Abstract
In the current changing environment, salt stress has become a major concern for plant growth and food production worldwide. Understanding the mechanisms of how plants function in saline environments is critical for initiating efforts to mitigate the detrimental effects of salt stress. Agricultural productivity is linked to nutrient availability, and it is expected that the judicious metabolism of mineral nutrients has a positive impact on alleviating salt-induced losses in crop plants. Nitrogen (N) is a macronutrient that contributes significantly to sustainable agriculture by maintaining productivity and plant growth in both optimal and stressful environments. Significant progress has been made in comprehending the fundamental physiological and molecular mechanisms associated with N-mediated plant responses to salt stress. This review provided an (a) overview of N-sensing, transportation, and assimilation in plants; (b) assess the salt stress-mediated regulation of N dynamics and nitrogen use- efficiency; (c) critically appraise the role of N in plants exposed to salt stress. Furthermore, the existing but less explored crosstalk between N and phytohormones has been discussed that may be utilized to gain a better understanding of plant adaptive responses to salt stress. In addition, the shade of a small beam of light on the manipulation of N dynamics through genetic engineering with an aim of developing salt-tolerant plants is also highlighted.
Keywords: crop productivity, genetic engineering, nitrogen metabolism, phytohormones, salt stress resilience
Introduction
Soil salinization is a detrimental ecological issue, limiting crop production, quality and global problem for more than half of all arable lands by 2050 (Qureshi and Daba, 2020; Naz et al., 2022). Due to the increased saline levels in the agricultural soils, nitrogen (N) status of soil depleted (Prasad et al., 1997), and it is predicted that up to 50% fertile land will be lost by the middle of the 21st century (Aslam et al., 2017). Mineral nutrient research provides an operative way for exploring plant acclimation to salt stress and improving plant growth and productivity (Torres Bazurto et al., 2017; Sousa et al., 2021). Nitrogen is an indispensable potent nutrient that mediates defense responses, along with a wide range of cellular, physiological and molecular responses critical to plant survival, and various signal transduction pathways associated with plant defense mechanisms and salt-stress resilience (Krapp, 2015; Zhang et al., 2020a; Zhang et al., 2020b). Research focus on sustaining N dynamics in managing salt stress responses gained attention. Salt stress has an impact on nitrification and ammonification in the soil as chloride (Cl-) competes with nitrate (NO3-) and causes ion toxicities and ionic disparities, which can explicitly restrict N uptake, transport and assimilation processes (Campbell, 1999; Zhao et al., 2020; Liu et al., 2022; Tzortzakis et al., 2022, Figure 1 ). Reductions in N uptake and metabolism have been reported in many plant species (Dalio et al., 2013; Singh et al., 2016; Shahzad et al., 2017; Fuertes-Mendizábal et al., 2020, Table 1 ). For instance, salt stress inhibited NO3 - and ammonium (NH4 +) uptake in Zea mays (maize) leaves (Fuertes-Mendizábal et al., 2020), and may decrease the proportion of N-transport amino acids: asparagine (Asn), glutamate (Glu), aspartate (Asp) and glutamine (Gln) in Cajanus cajan (pigeon pea; Dalio et al., 2013) and Oryza sativa (rice; Shahzad et al., 2017). Further, salt stress reduced water uptake, N content and nitrate reductase (NR) activity in Triticum aestivum (wheat; Ashfaque et al., 2014), inhibited activities of N-assimilation enzymes such as NR, nitrite reductase (NiR), glutamine synthetase (GS) and glutamate oxyglutarate aminotransferase cycle (GOGAT) in Cucumis sativus (cucumber) and Lycopersicon esculentum (tomato) seedlings (Shao et al., 2015; Singh et al., 2016, Figure 1 ). However, Meng et al. (2016) reported that salt stress has only minimal effect on NR and NiR activity while the substrate concentration (NO3 - supply) impact NR and NiR activity in Populus simonii (Chinese poplar), GS and GOGAT activity were significantly lower in salt-treated plants in comparision to control plants. They presume that this effect is due to decrease in NH4 + uptake via roots under salt stress. Salt stress also restricts NO3- uptake on the tonoplast membrane and causes an increase in NH4 + accumulation in shoots, disrupting N metabolism, and finally yield in rice seedlings (Thu Hoai et al., 2003, Figure 1 ). Thus, the detrimental effects of salt stress on plant metabolism could be attributed to changes in N uptake as well as the activities of various enzymes and genes involved in N metabolism (Chakraborty et al., 2018; Isayenkov and Maathuis, 2019).
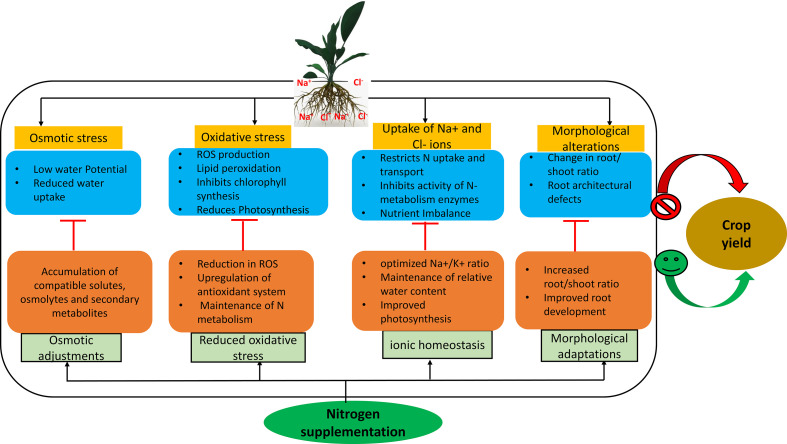
Figure 1.
N supplementation exerts a defensive response on plant physiological and morphological machinery during salt stress and helps in alleviating the osmotic stress, oxidative damage and morphological alterations. N also help in maintaining the ionic homeostasis along with N metabolism in plants during salt stress, which in turn helps in improving the plant performance during salt stress. Cl-, chloride; N, Nitrogen; Na+, sodium; K+, Potassium; ROS, reactive oxygen species.
Table 1.
Effect of salt stress on N metabolism and NUE in different plants.
| Plants | Salt concentration | Gene expression | Action | References |
|---|---|---|---|---|
| Rice (Oryza sativa) | 0.3% NaCl | OsNRT1.1, OsNRT1.5, OsNRT1.8, OsNRT2.1, OsAMT1.1, OsAMT1.2, OsNR1, OsGS1.2, OsNADH-GOGAT, OsFd-GOGAT, OsAS | Inhibition of NO3¯content, NR, GS and NADH-GOGAT activities however, NH4 + content, Fd-GOGAT, GDH and AspAT activities ↑, decreased expression of NR1 | Decui et al., 2020 |
| Maize (Zea mays) | 100 and 430 mM NaCl | GS1 and GS2 | Reduced NO3¯ content, N assimilation, free amino acids and soluble protein | Fuertes-Mendizábal et al., 2020 |
| Rice | 100 mM NaCl | AMT1.1, AMT1.2, NIA1, NIA2 | N content and accumulation ↓, NR and GS activities ↓, while, NH4 + uptake ↑ | Huang et al., 2020 |
| Tomato (Solanum lycopersicum and Solanum pimpinellifoilum) | 75 mM NaCl | SlNR, SlNiR, SlAMT1, AlAMT2, SlNRT1.2, SlGDH, SlGS | N-NO3¯, NR, NiR, GS, GDH activities and amino acid accumulation ↓, while, GOGAT activity ↑ | Lopez-Delacalle et al., 2020 |
| Beet root (Beta vulgaris) | NaCl and Na2SO4 (2:1M) | ALN, XDH | Down-regulation of ALN while, allantoin, betaine, L-citrulline and melatonin content ↑ and altered N metabolism | Liu et al., 2020 |
| Common bean (Phaseolous vulgaris) | 100 mM NaCl | NLP7, DREB2A | NLP7, NO3¯, NiR and GOGAT activity ↓, however, NH4 + content and NR activity ↑ | Ozfidan-Konakci et al., 2020 |
| Peanut (Arachis hypogaea) | 200 mM NaCl | NO3- transporter coding genes | Expression of NO3¯ transporter coding genes repressed thus, reduced N metabolism | Zhang et al., 2020b |
| Cucumber (Cucumis sativus) | 80 mM NaCl |
CsPIP1-2, CsPIP2-4 |
NO3¯ uptake, NR, GS, GOGAT, AspAT, AlaAT and IDH activity ↓, conversely NH4 + uptake and GDH activity ↑ | Li et al., 2019 |
| Cucumber | 25-75 mM NaCl | NR, NiR, GS, GOGAT, GPT, GDH | NO3¯ content, NR, NiR, and GOGAT activities ↓, while, GS and GDH activity ↑ | Ma et al., 2019 |
| Rice | 100, 200 and 300 mM NaCl | NR1, NR2, NR3 | Reduced NR activity in salt sensitive genotype | Rohilla and Yadav, 2019 |
| Common grass (Zostera marina) | 0.4 mM NaCl | NR, NiR, NAR, NRT2.1 | Up-regulation of NR and NiR gene, and increased NO3¯ assimilation | Lv et al., 2018 |
| Tomato (Solanum lycopersicon and Solanum pennelli) | 100 mM NaCl | NRT1.1, NRT1.2, AMT1.1, AMT1.2, GS1, NR | NO3¯content ↓, NH4 + content ↑, down-regulation of NRT1.1, NRT1.2, AMT1.2, GS1 and NR, while, up-regulation of AMT1.1 | Abouelsaad et al., 2016 |
| Chinese poplar (Populus simonii) | 75 mM NaCl | AMT1.2, AMT1.6, NRT1.1, NRT2.4a, NR, NiR, GS1.3, GS2, Fd-GOGAT, NADH-GOGAT | NH4 + uptake ↑, NO3¯ uptake and NH4 + assimilation ↓, weakened GS/GOGAT pathway | Meng et al., 2016 |
| Mustard (Brassica juncea) | 150 mM NaCl | NRT1.1, NRT1.5, NRT2.1, AMT1.2, AMT2, GS1.1, GDH1, ASN2, NiR1, GDH2 | Reduced NO3 - uptake and mobilization, down-regulation of N assimilation associated genes | Goel and Singh, 2015 |
| Cucumber | 84 mM NaCl | NR, GS, GOGAT, β-actin | NR, GS, GOGAT, GDH activity and soluble protein content↓, proline content↑ | Shao et al., 2015 |
| Arabidopsis (Arabidopsis thaliana) | 100 mM NaCl | NPF2.3, NPF7.3 | Reduced NO3¯ translocation from root to shoot | Taochy et al., 2015 |
| Sweet potato (Ipomoea batatas) | 100 mM NaCl | NR2, NiR2, GS2, NADH-GOGAT, NRT1.1, CLCc | NO3 - net influx↑, NH4 + assimilation ↑ | Yu et al., 2016 |
| Barley (Hordeum vulgare) | 150 mM NaCl | P5CS | P5CS activity and Ala accumulation↑, N metabolism↑ | Averina et al., 2014 |
| Maize | 25, 75 and 150 mM NaCl | ZmPAL1 | GS and GOGAT↓, proline accumulation↑, up-regulation of PAL1 | Ertani et al., 2013 |
| Rice | 100 mM NaCl | OsGS1, OsGS2, OsNR1, OsFd-GOGAT | Down-regulation of NH4 + assimilation genes, NO3 - deficiency | Wang et al., 2012a |
| Soybean (Glycine max) | 50, 100 and 200 mM NaCl | GS, GOGAT, HO-1 | Decreased leghemoglobin content, nitrogenase and GS/GOGAT activity while, NH4 + content ↑ | Zilli et al., 2008 |
| Tex-Mex tobacco (Nicotiana plumbaginifoia) | 300 mM NaCl | gdhA, gdhB | Increased NADH-GDH activity and gdhA level while, gdhB ↓ | Restivo, 2004 |
↑: increased; ↓: decreased; ALN, Allantoinase; XDH, Xanthine dehydrogenase; IDH, Iso-citrate dehydrogenase; PAL, Phenyl ammonia lyase; P5CS, Pyrroline-5-carboxylate synthase; PIP, Plasma membrane intrinsic protein; DREB, Dehydration-responsive element-binding protein; GPT, Glutamate pyruvate transaminase; NLP, Nodule inception like protein; NIA, Nitrate reductase; gdh, Glutamate dehydrogenase; HO, Heme oxygenase.
During green revolution years, usage of N fertilizers have been an effective way to increase crop performance and productivity. Therefore, optimizing N-metabolism through N-supplementation to saline soils may become as an effective practice for sustainable crop production facilitated by N-assimilation and cellular ion homeostasis (Hessini, 2022; Yin et al., 2022). Additionally, N is interrelated with phytohormones, and play a critical role in the modulation of plant responses to develop salt stress resilience (Ali et al., 2021; Chen et al., 2021). Further, there has been the possibility for significant improvements in nitrogen use efficiency (NUE) through genetic engineering approaches, which have made use of diversified germplasm responses to N supply, to sustain or improve their productivity while minimizing the use of N fertilizers.
The present review is an attempt to elucidate (a) a brief overview of N-sensing, transportation and assimilation in plants, (b) regulation of N metabolism and NUE under salt stress (c) N-supplementation mediated adaptive responses in plants under salt stress. Furthermore, the existing but not much explored crosstalk between N and phytohormones have been discussed to gain a better understanding of plant adaptive responses to salt stress. In addition, this also shades a small beam of light on manipulation of N dynamics through genetic engineering for sustainable agriculture.
Nitrogen sensing, transportation and assimilation in plants
Remarkable progress has been made in understanding the basic mechanisms of how plants sense and adapt to N circumstances. Nitrogen uptake and transportation occurs within and between the plant cells, requiring trans-membrane transporters to facilitate such response. NO3 - and NH4 + are the most prevalent forms of inorganic N available in the soil, and a vast number of NO3 - (NRTs) and NH4 + transporter families (AMTs) effectively support their absorption and allocation throughout the plant cells and tissues ( Figure 2 ). It is known that four families of transporters- NPF family [Nitrate Transporter 1 (NRT1)/Peptide Transporter (PTR) Family], NRT2 family [Nitrate Transporter 2], CLC family [Chloride Channel Family], and SLAC1/SLAH family [Slow Anion Associated Channel Homologs] - contribute to NO3 - uptake and transport in plants (Xia et al., 2015; Wei et al., 2021; Souza et al., 2022). However, the supply of NO3 - and the level of plant N nutrition tightly control the functioning of these transporters. For instance, the dual-affinity of NO3 - transporter like AtNRT1.1 (AtNPF6.3) and the low-affinity transporter-like AtNR1.2 (AtNPF4.6/CHL1) in Arabidopsis thaliana (Arabidopsis; Huang et al., 1999; Liu and Tsay, 2003) were reported to be influenced under high NO3 - conditions. Similarly, the dual-affinity behaviour of NO3 - transporter, MtNRT1.3 in Medicago truncatula (barrel medick; Morere-Le Paven et al., 2011), and the low-affinity NO3 - transporter ZmNPF7.9 (NRT1.5) in maize (Wei et al., 2021) and OsNPF8.9 (OsNRT1.1, Os3g13274, or AF140606), the OsNRT1.1 allele of OsNPF6.5 (OsNRT1.1B), OsNPF2.4 (OsNRT1.6), and a possible 6-transmembrane NO3 - transporter OsNRT1.1b (AK066920) in rice (Xia et al., 2015; Fan et al., 2016; Li et al., 2022a) all were shown to be implicated in the uptake of NO3 -. Contrarily, NRT2 family plays an imperative role in NO3 - acquisition under N deficient conditions (Zou et al., 2020; Li et al., 2022b; Du et al., 2022). For instance, NO3 - transporter-like NRT2.1, NRT2.2, NRT2.4, and NRT2.5 require a partner protein, NAR2.1 (Kiba et al., 2012; Lezhneva et al., 2014; Zou et al., 2020) in Arabidopsis to acclimatize under such conditions. In addition, other transporters such as CmNRT2.1 in Chrysanthemum indicum (Chrysanthemum; Gu et al., 2016), TaNRT2.1-6B in wheat (Li et al., 2022a), BnaZSNRT2s in Brassica napus (Canola; Du et al., 2022) and OsNRT2.1, OsNRT2.2, OsNRT2.4 and their partner protein OsNAR2.1 in rice (Naz et al., 2019; Souza et al., 2022) also function as NO3 - influx transporters ( Figure 2 ). Among all NO3 – transporters, the dual affinity NO3 – transporter NRT1.1 (AtNPF6.3) serve as a multifunctional protein with a critical role in both NO3 – acquisition and sensing. Hence, this transporter represents critical step towards understanding the molecular basis of NO3 – use in plants (Tsay et al., 1993).
