Age-associated B cells are heterogeneous with respect to expression of memory and plasmablast markers. Unlike resting memory, ABCs are chronically activated, with the capacity for self-renewal or differentiation. Depletion of CD11c+ ABCs improved renal disease in the MRL/lpr lupus model.
Abstract
Age-associated B cells (ABCs) are formed under inflammatory conditions and are considered a type of memory B cell (MBC) expressing the transcription factor T-bet. In SLE, ABC frequency is correlated with disease, and they are thought to be the source of autoantibody-secreting cells. However, in inflammatory conditions, whether autoreactive B cells can become resting MBCs is uncertain. Further, the phenotypic identity of ABCs and their relationship to other B cell subsets, such as plasmablasts, is unclear. Whether ABCs directly promote disease is untested. Here we report, in the MRL/lpr SLE model, unexpected heterogeneity among ABC-like cells for expression of the integrins CD11b and CD11c, T-bet, and memory or plasmablast markers. Transfer and labeling studies demonstrated that ABCs are dynamic, rapidly turning over. scRNA-seq identified B cell clones present in multiple subsets, revealing that ABCs can be plasmablast precursors or undergo cycles of reactivation. Deletion of CD11c-expressing B cells revealed a direct role for ABC-like B cells in lupus pathogenesis.
Introduction
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease characterized by the loss of self-tolerance to nucleic acid associated self-antigens. While several immune cell lineages are aberrantly activated in SLE, B cells are essential for both the initiation and propagation of disease (Chan et al., 1999; Marinov et al., 2021). Several immunotherapeutic approaches that broadly target B cells have been brought to the clinic in recent years, including B cell activating factor inhibition and B cell–specific depleting antibodies (Atisha-Fregoso et al., 2021; Lee et al., 2021). These approaches affect not only autoreactive B cells but also naive and pathogen-specific populations. A better understanding of B cell dysregulation in lupus could lead to more narrowly targeted therapeutics.
In addition to their role as the precursor to antibody-secreting cells (ASCs), B cells function as antigen-presenting cells, producers of inflammatory and regulatory cytokines, and as reservoirs of immune memory. In autoimmunity, B cells give rise to autoantibodies, present self-antigens to autoreactive T cells, and produce cytokines; however, it remains unclear to what extent resting autoreactive memory B cells (MBCs) can form in the presence of ubiquitous self-antigen.
Several groups have described a subset of B cells, sometimes termed “atypical memory B cells” in humans or “age-associated B cells” (ABCs) in mice, that is elevated in SLE patients and in murine lupus models, as well as in certain chronic bacterial or viral infections (Cancro, 2020; Mouat et al., 2022). These cells are proposed to be autoreactive memory B cells and are thought to be immediate precursors to autoantibody-secreting cells (Jenks et al., 2018; Rubtsov et al., 2011). In humans, DN2 atypical memory B cells, defined as CD21lowCD27−, are elevated in the peripheral blood lymphocytes of patients with SLE compared to healthy controls, and their frequency correlates with the severity of clinical disease manifestations, including nephritis (Jacobi et al., 2008; Wang et al., 2018; Wehr et al., 2004; Wei et al., 2007). In several murine models of SLE, an analogous population of ABCs is also increased in frequency compared to normal animals (Ricker et al., 2021; Rubtsov et al., 2011; Rubtsova et al., 2017). To date, much of the evidence of a pathogenic function for ABCs has been correlative, although it is clear that phenotypic murine ABCs and human DN2 B cells do include autoreactive cells, and that they are generated by an inflammatory cytokine environment (such as signaling by IFNγ and IL-21) combined with signaling through the B cell receptor (BCR) and endosomal TLRs (Manni et al., 2018; Naradikian et al., 2016; Rubtsov et al., 2011, 2013; Rubtsova et al., 2013; Zumaquero et al., 2019). These ABC-generating conditions are similar to those necessary for the extrafollicular plasmablast (PB) response.
A significant challenge to interpretation of studies of ABCs is that there has yet to be a consensus about the most important lineage-defining markers. Initial descriptions in mice described ABCs as CD19+ CD11c+ CD11b+ (Rubtsov et al., 2011) or as CD19+ CD43− CD93− CD21/35− CD23− (Hao et al., 2011). Subsequent studies focused on CD11c and T-bet coexpression as critical markers (Rubtsov et al., 2013; Rubtsova et al., 2013; Naradikian et al., 2016); however, the importance of T-bet for ABC formation and function has recently been called into question (Du et al., 2019b; Levack et al., 2020; Ricker et al., 2021). The situation is further complicated by potential differences between human DN2 B cells and mouse ABCs, which are generally isolated from different sites (i.e., peripheral blood lymphocytes vs. spleen), as well as the analysis of ABC-like cells in a variety of different disease states in both species. Although transcriptomic and phenotypic studies do suggest a considerable degree of overlap among these systems, they may not always be perfectly congruent, depending on the choice of lineage-defining markers and the details of the experimental model or disease state.
To better define the role of ABCs in a single, well-defined model of autoimmunity, we took advantage of several complimentary approaches in the MRL/lpr model of spontaneous SLE. We identify heterogeneity among ABC-like subsets, defined by CD11c and/or CD11b expression, with respect to the expression of T-bet, MBC markers, and PB markers. We show that CD11c+CD11b+ ABCs are proliferative and can self-renew or differentiate into PBs or germinal center (GC) B cells in vivo. In a mixed bone marrow chimera system in which CD11c+ B cells but not other CD11c-expressing cells are constitutively deleted, we show that CD11c+ ABCs contribute to lupus nephritis and T cell activation. Finally, using single-cell RNA sequencing (scRNA-seq) with VDJ analysis, we trace expanded B cell clones through multiple B cell subsets, finding evidence that the ABC population shares clonal mutations with the PB compartment. Together, this evidence suggests that, despite having memory-like features, ABCs are a highly dynamic and chronically reactivated population important for SLE pathogenesis, which may be an attractive target for future therapeutic approaches.
Results
ABC-like cells are phenotypically heterogeneous with respect to T-bet, ASC markers, and memory markers
Several groups have recently described the expansion of ABCs in murine models of lupus, including MRL/lpr (Aranburu et al., 2018; Manni et al., 2018; Ricker et al., 2021; Rubtsov et al., 2011; Rubtsova et al., 2017). A major challenge to interpreting and integrating murine studies is the varying definition of ABCs used by different groups, such that it is not always clear that the same population is being described across studies. ABCs have been defined variously as CD11c+, CD11c+CD11b+, CD11c+T-bet+, or CD43−CD93−CD21/35−CD23− B cells (Cancro, 2020; Mouat et al., 2022). In order to proceed with functional, genetic, and cell transfer studies of ABCs, it was important to define phenotypes and heterogeneity of ABC-like cells. We thus undertook a flow cytometric analysis of ABC-like populations in older MRL/lpr mice with an SLE-like disease.
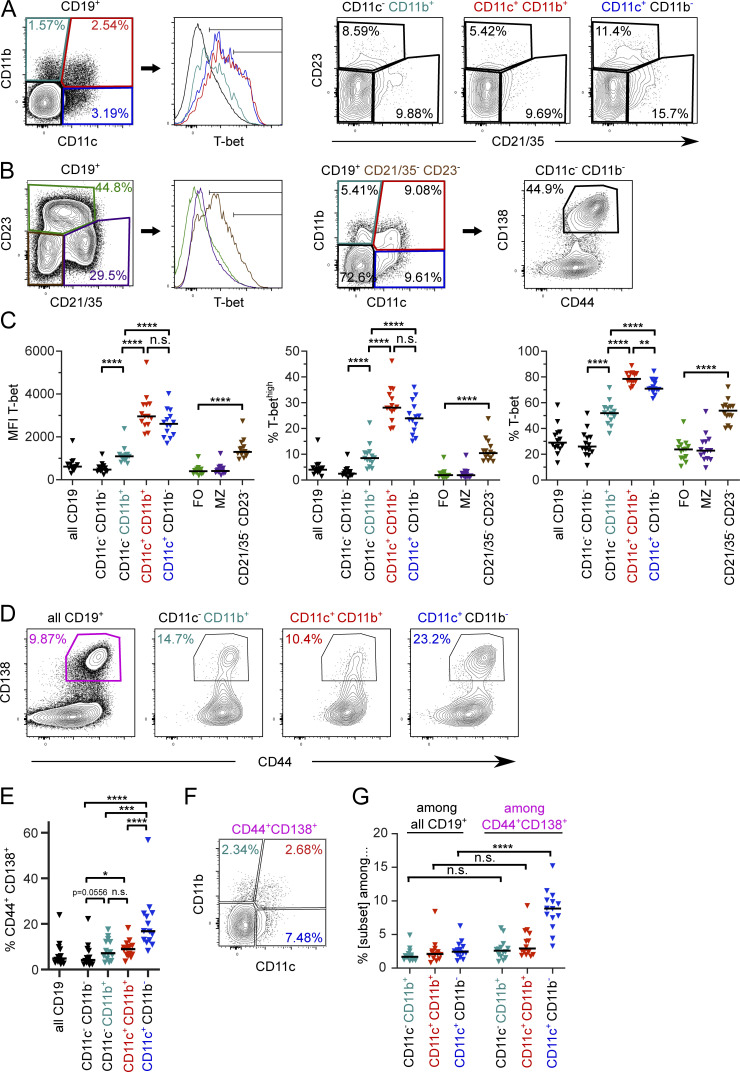
To determine whether T-bet expression was a reliable marker for ABCs in diseased MRL/lpr mice, we examined T-bet expression in the context of other B cell and putative ABC surface markers. A majority of CD19+ B cells that express CD11c also express T-bet by flow cytometry, with similar patterns and mean fluorescence intensity regardless of CD11b expression (Fig. 1, A and C). CD11b single-positive B cells were also T-bet positive in comparison to CD11c−CD11b− B cells, albeit with a lower mean fluorescence intensity and fewer T-bethigh cells than among CD11c+ B cells (Fig. 1, A and C).
Figure 1.
ABC-like cells are heterogeneous with respect to expression of T-bet and PB markers. Splenocytes from seven female and seven male MRL/lpr at 18 wk of age were stained as indicated. (A) Representative CD11c, CD11b, and T-bet staining in CD11c−CD11b+ (teal), CD11c+CD11b+ (red), CD11c+CD11b− (dark blue) or CD11c−CD11b− (black) CD19+ B cells. Representative CD21/35 versus CD23 staining from the subsets as gated in the leftmost plot. (B) Representative CD21/35, CD23, and T-bet staining in the same animal as A, indicating FO (green), MZ (purple), or CD21/35−CD23− (brown) T-bet expression. Representative CD11c vs. CD11b staining of CD21/35−CD23−cells and CD44 vs. CD138 staining of CD19+CD21/35−CD23−CD11c−CD11b− cells. (C) Mean fluorescence intensity (MFI) of T-bet staining in the populations shown in A and B. (D) Representative CD44+ CD138+ staining in total CD19+ B cells or indicated B cell subsets. (E) Frequency of CD44+ CD138+ cells among the indicated B cell subsets. (F) Representative CD11c and CD11b staining among all CD19+ CD44+ CD138+ PBs. (G) Frequency of CD11c and/or CD11b expression (as indicated) among total B cells or PBs. Horizontal lines indicate medians. Data are representative of at least three independent experiments. Statistical comparisons by two-tailed Mann-Whitney test; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
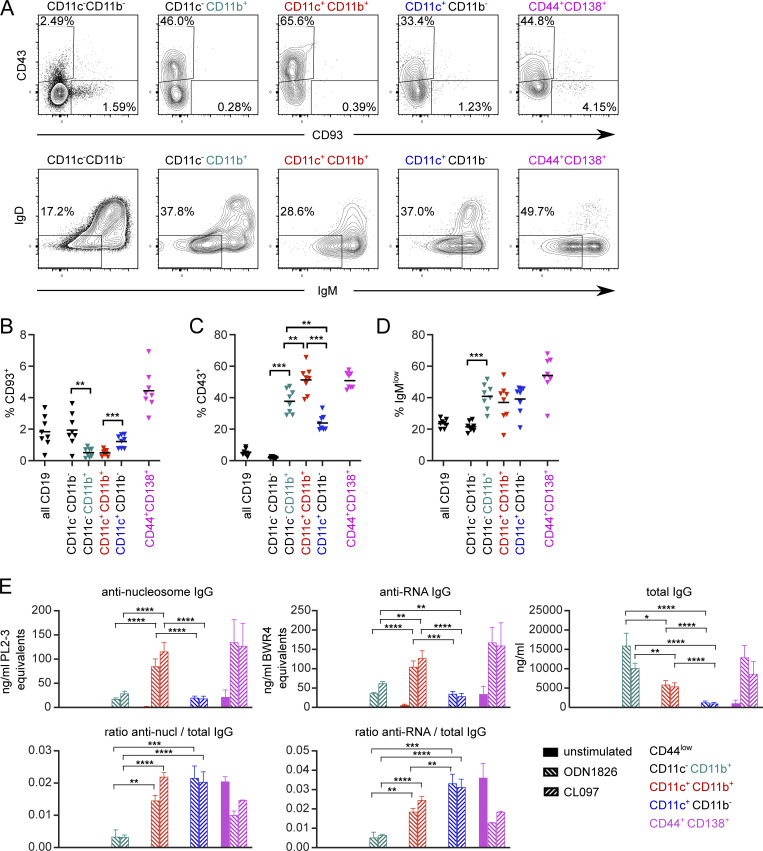
Other studies have defined ABCs as CD21/35−CD23−, often with prior CD93 and CD43 exclusion (Hao et al., 2011; Aranburu et al., 2018). We found that CD21/35−CD23− MRL/lpr B cells did indeed include a T-bet+ population, although a significant proportion of these cells did not express T-bet (Fig. 1, B and C). The majority of ABC-like cells defined by CD11c, CD11b, or CD11b and CD11c were CD21/35−CD23− (Fig. 1 A); however, the converse was not true. In aged MRL/lpr mice, the CD21/35−CD23− gate enriched for CD11c and/or CD11b expression, but a majority of CD21/35−CD23− cells lacked expression of those markers; about half of the remaining cells were PBs and the others, not defined in this staining panel, could include transitional or B-1 populations. Among CD11c and/or CD11b-expressing B cells, CD93 (a marker of transitional B cells) was not significantly expressed, but a high proportion of these cells were CD43+ and IgMlow (Fig. S1, A–D), suggesting that prior exclusion of CD43+ cells would have excluded some CD11c+T-bet+ ABC-like cells in the MRL/lpr model.
Figure S1.
