Abstract
Using hydrogen oxidising bacteria to produce protein and other food and feed ingredients is a form of industrial biotechnology that is gaining traction. The technology fixes carbon dioxide into products without the light requirements of agriculture and biotech that rely on primary producers such as plants and algae while promising higher growth rates, drastically less land, fresh water, and mineral requirements. The significant body of scientific knowledge on hydrogen oxidising bacteria continues to grow and genetic engineering tools are well developed for specific species. The scale‐up success of other types of gas‐ fermentation using carbon monoxide or methane has paved the way for scale‐up of a process that uses a mix of hydrogen, oxygen, and carbon dioxide to produce bacteria as a food and feed ingredients in a highly sustainable fashion.
Inspec keywords: biotechnology, fermentation, proteins, agriculture, genetic engineering, microorganisms, carbon compounds
Other keywords: feed ingredients, hydrogen oxidising bacteria, single‐cell protein, carbon dioxide, primary producers, food ingredients, industrial biotechnology, genetic engineering tools
1. Introduction
Bacteria have been part of the human diet for millennia as components of many traditional foods, such as yoghurt, cheese, fermented vegetables, and fermented fish [1, 2]. In the 1960s, initial steps were taken to directly harness bacterial biomass as a source of protein, fats, and vitamins, for which the term single‐cell protein (SCP) was subsequently coined [3, 4].
Today multiple factors coincide to drive market interest in SCP, most importantly the increased market demand for protein, and sustainability issues surrounding current protein production. Here, the authors will focus on one of the most sustainable forms of SCP: SCP produced with autotrophic bacteria that fix carbon dioxide (CO2) into their cellular biomass (Fig. 1).
Fig. 1.
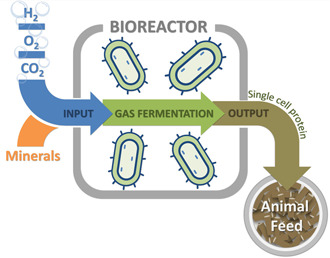
General process overview for SCP production using HOB and gaseous substrates
The United Nations has forecasted the world population to grow from 7.7 billion today to 9.7 billion by 2050 [5]. Inevitably, this has prompted concerns for future global food security, particularly as 2 billion individuals already experienced some level of food insecurity in 2019 [6]. The supply of animal‐derived protein is expected to double by 2050 to meet the persistent global consumption demand for high‐quality protein. Correspondingly, protein feed sources for livestock and aquaculture must also increase, despite land resources already being stretched [7, 8].
Agriculture currently utilises ∼43% of the land that is not a desert or covered by ice. This percentage will rise if agriculture is to meet the growing protein demand, which in turn will negatively affect biodiversity. Production of the type of SCP we focus on here requires much less land than agricultural protein (Fig. 2). To fix CO2 energy is required. The basis of our current food supply is formed by plants that use light as their energy source. The energy requirement for the most sustainably produced SCP, as discussed in this review, uses hydrogen (H2). This H2 can be produced by electrolysis using sustainably produced electricity. If the required electricity is based on solar power it requires 0.18–0.26 m2/kg of protein/year, which is considerably less than the 6–16 m2 of land/kg of protein per year needed for soybean. If wind power is used land‐use decreases to around 0.04 m2/kg of protein/year [9].
Fig. 2.
