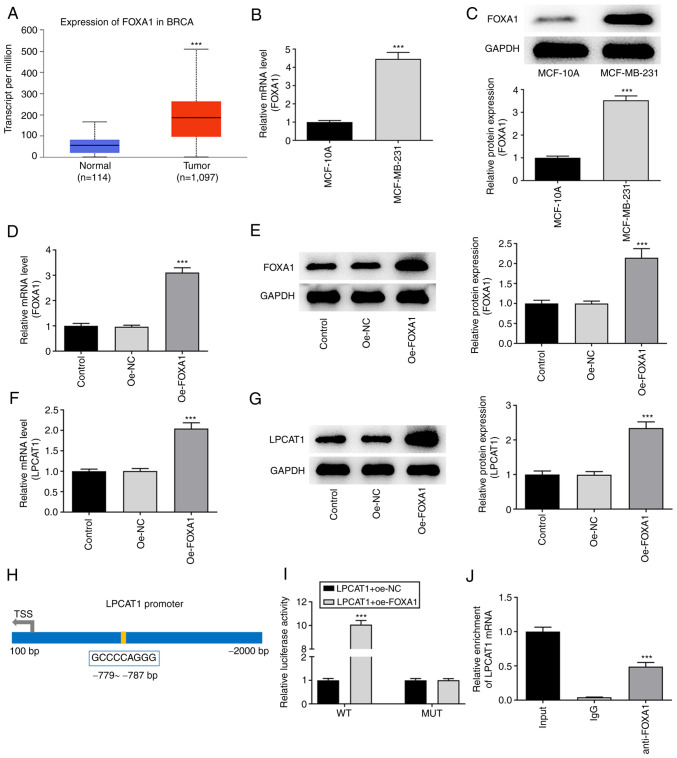
Analysis performed using the UALCAN database suggested that FOXA1 was also highly expressed in breast cancer tissues compared with normal breast tissues (Fig. 4A). The expression level of FOXA1 in MCF-10A and MDA-MB-231 cells was assessed using RT-qPCR (Fig. 4B) and western blotting (Fig. 4C). FOXA1 expression was significantly higher in MDA-MB-231 cells compared with MCF-10A cells. Following confirmation that FOXA1 was successfully overexpressed in the MDA-MB-231 cells transfected with oe-FOXA1 compared with those transfected with oe-NC (Fig. 4D and E), the levels of LPCAT1 were determined. FOXA1 overexpression boosted the increase of LPCAT1 in the MDA-MB-231 cells (Fig. 4F and G). The HumanTFDB website predicted binding sites between transcription factor FOXA1 and the LPCAT1 promoter (Fig. 4H). Thereafter, LPCAT1 promoter activity was determined using a luciferase reporter assay. The activity in the LPCAT1-WT + oe-FOXA1 group was significantly higher than that in the LPCAT1-WT + oe-NC group (Fig. 4I). The binding of FOXA1 to LPCAT1 was then evaluated using a ChIP assay (Fig. 4J). The relative enrichment of LPCAT1 in the anti-FOXA1 group was significantly higher compared with that in the lgG group.
Figure 4.
Association between FOXA1 and LPCAT1. (A) UALCAN database analysis based on The Cancer Genome Atlas data indicates that FOXA1 is highly expressed in breast cancer tissues. ***P<0.001 vs. normal. Expression levels of FOXA1 in MCF-10A and MDA-MB-231 cells were assessed using (B) RT-qPCR and (C) western blotting. ***P<0.001 vs. MCF-10A. Expression of FOXA1 in the MDA-MB-231 cells transfected with oe-FOXA1 was confirmed using (D) RT-qPCR and (E) western blotting. Expression of LPCAT1 in the transfected MDA-MB-231 cells was confirmed using (F) RT-qPCR and (G) western blotting. ***P<0.001 vs. oe-NC. (H) Binding sites for transcription factors FOXA1 and LPCAT1 promoters predicted using the HumanTFDB website. (I) LPCAT1 promoter activity was determined with a luciferase reporter assay. ***P<0.001 vs. LPCAT1 + oe-NC (WT). (J) Binding between FOXA1 and LPCAT1 was evaluated with a chromatin immunoprecipitation assay. ***P<0.001 vs. IgG. FOXA1, forkhead box A1; LPCAT1, lysophosphatidylcholine acyltransferase 1; RT-qPCR, reverse transcription-quantitative PCR; HumanTFDB, Human Transcription factor Database; oe, overexpression; NC, negative control; BRCA, breast invasive carcinoma; WT, wild type; MUT, mutant.
