Abstract
Collagen triple helix repeat containing 1 (CTHRC1) is a secreted glycoprotein that decreases the deposition of collagen matrix and accelerates tumor metastasis. However, the relationship between CTHRC1 and the outcomes of head and neck squamous cell carcinoma (HNSCC) and tumor-infiltrating lymphocytes remains unclear. In the present study, the transcriptional level of CTHRC1 and its association with overall survival (OS) and relapse-free survival (RFS) time in diverse cancer types were evaluated using The Cancer Genome Atlas, Tumor Immune Estimation Resource (TIMER), ONCOMINE and Kaplan-Meier plotter databases. The association of CTHRC1 expression level with the clinicopathological parameters of patients with HNSCC from The University of ALabama at Birmingham CANcer data analysis Portal (UALCAN) database were also evaluated. Enrichment analysis of CTHRC1 was carried out using gene set enrichment analysis software. CIBERSORT and TIMER databases were used to evaluate the relationship between the expression level of CTHRC1 and the proportion of tumor-infiltrating immune cells (TICs) in multiple cancer types. Moreover, immunohistochemistry was used to verify the expression of CTHRC1 in clinical samples of HNSCC. CTHRC1 was upregulated in HNSCC and high expression of CTHRC1 was associated with worsening clinicopathologic parameters and shorter OS and RFS times. There were eight HALLMARK gene sets, 1,231 immune signature gene sets and 14 KEGG gene sets significantly enriched in the high CTHRC1 expression group, while no gene set was enriched in the low CTHRC1 expression group. The expression of CTHRC1 was closely correlated with the proportion of TICs, where the expression of CTHRC1 was significantly positively correlated with the amount of infiltrated M0 and M2 macrophages, and significantly negatively associated with the levels of M1 macrophages. These findings suggest that CTHRC1 is an adverse prognostic marker and is associated with immune cell infiltration in HNSCC.
Keywords: collagen triple helix repeat containing 1, marker, immune infiltration, head and neck squamous cell carcinoma
Introduction
Head and neck squamous cell carcinoma (HNSCC) is one of the most common malignant tumors, accounting for ~90% of the total number of head and neck tumors (1), with >800,000 new cases globally every year (2). Multiple therapies, including surgery, radiotherapy, chemotherapy and systemic therapy, can be applied for HNSCC treatment (3); however, the 5-year survival rate is only 50% (4). HNSCC is an immunosuppressive disease which demonstrates impaired function of immune cells (5–7). The mechanisms of immune escape in HNSCC include upregulation of programmed death-ligand 1 in human papillomavirus (HPV)-positive tumors, downregulation of interferon regulatory factors and activation of the STAT1 signaling pathway (3), which contribute to the development of HNSCC. Previous studies have reported that Keytruda® and Opdivo® can efficiently prevent the immune escape state and improve the prognosis of patients due to their specific binding to PD-1 (8,9). Therefore, it is important to explore immune checkpoints in the development of targeted drugs to improve the prognosis of patients with HNSCC.
Collagen triple helix repeat containing 1 (CTHRC1) was first discovered in the injured arteries of rats (10). CTHRC1 serves an important role in wound repair (11), hepatocyte fibrosis (12), bone reconstruction (13) and adipose tissue formation (14). In previous studies, CTHRC1 has been reported to be abnormally expressed in colorectal and gastric cancer (15–17). Furthermore, CTHRC1 inhibits collagen deposition by mediating the Wnt/planar cell polarity (PCP), TGF-β/bone morphogenetic protein and ERK signaling pathways, and participates in the metastasis of tumors (18–20). CTHRC1 promotes the proliferation of HeLa cells via activation of the Wnt/PCP signaling pathway and promotion of the proliferation of breast cancer cells via a Linc00707-mediated competing endogenous RNA mechanism (21,22). Furthermore, upregulation of CTHRC1 results in a worse prognosis for patients with liver and epithelial ovarian cancers (19,23). Therefore, CTHRC1 can be considered a carcinogenic driving factor for the progression and metastasis of esophageal squamous cell carcinoma, and as a potential biomarker for prognosis and individualized treatment (20,24). It has been reported that CTHRC1 is involved in immune cell infiltration in the tumor microenvironment, has a pivotal role in the regulation of M2 macrophage polarization in ovarian tumors and is seen as a target for antitumor immunotherapy (25). However, the roles of CTHRC1 in HNSCC and its tumor immune microenvironment remain unclear. Therefore, in the present study, the Tumor Immune Estimation Resource (TIMER) and ONCOMINE databases were used to analyze the transcriptional levels of CTHRC1 in HNSCC. The University of ALabama at Birmingham CANcer data analysis Portal (UALCAN) and CIBERSORT websites were used to evaluate the association between CTHRC1 expression levels and clinical features or the immune microenvironment, to provide solid evidence for the significance of CTHRC1 in HNSCC.
Materials and methods
The cancer genome atlas (TCGA) database analysis
The RNA-seq data (workflow type, HTSeq-FPKM) and relevant clinical data for the ‘TCGA-HNSC’ cohort, including 502 tumor samples and 44 normal samples, were downloaded from the TCGA database (https://tcga-data.nci.nih.gov/tcga/) (26).
ONCOMINE database analysis
Expression levels of CTHRC1 were analyzed in multiple cancer types using the ONCOMINE database (https://www.oncomine.org/resource/main.html) (27), and the cut-off P-value used was 0.05, while the log fold change cut-off was equal to 1.
TIMER database analysis
The TIMER database (https://cistrome.shinyapps.io/timer/) (28) has incorporated 39 types of cancer in the TCGA database. Gene expression levels were presented as log2 transcripts per million. The TIMER database was used to evaluate the transcriptional level of CTHRC1 in multiple cancer types and its correlation with immune cell infiltration.
Kaplan-Meier (KM) plotter analysis
KM plotter contains data and clinical information from the Gene Expression Omnibus, TCGA and European Genome-Phenome Archive databases. The prognostic value of the mRNA expression levels of CTHRC1 in 21 cancer types was evaluated using the KM plotter (http://kmplot.com/analysis/). P<0.05 was considered to indicate a statistically significant difference.
UALCAN analysis
UALCAN (http://ualcan.path.uab.edu) (29) contains level 3 RNA-seq data and clinical information from 31 cancer types from the TCGA database and is an interactive and comprehensive web resource for analyzing cancer omics data. In the present study, UALCAN was used to investigate the relationship between the levels of CTHRC1 and clinicopathologic parameters, including age, TP53 mutation, nodal metastasis (N0, no regional lymph node metastasis; N1, metastases in 1–3 axillary lymph nodes; N2, metastases in 4–9 axillary lymph nodes; N3, metastases in ≥10 axillary lymph nodes), individual cancer stages, tumor grades (grade 1, well differentiated; grade 2, moderately grade; grade 3, poorly differentiated; grade 4, undifferentiated) (30,31) and HPV status. Unpaired student's t-test was used to assess transcriptional expression and P<0.05 was considered to indicate a statistically significant difference.
