Summary
A reproducible imaging protocol should include four main detailed sections. The first should describe the sample preparation and include details about the tissue and/or cell culture preparation, the staining procedure, the optical grade of the coverslip, and the type of mounting media used to mount the sample. The second section should describe the configuration and components of the microscope and include the type of stand, stage, illumination, and detector, as well as the emission (EM) and excitation (EX) filters, objective, and immersion medium specifications. Specialized microscopes may have other important components in the optical path to include. The third section should describe the settings used to acquire an image like the exposure and/or dwell time, final magnification and optical resolution, the pixel and field of view (FOV) sizes, time intervals for any time lapse, total power at the objective (i.e., directed at your sample) and number of planes and step size used to collect a 3-dimensional image, and order of operations used in multi-dimensional image acquisitions. The final section should include details about the image analysis workflow such as the image processing steps, segmentation and measurement methods used to extract information from the image, data size, and necessary computing hardware and networking requirements if data sets are >1 GB, as well as citations and versions for the software and code used to perform any of these steps. Every effort should be made to make an example dataset with accurate metadata available online. Finally, specifics about the type of replicates included in the experiment and details about the statistical analysis conducted are also necessary.
Graphical abstract
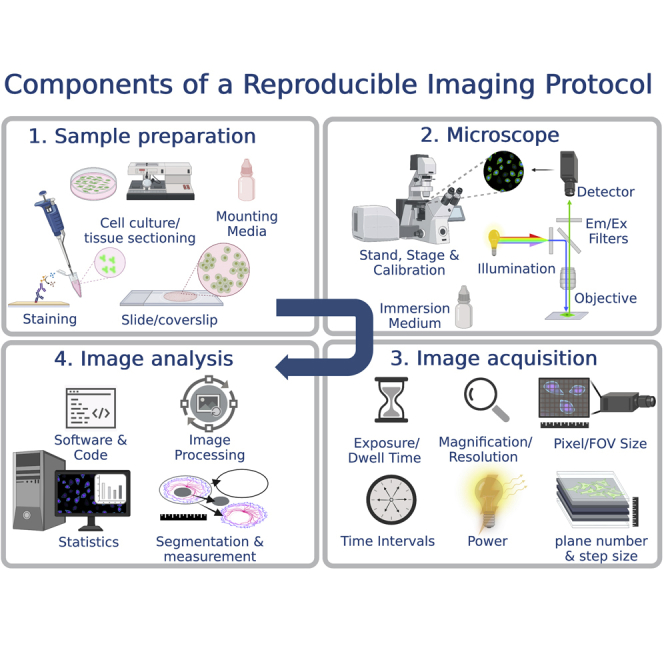
A reproducible imaging protocol should include four main detailed sections. The first should describe the sample preparation and include details about the tissue and/or cell culture preparation, the staining procedure, the optical grade of the coverslip, and the type of mounting media used to mount the sample. The second section should describe the configuration and components of the microscope and include the type of stand, stage, illumination, and detector, as well as the emission (EM) and excitation (EX) filters, objective, and immersion medium specifications. Specialized microscopes may have other important components in the optical path to include. The third section should describe the settings used to acquire an image like the exposure and/or dwell time, final magnification and optical resolution, the pixel and field of view (FOV) sizes, time intervals for any time lapse, total power at the objective (i.e., directed at your sample) and number of planes and step size used to collect a 3-dimensional image, and order of operations used in multi-dimensional image acquisitions. The final section should include details about the image analysis workflow such as the image processing steps, segmentation and measurement methods used to extract information from the image, data size, and necessary computing hardware and networking requirements if data sets are >1 GB, as well as citations and versions for the software and code used to perform any of these steps. Every effort should be made to make an example dataset with accurate metadata available online. Finally, specifics about the type of replicates included in the experiment and details about the statistical analysis conducted are also necessary.
Introduction
STAR Protocols strives toward making bench-ready protocols available to researchers. By having a detailed step-by-step protocol with expanded notes and conceptual background, both novice and expert users have access to details of techniques reported in published scientific articles.
