Abstract
The mechanism by which origin recognition complexes (ORCs) identify replication origins was investigated using purified Orc proteins from Schizosaccharomyces pombe. Orc4p alone bound tightly and specifically to several sites within S. pombe replication origins that are genetically required for origin activity. These sites consisted of clusters of A or T residues on one strand but were devoid of either alternating A and T residues or GC-rich sequences. Addition of a complex consisting of Orc1, -2, -3, -5, and -6 proteins (ORC-5) altered neither Orc4p binding to origin DNA nor Orc4p protection of specific sequences. ORC-5 alone bound weakly and nonspecifically to DNA; strong binding required the presence of Orc4p. Under these conditions, all six subunits remained bound to chromatin isolated from each phase of the cell division cycle. These results reveal that the S. pombe ORC binds to multiple, specific sites within replication origins and that site selection, at least in vitro, is determined solely by the Orc4p subunit.
Each time that a eukaryotic cell divides, it replicates its genome once and only once. In the budding yeast Saccharomyces cerevisiae, the mechanism that regulates this process begins with assembly of a single six-subunit origin recognition complex (ORC) at specific sites along the genome and DNA synthesis begins at or near the site where ORC binds (4). In S. cerevisiae, these DNA replication origins consist of 100 to 150 bp that can act as autonomously replicating sequences (ARS) and contain several noninterchangeable elements (34a). The A element is an 11-bp ARS consensus sequence consisting of (A/T)TTTA(T/C)(A/G)TTT(A/T) that is essential for origin function and ORC binding. Three B elements are located at the 3′ side of the T-rich strand of the A element. These elements act as auxiliary sequences; they facilitate replication but are dispensable under some conditions. B1 lies adjacent to the A element and facilitates ORC binding. B2 is located further away within an easily unwound DNA sequence referred to as a DNA unwinding element. B3 is found in some, but not all, replication origins, where it is a binding site for transcription factor Abf1. Once a functional ORC:DNA complex is established, it interacts sequentially with Cdc6 and Mcm2-7 proteins to form a prereplication complex. A novel Mcm loading factor, Cdt-1 protein, may also be involved (29). Prereplication complexes are then acted upon by the protein kinases Cdk1/Clb5, Clb6, and Cdc7/Dbf4 to allow Cdc45 to load DNA primase-DNA polymerase α and initiate DNA synthesis. Since homologues of all of these proteins are found throughout the eukaryotic kingdom and since most of them have been shown to be required for initiation of DNA replication in other organisms, the mechanism for initiation of DNA replication appears to be highly conserved among the eukaryotes (5, 22).
Two differences between S. cerevisiae and other eukaryotes have emerged that point to the interaction of ORC with DNA replication origins as a critical regulatory step in determining where and when DNA replication will occur. First, DNA replication origins in eukaryotes, such as the fission yeast Schizosaccharomyces pombe, Drosophila, and mammals, are much larger and less well defined in terms of their genetically required cis-acting sequences than those in S. cerevisiae (3, 10, 31, 37). In fact, the number and locations of replication origins can change during the early stages of development in both Drosophila (45) and Xenopus (20), demonstrating that epigenetic as well as genetic factors determine where initiation sites are established in the metazoans. Second, although the ORC in yeast appears to be stably bound throughout its cell division cycle, the stability of one or more ORC subunits in mammals, Drosophila, and Xenopus is cell cycle dependent (references 24 and 34 and references therein). Unfortunately, outside the S. cerevisiae paradigm, the mechanism by which an ORC recognizes its DNA binding site and establishes a functional ORC-DNA complex is not yet understood. One solution to this problem has been to extend the studies carried out on S. cerevisiae to a related but evolutionarily distant organism with more complex replication origins, such as S. pombe.
S. pombe, like S. cerevisiae, contains replication origins that can exhibit ARS activity (13, 32). However, S. pombe replication origins are 500 to 1,500 bp in size and in general are a very AT-rich sequence. Moreover, S. pombe origins do not contain a core consensus sequence that is essential for origin function (32), although they do contain more than one region that significantly affects origin activity (8, 12, 23, 37). Each of these regions is extremely AT rich (≥80%) and highly asymmetric, with adenines clustered in one strand and thymines in the other. These results indicate that S. pombe replication origins contain several functionally important DNA sequence elements whose functions are as yet unknown. The largeness and sequence complexity observed in mammalian replication origins, together with the presence of several AT-rich regions, suggest that S. pombe may provide a paradigm for understanding metazoan replication origins. In fact, initiation of DNA synthesis at Drosophila (21) and human (1) replication origins occurs in AT-rich sequences that are strikingly similar to those identified as critical elements in S. pombe replication origins. Moreover, S. pombe Orc4p is unique in that its N-terminal domain consists of nine copies of an AT-hook motif that binds preferentially to AT-rich sequences (7, 33) and that is required for binding of Orc4p to DNA (7). Thus, it appears that Orc4p may play a critical role in selecting initiation sites for DNA replication in S. pombe. However, the precise role of Orc4p, as well as the other ORC subunits, in origin recognition remains unknown.
In an effort to answer this question, we took advantage of the fact that it can be purified as an intact five-member complex (the ORC-5 complex) consisting of the Orc1, Orc2, Orc3, Orc5 and Orc6 protein subunits (33). Orc4 protein (Orc4p) is absent, because it remains bound to DNA under conditions in which the other five ORC subunits can be eluted, and it interacts weakly with the other five Orc subunits (33). Therefore, the Orc4 protein (Orc4p) was purified from insect cells expressing a cloned SpOrc4 gene. Using DNA band shift analysis, DNA affinity chromatography analysis, and DNase I footprinting analysis, we determined whether or not Orc4p and the ORC-5 complex, either alone or in combination, selected one or more specific sequences within S. pombe replication origins.
The results revealed a novel mechanism for selection of DNA replication initiation sites by a eukaryotic origin recognition complex. Only the Orc4p subunit bound strongly and specifically to several specific sequences within S. pombe replication origins. These sites were highly asymmetric AT-rich sequences that corresponded to critical regions detected by deletion analysis of S. pombe origins. In contrast, the ORC-5 complex bound only weakly and nonspecifically to the same DNA sequences and altered neither the selection of DNA binding sites by Orc4p nor the footprint that it produced. Moreover, ORC-5 binding to DNA was strongly dependent on the presence of Orc4p. Once bound to chromatin in vivo, all six Orc proteins remained bound to chromatin isolated from different phases of the cell cycle under the same salt conditions used to characterize interaction of these proteins with DNA in vitro. Thus, the properties of these purified proteins suggest that S. pombe Orc4p is solely responsible for selecting ORC binding sites on DNA and that Orc4p targets the remaining ORC subunits to replication origins.
MATERIALS AND METHODS
Purification of ORC-5 complex.