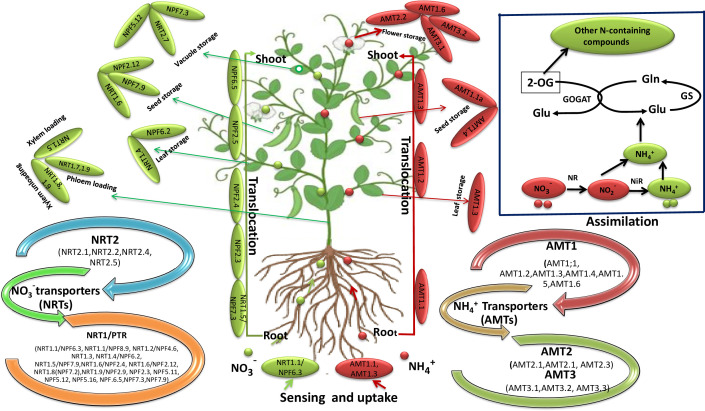
Figure 2.
Schematic diagram showing the routes of NO3¯ and NH4 + sensing, transportation and accumulation in plants. The green dots and arrows denote the transport of NO3¯ and the red dots and arrows indicates the transport of NH4 + ions in plants. In the uptake process, NO3¯ and NH4 + enter the plant roots via NRTs and AMTs transporters respectively. For example, NRT1.1/NPF6.3/CHL1, AMT1.1 and AMT1.3 involved in the sensing of NO3¯ and NH4 + respectively. These transporter are localized in the plasma membrane of root cells. After uptake, NO3¯ transported via NRT1.5/NPF7.3, NPF2.3, NPF2.4, NPF2.5 and NPF6.5 and NH4 + via AMT1.1, AMT1.2 and AMT1.3 to the shoot through xylem. NRT1.5 is involved in NO3¯ loading in the xylem, while as NRT1.8, NRT1.9 are involved in NO3¯ unloading from the xylem. However, NRT1.7, NRT1.9 transport NO3¯ into the phloem. Furthermore, NRT1.4/NPF6.2 and AMT1.3 mediates NO3¯ and NH4 + transport to the leaf/petiole. NO3¯ accumulation within the leaf vacuole is mediated by NPF7.3, NPF5.12, and NRT2.7. AMT1.1, AMT1.2 and AMT1.3 mediates NH4 + transoction from root to shoot. NRT1.6/NPF2.12, NPF7.9 and AMT1.1a, AMT1.4 is involved in the transportation of NO3¯ and NH4 + respectively in the seed where they accumulated in the vacuoles. NRT2.7 is a tonoplast transporter of embryo which regulates NO3¯accumulation within seed vacuole. AMTs, ammonium transporters; Gla, glutamine; Glu, glutamate; GOGAT, glutamate synthase; GS, glutamine synthetase; NH4 +, ammonium; NiR, nitrite reductase; NO3¯, nitrate; NR, nitrate reductase; NRTs, nitrate transporters; 2-OG, 2-Oxoglutarate.
The transport and distribution of NO3 - from root to shoot occurs via the xylary tissues ( Figure 2 ). Long-distance root to shoot transport in Arabidopsis is mediated by the low-affinity NO3 – transporter NPF7.3 (AtNRT1.5), which is essential for NO3 – loading from the root cytoplasm of pericycle cells into the xylem vessel (Sena Falero, 2022). AtNPF7.2 (NRT1.8) and AtNPF2.9 (NRT1.9), negative regulators of root to shoot NO3 – transport, have promising roles in xylem unloading (Wang and Tsay, 2011). Moreover, NO3 - is locally stored in vacuoles or assimilated in the cytoplasm of the leaves. According to recent evidences, tonoplast-localized NO3- transporters (AtNPF5.11, AtNPF5.12, and AtNPF5.16) could retrieve NO3 - from vacuoles and transport it to the cytosol, acting as star players in the regulation of NO3 - allocation between roots and shoots (He et al., 2017). According to de Los Rios et al. (2021), NPF6.2 (NRT1.4), a low-affinity NO3 -transporter, plays a key role in the regulation of leaf NO3 - homeostasis. TaNRT1.4/AtNPF6.2 constitutively expressed in the midribs and petiole of wheat leaves indicates the role of AtNRT1.4/AtNPF6.2 in the regulation of NO3- homeostasis in leaves (Chiu et al., 2004). Moreover, AtNRT1.6/AtNPF2.12 is involved in NO3 - delivery from maternal tissue to early developing embryos, which influences seed set and NO3 - acquisition (Almagro et al., 2008). AtNPF5.5, is activated in embryos and its knockout affects N import and accumulation, but apparently leads to unaffected seed development (Léran et al., 2015). AtNRT2.7 is a tonoplast transporter of embryo, up regulated in reproductive organs, which regulate NO3- accumulation in the seed vacuole, thus inducing seed germination (Chopin et al., 2007).
OsNPF2.4 and OsNPF7.9 play critical roles in rice, allow the roots to acquire low-affinity NO3 - by the root, translocation from the root to the shoot, and remobilization from old leaf (source) to young leaf and root (sink organs; Wei et al., 2018; Guan et al., 2022). OsNPF7.3 (OsPTR6) transporter, which is located on the vacuolar membrane, may transport the tripeptide Glycine-Glycine-Leucine (Gly-Gly-Leu) and the dipeptide Glycine-Histidine (Gly-His) (Ouyang et al., 2010). By increasing N absorption and GS activity, it increases rice growth, but when NH4 + levels are high, it decreases NUE (Ouyang et al., 2010). Additionally, ZmNPF7.9 in maize plays a role in transporting NO3 – from maternal tissues to the developing endosperm, which could provide valuable knowledge for maize genetic improvement (Wei et al., 2021). Furthermore, the low affinity transporter, VvNPF6.5 modulates NO3- uptake and translocation from roots to shoot in Vitis vinifera (grapevine) and is implicated in primary NO3- response (He et al., 2020).
Under salt stress, NO3- dependent transport systems facilitate the uptake and loading of sodium (Na+) ion into the xylem, and may represent a key pathway for the accumulation of Na+ in Arabidopsis shoots (Álvarez-Aragón and Rodríguez-Navarro, 2017). AtNPF2.4 was much less permeable to NO3 − and catalysed passive Cl− efflux out of cells. atnpf2.4 knockdown mutants showed decreased shoot Cl– accumulation, while overexpression of AtNPF2.4 increased shoot Cl– accumulation, suggesting that AtNPF2.4 might function to load Cl– into the xylem of Arabidopsis roots during salt stress (Li et al., 2016). Another permeable to Cl- transporter AtNPF2.5, the closest homolog to AtNPF2.4, has also been shown to serve as a pathway for Cl- efflux from the root, which contributes to the deprivation of Cl- from Arabidopsis shoots under salt stress (Li et al., 2017). Furthermore, under salt stress, AtNPF2.3 promotes NO3 - translocation from roots to shoots (Taochy et al., 2015). CLCs transporters are also essential for the absorption and allocate Cl- and NO3 – (Wei et al., 2019; Liu et al., 2020). Rice genome contains five CLC genes, but their underlying mechanisms in NO3 – translocation have not yet been identified (Diédhiou and Golldack, 2006). In contrast, the tonoplast-localized OsCLC1 and OsCLC2 proteins work to compartmentalize Cl- ions into the vacuole to prevent Cl– induced toxicity under situations of salt stress, and over-expression of OsCLC1 promotes salt tolerance, resulting in higher grain yield in rice (Nakamura et al., 2006; Diédhiou and Golldack, 2006). At the cellular and molecular research, more in-depth research is required to understand the function of various NO3 – related transporters in response to salt stress in various crop species, which will help to improve agricultural efficiency in the future.
NH4 + transport across membranes is facilitated by proteins from the NH4 + transporter/methyl ammonium permeases (AMT/MEP) family in a diverse array of crop species (Giehl et al., 2017; Wu et al., 2019; Bindel and Neuhäuser, 2021). AMT1;4 is primarily restricted to pollen, although four AMT family homologs - AMT1;1, AMT1;2, AMT1;3, and AMT1;5, and one MEP subfamily homolog - AMT2;1 - are expressed in roots of Arabidopsis (Bindel and Neuhäuser, 2021). The primary transporters for high-affinity NH4 + absorption into Arabidopsis are the root-expressed AMT1-type proteins AMT1;1, AMT1;2, AMT1;3, and AMT1;5 (Giehl et al., 2017; Sasaki and Kojima, 2018; Wu et al., 2019). About two-thirds of the high-affinity NH4 + absorption capability in roots is accounted for by two of these transporters, AMT1;1 and AMT1;3, which are mostly localized in rhizodermal and cortical cells, including root hairs (Sasaki and Kojima, 2018). It follows that AMT1;2 may mediate the uptake of NH4 + entering the root via the apoplastic transport channel (Giehl et al., 2017; Sasaki and Kojima, 2018). OsAMT1;1 is found in the root stele, epidermis, and mesophyll cells of rice, and knockdown of this gene significantly reduces the mobilization of NH4 + from the root to the shoot, indicating that OsAMT1;1 is associated with NH4 + translocation from the root to the shoot (Li et al., 2016). High affinity AMTRs are also highly expressed in the leaf, including NtAMT1.3 in Nicotiana tabacum (tobacco; Fan et al., 2017), ZmAMT1.1a and ZmAMT1.3 in maize (Gu et al., 2013), and GhAMT1.3 in Gossypium hirsutum (cotton; Sun et al., 2019). In Chinese poplar floral organs, PtrAMT1;5 is stamen-specific, whereas PtrAMT1;6 is female flower-specific (Couturier et al., 2007). In Sorghum bicolor (sorghum) floral organs, SbAMT1;1, SbAMT1;2, SbAMT2;1, SbAMT3;1, and SbAMT3;3 are identified in the pistil and stamen, while SbAMT2;2 and SbAMT3;2 are only found in the pistil (Koegel et al., 2013). LjAMT1;1-1;3 expression has also been found in Lotus japonicus flowers ((Birdsfoot trefoil; D`Apuzzo et al., 2004). ZmAMT1.1a transcripts have been found in seeds (Zhao et al., 2018). These AMTs might aid in delivering NH4 + nutrients to the reproductive organs. The exploration of NH4 + nutrition in plants is still in its early stages and is a largely uncharted area of study.
Ammonium transporter PsAMT1.2 mediates NH4 + uptake and metabolism under salt stress in Chinese poplar (Li et al., 2021). PsAMT1.2-overexpressing transgenic plants with better growth had relatively high ratio of K+/Na+ compared with the wild type (WT) under salt stress. Similarly, overexpressing the PutAMT1;1 gene (from Puccinellia tenuiflora, forage grass) in Arabidopsis greatly increases its ability to tolerate salt stress during the early stage of root development following seed germination and improved root to shoot NH4 + mobilization under salt stress (Bu et al., 2019). Zhu et al. (2020) demonstrated that overexpression of BcAMT1.2 (from Brassica juncea, mustard) in Arabidopsis increased the expression levels of N assimilation genes, indicating that an increase in BcAMT1.2 mRNA abundance could also affect NH4 + assimilation directly or indirectly.
Organic compounds in the soil can also aid plant N nutrition (Geisseler et al., 2022). Amino acids account for the majority of low molecular weight dissolved organic N in soil (Tsuzuki et al., 2022). Evidence suggests that Arabidopsis has a single target of rapamycin (TOR) kinase gene (AtTORC) which is known to be involved in the maintenance of N metabolism including accumulation of amino acids thereby, influencing plant growth and metabolism (Caldana et al., 2013; Dobrenel et al., 2016). It has been revealed that target of TOR is repressed in N-deprived seedlings, however, resupply of N sources such as NO3-, NH4 +, amino acids promptly reactivates TOR kinase (Liu et al., 2018). Further, it has been speculated that a number of amino acid permeases (AAP) family, including AAP1, AAP2, AAP3, AAP6, AAP7, AAP8, and AAP16, have a variety of roles in the transport of amino acids (Zhao et al., 2012; Cheng et al., 2016; Wang et al., 2022; Pereira et al., 2022). The families of cationic amino acid transporters (CAT), oligopeptide transporters (OPT), lysine-histidine like transporters (LHT), proline transporters (ProT), and aromatic and neutral amino acid transporters (ANT) have also been identified as having the potential to transport amino acids (Feng et al., 2018; Lin et al., 2019; Guo et al., 2020; Kurt and Filiz, 2022). Only a small subset of these transporters, though, has been essentially described for their function in transporting amino acids. For example, LHT1 aids in the efficient transport of proline and γ- amino butyric acid (GABA) during vegetative and reproductive growth, whereas ProT1 and ProT3 effectively deliver amino acids from vegetative to reproductive organs for grain yield, nutritional quality, and functioning (Lin et al., 2019; Guo et al., 2020).
The coordinated regulation of NO3 - transporter McNRT1 and the amino acid transporters McAAT1 and McAAT2 from Mesembryanthemum crystallinum (crystalline ice plant) mediates the uptake and distribution of nitrogenous compounds and amino acids under conditions of high salt in plants (Popova et al., 2003). Over-expression of AhProT (a proline transporter from Atriplex hortensis, mountain spinach) had increased proline content in root tips and showed salt tolerance in Arabidopsis (Shen et al., 2002). It is noted that, higher expression level of ProT1 in Arabidopsis, implying that ProT1 plays an important role in N distribution during salt stress (Rentsch et al., 1996).
In plants, GS and the GOGAT account for more than 95% of NH4 + assimilation (Liu et al., 2022). NH4 + can also be assimilated by glutamate dehydrogenase (GDH), which converts α-oxoglutarate to glutamate. These metabolic intermediates function as important signalling molecules or as significant amino donors for the formation of other amino acids and N-containing compounds, supporting plant growth and development as well as plant responses to stress conditions (Zhi et al., 2020).