Tumor infiltration cell (TIC) profile analysis
Based on the validated leukocyte gene signature matrix (LM22), the CIBERSORT (http://cibersort.stanford.edu/) computational method was used to analyze the infiltration ratio of 22 TICs in each HNSCC sample, and the ratios of all were equal to 1.
Barplot, corrplot, vioplot, Venn and scatter plot
R 64.4.0.0 software (https://www.r-project.org) was used to plot associations with TICs. The Barplot and corrplot were generated using the corrplot package (version 0.84, http://cran.r-project.org/src/contrib/Archive/corrplot/), while vioplot was produced using the BiocManager (version 3.12, http://www.bioconductor.org/install) and vioplot packages (version 0.3.4, http://cran.r-project.org/src/contrib/Archive/vioplot/). The Venn plot was generated using the Venn Diagram package (http://www.rdocumentation.org/packages/VennDiagram/versions/1.6.18). The scatter plot was generated using the ggplot2 (version 3.2.1, http://cran.r-project.org/src/contrib/Archive/ggplot2/), ggpubr (version 0.2.4, http://cran.r-project.org/src/contrib/Archive/ggpubr/) and ggExtra packages (version 0.9, http://cran.r-project.org/src/contrib/Archive/ggExtra/). All packages were used with standard settings.
Gene sets for enrichment analysis (GSEA)
GSEA were downloaded from the Broad Institute Website (version 4.0.2, http://software.broadinstitute.org/gsea/index.jsp). HALLMARK, Kyoto Encyclopedia of Genes and Genomes (KEGG) and IMMUNE SIGNATURE gene sets were obtained from the Molecular Signatures Database (http://www.gsea-msigdb.org/gsea/msigdb/index.jsp). The transcriptome of patients with HNSCC were assessed using GSEA software (version 4.0.2, http://software.broadinstitute.org/gsea/downloads.jsp) and all the samples with nominal (NOM) P<0.01 and false discovery rate (FDR) Q<0.06 were considered to indicate a statistically significant difference.
Tissue microarray (TMA) and ethical approval
From January 2020 to December 2020, 70 patients with primary HNSCC who underwent radical resection of tumors, did not receive preoperative radiotherapy or chemotherapy in the People's Hospital of Tongxu County (Kaifeng, China) and provided written informed consent were enrolled in the present study, and their tumor tissue samples were collected. A total of 11 patients with benign head and neck lesions were included, informed consent was signed and normal tissue samples adjacent to benign lesions were collected. Patients with HNSCC were between 32 and 79 years old, with a mean age of 59.9 years. Human tissue specimens were obtained from patients who had provided written informed consent and the present study and tissue collection were approved by the Ethics Supervision Committee of The People's Hospital of Tongxu County (Kaifeng, China; approval no. TX20NP003) and the National Human Genetic Resources Sharing Service Platform (2005DKA21300). The study protocol was approved by the Ethics Committee of Union Hospital, Tongji Medical College, Huazhong University of Science and Technology (Wuhan, China; approval no. 2020IEC-J050).
Immunohistochemistry (IHC)
The tissue samples were fixed using 10% formalin solution at room temperature for 24 h, dehydrated, embedded in paraffin and sectioned into 4 µm sections. The expression of CTHRC1 protein was assessed using the immunohistochemistry ultrasensitive TMS-P method. After the sections were deparaffinized, hydrated and washed, they were placed in EDTA repair solution (pH 9.0), and boiled using a microwave oven for 2 min, and then cooled to room temperature naturally. Then sections were blocked using 3% H2O2 solution for 15 min at room temperature in the dark room. The sections were washed and incubated overnight at 4°C with CTHRC1 antibodies (1:200; cat no. 16534-1-AP; Wuhan Sanying Biotechnology). After washing three times with phosphate buffered saline, the sections were stained with secondary antibodies using the Ready-to-use Ultrasensitive™ SP kit (cat. no. Kit-9720; Fuzhou Maxin Biotechnology Development Co., Ltd.) and incubated for 30 min in a wet box at 37°C. DAB horseradish peroxidase color development kit (cat no. P0202; Beyotime Institute of Biotechnology) was used for color development. The sections were re-stained using hematoxylin staining solution (cat no. C0107; Beyotime Institute of Biotechnology) for 2 min at room temperature, dehydrated using ethanol, sealed and imaged using a Pannoramic MIDI scanner (3DHISTECH, Ltd.) and ImageJ software (version 1.8.0, National Institutes of Health) was used for image analysis. The cells on the microarrays were scored according to the staining intensity of the marker, as follows: 0, no coloring; 1, light yellow; 2, brown-yellow; and 3, tan. The proportion of cells under each score was then recorded, and the H-score value of each specimen was calculated. H-scores were defined as: (1× percentage of cells staining at 1) + (2× percentage of cells staining at 2) + (3× percentage of cells staining at 3).
Statistical analysis
SPSS 26.0 software (IBM Corp.) was used for statistical analyses. Fisher's exact test was used to analyze the relationship between clinical characteristics and CTHRC1 mRNA expression levels. The Mann-Whitney U test was used for calculating statistical differences in the H-scores in TMA and the ratio differentiation level of 21 types of immune cells in high and low CTHRC1 expression groups in HNSCC. Pearson correlation coefficient analysis was used to assess the correlation value between two kinds of cells. P<0.05 was considered to indicate a statistically significant difference.