A 15-liter culture of S. pombe h− ura4-D18 Orp1-3HA expressing an N-terminal hemagglutinin-tagged Orc1 protein (18) was grown in yeast extract medium at 32°C (optical density at 595 nm [OD595] = 2 to 3). Cells were harvested and resuspended in 200 ml of buffer B (1.2 M sorbitol, 20 mM potassium phosphate [pH 6.8], and 5 mM dithiothreitol). Spheroplasts were prepared by addition to the culture of 0.2 mg each of lysoenzyme (Sigma) and Zymolyase (ICN) per ml and were incubated at 30°C with gentle swirling until most of the cells could be lysed in the presence of 1% sodium dodecyl sulfate (SDS). Spheroplasts were recovered by centrifugation (3,000 × g, 5 min, 4°C), washed once with 10 pellet volumes of ice-cold buffer B, resuspended in 100 ml of buffer A (50 mM HEPES-HCl [pH 7.5], 100 mM KCl, 5 mM magnesium acetate, 5 mM dithiothreitol, and 10% glycerol), and lysed by the addition of Triton X-100 to 1%. Chromatin was immediately recovered by centrifugation (30,000 × g, 20 min, 4°C), resuspended in 10 ml of buffer A containing 0.5 M KCl, and then separated into soluble and insoluble fractions by centrifugation (30,000 × g, 20 min, 4°C). The ORC-5 complex was purified from the soluble fraction, as previously described (33).
Purification of S. pombe Orc4 protein.
Using PCR and appropriate DNA primers, the S. pombe Orc4 gene was isolated from a S. pombe cDNA library (ATCC 87275) and cloned into Bluescript II SK(−) plasmid (Stratagene). Sequence analysis of both DNA strands from four independent clones confirmed the published sequence of the Orc4 gene (7). The Orc4 gene was cloned into pFastBac1 and overexpressed in insect cells according to the manufacturer's instructions (Life Technologies). Cells from a 2-liter culture were extracted with 30 ml of buffer A containing 1 M KCl. After centrifugation (30,000 × g, 30 min, 4°C), the supernatant was dialyzed against buffer A to reduce salt to 0.1 M. All steps were carried out at 4°C in buffer A. Any precipitate was removed by centrifugation, and the supernatant was adsorbed onto a Q-Sepharose column. Orc4p was eluted at 0.4 to 0.6 M KCl in linear gradient, dialyzed against buffer A, and then fractionated by gel filtration using Superose-12 (Pharmacia). Fractions containing Orc4p were adsorbed onto a double-stranded DNA–cellulose column, and Orc4p was eluted at 0.5 to 0.7 M KCl in linear gradient, dialyzed against buffer A, and then loaded onto SP-Sepharose (Pharmacia) and eluted at 0.2 to 0.3 M KCl. Fractions containing Orc4p were dialyzed against buffer A containing 40% glycerol.
DNA replication origins.
A plasmid containing the 363-bp S. pombe ARS3002 sequence (49) was kindly provided by J. Huberman and digested with HindIII and EcoRI to release the 363-bp ARS3002 sequence. The 1.2-kb S. pombe ARS1 DNA fragment was released from pESP-1 (Stratagene) by digestion with EcoRI restriction endonuclease. An 853-bp sequence encompassing the ARS1 core (8) was obtained using PCR and appropriate DNA primers. Control DNA consisted of a random sequence (688 bp; 50% AT rich) isolated by digestion of pBluescript II KS(−) plasmid with restriction enzymes SspI and EcoRI.
DNA band shift assay.
DNA fragments were radiolabeled at their 5′ ends using T4 polynucleotide kinase according to the manufacturer's instructions (New England BioLabs). A 20-μl reaction mixture contained 5 ng of [32P]DNA, 0.5 μg of dG-dC DNA (∼750 nucleotides), 50 mM HEPES-HCl (pH 7.5), 100 mM KCl, 5 mM MgCl2, 0.5 mg of bovine serum albumin/ml, 5 mM dithiothreitol, and the indicated amounts of Orc4p and ORC-5. Reaction mixtures were incubated at room temperature for 10 min before fractionating them by electrophoresis in a 1.2% agarose gel (Tris-borate-EDTA buffer) for 3 h at 5 V/cm and room temperature. The gel was then dried, and [32P]DNA was detected by autoradiography.
ORC-5 and Orc4p DNA binding assay.
A 935-bp DNA fragment (5′-AGTGCAC to GTCTTCA-3′) containing the ARS3002 origin (49) was prepared from the S. pombe genome using PCR and appropriate DNA primers and attached to cellulose (Sigma) (2). A longer ARS3002-containing DNA fragment was used to improve the affinity between DNA and cellulose. A 0.5-ml sample of ARS3002-cellulose was then incubated with 10 μg of Orc4p in buffer A, loaded into a column, and washed with 10 ml of buffer A. Then 5 μg of ORC-5 complex was passed through the column with a flow rate of 0.1 ml/min. After the column was washed with 5 ml of buffer A at the same flow rate, proteins were eluted with 1.5 ml of buffer A containing 0.9 M KCl and fractionated by electrophoresis in an SDS-polyacrylamide gel and individual proteins were identified by immunoblotting using appropriate antibodies.
Chromatin-associated ORC proteins.
A 500-ml culture of S. pombe (OD595 = 1) was harvested by centrifugation (2,000 × g, 5 min, 4°C), washed with water, resuspended in 10 ml of buffer B containing 0.2 mg each of lysing enzyme and Zymolyase per ml, and then incubated at 30°C with gentle swirling until the cells were sensitive to 1% SDS. The cells were then harvested and resuspended in 5 ml of buffer A plus a protease inhibitor set (Roche Molecular Biochemicals) and were lysed with 1% Triton X-100. Chromatin was immediately recovered by centrifugation (30,000 × g, 20 min, 4°C) and resuspended in loading buffer, and the proteins were fractionated by electrophoresis in an 8% polyacrylamide gel containing 0.1% SDS. Orc proteins were detected by immunoblotting (33) using antisera prepared against Orc2 and Orc6 proteins (33) that were kindly provided by J.-K. Lee and S. Raychaudhuri (Memorial Sloan-Kettering Cancer Center, New York, N.Y.). Antisera against Orc3, -4, and -5 were made by Cocalico Biologicals using peptides synthesized by Research Genetics. Hemagglutinin-tagged Orc1 was detected using the antihemagglutinin peptide (Roche Molecular Biochemicals).
In vitro footprinting assay.
A footprinting assay was carried out on two S. pombe ARS DNA fragments, ARS3002 and ARS1, according to a previously described method (25). The ARS3002 fragment has a length of 363 bp (see Fig. 8). The fragment can function as a weak ARS element and is able to give 30% of the transformation frequency compared to a longer ARS3002 fragment (49). The ARS1 fragment has a length of 853 bp with ends at 5′-CATAGTGACT and ATTCTGTCTC-3′. This ARS1 fragment is fully active (8). 32P labeling of these two DNA fragments at one end is described above. The DNA binding conditions for Orc4p and ORC-5 were the same as those described for gel shift experiments.
FIG. 8.
The 363-bp sequence for ARS3002 (49) was used for DNA band shift assays and for in vitro footprinting experiments. Regions A, B, and C, protected by Orc4p, are in boldfaced italics. Two regions required for ARS3002 activity (deletions 8 and 10 [49]) are doubly underlined. One region not required for ARS3002 activity (deletion 6) is wavily underlined. This region contains a block of alternating AT residues.
