Abstract
In Saccharomyces cerevisiae, four factors [cleavage factor I (CF I), CF II, polyadenylation factor I (PF I), and poly(A) polymerase (PAP)] are required for maturation of the 3′ end of the mRNA. CF I and CF II are required for cleavage; a complex of PAP and PF I, which includes CF II subunits, participates in polyadenylation, along with CF I. These factors are directed to the appropriate site on the mRNA by two sequences: one A-rich and one UA-rich. CF I contains five proteins, two of which, Rna15 and Hrp1, interact with the mRNA through RNA recognition motif-type RNA binding motifs. Previous work demonstrated that the UV cross-linking of purified Hrp1 to RNA required the UA-rich element, but the contact point of Rna15 was not known. We show here that Rna15 does not recognize a particular sequence in the absence of other proteins. However, in complex with Hrp1 and Rna14, Rna15 specifically interacts with the A-rich element. The Pcf11 and Clp1 subunits of CF I are not needed to position Rna15 at this site. This interaction is essential to the function of CF I. A mutant Rna15 with decreased affinity for RNA is defective for in vitro RNA processing and lethal in vivo, while an RNA with a mutation in the A-rich element is not processed in vitro and can no longer be UV cross-linked to the Rna15 subunit assembled into CF I. Thus, the recognition of the A-rich element depends on the tethering of Rna15 through an Rna14 bridge to Hrp1 bound to the UA-rich motif. These results illustrate that the yeast 3′ end is defined and processed by a mechanism surprisingly different from that used by the mammalian system.
The 3′ ends of most eukaryotic mRNAs undergo maturation before export of the transcript to the cytoplasm. This maturation occurs in two steps that are tightly coupled in vivo but can be experimentally uncoupled in vitro. First, the nascent transcript is cleaved at a specific site downstream of the translational stop codon, and then a poly(A) tail is added. Processing is performed by a multisubunit complex that provides the nuclease activity and also confers sequence specificity to a template-independent poly(A) polymerase (PAP) (43, 68, 71). This event regulates the efficiency of transcriptional termination (14, 29, 51), nuclear export (13, 30, 36), and translation (54) and contributes to the longevity of the RNA in the cytoplasm (46, 63, 70). The choice of poly(A) site can even change the amount of sequence found in the mRNA and is thus an additional mechanism for the control of gene expression (18).
An intricate dance involving the recognition of cis-acting signals in the pre-mRNA by multisubunit protein factors defines the cleavage site by directing the processing machinery to the appropriate location on the nascent transcript (25, 71). In mammals, cleavage of the nascent mRNA precursor occurs at a loosely conserved element called the poly(A) site, usually containing a CA dinucleotide. This site is flanked by three other sequences which control the exact location and efficiency of cleavage. These are called the A-rich element, the downstream element, and the upstream element. The A-rich element, most often the sequence AAUAAA, is 10 to 30 nucleotides upstream from the poly(A) site. This element is highly conserved within the animal, plant, and fungal kingdoms. The downstream element is U-rich or GU-rich and lies within 50 nucleotides downstream of the poly(A) site. Mutations in either element affect the efficiency of the 3′-end processing machinery, and the positioning of the cleavage site is affected by the distance between these two signals. Upstream elements, found in some viral and cellular genes, are not essential but stimulate the overall efficiency of processing.
Four multisubunit protein factors, plus RNA polymerase II, are required for cleavage of the mammalian mRNA precursor and have been extensively characterized biochemically. They are called cleavage-polyadenylation specificity factor (CPSF), cleavage stimulatory factor (CstF), and mammalian cleavage factors I and II (CF IM and CF IIM). Cleavage is most efficient in the presence of PAP and RNA polymerase II. Polyadenylation of the cleaved transcript requires CPSF, poly(A) binding protein II (PAB II), and PAP (43, 68, 71).
The actual 3′-end processing reaction occurs within a structure created through interactions of these factors with each other and with the pre-mRNA. The binding of CPSF to the A-rich sequence and of CstF to the downstream element is strengthened through protein-mediated interactions of the two factors (43, 71). Since CF IM and CF IIM are required for the cleavage step, these factors may be directed to the poly(A) site through interactions with bound CPSF or CstF (15) and through the binding of CF IM to the mRNA (52). The interaction of CPSF at the AAUAAA signal also gives specificity to the PAP, and the binding of PAB II to the nascent poly(A) tail stabilizes the polyadenylation complex (7).
In Saccharomyces cerevisiae a similar but not identical machinery is involved in 3′ end maturation. At least four different types of elements contribute to the efficiency and accuracy of yeast 3′-end processing (26). As in mammals, the poly(A) site is the location of endonucleolytic cleavage of the mRNA precursor, which then becomes the site of initiation for polymerization of the adenosine tail. Mutagenesis studies with the CYC1 and ADH1 genes (27, 53) have defined the poly(A) site as Py(A)n. As in mammals, the A-rich sequence, ca. 20 bases upstream of the cleavage site, is extremely well conserved (22, 66). In vivo, mutations in this element most often do not affect the overall efficiency of processing but instead cause a defocusing of the endonucleolytic activity through the activation of previously cryptic poly(A) sites. Because of this effect, this motif has often been referred to as the positioning element (25).
Two global sequence analyses detected a bias toward U-rich sequences immediately upstream and downstream of the poly(A) site (22, 66), and mutation of these sequences affects 3′-end formation in vitro and in vivo (16). However, additional downstream sequence does not appear to be very important for in vivo processing (3, 55), a finding consistent with the finding that precursors with only 7 to 10 nucleotides beyond the cleavage site are readily processed in vitro (10, 11).
Instead of a downstream element, yeast mRNAs contain a functionally analogous sequence which is UA-rich in nature and located upstream of the A-rich sequence. This sequence can function from up to 30 nucleotides away from the A-rich sequence and, in some genes, in both orientations. Mutations in this sequence reduce the overall efficiency but not the accuracy of the cleavage reaction on the mRNA in vivo (71).
The machinery required for processing of yeast mRNA 3′ ends consists of four factors, defined by chromatographic fractionation of whole-cell extract (11). CF I and II recognize the processing signals on the mRNA and perform the endonucleolytic cleavage, whereas CF I, polyadenylation factor I (PF I), and PAP are required for the polyadenylation step. A factor containing the CF II subunits combined with a cohort of polyadenylation-specific proteins can be isolated by affinity chromatography (50). This complex can provide both CF II and PF I activities and has been designated cleavage-polyadenylation factor (CPF). Unlike the mammalian system, much less is known about the means through which these factors recognize the cis elements contained in the pre-mRNA.
When purified CF II is used in cross-linking experiments, the Cft2 subunit cross-links to a model mRNA precursor only if the substrate contains the UA-rich element and sequence beyond the poly(A) site (72). A recent study using RNase H protection analysis has found that CPF, as well as recombinant Cft2, interacts with the region around the poly(A) site (16).
CF I contains two proteins with obvious RNP-type RNA-binding motifs: Rna15 (45) and Hrp1 (33). Hrp1 has been shown to require the UA-rich element for binding to mRNA (12, 33), and SELEX experiments with Hrp1 have selected (UA)3 sequences (65). Mutations in the RNA binding domains of Hrp1 generate a thermosensitive phenotype (33), implying that recognition of the UA-rich element is an essential step in mRNA processing. Rna15 contains a single RNA recognition motif (RRM) domain, but its binding has not been mapped to a particular site on the mRNA. In vivo, yeast carrying a thermosensitive mutation at the RNA15 locus display defocused polyadenylation (17), mimicking mutations at the A-rich element and implying a role for this protein in contributing to the specificity of the cleavage reaction.
In the study presented here, we use CF I reconstituted from separately expressed and purified proteins (23) to directly demonstrate the binding of Rna15 to the A-rich element. We show that Rna15 in the absence of other CF I proteins does not recognize a specific RNA sequence but gains this ability when assembled into a subcomplex of CF I containing Hrp1 and Rna14, a protein previously shown to bridge Rna15 and Hrp1 (23). Furthermore, through an Rna15 construct with attenuated RNA binding, we show that Rna15 recognition of the A-rich element is an essential step in mRNA 3′-end formation. Our identification of the RNA sequences recognized by CF I illustrates a central but previously unknown aspect of the mechanism by which the yeast 3′ end is defined and processed.
MATERIALS AND METHODS
Yeast strains, media, and plasmids.
Yeast strains LM31 (MATα RNA15::TRP1 ura3-1 trp1-1 ade2-1 leu2-3,112 his3-11) and rna15-2 (MATα rna15-2) were gifts from F. Lacroute (45). Yeast were transformed by the lithium acetate technique (21). For the high-copy suppressor and phenotype assays, yeast dropout medium and medium containing 5-fluoroorotic acid (5-FOA) or X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) were prepared as described elsewhere (59).