Results
CTHRC1 was upregulated in multiple human cancer types
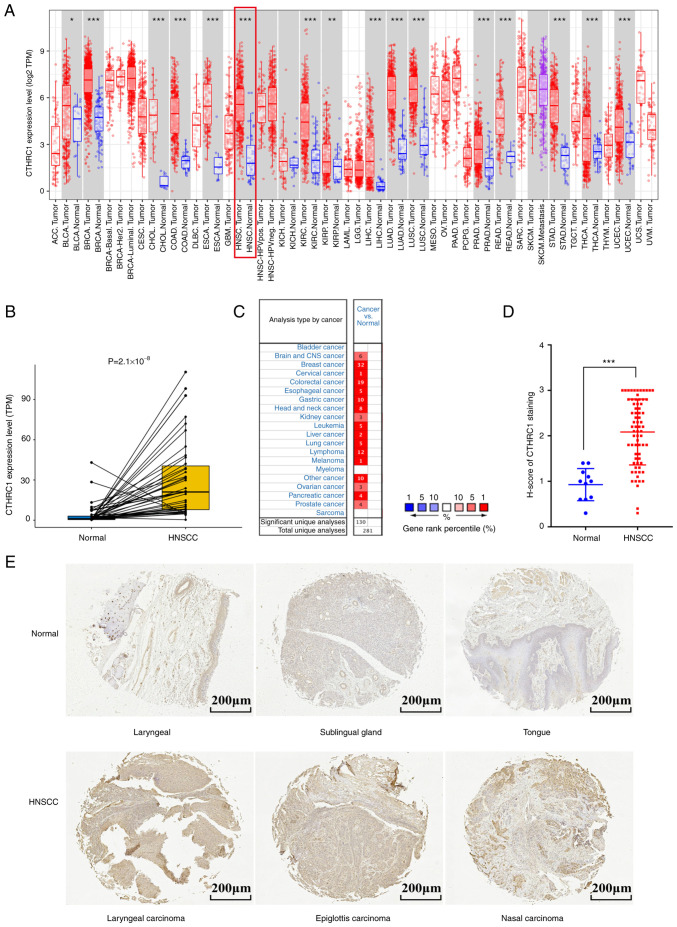
To evaluate the distinct prognostic and potential therapeutic value of CTHRC1 in HNSCC, the mRNA expression level of CTHRC1 was studied using the TCGA, ONCOMINE and TIMER databases. Data from the TCGA database showed that CTHRC1 was highly expressed in 16 cancer types, including HNSCC (Fig. 1A and B). Results from the ONCOMINE database also showed that the mRNA expression levels of CTHRC1 were significantly upregulated in multiple tumor tissues compared with normal tissues (Fig. 1C). Furthermore, the same result was demonstrated by HNSCC clinical tissues samples when compared with normal tissues (Fig. 1D and E). Taken together, these data indicated that CTHRC1 was enriched in multiple human cancer types.
Figure 1.
CTHRC1 expression levels in different types of human cancer. (A) The transcription levels of human CTHRC1 in different tumor types in the TCGA database were assessed using TIMER. (B) CTHRC1 transcriptional levels in adjacent and tumor tissues in patients with HNSCC from the TCGA database. (C) Expression level of CTHRC1 in different cancer datasets compared with normal tissues in the ONCOMINE database, the number indicated the number of data sets included by The National Center for Biotechnology Information that matched the conditions. (D) Scatter plot of CTHRC1 expression (using H-score) in HNSCC tissues compared with normal tissues. (E) Representative immunohistochemistry of CTHRC1 in clinical specimens. *P<0.05, **P<0.01, ***P<0.001. TCGA, The Cancer Genome Atlas; ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangio carcinoma; CNS, central nervous system; COAD, colon adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; HPV, human papillomavirus; neg, negative; pos, positive; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LGG, (brain) lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumor; THCA, thyroid carcinoma; THYM, thymoma, UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma; CTHRC1, collagen triple helix repeat containing 1; TPM, transcripts per million.
Association of CTHRC1 expression with clinicopathological parameters of patients with HNSCC
Since clinical pathology can determine the progression and prognosis of diseases, the association of transcriptional levels of CTHRC1 with clinicopathological parameters in patients with HNSCC was investigated using UALCAN. As presented in Fig. 2 and Table I, the transcriptional level of CTHRC1 was significantly associated with age (Fig. 2A), TP53 mutation (Fig. 2B), HPV status (Fig. 2C), tumor grade (Fig. 2D), nodal metastasis status (Fig. 2E) and individual cancer stages (Fig. 2F). The mRNA expression level of CTHRC1 increased markedly with age. Patients with TP53 mutations and those who were HPV-negative also had higher CTHRC1 mRNA expression levels. CTHRC1 mRNA expression levels increased with tumor progression and, in general, the more lymph node metastases in patients, the higher the mRNA expression level of CTHRC1. Among the patients, the transcriptional level of CTHRC1 in the N2 group (4–9 axillary lymph node metastasis) was lower than that of the N3 group (≥10 axillary lymph node metastasis), which may be due to the limited sample size. Additionally, the mRNA expression level of CTHRC1 in pathological stage IV patients was significantly higher than those in stage III or II, but markedly lower than that in patients with stage I, which may also be due to the difference in sample size. In summary, these results suggested that the mRNA expression level of CTHRC1 was closely related to the clinicopathological parameters of patients with HNSCC.
Figure 2.
Relationship between mRNA expression levels of CTHRC1 and the clinical characteristics of patients with HNSCC. Relationship between CTHRC1 mRNA expression level and (A) age, (B) TP53 mutation, (C) HPV status, (D) tumor grade, (E) nodal metastasis status and (F) cancer stages in HNSCC patients. *P<0.05, **P<0.01, ***P<0.001. CTHRC1, collagen triple helix repeat containing 1; HNSCC, head and neck squamous cell carcinoma; HPV, human papillomavirus; TCGA, The Cancer Genome Atlas; Yrs, years.
Table I.
Relationship between clinical characteristics and CTHRC1 expression in patients with head and neck squamous cell carcinoma.
| Characteristic | Low expression of CTHRC1, n=251 (%) | High expression of CTHRC1, n=250 (%) | P-value |
|---|---|---|---|
| Sex, n | 0.978 | ||
| Female | 67 (26.7) | 67 (26.8) | |
| Male | 184 (73.3) | 183 (73.2) | |
| Age, n | 0.072 | ||
| <60 years | 121 (48.4) | 100 (40.0) | |
| ≥60 years | 129 (51.6) | 150 (60.0) | |
| HPV | 0.009 | ||
| status, n | |||
| Negative | 196 (81.3) | 216 (90.0) | |
| Positive | 45 (18.7) | 24 (10.0) | |
| Tumor | 0.118 | ||
| stage, n | |||
| 1 | 20 (9.0) | 26 (11.7) | |
| 2 | 73 (32.7) | 59 (26.6) | |
| 3 | 54 (24.2) | 42 (18.9) | |
| 4 | 76 (34.1) | 95 (42.8) | |
| Node | 0.412 | ||
| stage, n | |||
| 0 | 93 (45.8) | 78 (38.0) | |
| 1 | 32 (15.8) | 33 (16.1) | |
| 2 | 75 (36.9) | 90 (43.9) | |
| 3 | 3 (1.5) | 4 (2.0) | |
| Metastasis | >0.999 | ||
| stage, n | |||
| 0 | 97 (99.0) | 89 (100.0) | |
| 1 | 1 (1.0) | 0 (0) | |
| Pathological | 0.070 | ||
| Stage, n | |||
| I | 10 (4.6) | 15 (6.9) | |
| II | 44 (20.4) | 26 (12.0) | |
| III | 41 (19.0) | 37 (17.1) | |
| IV | 121 (56.0) | 139 (64.1) | |
| Grade, n | 0.016 | ||
| 1 | 39 (16.2) | 22 (9.1) | |
| 2 | 150 (62.2) | 150 (62.2) | |
| 3 | 50 (20.7) | 69 (28.6) | |
| 4 | 2 (0.8) | 0 (0) | |
| Radiation | 0.985 | ||
| therapy, n | |||
| No | 77 (34.8) | 73 (34.9) | |
| Yes | 144 (65.2) | 136 (65.1) |
A total of 501 patients were included, including 251 patients in the low expression group and 250 patients in the high expression group, certain patients had incomplete clinical information available. In each characteristic, patients lacking that information were excluded. CTHRC1, collagen triple helix repeat containing 1; HPV, human papillomavirus.