RESULTS
Purification of ORC-5 complex and Orc4 protein subunit.
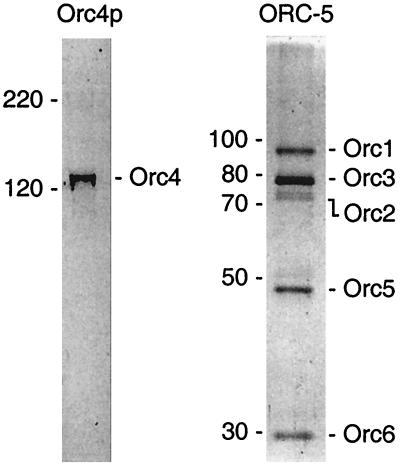
The ORC-5 complex, composed of the Orc1, -2, -3, -5, and -6 protein subunits, was isolated from S. pombe cells. Based on silver staining (Fig. 1), the complex was more than 98% pure. Based on immunoblotting (data not shown), the faint band (∼50 kDa) just above Orc5p resulted from degradation of Orc1p. Both Orc2p and Orc3p appeared as two closely migrating bands. Treatment of ORC-5 samples with λ-phosphatase converted the slower-migrating form in each case into the faster-migrating form (data not shown). Therefore, the slower-migrating form of both Orc2p and Orc3p was considered to be a phosphorylated form of these subunits. Orc2p was not as well stained by silver as were the other ORC subunits, but Coomassie blue staining confirmed that the ORC-5 complex contained an equimolar ratio of the five Orc subunits. In addition, ORC-5 exhibited ATPase activity (data not shown), as previously observed for S. cerevisiae ORC (27).
FIG. 1.
Purified S. pombe Orc4 protein and ORC-5 complex. Purified Orc4p and purified ORC-5 complex were fractionated by electrophoresis in SDS–8% polyacrylamide gels. Proteins in the Orc4p gel were stained with Coomassie blue, and those in the ORC-5 gel were stained with silver nitrate. Based on the migration of standard proteins (Bio-Rad) fractionated in parallel on the same gel, molecular masses were estimated as Orc1p (95 kDa), Orc2p (72 kDa), Orc3p (79 kDa), Orc4p (130 kDa), Orc5p (48 kDa), and Orc6p (29 kDa).
The Orc4p subunit was absent from the purified ORC-5 complex, because at 0.5 M KCl, most of the Orc4p remained bound to cellular DNA while all of the ORC-5 complex was eluted. Although higher salt concentrations also eluted Orc4p, a complete ORC was still not recovered, presumably due to weak protein-protein interactions. Therefore, Orc4p was purified separately by expressing the SpOrc4 gene in insect cells and by then eluting Orc4p from insect cell chromatin in 1 M KCl. SpOrc4p was then purified to apparent homogeneity (Fig. 1), as described in Materials and Methods.
Binding of ORC to replication origins is dependent on Orc4 protein.
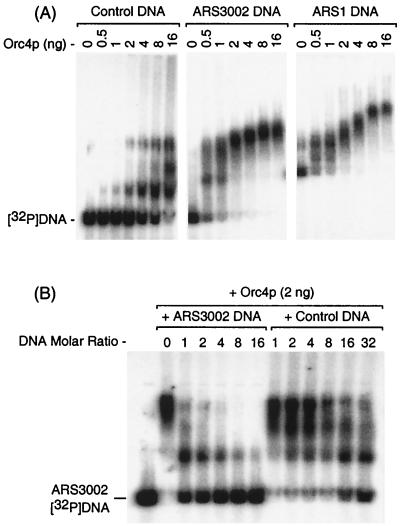
To determine whether or not purified Orc4p could bind specifically to S. pombe DNA replication origins, [32P]DNA fragments containing S. pombe ARS3002 or ARS1 were incubated with increasing amounts of Orc4p and were then fractionated by gel electrophoresis. ARS3002 was contained in a 363-bp DNA fragment that gives 30% of the transformation frequency observed with a longer DNA fragment (12). ARS1 was contained in a 853-bp DNA fragment that gives a 100% transformation frequency (8). Orc4p rapidly and tightly bound to both replication origins (Fig. 2A). As the amount of Orc4p was increased, all the free DNA formed protein-DNA complexes of increasing complexity, suggesting that at least two and as many as four or five Orc4p molecules were bound to each replication origin. ATP did not affect binding of Orc4p to either origin (data not shown). When the same experiment was carried out using a DNA fragment of average sequence composition that did not exhibit ARS activity in S. pombe, an equivalent level of Orc4p binding was not observed until the ratio of protein to DNA was increased at least 16-fold (Fig. 2A). Specificity of Orc4p for origin DNA could also be demonstrated with competitor DNA. For example, increasing the ratio of an unlabeled ARS3002 DNA fragment to the ARS3002 [32P]DNA fragment from 1 to 16 completely eliminated binding of Orc4p to ARS3002 [32P]DNA, whereas increasing the ratio of the control DNA fragment to the ARS3002 [32P]DNA fragment as much as 32-fold did not (Fig. 2B). These results reveal the existence of several Orc4p-specific binding sites in S. pombe ARS elements.
FIG. 2.
Orc4p binds specifically to S. pombe DNA replication origins. (A) DNA band shift assays were carried out by incubating the indicated amount of Orc4p with either 32P-labeled S. pombe replication origin ARS3002 (363 bp) or ARS1 (853 bp) or with a control DNA (688 bp) consisting of an average sequence. Samples were then fractionated by agarose gel electrophoresis, and [32P]DNA-Orc4p complexes were detected by autoradiography. (B) Competition between ARS3002 [32P]DNA (5 ng) and either unlabeled ARS3002 DNA or control DNA was detected by incubating the indicated molar ratio of DNA with 2 ng of Orc4p. Molar DNA ratios were that of unlabeled competitor DNA to [32P]DNA. Conditions were the same as for panel A.
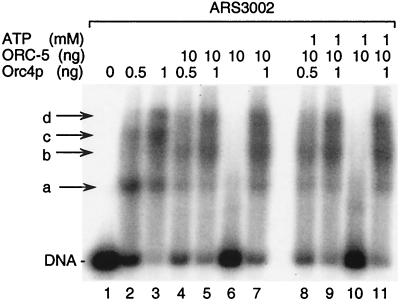
To determine whether or not Orc4p binding to S. pombe DNA replication origins was affected by the other ORC subunits, similar DNA band shift experiments were carried out with Orc4p in either the presence or absence of the ORC-5 complex using [32P]DNA fragments containing either S. pombe ARS3002 or ARS1 sequences. Low concentrations of Orc4p alone generated at least three protein-ARS3002 DNA complexes (Fig. 3, lanes 2 and 3, complexes a, c, and d). In contrast, ORC-5 had little affinity for origin DNA, regardless of the presence or absence of ATP (Fig. 3, lanes 6 and 10). Nevertheless, ORC-5 did alter the DNA binding pattern observed with Orc4p when Orc4p was either prebound (Fig. 3, lanes 4, 5, 8, and 9) or included with ORC-5 before addition of DNA (Fig. 3, lanes 7 and 11). Under these conditions, the fraction of [32P]DNA in DNA-protein complexes a and c was reduced, and a new DNA-protein complex (b) appeared. However, a single ORC-ARS3002 DNA complex should migrate more slowly than complex d, which is composed of at least three Orc4p molecules (390 kDa) per DNA molecule, because a single ORC is ∼450 kDa. Therefore, ORC-5 appeared to be weakly bound under the conditions of this DNA band shift assay. If it dissociated from the Orc4p-DNA complex during the beginning of electrophoresis, then the effect would be seen as a retardation in the migration of complex a, causing the appearance of b. This hypothesis was subsequently confirmed both by DNA affinity chromatography assays and by chromatin affinity assays described below.
