A genome-wide CRISPR/Cas9 knockout screen uncovered novel mechanisms of adaptive resistance to pharmacologic inhibition of SHP2.
Abstract
Allosteric SHP2 inhibitors are a novel class of compounds that target hyperactive Ras/Mitogen Activated Protein Kinase (MAPK) signaling. In this issue of JEM, Wei et al. (2023. J. Exp. Med. https://doi.org/10.1084/jem.20221563) report a genome-wide CRISPR/Cas9 knockout screen that uncovered novel mechanisms of adaptive resistance to pharmacologic inhibition of SHP2.
Hyperactive Ras/MAPK signaling is among the most compelling biochemical targets in cancer therapy. The canonical MAPK pathway, which includes Raf, MEK, and ERK, regulates key cell fate decisions in diverse tissue contexts and is aberrantly activated in most cancers and in “RASopathy” developmental disorders. In cancer, hyperactive Ras/MAPK signaling can be caused by either somatic Ras mutations or by alterations of proteins that modulate Ras signal output by increasing the ratio of active Ras-GTP to inactive Ras-GDP. The core components of the Ras molecular switch are guanine nucleotide exchange factors (GNEFs) such as SOS1 and SOS2, which increase Ras-GTP levels by promoting guanine nucleotide dissociation, and GTPase activating proteins (GAPs), which greatly enhance the slow intrinsic Ras GTPase activity. Accordingly, oncogenic Ras mutants encode proteins that accumulate in the GTP-bound conformation due to reduced intrinsic GTPase activity and resistance to GAPs. Interestingly, most of the mutant Ras proteins encoded by germline RASopathy mutations have similar, though less potent, biochemical consequences (Tajan et al., 2018).

Insights from Bogdan Popescu and Kevin Shannon.
Whereas oncogenic Ras proteins were regarded as “undruggable” for decades, the efficacy of potent and selective inhibitors of K-RasG12C in lung adenocarcinoma has overturned this assumption (Ostrem and Shokat, 2016). However, major challenges remain, including the almost inevitable emergence of “adaptive resistance” due to genetic and epigenetic mechanisms that result in re-activation of MAPK signaling (Punekar et al., 2022). Furthermore, inhibiting mutant Ras proteins such as K-RasG12D that lack a “covalent handle” (i.e., nucleophilic cystine residue) remains challenging. In many respects, the recent experience with K-RasG12C inhibitors has only further reinforced the hard lessons learned over many decades that advanced cancers are rarely—if ever—cured with even the most potent and selective single agents.
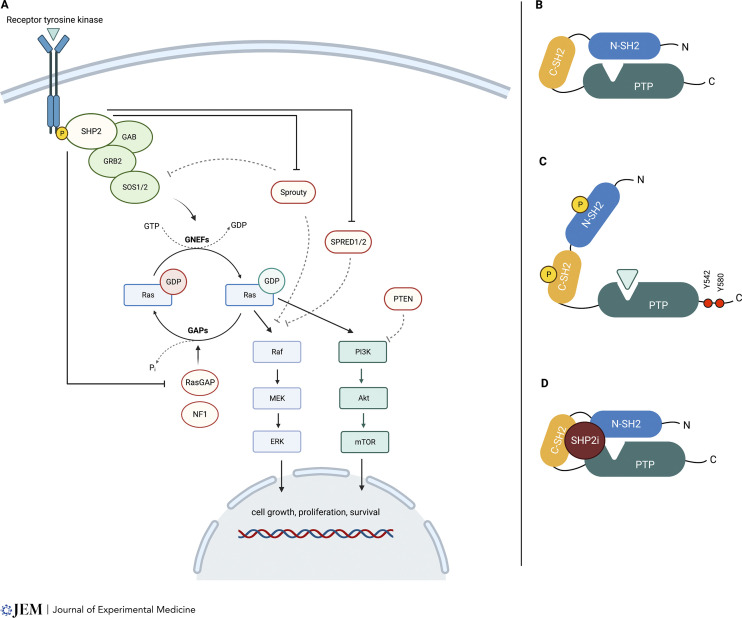
Mutations in PTPN11, which encodes the intracellular protein tyrosine phosphatase SHP2, illustrate how dysfunctional proteins that regulate the equilibrium between Ras-GDP and Ras-GTP cause human disease. Specifically, germline PTPN11 mutations are the most common cause of the RASopathy disorder Noonan syndrome (Tartaglia et al., 2001; Tajan et al., 2018). Somatic driver mutations occur in juvenile myelomonocytic leukemia, acute myeloid leukemia (AML), other hematologic malignancies, and, less frequently, in solid cancers (Chan et al., 2008). SHP2 is an intracellular protein tyrosine phosphatase that functions as a signal relay molecule in Ras/MAPK