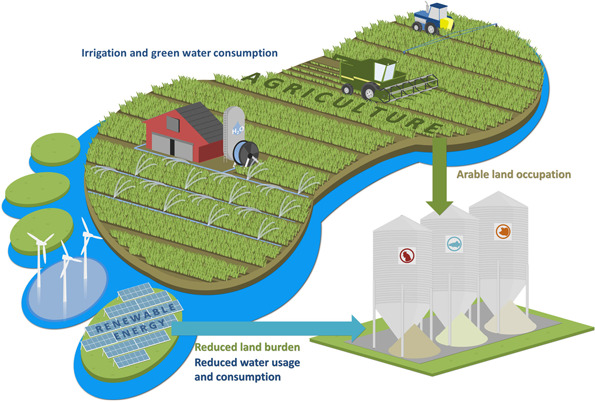
Illustrating the advantage of protein production using HOB: reduced land and freshwater use
2. Micro‐organisms
The metabolic diversity of bacteria allows SCP to be produced from a variety of substrates. Most biotechnology and SCP production are based on heterotrophic bacteria grown on substrates such as cheap carbohydrates, lipids or methane and methanol, which inherently release CO2. In light of climate change, ideally, our food and animal feed would be produced by primary producers capable of fixing CO2. The CO2 fixing abilities of autotrophic bacteria make them most suitable for truly sustainable SCP production [10].
The most attractive energy sources to enable CO2 fixation into SCP are light for photosynthetic cyanobacteria or H2 for certain chemolithoautotrophic bacteria. The cultivation of photosynthetic bacteria is limited by their light requirement. While natural light has the advantage of being a free energy source, its capture requires a large surface area, furthermore, it is only available half of the day and has a large seasonal variation in many highly populated parts of the world. Using artificial light is a possibility but this increases costs significantly.
As the main substrates for H2‐based autotrophy are gases, the technology is generally described as gas fermentation. Broadly speaking the two types of autotrophic gas fermentation are anaerobic and aerobic gas fermentation. Anaerobic metabolism, fed on H2 and CO2, is only feasible if 70–80% of the carbon ends up in reduced products such as acetate and roughly 20% as SCP. From the perspective of protein production, this is inefficient unless the produced acetate is used to feed an aerobic second stage process where acetate is used as a carbon source. This has recently been demonstrated on lab‐scale [11].
Aerobic H2‐based autotrophy has the capability to produce cell biomass without any significant by‐product. This is due to the energy released by the reaction of H2 with oxygen (O2). Indeed, a mixture of O2 and H2 under optimal conditions can easily react to producing an ear‐piercing bang or ‘Knall’ in German, where the gas mixture is known as Knallgas. Aerobic H2 autotrophs are therefore known as Knallgas bacteria or the H2 oxidising bacteria (HOB).
The generalised reaction of aerobic CO2 fixation via H2 oxidation can be given as:
If one assumes an overall process where H2 and O2 are produced via electrolysis the overall process is analogous to oxygenic photosynthesis:
Indeed, most of the industrially relevant HOB use the same metabolic pathway as plants to assimilate the CO2 into biomass although alternative routes are utilised by certain HOB, such as those of the Aquificae phylum [12].
HOB are found in many different bacterial phyla. Once a lab is established to feed mixtures of H2 and O2 to bacteria, HOB are fairly easy to isolate. They have been detected in many different environments, such as Antarctic subglacial lakes, temperate soils, and hot hydrothermal vents [13, 14, 15, 16, 17, 18].
Notwithstanding the wide range of HOB available only a handful are used and studied in detail. Interestingly many of these more prominent HOB have been mostly studied for reasons other than either their H2 oxidising capabilities or their quality as a food or feed source.
Examples of HOB with industrial potential that have received significant research attention are Rhodococcus opacus and Xantobacter autotrophicus.
R. opacus is an attractive HOB for lipid production for nutrition, biofuel, and bio‐commodity chemicals [19].
X. autotrophicus produces considerable amounts of zeaxanthin, a common caroteinoid used as a food dye.
X. autotrophicus's most prominent proposed biotechnological use is its capability to degrade chlorinated hydrocarbons [20].
By far the most studied HOB is Cupriavidus necator, which we will review in more detail as an example of the possibilities the HOB offer biotechnology.
Multiple studies have been performed on C. necator as a SCP source, but it is more widely known for its production of polyhydroxyalkanoate (PHA) bioplastics.
