Abstract
The immunopurification of the endogenous cytoplasmic murine histone deacetylase 6 (mHDAC6), a member of the class II HDACs, from mouse testis cytosolic extracts allowed the identification of two associated proteins. Both were mammalian homologues of yeast proteins known to interact with each other and involved in the ubiquitin signaling pathway: p97/VCP/Cdc48p, a homologue of yeast Cdc48p, and phospholipase A2-activating protein, a homologue of yeast UFD3 (ubiquitin fusion degradation protein 3). Moreover, in the C-terminal region of mHDAC6, a conserved zinc finger-containing domain named ZnF-UBP, also present in several ubiquitin-specific proteases, was discovered and was shown to mediate the specific binding of ubiquitin by mHDAC6. By using a ubiquitin pull-down approach, nine major ubiquitin-binding proteins were identified in mouse testis cytosolic extracts, and mHDAC6 was found to be one of them. All of these findings strongly suggest that mHDAC6 could be involved in the control of protein ubiquitination. The investigation of biochemical properties of the mHDAC6 complex in vitro further supported this hypothesis and clearly established a link between protein acetylation and protein ubiquitination.
An increasing number of histone deacetylases (HDACs) are being characterized in higher eukaryotes. These proteins have been grouped in distinct families according to the similarity of their sequence to a yeast founding member. Class I HDACs are homologous to yeast RPD3, while class II members are related to yeast HDA1 and class III members are related to yeast SIR2 deacetylase (13, 20). Class I HDACs are found in various nuclear multiprotein complexes containing either HDAC1/2 or HDAC3. Class II HDACs show the interesting property of being capable of a nucleocytoplasmic shuttling. Indeed, all of the class II HDACs, HDAC4, -5, -6, and -7, are subject to a regulated intracellular localization (20). Although there is evidence for a role for some of these HDACs in transcriptional repression, their possible function in the cytoplasm remains elusive (20, 21). Within these enzymes, the endogenous HDAC6 was found to be essentially cytoplasmic (2, 37). A fraction of the murine HDAC6 (mHDAC6) translocates, however, in the nucleus under specific circumstances, such as arrest of cell proliferation (37). In order to gain an insight into the function of cytoplasmic HDACs, cytosolic mHDAC6 was immunopurified from mouse testis cytosolic extracts. The identified mHDAC6-associated proteins showed striking sequence homology to yeast regulatory proteins involved in the control of protein ubiquitination. These proteins are the mammalian homologue of yeast UFD3, known as phospholipase A2-activating protein (PLAP) (12), as well as the homologue of yeast Cdc48p AAA ATPase (p97/VCP/Cdc48p) (11). The UFD pathway was discovered in yeast after the observation that a protein containing a nonremovable N-terminal ubiquitin (Ub) moiety had a short half-life (19). The protein degradation pathway involved was called UFD, for Ub fusion degradation. A genetic approach was used to dissect this pathway, and five genes termed UFD1 to UFD5 were discovered to be involved in the degradation of the substrate in vivo (12, 19). Evidence of the role of some of these proteins in Ub-dependent degradation of target substrates was later discovered. For instance, UFD2 (also known as E4) was shown to bind to Ub moieties of preformed conjugates and catalyze Ub chain assembly (22). UFD5 is a transcription factor regulating genes encoding proteosomal subunits (38). Interestingly, Cdc48p was shown to interact with both UFD2 (22) and UFD3 (12), but its role in the function of these UFD proteins has remained unclear.
In mammals, several proteins showing striking sequence homology with these yeast UFD proteins have been discovered. A mammalian homologue of yeast UFD1 has recently been identified and was shown to form a specific complex with the mammalian homologue of yeast Cdc48p, p97/VCP/Cdc48 (27). The gene encoding the mammalian homologue of UFD2 has been shown to fuse with another gene (named D4Cole1e) in the slow Wallerian degeneration mutant mouse (8). The chimeric mRNA is abundantly expressed in the nervous system and encodes a fusion protein containing the 40 N-terminal amino-acid region of UFD2. The homologue of UFD3 in mammals is PLAP (reference 12 and reported here) which seems to participate in the control of the cellular levels of prostaglandin and the activity of phospholipase A2 and phospholipases C and D (30). UFD4 is homologous to E6AP (19), a human protein possessing a Ub-ligase activity (34), interacting with the papillomavirus-encoded oncogene E6 and involved in the Ub-dependent degradation of p53 (17, 18).
The mammalian homologues of yeast UFD proteins therefore appear to be involved in a variety of functions. Here we show that one of them, the mammalian homologue of UFD3, is a member of the cytoplasmic mHDAC6 complex. It is also shown here that mHDAC6 can specifically interact with Ub and that the Ub-bound mHDAC6 maintains its deacetylase activity. Moreover p97/VCP/Cdc48, interacting with UFD proteins in yeast as well as in mammals, was also found in the mHDAC6 complex. That mHDAC6 is one of the major cytoplasmic Ub-binding proteins present in mouse testis cytosolic extracts and that most of the Ub-binding proteins identified in this work were involved in the Ub/proteasome-dependent regulatory pathways strongly suggest that the deacetylase mHDAC6 is also involved in the Ub signaling pathway. This conclusion was further supported by in vitro experiments with mouse testis cytosolic extracts.
MATERIALS AND METHODS
Fractionation of spermatogenic cells.
Spermatogenic cells of all stages were recovered from the testes of an adult mouse. Cell suspensions enriched in mouse spermatogenic cells at specific stages of their differentiation, were obtained by velocity sedimentation at unit gravity on a bovine serum albumin (BSA) gradient, according to a method described previously (4, 31). This technique allows germ cells to be separated according to their respective sizes and densities.
Cell fractionation and Western blotting.
Western blot analysis was performed by standard procedures. Cytoplasmic and nuclear extracts were obtained as follows. Cells were lysed in buffer D (15 mM NaCl, 60 mM KCl, 12% sucrose, 2 mM EDTA, 0.5 mM EGTA, 0.65 mM spermidine, 1 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.05% Triton X-100). Nuclei were pelleted by centrifugation, and the supernatant (cytoplasmic extract) was kept at −80°C. Nuclei were resuspended in a small volume of buffer D and layered over 11 ml of the same buffer and centrifuged at 1,000 × g for 5 min. The pellet was lysed directly in protein loading buffer and homogenized by sonication.
Large-scale purification of mHDAC6 and associated proteins.
Mouse testes were isolated, sliced, and homogenized in an ice-cold lysis buffer (500 μl/testis) containing 0.34 M sucrose, 60 mM KCl, 15 mM NaCl, 15 mM Tris-HCl (pH 7.4), 0.65 mM spermidine, 2 mM EDTA, 0.5 mM EGTA, 0.05% Triton X-100, 1 mM DTT, 0.5 mM PMSF. The homogenate was incubated for 30 min on ice and centrifuged at 16,000 × g for 30 min at 4°C. The supernatant was recovered and centrifuged for 1 h at 100,000 × g, at 4°C (cytoplasmic extract). Anti-mHDAC6 antibody (37) or peptide-blocked anti-mHDAC6 antibody (antibody preincubated for 30 min at 4°C with its target peptide) was incubated with the extract at 4°C for 1 h. Immunocomplexes were precipitated with protein G-Sepharose, washed three times in lysis buffer, and eluted by the target peptide (1 mg/ml) at 4°C overnight.
