Abstract
Both Saccharomyces and non‐Saccharomyces yeast strains are of great importance for the fermentation industry, especially with the flourishing of craft breweries, which are driving current innovations. Non‐conventional yeasts can produce novel beverages with attractive characteristics such as flavour, texture, and reduced alcohol content; however, they have been poorly explored. A new method for screening the fitness of conventional and non‐conventional yeast libraries utilising robotic platforms and solidified media representing industrial conditions is proposed. As proof of concept, a library formed of 6 conventional and 17 non‐conventional yeast strains was distributed in 96, 384 and 1536 arrays onto a YPD agar medium. Following this, the library was replicated in different conditions mimicking beer and cider fermentation conditions. The colony size was monitored over time, and fitness values measured in maximum pixels/h and maximum biomass were calculated. Significant differences in growth were observed in between the different strains and conditions. As examples, Candida milleri Y‐7245 displayed good performance in wort conditions, and Kazachstania yakushimaensis Y‐48837 stood out for its performance in apple juice. The method is proposed to be used as a pre‐screening step when studying vast yeast libraries. This would enable interested parties to discover potential hits for further study at a low initial cost. Furthermore, this method can be used in other applications where the desired screening media can be solidified.
Keywords: beer, high‐throughput, non‐Saccharomyces , screening, yeast
1. INTRODUCTION
Yeasts are intertwined with human history and still play a crucial role in the food industry in the present day [1]. They are key players in processes involved in the production of food such as bread, dairy products, cocoa and coffee as well as the vast majority of alcoholic beverages [2]. Some of these fermentative processes occur due to the spontaneous activity of wild yeasts, whereas others, such as beer production, typically make use of pure inocula.
Saccharomyces cerevisiae dominates both spontaneous and controlled fermentations and has traditionally been the species of choice for the production of alcoholic beverages [3]. However, non‐conventional yeasts (NCY) have gained interest in recent years [4, 5, 6]. The use of NCY has opened the door for innovation within the brewing application. These organisms, which were traditionally considered as spoilage, can produce beverages with improved characteristics such as aroma profile [7, 8], texture [9], and recently marketable low‐alcohol content [9, 10, 11, 12].
The use of NCY in brewing has grown significatively [5]—partially, thanks to the expansion of the craft brewing industry, which appears to be more open to innovation than large‐scale industrial brewing. Craft brewers are continuously looking for new ingredients, processes and yeast strains for manufacturing distinctive products [13, 14].
However, the biological knowledge of NCY is limited, with the exception of some organisms such as Kluyveromyces lactis [15] and Yarrowia lipolytica [16]. One way of overcoming this limitation is the use of high‐throughput screening (HTS).
HTS technologies have been employed in a multitude of research fields for more than 2 decades, being particularly prevalent in the pharmaceutical industry [17]. In the field of yeast research, this has been done primarily in whole‐genome studies making use of the S. cerevisiae deletion collection [18] and synthetic gene array (SGA) approaches [19, 20]. These studies have been based on screening different mutant strains of the same species, but a genera‐wide approach can also be employed. The suitability of different NCY for the brewing industry has been assessed before by using liquid media HTS approaches [7, 21, 22]. However, these methods are normally carried out at very low volumes, thus being questionably representative of real brewing conditions. The number of strains that can be simultaneously studied is another caveat in liquid medium experiments. These are typically performed in 96 or 384 well‐plate formats, whereas solid medium high‐density arrays can technically be as dense as 24,576 samples per plate [23].
In this study, we propose a new method for the rapid screening of conventional and non‐conventional yeast strains using arrays in the media mimicking industrial conditions. This could be used as an initial step for hit generation, identifying strains with potential use in the brewing industry. A yeast library composed of a vast number of wild isolates could be screened with our proposed method, allowing rapid selection for downstream analysis (Figure 1).
FIGURE 1.
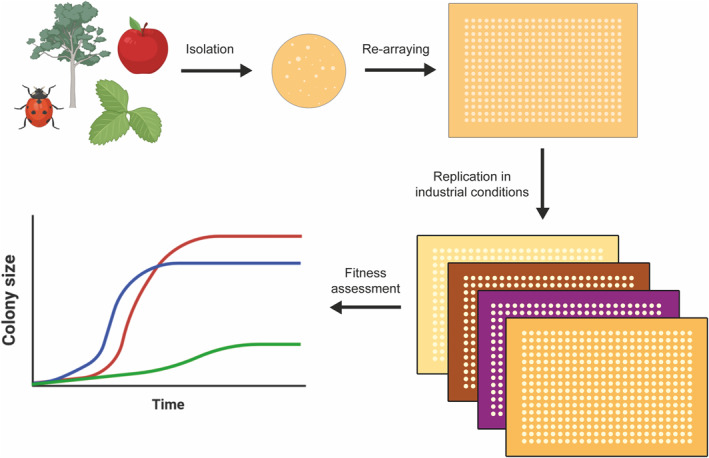
Schematic representation of the proposed workflow. A vast number of Saccharomyces and non‐Saccharomyces isolates can be re‐arrayed and replicated in solidified media representing industrial conditions (i.e.,: wort and apple juice). Fitness is assessed by monitoring colony size over time
2. MATERIALS AND METHODS
To prove the feasibility of the proposed method, we re‐arrayed a collection of conventional and non‐conventional yeast strains onto plates containing solidified media representing cider and beer production conditions. Growth performance was assessed by monitoring the colony size over time.