CO2 fixation in C. necator occurs via the Rubisco enzyme as a part of the Calvin‐cycle. The reducing power and energy needed to reduce CO2 and form biomass come from the oxidation of H2 via [NiFe]‐dependent hydrogenases. The nitrogen needed for biomass formation can be supplied in the form of ammonia, urea, or even as nitrogen gas.
Besides the capability of C. necator strains to grow on CO2 as the sole carbon‐source they can also grow on a large number of organic compounds found in soil, e.g. succinate, fumarate, and malate, while sugar metabolism is often restricted to fructose.
During growth under certain nutrient limitations, the cells accumulate PHA, which can constitute up to 90% of the cells. However, under controlled conditions, the PHA content can be minimised and 75% of the dry matter is protein and the biomass can be used for SCP. It is because of this capability to produce a high protein ratio that the strain was proposed as a protein generator for space stations in the 1960s [21]
The cells of C. necator H16 contain two chromosomes and one smaller megaplasmid with a total of 6543 genes [22]. A large number of encoded proteins reflect a diverse and robust metabolism needed to thrive in a complex environment. The key proteins needed for DNA and protein synthesis are located on chromosome I, while chromosome II encodes for many of the enzymes needed for utilising diverse substrates. Two highly similar gene clusters for the Calvin cycle pathway are present, one on chromosome II while the other is present on the megaplasmid. The NiFe‐dependent hydrogenase used for H2 oxidation is located on the megaplasmid.
The genomic data has guided the creation of a whole‐genome‐scale metabolic model, which contains 1391 reactions [23]. This model was used to design metabolic engineering strategies to improve the PHA producing potential of the strain and to devise strategies for the production of other chemicals.
The nutritional quality of many HOB is naturally already higher than many plant products however further improvements are desirable. Three routes to improvements in both product quality and process efficiency are process optimisation, directed evolution, and genome editing. The potential of the latter has improved significantly in the recent decade due to the increased knowledge of the genome and metabolism under various growth conditions.
Early studies of C. necator made use of Tn5 mutagenesis to develop single gene knock out mutants. This method was used to study the polyhydroxybutyrate (PHB) biosynthetic pathway, the soluble hydrogenase, post‐translational modifications to PHA synthase, and other metabolic pathways [24, 25, 26, 27]. The availability of suicide vectors employing the sacB counter‐selection mechanism allows targeted gene deletion and genomic integration via homologous recombination. This method has been exploited to study PHB biosynthesis, 3‐hydroxypropionate metabolism, and investigate the production of protein and cyanophycin [28, 29, 30, 31, 32].
Plasmid‐based expression in C. necator has largely relied on the broad‐host‐range plasmid pBBR1, which was used to introduce pathways for the production of ethyl ketones, fatty acids, isopropanol, and non‐native PHA [33, 34, 35, 36]. Efforts to further develop the genetic tools have included increasing the number of vectors suitable for use in C. necator [37], identifying inducible and constitutive promoters [38, 39], and increasing the stability of those vectors [40, 41]. The latter is important for maximising product formation, as plasmid‐based pathways offer a higher gene dosage than those integrated into the genome and plasmid instability is particularly problematic in long‐running fermentations with higher selection pressure on the cells. Plasmid addiction systems rely on the deletion of an essential gene from the chromosome, and expression of that gene on a plasmid which contains the desired product synthesis pathway. This method of improving plasmid stability has been adopted for the production of cyanophycin, PHA, and arginine [32, 42, 43].
Recent studies have shown the capability of C. necator to produce compounds from CO2 other than the well‐characterised PHB such as methyl ketones, terpenes, carotenoids, acetoin, and isopropanol [33, 44, 45, 46]. Furthermore, PHB has been produced using both syngas and real CO2‐rich industrial off‐gases emphasising the capability of C. necator to thrive and produce compounds in the presence of impure CO2 sources [47, 48].
