Abstract
The GGAs (Golgi-localized, gamma-ear-containing, ARF-binding proteins) are a family of multidomain adaptor proteins involved in protein sorting at the trans-Golgi network of eukaryotic cells. Here we present results from a functional characterization of the two Saccharomyces cerevisiae GGAs, Gga1p and Gga2p. We show that deletion of both GGA genes causes defects in sorting of carboxypeptidase Y (CPY) and proteinase A to the vacuole, vacuolar morphology, and maturation of α-factor. A structure-function analysis reveals a requirement of the VHS, GAT, and hinge for function, while the GAE domain is less important. We identify putative clathrin-binding motifs in the hinge domain of both yeast GGAs. These motifs are shown to mediate clathrin binding in vitro. While mutation of these motifs alone does not block function of the GGAs in vivo, combining these mutations with truncations of the hinge and GAE domains diminishes function, suggesting functional cooperation between different clathrin-binding elements. Thus, these observations demonstrate that the yeast GGAs play important roles in the CPY pathway, vacuole biogenesis, and α-factor maturation and identify structural determinants that are critical for these functions.
The GGAs (Golgi-localized, gamma-ear-containing, ARF-binding proteins) are ubiquitous adaptor-like proteins that associate with the cytoplasmic face of the trans-Golgi network (TGN) (4, 16, 22, 37, 47). Three GGAs exist in humans (GGA1, GGA2, and GGA3), one each exists in Drosophila melanogaster and Caenorhabditis elegans, and two exist in the budding yeast Saccharomyces cerevisiae (Gga1p and Gga2p). The GGAs are monomeric but have a multidomain structure consisting of VHS (Vps27, Hrs, and STAM), GAT (GGA and TOM), hinge, and GAE (gamma-adaptin ear) domains.
Biochemical and immunocytochemical analyses have revealed that each of the four GGA domains serves a specific function. The VHS domain of the human GGAs functions as a recognition module for acidic cluster-dileucine sorting signals contained within the cytosolic tails of sortilin (31) and the cation-independent (CI) and cation-dependent (CD) mannose 6-phosphate receptors (MPRs) that sort lysosomal hydrolases to lysosomes (38, 56). The GAT domain of human and yeast GGAs mediates interactions with the GTP-bound form of members of the ARF (ADP-ribosylation factor) family of proteins (4, 16, 55). GAT-ARF interactions are responsible for the regulated recruitment of the GGAs from the cytosol to the Golgi complex (4, 16, 39). The hinge domain of all the GGAs contains putative clathrin-binding motifs composed of acidic and bulky hydrophobic amino acids (16, 39). For the human GGAs, these motifs have been shown to mediate interactions with clathrin in vitro and to promote recruitment of clathrin to the TGN in vivo (39). Finally, the GAE domain of the human GGAs binds proteins such as γ-synergin and rabaptin-5, which may function to regulate assembly of GGA-containing coats or formation of coated vesicular carriers (22, 47). These properties of the human GGA domains indicate that they may mediate ARF-dependent recruitment of clathrin to the TGN in order to sort intracellular cargo receptors from the TGN to the endosomal system. Despite the detailed characterization of the properties of GGA domains, the significance of these properties for the function of the GGAs in vivo remains to be assessed.
The existence of two GGAs in yeast provides an opportunity to perform analyses of the physiological roles of the GGAs in an organism easily amenable to genetic manipulation. In yeast, biosynthetic protein sorting from the Golgi complex to the vacuole, the equivalent of the mammalian lysosome, is mediated by two principal routes (for reviews, see references 7, 12, 26, and 29). The alkaline phosphatase (ALP) pathway sorts vacuolar proteins such as ALP and the t-SNARE Vam3p from the Golgi complex to the vacuole directly. Formation of Golgi complex-derived carrier vesicles and transport through this pathway require the adaptor protein (AP) complex AP-3 (14, 46) and the VPS (vacuolar protein sorting) gene products Vps41p/Vam2p, Vps39p/Vam6p (30), and Vps1p (32, 49), a member of the dynamin family of proteins (49). In contrast, the carboxypeptidase Y (CPY) pathway sorts vacuolar proteins including CPY and proteinase A (PrA) and subunits of the vacuolar ATPase from the Golgi complex to the vacuole via a prevacuolar endosomal compartment (PVC). Sorting of both CPY and PrA through this pathway is mediated by interactions with the transmembrane receptor Vps10p, which delivers these proteins to the PVC prior to recycling back to the Golgi complex (13, 27). Vesicle formation and transport in the CPY pathway involve the coat-scaffolding protein clathrin (10, 45), the putative aminophospholipid translocase Drs2p (11), and the synaptojanin-like proteins Inp52p and Inp53p (1, 33), as well as numerous VPS gene products, including Vps41p/Vam2p, Vps39p/Vam6p (30), and Vps1p (1, 49). Mutations in factors operating in these respective pathways result, to differing degrees, in impaired vacuolar sorting and defects in vacuole biogenesis. The yeast GGAs appear to play important, redundant roles in biosynthetic sorting to the vacuole based on studies of a mutant gga1Δ gga2Δ strain containing disruptions of both yeast GGA genes. This mutant is defective for transport of pro-CPY to the vacuole and missorts pro-CPY to the periplasmic space (16, 22, 55). The gga1Δ gga2Δ mutant was also found to be defective in sorting of the syntaxin Pep12p from the Golgi complex to late endosomes (2). The presence of a vacuolar morphology defect in gga1Δ gga2Δ cells has, however, been debated, with one study reporting abnormal morphology (22) and another showing no morphological differences between wild-type and gga1Δ gga2Δ strains (55).
As is evident from the well-defined phenotypes arising from mutations in the yeast GGA genes, these proteins appear to play an important role in biosynthetic protein sorting. To expand our understanding of the GGAs' function and to assess the relative importance of the different GGA domains in vivo, we have performed a structure-function analysis of yeast Gga1p and Gga2p. First, we elaborate on the gga1Δ gga2Δ mutant phenotype by reporting defects in sorting of additional vacuolar proteins and abnormal vacuolar morphology and a strong defect in maturation of the mating pheromone α-factor. We then analyze the functional requirement of individual GGA domains and find that the VHS, GAT, and hinge domains are important for GGA-mediated pro-CPY sorting and pro-α-factor processing, while the GAE domain appears less important. In addition, we show that, like their human counterparts, the yeast GGAs are capable of binding clathrin via acidic-bulky-hydrophobic motifs in their hinge domains. We also present evidence that these clathrin-binding motifs contribute to GGA-mediated sorting in vivo. Finally, mutational analysis of the Gga2p VHS domain identifies a highly conserved sequence important for this protein's function.
MATERIALS AND METHODS
Media, strains, and antibodies.
S. cerevisiae cells were grown at 30 or 25°C in yeast extract-peptone-dextrose (YEPD) medium (rich medium) or synthetic complete (minimal) medium supplemented with amino acids and nucleotides where appropriate. Plasmids expressing prototrophic markers were maintained by growth of transformed strains in synthetic complete medium lacking the respective amino acid or nucleotide (selective medium). All media and supplements were obtained from Bio 101 (Vista, Calif.). Genotypes of yeast strains used in this study are listed in Table 1. Plasmid manipulations were performed in Escherichia coli strain DH5α (PGC Scientifics, Gaithersburg, Md.) using standard transformation protocols and media. Mouse monoclonal HA.11 antihemagglutinin (anti-HA) antibodies were obtained from Covance (Vienna, Va.). Mouse anti-CPY antibodies were purchased from Molecular Probes (Eugene, Oreg.). Mouse anti-α-factor (αf), anti-PrA, anti-clathrin heavy chain (Chc1p), anti-Kar2p, and rabbit anti-ALP antibodies were the generous gifts of Todd Graham, Carol Woolford, Sandra Lemmon, Mark Rose, and Tom Stevens, respectively.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| CMY119 | MATα gga1Δ1::TRP1 gga2Δ1::HIS3 ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | 16 |
| SEY6210 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | 42 |
| SEY6211 | MATaura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 ade2-101 suc2-Δ9 | 42 |
| RC634 | MATasst1-3 rme ade2-1 ura1 his6 met1 can1 cyh2 GAL | 9 |
| JHRY28-3A | MATα ura3-52 vps1a | Tom Stevens lab |
| BJ2407 | MATa/MATα prb1-1122/prb1-1122 prc1-407/prc1-407 pep4-3/pep4-3 leu2/leu2 trp1/trp1 ura3-52/ura3-52 gal2/gal2 | Neil Green lab |
The vps1 allele was also originally referred to by the designations vpl1-1 and vpt26.
Metabolic labeling, immunoprecipitation, and immunoblotting.
Metabolic labeling of yeast cells with 35S Express label (NEN Life Science Products, Boston, Mass.), pulse-chase analyses, immunoprecipitations, and preparation of whole-cell lysates were performed at 30°C according to published protocols (5). For α-factor experiments, cells were pulse-labeled at 25°C, as temperatures lower than 30°C have been used to visualize α-factor-processing intermediates (21). Immunoprecipitates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography. For immunoblotting, protein samples were resolved by SDS-PAGE and transferred to Protran nitrocellulose membranes (Schleicher and Schuell, Dassel, Germany), followed by incubation with appropriate primary antibodies, incubation with horseradish peroxidase-conjugated secondary antibodies, and visualization with enhanced chemiluminescence reagents (NEN Life Sciences Products, Inc.) according to the manufacturer's protocols.
Microscopy analysis of yeast vacuoles.
To visualize yeast vacuoles in vivo, cells were grown in YEPD to 1 to 3 units of optical density at 600 nm (OD600) per ml at 30°C and treated with vital dyes N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenylhexatrienyl) (FM4-64) and 5-(and -6)-carboxy-2′,7′-dichlorofluorescein diacetate (CDCFDA) (Molecular Probes) essentially as described previously (50) with slight modifications. In brief, cells were pelleted and resuspended to 30 units of OD600/ml in fresh YEPD to which FM4-64 (as a 16 mM stock in dimethyl sulfoxide) was added to a 40 μM concentration or in fresh YEPD containing 50 mM citric acid, pH 5.0, to which CDCFDA (as a 10 mM stock in dimethylformamide) was added to a 10 μM concentration. Cells with FM4-64 and CDCFDA were incubated with shaking for 15 and 20 min, respectively, at 30°C in a darkened incubator. Following incubation, cells were pelleted (700 × g for 3 min) and washed twice with YEPD. Cells with CDCFDA were resuspended in YEPD to 30 units of OD600/ml and examined immediately. Cells with FM4-64 were resuspended in YEPD to 15 units of OD600/ml and further incubated for 45 min at 30°C with shaking, after which time they were pelleted, resuspended in fresh YEPD to 30 units of OD600/ml, and examined immediately. Cell suspensions were viewed on standard glass slides using an Olympus (Sterling, Va.) PlanApo 100× objective with a 1.4-mm numerical aperture on an Olympus IX70 fluorescence microscope (excitation and emission wavelengths for dyes were as recommended by Molecular Probes). Cells were imaged using TILLvisION v3.3 software (T.I.L.L. Photonics GmbH, Martinsried, Germany).
