Highlights
-
•
Data from the European Prevention of Alzheimer’s Dementia (EPAD) cohort are analyzed.
-
•
CSF p-tau relates to gray matter connectivity alterations in preclinical AD.
-
•
Amyloid and tau show differential effects on network connectivity.
Keywords: Brain connectivity, Gray matter networks, Preclinical Alzheimer, Tau, Amyloid
Abstract
Gray matter networks are altered with amyloid accumulation in the earliest stage of AD, and are associated with decline throughout the AD spectrum. It remains unclear to what extent gray matter network abnormalities are associated with hyperphosphorylated-tau (p-tau). We studied the relationship of cerebrospinal fluid (CSF) p-tau181 with gray matter networks in non-demented participants from the European Prevention of Alzheimer’s Dementia (EPAD) cohort, and studied dependencies on amyloid and cognitive status. Gray matter networks were extracted from baseline structural 3D T1w MRI. P-tau181 and abeta were measured with the Roche cobas Elecsys System. We studied the associations of CSF biomarkers levels with several network’s graph properties. We further studied whether the relationships of p-tau 181 and network measures were dependent on amyloid status and cognitive stage (CDR). We repeated these analyses for network properties at a regional level, where we averaged local network values across cubes within each of 116 areas as defined by the automated anatomical labeling (AAL) atlas. Amyloid positivity was associated with higher network size and betweenness centrality, and lower gamma, clustering and small-world coefficients. Higher CSF p-tau 181 levels were related to lower betweenness centrality, path length and lambda coefficients (all p < 0.01). Three-way interactions between p-tau181, amyloid status and CDR were found for path length, lambda and clustering (all p < 0.05): Cognitively unimpaired amyloid-negative participants showed lower path length and lambda values with higher CSF p-tau181 levels. Amyloid-positive participants with impaired cognition demonstrated lower clustering coefficients in association to higher CSF p-tau181 levels.
Our results suggest that alterations in gray matter network clustering coefficient is an early and specific event in AD.
Introduction
Alzheimer’s Disease (AD) is a progressive and disabling neurodegenerative disease, and is estimated to be responsible for 60 % to 80 % of dementia cases [45]. Amyloid pathology is hypothesized to initiate the AD pathological cascade [59], [20] followed by the aggregation of intraneuronal hyperphosphorylated tau (p-tau) [30], [24]. Both amyloid and tau can provoke disruptions of neuronal connectivity and synaptic loss, even in early disease stages, when cognition is still normal [3], [40]. Progressive synaptic loss as the disease progresses shows a close correspondence with cognitive decline [49]. As such, imaging measures that can capture consequences of connectivity loss may help identify individuals at risk for cognitive decline for secondary prevention interventions.
One approach captures brain connectivity from patterns of covariation of gray matter volume between brain areas on structural magnetic resonance image (MRI) T1-weighted sequences [32], [56]. Such patterns of cortical similarity have been associated with coordinated growth during development [1], functional co-activation [2] and axonal connectivity [18]. Gray matter networks have been reported as being disrupted in AD [39], [55], [22], correlate with disease severity [57], [64], [39] and predict imminent cognitive decline [57]. In preclinical stages, they are associated with early amyloid accumulation [26], [54]. Another pathological hallmark of AD is tau neurofibrillary tangles. Tau is promoter of axonal stabilization, suggesting that individuals with higher tau levels will show greater gray matter network disruptions and that this would relate to amyloidosis. CSF p-tau 181is a sensitive marker for tau tangles [51] and so it can be hypothesized that this measure may show stronger associations with GM networks.
Here, we studied whether higher CSF p-tau 181 levels are associated with worse gray matter network disruptions in people without dementia from the European Prevention of Alzheimer’s Dementia (EPAD) cohort. We further tested whether relationships between p-tau 181 levels and gray matter network connectivity were dependent on amyloid and/or CDR status.
Methods
Study population
Data were drawn from the v1500.0 baseline data release from the European Prevention of Alzheimer’s Dementia (EPAD) multicenter study [42]. EPAD general inclusion criteria were age above 50 years and no diagnosis of dementia. Demographic, cognitive, neuroimaging, fluid biomarkers and genetic outcomes were collected [50]. For this study, we selected individuals who had cerebrospinal fluid (CSF), neuropsychological and structural MRI data available. Neuropsychological examination included the Mini-Mental State Evaluation (MMSE, [14]), and Clinical Dementia Rating (CDR) scale [33].
CSF analysis
CSF biomarkers were obtained using a harmonized pre-analytical protocol and analyses were performed on the fully automated Roche cobas Elecsys System at the Clinical Neurochemistry Laboratory, Mölndal, Sweden [50]. Concentrations of amyloid-beta (Aβ 1–42) and phosphorylated tau (p-tau181) were determined according to the manufacturer’s instructions. We used CSF Aβ1-42 levels < 1000 pg/mL to define amyloid positive (A+) subjects as validated previously [23]. Continuous p-tau181 measurements were used in the present analyses.
MRI acquisition and preprocessing
3D-T1 weighted structural MRI scans were acquired at baseline with seven different MRI scanners from 19 scanning sites using a harmonized scanning protocol [53]. ExploreASL [34], an SPM-based toolbox [37] designed to harmonize image processing in multimodal and multicenter studies, was used for segmentation of subject specific gray matter (GM), white matter (WM) and CSF from T1w images. Specifically, this step was performed using the Computational Anatomy Toolbox 12 (CAT12), which estimates and corrects the bias field inhomogeneity in 3D T1w images, and iteratively improves the non-linear registration to MNI standard space and the creation of partial volume maps of gray matter (GM), white matter (WM) and CSF [17]. GM segmentations were resliced into 2 × 2 × 2 mm3 voxels to reduce the total number of voxels. The Automated Anatomical Labeling atlas (AAL3, [58], [43]), including 116 regions of interest (ROI), was warped into each subject space in order to summarize local network properties. MRI and CSF biomarkers were acquired on the same day for each participant.
