Summary
Alternative splicing contributes to cancer development. Indeed, splicing analysis of cancer genome-wide association study (GWAS) risk variants has revealed likely causal variants. To systematically assess GWAS variants for splicing effects, we developed a prioritization workflow using a combination of splicing prediction tools, alternative transcript isoforms, and splicing quantitative trait locus (sQTL) annotations. Application of this workflow to candidate causal variants from 16 endometrial cancer GWAS risk loci highlighted single-nucleotide polymorphisms (SNPs) that were predicted to upregulate alternative transcripts. For two variants, sQTL data supported the predicted impact on splicing. At the 17q11.2 locus, the protective allele for rs7502834 was associated with increased splicing of an exon in a NF1 alternative transcript encoding a truncated protein in adipose tissue and is consistent with an endometrial cancer transcriptome-wide association study (TWAS) finding in adipose tissue. Notably, NF1 haploinsufficiency is protective for obesity, a well-established risk factor for endometrial cancer. At the 17q21.32 locus, the rs2278868 risk allele was predicted to upregulate a SKAP1 transcript that is subject to nonsense-mediated decay, concordant with a corresponding sQTL in lymphocytes. This is consistent with a TWAS finding that indicates decreased SKAP1 expression in blood increases endometrial cancer risk. As SKAP1 is involved in T cell immune responses, decreased SKAP1 expression may impact endometrial tumor immunosurveillance. In summary, our analysis has identified potentially causal endometrial cancer GWAS risk variants with plausible biological mechanisms and provides a splicing annotation workflow to aid interpretation of other GWAS datasets.
Keywords: endometrial cancer, GWAS, splicing, spliceogenic, sQTL, NF1, SKAP1, SpliceAI-10k calculator
We have developed a workflow to prioritize GWAS variants for splicing effects and applied this to the largest available GWAS of endometrial cancer risk. Our workflow reveals two candidate spliceogenic GWAS variants that appear to affect endometrial cancer risk through alternative splicing of NF1 and SKAP1.
Introduction
Genome-wide association studies (GWASs) have identified thousands of loci associated with complex traits and diseases.1 Most GWAS variants are located in noncoding regions and likely regulate gene expression. However, it is difficult to assign causality to variants and uncover the underlying target genes (reviewed by Tam et al.2), especially given the myriad of mechanisms that impact gene expression. Further, as genetic variants are correlated by linkage disequilibrium, it is challenging to disentangle statistically prioritized credible sets of correlated GWAS variants that contain the causal variant(s). Functional analyses are thus required to identify likely causal GWAS variants and their target genes. Expression quantitative trait locus (eQTL) analyses have succeeded in correlating GWAS variants with gene expression, revealing candidate causal genes at ∼20% of GWAS loci using currently available eQTL data.3 Splicing QTL (sQTL) analyses can identify variants associated with alternative transcript isoforms, associations that tend to be independent of eQTLs.4,5 Although sQTLs provide a functional mechanism for likely causal variants and genes at a smaller fraction of GWAS loci with available sQTL data (∼10%),3 sQTLs have been reported to have larger effects on traits than variants affecting only gene expression.6 However, GWAS variants are often not assessed for effects on splicing, possibly due to a lack of appropriate pipelines for analysis of common genetic variants. Alternative splicing dysregulation plays a role in cancer development and progression,7 and sQTL analyses have shown that alternative splicing is a mechanism through which GWAS variants may impact cancer risk.8,9,10 Splicing prediction analysis has yet to be integrated with GWAS data for many cancer types, including endometrial cancer (MIM: 608089).11
sQTL discovery is expected to increase as well-validated mapping methods are developed and long-read sequencing approaches are used. The incompleteness of current sQTL datasets means that some GWAS variants that affect splicing may not be revealed. To address this issue, in silico splicing predictors used to identify pathogenic variants for Mendelian disorders could be used in a complementary approach to analyze GWAS variants for splicing effects.6 Here, we have developed such a strategy to identify and prioritize endometrial cancer GWAS risk variants that alter splicing profiles (here termed spliceogenic variants). Firstly, we prioritized candidate causal endometrial cancer risk single-nucleotide polymorphisms (SNPs) that create or alter splicing motifs (i.e., 5′ and 3′ splice sites, polypyrimidine tracts, branchpoints, and splicing regulatory elements). Then, we leveraged large-scale catalogs of alternative transcript isoforms and tissue sQTLs to assess the predicted splicing events and provide supporting evidence for the predicted impact of spliceogenic variants. A flow diagram summarizing the workflow is shown in Figure S1.