FIG. 3.
ORC-5 selectively binds to Orc4p-DNA complexes. DNA band shift assays using [32P]ARS3002 were carried out as for Fig. 3. The amounts of Orc4p, ORC-5, and ATP added to each reaction are indicated. In lanes 4, 5, 8, and 9, Orc4p was allowed to bind to DNA for 5 min before addition of ORC-5. In lanes 7 and 11, Orc4p and ORC-5 were combined, kept on ice for 5 min, and then added to the reaction.
Taken together, the results from DNA band shift assays revealed that Orc4p bound strongly and specifically to DNA containing a S. pombe replication origin. Furthermore, these assays showed that the ORC-5 complex alone bound weakly to the same DNA. However, the ORC-5 complex did cause a detectable band shift of Orc4p-DNA complexes, indicating a physical interaction between Orc4p and the ORC-5 complex.
ORC-5 complex physically interacts with Orc4 protein.
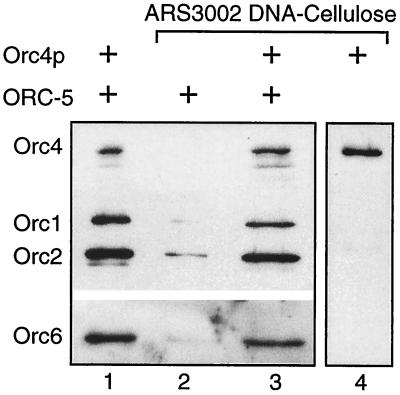
To demonstrate that the ORC-5 complex did physically interact with Orc4p under the conditions used in the DNA band shift assays, an ARS3002 DNA fragment was attached to DNA cellulose and tested for its ability to bind ORC-5 before and after Orc4p was bound. Salt conditions (buffer A which contains 0.1 M KCl) used in this assay were the same as those in the DNA band shift assays. When an aliquot of Orc4p was passed through this column, more than 95% of the protein was retained, consistent with a strong interaction between the Orc4p and origin DNA. In contrast, when an aliquot of the ORC-5 complex was passed through this column, only trace amounts of these proteins were bound to the resin (Fig. 4, lane 2). However, when an aliquot of the ORC-5 complex was passed through resin that had been preloaded with enough Orc4p to bind all of the ORC-5 proteins in the starting sample, about 45% of the ORC-5 proteins were retained (Fig. 4, lane 3). This represented at least a 10-fold increase in the affinity of ORC-5 proteins for DNA to which Orc4p was already bound. The fact that all of the ORC-5 proteins were not retained by the Orc4p/DNA resin suggests that the affinity between ORC-5 and Orc4p is weak. The results from these DNA affinity chromatography assays demonstrated that the ORC-5 complex physically interacted with the Orc4p-DNA complex under the conditions used above in DNA band shift assays. Together with the results from DNA band shift assays, they suggest that Orc4p binds to the origin DNA sequence and that the ORC-5 complex binds strongly to Orc4p, not to the DNA.
FIG. 4.
The ORC-5 complex binds to the Orc4p-origin DNA complex. An aliquot of the ORC-5 complex (lane 1) was passed through ARS3002 cellulose (lane 2) or ARS3002-Orc4p cellulose (lane 3) to which an aliquot of Orc4p (lane 1) had been prebound (lane 4). The column was washed with starting buffer (buffer A which contains 0.1 M KCl) to remove unbound proteins and then with 1 M KCl (buffer A + 0.9 M KCl) to elute bound ORC proteins. ORC proteins in the eluent were fractionated by electrophoresis in an SDS-polyacrylamide gel and were then detected by immunoblotting using antisera against Orc1, -2, -4, and -6 proteins.
All six ORC subunits bind to chromatin throughout cell division cycle.
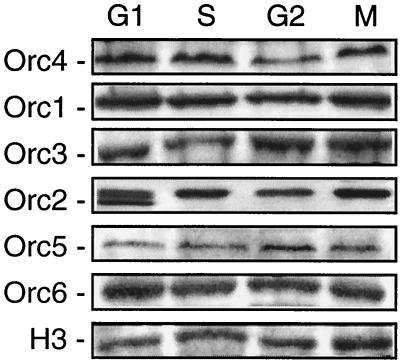
S. pombe is more closely related to higher eukaryotic cells than is S. cerevisiae (36). Therefore, since it has been reported that one or more of the ORC subunits in mammals and frogs do not remain stably bound to chromatin throughout the cell division cycle (references 24 and 34 and references therein), we considered the possibility that the same might be true in S. pombe. Therefore, S. pombe chromatin was extracted under the same salt conditions used in the DNA band shift and DNA affinity assays and was then analyzed for the presence of ORC proteins. Cells were arrested at specific times during their division cycle using temperature-sensitive mutations. Cells were arrested in the G1 phase by a nonfunctional cdc10 protein (required for transcription at Start [48]), in the S phase by a nonfunctional cdc22 protein (ribonucleotide reductase [14]), in the G2 phase by a nonfunctional cdc25 protein (activates cdk1-cyclin B [43]), or in metaphase by benomyl (an inhibitor of microtubule assembly [46]). Chromatin was isolated and chromatin-associated proteins were fractionated by electrophoresis in SDS polyacrylamide gels.
The results revealed that all six Orc proteins were associated with the chromatin fraction at equivalent concentrations during each phase of the cell cycle (Fig. 5). The only variation was the appearance of a novel form of Orc2 during the G1 phase, as previously reported for cycling S. pombe cells (30). Thus, both the ORC-5 complex and Orc4p remained bound to yeast chromatin in vivo throughout the yeast cell division cycle. In addition, all six S. pombe Orc proteins remained stably bound to chromatin under the same buffer conditions used to detect DNA band shifts and DNA binding affinity in vitro.
FIG. 5.
All six ORC subunits are bound to chromatin in 100 mM KCl. To get chromatin from different phases of cells, S. pombe strains JL197 ura4-D18 cdc10-129 (G1), JL202 ura4-D18 cdc22-M45 (S), and JL206 ura4-D18 cdc25-22 (G2) were grown to an OD595 of 0.5 at 26°C and were then shifted to 36.5°C for an additional 3.5 h. To arrest cells at the M phase, 25 μg of benomyl/ml was added to a cell culture with an OD595 of 0.5, and the culture was incubated for an additional 3.5 h. Chromatin was prepared and analyzed as described in Materials and Methods.
