Abstract
Background & Aims
Patterns of liver HBV antigen expression have been described but not quantified at single-cell resolution. We applied quantitative techniques to liver biopsies from individuals with chronic hepatitis B and evaluated sampling heterogeneity, effects of disease stage, and nucleos(t)ide (NUC) treatment, and correlations between liver and peripheral viral biomarkers.
Methods
Hepatocytes positive for HBV core and HBsAg were quantified using a novel four-plex immunofluorescence assay and image analysis. Biopsies were analysed from HBeAg-positive (n = 39) and HBeAg-negative (n = 75) participants before and after NUC treatment. To evaluate sampling effects, duplicate biopsies collected at the same time point were compared. Serum or plasma samples were evaluated for levels of HBV DNA, HBsAg, hepatitis B core-related antigen (HBcrAg), and HBV RNA.
Results
Diffusely distributed individual HBV core+ cells and foci of HBsAg+ cells were the most common staining patterns. Hepatocytes positive for both HBV core and HBsAg were rare. Paired biopsies revealed large local variation in HBV staining within participants, which was confirmed in a large liver resection. NUC treatment was associated with a >100-fold lower median frequency of HBV core+ cells in HBeAg-positive and HBeAg-negative participants, whereas reductions in HBsAg+ cells were not statistically significant. The frequency of HBV core+ hepatocytes was lower in HBeAg-negative participants than in HBeAg-positive participants at all time points evaluated. Total HBV+ hepatocyte burden correlated with HBcrAg, HBV DNA, and HBV RNA only in baseline HBeAg-positive samples.
Conclusions
Reductions in HBV core+ hepatocytes were associated with HBeAg-negative status and NUC treatment. Variation in HBV positivity within individual livers was extensive. Correlations between the liver and the periphery were found only between biomarkers likely indicative of cccDNA (HBV core+ and HBcrAg, HBV DNA, and RNA).
Impact and Implications
HBV infects liver hepatocyte cells, and its genome can exist in two forms that express different sets of viral proteins: a circular genome called cccDNA that can express all viral proteins, including the HBV core and HBsAg proteins, or a linear fragment that inserts into the host genome typically to express HBsAg, but not HBV core. We used new techniques to determine the percentage of hepatocytes expressing the HBV core and HBsAg proteins in a large set of liver biopsies. We find that abundance and patterns of expression differ across patient groups and even within a single liver and that NUC treatment greatly reduces the number of core-expressing hepatocytes.
Keywords: HBV, NUC, HBsAg, HBV core, Biomarkers, HBeAg
Abbreviations: ADV, adefovir; ALT, alanine aminotransferase; cccDNA, covalently closed circular DNA; CHB, chronic hepatitis B; CNN, convolutional neural network; dslDNA, double-stranded linear DNA; HBcrAg, hepatitis B core-related antigen; HBV core, hepatitis B core antigen; HCC, hepatocellular carcinoma; IF, immunofluorescence; Na+K+-ATPase, sodium–potassium ATPase; NUC, nucleo(t)side; QC, quality control; TDF, tenofovir disoproxil fumarate; HBeAg, Hepatitis B e antigen; HBV, Hepatitis B Virus; HBsAg, Hepatitis B surface antigen
Graphical abstract
Highlights
-
•
We applied a novel imaging technique to quantify HBsAg and HBV core antigen burden in liver biopsies from patients with CHB.
-
•
The frequency of HBV core+ hepatocytes was lower in HBeAg-negative participants than in HBeAg-positive participants.
-
•
NUC treatment was associated with a significant decline in HBV core+ cells but not in HBsAg.
-
•
Duplicate biopsies collected at the same time point revealed large local variation in HBV staining within participants.
-
•
HBV+ hepatocyte burden correlated with HBcrAg, HBV DNA, and HBV RNA only in HBeAg-positive participants at baseline.
Introduction
An estimated 260 million people worldwide are chronically infected with HBV, resulting in more than 800,000 annual deaths from cirrhosis, liver failure, and hepatocellular carcinoma (HCC).1 Current treatments for chronic hepatitis B (CHB) infection can efficiently suppress viral replication and improve patient outcome, but typically require life-long therapy and rarely lead to functional cure (defined as persistent HBsAg loss).2 Consequently, curative therapies for CHB are required.
During the HBV replication cycle, viral RNA is reverse-transcribed and processed into covalently closed circular DNA (cccDNA) that is transcribed to express all viral proteins.3 However, the viral RNA can also generate a double-stranded linear form (dslDNA) as a consequence of failed primer–template switching during reverse transcription.4 HBV dslDNA can integrate into the host genome,5 resulting in a chromosomal template that is transcribed to express HBsAg, but not the viral core protein (HBV core) or polymerase protein.4,6 These hepatocytes with integrated HBV sequences can clonally expand[7], [8], [9] and may contribute to the development of HCC.[10], [11], [12] The relative proportion of HBV in the form of cccDNA or integrated viral DNA varies at different stages of CHB natural history, with cccDNA more abundant in HBeAg-positive (HBeAg+) individuals whereas HBeAg-negative (HBeAg-) individuals have predominantly integrated HBV DNA.6,13,14 Sustained HBsAg loss in the periphery is the clinical marker of functional cure, so strategies that result in elimination of both cccDNA and integrated HBV are likely necessary to achieve this goal.
Current HBV cure strategies aim to induce immune-mediated clearance of HBV-positive hepatocytes to eliminate otherwise stable viral DNA.15 However, excessive immune-mediated hepatocyte killing could lead to potential adverse events in patients. Consequently, individuals with fewer HBV-positive cells may have a lower safety risk; however, there is no consensus on the fraction of HBV-positive hepatocytes in populations with CHB.13,15 Quantification of the percentage of HBV-positive hepatocytes across disease stages and pre- and post-nucleos(t)ide (NUC) treatment requires a method that can accurately identify and quantify HBV-positive cells in liver samples, a sufficiently large liver biopsy collection to draw meaningful conclusions in different patient groups, and a greater understanding of how well individual biopsies represent the entire liver. We developed a multiplex immunofluorescence (IF) assay and image analysis methods to describe the abundance of HBV antigen containing hepatocytes across a large liver biopsy collection to describe the HBV viral burden across different disease stages, as well as pre- and post-NUC treatment. We further evaluated correlations between HBV burden in the liver and well-established peripheral viral biomarkers.
Materials and methods
Liver biopsy collection
Liver biopsies were obtained from HBeAg+ and HBeAg- individuals enrolled in two randomised phase III clinical trials of tenofovir disoproxil fumarate (TDF): GS-US-174-0102 (ClinicalTrials.gov: NCT00117676) and GS-US-174-0103 (ClinicalTrials.gov: NCT00116805).16,17 HBeAg- (GS-US-174-0102, n = 375) and HBeAg+ (GS-US-174-0103, n = 266) participants with elevated alanine aminotransferase (ALT) were randomised (2:1) to receive TDF or adefovir dipivoxil (ADV) for 48 weeks (Fig. S1). Based on the current EASL guidelines,18 the patient populations were classified as HBeAg+ chronic hepatitis (formerly ‘immune active’) and HBeAg- chronic hepatitis. Participants were predominantly genotypes A, B, C, and D (Table 1), consistent with the geographic distribution of HBV genotypes at study sites. Within each genotype, a mixture of HBeAg+ and HBeAg- participants were present (Table 1).