Regulation of nitrogen metabolism and nitrogen use efficiency under salt stress
Regulation of N-metabolism is critical for salt tolerance, and the interaction between salt stress and N is a complex network that influences plant function (Yu et al., 2016; Tian et al., 2022; Table 1 ). Researchers have reported various physiological and molecular processes using a variety of tools ranging from agronomic to genetic approaches for developing salt tolerant plants (Xu et al., 2012; Reddy et al., 2017; Jha et al., 2019). It is known that cultivars that can sustain high NR and GS/GOGAT activities are more resistant to salt stress (Teh et al., 2016; de la Torre-González et al., 2020). On the other hand, species that can retain a higher NO3 − influx are more resilient to salt stress (Liu et al., 2022). Adequate N supply adjusts and rectifies nutritional disparities in salt-stressed plants. For instance, NO3 - fed rice and canola plants grew faster and had higher salt toxicity than NH4 +-supplied plants, as evidenced by restricted allocation of Na+ in leaf apoplasts (Gao et al., 2016). In contrast, NH4 + fertilization significantly ameliorated the salt-triggered growth inhibition in sorghum (Coelho et al., 2020). Thus, the effect of N forms on salt resilience varies depending on the plant species. Thus, understanding N metabolism in response to salt stress may be critical for salt tolerance research and will be an interesting research topic for salt stress physiology in the future. Genes involved in N uptake and metabolic processes also play key roles in plant resilience to salt stress. Expression of NO3- transporters were down-regulated, while the expression of NH4 + transporters were up-regulated in roots of salt-stressed- Chinese poplar (Zhang et al., 2014a). Similarly, genes associated with NO3- uptake, reduction and N metabolism were down-regulated whereas, NH4 + transporter genes (AMT1.2 and AMT1.6) were up-regulated in Chinese poplar in response to salt stress (Meng et al., 2016). Furthermore, salt stress up regulated the expression of OsNRT1.2 and OsAMT2.1, while, down-regulated the expression of OsNRT2.1, in old leaves of salt-stressed rice (Wang et al., 2012a). Salt stress reduced the expression of NRT1.1 and NRT1.2, which was associated with the reduced NO3- content and expression level of AMT1.2, GS1 and NR gene in two cultivars of tomato (Abouelsaad et al., 2016). The genes (OsGS1.2, OsGS2, OsNR1, and OsFd-GOGAT) involved in NH4 + assimilation were down-regulated in rice seedlings due to reduced NO3- uptake, resulting in the inhibition of N-assimilation under salt stress (Wang et al., 2012a). The combined action of NRT1.5/NPF7.3 and NRT1.8/NPF7.2 regulates the partition of NO3- from root to shoot. In response to salt stress, expression of NRT1.8/NPF7.2 were up-regulated while expression of NRT1.5/NPF7.3 were repressed in roots which mediates reallocation of NO3- back to the roots, and limits NO3- translocation to the shoot under stress conditions (Li et al., 2010; Zhang et al., 2014b).
For the world’s sustainable food production, there is an urgent need to improve NUE in agricultural farming systems. Improving NUE under salt stress is critical for plants, as they are increasingly confronted with two major environmental constraints: excessive N fertilization and soil salinization. Several studies have been conducted to assess NUE under saline conditions (Song et al., 2019; Guo et al., 2021; Phan et al., 2022; Table 1 ). It has been proposed that NUE can be determined by agronomic NUE (aNUE), physiological NUE (pNUE), internal NUE (iNUE) and N recovery efficiency (NRE) which gets affected by salt stress. Nitrogen fertilization enhance NUE in rice plants under salt stress as application of low N (0.36 mM) under high salt stress (113 mM) gave better aNUE and agNUE than a high N (2.86 mM) rate under moderate (56 mM) salt stress (Phan et al, 2022). Moreover, salt treatment reduces photosynthetic-NUE in Capsicum annuum (chilli pepper; Huez-Lópeze et al., 2011), mustard (Iqbal et al., 2015; Iqbal et al., 2017), and cucumber (Ma et al., 2019). Furthermore, NUE was significantly reduced in the salt-sensitive genotype of Avena sativa (oat) compared to tolerant genotype (Song et al., 2019). Recently, Guo et al. (2021) revealed that salt stress significantly reduced N metabolism, NUE, and grain yield in rice seedlings. Effective crop management practices and smart decision-making are required to ensure high crop yields under salt stress conditions and meet increasing global food security. Cultivation of salt-tolerant cultivars is an excellent strategy to increase NUE. Many researchers have found a significant link between NUE and salt resilience of cereal crops. The salt-tolerant genotype of wheat had a higher NUE than the salt-sensitive genotype (Borzouei et al., 2014). High salt stress resistance in maize was attributed to higher NUE attributes such as pNUE, NYE (nitrogen yield efficiency) and NHI (nitrogen harvest index) (Javed et al., 2021). According to Bonnave and Bertin (2018), higher NUE is also responsible for conferring salt resilience in rice. It has been revealed that salt-tolerant wheat lines retained higher yield and yield-related attributes, as well as the maximum NUE indexes, compared to salt-sensitive wheat lines, which could be accountable to the greater N uptake proficiency in the tolerant genotype (Lamichhane et al., 2021). Nitrogen application at the proper dose and timings may also minimize N losses and increase efficiency. It has been reported that moderately low N (2.0 mM) supplementation can help to reduce salt (50 mM, 100 mM)-triggered damage in Lolium multiflorum (annual ryegrass) and maximize NUE (Shao et al., 2020). Root stock grafting improves NUE of Citrullus lanatus (watermelon) by increasing nutrient uptake and activating NO3- and NO2- reductase genes (Nawaz et al., 2017). Furthermore, physiological traits that regulates elemental control and the synergy of N partitioning to photosynthesis and remobilized N to sink organs can also be useful for generating high NUE cultivars. Improving N transition to sink organs such as leaves or grains can optimize the NHI and thus the overall NUE (Phan et al., 2022). Salt-stress induced responses also include the accumulation of compatible solutes as well as changes in ion transport (such as uptake, extrusion and sequestration of ions), which may potentially result in the maintenance of redox homeostasis and detoxification, increased NUE and survival under salt stress.
Nitrogen use efficiency is a complex trait controlled by multiple genes. For instance, alanine aminotransferase (AlaAT) and aspartate aminotransferase (AspAT) are involved in the synthesis and degradation of alanine (Ala) and Asp respectively. The increased NUtE under salt stress was attributed to the overexpression of AlaAT and AspAT genes in canola and rice (Beatty et al., 2009; Decui et al., 2020). Accordingly, the activity of AlaAt and AspAT were shown to be increased in Morus alba (white mulberry) and Jatropha curcas (jatropha) under salt stress conditions (Gao et al., 2013; Ullah et al., 2019). The increased activity of AlaAT and AspAT enzymes may be attributed to high glutamate demands, which maintains the C: N ratio under salt stress conditions, thereby improving NUE (Naliwajski and Skłodowska, 2018). On the contrary, the activity of AlaAT and AspAt were found to be suppressed in wheat and cucumber under salt stress condition (Ahanger and Agarwal, 2017; Li et al., 2019).
Thus, improving NUE in crops is an important goal in agricultural research and our future food production capabilities. Understanding the physiological and molecular mechanisms regulating plant N uptake, assimilation, and redistribution within the cell is critical for enhancing NUE and achieving maximum crop growth, ensuring a better return on investment, and mitigating the adverse effects of salt stress.
Nitrogen supplementation mediated salt stress tolerance in plants
Nitrogen has been widely reported to be a multifunctional and the most regulating nutrient element for crop production, influencing the primary production of the agricultural system (Melino et al., 2022). The constructive regulatory roles of N-supply in plants to develop salt tolerance have been reported ( Figure 1 , Table 2 ). For instance, supplementation with 50 and 100 mg N kg− 1 soil alleviates the deleterious effects of salt stress by activating the antioxidant system and increasing the accumulation of osmolytes and secondary metabolites in wheat plants (Ahanger et al., 2019). Similarly, supplementation of 2.5 mmol L−1 N to two cotton genotypes with varying salt stress sensitivity was reported to augment the antioxidant system, accumulation of compatible solutes and ameliorate salt-induced toxicity (Sikder et al., 2020). This revealed the positive role of N by maintaining relative water content (RWC) and protecting the photosynthetic apparatus, especially in the salt-sensitive genotype. Nitrogen fertilizer (105 and 210 kg ha−1) applied to salt-stressed wheat plants was reported to have a positive effect on germination rates and seedling characteristics (Ibrahim et al., 2016). In another study, N (86 and 210 kg ha−1) significantly decreased the damaging impacts of salt stress and improved germination percentage in wheat seedlings (Ibrahim et al., 2016). In hybrid corn (AG 1051), N application (80 and 160 kg ha-1) can ameliorate the deleterious effects of salt stress and enhanced photosynthetic characteristics (Sousa et al., 2021). Furthermore, N application at moderately low levels (2.0 mM) could help in promoting plant growth during salt stress in rye grass by alleviating salt stress induced oxidative damage and reconstitution of N metabolism (Shao et al., 2020). Apart from ensuring physiological homeostasis, N supplementation also affects crop yield under salt stress. A significant improvement in wheat yield traits by application of N fertilizer during salt stress (Ibrahim et al., 2018). Supplementation of N at 200 mg N pot−1 ameliorates the detrimental effects of salt stress, which increased plant growth and yield through sustaining the integrity of the photosynthesis and chlorophyll fluorescence processes of oat plants (Song et al., 2019). NO3 - supplemented wheat and maize plants exhibited greater tolerance and performance as compared to the NH4 + fed plants under salt stress (Lewis et al., 1989). Additionally, NO3- fed Nerium oleander (oleander) grows faster with enhanced tolerance to salt stress than NH4 + fed plants. The higher salt tolerance of NO3 - fed plants was speculated to be due to decreased transport and accumulation of Na+ and Cl- in the shoots, however, accumulation of Na+ and Cl- in shoots in NH4 + fed plants may result in reduced plant growth. Furthermore, enhanced tolerance to salt stress has been observed in Chinese poplar exposed to NO3 - as compared to NH4 + as a source of N (Meng et al., 2016), possibly by influencing N uptake, metabolism and accumulation (amino acids, proteins and total N). On the contrary, Hessini et al. (2019) have revealed that NH4 + favoured the growth of maize plant under salt stress more than NO3 - by inducing the accumulation of inorganic salts and thereby improving the plant’s capacity to acclimatize to osmotic stress. Shaviv et al. (1990) reported that mixed NH4 +- NO3 - nutrition effectively mitigated the negative effects of salt stress in wheat, resulting in significantly higher dry matter, grain and protein yields as compared to NO3 - alone, which may be beneficial to growers desiring ways to maximize grain yield under salt stress conditions. Likewise, Flores et al. (2001) reported that combined NO3 -/NH4 + regime increase the rate of N assimilation, as well as the levels of Fe and chlorophyll in tomato plants, thereby mitigating salt injury. More research is required to understand the mechanism that regulates N responses. Despite the fact that omics approaches to understanding N-mediated salt stress acclimation responses have not been widely used, there are some reports indicating the interplay of N in plants during salt stress. For instance, transcriptomic analysis of cotton, soybean, alfalfa under salt stress revealed a number of differentially expressed genes (DEGs) related to N absorption (Postnikova et al., 2013; Bahieldin et al., 2015). Overexpression of Fd-NiR (ferredoxin-dependent assimilatory nitrite reductase) and GAD1 (glutamic acid decarboxylase) genes linked to N assimilation and glutamine/glutamine family of amino acids was significantly up-regulated in leaves of mulberry seedlings under salt stress, suggesting that these genes may be important mechanisms for mulberry to acclimatize to salt stress (Zhang et al., 2020a). Furthermore, omics (UPLC-MS and RNA-seq) analysis revealed that N metabolism related genes, encoding xanthin dehydrogenase (XDH), AspAT, AlaTA and allantoinase (ALN), and two hormones (melatonin and (S)-2-aminobutyric acid) in Beta vulgaris (sugar beet) were activated under salt stress, which might be attributed to the adaptive response of sugar beet to salt stress (Liu et al., 2020). These studies provides a valuable agronomic and molecular approaches for improving salt stress resistance in plants ( Table 2 ). Therefore, further research into the N mediated regulation of plant transcriptome, proteome as well as metabolome will aid in deciphering the N mediated molecular regulatory networks governing salt stress responses in plants, thereby providing new directions for developing salt stress tolerant plants.
Table 2.
Representative studies on exogenously- sourced nitrogen in regulating various plant processes for adaptation to salt stress.