FOXA1 regulates LPCAT1
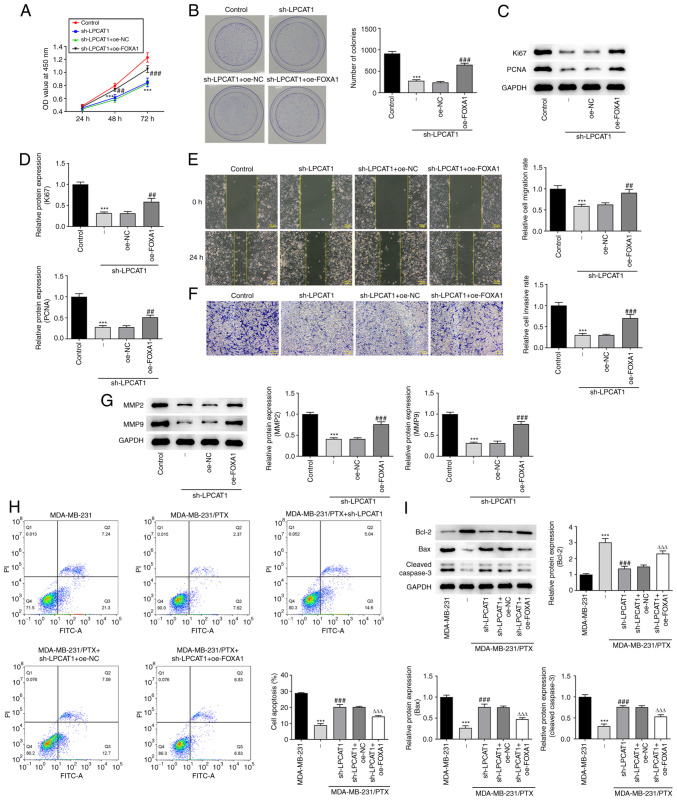
To evaluate the effect of FOXA1 overexpression on the regulatory role of LPCAT, the proliferation and colony formation ability of MDA-MB-231 cells co-transfected with sh-LPCAT1 and oe-FOXA1 were assessed. The co-transfection increased cell proliferation (Fig. 5A) and colony formation (Fig. 5B) and enriched the expression of Ki67 and PCNA proteins (Fig. 5C and D) compared with those in cells transfected with sh-LPCAT1 and oe-NC. These results indicate that FOXA1 overexpression attenuated the effects of LPCAT knockdown on cell proliferation. Furthermore, FOXA1 overexpression attenuated the effects of sh-LPCAT1 on cell migration (Fig. 5E) and invasion (Fig. 5F), which was accompanied by increased levels of MMP2 and MMP9 (Fig. 5G). The effect of FOXA1 overexpression on the drug resistance of the cells was also evaluated. The flow cytometry results indicated that the level of apoptosis was decreased in MDA-MB-231/PTX cells subjected to co-transfection with sh-LPCAT1 and oe-FOXA1 compared with that in cells transfected with sh-LPCAT1 plus oe-NC (Fig. 5H). In addition, the expression levels of Bcl-2 in the sh-LPCAT1 and oe-FOXA1 co-transfected cells were increased whereas those of Bax and cleaved caspase 3 were decreased compared with those in the sh-LPCAT1 plus oe-NC group (Fig. 5I).
Figure 5.