Table 1.
Baseline participant population.
| HBeAg+ (n = 39) | HBeAg- (n = 75) | Total cohort (N = 114) | |
|---|---|---|---|
| Baseline demographics | |||
| Age, years | 30 (24, 42) | 47 (39, 54) | 42 (31, 51) |
| Men | 34 (87) | 64 (85) | 98 (86) |
| Women |
5 (13) |
11 (15) |
16 (14) |
| HBV genotype | |||
| A | 6 (15) | 11 (15) | 17 (15) |
| B | 5 (13) | 8 (11) | 13 (11) |
| C | 10 (26) | 6 (8) | 16 (14) |
| D | 16 (41) | 48 (64) | 64 (56) |
| E | 1 (3) | 0 | 1 (1) |
| F | 1 (3) | 0 | 1 (1) |
| G | 0 | 1 (1) | 1 (1) |
| U |
0 |
1 (1) |
1 (1) |
| Baseline peripheral biomarker values | |||
| HBV DNA, log10 IU/ml | 8.0 (7.6, 8.7); n = 39 | 6.0 (5.4, 7.3); n = 69 | 7.1 (5.8, 8.0); n = 108 |
| HBsAg, l log10 IU/ml | 4.6 (4.1, 5.1); n = 39 | 3.8 (3.3, 4.1); n = 70 | 4.0 (3.6, 4.4); n = 109 |
| HBV RNA, log10 IU/ml | 7.0 (6.5, 7.7); n = 28 | 4.8 (4.2, 5.7); n = 45 | 5.7 (4.6, 6.7); n = 73 |
| HBcrAg, log10 IU/ml | 7.9 (7.3, 8.3); n = 27 | 5.1 (4.3, 5.8); n = 43 | 6.2 (4.9, 7.8); n = 70 |
| ALT, U/L | 123 (80, 179); n = 38 | 116 (81, 234); n = 71 | 119 (81, 202); n = 109 |
Data are n (%) or median (IQR). Statistical tests performed are as follows: Fisher’s exact test for categorical data and the Wilcoxon rank-sum test for continuous data. ALT, alanine aminotransferase; HBcrAg, hepatitis B core-related antigen.
Optional core needle liver biopsies were collected at baseline and Weeks 44–48 (referred to as ‘Week 48’). Participants who completed 48 weeks of treatment and provided a liver biopsy at Week 48 were given the option to begin (or continue) treatment with TDF for up to 7 additional years. A third optional liver biopsy was collected at Week 240 in these participants who continued treatment. A total of 431 liver biopsies were stained using multiplex IF assay. Participant samples analysed were collected from 12 countries (the USA, Great Britain, Germany, Canada, Greece, Bulgaria, Czech Republic, the Netherlands, France, Spain, Australia, and Italy). All participants signed an informed consent form before screening and in accordance with local regulatory and ethics committee requirements. The experimental protocol in these trials was approved by Gilead Sciences and all local regulatory agencies.
Multiplex immunofluorescence
An automated multiplex IF assay was performed on n = 431 core needle liver biopsies (n = 133 stained using duplex IF for HBsAg and HBV core only; n = 298 stained using four-plex) from the GS-US-174-0102 and GS-US-174-0103 studies. Unmasking of antigens in formalin-fixed, paraffin-embedded tissue sections was achieved by heat-induced epitope retrieval. The multiplex antibody panel for HBV core (Gilead Sciences 366-2,19 Rabbit IgG), HBsAg (Gilead Sciences XTL17, Mouse IgG2a), histone H3 (Abcam EPR16987, Rabbit IgG), and sodium–potassium ATPase (Na+K+-ATPase; Abcam EP1845Y, Rabbit IgG) as well as the HBsAg/HBV core duplex was developed using the same primary antibodies listed above, optimised and performed using the Opal technology workflow20 (Akoya Biosciences; Fig. S2) on a Bond RX autostainer (Leica Biosystem), and scanned on the Vectra Polaris (Akoya Biosciences). HBsAg detection antibody XTL17 has similar affinity for HBsAg of genotypes A, B, C, D, E, G, and H (0.3 nM by ELISA). The HBV core detection antibody 366-2 binds to a linear epitope that is conserved across genotypes and present only in HBV core antigen (not HBeAg).19 Spectral DAPI (Akoya Biosciences) was used to detect nuclei in both assays. Assay performance was tested for nuclear dropout (loss of nuclear counterstain signal by DAPI; Fig. S3A), optimal antibody order, and appropriate positive and negative controls.
Whole-slide image analysis
Immunofluorescent whole-slide images of liver biopsies were analysed using Visiopharm software (version 2020.05, Visiopharm Corporation). To automate and standardise the analysis, a customised multistep algorithm was developed: (1) a decision forest algorithm was trained to detect the tissue from the background glass slide at 4× magnification to limit the high-resolution analysis to only the relevant regions; and (2) a convolutional neural network (CNN; U-Net∗)21 was trained at 20× magnification to segment and classify (a) nuclei (for HBsAg/HBV core duplex stains, CNN trained on the nuclear DAPI channel) or (b) cells (for four-plex stains, CNN was trained on the membrane marker Na+K+-ATPase to detect cell outlines and nuclear histone H3 in addition to the DAPI channel to identify nuclei). Nuclear size exclusion was used in the duplex assay to distinguish hepatocytes from lymphocyte populations, whereas in the four-plex assay both cytoplasmic exclusion and nuclear size exclusion were used. Hepatocytes were classified as HBsAg+ or HBV core+ individually, as well as ‘total HBV+’ if they stained for either HBsAg or HBV core, or both viral antigens.
Peripheral biomarker analysis
Participant serum or plasma was collected longitudinally throughout the GS-US-174-0102 and GS-US-174-0103 clinical trials. HBV DNA and qualitative HBeAg were measured on-study at Labcorp (previously Covance Central Laboratories).16 In participants with sufficient residual sample volume at baseline, Week 48, and/or Week 240, the following viral biomarkers were evaluated retrospectively: HBV RNA22 (DDL Diagnostics), hepatitis B core-related antigen (HBcrAg; Lumipulse, Fujiribio; DDL Diagnostics), and quantitative HBsAg (Architect i2000SR, Abbot; Labcorp).