| Plant | Salinity level | Source and level of N applied | Plant responses | References | |
|---|---|---|---|---|---|
| Mung bean (Vigna radiata) | 75 mM | Nitrogen fertilizer | 60 kg ha-1 | Improved water relation, accumulation of nutrients and yield | Kabir et al., 2005 |
| Sorghum (Sorghum bicolor) | 0.6, 6, 8 dS m-1 | Urea and ammonium nitrate | 137 kg ha-1 | Increase uptake of N, P, Ca Mg and decreased Na+ and Cl- concentration | Esmaili et al., 2008 |
| Cotton (Gossypium hirsutum) | 7.7, 12.5 dS m-1 | Urea | 135 and 270 kg ha−1/ | Enhance N uptake and alleviates adverse effects of salinity | Chen et al., 2008 |
| Maize (Zea mays) | 12.4 dSm−1 | Urea and Nitrate | 100 kg ha−1 | Improved nitrogen use efficiency | Irshad et al., 2008 |
| Suaeda physo phora | 75 mmol L−1 | Nitrate | 10 mM L-1 | Increased root:shoot ratio, effective N uptake in shoots and lateral root development | Jun-Feng et al., 2010 |
| Seep weed (Suaeda salsa) | 42 g | Urea | 4.8 g | Increased the biomass of leaf, shoot and root, regulation of Na+, K+ and Cl- accumulation | Guan et al., 2011 |
| Wheat | 100, 150 and 200 mM | Nitrogen fertilizer | 86 and 210 kg ha−1 | Improved germination percentage, shoot and root length, dry weight, salt tolerance index, and seedling vigor index | Ibrahim et al., 2016 |
| Mustard (Brassica juncea) | 50 mM | Nitrogen fertilizer | 100 mM | Enhanced antioxidant system | Naveed et al., 2019 |
| Wheat (Triticum aestivum) | 1.4 and 2.5 dS m−1 | Urea | 86 kg ha-1 | Accumulation of antioxidant enzymes, improved seedling growth | Ibrahim et al., 2019 |
| Oat (Avena sativa) | 100 mM | Nitrogen fertilizer | 50,100, 200 kg ha−1 | Improved photosynthetic rates, grain yield and other yield components | Song et al., 2019 |
| Wheat | 100 mM | Urea | 50 and 100 mg kg− 1 soil | Increased osmolyte and secondary metabolite accumulation, and redox components in N supplemented plants regulated the ROS metabolism and salt tolerance | Ahanger et al., 2019 |
| Rice (Oryza sativa) | 2.5-3.5 dS m-1 | Urea | 300 kg ha-1 | Enhanced antioxidant system and solube sugars content, and improved grain yield | Zhu et al., 2020 |
| Regrass (Lolium multiflorum) | 50 and 100 mM | Nitrogen fertilizer | 2.0 mM | Promoted growth via regulating photosynthesis, alleviating ROS-induced damage and maintenance of N metabolism. | Shao et al., 2020 |
| Gossypium hirsutum (cotton) | 200 mm.L-1 | Nitrate | 2.5 mmol L−1 | Enhanced the accumulation of osmolytes, such as soluble sugars, soluble proteins, and free amino acids. | Sikder et al., 2020 |
| Hybrid corn (AG 1051), | 3.0 dS m-1 | Nitrogen fertilizer | 80 and 160 kg ha-1 | Increased plant height, leaf area, photosynthesis, transpiration, and conductance | Sousa et al., 2021 |
Crosstalk of nitrogen with phytohormones to induce salt stress tolerance
Phytohormones, besides controlling plant growth and development under normal conditions, may also play a critical role in how plants respond to salt stress, which could lead to increased plant growth and productivity (Yu et al., 2020). Both N and phytohormones interact in a complex system with diverse synergistic and antagonistic interactions, resulting in plant responses under normal and salt stress conditions ( Figure 3 ). For example, Vigna radiata (mungbean) plants sprayed with indole-3-acetic acid (IAA), gibberellic acid (GA) and kinetin (CK) ranging from 0.1-10 µM significantly increased the activity of key enzymes of N metabolism (GS, GOGAT and GDH) along with minimizing the effects of salt stress in plants (Chakrabarti and Muhkerji, 2003). Thus, it is possible that there is a complex crosstalk between N, IAA, GA and CK that modulates salt stress-induced mitigation responses in plants that has yet to be comprehended. Application of N (90 and 135 kg N ha-1) or GA3 (144.3 and 288.7 μM) alone or in combination reversed the inhibitory effect of salt stress in sorghum, making it a useful approach for maintaining cellular homeostasis, photosynthesis, lowering oxidative damage, and improving and managing overall crop productivity under salt stress (Ali et al., 2021). Similarly, application of N (40 mg N kg-1) or GA3 (10-5 M) alone or in combination proved to be a physiological remedy to enhance the resistance against the adverse effects of salt stress in mustard (Siddiqui et al., 2008), thus indicating that a more complex N–GA interaction may be involved in salt stress responses. The crosstalk of N with ethylene is also important in plant cellular metabolism, and it has the capability to deal with salt stress. In mustard, cumulative treatment of ethephon with split doses of N and sulphur (S) mitigated salt-induced damages by improving photosynthetic efficiency, assimilation of N and S, proline content and antioxidant defense system and decreased generation of oxidative stress markers (Jahan et al., 2021). In another study, foliar spraying of ethylene (2-chlorethyl-phosphonic acid) to salt- exposed Pennisetum typhoides (pearl millet) seedlings ameliorated salt- induced toxicity mainly by increasing the activity of several enzymes associated with N metabolism as well as improving protein synthesis and proline levels (Eder et al., 1977), which could play a significant role in sustaining crop productivity and nutritional quality. Moreover, ethephon have been found to act synergistically with N to improve photosynthesis in mustard by modulating the pNUE and antioxidant metabolism under salt stress conditions (Iqbal et al., 2017). An interaction between N and ethylene along with proline was also revealed in salt- exposed mustard, where N supplementation significantly increased photosynthetic efficiency mainly by regulating proline and ethylene production (Iqbal et al., 2015). More research is needed to gain a better understanding of N and ethylene-mediated salt stress alleviation. Furthermore, a functional interaction between abscisic acid (ABA) and N in mitigating salt stress resilience has been reported in mustard, where ABA (25 µM) in coordination with N (10 mM) ameliorates salt- inhibited photosynthetic efficiency by increasing the activity of antioxidant enzymes and accumulation of proline, which maintains osmotic balance and reduces oxidative stress (Majid et al., 2021). However, more researches are required to promote the differential roles of N and ABA under salt stress, which could aid in improving plant resilience traits. The cumulative treatment of N supplementation (3 mM) and seed priming with 24- epibrassinolide (24-EBL;100 mM) significantly enhanced salt stress tolerance in soybean plants by increasing antioxidant activity and osmolyte accumulation, resulting in improved photo-protection through maintenance of tissue water content (Soliman et al., 2020). Furthermore, the application of EBL (10-7 M and 0.5 × 10-9 M) to pigeon pea reduced the adverse effects of salt stress by increasing NO3 - uptake, which was associated with increased activity of NR along with increased levels of free amino acids and soluble proteins in salt- stressed plant roots, showing the significance of brassinosteroid (BR) synthesis for plants growing under salt stress (Dalio et al., 2013). In another report, co- application of EBL and sodium nitroprusside (SNP) along with N and ABA was more effective than their individual treatments in decreasing salt- induced oxidative stress in mustard (Gupta et al., 2017). The positive effect was accomplished by reducing Na+ acquisition, oxidative stress and increasing proline content. Thus, N appears to have an intriguing crosstalk with BRs in modulating salt stress amelioration. Salicylic acid (SA) and N are also implicated in an intriguing crosstalk that regulates plant metabolism and can mitigate the negative impacts of salt stress. For instance, SA and N prevented salt- induced adversities in two mungbean cultivars by improving photosynthetic efficiency, as well as modulating N, S and antioxidant metabolism (Nazar et al., 2011). It has been revealed that treatment of SA (0.5 mM) improved the efficacy of salt- stressed mungbean plants by increasing the activity of N assimilatory enzymes, proline accumulation and defense system, while decreasing the content of Na+ and Cl- ions, resulting in increased photosynthesis and growth (Hussain et al., 2021). Furthermore, Yang et al. (2019) explored the role of methyl jasmonate (MeJA) in mitigating salt- induced damages in Glycyrrhiza uralensis (Chinese liquorice) mainly by increasing the activity antioxidant defense system and modulating N-metabolism, which in turn regulates the survival of these plants under salt- stress conditions. Thus, N has gained popularity as an essential macro element capable of inducing salt stress-adaptation in plants. It mainly revitalises the reservoir of antioxidants, necessary to detoxify the toxic reactive oxygen species (ROS) produced under salt stress. Given the importance of N as a major nutrient in agricultural systems, and its crosstalk with different phytohormones, it can be further utilised to strengthen the production of more tolerant varieties to salt stress. To date, there is scant data that elucidates the potential crosstalk of N and phytohormones-triggered responses under salt stress. More in-depth and convincing research is needed at the cellular and molecular levels under salt stress, to explore the intrinsic signalling mechanism of N-phytohormone interactions that would provide new insights into the complex signalling pathways, which could be used to increase the yield and stress resilience under today’s changing environment. These findings will benefit not only plant scientists but also breeders and will open up new avenues of research into plant responses to salt stress.
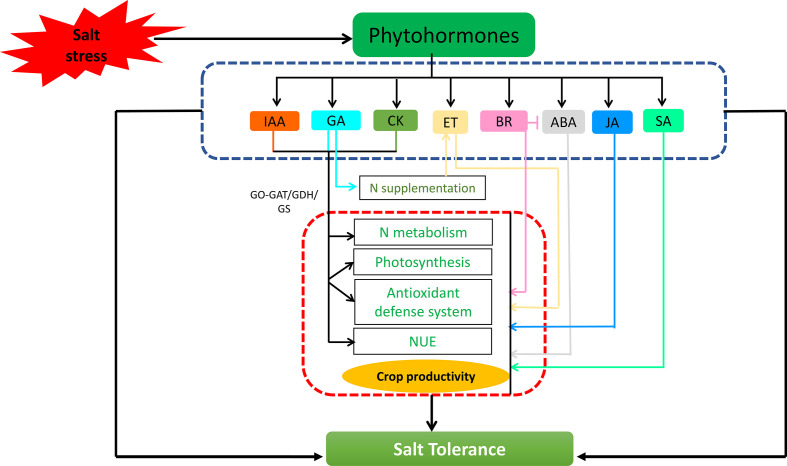
Figure 3.
Phytohormones have been found to regulate nitrogen metabolism during salt stress, either in conjugation with nitrogen sources or by interacting with other phytohormones leading to improved physio-biochemical attributes, and promoting plant growth and development under salt stress.
Manipulation of nitrogen dynamics through genetic engineering
Engineering metabolic pathways by reinforcement of gene identification associated with N metabolism and modifying their key genes expression have emerged as an important factor for ameliorating constraints from crop nutrition and for improving crop quality. Generating transgenic crops by modulating the expression of genes involved in uptake, transport and distribution of N can help in maintaining N homeostasis and promoting plant growth under normal or stressful conditions ( Figure 4 ). For instance, overexpression of OsAMT1;1 in transgenic rice greatly improved N uptake, translocation capacity, and aided in the maintenance of N homeostasis, resulting in higher NUE, amino acid levels, photosynthetic pigments, and sugars, as well as increased grain yield, particularly under both suboptimal and optimal N fertilizer environments (Ranathunge et al., 2014). Likewise, simultaneous activation of the OsAMT1;2 and glutamate synthetase 1 (OsGOGAT1) genes in rice serves as an efficient breeding strategy to improve plant growth, NUE, and grain yield, particularly under N limitation, by improving both N uptake and utilization (Lee et al., 2020). Overexpression of the high-affinity urea transporter gene, OsDUR3 (from rice) improves urea acquisition and allocation in transgenic Arabidopsis roots (Wang et al., 2012b). Under N-deficit environments, overexpression of maize Dof1, an activator of genes involved in organic acid metabolism improves plant growth and amino acid levels (Gln and Glu), decreases glucose levels, improves N-assimilation, and enhances N-content in Arabidopsis (Yanagisawa et al., 2004). ZmDof1 overexpression enhances N uptake, assimilation, and N and C levels in transgenic rice, resulting in increased photosynthetic rate and biomass in transgenic rice (Kurai et al., 2011). The high-affinity nitrate transporter gene (OsNRT2.1) has been widely used for the production and remodelling of transgenic rice with optimized NO3 - utilization, which is critical for maintaining crop yield under low N conditions (Katayama et al., 2009). Additionally, under both limited and sufficient N conditions, overexpression of TaNRT2.1-6B (dual-affinity NO3 - transporter) increased N influx, root growth, SPAD value, grain yield, and NUE in wheat, whereas gene silence lines had the inverse effects (Li et al., 2022a). Similarly, in Hordeum vulgare (barley), overexpression of the high affinity NO3 - transporter, OsNRT2.3b under the influence of ubiquitin promoter optimized yield, NUE, and regulation of nutrient uptake and homeostasis (Luo et al., 2020).
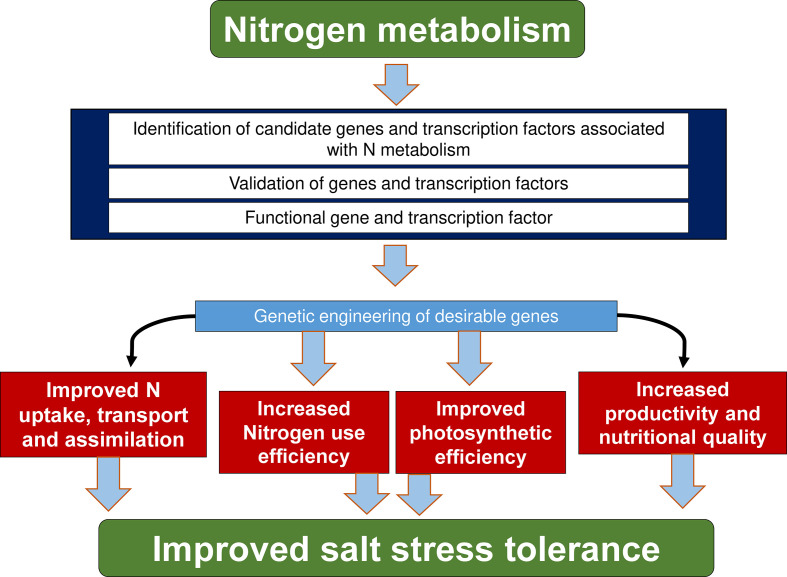
Figure 4.
Specific targets for manipulating nitrogen dynamics through genetic engineering. Nitrogen dynamics is mainly associated with higher expression of nitrate and ammonium transporter genes as well as transcription factors, and the overexpression of these genes in transgenic plants leads to increased nitrogen use efficiency, photosynthesis, crop productivity and nutritional quality, nitrate and ammonium uptake and salt-stress resistance.
Under salt stress condition, transgenic rice plant over-expressing GS2 (chloroplastic glutamine synthetase) had enhanced photorespiration capacity and salt tolerance. This study decoded the role of GS2 in maintaining intracellular K+ and Na+ homeostasis, which has important implications for plant response to salt stress (Hoshida et al., 2000). Transcriptomic studies revealed that genes associated with N metabolism (NR, NIR) were substantially up- regulated in seagrass (Zostera marina) under salt stress, suggesting this may be an adaptation for NO3- absorption, transport and assimilation in a high salt environment (Lv et al., 2018). In addition, the genes (NR) and metabolites (amino acids) related to N metabolism have been found to be strongly regulated by salt stress in tomato (Mellidou et al., 2021). Concurrent overexpression of OsGS1;1 and OsGS2 isoforms in rice improved photosynthetic and agronomic performance under salt stress during the reproductive stage (James et al., 2018). Furthermore, most NRTs were found to be overexpressed in Sophora japonica (Japanese pagoda tree) under salt stress, allowing the root to maintain high NO3-uptake ability (Tian et al., 2022). Moreover, BjNRT1.3, BjNRT1.4 and BjNRT1.8 were up-regulated under salt (1h, 24h) and osmotic (1h) stress in mustard while the down-regulation of BjNRT1.1, BjNRT1.5, BjNRT2.1, BjAMT1.2 and BjAMT2 after stress exposure, suggesting their involvement in plant tolerance mechanism to salt stress (Goel and Singh, 2015). Expression of splice variant of OsNRT2.3 (OsNRT2.3b) has been identified for mediating cytosol pH sensing and balancing the uptake of NO3- and NH4 + in response to external perturbations of N supplies (Fan et al., 2016). Surprisingly, the high affinity tomato (SlNRT2) gene has been found to be involved in facilitating adaptive responses in plants exposed to salt stress (Akbudak et al., 2022). It has been revealed that salt stress induces the expression levels of both GLN-1.1 and GLN-1.2 in Arabidopsis roots probably due to higher demand of GS or other N-metabolism related enzymes required by the plant as an adaptation against higher level of salt stress (Debouba et al., 2013). Recently, Sathee et al. (2021) have shown that rice genotypes differing in reproductive stage salt tolerance have different transcript abundance of NIA2, GLN2, GLN1.1 and GLN1.2, Fd-GOGAT and NADH-GOGAT genes; the expression levels was found higher in salt tolerant than salt sensitive cultivar, suggesting its protective role against salt-induced oxidative damage. Ectopic expression of MdNIA2 from Malus domestica (apple) in Arabidopsis and apple callus elevated the NUE and increased root hair elongation and formation, resulting in promoted plant growth and salt stress tolerance (Liu et al., 2023). Thus, using transgenic strategies to manipulate the N status of plants has aided in plant growth, increased N content, uptake, and remobilization for combating food insecurity, and improved productivity to foster sustainable agriculture programmes.
In recent times, researchers have uncovered a plethora of transcription factors (TFs) that regulate the transcription of targeted genes involved in absorption, redistribution, and assimilation of N. The majority of these TFs have been used to boost NUE in crop plants. The NAC superfamily is one of the most extensive and critical plant-specific TF families. For instance, NO3 −-inducible NAC TF (TaNAC2-5A) was identified in wheat, binding to the promoter region of genes involved in N transport and assimilation, its overexpression improved root growth, NO3 − influx rate, grain yield, and thus increased the root’s capacity to accumulate N under N- limited environments (He et al., 2015). Similarly, OsNAP (an apetala3/pistillata-activated NAC-like), a member of the NAC TF family, is regulated by ABA and mediates age-related senescence (Liang et al., 2014). This TF binds to the promoter region of the nutrient transporter and regulates N re-translocation for grain filling in rice leaves. Overexpression of OsNAP increased grain N content in rice, its knockdown reduced grain yields with impaired leaf senescence (Liang et al., 2014). Furthermore, salt stress induced the expression of NO3 - responsive transcript factor, OsNLP2 in rice during early stage of seed germination, which was accompanied by an increase in OsNR1/2 and NR activity, resulting in improved salt tolerance (Yi et al., 2022). Similarly, another nitrate responsive TF (OsMADS27), positively regulates salt tolerance in rice in a NO3 – dependent manner by regulating salt-responsive genes, maintaining nutrient homeostasis and modulating the expression of genes related to N uptake and assimilation (Alfatih et al., 2022). OsMYB305, a MYB transcription factor, is mainly expressed in young rice panicles, but it is also expressed in roots when N levels are low. Rice plants overexpressing this TF exhibited increased plant growth, more tiller number and enhanced N uptake and assimilation than WT. The increased NO3 - uptake in the overexpression lines, associated with the increased expression of N-related genes (OsNRT2.1, OsNRT2.2, OsNAR2.1, and OsNIR2) in roots suggests that OsMYB305 could be a potential candidate for rice NUE improvement (Wang et al., 2020). Higher NUE of indica than japonica is associated with a natural transformation of OsMYB61, whose expression is restrained by growth regulatory factor 4 (GRF4), a C/N metabolic regulator in rice (Gao et al., 2020). OsMYB61 expression was induced by a low supply of N, and this effect was more pronounced in the indica than in the japonica subspecies. Transcription factors belonging to basic leucine zipper (bZIP) family play an intrinsic part in the manipulation of N dynamics and resilience to abiotic stresses (Yang et al., 2019). Overexpression of TabZIP15 in transgenic wheat plants improves salt stress tolerance and root development (Bi et al., 2021). AtbZIP24 increased salt tolerance by maintaining osmotic balance, amino acid levels (Gln and Glu) and increased growth and development, involving homo- and heterodimerization, or post-transcriptional modification (Yang et al., 2009). Other TFs, rice Nin-Like Protein 1 (OsNLP1) have also been implicated in N utilization (Alfatih et al., 2020). Over-expression of this gene was found to be significant in improving rice yield and NUE under diverse N environments, whereas knockout of the gene with the CRISPR/Cas9 system exerted negative impacts on crop productivity at low as well as under normal N conditions, but no significant change was observed at high N levels. Overexpression of FtWRKY46 derived from Tartary buckwheat in Arabidopsis increased salt tolerance by regulating ROS detoxification and stress-related gene expression (Lv et al., 2020). Reaumuria trigyna derived RtWRKY1 confers tolerance to salt stress in transgenic Arabidopsis by regulating plant growth, N-metabolism, osmotic balance, Na+/K+ homeostasis, and the antioxidant system (Du et al., 2017). Thus, the aforementioned studies opens up new possibilities and fill a gap in our understanding of the dynamics of NUE and crop productivity. More convincing research is needed to support the differential role of genes and TFs encoding NH4+ and NO3- assimilation enzymes that regulates N metabolism in plants under salt stress.