CD11c and CD11b ABC-like cells and PBs include CD43+ but not CD93+ populations in MRL/lpr and are frequently switched. Spleens were stained from four male and four female 16-wk-old MRL/lpr mice and analyzed on a Cytek Aurora. (A) Plot of CD93 and CD43 expression (top) and IgM vs. IgD surface staining (bottom) among CD19+ B cells gated on indicated subsets. (B) Frequency of CD93-positive staining among indicated subsets. (C) Frequency of CD43-positive staining among indicated subsets. (D) Frequency of IgMlowIgD− cells among indicated subsets. (B–D) Horizontal lines indicate means. Data are representative of three experiments. (E) MRL/lpr splenocytes from 15- to 18-wk-old female mice were pooled, B-enriched, and sorted for CD44low cells (CD19+ CD11c− CD11b− CD44− CD138−; black bars; below limit of detection), CD11b+ ABCs (CD19+ CD11c− CD11b+ CD138−; teal bars), CD11b+CD11c+ ABCs (CD19+ CD11c+ CD11b+ CD138−; red bars), CD11c+ ABCs (CD19+ CD11c+ CD11b− CD138−; blue bars) or PBs (CD19+ CD44+ CD138+ CD11c− CD11b−; pink bars). Sorted cells were cultured for 6 d with media alone, 2 μg/ml ODN 1826 (TLR9 ligand), or 2 μg/ml CL097 (TLR7 ligand). Culture supernatants were harvested and secreted anti-nucleosome IgG, anti-RNA IgG, or total IgG were assessed by ELISA (top). The ratio of anti-nucleosome or anti-RNA to total IgG was calculated (bottom). Bars indicate mean + SEM. Data are pooled from two independent sorts with a total of 7–12 technical replicates (stimulation wells) except naive and CD44+CD138+ groups, which had 1–3 technical replicates. Statistical comparisons by two-tailed Mann-Whitney test; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
In sum, CD11c expression was a better predictor of T-bet expression, particularly among the T-bethighest populations, than CD11b or the absence of CD21/35 and CD23. We primarily focused on CD11c and CD11b expressing populations in subsequent experiments.
As CD11c expression has been observed on ASCs (Racine et al., 2008), we next asked whether ABC-like B cells defined by CD11c and/or CD11b expression also express ASC markers. CD11c+, CD11c+CD11b+, and CD11b+ B cells were all CD44high (Fig. 1 D). All three subsets included CD138+ cells, with CD11c+ single-positive B cells the most statistically enriched for CD138+ cells compared to CD11c−CD11b− B cells (Fig. 1, D and E). Nonetheless, the great majority of these populations were CD138−. Conversely, gating first on CD44+CD138+ PBs also enriched for CD11c single-positive B cells, but not CD11b single-positive or CD11b+CD11c+ cells (Fig. 1, F and G). Importantly, the majority of PBs did not express either CD11b or CD11c, indicating that ABC-like and ASC populations mainly do not overlap.
ABCs have been proposed to be a type of MBC that, in the context of autoimmune disease, are driven by self-antigen recognition (Naradikian et al., 2016). We therefore assessed the expression of MBC surface markers (Tomayko et al., 2010; Zuccarino-Catania et al., 2014) on CD11c and/or CD11b ABC-like subsets in MRL/lpr mice. CD80, PD-L2, and CD73 were indeed expressed, in heterogeneous fashion, among the B cell subsets defined by CD11b or CD11c expression. CD11c+CD11b+ B cells included the highest proportion of CD80+ and CD80+PD-L2+ cells (Fig. 2, A–D) and expressed the most CD73 (Fig. 2, E and F), suggesting that these are the most memory-like. CD11b single-positive B cells also included a high proportion of CD80+ B cells and some CD80+PD-L2+ cells, although at a lesser frequency than in the CD11c+CD11b+ gate. CD11c single-positive B cells were less likely to express either CD80 or PD-L2. Both CD11b and CD11c single-positive populations expressed intermediate amounts of CD73 compared to CD11b/c double-positive (DP) cells (Fig. 2, E and F). Interestingly, none of the ABC-like populations defined by CD11b and/or CD11c included a notable number of the PD-L2 single-positive MBCs (Fig. 2 B), a subset found in the T-dependent nitrophenol-chicken γ globulin immunization model (Tomayko et al., 2010; Zuccarino-Catania et al., 2014).
Figure 2.
ABC-like cell subsets are heterogeneous in expression of MBC markers and do not account for all CD80 and PD-L2 expression among B cells. (A) Representative CD11c and CD11b staining on CD19+ splenocytes from a 15.5-wk female MRL/lpr. (B) Representative staining of CD80 and PD-L2 among all CD19+ B cells (far right) or indicated CD11c and/or CD11b B cell subsets. (C) Quantification of CD80 single-positive B cells among indicated subsets. (D) Quantification of CD80+ PD-L2+ B cells among indicated subsets. (E) Representative staining of CD73 in indicated B cell subsets. (F) Quantification of CD73+ cells among indicated B cell subsets. (G) Representative staining of CD11c and CD11b among total CD80+PD-L2− (top) or CD80+PD-L2+ (bottom) B cells. Summary data are pooled from five (C and D) or seven (F) independent experiments and include both male and female mice. Horizontal lines indicate medians. Statistical comparisons by two-tailed Mann-Whitney; *P < 0.05; **P < 0.01; ****P < 0.0001.
Conversely, CD11c and/or CD11b expression was turned on in only a fraction of MRL/lpr phenotypic MBCs. Gating first on CD80+ or CD80+PD-L2+ populations out of total B cells (Fig. 2 B, far right), we found that of CD80+ B cells, more than half did not express either CD11c or CD11b (Fig. 2 G). Of CD80+PD-L2+ B cells, about two-thirds expressed CD11b and/or CD11c. Overall, this analysis revealed a remarkable amount of heterogeneity among ABC-like cells, T-bet–positive cells, and MBCs in older MRL/lpr mice. That said, the cells positive for both CD11b and CD11c were almost uniformly T-bet+, with most expressing the memory marker CD80; therefore, we generally focused on this CD11c+CD11b+ population for subsequent functional studies, while continuing to characterize other related populations when possible.
Anti-nucleic acid specificity drives ABC-like B cell generation in a TLR9-dependent fashion
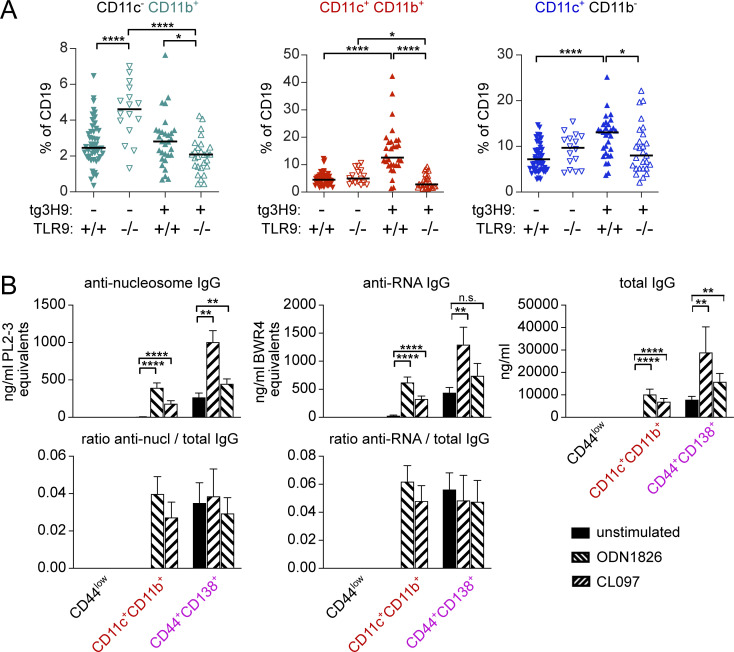
Several reports have described a role for endosomal TLRs, particularly TLR7, in the generation of ABCs (Jenks et al., 2018; Naradikian et al., 2016; Ricker et al., 2021; Rubtsov et al., 2011, 2013; Rubtsova et al., 2013; Zumaquero et al., 2019). However, whether the BCR itself is instructional in this process, or rather whether it is a passenger to non-specific TLR stimulation, has not been tested. We therefore asked whether an anti-DNA BCR would enhance ABC formation in an autoimmune context, and if so, whether TLR9 is necessary for this, using MRL/lpr mice expressing the 3H9 BCR heavy chain as a transgene (Nickerson et al., 2013). In these animals, pairing of the heavy chain with most endogenous light chains results in anti-nucleosome or anti-dsDNA reactivity (Ibrahim et al., 1995). The proportion of CD11c+CD11b+ and CD11c+ B cells, but not CD11b+ single-positive cells, was greater in 3H9+ mice than in nontransgenic controls (Fig. 3 A, first vs. third columns). Importantly, the proportion of all three ABC-like populations was diminished in 3H9 mice when TLR9 was absent (Fig. 3 A, third vs. fourth columns), a condition under which we have previously shown anti-nucleosome ASC differentiation is also blocked (Nickerson et al., 2013). In contrast, the absence of TLR9 did not have much effect on CD11c-expressing ABCs in MRL/lpr with an unrestricted BCR repertoire (tg3H9− groups), although the population of CD11b single-positive B cells was somewhat increased (Fig. 3 A, first vs. second columns). Thus, anti-DNA BCR specificity drives ABC formation, and this process is TLR9 dependent.
Figure 3.
ABC-like cells are enriched for anti-nucleic acid reactivity. (A) Proportion of B cells expressing CD11b alone (teal), CD11c and CD11b (red), or CD11c (blue) in mice of the indicated 3H9 and TLR9 genotypes. (B) MRL/lpr splenocytes from 14- to 18-wk-old male or female mice were B-enriched and sorted for CD44low cells (CD19+ CD44− CD138−), ABCs (CD19+ CD11c+ CD11b+ CD138−), or PBs (CD19+ CD44+ CD138+ CD11c− CD11b−). Sorted cells were cultured for 6 d with media alone, 2 μg/ml ODN 1826 (TLR9 ligand), or 2 μg/ml CL097 (TLR7 ligand). Culture supernatants were harvested and secreted anti-nucleosome IgG, anti-RNA IgG, or total IgG were assessed by ELISA (top). The ratio of anti-nucleosome or anti-RNA to total IgG was calculated (bottom). Data are pooled from two independent sorts with at least eight technical replicates (stimulation wells) per condition. Bars indicate mean + SD. Statistical comparisons by two-tailed Mann-Whitney; *P < 0.05; **P < 0.01; ****P < 0.0001.
ABCs are thought to be a source for autoantibodies, and to be poised to differentiate into PBs following endosomal TLR stimulation (Manni et al., 2018; Rubtsov et al., 2011). To test whether ABCs can differentiate to autoreactive PBs ex vivo, we pooled MRL/lpr splenocytes from several donors and enriched for total B cells by negative magnetic selection, followed by cell sorting for CD44low B cells (CD19+CD44lowCD11c−CD11b−CD138−), ABCs (CD19+CD44highCD11c+CD11b+CD138−), or PBs (CD19+CD44+CD11c−CD11b−CD138+). Equivalent numbers of sorted cells of each population were cultured for 6 d in the presence or absence of TLR7 or TLR9 ligands. The resulting cell culture supernatants were then tested for the presence of anti-nucleosome, anti-RNA, and total IgG (Fig. 3 B, top). As expected, CD44low B cells did not produce IgG under these conditions, and most cells died after 6 d in culture (not shown). In contrast, unstimulated CD11c+CD11b+ ABCs did not secrete Ig; however, following TLR7 or TLR9 stimulation they produced anti-nucleosome and anti-RNA Ig at titers comparable to unstimulated PBs. PBs secreted IgG, including anti-nucleosome and anti-RNA IgG, even without in vitro TLR stimulation (Fig. 3 B). Somewhat surprisingly, the total amount of IgG secreted by PBs was enhanced by the addition of TLR7 or TLR9 ligands. This interesting result suggests that PBs are still dynamically capable of responding to TLR signals, although it is not clear if this was an effect on survival, proliferation, or per-cell antibody production (Fig. 3 B, top right). The ratio of secreted autoreactive IgG of these specificities to total IgG was similar between ABCs and PBs (Fig. 3 B, bottom), suggesting that autoreactive specificities are similarly enriched in the two populations. In a follow-up experiment, we examined whether CD11b single-positive or CD11c single-positive cells also differentiate into autoreactive ASCs. Interestingly, CD11b single-positive cells produced the most IgG following TLR stimulation, but proportionally little of it was anti-RNA or anti-nucleosome (Fig. S1 E). CD11c single-positive cells produced less total IgG than CD11b+CD11c+ cells, but a similar fraction of that IgG was autoreactive (Fig. S1 E). Thus, the ABC population contains autoreactive cells capable of differentiation to autoreactive ASCs ex vivo following TLR ligation, but ABCs are not themselves ASCs.
ABCs are proliferative
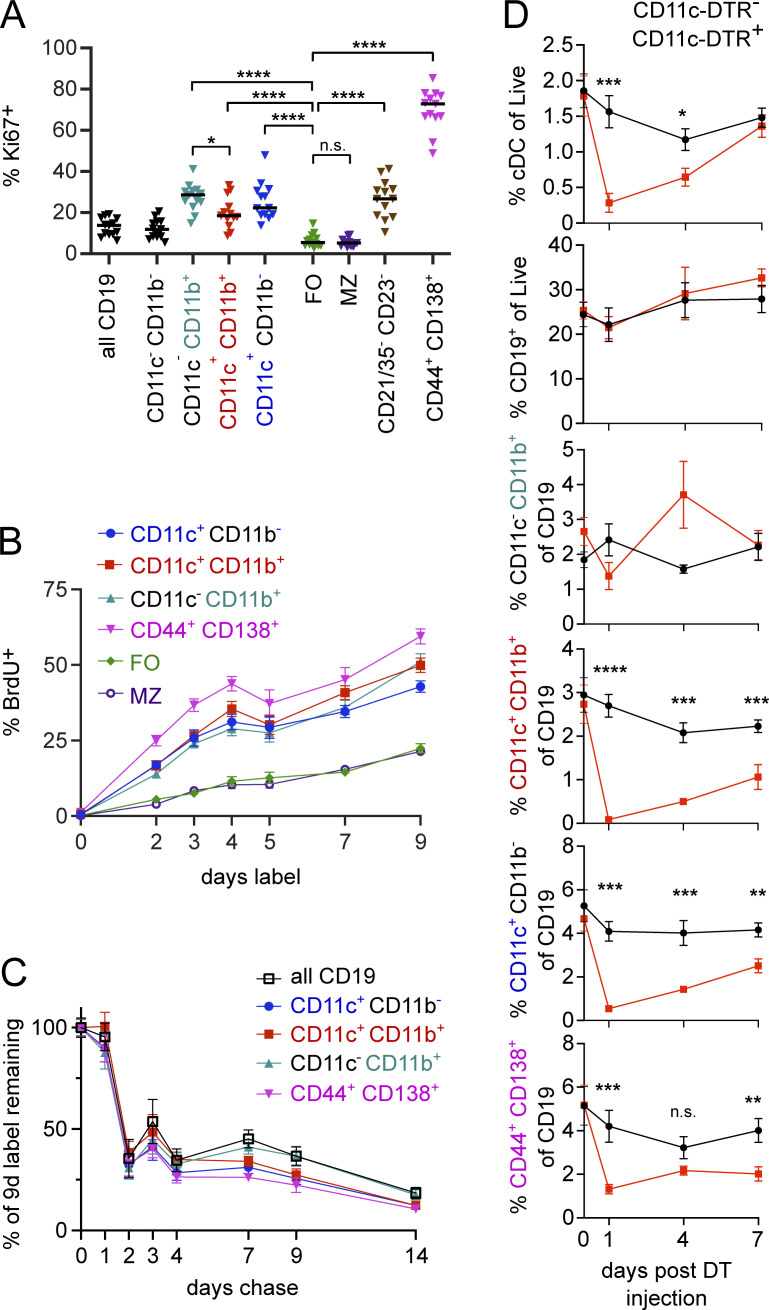
Conventional MBCs are non-cycling unless re-exposed to antigen; however, if ABCs are autoreactive, they may be chronically restimulated by ubiquitous self-antigens. To assess whether ABCs in mice with lupus are cycling in vivo, we performed Ki67 staining. 20–30% of ABC-like B cells were Ki67+, whether defined as CD11b+ and/or CD11c+ or CD21/35−CD23−, a significantly different frequency from follicular (FO) or marginal zone (MZ) B cells, which had <5% Ki67+ cells (Fig. 4 A). Nonetheless, the frequency of Ki67 positivity of ABC-like cells was substantially lower than that among PBs (Fig. 4 A) or GC B cells (not shown).