Plasmids expressing Rna14, Rna15, Pcf11, PAP, Hrp1, and Dp1 used in the two-hybrid and rescue assays were cloned as follows. For Lex-A translational fusions, the open reading frames (ORFs) were cloned into pEG202 (19), which also contains the HIS3 marker and 2μ origin, to create pEG202(Rna14), pEG202(Rna15), pEG202(Pcf11), pEG202(PAP), and pEG202(Hrp1). For Gal4-activation domain translational fusions, ORFs were cloned into pACT2 (Clontech, Palo Alto, Calif.), which also contains the LEU2 marker and 2μ origin, to create pACT2(Rna14), pACT2(Rna15), pACT2(Pcf11), pACT2(PAP), pACT2(Hrp1), and pACT2(DP1). High-copy suppressor and phenotype assays were performed by using the pEG202 and pACT2 series of vectors. For creation of the Rna15 F63,66A double point mutant, we utilized the overlap extension method of Mikaelian and Sergeant (41). For testing the ability of this mutant to rescue a genetic disruption in vivo at the RNA15 locus and for directed two-hybrid tests against other CF I proteins, it was substituted for the wild-type RNA15 ORF in plasmid pEG202(Rna15) (33). The Bac-to-Bac baculovirus kit (Gibco) was used for expression of the wild-type and mutant Rna15 proteins. The corresponding ORFs were cloned into plasmid pFASTBAC1 (Gibco) and expressed as described previously (23). Plasmid pJCGAL7-11, containing a mutation in the cleavage site from GUAAAUC to GGGCCC, and plasmid pJCGAL7-12, containing a mutation in the A-rich element from AAUAAU to AGAUCU, were created by site-specific mutagenesis on plasmid pJCGAL7-1 (11) by the method of Mikaelian and Sergeant (41). Plasmids for transcribing the other mutant substrates have been described by Chen and Moore (11).
Preparation of CF I and expression and purification of CF I and CF II subunits.
CF I was prepared as described earlier (11) by fractionation of yeast whole-cell extract on a Hi-Trap Q column (AP Biotech). The expression and purification of active Pcf11–glutathione S-transferase (GST), Rna14-His6, His6-Clp1, Rna15-His6, His6-Hrp1, and GST-Hrp1 have been described in detail elsewhere (23, 33).
RNA processing assays.
Capped, 32P-labeled RNAs were prepared by runoff transcription from plasmids pJCGAL7-1 for wild-type and pJCGAL7-12 for mutant substrates as described by Chen and Moore (11). RNAs containing sequences on both sides of the poly(A) site were designated full length and those with only an upstream sequence were described as precleaved. Each reaction was done in a volume of 10 μl, containing 1 mM ATP, 10 mM creatine phosphate, 1 mM magnesium acetate, 75 mM potassium acetate, 2% polyethylene glycol 8000 (PEG 8000), 1 mM dithiothreitol (DTT), 0.1 mg of bovine serum albumin (New England Biolabs)/ml, 0.4 U of RNasin (Promega), 10 nM radioactive RNA precursor (ca. 250,000 to 280,000 cpm), and 1 μl of yeast whole-cell extract, prepared as described previously (11). For experiments requiring the addition of recombinant protein to rescue the activity of yeast whole-cell extract, 100 ng of Rna15 or 1 μl of insect cell lysate (determined by visual inspection of a Coomassie blue-stained gel to contain approximately the same amount of Rna15) was used. Reactions were prepared on ice and incubated first at 4°C for 10 min and then at 30°C for 20 min. Reactions were stopped by the addition of proteinase K and sodium dodecyl sulfate (SDS) (11), brought to a volume of 30 μl with Tris-EDTA (pH 7.5) and extracted once with phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol). One-tenth of the reaction was resolved on a 5% acrylamide–8.3 M urea gel and then visualized by using a Storm 960 phosphorimager.
UV cross-linking for protein labeling.
CF I or subcomplexes of CF I were assembled by combining 100 ng of each recombinant protein under buffer conditions identical to the RNA processing assay and then maintaining the mixtures at 4°C for 2 h. Reactions were assembled on ice in a final volume of 15 μl containing 250,000 to 280,000 cpm of RNA prepared as described above (equivalent to a final concentration of 6.7 nM), 0.125 μg of yeast tRNA, 1 mM magnesium acetate, 75 mM potassium acetate, 10 mM HEPES-KOH (pH 7.0), 2% PEG 8000, and 1 mM DTT. After incubation for 10 min at 30°C, samples were returned to ice and irradiated with 1.2 mJ of energy from a UV Stratalinker model 2400 (Stratagene). RNA was then digested with 8 μg of Rnase A for 1 h at 37°C. The entire sample was resolved by SDS-polyacrylamide gel electrophoresis (PAGE), proteins were fixed and visualized with the Pierce Silver Stain kit or by Western blotting, and radioactively tagged proteins were identified with a Storm 960 phosphorimager. For cross-linking of CF I isolated from yeast extract, 10 15-μl reactions, each containing 10 μl of CF I, were combined.
UV cross-linking and primer extension.
The procedure was based on the approach previously described by Urlaub et al. (64). To obtain covalent protein-RNA complexes for mapping by primer extension, reactions were assembled as described above, except that 1 μg of unlabeled GAL7-1 RNA was substituted for radiolabeled precursor. After incubation and cross-linking, reactions were supplemented with 10 μl of 0.2 mM Ni(NO3)2 in 20 mM potassium acetate (pH 5.2) to sequester the EDTA in the buffer containing the proteins. Proteins displaying the His6-epitope were then captured by incubation with 20 μl of Talon beads (Clontech) in 200 μl of enrich buffer (0.1% SDS, 500 mM KCl, 20 mM Tris-Cl [pH 8.0]) for 1 h at 4°C. Unbound proteins were removed by three washes in 1 ml of enrich buffer, and then bound proteins were eluted in 20 μl of elution buffer (0.1% SDS, 500 mM KCl, 20 mM Tris-Cl [pH 8.0], 100 mM EDTA), transferred to another tube, and digested with 200 μl of digestion buffer (10 mM EDTA, 0.1% SDS, 1 mg of proteinase K/ml) for 20 min at 37°C. RNA containing bases modified by cross-linking to peptide residues was then recovered by extraction with phenol-chloroform-isoamyl alcohol (25:24:1) and ethanol precipitation in the presence of 20 μg of glycogen and 1 M sodium acetate. Precipitated RNA was then washed with 1 ml of 70% ethanol and vacuum dried.
For primer extension, 10 pmol of DNA oligomer 5′-GGAAAGGACCACATTACATAAC-3′ was labeled at the 5′ end by using [γ-32P]ATP as described previously (56). After the labeling step, the reaction was extracted with phenol-chloroform-isoamyl alcohol (25:24:1), ethanol precipitated, and resuspended in 5 μl of FA dye (80% formamide, 50 mM Tris-borate [pH 8.0], 1 mM EDTA, 0.1% xylene cyanol, 0.1% bromphenol blue). Full-length radiolabeled oligomer was then separated from prematurely terminated synthesis products on a 15% acrylamide (29:1, bis)–8 M urea gel and recovered by using the QiaEx II Kit (Qiagen) according to the manufacturer's instructions. The typical yield was 8 × 106 cpm/pmol in a volume of 20 μl.
Primer extension on the RNA prepared above was carried out by using Moloney murine leukemia virus reverse transcriptase (Gibco). The RNA pellet was resuspended in 9.5 μl of extension buffer (1× First Strand buffer [Gibco]; 10 mM DTT; 10 mM [each] dATP, dCTP, dGTP, and dTTP; 10,000 cpm of gel-purified, radiolabeled DNA primer) and preincubated for 2 min at 42°C before the addition of 0.5 μl of Moloney murine leukemia virus reverse transcriptase (2 U/μl). Extensions were carried out for 1 h at 42°C and then halted and visualized as described for the RNA processing assays. For mapping of the polymerase pause sites, extension products were resolved in parallel with DNA sequencing reactions by using the same oligomer as a primer to sequence the plasmid pGAL7-1 (11) with the Circumvent thermocycle DNA sequencing kit (AP Biotech).
GST pulldowns and immunoprecipitations.
To study the assembly of CF IA proteins, 10 μg of Pcf11-GST fusion protein was bound to 20 μl of glutathione-agarose beads in 200 μl of IP-150 buffer (150 mM KCl, 20 mM Tris-Cl [pH 8.0], 0.1% NP-40) for 1 h at 4°C (23). To minimize nonspecific interactions between components, the bead-GST-protein complexes were then blocked for an additional hour with 200 μl of IP-150 plus 10% (vol/vol) fetal calf serum. Unbound proteins were removed with three 5-min washes with 1 ml of IP-150 buffer, and 100 μg of each of the other recombinant CF IA proteins was added in 200 μl of IP-150 buffer for 1 h. After being washed with IP-150 buffer as described above, assembled complexes were eluted with IP-150 plus 50 mM glutathione and resolved by SDS-PAGE and Western blotting. Proteins were visualized by cutting the membrane into sections and probing them with antibodies to GST, the His6 epitope, or Rna15.