2.1. Strains
A total of 23 strains were used in the study (Table 1). Six of them were Saccharomyces and 17 non‐Saccharomyces. All the strains except for the lab strains used as control (BY4741 and ESM356‐1) were obtained from the NRRL culture collection (NRRL, ARS Culture Collection).
TABLE 1.
Strains utilised in the study
| Species | Abbreviation | Strain | Isolation source |
|---|---|---|---|
| Candida milleri | Cm | Y‐7245 | Sour dough, California |
| C. milleri | Cm | Y‐7248 | Sour dough, California |
| Cyberlindnera fabianii | Cf | Y‐6710 | Rice, Japan |
| C. meyerae | Cm | Y‐17236 | Insect, South Africa |
| Hanseniaspora vineae | Hv | Y‐17530 | Tree, Canada |
| Kazachstania yakushimaensis | Ky | Y‐48837 | Leaves, Japan |
| Kluyveromyces lactis | Kl | Y‐1140 | Cream, Illinois |
| Metschnikowia bicuspidata | Mb | Y‐17917 | Clothes, New Zealand |
| Nadsonia fulvescens | Nf | Y‐12791 | Unknown |
| Naumovozyma castellii | Nc | Y‐12630 | Soil, Finland |
| Pichia deserticola | Pd | Y‐12921 | Cactus, Haiti |
| P. scutulata | Ps | Y‐7663 | Tree, Hawaii |
| Rhodotorula kratochvilovae | Rk | Y‐1621 | Air, Japan |
| Saccharomyces cerevisiae | Sc | BY4741 | Lab strain, singer |
| S. cerevisiae | Sc | ESM356‐1 | Lab strain, singer |
| S. cerevisiae | Sc | Y‐11875 | Brewery, UK |
| S. cerevisiae | Sc | Y‐12632 | Brewery, UK |
| S. cerevisiae | Sc | Y‐2432 | Brewery, UK |
| S. kudriavzevii | Sk | Y‐27339 | Unknown |
| S. pastorianus | Sp | Y‐12693 | Brewery, Denmark |
| Starmera caribaea | Sc | Y‐17468 | Pear, Bahamas |
| Tetrapisispora blattae | Tb | Y‐10934 | Cockroach, Germany |
| Torulaspora microellipsoides | Tm | Y‐17060 | Tree, Hawaii |
2.2. Culture conditions
YPD broth (Foremedium), YPD agar (Foremedium), 12°P wort (Spraymalt, Brewferm,), malt (malt extract 100 g/L, Sigma Aldrich, Germany) [24], synthetic apple juice (102.3 g/L sucrose, 63.5 g/L glucose, 186 g/L fructose, 5/L g peptone, 5 g/L yeast extract, Sigma Aldrich, Germany) [25] and natural pasteurised apple juice (Torre Cider Farm) were used as the culture media. For generating the solidified media, 2% agar was added prior to autoclaving, with the exception of the apple juice in which the agar was added after pre‐heating it at 80°C in sterile conditions.
2.3. Phenotypic screening in solidified industrial conditions
An initial screen was carried out in the YPD agar plates at 30°C with a density of 96 colonies per plate. The strains were revived from −80°C in freshly prepared YPD broth +2% glucose (Foremedium) at 30°C with agitation of 120 rpms during 48 h. Then, the strains were grown in petri dishes containing YPD agar at 30°C until individual colonies were observed.
The PIXL colony picker robot (Singer Instruments) was used to re‐array the strains in a 96 array format using SBS PlusPlates (Singer Instruments) containing YPD agar. The first row of the plates was filled with the lab controls (6 replicas of each) and the remaining strains were arrayed in quadruplicate. The plates were sealed and incubated for 48 h at 30°C. Then, the ROTOR HDA (Singer Instruments) was used to replicate the previously generated arrays onto YPD agar, and the plates were sealed and incubated for 24 h at 30°C. These were used as source plates for the generation of the screen. This was done to ensure a homogeneous transfer of cellular material across all the positions.
The generated YPD source arrays were replicated in YPD agar using the ROTOR HDA (Singer Instruments). The plates were sealed and incubated for 120 h at 30°C. PhenoBooth (Singer Instruments) was used to image each plate every ≈ 6 h. The PhenoBooth web app was used to subtract the background and export a colony radius value measured in pixels for all strains and timepoints. The colony size values were then normalised, taking into account the plate, row, and column means.
The experiment was repeated using 384 and 1536 arrays. As described previously, PIXL (Singer Instruments) was used to re‐array the strains onto YPD agar. For the 384 format, four 384 arrays using a randomly distributed pattern were used. The same procedure was followed for the 1536 format, in this case, the plate edges (rows A and AF, and columns 1 and 48) were filled with the S. cerevisiae ESM356‐1 lab strain. This was done to reduce the colony–neighbour effect as much as possible. The ROTOR HDA (Singer Instruments) was used to generate and replicate the source arrays onto solidified, industrially relevant media in triplicate. These were wort, malt extract (ME), apple juice (AJ), and synthetic apple juice (SAJ) with the YPD medium included as a control. The plates were sealed and incubated for 120 h for the 384 arrays and 96 h for the 1536 arrays. Images were captured and analysed with the PhenoBooth (Singer Instruments) every ≈ 6 h as described above. To mimic ale brewing conditions, the experiment was performed at both 20 and 25°C. 16 and 59 in‐plate replicas per strain were used for the 384 and the 1536 screenings, respectively. Furthermore, all experiments were carried out in triplicate.