We selected intronic and exonic candidate causal SNPs from the largest endometrial cancer GWAS risk meta-analysis (12,906 cases and 108,979 controls), performed by the Endometrial Cancer Association Consortium.12 The reference allele, alternate allele, and chromosomal position of the selected SNPs were submitted to the Ensembl Variant Effect Predictor (VEP)13 online tool to generate the variant call format file and obtain the transcript annotations. All coordinates, nomenclature, and analyses were based on the GRCh38 assembly. Using the VEP-generated variant call format file as input, SpliceAI (v.1.3.1)14 was used to predict the probabilities of gain or loss of acceptor and donor splice sites indicated as delta scores in the output file. SpliceAI was shown as the best single splicing prediction strategy for variants in Mendelian disease genes in a comparative study of nine in silico methods.15 SpliceAI can evaluate up to 10 kb of a nucleotide sequence,14 making it suitable for the analysis of variant effect on splicing motifs located far from native splice sites. The distance parameter of the SpliceAI run was set at the maximum allowable for this tool, 4,999 bp flanking the variant to capture gain or loss of distant splice sites. Due to a design limitation of SpliceAI v.1.3.1, only variants in protein-coding genes were scored. The chromosomal coordinates and alleles with SpliceAI scores were then matched with VEP annotation to obtain the corresponding c. position based on the high-quality Matched Annotation from NCBI and EMBL-EBI (MANE) Select transcripts. The MANE Select transcript is considered here as the canonical transcript. Finally, SpliceAI delta scores were inputted into our SpliceAI-10k calculator16 to predict the type and size of mRNA aberrations (pseudoexonization, whole/partial intron retention, partial exon deletion, or exon skipping) and assess their effect on reading frame. The SpliceAI-10k calculator demonstrated high accuracy for predicting pseudoexons or alternative exons activated by deep intronic variants,16 which comprise the bulk of genic variants in our GWAS dataset. By design, the SpliceAI-10k calculator can analyze single-nucleotide substitutions only. The SpliceAI-10k calculator default thresholds were based on the analysis of rare high-risk variants. Considering the expected subtle splicing effects of GWAS SNPs, we arbitrarily adjusted the calculator threshold to the lowest score of 0.01 for acceptor and donor gain in deep intronic regions and a minimum score of 0.01 and maximum score of 0.05 for native acceptor and donor loss to increase sensitivity. Events predicted as pseudoexons were termed here as alternative exon inclusion to differentiate the potentially modest changes in alternative splicing caused by GWAS SNPs from severely abnormal splicing events caused by rare high-risk variants. We searched the Ensembl Genome Browser release 10617 for alternative transcript isoforms that harbor the alternative exons predicted by SpliceAI-10k calculator.
The functional consequence of alternative transcripts (i.e., in frame or frameshift) was derived from the amino acid sequence predicted by the SpliceAI-10k calculator. Predicted alternative transcript sequences were visualized in HEXplorer18 to identify the affected splicing motifs. These include the 3′ splice site indicated by MaxEntScan score, the 5′ splice site indicated by H-bond score, and splicing regulatory elements indicated by HEXplorer exon-intron Z score (HZEI).
For genes with predicted Ensembl-annotated alternative exon inclusion, we identified sQTLs (p < 1 × 10−5) from potentially relevant tissues (i.e., uterus, vagina, ovary, Epstein-Barr virus [EBV]-transformed lymphocytes, whole blood, subcutaneous adipose, and visceral omentum) from Genotype Tissue Expression (GTEx) Project v.8.19 sQTLs were intersected with the GWAS candidate causal SNPs located in genes with predicted alternative splicing effects. Each sQTL was reviewed to identify if the SNP location was consistent with the size and location of the event predicted by the SpliceAI-10k calculator. The sQTL intron ID, indicating the chromosomal positions of the excised intron boundaries (i.e., the 5′ and 3′ splice sites), was used to identify the differentially expressed alternative exon in Ensembl. Colocalization between GWAS signals and sQTL was assessed using the ezQTL20 web platform and the hypothesis prioritization for multitrait colocalization (HyprColoc) algorithm.21
Analysis of candidate causal variants from 16 endometrial cancer GWAS risk loci12 identified 209 exonic and intronic SNPs located in protein-coding genes. SpliceAI predictions were returned for 177 candidate causal SNPs at eight GWAS risk loci (Table S1). As some of the SNPs are located in overlapping genes, this corresponded to a greater number of gene-based SNP locations (i.e., 3 exonic and 184 intronic; Table S1). Seven candidate causal SNPs, at four GWAS risk loci, were predicted to alter splicing motifs of CYP19A1 (MIM: 107910), EIF2AK4 (MIM: 609280), NF1 (MIM: 613113), and SKAP1 (MIM: 604969) (Table 1). The Ensembl database had no record of alternative transcripts that harbor the predicted alternative exons in CYP19A1 and EIF2AK4, so these were not analyzed further. Splicing prediction results (Table 2; Figure S2) and Ensembl alternative transcript annotation (Table S2) provided evidence that three SNPs in NF1 and another in SKAP1 may modify splicing of these genes through effects on splicing motifs.
Table 1.