Statistical analysis
All statistical analyses were performed using GraphPad Prism v8.0 (GraphPad Software, San Diego, CA, USA) and the statistical software R (R Foundation for Statistical Computing, Vienna, Austria). Comparison of baseline characteristics between HBeAg+ and HBeAg- participants was performed using a Fisher’s exact test for categorical data and the Wilcoxon rank-sum test for continuous data. Comparisons of % HBV+ hepatocytes (%HBsAg+, %HBV core+, and % total HBV+ [HBsAg+ and/or HBV core+]) between time points and between HBeAg status were performed using a Mann–Whitney U test or Wilcoxon test for paired and unpaired samples, respectively. Pearson’s correlation analysis was performed between % HBV+ hepatocytes and peripheral viral biomarkers. The corresponding p values were corrected for multiple hypothesis testing using the Benjamini–Hochberg method.
Results
Participant characteristics
In total, 431 core needle liver biopsies (133 stained using duplex and 298 stained using four-plex) were subjected to the multiplex IF assay. Final analysis was performed on 220 liver biopsies analysed using the four-plex IF assay that passed stringent quality control (QC) criteria, which included absence of extensive tissue folds, mechanical disruption or tissue loss, strong autofluorescence in one or more of the signal channels; or dropout of nuclear DAPI/histone H3 signal and/or Na+K+-ATPase membrane marker in the majority of the biopsy. These 220 biopsies were obtained from 114 total individuals (75 HBeAg- and 39 HBeAg+) at baseline, Week 48, and/or Week 240 (Table 1 and Table S1). Statistically significant differences (p <0.001) in baseline characteristics were observed for age, HBV DNA, HBsAg, HBcrAg, and HBV RNA, but not ALT, between HBeAg- and HBeAg+ participants (Table 1). These differences are consistent with known differences between stages of HBV disease and the enrolment criteria for the studies.16,23
Development of a four-plex assay to accurately quantify biopsy HBV+ hepatocytes
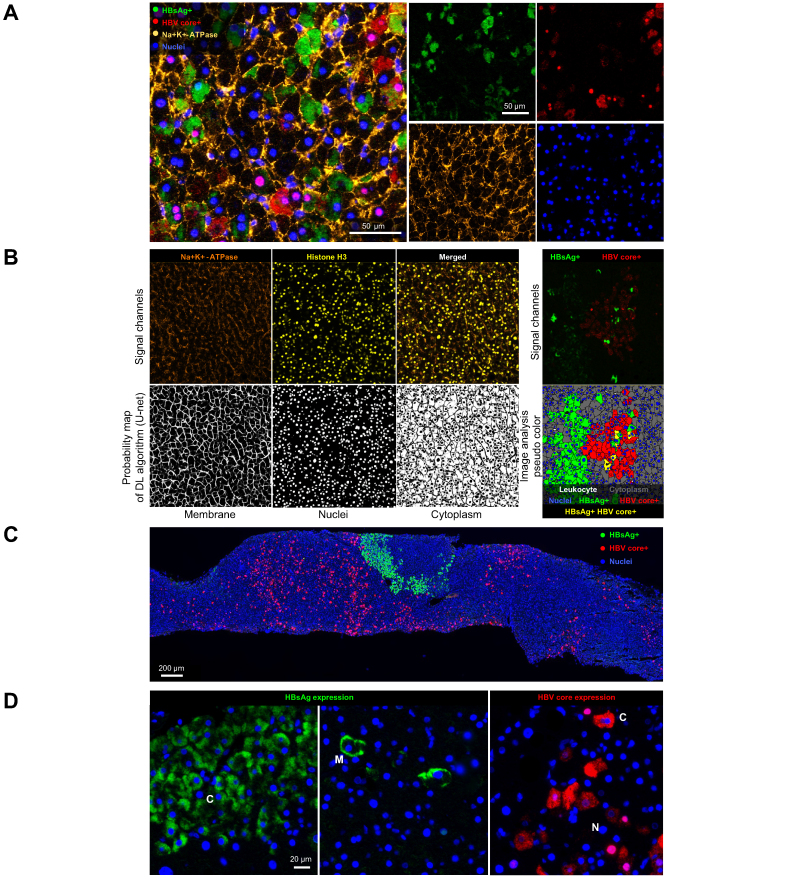
To assess HBV+ hepatocyte burden in core needle liver biopsies, we initially developed a duplex IF assay using antibodies against HBsAg and HBV core in the liver. We identified multiple pitfalls of the duplex assay that resulted in challenges for downstream analysis, including high numbers of biopsies that failed QC, difficulty assigning viral antigens to individual hepatocytes, and inability to consistently and accurately quantify the total number of hepatocytes. A major reason for QC failure was dropout of nuclear DAPI signal in almost 40% of samples, potentially caused by the archival nature of the biopsies in this study (Fig. S3A). We therefore expanded the multiplex panel to include histone H3, which detected hepatocyte nuclei in many samples where DAPI failed (Fig. 1A). In addition, we expanded our multiplex panel to include Na+K+-ATPase, a heterodimeric surface membrane protein complex, to accurately detect individual hepatocytes and enable assignment of HBsAg and HBV core staining to individual cells (Fig. 1A and Fig. S3B). As a result, we were able to reduce the QC failure rate from 38% with the duplex IF assay to 23% with the four-plex IF assay. We applied the four-plex IF assay and customised image analysis algorithm to a total of 298 core needle liver biopsies. Of those initially evaluated, 220 passed QC. Remaining causes of QC failure included extensive tissue folds, mechanical disruption or tissue loss, strong autofluorescence in one or more of the signal channels, or dropout of nuclear DAPI/histone H3 signal and/or Na+K+-ATPase membrane marker in the majority of the biopsy.
Fig. 1.
Development of a four-plex IF assay to accurately quantify liver HBV burden.
(A) Representative image of four-plex displayed as single channels (right) or overlayed image (left). (B) Representative images of signal channels for cell membrane (Na+K+-ATPase) and nuclear staining (histone H3) used to train CNN (U-Net; upper left) and corresponding probability maps to robustly segment cells (lower left); signal channels for HBV antigens (upper right) to accurately identify HBV-infected hepatocytes (lower right). (C) Representative image of multiplex IF showing diffuse HBcAg staining and foci of HBsAg staining. (D) Representative images of differential cell localisation of HBsAg and HBcAg expression. CNN, convolutional neural network; DL, deep learning; IF, immunofluorescence; Na+K+-ATPase, sodium–potassium ATPase.
A deep learning algorithm was developed to robustly detect cell membranes and nuclei (Fig. 1B). Compared with a threshold-based approach, the deep learning algorithm allowed for cell segmentation that was less affected by fluorescent intensity fluctuations. Nucleated and segmented cells were qualified as hepatocytes by their large size and were easily distinguished from leucocytes, which have significantly smaller nuclei and cytoplasm. Hepatocytes were classified as HBV+ if they stained for HBsAg and/or HBV core. This approach allowed for robust quantification of HBV positive cells as a percentage of total hepatocytes (Fig. 1B).