Microscopy techniques are under rapid and continuous development, while easier entry-level instrumentation and expanded accessibility also make these techniques more prominent in biological research. To ensure microscopy protocols are thoroughly reported and reproducible, in this review, we will cover key points for describing sample preparation, microscope and imaging modalities, image acquisition, and image analysis components of an imaging protocol. Such guidelines for reporting microscopy techniques are under development and are a topic of active discussion by the imaging community. More in-depth discussions of standards have previously been reported. Here, we highlight the necessary content and critical details required for the reproducibility of protocols published by STAR Protocols.
One consideration we emphasize throughout an imaging protocol is the need to be specific, as well as to clearly present the conceptual reasoning for each step. Specific details will be helpful to readers who have similar instrumentation and software packages and who can utilize platform-specific details, while conceptual reasoning enables those who have access to different instruments or software tools to be able to carry out conceptually similar steps. If the steps are presented without the reasoning behind them, it is difficult to translate the protocol to different platforms and it is not possible to ensure reproducibility. Microscope details can be complex and are often unknown or misunderstood by non-experts, so it is critical to integrate both levels of reporting in protocols. The same is true for image analysis software and computing hardware details.
Below is a description of each section to be included in an imaging protocol where we highlight commonly overlooked items of importance. We provide Key Resource Table examples for the main sections as well as fictitious excerpts of imaging protocol steps inspired by protocols submitted for publication to demonstrate how to review and identify if a protocol step is suitable (cyan comment bubble), incomplete (yellow comment bubble), and/or missing (magenta comment bubble) critical information for reproducibility. Common questions and answers are addressed.
Sample preparation
Sample culture/collection and staining steps are usually reported reasonably well, with all requisite components clearly identified. These are steps that scientists are most likely to record in detail in their laboratory notebooks (we recommend extending this practice to all the sections discussed in this article). However, the sample preparation section often ends abruptly with a simple “and then images were collected,” without describing how the sample is mounted, whether there was a coverslip used, and if so, what the thickness of the coverslip should be (the appropriate thickness will depend on the imaging objective), or the immersion medium. For some modalities, such as light sheet microscopy, it may be important to include information about other components of a mounting strategy that could be in the light path (such as FEP tubing or a cuvette). While these details may seem trivial, leaving them unspecified potentially introduces a significant amount of experimental variation that could otherwise be carefully controlled. Any components associated with the sample that you introduce into the optical path(s) of the microscope system should be reported.
Example 1. Example key resources table (excerpt from a protocol using human inducible stem cell culture, expressing EGFP-tagged protein, stained with live dyes and imaged live at Allen Institute for Cell Science)
| Reagent or resource | Source | Identifier |
|---|---|---|
| Experimental models: Cell lines | ||
| AICS-0014 cl. 6, nucleoli (DFC) | https://catalog.coriell.org/0/Sections/Search/Sample_Detail.aspx?Ref = AICS-0014&Product = iPSC | CVCL_JM17 |
| Chemicals, peptides, and recombinant proteins | ||
| mTeSR™1 Medium Without Phenol Red | STEMCELL Technologies | Cat#05876 |
| mTeSR™1 Medium | STEMCELL Technologies | Cat#85850 |
| DPBS (1X) | Thermo Fischer Scientific | Cat#14190144 |
| StemPro™ Accutase™ Cell Dissociation Reagent | Thermo Fischer Scientific | Cat#A1110501 |
| CellMaskTM Deep Red Plasma Membrane Stain | Thermo Fisher Scientific | Cat#C10046; Lot#1813792 (5X final concentration); Lot#1853335 and #1900978 (3X final concentration) |
| NucBlueTM Live ReadyProbesTM Reagent | Thermo Fisher Scientific | Cat#R37605 |
| Alexa Fluor® 647 Mouse anti-Human Nanog, Clone N31-355 | BD Biosciences | AB_10611718; Cat#561300 |
Example 2. Mounting media and coverslip thickness (excerpt from protocol for mounting sectioned tissue)
Q: Why is it important to provide information about the characteristics of the mounting media and coverslip?
A: Mismatch between sample refractive index, mounting media refractive index, or an objective’s specified coverslip thickness and refractive index produces aberrations that can change quantitative results at the micrometer level. Other important information to provide regarding the mounting media should include incompatibility with specific fluorophores, and the time needed for curing the mounting media and its effect on the refractive index or morphology of the sample.