Potential value of CTHRC1 mRNA expression level in assessment of the survival time of patients with HNSCC
Since CTHRC1 was differentially expressed in diverse cancer types, the relationship between CTHRC1 expression levels and the survival time of patients with HNSCC was next evaluated using the KM plotter. Patients were grouped, into CTHRC1 high and CTHRC1 low expression groups, using the auto select best cutoff function on the KM plotter website (32). The data indicated that the CTHRC1 high expression group had a worse OS (Fig. 3A) and RFS time (Fig. 3B). These results demonstrated that CTHRC1 had the potential to be a prognostic biomarker in HNSCC.
Figure 3.
KM survival analysis of CTHRC1 in head and neck squamous cell carcinoma from the KM Plotter database. (A) Overall survival and (B) relapse-free survival. CTHRC1, collagen triple helix repeat containing 1; HR, hazard ratio; KM, Kaplan-Meier.
CTHRC1 has a potential role in mediating tumor progression
In order to further investigate the potential function of CTHRC1, GSEA of the data from patients with HNSCC was conducted. A total of eight HALLMARK gene sets (Fig. 4A), 14 KEGG gene sets (Fig. 4B) and 1,231 immune signature gene sets (Fig. 4C) were significantly enriched in the high CTHRC1 expression groups (NOM P<0.01; FDR Q<0.06). These gene sets included angiogenesis, apical junction, coagulation, epithelial mesenchymal transformation (EMT), the KRAS signaling pathway, the notch signaling pathway, glycosaminoglycan biosynthesis, chondroitin sulfate and the TGF-β signaling pathway. Most of these gene sets serve pivotal roles in tumorigenesis. Therefore, CTHRC1 may mediate immune cell infiltration during cancer development, and it may be regarded as an important indicator for cancer progression.
Figure 4.
Gene sets for enrichment analysis for patients with head and neck squamous cell carcinoma with high or low CTHRC1 expression. Each row represents a specific set of genes with a unique color. Enriched gene sets with high expression of CTHRC1 in (A) HALLMARK, (B) KEGG and (C) IMMUNE SIGNITURE. Representative gene sets are shown. Nominal P<0.01, false discovery rate Q<0.06. CTHRC1, collagen triple helix repeat containing 1; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Association of CTHRC1 with immune cell infiltration in HNSCC
The immune system serves pivotal roles in the tumorigenesis of HNSCC (3). The aforementioned data showed that CTHRC1 may be involved in immune responses and tumor associated pathways. Therefore, the association between CTHRC1 expression and immune cell invasion in patients with HNSCC from the TIMER database was evaluated (Fig. 5A). The results indicated that CTHRC1 expression was significantly correlated with the degree of infiltration of B cells, CD4+ T cells, neutrophils and dendritic cells (DCs) in HPV negative patients with HNSCC (Fig. 5B). However, no significant correlation was demonstrated for these groups in HPV positive patients with HNSCC. The expression of CTHRC1 was significantly associated with the infiltration of macrophages in both HPV negative and positive patients with HNSCC (Fig. 5C). This suggested that the relationship between CTHRC1 expression and macrophage infiltration was not related to HPV infection. Collectively, these results demonstrated that CTHRC1 expression was significantly associated with immune cell infiltration in HNSCC.
Figure 5.
Correlation between tumor-infiltrating immune cell proportions and CTHRC1 expression in HNSCC in the Tumor Immune Estimation Resource database. (A) HNSCC, (B) HPVneg HNSCC and (C) HPVpos HNSCC. CTHRC1, collagen triple helix repeat containing 1; HNSCC, head and neck squamous cell carcinoma; HPV, human papillomavirus; neg, negative; pos, positive.
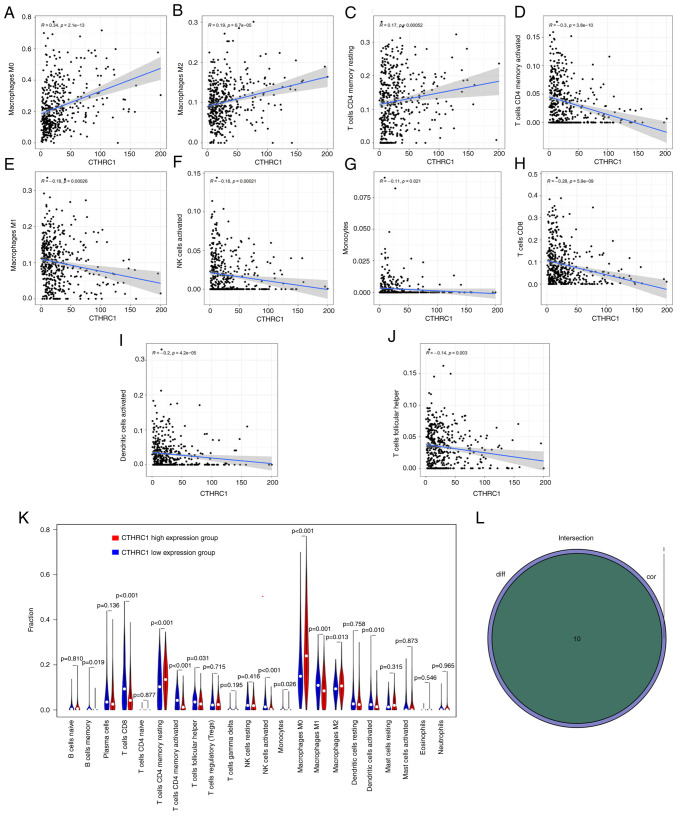
The correlation of CTHRC1 expression with the proportion and distribution of TICs in HNSCC
The infiltration of 22 TICs in each HNSCC sample and the CIBERSORT algorithm was used to further evaluate the correlation between the transcriptional levels of CTHRC1 and the immune microenvironment (Fig. 6A and B). Correlation analysis indicated that the transcriptional levels of CTHRC1 were significantly positively correlated with M0 macrophages (P=2.1×10−13; Fig. 7A), M2 macrophages (P=6.7×10−5; Fig. 7B) and resting CD4+ memory T cells (P=5.2×10−4; Fig. 7C), and were significantly negatively associated with the levels of activated CD4+ memory T cells (P=3.8×10−10; Fig. 7D), M1 macrophages (P=2.6×10−4; Fig. 7E), activated natural killer (NK) cells (P=2.1×10−4; Fig. 7F), monocytes (P=2.1×10−2; Fig. 7G), CD8+ T cells (P=5.9×10−9; Fig. 7H), activated DCs (P=4.2×10−5; Fig. 7I) and follicular helper T (Tfh) cells (P=3.0×10−3; Fig. 7J) (Table II). Analysis demonstrated that the transcriptional levels of CTHRC1 were correlated with the infiltration of 10 types of TICs, including resting CD4+ memory T cells, memory B cells, activated CD4+ memory T cells, CD8+ T cells, Tfh cells, M0 macrophages, activated NK cells, M1 macrophages, M2 macrophages and activated DCs (Fig. 7K and L). The high CTHRC1 expression group tended to have a larger proportion of resting CD4+ memory T cells, M0 macrophages and M2 macrophages compared with the low CTHRC1 expression group, whereas the low CTHRC1 expression group had a larger proportion of resting CD4+ memory T cells, memory B cells, activated CD4+ memory T cells, CD8+ T cells, Tfh cells, activated NK cells, M1 macrophages and activated DCs compared with the high CTHRC1 expression group.