activation downstream of activated receptor tyrosine kinases (RTKs). Despite decades of investigation, aspects of SHP2 biology remain enigmatic. This is, in part, due to its dual function as a scaffold for assembling signaling complexes that promote GNEF activation and as an enzyme that de-phosphorylates GAPs and other negative regulators of Ras, such as Sprouty proteins (see panel A of figure). SHP2 also modulates horizontal signaling pathways such as PI3 kinase (PI3K)/AKT and JAK/STAT and mediates interactions with the tumor microenvironment (Kerr et al., 2021). However, many of the studies investigating the functions of SHP2 relied on over-expression in cell line models, so may indicate what SHP2 can do but not what it does do in different primary cell lineages.
Ras/MAPK Activation by SHP2. (A) SHP2 modulates Ras signaling activation downstream of RTKs by functioning as a scaffold for SOS-mediated guanine nucleotide exchange and as an enzyme that dephosphorylates multiple substrates, including known negative regulators of Ras-GTP. (B) In the inactive conformation, SHP2 PTPase activity is autoinhibited by the N-SH2 domain. (C) Upon recruitment by activated RTKs, the SHP2 adopts an open (active) state, exposing the catalytic site and the scaffold docking sites. (D) Allosteric SHP2 inhibitors stabilize the closed conformation, hampering both the catalytic and the scaffold functions.
SHP2 is composed of tandem SH2 (C-SH2 and N-SH2) domains, a catalytic protein tyrosine phosphatase (PTPase) domain, and a C-terminal module containing two tyrosine residues (Y542 and Y580) that serve as docking sites for scaffold assembly. The basal state of SHP2 is a sterically closed, autoinhibited conformation, with the N-SH2 domain occluding the catalytic cleft. Upon activation, the N-SH2 domain undergoes a conformational shift that allows the PTPase to adopt an “open,” catalytically active conformation (see panels B and C of figure) and promotes phosphorylation of Y542/Y580 residues for the subsequent recruitment of GRB2 to the scaffold complex (Araki et al., 2003). Biochemical and functional analysis of mutant SHP2 proteins have shown that PTPN11E76K and other oncogenic N-SH2 mutations disrupt key autoinhibitory interactions and constitutively increase PTPase activity, which is essential for hyperactive MAPK signaling and myeloid transformation (LaRochelle et al., 2018). However, the key substrate(s) that is/are dephosphorylated remain unknown.
Allosteric SHP2 inhibitors stabilize the closed, inactive conformation of SHP2 like a “molecular glue,” cleverly inhibiting both the phosphatase and scaffold functions (see panel D of figure). This reduces Ras-GTP levels by suppressing GNEF activity in cancers driven by oncogenic RTKs and, compellingly, by some guanidine “fast-exchanging” mutant Ras proteins including K-RasG12C (Chen et al., 2016; Nichols et al., 2018). In vitro studies have shown that concurrent targeting of SHP2 and other Ras/MAPK pathway proteins can preempt adaptive resistance (Fedele et al., 2021), and several allosteric SHP2 inhibitors are currently undergoing evaluation in phase I or I/II clinical trials as either monotherapy or in vertical combinations with other inhibitors.