CPY colony blotting and α-factor secretion assays.
Colony blot assay for CPY mislocalization was performed essentially as previously described (41) with slight modifications. In brief, strains were grown to 1 to 1.5 units of OD600/ml at 30°C and concentrated to 0.2 units of OD600/ml and 5 μl was spotted on selective medium. Plates were incubated overnight at 30°C to allow cell growth, after which a nitrocellulose membrane (see above) was prewetted with distilled H2O and placed on colonies, and incubation was continued for an additional 12 to 14 h. Membranes were then removed, rinsed with distilled H2O, and incubated with anti-CPY antibodies (1:4,000 dilution), followed by incubation with horseradish peroxidase-conjugated secondary antibody and visualization with enhanced chemiluminescence reagents as described above. Densitometry measurements were derived using NIH Image version 1.62 software from multiple trials (average n = 6). Mean ± standard errors were calculated and are reported in Table 3. Densitometry measurement for gga1Δ gga2Δ mutant cells containing empty vector was performed as an internal control in each trial, weighed as 100% missorting, and used to calculate percentages of strains tested. These percentages of missorting were used to produce the scoring system presented in Fig. 3 and 6 (see legend to Fig. 3). For α-factor secretion assays, sst1-3 mutant strain RC634 was grown overnight at 30°C to stationary phase in 10 ml of YEPD. Cells were pelleted and resuspended in 10 ml of fresh medium. A 200-μl aliquot was removed and spread evenly on an agar plate containing YEPD which was then used immediately. Strains to be tested for α-factor secretion were grown to 1 to 1.5 units of OD600/ml at 30°C, concentrated to 1 unit of OD600/ml, and serially diluted in distilled H2O to concentrations indicated in the figure legends. Three microliters of each dilution was spotted on plates containing RC634 cells. Plates were incubated at 30°C for approximately 48 to 72 h to allow visualization of α-factor-induced RC634 growth inhibition (i.e., growth halo).
TABLE 3.
Percentages of CPY missorting in cells expressing wild-type and mutant GGA1 and GGA2-encoded proteins
| Strain (plasmid) | % ± SEMa | No. of trials |
|---|---|---|
| Wild type (vector) | 0.5 ± 0.3 | 17 |
| gga1Δ gga2Δ (vector) | 100.0 | |
| gga1Δ gga2Δ (GGA2) | 0.7 ± 0.2 | 12 |
| gga1Δ gga2Δ (gga2-ΔGAE) | 4.5 ± 1.3 | 7 |
| gga1Δ gga2Δ (gga2-Δhinge/GAE) | 34.7 ± 2.0 | 10 |
| gga1Δ gga2Δ (gga2-ΔVHS) | 72.5 ± 1.9 | 9 |
| gga1Δ gga2Δ (gga2-ΔVHS/GAT) | 101.4 ± 1.9 | 5 |
| gga1Δ gga2Δ (gga1-N215A) | 13.5 ± 2.2 | 10 |
| gga1Δ gga2Δ (gga2-N219A) | 25.9 ± 3.6 | 10 |
| Wild type (gga2-ΔVHS/GAT) | 1.4 ± 0.7 | 4 |
| Wild type (gga2-ΔVHS/GAT + CBS#1) | 1.6 ± 0.7 | 4 |
| gga1Δ gga2Δ (GGA1) | 0.6 ± 0.5 | 8 |
| gga1Δ gga2Δ (gga1-CBS#1-CBS#2) | 0.9 ± 0.5 | 3 |
| gga1Δ gga2Δ (gga1-ΔGAE) | 4.3 ± 0.3 | 4 |
| gga1Δ gga2Δ (gga1-ΔGAE + CBS#1-CBS#2) | 13.6 ± 2.3 | 5 |
| gga1Δ gga2Δ (gga2-CBS#1) | 0.9 ± 0.3 | 4 |
| gga1Δ gga2Δ (gga2-ΔGAE + CBS#1) | 13.6 ± 3.3 | 4 |
| gga1Δ gga2Δ (gga2-Δh[442–471]/GAE) | 4.3 ± 0.3 | 3 |
| gga1Δ gga2Δ (gga2-Δh[409–471]/GAE) | 2.0 ± 0.3 | 3 |
| gga1Δ gga2Δ (gga2-Δh[370–471]/GAE) | 5.2 ± 1.2 | 3 |
| gga1Δ gga2Δ (gga2-Δh[409–471]/GAE + CBS#1) | 11.7 ± 0.6 | 9 |
| gga1Δ gga2Δ (gga2-Δh[370–471]/GAE + CBS#1) | 28.6 ± 1.9 | 9 |
See Materials and Methods for method of calculating percent CPY missorting and standard error of the mean (SEM).
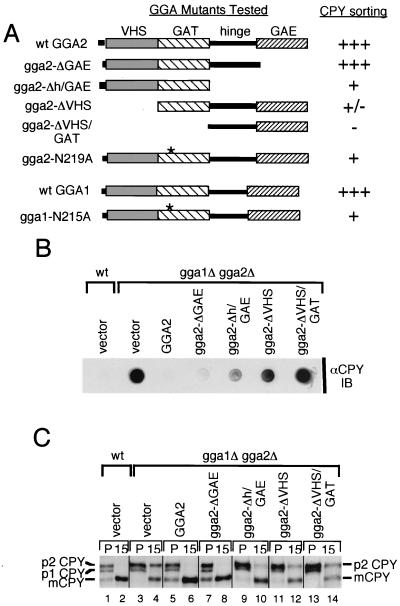
FIG. 3.
GGA domains are differentially required for vacuolar sorting. (A) Schematic representation of GGA1 and GGA2 mutants analyzed. Plasmids encoding wild-type Gga2p, Gga2p mutants containing truncations of one or more designated domains and GAT point mutant, wild-type Gga1p, and Gga1p GAT point mutant were generated (Table 2 has plasmid descriptions) and transformed into gga1Δ gga2Δ mutant cells. The scale indicates the ability of encoded proteins to complement the gga1Δ gga2Δ CPY missorting phenotype as determined by relative percentages of CPY detected (i.e., missorted) in the CPY colony blot below (values are listed in Table 3). Range employed: +++, ≤5% (i.e., similar to wild type); ++, 6 to 25%; +, 26 to 50%; +/−, 51 to 90%; −, 91 to 100% (i.e., similar to gga1Δ gga2Δ mutant). Asterisks represent N215A and N219A mutations for GGA1 and GGA2, respectively. (B) Complementation of gga1Δ gga2Δ CPY missorting by GGA2 truncations. Wild-type cells transformed with vector alone and gga1Δ gga2Δ mutant cells transformed with vector alone and plasmids expressing wild-type Gga2p and Gga2p mutants indicated were grown on SC-URA (selective) medium and blotted with a nitrocellulose membrane, followed by immunoblotting (IB) of the membrane with anti-CPY antibodies as described in Materials and Methods. (C) Complementation of gga1Δ gga2Δ CPY processing defect by GGA2 truncations. Wild-type cells transformed with vector alone (lanes 1 and 2) and gga1Δ gga2Δ mutant cells transformed with vector alone (lanes 3 and 4) and plasmids expressing wild-type Gga2p (lanes 5 and 6) and Gga2p mutants indicated (lanes 7 to 14) were pulse-labeled (P) for 10 min, and labeled proteins were chased for 15 min as described in Materials and Methods. Lysates were subjected to immunoprecipitation with anti-CPY antibodies. Immunoprecipitated proteins were resolved by SDS-PAGE, and forms of CPY are indicated as in Fig. 1A. (D) Complementation of gga1Δ gga2Δ CPY missorting by GGA1 and GGA2 GAT point mutants. Wild-type cells transformed with vector alone and gga1Δ gga2Δ cells transformed with vector alone and plasmids expressing wild-type GGA1 and GGA2 and GAT point mutants were examined for CPY mislocalization as described for panel B. (E) Test for possible dominant negative effect for gga2-ΔVHS/GAT mutant protein. Wild-type cells transformed with vector alone and plasmids expressing gga2-ΔVHS/GAT- and gga2-ΔVHS/GAT-containing point mutations in a putative clathrin-binding motif (CBS#1) (Table 2 and Materials and Methods) and gga1Δ gga2Δ mutant cells transformed with vector alone were analyzed as described for panel B for CPY mislocalization. wt, wild type.
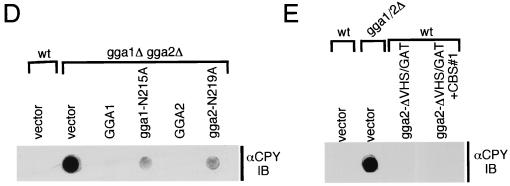
FIG. 6.
Functional analysis of GGA hinge, CBS motifs, and GAE domains in vivo. (A) Schematic representation of GGA mutants analyzed. Plasmids encoding wild-type Gga1p and Gga2p and Gga1p and Gga2p mutants containing mutations in their CBS motifs and/or domain truncations were generated (Table 2 shows plasmid descriptions). The relative amount of CPY missorting in gga1Δ gga2Δ transformants (as assessed in panels B and C) is indicated using the scale described for Fig. 3 and based on values in Table 3. The asterisks represent site-directed mutations in CBS#1 and CBS#2 of Gga1p and CBS#2 of Gga2p as noted in names of fusions to the left. Numbers in brackets indicate residues of hinge domains truncated from the respective mutants. (B) Complementation of gga1Δ gga2Δ CPY missorting and α-factor-secretion defects by GGA1 mutants. Indicated strains were examined for relative levels of CPY missorting and secretion of mature α-factor as described for Fig. 3. (C) Complementation of gga1Δ gga2Δ CPY missorting and α-factor-secretion defects by GGA2 mutants. Indicated strains were examined as described for panel B. WT and wt, wild type.
Preparation of GGA truncation-epitope tag and glutathione S-transferase (GST) fusion constructs, site-directed mutagenesis, and preparation of KEX2 epitope-tagged construct.