Example 3. Mounting live samples (excerpt from a light sheet imaging protocol)
Q: Why is it important to specify the type of mounting medium?
A: An incorrect mounting medium can harm the live sample or introduce undesirable background fluorescence. Besides keeping the tissue healthy and alive, the mounting medium and apparatus should also have similar refractive index to that of the tissue when performing fluorescence microscopy.
Q: For a multicellular organism, is the mounting orientation critical for the success of the experiment?
A: Yes, failure to specify how the anatomy of the sample should be oriented can reduce image resolution and contrast. Best results can be achieved when there is minimal scattering tissue between the anatomy of interest and the objective(s).
Microscope
No two microscopes are exactly alike. Even common models of commercial microscopes will be customized during installation with a unique combination of light sources, objectives, stages, filters, detectors, various specialized modules (for example a spinning disk scan head), and control software. For protocol reporting, which aims to improve reproducibility, it is necessary to provide sources and identifiers for each of these components. The key resources table is a good place to provide these details. Below is an example provided from the Allen Institute on how to fill out the key resources table for a spinning disk confocal microscope.
Example 4. Modified key resource table (excerpt from a protocol using Zeiss spinning disk confocal microscope at Allen Institute for Cell Science)
| Category | Reagent or resource | Source | Identifier |
|---|---|---|---|
| Stand | Observer.Z1 microscope stand | Zeiss | |
| Objectives | 10X/0.45 NA Plan-Apochromat objective, FWD = 2.1mm | Zeiss | Cat#420640-9900-000 |
| 100X/1.25 W C-Apochromat Korr UV Vis IR objective, FWD = 0.25mm | Zeiss | Cat#421797-9970-000 | |
| Advanced modality | Spinning disk head CSU-X with Primary dichroic RQFT 405/488/568/647 BP filter | Yokogawa | M1N-E/FBO/C101; 24V DC; P8X006; 95P900140 |
| Emission filters | NucBlue Live dye: BP filter 450/50 | Chroma | Cat#ET450/50m |
| mEGFP tag structure or bright field: BP filter 526/50 | SEMROCK | Cat#FF03-525/50 | |
| CMDR dye: BP filter 690/50 | Chroma | Cat#690/50m | |
| mTagRFP-T tag structure: BP filter 600/50 | Chroma | Cat#ET600/50m | |
| Bright field: BP filter 706/95 | Chroma | Cat#ET706/95m | |
| Fluorescence Excitation light source | Laser: LASOS 405 50mw | Zeiss | Part#400600-9011-000 |
| Laser: LASOS 488 100mw | Zeiss | Part#400600-9061-000 | |
| Laser: LASOS 561 75mw | Zeiss | Part#400600-9111-000 | |
| Laser: LASOS 638 75mw | Zeiss | Part#400600-9121-000 | |
| Transmitted light source | Transmitted light red; LAMBDA TLED+ with 740 nm wavelength LED | Sutter Instruments | |
| Transmitted light white; Attachment lamp VIS-LED (400–700 nm) with collector for laser system | Zeiss | Part#423053-9060 | |
| Detector | Orca Flash 4.0 V2+ cameras | Hamamatsu | Part#C1144-22CU |
| Stage components | Piezo z-drive: Prior NanoScan Z 100 mm piezo z-stage: NZ100ZM/a | Zeiss | Part#2802000 224 |
| PECON Incubator XLmulti S1; (heat and CO2 control: 37°C with 5% CO2) | Zeiss | Part#2802000 224 | |
| Stage insert: H201 k frame slim profile model | Okolab | Part#H201 k |
Q: What critical information about the objective should be included?
A: For objectives, it is important to include the numerical aperture of the lens (which in part determines the theoretical resolution limit of the imaging data), expected index of refraction at the lens interface (air, water, other immersion medium or clearing media), and type of corrections applied to the lens design. For example, 10X/0.45 NA Plan-Apochromat objective (Zeiss, Cat# 420640-9900-000).
Q: What critical information about the illumination should be included?
