Summary
Background
It has been widely recognized that a critical time window for neurodevelopment occurs in early life and the host's gut microbiome plays an important role in neurodevelopment. Following recent demonstrations that the maternal prenatal gut microbiome influences offspring brain development in murine models, we aim to explore whether the critical time window for the association between the gut microbiome and neurodevelopment is prenatal or postnatal for human.
Methods
Here we leverage a large-scale human study and compare the associations between the gut microbiota and metabolites from mothers during pregnancy and their children with the children's neurodevelopment. Specifically, using multinomial regression integrated in Songbird, we assessed the discriminating power of the maternal prenatal and child gut microbiome for children's neurodevelopment at early life as measured by the Ages & Stages Questionnaires (ASQ).
Findings
We show that the maternal prenatal gut microbiome is more relevant than the children's gut microbiome to the children's neurodevelopment in the first year of life (maximum Q2 = 0.212 and 0.096 separately using the taxa at the class level). Moreover, we found that Fusobacteriia is more associated with high fine motor skills in ASQ in the maternal prenatal gut microbiota but become more associated with low fine motor skills in the infant gut microbiota (rank = 0.084 and −0.047 separately), suggesting the roles of the same taxa with respect to neurodevelopment can be opposite at the two stages of fetal neurodevelopment.
Interpretation
These findings shed light, especially in terms of timing, on potential therapeutic interventions to prevent neurodevelopmental disorders.
Funding
This work was supported by the National Institutes of Health (grant numbers: R01AI141529, R01HD093761, RF1AG067744, UH3OD023268, U19AI095219, U01HL089856, R01HL141826, K08HL148178, K01HL146980), and the Charles A. King Trust Postdoctoral Fellowship.
Keywords: Childhood neurodevelopment, Maternal gut microbiome, Early-life gut microbiome, Ages and stages questionnaire
Research in context.
Evidence before this study
Neurodevelopment has been associated with gut microbial composition in animal models. It has been demonstrated that the maternal prenatal gut microbiome also influences offspring brain development in mouse model. Yet, for humans it is still unclear if the critical time window for the association between the gut microbiome and neurodevelopment is prenatal, postnatal or both. Thus, microbiome data in human populations are highly warranted, especially with longitudinal data allowing exploration of the vertical transmission from mother to child.
Added value of this study
The maternal prenatal gut microbiome is more relevant than the children's gut microbiome to the children's neurodevelopment. Although similar class-level taxa were associated with neurodevelopment among both the maternal prenatal and infant gut microbiota, those taxa may play different roles during pregnancy and in infancy.
Implications of all the available evidence
Investigation of the correlations between the ASQ and multi-omics data allows us to understand the association between the gut microbiome and neurodevelopment more comprehensively. Demonstrating the role of the gut microbes can be different, especially when the taxa are acquired or dominated at different stages of life or in the vertical transmission from mother to child, we argue that the role of the gut microbiota need to be separately investigated. Overall, our results underscore the importance of maternal prenatal gut microbiome in offspring neurodevelopment, which may guide us in designing potential interventions.
Introduction
Mounting evidence suggests that the gut microbiome influences not only the host's health status1 but also brain function and host behavior.2,3 In particular, neurodevelopment has been shown to be associated with gut microbial composition in animal models.4, 5, 6, 7, 8 For example, compared with normal mice, germ-free mice differ in social behavior, stress response, cognition, and other functions, providing solid evidence for the role of the microbiome in neurodevelopment.9 Some of these effects can be mitigated by exposure to microbes throughout adolescence, while others persist despite microbial colonization,10 suggesting that early life is a sensitive time period for the effects of microbial exposure on neurodevelopment.11 Since a human child's brain grows to 80–90% of its adult volume by two years of age,12 it is crucial to identify the critical time window for the association between the gut microbiome and neurodevelopment in the early years.
In addition to the relationship between the infant gut microbiome and neurodevelopment, previous studies in mice13, 14, 15 and humans16 suggest that the maternal prenatal microbiome also influences brain development in offspring. However, it is still unclear if the impact of the maternal microbiome is limited to events during pregnancy or occurs postnatally as a result of vertical transmission.17, 18, 19 A recent mouse study revealed that the maternal prenatal gut microbiome promotes fetal thalamocortical axonogenesis by signaling neurons in the developing brain of the offspring with microbially modulated metabolites.20 However, the relative importance of the association between maternal prenatal or infant gut microbiome and neurodevelopment in humans remains to be determined.
Although mouse studies are important in providing evidence supporting the concept that the gut microbiome is involved in neurodevelopment,4, 5, 6, 7, 8, 9, 10, 11,13, 14, 15,20,21 there are significant limitations to human translation of these findings.22 Microbiome data in human populations are clearly warranted, especially with longitudinal data allowing exploration of the vertical transmission from mother to child. In this study, we leveraged the gut microbiome and metabolome (microbiome for short) data from the VDAART,23 which assessed the effect of vitamin D supplementation for mothers in pregnancy on the development of asthma-related phenotypes in offspring. Specifically, we assessed associations between the maternal prenatal and child gut microbiome on children's neurodevelopment at year one, year two, and year three separately as measured by the Ages and Stages Questionnaire, third edition (ASQ for short hereafter)24 (see Methods) due to the complex trajectory of the ASQ for the first three years of life. Notably, the ASQ is one of the major screening tools recommended by The American Academy of Pediatrics. It has been widely used to assess risks for developmental delays,25 e.g., early life ASQ is strongly associated with long term outcomes such as IQ, school difficulties at age 5–6, and autism spectrum disorder.26, 27, 28 More specifically, we examined the discriminating power of (i) the maternal prenatal gut microbiome, (ii) the infant gut microbiome (3–6 months), and (iii) the gut microbiome of children at year one and year three on outcomes of children's neurodevelopment (Fig. 1). The comparison of relevance (as measured by their discriminating power for the ASQ) of maternal prenatal, infant and child gut microbiome allowed us to identify the earliest, most primary association between the gut microbiome and the neurodevelopment in humans. Interestingly, we found that the maternal prenatal gut microbiome is more relevant to their children's first-year neurodevelopment than the infant (months 3–6) gut microbiome or the gut microbiome at year one, while the discriminating power of maternal prenatal and infant gut microbiome gradually diminishes for neurodevelopment at year two. Neither the maternal prenatal nor children's gut microbiome has any discriminating power for the children's neurodevelopment at year three. Our results underscore the importance of the maternal prenatal gut microbiome in offspring neurodevelopment, which may guide us in designing potential interventions, particularly with regard to the timing of those interventions.
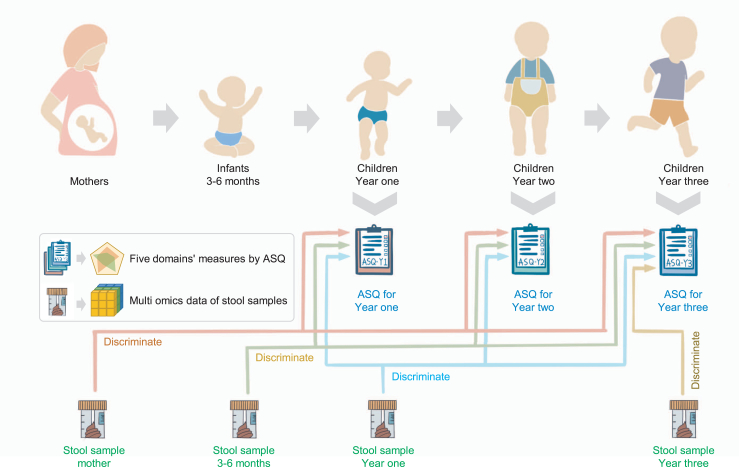
Fig. 1.