Gray matter network construction
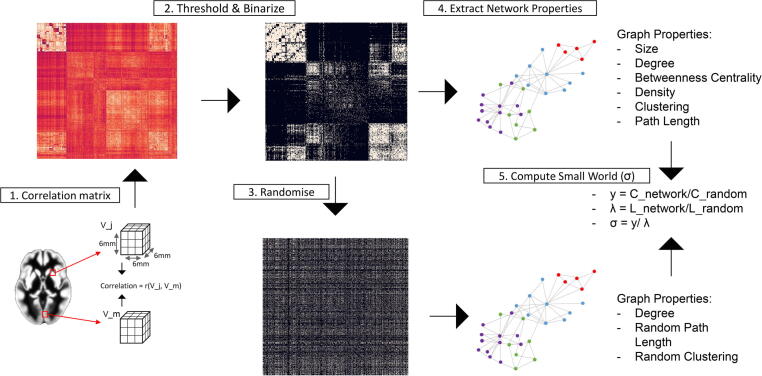
Single-subject gray matter networks were computed from gray matter segmentations, using an automated method previously described and publicly available [56]. Nodes were defined as 3 × 3 × 3 voxel cubes. Using cubes keeps the 3D structure of the cortex thereby using spatial information from the MRI scan in addition to GM density values. Connections are then determined between each pair of nodes by calculating Pearson’s correlations of gray matter density values across corresponding voxels between cube pairs. High correlations between cubes thus reflect similarity in both the local thickness and the folding structure of the cortex. This process results in an NxN similarity matrix for each participant, where N is the number of cubes extracted for that participant. Networks are then binarized using a subject-specific threshold determined with a random permutation method ensuring a similar chance for all individuals to include at most 5 % spurious correlations in the network [35]. From the resulting undirected and unweighted networks, the Brain Connectivity Toolbox (BCT, [44]) was used to extract graph features describing local and global network topology. GM network construction is shown in Fig. 1.
Fig. 1.
Gray Matter Network construction. After preprocessing, each GM segmentation is divided into 3 × 3 × 3 voxel cubes and 1) similarity between all N cubes within a scan was computed with Pearson’s correlation coefficient and stored as an N by N matrix; 2) the similarity matrix was binarized using a threshold that ensured 5 % chance of spurious connections (corresponding to a significance level of p-value = 0.05 FDR-corrected); 3) five random matrices with similar spatial degree distribution were generated; 4) network properties (size, density, degree, BC, clustering, path length) from the binarized matrix were calculated and 5) normalized properties (gamma, lambda and small-world coefficient) through the comparison with the random networks were computed.
Network metrics
We computed the following network metrics (see detailed explanation below): size, connectivity density, degree, betweenness centrality (BC), clustering coefficient (Cp) and characteristic path length (Lp). In addition, path length and clustering coefficient of each graph were compared with those extracted from 5 randomized reference graphs, generated by rearranging the edges while keeping identical size and degree distribution [31], to obtain normalized clustering (gamma, γ), normalized path length (lambda, λ) and small-world coefficient, as the ratio of γ and λ.
Connectivity density is computed as the fraction of present connections relative to possible connections, and gives an indication on the sparsity of the network. Degree is the number of a node’s connections, and the global degree is the average across all nodes. Networks showing disconnected nodes, i.e. nodes with degree equal to zero, were excluded from the analysis [4]. Betweenness centrality is defined as the number of all shortest paths in the network that pass through a given node [16]. Clustering coefficient is the fraction of a node’s neighbors which are also neighbors of each other [61], and characteristic path length is the length of all paths between all pairs of nodes in the network [5]. Degree, betweenness centrality, clustering and path length were first extracted for each GMN cube in the subject’s network and then averaged to obtain mean global topological descriptors. Regional values were computed as the mean across cubes within each of the 116 regions defined by the AAL atlas [58]. Global and local GM volume were also extracted in this step to be used as a covariate in subsequent statistical analyses. All global and regional GMN properties were z-scored to improve comparability and interpretability of results.
Statistical analyses
Chi-square test and paired samples t-test were initially used to compare demographics, clinical and network characteristics between A+ and A− participants.
We first studied associations of amyloid status and p-tau181 levels with GM network measures across the total group. For each of the network’s metrics, we ran separate linear mixed models with amyloid status and p-tau 181as main effects. A random intercept for the scanning site was included to adjust for potential center-specific effects. Models were further adjusted for CDR, age, sex, total GM volume and network size. Covariate selection was based on previous evidence showing that participant demographics, network size and disease status show an effect on gray matter network topology [9], [19]. Next, we tested whether the relationship between p-tau181 and GM network meaures were dependent on amyloid status or CDR by testing the effect of the three-way interaction between p-tau, amyloid status and CDR on network properties. We repeated these analyses at a regional level for all ROI included in the AAL atlas across the whole group. Regional associations were corrected for age, sex, regional GM volume, site and adjusted for multiple testing using false discovering rate (FDR) correction [62]. All variables were Z-scored before being used in linear models. An overview of the statistical model used is given in Table S1 of Supplementary materials.
Statistical significance was set at p-value < 0.05. All statistical analyses were performed in R, version 4.0.3.1
Results
Sample characteristics
From all 1500 baseline EPAD participants, a total of 1198 participants fulfilled the inclusion criteria for this study (Table 1). Mean age was 65.3 ± 7.14 and 673 (56.2 %) were female. All participants were without dementia at inclusion with an average MMSE of 28.7 (SD = 1.53), and 17 % had a CDR of 0.5. There were 384 (32.1 %) individuals who were CSF A+. Higher p-tau181 concentrations were found in A+ compared to A− participants, showing mean p-tau181 measurements of 21 pg/ml and 18.3 pg/ml respectively. No differences in total tau were observed based on amyloid status. Moreover, A+ participants showed lower network density, clustering, gamma and small-world coefficient, and higher path length values.
Table 1.
Demographic, clinical and network characteristics of amyloid positive and negative individuals.
| Characteristics | Amyloid negative (n = 814) |
Amyloid positive (n = 384) |
p-value |
|---|---|---|---|
| Female, N (%) | 475 (58.4) | 198 (51.6) | 0.032 |
| Age, Mean ± SD | 64.8 ± 6.96 | 66.5 ± 7.37 | <0.001 |
| MMSE, mean ± SD | 28.7 ± 1.42 | 28.4 ± 1.92 | <0.001 |
| CDR = 0.5, N (%) | 115 (14.1) | 97 (25.3) | <0.001 |
| Total tau, Mean ± SD | 218 ± 75.2 | 226 ± 126 | 0.182 |
| Phospho tau, Mean ± SD | 18.3 ± 7.28 | 21.0 ± 14.7 | <0.001 |
| Network Density, Mean ± SD | 0.20 ± 0.01 | 0.19 ± 0.01 | <0.001 |
| Network Size, Mean ± SD | 7226.76 ± 624.08 | 7247.38 ± 666.45 | 0.602 |
| Network Degree, Mean ± SD | 1454.00 ± 135.43 | 1436.94 ± 144.72 | 0.047 |
| Network Betweenness Centrality, Mean ± SD | 6177.08 ± 550.33 | 6222.36 ± 585.85 | 0.193 |
| Network Path Length, Mean ± SD | 1.85 ± 0.02 | 1.86 ± 0.02 | <0.001 |
| Network Lambda, Mean ± SD | 1.03 ± 0.01 | 1.03 ± 0.01 | 0.220 |
| Network Clustering, Mean ± SD | 0.49 ± 0.02 | 0.48 ± 0.02 | <0.001 |
| Network Gamma, Mean ± SD | 1.35 ± 0.04 | 1.34 ± 0.05 | <0.001 |
| Network Small-World Coefficient, Mean ± SD | 1.31 ± 0.04) | 1.30 ± 0.05 | <0.001 |
Abbreviations: MMSE = Mini-mental state evaluation; CDR = Clinical dementia rating scale. Statistical differences between amyloid positive and negative individuals are reported.