Mass spectrometry and protein identification.
After separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), the protein bands were excised from the Coomassie blue-stained gel and washed with 50% acetonitrile and 25 mM NH4HCO3. Gel pieces were dried in a vacuum centrifuge and reswollen in 20 μl of 25 mM NH4HCO3 containing 0.5 μg of trypsin (Promega, sequencing grade). After a 4-h incubation at 37°C, a 0.5-μl aliquot was removed for matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis and spotted onto the MALDI sample probe on top of a dried 0.5-μl mixture of a 4-volume solution of saturated α-cyano-4-hydroxy-trans-cinnamic acid in acetone and 3 volumes of nitrocellulose (10 mg/ml) dissolved in acetone-isopropanol (1:1 [vol/vol]). Samples were rinsed by placing a 5-μl volume of 0.1% (vol/vol) trifluoroacetic acid (TFA) on the matrix surface after the analyte solution had dried completely. After 2 min, the liquid was blown off by pressurized air. MALDI mass spectra of peptide mixtures were obtained by using a Bruker Biflex mass spectrometer (Bruker-Franzen Analytik). Monoisotopic peptide masses were assigned and used for database searching. When no consistent hit was found, protein identification was achieved by tandem mass spectrometry (MS/MS) analysis. After in-gel tryptic digestion, the gel pieces were extracted with 5% (vol/vol) formic acid solution and then with acetonitrile. The extracts were combined with the original digest, and the sample was evaporated to dryness in a vacuum centrifuge. The residues were dissolved in 0.1% (vol/vol) formic acid and desalted with a Zip Tip (Millipore). Elution of the peptides was performed with 5 to 10 μl of a 50:50:0.1 (vol/vol) acetonitrile-H2O-formic acid solution. The peptide solution was introduced into a glass capillary (Protana) for nanoelectrospray ionization. MS/MS experiments were carried out on a Q-TOF hybrid mass spectrometer (Micromass) in order to obtain sequence information. Collision-induced dissociation (CID) of selected precursor ions was performed with argon as the collision gas and with collision energies of 40 to 60 eV. MS/MS sequence information was used for database searching with the programs (i) MS-Edman located at the University of California San Francisco (http: //prospector.ucsf.edu/), (ii) BLAST located at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/), and (iii) FASTS or TFASTS located at the University of Virginia (http://fasta.bioch.virginia.edu /fasta/cgi/).
Ub-binding assays.
Twenty-five microliters of extracts was diluted with 25 μl of buffer A containing 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 2 mM ATP, and 0.2 mM DTT. Ten microliters of 50% Ub-agarose matrix (preincubated with 1 mg of BSA per ml) was added. After 1 h at room temperature, Ub-agarose was pelleted and washed in buffer A. Bound proteins were eluted and used for Western blot analysis. Glutathione S-transferase (GST) pull-downs were performed by the same protocol.
Identification of cytoplasmic Ub-binding proteins (by Ub-agarose pull-down).
Five hundred microliters of cytoplasmic extract from mouse testis was incubated with 500 μg of free Ub (10 μg/μl; Sigma) or 50 μl of buffer A containing 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 2 mM ATP, and 0.2 mM DTT for 30 min at 4°C. The pull-down was performed by addition of 200 μl of 50% Ub-agarose matrix (preincubated with 1 mg of BSA per ml). After 1 h at room temperature, Ub-agarose was pelleted and washed three times with buffer A. Bound proteins were eluted in buffer A containing free Ub. Eluted proteins were concentrated and analyzed by SDS-PAGE and mass spectrometry.
In vitro ubiquitination reaction.
Reactions were performed in a buffer containing 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, and 0.2 mM DTT in a total volume of 20 μl. Cytoplasmic extracts from mouse testis were incubated for 30 min at room temperature with various amounts of bacterially expressed and purified His C-terminal mHDAC6 or the same fragment from wild-type mHDAC6 or m2 mutant fused to GST or BSA. Ubiquitination assay was performed by addition of 4 mM ATP and incubation for 1 h at room temperature. Reactions were stopped by addition of SDS-PAGE sample buffer, and ubiquitinated proteins were analyzed by SDS-PAGE and Western blotting with an antibody against Ub.
In situ immunodetection procedure.
Anti-HDAC6 was a rabbit polyclonal antibody raised against a C-terminal peptide (37). For in situ immunofluorescence analysis, cells were fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for 5 min at room temperature and permeabilized by the addition of 0.1% Triton X-100. Incubation with primary antibodies was carried out overnight at 4°C in the PBS-milk solution. After the addition of the secondary antibodies, cells were washed and counterstained with Hoechst 33258. The preparations were then observed under an epifluorescence microscope (Zeiss Axiophot). Numeric digital image acquisitions were realized with a cooled charge-coupled device camera (C4880 Hamamatsu). Cryosections and immunolocalization were performed as described before (6).
Plasmid constructs.
Vectors expressing hemagglutinin (HA)-tagged mHDAC1, -4, -5, or -6 have been described previously (23). mHDAC6 deletion mutants were obtained by PCR and substitution mutants by using the QuickChange site-directed mutagenesis kit (Stratagene) and controlled by sequencing. Human SUMO-1 and Ub cDNAs were cloned in pGEX-5X3 vector (Pharmacia) after reverse transcription-PCR (RT-PCR) amplification. The C-terminal Ub-binding domain of the mHDAC6 wild type or m2 mutant (amino acids 824 to 1149) was PCR amplified and cloned in either the pET28b (Novagen) or pGEX-5X3 plasmid. The histidine-tagged and GST fusion proteins were then purified with, respectively, the Ni-nitrilotriacetic acid (NTA) system (Qiagen) or gluthatione-Sepharose beads (Pharmacia).
RESULTS
mHDAC6 is predominantly expressed in spermatogenic cells.
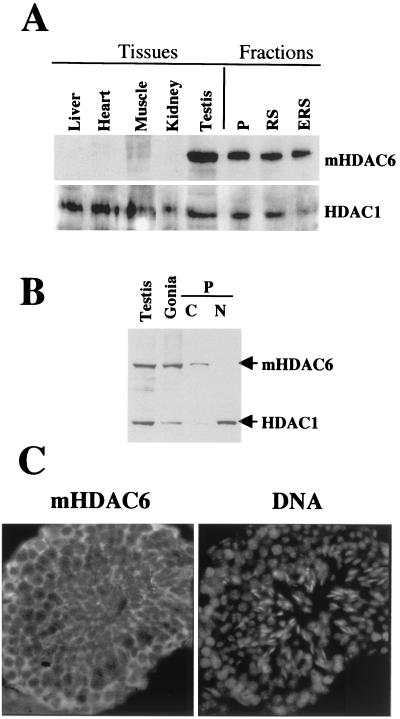
The expression level of mHDAC6 was first examined in various mouse tissues in order to find the most appropriate tissue for a large-scale purification of the endogenous protein. The use of highly-specific anti-mHDAC6 antibody produced in our laboratory (37) allowed us to show that mHDAC6 was predominantly expressed in the testis (Fig. 1A). We then looked at whether mHDAC6 was expressed in specific spermatogenic cell subpopulations and whether it was localized, as observed earlier in somatic cells (37), in the cell cytoplasm. For this purpose, spermatogenic cells were fractionated with a sedimentation chamber into three fractions enriched in pachytene spermatocytes (Fig. 1A, P), round spermatids (Fig. 1A, RS), and a mixture of round and elongated spermatids (Fig. 1A, ERS). We also prepared an extract from 6-day-old mice testes, which are known to contain essentially spermatogonia and Sertoli cells (3) (Fig. 1B, Gonia). Each of the cell populations examined contained considerable amounts of mHDAC6 (Fig. 1A, fractions). Moreover, as in tissue culture cells, mHDAC6 was localized predominantly in the cytoplasm (Fig. 1B). This conclusion was confirmed afterwards by in situ immunodetection of mHDAC6 on testis cryosections, in which mHDAC6 was found in the cytoplasm of the majority of cells (Fig. 1C). One should keep in mind that, although mHDAC6 is overexpressed in mouse testis, it is not encoded by a testis-specific gene, since the presence of mHDAC6 mRNA in various mouse tissues (36) was previously shown, as was the presence of the protein in four unrelated murine cell lines (37).