Figure 6.
TIC profiles and correlation analysis of HNSCC samples. (A) The proportion of 22 types of TICs in HNSCC tumor samples. (B) Heatmap showing the correlation between 22 kinds of TICs, the number in each box indicates the corresponding correlation value between two kinds of cells. CTHRC1, collagen triple helix repeat containing 1; HNSCC, with head and neck squamous cell carcinoma; TICs, tumor-infiltrating immune cells.
Figure 7.
Association between the percentage of each TIC and the CTHRC1 expression level. (A-J) The association of the percentage of 10 types of TICs with the transcriptional levels of CTHRC1. The linear model of immune cell proportional tendency and CTHRC1 expression was fitted by the gray line, and Pearson coefficient was used to test the correlation. (K) Violin plot indicating the ratio differentiation level of 21 types of immune cells in high and low CTHRC1 expression groups in head and neck squamous cell carcinoma tumor samples. (L) Venn plot displaying 10 kinds of TICs correlated with CTHRC1 expression codetermined by difference and correlation tests displayed in violin and scatter plots, respectively. CTHRC1, collagen triple helix repeat containing 1; NK, natural killer; TICs, tumor-infiltrating immune cells.
Table II.
Correlation analysis between collagen triple helix repeat containing 1 and related gene markers of tumor-infiltrating immune cells in Tumor Immune Estimation Resource.
| Nonea | Purityb | ||||
|---|---|---|---|---|---|
|
|
|
||||
| Cell type | Gene marker | Corc | P-value | Cor | P-value |
| CD8+ T cell | CD8A | −0.005 | 9.19×10−1 | −0.277 | 4.03×10−10 |
| CD8B | 0.048 | 2.90×10−1 | −0.242 | 5.06×10−8 | |
| T cell (general) | CD3D | 0.029 | 5.22×10−1 | −0.298 | 1.35×10−11 |
| CD3E | 0.110 | 1.49×10−2 | −0.299 | 1.17×10−11 | |
| CD2 | 0.116 | 9.77×10−3 | −0.285 | 1.15×10−10 | |
| B cell | CD19 | 0.028 | 5.36×10−1 | −0.261 | 4.33×10−9 |
| CD79A | 0.053 | 2.43×10−1 | −0.228 | 3.05×10−7 | |
| Monocyte | CD86 | 0.358 | 2.55×10−16 | −0.295 | 2.26×10−11 |
| CSF1R | 0.415 | 5.94×10−22 | −0.304 | 4.98×10−12 | |
| TAM | CCL2 | 0.394 | 1.07×10−19 | −0.258 | 5.98×10−9 |
| CD68 | 0.272 | 9.12×10−10 | −0.172 | 1.24×10−4 | |
| IL10 | 0.332 | 4.03×10−14 | −0.313 | 1.21×10−12 | |
| M1 Macrophage | NOS2 | 0.122 | 6.55×10−3 | 0.071 | 1.16×10−1 |
| IRF5 | 0.103 | 2.26×10−2 | −0.001 | 9.75×10−1 | |
| PTGS2 | −0.006 | 8.96×10−1 | 0.1 | 2.67×10−2 | |
| M2 Macrophage | CD163 | 0.399 | 3.28×10−20 | −0.286 | 1.02×10−10 |
| VSIG4 | 0.425 | 5.06×10−23 | −0.257 | 7.42×10−9 | |
| MS4A4A | 0.438 | 1.69×10−24 | −0.287 | 8.35×10−11 | |
| Neutrophils | CEACAM8 | 0.001 | 9.90×10−1 | 0.034 | 4.48×10−1 |
| ITGAM | 0.416 | 5.06×10−23 | −0.137 | 2.27×10−3 | |
| CCR7 | 0.194 | 1.53×10−5 | −0.322 | 2.23×10−13 | |
| Natural killer cell | KIR2DL1 | −0.006 | 8.93×10−1 | −0.093 | 3.81×10−2 |
| KIR2DL3 | −0.01 | 8.20×10−1 | −0.136 | 2.56×10−3 | |
| KIR2DL4 | −0.134 | 2.89×10−3 | −0.183 | 4.30×10−5 | |
| KIR3DL1 | −0.042 | 3.48×10−1 | −0.144 | 1.35×10−3 | |
| KIR3DL2 | 0.047 | 2.97×10−1 | −0.147 | 1.05×10−3 | |
| KIR3DL3 | −0.045 | 3.23×10−1 | −0.086 | 5.68×10−2 | |
| KIR2DS4 | 0.017 | 7.12×10−1 | −0.148 | 9.93×10−4 | |
| Dendritic cell | HLA-DPB1 | 0.223 | 6.05×10−7 | −0.302 | 7.78×10−12 |
| HLA-DQB1 | 0.178 | 7.19×10−5 | −0.228 | 3.28×10−7 | |
| HLA-DRA | 0.194 | 1.41×10−5 | −0.299 | 1.16×10−11 | |
| HLA-DPA1 | −0.209 | 7.58×10−2 | −0.52 | 2.00×10−6 | |
| BDCA-1 (CD1C) | −0.01 | 9.34×10−1 | −0.58 | 5.92×10−8 | |
| BDCA-4 (NRP1) | 0.455 | 5.15×10−5 | 0.065 | 5.80×10−1 | |
| CD11c (ITGAX) | −0.064 | 5.88×10−1 | −0.387 | 6.55×10−4 | |
| Th1 | T-bet (TBX21) | 0.166 | 1.59×10−1 | −0.617 | 4.68×10−9 |
| STAT4 | 0.133 | 2.64×10−1 | −0.496 | 7.06×10−6 | |
| STAT1 | 0.291 | 1.26×10−2 | 0.05 | 6.69×10−1 | |
| IFN-γ (IFNG) | 0.092 | 4.37×10−1 | −0.421 | 1.85×10−4 | |
| TNF-α (TNF) | 0.112 | 1.03×10−1 | −0.467 | 2.7×10−5 | |
| Th2 | GATA3 | 0.248 | 2.63×10−8 | −0.227 | 3.63×10−7 |
| STAT6 | 0.056 | 2.12×10−1 | 0.069 | 1.26×10−1 | |
| STAT5A | 0.188 | 2.81×10−5 | −0.132 | 3.28×10−3 | |
| IL13 | 0.071 | 1.15×10−1 | −0.154 | 5.78×10−4 | |
| Tfh | BCL6 | 0.192 | 1.03×10−1 | 0.25 | 3.19×10−2 |
| IL21 | NA | NA | NA | NA | |
| Th17 | STAT3 | 0.124 | 2.96×10−1 | 0.062 | 5.98×10−1 |
| IL17A | 0.236 | 4.42×10−2 | −0.238 | 4.09×10−2 | |
| Treg | FOXP3 | 0.312 | 7.25×10−3 | 0.009 | 9.37×10−1 |
| CCR8 | 0.129 | 2.77×10−1 | −0.33 | 4.11×10−3 | |
| STAT5B | 0.008 | 9.44×10−1 | −0.036 | 7.59×10−1 | |
| TGFβ (TGFB1) | 0.49 | 1.08×10−5 | −0.263 | 2.35×10−2 | |
| T cell exhaustion | PD-1 (PDCD1) | 0.109 | 3.58×10−1 | −0.581 | 5.89×10−8 |
| CTLA4 | 0.206 | 8.04×10−2 | −0.504 | 4.78×10−6 | |