CSF biomarkers relationship with global network measures
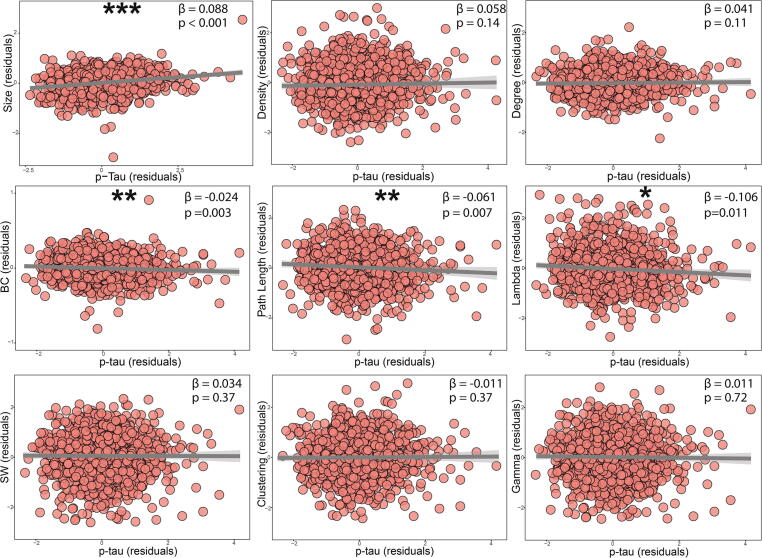
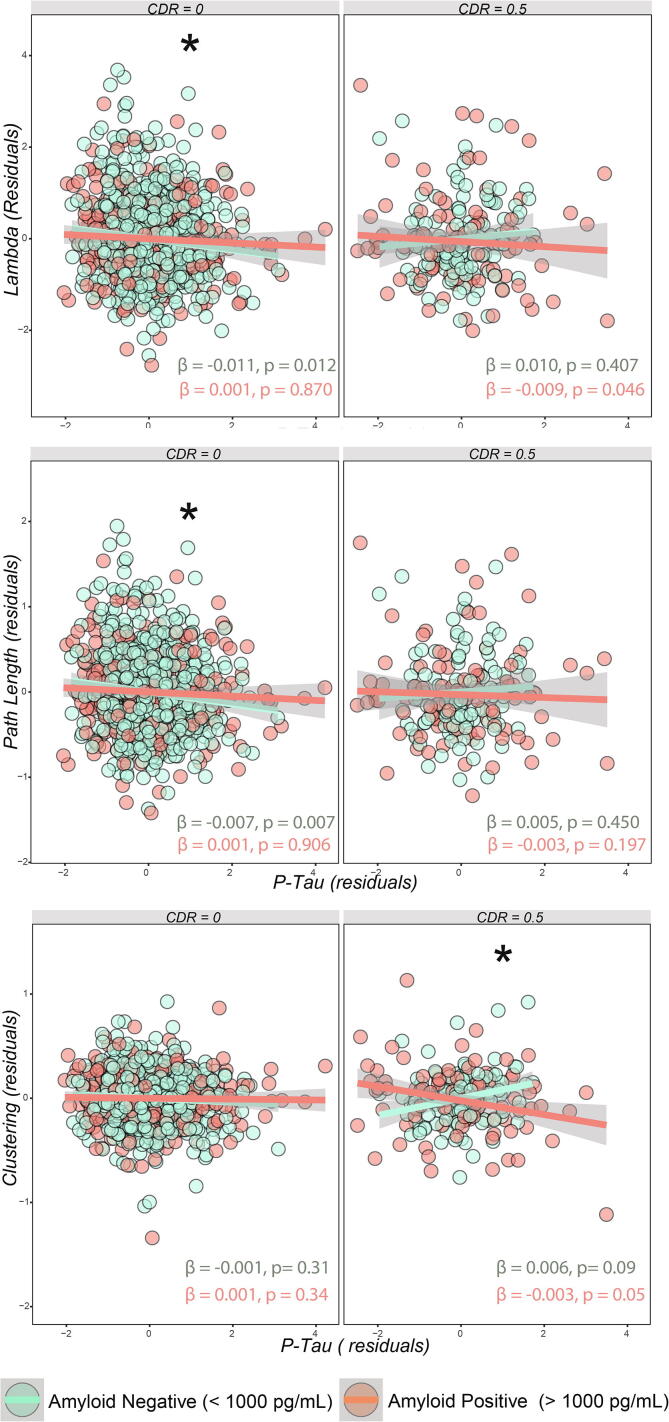
Amyloid positivity was associated with larger network size (β = 0.12, p < 0.001), which was used as a covariate in other models, lower gamma (β = −0.1, p < 0.05), lower small-world coefficient (β = −0.12, p < 0.05; Fig. S1). Higher p-tau181 levels were associated with a higher network size (β = 0.088, p < 0.001; Fig. 2), and lower betweenness centrality (β = −0.024, p = 0.003), lambda (β = −0.106, p = 0.011) and path length (β = −0.061, p = 0.007). A three way interaction of p-tauXabetaXCDR was significant for lambda, path length and clustering. For the participants in the CDR = 0 group, A− participants had higher path length and lambda values than A+, and only in A−, higher p-tau181 was associated with lower values for path length (for A−: β = −0.007, p = 0.007; for A+: β = 0.001, p = 0.906; p interaction = 0.04) and lambda (for A−: β = −0.011, p = 0.012; for A+: β = 0.001, p = 0.870; p interaction = 0.027). In participants with CDR = 0.5, A+ participants showed lower clustering coefficient compared to A−, and only in the A+ group higher p-tau181 concentrations were associated with lower clustering coefficient values (for A−: β = 0.006, p = 0.09; for A+: β = −0.003, p = 0.05; p interaction = 0.006). Significant three-way interactions are shown in Fig. 3. All model coefficients are shown in Tables S2–S4 of Supplementary materials. Beta values refers to standardized measures.