The most common pattern observed across these biopsies was mutually exclusive HBsAg and HBV core staining, with diffusely distributed individual HBV core+ cells and foci of HBsAg+ cells (Fig. 1C and D). HBsAg was localised to either the cell membrane or cytoplasm within the hepatocyte (Fig. 1D). HBV core was present either as strong nuclear staining or as cytoplasmic staining within the hepatocyte (Fig. 1D). Although HBV core and HBsAg staining were typically nonoverlapping, patches of mixed positive staining were observed in some samples, and a subset of hepatocytes that were dual positive for HBsAg and HBV core were identifiable. These dual-positive hepatocytes were infrequent, as previously reported[23], [24], [25], [26], [27], [28] (Fig. S4).
Analysis of duplicate liver biopsies identifies sampling heterogeneity with core needle biopsies
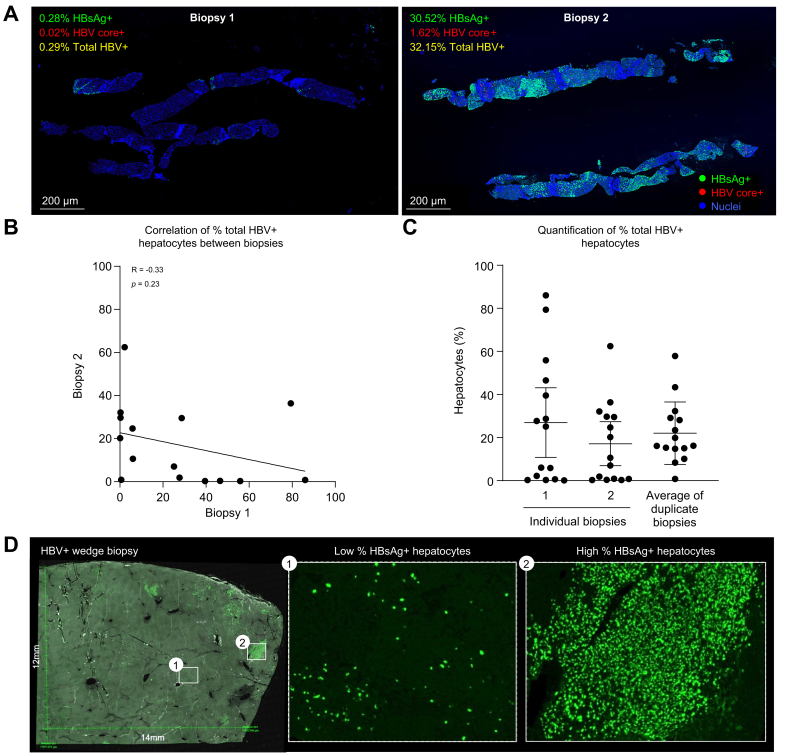
We identified 15 instances where two core needle biopsies were collected from different portions of the liver at the same time point, at either baseline or Week 48. This unique subset of samples allowed us to evaluate whether a single core needle biopsy is representative of the HBV burden across different areas of the liver.
Surprisingly, we found no correlation in total HBV+ hepatocyte burden between two liver biopsies collected from the same individual and time point (Fig. 2A and B). This result suggests that there is extensive heterogeneity in HBV positivity within individual livers and hence a large effect of random sampling on the frequency of HBV-positive hepatocytes detected in individual core needle liver biopsies. When HBsAg+ and HBV core+ hepatocytes were plotted separately, we found no correlation in HBsAg+ hepatocytes (Fig. S5A) and a statistically significant but poor correlation in HBV core+ hepatocytes (Fig. S5B) between two biopsies from the same participant and time point. To evaluate how biopsy sampling may impact analysis of a collection of individual biopsies, we plotted the total HBV+ hepatocyte burden from each of the two individual biopsies and compared it with the average of the duplicate biopsies (Fig 2C). Individual biopsy data produced a non-normal distribution of values, whereas the average of two biopsies approached a more normal distribution with fewer on the extreme high or low ends of the scale. This pattern suggested a ‘hot spot’ effect of core needle biopsy sampling, in which the measured value tends to be higher or lower than the true tissue average because local regions of the liver tend to have either very high or very low percentages of HBV+ hepatocytes. To better visualise the heterogeneity in HBV positivity, we analysed one large 12 mm × 14 mm wedge biopsy that was obtained from a single participant at Week 48 post NUC treatment (Fig. 2D). Staining in this sample was almost exclusively HBsAg+, with little to no HBV core+ hepatocytes. Heterogeneity in staining within this single biopsy was extensive, with large regions of no apparent HBV staining, areas with a small percentage of HBsAg+ hepatocytes, and focal regions of near-complete HBsAg positivity, consistent with observations above.
Fig. 2.
Analysis of duplicate liver biopsies suggests inherent sampling heterogeneity for viral antigens.
(A) Representative images from duplicate biopsies collected from the same participant and time point. Embedded legends indicate markers, corresponding colour, and percent hepatocytes positive for viral antigens. (B) Correlation between % total HBV+ hepatocytes (defined as HBsAg+ and/or HBV core+) in biopsy 1 vs. biopsy 2 from the same participant and time point using Pearson’s correlation analysis. (C) Quantification of total HBV burden when duplicate biopsies are plotted individually (left) or averaged for the same participant and time point (right). (D) Representative image of a large wedge biopsy with regions of low (embedded image 1) and high (embedded image 2) % of HBsAg+ hepatocytes.
Collectively, these data indicate that sampling heterogeneity has a large impact on the results obtained with single core needle biopsies, such that individual biopsies often skew toward very high or very low values that are likely not representative of the overall average across a patient’s liver.
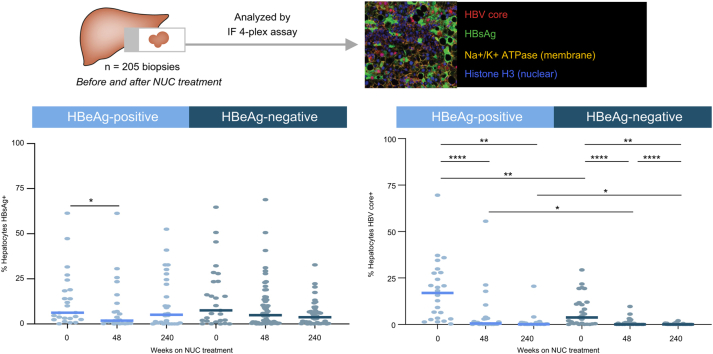
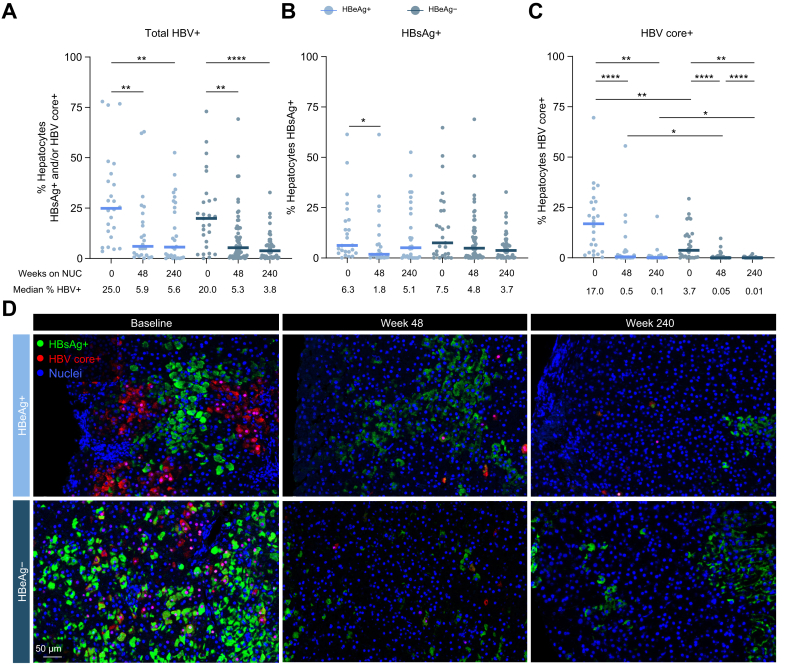
NUC-treated HBeAg- participants have the lowest HBV+ hepatocyte burden
As single core needle biopsies are not representative of the entire liver burden for individual patients, we focused on population-level analyses of HBV+ hepatocyte burden between HBeAg+ (n = 39) and HBeAg- (n = 75) participants with CHB at baseline and post NUC treatment. Participants for which two biopsies were collected at the same time point had their values averaged for analysis.