In this issue of JEM, Wei and colleagues performed a genome-wide CRISPR/Cas9 knockout screen to elucidate candidate mechanisms of resistance to the allosteric SHP2 inhibitor SHP-099 in the FLT3-mutant AML cell lines MOLM-13 and MV4-11 (Wei et al., 2023). This is a clinically relevant disease context as FLT3 mutations specify proteins with constitutively elevated RTK activity. Accordingly, the FDA-approved Flt3 kinase inhibitor gilteritinib prolongs survival in relapsed/refractory AML, although these patients invariably relapse due to the emergence of resistant clones that reactivate MAPK signaling with mutations in NRAS/KRAS and other Ras pathway genes, including PTPN11 (McMahon et al., 2019).
In addition to showing that disrupting genes encoding known negative regulators of Ras/MAPK signaling (e.g., NF1, SPRED) and proteins involved in parallel signaling pathways (e.g., PTEN) or cell cycle progression (e.g., CDKN1B, FBXW7, RB1) conferred SHP-099 resistance, Wei and colleagues identified novel candidate resistance genes such as INPPL1, MAP4K5, BIRC6, and LZTR1. The authors validated these hits in a subsequent mini-screen of a larger set of cell lines genetically dependent on SHP2, including AML lines driven by BCR-ABL for JAK2V617F and solid cancer cell lines driven by mutant EGFR proteins or RASG12C.
Wei et al. (2023) went on to characterize how disrupting INPPL1, MAP4K5, and LZTR1 induced resistance to SHP-099. INPPL1 encodes a phosphatidyl-inositide phosphatase that negatively regulates the PI3K/Akt pathway. As PI3K is a key downstream effector of Ras-GTP, a simple interpretation of this finding could be that cancer cells “switch” from MAPK dependency by hyperactivating this parallel signaling pathway. Surprisingly, the authors showed that INPPL1 inactivation restored MAPK pathway activation downstream of SHP2, resulting in resistance. Mutagenesis studies revealed that SHP-099 sensitivity is independent of the enzymatic activity of INPPL1 but requires the integrity of its NPXY motif. MAP4K5 is a MAP4K family member known to activate p38/JNK signaling. However, Wei and colleagues did not observe consistent changes in JNK or p38 phosphorylation in isogenic MAP4K5 knockout cell lines that were exposed to SHP-099, but instead detected higher levels of ERK phosphorylation. Notably, MAP4K5 kinase activity is essential for sensitivity to SHP-099. These data unexpectedly implicate MAP4K5 as a negative regulator of MAPK signaling. LTZR1 promotes ubiquitin-ligase degradation of Ras superfamily proteins, and germline loss-of-function mutations occur in some patients with RASopathies. The critical biochemical targets of LTZR1 are a matter of some controversy, and the study of Wei et al. (2023) sheds new light on this question. Specifically, the authors found that RIT1 was over-expressed across all LZTR1 knockout cell lines and made the key observation that disrupting RIT1 restored sensitivity to SHP-099. These data are consistent with the work of Castel et al. (2019) showing that LTZR1 normally suppresses MAPK signaling by modulating RIT1 protein levels. Although their primary goal was to identify SHP2 resistance genes, the rigorous biochemical and functional studies of Wei and colleagues raise new questions regarding how normal cells regulate Ras/MAPK signal output, such as what protein-protein interaction(s) link INPPL1 to MAPK signaling and what substrates of the MAP4K5 negatively regulate ERK phosphorylation directly or indirectly?