FIG. 1.
mHDAC6 is overexpressed in mouse testis. (A) Extracts prepared from the indicated mouse tissues were used to obtain a Western blot, which was successively probed with anti-HDAC6 and anti-HDAC1 antibodies. Fractions correspond to extracts prepared from spermatogenic cell populations enriched in the indicated cell types. P, pachytene; RS, round spermatids; RES, elongated and round spermatids. (B) Extracts from adult mouse testis (Testis) or from testis isolated from 6-day-old mice enriched in spermatogonia (Gonia), as well as cytoplasmic (C) and nuclear (N) extracts isolated from pachytene-enriched cells (P) were analyzed as in panel A. (C) A cryosection from adult mouse testis was used to immunodetect in situ mHDAC6 by using the anti-HDAC6 antibody (HDAC6 panel). The DNA panel shows corresponding Hoechst-labeled nuclei.
Immunopurification of mHDAC6-containing complex from testis cytosolic extracts.
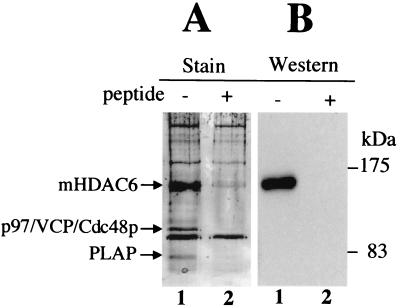
The preferential accumulation of mHDAC6 in the cytoplasm of mouse spermatogenic cells prompted us to use a testis cytoplasmic extract in order to immunoprecipitate mHDAC6 and identify the associated proteins. The immunopurified complex was eluted with the target immunogenic peptide. As a control, the procedure of immunopurification was carried out with an antibody preincubated with an excess of the immunogenic target peptide. A fraction of these immunoprecipitated materials (eluate from anti-mHDAC6 antibodies or peptide-blocked antibodies) was used to run a gel, which was then silver stained. Figure 2 clearly shows that mHDAC6 was very efficiently immunopurified with our anti-mHDAC6 antibody, while it was not with the same antibody blocked with the immunogenic peptide. The identity of mHDAC6 was confirmed by Western blot analysis of the materials described above. Interestingly, two bands, one migrating at 97 kDa and the other around 82 kDa, were specifically coimmunoprecipitated with mHDAC6 and were not present in eluate obtained from the peptide-blocked antibody. These proteins were then isolated from a preparative gel and digested with trypsin, and the fragments were analyzed by mass spectrometry. Comparison with the predicted tryptic fragments of protein databases identified one of these proteins as the mouse homologue of the yeast Cdc48p, p97/VCP/Cdc48 (11). The other protein was PLAP. A database search revealed that PLAP shows striking sequence homology to yeast UFD3 protein: 31 and 49% sequence identity and similarity, respectively, to the yeast protein (12; data not shown). Interestingly, it had been shown earlier in yeast that Cdc48p and UFD3 could form a specific complex (12). Our findings added to these data suggest that this interaction between Cdc48p and UFD3 would be conserved during evolution and therefore have important functional significance. Moreover, the interaction of mHDAC6 and p97/VCP/Cdc48 is not specific to testis, since we also observed the presence of both proteins in a complex in murine erythroleukemia cells by immunoprecipitating p97/VCP/Cdc48 (data not shown).
FIG. 2.
p97/VCP/Cdc48p and PLAP are associated with mHDAC6. (A) Mouse testis cytoplasmic extracts were used to immunoprecipitate mHDAC6 with an anti-mHDAC6 antibody (lane −) or the same antibody preincubated with its target peptide (lane +). Bound proteins were eluted and analyzed by SDS-PAGE and revealed after silver staining. The indicated bands were excised and identified by mass spectrometry (MALDI-TOF analysis and MS/MS). (B) The identity of mHDAC6 in the immunoprecipitates was confirmed by Western blotting.
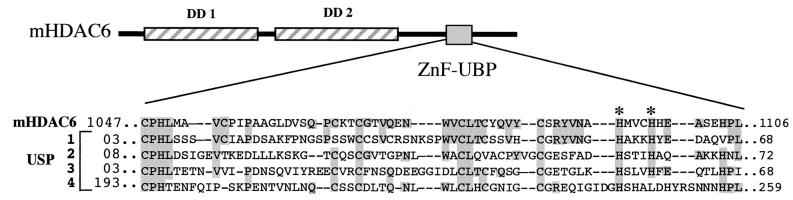
Sequence homology between several Ub-specific proteases and the C-terminal region of mHDAC6.
A search in the databank with the nondeacetylase regions of mHDAC6 showed a significant sequence homology between a limited region of mHDAC6 located in its C-terminal domain and particular members of Ub-specific proteases (UBPs) from different species (Fig. 3). Indeed, this region is cysteine and histidine rich, and both PFAM and SMART protein prediction web servers (33) identified it as the “ubiquitin carboxyl-terminal hydrolase-like zinc finger” (ZnF-UBP) domain. Enzymes involved in the cleavage of Ub fall into two families of cysteine proteases, UBPs and UCHs (Ub C-terminal hydrolases). These enzymes are capable of interacting with Ub, and in some of them, a domain known as UBA (Ub associated) has been shown to mediate this interaction (5, 15). However, since UBA is not present in all the UBPs and UCHs, we wondered whether the ZnF-UBP domain could be another Ub-binding domain present in UBA-less proteins. Accordingly, the Ub-binding activity of the mHDAC6 ZnF-UBP domain was then investigated.
FIG. 3.
Homology between the mHDAC6 C-terminal domain and Ub-specific proteases (USPs). The sequence of the C-terminal region of mHDAC6 was aligned with that of several USPs from different species. Identical amino acids are boxed. The USP sequences are as follows: 1, human USP3, AF073344; 2, human USP20, AB023220; 3, Schizosaccharomyces pombe USP, AL021838; 4, Saccharomyces cerevisiae USP, P38237. Asterisks indicate histidines replaced by alanines described in a further experiment.
mHDAC6 is a Ub-binding protein.