Full-length and truncated open reading frame (ORF) sequences for yeast GGA1 and GGA2 were amplified from cDNAs (16) through PCR using an Advantage cDNA PCR kit (Clontech, Palo Alto, Calif.) and oligonucleotide primers containing BamHI (5′ primer) and AvaI (3′ primer) restriction sites. PCR products were cloned directionally using standard molecular biology methods into the multiple cloning site of yeast expression vector pYX112 (R & D Systems, Minneapolis, Minn.) to produce an in-frame fusion at the 3′ terminus with the HA epitope coding sequence (plasmids used are listed in Table 1). Sequences encoding Gga2p hinge truncation mutants were amplified using the above 5′ primer and a 3′ primer containing a STOP sequence (TAA) and a HindIII restriction site for directional cloning into pYX112. The DNA sequence of fragments encoding all Gga1p and Gga2p truncations may be inferred through analyzing yeast GGA1 (ORF YDR358w) and yeast GGA2 (ORF YHR108w) coding sequences relative to the corresponding amino acids presented in the text and tables. To generate GGA1 and GGA2 GAT point mutants, site-directed mutagenesis was performed using a QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) according to the manufacturer's instructions. Oligonucleotide primers were designed to contain nucleotide changes that generate alanine substitutions for asparagines 215 and 219 in Gga1p and Gga2p, respectively. Mutant proteins were cloned into pYX112 as described above. Western blot analyses demonstrate that wild-type and mutant GGA proteins are expressed at similar levels (data not shown). The Kex2p ORF was amplified from yeast genomic DNA using the Advantage cDNA PCR kit (Clontech) and oligonucleotide primers containing BamHI (5′ primer) and SalI (3′ primer) restriction sites. The resultant product was cloned directionally into pYX112 to produce an in-frame fusion at the 3′ terminus with the HA epitope coding sequence. Kex2p containing an HA epitope at the C terminus is known to functionally replace endogenous Kex2p in vivo (32). To generate GST fusion constructs, DNA sequences encoding Gga1p VHS/GAT (amino acids [aa] 1 to 331) and hinge/GAE (aa 332 to 557) domains and Gga2p VHS/GAT (aa 1 to 336), hinge/GAE (aa 337 to 585), Δhinge/GAE (aa 357 to 585), and GAE (aa 472 to 585) domains were amplified as described above using oligonucleotide primers containing BamHI (5′ and 3′ primers) restriction sites for GGA1 fragments and BamHI (5′ primer) and XhoI (3′ primer) restriction sites for GGA2 fragments. PCR products were cloned directionally into bacterial GST expression vector pGEX-5x-1 (Pharmacia Biotech, Piscataway, N.J.) to create in-frame fusions at the 5′ terminus with the GST coding sequence. Site-directed mutagenesis was performed using a QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. Oligonucleotide primers were designed to contain nucleotide changes that generate alanine substitutions for the first two residues of putative clathrin-binding motifs in Gga1p (LIDFD changed to AADFD; LLDFD changed to AADFD) and Gga2p (LIDFN changed to AADFN). Further details of plasmid construction and primer sequences used in PCR methods and site-directed mutagenesis reactions are available upon request.
Preparation of yeast cytosolic protein extract and clathrin-binding assay.
For isolation of cytosolic proteins, protease-deficient yeast strain BJ2407 was grown at 30°C to 1 to 1.5 units of OD600/ml. Three liters of cells was pelleted (5,000 × g for 10 min), washed once in distilled H2O, and resuspended in a total of 10 ml of ice-cold lysis buffer (20 mM Tris-HCl [pH 7.5], 1 mM EDTA, 1 mM 2-mercaptoethanol, 50 mM NaCl, and 0.05% [wt/vol] NaN3) with protease inhibitors leupeptin (0.5 μg/ml), pepstatin (0.7 μg/ml), aprotinin (1 μg/ml), and 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride (0.25 mM). Acid-washed glass beads were added to cell slurry to a one-half volume. Cells were vortexed at top speed at 4°C for 30 s and chilled on ice for 30 s; this was repeated 10 times. The lysate was removed and centrifuged (5,000 × g for 15 min) at 4°C to remove unbroken cells and debris. The supernatant was decanted and subjected to a high-speed spin (110,000 × g for 1 h) at 4°C. The resulting supernatant (S100) representing the cytosolic protein fraction was removed, and the protein concentration was determined using the Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, Calif.). Sterile glycerol was added to a 10% concentration to the extract, which was then divided into 1-ml aliquots (2.5 mg/ml) and frozen at −70°C until used.
For use in the clathrin-binding assay, GST fusion proteins were isolated from bacteria using glutathione-Sepharose (Pharmacia Biotech) according to the manufacturer's directions. One hundred micrograms of each isolated GST fusion was then bound to 50 μl of glutathione-Sepharose in phosphate-buffered saline (PBS) with protease inhibitors (as above for lysis buffer) in a total of 1 ml by rotation for 30 min at room temperature. Bound GST fusions were then pelleted (12,000 × g for 15 s) and washed once with PBS. Aliquots of the S100 cytosolic protein extract (see above) were thawed on ice and centrifuged at 4°C (12,000 × g for 10 min) to remove aggregates, and supernatant was added to each glutathione-GST fusion complex. Incubation proceeded for 4 h at 4°C by rotation. Following incubation, glutathione-GST fusion complexes with bound cytosolic proteins were pelleted (100 × g for 2 min) at 4°C. The pellet was washed once with PBS containing protease inhibitors, resuspended in 50 μl of 4× protein sample buffer (5), and heated at 95°C for 5 min, and aliquots representing 20 μg were analyzed by SDS-PAGE followed by immunoblotting as described above. To test for clathrin depletion, volumes representing 50 μg of protein were removed from post-pull-down supernatants and precipitated with trichloroacetic acid. Precipitated proteins were resuspended in 4× protein sample buffer, heated at 95°C for 5 min, and analyzed by SDS-PAGE and immunoblotting as described above. An aliquot representing 2% (50 μg) of the original cytosolic extract was also analyzed in this manner. To estimate the percentage of clathrin bound by GST fusions, densitometry was performed on multiple trials of anti-Chc1p immunoblots of GST pull-down (n = 2 or 3) using NIH Image software, version 1.62. Values obtained were corrected for loading, averaged, and presented as percentages of clathrin bound to GST fusion relative to clathrin input (i.e., 100%).
RESULTS
A double mutant of yeast GGA1 and GGA2 displays defective sorting of multiple vacuolar hydrolases and altered vacuolar morphology.
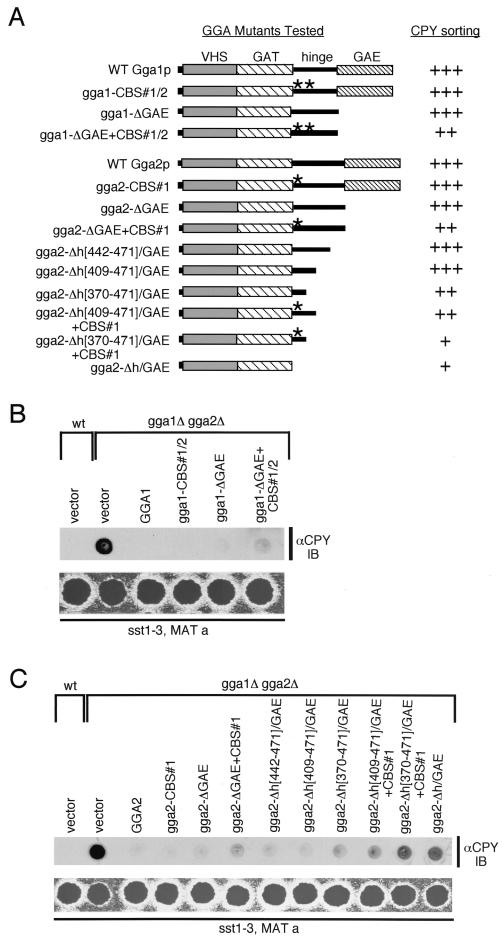
Yeast Gga1p and Gga2p share homology with various components of the protein trafficking machinery and play an apparently redundant role in efficient sorting of the vacuolar hydrolase CPY (16, 22, 55). Here we further elaborate on the requirement of the yeast GGAs in biosynthetic sorting from the Golgi complex to the vacuole by examining the processing of additional vacuolar proteins. We subjected wild-type and gga1Δ gga2Δ mutant cells (Table 1) to pulse-chase analyses followed by immunoprecipitation (Materials and Methods) with antibodies to vacuolar hydrolases CPY and PrA, substrates of the CPY sorting pathway, and ALP, a substrate of the ALP sorting pathway. As seen in Fig. 1A, gga1Δ gga2Δ mutant cells (lanes 5 to 8) exhibited a delayed and incomplete maturation of CPY relative to that of wild type (lanes 1 to 4), presumably due to impaired sorting of the 69-kDa Golgi complex-modified form of CPY (p2 CPY) to the vacuole, where final processing occurs, yielding the 61-kDa mature form (mCPY). The gga1Δ gga2Δ mutant also displayed a slower processing of the 52-kDa Golgi complex-modified form of PrA (p2 PrA) to the 42-kDa vacuolar, mature form (mPrA), with p2 PrA visible even after 30 min into the chase (lane 8). Interestingly, a slight ALP processing defect was also reproducibly seen in the gga1Δ gga2Δ mutant. Here, more of the 74-kDa Golgi complex-modified form of ALP (pALP) appeared present in the pulse (lane 5), with slower processing to the 72-kDa vacuolar, mature form (mALP) over the first 5 min of the chase (lane 6), relative to wild-type cells (lanes 1 and 2). These results suggest that the yeast GGAs participate in the CPY pathway for sorting of multiple vacuolar hydrolases and may also participate to a small degree in sorting via the ALP pathway.
FIG. 1.
The gga1Δ gga2Δ mutant displays a sorting defect for multiple vacuolar hydrolases and exhibits abnormal vacuolar morphology. (A) Vacuolar hydrolase processing in wild-type and gga1Δ gga2Δ mutant cells. Wild-type (lanes 1 to 4) and gga1Δ gga2Δ mutant (lanes 5 to 8) cells were pulse-labeled (P) for 10 min, and labeled proteins were chased for the times indicated (in minutes). Sequential immunoprecipitations from lysates were performed using anti-CPY, anti-PrA, and anti-ALP antibodies as described in Materials and Methods. Immunoprecipitated proteins were resolved by SDS-PAGE (8% acrylamide gels) and analyzed by fluorography. p1 CPY, p2 CPY, and mCPY refer to ER-glycosylated pro-CPY (67 kDa), Golgi complex-glycosylated pro-CPY (69 kDa), and mature (vacuolar) CPY (61 kDa), respectively. p2 PrA and mPrA refer to Golgi complex-glycosylated pro-PrA (52 kDa) and mature (vacuolar) PrA (mPrA) (42 kDa), respectively. pALP and mALP refer to membrane-bound, Golgi complex-glycosylated pro-ALP (74 kDa) and soluble, mature (vacuolar) ALP (mALP) (72 kDa), respectively. The asterisk indicates the position of a pseudomature form of CPY. (B) Analysis of vacuolar morphology of wild-type and gga1Δ gga2Δ and vps1 mutant cells by light microscopy. Wild-type, gga1Δ gga2Δ, and vps1 cells were incubated with vacuolar membrane-specific dye FM4-64 and vacuolar lumen-specific dye CDCFDA as described in Materials and Methods. Vacuoles were visualized by fluorescence microscopy using a small amount of transmitted light to reveal cell size and shape.