Orc4 protein binds to specific sites within replication origins.
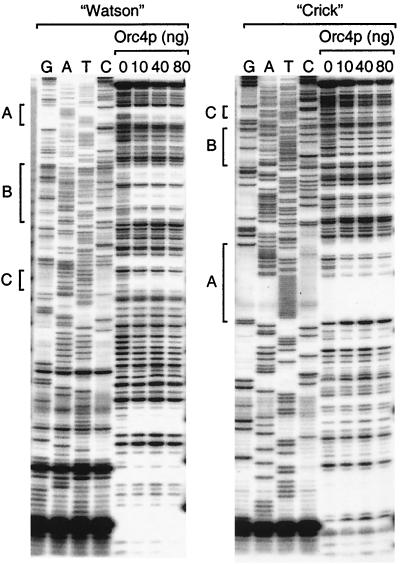
To determine whether or not the S. pombe ORC binds to specific sites within S. pombe replication origins, a 363-bp DNA fragment containing ARS3002 was radiolabeled at one of its 5′ ends and was then incubated with increasing amounts of Orc4p before digestion of the DNA with DNase I. The results revealed three specific sites that were protected by Orc4p on both strands of ARS3002 (Fig. 6). These sites were AT-rich sequences within ARS3002 with A clustered in one strand and T in the other (See Fig. 8).
FIG. 6.
Orc4 protein binds to specific sites in ARS3002. A 363-bp DNA fragment containing S. pombe ARS3002, labeled at one of its 5′ ends, was incubated with the indicated amount of Orc4p and then digested with DNase I (0.2 U for bare DNA, 0.5 U for 10 ng of Orc4p, and 2 U for 40 to 80 ng of Orc4p) as previously described (25). The DNA binding conditions for Orc4p and ORC-5 complex were the same as described for DNA band shift experiments. Lanes G, A, T, and C refer to DNA sequencing obtained using the dideoxyribonucleotide method according to the manufacturer's instructions. A DNA primer 25 nucleotides long was annealed to the 3′ end of the strand that was complementary to the 5′ 32P-labeled strand in order to reveal the sequence of the 5′ 32P-labeled strand. Sequences protected by Orc4p are indicated as A, B, and C.
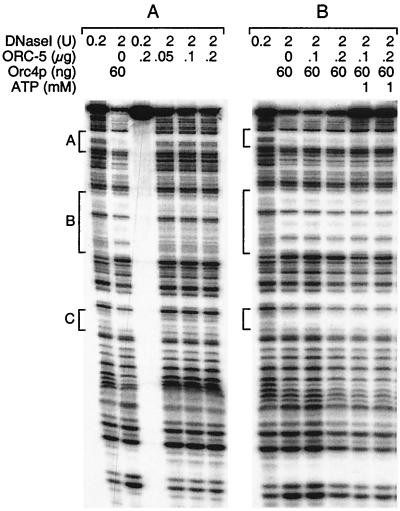
In contrast to Orc4p, the ORC-5 complex did not bind to specific sites on ARS3002, although it did bind this DNA weakly. Low concentrations of DNase I that were used to digest naked ARS3002 [32P]DNA (Fig. 7A, lane 1) failed to digest this DNA in the presence of ORC-5 (Fig. 7A, lane 3). However, the higher concentrations of DNase I that revealed three specific Orc4p binding sites (Fig. 7A, lane 2) failed to reveal any specific DNA binding sites for ORC-5 (Fig. 7A, lanes 4 to 6). The failure to observe specific DNA binding sites for ORC-5 did not result from overdigestion of the DNA, because [32P]DNA in the presence of ORC-5 complex was not digested to a greater extent than the same DNA in the presence of Orc4p. Therefore, ORC-5 complex protected DNA from digestion by binding “randomly” along the DNA. The ORC-5 complex also did not alter Orc4p binding specificity. The three footprints created by Orc4p on ARS3002 DNA remained unchanged in the presence of either ORC-5 or both ORC-5 and ATP (Fig. 7B).
FIG. 7.
The ORC-5 complex neither binds to specific sites of its own nor alters the binding pattern of Orc4p. The experiment described for Fig. 6 (Watson strand) was carried out in the presence of the indicated amounts of Orc4p, ORC-5 complex, and ATP using the amounts of DNase I indicated. When both Orc4p and ORC-5 were used, they were mixed together before addition of [32P]DNA. Protected regions A, B, and C are indicated.
Similar experiments were carried out with an 853-bp DNA fragment (5′-CATAGTGACT to ATTCTGTCTC-3′) containing S. pombe ARS1 in order to determine whether similar sequences were protected in other replication origins. As with ARS3002, Orc4p was bound to specific sites within ARS1 that were similar to those identified in ARS3002 (Table 1). Moreover, protected region D corresponded to the rightmost 30 bp of segment 1 identified by internal deletion or substitution studies as having a strong effect on ARS1 activity (8).
TABLE 1.
Sites within the S. pombe ARS1 sequence that are protected by Orc4pa
| Designation | Sequence |
|---|---|
| A | 243-TTTTTATTGAATAATATTTTA-263 |
| B | 368-AAAAATTAAATAAT-381 |
| C | 542-AAATTAAAAAA-552 |
| D | 875-TTACAAAAAATACAAATT-892 |
ARS1 is nucleotides 71 to 1264 in pESP-1 (GenBank accession no. U67875).
DISCUSSION
ORC specific DNA binding sites in S. pombe replication origins.
Using purified Orc proteins from S. pombe, the results presented here demonstrate for the first time that S. pombe Orc proteins interact with specific sites within S. pombe DNA replication origins. These sites are recognized specifically by the Orc4p subunit, and they are characterized by clusters of A on one strand and T on another [(AAAA)n, (AAAT)n, or (AATAAT)n, with n ≥ 2]. This binding was clearly site specific, because Orc4p did not bind either to alternating (AT)n sequences or to GC-rich sequences present in both ARS3002 and ARS1. Since the presence of a protein complex containing the remaining five ORC subunits (ORC-5) altered neither the selection of DNA binding sites by Orc4p nor the footprint it produced, site-specific DNA binding of the S. pombe ORC, at least in vitro, is determined solely by its Orc4p subunit.
The Orc4p DNA binding sites identified here corresponded to the essential regions identified by internal deletion studies. For example, Orc4p binding sites A and B in ARS3002 corresponded to DNA sequences 10 and 8, respectively, that are required for efficient transformation (Fig. 8). On the other hand, the smaller, and presumably weaker, Orc4p binding site C lay within DNA sequence 6, a region that is not required for ARS3002 activity. Multiple ORC binding sites within a single origin would account for the observation that full activity for S. pombe replication origins requires a larger sequence than observed with S. cerevisiae (8, 12, 37).