Analysis design. Conceptual diagram illustrating the analysis design and collection of stool samples and ASQ questionnaires. The gut microbial compositions of mothers (prenatal), infants (months 3–6), and children (year one and year three) were separately used to discriminate neurodevelopmental outcomes as measured by ASQ at different ages (ASQ-Y1, ASQ-Y2, and ASQ-Y3) to quantify the associations and relevance between the gut microbiome and neurodevelopment.
Methods
Ethics statement
Subjects were offspring of participants in VDAART, a multi-site randomized, double-blind, placebo-controlled trial of Vitamin D supplementation during pregnancy for prevention of asthma and other allergic diseases in offspring conducted in the United States (NCT00920621)23 The study protocol was approved by the institutional review boards at each participating institution and at Brigham and Women's Hospital. All participants provided written informed consent. Pregnant women (n = 876) were recruited during the first trimester of pregnancy from three sites across the United States: Boston Medical Center, Washington University in St. Louis, and Kaiser Permanente Southern California Region in San Diego.
VDAART study design and sample collection
After delivery, 806 children were followed using an over-the-phone quarterly health questionnaire and annual in-person visits. Stool samples were collected from mothers in the third trimester, and from their children at months 3–6, age one, and age three years. Stool was not collected if the mother or child had used antibiotics in the past 7 days. Then metagenomic sequencing, metabolomic profiling, and measurement of short chain fatty acids were conducted on the collected stool samples as detailed below (see Supplementary files: Fig. S1 for the detailed sample size of each omics dataset).
Ages and stages questionnaire
At the end of the first, second, and third year, study researchers administered the Ages and Stages Questionnaire (third edition) to the VDAART primary caregivers during a study clinic visit or phone interview. The ASQ assesses 5 developmental domains (gross motor skills, fine motor skills, problem-solving ability, communication, and personal and social skills) at different ages (year one, n = 99; year two, n = 102; and year three, n = 110). Each domain is assessed by 6 questions ascertaining achievement of relevant skills and answered as yes (10 points), sometimes (5 points), or not yet (0 points). Scores for individual items (within the same domains) are summed to give an overall continuous score for each of the 5 domains (possible range, 0–60).
Sequencing and profiling of bacterial 16S rRNA
Metagenomic sequencing and profiling were performed on stool samples by sequencing the 16S rRNA hypervariable region 4 (V4) on the Illumina MiSeq platform, as previously described.29 Raw reads were demultiplexed and processed (e.g., quality filtering and trimming) using tools available in QIIME 2 (version 2020.8).30 Clean reads (averagely 51,935 ± 24,962 ranging from 23,983 to 179,820) were then denoised and clustered with dada2 and classified using a pre-trained machine-learning-based classifier of the Silva database.
Measurement of short chain fatty acids (SCFAs)
Eight SCFAs were measured by liquid chromatography with tandem mass spectrometry by Metabolon, Inc (Morrisville, NC): acetic acid, propionic acid, butyric acid, isobutyric acid, 2-methyl-butyric acid, isovaleric acid, valeric acid, and hexanoic acid. Sample preparation was conducted according to previously described methods.31 In brief, the reaction mixture was diluted, and an aliquot injected onto an Agilent 1290/AB Sciex QTrap 5500 liquid chromatography with tandem mass spectrometry system equipped with a C18 reversed phase ultra-high performance liquid chromatography column. Raw data were collected and processed using AB SCIEX software Analyst 1.6.2.
Metabolomics profiling
Stool samples taken from the selected paired mothers and children (Fig. 1) were used for metabolomic profiling. Sample preparation was conducted according to previously described methods.32 Nontargeted global metabolomic profiles were generated at Metabolon Inc. by using ultra-performance liquid chromatography–tandem mass spectroscopy (UPLC-MS/MS). Briefly, four platforms (UPLC-MS/MS under positive ionization, UPLC-MS/MS under negative ionization, UPLC-MS/MS polar platform, and gas chromatography–MS) were used to detect a comprehensive list of metabolites throughout the metabolome. Metabolites were identified by their m/z, retention time, and through comparison to library entries of purified known standards. For the data preparation and quality control, metabolites with CV >25%, missingness >10%, or no variability based on the IQR we removed in the quality control samples. Metabolon's authentic standard library contains identifying ion and chromatographic features of over 4000 known metabolites (and an additional 7000 entries for unnamed compounds) present in metabolic pathways.
Statistics
The Bray–Curtis dissimilarity matrices (based on ASVs, metabolites, and SCFAs) were calculated by QIIME2. PERMANOVA (Adonis) was then used to test the effect size of host factors such as recruitment site, education level, marital status, family income, gestational age (when maternal stool samples were collected since pregnancy), Vitamin D treatment, mothers' health status, and children's ASQ measures on the multi-omics data. We chose not to use classic multiple comparison correction techniques (e.g., Bonferroni) for two reasons: (1) these would be too conservative, given the correlated nature of the outcomes and the compositionality of microbiome and metabolome data; and (2) these analyses were performed for the purposes of covariate selection, instead of causal inference.
Songbird33 was used to perform the multinomial regression (including null model construction), which is specially designed to process compositional data such as metagenomic and metabolomic profiling results. Songbird leverages the ratios between any two taxa/metabolites instead of the relative abundance as features to deal with the compositionality issue. In the Songbird workflow, once a good fit is created for the model, a comparison with a null model (which is made without any metadata input) is created to make sure the model is more predictive than the null model. This will allow us to see how strongly the covariates in the formula can be associated with the features of the model, with respect to random chance (Supplementary files: Fig. S2). We included as covariates all subject characteristics that were associated with gut microbiota and metabolites (PERMANOVA test P-value <0.05 or F value >2, Table S1). The model with these covariates was compared with a null model containing no covariates to estimate a Q2 adapted from the partial least squares literature. As for the statistic of Songbird, Q2 is denoted as 1 – m1/m2, where m1 indicates the average absolute model error and m2 indicates the average absolute null model error. Q2 values close to 1 indicate a high discriminant accuracy on cross-validation samples. Q2 values that are low or below zero indicate poor discriminant accuracy, suggesting possible overfitting.
The output from Songbird also includes differentials that describe the log-fold change of microbes/metabolites with respect to ASQ measures. The most important aspect of these differentials are ranks (Songbird does not generate P-values but instead generates taxa's or metabolites' ranks because when using a reference-frame-based approach, there is no reasonable null distribution for the generation of test statistics), which are obtained by sorting a column of differentials from lowest to highest. These ranks give information on the relative associations of features (microbes and metabolites) with a given covariate, e.g., the features with the most negative differential ranking values will be more associated with low ASQ samples, whereas the features with the most positive differential ranking values will be more associated with high ASQ samples.