FOXA1 regulates LPCAT1. (A) Proliferation and (B) colony formation of MDA-MB-231 cells co-transfected with sh-LPCAT1 and oe-FOXA1 was assessed. (C and D) Expression levels of Ki67 and PCNA were determined using western blotting. (C) Representative images and (D) densitometrically quantified results are presented. (E) Cell migration and (F) invasion potential were assessed using wound healing and Transwell assays, respectively. Scale bar, 100 µm. (G) Expression levels of MMP2 and MMP9 were determined using western blotting. ***P<0.001 vs. control; ##P<0.01 and ###P<0.001 vs. sh-LPCAT1 + oe-NC. (H) Apoptosis after 4 nM PTX treatment was assessed by flow cytometry. (I) Enrichment of apoptosis-associated proteins in the co-transfected cells was determined using western blotting. ***P<0.001 vs. MDA-MB-231; ###P<0.001 vs. MDA-MB-231/PTX + sh-NC; ΔΔΔP<0.001 vs. sh-LPCAT1 + oe-NC. FOXA1, forkhead box A1; LPCAT1, lysophosphatidylcholine acyltransferase 1; sh, short hairpin; oe, overexpression; NC, negative control; PCNA, proliferating cell nuclear antigen; OD, optical density; PI, propidium iodide; FITC-A, fluorescein isothiocyanate-Annexin V.
Discussion
PTX has been extensively known for its antitumor activity. It has a broad range of anticancer properties and can be employed in the chemotherapy of various solid tumors (20), including non-small cell lung cancer, ovarian cancer and esophageal cancer. At present, it is the first-line drug in the chemotherapy of breast cancer (21). Resistance to chemotherapy drugs is a serious issue in cancer treatment, and recurrence and metastasis are the predominant causes of mortality in patients with breast cancer (22). Primary resistance is resistance that is present prior to therapy (23), whereas acquired resistance develops over time after drug usage. As a consequence, the long-term effects of PTX use may be unsatisfactory (24). The mechanisms underlying chemotherapy resistance have not been fully elucidated and require additional investigation. The key to improving the prognosis of breast cancer is the effective control of metastases and treatment resistance (25). Therefore, investigating the molecular mechanisms of breast cancer invasion, metastasis and drug resistance, as well as identifying specific targets for the reversal of chemotherapy resistance (25), may provide new research and development options for gene-targeted breast cancer therapy.
The findings of the present study indicate that LPCAT1 modulates breast cancer cell proliferation, metastatic potential and drug resistance. Given the previous findings of LPCAT1 in various tumors (26), it may be inferred that LPCAT1 is a novel target that is prevalent in a wide range of cancers. Furthermore, it is considered to be an enzyme that is associated with genetic and metabolic anomalies in cancer cells (11,27), contributing to aggressive tumor growth. A novel finding of the present study is that LPCAT1 is implicated in chemoresistance. Furthermore, FOXA1 was discovered to alter the function of LPCAT1 in breast cancer through rescue experiments.
The role of FOXA1 in breast cancer has been reported in previous studies. For example, one study showed that microRNA (miR)-100 inhibits the proliferation, migration and invasion of breast cancer cells by targeting FOXA1 (28). Another study demonstrated that by sponging miR-23a-3p and thereby promoting FOXA1, the long non-coding RNA NEAT1 enhances drug resistance in breast cancer cells (29). Furthermore, somatic point mutations in FOXA1 occur at a rate of 4–8%, and FOXA1 mutations boost cancer progression by reprogramming functions such as the androgen receptor (AR) (30,31). The AR is the therapeutic target in some types of breast cancer, which indicates that FOXA1 mutation may result in the failure of targeted AR therapy (32). According to recent research, increased levels of FOXA1 are associated with reduced interferon activity and T-cell infiltration in patients with estrogen-positive luminal breast cancer treated with neoadjuvant chemotherapy, indicating that the role of FOXA1 in the cancer immune response contributes to immune evasion and therapeutic resistance (33). Nevertheless, it is uncertain if LPCAT1 also mediates immune responses in breast cancer, which necessitates further research in the future.