NUC treatment for 48 weeks significantly reduced total HBV+ hepatocyte burden in both HBeAg+ and HBeAg- participants (Fig. 3A and D). Comparable results were obtained in participants who received TDF or ADV (Fig. S6A). This decline in total HBV+ hepatocyte burden with treatment was driven by large decreases in HBV core+ hepatocytes (Fig. 3C), whereas minimal changes in HBsAg+ hepatocytes were observed over time (Fig. 3B). HBeAg status also impacted HBV core positivity, as the frequency of HBV core+ hepatocytes was lower in HBeAg- participants than in HBeAg+ participants at all time points evaluated (Fig. 3C). As previously mentioned, HBsAg+ HBV core+ cells were rare and only found in HBeAg+ participants at baseline (Fig. S4). HBeAg-, NUC-treated participants had the lowest HBV+ hepatocyte burden amongst the populations tested. A subset of participants within our cohort had a complete longitudinal time course available before and after NUC treatment (Table S1). Paired analysis of this biopsy subset showed trends similar to the results observed with the entire cohort (Fig. S6B–D).
Fig. 3.
HBeAg status and NUC treatment impact HBV+ hepatocyte burden.
Quantification of (A) % total HBV+ hepatocytes (defined as HBsAg+ and/or HBV core+), (B) % HBsAg+ hepatocytes, and (C) % HBV core+ hepatocytes before and after NUC treatment in HBeAg+ and HBeAg- participants. (D) Representative images from HBeAg+ and HBeAg- participants throughout NUC treatment. Embedded legend indicates markers and corresponding colour. ∗p <0.05, ∗∗p <0.01, ∗∗∗p <0.001, ∗∗∗∗p <0.0001, Mann–Whitney U test. Only statistically significant differences are shown. NUC, nucleo(t)side.
Correlations between HBV+ hepatocyte burden and peripheral biomarkers are dependent on stage of infection and treatment status
Finally, we sought to understand if any correlations existed between the frequency of HBV+ hepatocytes and well-established peripheral viral biomarkers. In a subset of samples with available paired serum or plasma, we evaluated HBsAg, HBcrAg, HBV DNA, and HBV RNA (Fig. S7A) and performed correlation analyses of liver biopsy vs. peripheral biomarker data, stratified by HBeAg status.
In HBeAg+ participants, we identified statistically significant correlations between total HBV+ hepatocyte burden and peripheral HBcrAg, HBV DNA, and HBV RNA at baseline, before NUC treatment (Table 2 and Fig. S8). Upon NUC treatment, however, most correlations between peripheral viral biomarkers and total HBV+ hepatocyte burden were lost. Correlations between peripheral biomarkers and HBsAg+ hepatocytes or HBV core+ hepatocytes were inconsistent when analysed independently (Table S2).
Table 2.
Correlation between peripheral viral biomarkers and total HBV+ liver burden.
| HBeAg status | Peripheral viral biomarker | Baseline |
Week 48 |
Week 240 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| R value | p value | Adjusted p value | R value | p value | Adjusted p value | R value | p value | Adjusted p value | ||
| HBeAg+ | HBsAg | 0.195 | 0.361 | 0.629 | 0.116 | 0.589 | 0.748 | 0.279 | 0.15 | 0.355 |
| HBcrAg | 0.638 | 0.002 | ∗0.027 | 0.349 | 0.202 | 0.396 | 0.05 | 0.815 | 0.896 | |
| HBV DNA | 0.654 | 0.001 | ∗0.018 | 0.619 | 0.032 | 0.139 | NR | – | – | |
| HBV RNA | 0.651 | 0.001 | ∗0.025 | 0.519 | 0.084 | 0.258 | −0.016 | 0.949 | 0.971 | |
| HBeAg- | HBsAg | 0.279 | 0.168 | 0.371 | 0.103 | 0.435 | 0.694 | 0.433 | 0.004 | 0.033∗ |
| HBcrAg | −0.101 | 0.682 | 0.835 | −0.111 | 0.552 | 0.721 | −0.008 | 0.971 | 0.97 | |
| HBV DNA | −0.118 | 0.557 | 0.721 | NR | – | – | NR | – | – | |
| HBV RNA | −0.163 | 0.48 | 0.719 | −0.398 | 0.142 | 0.335 | 0.046 | 0.932 | 0.97 | |
Bold text indicates statistical significance with adjusted p value. ∗Adjusted p <0.05 (Pearson’s correlation analysis; p values adjusted for false discovery rate using the Benjamini–Hochberg procedure). HBcrAg, hepatitis B core-related antigen; NR, not reported (owing to insufficient [<5] data points for analysis).
Discussion
Here, we apply a novel four-plex IF assay to quantify HBV antigens in a large cohort of HBV-infected individuals and provide insight into viral antigen burden and distribution over time. Use of histone H3/DAPI and Na+K+-ATPase allowed us to clearly define nuclei and cell membranes, which enabled training of a CNN to robustly segment cell membranes and nuclei. We identified population-level reductions in HBV core+ hepatocytes associated with NUC treatment and lower frequencies of HBV core+ hepatocytes in HBeAg+ vs. HBeAg- participants. Importantly, we demonstrated extensive regional variation in HBV positivity in liver samples. Significant correlations between peripheral biomarkers and total HBV+ hepatocyte burden were identified in untreated HBeAg+ participants, with few significant correlations observed in HBeAg- and/or NUC-treated participants.