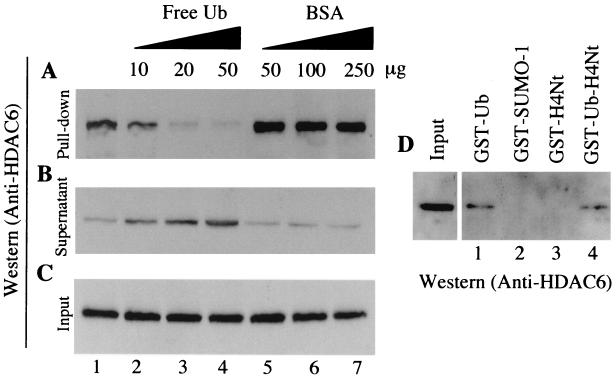
An extract from mouse testis was incubated with Ub-agarose, and a pull-down experiment was carried out. Figure 4A shows that the endogenous mHDAC6, present in the extract, was able to interact efficiently with Ub (lane 1). As a control, prior to the addition of the Ub-agarose, the extract was incubated with increasing amounts of free Ub (lanes 2 to 4) or BSA (lanes 5 to 7). The addition of free Ub abolished the binding of mHDAC6 to the Ub-agarose matrix, while the addition of the same number of BSA molecules had no effect on the mHDAC6-Ub interaction. The interaction with Ub was specific, since, while mHDAC6 in the extract could efficiently interact with GST-Ub (Fig. 4D, lane 1), no interaction was observed between mHDAC6 and GST-SUMO1, a Ub-like protein (lane 2). We also compared the ability of a fusion protein containing GST, Ub, and the N-terminal tail of histone H4, with that of the same fusion protein without the Ub part, to bind mHDAC6. Again, mHDAC6 was efficiently retained on the fusion protein containing Ub (Fig. 4D, lane 4), whereas no interaction was observed with GST-H4 N-terminal region (lane 3).
FIG. 4.
mHDAC6 specifically interacts with Ub. (A) Mouse testis cytoplasmic extracts were incubated with an Ub-agarose matrix (lane 1). In control experiments, the indicated amounts of free Ub (lanes 2 to 4) or BSA (lanes 5 to 7) were added to the extract before the addition of the Ub-agarose. After the pull-down, bound proteins were eluted and analyzed by Western blotting with an anti-mHDAC6 antibody. (B and C) Ten microliters of supernatant after the pull-down (B) and the same amounts of the input materials (C) were used to monitor the presence of mHDAC6. (D) mHDAC6 interacts with Ub-fusion proteins, but not with the GST–SUMO-1 fusion. The pull-down was performed as described above with the immobilized indicated GST-fusion proteins. Bound proteins were analyzed as described above with an anti-mHDAC6 antibody. H4Nt, the N-terminal H4 histone tail.
The mHDAC6 C-terminal ZnF-UBP domain mediates the binding of Ub.
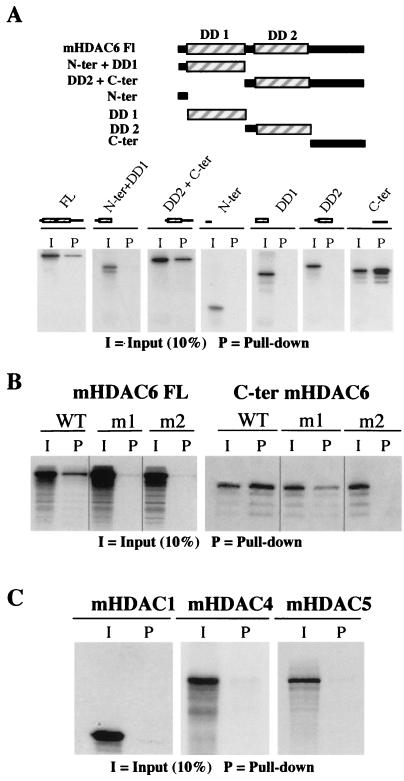
We then tried to identify the Ub-binding site of mHDAC6. Several deletion mutants of the protein were used to produce 35S-labeled proteins in vitro. Figure 5A shows that the C-terminal region of mHDAC6 is capable of efficient interaction with Ub (C-terminal panel). In agreement with this observation, all mHDAC6 fragments containing the C-terminal domain could also interact with Ub, while no interaction was observed between mHDAC6 truncation mutants lacking the C-terminal domain and Ub (Fig. 5A). In order to confirm the direct involvement of the mHDAC6 ZnF-UBP domain in Ub binding, histidines supposedly important in the formation of the zinc finger (Fig. 3) were mutated. In the full-length protein, the replacement of either one (m1 mutant) or two (m2 mutant) histidines drastically affected the Ub-binding capacity of mHDAC6 (Fig. 5B, left panel). The isolated 325-amino-acid C-terminal region of HDAC6 has a strong Ub binding activity (Fig. 5A, C-terminal panel, and 5B, right panel, wild type [WT]). The replacement of two histidines by alanines was necessary to completely abolish the binding to Ub by this fragment (Fig. 5B, m2 panel).
FIG. 5.
ZnF-UBP is the Ub-binding site of mHDAC6. (A) The indicated regions of mHDAC6 were cloned in an expression vector and used to generate 35S-labeled proteins in vitro. The pull-down was performed with Ub-agarose. (B) 35S-labeled mHDAC6 (full length or the C-terminal domain as indicated) bearing mutations in the ZnF-UBP (in m1, histidine 1098 is replaced by alanine, and in m2, histidines 1094 and 1098 are replaced [Fig. 3]) was produced and used in a Ub-agarose pull-down assay as described above. (C) mHDAC1, mHDAC4, and mHDAC5 were labeled in vitro and used in Ub pull-down assays as described above.
This Ub-binding capacity of mHDAC6 was not shared with other HDACs, since two other members of the class II HDACs, mHDAC4 and mHDAC5, and one member of the class I HDACs, mHDAC1, did not interact with Ub (Fig. 5C).
mHDAC6 is one of the major Ub-binding proteins present in the mouse testis cytosolic extract.
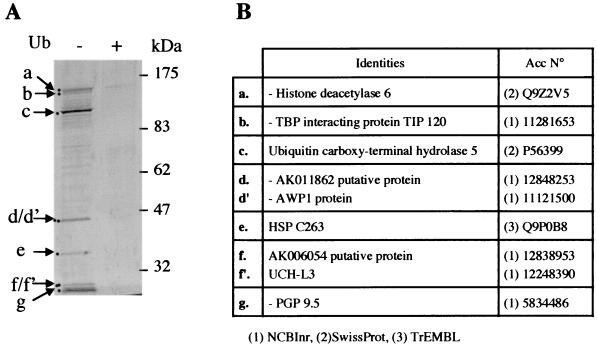
In order to approach the complexity and the nature of the Ub-binding proteins in the mouse testis cytosolic extracts, a systematic identification of Ub-binding proteins present in these extracts was performed. Nine proteins efficiently interacting with Ub-agarose were identified (Fig. 6). The binding of all of them to Ub-agarose was severely affected by the addition of free Ub to the extracts prior to the Ub-agarose pull-down experiment (Fig. 6, compare Ub− and Ub+ lanes). Interestingly, three of these proteins were members of Ub carboxy-terminal hydrolases (Ub carboxy-terminal hydrolase 5, UCH-L3, and PGP9.5) and one, Ub carboxy-terminal hydrolase 5, contained a ZnF-UBP. TIP120, a TATA-binding protein (TBP)-interacting protein that appears to be involved in the activation of transcription, was also found to be present in a complex containing several proteasomal ATPases (24). Four proteins with unknown function were also present in the pulled-down materials, AK011862, AWP1, HSP C263, and AK006054. The first two contained a Ub-like domain (not shown).