An average colony size for each time point and condition was calculated using the ggplot2 R package [26]. Statistical significance in between colony sizes was assessed via the two‐way ANOVA analysis in GraphPad Prism 8 (GraphPad Software). A growth value measured in pixels/h was calculated for each time frame (0–6 h, 6–12 h, 12–24 h, 24–48 h, 48–72 h, 72–96 h, 96–120 h), strain and condition. The maximum growth rate (MGR) and final biomass values were normalised for each condition and represented in a heatmap generated with GraphPad Prism 8 (GraphPad Software).
To validate the solid media screening data, growth curves were performed for all the strains in YPD broth at 30°C. Overnight YPD broth cultures were used to inoculate a 96‐MWP containing YPD broth at an initial OD600 of 0.1; this was performed in triplicate. A SPECTROstar Nano plate reader (BMG) was used to incubate, shake and monitor the OD600 absorbance at 30°C every 30 min for 55.5 h. Growth curves were plotted using GraphPad Prism 8 (GraphPad Software).
3. RESULTS
The fitness of different conventional and non‐conventional yeast strains was measured using the colony size over time in different solidified media representing industrial conditions. Three different array formats were tested—96, 384, and 1536 colonies per plate.
3.1. Feasibility of the method
On the 96‐format array screening, the colony size was significatively different (Dunnett's multiple comparisons test adjusted p‐value <0.0001) from the control strain (Sc ESM356‐1) in 17 of the strains after 96 h of growth. As in 96 format, statistically significant differences with the control strain were observed in both 384 and 1536 screenings after 96 h of incubation. The number of strains that showed differential growth compared to the control strain was over 17, for 19 out of the 20 differing conditions. In the ME at 25°C, 15 strains showed differential growth, accounting for 68% of the total strains studied. Overall, statistically significant differential growth was observed in 89% of cases.
To validate the solid screening data, the growth behaviour in YPD broth at 30°C was also studied. Results were comparable in between solid and liquid media for the majority of strains (Figure S1): only Cm Y‐7248 and Hv Y‐17530 displayed marked differences in growth kinetics between solid and liquid experiments.
3.2. Growth behaviour of the strains
In the proof‐of‐concept screening carried out in YPD at 30°C using 96 arrays, the strains that showed the highest MGR were Pd Y‐12921, Tm Y‐17060, Kl Y‐1140, Nf Y‐12791, and Sca Y‐17468 (Table 2). The strain with the lowest MGR was Mb Y‐17917 (Table 2). The fitness in terms of maximum biomass (Table S1) was comparable to the MGR in all cases with the following exceptions: on the 384‐array screening, the relative maximum biomass was lower than the relative MGR in Sc ESM356‐1 and Sc BY4741 across all the conditions (except wort at 20°C, in which the maximum biomass was higher) and the same trend was observed in Sk Y‐27339 and Sca Y‐17468 in wort at 20°C and 25°C, respectively; furthermore, on the 1536‐array screening, Mb Y‐17917 showed a lower relative maximum biomass compared with the relative MGR as well; in this case only in the ME at 25°C.
TABLE 2.
Maximum growth rate (pixels/h) for all the studied strains in the different formats and conditions
| 96 YPD 30°C | 384 YPD 20°C | 384 YPD 25°C | 384 ME 20°C | 384 ME 25°C | 384 wort 20°C | 384 wort 25°C | 384 SAJ 20°C | 384 SAJ 25°C | 384 AJ 20°C | 384 AJ 25°C | 1536 YPD 20°C | 1536 YPD 25°C | 1536 ME 20°C | 1536 ME 25°C | 1536 wort 20°C | 1536 wort 25°C | 1536 SAJ 20°C | 1536 SAJ 25°C | 1536 AJ 20°C | 1536 AJ 25°C | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sc ESM356‐1 | 9.39 | 5.98 | 5.87 | 4.16 | 7.32 | 4.25 | 5.42 | 2.12 | 3.06 | 0.41 | 0.60 | 1.47 | 2.00 | 1.32 | 1.90 | 2.52 | 2.76 | 1.23 | 1.47 | 0.33 | 0.48 |