S. pombe Orc4p is unique among Orc proteins because its N terminus is extended by ∼60 kDa and contains nine AT-hook motifs (7, 33). These AT-hook motifs are required for DNA binding, because only the N-terminal half of Orc4p binds to origin DNA in vitro, not the C-terminal half (7). Furthermore, overexpression of either the N-terminal or C-terminal half of Orc4p, as well as of intact Orc4p, markedly inhibits growth of S. pombe. In view of the results presented here showing that site-specific binding of S. pombe ORC proteins to replication origins depends solely on Orc4p, overexpression of the N-terminal domain would be expected to interfere with Orc4p binding to replication origins in vivo. Moreover, the results presented here showing that ORC-5 does not bind to DNA in the absence of Orc4p suggest that the C-terminal half of Orc4p binds to one or more of the remaining ORC subunits. In fact, the C-terminal domain alone can form a complex with the other Orc subunits, and the ability to immunoprecipitate this complex without prior digestion by DNase I reveals that it is not bound to DNA in cell extracts (33). Therefore, overexpression of the C-terminal half of Orc4p would be expected to interfere with ORC assembly in vivo. If ORC-5 and Orc4p exist as a stable ORC in vivo, then overexpression of intact Orc4p would compete with ORC for binding to replication origins. If not, then overexpression of Orc4p may interfere with cell growth by causing ORC-5 to bind nonproductively to non-origin, AT-rich DNA sequences.
Taken together, these results provide compelling evidence that S. pombe employs a novel mechanism for site-specific initiation of DNA replication in which a single ORC subunit determines where on the genome an ORC is assembled. Although other eukaryotes lack an ORC subunit that contains multiple AT-hook motifs, such a model may apply if they contain an AT-hook protein that binds to one of their ORC subunits.
ORC DNA binding specificity.
Previous results have shown that the six S. pombe Orc subunits can be coimmunoprecipitated when their genes are transcribed and translated in vitro (33), suggesting that all six subunits are bound to chromatin as a single protein complex. The results presented here (diagrammed in Fig. 9) clearly show that the ability of this complex to recognize DNA replication origins is dependent solely on the Orc4p subunit. First, a complex of ORC subunits 1, 2, 3, 5, and 6 (the ORC-5 complex) exhibited very weak binding to DNA in vitro. In the presence of 0.1 M KCl, it altered the DNA band shift pattern of only a small amount of the [32P]DNA probe (compare lanes 1 with lanes 6 and 10 in Fig. 3) and provided only slight protection of [32P]DNA against DNase I (compare lanes 3 and 6 in Fig. 7A). Second, since neither the presence nor absence of the ORC-5 complex made any difference in the DNase I digestion pattern (Fig. 7), ORC-5 was randomly associated along the DNA sequence. Therefore, since S. pombe initiates DNA replication at specific DNA sequences and since S. pombe Orc proteins are required for DNA replication and cell cycle progression (7, 18), interaction of the S. pombe ORC with DNA appeared to be determined solely by the Orc4p subunit.
FIG. 9.
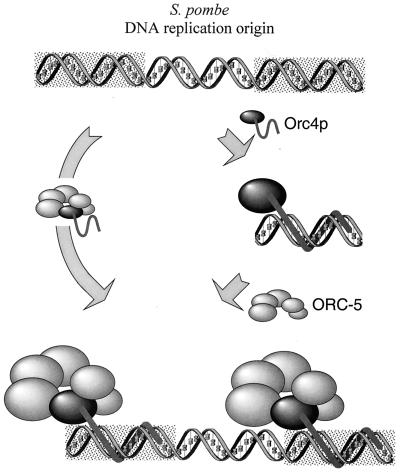
Site-specific DNA binding by the S. pombe ORC is determined solely by the Orc4p subunit, which directs ORC to several specific sites (rectangular boxes in corners) within S. pombe DNA replication origins. In vitro, purified ORC-5 complex did not bind to replication origins in the absence of purified Orc4p, but in vivo, Orc4p and ORC-5 may exist as a single protein complex that binds to newly replicated origins in a single step.
This hypothesis is supported directly by six lines of biochemical evidence. First, only Orc4p bound tightly and specifically to origin DNA (Fig. 2 and 3). Second, only Orc4p produced site-specific protection against DNase I (Fig. 6 and 7). Third, only in the presence of Orc4p did the ORC-5 complex alter the DNA band shift pattern on ARS3002 (Fig. 3). Fourth, only in the presence of Orc4p did the ORC-5 complex bind to DNA cellulose containing an S. pombe replication origin (Fig. 4). Fifth, all six ORC subunits remained on S. pombe chromatin under the same ionic conditions (0.1 M KCl) used to measure Orc protein binding to DNA. Extraction of the complete six-subunit complex from chromatin requires high-salt or DNase I treatment, presumably reflecting the unique DNA binding properties of the Orc4p subunit (33). Finally, whereas ATP affects binding of S. cerevisiae ORC to DNA (27), ATP did not affect binding of S. pombe Orc4p (± ORC-5) to DNA. Thus, unlike the S. cerevisiae ORC in which several subunits make specific contacts with origin DNA (26), the S. pombe ORC appears to use only the large N-terminal, AT-hook domain of its Orc4 subunit to bind origin DNA.
The existence of multiple ORC binding sites in a single S. pombe replication origin of 500 to 1,500 bp differs strikingly from what was found in budding yeast. S. cerevisiae replication origins each bind a single ORC molecule and initiate replication at a single nucleotide site (4). Replication origins in S. pombe require a much larger sequence for a full ARS activity (8, 12, 23, 37), consistent with the hypothesis that several SpORC molecules must bind to a single origin for efficient initiation of DNA replication. Multiple, closely spaced ORCs may facilitate recruitment of S. pombe Cdc18 and Cdt-1 proteins in the assembly of a prereplication complex.
How does Orc4p recognize specific sequences? The AT-hook motif was first discovered in the HMG-I(Y) proteins, a family of proteins ∼10 kDa in size that usually contain three AT-hook motifs (39). These hook motifs bind to the narrow groove of AT-rich sequences where each motif covers approximately 5 or 6 bp and collectively can alter DNA conformation such as bends and supercoils (35). Although HMG-I(Y) proteins can bind to any stretch of 6 AT bp in duplex DNA, its affinity may differ among those AT sequences due either to the length of the AT sequence or of their flanking sequences (44). Such parameters could allow the AT-hook motifs in S. pombe Orc4p to identify specific sequences.
S. pombe Orc4p DNA binding differs from that of HMG-I(Y) proteins in several important ways. SpOrc4p binds to highly asymmetric AT-rich sequences with A in one strand and T in the other but not to alternating AT rich sequences. SpOrc4p is eluted from chromatin by ≥0.6 M KCl, while HMG-I(Y) proteins are eluted by 0.35 M KCl (6). Therefore, consistent with its larger number of AT-hook motifs, SpOrc4p binds more tightly to chromatin than HMG-I(Y)p. Finally, in contrast to HMG-I(Y) proteins, we did not detect DNA bending by SpOrc4p alone or in the presence of ORC-5 complex (data not shown).