A: For illumination sources, it is important to indicate the type of light (arc lamp, LED, or laser for example), wavelength characteristics such as wavelength peaks and/or range, and maximum output power. Without this information, there is no starting point to begin to understand what “10% laser power” means (which will still be an incomplete characterization due to other variables in the light path that will affect power at the sample, thus protocols for better characterization of power at the sample are under development). Pulsed light sources should include details about the pulse duration and repetition rate and the appropriateness of the light source for the fluorophores being used.
Q: What critical information about the filters should be included?
A: For filters, it is important to include the specifications that convey the spectral characteristics of the wavelengths that contribute to the data. In the case of short or long-pass filters, usually, a single cut-off frequency is specified while bandpass filters include a central wavelength and the width of the band or top and bottom cut-offs. Such information allows for reproducibility with optimal conditions avoiding cross-talks and bleed-through between different channels.
Q: What critical information about the detector should be included?
A: For the detector, it is helpful to provide the type (PMT, HyD, APD, CCD, sCMOS, etc.) as well as the make and model so the relative sensitivity is understood, and low-signal regions aren’t missed because of readers attempting to use a detector with insufficient limits of detection. It should also be noted if the detector is run in a particular mode which change the readout of the signal (e.g., line or frame averaging for photon counting mode (PMTs) or binning, pixel size or noise-reduction mode for cameras) and therefore, the noise characteristics of the resulting image.
Q: What additional information is critical for reproducibility?
A: For all components, manufacturer and parts number are critical so that further specifications can be found as needed. While this type of list may seem difficult to create, if you have used an imaging facility for your experiments, facility imaging scientists will be able to contribute to this portion of the protocol. Application specialists from the manufacturer may be helpful. In the case of collaborating with a colleague who has a custom-built microscope, they should have such details available.
Image acquisition
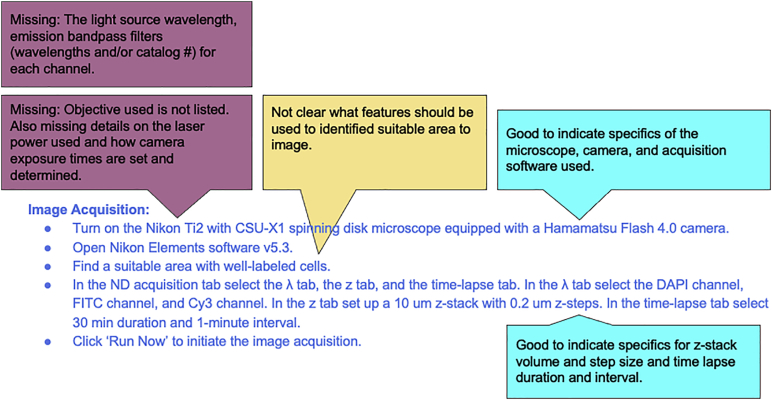
From the available configurations of a given microscope, it is important to provide a detailed description of what components and settings are used during image data acquisition. Image acquisition steps in a protocol often lack sufficient detail about microscope illumination and detection paths, rationale for acquisition settings, and details about microscope control software. As previously mentioned, details about immersion or imaging media are also often missing. Software and experiment settings for acquisition (and processing) should be included in the Key Resource Table as shown below.
Example 5. Key resource table (software packages utilized for image acquisition)
| Reagent or resource | Source | Identifier |
|---|---|---|
| Software and algorithms | ||
| Open access acquisition software example | ||
| Micro-Manager 2.0.1 | J. Edelstein, et al., J. Biol. Methods 1, e10 | https://micro-manager.org/ |
| Commercial acquisition software examples | ||
| LAS version 4.5.22 | Leica | |
| Nikon Elements AR version 5.20.02 Build 1453 | Nikon | |
| cellSens v4.1 | Olympus | |
| Zen 2.3 (blue edition); version 23.69.1003; service pack 2.3.69.01000; hotfix 2.3.69.01003 | Zeiss | |
| Custom acquisition code or software example | ||
| Code Package Name | https://github.com/RepositoryName | Citation if available |
Q: What are critical and often omitted information about software packages?
A: Always include details on the version of the software used. Depending on the source this can include build numbers, service pack numbers, and other identifiers. For open access software, cite the publication for the software and link to this valuable community resource to your reader. Often, the software website will include reference instructions. Image processing and analysis software should be treated the same.