For immunoprecipitations, 0.5 μl of anti-Rna15, 0.5 μl of anti-Rna14, or 0.5 μl of anti-Hrp1 polyclonal antiserum was bound to 10 μl of protein A-agarose beads (Gibco) for 1 h in 1 ml of IP-150, blocked with fetal calf serum, and incubated with the CF I fraction containing proteins photo-cross-linked to radiolabeled RNA precursor for 1 h at 4°C. Complexes were eluted by resuspending the beads in 20 μl of SDS buffer and heating the mixtures at 65°C for 5 min before resolution by SDS-PAGE. For experiments in which precipitation of a particular protein was desired in the absence of other CF I proteins, the complex was disrupted by substituting the more stringent IP-250 buffer (250 mM KCl, 20 mM Tris-Cl [pH 8.0], 0.1% NP-40) for IP-150 buffer.
Poly(U) binding assays.
Poly(U) Sepharose beads (Pharmacia) were washed and prepared according to manufacturer's instructions prior to use in order to remove unbound poly(U). Then, 20 μl of beads per sample was washed three times with 1 ml of PU buffer (20 mM Tris-Cl [pH 8.0], 0.1% NP-40) supplemented with either 50, 100, 150, 250, or 500 mM (final concentration) KCl. Aliquots of insect cell lysate containing wild-type or mutant Rna15 were then mixed with the beads, brought to a final volume of 1.5 ml, and incubated at 4°C for 1 h. Unbound proteins were then removed with three 1-ml washes with PU buffer supplemented to the same concentration of KCl. Bound proteins were removed by boiling them for 5 min in 20 μl of SDS buffer prior to loading on the gel.
RESULTS
Two proteins in CF I independently contact the mRNA precursor.
Two proteins of the CF I complex, Hrp1 and Rna15, contain the well-conserved RRM found in proteins known to bind single-stranded RNA (5). Rna15 contains a single copy of this motif (45), and recombinant Rna15 purified from Escherichia coli has been shown to bind to poly(U) RNA in the absence of other yeast proteins (8). Hrp1 contains two repeats of the RRM, and purified recombinant Hrp1 has also been shown to bind polyadenylation precursor (33). The other three subunits of CF I (Clp1, Rna14, and Pcf11) do not have any obvious RNA-binding motifs (2, 15, 45).
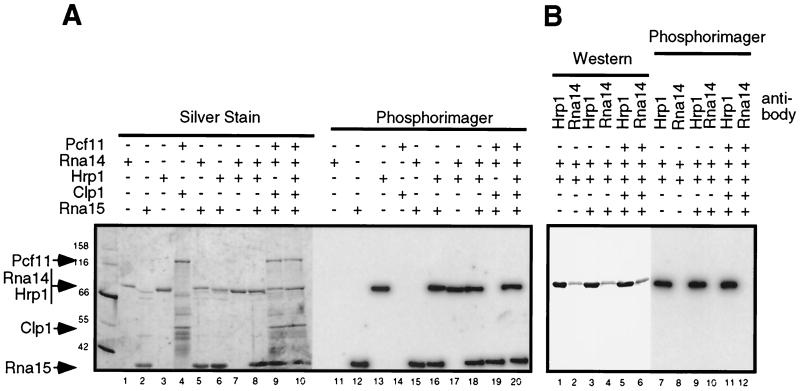
We assembled subcomplexes of CF I from previously characterized active subunits (23), bound them to a radioactively labeled polyadenylation precursor RNA containing the GAL7 3′ untranslated region (3′-UTR), and subjected them to photochemical cross-linking. The photochemical conjugation of a radioactive nucleotide to the protein of interest is an indication of the protein's proximity to the RNA (57). The results of these experiments are presented in Fig. 1.
FIG. 1.
Cross-linking of CF I proteins to GAL7 3′-UTR in vitro demonstrates the binding of Rna15 and Hrp1. Subcomplexes or complete CF I were assembled from purified proteins as described in Materials and Methods and cross-linked to the GAL7-1 RNA precursor by using UV irradiation. The presence of a plus sign indicates the addition of that protein to the assembly reaction. (A) After cross-linking and digestion of the RNA, proteins were resolved by SDS-PAGE and visualized by silver staining, and radioactively tagged proteins were identified by PhosphorImager analysis. (B) Determination of the identity of the 70-kDa radioactively tagged protein. After photo-cross-linking, complexes containing both Hrp1 and Rna14 were disrupted, and proteins were immunoprecipitated by using either anti-Hrp1 or anti-Rna14 antisera, indicated by the Hrp1 or Rna14 labels. Captured proteins were released by boiling in SDS buffer and identified by Western blotting by using antibodies to the His6 epitope found on both proteins.
The two RRM-containing proteins, Rna15 and Hrp1, are cross-linked to the RNA (Fig. 1A, lanes 12 and 13), whereas Rna14, the protein that bridges them in the CF I complex (23), is not (Fig. 1A, lane 11). Pcf11-GST and His6-Clp1 are also not labeled (Fig. 1A, lane 14). When Rna15-His6 was combined with His6-Rna14 or His6-Hrp1, the results were identical to those obtained with these proteins alone (Fig. 1A, lanes 15 and 16). When His6-Hrp1 was combined with His6-Rna14 alone or in conjunction with Rna15-His6, only His6-Hrp1 and Rna15-His6 bound the RNA (Fig. 1A, lanes 17 and 18). In the CF IA complex (Rna14, Rna15, Pcf11, and Clp1), as well as in the complete CF I (CF IA plus Hrp1), only Rna15-His6 and His6-Hrp1 were radioactively labeled (Fig. 1A, lanes 19 and 20). Since His6-Rna14 could not be resolved from His6-Hrp1 under these conditions (Fig. 1A, lanes 1 and 3), reactions in which His6-Rna14 and His6-Hrp1 were both present were split in half, and proteins were immunoprecipitated with either anti-Hrp1 or anti-Rna14 antiserum under conditions which disrupted the CF I complex. Precipitated proteins were visualized by Western blotting with anti-His5 antibody, and the membrane was subjected to autoradiography to determine the identity of the radioactively labeled protein. None of the conditions tested resulted in the labeling of Rna14 (Fig. 1B). These results show that Rna15 and Hrp1 are probably the only two proteins in CF I which contact the RNA and that their efficiency of photo-cross-linking is not modified by the presence or absence of other CF I subunits.
Rna15 alone does not display sequence specificity.
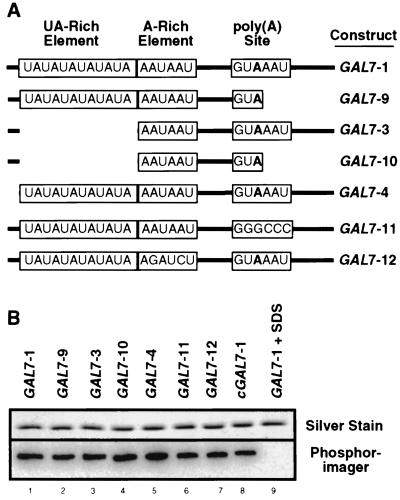
We next attempted to determine the RNA sequence to which Rna15 bound. While Hrp1 alone and in nuclear extracts has been shown to have some specificity for the UA-repeat element (12, 33), little is known about the sequence recognized by Rna15. Selection-amplification experiments with recombinant Rna15 failed to evolve a recognizable yeast polyadenylation signal, although the identical procedure with the mammalian homolog, CstF64, yielded sequences which were functional in reconstituted mRNA 3′-end processing assays (60). We utilized full-length or precleaved radioactive RNA precursors derived from the GAL7 3′-UTR (11) in which various deletions or mutations had been introduced (Fig. 2A) and tested whether these mutants were photochemically cross-linked to the recombinant protein.
FIG. 2.
Rna15 alone does not recognize a specific RNA sequence in the absence of other CF I proteins. (A) Schematic of GAL7 3′-UTR and mutant derivatives utilized in the cross-linking study. RNA is indicated by a solid line, deletions are indicated by the absence of the line, conserved sequences are boxed, and the cleavage site is indicated by the boldface adenosine residue immediately upstream. (B) Silver stain and phosphorimager scan from gel containing purified Rna15 after UV cross-linking to the RNAs indicated. cGAL7-1, transcript derived from the complementary strand to the GAL7 3′-UTR; GAL7-1 + SDS, cross-linking reaction was done under denaturing conditions.
Rna15 cross-linked to both full-length and precleaved RNA (Fig. 2B, lanes 1 and 2), befitting the necessity of CF I in both the cleavage and the polyadenylation steps (11). Deletion of the UA-rich sequence did not affect the binding of Rna15-His6 to either the full-length or precleaved precursor (Fig. 2B, lanes 3 and 4). Mutation of the poly(A) site from GUAAAU to GGGCCC or mutation of the A-rich sequence from AAUAAU to AGAUCU also did not alter binding (Fig. 2B, lanes 6 and 7). In spite of its propensity to bind pyrimidine-rich sequences (60), Rna15 still bound well to an RNA from which a nonessential stretch upstream of the UA-rich signal, containing three repeats of (U)3-6 had been removed (Fig. 2B, lane 5). To confirm that Rna15 was not interacting with previously uncharacterized sequences still present in the RNAs analyzed, we also demonstrated binding to RNA transcribed from the complementary strand of the GAL7 3′-UTR (Fig. 2B, lane 8), a substrate not processed in vitro (1). Binding of Rna15 to the RNA was abolished when the cross-linking reaction was performed with protein pretreated with SDS (Fig. 2B, lane 9).