Predicted spliceogenic candidate causal GWAS SNPs and their predicted functional consequences
| SNP | Effect allele frequency | HGVS (MANE select transcript) | SpliceAI max delta score | Predicted mRNA splicing effecta | Predicted functional consequenceb | Ensembl- annotated alternative exon |
|---|---|---|---|---|---|---|
| rs7177179 | 0.25 | ENST00000263791.10(EIF2AK4):c.2767–1183T>C | 0.17 | 107 bp alternative exon | p.(Lys923fs) | no |
| rs7173595 | 0.69 | ENST00000396402.6(CYP19A1):c.145+1229G>A | 0.01 | 100 bp alternative exon | p.(Gly49fs) | no |
| rs28518777 | 0.34 | ENST00000396402.6(CYP19A1):c.–38–18360C>T | 0.02 | 199 bp alternative exon | 5′ UTR insertion | no |
| rs35888506 | 0.45 | ENST00000358273.9(NF1):c.4836–1609C>T | 0.11 | 97 bp alternative exon | p.(Phe1613fs) | yes |
| rs2854320 | 0.50 | ENST00000358273.9(NF1):c.8377+6342C>A | 0.01 | 54 bp alternative exon | p.(Pro2792_Gly2793ins18) | yes |
| rs7502834 | 0.45 | ENST00000358273.9(NF1):c.8377+1709G>A | 0.02 | 77 bp alternative exon | p.(Gly2793fs) | yes |
| rs2278868 | 0.56 | ENST00000336915.11(SKAP1):c.481G>A | 0.07 | 125 bp exon skipping | p.(Ser148fs) | yes |
HGVS, Human Genome Variation Society; MANE, Matched Annotation from NCBI and EMBL-EBI.
Predicted by the SpliceAI-10k calculator.
Predicted consequence for the canonical protein isoforms were derived from the results of the SpliceAI-10k calculator.
Table 2.
SNP-affected alternative exons and bioinformatic scores of relevant splicing motifs
| SNP | Alternative exon and location | SNP position relative to 3′ ss | SNP position relative to 5′ ss | 3′ ss MES Ref score | 5′ ss H-bond Ref score | ΔHZEI score | SNP effect on splicing motif |
|---|---|---|---|---|---|---|---|
| rs35888506 | ENSE00003938169 (NF1) chr17:31,324,113–31,324,209 | – | +2 | 4.73 | 0 | N/A | strengthening of 5′ ss (H-bond = 17.5); donor gain (GC 5′ ss → GT 5′ ss) |
| rs2854320 | ENSE00001657839 (NF1) chr17:31,367,225–31,367,278 | −180 | – | 6.19 | 12.3 | 6.41 | ISE loss; branchpoint gain (TCTCT → TCTAT) |
| rs7502834 | ENSE00003966146 (NF1) chr17:31,362,288–31,362,364 | – | +48 | 8.10 | 15.8 | −1.66 | ISE gain |
| rs2278868 | ENSE00003557988 (SKAP1) chr17:48,184,847–48,184,723 | +38 | −86 | 8.57 | 14 | −2.19 | ESE loss |
ESE, exonic splicing enhancer; H-bond, hydrogen bond; HZEI, HEXplorer exon-intron Z score; ISE, intronic splicing enhancer; MES, MaxEntScan; ss, splice site.
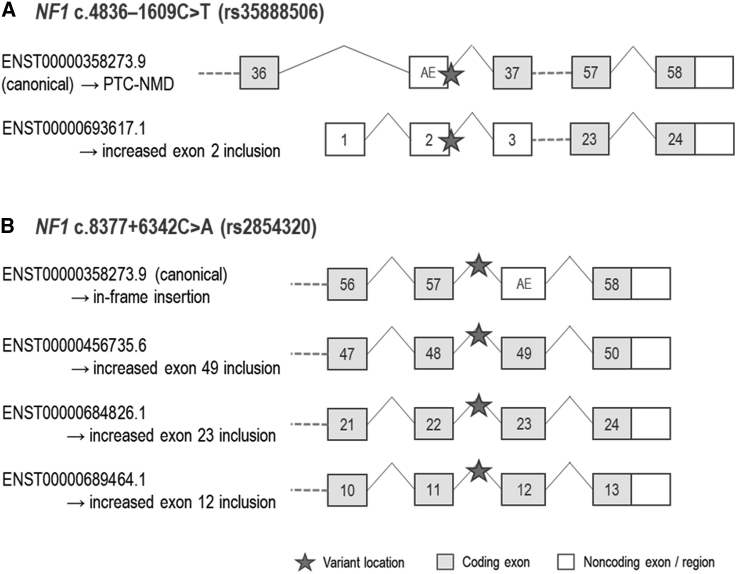
The protective allele of rs35888506 (T), located in intron 36 of the NF1 canonical transcript, is predicted to activate an alternative 97 bp exon (Figure 1A) by conversion of the pre-existing GC 5′ splice site into a stronger GT 5′ splice site (Table 2; Figure S2A). We anticipate that the resultant out-of-frame transcript, which is not present in the Ensembl database, would be subject to nonsense-mediated decay (NMD) (Figure 1A). The same alternative exon (exon 2; Figure 1A) is present in an Ensembl-annotated alternative transcript and is predicted to encode an N-terminal truncated 1,027 amino acid NF1 protein. Thus, splicing analysis indicates that the T allele would increase expression of both alternative transcripts.
Figure 1.
rs35888506 and rs2854320 are predicted to affect NF1 splicing
(A) and (B) show the predicted splicing events for rs35888506 and rs2854320 (locations denoted by the star symbols), respectively. For each panel, vertically aligned exons have identical chromosomal locations, although the positions of stop codons at the last exons and the end of the 3′ untranslated regions may vary. AE, alternative exon mapped to the canonical transcript; PTC-NMD, premature terminating codon-nonsense-mediated decay).