Vacuolar protein sorting (vps) mutants are often abnormal for vacuole number and structure, a finding that has led to grouping of these mutants into six classes (A to F) according to their vacuole morphology (40). Recent studies examining gga1Δ gga2Δ cells have come to somewhat different conclusions concerning their vacuole morphology (22, 55). Here, we examined the requirement of the yeast GGAs in maintenance of proper vacuole morphology through staining vacuoles with vital dyes specific for vacuolar membranes (FM4-64) and the vacuole lumen (CDCFDA) (50). As shown in Fig. 1B, wild-type cells exhibited normal vacuole number and morphology. In contrast, vacuoles in gga1Δ gga2Δ mutant cells appeared enlarged, with surrounding smaller, possibly fragmented, vacuolar structures. This vacuole morphology is similar to the class F phenotype seen for the vps1 mutant (Fig. 1B), which is defective in a dynamin-like GTPase associated with Golgi membranes (49, 53). Such class F mutants typically contain a large, central vacuole surrounded by smaller class B-like vacuolar structures (40). This finding confirms a vacuolar morphology defect in the gga1Δ gga2Δ mutant as being similar to class F mutants and establishes that the yeast GGAs are required for proper vacuole biogenesis.
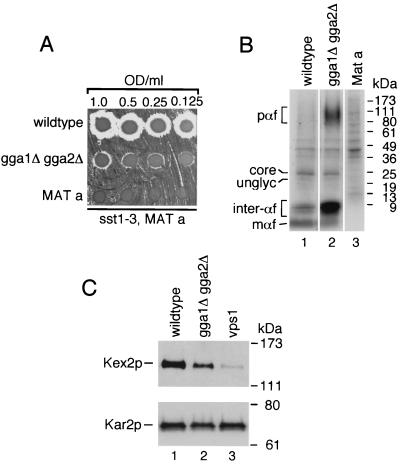
The gga1Δ gga2Δ mutant displays a defect in α-factor maturation and a decrease in Kex2p levels.
To further define the requirement of the GGAs in sorting from the Golgi complex, we analyzed α-factor maturation. The mating pheromone α-factor is synthesized as a pre-pro-form that is subject to endoplasmic reticulum (ER) and Golgi complex glycosylation and cleavage by the Golgi complex-localized endoprotease Kex2p (20). This proteolytic processing produces mature α-factor peptides that are secreted and act to prepare MATa cells for mating. To examine secretion of mature α-factor, wild-type (MATα), gga1Δ gga2Δ mutant (MATα), and wild-type (MATa) cells, as a negative control, were spotted on a lawn of a MATa strain which undergoes G1 arrest upon exposure to even low levels of the pheromone (9). This assay revealed diminished levels of mature α-factor secretion from the gga1Δ gga2Δ mutant as measured by the smaller zone of inhibition of the tester strain relative to that seen around wild-type cells (Fig. 2A). To define the block in α-factor maturation producing this secretion defect, the above strains were subjected to pulse-labeling and immunoprecipitation analysis with anti-α-factor antibodies (Materials and Methods). This analysis revealed an accumulation of high-molecular-weight α-factor species in the gga1Δ gga2Δ mutant. The size of these species (ranging from ∼90 to 150 kDa) suggests that they are α1-3-linked mannose-modified forms of pro-α-factor generated in the Golgi complex. Additional species with molecular masses of ∼6 to 8 kDa were also seen to accumulate in the gga1Δ gga2Δ mutant. These may represent late-Golgi complex species in various states of processing but with the highly glycosylated prosequence removed.
FIG. 2.
The gga1Δ gga2Δ mutant is defective for α-factor maturation and shows a decrease in Kex2p levels. (A) Secretion of mature α-factor in the gga1Δ gga2Δ mutant. Wild-type (MATα), gga1Δ gga2Δ (MATα) mutant, and wild-type (MATa) (negative control) cells were serially diluted to concentrations indicated, and equal amounts were spotted on rich medium (YEPD) containing a freshly spread lawn of α-factor-supersensitive mutant strain RC634 (MATa sst1-3). The agar plate was then incubated at 30°C to assess α-factor secretion of test strains as indicated by relative growth inhibition of sst1-3 mutant cells (halo). (B) Processing of α-factor in the gga1Δ gga2Δ mutant. Strains described for panel A were pulse-labeled at 25°C for 7.5 min as described in Materials and Methods. Various α-factor Golgi-processing intermediates and mature α-factor were immunoprecipitated with anti-α-factor antibodies, resolved by SDS-PAGE (4 to 20% acrylamide gels), and analyzed by fluorography. pαf (pro-α-factor) denotes a range of high-molecular-mass α-factor intermediates (∼90 to 150 kDa) containing the pro-region and Golgi complex-derived, complex glycosylation. unglyc and core refer to unglycosylated (∼20-kDa) and ER-localized, core-glycosylated (∼26-kDa) forms of pro-α-factor, respectively. inter-αf and mαf represent a range of low-molecular-mass α-factor intermediates (probably the α-factor tetrapeptide in various states of processing but with the prosequence removed) (∼6 to 8 kDa) and the 13-aa terminally processed (mature-secreted) form (∼2 kDa), respectively. Relative positions of molecular mass markers are indicated on the right. (C) Steady-state levels of Kex2p in the gga1Δ gga2Δ mutant. Whole-cell lysates were prepared from wild-type, gga1Δ gga2Δ, and vps1 cells expressing epitope-tagged Kex2p. Proteins were resolved by SDS-PAGE (8% acrylamide gels) and analyzed by immunoblotting using anti-HA and anti-Kar2p (to assess relative protein loading) antibodies as described in Materials and Methods. Positions of Kex2p and Kar2p and molecular mass size markers are indicated.
In vps1 cells, proteins that normally cycle between the late-Golgi complex and endosomal compartments, such as the CPY receptor Vps10p and Kex2p, as well as proteins destined for the vacuole, such as ALP, are mislocalized to the plasma membrane (32). This mislocalization precedes transport of Vps10p and Kex2p to the vacuole (32). Based on the observed defect in pro-α-factor processing in the Golgi complex, we sought to test the steady-state levels of Kex2p in our GGA-deficient mutant. For this purpose, we examined wild-type, gga1Δ gga2Δ, and vps1 mutant cells expressing an epitope-tagged Kex2p (Table 2 lists plasmids used). We observed a decrease in Kex2p steady-state levels in the gga1Δ gga2Δ mutant (Fig. 2C, lane 2) relative to wild type (lane 1). However, the decrease was not as severe as was seen in vps1 mutant cells (lane 3).
TABLE 2.
Plasmids used in this study
| Plasmid | Descriptiona | Reference |
|---|---|---|
| pCM54B4 | CEN-URA plasmid encoding Gga1p-HA (pYX112 based) | This study |
| pCM89B2 | CEN-URA plasmid encoding Gga1p-HA containing CBS#1 and CBS#2 mutations (pYX112 based) | This study |
| pCM123B3 | CEN-URA plasmid encoding Gga1p-ΔGAE (aa 445–557)-HA (pYX112 based) | This study |
| pCM124B5 | CEN-URA plasmid encoding Gga1p-ΔGAE (aa 445–557)-HA containing CBS#1 and CBS#2 mutations (pYX112 based) | This study |
| pCM53-3 | CEN-URA plasmid encoding Gga2p-HA (pYX112 based) | This study |
| pCM90-3 | CEN-URA plasmid encoding Gga2p-HA containing CBS#1 mutation (pYX112 based) | This study |
| pCM62-2 | CEN-URA plasmid encoding Gga2p-ΔGAE (aa 472–585)-HA (pYX112 based) | This study |
| pCM126-4 | CEN-URA plasmid encoding Gga2p-ΔGAE (aa 472–585)-HA containing CBS#1 mutation (pYX112 based) | This study |
| pCM63-1 | CEN-URA plasmid encoding Gga2p-Δhinge/GAE (aa 337–585)-HA (pYX112 based) | This study |
| pCM64-1 | CEN-URA plasmid encoding Gga2p-ΔVHS (aa 1–169)-HA (pYX112 based) | This study |
| pCM65-2 | CEN-URA plasmid encoding Gga2p-ΔVHS/GAT (aa 1–336)-HA (pYX112 based) | This study |
| pCM135-1 | CEN-URA plasmid encoding Gga2p-ΔVHS (aa 1–39)-HA (pYX112 based) | This study |
| pCM136-3 | CEN-URA plasmid encoding Gga2p-ΔVHS (aa 1–77)-HA (pYX112 based) | This study |
| pCM137-2 | CEN-URA plasmid encoding Gga2p-ΔVHS (aa 1–97)-HA (pYX112 based) | This study |
| pCM138-1 | CEN-URA plasmid encoding Gga2p-ΔVHS (aa 1–132)-HA (pYX112 based) | This study |
| pCM139-1 | CEN-URA plasmid encoding Gga2p-ΔVHS (aa 1–155)-HA (pYX112 based) | This study |
| pCM142B-3 | CEN-URA plasmid encoding Gga2p-Δhinge/GAE (aa 442–585) (pYX112 based) | This study |
| pCM143B-9 | CEN-URA plasmid encoding Gga2p-Δhinge/GAE (aa 409–585) (pYX112 based) | This study |
| pCM144B-4 | CEN-URA plasmid encoding Gga2p-Δhinge/GAE (aa 370–585) (pYX112 based) | This study |
| pCM146-16 | CEN-URA plasmid encoding Gga2p-Δhinge/GAE (aa 409–585) containing CBS#1 mutation (pYX112 based) | This study |
| pCM147-8 | CEN-URA plasmid encoding Gga2p-Δhinge/GAE (aa 370–585) containing CBS#1 mutation (pYX112 based) | This study |
| pCM85-1 | CEN-URA plasmid encoding Gga1-N215A (pYX112 based) | This study |
| pCM86-5 | CEN-URA plasmid encoding Gga2-N219A (pYX112 based) | This study |
| pCM147-8 | CEN-URA plasmid encoding Gga2p-Δhinge/GAE (aa 370–585) containing CBS#1 mutation (pYX112 based) | This study |
| pCM146-1 | CEN-URA plasmid encoding Gga2p-hinge/GAE (aa 337–585) containing CBS#1 mutation (pYX112 based) | This study |
| pCM133-2 | CEN-URA plasmid encoding Kex2p-HA (pYX112 based) | This study |
| pCM102-2 | GST fusion vector pGEX-5X-1 encoding GST-Gga1p-VHS/GAT (aa 1–331) | This study |
| pCM24-1 | GST fusion vector pGEX-5X-1 encoding GST-Gga1p-hinge/GAE (aa 332–557) | This study |
| pCM3-1 | GST fusion vector pGEX-5X-1 encoding GST-Gga1p-hinge/GAE (aa 332–557) containing CBS#1 mutation | This study |
| pCM8-3 | GST fusion vector pGEX-5X-1 encoding GST-Gga1p-hinge/GAE (aa 332–557) containing CBS#2 mutation | This study |
| pCM9-2 | GST fusion vector pGEX-5X-1 encoding GST-Gga1p-hinge/GAE (aa 332–557) containing CBS#1 and CBS#2 mutations | This study |
| pCM103-3 | GST fusion vector pGEX-5X-1 encoding GST-Gga2p-VHS/GAT (aa 1–336) | This study |
| pCM43-5 | GST fusion vector pGEX-5X-1 encoding GST-Gga2p-hinge/GAE (aa 337–585) | This study |
| pCM98-1 | GST fusion vector pGEX-5X-1 encoding GST-Gga2p-hinge/GAE (aa 337–585) containing CBS#1 mutation | This study |
| pCM151B-1 | GST fusion vector pGEX-5X-1 encoding GST-Gga2p-GAE (aa 472–585) | This study |
| pCM152B-1 | GST fusion vector pGEX-5X-1 encoding GST-Gga2p-Δhinge/GAE (aa 357–585) | This study |
Plasmids are described relative to full-length or mutant proteins that they encode. Amino acid residues indicated in parentheses for plasmids encoding Gga1p and Gga2p truncations indicate the residues deleted, while those indicated in parentheses for GST fusion constructs indicate the residues of Gga1p or Gga2p fused to GST. .