Rna15 recognizes the A-rich element in complex with Rna14 and Hrp1.
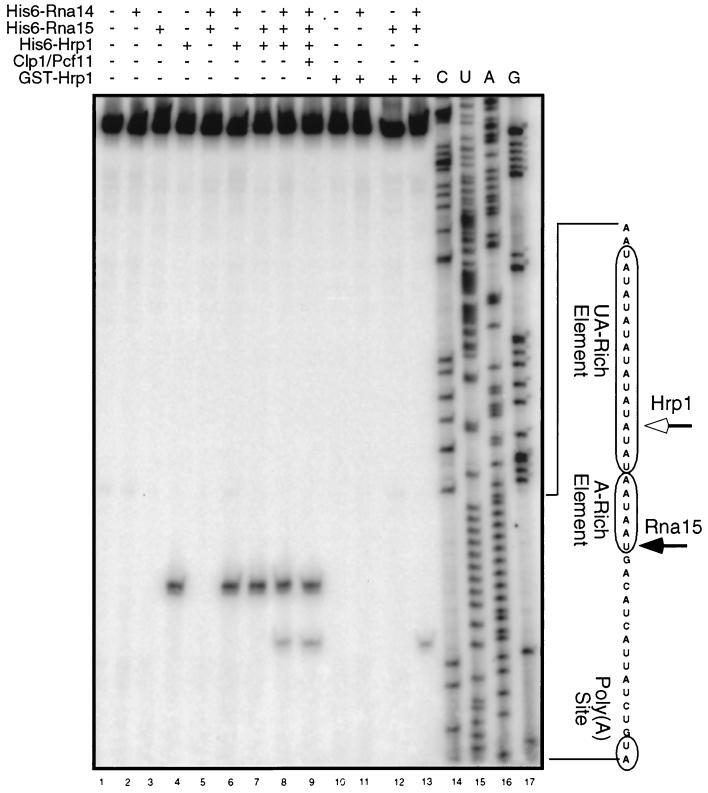
To map the signals recognized by the subunits of CF I that bind the mRNA, we exploited the propensity of polymerases to pause one nucleotide upstream of the site of cross-linking to peptide moieties (64). This pausing is due to the template's retention of short peptide fragments refractory to proteolysis. We first cross-linked intact CF I or various subassemblies to unlabeled RNA precursor. The RNA-binding proteins Rna15 and Hrp1 contained the His6 epitope tag, thus permitting their precipitation by metal chelate affinity capture (MCAC) for the enrichment of cross-linked RNAs. After elution from the beads, the proteins were removed by protease digestion, and the RNA was utilized as a template for reverse transcription. We designed an oligonucleotide complementary to sequence downstream of the poly(A) site as a primer to direct the polymerase upstream toward the cis elements. Polymerase pause sites, revealed as novel bands in the extension reaction, were mapped by comparison to a DNA sequencing ladder utilizing the same primer bound to the DNA template used for transcription of the RNA precursor.
No major pauses were observed when this procedure was carried out upon naked RNA (Fig. 3, lane 1) or in the presence of His6-Rna14, which was shown in the previous experiment not to cross-link to RNA (Fig. 3, lane 2). When His6-Hrp1 was cross-linked to the RNA, a pause is observed at an adenosine base within the UA-rich element (Fig. 3, lane 4) corresponding to a cross-link at the uracil base immediately upstream. This result is consistent with previously published work indicating recognition of the UA-rich element by Hrp1 (12, 33, 65).
FIG. 3.
Mapping of the binding sites of Hrp1 and Rna15. Subcomplexes or complete CF I was assembled from purified active proteins and photo-cross-linked to GAL7-1 RNA; a plus sign indicates the addition of a particular protein to the assembly reaction. Cross-linking of Hrp1 to the UA-rich element creates a polymerase pause site at the 14th nucleotide of the UA-rich element (open arrow). Specific binding of Rna15 in complex with Hrp1 and Rna14 creates a polymerase pause site at the sixth nucleotide of the A-rich element (closed arrow).
Interestingly, while Rna15 can bind RNA in the absence of other yeast proteins (Fig. 1) (8), no pause was observed when this protein was cross-linked alone (Fig. 3, lane 3). The addition of Rna15-His6 to His6-Hrp1 resulted in a pattern indistinguishable from that observed from His6-Hrp1 alone (Fig. 3, lanes 4 and 7). However, when a trimer of His6-Hrp1, His6-Rna14, and Rna15-His6 was added to the RNA, a new pause site was observed, corresponding to a uracil residue in the A-rich element, suggesting the presence of a cross-link at the adenosine residue immediately upstream (Fig. 3, lane 8). The appearance of this second pause site was dependent on the presence of all three proteins, for it was not observed with a dimer of His6-Rna14 and Rna15-His6 (Fig. 3, lane 5). The addition of His6-Clp1 and Pcf11-GST to assemble a complete CF I did not change the position or the intensity of the observed pauses (Fig. 3, lane 9).
To confirm that the pause within the A-rich element was due to the binding of His6-Rna15 to the RNA and not to a second contact by His6-Hrp1, we utilized an alternative form of recombinant Hrp1. Instead of the His6-Hrp1 used above, we substituted a GST-Hrp1 that is functionally identical (33). This form of the protein would thus be present for the cross-linking reaction but RNAs covalently bound to it would not be precipitated by MCAC for entry into the reverse transcriptase template pool. No pauses were observed upon the cross-linking of this protein alone or in combination with Rna15-His6 or His6-Rna14 (Fig. 3, lanes 10 to 12). However, when a trimer of GST-Hrp1, His6-Rna14, and Rna15-His6 was photo-cross-linked to the RNA, a pause corresponding to the A-rich element was observed (Fig. 3, lane 13). This pause site could only be derived from a pool of RNAs to which His6-Rna15 had been cross-linked. No pauses were observed in sequence between the UA-rich element and the 5′ end. Furthermore, none of the conditions tested generated a pause site in the vicinity of the poly(A) site (data not shown). These results demonstrate the binding of Rna15 to the A-rich element in a subassembly of CF I containing Hrp1 and Rna14.
In summary, the cross-linking of Hrp1 results in a single polymerase pause site within the UA-rich element. However, under conditions in which Rna15 alone cross-links to the RNA, no unique pause is created in the presence of Rna15. The creation of an Rna15-dependent pause site in combination with Rna14 and Hrp1 implies that Hrp1 directs Rna15 to the A-rich element by means of an Rna14 bridge. In the absence of these two proteins, Rna15 binds RNA nonspecifically. Pcf11 and Clp1 do not appear to play a role in influencing the binding or location of either Rna15 or Hrp1.
Targeted mutagenesis of the Rna15 RRM.
To determine whether Rna15 recognition of the A-rich element is essential for mRNA 3′-end processing, we decided to attenuate the RNA-binding property of this protein by replacing conserved amino acids with alanine, which is believed to be functionally quiescent and noncontributory in terms of structure (69). The RNA-binding domain of Rna15 is highly homologous to the classical bipartite RNP2/RNP1 motif (45). The RNP1 motif has been derived by pattern-matching from the SWISS-PROT database as (RKLND)G(YF)(GA)(FY)(IV)(QNDETHK)(FYRM) (35). We decided to replace phenylalanines 63 and 65 in Rna15, changing the RNP1 motif from KGYAFIEF to KGYAAIEA. These amino acids were chosen because of their high degree of conservation and their involvment in base-stacking interactions with the RNA (67). The resultant mutant was thus named Rna15 F63,66A.
The Rna15 F63,66A protein cannot substitute for wild-type Rna15 protein in vivo.
To assay the effect of this mutation on the in vivo function of the Rna15 protein, we next tested the ability of a plasmid expressing this fusion protein to rescue an otherwise lethal disruption at the RNA15 locus. The yeast strain LM31 contains ura3, his3, and leu2 mutations, as well as a disruption of the RNA15 locus rescued in trans by a genomic copy of the RNA15 gene on a URA3 plasmid (45). We transformed LM31 with one of the following plasmids: pEG202(Rna15), pEG202(Rna15 F63,66A), or pEG202(Rna14). After selection for uptake of these plasmids, transformants were counterselected on medium containing 5-FOA, an additive lethal to uridine prototrophs. This permitted identification of cells which had received a gene capable of substituting for the wild-type RNA15 encoded on the URA3 covering plasmid. Cells which were transformed with a wild-type copy of RNA15 were viable on medium containing 5-FOA, but those which received the mutant RNA15 F63,66A or RNA14 were not (data not shown). Thus, the Rna15 F63,66A mutant cannot substitute for the wild-type protein in vivo.