The number of AT-hook motifs utilized by S. pombe Orc4p may differ from one DNA site to another, because the protected regions A and B in ARS3002 were only 20 to 30 bp (Fig. 6 and 8), consistent with four or five bound AT-hook motifs, while protected region C was only 5 to 10 bp, consistent with only one or two bound AT-hook motifs. The presence of multiple DNA binding sites in replication origins that differ in their affinity for Orc4p would account for the observation that increasing the ratio of Orc4p to origin DNA increased the number of protein-DNA complexes detected in DNA band shift assays (Fig. 2 and 3).
Relationship between S. pombe and metazoans.
Replication origins in the metazoans are more similar in size and complexity to those in S. pombe than to those in S. cerevisiae (10). In addition, metazoan ORC proteins are more homologous to S. pombe ORC proteins in size and amino acid composition, with the exception of Orc4p, which in S. pombe contains an additional 60 kDa consisting of nine AT-hook motifs (33). Since both the human lamin B2 origin (1) and the S. pombe ARS1 origin (17) appear to initiate leading-strand DNA synthesis at a single specific nucleotide location, it is possible that mammalian replication origins also contain multiple ORC binding sites. Moreover, Drosophila replication origins appear to consist of multiple AT-rich discrete initiation sites similar to those found in S. pombe (21). Missing so far in the metazoans is a protein containing multiple AT-hook motifs that interacts with ORC and guides it to specific AT-rich sequences in metazoan replication origins. One candidate for such a protein is the Drosophila D1 protein, which contains seven AT-hook motifs (47). Its distribution along the chromosomes (40) is similar to that of DmOrc2p (38).
None of the S. pombe ORC subunits dissociated from S. pombe chromatin during the cell division cycle (Fig. 5), confirming a previous report that S. pombe Orc1, Orc2 and Orc5 are bound to chromatin throughout the cell cycle (30), and extending it to include Orc3, Orc4 and Orc6. Thus, in this respect, S. pombe is similar to S. cerevisiae. S. cerevisiae ORC is stably bound to chromatin throughout the cell cycle (11, 16, 28), while Xenopus ORC appears to be released from chromatin during mitosis (9, 15, 19, 41, 42). In mammals, the Orc1p subunit becomes unstably bound during the S-to-M transition and then rebinds stably to chromatin during the M-to-G1 transition (24, 34; J.-K. Li and M. L. DePamphilis, unpublished results). Therefore, despite the fact that origin complexity in S. pombe resembles that in the metazoans, regulation of ORC activity in S. pombe appears to differ from that in multicellular animals.
ACKNOWLEDGMENTS
We thank Walter Stünkel (NICHD/NIH) for his helpful advice on band shift and footprinting analysis, Joon-Kyu Lee for S. pombe strains JL197, JL202, and JL206, and Joon-Kyu Lee and Santanu Raychaudhuri (Memorial Sloan-Kettering Cancer Center) for antisera against S. pombe Orc2 and Orc6 proteins.
REFERENCES
- 1.Abdurashidova G, Deganuto M, Klima R, Riva S, Biamonti G, Giacca M, Falaschi A. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science. 2000;287:2023–2026. doi: 10.1126/science.287.5460.2023. [DOI] [PubMed] [Google Scholar]
- 2.Alberts B, Herrick G. DNA cellulose chromatography. Methods Enzymol. 1971;21:198–217. [Google Scholar]
- 3.Altman A L, Fanning E. The Chinese hamster dihydrofolate reductase replication origin beta is active at multiple ectopic chromosomal locations and requires specific DNA sequence elements for activity. Mol Cell Biol. 2001;21:1098–1110. doi: 10.1128/MCB.21.4.1098-1110.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bielinsky A K, Gerbi S A. Chromosomal ARS1 has a single leading strand start site. Mol Cell. 1999;3:477–486. doi: 10.1016/s1097-2765(00)80475-x. [DOI] [PubMed] [Google Scholar]
- 5.Bogan J A, Natale D A, DePamphilis M L. Initiation of eukaryotic DNA replication: conservative or liberal? J Cell Physiol. 2000;184:139–150. doi: 10.1002/1097-4652(200008)184:2<139::AID-JCP1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 6.Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 7.Chuang R Y, Kelly T J. The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc Natl Acad Sci USA. 1999;96:2656–2661. doi: 10.1073/pnas.96.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clyne R K, Kelly T J. Genetic analysis of an ARS element from the fission yeast Schizosaccharomyces pombe. EMBO J. 1995;14:6348–6357. doi: 10.1002/j.1460-2075.1995.tb00326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coleman T R, Carpenter P B, Dunphy W G. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- 10.DePamphilis M L. Replication origins in metazoan chromosomes: fact or fiction? Bioessays. 1999;21:5–16. doi: 10.1002/(SICI)1521-1878(199901)21:1<5::AID-BIES2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 11.Diffley J F, Cocker J H, Dowell S J, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- 12.Dubey D D, Kim S M, Todorov I T, Huberman J A. Large, complex modular structure of a fission yeast DNA replication origin. Curr Biol. 1996;6:467–473. doi: 10.1016/s0960-9822(02)00514-6. [DOI] [PubMed] [Google Scholar]
- 13.Dubey D D, Zhu J, Carlson D L, Sharma K, Huberman J A. Three ARS elements contribute to the ura4 replication origin region in the fission yeast, Schizosaccharomyces pombe. EMBO J. 1994;13:3638–3647. doi: 10.1002/j.1460-2075.1994.tb06671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandez Sarabia M J, McInerny C, Harris P, Gordon C, Fantes P. The cell cycle genes cdc22+ and suc22+ of the fission yeast Schizosaccharomyces pombe encode the large and small subunits of ribonucleotide reductase. Mol Gen Genet. 1993;238:241–251. doi: 10.1007/BF00279553. [DOI] [PubMed] [Google Scholar]
- 15.Findeisen M, El-Denary M, Kapitza T, Graf R, Strausfeld U. Cyclin A-dependent kinase activity affects chromatin binding of ORC, Cdc6, and MCM in egg extracts of Xenopus laevis. Eur J Biochem. 1999;264:415–426. doi: 10.1046/j.1432-1327.1999.00613.x. [DOI] [PubMed] [Google Scholar]
- 16.Fujita M, Hori Y, Shirahige K, Tsurimoto T, Yoshikawa H, Obuse C. Cell cycle dependent topological changes of chromosomal replication origins in Saccharomyces cerevisiae. Genes Cells. 1998;3:737–749. doi: 10.1046/j.1365-2443.1998.00226.x. [DOI] [PubMed] [Google Scholar]
- 17.Gomez M, Antequera F. Organization of DNA replication origins in the fission yeast genome. EMBO J. 1999;18:5683–5690. doi: 10.1093/emboj/18.20.5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grallert B, Nurse P. The ORC1 homolog orp1 in fission yeast plays a key role in regulating onset of S phase. Genes Dev. 1996;10:2644–2654. doi: 10.1101/gad.10.20.2644. [DOI] [PubMed] [Google Scholar]