In combination, these results indicate that the yeast GGAs are required for efficient Golgi complex processing of pro-α-factor and that the observed defect in α-factor maturation seen in the gga1Δ gga2Δ mutant is possibly due to missorting of the Golgi endoprotease Kex2p.
Yeast GGA domains are differentially required for protein function in vivo.
The yeast GGAs, like their mammalian homologs, display a modular organization consisting of an amino-terminal VHS domain, a GAT domain, a variable hinge region, and a carboxy-terminal GAE domain (4, 16, 22, 43, 47). To address the relative importance of these domains for GGA function in vivo, a series of truncation mutants of Gga2p were produced (Fig. 3A) and expressed in the gga1Δ gga2Δ mutant.
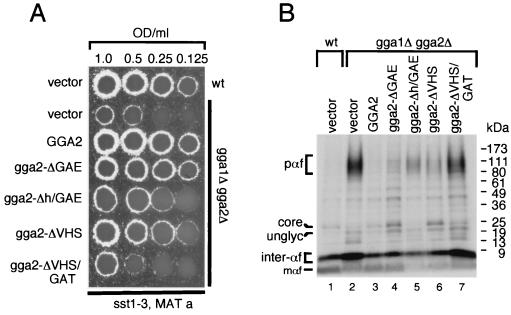
The gga1Δ gga2Δ mutant displays a defect in CPY sorting with a substantial percentage of CPY missorted to the periplasmic space (16, 22, 55). Missorted CPY is easily detectable through immunoblotting of colonies using anti-CPY antibodies (Materials and Methods). Initially, we examined the above truncations for their ability to complement this phenotype (Fig. 3B) (scoring of complementation is noted in Fig. 3A). We found the gga2-ΔGAE mutant complemented to a level similar to that of wild-type Gga2p with ≤5% CPY missorting (see Materials and Methods and Fig. 3 legend for method of calculating percent CPY missorting and Table 3 for summary of relative percent CPY missorted in strains tested) detected in blot assay. The gga2-Δh/GAE and gga2-ΔVHS mutants complemented much less, and the gga2-ΔVHS/GAT mutant provided little or no apparent complementation relative to controls.
The ability of these truncations to complement the gga1Δ gga2Δ mutant CPY processing defect was then assessed through pulse-chase and immunoprecipitation analysis (Materials and Methods). As shown in Fig. 3C, results paralleled those from the CPY missorting analysis, with truncation mutants displaying different degrees of complementation. CPY processing kinetics ranged from apparently wild-type level in mutant cells expressing the gga2-ΔGAE mutant (lanes 7 and 8) to similar to vector alone in cells expressing the gga2-ΔVHS/GAT mutant (lanes 13 and 14). Mutant cells expressing the gga2-Δh/GAE (lanes 9 and 10) and gga2-ΔVHS (lanes 11 and 12) mutants displayed intermediate levels of CPY processing relative to controls. To further address the importance of the GAT domain to GGA function, we produced GGA1 and GGA2 mutants containing N215A and N219A point mutations, respectively. This asparagine residue is conserved in mammalian and yeast GGA proteins and has been demonstrated to be important for Golgi localization of mammalian GGA3 (39). gga1Δ gga2Δ cells expressing wild-type and GAT mutant proteins were examined for CPY missorting as described above. Results indicated a reduced ability to complement the CPY missorting defect by both mutant proteins, though the effect was stronger for the GGA2 mutant (Fig. 3D and Table 3).
To eliminate the possibility that our GGA mutant gga2-ΔVHS/GAT acts as a dominant negative, we expressed this mutant and a second ΔVHS/GAT mutant containing a mutation in a putative clathrin-binding motif (described in the following section) in wild-type cells and assayed for any increase in CPY mislocalization. We detected no appreciable increase in CPY mislocalization relative to vector alone (Fig. 3E and Table 3), indicating that these fusions do not act in a dominant manner at these expression levels. This was important to examine based on findings described in the following section which show strong clathrin-binding activity by the h/GAE region of GGA2.
As the gga1Δ gga2Δ mutant also displayed a defect in α-factor maturation, we sought to examine the ability of the Gga2p mutants to complement this phenotype. Mutant cells expressing Gga2p truncations were assayed for secretion of mature α-factor (Fig. 4A) and for pro-α-factor processing (Fig. 4B). Both assays revealed findings similar to the above analyses of CPY processing-sorting with the gga2-ΔGAE truncation complementing the α-factor secretion and pro-α-factor processing defects to the greatest level relative to other truncations. Again, the gga2-Δh/GAE and gga2-ΔVHS mutants provided intermediate levels of complementation while the gga2-ΔVHS/GAT mutant provided little or no apparent complementation of the gga1Δ gga2Δ mutant phenotypes. The ability of the gga2-ΔGAE truncation to complement the CPY and α-factor maturation phenotypes is not attributable to high expression levels, as identical findings were seen with clones utilizing the endogenous GGA2 promoter (data not shown). Together, these findings reveal differential requirements for GGA domains for function in vivo. Here, the GAE domain appears to be largely nonessential for function in the assays employed. Interestingly, the combined removal of the GAE and hinge domains significantly reduced the ability to complement the gga1Δ gga2Δ mutant phenotypes examined, indicating that the hinge domain is important for function. Also, the VHS domain appears important for function, as the ΔVHS truncation largely failed to functionally replace wild-type Gga2p. Finally, removal of VHS and GAT, the region implicated in ARF binding in yeast and mammalian cells, domains completely eliminated the function of the protein.
FIG. 4.
GGA domains are differentially required for α-factor maturation. (A) Complementation of gga1Δ gga2Δ α-factor secretion defect by GGA2 truncations. Strains described for Fig. 3 were spotted on a lawn of tester strain RC634 (MATa sst1-3). Relative growth inhibition of tester lawn is indicative of level of mature α-factor secretion. (B) Complementation of gga1Δ gga2Δ α-factor-processing defect by GGA2 truncations. Strains described for panel A were pulse-labeled, and processed and mature forms of α-factor were immunoprecipitated using anti-α-factor antibodies. Immunoprecipitated proteins were resolved by SDS-PAGE and analyzed, and forms of α-factor and molecular mass standards are indicated as in Fig. 2B.
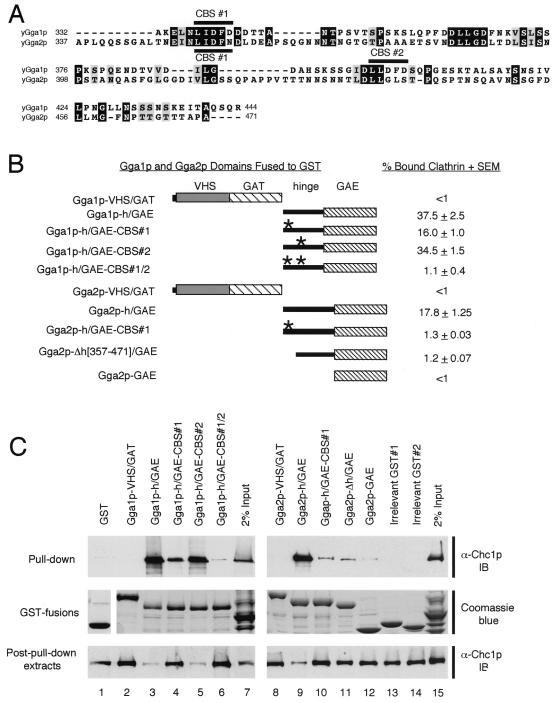
Gga1p and Gga2p interact with clathrin via consensus clathrin-binding motifs.
The requirement of the hinge domain for function, even in the absence of the GAE domain (Fig. 3 and 4), suggests that the hinge fulfills a role other than just linking the VHS/GAT and GAE domains of the GGAs. Interestingly, sequence analyses of the yeast GGAs revealed the presence of potential clathrin-binding motifs within their hinge domains. These motifs resemble a five-residue L(L, I)(D, E, N)(L, F)(D, E) consensus sequence found in a number of clathrin-binding proteins, including human GGA1 and GGA2; the β1, β2, and β3 subunits of mammalian AP complexes; and β-arrestin (15, 16, 25, 48; for reviews, see references 24 and 48). Gga1p contains two such motifs, LIDFD and LLDFD, while Gga2p contains one motif, LIDFN, which matches the consensus sequence only for the first four residues (Fig. 5A). These sequences are herein referred to as CBSs (for clathrin-binding sites). To test for GGA interactions with clathrin and to assess the possible contribution of the CBS motifs to this binding, a series of GST fusions containing sequences of Gga1p and Gga2p were produced (Fig. 5B). GST fusion proteins were incubated with a yeast cytosolic extract, and bound proteins were recovered and analyzed by SDS-PAGE followed by immunoblot analysis using anti-Chc1p antibodies (Materials and Methods) (Fig. 5C). This analysis revealed strong clathrin binding by the Gga1p and Gga2p hinge/GAE domains with little or no apparent binding by the VHS/GAT domains (Fig. 5B shows estimations of relative binding strengths of GST-GGA fusions). Mutation of the Gga1p CBS#1 and, to a lesser degree, CBS#2 reduced clathrin binding relative to the wild-type hinge/GAE. Mutation of both CBS#1 and CBS#2 eliminated most clathrin binding, though some residual binding was reproducibly observed for this fusion. Similarly, mutation of the CBS#1 present in Gga2p greatly reduced, but did not completely eliminate, clathrin binding. These results were suggestive that hinge and GAE sequences other than the CBS motifs may contribute to clathrin binding. To examine this, we tested Gga2p hinge (minus the region containing the CBS#1 site)/GAE (construct Gga2p-Δh[357-471]/GAE) and Gga2p GAE (construct Gga2p-GAE) sequences for binding. Results showed that the GGA2 Δh[357-471]/GAE region is capable of binding clathrin at levels above GST alone, the VHS/GAT sequence, and two irrelevant GST fusions. Also, the GGA2 GAE domain is capable of reproducibly binding a slight, but detectable, amount of clathrin.
FIG. 5.