Expression of the RNA15 gene from a high-copy plasmid is able to suppress the temperature sensitivity of strains bearing the rna14-1 mutation (45). We next investigated the ability of overexpression of proteins known to contact Rna15 (23) to rescue the lethality of the Rna15 F63,66A mutant. The yeast strain LM31 described above was transformed with pEG202(Rna15 F63,66A) so that it now contained two plasmids coding for Rna15. This resultant strain was transformed with one of the following high-copy plasmids: pACT2(Rna14), pACT2(Rna15), pACT2(Pcf11), pACT2(Hrp1), pACT2(PAP), or pACT2(Pcf11). Transformants capable of losing the URA3 covering plasmid were selected by 5-FOA as described above. Only cells which had received a plasmid expressing wild-type Rna15 were viable, indicating that overexpression of Rna14 or Pcf11 is not sufficient for in vivo function of Rna15 F63,66A (data not shown).
The Rna15 F63,66A protein retains in vivo interactions with other CF I components.
To determine whether the alanine changes had disrupted the overall structure of the protein, we used the pEG202(Rna15 F63,66A) plasmid, which expresses the protein as a fusion to the LexA binding domain, to conduct a two-hybrid test against Pcf11 and Rna14, two proteins known to directly interact with Rna15 (23). In this system, positive interaction in vivo between two proteins results in the production of β-galactosidase, which can be observed as the development of “blue” colonies when cells are plated on medium containing X-Gal. The wild-type LexA-Rna15 fusion interacted with activation domain fusions to Rna14, Pcf11, and Hrp1. No interaction was seen with yeast PAP or with a fusion to the Drosophila melanogaster transcription factor Dp1. The mutant fusion protein LexA-Rna15 F63,66A maintained the same interactions, with similar efficiency, as seen with the wild-type protein (data not shown). These results also confirmed the stable expression of the mutant protein in the host cell. In experiments described below, we also demonstrate in vitro incorporation of this mutant protein into the CF IA complex.
The Rna15 F63,66A mutant does not disrupt the assembly of the CF IA complex in vitro.
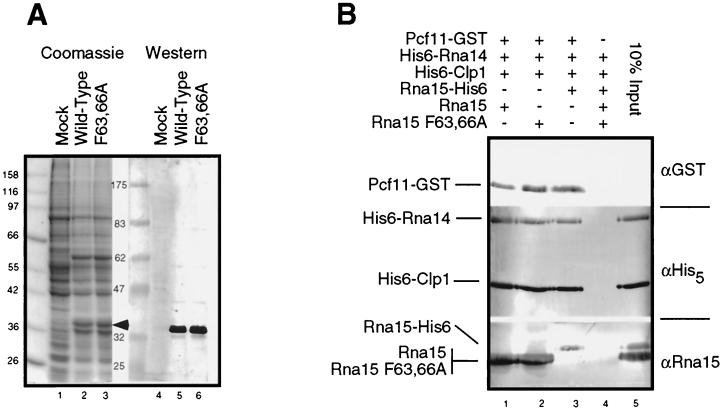
The Rna15 and Rna15 F63,66A proteins were expressed in insect cells as described in Materials and Methods. Both proteins were expressed at similar levels (Fig. 4A, compare lanes 2 and 3) and were recognized by anti-Rna15 antibodies (Fig. 4A, lanes 5 and 6). To confirm that the mutation does not affect the ability of the protein to participate in protein-protein interactions with other members of the CF IA complex, we assembled CF IA in vitro by using the Rna15F63,66A mutant protein (Fig. 4B). A mixture of Pcf11-GST, His6-Rna14, His6-Clp1, and lysate from insect cells expressing either the wild-type Rna15 or the Rna15 F63,66A mutant or purified, recombinant Rna15-His6 expressed in E. coli was incubated with glutathione-Sepharose beads. Unbound proteins were removed by extensive washes, the assembled complexes were eluted from the beads, and their composition was determined by Western blotting.
FIG. 4.
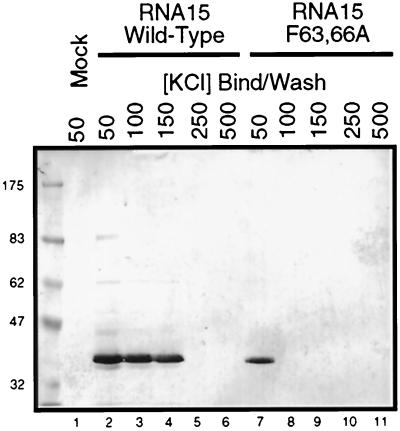
The Rna15 F63,66A mutant does not disrupt assembly of the CF IA complex. (A) Clarified whole-cell lysates of insect cells either mock infected (Mock) or infected with recombinant baculoviruses expressing either wild-type or the F63,66A mutant Rna15 proteins (indicated by the arrow). The protein immediately below the Rna15 is a 32-kDa baculovirus structural protein. (B) GST-Pcf11 bound to glutathione-Sepharose beads was used to nucleate the assembly of CF IA incorporating either Rna15-His6 purified from E. coli or insect cell lysates from cells expressing wild-type or mutant Rna15. The plus symbol indicates the presence of a particular protein in the assembly reaction. Proteins bound to the beads through interaction with GST-Pcf11 were released by the addition of glutathione and visualized by Western blotting with the indicated antisera. The “10% input” lane represents direct loading on the gel of a quantity of protein equal to 10% of the amount added to the complex assembly reaction.
In the absence of Pcf11-GST, no complex was assembled (Fig. 4B, lane 4). As shown previously (23), in the presence of Pcf11-GST, the other three CF IA subunits (Rna14, Rna15, and Clp1) were incorporated into the CF IA complex (Fig. 4B, lanes 1 to 3). The wild-type Rna15 and the Rna15 F63,66A proteins in the insect cell lysates were able to assemble into CF IA with equal efficiencies (Fig. 4B, lanes 1 and 2). This result confirms that the inability of the Rna15 F63,66A mutant to function in vivo is not due to a disruption of protein-protein interactions with Rna14 or Pcf11.
Rna15 F63,66A displays decreased binding affinity for RNA.
To determine whether the mutant protein had decreased affinity for RNA, we employed a poly(U) pulldown assay. When insect cell lysate from mock-infected cells was incubated with poly(U) Sepharose at 50 mM KCl, no proteins reactive to the Rna15 antibody were bound to the beads (Fig. 5, lane 1). Rna15 binds well at 50, 100, and 150 mM KCl but does not bind at 250 mM (Fig. 5, lanes 2 to 5). This salt concentration is consistent with the elution of CF IA from a poly(U) Sepharose column at 250 mM KCl (34). In contrast, the Rna15 F63,66A mutant, which bound at 50 mM KCl, could not bind at 100 mM, suggesting that the mutant is greatly impaired in its ability to interact with RNA (Fig. 5, lanes 7 to 8).
FIG. 5.
The Rna15 F63,66A mutant displays reduced affinity for RNA, as demonstrated by a poly(U) binding assay with wild-type and mutant Rna15 proteins. Proteins in the lysates from Fig. 4A were assayed for the ability to bind to poly(U) Sepharose beads in the presence of increasing concentrations of KCl and then detected by Western blotting with Rna15 antibody.
Failure of Rna15 to bind to the A-rich element prevents RNA 3′-end formation in vitro.
To determine whether the inability of the Rna15 F63,66A mutation to rescue a disruption of the RNA15 locus in vivo was due to a defect in mRNA 3′-end formation, we assayed its activity in an in vitro processing assay. Yeast whole-cell extract made from cells with the rna15-2 mutation are defective for cleavage and polyadenylation of an RNA precursor in this assay (44), but the 3′-end maturation activity can be rescued with the addition of functional Rna15 (23). We assayed rescue of the rna15-2 extract with insect cell lysate made from cells expressing Rna15 F63,66A.
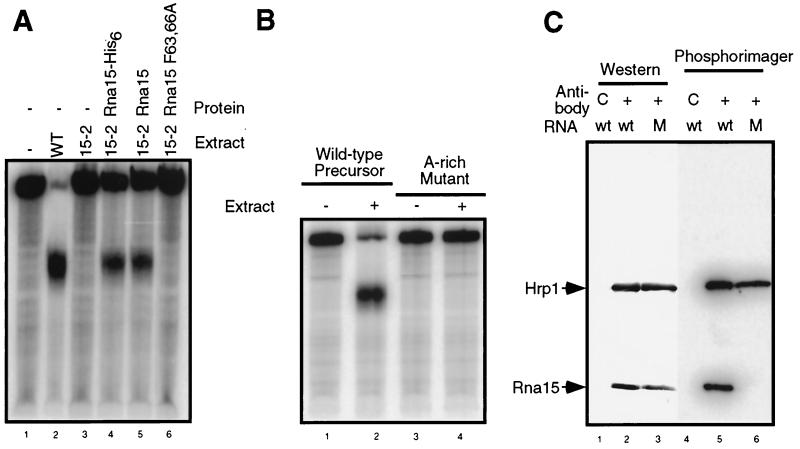
The RNA precursor is efficiently cleaved and polyadenylated by whole-cell extract made from wild-type cells but not by extract made from rna15-2 cells (Fig. 6A, compare lanes 2 and 3). The processing activity of the rna15-2 extract is restored to ca. 50% of wild-type levels by the addition of purified recombinant Rna15-His6 or lysate from insect cells expressing Rna15. However, lysate from insect cells expressing the Rna15 F63,66A mutant does not rescue the processing activity of the rna15-2 extract (Fig. 6A, lane 6). This result suggests that, even though the mutant protein maintains appropriate protein-protein interactions with other CF I proteins, its decreased affinity for RNA prevents its participation in mRNA 3′-end formation.