| LAG3 | 0.195 | 9.89×10−2 | −0.259 | 2.61×10−2 | |
| TIM-3 (HAVCR2) | −0.001 | 9.94×10−1 | −0.552 | 3.34×10−7 | |
| GZMB | 0.062 | 6.05×10−1 | −0.406 | 3.33×10−4 | |
None, correlation without adjustment.
Purity, correlation adjusted by purity.
Cor, R value of Spearman's correlation. TAM, tumor-associated macrophage; Th, T helper cell; Tfh, follicular helper T cell; Treg, regulatory T cell; NA, not available in database.
Discussion
HNSCC is among the six most common types of human tumor. Despite the development of medical technology the survival rate of patients with HNSCC is still low (33). Abnormal gene expression or mutations may be closely related to the occurrence, development and prognosis of tumors. However, the molecular mechanisms of HNSCC still need to be investigated. In the present study, it was found that CTHRC1 was abnormally upregulated in HNSCC and was significantly associated with age, TP53 mutation, nodal metastasis status, individual cancer stages and HPV status. Furthermore, high expression levels of CTHRC1 were significantly associated with shorter OS and RFS times in patients with HNSCC. These results demonstrated that CTHRC1 may function as a pro-oncogene in HNSCC.
CTHRC1 is a secretory glycoprotein which negatively regulates the deposition of collagen matrix and is involved in vascular remodeling and cell migration (10). In previous studies, the mRNA and protein expression levels of CTHRC1 in oral squamous cell carcinoma samples were found to be higher than those in normal specimens (34), and were associated with metastasis in tongue squamous cell carcinoma (35). Therefore, it could be hypothesized that CTHRC1 may be involved in the development of head and neck tumors.
In the present study, analysis indicated that upregulation of CTHRC1 was mainly involved in tumor and immune-related pathways, such as angiogenesis, apical junction, EMT and the KRAS and TGF-β signaling pathways. The EMT signaling pathway is associated with visibility, acquisition of mobility and self-renewal (36). The abnormal activation of the KRAS (37), notch (38) and TGF-β (39) signaling pathways are closely related to tumorigenesis. In the present study, upregulation of CTHRC1 was indicated in 1,231 immune related pathways, which suggested a potential regulatory role of CTHRC1 in the tumor immune microenvironment. Furthermore, the results also showed that CTHRC1 was significantly positively associated with M0 macrophages, M2 macrophages and resting CD4+ memory T cells, and significantly negatively associated with the levels of activated CD4+ memory T cells, activated NK cells, M1 macrophages, CD8+ T cells, monocytes, activated DCs and Tfh cells. Moreover, the high CTHRC1 expression group tended to have a larger proportion of M0 macrophages and M2 macrophages compared with the low CTHRC1 expression group, while the low CTHRC1 expression group had a larger proportion of M1 macrophages compared with the high CTHRC1 expression group. A previous study reported that CTHRC1 can activate the STAT6 signaling pathway, induce the M2-like macrophage phenotype in a dose-dependent manner, and improve the migration and invasion ability of ovarian cancer cells (40). Therefore, CTHRC1 may mediate the occurrence and development of HNSCC by mediating macrophage polarization, which leads to poor prognosis.
There are several limitations and challenges in the present study. Firstly, the sample size is small. Larger studies in HNSCC are needed to confirm the results. Secondly, CTHRC1 expression may not be a highly specific diagnostic and prognostic biomarker of HNSCC in humans, but be a shared diagnostic and prognostic biomarker of survival in different human cancers (23,24). These may limit its use in clinical diagnosis and prognosis.
In conclusion, the upregulation of CTHRC1 in patients with HNSCC is related to the pathological grade, Tumor-Node-Metastasis stage, lymphatic metastasis, HPV status, TP53 mutation and TICs, which may lead to poor prognosis in patients with HNSCC. In addition, upregulation of CTHRC1 may activate tumor and immune related pathways, leading to the infiltration of immune cells and macrophage polarization in the tumor microenvironment in HNSCC. The present study identified a potential role of CTHRC1 in immunology and its adverse prognostic value in HNSCC. CTHRC1 should therefore be considered as a prognostic marker and therapeutic target for HNSCC.
Acknowledgements
Not applicable.
Funding Statement
This work was financially supported by the Hubei Provincial Natural Science Foundation of China (grant no. 2019CFB470) and Hubei Key Laboratory of Biological Targeted Therapy, China (grant no. 2020swbx014).