Example 6. Platform-specific acquisition (excerpt without enough quantitative details about excitation lines and emission filters)
Q: Are detailed step-by-step methods for specific software packages enough?
A: The detailed step-by-step methods for specific software packages are very useful for other groups using the same software and equipment; however, make sure you provide enough general information so that your settings can be easily translated to other platforms. Carefully identify and be sure to highlight key details that will be required to replicate your methods.
Q: Why is reporting the channels used not enough for the protocol?
A: As further illustrated below, you need to provide details about the excitation laser lines and emission filter details or light sources and filter combinations utilized in your settings, as these directly impact the fluorescence measurements taken during imaging. The order in which multi-channels are acquired and if they are acquired sequentially (z-stack of channel 1 then of channel 2, etc…) or interwoven (all channels are imaged at a focal point before moving to the next focal point until the z-stack is completed) for a z-stack acquisition is also very important for the reproducibility of an experiment. This is particularly important when doing live imaging since small movements may affect the alignment of the different channels and different settings produce different local time steps.
Example 7. Unclear acquisition settings for measurement and display (excerpt from multi-color fluorescence imaging protocol)
Q: When would this information be sufficient and when would it not?
A: While the resulting image may only show the different markers and probes in a selected color, identifying a channel by a color alone, especially if there is more than one color used, is not sufficient for the reproducibility of the image itself. Since channel names are generic, for each channel, always provide the exact name of the fluorophore used as a dye or secondary conjugated antibody (including the product number in the key resources table) or the specific fluorescent protein used (e.g., EGFP), the wavelength of the laser line or light source, the excitation and emission bandpass filters and dichroic mirror(s). Always identify all units, i.e., nm for each wavelength and Watt or mW, for each excitation line used (see above question on reporting illumination).
Q: What display color choices are accessible to colorblind people?
A: Avoid red and green as those two colors cannot be distinguished by people with red-green colorblindness. Magenta and teal are good replacement colors and many tools exist to help you select appropriate color palettes for your figures and images. This applies to both the image and its annotations and more complex lookup tables. Make sure to identify clearly if the annotation name refers to the structure, dye, or channel name to avoid confusion.
Q: How are bandpass filter information and fluorophore information used?
A: By knowing the excitation and emission spectrum of your fluorophore, you can select filters that will limit the range of wavelength of light to better excite and collect the emission light of a fluorophore. Using the wrong combination of filters, (too narrow) can reduce the intensity of your fluorophore or (too wide) may include non-specific signal coming from other sources of fluorescence (another fluorophore or non-specific autofluorescence from your specimen).
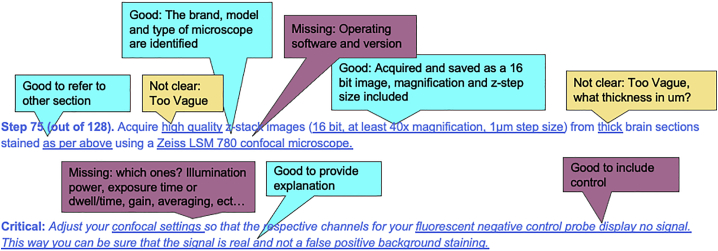
Example 8. Oversimplified acquisitions settings, missing objective, and pixel size parameters (excerpt from confocal imaging protocol)
Q: When would this information be sufficient and when would it not?
A: Minimal information is sufficient if only qualitative information is extracted from an image, i.e., cell confluency from a bright field image or when assessing whether there is positive expression of fluorescence protein in transfected cells. However, the information is insufficient for reproducibility of an image from which quantitative information needs to be extracted. Although similar images can be acquired on a different microscope, it would be valuable for the author to include their own process and to include as much details as possible about the system used so that imaging can be reproduced accurately (light path, objective full description including N.A., working distance, immersion medium and correction type, light source, and detectors). Additional tips can be provided for imaging on a different system.
Q: What specific information is missing?