FIG. 6.
Disruption of the interaction between the A-rich element and Rna15 prevents RNA 3′-end processing in vitro. (A) Rna15 F63,66A cannot rescue the processing activity of rna15-2 mutant extract. Full-length radioactive GAL7-1 RNA was incubated with extract made from wild-type cells (WT) or from cells carrying the rna15-2 mutation (15-2) under standard processing conditions. As indicated, reactions were supplemented with Rna15-His6 protein purified from E. coli or with lysates of insect cells expressing wild-type or mutant Rna15. RNAs recovered from the reactions were separated by electrophoresis through a 5% acrylamide-bisacrylamide (19:1) gel supplemented with 8 M urea. Lane 1 shows unreacted precursor. (B) The A-rich element is essential for 3′-end processing of RNA in vitro. The addition of yeast whole-cell extract (indicated by a plus sign) results in the cleavage and polyadenylation of the GAL7-1 precursor. When this sequence is mutated from AAUAAU to AGAUCU in GAL7-12, this processing is abolished. (C) Mutation of the A-rich element prevents Rna15 binding to the RNA. Proteins in a Q-Sepharose fraction containing CF I activity were photo-cross-linked to either wild-type (wt) RNA or RNA containing a mutation in the A-rich sequence (M). After digestion of the RNA, samples were immunoprecipitated by using combined anti-Rna15 and anti-Hrp1 antiserum or a control antibody (C) as indicated. Proteins were resolved by SDS-PAGE, transferred to membrane and then detected by Western blotting or autoradiography.
To confirm that the RNA sequence to which Rna15 was shown to bind in Fig. 3 was critical for mRNA 3′-end formation, we tested the processing of a mutation in the A-rich sequence in our in vitro system. The AAUAAU hexanucleotide in the GAL7 precursor was mutated to AGAUCU. Precursor RNAs transcribed from this template showed no detectable processing in vitro (Fig. 6B), demonstrating the importance of this sequence.
Finally, we confirmed that the in vitro processing defect observed in the A-rich element mutant was due to the inability of Rna15 to bind at this location. Aliquots of a Q-Sepharose fraction containing only CF I activity (23) were photo-cross-linked to the full-length wild-type or mutant RNA precursor. After digestion of the RNA and disruption of the CF I complex, Hrp1 and Rna15 were precipitated from the reaction by using a mixture of antibodies bound to beads and then resolved by SDS-PAGE. The proteins were identified by Western blotting and subjected to autoradiography in order to determine the ability of these proteins to bind the mutant and wild-type RNAs. Both proteins were precipitated from the reaction by using this procedure (Fig. 6C, lanes 2 and 3) but not when a control antibody raised against Rna14 was used instead (Fig. 6C, lane 1). Both Rna15 and Hrp1 were cross-linked to the wild-type precursor, but with the mutant RNA only Hrp1 cross-linking could be detected (Fig. 6C, compare lanes 5 and 6). Thus, Hrp1 interaction is not affected by this mutation in the RNA. However, interaction of Rna15 when it is a component of CF I requires the A-rich element.
DISCUSSION
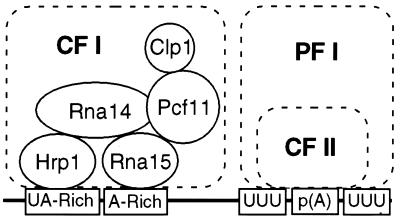
We had previously defined a minimal cohort of proteins required for reconstitution of CF I activity in vitro and described the protein-protein interactions responsible for assembly of the complex (23). The experiments described here illustrate the functional significance of this architecture as a means to assemble a complex capable of recognizing a specific RNA sequence (Fig. 7). In this report, we have focused on the roles of the Rna15, Hrp1, and Rna14 subunits of CF I. Although Rna15 had been shown to interact with RNA and was a necessary component of the processing machinery, its binding site on the precursor had not been characterized. We show that the ability of Rna15 to bind RNA is essential for its function in vivo and in vitro but that Rna15 alone does not recognize a specific RNA sequence. However, when Rna15 was in complex with Hrp1 and Rna14, it specifically recognizes the A-rich element, a critical sequence in yeast which until now had no occupant. Disruption of the interaction of Rna15 with the A-rich element, either by mutating the RRM of Rna15 or by altering the sequence of the RNA substrate, prevents CF I from functioning.
FIG. 7.
Model for interaction of the polyadenylation complex with conserved sequence elements in the pre-mRNA. The CF I architecture is taken from the protein-protein interaction studies of Gross and Moore (23).
In contrast to Rna15, prior work had suggested that Hrp1 recognized the UA-rich element in the absence of other proteins (12, 33, 65). Deletion of this element in the GAL7 precursor prevents 3′-end processing in vitro and in vivo (31), a finding consistent with the requirement for Hrp1 in the in vitro cleavage and polyadenylation of this substrate (23, 33). By primer extention analysis, we now precisely map the UA-rich motif as the Hrp1 binding site and show that this interaction is maintained when Hrp1 is incorporated into CF I. It has been suggested that the contribution of Hrp1 to accurate cleavage was to mask cryptic sites from the processing machinery (42). Our data demonstrate a much more direct role for Hrp1 in specifying the authentic cleavage site, in agreement with the strong interaction it exhibits with Rna14 (23). These results support a model for assembly of a cleavage complex in which Hrp1 binding at the UA-rich element directs Rna15 to the A-rich sequence through an Rna14 bridge (Fig. 7). CF II recognizes the U-rich tracts flanking the poly(A) site, and cross-factor interactions lock the complex on the correct cleavage site (16). Most of the RNA interactions would be maintained after cleavage, ensuring a stable association of the polyadenylation complex with the RNA. This scenario suggests that, in spite of the sequence conservation between protein subunits, the mechanism by which yeast defines the poly(A) site is significantly different from that of mammals, in which the A-rich element is recognized by the CF II homologue and the U-rich element is recognized by the CF I counterpart (6, 47).
Common themes have been preserved in protein-protein interactions and are exemplified by Rna14, which appears to serve three functions as a component of CF I. The first function is to facilitate the assembly of a multiprotein complex containing three RRMs: two contributed by Hrp1 (33) and one contributed by Rna15 (45). This subcomplex gives CF I specificity for the two parts of the polyadenylation signal farthest away from the poly(A) site. The concatenation of multiple RNA-binding domains is often a prerequisite for the specific binding of a protein or complex of proteins. For example, nucleolin is a protein that regulates ribosome biogenesis through binding to a specific sequence in the pre-rRNA (20). Nucleolin contains four RNP1 sequences, but individual domains alone lose the ability to discriminate between specific and nonspecific sequences (24). An analogous situation is seen with the hnRNP A1 splicing factor, where two binding domains are required for specificity (9, 39, 40, 58). In both of these cases, isolated domains behave like the single-RRM-containing Rna15, binding RNA irrespective of sequence, while the intact protein recognizes a specific sequence like the three-RRM-containing CF I.
The second function Rna14 may have is to serve as a bridge between the core proteins of CF I and the CF II/PF I complex. Rna14 interacts with Pcf11, which also simultaneously binds Rna15 and Clp1 (23). Rna14 also interacts with Pfs2, a PF I protein, which in turn interacts with the PF I subunits Fip1 and Brr5/Ysh1 (50). Pfs2 contains several repeats of the WD-40 sequence, a motif involved in protein-protein interactions, and may act as the central organizing component of PF I (49). Therefore, Rna14 may connect the two major organizing centers of CF I and PF I: Pcf11 and Pfs2.
The third function Rna14 may perform is a mechanism for signaling completion of the cleavage reaction to the waiting polyadenylation machinery. Rna14 also interacts with Fip1, a PF I protein which binds to PAP and regulates its activity (73). Through its interaction with Pcf11, Rna14 may sense the completion of the cleavage reaction and transmit this information to Fip1. Fip1 would then relax its inhibition of PAP activity (28), permitting polyadenylation to occur.
Two of Rna14's functions, the bridging of two RNA-binding proteins to confer specificity to the Rna15 homolog and the linking of two different processing factors, are conserved in the mammalian homolog of Rna14, CstF77. The mammalian homolog of Rna15, CstF64, also binds mRNA nonspecifically in the absence of other proteins (37) but becomes specific in the complex. Processing specificity in mammals is provided by CPSF, whose p160 subunit binds the AAUAAA signal (32). CstF64 is connected to CPSF160 via CstF77. In this assembly, CstF specifically binds the downstream element (60). The result of this assembly is that CPSF and CstF together have a higher affinity for the mRNA than does either factor alone (48). CstF77 also binds the mammalian homolog of Pfs2, CstF50, although the significance of this interaction and any contribution to CPSF-CstF bridging is unknown (62).