Consistent with existing literature, we observed strong HBV core and HBsAg staining in baseline liver samples from HBeAg+ participants, whereas the HBeAg- liver samples demonstrated high HBsAg staining but little to no HBV core staining.23,24,[27], [28], [29], [30] We observed relatively few cells in which both HBV core and HBsAg were detected at the same time, as previously reported.[24], [25], [26], [27], [28] The presence of HBsAg+/HBV core-cells is consistent with literature characterising hepatocytes containing integrated HBV DNA.4,14 HBV dslDNA has been shown to express HBsAg, but not HBV core,[31], [32], [33] and targeted long-read sequencing showed that many HBV integrations are clonally expanded, consistent with the foci of HBsAg+/HBV core-cells.6,8 In addition, HBsAg+/HBV core-cells were dominant in HBeAg- individuals, where most HBV transcripts derive from integrated DNA.6
In contrast, HBsAg-/HBV core+ staining is less well defined, as infected hepatocytes containing cccDNA should be able to co-express HBV core and HBsAg. However, the fact that HBsAg-/HBV core+ cells were most common in untreated HBeAg+ participants is informative, as most HBV transcripts in this population have been shown to derive from cccDNA.6 We similarly observed that HBeAg+ participants contained significantly more HBV core+ hepatocytes than HBeAg- participants at all time points. Consequently, it is likely that HBsAg-/HBV core+ cells represent cccDNA-containing cells and the level of intracellular HBsAg simply falls below the limit of assay detection in many cases, as previously reported.34 This would imply that the amount of intracellular HBsAg is lower in most cccDNA+ cells than in those with integrated HBV. This could occur if cccDNA+ cells produce lower amounts of HBsAg than cells with integrated HBV and/or if HBsAg is more efficiently secreted from cccDNA+ cells. Indeed, HBV integration has been reported to change the ratios of large, medium, and small HBsAg production, and thereby disrupt efficient particle secretion and cause HBsAg accumulation in the cytoplasm.35 A subset of cells with cccDNA may have sufficient HBsAg for detection, explaining the occasional presence of dual HBsAg+/HBV core+ hepatocytes, as well as their disappearance upon NUC treatment. This scenario is consistent with all the staining patterns observed here and with the literature cited above.
We determined that individual core needle biopsies are typically not representative of the entire liver; hence, caution is needed in extrapolating biopsy measurements to the entire organ.14 In 15 individuals who had two biopsies collected from different parts of the liver at the same time point, there were poor correlations between HBV antigen staining in the two biopsies. Many samples contained ‘hot spots’ of HBsAg+ hepatocytes, presumed to be clonal expansions harbouring integrated HBV DNA. The associated focal pattern of HBsAg staining could explain the tendency observed here for needle biopsies to yield liver HBV antigen measurements that were frequently much higher or lower than the average burden. This conclusion is supported by the heterogeneous distribution in HBV antigen staining in a single large wedge biopsy. These data suggest the need for caution when interpreting biopsy results for individual patients. However, in our subgroup analysis, the median frequency of HBV+ hepatocytes was similar whether single biopsy values or averages of two biopsies were used to measure liver HBV positivity, suggesting that comparisons of population medians is reliable despite the observed liver sampling effects on an individual patient level. Consequently, we focused our analyses on population-level comparisons, rather than on individual-patient comparisons.
A major finding of this study is that NUC treatment resulted in large reductions in HBV core+, but not HBsAg+, hepatocytes in both HBeAg+ and HBeAg- participants. To our knowledge, only a single previous report has documented this pattern of NUC effects in the liver and was semiquantitative.36 We confirm and extend that observation with quantitative analysis of a much larger biopsy set. Our results suggest that NUC treatment reduces HBV core burden (likely cccDNA+) by reducing HBV replication (and hence viral spread) but does not significantly impact antigen expression from pre-existing integrated HBV DNA. The recently reported effect of NUCs on reducing integration burden37 likely results from preventing formation of new integrations, rather than direct effects on existing integrations. Accordingly, in the periphery, we observed a multi-log10 decrease in HBV DNA with NUC treatment vs. a half-log10 median HBsAg decline,16 consistent with integrated HBV DNA maintaining HBsAg production during NUC treatment.38 Collectively, these results suggest a moderate half-life of cccDNA+ hepatocytes, as the HBV core+ hepatocyte population is reduced by >95% over 48 weeks of treatment. This occurs even though NUC treatment does not completely block viral replication,39 indicating that the level of residual infectious virus during NUC treatment is too low to compensate for natural hepatocyte turnover and/or immune-mediated pressure on HBV core+ cells. A moderate cccDNA half-life is also supported by previous reports that showed that a year of NUC treatment eliminates over 80% of cccDNA40 and that, in cases of lamivudine resistance, a majority of cccDNA can be replaced with resistant genomes in a span of approximately 6 months.41
Statistically significant correlations between total detected HBV+ hepatocyte frequency and the peripheral biomarkers HBcrAg, HBV DNA, and HBV RNA were present only in HBeAg+ participants, where a large portion of HBV+ hepatocytes represent cccDNA+ infected cells that may distribute more evenly in the liver than clonal integrations. In contrast, correlations between detected HBV+ hepatocyte frequency and peripheral viral biomarkers were largely absent in HBeAg- and NUC-treated participants, where the large majority of HBV antigen staining in biopsies represents integrations8,13,23 that do not produce HBV DNA or HBcrAg. As previously reported, correlations between biopsy HBsAg and peripheral HBsAg were largely absent across patient groups,42 likely owing to the biopsy sampling heterogeneity and foci of HBsAg+ hepatocytes demonstrated in our study. The potential presence of HBV+ hepatocytes below the limits of detection by immunohistochemistry could also have impacted correlation analyses.
We also found inconsistent correlations between peripheral biomarkers and either HBsAg+ hepatocytes or HBV core+ hepatocytes analysed individually. Earlier efforts to identify such correlations have also yielded conflicting results. We identified a significant correlation between HBsAg+ hepatocytes and peripheral HBV DNA in untreated HBeAg+ but not HBeAg- individuals. One previous report found the same,43 another found no correlations in either group,29 and a third found an inverse correlation between intrahepatic and serum HBsAg.42 Similarly, we identified no clear patterns of correlation between peripheral markers and HBV core+ hepatocytes, despite a previous report correlating liver HBV core staining with serum HBV DNA.29 Notably, our analysis algorithm did not differentiate between nuclear HBV core and cytoplasmic HBV core, which vary based on HBeAg status and inflammation, or between membranous HBsAg and cytoplasmic HBsAg, which may differ between cccDNA and integrated HBV.27,44 These variables, differences in patient groups, and differences in methods for quantifying viral antigens may explain the disparate results of correlation analyses here and in the literature.
An accurate understanding of HBV+ hepatocyte burden across disease stage and treatment status may help inform clinical development of novel HBV cure approaches. Many HBV cure strategies now aim to achieve immune-mediated elimination of the HBV-positive cells, and safety may be linked to the number of hepatocytes that need to be killed and the speed with which they are eliminated. Identification of individuals with a low HBV+ hepatocyte burden would be the safest way to initiate such studies. Our current study suggests that at a population level, HBeAg- individuals on NUC treatment have the lowest HBV+ hepatocyte burden and may represent the best starting population for clinical development of some HBV cure agents. In our study cohort, serum viral biomarkers were not successful in identifying a population with lower HBV+ hepatocyte burden than HBeAg– NUC-treated individuals overall. Our results also suggest that cytolytic mechanisms capable of killing cells expressing HBsAg are likely to be required to achieve cure, as HBV core+ (likely cccDNA+) cells represent only a small fraction of the HBV+ hepatocytes in NUC-treated individuals.