The protective allele of rs2854320 (A), located in intron 57 of the NF1 canonical transcript, is predicted to create a branchpoint motif (Table 2) that would be expected to result in inclusion of an alternative exon downstream in an alternative transcript (Figures 1B and S2B), which is not present in the Ensembl database. We project that translation of this transcript would insert 18 amino acids (in frame) at the C terminus of the canonical NF1 protein. The same exon is the penultimate exon of three NF1 Ensembl-annotated alternative transcript isoforms, and thus the A allele is also predicted to increase the expression of these isoforms (Figure 1B). Although all three transcripts are predicted to encode truncated protein isoforms, there is only evidence of protein expression from ENST00000456735.6 (a 2,502 amino acid isoform (PDB:H0Y465), ProteomicsDB, accessed June 1, 2022).
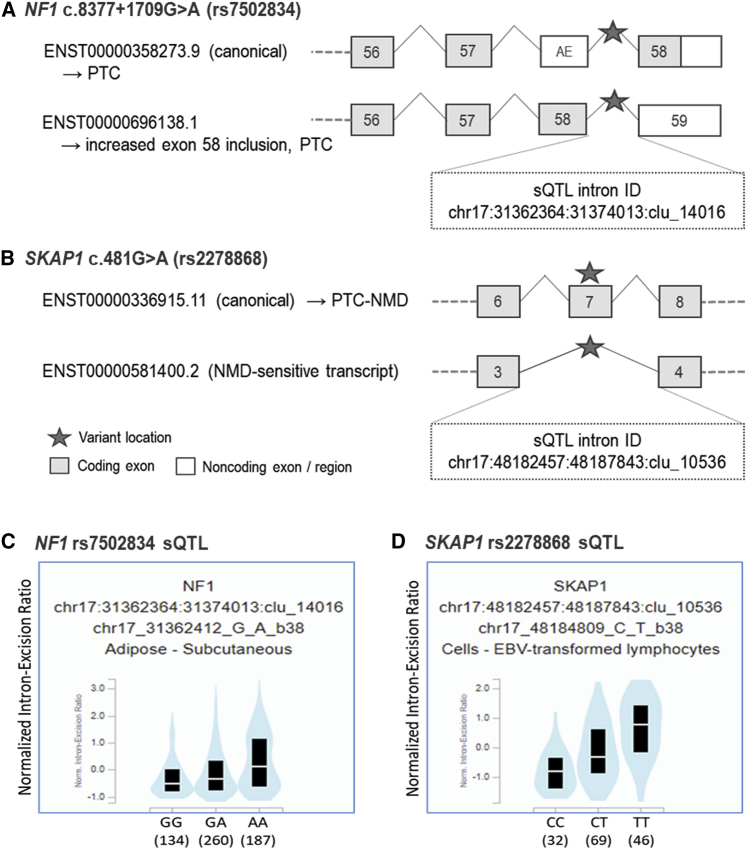
For the remaining two candidate spliceogenic SNPs, the predicted splicing was supported by evidence from both Ensembl annotations and sQTL data. The protective allele of rs7502834 (A), located in intron 57 of the NF1 canonical transcript, is predicted to lead to the inclusion of a 77 bp alternative exon (exon 58; Figure 2A) through strengthening of an intronic spicing enhancer motif (ΔHZEI = −1.66; Table 2) downstream of the exon 5′ splice site (Figure S2C). Inclusion of this alternative exon generates an Ensembl-annotated alternative transcript (Figure 2A), and a termination codon near the 3′ end of this alternative exon is predicted to truncate 32 amino acids from the C terminus of NF1. Consistent with the splicing prediction, sQTL data show that the protective allele of rs7502834 is associated with inclusion of alternative exon 58 in NF1 transcripts expressed in subcutaneous adipose tissue (p = 7.8 × 10−7; Figure 2C). Furthermore, we found evidence for colocalization between the sQTL and endometrial cancer risk signal (Figure S3), with a posterior probability of 0.89, providing evidence that this NF1 splicing event may explain the genetic association with endometrial cancer risk.
Figure 2.
rs7502834 (NF1) and rs2278868 (SKAP1) are predicted to affect splicing, and sQTL data demonstrate associations with corresponding splicing events
(A) and (B) show the predicted splicing events for rs7502834 and rs2278868 (denoted by the star symbols), respectively, with the corresponding intron IDs for the sQTLs. Vertically aligned exons have identical chromosomal locations, although the end of the 3′ untranslated regions may vary.
(C) and (D) show sQTL violin plots of normalized intron-exclusion ratios (GTEx v.8) for rs7502834 and rs2278868, respectively (see Table S3 for further details). Black boxes indicate interquartile ranges and the white lines show median values for each genotype. AE, alternative exon mapped to the canonical transcript; NMD, nonsense-mediated decay; PTC-NMD, premature terminating codon-nonsense-mediated decay; sQTL, splicing quantitative trait locus.