FIG. 6.
Identification of the major Ub-binding proteins present in mouse testis cytosolic extracts. (A) Proteins in mouse cytosolic testis extract retained on Ub-agarose beads were visualized on silver-stained SDS gel and compared with proteins retained on Ub-agarose beads when the pull-down was performed in the presence of an excess of free Ub (lane +). (B) The indicated bands were excised from a Coomassie blue-stained preparative SDS gel and identified by mass spectrometry as in Fig. 2. The table shows the identity of the excised bands. The right column shows the accession number and the corresponding database.
Involvement of mHDAC6 in the control of protein ubiquitination.
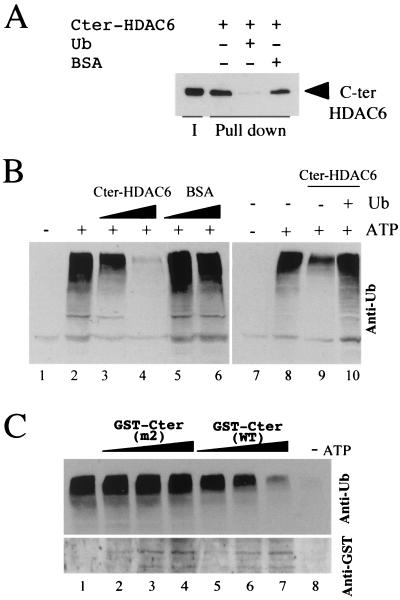
Data obtained so far strongly suggest that mHDAC6 may have a role in protein ubiquitination. A direct approach to investigate this issue would have been the use of an in vitro ubiquitination system. However, such a system would have hardly allowed the study of the role of mHDAC6 activity in protein ubiquitination. Indeed, if using an extract, putative acetylated substrate proteins could be nonspecifically deacetylated due to the presence of free and active deacetylases. If use of a purified system containing E1, E2 and E3 (see for instance, reference 22) had been chosen, one would first have to find an acetylable substrate, find an appropriate acetylase to acetylate it, and then evaluate the role of mHDAC6 and its deacetylase activity in protein ubiquitination. Considering all these difficulties, we chose to look at the influence of the Ub-binding 325-amino-acid C-terminal domain of mHDAC6 on in vitro protein ubiquitination. For this purpose, the histidine-tagged C-terminal domain of mHDAC6 was expressed in bacteria, and the purified protein was used to show its ability to directly bind to Ub (Fig. 7A). We also showed that the mouse cytosolic extract was very efficient to direct protein ubiquitination upon the addition of ATP (Fig. 7B, compare lanes 1 and 2). The addition of the increasing amounts of mHDAC6 C-terminal domain interfered severely with protein ubiquitination in the extract (lanes 3 and 4). This inhibiting effect was relieved by the presence of free Ub (lane 10). These data confirmed the ability of mHDAC6 to specifically interact with Ub in the extract. They also showed that the interaction between mHDAC6 C-terminal domain and Ub severely interfered with the in vitro ubiquitination of proteins in the extract. In order to confirm these results, recombinant proteins made of GST fused to the C-terminal region of mHDAC6 either from the m2 mutant unable to bind to Ub (Fig. 5B) or from the wild type protein were also expressed in bacteria and purified.
FIG. 7.
Ub binding by mHDAC6 C-terminal region interferes with in vitro ubiquitination of proteins present in mouse testis cytosolic extract. (A) mHDAC6 binds directly to Ub. mHDAC6 C-terminal end (amino acids 824 to 1149) was cloned in an expression vector (pET-28a+; NOVAGEN). His-HDAC6 C-terminal region was produced and purified and used to perform Ub-agarose pull-down experiments in the absence (−) or presence (+) of free Ub or BSA as indicated. After the pull-down, bound proteins were eluted and analyzed by Western blotting with an anti-mHDAC6 antibody. I, indicates 50% of the input material; lanes labeled Pull down show materials eluted after Ub-agarose pull-down. (B) Mouse testis cytosolic extract is capable of directing an in vitro ubiquitation of extract proteins in an ATP-dependent manner. Cytosolic testis extracts were incubated in the absence (−) or presence (+) of ATP and increasing amounts of the purified mHDAC6 C-terminal fragment (lanes 3 and 4) and the same number of molecules of BSA (lanes 5 and 6). After incubation, a fraction of the extract was used to monitor protein ubiquitination with a Western blot and an anti-Ub antibody (Santa Cruz). The right panel shows that the addition of free Ub relieves the mHDAC6 C-terminal-mediated repression of protein ubiquitination (compare lanes 9 and 10). (C) The C-terminal region of mHDAC6 (the same fragment described above) from the wild-type protein or the m2 mutants (Fig. 3 and 5B) was fused to GST, expressed in bacteria, and purified (GST-Cter WT and GST-Cter m2, respectively). Increasing amounts of recombinant proteins were added to the extract as described above, and protein ubiquitination was monitored (upper panel). The lower panel shows the amount of GST-fusion proteins added to the extract in this experiment by analyzing a fraction of the samples with an anti-GST antibody. Lane 1 shows a standard ubiquitination reaction, and lane 8 shows the same reaction without ATP.
The influence of these recombinant proteins on in vitro protein ubiquitination was monitored as described above. Figure 7C shows that, while the GST containing the wild-type C-terminal region of mHDAC6 severely interfered with protein ubiquitination (lanes 5 to 7), the mutated version of this protein unable to bind to Ub did not affect protein ubiquitination in the extract (lanes 2 to 4).
We therefore suggested that mHDAC6 recruited by ubiquitinated proteins would have to be removed before the addition of other Ub moieties and multi-Ub chain assembly. In agreement with this hypothesis, we found that Ub binding led to the dissociation of the mHDAC6-containing complex. This implies that the mHDAC6-interacting proteins present in the complex could play a role in dissociating Ub-mHDAC6 (see below).
Ub binding dissociates the mHDAC6-containing complex.