| Sc BY4741 | 8.86 | 6.07 | 6.23 | 4.95 | 7.32 | 4.20 | 6.39 | 2.21 | 3.18 | 0.94 | 0.70 | 1.28 | 1.87 | 0.83 | 1.39 | 1.15 | 1.98 | 1.12 | 1.28 | 0.35 | 0.41 |
| Ps Y‐7663 | 7.59 | 6.01 | 6.31 | 5.86 | 7.88 | 7.23 | 6.51 | 2.30 | 1.26 | 3.67 | 5.53 | 0.29 | 2.02 | 2.39 | 1.76 | 4.02 | 3.37 | 0.25 | 0.30 | 1.96 | 2.07 |
| Rk Y‐1621 | 4.81 | 2.52 | 2.41 | 2.38 | 3.85 | 6.79 | 2.22 | 0.25 | 0.24 | 1.11 | 1.32 | 0.19 | 1.92 | 0.27 | 0.23 | 0.37 | 2.47 | 0.11 | 0.19 | 0.34 | 0.31 |
| Sk Y‐27339 | 3.72 | 1.74 | 2.88 | 1.29 | 2.61 | 9.78 | 2.64 | 1.08 | 1.80 | 1.90 | 4.35 | 0.49 | 1.89 | 0.57 | 0.33 | 0.74 | 1.82 | 0.64 | 0.49 | 0.77 | 0.75 |
| Sca Y‐17468 | 13.51 | 4.93 | 6.46 | 4.22 | 6.43 | 6.27 | 7.30 | 1.57 | 0.36 | 3.58 | 3.54 | 0.27 | 2.46 | 1.27 | 1.36 | 2.21 | 1.87 | 0.20 | 0.27 | 1.22 | 1.39 |
| Tm Y‐17060 | 14.88 | 5.48 | 5.44 | 3.76 | 6.31 | 3.45 | 5.14 | 1.37 | 0.21 | 2.17 | 3.91 | 0.28 | 2.23 | 1.19 | 1.50 | 1.45 | 1.73 | 0.17 | 0.28 | 0.89 | 1.24 |
| Sp Y‐12693 | 4.08 | 1.90 | 3.89 | 0.87 | 2.43 | 11.61 | 0.71 | 0.77 | 1.19 | 0.17 | 0.25 | 0.91 | 1.95 | 2.04 | 1.85 | 3.53 | 3.36 | 0.95 | 0.91 | 2.20 | 2.17 |
| Sc Y‐12632 | 8.63 | 5.05 | 5.21 | 4.53 | 6.34 | 3.95 | 6.26 | 3.33 | 4.01 | 4.80 | 7.77 | 1.83 | 1.82 | 1.77 | 1.58 | 2.78 | 2.58 | 1.65 | 1.83 | 1.77 | 1.87 |
| Sc Y‐11875 | 7.71 | 4.90 | 5.48 | 4.66 | 6.03 | 5.58 | 5.98 | 3.21 | 4.04 | 4.60 | 7.44 | 0.92 | 1.51 | 0.87 | 0.81 | 1.21 | 1.56 | 0.68 | 0.92 | 0.80 | 0.92 |
| Sc Y‐2432 | 10.43 | 4.49 | 5.10 | 4.48 | 7.60 | 8.19 | 6.36 | 2.85 | 3.26 | 3.57 | 5.79 | 2.10 | 2.20 | 2.02 | 2.06 | 3.32 | 3.26 | 1.88 | 2.10 | 1.91 | 2.15 |
| Cm Y‐7245 | 11.14 | 10.14 | 9.30 | 6.09 | 8.64 | 8.43 | 8.09 | 2.93 | 3.50 | 4.12 | 6.77 | 1.08 | 3.24 | 1.59 | 2.16 | 2.86 | 3.29 | 1.05 | 1.08 | 1.77 | 2.23 |
| Cm Y‐7248 | 9.95 | 7.41 | 9.90 | 5.75 | 7.37 | 7.02 | 7.87 | 2.92 | 2.99 | 3.52 | 6.74 | 1.77 | 2.42 | 1.51 | 2.02 | 2.99 | 3.22 | 1.52 | 1.77 | 1.94 | 2.82 |
| Hv Y‐17530 | 6.76 | 4.80 | 6.44 | 4.36 | 6.42 | 6.13 | 6.19 | 2.16 | 2.54 | 2.76 | 5.10 | 0.96 | 1.54 | 0.81 | 0.71 | 1.24 | 1.49 | 0.65 | 0.96 | 1.99 | 2.39 |
| Ky Y‐48837 | 10.28 | 6.88 | 8.48 | 5.94 | 7.55 | 4.45 | 7.84 | 2.82 | 1.48 | 6.70 | 10.43 | 0.33 | 2.68 | 1.61 | 2.10 | 3.42 | 3.78 | 0.39 | 0.33 | 2.10 | 2.98 |
| Tb Y‐10934 | 6.39 | 2.52 | 2.93 | 1.89 | 2.88 | 7.15 | 3.08 | 0.94 | 0.52 | 0.67 | 1.03 | 0.28 | 1.58 | 0.56 | 0.50 | 1.10 | 1.27 | 0.15 | 0.28 | 0.31 | 0.40 |
| Kl Y‐1140 | 14.69 | 6.82 | 9.83 | 4.87 | 7.53 | 5.48 | 6.99 | 2.52 | 3.70 | 2.45 | 2.87 | 1.11 | 4.23 | 1.94 | 1.97 | 2.70 | 2.49 | 1.38 | 1.11 | 1.38 | 1.39 |
| Cf Y‐6710 | 11.06 | 4.49 | 6.46 | 4.42 | 6.60 | 3.72 | 6.73 | 2.17 | 2.39 | 2.89 | 4.89 | 0.44 | 2.76 | 1.82 | 1.83 | 2.54 | 2.33 | 0.49 | 0.44 | 1.43 | 1.83 |
| Cm Y‐17236 | 12.65 | 5.30 | 5.64 | 4.04 | 6.45 | 4.86 | 4.86 | 1.41 | 0.47 | 2.21 | 4.07 | 0.28 | 2.59 | 1.28 | 1.54 | 1.55 | 1.65 | 0.19 | 0.28 | 0.93 | 1.26 |
| Mb Y‐17917 | 3.65 | 6.15 | 4.14 | 1.64 | 2.96 | 7.67 | 3.17 | 1.04 | 1.18 | 1.58 | 1.38 | 0.28 | 2.27 | 0.18 | 2.10 | 0.15 | 2.86 | 0.00 | 0.34 | 0.43 | 0.37 |
| Nf Y‐12791 | 14.33 | 5.25 | 6.00 | 3.94 | 6.05 | 5.23 | 4.88 | 1.33 | 0.19 | 2.26 | 3.99 | 0.25 | 2.14 | 1.23 | 1.55 | 1.55 | 1.88 | 0.21 | 0.25 | 0.90 | 1.28 |
| Nc Y‐12630 | 11.04 | 5.61 | 7.17 | 4.65 | 6.58 | 5.57 | 5.71 | 2.94 | 3.22 | 3.58 | 5.62 | 0.68 | 2.52 | 1.31 | 1.38 | 3.04 | 2.73 | 0.60 | 0.68 | 1.44 | 1.53 |