Role of funding source
The Funders do not play any roles in study design, data collection, data analyses, interpretation, or writing of report.
Results
Participants and data collection
In the VDAART cohort, 876 pregnant women were recruited from three sites across the United States between 2009 and 2015, and their offspring continue to be followed (Methods). In this study, 116 mother-child pairs were selected for their relatively complete multi-omics datasets and ASQ questionaries (Methods, Supplementary files: Fig. S1): (i) The stool metagenomic sequencing data are accessible for 116 mothers (prenatal), 85 infants at months 3–6, 105 children at year one, and 80 children at year three; (ii) The stool short-chain fatty acid (SCFA) measurements are accessible for 113 mothers (prenatal), 73 infants at months 3–6, 68 children at year one, and 65 children at year three; (iii) The stool metabolomic profiles are available for 111 mothers (prenatal), 114 infants at months 3–6, 75 children at year one, and 70 children at year three. As for the ASQ measures of the children, completed ASQ data are available for 99 children at year one (ASQ-Y1), 102 children at year two (ASQ-Y2), and 110 children at year three (ASQ-Y3). Even though the sample size in our study declines with time, it remains top in longitudinal mother-infant microbiome studies.
The maternal participants (n = 116) are ethnically diverse (22% Hispanic or Latino and 78% not Hispanic or Latino) and racially diverse (38% white, 49% black, and 13% other, Table 1). They also exhibit a representative range of family incomes (36% less than 30 k/year, 30% 30 k–100 k/year, and 9% more than 100 k/year), pre-pregnancy BMI (58% overweight), and education levels (62% attended at least some college). In terms of health status, 35% of mothers had asthma, 28% had eczema, 56% had a fever, and one mother had gestational diabetes during pregnancy. Most of their children (65%) were delivered vaginally, and more than half were breastfed (52%) in the first six months of life. In addition, 48% of mothers were randomized to receive high-dose vitamin D during pregnancy. All the above clinical covariates that might be associated with the gut microbiota or neurocognitive development were considered potential confounders in the downstream statistical analysis.
Table 1.
Characteristics of mother participants in this study.
| Characteristic and mothers # (n = 116) | |
|---|---|
| Recruitment site | |
| Boston | 52 (45%) |
| St Louis | 49 (42%) |
| San Diego | 15 (13%) |
| Maternal education level | |
| College graduate or graduate school | 37 (32%) |
| Some college | 35 (30%) |
| High school or technical school | 24 (21%) |
| Less than high school | 20 (17%) |
| Family income | |
| >100 K/year | 10 (9%) |
| 30K–100 K/year | 35 (30%) |
| <30 K/year | 42 (36%) |
| Unknown | 29 (25%) |
| Marital status | |
| Single | 64 (55%) |
| Married | 47 (41%) |
| Divorced | 5 (4%) |
| Maternal ethnicity | |
| Hispanic or Latino | 26 (22%) |
| Not Hispanic or Latino | 90 (78%) |
| Maternal race | |
| Black or African American | 57 (49%) |
| White | 44 (38%) |
| Other | 15 (13%) |
| Gestation days | |
| 30–33 weeks | 57 (49%) |
| 34–37 weeks | 59 (51%) |
| Pre-pregnancy BMI | |
| Overweight | 67 (58%) |
| Normal | 36 (31%) |
| Unknown | 13 (11%) |
| Eczema | |
| Yes | 32 (28%) |
| No | 84 (72%) |
| Asthma | |
| Yes | 41 (35%) |
| No | 75 (65%) |
| High fever | |
| Yes | 65 (56%) |
| No | 51 (44%) |
| Treatment | |
| Vitamin D | 61 (53%) |
| Blank | 55 (47%) |
| Delivery mode | |
| Cesarean section | 41 (35%) |
| Vaginal birth | 75 (65%) |
| Child gender | |
| Female | 52 (45%) |
| Male | 64 (55%) |
| Child ethnicity | |
| Hispanic or Latino | 30 (26%) |
| Not Hispanic or Latino | 86 (74%) |
| Child race | |
| Black or African American | 66 (57%) |
| White | 36 (31%) |
| Other | 14 (12%) |
| Breast feeding until month 6/12 | |
| Yes | 60 (52%)/35 (30%) |
| No | 56 (48%)/81 (70%) |
| Formular used until month 6/12 | |
| Yes | 87 (75%)/88 (75%) |
| No | 26 (22%)/25 (22%) |
| Unknown | 3 (3%)/3 (3%) |
Data are given as number (and percentage) of individuals. Besides mothers' characteristics, their children's information such as gender, ethnicity, and race are also included. Not all metagenomic and metabolomic data are available for all 116 mother-child pairs, please see Supplementary files: Fig. S1 for detailed information.
Assessment of children's neurodevelopment by ASQ and profiling of the gut microbiome at different stages
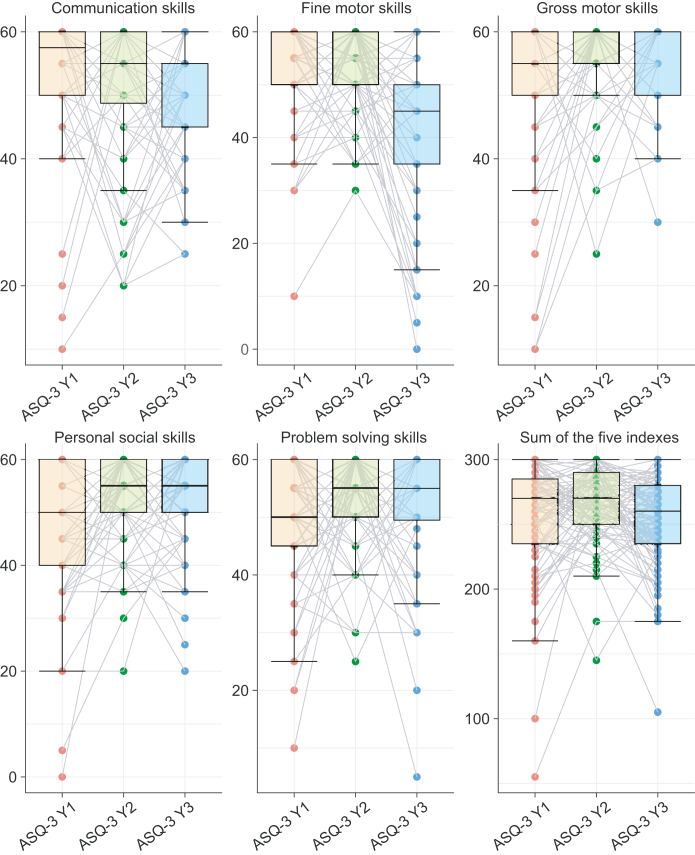
To evaluate children's neurodevelopment, ASQ measures at year one, year two, and year three were analyzed. Scores within the domains of communication skills (COM), gross motor skills (GM), fine motor skills (FM), problem-solving ability (ProS), and personal social skills (PerS) were summed to give an overall continuous measurement for each of the five domains (see Methods). We found that the ASQ scores over the three years measured were unstable and unpredictable whether separated into individual domains or combined in a single summary score (Fig. 2), which is in line with previous studies.34 For example, subjects with lower scores at year one (ASQ-Y1) may have higher scores at year two (ASQ-Y2) and may again have lower scores at year three (ASQ-Y3). Thus, it is inappropriate to categorize the children based on the irregular change of the three years' ASQ measures (within person ASQ through three years). Hence, the association between the gut microbiota and ASQ measures was separately analyzed year by year in this study.