While SHP2 is an attractive therapeutic target in cancers that are dependent on Ras/MAPK signaling, the driver mutations and disease settings that are most likely to respond to these therapies are unknown. In some contexts, allosteric SHP2 inhibitors are ineffective ab initio. Cancers harboring oncogenic PTPN11 mutations such as PTPN11E76K or PTPN11D61V that bias SHP2 toward a hyperactive conformation are insensitive to allosteric inhibitors that bind the closed conformation of the protein. Developing “next generation” SHP2 inhibitors with activity against cancer-associated PTPN11 mutations is, in this respect, an appealing therapeutic strategy. Furthermore, the susceptibility of mutant Ras proteins to allosteric SHP2 inhibitors tracks with their dependence on SOS-mediated nucleotide exchange (Gebregiworgis et al., 2021). Interestingly, Wei and colleagues identified NF1 as a quasi-universal resistance gene. This contrasts with a previous report showing that NF1-deficient cell lines are sensitive to the SHP2 inhibitor RMC-4550 (Nichols et al., 2018), and may reflect the high kinase output of mutant Flt3 proteins. Indeed, it should be noted that the authors performed most of their assays in AML cell lines, and it is therefore uncertain if solid malignancies will exhibit similar mechanisms of resistance. Moreover, resistance mechanisms identified in cell line screens are cell-intrinsic and do not account for the potential effects of SHP2 inhibition on immune reprogramming of the tumor microenvironment.
Ongoing clinical trials of allosteric SHP2 inhibitors will shape future clinical development (Kerr et al., 2021). In the interim, the work of Wei et al. (2023) identifies candidate resistance genes that predominantly converge on MAPK pathway reactivation. The authors’ data showing that disrupting INPPL1, MAP4K5, or LZTR1 confers cross-resistance to gilteritinib and dasatinib in AML cell lines driven by mutant Flt3 or BCR-ABL, respectively, are consistent with SHP2 functioning downstream of both oncoproteins. This observation raises the question whether drug combinations that target SHP2 and either mutant Flt3 or BCR-ABL might improve efficacy and reduce the likelihood of clinical resistance due to deeper MAPK pathway inhibition and if a similar strategy of combining SHP2 and other vertical inhibitors of Ras/MAPK might be efficacious in some solid cancers. However, this approach might be associated with intolerable toxicities. Nevertheless, the findings reported by Wei and colleagues underscore that cancer cells that are exposed to SHP2 inhibitor monotherapy develop adaptive resistance by multiple genetic mechanisms and emphasize the need to ultimately investigate the efficacy and safety of drug combinations to realize maximal clinical benefit.
References
- Araki, T., Nawa H., and Neel B.G.. 2003. J. Biol. Chem. 10.1074/jbc.M306461200 [DOI] [PubMed] [Google Scholar]
- Castel, P., et al. 2019. Science. 10.1126/science.aav1444 [DOI] [Google Scholar]
- Chan, G., et al. 2008. Cancer Metastasis Rev. 10.1007/s10555-008-9126-y [DOI] [PubMed] [Google Scholar]
- Chen, Y.N., et al. 2016. Nature. 10.1038/nature18621 [DOI] [Google Scholar]
- Fedele, C., et al. 2021. J. Exp. Med. 10.1084/jem.20201414 [DOI] [Google Scholar]
- Gebregiworgis, T., et al. 2021. Nat. Commun. 10.1038/s41467-021-26526-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr, D.L., et al. 2021. Curr. Opin. Chem. Biol. 10.1016/j.cbpa.2020.11.007 [DOI] [PubMed] [Google Scholar]
- LaRochelle, J.R., et al. 2018. Nat. Commun. 10.1038/s41467-018-06823-9 [DOI] [Google Scholar]
- McMahon, C.M., et al. 2019. Cancer Discov. 10.1158/2159-8290.CD-18-1453 [DOI] [Google Scholar]
- Nichols, R.J., et al. 2018. Nat. Cell Biol. 10.1038/s41556-018-0169-1 [DOI] [Google Scholar]
- Ostrem, J.M.L., and Shokat K.M.. 2016. Nat. Rev. Drug Discov. 10.1038/nrd.2016.139 [DOI] [PubMed] [Google Scholar]
- Punekar, S.R., et al. 2022. Nat. Rev. Clin. Oncol. 10.1038/s41571-022-00671-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajan, M., et al. 2018. Endocr. Rev. 10.1210/er.2017-00232 [DOI] [PubMed] [Google Scholar]
- Tartaglia, M., et al. 2001. Nat. Genet. 10.1038/ng772 [DOI] [PubMed] [Google Scholar]
- Wei, W., et al. 2023. J. Exp. Med. 10.1084/jem.20221563 [DOI] [Google Scholar]