The most obvious targets of genetic engineering of strains for SCP use are higher protein content or improved amino acid profiles, but the value can be added to the strains via their production of specific functional proteins, vitamins, valuable fatty acids, carotenoids, terpenoids, flavours, and food dyes or non‐toxic valuable co‐products. This does require acceptance of the use of genetically modified organisms in addition to the acceptance of SCP. In the last few years, there are trends of growing public and legislative support provided the product is more sustainable. This is illustrated by the popularity of the impossible burger that contains haeme produced by genetically modified organism (GMO) microbes [49]. Public discourse seems to be positive about the idea of incorporating the sustainable versions of biotechnology into our food supply chain [50].
3. Commercialisation of HOB
In recent years, several companies have undertaken the engineering challenge of feeding the highly energetic H2, O2, and CO2 mixture to the HOB to produce food and feed ingredients.
The California‐based company Kiverdi, founded in 2011, produces the SCP products Air Protein™ & CO2 Aquafeed as well as oils and bioplastics [51]. Also based in California is Novonutrients that tailor their SCP to supply aquaculture. Novonutrients’ initial feed trials demonstrated the superiority of their SCP compared to control diets that included soy and algae, with respect to fish growth rate [52]. Founded in 2017, the Finnish company Solar Foods produces Solein, their SCP for human food pending EU novel food license approval [53]. The Belgium company Avecom developed several SCPs from low‐value substrates. In their Power to Protein project, a collaboration with the Dutch KWR, HOB were used to produce SCP. This project experimented with using ammonium from waste‐water as a nitrogen source [54]. Deep Branch Biotechnology was founded in Nottingham, UK in 2018 by a team that has a background in both gas fermentation and synthetic biology. Their first SCP produced by HOB, Proton™ is a protein source with a tailored amino acid profile. In 2019, Deep Branch Biotechnology partnered with the Drax Group to pilot their SCP production technology using CO2 from Drax flue gas [55].
4. Other commercial gas fermentation
In this review, we have focused on gas fermentation using H2, O2, and CO2 as growth substrates for bacteria. Other types of gas fermentation have been successfully scaled from lab to industrial scale by a handful of companies. Since the engineering challenges are similar, the success of these companies has paved the way for companies utilising HOB.
After the 1990s discovery that some acetogenic bacteria can produce considerable amounts of ethanol when fed carbon monoxide [56], several companies sprung up to use that process, including Lanzatech which remains the most successful example [57]. Lanzatech overcame many technical challenges in gas fermentation. They focus on biofuel and chemical production, however, they also have a patent on using their biomass by‐product as SCP.
The German company Electrochaea uses hydrogenotrophic methanogens to produce methane from CO2 and H2. Their process is anaerobic and thus does not have flammability issues inside the reactor, however, the large H2 demands necessitates similar safety precautions as HOB‐based biotechnology. Their use of a pure archaeal strain in a biotechnological process is unique [58].
Two examples of companies that have scaled SCP production using gas fermentation are Calysta and Unibio [59, 60]. Both companies use a flammable gas mixture of methane and oxygen as a substrate. This type of gas‐fermentation technology has been developing ever since its initial conception in the 1960s. Although the methanotrophic process inherently releases CO2 while HOB‐based gas fermentation captures it, these commercialised technologies are similar enough to HOB fermentation to prove that scaling up gas fermentation processes for commercial SCP production is achievable.
5. Conclusion
Feeding H2, O2, and CO2 to HOB to produce SCP and added value food and feed ingredients is a promising form of biotechnology as is demonstrated by the range of start‐up companies involved. There are still process efficiency and scale‐up challenges to overcome but both industry and academia are highly motivated. Furthermore, the increased need for sustainability and recent success stories in adjacent technologies suggest that this powerful application of HOB technology will be realised in the short‐ to medium‐term.