The risk allele (A) of rs2278868 is a missense variant p.(Gly161Ser) that is predicted to lead to exon skipping through exonic splicing enhancer loss (ΔHZEI = −2.19; Table 2) in exon 7 of the canonical SKAP1 transcript (Figures 2B and S2D). Skipping of exon 7 will produce an Ensembl-annotated out-of-frame alternative transcript that is predicted to be subject to NMD (Figure 2B). sQTL data again support the predicted splicing, with the risk allele of rs2278868 associated with skipping of exon 7 in EBV-transformed lymphocytes (p = 1.80 × 10−10; Figure 2D). Colocalization analysis demonstrated that the sQTL and corresponding GWAS risk signal overlapped (posterior probability = 0.92; Figure S3), again supporting a causal role for variant-induced splicing in endometrial cancer risk.
Our prioritization workflow identified seven candidate causal endometrial cancer risk SNPs, with potential effects on splicing at four of the 16 established endometrial cancer risk loci12: 15q15.1 (EIF2AK4), 15q21.2 (CYP19A1), 17q11.2 (NF1), and 17q21.32 (SKAP1). Notably, genetically predicted expression of these four genes had recently been associated with endometrial cancer risk in a transcriptome-wide association study (TWAS). Further analysis provided evidence that EIF2AK4, CYP19A1, and SKAP1 expression may have causal effects on endometrial cancer risk, but the association with genetically predicted NF1 expression did not pass a multiple-testing threshold and was not evaluated for causality.22 The current study supports the TWAS findings and bolsters the hypothesis that altered NF1 expression affects endometrial cancer risk. Moreover, we identify splicing mechanisms that may explain the TWAS associations and prioritize two candidate spliceogenic SNPs that appear to mediate their effects on endometrial cancer risk through NF1 and SKAP1 isoform expression changes.
This study demonstrates the utility of our approach to detect GWAS variants with subtle effects on splicing, highlighting potential causal genes. Moreover, the SpliceAI-10k calculator can be implemented in R to analyze large variant datasets, facilitating the selection of candidate spliceogenic SNPs.16 This method can also detect branchpoints outside the common branchpoint window (−18 to −44 bp from the 3′ splice site)23 that are less likely to be picked up by most splicing prediction tools. There are multiple examples of distal branchpoints associated with alternative splicing.24,25,26 We have previously annotated an experimentally inferred noncanonical TCTAT branchpoint motif 179 bp upstream of exon 19 of BLM27 and note that the putative noncanonical TCTAT branchpoint motif created by rs2854320 (NF1) is located 180 bp upstream of the 54 bp alternative exon.
Of the three predicted spliceogenic risk SNPs located in NF1, the effect of rs7502834 was supported by sQTL data that showed that the protective allele was associated with inclusion of the corresponding alternative exon in NF1 transcripts in subcutaneous adipose tissue. Importantly, the sQTL and endometrial cancer GWAS risk signals colocalized at the NF1 locus, suggesting that this splicing event may have a protective effect on endometrial cancer risk by reducing canonical NF1 transcript expression. This effect is consistent with a nominally significant association between decreased NF1 subcutaneous adipose expression and decreased endometrial cancer risk in our recent TWAS.22
NF1 encodes neurofibromin (NF1), a large multifunctional tumor-suppressor protein that is involved in several cell signaling pathways and regulates many cellular processes such as proliferation and migration.28 NF1 is also associated with neurofibromatosis type 1 (MIM: 162200), a Mendelian disease characterized by fibromatous skin tumors. Given that NF1 is a tumor suppressor, one may hypothesize that the protective alleles of the endometrial cancer risk SNPs would increase NF1 expression. However, NF1 regulates the mammalian target of rapamycin (mTOR) pathway,29 which is implicated in obesity and type 2 diabetes (MIM: 125953).30 Obesity is a well-established risk factor for endometrial cancer,31 and Mendelian randomization analyses have shown that increased body mass index and insulin levels are causally associated with endometrial cancer risk.11 In contrast, individuals with neurofibromatosis type 1 have a lower incidence of diabetes than healthy controls.32,33 Studies in model organisms have also suggested that NF1 loss protects against obesity: increasing the metabolic rate in Drosophila34; and reducing visceral and subcutaneous fat mass, and conferring protection from diet-induced obesity and hyperglycemia in mice.35 Thus, these findings indicate that decreased NF1 expression may reduce endometrial cancer risk through protecting against obesity and its sequelae.
We predicted that the risk allele of rs2278868 generates a SKAP1 NMD-sensitive transcript, an association supported by sQTL data from EBV-transformed lymphocytes. Again, we found evidence for colocalization of sQTL and GWAS risk signals, indicating that reduced expression of the canonical SKAP1 transcript in lymphocytes may increase endometrial cancer risk. Consistent with this finding, our previous endometrial cancer TWAS provided evidence that decreased expression of SKAP1 in whole blood was causally associated with endometrial cancer risk.22 SKAP1 encodes Src kinase associated phosphoprotein 1, which has multiple roles in T cell function related to immune responses. For example, SKAP1 is involved in antigen activation of the T cell receptor through binding of antigen-presenting cells36 and is necessary for efficient T cell cycling,37 an important feature of T cell clonal expansion in response to pathogens and cancer neoantigens. Given these functions, our findings suggest that decreased SKAP1 expression may impair T cell tumor responses, resulting in decreased tumor immunosurveillance and increased endometrial cancer risk.