Figure 4.
ABC-like cells are proliferative. (A) B cells of the indicated subsets were stained for Ki67 by flow cytometry. (B) MRL/lpr mice were provided BrdU in drinking water for the indicated durations and splenocytes were analyzed for incorporation of label. Data are pooled from four experiments. (C) MRL/lpr mice were provided BrdU in drinking water for 9 d and returned to normal drinking water for the indicated durations. BrdU incorporation was normalized to the average amount of label observed on d9 within each experiment for each subset. Data are pooled from two experiments. (D) CD11c-DTR+ MRL/lpr (red squares) or littermate controls (black circles) were given 8 ng/kg DT i.p. at 7, 4, or 1 d prior to analysis or left untreated. Splenocytes were analyzed for the presence of indicated cell types by flow cytometry. Statistics in A and D by two-tailed Mann-Whitney; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To quantitate the proliferative activity of ABCs, we performed a BrdU labeling-up analysis. About 25% of ABCs incorporated BrdU within 3 d and 40–50% incorporated BrdU after 9 d of labeling (Fig. 4 B); the labeling kinetics were similar in CD11c+, CD11c+CD11b+, and CD11b+ subsets. The rate of labeling in ABC-like cells was slightly less than that of PBs but was more rapid than that in FO or MZ B cells. These data are indicative of a dynamically renewing population; either ABCs themselves are proliferative or they rapidly turn over and are regenerated from a proliferating precursor population.
To further assess the turnover of ABCs, we performed labeling for 9 d followed by a chase period of up to 2 wk. We found that the majority of cells that acquired BrdU during the labeling period were BrdU negative within 2–4 d after the mice were switched back to regular drinking water, indicating that they had continued to proliferate and thus diluted out the BrdU label (Fig. 4 C). About 25% of ABCs that acquired label during the labeling window retained detectable label from 4 to at least 9 d after label was removed, suggesting that the ABC-like populations contain a mixture of cells, with the majority that rapidly dilute BrdU but possibly a subset of a more stable population that, once labeled, undergoes little further proliferation for at least 9 d.
If ABCs are rapidly turning over, then they should reconstitute quickly after selective depletion. To test this, we used CD11c-DTR MRL/lpr mice, which express a transgene encoding the receptor for diphtheria toxin (DT) under control of the CD11c promoter, such that cells expressing CD11c are eliminated when exposed to DT. We injected CD11c-DTR MRL/lpr or transgene-negative littermate controls i.p. with a single dose of DT at varying times prior to analysis. As expected, conventional splenic CD11c+MHCII+ dendritic cells were depleted by DT under these conditions but recovered to their typical proportion out of total splenocytes within 7 d (Fig. 4 D). DT treatment had no effect on the frequency of total B cells in the spleen, but specifically depleted CD11c+ and CD11c+CD11b+ B cells. Within 7 d, these populations recovered to about one third to one half of their frequency in untreated controls. This suggested a turnover half-life in the compartment that is consistent with that estimated by BrdU labeling. Interestingly, the PB compartment was also partially depleted by DT treatment, and to a greater extent than might have been expected based on the frequency of PBs that are CD11c+ by flow cytometry (Fig. 1 G). This could be explained if a fraction of the PB compartment is generated over the short time course of this experiment from ABC precursors, which in turn had been depleted.
ABCs self-renew and give rise to PBs or GCs
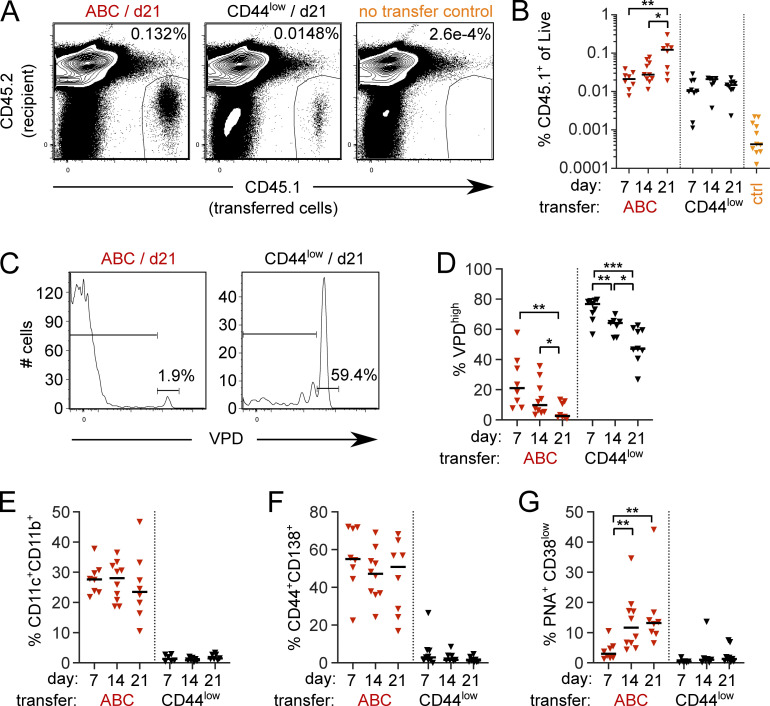
To formally demonstrate the self-renewal capacity and differentiation potential of ABCs in vivo, we performed a precursor-product analysis using an adoptive transfer design. Pooled splenocytes from aged CD45.1+ MRL/lpr mice were B cell enriched by negative magnetic selection and sorted for naive-like CD44low B cells (CD19+CD44lowCD11c−CD11b−CD138−) or ABCs (CD19+CD44highCD11c+CD11b+CD138−) populations. Violet Proliferation Dye (VPD)–labeled ABCs or control naive-like B cells were then separately transferred into congenic, pre-autoimmune CD45.2 MRL/lpr recipients. At 7, 14, or 21 d following transfer, recipient mice were analyzed for the presence and phenotype of the transferred cells. By day 21 (d21) after transfer, the frequency of ABC progeny had increased by about 10-fold compared to d7 recipients, while the frequency of control naive-like B cell progeny did not increase over time after transfer (Fig. 5, A and B). About 80% of the ABC progeny group had diluted VPD within 7 d, although a small proportion of the transferred cells did retain VPD label as late as 21 d after transfer (Fig. 5, C and D). In contrast, on average ∼50% of transferred naive-like B cells were in the VPD highest peak at d21. Together these data indicated that after transfer, most ABC proliferated, and their progeny was preserved, while most naive-like B cells exhibited little or no proliferation.
Figure 5.
CD11c+CD11b+ ABCs self-renew or differentiate. Sorted ABCs (CD19+ CD11c+ CD11b+ CD138−) or CD44low B cells (CD19+ CD44low CD138− CD11c− CD11b−) isolated from spleens of CD45.1+ MRL/lpr females at 14–20 wk of age were VPD labeled and adoptively transferred into preautoimmune female CD45.2 MRL/lpr recipients. Recipient spleens were analyzed at d7, d14, or d21 following transfer. (A) Representative staining of total live splenocytes at d21 after transfer from mice receiving ABCs (left), CD44low B cells (middle), or no transfer controls (right). (B) Frequency of transferred CD45.1+ ABCs (red) or CD44low B cells (black) in recipient spleens at indicated times after transfer. (C) VPD staining from mice in A gated on CD45.1+ transferred cells. (D) Proportion of VPD undiluted cells among transferred cells. (E) Proportion of CD11c+CD11b+ ABCs among transferred cells. (F) Proportion of CD44+CD138+ PBs among transferred cells. (G) Proportion of PNA+CD38low GC B cells among transferred cells. Data are pooled from five independent sorts/transfers with 3–4 experiments per timepoint. Statistical comparisons by two-tailed Mann-Whitney; *P < 0.05; **P < 0.01; ***P < 0.001.
25–30% of the sorted and transferred CD11c+CD11b+ cells retained CD11c and CD11b surface expression following transfer even at d21 (Fig. 5 E), consistent with the notion that ABCs have the ability to self-renew while maintaining the ABC phenotype. Further, many of the CD138− ABCs that were transferred differentiated to CD138+ PBs within the first 7 d after transfer (Fig. 5 F), directly demonstrating a precursor-product relationship between ABCs and PBs, and suggesting that ABCs readily undergo such differentiation in vivo in this autoimmune setting. Interestingly, some transferred ABCs were able to adopt a GC phenotype at the later d14 and d21 timepoints (Fig. 5 G). This is consistent with a memory-like nature for CD11c+CD11b+ ABCs, in that they can give rise to both ASCs and secondary GCs. In addition, these data directly corroborate the rapid turnover of most ABCs that was inferred from both the BrdU labeling and transient depletion experiments.
ABCs contribute to renal disease and T cell activation
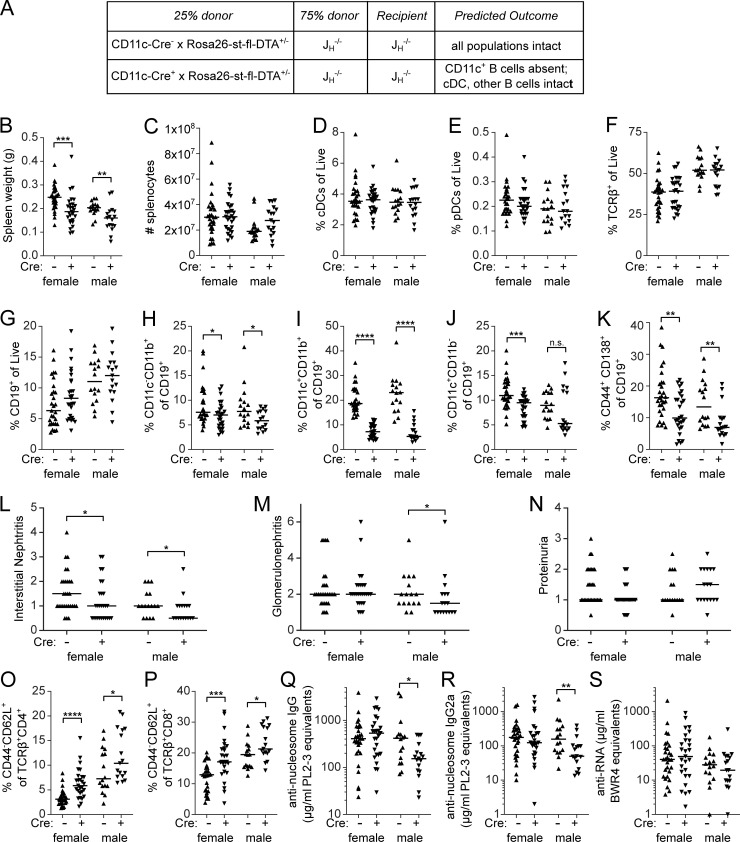
To evaluate whether ABCs are important in MRL/lpr disease outcomes, we needed a system to specifically deplete ABCs without affecting other CD11c-expressing populations. To do this, we generated mixed bone marrow chimeras comprised of 75% JH−/− MRL/lpr bone marrow and 25% either CD11c-Cre+ Rosa26-loxP-eGFP-stop-loxP-DTA (R26-DTA) MRL/lpr marrow or control CD11c-Cre− R26-DTA marrow, transferred into irradiated JH−/− MRL/lpr recipients (Fig. 6 A). JH−/− MRL/lpr bone marrow does not generate B cells, including ABCs, but is capable of reconstituting all other hematopoietic compartments including CD11c-expressing non-B cells such as conventional and plasmacytoid dendritic cells (cDCs and pDCs). The R26-DTA MRL/lpr donor is capable of generating all hematopoietic compartments, including B cells; when CD11c-Cre is also present, any cell (including ABCs, cDCs, or pDCs) that turns on the expression of the CD11c promoter should be deleted by DTA (DT subunit A) soon after CD11c promoter activity is induced (Teichmann et al., 2010). In these mixed chimeras, cDCs and pDCs will be provided by the JH−/− marrow while CD11c-expressing ABCs should be deleted.
Figure 6.
Constitutive genetic depletion of CD11c+ B cells improves renal disease and reduces T cell activation (A) Mixed bone marrow chimeras were generated in JH−/− MRL/lpr recipients with indicated bone marrow. Data are pooled from three independently generated cohorts of chimeras. (B–S) Recipients were assessed at 24 wk after chimerism for (B) spleen weight; (C) spleen cell number; proportion of (D) CD11c+MHCII+ conventional dendritic cells of live, (E) CD317+ SiglecH+ plasmacytoid dendritic cells of live, (F) TCRβ+ T cells of live, (G) CD19+ B cells of live, (H) CD11c− CD11b+ ABCs among B cells, (I) CD11c+CD11b+ ABCs among B cells, (J) CD11c+CD11b− ABCs among B cells, (K) CD44+ CD138+ PBs among B cells. (L–M) H&E-stained kidney sections were assessed for (L) interstitial infiltrates and (M) glomerulonephritis. (N) Urine was assessed for proteinuria by albustix. (O and P) Frequency of CD44lowCD62L+ (naive) cells among (O) CD4 T cells or (P) CD8 T cells. (Q) anti-nucleosome IgG, (R) anti-nucleosome IgG2a, and (S) anti-RNA IgG were measured by ELISA. Statistical comparisons by one-tailed Mann-Whitney; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Mixed bone marrow chimeric mice were evaluated for clinical disease and splenic immune populations at 24 wk after generation. Chimeric mice in which the R26-DTA donor was CD11c-Cre+ had slightly smaller spleens than CD11c-Cre− R26-DTA controls, although the total number of nucleated splenocytes was not significantly different between the groups (Fig. 6, B and C). We did not observe a difference in the relative proportion of cDCs, pDCs, T cells, or total B cells between the two groups (Fig. 6, D–G).