6. Acknowledgments
We would like to thank Ying Zhang, the academic supervisor of Zahara Mortimer, and Katalin Kovacs who is the academic supervisor of Callum McGregor. We also extend our gratitude to Nigel Minton and the SBRC Nottingham for their advice and support. Deep Branch Biotechnology Ltd is supported by Innovate UK grant number 105259.
7 References
- 1. Campbell-Platt, G. : ‘Fermented foods — a world perspective’, Food Res. Int., 1994, 27, (3), pp. 253–257 [Google Scholar]
- 2. Boethius, A. : ‘Something rotten in Scandinavia: the world's earliest evidence of fermentation’, J. Archaeol. Sci., 2016, 66, pp. 169–180 [Google Scholar]
- 3. Matelbs, R.I. , Tannenbaum, S.E. : ‘Single-cell protein’, Econ. Bot., 1968, 22, (1), pp. 42–50 [Google Scholar]
- 4. Snyder, H.E. : ‘Microbial sources of protein’, Adv. Food Res., 1970, 18, pp. 85–140 [DOI] [PubMed] [Google Scholar]
- 5.United Nations, Department of Economic and Social Affairs, Population Division, ‘World Population Prospects 2019: Highlights (ST/ESA/SER.A/423)’, 2019.
- 6. Alexandratos, N. , Bruinsma, J. : ‘World agriculture towards 2030/2050’, 2012.
- 7. Boland, M.J. , Rae, A.N. , Vereijken, J.M. , et al.,: ‘The future supply of animal-derived protein for human consumption’, Trends Food Sci. Technol., 2013, 29, (1), pp. 62–73 [Google Scholar]
- 8. Henchion, M. , Hayes, M. , Mullen, A. , et al.,: ‘Future protein supply and demand: strategies and factors influencing a sustainable equilibrium’, Foods, 2017, 6, (7), p. 53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sillman, J. , Nygren, L. , Kahiluoto, H. , et al.,: ‘Bacterial protein for food and feed generated via renewable energy and direct air capture of CO2: can it reduce land and water use?’, Glob. Food Sec., 2019, 22, pp. 25–32 [Google Scholar]
- 10. Bogdahn, I. : ‘Agriculture-independent, sustainable, fail-safe and efficient food production by autotrophic single-cell protein’, 2015, PeerJ PrePrints 3:e1279v2 10.7287/peerj.preprints.1279v2 [DOI]
- 11. Molitor, B. , Mishra, A. , Angenent, L.T. : ‘Power-to-protein: converting renewable electric power and carbon dioxide into single cell protein with a two-stage bioprocess’, Energy Environ. Sci., 2019, 12, (12), pp. 3515–3521 [Google Scholar]
- 12. Berg, I.A. : ‘Ecological aspects of the distribution of different autotrophic CO2 fixation pathways’, Appl. Environ. Microbiol., 2011, 77, (6), pp. 1925–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Götz, D. , Banta, A. , Beveridge, T.J. , et al.,: ‘ Persephonella marina gen. nov., sp. nov. and Persephonella guaymasensis sp. nov., two novel, thermophilic, hydrogen-oxidizing microaerophiles from deep-sea hydrothermal vents’, Int. J. Syst. Evol. Microbiol., 2002, 52, (4), pp. 1349–1359 [DOI] [PubMed] [Google Scholar]
- 14. Ohi, K. , Komemushi, S. , Okazaki, M. , et al.,: ‘A new species of hydrogen-utilizing bacterium’, J. Gen. Appl. Microbiol., 1979, 25, (1), pp. 53–58 [Google Scholar]
- 15. Wilde, E. : ‘Untersuchungen über Wachstum und Speicherstoffsynthese von hydrogenomonas’, Arch. Mikrobiol., 1962, 43, (2), pp. 109–137 [PubMed] [Google Scholar]
- 16. Lavire, C.. , Normand, P. , Alekhina, I. , et al.,: ‘Presence of Hydrogenophilus thermoluteolus DNA in accretion ice in the subglacial lake vostok, Antarctica, assessed using rrs, cbb and hox’, Environ. Microbiol., 2006, 8, (12), pp. 2106–2114 [DOI] [PubMed] [Google Scholar]