Gga1p and Gga2p interact with clathrin in vitro. (A) Alignment of Gga1p and Gga2p hinge domains. Amino acid sequences representing hinge domains of Gga1p and Gga2p are aligned, and positions of putative Gga1p CBSs (CBS#1, LIDFD, and CBS#2, LLDFD) and a putative Gga2p CBS (CBS#1, LIDFN) are indicated. Black and gray shading represents amino acid identity and similarity, respectively. (B) Schematic representation of GST-GGA domain fusions tested for clathrin binding. GST-Gga1p and GST-Gga2p fusions (Table 2 shows details of GST expression constructs) are presented relative to their percent clathrin binding in vitro (as determined from panel C and calculated as described in Materials and Methods). The asterisk represents alanine substitution mutations in CBS#1 and/or CBS#2 of Gga1p and CBS#2 of Gga2p (Materials and Methods) as noted in names of fusions to the left. (C) Assay for in vitro clathrin binding. GST alone, GST fusions presented in panel B, and irrelevant GST fusions (as negative controls) were expressed in bacteria, isolated, and incubated with a yeast cytosolic extract, and fusion-bound protein complexes were recovered as described in Materials and Methods. Pull-down samples and trichloroacetic acid precipitants of input lysates and post-pull-down extracts were resolved by SDS-PAGE (8% acrylamide gels) and analyzed by immunoblotting (IB) using anti-Chc1p antibodies (Materials and Methods). Aliquots of GST and GST fusions were resolved by SDS-PAGE and visualized by Coomassie blue staining as a control for levels of fusions used in pull-down experiments.
Together, these results demonstrate that the yeast GGAs bind clathrin mainly via consensus CBS motifs located in their hinge domains. For Gga1p, it appears that two such motifs may act together to bind clathrin, while for Gga2p, it appears that only one such site is required for binding in vitro. Also, this study reveals that other sequences in the hinge and ear also contribute to interactions with clathrin in vitro.
Functional analysis of GGAs with CBS mutations and GAE and hinge domain truncations in vivo.
Next, we tested the in vivo requirement of the identified CBS sequences for GGA function in the context of the full-length proteins as well as in a series of GAE and hinge truncation mutants (Fig. 6A shows a schematic of GGA Δhinge/CBS/ΔGAE mutants). Mutant Gga1p and Gga2p were expressed in gga1Δ gga2Δ mutant cells, and their function was assessed through assays for CPY sorting and α-factor maturation (as described for Fig. 3 and 4 and in Materials and Methods). Surprisingly, we found that mutation of CBS sequences did not affect the function of Gga1p (Fig. 6B) and Gga2p (Fig. 6C) in vivo (Fig. 6A shows a summary of results, and Table 3 shows percent CPY missorting values). Recent studies, however, have shown that even though canonical CBS motifs account for most of the clathrin-binding activity in vitro, other sequences cooperate to yield maximal binding, including the GAE domain of human GGA1 (39) and the related ear domains of gamma-adaptin (18) and β2-adaptin (34). Moreover, findings in this study demonstrate that GGA2 hinge and GAE sequences not containing consensus CBS motifs can interact, albeit weakly, with clathrin in vitro (Fig. 5). These observations prompted us to test the effect of CBS mutations in the context of GGAs with truncations of the GAE and parts of the hinge domains. We observed that combining the CBS mutations with deletion of the GAE domain resulted in a partial loss of complementation for mutants of both Gga1p (Fig. 6B) and Gga2p (Fig. 6C) (Fig. 6A shows a summary of results). Estimations of the CPY missorting in these transformants using densitometry (Materials and Methods) revealed an approximately 14% defect (Table 3) relative to gga1Δ gga2Δ mutant cells transformed with vector alone (considered 100%). Serial truncations of the hinge region of Gga2p revealed a gradual loss of the ability to complement (Fig. 6C). Removal of hinge residues 370 to 409 (compare gga2-Δh[409–471]/GAE with gga2-Δh[370–471]/GAE) caused a particularly noticeable decrease in activity (Fig. 6C), while further removal of hinge residues 337 to 369, containing the CBS (compare gga2-Δh[370–471]/GAE with gga2-Δh/GAE), caused an even greater loss of activity (Fig. 6C). These observations suggested that the 370–408 segment containing an unidentified element and the 337–369 segment containing the CBS are important for function. Combining either the gga2-Δh[409–471]/GAE or gga2-Δh[370–471]/GAE hinge truncations with mutation of the CBS diminished the ability of both constructs to complement. In fact, the gga2-Δh[370–471]/GAE hinge construct with a mutation of the CBS was as ineffective as the gga2-Δh/GAE construct in complementing the CPY missorting defect. Both of these constructs exhibited ∼29 to 35% CPY missorting relative to mutant cells transformed with vector alone. The lowered ability of these Gga2p hinge and hinge-CBS mutants to complement can also be seen in the α-factor maturation assay, although this assay is somewhat less sensitive than the CPY blotting assay employed.
Taken together, these observations suggest that the CBS motifs in Gga1p and Gga2p contribute to Ggap function in vivo. Evidence for this is seen from the above analyses in which combining mutations in the CBS motifs with deletions of the hinge and/or GAE domain results in diminished activity relative to the hinge and GAE deletions alone. One explanation for the synthetic nature of these mutations is that the CBS motifs and other sequences in the hinge and GAE domains cooperate for GGA function.
Functional analysis of GGAs with VHS domain truncations in vivo.
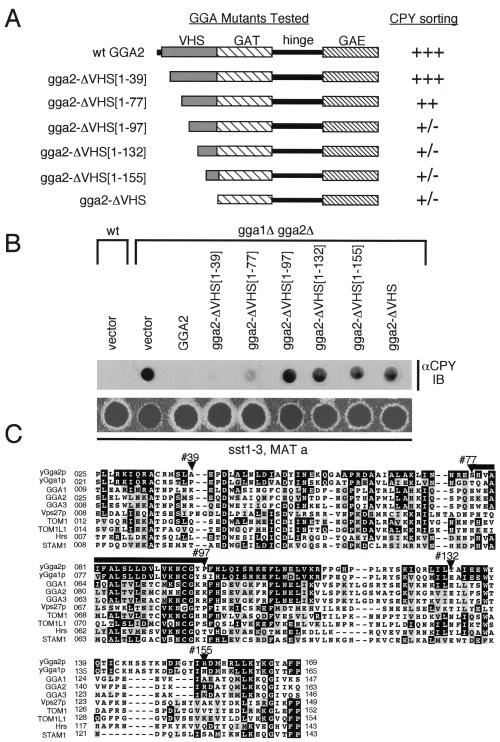
Recent studies have demonstrated that the mammalian GGA VHS domains interact with sorting signals in the cytosolic domains of sortilin (31), as well as the CD- and CI-MPRs (38, 56). This suggests that VHS domains may be generally involved in cargo recognition. To further examine the functional importance of the VHS domain of the yeast GGAs, a series of Gga2p VHS truncation mutants were prepared (Fig. 7A) and expressed in gga1Δ gga2Δ mutant cells. These mutants were then assessed for their ability to complement the gga1Δ gga2Δ phenotype using the CPY immunoblot and α-factor secretion assays (as described for Fig. 6 and Materials and Methods). Results in Fig. 7B show that removal of the first 39 residues produced little effect on the protein's function while truncation to residue 77 led to partial loss of complementation (the gga2-ΔVHS[1–77] mutant) (Fig. 7A shows a summary of results). However, further removal of residues 77 to 97 (the gga2-ΔVHS[1–97] mutant) resulted in a loss of function apparently equal to complete removal of the VHS domain (the gga2-ΔVHS mutant). These results suggest that residues 77 to 97 delineate a region of the Gga2p VHS domain critical for function. This region is highly conserved among GGA family members and other proteins containing the VHS domain (Fig. 6C). Future studies will attempt to characterize the role of the Ggap VHS domains and identify key residues within this conserved block critical to this function.
FIG. 7.
Functional analysis of the GGA VHS domain. (A) Schematic representation of GGA2 VHS mutants analyzed. Plasmids encoding wild-type Gga2p and Gga2p mutants containing serial truncations of the VHS domain were generated (Table 2 shows plasmid descriptions). The relative ability of the encoded proteins to complement the gga1Δ gga2Δ defects in CPY sorting (as assessed in panel B) is indicated using the scale described for Fig. 3. Numbers in brackets indicate residues truncated from the respective Gga2p mutants. (B) Complementation of gga1Δ gga2Δ CPY missorting and α-factor secretion defects by GGA2 mutants. Wild-type cells transformed with vector alone and gga1Δ gga2Δ cells transformed with vector alone and plasmids encoding wild-type Gga2p and mutants indicated were examined for relative levels of CPY missorting and secretion of mature α-factor as described for Fig. 3. (C) Alignment of yeast and mammalian VHS domains. VHS domains from yeast and mammalian GGA family members and VHS domains found in additional proteins are aligned relative to yeast Gga2p. Black and gray shading represents amino acid identity and similarity, respectively. Residues marking sites of serial Gga2p VHS truncations are indicated. The region of the Gga2p VHS domain (amino acids 77 to 97) demonstrated as important for protein function is indicated with a bar. wt, wild type.
DISCUSSION
Two pathways for the biosynthetic transport of vacuolar proteins from the Golgi complex to the vacuole have been described for S. cerevisiae. The first pathway, known as the ALP pathway, involves direct transport from the late Golgi complex to the vacuole and requires the AP-3 complex as well as a number of VPS gene products. The second pathway, known as the CPY pathway, involves sorting from the late Golgi complex to the PVC prior to transport to the vacuole and involves numerous VPS gene products and clathrin. In this study, we present evidence that the yeast GGAs function primarily in the CPY pathway of vacuolar sorting. We demonstrate that disruption of the two GGA genes impairs processing of CPY as well as PrA, both substrates of the CPY pathway. In contrast, processing of alkaline phosphatase, a substrate of the ALP pathway, is only slightly delayed in gga1Δ gga2Δ mutant cells. This effect on ALP maturation may be indirect, perhaps due to impaired sorting of the vacuolar hydrolase PrA, which processes pro-ALP in the vacuole.
In addition to the defects in vacuolar sorting, we observe a defect in vacuolar morphology in the gga1Δ gga2Δ mutant. Mutant cells exhibit enlarged vacuoles with, in some cases, surrounding smaller vacuolar structures. This phenotype is analogous to the class F vps mutants (40) vps1 and vps26, the latter of which contains a defect in the p50 subunit of the retromer complex (44). The phenotype detailed here is therefore similar to that reported previously by Hirst et al. (22) for cells deficient in the GGAs. The absence of a morphological defect in GGA mutant cells reported in another study (55) may have been due to a different genetic background or the growth conditions used in that study. The class F vacuole morphology is suggestive of abnormal accumulation of vacuolar material, possibly due to increased delivery of plasma membrane-derived vesicles containing cargo missorted from the Golgi complex, as has been suggested for the vps1 mutant (32). Alternatively, vacuolar material may accumulate due to defective retrieval of late-endosomal–vacuolar proteins, shown to occur in mutants of the retromer subunits including Vps26p (44) and the vacuolar inheritance factor Vac7p (6).