Based on our results, many changes in the function of the polyadenylation machinery may be explained as resulting from an alteration of the Hrp1-Rna14-Rna15 or CPSF160-CstF77-CstF64 subassembly essential for connecting the catalytic machinery to the mRNA. When we created an Rna15 protein that had a greatly reduced affinity for RNA through mutations in the RRM but maintained protein-protein interactions with other CF I components, this protein could not provide wild-type function in vitro or in vivo. A lethal defect in mRNA processing due to mutations in Rna14, such as that caused by the rna14-1 allelle, can be rescued by overexpression of Rna15, possibly by promoting the formation of a greater number of Hrp1-Rna14-Rna15 assemblages in the cell (45). Increased levels of CstF64, the mammalian homolog of Rna15, promote the use of more upstream poly(A) sites in B cells (61), probably through the more efficient assembly of the specificity complex in these cells. Conversely, in Drosophila, mutations in suppressor of forked, the Rna14 homolog, result in the skipping of weak poly(A) sites (4), possibly due to destabilization of the specificity mechanism. This phenomenon is also observed in yeast, where rna14 and rna15 mutants exhibit preferential use of the most distal poly(A) site of the ACT1 gene at restrictive temperatures (38).
The contributions of Pcf11 and Clp1 toward CF I activity are still unclear. Since Pcf11 interacts simultaneously with Rna14 and Rna15, its role in vivo may also be to stabilize their interaction. While our results did not reveal any augmentation of Rna15 RNA-binding specificity by these proteins, their requirement for both the cleavage and the polyadenylation activities of CF I (23) suggests that they may provide catalytic functions or bridges to the CF II/PF I complexes. Interestingly, their mammalian homologues are components of CF IIM and are only required for the cleavage step (15).
We have now defined two critical contact points between CF I and the pre-mRNA, and we have shown how the architecture of CF I contributes to its essential function of recognizing the nascent 3′ end. However, the roles of CF II in recognition and processing are still unclear. In the future, similar experiments with highly purified CF II may permit the determination of the arrangement of proteins within the complex, the mapping of connection points to CF I and the substrate mRNA, and the identification of the protein or proteins responsible for the endonucleolytic cleavage.
ACKNOWLEDGMENTS
We are grateful to all members of the Moore lab for helpful discussions and technical advice and to K. Sparks and A. Zhelkovsky for their assistance with preparation of the manuscript. We also acknowlege the essential support of A. E. Gross at all stages of this work.
This work was supported by grant GM41752 to C.L.M.
REFERENCES
- 1.Abe A, Hiraoka Y, Fukasawa T. Signal sequence for generation of mRNA 3′ ends in the Saccharomyces cerevisiae GAL7 gene. EMBO J. 1990;9:3691–3697. doi: 10.1002/j.1460-2075.1990.tb07581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amrani N, Minet M, Wyers F, Dufour M E, Aggerbeck L P, Lacroute F. PCF11 encodes a third protein component of yeast cleavage and polyadenylation factor I. Mol Cell Biol. 1997;17:1102–1109. doi: 10.1128/mcb.17.3.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aranda A, Moore C, Perez-Ortin J, del Olmo M. Transcription termination downstream of the Saccharomyces cerevisiae FBP1 poly(A) site does not depend on efficient 3′ end processing. RNA. 1998;4:303–318. [PMC free article] [PubMed] [Google Scholar]
- 4.Audibert A, Simonelig M. Autoregulation at the level of mRNA 3′ end formation of the suppressor of forked gene of Drosophila melanogaster is conserved in Drosophila virilis. Proc Natl Acad Sci USA. 1998;95:14302–14307. doi: 10.1073/pnas.95.24.14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bandziulis R J, Swanson M S, Dreyfuss G. RNA-binding proteins as developmental regulators. Genes Dev. 1989;3:431–437. doi: 10.1101/gad.3.4.431. [DOI] [PubMed] [Google Scholar]
- 6.Beyer K, Dandekar T, Keller W. RNA ligands selected by cleavage stimulation factor contain distinct sequence motifs that function as downstream elements in 3′-end processing of pre-mRNA. J Biol Chem. 1997;272:26769–26779. doi: 10.1074/jbc.272.42.26769. [DOI] [PubMed] [Google Scholar]
- 7.Bienroth S, Keller W, Wahle E. Assembly of a processive messenger RNA polyadenylation complex. EMBO J. 1993;12:585–594. doi: 10.1002/j.1460-2075.1993.tb05690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birse C E, Minvielle-Sebastia L, Lee B A, Keller W, Proudfoot N J. Coupling termination of transcription to messenger RNA maturation in yeast. Science. 1998;280:298–301. doi: 10.1126/science.280.5361.298. [DOI] [PubMed] [Google Scholar]
- 9.Burd C, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butler J S, Sadhale P, Platt T. RNA processing in vitro produces mature 3′ ends of a variety of Saccharomyces cerevisiae mRNAs. Mol Cell Biol. 1990;10:2599–2605. doi: 10.1128/mcb.10.6.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Moore C L. Separation of factors required for cleavage and polyadenylation of yeast pre-mRNA. Mol Cell Biol. 1992;12:3470–3481. doi: 10.1128/mcb.12.8.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S, Hyman L E. A specific RNA-protein interaction at yeast polyadenylation efficiency elements. Nucleic Acids Res. 1998;26:4965–4974. doi: 10.1093/nar/26.21.4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z, Krug R. Selective nuclear export of viral RNAs in influenza virus-infected cells. Trends Microbiol. 2000;8:376–383. doi: 10.1016/s0966-842x(00)01794-7. [DOI] [PubMed] [Google Scholar]
- 14.Cramer P, Srebrow A, Kadener S, Werbajh S, de la Mata M, Melen G, Nogues G, Kornblihtt A R. Coordination between transcription and pre-mRNA processing. FEBS Lett. 2001;498:179–182. doi: 10.1016/s0014-5793(01)02485-1. [DOI] [PubMed] [Google Scholar]
- 15.de Vries H, Ruegsegger U, Hubner W, Friedlein A, Lengen H, Keller W. Human pre-mRNA clevage factor IIm contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J. 2000;19:5895–5904. doi: 10.1093/emboj/19.21.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dichtl B, Keller W. Recognition of polyadenylation sites in yeast pre-mRNAs by cleavage and polyadenylation factor. EMBO J. 2001;20:3197–3209. doi: 10.1093/emboj/20.12.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duvel K, Braus G. Different positioning elements select poly(A) sites at the 3′ end of GCN4 mRNA in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1999;27:4751–4758. doi: 10.1093/nar/27.24.4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwalds-Gilbert G, Veraldi K L, Milcarek C. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Estojak J, Brent R, Golemis E. Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghisolfi-Nieto L, Joseph G, Puvion-Dutilleul F, Amalric F, Bouvet P. Nucleolin is a sequence-specific RNA-binding protein: characterization of targets on pre-ribosomal RNA. J Mol Biol. 1996;260:34–53. doi: 10.1006/jmbi.1996.0380. [DOI] [PubMed] [Google Scholar]
- 21.Gietz D, St. Jean A, Woods R, Schiestl R. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graber J, Cantor C, Mohr S, Smith T. Genomic detection of new yeast pre-mRNA 3′-end processing signals. Nucleic Acids Res. 1999;27:888–894. doi: 10.1093/nar/27.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gross S, Moore C. Five subunits are required for reconstitution of the cleavage and polyadenylation activities of Saccharomyces cerevisiae cleavage factor I. Proc Natl Acad Sci USA. 2001;98:6080–6085. doi: 10.1073/pnas.101046598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guillaume S, Joseph G, Ghisolfi-Nieto L, Bauzan M, Erard M, Amalric F, Bouvet P. Two RNA-binding domains determine the RNA-binding specificity of nucleolin. J Biol Chem. 1997;272:13109–13116. doi: 10.1074/jbc.272.20.13109. [DOI] [PubMed] [Google Scholar]
- 25.Guo Z, Sherman F. 3′-end forming signals of yeast mRNA. Trends Biochem Sci. 1996;21:477–481. doi: 10.1016/s0968-0004(96)10057-8. [DOI] [PubMed] [Google Scholar]
- 26.Guo Z, Sherman F. Signals sufficient for 3′-end formation of yeast mRNA. Mol Cell Biol. 1996;16:2772–2776. doi: 10.1128/mcb.16.6.2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heidmann S, Obermaier B, Vogel K, Domdey H. Identification of pre-mRNA polyadenylation sites in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:4215–4229. doi: 10.1128/mcb.12.9.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helmling S, Zhelkovsky A, Moore C. Fip1 regulates the activity of poly(A) polymerase through multiple interactions. Mol Cell Biol. 2001;21:2036–2037. doi: 10.1128/MCB.21.6.2026-2037.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirose Y, Manley J L. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 30.Huang Y, Carmichael G. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol Cell Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hyman L E, Moore C L. Termination and pausing of RNA polymerase II downstream of yeast polyadenylation sites. Mol Cell Biol. 1993;13:5159–5167. doi: 10.1128/mcb.13.9.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanneganti G K M, Manley J L. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev. 1995;9:2672–2683. doi: 10.1101/gad.9.21.2672. [DOI] [PubMed] [Google Scholar]