- 17. Matassa, S. , Verstraete, W. , Pikaar, I. , et al.,: ‘Autotrophic nitrogen assimilation and carbon capture for microbial protein production by a novel enrichment of hydrogen-oxidizing bacteria’, Water Res.., 2016, 101, pp. 137–146 [DOI] [PubMed] [Google Scholar]
- 18. Dou, J. , Huang, Y. , Ren, H. , et al.,: ‘Autotrophic, heterotrophic, and mixotrophic nitrogen assimilation for single-cell protein production by two hydrogen-oxidizing bacterial strains’, Appl. Biochem. Biotechnol., 2019, 187, (1), pp. 338–351 [DOI] [PubMed] [Google Scholar]
- 19. Holder, J.W. , Ulrich, J.C. , DeBono, A.C. , et al.,: ‘Comparative and functional genomics of Rhodococcus opacus PD630 for biofuels development’, PLOS Genet., 2011, 7, (9), p. e1002219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Meusel, M. , Rehm, H.-J. : ‘Biodegradation of dichloroacetic acid by freely suspended and adsorptive immobilized Xanthobacter autotrophicus GJ10 in soil’, Appl. Microbiol. Biotechnol., 1993, 40, (1), pp. 165–171 [Google Scholar]
- 21. Foster, J.F. , Litchfield, J.H. : ‘A continuous culture apparatus for the microbial utilization of hydrogen produced by electrolysis of water in closed-cycle space systems’, Biotechnol. Bioeng., 1964, 6, (4), pp. 441–456 [Google Scholar]
- 22. Little, G.T. , Ehsaan, M. , Arenas-López, C. , et al.,: ‘Complete genome sequence of Cupriavidus necator H16 (DSM 428)’, Microbiol. Resour. Announc., 2019, 8, p. 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park, J. , Kim, T. , Lee, S. : ‘Genome-scale reconstruction and in silico analysis of the Ralstonia eutropha H16 for polyhydroxyalkanoate synthesis, lithoautotrophic growth, and 2-methyl citric acid production’, BMC Syst. Biol., 2011, 5, (1), p. 101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schubert, P. , Steinbüchel, A. , Schlegel, H.G. : ‘Cloning of the Alcaligenes eutrophus genes for synthesis of poly-beta-hydroxybutyric acid (PHB) and synthesis of PHB in Escherichia coli ’, J. Bacteriol., 1988, 170, (12), pp. 5837–5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoppensack, A. , Rehm, B.H.A. , Steinbüchel, A. : ‘Analysis of 4-phosphopantetheinylation of polyhydroxybutyrate synthase from Ralstonia eutropha: generation of β-alanine auxotrophic Tn5 mutants and cloning of the panD gene region’, J. Bacteriol., 1999, 181, (5), pp. 1429–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang, Z.-X. , Brämer, C.O. , Steinbüchel, A. : ‘The glyoxylate bypass of Ralstonia eutropha ’, FEMS Microbiol. Lett., 2003, 228, (1), pp. 63–71 [DOI] [PubMed] [Google Scholar]
- 27. Brämer, C.O. , Steinbüchel, A. : ‘The methylcitric acid pathway in Ralstonia eutropha: new genes identified involved in propionate metabolism the GenBank accession numbers for the nucleotide sequences of the prp gene cluster are AF325554 and AF331923’, Microbiology, 2001, 147, (8), pp. 2203–2214 [DOI] [PubMed] [Google Scholar]