Availability of data and materials
The data used in the present study may be accessed from the TCGA (https://tcga-data.nci.nih.gov/tcga/, head and neck squamous cell carcinoma, the RNA-seq data ‘workflow type, HTSeq-FPKM’ and relevant clinical data for the ‘TCGA-HNSC’ cohort), oncomine (https://www.oncomine.org/resource/main.html, Gene, CTHRC1; cut-off P-value, 0.05; cut-off fold change, 1), KM plotter (http://kmplot.com/analysis/, head and neck squamous cell carcinoma, KM plotter contains data and clinical information from the Gene Expression Omnibus, TCGA and European Genome-Phenome Archive databases), UALCAN (http://ualcan.path.uab.edu, TCGA-head and neck squamous cell carcinoma, contains level 3 RNA-seq data and clinical information from the TCGA database and is an interactive and comprehensive web resource for analyzing cancer omics data) and Timer (https://cistrome.shinyapps.io/timer/, head and neck squamous cell carcinoma, gene expression levels were presented as log2 transcripts per million) databases. The remaining datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LW and JHZ proposed the idea for and designed the study. RLZ, MLY and RZ completed the data analysis work, RLZ and MLY drew the figures, MLY and LW drafted the manuscript and JHZ reviewed the manuscript. RLZ, MLY and LW confirm the authenticity of all the raw data. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Human tissue specimens were obtained from patients who provided written informed consent and the present study and tissue collection was approved by the Ethics Supervision Committee of The People's Hospital of Tongxu County (Kaifeng, China; approval no. TX20NP003) and the National Human Genetic Resources Sharing Service Platform (2005DKA21300). The study protocol was approved by the Ethics Committee of Union Hospital, Tongji Medical College, Huazhong University of Science and Technology (Wuhan, China; approval no. 2020IEC-J050).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Wyss A, Hashibe M, Chuang SC, Lee YC, Zhang ZF, Yu GP, Winn DM, Wei Q, Talamini R, Szeszenia-Dabrowska N, et al. Cigarette, cigar, and pipe smoking and the risk of head and neck cancers: Pooled analysis in the international head and neck cancer epidemiology consortium. Am J Epidemiol. 2013;178:679–690. doi: 10.1093/aje/kwt029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Ferris RL. Immunology and immunotherapy of head and neck cancer. J Clin Oncol. 2015;33:3293–3304. doi: 10.1200/JCO.2015.61.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 5.Kuss I, Hathaway B, Ferris RL, Gooding W, Whiteside TL. Decreased absolute counts of T lymphocyte subsets and their relation to disease in squamous cell carcinoma of the head and neck. Clin Cancer Res. 2004;10:3755–3762. doi: 10.1158/1078-0432.CCR-04-0054. [DOI] [PubMed] [Google Scholar]
- 6.Bauernhofer T, Kuss I, Henderson B, Baum AS, Whiteside TL. Preferential apoptosis of CD56dim natural killer cell subset in patients with cancer. Eur J Immunol. 2003;33:119–124. doi: 10.1002/immu.200390014. [DOI] [PubMed] [Google Scholar]
- 7.Ferris RL. Progress in head and neck cancer immunotherapy: Can tolerance and immune suppression be reversed? ORL J Otorhinolaryngol Relat Spec. 2004;66:332–340. doi: 10.1159/000081891. [DOI] [PubMed] [Google Scholar]
- 8.Burtness B, Harrington KJ, Greil R, Soulières D, Tahara M, de Castro G, Jr, Psyrri A, Basté N, Neupane P, Bratland Å, et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): A randomised, open-label, phase 3 study. Lancet. 2019;394:1915–1928. doi: 10.1016/S0140-6736(19)32591-7. [DOI] [PubMed] [Google Scholar]
- 9.Ferris RL, Blumenschein G, Jr, Fayette J, Guigay J, Colevas AD, Licitra L, Harrington K, Kasper S, Vokes EE, Even C, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. New Engl J Med. 2016;375:1856–1867. doi: 10.1056/NEJMoa1602252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pyagay P, Heroult M, Wang Q, Lehnert W, Belden J, Liaw L, Friesel RE, Lindner V. Collagen triple helix repeat containing 1, a novel secreted protein in injured and diseased arteries, inhibits collagen expression and promotes cell migration. Circ Res. 2005;96:261–268. doi: 10.1161/01.RES.0000154262.07264.12. [DOI] [PubMed] [Google Scholar]
- 11.Duan X, Yuan X, Yao B, Song W, Li Z, Enhejirigala, Kong Y, Wang Y, Fu X, Huang S. The role of CTHRC1 in promotion of cutaneous wound healing. Signal Transduct Target Ther. 2022;7:183. doi: 10.1038/s41392-022-01008-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bian Z, Miao Q, Zhong W, Zhang H, Wang Q, Peng Y, Chen X, Guo C, Shen L, Yang F, et al. Treatment of cholestatic fibrosis by altering gene expression of Cthrc1: Implications for autoimmune and non-autoimmune liver disease. J Autoimmun. 2015;63:76–87. doi: 10.1016/j.jaut.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeshita S, Fumoto T, Matsuoka K, Park KA, Aburatani H, Kato S, Ito M, Ikeda K. Osteoclast-secreted CTHRC1 in the coupling of bone resorption to formation. J Clin Invest. 2013;123:3914–3924. doi: 10.1172/JCI69493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stohn JP, Wang Q, Siviski ME, Kennedy K, Jin YR, Kacer D, DeMambro V, Liaw L, Vary CP, Rosen CJ, et al. Cthrc1 controls adipose tissue formation, body composition, and physical activity. Obesity (Silver Spring) 2015;23:1633–1642. doi: 10.1002/oby.21144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ni SJ, Ren F, Xu M, Tan C, Weng W, Huang Z, Sheng W, Huang D. CTHRC1 overexpression predicts poor survival and enhances epithelial-mesenchymal transition in colorectal cancer. Cancer Med. 2018;7:5643–5654. doi: 10.1002/cam4.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding X, Huang R, Zhong Y, Cui N, Wang Y, Weng J, Chen L, Zang M. CTHRC1 promotes gastric cancer metastasis via HIF-1α/CXCR4 signaling pathway. Biomed Pharmacother. 2020;123:109742. doi: 10.1016/j.biopha.2019.109742. [DOI] [PubMed] [Google Scholar]
- 17.Yan L, Yu J, Tan F, Ye GT, Shen ZY, Liu H, Zhang Y, Wang JF, Zhu XJ, Li GX. SP1-mediated microRNA-520d-5p suppresses tumor growth and metastasis in colorectal cancer by targeting CTHRC1. Am J Cancer Res. 2015;5:1447–1459. [PMC free article] [PubMed] [Google Scholar]