Conclusion and future prospective
In conclusion, salt stress poses a serious threat to soil fertility and productivity of agriculture. Correct plant mineral nutrition has often been shown to have a protective role in bestowing salt stress tolerance and in sustaining crop production. Here, we provided convincing evidence for N-mediated salt stress tolerance in plants by up-regulating salt stress tolerance genes. The use of N as fertilizer or soil supplementation with N-containing compounds in agriculture system may emerge as a useful tool for combating salt-induced effects and regulating N metabolism and NUE under salt stress conditions. There are still some gaps in our understanding of crosstalk of N with phytohormones that need to be filled in order to improve salt stress tolerance. Despite significant progress, there is an urgent need to develop more understanding of N-dynamics in the crop improvement against salt stress. The identification and analysis of key genes would be effective in N-mediated salt tolerance, providing a new framework for developing crop varieties with improved NUE to meet the requirements for salt stress tolerance and sustainable agriculture. Furthermore, the advent of genetic engineering technology holds significant promise for crop improvement and stress management.
Author contributions
Conceptualization, MIRK. Software, FN and FA. Writing- Original draft preparation, FN, FA, HC and PC. Writing – Review and editing, MIRK, FN, MM, SK and MA. All authors contributed to the article and approved the submitted version.
Acknowledgments
MIRK is gratefully acknowledging the SERB-DST grant (SRG/2020/001004). FN acknowledges DBT –RA (No. DBT-RA/2022/January/N/1186).
Glossary
| AAPs | amino acid permeases |
| AMT | ammonium transporter |
| APX | ascorbate peroxidase |
| BABA | β-aminobutyric acid |
| CLC | chloride channel family |
| DEG | differentially expressing genes |
| GOGAT | glutamate synthase |
| GS | glutamine synthetase |
| G6PHD | glucose-6-phosphate dehydrogenase |
| LHTs | lysine/histidine transporters |
| NAC | NAM (no apical meristem), Petunia, ATAF1–2 (Arabidopsis thaliana activating factor), and CUC2 (cup-shaped cotyledon, Arabidopsis) |
| NCC | nitrogen containing compounds |
| NH4 + | ammonia |
| NiR | nitrite reductase |
| NO2 - | nitrite |
| NO3 - | nitrate |
| NR | nitrate reductase |
| NRT2 | nitrate transporter 2 |
| NUE | nitrogen use efficiency |
| NRE | nitrogen recovery efficiency |
| ProTs | proline transporters, GBTs, glycine betaine transporters |
| PTR | peptide transporter family |
| QAC | quaternary ammonium compounds |
| SLAC1 | slow anion channel homologs |
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Abouelsaad I., Weihrauch D., Renault S. (2016). Effects of salt stress on the expression of key genes related to nitrogen assimilation and transport in the roots of the cultivated tomato and its wild salt-tolerant relative. Sci. Hortic. 211, 70–78. doi: 10.1016/j.scienta.2016.08.005 [DOI] [Google Scholar]
- Ahanger M. A., Agarwal R. M. (2017). Salinity stress induced alterations in antioxidant metabolism and nitrogen assimilation in wheat (Triticum aestivum l.) as influenced by potassium supplementation. Plant Physiol.Biochem. 115, 449–460. doi: 10.1016/j.plaphy.2017.04.017 [DOI] [PubMed] [Google Scholar]
- Ahanger M. A., Qin C., Begum N., Maodong Q., Dong X. X., El-Esawi M., et al. (2019). Nitrogen availability prevents oxidative effects of salinity on wheat growth and photosynthesis by up-regulating the antioxidants and osmolytes metabolism, and secondary metabolite accumulation. BMC Plant Biol.19 1), 1–12. doi: 10.1186/s12870-019-2085-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbudak M. A., Filiz E., Çetin D. (2022). Genome-wide identification and characterization of high-affinity nitrate transporter 2 (NRT2) gene family in tomato (Solanum lycopersicum) and their transcriptional responses to drought and salinity stresses. J. Plant Physiol. 272, 153684. doi: 10.1016/j.jplph.2022.153684 [DOI] [PubMed] [Google Scholar]
- Alfatih A., Wu J., Zhang Z. S., Xia J. Q., Jan S. U., Yu L. H., et al. (2020). Rice NIN-LIKE PROTEIN 1 rapidly responds to nitrogen deficiency and improves yield and nitrogen use efficiency. J. Exp. Bot. 71 (19), 6032–6042. doi: 10.1093/jxb/eraa292 [DOI] [PubMed] [Google Scholar]
- Alfatih A., Zhang J., Song Y., Jan S. U., Zhang Z. S., Xia J. Q., et al. (2022). Nitrate-responsive OsMADS27 promotes salt tolerance in rice. Plant Commun. 100458. doi: 10.1016/j.xplc.2022.100458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali A. Y. A., Ibrahim M. E. H., Zhou G., Nimir N. E. A., Elsiddig A. M. I., Jiao X., et al. (2021). Gibberellic acid and nitrogen efficiently protect early seedlings growth stage from salt stress damage in sorghum. Sci. Rep.11 1), 1–11. doi: 10.1038/s41598-021-84713-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almagro A., Lin S. H., Tsay Y. F. (2008). Characterization of the arabidopsis nitrate transporter NRT1. 6 reveals a role of nitrate in early embryo development. Plant Cell 20 (12), 3289–3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Aragón R., Rodríguez-Navarro A. (2017). Nitrate-dependent shoot sodium accumulation and osmotic functions of sodium in arabidopsis under saline conditions. Plant J. 91 (2), 208–219. doi: 10.1111/tpj.13556 [DOI] [PubMed] [Google Scholar]
- Ashfaque F., Khan M. I. R., Khan N. A. (2014). Exogenously applied H2O2 promotes proline accumulation, water relations, photosynthetic efficiency and growth of wheat (Triticum aestivum l.) under salt stress. Annu. Res. Rev. 4, 105–120. doi: 10.9734/ARRB/2014/5629 [DOI] [Google Scholar]
- Aslam M., Ahmad K., Akhtar M. A., Maqbool M. A. (2017). Salinity stress in crop plants: effects of stress, tolerance mechanisms and breeding strategies for improvement. J. Agric. Basic Sci. 2 (1), 2518–4210. [Google Scholar]
- Averina N. G., Beyzaei Z., Shcherbakov R. A., Usatov A. V. (2014). Role of nitrogen metabolism in the development of salt tolerance in barley plants. Russ. J. Plant Physiol. 61 (1), 97–104. doi: 10.1134/S1021443713060022 [DOI] [Google Scholar]
- Bahieldin A., Atef A., Sabir J. S., Gadalla N. O., Edris S., Alzohairy A. M., et al. (2015). RNA-Seq analysis of the wild barley (H. spontaneum) leaf transcriptome under salt stress. C.R. Biol. 338 (5), 285–297. [DOI] [PubMed] [Google Scholar]
- Beatty P. H., Shrawat A. K., Carroll R. T., Zhu T., Good A. G. (2009). Transcriptome analysis of nitrogen-efficient rice over-expressing alanine aminotransferase. Plant Biotech. 7 (6), 562–576. doi: 10.1111/j.1467-7652.2009.00424.x [DOI] [PubMed] [Google Scholar]
- Bi C., Yu. Y., Dong C., Yang Y., Zhai Y., Du F., et al. (2021). The bZIP transcription factor TabZIP15 improves salt stress tolerance in wheat. Plant Biotechnol. J. 19 (2), 209. doi: 10.1111/pbi.13453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindel N., Neuhäuser B. (2021). High-affinity ammonium transport by Arabidopsis thaliana AMT1; 4. Acta Physiol. Plant 43 (4), 1–5. [Google Scholar]
- Bonnave M., Bertin P. (2018) Evaluation of nitrogen use efficiency (NUE) of rice (O. sativa) under saline conditions. Available at: http://hdl.handle.net/2078.1/.
- Borzouei A., Eskandari A., Kafi M., Mousavishalmani A., Khorasani A. (2014). Wheat yield, some physiological traits and nitrogen use efficiency response to nitrogen fertilization under salinity stress. Indian J. Plant Physiol. 19 (1), 21–27. doi: 10.1007/s40502-014-0064-0 [DOI] [Google Scholar]
- Bu Y., Takano T., Liu S. (2019). The role of ammonium transporter (AMT) against salt stress in plants. Plant Signal. Behav.14 8), 1625696. doi: 10.1080/15592324.2019.1625696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldana C., Li Y., Leisse A., Zhang Y., Bartholomaeus L., Fernie A. R., et al. (2013). Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana . Plant J. 73 (6), 897–909. doi: 10.1111/tpj.12080 [DOI] [PubMed] [Google Scholar]
- Campbell W. H. (1999). Nitrate reductase structure, function and regulation: Bridging the gap between biochemistry and physiology. Annu. Rev. Plant Biol. 50 (1), 277–303. doi: 10.1146/annurev.arplant.50.1.277 [DOI] [PubMed] [Google Scholar]
- Chakrabarti N., Mukherji S. (2003). Effect of phytohormone pretreatment on nitrogen metabolism in Vigna radiata under salt stress. Biol.Plant. 46 (1), 63–66. doi: 10.1023/A:1022358016487 [DOI] [PubMed] [Google Scholar]
- Chakraborty K., Basak N., Bhaduri D., Ray S., Vijayan J., Chattopadhyay K., et al. (2018). “Ionic basis of salt tolerance in plants: Nutrient homeostasis and oxidative stress tolerance,” in Plant nutrients and abiotic stress tolerance (Singapore: Springer; ), 325–362. [Google Scholar]
- Chen H., Zhang Q., Wang X., Zhang J., Ismail A. M., Zhang Z. (2021). Nitrogen form- mediated ethylene signal regulates root-to-shoot k+ translocation via NRT1. 5. Plant Cell Environ. 44 (12), 3806–3818. doi: 10.1111/pce.14182 [DOI] [PubMed] [Google Scholar]
- Chen Y., Schmelz E. A., Wäckers F., Ruberson J. R. (2008). Cotton plant, Gossypium hirsutum l., defense in response to nitrogen fertilization. J.Chem.Ecol. 34 (12), 1553- 1564. [DOI] [PubMed] [Google Scholar]
- Cheng L., Yuan H. Y., Ren R., Zhao S. Q., Han Y. P., Zhou Q. Y., et al. (2016). Genome-wide identification, classification, and expression analysis of amino acid transporter gene family in Glycine max . Front. Plant Sci. 7, 515. doi: 10.3389/fpls.2016.00515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C. C., Lin C. S., Hsia A. P., Su R. C., Lin H. L., Tsay Y. F. (2004). Mutation of a nitrate transporter, AtNRT1: 4, results in a reduced petiole nitrate content and altered leaf development. Plant Cell Physiol. 45 (9), 1139–1148. doi: 10.1093/pcp/pch143 [DOI] [PubMed] [Google Scholar]
- Chopin F., Orsel M., Dorbe M. F., Chardon F., Truong H. N., Miller A. J., et al. (2007). The arabidopsis ATNRT2. 7 nitrate transporter controls nitrate content in seeds. Plant Cell. 19 (5), 1590–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho D. G., Miranda R. D. S., Paula-Marinho S. D. O., de Carvalho H. H., Prisco J. T., Gomes-Filho E. (2020). Ammonium nutrition modulates k+ and n uptake, transport and accumulation during salt stress acclimation of sorghum plants. Arch. Agron. Soil Sci. 66 (14), 1991–2004. doi: 10.1080/03650340.2019.1704736 [DOI] [Google Scholar]
- Couturier J., Montanini B., Martin F., Brun A., Blaudez D., Chalot M. (2007). The expanded family of ammonium transporters in the perennial poplar plant. New Phytol. 174 (1), 137–150. doi: 10.1111/j.1469-8137.2007.01992.x [DOI] [PubMed] [Google Scholar]
- D'Apuzzo E., Rogato A., Simon-Rosin U., El Alaoui H., Barbulova A., Betti M., et al. (2004). Characterization of three functional high-affinity ammonium transporters in Lotus japonicus with differential transcriptional regulation and spatial expression. Plant Physiol. 134 (4), 1763–1774. doi: 10.1104/pp.103.034322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalio R. J. D., Pinheiro H. P., Sodek L., Haddad C. R. B. (2013). 24-epibrassinolide restores nitrogen metabolism of pigeon pea under saline stress. Bot. Stud. 54 (1), 1–7. doi: 10.1186/1999-3110-54-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debouba M., Dguimi H. M., Ghorbel M., Gouia H., Suzuki A. (2013). Expression pattern of genes encoding nitrate and ammonium assimilating enzymes in Arabidopsis thaliana exposed to short term NaCl stress. J. Plant Physiol.170 2), 155–160. doi: 10.1016/j.jplph.2012.09.011 [DOI] [PubMed] [Google Scholar]
- Decui Y., Qing X., Fanggui Z., Mian L., Xin L. (2020). Effects of NaCl stress on nitrogen metabolism and related gene expression of rice with different resistance. Acta Agric. Bor. Sin. 35, 187–195. [Google Scholar]
- de la Torre-González A., Navarro-León E., Blasco B., Ruiz J. M. (2020). Nitrogen and photorespiration pathways, salt stress genotypic tolerance effects in tomato plants (Solanum lycopersicum l.). Acta Physiol. Plant 42 (1), 1–8. [Google Scholar]
- de Los Ríos L. M., Corratgé-Faillie C., Raddatz N., Mendoza I., Lindahl M., de Angeli A., et al. (2021). The arabidopsis protein NPF6. 2/NRT1. 4 is a plasma membrane nitrate transporter and a target of protein kinase CIPK23. Plant Physiol. Biochem. 168, 239–251. doi: 10.1016/j.plaphy.2021.10.016 [DOI] [PubMed] [Google Scholar]
- Diédhiou C. J., Golldack D. (2006). Salt-dependent regulation of chloride channel transcripts in rice. Plant Sci.170 4), 793–800. doi: 10.1016/j.plantsci.2005.11.014 [DOI] [Google Scholar]
- Dobrenel T., Caldana C., Hanson J., Robaglia C., Vincentz M., Veit B., et al. (2016). TOR signaling and nutrient sensing. Annu. Rev. Plant Biol. 67, 261–285. doi: 10.1146/annurev-arplant-043014-114648 [DOI] [PubMed] [Google Scholar]
- Du C., Zhao P., Zhang H., Li N., Zheng L., Wang Y. (2017). The reaumuria trigyna transcription factor RtWRKY1 confers tolerance to salt stress in transgenic arabidopsis. J. Plant Physiol. 215, 48–58. doi: 10.1016/j.jplph.2017.05.002 [DOI] [PubMed] [Google Scholar]
- Du R. J., Wu Z. X., Yu Z. X., Li P. F., Mu J. Y., Zhou J., et al. (2022). Genome- wide characterization of high-affinity nitrate transporter 2 (NRT2) gene family in Brassica napus . Int. J. Mol. Sci. 23 (9)4965. doi: 10.3390/ijms23094965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder A., Huber W., Sankhla N. (1977). Interaction between salinity and ethylene in nitrogen metabolism of Pennisetum typhoides seedlings. Biochem. Physiol. Pflanz. 171, 93–100. doi: 10.1016/S0015-3796(17)30279-2 [DOI] [Google Scholar]