In summary, the present study reveals that the presence of LPCAT1 contributes to breast cancer cell proliferation, metastatic potential and PTX resistance. Moreover, LPCAT1 is transcriptionally regulated by FOXA1, and the identification of this signaling pathway in PTX resistance suggests a new potential target for the alleviation of chemotherapy resistance. Follow-up experiments using animals with gene overexpression or knockdown are planned to verify this discovery, and new therapies may become available to patients in terms of this potential novel target.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HZ and YZ contributed to the study concept, experiments and analysis. HZ drafted the manuscript. HZ and YZ confirm the authenticity of all the raw data. Both authors read and approved the final version of the manuscript
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Fisusi FA, Akala EO. Drug combinations in breast cancer therapy. Pharm Nanotechnol. 2019;7:3–23. doi: 10.2174/2211738507666190122111224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDonald ES, Clark AS, Tchou J, Zhang P, Freedman GM. Clinical diagnosis and management of breast cancer. J Nucl Med. 2016;57((Suppl 1)):9S–16S. doi: 10.2967/jnumed.115.157834. [DOI] [PubMed] [Google Scholar]
- 4.Sharma R, Sharma R, Khaket TP, Dutta C, Chakraborty B, Mukherjee TK. Breast cancer metastasis: Putative therapeutic role of vascular cell adhesion molecule-1. Cell Oncol (Dordr) 2017;40:199–208. doi: 10.1007/s13402-017-0324-x. [DOI] [PubMed] [Google Scholar]
- 5.Rashid NS, Grible JM, Clevenger CV, Harrell JC. Breast cancer liver metastasis: Current and future treatment approaches. Clin Exp Metastasis. 2021;38:263–277. doi: 10.1007/s10585-021-10080-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugie T. Immunotherapy for metastatic breast cancer. Chin Clin Oncol. 2018;7:28. doi: 10.21037/cco.2018.05.05. [DOI] [PubMed] [Google Scholar]
- 7.Abu Samaan TM, Samec M, Liskova A, Kubatka P, Büsselberg D. Paclitaxel's mechanistic and clinical effects on breast cancer. Biomolecules. 2019;9:789. doi: 10.3390/biom9120789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chi Y, Xue J, Huang S, Xiu B, Su Y, Wang W, Guo R, Wang L, Li L, Shao Z, et al. CapG promotes resistance to paclitaxel in breast cancer through transactivation of PIK3R1/P50. Theranostics. 2019;9:6840–6855. doi: 10.7150/thno.36338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.AlFakeeh A, Brezden-Masley C. Overcoming endocrine resistance in hormone receptor-positive breast cancer. Curr Oncol. 2018;25((Suppl 1)):S18–S27. doi: 10.3747/co.25.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia ZH, Wang XG, Zhang H. Overcome cancer drug resistance by targeting epigenetic modifications of centrosome. Cancer Drug Resist. 2019;2:210–224. doi: 10.20517/cdr.2018.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bi J, Ichu TA, Zanca C, Yang H, Zhang W, Gu Y, Chowdhry S, Reed A, Ikegami S, Turner KM, et al. Oncogene amplification in growth factor signaling pathways renders cancers dependent on membrane lipid remodeling. Cell Metab. 2019;30:525–538.e8. doi: 10.1016/j.cmet.2019.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y, Wang Y, Wang Y, Wang N, Duan Q, Wang S, Liu M, Bilal MA, Zheng Y. LPCAT1 promotes cutaneous squamous cell carcinoma via EGFR-mediated protein kinase B/p38MAPK signaling pathways. J Invest Dermatol. 2022;142:303–313.e9. doi: 10.1016/j.jid.2021.07.163. [DOI] [PubMed] [Google Scholar]
- 13.He RQ, Li JD, Du XF, Dang YW, Yang LJ, Huang ZG, Liu LM, Liao LF, Yang H, Chen G. LPCAT1 overexpression promotes the progression of hepatocellular carcinoma. Cancer Cell Int. 2021;21:442. doi: 10.1186/s12935-021-02130-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao T, Zhang Y, Ma X, Wei L, Hou Y, Sun R, Jiang J. Elevated expression of LPCAT1 predicts a poor prognosis and is correlated with the tumour microenvironment in endometrial cancer. Cancer Cell Int. 2021;21:269. doi: 10.1186/s12935-021-01965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45((W1)):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu H, Miao YR, Jia LH, Yu QY, Zhang Q, Guo AY. AnimalTFDB 3.0: A comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 