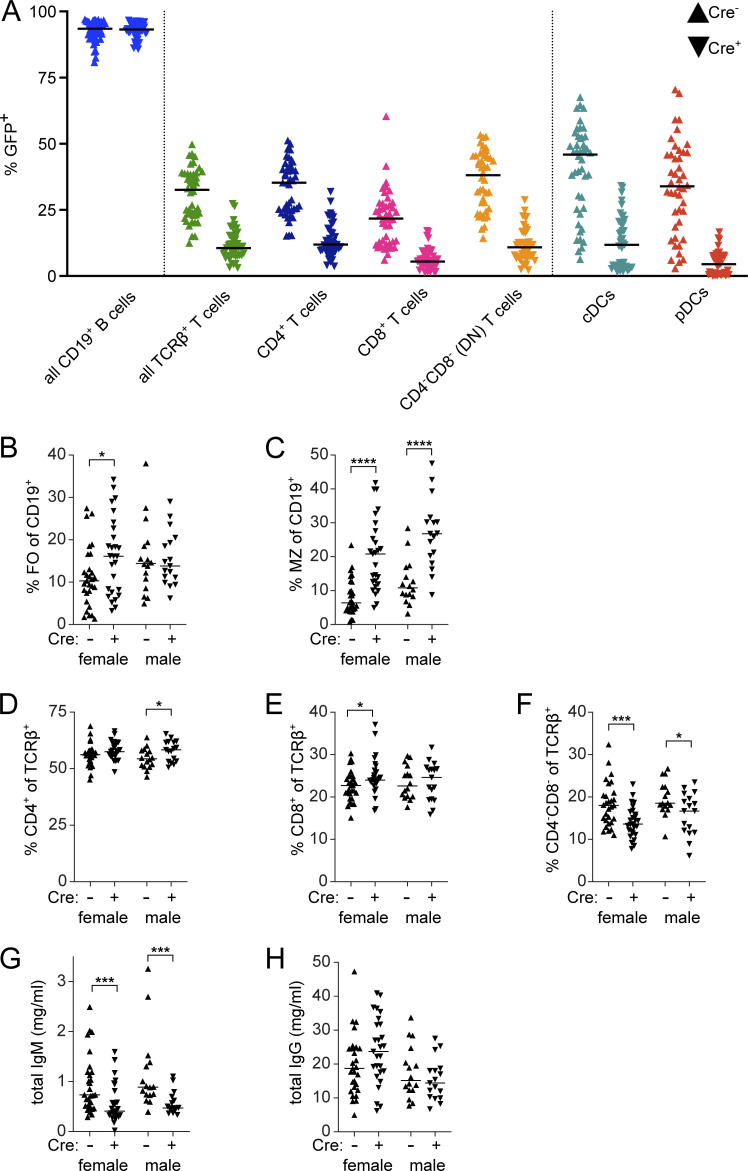
Using eGFP as a marker of the contribution made by the R26-DTA donor to any particular compartment, we evaluated the degree of chimerism in various populations (Fig. S2 A). As expected, all B cells were derived from the R26-DTA (eGFP+) donor, not the JH−/− donor, in both Cre+ and Cre− mice. Similarly, cDCs and pDCs were derived primarily from the JH−/− (non-GFP) donor when CD11c-Cre was present, but were produced from both donors when CD11c-Cre was not present. Unexpectedly, however, CD11c-Cre expression significantly limited the contribution from the non-JH−/− donor, even in compartments in which CD11c should not be expressed, including T cells. This is consistent with a recent report (Hou et al., 2020) describing a low level of CD11c expression in hematopoietic stem cells, particularly under inflammatory conditions, as found in aged MRL/lpr mice. This may have reduced the overall contribution from the Cre+ donor bone marrow, due to cell death at an earlier developmental stage than intended. Nonetheless, likely due to homeostatic mechanisms that govern individual compartments, the overall size of the various immune compartments was not different between the two groups; thus, this effect would not confound the interpretation of the disease phenotype comparisons.
Figure S2.
Constitutive deletion of CD11c+ B cells in bone marrow chimeras. (A) The extent to which the R26-DTA donor contributed to indicated subsets in recipient mice at 24 wk after chimerism was assessed by flow cytometry for presence of GFP in CD11c-Cre− (up-facing triangles) and CD11c-Cre+ mice (down-facing triangles). Data are pooled from males and females across all cohorts. (B–F) Proportion of (B) CD23+CD21/35− FO or (C) CD21/35+CD23− MZ B cells of CD19+ cells or (D) CD4+, (E) CD8+, or (F) CD4−CD8− (DN) cells among TCRβ+ T cells. (G and H) Total IgM (G) and total IgG (H) were measured by ELISA.
As designed, CD11c+CD11b+ ABCs were strongly affected, with a four- to fivefold reduction as a proportion of B cells in both genders (Fig. 6 I) in the frequency of ABC-like B cells in the CD11c-Cre+ group (Fig. 6, H–J). Depletion of CD11b+ B cells, though statistically significant, was modest (Fig. 6 H). Interestingly, CD11c+ single-positive B cells, which tend to have lower expression of CD11c than do CD11c+CD11b+ cells (Fig. 1 A), were also less affected (Fig. 6 J). It is important to recognize that, despite substantial reduction, the CD11c+CD11b+ compartment was not completely eliminated, as it still comprised 5–7% of B cells in the Cre+ mice. Of note, the PB compartment was also reduced by about 50% when CD11c-driven Cre was present (Fig. 6 K), similar to the degree of depletion of PBs in acutely DT-treated CD11c-DTR MRL/lpr mice (Fig. 4 D), which suggests that some but not all PBs are of ABC origin. The reduction in ABC frequency was accompanied by a relative increase in the proportion of follicular B cells in female chimeras and MZ B cells in both genders (Fig. S2, B and C).
Deletion of ABCs in these chimeras was accompanied by a decrease in histologic renal disease. Interstitial nephritis was significantly reduced in female and male chimeras in which ABCs were partially depleted (Fig. 6 L), and glomerulonephritis was reduced in the same males (Fig. 6 M). Proteinuria, which was mild in these radiation chimeras, was not further improved by partial depletion of ABCs (Fig. 6 N). Targeting ABCs resulted in a modest reduction in CD4−CD8− (double-negative [DN]) T cells and a proportionate increase in CD8 or CD4 T cells (Fig. S2, D–F). Within the CD4 and CD8 T cell populations, we observed an increase in CD44lowCD62Lhigh naive populations (Fig. 6, O and P), indicative of less B cell–driven immune activation and consistent with reports that ABCs can be effective APCs (Hao et al., 2011; Rubtsov et al., 2015). Anti-nucleosome IgG and IgG2a autoantibodies were reduced in male but not female chimeras in which ABCs were deleted (Fig. 6, Q–R) but anti-RNA IgG was unaffected (Fig. 6 S). Total serum IgM but not IgG was reduced in the CD11c-Cre+ group (Fig. S2, G and H). Therefore, genetic depletion of ABCs and CD11c+ ASCs reduced renal disease and splenic T cell activation with only modest effects on autoantibody titers, despite the reduction in PB frequency. It is likely that the observed escape from deletion served to limit the effects, such that the effects of ABCs on disease were underestimated by the system.
ABCs and PBs contain related, expanded B cell clones
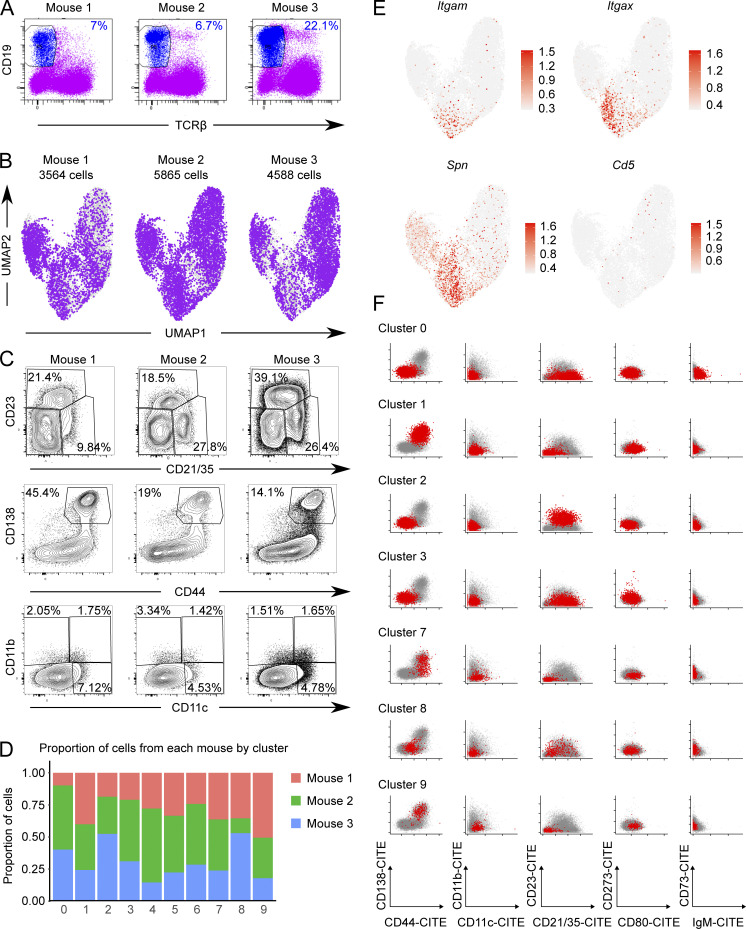
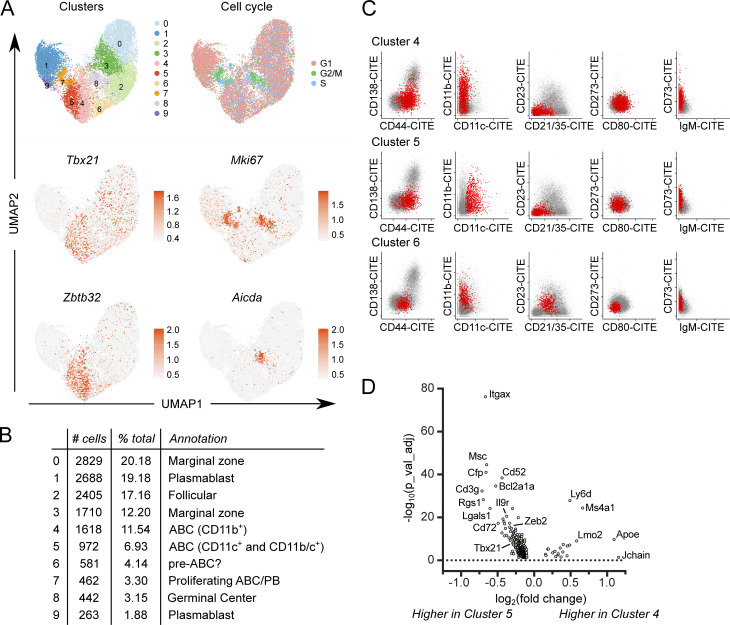
To elucidate the relationships of ABCs to other B cell subsets, including PBs, we performed scRNA-seq along with V region sequencing on total MRL/lpr CD19+ B cells. Total B cells (DAPI− CD19+ TCRβ−; Fig. S3 A) were sorted from the spleens of three 18-wk-old MRL/lpr females and stained with oligo-tagged sample hashing and surface feature antibodies, then pooled for scRNA-seq with V(D)J analysis. A total of 14,017 cells were recovered (Fig. 7 A and Fig. S3, B and D). Weighted nearest-neighbor clustering was performed on the basis of the gene expression and a 10 marker surface feature library, with cluster annotations determined based on differential gene and marker expression among clusters (Fig. 7, Fig. S3, and Fig. S4).
Figure S3.
Isolation and characterization of CD19+ MRL/lpr B cells by scRNA-seq. (A) CD19+ TCRβ− sort gates are indicated. Sorted B cells were stained with oligo-tagged sample hashing and surface feature antibodies prior to being pooled and processed via the 10× Chromium Next GEM Single Cell V(D)J v1.1 protocol per the manufacturer’s instructions. (B) Purple dots indicate contribution of cells from each donor mouse to the overall clustering UMAP. (C) A separate aliquot of total splenocytes from the same mice in A were stained with the indicated markers for conventional flow cytometry. (D) Summary of the contribution of each mouse to the clusters in Fig. 7 A as determined by the sample-hashing oligo-tagged anti-CD45/anti-β2m antibodies included in the surface feature library. (E) Expression of Itgam (CD11b), Itgax (CD11c), Spn (CD43), and Cd5 genes overlaid on the cluster UMAP. (F) Expression of CITE-seq mapped surface markers in scRNA-seq. Pairwise plots of normalized counts from oligo-tagged surface feature antibodies for B cell clusters not included in Fig. 7 C.
Figure 7.
scRNA-seq identifies multiple ABC-like clusters. (A) WNN clustering was performed on CD19+ B cells from 18-wk MRL/lpr females using Seurat 4.0.0 on the basis of gene expression and surface feature libraries. Expression of cell cycle–related gene signatures is shown (top right). Normalized expression of indicated genes is shown (middle and bottom rows). (B) Cluster size, frequency as a percentage of total, and annotations are indicated. (C) Pairwise plots of normalized counts from the oligo-tagged surface feature antibody library for ABC clusters 4, 5, and 6 are shown. Plots for other clusters are available in Fig. S3. (D) Volcano plot of DEGs between ABC-like clusters 4 and 5 showing 335 genes with log2(fold change) >0.1 and false discovery rate < 0.05.
Figure S4.