We note several caveats to our study. SpliceAI, trained on GENCODE v.24 and the GRCh37 reference assembly,14 has incomplete coverage of protein-coding regions, as evidenced by genic endometrial cancer GWAS risk SNPs that had no scores. Although our SpliceAI-based approach can detect variants that alter splicing, these are limited to exonic and intronic SNPs predicted to create or modify splice sites, the polypyrimidine tract, branchpoints, and cis-acting splicing regulatory elements. SNPs that influence alternative splicing by modifying trans-acting RNA-binding proteins, mRNA secondary structure, and factors outside of splicing motif sequence alteration38,39 have not been analyzed. The sQTL analysis of predicted spliceogenic variants is constrained by the current mapping of transcript isoforms from short-read sequencing and the relatively small sample sizes of the GTEx tissue datasets. Data from long-read sequencing approaches and larger datasets will provide further sQTLs to support candidate spliceogenic variants. Furthermore, functional studies are needed to assess the effects of altered NF1 and SKAP1 isoform expression in relevant models.
Other limitations of this study relate to the underlying endometrial cancer risk GWAS. This GWAS was performed using individuals with European ancestry, and thus the relevance of the current findings to other ancestry groups is unknown. Another limitation is the statistical power of the GWAS, with a larger GWAS dataset likely to refine candidate causal variants at risk loci and reveal further risk loci for splicing analysis.
In conclusion, our findings suggest causal endometrial cancer GWAS risk SNPs and indicate molecular mechanisms for the regulation of NF1 and SKAP1 in the development of endometrial cancer. We have also identified plausible biological pathways through which these genes may impact endometrial cancer risk, but further studies are needed to assess these. Lastly, given the likely contribution of variant-induced splicing to the risk of other common diseases, our workflow could facilitate the systematic identification of likely causal SNPs and genes for other GWAS.
Ethics declaration
This research was performed under QIMR Berghofer Project P1051, which has been approved by QIMR Berghofer’s Human Research Ethics Committee. Informed consent was not required because human participants were not involved in the current study.
Acknowledgments
D.M.C. was supported by a QIMR Berghofer Ailsa Zinns PhD Scholarship, QIMR Berghofer HDC Top Up Scholarship, and University of Queensland Research Training Tuition Fee Offset. A.B.S. was supported by National Health and Medical Research Council of Australia Investigator Fellowship funding (APP1177524). T.A.O’M. was supported by National Health and Medical Research Council of Australia Investigator Fellowship funding (APP1173170). We thank the many women who participated in the Endometrial Cancer Association Consortium (ECAC), and the numerous institutions and their staff who supported recruitment. We thank the efforts of Deborah Thompson for her contribution to ECAC. The ECAC GWASs were supported by the National Health and Medical Research Council of Australia (APP552402, APP1031333, APP110`9286, APP1111246, and APP1061779); the US National Institutes of Health (R01-CA134958); European Research Council (EU FP7 Grant); Wellcome Trust Center for Human Genetics (090532/Z/09Z); and Cancer Research UK. OncoArray genotyping of ECAC cases was performed with the generous assistance of the Ovarian Cancer Association Consortium (OCAC), which was funded through grants from the US National Institutes of Health (CA1X01HG007491-01 (C.I. Amos), U19-CA148112 (T.A. Sellers), R01-CA149429 (C.M. Phelan), and R01-CA058598 (M.T. Goodman); Canadian Institutes of Health Research (MOP-86727 [L.E. Kelemen]); and the Ovarian Cancer Research Fund (A. Berchuck). We particularly thank the efforts of Cathy Phelan. OncoArray genotyping of the BCAC controls was funded by Genome Canada Grant GPH-129344, US National Institutes of Health Grant U19 CA148065, and Cancer Research UK Grant C1287/A16563. All studies and funders are listed in O’Mara et al.12
Author contributions
Conceptualization, D.M.C., A.B.S., and D.M.G.; data curation, D.M.C., D.M.G., and T.A.O’M.; formal analysis, D.M.C. and T.A.O’M.; funding acquisition, A.B.S. and T.A.O'M; methodology, D.M.C.; supervision, A.B.S., D.M.G., and T.A.O’M.; visualization, D.M.C.; writing – original draft, D.M.C. and D.M.G.; writing – review & editing, all authors.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2023.100185.
Web resources
Ensembl Genome Browser, https://asia.ensembl.org/
Ensembl Variant Effect Predictor, https://asia.ensembl.org/info/docs/tools/vep/
ezQTL web platform, https://analysistools.cancer.gov/ezqtl
Genotype Tissue Expression Project, https://www.gtexportal.org/
Online Mendelian Inheritance in Man, http://www.omim.org
ProteomicsDB, https://www.proteomicsdb.org/
Supplemental information
Data and code availability
The R code for SpliceAI-10k calculator implementation can be accessed at https://github.com/adavi4/SAI-10k-calc. Endometrial cancer GWAS summary statistics are available from the NHGRI-EBI GWAS catalog (https://www.ebi.ac.uk/gwas/studies/GCST006464). GTEx sQTL data are available from https://www.gtexportal.org/. All other data that support the findings of this publication are available in the supplemental information of this report.