Fig. 2.
The change of the five domains in ASQ from year one to year three. The boxplots illustrate the comparison of the five ASQ measures and their sum score across the first three years of life, with grey lines connecting the same individual. ASQ-Y1 (orange boxes) refers to the ASQ at year one, ASQ-Y2 (green boxes) refers to the ASQ at year two, and ASQ-Y3 (blue boxes) refers to the ASQ at year three.
We analyzed the maternal prenatal, infant (months 3–6), and children's year one and year three gut microbial compositions. We found that the taxonomic diversity increased along with the children's age. In particular, we identified 3770 ASVs (Amplicon Sequence Variants) in the maternal prenatal gut microbiome, 1440 ASVs in the infant gut microbiome, 2376 ASVs in child gut microbiome at year one, and 2360 ASVs in child gut microbiome at year three (please see Supplementary files: Fig. S3 for an overall picture (PCoA) of the microbiome and metabolome data at different stages). Notably, compared to the gut microbiota, the maternal factors (Table 1) have a relatively smaller effect size on the neurodevelopment of their children (Supplementary files: Fig. S4).
The prenatal period may be the critical time window for the association between the gut microbiome and neurodevelopment
To further identify associations of the maternal prenatal gut microbiome, the infant gut microbiome (months 3–6), and the child gut microbiome (year one and year three) with neurodevelopment (ASQ-Y1, ASQ-Y2, and ASQ-Y3), we performed multinomial regression using reference frames33 to explicitly address the compositionality issue in microbiome data analysis. Specifically, we compared null models with a series of multinomial regression models built for discriminating the neurodevelopment of different ages (ASQ-Y1, ASQ-Y2, and ASQ-Y3) based on the gut microbiota and metabolites of different stages (mother prenatal, infant, child year one, and child year three). The Q2 score (adapted from the partial least squares literature) allows us to see how strongly the ASQ measures are associated with the gut microbiome at different stages, as compared to random chance (Methods). Higher Q2 (positive) indicates a higher discriminant accuracy (with values of 1 corresponding to 100% accuracy) on cross-validation and suggests a stronger relationship between the gut microbiome and neurodevelopment, while negative Q2 indicates poor discriminant accuracy and irrelevance.
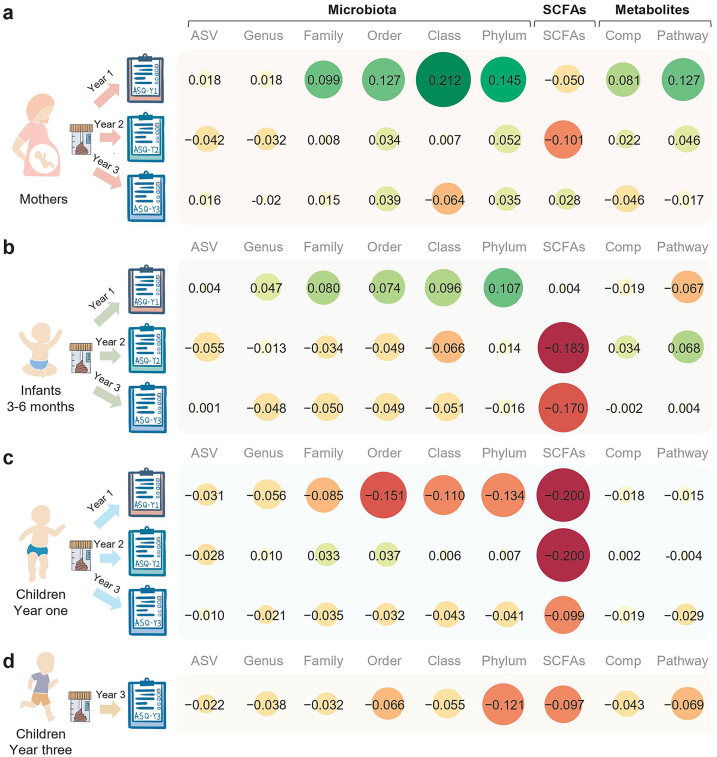
When using the maternal prenatal gut microbiome to discriminate the ASQ measures (Fig. 3a, Supplementary files: Fig. S5a), we found that the best performance of discriminating the five domains of ASQ-Y1 by the maternal prenatal gut microbiota is at the class-level (with average Q2∼0.21 compared to 0.145, 0.127, 0.099, 0.018, and 0.018 at the phylum, order, family, genus and ASV level, respectively). Moreover, both the maternal prenatal gut microbiota and metabolites are significantly associated with the five domains in children's ASQ-Y1, with Q2 ranging from 0.190 to 0.233 for microbiota at the class level and Q2 ranging from 0.108 to 0.157 for metabolites at the pathway level (Methods). Of note, the performance of discriminating ASQ-Y2 (or ASQ-Y3) measures by the maternal prenatal gut microbiome is much lower than that of discriminating ASQ-Y1 measures (with Q2 values less than 0.052 and 0.039 for ASQ-Y2 and ASQ-Y3).
Fig. 3.
Average discriminating power (Q2) of the gut microbiome for the ASQ in five domains. (a) The discriminating power of the maternal prenatal gut microbiome for child neurodevelopment. (b) The discriminating power of the infant gut microbiome in early life (months 3–6) for child neurodevelopment. (c) The discriminating power of the child gut microbiome at year one for child neurodevelopment. (d) The discriminating power of the child gut microbiome at year three for child neurodevelopment at the same time point. Red pies indicate negative Q2 and green pies indicate positive Q2. The number in each pie chart is the average Q2 of communication skills, fine motor skills, gross motor skills, personal social skills, and problem-solving skills. The higher Q2 (positive) indicates a higher discriminant accuracy for ASQ, while negative Q2 indicates poor discriminant accuracy or overfitting of the multinomial regression models for ASQ measures. The value less than −0.2 will be trimmed to −0.2 for visualization, and comp is short for compound (metabolite).
When using the infant gut microbiome to discriminate the ASQ measures (Fig. 3b, Supplementary files: Fig. S5b), we found that the infant gut microbiota is only relevant to ASQ-Y1 (Q2 ranges from 0.076 to 0.128 at the class level) while the metabolites are only relevant to ASQ-Y2 (Q2 ranges from 0.036 to 0.094 at the pathway level), and the SCFAs showed limited relevance to FM, GM, and ProS of ASQ-Y1. Of note, the discriminant of ASQ-Y1 measures using the infant gut microbiota (with average Q2∼0.096) is much weaker than that of using the maternal prenatal gut microbiota (with average Q2∼0.212). The performance of discriminating ASQ-Y3 measures using the infant gut microbiome is very low (with Q2 values less than 0.004).