| Pd Y‐12921 | 19.58 | 4.64 | 10.17 | 4.84 | 10.21 | 4.92 | 7.59 | 1.37 | 0.14 | 2.57 | 7.58 | 0.20 | 3.60 | 1.41 | 1.23 | 3.15 | 3.23 | 0.19 | 0.20 | 1.54 | 2.21 |
When introducing more nutrient and temperature variables on the 384 screening, the effect of these on the growth behaviour was evident. For example, in the 384‐array screening at 25°C, the growth of the strains Pd Y‐12921 and Ky Y‐48837 was greatly influenced by the nutrient conditions. These strains showed a high MGR in YPD, the ME and wort at 25°C but a very low MGR in SAJ. Another good example of the influence of the nutrient conditions can be observed in Ps Y‐7663, which showed high MGR values across all media and temperatures on the 1536 screening except in SAJ and YPD at 20°C (Table 2, Figure 2). A similar scenario can be observed in the lab strains Sc ESM356‐1 and Sc BY4741, which grew relatively well in all the conditions except AJ. Ky Y‐48837 was the best performing strain in AJ in terms of both MGR and maximum biomass.
FIGURE 2.
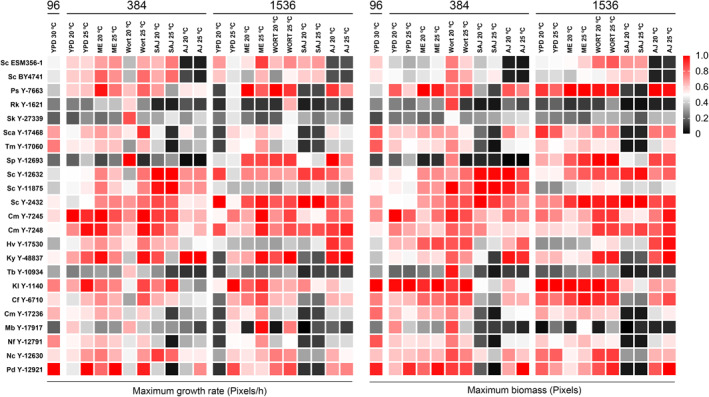
Heatmap representing the relative maximum growth rate (MGR) and the relative maximum biomass for each of the studied strains. Black represents the lowest values and red the highest. For raw pixels/h data and maximum biomass see Tables 2 and S1 respectively
On the other hand, strains such as Sc Y‐11875 and Sc Y‐12632 did show a very similar growth behaviour across all the nutrient conditions, as well as the C. milleri strains (Cm Y‐7245, Cm Y‐7248), which consistently displayed good growth kinetics. Contrarily, Tb Y‐10934 showed defective growth in all conditions except in 20°C wort on the 384‐array screening.
As expected, the temperature had an effect on fitness as well. It did play an especially significant role in the growth of Sp Y‐12693 on the 384 screening, and namely, in wort, at 20°C; its MGR was 11.61 pixels/h, the highest of all the studied strains in that condition. On the contrary, its MGR was 0.71 pixels/h at 25°C, the lowest one of all the strains tested. Interestingly, this behaviour was not observed in the 1536 array.
In the 384 and 1536 format screening, the MGR was also overall comparable with the final biomass (Figure 2), with only a few exceptions. The most notable ones were Pd Y‐1292 and Sca Y‐17468 at YPD 20°C in the 1536 density array, which showed a low MGR value but were able to reach a considerable colony size at the endpoints of the screening.
The results obtained with the 1536 screening were overall comparable with those obtained from the less dense formats (Figure 2), with some interesting exceptions such as Sp Y‐12663 and Sc‐11875.
4. DISCUSSION
In this study, a new approach for assessing the fitness of Saccharomyces and non‐Saccharomyces yeast strains employing high‐throughput robotic platforms was investigated. The method can be summarised in 3 steps: (1) re‐arraying a yeast library in a solid medium in the desired array density (96, 384 or 1536); (2) replica‐plating the array onto the industrial conditions of choice; (3) measuring the colony size increment over time for each of the strains.
As a proof of concept, we created a library of 23 yeast strains belonging to different genera and studied their growth rates in solidified media representing beer and cider brewing conditions. The feasibility of the method was assessed using a 96‐array format and standard lab conditions (YPD at 30°C). Since significant differences were observed in between the growth of the studied strains, the experiment was expanded to higher array densities and media mimicking industrial conditions.