References
- 1.Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E., et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gky1120. D1005-d1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tam V., Patel N., Turcotte M., Bossé Y., Paré G., Meyre D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019;20:467–484. doi: 10.1038/s41576-019-0127-1. [DOI] [PubMed] [Google Scholar]
- 3.Barbeira A.N., Bonazzola R., Gamazon E.R., Liang Y., Park Y., Kim-Hellmuth S., Wang G., Jiang Z., Zhou D., Hormozdiari F., et al. Exploiting the GTEx resources to decipher the mechanisms at GWAS loci. Genome Biol. 2021;22:49. doi: 10.1186/s13059-020-02252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y.I., Knowles D.A., Humphrey J., Barbeira A.N., Dickinson S.P., Im H.K., Pritchard J.K. Annotation-free quantification of RNA splicing using LeafCutter. Nat. Genet. 2018;50:151–158. doi: 10.1038/s41588-017-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y.I., van de Geijn B., Raj A., Knowles D.A., Petti A.A., Golan D., Gilad Y., Pritchard J.K. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352:600–604. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garrido-Martín D., Borsari B., Calvo M., Reverter F., Guigó R. Identification and analysis of splicing quantitative trait loci across multiple tissues in the human genome. Nat. Commun. 2021;12:727. doi: 10.1038/s41467-020-20578-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y., Qian J., Gu C., Yang Y. Alternative splicing and cancer: a systematic review. Signal Transduct. Target. Ther. 2021;6:78. doi: 10.1038/s41392-021-00486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo Z., Zhu H., Xu W., Wang X., Liu H., Wu Y., Wang M., Chu H., Zhang Z. Alternative splicing related genetic variants contribute to bladder cancer risk. Mol. Carcinog. 2020;59:923–929. doi: 10.1002/mc.23207. [DOI] [PubMed] [Google Scholar]
- 9.Caswell J.L., Camarda R., Zhou A.Y., Huntsman S., Hu D., Brenner S.E., Zaitlen N., Goga A., Ziv E. Multiple breast cancer risk variants are associated with differential transcript isoform expression in tumors. Hum. Mol. Genet. 2015;24:7421–7431. doi: 10.1093/hmg/ddv432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian J., Chen C., Rao M., Zhang M., Lu Z., Cai Y., Ying P., Li B., Wang H., Wang L., et al. Aberrant RNA splicing is a primary link between genetic variation and pancreatic cancer risk. Cancer Res. 2022;82:2084–2096. doi: 10.1158/0008-5472.CAN-21-4367. [DOI] [PubMed] [Google Scholar]
- 11.Wang X., Glubb D.M., O'Mara T.A. 10 Years of GWAS discovery in endometrial cancer: aetiology, function and translation. EBioMedicine. 2022;77:103895. doi: 10.1016/j.ebiom.2022.103895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Mara T.A., Glubb D.M., Amant F., Annibali D., Ashton K., Attia J., Auer P.L., Beckmann M.W., Black A., Bolla M.K., et al. Identification of nine new susceptibility loci for endometrial cancer. Nat. Commun. 2018;9:3166. doi: 10.1038/s41467-018-05427-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F. The Ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B., et al. Predicting splicing from primary sequence with deep learning. Cell. 2019;176:535–548.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 15.Rowlands C., Thomas H.B., Lord J., Wai H.A., Arno G., Beaman G., Sergouniotis P., Gomes-Silva B., Campbell C., Gossan N., et al. Comparison of in silico strategies to prioritize rare genomic variants impacting RNA splicing for the diagnosis of genomic disorders. Sci. Rep. 2021;11:20607. doi: 10.1038/s41598-021-99747-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Canson D.M., Davidson A.L., de la Hoya M., Parsons M.T., Glubb D.M., Kondrashova O., Spurdle A.B. SpliceAI-10k calculator for the prediction of pseudoexonization, intron retention, and exon deletion. bioRxiv. 2022 doi: 10.1101/2022.07.30.502132. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunningham F., Allen J.E., Allen J., Alvarez-Jarreta J., Amode M., Armean I.M., Austine-Orimoloye O., Azov A.G., Barnes I., Bennett R., et al. Ensembl 2022. Nucleic Acids Res. 2021;50:D988–D995. doi: 10.1093/nar/gkab1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erkelenz S., Theiss S., Otte M., Widera M., Peter J.O., Schaal H. Genomic HEXploring allows landscaping of novel potential splicing regulatory elements. Nucleic Acids Res. 2014;42:10681–10697. doi: 10.1093/nar/gku736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang T., Klein A., Sang J., Choi J., Brown K.M. ezQTL: a web platform for interactive visualization and colocalization of quantitative trait loci and GWAS. Dev. Reprod. Biol. 2022;20:541–548. doi: 10.1016/j.gpb.2022.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foley C.N., Staley J.R., Breen P.G., Sun B.B., Kirk P.D.W., Burgess S., Howson J.M.M. A fast and efficient colocalization algorithm for identifying shared genetic risk factors across multiple traits. Nat. Commun. 2021;12:764. doi: 10.1038/s41467-020-20885-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kho P.F., Wang X., Cuéllar-Partida G., Dörk T., Goode E.L., Lambrechts D., Scott R.J., Spurdle A.B., O'Mara T.A., Glubb D.M. Multi-tissue transcriptome-wide association study identifies eight candidate genes and tissue-specific gene expression underlying endometrial cancer susceptibility. Commun. Biol. 2021;4:1211. doi: 10.1038/s42003-021-02745-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Signal B., Gloss B.S., Dinger M.E., Mercer T.R. Machine learning annotation of human branchpoints. Bioinformatics. 2018;34:920–927. doi: 10.1093/bioinformatics/btx688. [DOI] [PubMed] [Google Scholar]