- Ertani A., Schiavon M., Muscolo A., Nardi S. (2013). Alfalfa plant-derived biostimulant stimulate short-term growth of salt stressed Zea mays l. plants. Plant Soil. 364 (1), 145- 158. doi: 10.1007/s11104-012-1335-z [DOI] [Google Scholar]
- Esmaili E., Kapourchal S. A., Malakouti M. J., Homaee M. (2008). Interactive effect of salinity and two nitrogen fertilizers on growth and composition of sorghum. Plant Soil Environ. 54 (12), 537–546. doi: 10.17221/425-PSE [DOI] [Google Scholar]
- Fan T. F., Cheng X. Y., Shi D. X., He M. J., Yang C., Liu L., et al. (2017). Molecular identification of tobacco NtAMT1. 3 that mediated ammonium root-influx with high affinity and improved plant growth on ammonium when overexpressed in arabidopsis and tobacco. Plant Sci. 264, 102–111. doi: 10.1016/j.plantsci.2017.09.001 [DOI] [PubMed] [Google Scholar]
- Fan X., Tang Z., Tan Y., Zhang Y., Luo B., Yang M., et al. (2016). Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Procd. Natl. Acad. Sci. 113 (26), 7118–7123. doi: 10.1073/pnas.1525184113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L., Yang T., Zhang Z., Li F., Chen Q., Sun J., et al. (2018). Identification and characterization of cationic amino acid transporters (CATs) in tea plant (Camellia sinensis). Plant Growth Regul. 84 (1), 57–69. doi: 10.1007/s10725-017-0321-0 [DOI] [Google Scholar]
- Flores P., Carvajal M., Cerdá A., Martínez V. (2001). Salinity and ammonium/nitrate interactions on tomato plant development, nutrition, and metabolites. J. Plant Nutr. 24 (10), 1561–1573. doi: 10.1081/PLN-100106021 [DOI] [Google Scholar]
- Fuertes-Mendizábal T., Bastías E. I., González-Murua C., González-Moro M. B. (2020). Nitrogen assimilation in the highly salt-and boron-tolerant ecotype Zea mays l. amylacea. Plants 9 (3), 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Liu M., Wang M., Shen Q., Guo S. (2016). Enhanced salt tolerance under nitrate nutrition is associated with apoplast na+ content in canola (Brassica napus l.) and rice (Oryza sativa l.) plants. Plant Cell Physiol. 57 (11), 2323–2333. [DOI] [PubMed] [Google Scholar]
- Gao S., Liu K. T., Chung T. W., Chen F. (2013). The effects of NaCl stress on jatropha cotyledon growth and nitrogen metabolism. J. Soil Sci. Plant Nutr.13 1), 99–113. doi: 10.4067/S0718-95162013005000010 [DOI] [Google Scholar]
- Gao Y., Xu Z., Zhang L., Li S., Wang S., Yang H., et al. (2020). MYB61 is regulated by GRF4 and promotes nitrogen utilization and biomass production in rice. Nat. Commun. 11 (1), 1–12. doi: 10.1038/s41467-020-19019-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisseler D., Ortiz R. S., Diaz J. (2022). Nitrogen nutrition and fertilization of onions (Allium cepa l.)–a literature review. Sci. Hortic. 291, 110591. doi: 10.1016/j.scienta.2021.110591 [DOI] [Google Scholar]
- Giehl R. F., Laginha A. M., Duan F., Rentsch D., Yuan L., von Wirén N. (2017). A critical role of AMT2; 1 in root-to-shoot translocation of ammonium in arabidopsis. Mol. Plant 10 (11), 1449–1460. doi: 10.1016/j.molp.2017.10.001 [DOI] [PubMed] [Google Scholar]
- Goel P., Singh A. K. (2015). Abiotic stresses downregulate key genes involved in nitrogen uptake and assimilation in Brassica juncea l. PloS One.10 11), e0143645. doi: 10.1371/journal.pone.0143645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C., Song A., Zhang X., Wang H., Li T., Chen Y., et al. (2016). Cloning of chrysanthemum high-affinity nitrate transporter family (CmNRT2) and characterization of CmNRT2. 1. Sci. Rep. 6 (1), 1–9. doi: 10.1038/srep23462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu R., Duan F., An X., Zhang F., von Wirén N., Yuan L. (2013). Characterization of AMT-mediated high-affinity ammonium uptake in roots of maize (Zea mays l.). Plant Cell Physiol. 54 (9), 1515–1524. doi: 10.1093/pcp/pct099 [DOI] [PubMed] [Google Scholar]
- Guan B., Yu J., Chen X., Xie W., Lu Z. (2011). Effects of salt stress and nitrogen application on growth and ion accumulation of suaeda salsa plants (International Conference on Remote Sensing, Environment and Transportation Engineering (IEEE; ), Nanjing, 8268–8272. [Google Scholar]
- Guan Y., Liu D. F., Qiu J., Liu Z. J., He Y. N., Fang Z. J., et al. (2022). The nitrate transporter OsNPF7. 9 mediates nitrate allocation and the divergent nitrate use efficiency between indica and japonica rice. Plant Physiol. 189 (1), 215–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Lu Y., Bao S., Zhang Q., Geng Y., Shao X. (2021). Carbon and nitrogen metabolism in rice cultivars affected by salt-alkaline stress. Crop Pasture Sci. 72 (5), 372–382. doi: 10.1071/CP20445 [DOI] [Google Scholar]
- Guo N., Hu J., Yan M., Qu H., Luo L., Tegeder M., et al. (2020). Oryza sativa lysine- histidine-type transporter 1 functions in root uptake and root-to-shoot allocation of amino acids in rice. Plant J. 103 (1), 395–411. doi: 10.1111/tpj.14742 [DOI] [PubMed] [Google Scholar]
- Gupta P., Srivastava S., Seth C. S. (2017). 24-epibrassinolide and sodium nitroprusside alleviate the salinity stress in Brassica juncea l. cv. varuna through cross talk among proline, nitrogen metabolism and abscisic acid. Plant Soil. 411 (1), 483–498. [Google Scholar]
- He X., Qu B., Li W., Zhao X., Teng W., Ma W., et al. (2015). The nitrate-inducible NAC transcription factor TaNAC2-5A controls nitrate response and increases wheat yield. Plant Physiol. 169 (3), 1991–2005. doi: 10.1104/pp.15.00568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Xi X., Zha Q., Lu Y., Jiang A. (2020). Ectopic expression of a grape nitrate transporter VvNPF6. 5 improves nitrate content and nitrogen use efficiency in arabidopsis. BMC Plant Biol. 20 (1), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y. N., Peng J. S., Cai Y., Liu D. F., Guan Y., Yi H. Y., Gong J. (2017). Tonoplast-localized nitrate uptake transporters involved in vacuolar nitrate efflux and reallocation in arabidopsis. M. Sci. Rep.7 1), 1–9. doi: 10.1038/s41598-017-06744-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessini K. (2022). Nitrogen form differently modulates growth, metabolite profile, and antioxidant and nitrogen metabolism activities in roots of Spartina alterniflora in response to increasing salinity. Plant Physiol. Biochem. 174, 35–42. doi: 10.1016/j.plaphy.2022.01.031 [DOI] [PubMed] [Google Scholar]
- Hessini K., Issaoui K., Ferchichi S., Saif T., Abdelly C., Siddique K. H., et al. (2019). Interactive effects of salinity and nitrogen forms on plant growth, photosynthesis and osmotic adjustment in maize. Plant Physiol. Biochem. 139, 171- 178. doi: 10.1016/j.plaphy.2019.03.005 [DOI] [PubMed] [Google Scholar]
- Hoshida H., Tanaka Y., Hibino T., Hayashi Y., Tanaka A., Takabe T., et al. (2000). Enhanced tolerance to salt stress in transgenic rice that overexpresses chloroplast glutamine synthetase. Plant Mol.Biol. 43 (1), 103–111. doi: 10.1023/A:1006408712416 [DOI] [PubMed] [Google Scholar]
- Huang J., Zhu C., Hussain S., Huang J., Liang Q., Zhu L., et al. (2020). Effects of nitric oxide on nitrogen metabolism and the salt resistance of rice (Oryza sativa l.) seedlings with different salt tolerances. Plant Physiol.Biochem 155, 374–383. doi: 10.1016/j.plaphy.2020.06.013 [DOI] [PubMed] [Google Scholar]
- Huang N. C., Liu K. H., Lo H. J., Tsay Y. F. (1999). Cloning and functional characterization of an arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11 (8), 1381–1392. doi: 10.1105/tpc.11.8.1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huez-López M. A., Ulery A. L., Samani Z., Picchioni G., Flynn R. P. (2011). Response of chile pepper (Capsicum annuum l.) to salt stress and organic and inorganic nitrogen sources: II. nitrogen and water use efficiencies, and salt tolerance. Trop. Subtrop. Ecosystems. 14 (3), 757–763. [Google Scholar]
- Hussain S. J., Khan N. A., Anjum N. A., Masood A., Khan M. I. R. (2021). Mechanistic elucidation of salicylic acid and sulphur-induced defence systems, nitrogen metabolism, photosynthetic, and growth potential of mungbean (Vigna radiata) under salt stress. J.Plant Growth Regul.40 3), 1000–1016. doi: 10.1007/s00344-020-10159-4 [DOI] [Google Scholar]
- Ibrahim M. E. H., Zhu X., Zhou G., Abidallhaa E. H. M. A. (2016). Effects of nitrogen on seedling growth of wheat varieties under salt stress. J. Agric. Sci. 8, 131–146. doi: 10.5539/jas.v8n10p131 [DOI] [Google Scholar]
- Ibrahim M. E. H., Zhu X., Zhou G., Ali A. Y. A., Ahmad I., Elsiddig A. M. I. (2018). Nitrogen fertilizer reduces the impact of sodium chloride on wheat yield. J.Agron.110 5), 1731–1737. doi: 10.2134/agronj2017.12.0742 [DOI] [Google Scholar]
- Ibrahim M. E. H., Zhu X., Zhou G., Ali A. Y. A., Ahmad I., Elsiddig A. M. I., et al. (2019). Promoting salt tolerance in wheat seedlings by application of nitrogen fertilizer. Pak. J. Bot. 51 (6), 1995–2002. doi: 10.30848/PJB2019-6(29) [DOI] [Google Scholar]
- Iqbal N., Umar S., Khan N. A. (2015). Nitrogen availability regulates proline and ethylene production and alleviates salinity stress in mustard (Brassica juncea). J. Plant Physiol. 178, 84–91. doi: 10.1016/j.jplph.2015.02.006 [DOI] [PubMed] [Google Scholar]
- Iqbal N., Umar S., Per T. S., Khan N. A. (2017). Ethephon increases photosynthetic- nitrogen use efficiency, proline and antioxidant metabolism to alleviate decrease in photosynthesis under salinity stress in mustard. Plant Signal.Behav. 12 (5), e1297000. doi: 10.1080/15592324.2017.1297000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irshad M., , Eneji A. E., Yasuda H. (2008). Comparative effect of nitrogen sources on maize under saline and non-saline conditions. J. Agron. Crop Sci. 194 (4), 256–261. doi: 10.1111/j.1439-037X.2008.00310.x [DOI] [Google Scholar]
- Isayenkov S. V., Maathuis F. J. (2019). Plant salinity stress: Many unanswered questions remain. Front. Plant Sci. 10, 80. doi: 10.3389/fpls.2019.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan B., Iqbal N., Fatma M., Sehar Z., Masood A., Sofo A., et al. (2021). Ethylene supplementation combined with split application of nitrogen and sulfur protects salt-inhibited photosynthesis through optimization of proline metabolism and antioxidant system in mustard (Brassica juncea l.). Plants 10 (7), 1303. doi: 10.3390/plants10071303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James D., Borphukan B., Fartyal D., Ram B., Singh J., Manna M., et al. (2018). Concurrent overexpression of OsGS1; 1 and OsGS2 genes in transgenic rice (Oryza sativa l.): impact on tolerance to abiotic stresses. Front. Plant Sci. 9, 786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed S. A., Arif M. S., Shahzad S. M., Ashraf M., Kausar R., Farooq T. H., et al. (2021). Can different salt formulations revert the depressing effect of salinity on maize by modulating plant biochemical attributes and activating stress regulators through improved n supply? Sustainability. 13 (14), 8022. doi: 10.3390/su13148022 [DOI] [Google Scholar]
- Jha U. C., Bohra A., Jha R., Parida S. K. (2019). Salinity stress response and ‘omics’ approaches for improving salinity stress tolerance in major grain legumes. Plant Cell Rep. 38, 255–277. doi: 10.1007/s00299-019-02374-5 [DOI] [PubMed] [Google Scholar]
- Jun-Feng Y. U. A. N., Gu F. E. N. G., Hai-Yan M. A., Chang-Yan T. I. A. N. (2010). Effect of nitrate on root development and nitrogen uptake of Suaeda physophora under NaCl salinity. Pedosphere 20 (4), 536–544. [Google Scholar]
- Kabir E., Hamid A., Haque M., Nawata E., Karim A. (2005). Effect of nitrogen fertilizer on salinity tolerance of mungbean (Vigna radiata l. wilczek). Japanese J. Tropic. 49 (2), 119–125. [Google Scholar]
- Katayama H., Mori M., Kawamura Y., Tanaka T., Mori M., Hasegawa H. (2009). Production and characterization of transgenic rice plants carrying a high-affinity nitrate transporter gene (OsNRT2. 1). Breed. Sci. 59 (3), 237–243. doi: 10.1270/jsbbs.59.237 [DOI] [Google Scholar]
- Kiba T., Feria-Bourrellier A. B., Lafouge F., Lezhneva L., Boutet-Mercey S., Orsel M., et al. (2012). The arabidopsis nitrate transporter NRT2. 4 plays a double role in roots and shoots of nitrogen-starved plants. Plant Cell 24 (1), 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koegel S., Ait Lahmidi N., Arnould C., Chatagnier O., Walder F., Ineichen K., et al. (2013). The family of ammonium transporters (AMT) in Sorghum bicolor: Two AMT members are induced locally, but not systemically in roots colonized by arbuscular mycorrhizal fungi. New Phytol. 198 (3), 853–865. doi: 10.1111/nph.12199 [DOI] [PubMed] [Google Scholar]
- Krapp A. (2015). Plant nitrogen assimilation and its regulation: A complex puzzle with missing pieces. Curr. Opin. Plant Biol. 25, 115–122. doi: 10.1016/j.pbi.2015.05.010 [DOI] [PubMed] [Google Scholar]
- Kurai T., Wakayama M., Abiko T., Yanagisawa S., Aoki N., Ohsugi R. (2011). Introduction of the ZmDof1 gene into rice enhances carbon and nitrogen assimilation under low-nitrogen conditions. Plant Biotechnol. J. 9 (8), 826–837. doi: 10.1111/j.1467-7652.2011.00592.x [DOI] [PubMed] [Google Scholar]
- Kurt F., Filiz E. (2022). “Functional divergence in oligopeptide transporters in plants,” in Cation transporters in plants (Academic Press: Elsevier; ), 167–183. [Google Scholar]
- Lamichhane S., Murata C., Griffey C. A., Thomason W. E., Fukao T. (2021). Physiological and molecular traits associated with nitrogen uptake under limited nitrogen in soft red winter wheat. Plants. 10 (1), 165. doi: 10.3390/plants10010165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Marmagne A., Park J., Fabien C., Yim Y., Kim S. J., et al. (2020). Concurrent activation of OsAMT1; 2 and OsGOGAT1 in rice leads to enhanced nitrogen use efficiency under nitrogen limitation. Plant J. 103 (1), 7–20. doi: 10.1111/tpj.14794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léran S., Garg B., Boursiac Y., Corratgé-Faillie C., Brachet C., Tillard P., et al. (2015). AtNPF5. 5, a nitrate transporter affecting nitrogen accumulation in arabidopsis embryo. Sci. Rep. 5 (1), 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis O. A. M., Leidi E. O., Lips S. H. (1989). Effect of nitrogen source on growth response to salinity stress in maize and wheat. New Phytol.111 2), 155–160. doi: 10.1111/j.1469-8137.1989.tb00676.x [DOI] [PubMed] [Google Scholar]