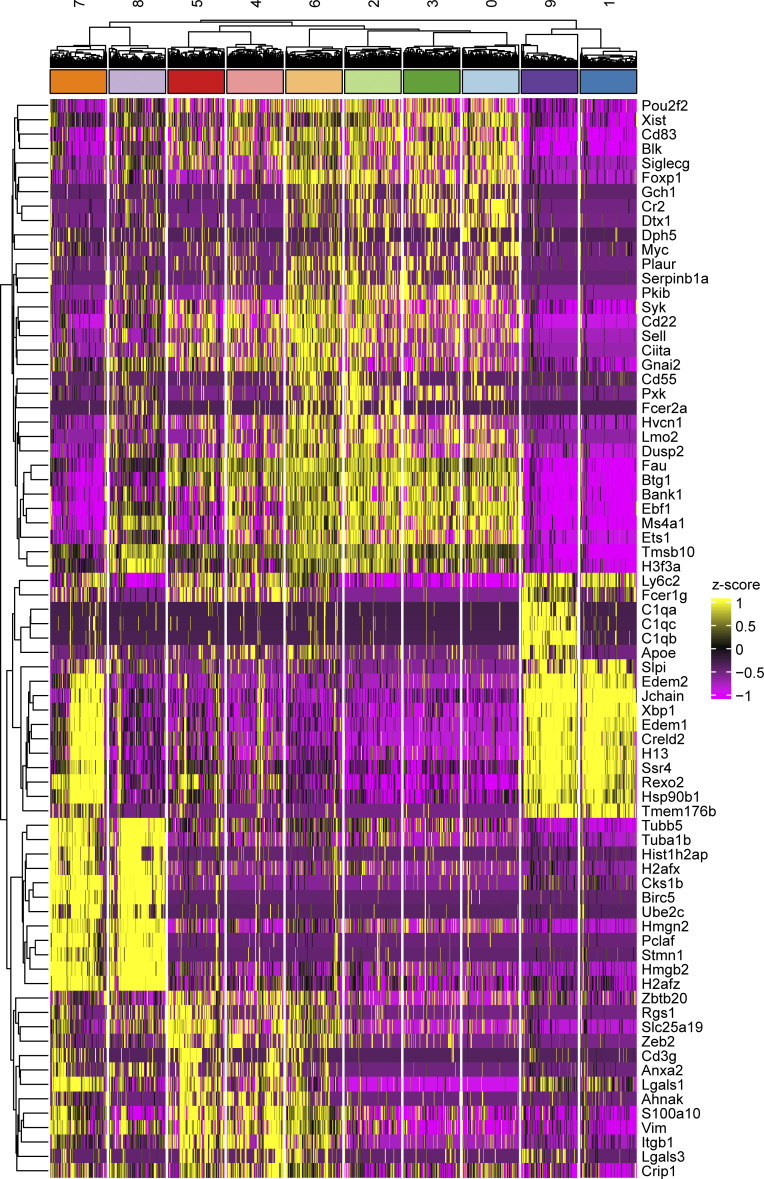
DEGs in scRNA-seq clusters. Heatmap showing z-scored expression of the top 10 most differentially expressed genes per cluster. Clusters are grouped based on unbiased hierarchical clustering.
ABCs resolved as a distinct B cell lineage that separated into three related clusters, clusters 4–6, primarily on the basis of CD11c vs. CD11b surface marker expression (Fig. 7, B and C; Fig. S3, E and F; and Fig. S4). Consistent with ABC surface marker expression by conventional flow cytometry (Figs. 1 and 2), the ABC clusters 4 and 5 were CD44high, negative for CD21 and CD23 expression, and expressed varying amounts of the B cell memory markers CD80, PD-L2, and CD73 as measured by Cellular Indexing of Transcripts and Epitopes by Sequencing (CITE-seq) tags (Fig. 7 C), as well as expressing transcripts for Tbx21 and Zbtb32, transcription factors associated with the ABC lineage (Fig. 7 A). ABC clusters 4 and 5 were transcriptionally similar to one another, although cluster 5 expressed higher levels of Itgax transcript and slightly higher levels of Tbx21 and Zeb2 than cluster 4 (Fig. 7 D and Fig. S3 E). Nearby cluster 6 expressed intermediate levels of surface CD44, CD21, and CD23, and was negative for CD80 and PD-L2, but was CD73int and CD11b+ (Fig. 7 C). Cluster 6 was also Tbx21 and Zbtb32 intermediate (Fig. 7 A) and expressed transcripts found in FO/MZ clusters (0, 2, and 3) as well as ABC transcripts (clusters 4 and 5; Fig. S4). Thus, cluster 6 may represent an early activated cell transitioning into the ABC compartment (labeled “pre-ABC?” in subsequent figures).
There were two proliferative clusters (7 and 8) with the highest G2/M gene signatures and Mki67 gene expression (Fig. 7 A). Of these, many cells in cluster 8 expressed Aicda transcript and GC-associated transcripts such as Gcsam (Fig. 7 A and Fig. S4). Though histologic GCs are rarely detected in older MRL/lpr mice (Luzina et al., 2001; William et al., 2002), based on transcriptomics this cluster was annotated as “GC,” although this cluster could also include B cells undergoing extrafollicular somatic mutation and proliferation. Cluster 7 included proliferating cells that were CD44high, CD11c+ but CD11b−, CD21−CD23−, CD80+CD73+ with varying levels of CD138 expression (Fig. S3 F). These cells were annotated as proliferating ABCs and PBs (“prolif. ABC/PB”) and likely represent the overlap between the two populations (Fig. 1, D–G).
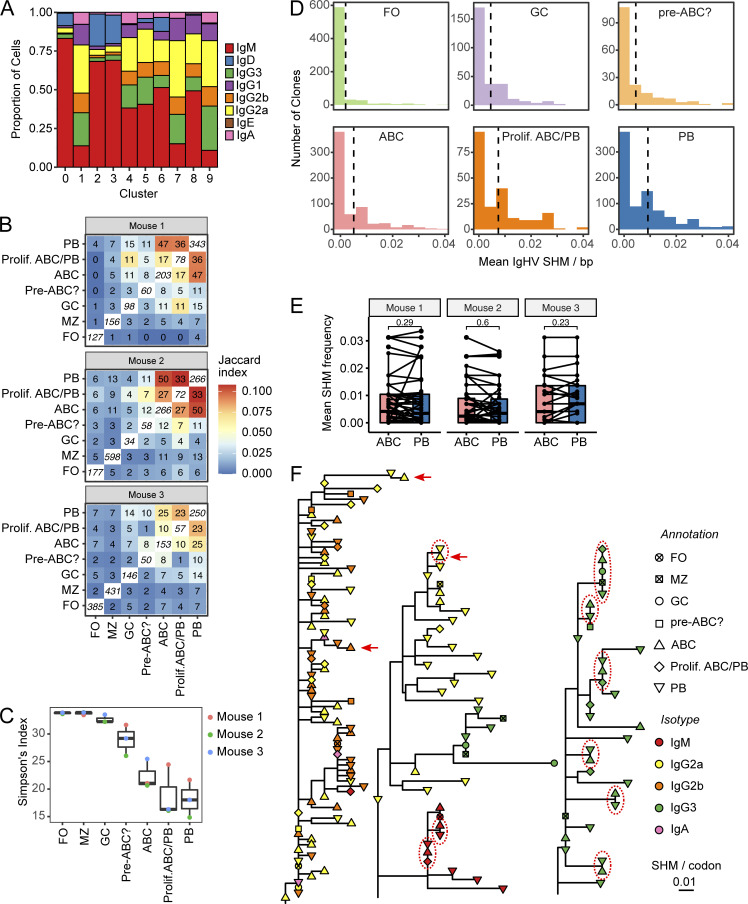
The extent of class switching observed in the 5′ VDJ library was greatest in the PB (clusters 1 and 9) and proliferating ABC/PB clusters (cluster 7), with intermediate amounts of class switch observed in the ABC (clusters 4 and 5), pre-ABC (cluster 6), and GC (cluster 8; Fig. 8 A). BCR sequence lineage tracing revealed significant clonal overlap among ABCs, proliferating ABC/PB, and PBs (Fig. 8 B). Commensurately, clonal diversity was lowest in the PB and proliferating ABC/PB clusters and intermediate in the ABC and pre-ABC clusters, indicating expansion of selected clones in these populations (Fig. 8 C). Interestingly, clonal diversity in the GC cluster was more similar to that of naive FO and MZ clusters than the PB cluster (Fig. 8 C), consistent with previous observations that spontaneous GCs in aged MRL/lpr are small and most PBs are generated by the extrafollicular pathway (William et al., 2002). The average BCR mutation frequency of ABC clones was higher than that of FO B cell clones but less than in the proliferating ABC/PB or PB clusters (Fig. 8 D and Fig. S5), consistent with the inferred differentiation of ABCs to terminally differentiated PBs. However, B cells from individual clones that were observed in both ABC and PB compartments had similar mutational content in both compartments (Fig. 8 E).
Figure 8.
ABCs and PBs share related expanded B cell clones with similar mutational burden. (A) Proportion of cells in each cluster with the indicated heavy chain isotype, based on 5′ VDJ library sequencing. (B) Normalized clonal overlap for indicated B cell subsets. Italicized numbers along diagonals indicate number of identified unique clones in each subset. Off-diagonal numbers indicate the number of overlapping clones between the subsets in the row and the column. Color of each box indicates the Jaccard similarity coefficient for each row and column. (C) Simpson’s diversity index was calculated for the indicated subsets from each of the three donor mice. (D) For each clone, the mean number of mutations per V segment was determined for each subset as indicated. The mean of the distribution for each subset is indicated by vertical dotted lines. Note that for clarity, only clones with at most 4% mutation frequency are included; for distributions including the full range of mutation frequency, see Fig. S5. (E) Mean number of mutations per V segment was compared for clones that were detected in both ABC and PB subsets. (F) Largest B cell lineage trees in each mouse. Horizontal distance indicates mutations per codon between tree nodes (see scale bar). Shape indicates annotated subset for each tip, while symbol color indicates Ig isotype. A few examples of identical clones in ABC and PB subsets are highlighted with red dotted circles; a few examples of an ABC with more mutations than a related PB are indicated by red arrows.
Figure S5.
Mutation frequency in MRL B cell subsets. (A) Distribution of mean number of mutations per V segment for each subset. (B) Distribution of mean number of mutations per V segment for each cluster.
We constructed clonal lineage trees from the most highly expanded B cell clones in each mouse and observed numerous examples of ABCs, proliferating ABC/PB, and PBs bearing identical, mutated BCRs (Fig. 8 F, dotted circles), as well as examples of ABCs with more mutations than a related PB (Fig. 8 F, red arrows). These data, together with the earlier data showing ABC proliferation, self-renewal, and differentiation capacity, support a model in which ABCs can both be generated by and be precursors of proliferating and mutating B cells as well as PBs.
Discussion
In this work, we have, in a genetically driven system of spontaneous lupus-like disease, defined a group of ABC-like cells using a combination of flow cytometry, functionality, development and scRNA-seq, providing new insights into the heterogeneity of these cells. We further investigated the ontogeny of ABCs, demonstrating that an anti-nuclear BCR instructs ABC development in a TLR9-dependent manner. Transfer and labeling studies further showed that, although many ABCs have MBC markers, they are a rapidly turning over population in the context of lupus. Single-cell V region sequencing combined with CITE-seq markers allowed us to track clones through various post-activation B cell compartments, revealing that ABCs can be direct PB precursors and that B cells can undergo cycles of activation and then differentiation back into ABCs, without entering a stable, long-term memory compartment. This conclusion was also supported by our transfer experiments, which demonstrated direct precursor-product relationships between ABCs and PBs. Perhaps most importantly, although B cell–expressed T-bet was shown in another study to be important for pathogenesis in different lupus models (Rubtsova et al., 2017), by taking a genetic cell depletion approach we could demonstrate that ABCs per se directly contribute to pathogenesis. Hence, overall, this work substantially enhances our understanding of the origins, biology, and functional impact of ABC-like cells.
ABC-like cells can differ across biological systems, and their various phenotypes and functions may be context dependent. Within a defined murine model, and in fact within a single mouse, we were able to define multiple populations of CD11c+, CD11b+ and CD11c+CD11b+ (DP) cells that had ABC-like qualities. Importantly, we observed that not all of these cells expressed high levels of T-bet. Thus, while T-bet has been proposed as a defining transcription factor of ABCs (Rubtsov et al., 2013; Rubtsova et al., 2013), our data suggest its expression is associated with ABC-like B cells but not causal of or directly linked to ABC development. Indeed, ABCs can develop in the absence of T-bet, and ABCs that have been induced can still function apparently normally in a recall response when T-bet has been deleted (Du et al., 2019b; Levack et al., 2020; Ricker et al., 2021).
The functional correlates of ABC-like phenotypic heterogeneity have been unclear. In MRL/lpr mice, we were able to tease apart specific functional and ontogenetic differences among CD11b and CD11c single-positive vs. DP populations. These included differences in the frequency of PB-like cells and classical MBC subsets, although all CD11b/c subsets were more proliferative than FO or MZ B cells. Within each of the ABC-like subsets, there was heterogeneity in terms of classic murine MBC subset markers: CD80, PD-L2, and CD73. Thus, if the documented functional differences between MBCs that lack both CD80 and PD-L2 (DN MBCs) or express both markers (DP MBCs) apply (Zuccarino-Catania et al., 2014), then each type of ABC-like cell itself could contain multiple, functionally distinct subsets. This further suggests that the ABC program may be considered as an overlay on top of the pathways that govern development of DN and DP classical MBC subsets (or vice versa), perhaps reflecting the quality or duration of local inflammatory conditions at the time of initial B cell activation, or the history of the cell with respect to reactivation. Notably, even among B cells with a restricted BCR specific for DNA, the resulting ABC-like cells are heterogeneous, suggesting that factors other than just specificity can control ABC subset differentiation. TLR9 expression altered the distribution of B cells among the ABC-like subsets in vivo with or without anti-DNA BCR restriction, extending prior work showing that TLR7 contributes to ABC formation (Rubtsov et al., 2011).
ABCs have been considered a type of MBC (Naradikian et al., 2016). In other systems, ABCs (and/or T-bet+ B cells) clearly have MBC markers (Du et al., 2019a; Johnson et al., 2020; Trivedi et al., 2019; Yates et al., 2013). In systems where there is no ongoing infection or inflammatory process, T-bet+ ABCs/MBCs seem to be resting cells with a relatively longer lifespan (Johnson et al., 2020; Naradikian et al., 2016; Trivedi et al., 2019). It is widely thought that MBCs can only form after cells disengage from cognate antigen; this seems unlikely to be the case for self-reactive B cells during ongoing autoimmunity. Although ABCs have been called MBCs in models of murine lupus, their actual dynamics (whether resting and stable or dynamic and turning over) has not been clarified. Thus, an important contribution of our work is the use of labeling to characterize the turnover of ABCs in situ, showing that in autoimmune mice they are a highly dynamic population, and not simply resting B cells. Combined with the scRNA-seq data in which we track clonal lineages, our data reveal a compartment in continuous flux and evolution, with some of the cells having a memory phenotype, but these being reactivated in a cyclic fashion. Hence, ABC-like cells are continually contributing to ongoing autoimmune activation.
These data also help to explain the relatively high V region mutation frequency of autoantibodies found in both murine models and humans, despite that many of them likely were not produced in GC, but rather through extrafollicular responses (Elsner and Shlomchik, 2020; Jenks et al., 2018; William et al., 2002). That such extrafollicular responses do undergo both somatic hypermutation (SHM) and isotype switch is no longer in dispute, but it does seem that the proliferative phases of these responses are transient and that either mutation rates among extrafollicular blasts are lower or that B cells in the responses only remain in the hypermutable state for relatively short periods. However, if such responses spawn ABC-like MBCs, which are then drawn back into proliferative phases in a cyclic fashion, high numbers of mutations could accumulate and selection on them could take place. Our data—particularly the clonal analysis of the scRNA-seq—provide direct support for this model.