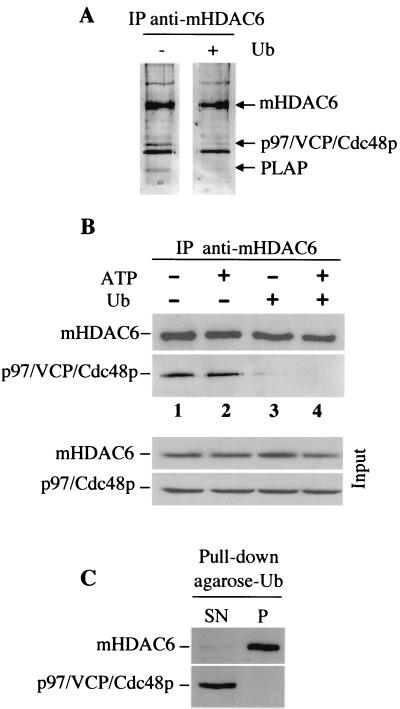
Interestingly, the analysis of the proteins interacting with Ub in the testis cytosolic extract (Fig. 6) did not show the presence of either p97/VCP/Cdc48 or PLAP. This observation suggests either that the mHDAC6 fraction present in the complex does not interact with Ub or that Ub binding dissociates the complex. In order to investigate this issue, the testis cytosolic extract was incubated with free Ub and mHDAC6 was immunoprecipitated with the anti-mHDAC6 antibody. The immunoprecipitated materials were then eluted with the immunogenic peptide and visualized on a silver-stained SDS-PAGE gel. This experiment, performed at the same time as that described in the legend to Fig. 2, showed that the coimmunoprecipitation of p97/VCP/Cdc48 and PLAP was lost when free Ub had been added to the extract (Fig. 8A, compare lanes − and +). In order to confirm these findings, the immunoprecipitation of mHDAC6 was performed under different conditions, and the presence of p97/VCP/Cdc48 and mHDAC6 was monitored with specific antibodies. Figure 8B shows that, while p97/VCP/Cdc48p coimmunoprecipitated with mHDAC6 in the absence of Ub (Fig. 8B, lanes 1 and 2), p97/VCP/Cdc48p was found in a very small amount in the immunoprecipitated material from Ub-containing extracts (lanes 3 and 4). Since p97/VCP/Cdc48p is an ATPase, we also tested the role of ATP in this dissociation process and found that the release of p97/Cdc48p was independent of ATP hydrolysis. Finally, we performed Ub pull-down assays with Ub-agarose as described above and analyzed a fraction of Ub-bound and unbound materials with anti-mHDAC6 and anti-p97/VCP/Cdc48p antibodies. Figure 8C shows that, while almost all of the mHDAC6 present in the extract could be removed by using Ub-agarose beads, all of the p97/VCP/Cdc48p was found in the unbound material (Fig. 8C, SN lane). No p97/VCP/Cdc48p was found in the bound materials (lane P).
FIG. 8.
Ub binding by mHDAC6 induces the release of p97/VCP/Cdc48p and PLAP. mHDAC6 from mouse cytosolic extracts was immunoprecipitated (IP)as described in the legend to Fig. 2 in the presence or absence of free Ub (+ and − lanes, respectively). Immunoprecipitated proteins were eluted with the immunogenic peptide and analyzed on a silver-stained gel. (This experiment was performed in parallel with that shown in Fig. 2A, and Fig. 2A and 8A are from the same gel.) The positions of mHDAC6, p97/VCP/Cdc48p, and PLAP are indicated. (B) Prior to immunoprecipitations of mHDAC6, the mouse testis extracts were preincubated with or without ATP (2 mM) and/or Ub as indicated. The presence of mHDAC6 and p97/VCP/Cdc48p in the immunoprecipitates was then determined by Western blotting with the corresponding antibodies. The input panel represents 2% of the amount of extract used in the immunoprecipitation. (C) Mouse cytosolic extracts were subjected to Ub pull-down with Ub-agarose beads as described in the legends to Fig. 4 and 6. Unbound materials present in the supernatant (SN) after the pull-down and bound materials retained on Ub-agarose beads (P) were used to monitor the presence of p97/VCP/Cdc48p and mHDAC6 with the corresponding antibodies.
Ub-bound mHDAC6 maintains its deacetylase activity.
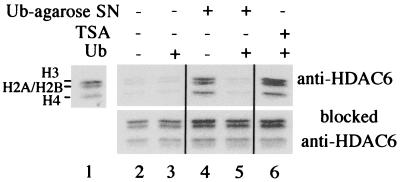
Since Ub binding could modulate the nature of the mHDAC6 complex, we wanted to know whether mHDAC6, when recruited by ubiquitinated proteins, would maintain its deacetylase activity and could therefore deacetylate critical lysines on the ubiquitinated protein itself or on other interacting proteins. The immunopurified mHDAC6 complex (eluate from anti-mHDAC6 antibodies or peptide-blocked antibodies) was used to deacetylate 3H-labeled hyperacetylated histones. Autoradiography allows monitoring of histone deacetylation, which results in the removal of the radioactive label from histones. Figure 9 shows that the binding of Ub by mHDAC6 did not modify its deacetylase activity (compare lanes 2 and 3), which remained trichostatin A (TSA) sensitive (lane 6). In order to show the Ub-binding ability of this purified mHDAC6 complex in these assays, the purified complex was incubated with the Ub-agarose matrix, in the presence or absence of free Ub, and centrifuged, and the supernatant was used to perform the deacetylase assay (Ub-agarose SN). Our data showed that Ub-agarose could deplete the deacetylase activity in the supernatant (lane 4) and that the presence of free Ub inhibited this depletion (lane 5). These experiments show that Ub can recruit active mHDAC6, which suggests that ubiquitinated proteins would also recruit the active deacetylase. Moreover, dissociation of Ub-bound mHDAC6 by histones during the reaction period was ruled out, since the incubation of mHDAC6-containing Ub-agarose beads with acetylated histones during various times did not lead to the release of mHDAC6 (not shown).
FIG. 9.
Ub recruits an active mHDAC6. Twenty microliters of immunopurified mHDAC6 (obtained as in Fig. 2) was incubated with 3H-labeled acetylated histones in the absence (lane 2) or in the presence of free Ub (lane 3). Lane 6 shows the inhibition of the deacetylase activity of the Ub-bound mHDAC6 by 100 ng of TSA per ml. The reaction mixtures containing or not containing free Ub were incubated with Ub-agarose and centrifuged, and labeled histones were added to the supernatant (Ub-agarose SN) to measure the deacetylase activity (lanes 4 and 5, respectively). Lane 1 shows the input of labeled histones. (Lower panels) As controls, the same experiments as those described above were performed, except that eluates from peptide-blocked anti-mHDAC6, obtained as described in the legend to Fig. 2, were analyzed.
DISCUSSION
Two lines of evidence suggest an involvement of mHDAC6 in the Ub-dependent signaling processes. First, two proteins copurified with mHDAC6 showed striking sequence homology to yeast proteins involved in Ub-dependent protein degradation. The first one, p97/VCP/Cdc48p, an AAA-ATPase, was shown to participate in different functions, depending on its partner proteins (29). Its role in Ub-dependent protein degradation was first suggested in yeast, where it was found in a complex with UFD3 (12) and then with E4/UFD2 (22). Moreover, p97/VCP/Cdc48p was recently shown to form a complex with the mammalian homologue of yeast UFD1 (27). Finally, p97/VCP/Cdc48 was found to specifically associate with the ubiquitinated forms of IκBα and to probably control its proteasome-dependent degradation (10). The second protein, PLAP, is a mammalian homologue of yeast UFD3. PLAP is involved in the activation of several phospholipases (30). In yeast, UFD3 seems to play a role in the control of the concentration of free Ub in cells (19). Indeed, UFD3 mutants have reduced levels of intracellular free Ub. The degradation of the UFD substrates in these mutants could be restored by overexpressing Ub. It is not clear how UFD3 may control the concentration of cellular Ub in yeast. Since UFD3/PLAP is capable of activating phospholipases, specifically phospholipase A2 (PLA2) (30), one may expect the involvement of these enzymes in the control of the free cellular Ub level. Interestingly, there is one hint in the literature regarding the participation of PLA2 in the control of Ub concentration. Indeed, it has been shown that a membrane-bound form of Ub was associated with budded virions of baculovirus and possessed a phospholipid anchor, which could be removed by PLA2 treatment (14). Therefore, PLA2 could release Ub from the membrane. Although the massive presence of cellular Ub in a membrane-bound form has not been evidenced, it is possible that PLAP/UFD3 participates in the release of this putative pool through the activation of PLA2. In the yeast UFD3 mutant, the absence of this PLAP-like activity might be responsible for the observed decrease in the amount of intracellular free Ub.