Financial support
This work was funded by Gilead Sciences, Inc.
Authors’ contributions
Designed the study: AA, PO, LD, BF, SB. Coordinated sample inventories and databases: MA, SC, ST, TT. Designed and performed the experiments: CM, NvB. Analysed the data: AA, PO, JB, NvB, SS. Wrote the manuscript: AA, PO, NvB, SPF, SB. Coordinated the clinical trial and biopsy collection: VS, PM, MB, AG. Approved the final version of the manuscript: all authors.
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary information.
Conflicts of interest
At the time this study was conducted, AA, PO, JB, NvB, CM, SC, MVA, VS, TT, ST, AG, SF, LD, BF, and SB were employees and stockholders of Gilead Sciences, Inc.
Please refer to the accompanying ICMJE disclosure forms for further details.
Acknowledgements
We would like to thank Sarah Tse from Bioscience Communications for the high-quality figures.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2022.100664.
Supplementary data
The following are the supplementary data to this article:
References
- 1.World Health Organization . Global hepatitis report. World Health Organization; Geneva: 2017. [Google Scholar]
- 2.Kim G.-A., Lim Y.-S., An J., Lee D., Shim J.H., Kim K.M., et al. HBsAg seroclearance after nucleoside analogue therapy in patients with chronic hepatitis B: clinical outcomes and durability. Gut. 2014;63:1325–1332. doi: 10.1136/gutjnl-2013-305517. [DOI] [PubMed] [Google Scholar]
- 3.Tong S., Revill P. Overview of hepatitis B viral replication and genetic variability. J Hepatol. 2016;64(Suppl. 1):S4–S16. doi: 10.1016/j.jhep.2016.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tu T., Budzinska M.A., Shackel N.A., Urban S. HBV DNA integration: molecular mechanisms and clinical implications. Viruses. 2017;9:75. doi: 10.3390/v9040075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pollicino T., Caminiti G. HBV-integration studies in the clinic: role in the natural history of infection. Viruses. 2021;13:368. doi: 10.3390/v13030368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Buuren N., Ramirez R., Soulette C., Suri V., Han D., May L., et al. Targeted long-read sequencing reveals clonally expanded HBV-associated chromosomal translocations in patients with chronic hepatitis B. JHEP Rep. 2022;4 doi: 10.1016/j.jhepr.2022.100449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang Z., Jhunjhunwala S., Liu J., Haverty P.M., Kennemer M.I., Guan Y., et al. The effects of hepatitis B virus integration into the genomes of hepatocellular carcinoma patients. Genome Res. 2012;22:593–601. doi: 10.1101/gr.133926.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mason W.S., Gill U.S., Litwin S., Zhou Y., Peri S., Pop O., et al. HBV DNA integration and clonal hepatocyte expansion in chronic hepatitis B patients considered immune tolerant. Gastroenterology. 2016;151:986–998.e4. doi: 10.1053/j.gastro.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Budzinska M.A., Shackel N.A., Urban S., Tu T. Sequence analysis of integrated hepatitis B virus DNA during HBeAg-seroconversion. Emerg Microbes Infect. 2018;7:142. doi: 10.1038/s41426-018-0145-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sung W.-K., Zheng H., Li S., Chen R., Liu X., Li Y., et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet. 2012;44:765–769. doi: 10.1038/ng.2295. [DOI] [PubMed] [Google Scholar]
- 11.Zhao L.-H., Liu X., Yan H.-X., Li W.-Y., Zeng X., Yang Y., et al. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat Commun. 2016;7 doi: 10.1038/ncomms12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Péneau C., Imbeaud S., La Bella T., Hirsch T.Z., Caruso S., Calderaro J., et al. Hepatitis B virus integrations promote local and distant oncogenic driver alterations in hepatocellular carcinoma. Gut. 2022;71:616–626. doi: 10.1136/gutjnl-2020-323153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wooddell C.I., Yuen M.F., Chan H.L., Gish R.G., Locarnini S.A., Chavez D., et al. RNAi-based treatment of chronically infected patients and chimpanzees reveals that integrated hepatitis B virus DNA is a source of HBsAg. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aan0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meier M.A., Calabrese D., Suslov A., Terracciano L.M., Heim M.H., Wieland S. Ubiquitous expression of HBsAg from integrated HBV DNA in patients with low viral load. J Hepatol. 2021;75:840–847. doi: 10.1016/j.jhep.2021.04.051. [DOI] [PubMed] [Google Scholar]
- 15.Loglio A., Viganò M., Lampertico P. Novel therapies that may cure chronic hepatitis B virus. Clin Liver Dis. 2021;25:875–899. doi: 10.1016/j.cld.2021.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Marcellin P., Heathcote E.J., Buti M., Gane E., de Man R.A., Krastev Z., et al. Tenofovir disoproxil fumarate versus adefovir dipivoxil for chronic hepatitis B. N Engl J Med. 2008;359:2442–2455. doi: 10.1056/NEJMoa0802878. [DOI] [PubMed] [Google Scholar]
- 17.Gordon S.C., Krastev Z., Horban A., Petersen J., Sperl J., Dinh P., et al. Efficacy of tenofovir disoproxil fumarate at 240 weeks in patients with chronic hepatitis B with high baseline viral load. Hepatology. 2013;58:505–513. doi: 10.1002/hep.26277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.European Association for the Study of the Liver EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol. 2017;67:370–398. doi: 10.1016/j.jhep.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 19.Hong X., Luckenbaugh L., Mendenhall M., Walsh R., Cabuang L., Soppe S., et al. Characterization of hepatitis B precore/core-related antigens. J Virol. 2021;95 doi: 10.1128/JVI.01695-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taube J.M., Roman K., Engle E.L., Wang C., Ballesteros-Merino C., Jensen S.M., et al. Multi-institutional TSA-amplified multiplexed immunofluorescence reproducibility evaluation (MITRE) study. J Immunother Cancer. 2021;9 doi: 10.1136/jitc-2020-002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ronneberger O., Fischer P., Brox T. In: Medical image computing and computer-assisted intervention – MICCAI 2015. Navab N., Hornegger J., Wells W.M., Frangi A.F., editors. Springer International Publishing; Cham: 2015. U-Net: convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 22.van Bömmel F., Bartens A., Mysickova A., Hofmann J., Krüger D.H., Berg T., et al. Serum hepatitis B virus RNA levels as an early predictor of hepatitis B e antigen seroconversion during treatment with polymerase inhibitors. Hepatology. 2015;61:66–76. doi: 10.1002/hep.27381. [DOI] [PubMed] [Google Scholar]