A: Other potentially relevant information about the image acquisition should include if the microscope is upright or inverted and the operating software and version used. Ideally, the full light path is described so that the image acquisition can be reproduced using a different system. This should include information about the objective, the different tube lens (impacts overall magnification, may be fixed or variable zoom), filters, and dichroic mirrors used. It should also include information about the light source and power as well as the type and characteristics of the detectors and its settings (i.e., exposure time or dwell time, averaging). Since this is a 3D image acquisition, information about the number of z-planes to be acquired in addition to the z-step size need to be provided. If multi-channels are used, you should also specify if the acquisition was done in series (one channel at a time) or interwoven (each channel imaged at each z-plane). Including acquisition settings in a table at the appropriate step may be helpful.
Q: What information is critical for image processing and analysis?
A: The resulting image file information is critical for reproducible analysis. This information is often included in the file format but not always. It includes the size of the image or field of view, the pixel size, dynamic range (given in bits), offset, and ideally the signal to noise ratio (SNR). Including the settings and requirements for successful positive and negative control images are also highly encouraged since these can be used to determine the intensity level of background noise and intensity level of the signal of interest.
Image processing and analysis
By this point in the protocol, much ground has already been covered and the level of detail is often hyper-focused, overly platform-specific and missing the conceptual background to extend the approach to other software platforms. “Click this button” and “Navigate to this menu” type instructions are useful only to those with the same acquisition software package and version. While it is helpful for individual labs to have such a manual for their pipelines within their groups, this approach often does not include the rationale for each step, so it is difficult to attempt to translate the approach to other software packages or deal with newer versions of the same software. Providing the rationale for each step is critical.
Despite detail present in some parts of the image processing and analysis protocols, vague descriptions of other aspects of this section may also be problematic. For example, important parameter choices in individual steps in the processing and analysis pipeline, data size, software versions, computational resources utilized, and imprecise reporting on image quality and types of statistical replicates makes reproducing results difficult.
Example 9: Imprecise language for image quality and sample quality metrics (excerpt from cell-tracking protocol)
Q: How do you define a “good-quality” image?
A: Image quality should be as objective a metric as possible, so readers will not have to subjectively assess whether an image is suitable for analysis. Low resolution, optical aberrations, blur from motion or defocus, and poor signal are common reasons that images are of insufficient quality to process. Common quality metrics include SNR and contrast ratio for tasks that involve segmentation, while bleaching rate or blur may be more suitable for tracking experiments. Relevant metrics can be chosen from the literature or devised independently, provided the calculation is clearly defined in the protocol. Different fields and research groups may define common metrics (e.g., SNR) differently, so it is critical to provide a simple and clear definition. It is also helpful to provide representative examples of poor-quality and good-quality images for control and experimental groups, as well as examples of how further analysis is compromised by use of poor-quality images.
Q. What is a “healthy enough” cell?
A. Control experiments should be designed to assess cell or organism health after imaging is performed, especially for time-lapse imaging. For example, to assess whether time-lapse experiments perturb normal embryonic development and morphogenesis, one can compare the endpoint image of a time lapse to images acquired from cells or embryos imaged just once at the equivalent time point. Depending on the biological system, it should also be possible to design an assay for molecular markers or structural indicators of cellular health that can be performed after imaging (e.g., antibody staining for stress response proteins or assessment of cell membrane integrity).
Q. What number of replicates is best?
A: Selection of appropriate statistical methods for different imaging experiments is beyond the scope of this discussion, but the “best” number of replicates is a number appropriate to provide statistical power to the biological question at hand. The problem we would like to address is the lack of detail surrounding the reporting. In this example the number of samples is simply reported (n≥3). It is not clear if n refers to number of cell cultures, number of cells, or number of organelles within cells, number of regions within a single organelle, or even repeated measurements of the same image, for example. Whatever has been chosen as the appropriate sets of replicates (biological, technical) should be clearly described.
Q: Should I include example data?
A: Yes, it is helpful to reviewers and readers for example data (raw, unprocessed images) to be provided by way of easy download (from an online data repository if possible). If the results of image processing and analysis are included for the example data, then it is simple to check if the protocol steps are being followed correctly, provided the requisite software is available.
Example 10: Not breaking down individual steps and parameter selection (excerpt from cell-tracking protocol)
Q: How to reference specific software?