Fig. 2.
Association of CSF p-tau181 with global gray matter network properties. Adjusted variable plots showing network metrics relationship with p-tau 181after correcting for age, sex, GM volume, size of the network and scanning site (as random intercept) are shown. * p < 0.05; ** p < 0.01. *** p < 0.001. Abbreviations: SW = Small world Coefficient; BC = Betweenness Centrality.
Fig. 3.
Interactions between p-tau181, amyloid status, and CDR. To facilitate interpretation, the residuals of the model after correcting for age, sex, GM volume, size of the network and scanning site (as random intercept) are shown; *Indicates significant three-way interaction after covariates correction with a p-value < 0.05.
CSF biomarkers relationship with local network measures
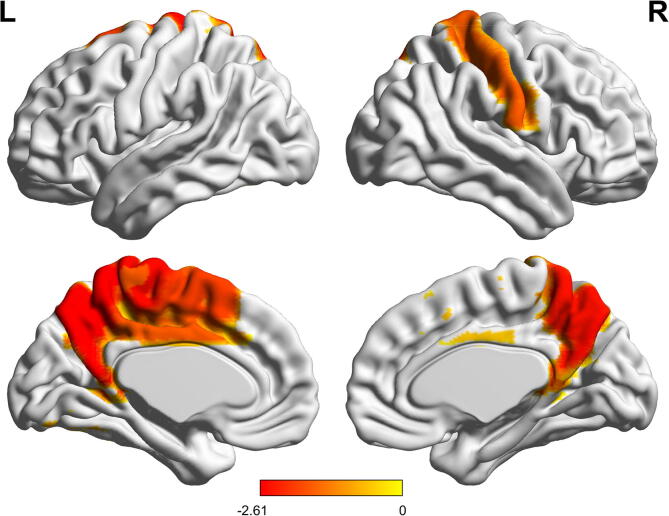
We then studied whether the associations observed in the global analysis were specific to a particular anatomical area. Amyloid status did not show significant association with regional network properties after adjusting for covariates and FDR correction. Across the whole group, we found that higher p-tau181 levels were related to lower path length values in the precuneus bilaterally; left paracentral lobule, left supplementary motor area and left middle cingulate cortex; and right postcentral gyrus (Fig. 4). Associations of p-tau181 with regional clustering, degree and betweenness centrality did not survive FDR multiple comparison correction (unadjusted relationships are shown in Fig. S1 of Supplementary materials). There was no significant effect of the interaction term on regional properties.
Fig. 4.
Associations of regional path length with p-tau181. Surface Plot of standardized beta-values of the relationship between p-tau181 and regional path length are shown for statistically significant regions after covariates correction (p-value < 0.05 FDR-corrected). Upper one is the lateral view, lower row is the medial view, for the left hemisphere (left column) and right hemisphere (right column), respectively.
Discussion
In this study, we found that CSF p-tau 181levels showed assocations with GM network measures that depended on amyloid and CDR status in a non-demented cohort. These results suggest that alterations in specific gray matter network properties reflect either amyloid and p-tau deposition in early stages of AD pathology, or are related to p-tau 181only, suggesting distinct pathological underlying mechanisms. An overview of our findings for each investigated graph properties, and comparison with previous studies, is reported in Table S5 of Supplementary materials.
Previous studies have shown that reduced clustering and decreased gamma in GM networks are associated with Aβdeposition in cognitively normal participants [26], [52]. Here, we also observed lower clustering values in participants with abnormal Aβonly. Possibly, a reduced clustering coefficient may reflect local synaptic dysfunction triggered by early Aβdeposition. This may introduce dissimilarities in gray matter morphology at a regional level, subsequently reducing clustering and gamma values. Similar to our findings, [54] have shown higher path length values in relation to lower Aβ42 CSF levels in cognitively unimpaired individuals. However, decreased lambda values have been observed in relation with Aβdeposition in patients with AD [63], [27], [55]. These discrepancies may be explained by non-monotonic path length changes during disease progression, with an increase due to the initial loss of local connections and reflecting asynchronous atrophy patterns, and an eventual decrease when the networks become more sparse in later disease phases [8].
We further extend on previous findings by showing that early CSF p-tau 181 measurements are also related to GM network alterations already from the preclinical and prodromal stages of AD. We found lower network betweenness centrality in relation to higher p-tau181 CSF concentrations, possibly reflecting a synchronous widespread disruption of connectivity following tau deposition. In a previous study, individuals with prodromal AD were found to have lower degree centrality values relating to higher levels of CSF total tau, with faster degree decline over time [10]. Similar findings have been reported using different MRI modalities. A fMRI-PET study [48] demonstrated that regions with high tau levels show lower functional connectivity as measured by clustering and degree in individuals with atypical AD, namely posterior cortical atrophy or logopenic progressive aphasia. Together, these findings suggest an additional effect of tau pathology in accelerating the synaptic loss initiated with Aβ.
In addition to the present literature, we further observed that the relationship between p-tau181and specific global network measures depended on both CDR and amyloid status. Amyloid positive participants with impaired cognition (CDR = 0.5) demonstrated lower clustering coefficients in association to higher CSF p-tau181 levels. Lower values of path length and lambda were associated with higher p-tau181 measurements and this relationship was stronger in A− CDR = 0 participants, which suggests potential primary tau-mediated effects on gray matter networks integration. A similar association of gray matter networks and tau biomarkers has previously been reported in Dicks et al. [10], where CSF total tau was related to a faster longitudinal decline in lambda showing local early regional disruptions comparable to our results. Regional alterations of path length in relationship with tau were found in both early amyloid related areas, such as precuneus and cingulate cortex, but also in regions which are only involved in the late stages of the disease, such as motor areas. These regional associations were however independent of amyloid or cognitive (CDR) status, and observed on the whole group. Similar patterns have been previously found in studies comparing tau and GM networks spatial distribution and might presumably be due to large-scale network-wide disruptions and to a lack of stimulation and/or neurotrophic factors from connecting regions [36]. One explanation might be that some individuals may have primary age-related tauopathy (PART), which is a neuropathological concept emerged to characterize individuals with altered levels of neurofibrillary tangles in the absence of amyloid pathology [7]. However, whether this condition represents an AD-independent, and what are the typical CSF profiles, remains unclear [28], [12].