Like other mutants affecting the CPY pathway (1, 10, 42), including clathrin (35, 45) and vps1 (53) mutants, the gga1Δ gga2Δ mutant exhibits reduced processing of pro-α-factor. This phenotype is likely due to missorting of Kex2p, the late-Golgi network subtilisin-like protease that converts pro-α-factor to mature α-factor (19, 28), to the vacuole possibly via the plasma membrane. In support of this scenario is the mislocalization of Kex2p in vps1 (32) and clathrin (35) mutants as well as the decreased steady-state levels of Kex2p observed in our GGA mutant (Fig. 2C) and vps1 (53) cells. In an apparently analogous manner, the CPY receptor Vps10p is also missorted and unstable in vps1 cells (8) as well as in other vps mutants (8, 43). However, Vps10p localization has been reported as similar to wild type for GGA mutant cells (22). In the GGA mutant, as well as other vesicle formation-transport mutants, inefficient incorporation of proteins into transport vesicles destined for the PVC may induce default transport to the vacuole, thus leading to the observed phenotypes. However, the possibility that the GGAs also function in the recycling of Kex2p and Vps10p from endosomes back to the Golgi complex cannot be ruled out.
Compared to other adaptor-type proteins, a unique aspect of the GGA family is their modular organization consisting of distinct domains termed VHS, GAT, hinge, and GAE. Here we demonstrate that these domains are differentially required for yeast GGA function in the CPY pathway and in α-factor maturation. Our results indicate that the VHS, GAT, and hinge domains play important roles in GGA function while the GAE domain appears less important in the assays used. These in vivo findings are especially interesting in light of recent studies that have addressed the function of individual mammalian GGA domains in vitro. The VHS domains of the human GGAs were recently shown to bind to acidic cluster-dileucine signals of three receptors that traffic between the TGN and endosomes: sortilin (31) and the CI- and CD-MPRs (38, 56). Overexpression of a truncated GGA1 construct lacking the hinge and GAE domains blocked transport of both MPRs from the TGN (38), consistent with a role for GGA1 in TGN-to-endosome sorting. By analogy, it is tempting to speculate that the VHS domain of the yeast GGAs may be involved in the recognition of sorting signals in the cytosolic tails of proteins that cycle between the late Golgi network and endosomes. Yeast Kex2p, like its mammalian ortholog furin, and Vps10p require sequences in their cytosolic tails for proper localization (8, 19). However, neither the Kex2p nor the Vps10p tail contains the exact combination of acidic and dileucine residues important for interactions between the above mammalian receptors and the mammalian GGAs. The yeast prevacuolar syntaxin Pep12p contains an FSDSPEF signal in its cytosolic tail that mediates sorting from the late Golgi complex to late endosomes. This sorting is dependent on the GGAs, although interactions of the FSDSPEF signal with the GGAs could not be demonstrated (2). Despite these facts, the importance of the yeast GGA VHS domain to function in vivo is apparent in analyses presented here. We show that deletion of the VHS domain severely reduces the ability of GGA2 to function in vivo (Fig. 3 and Table 3). However, it remains to be determined whether this domain functions in binding sorting signals in yeast. Also, point mutations of conserved residues of the GGA1 and GGA2 GAT domains lead to reduced activity (Fig. 3 and Table 3). The requirement of the GAT domain is likely due to its ability to interact with the GTP-bound form of ADP-ribosylation factor (ARF) proteins, a family of ras-like GTPases that regulate formation of transport vesicles (for reviews, see references 3 and 17). This has been shown for both the mammalian (16, 39) and the yeast (55) GGAs. Deletion of the GAT domain of the yeast GGAs may thus prevent the proteins from associating with the Golgi complex in vivo. The GAE domain is the least important for the functions of the yeast GGAs tested here. Expression of a gga2-ΔGAE mutant protein appears to fully complement sorting phenotypes seen in the gga1Δ gga2Δ mutant, independently of expression levels (Fig. 2 and data not shown). However, the strong conservation of this domain throughout evolution implies that it must play an important, though as yet undetermined, role in GGA function.
Recruitment of cytosolic clathrin to donor membranes drives the formation of secretory vesicles (for reviews, see references 24 and 36). The functional clathrin molecule, the triskelion, is composed of three heavy chains (Chc) and three light chains (Clc). Deletion of the yeast CHC or CLC genes results in extremely poor growth, defective endocytosis, and missorting of Golgi proteins, indicating the importance of clathrin in vesicle-mediated protein transport. Clathrin recruitment to membranes is believed to be mediated by adaptor proteins, such as the AP complexes. In mammalian cells, this is a function of the AP-1, AP-2, and AP-3 complexes. In yeast, however, only the AP-1 complex has been shown to associate physically with clathrin (54). In addition, null alleles of genes encoding AP-1 subunits, but not other AP genes, exhibit synthetic interactions with clathrin mutants (54). Surprisingly, deletion of genes encoding AP-1 subunits alone or in combination with subunits of AP-2 and AP-3, as well as mutations in yeast AP180, which binds clathrin in vitro (51), does not result in the clathrin-minus phenotypes seen in clathrin deletion mutants (23, 54). Furthermore, clathrin-coated vesicles which are indistinguishable from those in wild-type cells can be isolated from strains carrying multiple deletions of these genes (23, 54). This has led to the proposal that other adaptor proteins must exist that possess a redundant function in linking clathrin to membranes.
Our observations indicate that the yeast GGAs may act as clathrin adaptors in the CPY pathway. Upon initial sequence analysis of the GGA family (16), we observed that their hinge regions contained variants of the 5-aa L(L,I)(D,E,N)(L,F)(D,E) clathrin-binding motif previously identified in a host of mammalian proteins. These include the β-adaptin subunits of AP-1, AP-2, and AP-3; β-arrestins I and II (arrestin 3); and amphiphysins I and II and epsin I (reference 48 and references therein). Yeast β1-adaptin (54) and yeast AP180 (51), as well as the two yeast epsins (52), also contain variants of this motif. For some clathrin-binding proteins, the interaction has been defined at the atomic level. For example, the clathrin-binding motifs of β3A-adaptin and β-arrestin II bind to a groove between blades 1 and 2 of the seven-β-propeller structure of the clathrin amino-terminal domain (48). Despite extensive evidence for interactions of this motif with clathrin in vitro, however, the physiological roles of these interactions have yet to be demonstrated. In a previous study, we showed that the human GGAs interacted in vitro with clathrin mainly via the clathrin-binding motifs in their hinge domains, although the GAE domain also contributed to the binding to GGA1 (39). Here we show that clathrin-binding motifs in the hinge domains of yeast Gga1p and Gga2p also mediate interactions with clathrin in vitro. Mutation of these clathrin-binding motifs in the context of the full-length Gga1p and Gga2p had no effect on CPY sorting and pro-α-factor processing. However, combining mutations in the clathrin-binding motifs with removal of the GAE domain and the carboxy-terminal two-thirds of hinge (for Gga2p) and, to a lesser degree, with removal of the GAE domain alone (for Gga1p and Gga2p) uncovered a requirement for the clathrin-binding motifs in GGA-mediated CPY sorting and pro-α-factor processing. These observations indicate that interactions of the GGAs with clathrin via the canonical clathrin-binding motif contribute to GGA function in sorting through the CPY pathway and the Golgi complex maturation of α-factor. Because this contribution is apparent only in the context of hinge and GAE deletions, however, our results suggest that the hinge and GAE contribute additional determinants for direct or indirect attachment to clathrin. This belief is supported by findings in Fig. 5 which show that the GGA2 h/GAE domains missing the region containing the canonical clathrin-binding motif can still bind clathrin in vitro. This is in line with the emerging notion that optimal clathrin binding in vitro involves cooperative interactions with two or more clathrin-binding sites. Indeed, this has been demonstrated for mammalian β2-adaptin (34), GGA1 (39), and gamma-adaptin (18), all of which appear to interact with clathrin via the hinge and GAE (ear) domains.
In conclusion, we have demonstrated that the VHS, GAT, and hinge domains of the yeast GGAs fulfill essential functions in vivo. In addition, we show that the hinge domains of the yeast GGAs bind clathrin by virtue of canonical clathrin-binding motifs and that this ability is important for the function of the GGAs in the CPY pathway of Golgi complex-to-endosome-vacuole sorting and in α-factor maturation.
ACKNOWLEDGMENTS
We thank Xiaolin Zhu for excellent technical assistance; Todd Graham, Sandra Lemmon, Tom Stevens, Carol Woolford, and Mark Rose for generous gifts of antibodies; and Tom Stevens and Neil Green for yeast strains. We thank Jose Martina for the gift of negative control GST fusions. We thank Cathy Jackson and Cecilia Bonangelino for critical reading of the manuscript.
C.M. is supported by a National Research Council Post-Doctoral Associateship.