A: Both commercial and open-source software are constantly revised to fix bugs, modify existing functionality, and to add new functionality, with updates frequently released. In a protocol, it is imperative to provide the version number for the base software (e.g., FIJI/ImageJ v1.53q), and specific packages (e.g., TrackMate 7.7.2). Failure to provide these details can lead to confusion when trying to apply the protocol to a newer or older version of the software. Additionally, it is important to include links to where the software can be downloaded online (e.g., https://imagej.net/software/fiji/downloads, https://imagej.net/plugins/trackmate/) and citations for open-source software packages that have been reported in literature (e.g., for base software FIJI [Schindelin et al., Nat. Methods 9, 676–682] and recent release of Trackmate, [Ershov, et al., Nat. Methods 19, 829–832]. Often the website where the software is available will list the appropriate publications to cite. These details should be provided in the key resources table.
Q: Should I include data size and computing requirements?
A: Yes. It is helpful to provide nominal data size, as well as the details or minimum requirements for computing hardware and expected performance (how long the calculations should take on specific hardware). Different software are more or less equipped to scale up an image analysis pipeline, so these details are helpful to avoid storage/memory issues or wasting time trying to adapt software to a dataset that it won’t be able to handle. Additionally, providing these details helps readers get access to the requisite computing environment before attempting to apply the same steps.
Q. How much detail is helpful in the image processing analysis protocol?
A. As with bench work, each step in the protocol should provide enough detail for the work to be replicated without the reader needing to guess at specifics, with enough conceptual background for the reader to be able to extend the method to a different brand of equipment (in this case, different types of data sets or different processing software). Take care to enumerate every modification from the acquired data, including compression, file conversion, image pre-processing (e.g., filtering, contrast adjustments), processing (e.g., segmentation, object identification), and measurement (e.g., volume, velocity, diffusion calculations), as well as every parameter choice for each step. In this example case, TrackMate offers a variety of automatic and semi-automatic spot detection, segmentation, and tracking functionality. For reproducibility, the text should specify the selected algorithms, including any user-defined parameter values, which part of the process is semi-automated, and what constitutes a quality track. Qualitative descriptors like “accuracy” should be explained, for example “inspecting for accuracy” could be improved by saying “inspecting that tracks are correct for 9/10 objects on average over 5 time frames.”
Q. Should I provide code, macros?
A. Yes, providing code on github or other execution files that can help speed readers’ application of your image processing and analysis is helpful. Additionally, reviewers will need access to these during the publication process to be able to provide peer feedback.
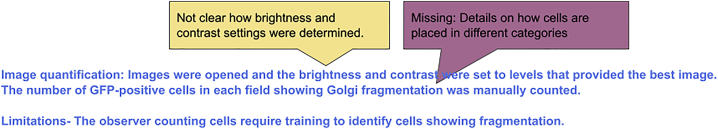
Example 11: Missing criteria for manual object identification (excerpt from fluorescence imaging protocol)
Q: Is manual counting for quantification acceptable?
A: In some cases, difficulties in automating analysis can result in using manual methods. If it has not been possible to automate or semi-automate the analysis, this limitation should be stated. Manual image analysis is prone to bias and possible mistakes. If reporting a manual method, be sure to include clear reasoning and criteria the observer is using for separating groups and any reasons for excluding data. State if any other methods to reduce bias are used, such as using an observer blind to treatments. Providing example images for how you are discriminating between groups can be useful to ensure similar classifications will be used across groups. Providing a clear description of qualitative terms like “best” is necessary, or alternatively examples of images and histograms that are representative of “best” results.
Q: What types of image processing are allowed and how should that be disclosed?
A: Any image processing steps used must be disclosed. Simple adjustments, such as changing the brightness and contrast of the image should be applied to the entire image. All images collected should have identical processing steps performed. Further discussion on reporting image processing can be found in other references.
Acknowledgments
N.G’s work was sponsored by the Allen Institute for Cell Science. The authors wish to thank the Allen Institute for Cell Science founder, Paul G. Allen, for his vision, encouragement, and support. H.C.G.’s work was supported by the Silicon Valley Community Foundation (CZI Imaging Scientist program 2019–198168). D.D.L.’s work was supported by Kavli Institute for Fundamental Neuroscience at UCSF. We would also like to thank the active community discussions hosted by BioImaging North America and QUAREP-LiMi.