In addition to the basic knowledge about ABC-like biology in the context of autoimmunity, this model provided the opportunity to directly test whether ABC-like CD11c+ B cells were pathogenic. This issue was partly addressed in prior work in which deletion of T-bet in B cells resulted in reductions, albeit somewhat modest, in disease in several lupus models (Rubtsova et al., 2017). At the time, it was not appreciated that T-bet is not needed for ABC-like cell development, nor for at least many of their functions (Du et al., 2019b; Levack et al., 2020; Ricker et al., 2021). Here we engineered the direct depletion of CD11c+ B cells without affecting CD11c+ myeloid cells. Disease was indeed ameliorated, demonstrating that depletion of ABCs can prevent lupus disease and thus that these cells actively promote such disease. That said, disease amelioration was rather limited compared to that of complete B cell deletion (Chan et al., 1999). This could be because B cells other than ABCs can substitute for them, suggesting that in reality, ABCs play only a limited role. However, more likely, the reduced disease is a minimal reflection of the true role of ABCs, as despite the genetic approach, some CD11c+ ABCs remained in the experimental animals, in addition to the CD11b+ population of ABC-like cells. The presence of residual CD11c+ and CD11b+CD11c+ B cells could reflect positive selection of autoreactive B cells that escape deletion, due to the inevitable inefficiency of Cre-based strategies. This would, in contrast, suggest an important role for ABCs. Similarly, deletion of MHCII in all B cells, which would impair B cell APC function directly, had a limited effect on disease, due to expansion of residual MHCII-positive B cells (Giles et al., 2015). These reductions in disease when targeting ABCs, even with technical limitations, along with the dynamic role of ABCs in contributing to autoantibodies and autoreactive plasmablasts, as well as potential APC function (Hao et al., 2011; Rubtsov et al., 2015), further support the development of specific ABC-like targeting approaches to treat lupus and other autoimmune diseases. The elucidation of the basic ABC-like cell biology in a relevant lupus model presented here will aid such efforts.
Materials and methods
Mice
MRL/lpr mice (#000485; Jackson Laboratory stock) were bred locally from commercial stocks. CD45.2 MRL/lpr were isolated during the backcrossing of IL-10flox MRL/lpr (Teichmann et al., 2012) after more than eight backcross generations by identifying founders in which the Il10 and Ptprc traits were unlinked, and progeny were bred to homozygosity for the CD45.2 allele. CD11c-DTR MRL/lpr were generated by backcrossing BALB/c FVB-Tg(Itgax-DTR/EGFP)57Lan/J (#004512; Jackson Laboratory stock) onto the MRL/lpr background for more than eight backcross generations. CD11c-Cre MRL/lpr, Rosa-flox-stop-flox-DTA MRL/lpr, JH−/− MRL/lpr, tg3H9 MRL/lpr, and Tlr9−/− MRL/lpr have been previously described (Chan et al., 1999; Nickerson et al., 2013; Teichmann et al., 2010). Male and female mice were used for all experiments unless otherwise noted in legends. Mice were housed under specific pathogen–free conditions, and all experiments were performed with the approval and oversight of the University of Pittsburgh Institutional Animal Care and Use Committee.
Flow cytometry and cell sorting
Splenocytes were prepared by manual disruption of spleens into cold staining medium (1× PBS + 3% calf serum + 5 mM EDTA + 0.05% sodium azide) followed by red blood cell lysis with ammonium chloride/potassium chloride (ACK) solution. Single-cell suspensions were washed with PBS and labeled with Ghost 510 live/dead reagent (Tonbo). Washed splenocytes were blocked with unconjugated anti-CD16/32 (clone 2.4G2) in staining medium and then stained with surface antibody cocktails on ice. Following washing and secondary staining with streptavidin-conjugated fluorophores where necessary, cells were washed in staining medium and fixed with 1% paraformaldehyde in 1× PBS. For T-bet intracellular staining, cells were permeablized with 0.1% Triton-X 100 in staining medium for 20 min on ice, then washed and blocked with staining medium containing 10% mouse serum and incubated with anti–T-bet antibody overnight. For Ki67 intracellular staining, cells were first fixed in 2% paraformaldehyde in 1× PBS then permeabilized with 1× Foxp3 Perm/Wash buffer (eBiosciences) for 30 min on ice, washed with Foxp3 Perm/Wash buffer, blocked with Foxp3 Perm/Wash buffer supplemented with 10% rat serum, then stained with anti-Ki67 antibody overnight. Following overnight incubations, cells were washed twice with Perm/Wash then twice with staining medium. Data were collected on a Becton Dickinson LSR II or Fortessa and analyzed in FlowJo.
Antibodies and flow cytometry reagents used in this study were: CD8-Alexa647 (TIB105), CD11c-Alexa488 (N418), CD11c-Alexa647 (N418), CD21/35-Alexa647 (7G6), CD44-Alexa488 (Pgp1), CD44-Alexa647 (Pgp1), CD45.1-Alexa647 (A20), CD45.2-Alexa488 (104), CD317-Alexa647 (eBio927)—lab grown from hybridoma and fluorophore conjugated; PNA-Alexa647 (Vector labs)—lab conjugated; CD4-BV421 (GK1.5), CD4-PE (GK1.5), CD8a-APC-Cy7 (53-6.7), CD11c-PE-Cy7 (N418), CD11b-APC-Cy7 (M1/70), CD11b-PE (M1/70), CD21/35-PerCP-Cy5.5 (7E9), CD23-PE-Cy7 (B3B4), CD38-PE (90), CD44-APCCy7 (IM7), CD62L-APC-eFluor780 (Mel-14), CD93-PE (AA4.1), CD138-BV605 (281-2), CD138-PE (281-2), I-A/I-E-BV605 (M5/114.15.2), T-bet-PE (4B10), TCRβ-PerCP-Cy5.5 (H57-597), TCRβ-BV421 (H57-597)—Biolegend; CD11b-BUV737 (M1/70), CD19-BUV395 (1D3), CD44-BV605 (1M7), CD73-PE (TY/11.8), CD80-BV421 (16-10A1), CD138-BV605 (281-2), CD273-biotin (TY25), SiglecH-BUV737 (440c), Streptavidin-BUV737—Becton Dickinson; CD45.1-APC-eFluor780 (A20), Ki67-FITC (SolA15), BrdU-Alexa647 (Mobu1)—eBiosciences/Invitrogen. All antibodies were titrated prior to use.
BrdU labeling and detection
Mice were provided ad libitum with drinking water containing a final concentration of 0.8 mg/ml BrdU and 1% sucrose; water bottles were changed daily during the labeling period. Splenocytes were isolated and surface stained as described above. After surface staining, cells were washed and resuspended in 0.6 ml cold 0.15 M sodium chloride. 1.2 ml cold 100% ethanol was added dropwise to each tube with vortexing and cells were incubated on ice for at least 30 min. Cells were then pelleted, washed with staining medium, and incubated overnight in 1% paraformaldehyde + 0.05% Tween-20 in 1× PBS. The next day, cells were pelleted and resuspended in a solution containing 100 Kunitz Units per ml DNase in 0.15 mM sodium chloride and 4.2 mM magnesium chloride in water. Cells were incubated for 30 min at room temperature, then washed with staining medium and transferred to 96-well plates. Cells were blocked in 10% mouse serum in staining medium then stained overnight with anti-BrdU antibody. The next day, cells were washed twice in staining medium and analyzed on a flow cytometer as above.
DT treatment
CD11c-DTR MRL/lpr mice and transgene-negative littermate controls were i.p. injected with a single dose of DT (List Biological Labs) at a concentration of 8 ng/kg body weight in PBS at 1, 4, or 7 d prior to experimental analysis.
Cell sorting, culture, and ELISA
MRL/lpr splenocytes were aseptically prepared by mechanical disruption into RPMI containing 2.5% FetalPlex + 1% Penn/Strep + 1% L-glutamine + 10 mM Hepes + 50 mM β-mercaptoethanol + 0.25 mg/ml gentamycin. Pooled splenocytes were RBC-lysed with ACK solution (Lonza) stopped with an excess of RPMI. Cells were pelleted and resuspended in separation media (1× PBS + 2% Fetalplex + 5 mM EDTA) and filtered through a 70-μm strainer. Cells were counted and the volume adjusted to yield a concentration of 2 × 108 cells/ml in separation medium supplemented with rat serum and unconjugated anti-CD16/32 (2.4G2; lab made). Cells were then stained by adding an equal volume of a 2× cocktail of biotinylated antibodies directed against Gr1 (RB6.8C5; Biolegend), CD49b (DX5; Biolegend), CD4 (GK1.5; lab made), CD8 (TIB105; lab made), CD90.2 (30H12; lab made), and Ter119 (Ter119; lab made) for 15–20 min on ice. Cells were washed in separation media and then resuspended at a concentration of 2 × 108 cells, to which a 2× solution containing iMag streptavidin-coated magnetic beads (Becton Dickinson, final concentration 80 μl iMag beads per ml) was added. After 5 min, cells were split in up to 8 ml aliquots in 14 ml polystyrene round-bottomed tubes, which were loaded into EasySep magnets. After 3 min, supernatants were poured into collection tubes. Tubes were removed from the magnets, washed with separation medium, and reloaded into the magnets to recover additional unbound cells. B cell enriched supernatants were washed, counted, and resuspended at a final concentration of 2 × 108 then stained by adding an equal volume of 2× solution containing fluorophore-conjugated antibodies against CD19, TCRβ, CD11c, CD11b, CD44, and CD138. Cells were washed and resuspended in 0.3 μM DAPI immediately prior to sorting. Cells were sorted on a BD FACSAria to isolate the indicated populations.
For cell culture, sorted cells were washed and resuspended in RPMI containing 10% FetalPlex + 1% Penn/Strep + 1% L-glutamine + 0.1 mM Hepes + 50 μM β-mercaptoethanol + 0.5 μg/ml gentamycin. 4 × 105 cells were cultured per well in 24-well plates containing media alone or media containing 2 μg/ml CL097 (TLR7 ligand) or 2 μg/ml ODN 1826 (TLR9 ligand). After 6 d in culture, cells and media were transferred to 1.5 ml Eppendorf tubes, centrifuged, and supernatants were transferred to fresh tubes. Total IgG, anti-nucleosome, and anti-RNA ELISAs were performed on cell culture supernatants diluted at 1:100 (total IgG) or undiluted (anti-nucleosome and anti-RNA) as described below.
Adoptive transfers
MRL/lpr splenic ABCs and naive B cells were isolated by sequential negative magnetic selection and cell sorting as described above. Sorted populations were washed in PBS and labeled with 1 μM VPD (Becton Dickinson) at 37°C for 10 min per the manufacturer’s instructions. Cells were washed with transfer buffer (1× PBS + 0.1 mM Hepes + 0.5% Penn/Strep + 0.625% ACD-A) and between 2 × 105 - 4 × 105 cells were transferred to 5–8-wk-old CD45.2 MRL/lpr through injection into the lateral tail vein in 200 μl transfer buffer.
Mixed bone marrow chimeras
Bone marrow from donor CD11c-Cre+ Rosa26-flox-stop-flox-DTA MRL/lpr, CD11c-Cre− Rosa26-flox-stop-flox-DTA MRL/lpr, and JH−/− MRL/lpr mice was isolated from tibias, femurs, and humeri by flushing the interior of the bone shafts with RPMI using a 27G1/2 needle and 1 ml syringe. Marrow was pooled by genotype and gender, and single-cell suspensions made by mechanical pipetting and passage through a 100-μm strainer. Pelleted cells were resuspended in ACK (Lonza) to lyse red blood cells. Marrow was washed with PBS, passed through a 40-μm cell strainer, counted, and mixed at a ratio of one part (CD11c-Cre+ or CD11c-Cre−) Rosa26-flox-stop-flox-DTA MRL/lpr marrow to three parts JH−/− MRL/lpr marrow in transfer buffer. 5 × 106 total bone marrow cells were introduced into recipient JH−/− MRL/lpr mice through injection into the lateral tail vein. Recipient mice were subject to whole body irradiation as two doses of 450 rads separated by 2–4 h, with an additional 1–2 h of rest, prior to receipt of donor marrow. Recipient mice were provided autoclaved drinking water and food containing 275 ppm trimethoprim and 1,365 ppm sulfadiazine (Uniprim, Envigo) ad libitum for 2–3 wk following irradiation and bone marrow transplant. Mice were analyzed at 24 wk after chimerism. Proteinuria was evaluated by Albustix (Siemens). Kidneys were bisected and fixed in 10% formalin. H&E slides were prepared by StageBio and scored by an observer blinded to the experimental group. Glomerulonephritis was scored on a scale of 1 to 6 (1, normal kidney; 2, mesangial expansion and increased mesangial cellularity and patent capillary loops; 3, enlarged glomeruli with moderate endocapillary hypercellularity; 4, 3+ with marked endocapillary hypercellularity and loss of patency of most capillary loops; 5, few glomeruli with necrosis [karyorrhexis] or few active [cellular or fibrocellular] or organized [fibrous] crescents; 6, many active [cellular or fibrocellular] or organized [fibrous] crescents, necrosis [karyorrhexis], obliteration of glomerular architecture, segmental/global sclerosis). Interstitial nephritis was scored on a scale of 1 to 4 in a blinded manner (1, minimal inflammation [lymphocytes and plasma cells] confined to the perivascular area; 2, expansion of inflammation throughout the interstitial space but maintained in discrete area; 3, diffuse infiltrates in over 40% of high-powered fields; 4, diffuse infiltrate throughout the entire interstitial space).
ELISA
Anti-nucleosome ELISAs were performed by coating Immulon 2HB plates with 10 μg/ml poly-L-lysine (Sigma-Aldrich) in PBS. Plates were washed and coated with 15 μg/ml dsDNA prepared by digestion of calf thymus DNA (Sigma-Aldrich) with S1 nuclease (Promega) for 30 min at 37 deg followed by ethanol precipitation. Plates were subsequently washed and coated with 10 μg/ml calf thymus histones type IIAS (Sigma-Aldrich). Plates were blocked with ELISA buffer (1× PBS 1% BSA 0.05% sodium azide) and serum samples diluted 1:200 in the same buffer were applied to the top row and diluted threefold down the plate down to 1:5,400. Bound antibody was detected with alkaline-phosphatase conjugated goat anti-mouse IgG (Southern Biotech) or goat anti-mouse IgG2a (Southern Biotech) and developed with pNPP (Sigma-Aldrich). Autoantibody concentrations were determined relative to PL2-3 anti-nucleosome monoclonal antibody standard using DeltaSoft 2.8.11 software (Biometallics). Anti-RNA ELISAs were performed in a similar fashion, except plates were coated first with poly-L-lysine then with 15 μg/ml total yeast RNA (Sigma-Aldrich) before blocking and concentrations were determined relative to BWR4 standard. Total IgG and IgM ELISAs were performed by coating plates with unconjugated goat anti-mouse IgG or IgM antibody (Southern Biotech), followed by serum sample diluted 1:10,000 (IgM) or 1:50,000 (IgG) in the top row, followed by goat anti-mouse IgG-AP or IgM-AP (Southern Biotech), relative to purified IgG or IgM standards.
scRNA-seq
Splenocytes from three 18-wk-old female MRL/lpr mice were isolated and blocked with 2.4G2. Cells were stained with anti-CD19 and anti-TCRβ antibodies, and sorted in the presence of 0.3 μM DAPI on a FACSAria II cell sorter. Total CD19+ cells were stained with a cocktail of oligo-tagged TotalSeq-C antibodies as indicated including sample hashing anti-CD45/β2m antibodies. Cell concentrations were normalized and samples were pooled then run through a 10× Genomics Chromium following the manufacturer’s protocol for the Chromium Next GEM Single Cell V(D)J Reagent Kit v1.1 with Feature Barcoding Technology for Cell Surface Protein. Sample libraries were pooled and sequenced on the Illumina HiSeq PE150 platform by Novogene. Sequencing data were demultiplexed and FASTQ files were generated using the “mkfastq” Cell Ranger pipeline (v5.0.0, 10× Genomics). Cell Ranger “count” was used to align reads to the mm10 reference genome, and mRNA transcript, and hashtag oligonucleotide (HTO) unique molecular identifier (UMI) quantification tables were generated. The raw barcode matrix files generated from the Cell Ranger pipeline were further utilized for downstream analysis using the Seurat package (v4.0.0; Butler et al., 2018) https://github.com/satijalab/seurat in R (v3.4.3).