2019;47(D1):D33–D38. doi: 10.1093/nar/gky822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Wu N, Zhang J, Wang H, Men X. MiR-153-5p enhances the sensitivity of triple-negative breast cancer cells to paclitaxel by inducing G2M phase arrest. Onco Targets Ther. 2020;13:4089–4097. doi: 10.2147/OTT.S241640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Verco S, Maulhardt H, Baltezor M, Williams E, Iacobucci M, Wendt A, Verco J, Marin A, Campbell S, Dorman P, diZerega G. Local administration of submicron particle paclitaxel to solid carcinomas induces direct cytotoxicity and immune-mediated tumoricidal effects without local or systemic toxicity: Preclinical and clinical studies. Drug Deliv Transl Res. 2021;11:1806–1817. doi: 10.1007/s13346-020-00868-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dan VM, Raveendran RS, Baby S. Resistance to intervention: Paclitaxel in breast cancer. Mini Rev Med Chem. 2021;21:1237–1268. doi: 10.2174/1389557520999201214234421. [DOI] [PubMed] [Google Scholar]
- 22.Mishra A, Srivastava A, Pateriya A, Tomar MS, Mishra AK, Shrivastava A. Metabolic reprograming confers tamoxifen resistance in breast cancer. Chem Biol Interact. 2021;347:109602. doi: 10.1016/j.cbi.2021.109602. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z, Li Z, Deng M, Liu B, Xin X, Zhao Z, Zhang Y, Lv Q. Downregulation of GPSM2 is associated with primary resistance to paclitaxel in breast cancer. Oncol Rep. 2020;43:965–974. doi: 10.3892/or.2020.7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ge X, Cao Z, Gu Y, Wang F, Li J, Han M, Xia W, Yu Z, Lyu P. PFKFB3 potentially contributes to paclitaxel resistance in breast cancer cells through TLR4 activation by stimulating lactate production. Cell Mol Biol (Noisy-le-grand) 2016;62:119–125. [PubMed] [Google Scholar]
- 25.Nedeljković M, Damjanović A. Mechanisms of chemotherapy resistance in triple-negative breast cancer-how we can rise to the challenge. Cells. 2019;8:957. doi: 10.3390/cells8090957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang B, Tontonoz P. Phospholipid remodeling in physiology and disease. Annu Rev Physiol. 2019;81:165–188. doi: 10.1146/annurev-physiol-020518-114444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tao M, Luo J, Gu T, Yu X, Song Z, Jun Y, Gu H, Han K, Huang X, Yu W, et al. LPCAT1 reprogramming cholesterol metabolism promotes the progression of esophageal squamous cell carcinoma. Cell Death Dis. 2021;12:845. doi: 10.1038/s41419-021-04132-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie H, Xiao R, He Y, He L, Xie C, Chen J, Hong Y. MicroRNA-100 inhibits breast cancer cell proliferation, invasion and migration by targeting FOXA1. Oncol Lett. 2021;22:816. doi: 10.3892/ol.2021.13077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu L, Wang F, Fan W, Jin Z, Teng C, Zhang J. lncRNA NEAT1 promotes the Taxol resistance of breast cancer via sponging the miR-23a-3p-FOXA1 axis. Acta Biochim Biophys Sin (Shanghai) 2021;53:1198–1206. doi: 10.1093/abbs/gmab098. [DOI] [PubMed] [Google Scholar]
- 30.Adams EJ, Karthaus WR, Hoover E, Liu D, Gruet A, Zhang Z, Cho H, DiLoreto R, Chhangawala S, Liu Y, et al. FOXA1 mutations alter pioneering activity, differentiation and prostate cancer phenotypes. Nature. 2019;571:408–412. doi: 10.1038/s41586-019-1318-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parolia A, Cieslik M, Chu SC, Xiao L, Ouchi T, Zhang Y, Wang X, Vats P, Cao X, Pitchiaya S, et al. Distinct structural classes of activating FOXA1 alterations in advanced prostate cancer. Nature. 2019;571:413–418. doi: 10.1038/s41586-019-1347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arruabarrena-Aristorena A, Maag JLV, Kittane S, Cai Y, Karthaus WR, Ladewig E, Park J, Kannan S, Ferrando L, Cocco E, et al. FOXA1 mutations reveal distinct chromatin profiles and influence therapeutic response in breast cancer. Cancer Cell. 2020;38:534–550.e9. doi: 10.1016/j.ccell.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He Y, Wang L, Wei T, Xiao YT, Sheng H, Su H, Hollern DP, Zhang X, Ma J, Wen S, et al. FOXA1 overexpression suppresses interferon signaling and immune response in cancer. J Clin Invest. 2021;131:e147025. doi: 10.1172/JCI147025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.