- 24.Corvelo A., Hallegger M., Smith C.W.J., Eyras E. Genome-wide association between branch point properties and alternative splicing. PLoS Comput. Biol. 2010;6:e1001016. doi: 10.1371/journal.pcbi.1001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taggart A.J., Lin C.-L., Shrestha B., Heintzelman C., Kim S., Fairbrother W.G. Large-scale analysis of branchpoint usage across species and cell lines. Genome Res. 2017;27:639–649. doi: 10.1101/gr.202820.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pineda J.M.B., Bradley R.K. Most human introns are recognized via multiple and tissue-specific branchpoints. Genes Dev. 2018;32:577–591. doi: 10.1101/gad.312058.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Canson D.M., Dumenil T., Parsons M.T., O’Mara T.A., Davidson A.L., Okano S., Signal B., Mercer T.R., Glubb D.M., Spurdle A.B. The splicing effect of variants at branchpoint elements in cancer genes. Genet. Med. 2022;24:398–409. doi: 10.1016/j.gim.2021.09.020. [DOI] [PubMed] [Google Scholar]
- 28.Ratner N., Miller S.J. A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat. Rev. Cancer. 2015;15:290–301. doi: 10.1038/nrc3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bergoug M., Doudeau M., Godin F., Mosrin C., Vallée B., Bénédetti H. Neurofibromin structure, functions and regulation. Cells. 2020;9:2365. doi: 10.3390/cells9112365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dann S.G., Selvaraj A., Thomas G. mTOR Complex1–S6K1 signaling: at the crossroads of obesity, diabetes and cancer. Trends Mol. Med. 2007;13:252–259. doi: 10.1016/j.molmed.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 31.Raglan O., Kalliala I., Markozannes G., Cividini S., Gunter M.J., Nautiyal J., Gabra H., Paraskevaidis E., Martin-Hirsch P., Tsilidis K.K., Kyrgiou M. Risk factors for endometrial cancer: an umbrella review of the literature. Int. J. Cancer. 2019;145:1719–1730. doi: 10.1002/ijc.31961. [DOI] [PubMed] [Google Scholar]
- 32.Martins A.S., Jansen A.K., Rodrigues L.O.C., Matos C.M., Souza M.L.R., de Souza J.F., Diniz M.d.F.H.S., Barreto S.M., Diniz L.M., de Rezende N.A., Riccardi V.M. Lower fasting blood glucose in neurofibromatosis type 1. Endocr. Connect. 2016;5:28–33. doi: 10.1530/EC-15-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kallionpää R.A., Peltonen S., Leppävirta J., Pöyhönen M., Auranen K., Järveläinen H., Peltonen J. Haploinsufficiency of the NF1 gene is associated with protection against diabetes. J. Med. Genet. 2021;58:378–384. doi: 10.1136/jmedgenet-2020-107062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Botero V., Stanhope B.A., Brown E.B., Grenci E.C., Boto T., Park S.J., King L.B., Murphy K.R., Colodner K.J., Walker J.A., et al. Neurofibromin regulates metabolic rate via neuronal mechanisms in Drosophila. Nat. Commun. 2021;12:4285. doi: 10.1038/s41467-021-24505-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tritz R., Benson T., Harris V., Hudson F.Z., Mintz J., Zhang H., Kennard S., Chen W., Stepp D.W., Csanyi G., et al. Nf1 heterozygous mice recapitulate the anthropometric and metabolic features of human neurofibromatosis type 1. Transl. Res. 2021;228:52–63. doi: 10.1016/j.trsl.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dadwal N., Mix C., Reinhold A., Witte A., Freund C., Schraven B., Kliche S. The multiple roles of the cytosolic adapter proteins ADAP, SKAP1 and SKAP2 for TCR/CD3 -mediated signaling events. Front. Immunol. 2021;12:703534. doi: 10.3389/fimmu.2021.703534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raab M., Strebhardt K., Rudd C.E. Immune adaptor SKAP1 acts a scaffold for Polo-like kinase 1 (PLK1) for the optimal cell cycling of T-cells. Sci. Rep. 2019;9:10462. doi: 10.1038/s41598-019-45627-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu X.-D., Ares M., Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 2014;15:689–701. doi: 10.1038/nrg3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen M., Manley J.L. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The R code for SpliceAI-10k calculator implementation can be accessed at https://github.com/adavi4/SAI-10k-calc. Endometrial cancer GWAS summary statistics are available from the NHGRI-EBI GWAS catalog (https://www.ebi.ac.uk/gwas/studies/GCST006464). GTEx sQTL data are available from https://www.gtexportal.org/. All other data that support the findings of this publication are available in the supplemental information of this report.