Declaration of interests
The authors declare no competing interests.
Recommended Reading
- Aaron J., Chew T.-L. A guide to accurate reporting in digital image processing – can anyone reproduce your quantitative analysis? J. Cell Sci. 2021;134:jcs254151. doi: 10.1242/jcs.254151. [DOI] [PubMed] [Google Scholar]
- Blainey P., Krzywinski M., Altman N. Replication. Nat. Methods. 2014;11:879–880. doi: 10.1038/nmeth.3091. [DOI] [PubMed] [Google Scholar]
- Boehm U., Nelson G., Brown C.M., Bagley S., Bajcsy P., Bischof J., Dauphin A., Dobbie I.M., Eriksson J.E., Faklaris O., et al. QUAREP-LiMi: a community endeavor to advance quality assessment and reproducibility in light microscopy. Nat. Methods. 2021;18:1423–1426. doi: 10.1038/s41592-021-01162-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo J.C., Cooper S., Heigwer F., Warchal S., Qiu P., Molnar C., Vasilevich A.S., Barry J.D., Bansal H.S., Kraus O., et al. Data-analysis strategies for image-based cell profiling. Nat. Methods. 2017;14:849–863. doi: 10.1038/nmeth.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromey D.W. Digital images are data: and should Be treated as such. Methods Mol. Biol. 2013;931:1–27. doi: 10.1007/978-1-62703-056-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A.D., Tsuchida M.A., Amodaj N., Pinkard H., Vale R.D., Stuurman N. Advanced methods of microscope control using μManager software. J. Biol. Methods. 2014;1:e10. doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ershov D., Phan M.S., Pylvänäinen J.W., Rigaud S.U., Le Blanc L., Charles-Orszag A., Conway J.R.W., Laine R.F., Roy N.H., Bonazzi D., et al. TrackMate 7: integrating state-of-the-art segmentation algorithms into tracking pipelines. Nat. Methods. 2022;19:829–832. doi: 10.1038/s41592-022-01507-1. [DOI] [PubMed] [Google Scholar]
- Hammer M., Huisman M., Rigano A., Boehm U., Chambers J.J., Gaudreault N., North A.J., Pimentel J.A., Sudar D., Bajcsy P., et al. Towards community-driven metadata standards for light microscopy: tiered specifications extending the OME model. Nat. Methods. 2021;18:1427–1440. doi: 10.1038/s41592-021-01327-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddleston J.M., Aaron J.S., Khuon S., Chew T.-L. A guide to accurate reporting in digital image acquisition – can anyone replicate your microscopy data? J. Cell Sci. 2021;134:jcs254144. doi: 10.1242/jcs.254144. [DOI] [PubMed] [Google Scholar]
- Katsnelson A. Colour me better: fixing figures for colour blindness. Nature. 2021;598:224–225. doi: 10.1038/d41586-021-02696-z. [DOI] [PubMed] [Google Scholar]
- Koho S., Fazeli E., Eriksson J.E., Hänninen P.E. Image quality Ranking method for microscopy. Sci. Rep. 2016;6 doi: 10.1038/srep28962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero Llopis P., Senft R.A., Ross-Elliott T.J., Stephansky R., Keeley D.P., Koshar P., Marqués G., Gao Y.-S., Carlson B.R., Pengo T., et al. Best practices and tools for reporting reproducible fluorescence microscopy methods. Nat. Methods. 2021;18:1463–1476. doi: 10.1038/s41592-021-01156-w. [DOI] [PubMed] [Google Scholar]
- Nelson G., Boehm U., Bagley S., Bajcsy P., Bischof J., Brown C.M., Dauphin A., Dobbie I.M., Eriksson J.E., Faklaris O., et al. QUAREP-LiMi: a community-driven initiative to establish guidelines for quality assessment and reproducibility for instruments and images in light microscopy. J. Microsc. 2021;284:56–73. doi: 10.1111/jmi.13041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saify F., Tiwari N. Mounting Media - An Untouched Aspect. Oral Maxillofac. Pathol. J. 2020;11:20–24. [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]