Cells expressing <200 genes or with >10% of UMIs mapping to mitochondrial DNA were filtered out. The HTO tables and antibody-derived tag (ADT) protein expression values were added to the dataset and normalized using the centered-log ratio transformation method using the “NormalizeData” function in Seurat. The normalized HTO count was used to determine whether each gel bead-in-emulsion contained a single cell using the Seurat “MULTIseqDemux” function to consider as “singlet,” otherwise “doublet.” Gene expression values for each cell were log2-normalized using the “NormalizeData” function, where expression of a gene was normalized to total expression of all genes in that cell and scaled by a factor of 10,000. UMI, mitochondrial content, hemoglobulin gene, Ig genes, and ribosomal gene content scores were “regressed-out” using Seurat’s “ScaleData” function.
Variable genes were detected using the “mean.var.plot” method in the “FindVariableFeatures” function with default cutoff, which computes the mean expression and dispersion (log(variance/mean)) per gene, followed by grouping the data into 20 bins based on their mean expression. Variable genes were then selected based on z-scored dispersion within each bin. These variable genes were used for dimensionality reduction based on principal component analysis (PCA) for gene expression and “APCA” for protein expression (ADT) data using the “RunPCA” function. For each cell, the weighted nearest neighbor (WNN) graph was calculated based on a weighted combination of RNA and ADT protein similarities using “FindMultiModalNeighbors” function. The WNN based on a weighted combination of RNA and protein data (ADT) were selected for further Uniform Manifold Approximation and Projection (UMAP) dimensional reduction and clustering analysis.
In order to identify distinct groups of cells, unsupervised clustering was performed using the “FindClusters” function, which calculates the k-nearest neighbors according to variable gene expression in all cells, thereby constructing a shared nearest neighbor graph using the Louvain algorithm. To avoid over clustering, we tested different resolution parameters, ranging from 0.1 to 2 in increments of 0.1, and the clustering progression was assessed and visualized using “Clustree” (v0.4.3; Zappia and Oshlack, 2018). Optimal resolution was determined based on continued separation prior to “overclustering” as observed by the increasing crossover arrows between clusters. Based on these observations, we chose the resolution 0.6 for clustering the data. Cell clusters were visualized using WNN-UMAP-dimensional reduction plots.
The “FindAllMarkers” function with default settings was utilized to find differentially expressed genes (DEGs) in each cluster, in comparison with all other clusters, using the Wilcoxon Rank Sum test with genes detected in a minimum of 10% of cells, a minimum of 0.25 average log-fold change, and a minimum of 0.01 Bonferroni-adjusted P value. The “CellCycleScoring” function was utilized to calculate the G1, G2/M, and S phase marker expression score in each cell (Nestorowa et al., 2016). Barplot, dotplot, and PCA plots were constructed using ggplot2 in R. Heatmaps were generated using the “pheatmap” function in R. All analyses and visualizations were performed in R. Computer code is available upon request. scRNA-seq data have been deposited in the NCBI Gene Expression Omnibus as GSE202110.
Inference of MRL/lpr germline IGHV gene sequences
Accurate somatic hypermutation analysis requires an accurate germline immunoglobulin heavy chain V region (IGHV) reference database. Because mouse strains have distinct genomes, we developed a de novo germline IGHV reference database for MRL/MpJ and MRL/lpr.
A whole dissected spleen, preserved in RNAlater (cat. no. AM7020; Thermo Fisher Scientific), was obtained from a female MRL/MpJ mouse from Jackson Laboratory (Jackson Laboratory stock #000486). We extracted total RNA from 30 mg of spleen using the RNeasy Mini kit (cat. no. 74104; Qiagen). An IgM 5′RACE AIRR-seq library was generated using the SMARTer Mouse BCR Profiling Kit (cat. no. 634422; Takara Bio), with custom IgM primers positioned in the CH3 and CH1 regions of IGHM (first round PCR, 5′-CAGATCCCTGTGAGTCACAGTACAC-3′, 12 µM; second round PCR, 5′-AATGATACGGCGACCACCGAGATCTACACTATAGCCTACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNNNNNGGGAAGACATTTGGGAAGGACTGAC-3′, 12.5 µM). The library was assessed using the Agilent 2100 Bioanalyzer High Sensitivity DNA Assay Kit (cat. no. 5067-4626; Agilent) and the Thermofisher Qubit 3.0 Fluorometer dsDNA High Sensitivity Assay Kit (cat. no. Q32851; Thermo Fisher Scientific). The library was diluted to 10 nM and sequenced on the Illumina MiSeq platform using the 600-cycle MiSeq Reagent Kit v3 (2 × 300 bp, paired-end; Illumina, cat. no. MS-102-3003). 47827 IgM read pairs were generated and used for downstream gene inference.
IgDiscover v0.12 (Corcoran et al., 2016) was used to construct a germline IGHV gene database for MRL/MpJ. Raw FASTQ reads were processed using the following IgDiscover parameters: (1) “barcode consensus” set to “false” because the sample did not contain a barcode; (2) “race_g” set to “true” to account for a run of G nucleotides at the start of the sequence; (3) “stranded” set to “true” since the forward primer was always at the 5′ end of the sequence; (4) “limit” set to “false” to process all reads; (5) “merge_program” set to “flash”; and (6) “ignore_j” set to “true” to ignore whether a joining (J) gene had been assigned to an inferred IGHV gene. We used IGHV mouse sequences downloaded from the ImMunoGeneTics Information System (IMGT; Lefranc, 2001; downloaded August 2021) as the starting database for IGHV inference. 53 IGHV sequences were inferred and used for downstream analysis.
To place these initial 53 MRL/lpr reference sequences into IMGT-gapped alignment, sequences were matched to the mouse IMGT GENE-DB reference database v3.1.37 (accessed 7/25/22; Giudicelli et al., 2005) using IgBlast v1.18.0 (Ye et al., 2013). These IMGT-aligned MRL/lpr sequences constituted our initial IGHV gene germline reference database. IMGT IGHV reference sequences were not included in this database.
To detect further MRL/lpr germline alleles, we performed single-cell BCR sequencing on B cells from spleen and bone marrow samples from two 4-wk-old female MRL/lpr mice, sorted on DAPI− CD45R+ CD93+ IgM+ Fraction E bone marrow and DAPI− TCRβ− CD19+ CD23+ CD21/35− CD138− CD11c− CD11b− FO splenocytes. Cells were stained with TotalSeq C sample hashing antibodies and run through the 10× Chromium Next GEM Single Cell V(D)J Reagent Kit v1.1 with Feature Barcoding Technology for Cell Surface Protein protocol. Surface feature and VDJ libraries were pooled at 1:1 and sequenced by Novogene on the NovaSeq platform.
BCR sequences were aligned to the initial IGHV MRL/lpr germline reference database using IgBlast. Murine IGHD and IGHJ reference databases from IMGT GENE-DB were also used. Cells which could not be definitively assigned to a particular mouse through hashtag oligo demultiplexing were removed. Unmutated germline sequences were reconstructed using createGermlines function from dowser v1.0.0 (Hoehn et al., 2022). We then calculated the Hamming distance from each sequence to its predicted germline ancestor. Because our sequencing used sorted naive B cells, we expected the overwhelming number of IGH V gene sequences to be in their germline state. Thus, IGHV gene sequences that differ from the germline reference represent novel alleles. However, it is possible some mutated B cells were included. To reduce the likelihood that spurious novel alleles were included, we applied an additional filter: each IMGT-gapped sequence was truncated to positions 1–312 (IMGT V-gene), and sequences were retained as novel IGHV alleles if an exact match was found in both 4-wk-old female mice, was not found in the initial germline reference database, and was paired with an IgM or IgD constant region. The logic behind this filter is that SHM introduces stochastic variation into V(D)J sequences; thus, IGHV sequences from IgM/IgD B cells that exactly match between inbred mice strains are more likely to be true novel alleles in the germline state than convergent mutated sequences. As an additional filter, we further required that each novel V gene sequence be paired with multiple J gene sequences. Sequences containing premature stop codons were also excluded. These filtering steps resulted in identification of 28 additional inferred MRL/lpr IGHV-gene alleles. These 28 additional genes were combined with the initial MRL/lpr germline database for a total of 81 inferred MRL/lpr IGHV germline sequences. Using a similar process as above, IMGT gaps were added based on alignment of each sequence to its closest matching IGHV sequence in the IMGT GENE-DB murine reference database.
scRNA-seq analysis of MRL/lpr B cell receptors
Starting with CellRanger output, V(D)J genes for each sequence were aligned to the 81 inferred MRL/lpr IGHV gene sequences, as well as IGHD and IGHJ reference sequences from IMGT GENE-DB v3.1.37 (Giudicelli et al., 2005), using IgBlast v1.17.0 (Ye et al., 2013). Cells that could not be definitively assigned to a particular mouse through hashtag oligo demultiplexing were removed. Nonproductive sequences were removed. Cells with multiple heavy chains were filtered by selecting only the heavy chain sequence with the most associated UMIs. Ties were resolved by selecting the first identified heavy chain associated with the cell barcode. Within each sample, sequences were grouped into clonal clusters, which contain B cells that relate to each other by somatic hypermutations from a common V(D)J ancestor. Sequences were first grouped by common IGHV gene annotations, IGHJ gene annotations, and junction lengths. Using the hierarchicalClones function of scoper v1.2.0 (Nouri and Kleinstein, 2018), sequences within these groups differing by a length-normalized Hamming distance of 0.1 within the junction region were defined as clones using single-linkage hierarchical clustering (Gupta et al., 2017). This threshold was determined by manual inspection of distance to nearest neighbor plots using shazam v1.1.1 (Yaari et al., 2013). Within each clone, germline sequences were reconstructed with D segment and N/P regions masked (replaced with “N” nucleotides) using the createGermlines function within dowser v1.0.0 (Hoehn et al., 2022). All BCR analyses used R v4.1.3.
SHM analysis, clonal overlap, and lineage tree analysis
V-gene SHM was calculated as the Hamming distance between each sequence and its predicted clonal germline sequence along IMGT positions 1–312 using shazam v1.1.1. Clonal overlap among each pair of cell types was determined by the Jaccard overlap (intersect/union) of clones between the two cell types. To infer initial lineage trees, we estimated tree topologies, branch lengths, and mouse-wide substitution model parameters using maximum likelihood under the GY94 model (Hoehn et al., 2019; Nielsen and Yang, 1998). We then estimated tree topologies, branch lengths and mouse-wide parameter values under the HLP19 model in IgPhyML v1.1.4 (Hoehn et al., 2019). Trees were visualized using dowser v1.0.0 (Hoehn et al., 2022) and ggtree v3.3.1.901 (Yu et al., 2016).
Online supplemental material
Fig. S1 shows expression of CD93, CD43, IgM, and IgD in ABC-like subsets and plasmablasts, and autoantibody and total Ig production from sorted, TLR-stimulated ABC subsets related to Fig. 3. Fig. S2 shows the degree of mixed chimerism and additional data in mice from Fig. 6. Fig. S3 shows the sorting strategy, conventional flow cytometric analysis of splenocytes from donor mice, and contribution of each donor mouse to the clusters, related to Fig. 7 and Fig. 8. Fig. S3 also shows expression of selected additional genes plotted on the cluster UMAP, and pairwise plots of normalized counts from oligo-tagged surface feature antibodies for B cell clusters not included in Fig. 7 C. Fig. S4 shows a heatmap of the top 10 DEGs by cluster related to Fig. 7. Fig. S5 shows the full range of mutation frequencies and the per-cluster mutation frequencies of data summarized in Fig. 8 D. Data S1 includes the full list of DEGs output by Seurat for the clusters described in Fig. 7 and Fig. S3. Data S2 contains the inferred germline MRL/lpr V region sequences used for BCR mutation analysis in Fig. 8.
Supplementary Material
includes the full list of DEGs output by Seurat for the clusters described in Fig. 7 and Fig. S3.
contains the inferred germline MRL/lpr V region sequences used for BCR mutation analysis in Fig. 8.
Acknowledgments
The authors would like to thank the University of Pittsburgh Division of Laboratory Animal Research for excellent animal husbandry. The authors also thank the Flow Cytometry Core.
This work was supported in part by National Institutes of Health (NIH) R21AI141938 and a Novel Research Grant from the Lupus Research Alliance (to K.M. Nickerson), NIH R37AI118841 (to M.J. Shlomchik), and NIH R01AI104739 (to S.H. Kleinstein).
Author contributions: K.M. Nickerson and M.J. Shlomchik conceptualized the research project and wrote the manuscript. K.M. Nickerson, A.D. Marinov, K.B. Thomas, and Y. Yang performed experiments. S. Smita and K.B. Hoehn (under the supervision of S.H. Kleinstein) performed bioinformatic analysis and contributed to experimental design. S.I. Bastacky evaluated renal pathology. J.T. Kos and C.T. Watson identified MRL/lpr germline V region sequences. All authors reviewed and commented upon the manuscript.