REFERENCES
- 1.Bensen E S, Costaguta G, Payne G S. Synthetic genetic interactions with temperature-sensitive clathrin in Saccharomyces cerevisiae. Roles for synaptojanin-like Inp53p and dynamin-related Vps1p in clathrin-dependent protein sorting at the trans-Golgi network. Genetics. 2000;154:83–97. doi: 10.1093/genetics/154.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black M W, Pelham H R. A selective transport route from Golgi to late endosomes that requires the yeast GGA proteins. J Cell Biol. 2000;151:587–600. doi: 10.1083/jcb.151.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boman A L, Kahn R A. Arf proteins: the membrane traffic police? Trends Biochem Sci. 1995;20:147–150. doi: 10.1016/s0968-0004(00)88991-4. [DOI] [PubMed] [Google Scholar]
- 4.Boman A L, Zhang C, Zhu X, Kahn R A. A family of ADP-ribosylation factor effectors that can alter membrane transport through the trans-Golgi. Mol Biol Cell. 2000;11:1241–1255. doi: 10.1091/mbc.11.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonifacino J S, Dasso M, Harford J B, Lippincott-Schwartz J, Yamada K M, editors. Current protocols in cell biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1998. [Google Scholar]
- 6.Bryant N J, Piper R C, Weisman L S, Stevens T H. Retrograde traffic out of the yeast vacuole to the TGN occurs via the prevacuolar/endosomal compartment. J Cell Biol. 1998;142:651–663. doi: 10.1083/jcb.142.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryant N J, Stevens T H. Vacuole biogenesis in Saccharomyces cerevisiae: protein transport pathways to the yeast vacuole. Microbiol Mol Biol Rev. 1998;62:230–247. doi: 10.1128/mmbr.62.1.230-247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cereghino J L, Marcusson E G, Emr S D. The cytoplasmic tail domain of the vacuolar protein sorting receptor Vps10p and a subset of VPS gene products regulate receptor stability, function, and localization. Mol Biol Cell. 1995;6:1089–1102. doi: 10.1091/mbc.6.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan R K. Recovery of Saccharomyces cerevisiae mating-type a cells from G1 arrest by α factor. J Bacteriol. 1977;130:766–774. doi: 10.1128/jb.130.2.766-774.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C Y, Graham T R. An arf1Δ synthetic lethal screen identifies a new clathrin heavy chain conditional allele that perturbs vacuolar protein transport in Saccharomyces cerevisiae. Genetics. 1998;150:577–589. doi: 10.1093/genetics/150.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C Y, Ingram M F, Rosal P H, Graham T R. Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J Cell Biol. 1999;147:1223–1236. doi: 10.1083/jcb.147.6.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conibear E, Stevens T H. Multiple sorting pathways between the late Golgi and the vacuole in yeast. Biochim Biophys Acta. 1998;1404:211–230. doi: 10.1016/s0167-4889(98)00058-5. [DOI] [PubMed] [Google Scholar]
- 13.Cooper A A, Stevens T H. Vps10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J Cell Biol. 1996;133:529–541. doi: 10.1083/jcb.133.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowles C R, Odorizzi G, Payne G S, Emr S D. The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell. 1997;91:109–118. doi: 10.1016/s0092-8674(01)80013-1. [DOI] [PubMed] [Google Scholar]
- 15.Dell'Angelica E C, Klumperman J, Stoorvogel W, Bonifacino J S. Association of the AP-3 adaptor complex with clathrin. Science. 1998;280:431–434. doi: 10.1126/science.280.5362.431. [DOI] [PubMed] [Google Scholar]
- 16.Dell'Angelica E C, Puertollano R, Mullins C, Aguilar R C, Vargas J D, Hartnell L M, Bonifacino J S. GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J Cell Biol. 2000;149:81–94. doi: 10.1083/jcb.149.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donaldson J G, Jackson C L. Regulators and effectors of the ARF GTPases. Curr Opin Cell Biol. 2000;12:475–482. doi: 10.1016/s0955-0674(00)00119-8. [DOI] [PubMed] [Google Scholar]
- 18.Doray B, Kornfeld S. The γ-subunit of AP-1 binds clathrin—implications for cooperative binding in coated vesicle assembly. Mol Biol Cell. 2001;12:1925–1935. doi: 10.1091/mbc.12.7.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuller R S, Brake A J, Thorner J. Intracellular targeting and structural conservation of a prohormone-processing endoprotease. Science. 1989;246:482–486. doi: 10.1126/science.2683070. [DOI] [PubMed] [Google Scholar]
- 20.Fuller R S, Sterne R E, Thorner J. Enzymes required for yeast prohormone processing. Annu Rev Physiol. 1988;50:345–362. doi: 10.1146/annurev.ph.50.030188.002021. [DOI] [PubMed] [Google Scholar]
- 21.Graham T R, Emr S D. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18 (NSF) mutant. J Cell Biol. 1991;114:207–218. doi: 10.1083/jcb.114.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirst J, Lui W W, Bright N A, Totty N, Seaman M N, Robinson M S. A family of proteins with gamma-adaptin and VHS domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome. J Cell Biol. 2000;149:67–80. doi: 10.1083/jcb.149.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang K M, D'Hondt K, Riezman H, Lemmon S K. Clathrin functions in the absence of heterotetrameric adaptors and AP180-related proteins in yeast. EMBO J. 1999;18:3897–3908. doi: 10.1093/emboj/18.14.3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirchhausen T. Clathrin. Annu Rev Biochem. 2000;69:699–727. doi: 10.1146/annurev.biochem.69.1.699. [DOI] [PubMed] [Google Scholar]
- 25.Krupnick J G, Goodman O B, Jr, Keen J H, Benovic J L. Arrestin/clathrin interaction. Localization of the clathrin binding domain of nonvisual arrestins to the carboxy terminus. J Biol Chem. 1997;272:15011–15016. doi: 10.1074/jbc.272.23.15011. [DOI] [PubMed] [Google Scholar]
- 26.Lemmon S K, Traub L M. Sorting in the endosomal system in yeast and animal cells. Curr Opin Cell Biol. 2000;12:457–466. doi: 10.1016/s0955-0674(00)00117-4. [DOI] [PubMed] [Google Scholar]
- 27.Marcusson E G, Horazdovsky B F, Cereghino J L, Gharakhanian E, Emr S D. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- 28.Mizuno K, Nakamura T, Ohshima T, Tanaka S, Matsuo H. Yeast KEX2 genes encode an endopeptidase homologous to subtilisin-like serine proteases. Biochem Biophys Res Commun. 1988;156:246–254. doi: 10.1016/s0006-291x(88)80832-5. [DOI] [PubMed] [Google Scholar]
- 29.Mullins C, Bonifacino J S. The molecular machinery for lysosome biogenesis. Bioessays. 2001;23:333–343. doi: 10.1002/bies.1048. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura N, Hirata A, Ohsumi Y, Wada Y. Vam2/Vps41p and Vam6/Vps39p are components of a protein complex on the vacuolar membranes and involved in the vacuolar assembly in the yeast Saccharomyces cerevisiae. J Biol Chem. 1997;272:11344–11349. doi: 10.1074/jbc.272.17.11344. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen M S, Madsen P, Christensen E I, Nykjaer A, Gliemann J, Kasper D, Pohlmann R, Petersen C M. The sortilin cytoplasmic tail conveys Golgi-endosome transport and binds the VHS domain of the GGA2 sorting protein. EMBO J. 2001;20:2180–2182. doi: 10.1093/emboj/20.9.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nothwehr S F, Conibear E, Stevens T H. Golgi and vacuolar membrane proteins reach the vacuole in vps1 mutant yeast cells via the plasma membrane. J Cell Biol. 1995;129:35–46. doi: 10.1083/jcb.129.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ooms L M, McColl B K, Wiradjaja F, Wijayaratnam A P, Gleeson P, Gething M J, Sambrook J, Mitchell C A. The yeast inositol polyphosphate 5-phosphatases Inp52p and Inp53p translocate to actin patches following hyperosmotic stress: mechanism for regulating phosphatidylinositol 4,5-bisphosphate at plasma membrane invaginations. Mol Cell Biol. 2000;20:9376–9390. doi: 10.1128/mcb.20.24.9376-9390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Owen D J, Vallis Y, Pearse B M, McMahon H T, Evans P R. The structure and function of the β2-adaptin appendage domain. EMBO J. 2000;19:4216–4227. doi: 10.1093/emboj/19.16.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Payne G S, Schekman R. Clathrin: a role in the intracellular retention of a Golgi membrane protein. Science. 1989;245:1358–1365. doi: 10.1126/science.2675311. [DOI] [PubMed] [Google Scholar]
- 36.Pishvaee B, Payne G S. Clathrin coats—threads laid bare. Cell. 1998;95:443–446. doi: 10.1016/s0092-8674(00)81611-6. [DOI] [PubMed] [Google Scholar]
- 37.Poussu A, Lohi O, Lehto V P. Vear, a novel Golgi-associated protein with VHS and γ-adaptin “ear” domains. J Biol Chem. 2000;275:7176–7183. doi: 10.1074/jbc.275.10.7176. [DOI] [PubMed] [Google Scholar]
- 38.Puertollano R, Aguilar R C, Gorshkova I, Crouch R J, Bonifacino J S. Sorting of mannose 6-phosphate receptors mediated by the GGAs. Science. 2001;292:1712–1716. doi: 10.1126/science.1060750. [DOI] [PubMed] [Google Scholar]
- 39.Puertollano R, Randazzo P A, Presley J F, Hartnell L M, Bonifacino J S. The GGAs promote ARF-dependent recruitment of clathrin to the TGN. Cell. 2001;105:93–102. doi: 10.1016/s0092-8674(01)00299-9. [DOI] [PubMed] [Google Scholar]
- 40.Raymond C K, Howald-Stevenson I, Vater C A, Stevens T H. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts C J, Raymond C K, Yamashiro C T, Stevens T H. Methods for studying the yeast vacuole. Methods Enzymol. 1991;194:644–661. doi: 10.1016/0076-6879(91)94047-g. [DOI] [PubMed] [Google Scholar]
- 42.Robinson J S, Graham T R, Emr S D. A putative zinc finger protein, Saccharomyces cerevisiae Vps18p, affects late Golgi functions required for vacuolar protein sorting and efficient α-factor prohormone maturation. Mol Cell Biol. 1991;11:5813–5824. doi: 10.1128/mcb.11.12.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seaman M N, Marcusson E G, Cereghino J L, Emr S D. Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35 gene products. J Cell Biol. 1997;137:79–92. doi: 10.1083/jcb.137.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seaman M N, McCaffery J M, Emr S D. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J Cell Biol. 1998;142:665–681. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seeger M, Payne G S. A role for clathrin in the sorting of vacuolar proteins in the Golgi complex of yeast. EMBO J. 1992;11:2811–2818. doi: 10.1002/j.1460-2075.1992.tb05348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stepp J D, Huang K, Lemmon S K. The yeast adaptor protein complex, AP-3, is essential for the efficient delivery of alkaline phosphatase by the alternate pathway to the vacuole. J Cell Biol. 1997;139:1761–1774. doi: 10.1083/jcb.139.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takatsu H, Yoshino K, Nakayama K. Adaptor γ-ear homology domain conserved in γ-adaptin and GGA proteins that interact with γ-synergin. Biochem Biophys Res Commun. 2000;271:719–725. doi: 10.1006/bbrc.2000.2700. [DOI] [PubMed] [Google Scholar]
- 48.ter Haar E, Harrison S C, Kirchhausen T. Peptide-in-groove interactions link target proteins to the β-propeller of clathrin. Proc Natl Acad Sci USA. 2000;97:1096–1100. doi: 10.1073/pnas.97.3.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vater C A, Raymond C K, Ekena K, Howald-Stevenson I, Stevens T H. The VPS1 protein, a homolog of dynamin required for vacuolar protein sorting in Saccharomyces cerevisiae, is a GTPase with two functionally separable domains. J Cell Biol. 1992;119:773–786. doi: 10.1083/jcb.119.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vida T A, Emr S D. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wendland B, Emr S D. Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J Cell Biol. 1998;141:71–84. doi: 10.1083/jcb.141.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wendland B, Steece K E, Emr S D. Yeast epsins contain an essential N-terminal ENTH domain, bind clathrin and are required for endocytosis. EMBO J. 1999;18:4383–4393. doi: 10.1093/emboj/18.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilsbach K, Payne G S. Vps1p, a member of the dynamin GTPase family, is necessary for Golgi membrane protein retention in Saccharomyces cerevisiae. EMBO J. 1993;12:3049–3059. doi: 10.1002/j.1460-2075.1993.tb05974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yeung B G, Phan H L, Payne G S. Adaptor complex-independent clathrin function in yeast. Mol Biol Cell. 1999;10:3643–3659. doi: 10.1091/mbc.10.11.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhdankina O, Strand N L, Redmond J M, Boman A L. Yeast GGA proteins interact with GTP-bound Arf and facilitate transport through the Golgi. Yeast. 2001;18:1–18. doi: 10.1002/1097-0061(200101)18:1<1::AID-YEA644>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 56.Zhu Y, Doray B, Poussu A, Lehto V-P, Kornfeld S. The VHS domain of GGA2 binds the lysosomal enzyme sorting motif of the Man-6-P/IGF II receptor. Science. 2001;292:1716–1718. doi: 10.1126/science.1060896. [DOI] [PubMed] [Google Scholar]