When using the child gut microbiome (year one and year three, Fig. 3c and d, Supplementary files: Fig. S5c and d) to discriminate the ASQ measures, we found that the year one child gut microbiome (including SCFAs) is not relevant for discriminating ASQ-Y1 but slightly relevant for discriminating ASQ-Y2 (Q2 ranges from to 0.011 to 0.065 for the five domains in ASQ-Y2 at the order level). We also found that year-three child gut microbiome (including SCFAs) cannot be associated with ASQ-Y3 measures (with Q2 values less than −0.02).
A high consistency finding by metagenomic and metabolomic data indicates that the maternal prenatal gut microbiome is much more relevant for discriminating the offspring year-one neurodevelopment (assessed by ASQ-Y1 measures) than the infant (months 3–6) and the child gut microbiome (year one). Also, the discriminating power of maternal prenatal and infant gut microbiome gradually diminishes with respect to neurodevelopment at year two (assessed by ASQ-Y2) and neither the maternal prenatal nor child microbiome has any discriminating power for the neurodevelopment at year three (assessed by ASQ-Y3), indicating the little impact of the maternal prenatal and early child microbiome on long term neurodevelopment and neurodevelopmental outcomes.
The different roles of the maternal and infant gut microbiota in neurodevelopment
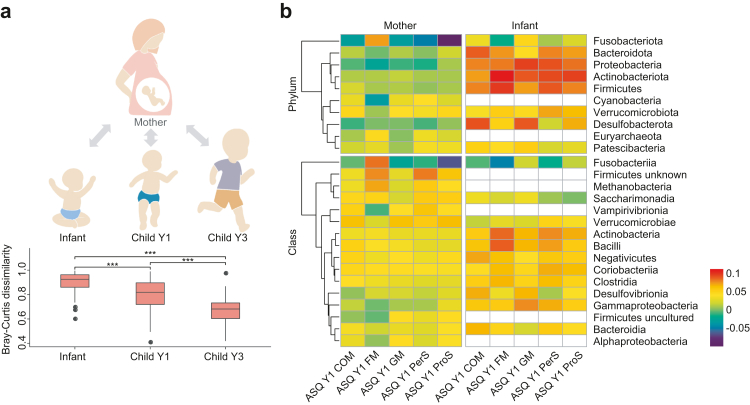
To further explore whether the discriminating power of the infant gut microbiome for neurodevelopment is a consequence of vertical transmission from mothers to their children, we analyzed the similarity of the gut microbiota between paired mothers (prenatal) and children (Fig. 4a). We found that the similarity of the infant gut microbiota with their mothers’ prenatal gut microbiota increased along with age. For example, the average Bray–Curtis dissimilarity of the gut microbiota between infant and their mother is 0.904, while for the child gut microbiota at year one and year three, at which points the microbiome was poorly discriminative of ASQ-Y1 (or ASQ-Y3, Fig. 3c), the number is 0.806 and 0.678, respectively. This suggests that the relevance between the infant gut microbiota and ASQ measures is not associated with the similarity with the maternal prenatal gut microbiota. Thus, the roles of the maternal prenatal and infant gut microbiomes in neurodevelopment are potentially different.
Fig. 4.
Different roles of maternal and infant gut microbiota in neurodevelopment. (a) Comparison of BC dissimilarity between paired maternal prenatal and infant gut microbiota. Red boxes refer to the BC dissimilarity between paired mothers and their children (n = 85, 105, 80 for infant, Child Y1, and Child Y3). (b) The heatmap shows the association ranks of classes from maternal prenatal, infant, and children gut microbiota against ASQ scores. The taxa with negative differential ranks are more associated with low-ASQ samples, whereas the taxa with positive differential ranks are more associated with high-ASQ samples. Since the multinomial regressions were separately performed at each stage, only the sign (positive or negative) of values and comparisons within stages are reasonable.
To further understand the role of the gut microbiota in neurodevelopment, we compared the differentials (the term “differential” refers to the logarithm of the fold change in abundance of taxa with respect to ASQ measures,33 see Methods) with respect to the five domains in ASQ-Y1 from the maternal prenatal and infant gut microbiota. Specifically, since the microbial taxa at the class and phylum level from these two stages are most discriminative of ASQ-Y1, we compared the ranks of differential classes and phyla in multinomial regression models, which give information on the relative associations of taxonomic classes and phyla with ASQ-Y1 (Methods). Although the differential taxa in the maternal and infant gut with respect to ASQ-Y1 exhibit high overlap (e.g., there are 11/16 and 8/10 ASQ-Y1 related classes and phyla shared between the two stages, Fig. 4b), our results demonstrated that the same taxa in the maternal prenatal and infant gut microbiota do not necessarily play the same role in neurodevelopment. For example, the class Fusobacteriia is more associated with high fine motor skills in ASQ-Y1 (rank = 0.084) in the maternal prenatal gut microbiota, but becomes more associated with low fine motor skills (rank = −0.047) in the infant gut microbiota (the relative abundance of Fusobacteriia is 0.02% and 0.03% in the maternal prenatal and infant gut microbiota, Fig. 4b). Moreover, in the maternal prenatal gut microbiota, the phylum Fusobacteriota is mostly associated with high scores in the five domains in ASQ-Y1 (rank for COM, FM, GM, PerS, and ProS are −0.030, 0.073, −0.031, −0.049, and −0.099 respectively) while Fusobacteriota exhibits opposite associations (0.033, −0.017, 0.048, 0.009, and 0.024) in the infant gut microbiota for neurodevelopment.
Similarly, to investigate the role of metabolites in neurodevelopment, we compared the differentials with respect to the five domains in ASQ-Y1 and ASQ-Y2 from the maternal prenatal and infant gut metabolome separately. Specifically, since the metabolites and pathways from these two stages are most discriminative of ASQ-Y1 and ASQ-Y2 separately (Fig. 3), we compared top-20 differential metabolites and pathways that are most relative to neurodevelopment (Supplementary files: Fig. S6). Then we found that there is no overlap between the top 20 metabolites between the two stages. However, 7 out of the top 20 ASQ associated pathways are shared between mother and infant, in which both support that Acetylated Peptides is the pathway most relevant to neurodevelopment (e.g., its rank for COM, FM, GM, PerS, and ProS are −0.743, −0.749, −0.756, −0.802, and −0.807 in maternal and −0.533, −0.496, −0.510, −0.488, and −0.497 in infant respectively).
Discussion
Mounting evidence linking offspring neurodevelopment with the maternal prenatal gut microbiota comes from mouse studies.4, 5, 6, 7, 8, 9, 10, 11,13, 14, 15,20,21 To date, only one human study has examined the role of the maternal prenatal gut microbiota in offspring neurodevelopment.16 More studies are certainly needed to address foundational questions about the maternal prenatal gut microbiota and children's neurodevelopment. In this work, we leveraged a human study to compare the ability of the maternal prenatal and child gut microbiomes to discriminate child neurodevelopment. Interestingly, the maternal prenatal gut microbiome exhibited stronger associations with child neurodevelopment at year one than the infant (months 3–6) or the first-year child gut microbiomes. Further analysis illustrated that similar class-level taxa were associated with neurodevelopment among both the maternal prenatal and infant gut microbiota, but those taxa may play different roles during pregnancy and in infancy.