The employment of high array densities permitted the use of a vast number of in‐plate replicas per strain (i.e., 59 in the 1536 array screening), generating a robust dataset. Importantly, we were able to detect statistically significant differences between the colony sizes of the various strains, in spite of the spread of the standard deviation observed in their replica in some cases (Table S1). This spread was mainly caused by the neighbour effect and the aberrant colony morphologies characteristic of some of the non‐Saccharomyces species. As expected, the overall colony size was reduced when the array density was increased. This was likely due to a higher neighbour effect brought about by the increased array density, limiting the amount of nutrients that each colony is able to access.
It is worth noting that we observed significant differences in the growth behaviour of Sp Y‐12693 (and Sc Y‐11875 and Hv Y‐17530 to a lesser extent) between the screenings performed employing 384 and 1536 arrays (Figure 2). This is intriguing and we cannot provide a clear explanation for why these strains are so affected by the array density. Given that the strains neighbouring Sp Y‐12693 in the 384 and 1536 arrays are different, it is possible that there was an effect due to the different competitive pressure between the two array types. One‐to‐one competition experiments with the relevant strains should be able to address this hypothesis in the future.
Interestingly, some strains were identified as vigorous growers only in a specific condition, whereas others displayed an overall good growth performance across all the conditions. For example, the growth of Sp Y‐12693 and Sk Y‐27339 was highly condition‐dependent and, on the other hand, the Cm and Sc brewing strains displayed a high MGR in virtually every condition (Figure 2). The growth kinetics of a strain in a specific condition can be influenced by numerous factors—nutrient uptake, pH and osmotic pressure resistance, temperature and ability to cope with toxic compounds, just to name a few. For example, as observed on the 384 screening, the growth of Tb Y‐10934 is clearly diminished in the SAJ and AJ media. This might be caused due to the inability to metabolise all the sugars present in the juice, together with a low resistance to the high osmotic pressure that takes place in this condition due to the very high concentrations of sugars as well as low pH. It is important to consider as well that adaptation for growing in a certain condition can be due to natural or artificial selection; for example, human selection for lager brewing. This is likely the case of S. pastorianus, which has been intentionally grown in wort and low‐temperature conditions for centuries [27]. Sp Y‐12693 showed a clear adaptation to low temperature in the 384 screening; however, that was not the only case. Other strains such as Sk Y‐27339 and Tb Y‐10934 also displayed a higher MGR at low temperature when using 384 arrays (Figure 2).
This highlights the importance of using adequate temperature when screening yeast for the brewing industry, since for some strains, it is a factor that greatly impacts fitness. Normally, fitness screenings for brewing industry purposes are performed at high temperature (25°C–30°C) [21, 28, 29, 30, 31]. However, lower temperatures are frequently used in wine [32] and beer production [33]. Limiting the fitness screenings to a single temperature condition constrains the discovery of new strains with brewing potential. It is understandable that increasing the number of conditions when handling vast libraries in liquid media can be troublesome. This is where this proposed method excelled; the use of solid media can facilitate the use of more temperature and nutrient conditions.
Adaptation to a specific growth condition was not generally detected in the non‐conventional species studied here. This could be due to a lack of artificially driven selection, as these species have never been used in an industrial manner. However, the S. cerevisiae ale strains, which have been used industrially in brewing for many years, also showed an overall good performance across all the conditions. This indicates that these strains were probably already very efficient in the uptake and metabolism of all the main sugars present in apple juice and wort. Wild fermentations are typically dominated by S. Cerevisiase, suggesting that naturally driven selection already provided S. Cerevisiase with the ability to ferment and live in high sugar content environments. Humans have taken advantage of this, enhancing it via domestication.
Additionally, fitness assessment of yeast isolates is normally performed in liquid medium [21, 28, 29, 30, 31, 32]. This is sensible for strains that will be used in the brewing industry; however, the use of liquid medium in the initial steps of the screening often involves a huge effort when dealing with vast libraries and a matrix of conditions: OD measurements need to be taken for each sample, with a limited factor of 384 samples per multi‐well plate. For example, in a study carried out by Wei and colleagues in 2019, they screened 236 yeast isolates in duplicate, using falcon tubes and 40 ml of apple juice per sample [7]. Our proposed method would have fit perfectly in the aforementioned Wei and colleagues' workflow, as a pre‐screening step. While fitness in solid and liquid conditions can vary in some cases, here we show that growth in solid media can be used as an accurate proxy for estimating the fitness in liquid media for the majority of the strains studied here (Figure S1). Additionally, being able to simultaneously screen 1536 samples per plate provides a clear high‐throughput advantage. Our proposed solid media screening approach would easily discard the less competent isolates, reducing the size of the library and the amount of human labour needed for the subsequent liquid small‐scale fermentation and downstream analysis experiments. Moreover, we believe that the workflow can be used with virtually any solid medium and organism that forms uniform colonies on agar, including bacteria.
5. CONCLUSIONS
The measurement of the colony size over time in solid media allows us to obtain growth kinetic values of potentially thousands of yeast strains in different industrially relevant conditions. We propose the use of the described method for the screening of vast yeast libraries formed by unknown isolates, with the aim of hijacking strains from nature for their use in the brewing industry.
The method could be used as a first filtering step when screening massive libraries. This would reduce time and effort on the subsequent fermentation performance and aroma profile analyses by reducing the number of yeast candidates that are worthy of studying.