The observed divergence between amyloid- and tau-mediated connectivity alterations has been previously described with other methodologies. In [21], the authors found a correlation between Aβ and cortical thickness that showed opposite directions in tau positive and negative participants, in cognitively unimpaired individuals. In other fMRI functional connectivity (FC) studies [29], [47], tau has often been linked to a clear FC decrease while both hypo- and hyper-connectivity of default mode networks nodes are observed with Aβdeposition [29], [47], [46]. Here, we found this discrepancy in relation to gray matter network integration properties. While clustering was associated with both amyloid status and tau, suggesting AD related changes, path length and lamba were specifically related to only tau. A possible explanation, that could also elucidate the stronger effect of tau found in unimpaired (A− CDR = 0) individuals, is that amyloid and tau could have a different effect on local and distant connectivity, suggesting two different underlying mechanisms. A recent study [38], showed distinct associations of Aβand tau pathology on synaptic and axonal connectivity using presynaptic (synaptosomal-associated protein 25; growth-associated protein 43), postsynaptic (neurogranin) and axonal (neurofilament light chain) CSF markers of synaptic and neuronal integrity. Early amyloid pathology was found to be associated with synaptic disruptions, i.e. elevated levels of pre- and postsynaptic markers. In contrast, tauopathy correlated with higher levels of neurofilament light chain, proxy of axonal damage. Similarly, other studies showed that Aβdeposition is associated with both pre- and post-synaptic changes in cognitively unimplaired individuals [41].The differential effect that Aβand tau exert on synaptic and axonal connectivity may be a possible explanation of our distinct GM networks findings and of the stronger tau effects found in A− CDR = 0 participants. Recent models have proposed that the Aβ-induced synaptic damage along the length of the axon might lead to dissociation of tau from microtubules and eventually result in tau-related axonal degeneration [13]. Our results are in line with this hypothesis of initial damage to synapses close to Aβ plaques producing local synaptic dysfunction and GM dissimilarity, thus showing low network segregation. Subsequent phosphorylation of tau and formation of tangles would eventually trigger axon loss and distal atrophy patterns over the entire brain, impairing network integration. As proposed in [6], tau would therefore play a bidirectional role in the process, with increased connectivity first accelerating tau spreading through trans-neuronal propagation, but later promoting network disruptions. However, it is not possible to disentangle to what extent this could possibly be solely due to the different spatial distribution of amyloid and tau deposition, resulting in different changes in both magnitude and direction of network effects. Future studies relating GM networks alteration to other synaptic CSF biomarkers will be needed to further explore this hypothesis.
A potential limitation of this study is that EPAD is a research cohort of healthy participants that were specifically selected for their elevated risk for AD, which may not be representative of the general early-phase AD population. Moreover, the groups we analyzed differed in size, and so analyses for smaller groups may have been underpowered (e.g. small percentage of A− CDR = 0.5 participants). Future studies should aim to collect more data on those specific groups. Previous studies have shown higher sensitivity of the ratio between Aβ42 and Aβ40 for defining amyloid positivity [25], however Aβ40 was not measured in EPAD CSF samples. Nonetheless, a strength of the EPAD cohort is that it is well-phenotyped. The multi-center aspect of EPAD is both a strength and a potential weakness. While the pan-European distribution of the EPAD centers enhances the generalizability of our findings and allows for high sample-size, between-scanner differences could have lowered our statistical power compared to a similar sample size acquired at a single scanner. Our harmonized scan protocol and centralized MRI processing may have partially accounted for this effect. Other harmonization strategies have been recently proposed to remove the effect of scanning site while keeping the variability of other biological variables [15], but their implementation on gray matter networks and derived graph measures has so far not been tested. Another potential limitation of our study is that we used CSF p-tau181 to specifically investigate AD-related changes in tau pathology, but this measuredoes not carry information about the spatial distribution of tau deposition across the brain. Moreover, although abnormal tau levels are often observed in amyloid positive participants, we did not find a difference in total tau between A groups. Recent evidence has shown that up to 30 % of individuals with AD can show normal CSF total tau levels (Disease Neuroimaging [11], even in the presence of tangles, possibly due to a different subtype of AD pathology [60]. Future studies may attempt to reproduce our findings with tau PET and non AD-specific markers of tau pathology, such as CSF total-tau. Finally, these findings were based on cross-sectional data and future longitudinal studies may confirm our cross-sectional findings, and further investigate the association of gray matter network properties with cognitive decline.
Conclusion
We found early GM networks disruptions in relationship to CSF p-tau181 levels, showing distinct characteristics depending on amyloid and cognitive status. Our results suggest that amyloid and tau deposition result in differential patterns of early GM networks alterations.
Funding
EPAD is supported by the EU/EFPIA Innovative Medicines Initiative (IMI) grant agreement 115736.
AM Wink, HM and FB are supported by AMYPAD (IMI 115952).
HM is supported by the Dutch Heart Foundation (2020T049), by the Eurostars-2 joint programme with co-funding from the European Union Horizon 2020 research and innovation programme, provided by the Netherlands Enterprise Agency (RvO), and by the EU Joint Program for Neurodegenerative Disease Research, provided by the Netherlands Organisation for health Research and Development and Alzheimer Nederland.
KB is supported by the Swedish Research Council (#2017-00915), the Alzheimer Drug Discovery Foundation (ADDF), USA (#RDAPB-201809-2016615), the Swedish Alzheimer Foundation (#AF-742881), Hjärnfonden, Sweden (#FO2017-0243), the Swedish state under the agreement between the Swedish government and the County Councils, the ALF-agreement (#ALFGBG-715986), the European Union Joint Program for Neurodegenerative Disorders (JPND2019-466-236), and the National Institute of Health (NIH), USA, (grant #1R01AG068398-01).
JMW is supported by the UK Dementia Research Institute (Foundation Chair) which receives its funding from DRI Ltd, funded by the UK Medical Research Council, Alzheimer’s Society and Alzheimer’s Research UK.
Dr. J.D. Gispert holds a ‘Ramón y Cajal’ fellowship (RYC-2013-13054) from the Spanish Ministry of Economy and Competitiveness, has received research support from the EU/EFPIA Innovative Medicines Initiative Joint Undertaking AMYPAD grant agreement n° 115952, and from Ministerio de Ciencia y Universidades (grant agreement RTI2018-102261).