References
- Aranburu, A., Höök N., Gerasimcik N., Corleis B., Ren W., Camponeschi A., Carlsten H., Grimsholm O., and Mårtensson I.L.. 2018. Age-associated B cells expanded in autoimmune mice are memory cells sharing H-CDR3-selected repertoires. Eur. J. Immunol. 48:509–521. 10.1002/eji.201747127 [DOI] [PubMed] [Google Scholar]
- Atisha-Fregoso, Y., Toz B., and Diamond B.. 2021. Meant to B: B cells as a therapeutic target in systemic lupus erythematosus. J. Clin. Invest. 131:e149095. 10.1172/JCI149095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, A., Hoffman P., Smibert P., Papalexi E., and Satija R.. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36:411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancro, M.P. 2020. Age-associated B cells. Annu. Rev. Immunol. 38:315–340. 10.1146/annurev-immunol-092419-031130 [DOI] [PubMed] [Google Scholar]
- Chan, O.T., Madaio M.P., and Shlomchik M.J.. 1999. B cells are required for lupus nephritis in the polygenic, Fas-intact MRL model of systemic autoimmunity. J. Immunol. 163:3592–3596. 10.4049/jimmunol.163.7.3592 [DOI] [PubMed] [Google Scholar]
- Corcoran, M.M., Phad G.E., Vázquez Bernat N., Stahl-Hennig C., Sumida N., Persson M.A.A., Martin M., and Karlsson Hedestam G.B.. 2016. Production of individualized V gene databases reveals high levels of immunoglobulin genetic diversity. Nat. Commun. 7:13642. 10.1038/ncomms13642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, S.W., Arkatkar T., Al Qureshah F., Jacobs H.M., Thouvenel C.D., Chiang K., Largent A.D., Li Q.Z., Hou B., Rawlings D.J., and Jackson S.W.. 2019a. Functional characterization of CD11c+ age-associated B cells as memory B cells. J. Immunol. 203:2817–2826. 10.4049/jimmunol.1900404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, S.W., Arkatkar T., Jacobs H.M., Rawlings D.J., and Jackson S.W.. 2019b. Generation of functional murine CD11c+ age-associated B cells in the absence of B cell T-bet expression. Eur. J. Immunol. 49:170–178. 10.1002/eji.201847641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsner, R.A., and Shlomchik M.J.. 2020. Germinal center and extrafollicular B cell responses in vaccination, immunity, and autoimmunity. Immunity. 53:1136–1150. 10.1016/j.immuni.2020.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles, J.R., Kashgarian M., Koni P.A., and Shlomchik M.J.. 2015. B cell-specific MHC class II deletion reveals multiple nonredundant roles for B cell antigen presentation in murine lupus. J. Immunol. 195:2571–2579. 10.4049/jimmunol.1500792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudicelli, V., Chaume D., and Lefranc M.P.. 2005. IMGT/GENE-DB: A comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 33:D256–D261. 10.1093/nar/gki010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, N.T., Adams K.D., Briggs A.W., Timberlake S.C., Vigneault F., and Kleinstein S.H.. 2017. Hierarchical clustering can identify B cell clones with high confidence in Ig repertoire sequencing data. J. Immunol. 198:2489–2499. 10.4049/jimmunol.1601850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, Y., O’Neill P., Naradikian M.S., Scholz J.L., and Cancro M.P.. 2011. A B-cell subset uniquely responsive to innate stimuli accumulates in aged mice. Blood. 118:1294–1304. 10.1182/blood-2011-01-330530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehn, K.B., Pybus O.G., and Kleinstein S.H.. 2022. Phylogenetic analysis of migration, differentiation, and class switching in B cells. PLoS Comput. Biol. 18:e1009885. 10.1371/journal.pcbi.1009885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehn, K.B., Vander Heiden J.A., Zhou J.Q., Lunter G., Pybus O.G., and Kleinstein S.H.. 2019. Repertoire-wide phylogenetic models of B cell molecular evolution reveal evolutionary signatures of aging and vaccination. Proc. Natl. Acad. Sci. U S A. 116:22664–22672. 10.1073/pnas.1906020116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, L., Voit R.A., Sankaran V.G., Springer T.A., and Yuki K.. 2020. CD11c regulates hematopoietic stem and progenitor cells under stress. Blood Adv. 4:6086–6097. 10.1182/bloodadvances.2020002504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim, S.M., Weigert M., Basu C., Erikson J., and Radic M.Z.. 1995. Light chain contribution to specificity in anti-DNA antibodies. J. Immunol. 155:3223–3233. 10.4049/jimmunol.155.6.3223 [DOI] [PubMed] [Google Scholar]
- Jacobi, A.M., Reiter K., Mackay M., Aranow C., Hiepe F., Radbruch A., Hansen A., Burmester G.R., Diamond B., Lipsky P.E., and Dörner T.. 2008. Activated memory B cell subsets correlate with disease activity in systemic lupus erythematosus: Delineation by expression of CD27, IgD, and CD95. Arthritis Rheum. 58:1762–1773. 10.1002/art.23498 [DOI] [PubMed] [Google Scholar]
- Jenks, S.A., Cashman K.S., Zumaquero E., Marigorta U.M., Patel A.V., Wang X., Tomar D., Woodruff M.C., Simon Z., Bugrovsky R., et al. 2018. Distinct effector B cells induced by unregulated toll-like receptor 7 contribute to pathogenic responses in systemic lupus erythematosus. Immunity. 49:725–739.e6. 10.1016/j.immuni.2018.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, J.L., Rosenthal R.L., Knox J.J., Myles A., Naradikian M.S., Madej J., Kostiv M., Rosenfeld A.M., Meng W., Christensen S.R., et al. 2020. The transcription factor T-bet resolves memory B cell subsets with distinct tissue distributions and antibody specificities in mice and humans. Immunity. 52:842–855.e6. 10.1016/j.immuni.2020.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D.S.W., Rojas O.L., and Gommerman J.L.. 2021. B cell depletion therapies in autoimmune disease: Advances and mechanistic insights. Nat. Rev. Drug Discov. 20:179–199. 10.1038/s41573-020-00092-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc, M.P. 2001. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 29:207–209. 10.1093/nar/29.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levack, R.C., Newell K.L., Popescu M., Cabrera-Martinez B., and Winslow G.M.. 2020. CD11c+ T-bet+ B cells require IL-21 and IFN-γ from type 1 T follicular helper cells and intrinsic Bcl-6 expression but develop normally in the absence of T-bet. J. Immunol. 205:1050–1058. 10.4049/jimmunol.2000206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzina, I.G., Atamas S.P., Storrer C.E., daSilva L.C., Kelsoe G., Papadimitriou J.C., and Handwerger B.S.. 2001. Spontaneous formation of germinal centers in autoimmune mice. J. Leukoc. Biol. 70:578–584. 10.1189/jlb.70.4.578 [DOI] [PubMed] [Google Scholar]
- Manni, M., Gupta S., Ricker E., Chinenov Y., Park S.H., Shi M., Pannellini T., Jessberger R., Ivashkiv L.B., and Pernis A.B.. 2018. Regulation of age-associated B cells by IRF5 in systemic autoimmunity. Nat. Immunol. 19:407–419. 10.1038/s41590-018-0056-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinov, A.D., Wang H., Bastacky S.I., van Puijenbroek E., Schindler T., Speziale D., Perro M., Klein C., Nickerson K.M., and Shlomchik M.J.. 2021. The type II anti-CD20 antibody Obinutuzumab (GA101) is more effective than rituximab at depleting B cells and treating disease in a murine lupus model. Arthritis Rheumatol. 73:826–836. 10.1002/art.41608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouat, I.C., Goldberg E., and Horwitz M.S.. 2022. Age-associated B cells in autoimmune diseases. Cell Mol. Life Sci. 79:402. 10.1007/s00018-022-04433-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naradikian, M.S., Myles A., Beiting D.P., Roberts K.J., Dawson L., Herati R.S., Bengsch B., Linderman S.L., Stelekati E., Spolski R., et al. 2016. Cutting edge: IL-4, IL-21, and IFN-γ interact to govern T-bet and CD11c expression in TLR-activated B cells. J. Immunol. 197:1023–1028. 10.4049/jimmunol.1600522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestorowa, S., Hamey F.K., Pijuan Sala B., Diamanti E., Shepherd M., Laurenti E., Wilson N.K., Kent D.G., and Göttgens B.. 2016. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood. 128:e20-31. 10.1182/blood-2016-05-716480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson, K.M., Christensen S.R., Cullen J.L., Meng W., Luning Prak E.T., and Shlomchik M.J.. 2013. TLR9 promotes tolerance by restricting survival of anergic anti-DNA B cells, yet is also required for their activation. J. Immunol. 190:1447–1456. 10.4049/jimmunol.1202115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, R., and Yang Z.. 1998. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 148:929–936. 10.1093/genetics/148.3.929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouri, N., and Kleinstein S.H.. 2018. A spectral clustering-based method for identifying clones from high-throughput B cell repertoire sequencing data. Bioinformatics. 34:i341–i349. 10.1093/bioinformatics/bty235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racine, R., Chatterjee M., and Winslow G.M.. 2008. CD11c expression identifies a population of extrafollicular antigen-specific splenic plasmablasts responsible for CD4 T-independent antibody responses during intracellular bacterial infection. J. Immunol. 181:1375–1385. 10.4049/jimmunol.181.2.1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricker, E., Manni M., Flores-Castro D., Jenkins D., Gupta S., Rivera-Correa J., Meng W., Rosenfeld A.M., Pannellini T., Bachu M., et al. 2021. Altered function and differentiation of age-associated B cells contribute to the female bias in lupus mice. Nat. Commun. 12:4813. 10.1038/s41467-021-25102-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov, A.V., Rubtsova K., Fischer A., Meehan R.T., Gillis J.Z., Kappler J.W., and Marrack P.. 2011. Toll-like receptor 7 (TLR7)-driven accumulation of a novel CD11c⁺ B-cell population is important for the development of autoimmunity. Blood. 118:1305–1315. 10.1182/blood-2011-01-331462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov, A.V., Rubtsova K., Kappler J.W., Jacobelli J., Friedman R.S., and Marrack P.. 2015. CD11c-Expressing B cells are located at the T cell/B cell Border in spleen and are potent APCs. J. Immunol. 195:71–79. 10.4049/jimmunol.1500055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov, A.V., Rubtsova K., Kappler J.W., and Marrack P.. 2013. TLR7 drives accumulation of ABCs and autoantibody production in autoimmune-prone mice. Immunol. Res. 55:210–216. 10.1007/s12026-012-8365-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsova, K., Rubtsov A.V., Thurman J.M., Mennona J.M., Kappler J.W., and Marrack P.. 2017. B cells expressing the transcription factor T-bet drive lupus-like autoimmunity. J. Clin. Invest. 127:1392–1404. 10.1172/JCI91250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsova, K., Rubtsov A.V., van Dyk L.F., Kappler J.W., and Marrack P.. 2013. T-box transcription factor T-bet, a key player in a unique type of B-cell activation essential for effective viral clearance. Proc. Natl. Acad. Sci. U S A. 110:E3216–E3224. 10.1073/pnas.1312348110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichmann, L.L., Kashgarian M., Weaver C.T., Roers A., Müller W., and Shlomchik M.J.. 2012. B cell-derived IL-10 does not regulate spontaneous systemic autoimmunity in MRL.Fas(lpr) mice. J. Immunol. 188:678–685. 10.4049/jimmunol.1102456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichmann, L.L., Ols M.L., Kashgarian M., Reizis B., Kaplan D.H., and Shlomchik M.J.. 2010. Dendritic cells in lupus are not required for activation of T and B cells but promote their expansion, resulting in tissue damage. Immunity. 33:967–978. 10.1016/j.immuni.2010.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomayko, M.M., Steinel N.C., Anderson S.M., and Shlomchik M.J.. 2010. Cutting edge: Hierarchy of maturity of murine memory B cell subsets. J. Immunol. 185:7146–7150. 10.4049/jimmunol.1002163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi, N., Weisel F., Smita S., Joachim S., Kader M., Radhakrishnan A., Clouser C., Rosenfeld A.M., Chikina M., Vigneault F., et al. 2019. Liver is a generative site for the B cell response to Ehrlichia muris. Immunity. 51:1088–1101.e5. 10.1016/j.immuni.2019.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., Wang J., Kumar V., Karnell J.L., Naiman B., Gross P.S., Rahman S., Zerrouki K., Hanna R., Morehouse C., et al. 2018. IL-21 drives expansion and plasma cell differentiation of autoreactive CD11c(hi)T-bet(+) B cells in SLE. Nat. Commun. 9:1758. 10.1038/s41467-018-03750-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehr, C., Eibel H., Masilamani M., Illges H., Schlesier M., Peter H.H., and Warnatz K.. 2004. A new CD21low B cell population in the peripheral blood of patients with SLE. Clin. Immunol. 113:161–171. 10.1016/j.clim.2004.05.010 [DOI] [PubMed] [Google Scholar]
- Wei, C., Anolik J., Cappione A., Zheng B., Pugh-Bernard A., Brooks J., Lee E.H., Milner E.C.B., and Sanz I.. 2007. A new population of cells lacking expression of CD27 represents a notable component of the B cell memory compartment in systemic lupus erythematosus. J. Immunol. 178:6624–6633. 10.4049/jimmunol.178.10.6624 [DOI] [PubMed] [Google Scholar]
- William, J., Euler C., Christensen S., and Shlomchik M.J.. 2002. Evolution of autoantibody responses via somatic hypermutation outside of germinal centers. Science. 297:2066–2070. 10.1126/science.1073924 [DOI] [PubMed] [Google Scholar]
- Yaari, G., Vander Heiden J.A., Uduman M., Gadala-Maria D., Gupta N., Stern J.N.H., O’Connor K.C., Hafler D.A., Laserson U., Vigneault F., and Kleinstein S.H.. 2013. Models of somatic hypermutation targeting and substitution based on synonymous mutations from high-throughput immunoglobulin sequencing data. Front. Immunol. 4:358. 10.3389/fimmu.2013.00358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates, J.L., Racine R., McBride K.M., and Winslow G.M.. 2013. T cell-dependent IgM memory B cells generated during bacterial infection are required for IgG responses to antigen challenge. J. Immunol. 191:1240–1249. 10.4049/jimmunol.1300062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, J., Ma N., Madden T.L., and Ostell J.M.. 2013. IgBLAST: An immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 41:W34–W40. 10.1093/nar/gkt382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G., Smith D.K., Zhu H., Guan Y., Lam T.T.Y., and McInerny G.. 2016. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8:28–36. 10.1111/2041-210X.12628 [DOI] [Google Scholar]
- Zappia, L., and Oshlack A.. 2018. Clustering trees: A visualization for evaluating clusterings at multiple resolutions. Gigascience. 7:giy083. 10.1093/gigascience/giy083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccarino-Catania, G.V., Sadanand S., Weisel F.J., Tomayko M.M., Meng H., Kleinstein S.H., Good-Jacobson K.L., and Shlomchik M.J.. 2014. CD80 and PD-L2 define functionally distinct memory B cell subsets that are independent of antibody isotype. Nat. Immunol. 15:631–637. 10.1038/ni.2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumaquero, E., Stone S.L., Scharer C.D., Jenks S.A., Nellore A., Mousseau B., Rosal-Vela A., Botta D., Bradley J.E., Wojciechowski W., et al. 2019. IFNγ induces epigenetic programming of human T-bethi B cells and promotes TLR7/8 and IL-21 induced differentiation. eLife. 8:e41641. 10.7554/eLife.41641 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
includes the full list of DEGs output by Seurat for the clusters described in Fig. 7 and Fig. S3.
contains the inferred germline MRL/lpr V region sequences used for BCR mutation analysis in Fig. 8.