A previous study found structural differences in the maternal prenatal gut microbiota between the mothers of children with behavioral problems (n = 20) in comparison to mothers of normative behavior children (n = 195),16 consistent with our finding of a significant effect of the maternal gut microbiota on ASQ measures (including problem solving skills and personal social skills). In our study, instead of using the Childhood Behavior Checklist at age two (which may lead to unbalanced case–control sample size and subsequent problematic differential abundance analysis), we measured neurodevelopment by ASQ in the first three years of life (ASQ-Y1, ASQ-Y2, and ASQ-Y3) and then quantitatively associated its five domain scores with maternal prenatal and child gut microbiome features at different time points (months 3–6, year one, and year three), allowing us to explore associations of the gut microbiome with neurodevelopment more comprehensively.
As both 16S rRNA and whole metagenome sequencing data in microbiome studies are inherently compositional, differential abundance analysis is challenging. Indeed, analyzing relative abundance data with inappropriate statistical methods can lead to 100% false discovery rates.33,35,36 Comparing ratios of taxa as relative differentials (logarithm of the fold change in abundance of taxa between two conditions) can circumvent this issue. Compared with previous studies about the association between maternal prenatal gut microbiota and child neurodevelopment, we applied reference frames to address the compositionality inherent in microbiome data.33 Specifically, we estimated the relative differentials directly using multinomial regression,37, 38, 39 then compared our model with a null model to quantify the relevance between features (the profiling results of the gut microbiota) and factors (ASQ measures), and the coefficients from multinomial regression analysis were ranked to determine which taxa varied the most along with ASQ measures.
In previous studies, Fusobacterium, Fusobacteriaceae, Fusobacteriales, Fusobacteria, and Fusobacteriota have been reported to be associated with brain and nervous system disorders like major depressive disorder40 and multiple sclerosis.41 Specifically, previous studies have associated the neuroinflammation (and intestinal inflammation) and Fusobacterium, with evidence that Fusobacterium releases outer membrane vesicles which activate TLR4 and NF-κB to stimulate proinflammatory signals and promote proinflammatory signaling cascades in the context of a depleted intestinal microbiome.42,43 In addition, the relative abundance of Fusobacteria was found to be different in rats who are vulnerable to early life stress.44 However, although validation and mechanistic studies are needed to confirm our findings, we report a potential two-sided role of Fusobacteria and Fusobacteriota in neurodevelopment with divergent associations with ASQ-Y1 when measured in pregnant mothers compared to during infancy. Further investigation at the ASV level reveals that 7 Fusobacteriota ASVs are shared in the maternal and infant gut microbiome, taking approximately 35% and 95% relative abundance to all Fusobacteriota ASVs separately. This suggests that the differential colonization of Fusobacteriota after the vertical transmission may lead to the different roles of Fusobacteriota in the maternal and infant gut microbiome on neurodevelopment. When establishing the gut microbiota in the first years of life, a healthy breastfeeding infant should have a microbial community that is distinct from an adult composition,45,46 which is mainly determined by environmental exposures47 such as delivery mode, breast (versus formula) feeding, and antibiotics.48 For example, breastmilk is an essential factor in modifying the infant gut microbiome and confers significant nutritional and immunological benefits to the infant, which ensures the vertically transmitting of the maternal gut microbiome and has also been associated with improved neurodevelopmental outcomes.49 Although infants have a markedly different gut microbial community from adults, the idiosyncratic microbial ecosystems in each baby will gradually converge toward a profile characteristic of the adult gastrointestinal tract in the first few years.50 Considering the role of the gut microbes can be different, especially if they are taxa that would be acquired or dominated at different stages of life,51 we believe the role of the gut microbiota should be separately investigated after vertical transmission.
As for the rest of the shared differential classes between the maternal prenatal and infant gut microbiota with respect to the five domains in ASQ-Y1, many of them, including Desulfovibrionia, Gammaproteobacteria, Negativicutes, Bacteriodia, Verrucomicrobiae, Clostridia, Actinobacteria, and Coribacteria, have been related to neurodevelopment in previous studies,52, 53, 54, 55, 56, 57, 58 which supports the accuracy of our overall findings (Table S2). For example, Desulfovibrionia and Coribacteria were found to be associated with brain aging and Alzheimer's disease; Gammaproteobacteria and Clostridia were found to be associated with major depressive disorder and autism spectrum disorders; Negativicutes, Bacteroides, Verrucomicrobiae, and Actinobacteria were found to be associated with neurodevelopment. Moreover, as the most ASQ-associated pathway supported by both maternal and infant metabolomic data in our study, Acetylated Peptides has been well acknowledged in brain development and function.59 Notably, altered histone deacetylation60 has been strongly associated with multiple neurological disorders such as neurodegenerative diseases,61 psychiatric disorders,62 acute brain injury,63 and Schizophrenia.64 This suggests that the targeted interventions for these diseases should begin as early as possible, for example, in the fetal stage during pregnancy, which may greatly increase the treatment effect.
Although the mothers in the VDAART cohort were supplemented with vitamin D during pregnancy, it has been proved to have a limited effect on the gut microbiome (Table S1) and thus would not influence the identified associations in this study. However, there are several limitations in this study. For example, larger studies may yield higher resolution insight into the relationship between the maternal gut microbiome composition during the third trimester of pregnancy and offspring neurodevelopmental outcomes, and more time points evaluated during earlier pregnancy and postnatal (first or second trimester and infant stool samples) are warranted to narrow down the critical time window for offspring neurodevelopment and to further validate the observation (because the maternal gut microbiome undergoes substantial remodeling throughout pregnancy). Moreover, the relative abundance of Fusobacteriia is extremely low in this study, further validation of its two-sided role by qPCR or digital PCR in a larger population is highly warranted. In addition, future research utilizing multi-omics data (e.g., whole metagenomic sequencing which would be useful in understanding the potential functions of the microbiome and decoding its taxonomic structure at the strain level) and systematic experiments are further warranted to deepen our understanding of the mechanism of prenatal and postnatal influence on neurodevelopment by the gut microbiome.
Contributors
ZS and YYL designed the project. ZS analyzed all the data with assistance from KLS and RK. ZS and YYL interpreted the results. ZS wrote the manuscript. YYL, KLS, RK, and SW edited the manuscript. JLS and AAL reviewed the manuscript. All authors approved the manuscript. YYL supervised the study.
Data sharing statement
Microbiome sequencing and metabolome profiling data from VDAART are part of the ECHO consortium and ECHO consortium members can obtain the data directly from the ECHO DCC or for those not part of ECHO directly from the authors. All other relevant data are available from the authors upon reasonable requests.
Declaration of interests
Scott. T. Weiss receives author royalty from UpToDate, and is on the Board of Directors of Histolix. Jessica Lasky-Su is a consultant for TruDiagnositc, and is on the Scientific Advisory Board of Precion. Augusto A. Litonjua is on the Data Safety Monitoring Board of PreCISE Network, and receives author royalty from UpToDate. No potential competing interest was reported by the other authors.