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: KB has served as a consultant, at advisory boards, or at data monitoring committees for Abcam, Axon, Biogen, JOMDD/Shimadzu. Julius Clinical, Lilly, MagQu, Novartis, Prothena, Roche Diagnostics, and Siemens Healthineers, and is a co-founder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program (all outside the work presented in this paper). JDG has received speaker’s fees from Biogen and Philips.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nbas.2022.100054.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Alexander-Bloch, Aaron, Jay N. Giedd, and Ed Bullmore. “Imaging Structural Co-Variance between Human Brain Regions.” Nat Rev. Neurosci 2013;14(5):322–36. [DOI] [PMC free article] [PubMed]
- 2.Alexander-Bloch, Aaron, Armin Raznahan, Ed Bullmore, and Jay Giedd. The Convergence of Maturational Change and Structural Covariance in Human Cortical Networks. J Neurosci 2013;33(7):2889–99. [DOI] [PMC free article] [PubMed]
- 3.Bero A.W., Bauer A.Q., Stewart F.R., White B.R., Cirrito J.R., Raichle M.E., et al. Bidirectional Relationship between Functional Connectivity and Amyloid-β Deposition in Mouse Brain. J Neurosci. 2012;32(13):4334–4340. doi: 10.1523/JNEUROSCI.5845-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bettstetter, Christian. On the Minimum Node Degree and Connectivity of a Wireless Multihop Network. In Proceedings of the 3rd ACM International Symposium on Mobile Ad Hoc Networking & Computing, 80–91. MobiHoc ’02. New York, NY, USA: Association for Computing Machinery; 2002.
- 5.Bullmore, Ed, Olaf Sporns. Complex Brain Networks: Graph Theoretical Analysis of Structural and Functional Systems. Nat Rev Neurosci 2009;10(3):186–98. [DOI] [PubMed]
- 6.Cope T.E., Rittman T., Borchert R.J., Simon Jones P., Vatansever D., Allinson K., et al. Tau Burden and the Functional Connectome in Alzheimer’s Disease and Progressive Supranuclear Palsy. Brain. 2018 doi: 10.1093/brain/awx347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crary J.F., Trojanowski J.Q., Schneider J.A., Abisambra J.F., Abner E.L., Alafuzoff I., et al. Primary Age-Related Tauopathy (PART): A Common Pathology Associated with Human Aging. Acta Neuropathol. 2014;128(6):755–766. doi: 10.1007/s00401-014-1349-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dicks E., van der Flier W.M., Scheltens P., Barkhof F., Tijms B.M. Single-Subject Gray Matter Networks Predict Future Cortical Atrophy in Preclinical Alzheimer’s Disease. Neurobiol Aging. 2020 doi: 10.1016/j.neurobiolaging.2020.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Dicks E., Tijms B.M., Ten Kate M., Gouw A.A., Benedictus M.R., Teunissen C.E., et al. Gray Matter Network Measures Are Associated with Cognitive Decline in Mild Cognitive Impairment. Neurobiol Aging. 2018;61(January):198–206. doi: 10.1016/j.neurobiolaging.2017.09.029. [DOI] [PubMed] [Google Scholar]
- 10.Dicks E., Vermunt L., van der Flier W.M., Barkhof F., Scheltens P., Tijms B.M., et al. Grey Matter Network Trajectories across the Alzheimer’s Disease Continuum and Relation to Cognition. Brain Commun. 2020 doi: 10.1093/braincomms/fcaa177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Disease Neuroimaging Initiative, Alzheimer’s. 2009. “Cerebrospinal Fluid Biomarker Signature in Alzheimer’s Disease Neuroimaging Initiative Subjects.” Annals of. https://onlinelibrary.wiley.com/doi/abs/10.1002/ana.21610?casa_token=_4vSEhs9fvYAAAAA:5Z6ElHUlCH61ybpscRUMTGCl62yEps1y1FufU9msBX6QmCLBo77A1gERu87UoQP1OaJEDiHuW5Mp4nQ.
- 12.Duyckaerts C., Braak H., Brion J.-P., Buée L., Del Tredici K., Goedert M., et al. PART Is Part of Alzheimer Disease. Acta Neuropathol. 2015;129(5):749–756. doi: 10.1007/s00401-015-1390-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards F.A. A Unifying Hypothesis for Alzheimer’s Disease: From Plaques to Neurodegeneration. Trends Neurosci. 2019 doi: 10.1016/j.tins.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Folstein M.F., Folstein S.E., McHugh P.R. ‘Mini-Mental State’: A Practical Method for Grading the Cognitive State of Patients for the Clinician. J Psychiatr Res. 1975;12(3):189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 15.Fortin J.-P., Cullen N., Sheline Y.I., Taylor W.D., Aselcioglu I., Cook P.A., et al. Harmonization of Cortical Thickness Measurements across Scanners and Sites. NeuroImage. 2018;167(February):104–120. doi: 10.1016/j.neuroimage.2017.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freeman L.C. A Set of Measures of Centrality Based on Betweenness. Sociometry. 1977 doi: 10.2307/3033543. [DOI] [Google Scholar]
- 17.Gaser C., Dahnke R. CAT-a Computational Anatomy Toolbox for the Analysis of Structural MRI Data. Hbm. 2016;2016:336–348. doi: 10.1093/gigascience/giae049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong G., He Y., Chen Z.J., Evans A.C. Convergence and Divergence of Thickness Correlations with Diffusion Connections across the Human Cerebral Cortex. NeuroImage. 2012;59(2):1239–1248. doi: 10.1016/j.neuroimage.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 19.Hafkemeijer A., Altmann-Schneider I., de Craen A.J.M., Slagboom P.E., van der Grond J., Rombouts Serge A.R.B. Associations between Age and Gray Matter Volume in Anatomical Brain Networks in Middle-Aged to Older Adults. Aging Cell. 2014;13(6):1068–1074. doi: 10.1111/acel.12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardy J., Selkoe D.J. The Amyloid Hypothesis of Alzheimer’s Disease: Progress and Problems on the Road to Therapeutics. Science. 2002;297(5580):353–436. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 21.Harrison, Theresa M, Richard Du, Giuliana Klencklen, Suzanne L. Baker, and William J. Jagust. Distinct Effects of Beta-Amyloid and Tau on Cortical Thickness in Cognitively Healthy Older Adults. Alzheimer’s & Dementia: The Journal of the Alzheimer's Association; 2020. https://alz-journals.onlinelibrary.wiley.com/doi/abs/10.1002/alz.12249. [DOI] [PMC free article] [PubMed]