- 18.Ma MZ, Zhuang C, Yang XM, Zhang ZZ, Ma H, Zhang WM, You H, Qin W, Gu J, Yang S, et al. CTHRC1 acts as a prognostic factor and promotes invasiveness of gastrointestinal stromal tumors by activating Wnt/PCP-Rho signaling. Neoplasia. 2014;16:265–278. 278, e1–e13. doi: 10.1016/j.neo.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou MZ, Cheng ZQ, Shen HW, He S, Li Y, Pan Y, Feng C, Chen X, Zhang Y, Lin M, et al. High expression of CTHRC1 promotes EMT of epithelial ovarian cancer (EOC) and is associated with poor prognosis. Oncotarget. 2015;6:35813–35829. doi: 10.18632/oncotarget.5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C, Li Z, Shao F, Yang X, Feng X, Shi S, Gao Y, He J. High expression of collagen triple helix repeat containing 1 (CTHRC1) facilitates progression of oesophageal squamous cell carcinoma through MAPK/MEK/ERK/FRA-1 activation. J Exp Clin Canc Res. 2017;36:84. doi: 10.1186/s13046-017-0555-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng M, Zhou Q, Liu X, Wang C, Liu G. CTHRC1 overexpression promotes cervical carcinoma progression by activating the Wnt/PCP signaling pathway. Oncol Rep. 2019;41:1531–1538. doi: 10.3892/or.2019.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan RX, Bao D, Zhang Y. Linc00707 promotes cell proliferation, invasion, and migration via the miR-30c/CTHRC1 regulatory loop in breast cancer. Eur Rev Med Pharmaco. 2020;24:4863–4872. doi: 10.26355/eurrev_202005_21175. [DOI] [PubMed] [Google Scholar]
- 23.Zhou H, Su L, Liu C, Li B, Li H, Xie Y, Sun D. CTHRC1 may serve as a prognostic biomarker for hepatocellular carcinoma. Onco Targets Ther. 2019;12:7823–7831. doi: 10.2147/OTT.S219429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sial N, Ahmad M, Hussain MS, Iqbal MJ, Hameed Y, Khan M, Abbas M, Asif R, Rehman JU, Atif M, et al. CTHRC1 expression is a novel shared diagnostic and prognostic biomarker of survival in six different human cancer subtypes. Sci Rep. 2021;11:19873. doi: 10.1038/s41598-021-99321-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bai Y, Yin K, Su T, Ji F, Zhang S. CTHRC1 in ovarian cancer promotes M2-like polarization of tumor-associated macrophages via regulation of the STAT6 signaling pathway. Onco Targets Ther. 2020;13:5743–5753. doi: 10.2147/OTT.S250520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Jensen MA, Zenklusen JC. A practical guide to the cancer genome atlas (TCGA) Methods Mol Biol. 2016;1418:111–141. doi: 10.1007/978-1-4939-3578-9_6. [DOI] [PubMed] [Google Scholar]
- 27.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/1538-7445.AM2017-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reis-Filho JS, Tutt ANJ. Triple negative tumours: A critical review. Histopathology. 2008;52:108–118. doi: 10.1111/j.1365-2559.2007.02889.x. [DOI] [PubMed] [Google Scholar]
- 31.Shien T, Shimizu C, Seki K, Shibata T, Hojo T, Ando M, Kohno T, Katsumata N, Akashi-Tanaka S, Kinoshita T, Fujiwara Y. Comparison among different classification systems regarding the pathological response of preoperative chemotherapy in relation to the long-term outcome. Breast Cancer Res Treat. 2008;113:307–313. doi: 10.1007/s10549-008-9935-2. [DOI] [PubMed] [Google Scholar]
- 32.Lánczky A, Győrffy B. Web-based survival analysis tool tailored for medical research (KMplot): Development and implementation. J Med Internet Res. 2021;23:e27633. doi: 10.2196/27633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 34.Lee CE, Vincent-Chong VK, Ramanathan A, Kallarakkal TG, Karen-Ng LP, Ghani WM, Rahman ZA, Ismail SM, Abraham MT, Tay KK, et al. Collagen triple helix repeat containing-1 (CTHRC1) expression in oral squamous cell carcinoma (OSCC): Prognostic value and clinico-pathological implications. Int J Med Sci. 2015;12:937–945. doi: 10.7150/ijms.11605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu GL, Sengupta PK, Jamal B, Yang HY, Bouchie MP, Lindner V, Varelas X, Kukuruzinska MA. N-glycosylation induces the CTHRC1 protein and drives oral cancer cell migration. J Biol Chem. 2013;288:20217–20227. doi: 10.1074/jbc.M113.473785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheel C, Eaton EN, Li SHJ, Chaffer CL, Reinhardt F, Kah KJ, Bell G, Guo W, Rubin J, Richardson AL, Weinberg RA. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145:926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weidhaas JB, Harris J, Schaue D, Chen AM, Chin R, Axelrod R, El-Naggar AK, Singh AK, Galloway TJ, Raben D, et al. The KRAS-variant and cetuximab response in head and neck squamous cell cancer: A secondary analysis of a randomized clinical trial. JAMA Oncol. 2017;3:483–491. doi: 10.1001/jamaoncol.2016.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loganathan SK, Schleicher K, Malik A, Quevedo R, Langille E, Teng K, Oh RH, Rathod B, Tsai R, Samavarchi-Tehrani P, et al. Rare driver mutations in head and neck squamous cell carcinomas converge on NOTCH signaling. Science. 2020;367:1264–1269. doi: 10.1126/science.aax0902. [DOI] [PubMed] [Google Scholar]
- 39.Oshimori N, Oristian D, Fuchs E. TGF-β promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell. 2015;160:963–976. doi: 10.1016/j.cell.2015.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo B, Yan H, Li L, Yin K, Ji F, Zhang S. Collagen triple helix repeat containing 1 (CTHRC1) activates Integrin β3/FAK signaling and promotes metastasis in ovarian cancer. J Ovarian Res. 2017;10:69. doi: 10.1186/s13048-017-0358-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used in the present study may be accessed from the TCGA (https://tcga-data.nci.nih.gov/tcga/, head and neck squamous cell carcinoma, the RNA-seq data ‘workflow type, HTSeq-FPKM’ and relevant clinical data for the ‘TCGA-HNSC’ cohort), oncomine (https://www.oncomine.org/resource/main.html, Gene, CTHRC1; cut-off P-value, 0.05; cut-off fold change, 1), KM plotter (http://kmplot.com/analysis/, head and neck squamous cell carcinoma, KM plotter contains data and clinical information from the Gene Expression Omnibus, TCGA and European Genome-Phenome Archive databases), UALCAN (http://ualcan.path.uab.edu, TCGA-head and neck squamous cell carcinoma, contains level 3 RNA-seq data and clinical information from the TCGA database and is an interactive and comprehensive web resource for analyzing cancer omics data) and Timer (https://cistrome.shinyapps.io/timer/, head and neck squamous cell carcinoma, gene expression levels were presented as log2 transcripts per million) databases. The remaining datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.