Acknowledgements
This work was supported by the National Institutes of Health (grant numbers: R01AI141529, R01HD093761, RF1AG067744, UH3OD023268, U19AI095219, U01HL089856, R01HL141826, K08HL148178, K01HL146980), and the Charles A. King Trust Postdoctoral Fellowship.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104491.
Appendix A. Supplementary data
References
- 1.Clemente J.C., Ursell L.K., Parfrey L.W., Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148(6):1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sherwin E., Bordenstein S.R., Quinn J.L., Dinan T.G., Cryan J.F. Microbiota and the social brain. Science. 2019;366(6465) doi: 10.1126/science.aar2016. [DOI] [PubMed] [Google Scholar]
- 3.Collins S.M., Surette M., Bercik P. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol. 2012;10(11):735–742. doi: 10.1038/nrmicro2876. [DOI] [PubMed] [Google Scholar]
- 4.Braniste V., Al-Asmakh M., Kowal C., et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci Transl Med. 2014;6(263):263ra158. doi: 10.1126/scitranslmed.3009759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu C., Murdock M.H., Jing D.Q., et al. The microbiota regulate neuronal function and fear extinction learning. Nature. 2019;574(7779):543–548. doi: 10.1038/s41586-019-1644-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erny D., de Angelis A.L.H., Jaitin D., et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci. 2015;18(7):965–977. doi: 10.1038/nn.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoban A.E., Stilling R.M., Ryan F.J., et al. Regulation of prefrontal cortex myelination by the microbiota. Transl Psychiatry. 2016;6:e774. doi: 10.1038/tp.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogbonnaya E.S., Clarke G., Shanahan F., Dinan T.G., Cryan J.F., O'Leary O.F. Adult hippocampal neurogenesis is regulated by the microbiome. Biol Psychiatry. 2015;78(4):E7–E9. doi: 10.1016/j.biopsych.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 9.Cryan J.F., Dinan T.G. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13(10):701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 10.Clarke G., Grenham S., Scully P., et al. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol Psychiatry. 2013;18(6):666–673. doi: 10.1038/mp.2012.77. [DOI] [PubMed] [Google Scholar]
- 11.Sudo N., Chida Y., Aiba Y., et al. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J Physiol. 2004;558(Pt 1):263–275. doi: 10.1113/jphysiol.2004.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heijtz R.D. Fetal, neonatal, and infant microbiome: perturbations and subsequent effects on brain development and behavior. Semin Fetal Neonatal Med. 2016;21(6):410–417. doi: 10.1016/j.siny.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 13.Buffington S.A., Di Prisco G.V., Auchtung T.A., Ajami N.J., Petrosino J.F., Costa-Mattioli M. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell. 2016;165(7):1762–1775. doi: 10.1016/j.cell.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu L., Zhong X., He Y., Shi Y. Butyrate, but not propionate, reverses maternal diet-induced neurocognitive deficits in offspring. Pharmacol Res. 2020;160 doi: 10.1016/j.phrs.2020.105082. [DOI] [PubMed] [Google Scholar]
- 15.Kim S., Kim H., Yim Y.S., et al. Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature. 2017;549(7673):528–532. doi: 10.1038/nature23910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dawson S.L., O'Hely M., Jacka F.N., et al. Maternal prenatal gut microbiota composition predicts child behaviour. eBioMedicine. 2021;68 doi: 10.1016/j.ebiom.2021.103400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meckel K.R., Kiraly D.D. Maternal microbes support fetal brain wiring. Nature. 2020;586(7828):203–205. doi: 10.1038/d41586-020-02657-y. [DOI] [PubMed] [Google Scholar]
- 18.Jasarevic E., Howard C.D., Morrison K., et al. The maternal vaginal microbiome partially mediates the effects of prenatal stress on offspring gut and hypothalamus. Nat Neurosci. 2018;21(8):1061–1071. doi: 10.1038/s41593-018-0182-5. [DOI] [PubMed] [Google Scholar]
- 19.Tochitani S. Vertical transmission of gut microbiota: points of action of environmental factors influencing brain development. Neurosci Res. 2021;168:83–94. doi: 10.1016/j.neures.2020.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Vuong H.E., Pronovost G.N., Williams D.W., et al. The maternal microbiome modulates fetal neurodevelopment in mice. Nature. 2020;586(7828):281–286. doi: 10.1038/s41586-020-2745-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luczynski P., Neufeld K.A.M., Oriach C.S., Clarke G., Dinan T.G., Cryan J.F. Growing up in a bubble: using germ-free animals to assess the influence of the gut microbiota on brain and behavior. Int J Neuropsychopharmacol. 2016;19(8):1–17. doi: 10.1093/ijnp/pyw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walter J., Armet A.M., Finlay B.B., Shanahan F. Establishing or exaggerating causality for the gut microbiome: lessons from human microbiota-associated rodents. Cell. 2020;180(2):221–232. doi: 10.1016/j.cell.2019.12.025. [DOI] [PubMed] [Google Scholar]
- 23.Litonjua A.A., Lange N.E., Carey V.J., et al. The Vitamin D Antenatal Asthma Reduction Trial (VDAART): rationale, design, and methods of a randomized, controlled trial of vitamin D supplementation in pregnancy for the primary prevention of asthma and allergies in children. Contemp Clin Trials. 2014;38(1):37–50. doi: 10.1016/j.cct.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Granana N., Romero Otalvaro A.M. In: Diagnosis, management and modeling of neurodevelopmental disorders. Martin C.R., Preedy V.R., Rajendram R., editors. Academic Press; 2021. Chapter 28–neurodevelopment and the ages and stages questionnaire, third edition (ASQ-3) pp. 319–328. [Google Scholar]
- 25.Chen J., Dueker G., Cowling C. Profiles and predictors of risk for developmental delay: insights gained from a community-based universal screening program. Early Hum Dev. 2018;127:21–27. doi: 10.1016/j.earlhumdev.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Charkaluk M.-L., Rousseau J., Calderon J., et al. Ages and stages questionnaire at 3 Years for predicting IQ at 5–6 years. Pediatrics. 2017;139(4) doi: 10.1542/peds.2016-2798. [DOI] [PubMed] [Google Scholar]
- 27.Halbwachs M., Muller J.B., Nguyen The Tich S., et al. Predictive value of the parent-completed ASQ for school difficulties in preterm-born children <35 weeks' GA at five years of age. Neonatology. 2014;106(4):311–316. doi: 10.1159/000363216. [DOI] [PubMed] [Google Scholar]