- 22.He Y., Chen Z., Evans A. Structural Insights into Aberrant Topological Patterns of Large-Scale Cortical Networks in Alzheimer’s Disease. J Neurosci. 2008;28(18):4756–4766. doi: 10.1523/JNEUROSCI.0141-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ingala S., De Boer C., Masselink L.A., Vergari I., Lorenzini L., Blennow K., et al. Application of the ATN Classification Scheme in a Population without Dementia: Findings from the EPAD Cohort. Alzheimer’s & Dementia. 2021 doi: 10.1002/alz.12292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jack C.R., Jr, Bennett D.A., Blennow K., Carrillo M.C., Feldman H.H., Frisoni G.B., et al. A/T/N: An Unbiased Descriptive Classification Scheme for Alzheimer Disease Biomarkers. Neurology. 2016;87(5):539–547. doi: 10.1212/WNL.0000000000002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janelidze S., Pannee J., Mikulskis A., Chiao P., Zetterberg H., Blennow K., et al. Concordance between Different Amyloid Immunoassays and Visual Amyloid Positron Emission Tomographic Assessment. JAMA Neurol. 2017;74(12):1492–1501. doi: 10.1001/jamaneurol.2017.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ten Kate M., ten Kate M., Visser P.J., Bakardjian H., Barkhof F., Sikkes S.A.M., et al. Gray Matter Network Disruptions and Regional Amyloid Beta in Cognitively Normal Adults. Front Aging Neurosci. 2018 doi: 10.3389/fnagi.2018.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim H.-J., Shin J.-H., Han C.E., Kim H.J., Na D.L., Seo S.W., et al. Using Individualized Brain Network for Analyzing Structural Covariance of the Cerebral Cortex in Alzheimer’s Patients. Front Neurosci. 2016;10(September):394. doi: 10.3389/fnins.2016.00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lafirdeen A.S., Mohamed E.C., Sabia S., Hourregue C., Lilamand M., Dugravot A., et al. Biomarker Profiles of Alzheimer’s Disease and Dynamic of the Association between Cerebrospinal Fluid Levels of β-Amyloid Peptide and Tau. PLoS ONE. 2019;14(5):e0217026. doi: 10.1371/journal.pone.0217026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malpas C.B., Saling M.M., Velakoulis D., Desmond P., O’Brien T.J. Differential Functional Connectivity Correlates of Cerebrospinal Fluid Biomarkers in Dementia of the Alzheimer’s Type. Neuro-Degenerative Diseases. 2016;16(3–4):147–151. doi: 10.1159/000438924. [DOI] [PubMed] [Google Scholar]
- 30.Mandelkow E.M., Mandelkow E. Tau in Alzheimer’s Disease. Trends Cell Biol. 1998;8(11):425–447. doi: 10.1016/s0962-8924(98)01368-3. [DOI] [PubMed] [Google Scholar]
- 31.Maslov S., Sneppen K. Specificity and Stability in Topology of Protein Networks. Science. 2002;296(5569):910–993. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- 32.Mechelli A., Friston K.J., Frackowiak R.S., Price C.J. Structural Covariance in the Human Cortex. J Neurosci. 2005;25(36):8303–8310. doi: 10.1523/JNEUROSCI.0357-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris J.C. The Clinical Dementia Rating (CDR): Current Version and Scoring Rules. Neurology. 1993;43(11):2412–3244. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- 34.Mutsaerts H.J.M.M., Petr J., Groot P., Vandemaele P., Ingala S., Robertson A.D., et al. ExploreASL: An Image Processing Pipeline for Multi-Center ASL Perfusion MRI Studies. NeuroImage. 2020;219(October) doi: 10.1016/j.neuroimage.2020.117031. [DOI] [PubMed] [Google Scholar]
- 35.Noble W.S. How Does Multiple Testing Correction Work? Nat Biotechnol. 2009;27(12):1135–2117. doi: 10.1038/nbt1209-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pelkmans W., Ossenkoppele R., Dicks E., Strandberg O., Barkhof F., Tijms B.M., et al. Tau-Related Grey Matter Network Breakdown across the Alzheimer’s Disease Continuum. Alzheimer’s Res Ther. 2021 doi: 10.1186/s13195-021-00876-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Penny W.D., Friston K.J., Ashburner J.T., Kiebel S.J., Nichols T.E. Elsevier; 2011. Statistical Parametric Mapping: The Analysis of Functional Brain Images. [Google Scholar]
- 38.Pereira J.B., Janelidze S., Ossenkoppele R., Kvartsberg H., Brinkmalm A., Mattsson-Carlgren N., et al. Untangling the Association of Amyloid-β and Tau with Synaptic and Axonal Loss in Alzheimer’s Disease. Brain: J Neurol. 2020 doi: 10.1093/brain/awaa395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pereira J.B., Mijalkov M., Kakaei E., Mecocci P., Vellas B., Tsolaki M., et al. Disrupted Network Topology in Patients with Stable and Progressive Mild Cognitive Impairment and Alzheimer’s Disease. Cereb Cortex. 2016;26(8):3476–3493. doi: 10.1093/cercor/bhw128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pooler, Amy M, Wendy Noble, and Diane P. Hanger. A Role for Tau at the Synapse in Alzheimer’s Disease Pathogenesis. Neuropharmacology 2014;76 Pt A (January):1–8. [DOI] [PubMed]
- 41.Potter P.E., Rauschkolb P.K., Pandya Y., Sue L.I., Sabbagh M.N., Walker D.G., et al. Pre- and Post-Synaptic Cortical Cholinergic Deficits Are Proportional to Amyloid Plaque Presence and Density at Preclinical Stages of Alzheimer’s Disease. Acta Neuropathol. 2011;122(1):49–60. doi: 10.1007/s00401-011-0831-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ritchie C.W., Muniz-Terrera G., Kivipelto M., Solomon A., Tom B., Molinuevo J.L. The European Prevention of Alzheimer’s Dementia (EPAD) Longitudinal Cohort Study: Baseline Data Release V500.0. J Prevent Alzheimer’s Disease. 2020;7(1):8–13. doi: 10.14283/jpad.2019.46. [DOI] [PubMed] [Google Scholar]