CONFLICT OF INTEREST
The company in which Dr. Oliver Jack Severn is employed, Singer Instruments, profits from the sale of the robotics platforms used within this study. This has been mitigated against via our academic collaboration with the University of Manchester, we are professionally confident in the studies integrity. In addition, it is made clear from the institutional affiliations of Oliver Severn to any reader.
PERMISSION TO REPRODUCE MATERIALS FROM OTHER SOURCES
None.
Supporting information
Supplementary Material 1
ACKNOWLEDGEMENTS
JEA‐C is supported by the European Commission (grant H2020‐MSCA‐ITN‐2017; number 764364). The authors wish to thank the wider Aromagenesis consortium for discussions and academic stimulation and, the Torre Cider Co Ltd for contributing materials.
Aguiar‐Cervera, J.E. , Delneri, D. , Severn, O. : A high‐throughput screening method for the discovery of Saccharomyces and non‐Saccharomyces yeasts with potential in the brewing industry. Eng. Biol. 5(3), 72–80 (2021). 10.1049/enb2.12013
DATA AVAILABILITY STATEMENT
All data is available from the corresponding author upon reasonable request, and in due course will be made available in accordance to the Pilot on Open Research Data in Horizon 2020.
REFERENCES
- 1. Gallone, B. , et al.: Domestication and divergence of Saccharomyces cerevisiae beer yeasts. Cell [Internet]. 166(6), 1397–1410 (2016). 10.1016/j.cell.2016.08.020. e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rai, A.K. , Jeyaram, K. : Role of yeasts in food fermentation. In: Satyanarayana, T. , Kunze, G. (eds.) Yeast diversity in human welfare [Internet], pp. 83–113. Springer, Singapore: (2017). 10.1007/978-981-10-2621-8 [DOI] [Google Scholar]
- 3. Lodolo, E.J. , et al.: The yeast Saccharomyces cerevisiae– the main character in beer brewing. FEMS Yeast Res [Internet]. 8(7), 1018–1036 (2008). 10.1111/j.1567-1364.2008.00433.x [DOI] [PubMed] [Google Scholar]
- 4. Jolly, N.P. , Augustyn, O.P.H. , Pretorius, I.S. : The role and use of non‐Saccharomyces yeasts in wine production. South Afr. J. Enol. Vitic. 27(1), 15–39 (2006). 10.21548/27-1-1475 [DOI] [Google Scholar]
- 5. Basso, R.F. , Alcarde, A.R. , Portugal, C.B. : Could non‐Saccharomyces yeasts contribute on innovative brewing fermentations? Food. Res. Int. 86, 112–120 (2016). 10.1016/j.foodres.2016.06.002 [DOI] [Google Scholar]
- 6. Gschaedler, A. : Contribution of non‐conventional yeasts in alcoholic beverages. Curr. Opin. Food. Sci. 13, 73–77 (2017). 10.1016/j.cofs.2017.02.004 [DOI] [Google Scholar]
- 7. Wei, J. , et al.: Characterization and screening of non‐Saccharomyces yeasts used to produce fragrant cider. LWT (Lebensm‐Wiss & Technol), vol. 107, pp. 191–198. (2019). 10.1016/j.lwt.2019.03.028 [DOI] [Google Scholar]
- 8. Holt, S. , et al.: Bioflavoring by non‐conventional yeasts in sequential beer fermentations. Food. Microbiol. 72, 55–66 (2018). 10.1016/j.fm.2017.11.008 [DOI] [PubMed] [Google Scholar]
- 9. Sannino, C. , et al.: Non‐conventional yeasts for producing alternative beers. In: Sibirny, A. (ed.) Non‐conventional Yeasts: from Basic Research to Application [Internet], pp. 361–388. Springer International Publishing, Cham: (2019). 10.1007/978-3-030-21110-311 [DOI] [Google Scholar]
- 10. Gamero, A. , et al.: Development of a low‐alcoholic fermented beverage employing cashew apple juice and non‐conventional yeasts. Fermentation. 5(3), 71 (2019). 10.3390/fermentation5030071 [DOI] [Google Scholar]
- 11. Bellut, K. , et al.: Screening and application of cyberlindnera yeasts to produce a fruity, non‐alcoholic beer. Fermentation, vol. 5(4), pp. 103. (2019). 10.3390/fermentation5040103 [DOI] [Google Scholar]
- 12. De Francesco, G. , et al.: Mrakia gelida in brewing process: an innovative production of low alcohol beer using a psychrophilic yeast strain. Food. Microbiol. 76, 354–362 (2018). 10.1016/j.fm.2018.06.018 [DOI] [PubMed] [Google Scholar]
- 13. Cabras, I. , Bamforth, C. : From reviving tradition to fostering innovation and changing marketing: the evolution of micro‐brewing in the UK and US, 1980‐2012. Bus. Hist. vol. 58(5). pp. 625–646. (2016). 10.1080/00076791.2015.1027692 [DOI] [Google Scholar]
- 14. Giannakou, K. , et al.: Biotechnological exploitation of Saccharomyces jurei and its hybrids in craft beer fermentation uncovers new aroma combinations. Food. Microbiol. 100, 103838 (2021). 10.1016/j.fm.2021.103838 [DOI] [PubMed] [Google Scholar]
- 15. Van Ooyen, A.J.J. , et al.: Heterologous protein production in the yeast Kluyveromyces lactis . FEMS Yeast. Res. 6(3), 381–392 (2006). 10.1111/j.1567-1364.2006.00049.x [DOI] [PubMed] [Google Scholar]