- 19.Hua X H, Newport J. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J Cell Biol. 1998;140:271–281. doi: 10.1083/jcb.140.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hyrien O, Maric C, Mechali M. Transition in specification of embryonic metazoan DNA replication origins. Science. 1995;270:994–997. doi: 10.1126/science.270.5238.994. [DOI] [PubMed] [Google Scholar]
- 21.Ina S, Sasaki T, Yokota Y, Shinomiya T. A broad replication origin of Drosophila melanogaster, oriDalpha, consists of AT-rich multiple discrete initiation sites. Chromosoma. 2001;109:551–564. doi: 10.1007/s004120000112. [DOI] [PubMed] [Google Scholar]
- 22.Kelly T J, Brown G W. Regulation of chromosome replication. Annu Rev Biochem. 2000;69:829–880. doi: 10.1146/annurev.biochem.69.1.829. [DOI] [PubMed] [Google Scholar]
- 23.Kim S M, Huberman J A. Multiple orientation-dependent, synergistically interacting, similar domains in the ribosomal DNA replication origin of the fission yeast, Schizosaccharomyces pombe. Mol Cell Biol. 1998;18:7294–7303. doi: 10.1128/mcb.18.12.7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kreitz S, Ritzi M, Baack M, Knippers R. The human origin recognition complex protein 1 dissociates from chromatin during S phase in HeLa cells. J Biol Chem. 2001;276:6337–6342. doi: 10.1074/jbc.M009473200. [DOI] [PubMed] [Google Scholar]
- 25.Leblanc B, Moss T. DNase I footprinting. In: Kneale G G, editor. DNA-protein interactions: principles and protocols. Vol. 30. Totowa, N.J: Humana Press; 1994. pp. 1–10. [Google Scholar]
- 26.Lee D G, Bell S P. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol Cell Biol. 1997;17:7159–7168. doi: 10.1128/mcb.17.12.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee D G, Makhov A M, Klemm R D, Griffith J D, Bell S P. Regulation of origin recognition complex conformation and ATPase activity: differential effects of single-stranded and double-stranded DNA binding. EMBO J. 2000;19:4774–4782. doi: 10.1093/emboj/19.17.4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang C, Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lygerou Z, Nurse P. Cell cycle. License withheld—geminin blocks DNA replication. Science. 2000;290:2271–2273. doi: 10.1126/science.290.5500.2271. [DOI] [PubMed] [Google Scholar]
- 30.Lygerou Z, Nurse P. The fission yeast origin recognition complex is constitutively associated with chromatin and is differentially modified through the cell cycle J. Cell Sci. 1999;112:3703–3712. doi: 10.1242/jcs.112.21.3703. [DOI] [PubMed] [Google Scholar]
- 31.Malott M, Leffak M. Activity of the c-myc replicator at an ectopic chromosomal location. Mol Cell Biol. 1999;19:5685–5695. doi: 10.1128/mcb.19.8.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maundrell K, Hutchison A, Shall S. Sequence analysis of ARS elements in fission yeast. EMBO J. 1988;7:2203–2209. doi: 10.1002/j.1460-2075.1988.tb03059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moon K Y, Kong D, Lee J K, Raychaudhuri S, Hurwitz J. Identification and reconstitution of the origin recognition complex from Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1999;96:12367–12372. doi: 10.1073/pnas.96.22.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Natale D A, Li C J, Sun W H, DePamphilis M L. Selective instability of Orc1 protein accounts for the absence of functional origin recognition complexes during the M-G(1) transition in mammals. EMBO J. 2000;19:2728–2738. doi: 10.1093/emboj/19.11.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Newlon C S. DNA replication in yeast. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 873–914. [Google Scholar]
- 35.Nissen M S, Reeves R. Changes in superhelicity are introduced into closed circular DNA by binding of high mobility group protein I/Y. J Biol Chem. 1995;270:4355–4360. doi: 10.1074/jbc.270.9.4355. [DOI] [PubMed] [Google Scholar]
- 36.Nurse P. Yeast aids cancer research. Nature. 1985;313:631–632. doi: 10.1038/313631a0. [DOI] [PubMed] [Google Scholar]
- 37.Okuno Y, Satoh H, Sekiguchi M, Masukata H. Clustered adenine/thymine stretches are essential for function of a fission yeast replication origin. Mol Cell Biol. 1999;19:6699–6709. doi: 10.1128/mcb.19.10.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pak D T, Pflumm M, Chesnokov I, Huang D W, Kellum R, Marr J, Romanowski P, Botchan M R. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell. 1997;91:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- 39.Reeves R, Nissen M S. Cell cycle regulation and functions of HMG-I(Y) Prog Cell Cycle Res. 1995;1:339–349. doi: 10.1007/978-1-4615-1809-9_28. [DOI] [PubMed] [Google Scholar]
- 40.Rodriguez Alfageme C, Rudkin G T, Cohen L H. Isolation, properties and cellular distribution of D1, a chromosomal protein of Drosophila. Chromosoma. 1980;78:1–31. doi: 10.1007/BF00291907. [DOI] [PubMed] [Google Scholar]
- 41.Romanowski P, Madine M A, Rowles A, Blow J J, Laskey R A. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr Biol. 1996;6:1416–1425. doi: 10.1016/s0960-9822(96)00746-4. [DOI] [PubMed] [Google Scholar]
- 42.Rowles A, Tada S, Blow J J. Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J Cell Sci. 1999;112:2011–2018. doi: 10.1242/jcs.112.12.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russell P, Nurse P. Negative regulation of mitosis by wee1+, a gene encoding a protein kinase homolog. Cell. 1987;49:559–567. doi: 10.1016/0092-8674(87)90458-2. [DOI] [PubMed] [Google Scholar]
- 44.Russnak R H, Candido E P, Astell C R. Interaction of the mouse chromosomal protein HMG-I with the 3′ ends of genes in vitro. J Biol Chem. 1988;263:6392–6399. [PubMed] [Google Scholar]
- 45.Sasaki T, Sawado T, Yamaguchi M, Shinomiya T. Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolα-dE2F locus of Drosophila melanogaster. Mol Cell Biol. 1999;19:547–555. doi: 10.1128/mcb.19.1.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Svoboda A, Bahler J, Kohli J. Microtubule-driven nuclear movements and linear elements as meiosis-specific characteristics of the fission yeasts Schizosaccharomyces versatilis and Schizosaccharomyces pombe. Chromosoma. 1995;104:203–214. doi: 10.1007/BF00352185. [DOI] [PubMed] [Google Scholar]
- 47.Wagner C R, Hamana K, Elgin S C. A high-mobility-group protein and its cDNAs from Drosophila melanogaster. Mol Cell Biol. 1992;12:1915–1923. doi: 10.1128/mcb.12.5.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitehall S, Stacey P, Dawson K, Jones N. Cell cycle-regulated transcription in fission yeast: Cdc10-Res protein interactions during the cell cycle and domains required for regulated transcription. Mol Biol Cell. 1999;10:3705–3715. doi: 10.1091/mbc.10.11.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu J, Carlson D L, Dubey D D, Sharma K, Huberman J A. Comparison of the two major ARS elements of the ura4 replication origin region with other ARS elements in the fission yeast, Schizosaccharomyces pombe. Chromosoma. 1994;103:414–422. doi: 10.1007/BF00362286. [DOI] [PubMed] [Google Scholar]