- 43.Rolls E.T., Huang C.-C., Lin C.-P., Feng J., Joliot M. Automated Anatomical Labelling Atlas 3. NeuroImage. 2020;206(February) doi: 10.1016/j.neuroimage.2019.116189. [DOI] [PubMed] [Google Scholar]
- 44.Rubinov M., Kötter R., Hagmann P., Sporns O. Brain Connectivity Toolbox: A Collection of Complex Network Measurements and Brain Connectivity Datasets. NeuroImage. 2009 doi: 10.1016/s1053-8119(09)71822-1. [DOI] [Google Scholar]
- 45.Scheltens P., Blennow K., Breteler M.M.B., de Strooper B., Frisoni G.B., Salloway S., et al. Alzheimer’s Disease. Lancet. 2016;388(10043):505–517. doi: 10.1016/S0140-6736(15)01124-1. [DOI] [PubMed] [Google Scholar]
- 46.Sepulcre J., Sabuncu M.R., Li Q., El Fakhri G., Sperling R., Johnson K.A. Tau and Amyloid β Proteins Distinctively Associate to Functional Network Changes in the Aging Brain. Alzheimer’s Dementia. 2017;13(11):1261–2129. doi: 10.1016/j.jalz.2017.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheline Y.I., Raichle M.E. Resting State Functional Connectivity in Preclinical Alzheimer’s Disease. Biol Psychiatry. 2013;74(5):340–437. doi: 10.1016/j.biopsych.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sintini I., Graff-Radford J., Jones D.T., Botha H., Martin P.R., Machulda M.M., et al. Tau and Amyloid Relationships with Resting-State Functional Connectivity in Atypical Alzheimer’s Disease. Cereb Cortex. 2020 doi: 10.1093/cercor/bhaa319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skaper S.D., Facci L., Zusso M., Giusti P. Synaptic Plasticity, Dementia and Alzheimer Disease. CNS Neurol Disord: Drug Targets. 2017;16(3):220–233. doi: 10.2174/1871527316666170113120853. [DOI] [PubMed] [Google Scholar]
- 50.Solomon, Alina, Miia Kivipelto, José Luis Molinuevo, Brian Tom, Craig W. Ritchie, and EPAD Consortium. European Prevention of Alzheimer’s Dementia Longitudinal Cohort Study (EPAD LCS): Study Protocol. BMJ Open 2019;8(12):e021017. [DOI] [PMC free article] [PubMed]
- 51.Tapiola T., Alafuzoff I., Herukka S.-K., Parkkinen L., Hartikainen P., Soininen H., et al. Cerebrospinal Fluid β-Amyloid 42 and Tau Proteins as Biomarkers of Alzheimer-Type Pathologic Changes in the Brain. Arch Neurol. 2009;66(3):382–439. doi: 10.1001/archneurol.2008.596. [DOI] [PubMed] [Google Scholar]
- 52.Teipel S.J., Cavedo E., Weschke S., Grothe M.J., Rojkova K., Fontaine G., et al. Cortical Amyloid Accumulation Is Associated with Alterations of Structural Integrity in Older People with Subjective Memory Complaints. Neurobiol Aging. 2017;57(September):143–152. doi: 10.1016/j.neurobiolaging.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 53.Kate T., Mara S.I., Schwarz A.J., Fox N.C., Chételat G., van Berckel B.N.M., et al. Secondary Prevention of Alzheimer’s Dementia: Neuroimaging Contributions. Alzheimer’s Res Ther. 2018;10(1):112. doi: 10.1186/s13195-018-0438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tijms B.M., ten Kate M., Wink A.M., Visser P.J., Ecay M., Clerigue M., et al. Gray Matter Network Disruptions and Amyloid Beta in Cognitively Normal Adults. Neurobiol Aging. 2016 doi: 10.1016/j.neurobiolaging.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 55.Tijms B.M., Möller C., Vrenken H., Wink A.M., de Haan W., van der Flier W.M., et al. Single-Subject Grey Matter Graphs in Alzheimer’s Disease. PLoS ONE. 2013;8(3):e58921. doi: 10.1371/journal.pone.0058921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tijms B.M., Seriès P., Willshaw D.J., Lawrie S.M. Similarity-Based Extraction of Individual Networks from Gray Matter MRI Scans. Cereb Cortex. 2012;22(7):1530–1541. doi: 10.1093/cercor/bhr221. [DOI] [PubMed] [Google Scholar]
- 57.Tijms B.M., Yeung H.M., Sikkes S.A.M., Möller C., Smits L.L., Stam C.J., et al. Single-Subject Gray Matter Graph Properties and Their Relationship with Cognitive Impairment in Early- and Late-Onset Alzheimer’s Disease. Brain Connect. 2014;4(5):337–346. doi: 10.1089/brain.2013.0209. [DOI] [PubMed] [Google Scholar]
- 58.Tzourio-Mazoyer N., Landeau B., Papathanassiou D., Crivello F., Etard O., Delcroix N., et al. Automated Anatomical Labeling of Activations in SPM Using a Macroscopic Anatomical Parcellation of the MNI MRI Single-Subject Brain. NeuroImage. 2002;15(1):273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- 59.Villemagne V.L., Burnham S., Bourgeat P., Brown B., Ellis K.A., Salvado O., et al. Amyloid β Deposition, Neurodegeneration, and Cognitive Decline in Sporadic Alzheimer’s Disease: A Prospective Cohort Study. Lancet Neurol. 2013;12(4):357–367. doi: 10.1016/S1474-4422(13)70044-9. [DOI] [PubMed] [Google Scholar]
- 60.Visser P.J., Reus L.M., Gobom J., Jansen I., Dicks E., van der Lee S.J., et al. Cerebrospinal Fluid Tau Levels Are Associated with Abnormal Neuronal Plasticity Markers in Alzheimer’s Disease. Mol Neurodegener. 2022;17(1):27. doi: 10.1186/s13024-022-00521-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watts D.J., Strogatz S.H. Collective Dynamics of ‘small-World’networks. Nature. 1998;393(6684):440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 62.Weisstein, Eric W. Bonferroni Correction. https://mathworld.Wolfram.Com/. http://mathworld.wolfram.com/BonferroniCorrection.html.
- 63.Xie T., He Y. Mapping the Alzheimer’s Brain with Connectomics. Front Psychiatry / Front Res Found. 2011;2:77. doi: 10.3389/fpsyt.2011.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yao Z., Zhang Y., Lin L., Zhou Y., Cunlu X.u., Jiang T., et al. Abnormal Cortical Networks in Mild Cognitive Impairment and Alzheimer’s Disease. PLoS Comput Biol. 2010;6(11):e1001006. doi: 10.1371/journal.pcbi.1001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.