The second line of evidence in favor of the involvement of mHDAC6 in protein ubiquitination relies on the analysis of the structure of mHDAC6 itself. Indeed, a Ub carboxyl-terminal hydrolase-like zinc finger (ZnF-UBP) domain was found in the C-terminal region of mHDAC6. This particular domain with unknown function is shared with a number of Ub-specific proteases (1). Here we showed that this finger specifically mediated the interaction of mHDAC6 with Ub. Besides mHDAC6, we have also identified eight other Ub-binding proteins in the mouse testis cytosolic extract. Several are Ub C-terminal hydrolases, which very probably directly interact with Ub. One of them, the Ub carboxy-terminal hydrolase 5, also contained a ZnF-UBP domain (not shown). Four noncharacterized proteins were also on the list, and two of them showed a specific structural motif suggesting a relationship with Ub signaling pathways. Indeed, both AK011826 and AWPI possess a Ub-like domain (not shown), which is also found in several proteins containing a small region of limited sequence identity to Ub (9).
Most interestingly, we also found TIP120 protein among the Ub-associated proteins. TIP120, a TBP-interacting protein, activates the basal level of transcription from all three classes (I, II, and III) of promoters (25). TIP120 is also part of a complex containing several proteasomal ATPases (24). TIP120 is therefore thought to participate in both transcriptional regulation and proteasome-dependent protein degradation. Here we report a novel property of TIP120: its ability to interact with Ub.
In the literature, besides enzymes directly involved in protein ubiquitination, other proteins with Ub-binding activity have been described. Indeed, several proteins involved in diverse functions share a potential Ub-binding domain termed UBA. Two plant de novo methyltransferases (7), as well as Rad23, involved in DNA repair (32), and the mammalian protein, p62 (35), are among these proteins. The UBA domain very probably links the activity of these proteins to that of the Ub-dependent signaling pathway. Indeed, the UBA domain was originally defined in various components of the Ub-dependent protein degradation pathway, such as Ub C-terminal hydrolases (UCHs), Ub-conjugating enzymes (E2), and Ub protein ligases (E3) (16). In the case of Rad23, it has recently been shown that UBA mediated the specific binding of Ub (5) and that this interaction inhibited multi-Ub chain formation (28).
Here we found that the interaction of mHDAC6 with Ub led to the dissociation of the mHDAC6 complex and notably to the release of p97/VCP/Cdc48. Interestingly, it has been reported that Ub binding by yeast E4/UFD2 resulted in the release of Cdc48p (22). Therefore, mHDAC6 shares three properties with the yeast E4/UFD2: (i) Ub binding, (ii) interaction with p97/VCP/Cdc48, and (iii) release of this partner after the interaction with Ub. However, it is not clear how mHDAC6 participates in the control of protein ubiquitination. Our data show that free Ub, and even a Ub sequence inserted in the middle of a protein (GST-Ub-H4) (Fig. 4), can recruit mHDAC6. The latter suggests that once a protein is monoubiquitinated, it becomes a potential target for mHDAC6. After the recruitment of mHDAC6 by a monoubiquitinated protein, at least two scenarios can be considered. First, mHDAC6 would deacetylate critical lysines on the substrate protein (and/or partner proteins) and to allow their ubiquitination. Indeed, the deacetylation of specific lysines may allow their subsequent ubiquitination. Second, mHDAC6 would control the activity of the ubiquitination machinery by deacetylating them. Our in vitro assays suggested that the Ub-mHDAC6 complex would have to be dissociated for the ubiquitination of substrate proteins to take place. Since the Ub-mHDAC6 interaction seems to dissociate the mHDAC6-p97/VCP/Cdc48p complex, one may assume that p97/VCP/Cdc48p could play a reverse role and would help mHDAC6 to release Ub. In agreement with this hypothesis, it has been suggested that the basic activity of p97/VCP/Cdc48p would be protein unfolding or disassembly of protein complexes (29).
Our experiments also suggest another property of mHDAC6. Indeed, the isolated C-terminal domain of mHDAC6 seems to have a better Ub-binding capacity than the full-length protein (Fig. 5A and B). This may suggest that the Ub-binding activity of mHDAC6 could be dependent on its conformation, which could itself be controlled by posttranslational modifications or its interaction with other partner proteins.
Unfortunately, there is no hint in the literature about a possible mechanism linking protein acetylation to the Ub signaling pathway. However, a relationship between protein acetylation and stability has recently been demonstrated in the case of E2F, which shows an increased half-life when acetylated (26). Our work therefore strongly suggests a link between two key posttranslational protein modifications—acetylation and ubiquitination—both modifying lysine residues.
ACKNOWLEDGMENTS
We are grateful to Jean-Jacques Lawrence for encouraging this work and N. Tonks for providing the anti-p97/Cdc48p antibody. We thank Marie-Paule Brocard for technical assistance.
D.S.-B. is a recipient of a postdoctoral fellowship from the Association pour la Recherche sur le Cancer, and A.V. is a recipient of a Ph.D. fellowship from the Ligue Nationale Contre le Cancer, Comité de la Haute Savoie. This work was supported by the Association pour la Recherche sur le Cancer (ARC).