- 28.Kelly R.S., Boulin A., Laranjo N., et al. Metabolomics and communication skills development in children; evidence from the ages and stages questionnaire. Metabolites. 2019;9(3):42. doi: 10.3390/metabo9030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee-Sarwar K.A., Kelly R.S., Lasky-Su J., et al. Integrative analysis of the intestinal metabolome of childhood asthma. J Allergy Clin Immunol. 2019;144(2):442–454. doi: 10.1016/j.jaci.2019.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolyen E., Rideout J.R., Dillon M.R., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37(8):852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee-Sarwar K.A., Kelly R.S., Lasky-Su J., et al. Fecal short-chain fatty acids in pregnancy and offspring asthma and allergic outcomes. J Allergy Clin Immunol Pract. 2020;8(3):1100–1102.e13. doi: 10.1016/j.jaip.2019.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelly R.S., Dahlin A., McGeachie M.J., et al. Asthma metabolomics and the potential for integrative omics in research and the clinic. Chest. 2017;151(2):262–277. doi: 10.1016/j.chest.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morton J.T., Marotz C., Washburne A., et al. Establishing microbial composition measurement standards with reference frames. Nat Commun. 2019;10(1):2719. doi: 10.1038/s41467-019-10656-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landa R.J., Gross A.L., Stuart E.A., Bauman M. Latent class analysis of early developmental trajectory in baby siblings of children with autism. J Child Psychol Psychiatry. 2012;53(9):986–996. doi: 10.1111/j.1469-7610.2012.02558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mandal S., Van Treuren W., White R.A., Eggesbo M., Knight R., Peddada S.D. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis. 2015;26 doi: 10.3402/mehd.v26.27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morton J.T., Sanders J., Quinn R.A., et al. Balance trees reveal microbial niche differentiation. mSystems. 2017;2(1) doi: 10.1128/mSystems.00162-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silverman J.D., Durand H.K., Bloom R.J., Mukherjee S., David L.A. Dynamic linear models guide design and analysis of microbiota studies within artificial human guts. Microbiome. 2018;6(1):202. doi: 10.1186/s40168-018-0584-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aijo T., Muller C.L., Bonneau R. Temporal probabilistic modeling of bacterial compositions derived from 16S rRNA sequencing. Bioinformatics. 2018;34(3):372–380. doi: 10.1093/bioinformatics/btx549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia F., Chen J., Fung W.K., Li H. A logistic normal multinomial regression model for microbiome compositional data analysis. Biometrics. 2013;69(4):1053–1063. doi: 10.1111/biom.12079. [DOI] [PubMed] [Google Scholar]
- 40.Cheung S.G., Goldenthal A.R., Uhlemann A.C., Mann J.J., Miller J.M., Sublette M.E. Systematic review of gut microbiota and major depression. Front Psychiatry. 2019;10:34. doi: 10.3389/fpsyt.2019.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katz Sand I., Zhu Y., Ntranos A., et al. Disease-modifying therapies alter gut microbial composition in MS. Neurol Neuroimmunol Neuroinflamm. 2019;6(1):e517. doi: 10.1212/NXI.0000000000000517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Engevik M.A., Danhof H.A., Ruan W., et al. Fusobacterium nucleatum secretes outer membrane vesicles and promotes intestinal inflammation. mBio. 2021;12(2):e02706–e02720. doi: 10.1128/mBio.02706-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barandouzi Z.A., Starkweather A.R., Henderson W.A., Gyamfi A., Cong X.S. Altered composition of gut microbiota in depression: a systematic review. Front Psychiatry. 2020;11:541. doi: 10.3389/fpsyt.2020.00541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.El Aidy S., Ramsteijn A.S., Dini-Andreote F., et al. Serotonin transporter genotype modulates the gut microbiota composition in young rats, an effect augmented by early life stress. Front Cell Neurosci. 2017;11:222. doi: 10.3389/fncel.2017.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka M., Nakayama J. Development of the gut microbiota in infancy and its impact on health in later life. Allergol Int. 2017;66(4):515–522. doi: 10.1016/j.alit.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 46.Niu J., Xu L., Qian Y., et al. Evolution of the gut microbiome in early childhood: a cross-sectional study of Chinese children. Front Microbiol. 2020;11:439. doi: 10.3389/fmicb.2020.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palmer C., Bik E.M., DiGiulio D.B., Relman D.A., Brown P.O. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5(7):e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Backhed F., Roswall J., Peng Y., et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17(5):690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 49.Granger C.L., Embleton N.D., Palmer J.M., Lamb C.A., Berrington J.E., Stewart C.J. Maternal breastmilk, infant gut microbiome and the impact on preterm infant health. Acta Paediatr. 2021;110(2):450–457. doi: 10.1111/apa.15534. [DOI] [PubMed] [Google Scholar]
- 50.Vatanen T., Franzosa E.A., Schwager R., et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature. 2018;562(7728):589–594. doi: 10.1038/s41586-018-0620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rinninella E., Raoul P., Cintoni M., et al. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. 2019;7(1):14. doi: 10.3390/microorganisms7010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hughes H.K., Rose D., Ashwood P. The gut microbiota and dysbiosis in autism spectrum disorders. Curr Neurol Neurosci Rep. 2018;18(11):81. doi: 10.1007/s11910-018-0887-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bezawada N., Phang T.H., Hold G.L., Hansen R. Autism spectrum disorder and the gut microbiota in children: a systematic review. Ann Nutr Metab. 2020;76(1):16–29. doi: 10.1159/000505363. [DOI] [PubMed] [Google Scholar]
- 54.Zhang W., Sun Z., Zhang Q., et al. Preliminary evidence for an influence of exposure to polycyclic aromatic hydrocarbons on the composition of the gut microbiota and neurodevelopment in three-year-old healthy children. BMC Pediatr. 2021;21(1):86. doi: 10.1186/s12887-021-02539-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsiao E.Y., McBride S.W., Hsien S., et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155(7):1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kelly J.R., Minuto C., Cryan J.F., Clarke G., Dinan T.G. Cross talk: the microbiota and neurodevelopmental disorders. Front Neurosci. 2017;11:490. doi: 10.3389/fnins.2017.00490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ding X., Xu Y., Zhang X., et al. Gut microbiota changes in patients with autism spectrum disorders. J Psychiatr Res. 2020;129:149–159. doi: 10.1016/j.jpsychires.2020.06.032. [DOI] [PubMed] [Google Scholar]
- 58.Qiao Y., Zhao J., Li C., et al. Effect of combined chronic predictable and unpredictable stress on depression-like symptoms in mice. Ann Transl Med. 2020;8(15):942. doi: 10.21037/atm-20-5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park J., Lee K., Kim K., Yi S.J. The role of histone modifications: from neurodevelopment to neurodiseases. Signal Transduct Target Ther. 2022;7(1):217. doi: 10.1038/s41392-022-01078-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Contestabile A., Sintoni S. Histone acetylation in neurodevelopment. Curr Pharm Des. 2013;19(28):5043–5050. doi: 10.2174/1381612811319280003. [DOI] [PubMed] [Google Scholar]
- 61.Chuang D.M., Leng Y., Marinova Z., Kim H.J., Chiu C.T. Multiple roles of HDAC inhibition in neurodegenerative conditions. Trends Neurosci. 2009;32(11):591–601. doi: 10.1016/j.tins.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abel T., Zukin R.S. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr Opin Pharmacol. 2008;8(1):57–64. doi: 10.1016/j.coph.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang H., Ni W., Wei P., et al. HDAC inhibition reduces white matter injury after intracerebral hemorrhage. J Cereb Blood Flow Metab. 2021;41(5):958–974. doi: 10.1177/0271678X20942613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Farrelly L.A., Zheng S., Schrode N., et al. Chromatin profiling in human neurons reveals aberrant roles for histone acetylation and BET family proteins in schizophrenia. Nat Commun. 2022;13(1):2195. doi: 10.1038/s41467-022-29922-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.