- 28. Lindenkamp, N. , Peplinski, K. , Volodina, E. , et al.,: ‘Impact of multiple β-ketothiolase deletion mutations in Ralstonia eutropha H16 on the composition of 3-mercaptopropionic acid-containing copolymers’, Appl. Environ. Microbiol., 2010, 76, (16), pp. 5373–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Budde, C.F. , Mahan, A.E. , Lu, J. , et al.,: ‘Roles of multiple acetoacetyl coenzyme A reductases in polyhydroxybutyrate biosynthesis in Ralstonia eutropha H16’, J. Bacteriol., 2010, 192, (20), pp. 5319–5328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arenas-López, C. , Locker, J. , Orol, D. , et al.,: ‘The genetic basis of 3- hydroxypropanoate metabolism in Cupriavidus necator H16’, Biotechnol. Biofuels, 2019, 12, (1), p. 150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Srinivasan, S. , Barnard, G.C. , Gerngross, T.U. : ‘Production of recombinant proteins using multiple-copy gene integration in high-cell-density fermentations of Ralstonia eutropha ’, Biotechnol. Bioeng., 2003, 84, (1), pp. 114–120 [DOI] [PubMed] [Google Scholar]
- 32. Voss, I. , Steinbüchel, A. : ‘Application of a KDPG-aldolase gene-dependent addiction system for enhanced production of cyanophycin in Ralstonia eutropha strain H16’, Metab. Eng., 2006, 8, (1), pp. 66–78 [DOI] [PubMed] [Google Scholar]
- 33. Müller, J. , MacEachran, D. , Burd, H. , et al.,: ‘Engineering of Ralstonia eutropha H16 for autotrophic and heterotrophic production of methyl ketones’, Appl. Environ. Microbiol., 2013, 79, (14), pp. 4433–4439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen, J.S. , Colón, B. , Dusel, B. , et al.,: ‘Production of fatty acids in Ralstonia eutropha H16 by engineering β-oxidation and carbon storage’, PeerJ, 2015, 3, p. e1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Grousseau, E. , Lu, J. , Gorret, N. , et al.,: ‘Isopropanol production with engineered Cupriavidus necator as bioproduction platform’, Appl. Microbiol. Biotechnol., 2014, 98, (9), pp. 4277–4290 [DOI] [PubMed] [Google Scholar]
- 36. Fukui, T. , Suzuki, M. , Tsuge, T. , et al.,: ‘Microbial synthesis of poly((R)-3-hydroxybutyrate-co-3-hydroxypropionate) from unrelated carbon sources by engineered Cupriavidus necator ’, Biomacromolecules, 2009, 10, (4), pp. 700–706 [DOI] [PubMed] [Google Scholar]
- 37. Bi, C. , Su, P. , Müller, J. , et al.,: ‘Development of a broad-host synthetic biology toolbox for Ralstonia eutropha and its application to engineering hydrocarbon biofuel production’, Microb. Cell Fact., 2013, 12, (1), p. 107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Alagesan, S. , Hanko, E.K.R. , Malys, N. , et al.,: ‘Functional genetic elements for controlling gene expression in Cupriavidus necator H16’, Appl. Environ. Microbiol., 2018, 84, (19), pp. e00878–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Johnson, A.O. , Gonzalez-Villanueva, M. , Tee, K.L. , et al.,: ‘An engineered constitutive promoter set with broad activity range for Cupriavidus necator H16’, ACS Synth. Biol., 2018, 7, (8), pp. 1918–1928 [DOI] [PubMed] [Google Scholar]
- 40. Gruber, S. , Hagen, J. , Schwab, H. , et al.,: ‘Versatile and stable vectors for efficient gene expression in Ralstonia eutropha H16’, J. Biotechnol., 2014, 186, pp. 74–82 [DOI] [PubMed] [Google Scholar]
- 41. Sydow, A. , Pannek, A. , Krieg, T. , et al.,: ‘Expanding the genetic tool box for Cupriavidus necator by a stabilized L-rhamnose inducible plasmid system’, J. Biotechnol., 2017, 263, pp. 1–10 [DOI] [PubMed] [Google Scholar]