- Lezhneva L., Kiba T., Feria-Bourrellier A. B., Lafouge F., Boutet-Mercey S., Zoufan P., et al. (2014). The arabidopsis nitrate transporter NRT 2.5 plays a role in nitrate acquisition and remobilization in nitrogen- starved plants. Plat J. 80 (2), 230–241. [DOI] [PubMed] [Google Scholar]
- Li B., Byrt C., Qiu J., Baumann U., Hrmova M., Evrard A., et al. (2016). Identification of a stelar- localized transport protein that facilitates root-to-shoot transfer of chloride in arabidopsis. Plant Physiol. 170 (2), 1014–1029. doi: 10.1104/pp.15.01163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Qiu J., Jayakannan M., Xu B., Li Y., Mayo G. M., et al. (2017). AtNPF2. 5 modulates chloride (Cl–) efflux from roots of arabidopsis thaliana. Front. Plant Sci. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Tang Z., Wei J., Qu H., Xie Y., Xu G. (2016). The OsAMT1. 1 gene functions in ammonium uptake and ammonium–potassium homeostasis over low and high ammonium concentration ranges. J. Genet. Genomics 43 (11), 639–649. [DOI] [PubMed] [Google Scholar]
- Li J. Y., Fu Y. L., Pike S. M., Bao J., Tian W., Zhang Y., et al. (2010). The arabidopsis nitrate transporter NRT1. 8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22 (5), 1633–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Wang T., Zhang H., Liu S., Li W., Abou Elwafa S. F., et al. (2022. a). Tanrt2. 1-6b is a dual-affinity nitrate transporter contributing to nitrogen uptake in bread wheat under both nitrogen deficiency and sufficiency. Crop J. 10, 993–1005. doi: 10.1016/j.cj.2021.11.012 [DOI] [Google Scholar]
- Li Q., Lu X., Wang C., Shen L., Dai L., He J., et al. (2022. b). Genome-wide association study and transcriptome analysis reveal new QTL and candidate genes for nitrogen deficiency tolerance in rice. Crop J. 10, 942–951. doi: 10.1016/j.cj.2021.12.006 [DOI] [Google Scholar]
- Li S., Li Y., He X., Li Q., Liu B., Ai X., et al. (2019). Response of water balance and nitrogen assimilation in cucumber seedlings to CO2 enrichment and salt stress. Plant Physiol. Biochem. 139, 256–263. doi: 10.1016/j.plaphy.2019.03.028 [DOI] [PubMed] [Google Scholar]
- Li W., Feng Z., Zhang C. (2021). Ammonium transporter PsAMT1. 2 from Populus simonii functions in nitrogen uptake and salt resistance. Tree Physiol. 41 (12), 2392- 2408. [DOI] [PubMed] [Google Scholar]
- Liang C., Wang Y., Zhu Y., Tang J., Hu B., Liu L., et al. (2014). OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Procd. Natnl Sci. Acad. 111 (27), 10013–10018. doi: 10.1073/pnas.1321568111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. H., Xu Z. J., Peng J. S., Zhao J., Zhang G. B., Xie J., et al. (2019). OsProT1 and OsProT3 function to mediate proline- and γ-aminobutyric acid-specific transport in yeast and are differentially expressed in rice (Oryza sativa l.). Rice.12 1), 1–10. doi: 10.1186/s12284-019-0341-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K. H., Tsay Y. F. (2003). Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 22 (5), 1005–1013. doi: 10.1093/emboj/cdg118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang B., Liu D., Zou C., Wu P., Wang Z., et al. (2020). Transcriptomic and metabolomic analyses reveal mechanisms of adaptation to salinity in which carbon and nitrogen metabolism is altered in sugar beet roots. BMC Plant Biol.20 1), 1–21. doi: 10.1186/s12870-020-02349-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Zhao Y., Liu X., Korpelainen H., Li C. (2022). Ammonium and nitrate affect sexually different responses to salt stress in Populus cathayana . Physiol. Plant.174 1), e13626. doi: 10.1111/ppl.13626 [DOI] [PubMed] [Google Scholar]
- Liu R. X., Li H. L., Rui L., Liu G. D., Wang T., Wang X. F., et al. (2023). An apple NITRATE REDUCTASE 2 gene positively regulates nitrogen utilization and abiotic stress tolerance in arabidopsis and apple callus. Plant Physiol. Biochem. 196, 23–32. doi: 10.1016/j.plaphy.2023.01.026 [DOI] [PubMed] [Google Scholar]
- Liu Y., Wang Y., Xiong Y. (2018). “Nitrogen–TOR signaling in shoot apex activation,” in EMBO workshop. target of rapamycin (TOR) signaling in photosynthetic organisms (Programme and Abstract Book; ), 96. [Google Scholar]
- Lopez-Delacalle M., Camejo D. M., García-Martí M., Nortes P. A., Nieves-Cordones M., Martínez V., et al. (2020). Using tomato recombinant lines to improve plant tolerance to stress combination through a more efficient nitrogen metabolism. Front. Plant Sci. 10, 1702. doi: 10.3389/fpls.2019.01702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B., Xu M., Zhao L., Xie P., Chen Y., Harwood W., et al. (2020). Overexpression of the high-affinity nitrate transporter OsNRT2. 3b driven by different promoters in barley improves yield and nutrient uptake balance. Int. J. Mol. Sci. 21 (4), 1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv B., Wu Q., Wang A., Li Q., Dong Q., Yang J., et al. (2020). A WRKY transcription factor, FtWRKY46, from tartary buckwheat improves salt tolerance in transgenic Arabidopsis thaliana . Plant Physiol. Biochem. 147, 43–53. doi: 10.1016/j.plaphy.2019.12.004 [DOI] [PubMed] [Google Scholar]
- Lv X., Yu P., Deng W., Li Y. (2018). Transcriptomic analysis reveals the molecular adaptation to NaCl stress in Zostera marina l. Plant Physiol. Biochem. 130, 61–68. doi: 10.1016/j.plaphy.2018.06.022 [DOI] [PubMed] [Google Scholar]
- Ma S., Guo S., Chen J., Sun J., Wang Y., Shu S. (2019). Enhancement of salt-stressed cucumber tolerance by application of glucose for regulating antioxidant capacity and nitrogen metabolism. Can. J. Plant Sci. 100 (3), 253–263. [Google Scholar]
- Majid A., Rather B. A., Masood A., Sehar Z., Anjum N. A., Khan N. A. (2021). Abscisic acid in coordination with nitrogen alleviates salinity-inhibited photosynthetic potential in mustard by improving proline accumulation and antioxidant activity. Stresses 1 (3), 162–180. doi: 10.3390/stresses1030013 [DOI] [Google Scholar]
- Melino V. J., Tester M. A., Okamoto M. (2022). Strategies for engineering improved nitrogen use efficiency in crop plants via redistribution and recycling of organic nitrogen. Curr. Opin. Biotechnol. 73, 263–269. doi: 10.1016/j.copbio.2021.09.003 [DOI] [PubMed] [Google Scholar]
- Mellidou I., Ainalidou A., Papadopoulou A., Leontidou K., Genitsaris S., Karagiannis E., et al. (2021). Comparative transcriptomics and metabolomics reveal an intricate priming mechanism involved in PGPR-mediated salt tolerance in tomato. Front . Plant Sci. 12, 713984. doi: 10.3389/fpls.2021.713984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng S., Su L., Li Y., Wang Y., Zhang C., Zhao Z. (2016). Nitrate and ammonium contribute to the distinct nitrogen metabolism of Populus simonii during moderate salt stress. PloS One 11 (3), e0150354. doi: 10.1371/journal.pone.0150354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morere-Le Paven M. C., Viau L., Hamon A., Vandecasteele C., Pellizzaro A., Bourdin C., et al. (2011). Characterization of a dual-affinity nitrate transporter MtNRT1. 3 in the model legume Medicago truncatula . J. Exp. Bot.62 15), 5595–5605. [DOI] [PubMed] [Google Scholar]
- Nakamura A., Fukuda A., Sakai S., Tanaka Y. (2006). Molecular cloning, functional expression and subcellular localization of two putative vacuolar voltage-gated chloride channels in rice (Oryza sativa l.). Plant Cell Physiol. 47 (1), 32–42. doi: 10.1093/pcp/pci220 [DOI] [PubMed] [Google Scholar]
- Naliwajski M. R., Skłodowska M. (2018). The relationship between carbon and nitrogen metabolism in cucumber leaves acclimated to salt stress. PeerJ 6, e6043. doi: 10.7717/peerj.6043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveed N., Mir M. Y., Hamid S. (2019). Effect of nitrogen and phosphorus on the growth and redox homeostasis of salt-stressed mustard plants. Asian J. Plants Sci. 18, 52–59. doi: 10.3923/ajps.2019.52.59 [DOI] [Google Scholar]
- Nawaz M. A., Wang L., Jiao Y., Chen C., Zhao L., Mei M., et al. (2017). Pumpkin rootstock improves nitrogen use efficiency of watermelon scion by enhancing nutrient uptake, cytokinin content, and expression of nitrate reductase genes. Plant Growth Regul. 82 (2), 233–246. doi: 10.1007/s10725-017-0254-7 [DOI] [Google Scholar]
- Naz M., Luo B., Guo X., Li B., Chen J., Fan X. (2019). Overexpression of nitrate transporter OsNRT2. 1 enhances nitrate-dependent root elongation. Genes 10 (4), 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naz N., Fatima S., Hameed M., Ashraf M., Ahmad M. S. A., Ahmad F., et al. (2022). Contribution of structural and functional adaptations of hyper-accumulator suaeda vera forssk. ex JF gmel. for adaptability across salinity gradients in hot desert. Environ. Sci. pollut. Res. 29 (42), 64077–64095. [DOI] [PubMed] [Google Scholar]
- Nazar R., Iqbal N., Syeed S., Khan N. A. (2011). Salicylic acid alleviates decreases in photosynthesis under salt stress by enhancing nitrogen and sulfur assimilation and antioxidant metabolism differentially in two mungbean cultivars. J. Plant Physiol. 168 (8), 807–815. doi: 10.1016/j.jplph.2010.11.001 [DOI] [PubMed] [Google Scholar]
- Ouyang J., Cai Z., Xia K., Wang Y., Duan J., Zhang M. (2010). Identification and analysis of eight peptide transporter homologs in rice. Plant Sci.179 4), 374–382. doi: 10.1016/j.plantsci.2010.06.013 [DOI] [Google Scholar]
- Ozfidan-Konakci C., Yildiztugay E., Alp F. N., Kucukoduk M., Turkan I. (2020). Naringenin induces tolerance to salt/osmotic stress through the regulation of nitrogen metabolism, cellular redox and ROS scavenging capacity in bean plants. Plant Physiol. Biochem. 157, 264–275. doi: 10.1016/j.plaphy.2020.10.032 [DOI] [PubMed] [Google Scholar]
- Pereira E. G., Bucher C. P. C., Bucher C. A., Santos L. A., Lerin J., Catarina C. S., et al. (2022). The amino acid transporter OsAAP1 regulates the fertility of spikelets and the efficient use of n in rice. Plant Soil 480 (1-2), 507–521. doi: 10.1007/s11104-022-05598-9 [DOI] [Google Scholar]
- Phan N. T. H., Heymans A., Bonnave M., Lutts S., Pham C. V., Bertin P. (2022). Nitrogen use efficiency of rice cultivars (Oryza sativa l.) under salt stress and low nitrogen conditions. J. Plant Growth Regul. 1-15. doi: 10.1007/s00344-022-10660-y [DOI] [Google Scholar]
- Popova O. V., Dietz K. J., Golldack D. (2003). Salt-dependent expression of a nitrate transporter and two amino acid transporter genes in Mesembryanthemum crystallinum . Plant Mol. Biol.52 3), 569–578. doi: 10.1023/A:1024802101057 [DOI] [PubMed] [Google Scholar]
- Postnikova O. A., Shao J., Nemchinov L. G. (2013). Analysis of the alfalfa root transcriptome in response to salinity stress. Plant Cell Physiol. 54 (7), 1041–1055. doi: 10.1093/pcp/pct056 [DOI] [PubMed] [Google Scholar]
- Prasad A., Patra D. D., Anwar M., Singh D. V. (1997). Interactive effects of salinity and nitrogen on mineral n status in soil and growth and yield of German chamomile (Matricaria chamomilla). J. Indian Soc Soil Sci. 45 (3), 537–541. [Google Scholar]
- Qureshi A. S., Daba A. W. (2020). Evaluating growth and yield parameters of five quinoa (Chenopodium quinoa w.) genotypes under different salt stress conditions. J. Agric. Sci. 12, 128. [Google Scholar]
- Ranathunge K., El-Kereamy A., Gidda S., Bi Y. M., Rothstein S. J. (2014). AMT1; 1 transgenic rice plants with enhanced NH4 + permeability show superior growth and higher yield under optimal and suboptimal NH4 + conditions. J. Exp. Bot. 65 (4), 965–979. doi: 10.1093/jxb/ert458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy I. N. B. L., Kim B. K., Yoon I. S., Kim K. H., Kwon T. R. (2017). Salt tolerance in rice: Focus on mechanisms and approaches. Rice Sci. 24 (3), 123–144. doi: 10.1016/j.rsci.2016.09.004 [DOI] [Google Scholar]
- Rentsch D., Hirner B., Schmelzer E., Frommer W. B. (1996). Salt stress-induced proline transporters and salt stress-repressed broad specificity amino acid permeases identified by suppression of a yeast amino acid permease-targeting mutant. Plant Cell 8 (8), 1437–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restivo F. M. (2004). Molecular cloning of glutamate dehydrogenase genes of Nicotiana plumbaginifolia: Structure analysis and regulation of their expression by physiological and stress conditions. Plant Sci. 166 (4), 971–982. doi: 10.1016/j.plantsci.2003.12.011 [DOI] [Google Scholar]
- Rohilla P., Yadav J. P. (2019). ). acute salt stress differentially modulates nitrate reductase expression in contrasting salt responsive rice cultivars. Protoplasma 256 (5), 1267- 1278. [DOI] [PubMed] [Google Scholar]
- Sasaki K., Kojima S. (2018). Identification of genomic regions regulating ammonium- dependent inhibition of primary root length in Arabidopsis thaliana . Soil Sci. Plant Nutr. 64 (6), 746–751. doi: 10.1080/00380768.2018.1524268 [DOI] [Google Scholar]
- Sathee L., Jha S. K., Rajput O. S., Singh D., Kumar S., Kumar A. (2021). Expression dynamics of genes encoding nitrate and ammonium assimilation enzymes in rice genotypes exposed to reproductive stage salinity stress. Plant Physiol. Biochem. 165, 161–172. doi: 10.1016/j.plaphy.2021.05.013 [DOI] [PubMed] [Google Scholar]