- 16. Blazeck, J. , et al.: Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat. Commun. 5(1), 3131 (2014). 10.1038/ncomms4131 [DOI] [PubMed] [Google Scholar]
- 17. Macarron, R. , et al.: Impact of high‐throughput screening in biomedical research. Nat. Rev. Drug. Discov. 10(3), 188–195 (2011). 10.1038/nrd3368 [DOI] [PubMed] [Google Scholar]
- 18. Giaever, G. , et al.: Functional profiling of the Saccharomyces cerevisiae genome. Nature. 418(6896), 387–391 (2002). 10.1038/nature00935 [DOI] [PubMed] [Google Scholar]
- 19. Baryshnikova, A. , et al.: Synthetic genetic array (SGA) analysis in Saccharomyces cerevisiae and Schizosaccharomyces pombe . In: Weissman, J. , Guthrie, C. , Fink, G. (eds.) Guide to Yeast Genetics: Functional Genomics, Proteomics, and Other Systems Analysis [Internet], (2nd ed). pp. 145–179. Academic Press, San Diego: (2010). https://www.sciencedirect.com/bookseries/methods‐in‐enzymology/vol/470/suppl/C [DOI] [PubMed] [Google Scholar]
- 20. Costanzo, M. , Boone, C. : SGAM: an array‐based approach for high‐resolution genetic mapping in Saccharomyces cerevisiae . Methods Mol. Biol. 548, 37–53 (2009). 10.1007/978-1-59745-540-43 [DOI] [PubMed] [Google Scholar]
- 21. Gamero, A. , et al.: High‐throughput screening of a large collection of non‐conventional yeasts reveals their potential for aroma formation in food fermentation. Food. Microbiol. 60, 147–159 (2016). 10.1016/j.fm.2016.07.006 [DOI] [PubMed] [Google Scholar]
- 22. Gutiérrez, A. , et al.: Evaluation of non‐Saccharomyces yeasts in the fermentation of wine, beer and cider for the development of new beverages. J. Inst. Brew. (2018). (June). http://doi.wiley.com/10.1002/jib.512 [Google Scholar]
- 23. Bean, G.J. , et al.: Development of ultra‐high‐density screening tools for microbial “omics”. PLoS One [Internet]. 9(1), e85177 (2014). 10.1371/journal.pone.0085177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Denby, C.M. , et al.: Industrial brewing yeast engineered for the production of primary flavor determinants in hopped beer. Nat Commun [Internet]. 9(1), 965 (2018). 10.1038/s41467-018-03293-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pando Bedriñana, R. , Mangas Alonso, J.J. , Suárez Valles, B. : Evaluation of autochthonous Saccharomyces bayanus strains under stress conditions for making ice ciders. LWT ‐ Food Sci Technol [Internet]. 81, 217–225 (2017). 10.1016/j.lwt.2017.03.055 [DOI] [Google Scholar]
- 26. Wickham, H. In: Gentleman, R. , Hornik, K. , Parmigiani, G. (Eds.) ggplot2: Elegant Graphics for Data Analysis, (2nd ed) pp. 182. Springer Nature; (2016) [Google Scholar]
- 27. Dunn, B. , Sherlock, G. : Reconstruction of the genome origins and evolution of the hybrid lager yeast Saccharomyces pastorianus . Genome. Res. 18(10), 1610–1623 (2008). 10.1101/gr.076075.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tian, T. , et al.: A multiple‐step strategy for screening Saccharomyces cerevisiae strains with improved acid tolerance and aroma profiles. Appl. Microbiol. Biotechnol. 104(7), 3097–3107 (2020). 10.1007/s00253-020-10451-z [DOI] [PubMed] [Google Scholar]
- 29. Simon Amoikon, T.L. , et al.: A study on the potential of yeasts isolated from palm wines to produce flavouring compounds, LWT. vol. 128, pp. 109506. (2020). 10.1016/j.lwt.2020.109506 [DOI] [Google Scholar]
- 30. De Francesco, G. , et al.: Screening of new strains of Saccharomycodes ludwigii and Zygosaccharomyces rouxii to produce low‐alcohol beer. J. Inst. Brew. 121(1), 113–121 (2015). 10.1002/jib [DOI] [Google Scholar]
- 31. Colomer, M.S. , et al.: Assessing population diversity of Brettanomyces yeast species and identification of strains for brewing applications. Front. Microbiol. 11, 637 (2020). 10.3389/fmicb.2020.00637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. García‐Ríos, E. , et al.: The genetic architecture of low‐temperature adaptation in the wine yeast Saccharomyces cerevisiae. BMC. Genomics. 18(1), 159 (2017). 10.1186/s12864-017-3572-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lasanta, C. , et al.: Influence of fermentation temperature and yeast type on the chemical and sensory profile of handcrafted beers. J. Sci. Food. Agric. 101(3), 1174–1181 (2021). 10.1002/jsfa.10729 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1
Data Availability Statement
All data is available from the corresponding author upon reasonable request, and in due course will be made available in accordance to the Pilot on Open Research Data in Horizon 2020.