- 23.Montanari N.R., Ramírez R., Aggarwal A., van Buuren N., Doukas M., Moon C., et al. Multi-parametric analysis of human livers reveals variation in intrahepatic inflammation across phases of chronic hepatitis B infection. J Hepatol. 2022;77:332–343. doi: 10.1016/j.jhep.2022.02.016. [DOI] [PubMed] [Google Scholar]
- 24.Hsu H.C., Lai M.Y., Su I.J., Chen D.S., Chang M.H., Yang P.M., et al. Correlation of hepatocyte HBsAg expression with virus replication and liver pathology. Hepatology. 1988;8:749–754. doi: 10.1002/hep.1840080408. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X., Lu W., Zheng Y., Wang W., Bai L., Chen L., et al. In situ analysis of intrahepatic virological events in chronic hepatitis B virus infection. J Clin Invest. 2016;126:1079–1092. doi: 10.1172/JCI83339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naumov N.V., Mondelli M., Alexander G.J., Tedder R.S., Eddleston A.L., Williams R. Relationship between expression of hepatitis B virus antigens in isolated hepatocytes and autologous lymphocyte cytotoxicity in patients with chronic hepatitis B virus infection. Hepatology. 1984;4:63–68. doi: 10.1002/hep.1840040111. [DOI] [PubMed] [Google Scholar]
- 27.Hsu H.C., Su I.J., Lai M.Y., Chen D.S., Chang M.H., Chuang S.M., et al. Biologic and prognostic significance of hepatocyte hepatitis B core antigen expressions in the natural course of chronic hepatitis B virus infection. J Hepatol. 1987;5:45–50. doi: 10.1016/s0168-8278(87)80060-0. [DOI] [PubMed] [Google Scholar]
- 28.van Buuren N., Ramirez R., Turner S., Chen D., Suri V., Aggarwal A., et al. Characterization of the liver immune microenvironment in liver biopsies from patients with chronic HBV infection. JHEP Rep. 2022;4 doi: 10.1016/j.jhepr.2021.100388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramakrishna B., Mukhopadhya A., Kurian G. Correlation of hepatocyte expression of hepatitis B viral antigens with histological activity and viral titer in chronic hepatitis B virus infection: an immunohistochemical study. J Gastroenterol Hepatol. 2008;23:1734–1738. doi: 10.1111/j.1440-1746.2008.05416.x. [DOI] [PubMed] [Google Scholar]
- 30.Safaie P., Poongkunran M., Kuang P.P., Javaid A., Jacobs C., Pohlmann R., et al. Intrahepatic distribution of hepatitis B virus antigens in patients with and without hepatocellular carcinoma. World J Gastroenterol. 2016;22:3404–3411. doi: 10.3748/wjg.v22.i12.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang W., Summers J. Integration of hepadnavirus DNA in infected liver: evidence for a linear precursor. J Virol. 1999;73:9710–9717. doi: 10.1128/jvi.73.12.9710-9717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freitas N., Abe K., Cunha C., Menne S., Gudima S.O. Support of the infectivity of hepatitis delta virus particles by the envelope proteins of different genotypes of hepatitis B virus. J Virol. 2014;88:6255–6267. doi: 10.1128/JVI.00346-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Budzinska M.A., Shackel N.A., Urban S., Tu T. Cellular genomic sites of hepatitis B virus DNA integration. Genes (Basel) 2018;9:365. doi: 10.3390/genes9070365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gowans E.J., Burrell C.J. Widespread presence of cytoplasmic HBcAg in hepatitis B infected liver detected by improved immunochemical methods. J Clin Pathol. 1985;38:393–398. doi: 10.1136/jcp.38.4.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang H.-Y., Tsai H.-W., Teng C.-F., Hui-Ching Wang L., Huang W., Su I.-J. Ground glass hepatocytes provide targets for therapy or prevention of hepatitis B virus-related hepatocellular carcinoma. AIMS Med Sci. 2018;5:90–101. [Google Scholar]
- 36.Tsai H.-W., Lin Y.-J., Wu H.-C., Chang T.-T., Wu I.-C., Cheng P.-N., et al. Resistance of ground glass hepatocytes to oral antivirals in chronic hepatitis B patients and implication for the development of hepatocellular carcinoma. Oncotarget. 2016;7:27724–27734. doi: 10.18632/oncotarget.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu Y.-C., Suri V., Nguyen M.H., Huang Y.-T., Chen C.-Y., Chang I.-W., et al. Inhibition of viral replication reduces transcriptionally active distinct hepatitis B virus integrations with implications on host gene dysregulation. Gastroenterology. 2022;162:1160–11670.e1. doi: 10.1053/j.gastro.2021.12.286. [DOI] [PubMed] [Google Scholar]
- 38.Grudda T., Hwang H.S., Taddese M., Quinn J., Sulkowski M.S., Sterling R.K., et al. Integrated hepatitis B virus DNA maintains surface antigen production during antivirals. J Clin Invest. 2022;132 doi: 10.1172/JCI161818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burdette D.L., Lazerwith S., Yang J., Chan H.L.Y., Delaney W.E.I.V., Fletcher S.P., et al. Ongoing viral replication and production of infectious virus in patients with chronic hepatitis B virus suppressed below the limit of quantitation on long-term nucleos(t)ide therapy. PLoS One. 2022;17 doi: 10.1371/journal.pone.0262516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Werle-Lapostolle B., Bowden S., Locarnini S., Wursthorn K., Petersen J., Lau G., et al. Persistence of cccDNA during the natural history of chronic hepatitis B and decline during adefovir dipivoxil therapy. Gastroenterology. 2004;126:1750–1758. doi: 10.1053/j.gastro.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 41.Huang Q., Zhou B., Cai D., Zong Y., Wu Y., Liu S., et al. Rapid turnover of hepatitis B virus covalently closed circular DNA indicated by monitoring emergence and reversion of signature-mutation in treated chronic hepatitis B patients. Hepatology. 2021;73:41–52. doi: 10.1002/hep.31240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lau J.Y., Bain V.G., Davies S.E., Alexander G.J., Williams R. Export of intracellular HBsAg in chronic hepatitis B virus infection is related to viral replication. Hepatology. 1991;14:416–421. [PubMed] [Google Scholar]
- 43.Thompson A.J., Nguyen T., Iser D., Ayres A., Jackson K., Littlejohn M., et al. Serum hepatitis B surface antigen and hepatitis B e antigen titers: disease phase influences correlation with viral load and intrahepatic hepatitis B virus markers. Hepatology. 2010;51:1933–1944. doi: 10.1002/hep.23571. [DOI] [PubMed] [Google Scholar]
- 44.Chu C.-M., Liaw Y.-F. Intrahepatic distribution of hepatitis B surface and core antigens in chronic hepatitis B virus infection. Hepatocyte with cytoplasmic/membranous hepatitis B core antigen as a possible target for immune hepatocytolysis. Gastroenterology. 1987;92:220–225. doi: 10.1016/0016-5085(87)90863-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary information.