- Sena Falero M. F. (2022). Role of the nitrate/potassium transporter, NRT1. 5/NPF7. 3, in the regulatory network of ion homeostasis in Arabidopsis thaliana (Doctoral dissertation). Germany: University of Berlin. doi: 10.17169/refubium-31049 [DOI] [Google Scholar]
- Shahzad R., Khan A. L., Bilal S., Waqas M., Kang S. M., Lee I. J. (2017). Inoculation of abscisic acid-producing endophytic bacteria enhances salinity stress tolerance in Oryza sativa . Environ. Exp. Bot. 136, 68–77. doi: 10.1016/j.envexpbot.2017.01.010 [DOI] [Google Scholar]
- Shao A., Sun Z., Fan S., Xu X., Wang W., Amombo E., et al. (2020). Moderately low nitrogen application mitigate the negative effects of salt stress on annual ryegrass seedlings. Peer J. 8, e10427. doi: 10.7717/peerj.10427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Q. S., Shu S., Du J., Xing W. W., Guo S. R., Sun J. (2015). Effects of NaCl stress on nitrogen metabolism of cucumber seedlings. Russ. J. Plant Physiol. 62 (5), 595–603. doi: 10.1134/S1021443715050155 [DOI] [Google Scholar]
- Shaviv A., Hazan O., Neumann P. M., Hagin J. (1990). Increasing salt tolerance of wheat by mixed ammonium nitrate nutrition. J. Plant Nutt. 13 (10), 1227–1239. doi: 10.1080/01904169009364148 [DOI] [Google Scholar]
- Shen YG, Zhang W.K., Yan D.Q., Du B.X., Zhang J.S., Chen S.Y. (2002). Overexpression of proline transporter gene from Halophyte confers tolerance in Arabidopsis. Acta Bot. Sin. 44, 956–962. doi: 10.1111/j.1439-037X.2008.00310.x [DOI] [Google Scholar]
- Siddiqui M. H., Khan M. N., Mohammad F., Khan M. M. A. (2008). Role of nitrogen and gibberellin (GA3) in the regulation of enzyme activities and in osmoprotectant accumulation in Brassica juncea l. under salt stress. J. Agron. Crop Sci.194 3), 214–224. [Google Scholar]
- Sikder R. K., Wang X., Zhang H., Gui H., Dong Q., Jin D., et al. (2020). Nitrogen enhances salt tolerance by modulating the antioxidant defense system and osmoregulation substance content in Gossypium hirsutum . Plants 9 (4), 450. doi: 10.3390/plants9040450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M., Singh V. P., Prasad S. M. (2016). Responses of photosynthesis, nitrogen and proline metabolism to salinity stress in Solanum lycopersicum under different levels of nitrogen supplementation. Plant Physiol.Biochem 109, 72–83. doi: 10.1016/j.plaphy.2016.08.021 [DOI] [PubMed] [Google Scholar]
- Soliman M., Elkelish A., Souad T., Alhaithloul H., Farooq M. (2020). Brassinosteroid seed priming with nitrogen supplementation improves salt tolerance in soybean. Physiol. Mol. Biol. 26 (3), 501–511. doi: 10.1007/s12298-020-00765-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Zhou G., Ma B. L., Wu W., Ahmad I., Zhu G., et al. (2019). Nitrogen application improved photosynthetic productivity, chlorophyll fluorescence, yield and yield components of two oat genotypes under saline conditions. Agronomy 9 (3), 115. doi: 10.3390/agronomy9030115 [DOI] [Google Scholar]
- Sousa H. C., Sousa G. G. D., Lessa C. I., Lima A. F. D. S., Ribeiro R. M., Rodrigues F. (2021). Growth and gas exchange of corn under salt stress and nitrogen doses. H. D. C. Rev. Bras. Eng. Agríc. Ambient. 25, 174–181. doi: 10.1590/1807-1929/agriambi.v25n3p174-181 [DOI] [Google Scholar]
- Souza A. F. F., Bucher C. A., Arruda L. N., Rangel R. P., Santos L. A., Fernandes M. S., et al. (2022). Knockdown of OsNRT2. 4 modulates root morphology and alters nitrogen metabolism in response to low nitrate availability in rice. Mol. Plant Breed. 42 (1), 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. C., Sheng S., Fan T. F., Liu L., Ke J., Wang D. B., et al. (2019). Molecular identification and functional characterization of GhAMT1. 3 in ammonium transport with a high affinity from cotton (Gossypium hirsutum l.). Physiol. Plant 167 (2), 217–231. [DOI] [PubMed] [Google Scholar]
- Taochy C., Gaillard I., Ipotesi E., Oomen R., Leonhardt N., Zimmermann S., et al. (2015). The arabidopsis root stele transporter NPF2. 3 contributes to nitrate translocation to shoots under salt stress. Plant J. 83 (3), 466–479. [DOI] [PubMed] [Google Scholar]
- Teh C. Y., Shaharuddin N. A., Ho C. L., Mahmood M. (2016). Exogenous proline significantly affects the plant growth and nitrogen assimilation enzymes activities in rice (Oryza sativa) under salt stress. Acta Physiol.Plant. 38 (6), 1–10. doi: 10.1007/s11738-016-2163-1 [DOI] [Google Scholar]
- Thu Hoai N. T., Shim I. S., Kobayashi K., Kenji U. (2003). Accumulation of some nitrogen compounds in response to salt stress and their relationships with salt tolerance in rice (Oryza sativa l.) seedlings. Plant Growth Rgul. 41 (2), 159–164. doi: 10.1023/A:1027305522741 [DOI] [Google Scholar]
- Tian J., Pang Y., Yuan W., Peng J., Zhao Z. (2022). Growth and nitrogen metabolism in Sophora japonica (L.) as affected by salinity under different nitrogen forms. Plant Sci. 322, 111347. [DOI] [PubMed] [Google Scholar]
- Torres Bazurto J., Sanchez J. D., Cayón Salinas D. G. (2017). Nutrient accumulation models in the banana (Musa AAA simmonds cv. williams) plant under nitrogen doses. Acta Agron. 66 (3), 391–396. [Google Scholar]
- Tsay Y. F., Schroeder J. I., Feldmann K. A., Crawford N. M. (1993). The herbicide sensitivity gene CHL1 of arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72 (5), 705–713. doi: 10.1016/0092-8674(93)90399-B [DOI] [PubMed] [Google Scholar]
- Tsuzuki C., Hachisu M., Iwabe R., Nakayama Y., Nonaga Y., Sukegawa S., et al. (2022). An amino acid ester of menthol elicits defense responses in plants. Plant Mol.Biol.109 4), 523–531. doi: 10.1007/s11103-021-01150-y [DOI] [PubMed] [Google Scholar]
- Tzortzakis N., Pitsikoulaki G., Stamatakis A., Chrysargyris A. (2022). Ammonium to total nitrogen ratio interactive effects with salinity application on Solanum lycopersicum growth, physiology, and fruit storage in a closed hydroponic system. Agronomy 12 (2), 386. doi: 10.3390/agronomy12020386 [DOI] [Google Scholar]
- Ullah A., Li M., Noor J., Tariq A., Liu Y., Shi L. (2019). Effects of salinity on photosynthetic traits, ion homeostasis and nitrogen metabolism in wild and cultivated soybean. Peer J. 7, e8191. doi: 10.7717/peerj.8191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Xu T., Yin Z., Wu W., Geng H., Li L., et al. (2020). Overexpression of OsMYB305 in rice enhances the nitrogen uptake under low-nitrogen condition. Front. Plant Sci. 11, 369. doi: 10.3389/fpls.2020.00369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhang M., Guo R., Shi D., Liu B., Lin X., et al. (2012. a). Effects of salt stress on ion balance and nitrogen metabolism of old and young leaves in rice (Oryza sativa l.). BMC Plant Biol. 12 (1), 1–11. doi: 10.1186/1471-2229-12-194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Li Y., Huang Y., Zhao X., Dong Z., Jin W., et al. (2022). Amino acid permease 6 regulates grain protein content in maize. Crop J. 10 (6), 1536–1544. doi: 10.1016/j.cj.2022.03.007 [DOI] [Google Scholar]
- Wang W. H., Köhler B., Cao F. Q., Liu G. W., Gong Y. Y., Sheng S., et al. (2012. b). Rice DUR3 mediates high-affinity urea transport and plays an effective role in improvement of urea acquisition and utilization when expressed in arabidopsis. New Phytol. 193 2), 432–444. doi: 10.1111/j.1469-8137.2011.03929.x [DOI] [PubMed] [Google Scholar]
- Wang Y. Y., Tsay Y. F. (2011). Arabidopsis nitrate transporter NRT1. 9 is important in phloem nitrate transport. Plant Cell 23 (5), 1945–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Zheng Y., Feng H., Qu H., Fan X., Yamaji N., et al. (2018). OsNRT2. 4 encodes a dual-affinity nitrate transporter and functions in nitrate-regulated root growth and nitrate distribution in rice. J. Exp. Bot. 69 (5), 1095–1107. [DOI] [PubMed] [Google Scholar]
- Wei P., Che B., Shen L., Cui Y., Wu S., Cheng C., et al. (2019). Identification and functional characterization of the chloride channel gene, GsCLC-c2 from wild soybean. BMC Plant Biol. 19 (1), 1–15. doi: 10.1186/s12870-019-1732-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y. M., Ren Z. J., Wang B. H., Zhang L., Zhao Y. J., Wu J. W., et al. (2021). A nitrate transporter encoded by ZmNPF7. 9 is essential for maize seed development. Plant Sci. 308, 110901. [DOI] [PubMed] [Google Scholar]
- Wu X., Liu T., Zhang Y., Duan F., Neuhäuser B., Ludewig U., et al. (2019). Ammonium and nitrate regulate NH4 + uptake activity of arabidopsis ammonium transporter AtAMT1; 3 via phosphorylation at multiple c-terminal sites. J. Exp. Bot.70 18), 4919–4930. doi: 10.1093/jxb/erz230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X., Fan X., Wei J., Feng H., Qu H., Xie D., et al. (2015). Rice nitrate transporter OsNPF2. 4 functions in low-affinity acquisition and long-distance transport. J. Exp. Bot. 66 (1), 317–331. [DOI] [PubMed] [Google Scholar]
- Xu G., Fan X., Miller A. J. (2012). Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 63, 153–182. doi: 10.1146/annurev-arplant-042811-105532 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S., Akiyama A., Kisaka H., Uchimiya H., Miwa T. (2004). Metabolic engineering with Dof1 transcription factor in plants: Improved nitrogen assimilation and growth under low-nitrogen conditions. Procd. Nat. Acad. Sci. 101 (20), 7833–7838. doi: 10.1073/pnas.0402267101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang O., Popova O. V., Süthoff U., Lüking I., Dietz K. J., Golldack D. (2009). The arabidopsis basic leucine zipper transcription factor AtbZIP24 regulates complex transcriptional networks involved in abiotic stress resistance. Gene 436 (1-2), 45–55. doi: 10.1016/j.gene.2009.02.010 [DOI] [PubMed] [Google Scholar]
- Yang S., Xu K., Chen S., Li T., Xia H., Chen L., et al. (2019). A stress- responsive bZIP transcription factor OsbZIP62 improves drought and oxidative tolerance in rice. BMC Plant Biol. 19 (1), 1–15. doi: 10.1186/s12870-019-1872-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X. M., Long X. H., Shao T. Y. (2022). Effect of inorganic nitrogen and phosphorous on morphology, ion uptake and photosynthesis activity in Jerusalem artichoke plants under salt stress. J. Plant Nutr. 45 (9), 1378–1392. doi: 10.1080/01904167.2021.2020828 [DOI] [Google Scholar]
- Yi Y., Peng Y., Song T., Lu S., Teng Z., Zheng Q., et al. (2022). NLP2-NR module associated NO is involved in regulating seed germination in rice under salt stress. Plants 11 (6), 795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Xu T., Li X., Tang J., Ma D., Li Z., et al. (2016). NaCl-Induced changes of ion homeostasis and nitrogen metabolism in two sweet potato (Ipomoea batatas l.) cultivars exhibit different salt tolerance at adventitious root stage. Environ. Exp. Bot. 129, 23–36. [Google Scholar]
- Yu Z., Duan X., Luo L., Dai S., Ding Z., Xia G. (2020). How plant hormones mediate salt stress responses. Trends Plant Sci. 25 (11), 1117–1130. [DOI] [PubMed] [Google Scholar]
- Zhang C., Meng S., Li Y., Zhao Z. (2014. a). Net NH4 + and NO3– fluxes, and expression of NH4 + and NO3– transporter genes in roots of Populus simonii after acclimation to moderate salinity. Trees 28 (6), 1813–1821. [Google Scholar]
- Zhang G. B., Yi H. Y., Gong J. M. (2014. b). The arabidopsis ethylene/jasmonic acid- NRT signaling module coordinates nitrate reallocation and the trade-off between growth and environmental adaptation. Plant Cell 26 (10), 3984–3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Huo Y., Xu Z., Guo K., Wang Y., Zhang X., et al. (2020. a). Physiological and proteomics responses of nitrogen assimilation and glutamine/glutamine family of amino acids metabolism in mulberry (Morus alba l.) leaves to NaCl and NaHCO3 stress. Plant Signal. Behav. 15 (10), 1798108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhao X., Sun Q., Yan C., Wang J., Yuan C., et al. (2020. b). Comparative transcriptome analysis reveals molecular defensive mechanism of Arachis hypogaea in response to salt stress. Int. J. Genom. 2020, 1–13. doi: 10.1155/2020/6524093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Zhang H., Song C., Zhu J. K., Shabala S. (2020). Mechanisms of plant responses and adaptation to soil salinity. Innov. 1 (1), 100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Ma H., Yu L., Wang X., Zhao J. (2012). Genome-wide survey and expression analysis of amino acid transporter gene family in rice (Oryza sativa l.). PloS One 7 (11), e49210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Liu Z., Duan F., An X., Liu X., Hao D., et al. (2018). Overexpression of the maize ZmAMT1; 1a gene enhances root ammonium uptake efficiency under low ammonium nutrition. Plant Biotechnol. Rep. 12(1), 47- 56. [Google Scholar]
- Zhi S., Tang X., Zheng Z. N., Xu F. X., Ren Y. H., Wang X. L. (2020). Expression of glutamate dehydrogenase genes of mulberry in Morus alba l. and transgenic tobacco in relation to biotic and abiotic stresses. Russ. J. Plant Physiol. 67 (4), 703–714. [Google Scholar]
- Zhu Y., Huang X., Hao Y., Su W., Liu H., Sun G., et al. (2020). Ammonium transporter (BcAMT1. 2) mediates the interaction of ammonium and nitrate in Brassica campestris . Front. Plant Sci. 10 1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilli C. G., Balestrasse K. B., Yannarelli G. G., Polizio A. H., Santa-Cruz D. M., Tomaro M. L. (2008). Heme oxygenase up-regulation under salt stress protects nitrogen metabolism in nodules of soybean plants. Environ. Expt. Bot. 64 (1), 83–89. doi: 10.1016/j.envexpbot.2008.03.005 [DOI] [Google Scholar]
- Zou X., Liu M. Y., Wu W. H., Wang Y. (2020). Phosphorylation at Ser28 stabilizes the arabidopsis nitrate transporter NRT2. 1 in response to nitrate limitation. J. Integ. Plant Biol. 62 (6), 865–876. doi: 10.1111/jipb.12858 [DOI] [PubMed] [Google Scholar]