- 33.Kessler M, Henry M, Gross S, Shen E, Zhao J, Silver P, Moore C. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 1997;11:2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kessler M M, Zhao J, Moore C L. Purification of the Saccharomyces cerevisiae cleavage/polyadenylation factor I. J Biol Chem. 1996;271:27167–27175. doi: 10.1074/jbc.271.43.27167. [DOI] [PubMed] [Google Scholar]
- 35.Landsman D. RNP-1, an RNA-binding motif, is conserved in the DNA-binding cold shock domain. Nucleic Acids Res. 1992;20:2861–2864. doi: 10.1093/nar/20.11.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Long R M, Elliott D J, Stutz F, Rosbash M, Singer R H. Spatial consequences of defective processing of specific yeast mRNAs revealed by fluorescent in situ hybridization. RNA. 1995;1:1071–1078. [PMC free article] [PubMed] [Google Scholar]
- 37.MacDonald C C, Wilusz J, Shenk T. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol Cell Biol. 1994;14:6647–6654. doi: 10.1128/mcb.14.10.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandart E, Parker R. Effects of mutations in the S. cerevisiae RNA14, RNA15, and PAP1 genes on polyadenylation in vivo. Mol Cell Biol. 1995;15:6979–6986. doi: 10.1128/mcb.15.12.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayeda A, Munroe S, Caceres J, Krainer A. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 1994;13:5483–5495. doi: 10.1002/j.1460-2075.1994.tb06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayeda A, Munroe S, Xu R, Krainer A. Distinct functions of the closely related tandem RNA-recognition motifs of hnRNP A1. RNA. 1998;4:1111–1123. doi: 10.1017/s135583829898089x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mikaelian I, Sergeant A. A general and fast method to generate multiple site-directed mutations. Nucleic Acids Res. 1992;20:376. doi: 10.1093/nar/20.2.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Minvielle-Sebastia L, Beyer K, Krecic A M, Hector R E, Swanson M S, Keller W. Control of cleavage site selection during mRNA 3′ end formation by a yeast hnRNP. EMBO J. 1998;17:7454–7468. doi: 10.1093/emboj/17.24.7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minvielle-Sebastia L, Keller W. mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr Opin Cell Biol. 1999;11:352–357. doi: 10.1016/S0955-0674(99)80049-0. [DOI] [PubMed] [Google Scholar]
- 44.Minvielle-Sebastia L, Preker P J, Keller W. RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3′-end processing factor. Science. 1994;266:1702–1705. doi: 10.1126/science.7992054. [DOI] [PubMed] [Google Scholar]
- 45.Minvielle-Sebastia L, Winsor B, Bonneaud N, Lacroute F. Mutations in the yeast RNA14 and RNA15 genes result in an abnormal mRNA decay rate: sequence analysis reveals an RNA-binding domain in the Rna15 protein. Mol Cell Biol. 1991;11:3075–3087. doi: 10.1128/mcb.11.6.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitchell P, Tollervey D. mRNA turnover. Curr Opin Cell Biol. 2001;13:320–325. doi: 10.1016/s0955-0674(00)00214-3. [DOI] [PubMed] [Google Scholar]
- 47.Murthy K G, Manley J L. Characterization of the multisubunit cleavage-polyadenylation specificity factor. J Biol Chem. 1992;267:14804–14811. [PubMed] [Google Scholar]
- 48.Murthy K G K, Manley J L. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′end formation. Genes Dev. 1995;9:2672–2683. doi: 10.1101/gad.9.21.2672. [DOI] [PubMed] [Google Scholar]
- 49.Neer E, Schmidt C, Nambudripad R, Smith T. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 50.Ohnacker M, Barabino S, Preker P, Keller W. The WD-repeat protein Pfs2p bridges two essential factors within the yeast pre-mRNA 3′-end-processing complex. EMBO J. 2000;19:47–57. doi: 10.1093/emboj/19.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Proudfoot N. Connecting transcription to messenger RNA processing. Trends Biochem Sci. 2000;25:290–293. doi: 10.1016/s0968-0004(00)01591-7. [DOI] [PubMed] [Google Scholar]
- 52.Ruegsegger U, Beyer K, Keller W. Purification and characterization of human cleavage factor IM involved in the 3′ end processing of messenger RNA precursors. J Biol Chem. 1996;271:6107–6113. doi: 10.1074/jbc.271.11.6107. [DOI] [PubMed] [Google Scholar]
- 53.Russo P, Li W, Guo Z, Sherman F. Signals that produce 3′ termini in CYC1 mRNA of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:7836–7849. doi: 10.1128/mcb.13.12.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sachs A, Varani G. Eukaryotic translation initiation: there are (at least) two sides to every story. Nat Struct Biol. 2000;7:356–361. doi: 10.1038/75120. [DOI] [PubMed] [Google Scholar]
- 55.Sadhale P P, Platt T. Unusual aspects of in vitro RNA processing in the 3′ regions of the GAL1, GAL7, and GAL10 genes in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:4262–4270. doi: 10.1128/mcb.12.10.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sambrook J, Russel D. Molecular cloning: a laboratory manual. 4th ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 57.Setyono B, Greenberg J R. Proteins associated with poly(A) and other regions of mRNA and hnRNA molecules as investigated by crosslinking. Cell. 1981;24:775–783. doi: 10.1016/0092-8674(81)90103-3. [DOI] [PubMed] [Google Scholar]
- 58.Shamoo Y, Abdul-Manan N, Patten A, Crawford J, Pellegrini M, Williams K. Both RNA-binding domains in heterogenous nuclear ribonucleoprotein A1 contribute toward single-stranded RNA binding. Biochemistry. 1994;33:8272–8281. doi: 10.1021/bi00193a014. [DOI] [PubMed] [Google Scholar]
- 59.Sherman F, Fink G R, Hicks J B. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 60.Takagaki Y, Manley J. RNA recognition by the human polyadenylation factor CstF. Mol Cell Biol. 1997;17:3907–3914. doi: 10.1128/mcb.17.7.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takagaki Y, Manley J L. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol Cell. 1998;2:761–771. doi: 10.1016/s1097-2765(00)80291-9. [DOI] [PubMed] [Google Scholar]
- 62.Takagaki Y, Manley J L. A polyadenylation factor subunit is the human homologue of the Drosophila suppressor of forked protein. Nature. 1994;372:471–474. doi: 10.1038/372471a0. [DOI] [PubMed] [Google Scholar]
- 63.Tucker M, Parker R. Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae. Annu Rev Biochem. 2000;69:571–595. doi: 10.1146/annurev.biochem.69.1.571. [DOI] [PubMed] [Google Scholar]
- 64.Urlaub H, Hartmouth K, Kosta S, Grelle G, Luhrmann R. A general approach for identification of RNA-protein cross-linking sites within native human spliceosomal small nuclear ribonucleoproteins (snRNPs) J Biol Chem. 2000;275:4158–5168. doi: 10.1074/jbc.M007434200. [DOI] [PubMed] [Google Scholar]
- 65.Valentini S, Weiss V, Silver P. Arginine methylation and binding of Hrp1p to the efficiency element for mRNA 3′-end formation. RNA. 1999;5:272–280. doi: 10.1017/s1355838299981633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Helden J, del Olmo M, Perez-Ortin J. Statistical analysis of yeast genomic downstream sequences reveals putative polyadenylation signals. Nucleic Acids Res. 2000;28:1000–1010. doi: 10.1093/nar/28.4.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Varani G, Nagai K. RNA recognition by RNP proteins during RNA processing. Annu Rev Biophys Biomol Struct. 1998;27:407–445. doi: 10.1146/annurev.biophys.27.1.407. [DOI] [PubMed] [Google Scholar]
- 68.Wahle E, Ruegsegger U. 3′-end processing of pre-mRNA in eukaryotes. FEMS Microbiol Rev. 1999;648:1–18. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 69.Wells J. Systemic mutational analyses of protein-protein interfaces. Methods Enzymol. 1991;202:390–411. doi: 10.1016/0076-6879(91)02020-a. [DOI] [PubMed] [Google Scholar]
- 70.Wilusz C J, Wormington M, Peltz S W. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 71.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao J, Kessler M M, Helmling S, Moore C L. Pta1, a component of yeast CF II, is required for both cleavage and poly(A) addition reactions of mRNA 3′ end formation. Mol Cell Biol. 1999;19:7733–7740. doi: 10.1128/mcb.19.11.7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhelkovsky A, Helmling S, Moore C. Processivity of the Saccharomyces cerevisiae poly(A) polymerase requires interactions at the carboxyl-terminal RNA binding domain. Mol Cell Biol. 1998;18:5942–5951. doi: 10.1128/mcb.18.10.5942. [DOI] [PMC free article] [PubMed] [Google Scholar]