- 42. Sato, S. , Maruyama, H. , Fujiki, T. , et al.,: ‘Regulation of 3-hydroxyhexanoate composition in PHBH synthesized by recombinant Cupriavidus necator H16 from plant oil by using butyrate as a co-substrate’, J. Biosci. Bioeng., 2015, 120, (3), pp. 246–251 [DOI] [PubMed] [Google Scholar]
- 43. Lütte, S. , Pohlmann, A. , Zaychikov, E. , et al.,: ‘Autotrophic production of stable-isotope- labeled arginine in Ralstonia eutropha strain H16’, Appl. Environ. Microbiol., 2012, 78, (22), pp. 7884–7890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Krieg, T. , Sydow, A. , Faust, S. , et al.,: ‘CO2 to terpenes: autotrophic and electroautotrophic α-humulene production with Cupriavidus necator ’, Angew. Chem. Int. Ed., 2018, 57, (7), pp. 1879–1882 [DOI] [PubMed] [Google Scholar]
- 45. Garrigues, L. , Maignien, L. , Lombard, E. , et al.,: ‘Isopropanol production from carbon dioxide in Cupriavidus necator in a pressurized bioreactor’, New Biotechnol., 2020, 56, pp. 16–20 [DOI] [PubMed] [Google Scholar]
- 46. Windhorst, C. , Gescher, J. : ‘Efficient biochemical production of acetoin from carbon dioxide using Cupriavidus necator H16’, Biotechnol. Biofuels, 2019, 12, (1), p. 163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Heinrich, D. , Raberg, M. , Steinbüchel, A. : ‘Studies on the aerobic utilization of synthesis gas (syngas) by wild type and recombinant strains of Ralstonia eutropha H16’, Microb. Biotechnol., 2018, 11, (4), pp. 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Garcia-Gonzalez, L. , De Wever, H. : ‘Valorisation of CO2-rich off-gases to biopolymers through biotechnological process’, FEMS Microbiol. Lett., 2017, 364, (20), 10.1093/femsle/fnx196 [DOI] [PubMed] [Google Scholar]
- 49.‘Does it contain genetically modified ingredients? – Impossible foods’ [Online] . Available at https://faq.impossiblefoods.com/hc/en-us/articles/360023038894-Does-it-contain-genetically-modified-ingredients (Accessed 26 February 2020)
- 50. Monbiot, G. : ‘Lab-grown food will soon destroy farming – and save the planet’, The Guardian, 2019.
- 51. Kiverdi, Inc.: Available at https://www.kiverdi.com/ (accessed 27 February 2020)
- 52. NovoNutrients : Available at https://www.novonutrients.com/ (accessed 27 February 2020)
- 53. Home-Solar Foods Ltd : Available at https://solarfoods.fi/ (accessed 27 February 2020)
- 54. Oesterholt, F. , Broeders, E. , Zamalloa, C. : ‘Power-to- protein: eiwitproductie in een circulaire economie’, KWR 2018.078, 2019.
- 55.‘Deep Branch Biotechnology – Transforming the polluters of today into the producers of tomorrow’ . Available at https://deepbranchbio.com/ (accessed 27 February 2020)
- 56. Abrini, J. , Naveau, H. , Nyns, E.-J. : ‘ Clostridium autoethanogenum sp. Nov., an anaerobic bacterium that produces ethanol from carbon monoxide’, Arch. Microbiol., 1994, 161, (4), pp. 345–351 [Google Scholar]
- 57. LanzaTech : Available at http://www.lanzatech.com/ (accessed 1 June 2017)
- 58. Electrochaea GmbH – Power-to-Gas Energy Storage | Technology : Available at http://www.electrochaea.com/technology/ (accessed 10 May2017)
- 59. Home – Unibio : Available at https://www.unibio.dk/ (accessed 27 February 2020)
- 60. Calysta – More From Less : Available at http://calysta.com/ (accessed 27 February 2020)