REFERENCES
- 1.Amerik A Y, Li S J, Hochstrasser M. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol Chem. 2000;381:981–992. doi: 10.1515/BC.2000.121. [DOI] [PubMed] [Google Scholar]
- 2.Barlow A L, van Drunen C M, Johnson C A, Tweedie S, Bird A, Turner B M. dSIR2 and dHDAC6: two novel, inhibitor-resistant deacetylases in Drosophila melanogaster. Exp Cell Res. 2001;265:90–103. doi: 10.1006/excr.2001.5162. [DOI] [PubMed] [Google Scholar]
- 3.Bellvé A R, Cavicchia J C, Millette C F, O'Brien D A, Bhatnagar Y M, Dym M. Spermatogenic cells of the prepubertal mouse. Isolation and morphological characterization. J Cell Biol. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellvé A R. Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol. 1993;225:84–113. doi: 10.1016/0076-6879(93)25009-q. [DOI] [PubMed] [Google Scholar]
- 5.Bertolaet B L, Clarke D J, Wolff M, Watson M H, Henze M, Divita G, Reed S I. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat Struct Biol. 2001;8:417–422. doi: 10.1038/87575. [DOI] [PubMed] [Google Scholar]
- 6.Callanan M, Kudo N, Gout S, Brocard M P, Yoshida M, Dimitrov S, Khochbin S. Developmentally regulated activity of CRM1/XPO1 during xenopus embryogenesis. J Cell Sci. 2000;113:451–459. doi: 10.1242/jcs.113.3.451. [DOI] [PubMed] [Google Scholar]
- 7.Cao X, Springer N M, Muszynski M G, Phillips R L, Kaeppler S, Jacobsen S E. Conserved plant genes with similarity to mammalian de novo DNA methyltransferases. Proc Natl Acad Sci USA. 2000;97:4979–4984. doi: 10.1073/pnas.97.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conforti L, Tarlton A, Mack T G, Mi W, Buckmaster E A, Wagner D, Perry V H, Coleman M P. A Ufd2/D4Cole1e chimeric protein and overexpression of Rbp7 in the slow Wallerian degeneration (WldS) mouse. Proc Natl Acad Sci USA. 2000;97:11377–11382. doi: 10.1073/pnas.97.21.11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conklin D, Holderman S, Whitmore T E, Maurer M, Feldhaus A L. Molecular cloning, chromosome mapping and characterization of UBQLN3 a testis-specific gene that contains an ubiquitin-like domain. Gene. 2000;249:91–98. doi: 10.1016/s0378-1119(00)00122-0. [DOI] [PubMed] [Google Scholar]
- 10.Dai R M, Chen E, Longo D L, Gorbea C M, Li C C. Involvement of valosin-containing protein, an ATPase co-purified with IkappaBalpha and 26 S proteasome, in ubiquitin-proteasome-mediated degradation of IkappaBalpha. J Biol Chem. 1998;273:3562–3573. doi: 10.1074/jbc.273.6.3562. [DOI] [PubMed] [Google Scholar]
- 11.Frohlich K U, Fries H W, Rudiger M, Erdmann R, Botstein D, Mecke D. Yeast cell cycle protein CDC48p shows full-length homology to the mammalian protein VCP and is a member of a protein family involved in secretion, peroxisome formation, and gene expression. J Cell Biol. 1991;114:443–453. doi: 10.1083/jcb.114.3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghislain M, Dohmen R J, Levy F, Varshavsky A. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 1996;15:4884–4899. [PMC free article] [PubMed] [Google Scholar]
- 13.Gray S G, Ekström T J. The human histone deacetylase family. Exp Cell Res. 2001;262:75–83. doi: 10.1006/excr.2000.5080. [DOI] [PubMed] [Google Scholar]
- 14.Guarino L A, Smith G, Dong W. Ubiquitin is attached to membranes of baculovirus particles by a novel type of phospholipid anchor. Cell. 1995;80:301–309. doi: 10.1016/0092-8674(95)90413-1. [DOI] [PubMed] [Google Scholar]
- 15.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann K, Bucher P. The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem Sci. 1996;21:172–173. [PubMed] [Google Scholar]
- 17.Huibregtse J M, Scheffner M, Howley P M. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol Cell Biol. 1993;13:4918–4927. doi: 10.1128/mcb.13.8.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huibregtse J M, Scheffner M, Howley P M. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol Cell Biol. 1993;13:775–784. doi: 10.1128/mcb.13.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson E S, Ma P C, Ota I M, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- 20.Khochbin S, Verdel A, Lemercier C, Seigneurin-Berny D. Functional significance of histone deacetylase diversity. Curr Opin Genet Dev. 2001;11:162–166. doi: 10.1016/s0959-437x(00)00174-x. [DOI] [PubMed] [Google Scholar]
- 21.Khochbin S, Kao H Y. Histone deacetylase complexes: functional entities or molecular reservoirs. FEBS Lett. 2001;494:141–144. doi: 10.1016/s0014-5793(01)02327-4. [DOI] [PubMed] [Google Scholar]
- 22.Koegl M, Hoppe T, Schlenker S, Ulrich H D, Mayer T U, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 23.Lemercier C, Verdel A, Galloo B, Curtet S, Brocard M P, Khochbin S. mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity. J Biol Chem. 2000;275:15594–15599. doi: 10.1074/jbc.M908437199. [DOI] [PubMed] [Google Scholar]
- 24.Makino Y, Yoshida T, Yogosawa S, Tanaka K, Muramatsu M, Tamura T A. Multiple mammalian proteasomal ATPases, but not proteasome itself, are associated with TATA-binding protein and a novel transcriptional activator, TIP120. Genes Cells. 1999;4:529–539. doi: 10.1046/j.1365-2443.1999.00277.x. [DOI] [PubMed] [Google Scholar]
- 25.Makino Y, Yogosawa S, Kayukawa K, Coin F, Egly J M, Wang Z, Roeder R G, Yamamoto K, Muramatsu M, Tamura T. TATA-binding protein-interacting protein 120, TIP120, stimulates three classes of eukaryotic transcription via a unique mechanism. Mol Cell Biol. 1999;19:7951–7960. doi: 10.1128/mcb.19.12.7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-Balbas M A, Bauer U M, Nielsen S J, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19:662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer H H, Shorter J G, Seemann J, Pappin D, Warren G. A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 2000;19:2181–2192. doi: 10.1093/emboj/19.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ortolan T G, Tongaonkar P, Lambertson D, Chen L, Schauber C, Madura K. The DNA repair protein Rad23 is a negative regulator of multi-ubiquitin chain assembly. Nat Cell Biol. 2000;2:601–608. doi: 10.1038/35023547. [DOI] [PubMed] [Google Scholar]
- 29.Patel S, Latterich M. The AAA team: related ATPases with diverse functions. Trends Cell Biol. 1998;8:65–71. [PubMed] [Google Scholar]
- 30.Ricardo D A, Crowe S E, Kuhl K R, Peterson J W, Chopra A K. Prostaglandin levels in stimulated macrophages are controlled by phospholipase A2-activating protein and by activation of phospholipase C and D. J Biol Chem. 2001;276:5467–5475. doi: 10.1074/jbc.M006690200. [DOI] [PubMed] [Google Scholar]
- 31.Romrell L J, Bellve A R, Fawcett D W. Separation of mouse spermatogenic cells by sedimentation velocity. Dev Biol. 1976;49:119–131. doi: 10.1016/0012-1606(76)90262-1. [DOI] [PubMed] [Google Scholar]
- 32.Schauber C, Chen L, Tongaonkar P, Vega I, Lambertson D, Potts W, Madura K. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature. 1998;391:715–718. doi: 10.1038/35661. [DOI] [PubMed] [Google Scholar]
- 33.Schultz J, Milpetz F, Bork P, Ponting C P. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwarz S E, Rosa J L, Scheffner M. Characterization of human hect domain family members and their interaction with UbcH5 and UbcH7. J Biol Chem. 1998;273:12148–12154. doi: 10.1074/jbc.273.20.12148. [DOI] [PubMed] [Google Scholar]
- 35.Vadlamudi R K, Joung I, Strominger J L, Shin J. p62, a phosphotyrosine-independent ligand of the SH2 domain of p56lck, belongs to a new class of ubiquitin-binding proteins. J Biol Chem. 1996;271:20235–20237. doi: 10.1074/jbc.271.34.20235. [DOI] [PubMed] [Google Scholar]
- 36.Verdel A, Khochbin S. Identification of a new family of higher eukaryotic histone deacetylases. J Biol Chem. 1999;274:2440–2445. doi: 10.1074/jbc.274.4.2440. [DOI] [PubMed] [Google Scholar]
- 37.Verdel A, Curtet S, Brocard M P, Rousseaux S, Lemercier C, Yoshida M, Khochbin S. Active maintenance of mHDA2/mHDAC6 histone-deacetylase in the cytoplasm. Curr Biol. 2000;10:747–749. doi: 10.1016/s0960-9822(00)00542-x. [DOI] [PubMed] [Google Scholar]
- 38.Xie Y, Varshavsky A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback. Proc Natl Acad Sci USA. 2001;98:3056–3061. doi: 10.1073/pnas.071022298. [DOI] [PMC free article] [PubMed] [Google Scholar]