Abstract
Purpose:
Primary open-angle glaucoma (POAG) is a degenerative eye disease for which early treatment is critical to mitigate visual impairment and, in severe cases, irreversible blindness. POAG-associated loci individually confer incremental risk. Genetic risk score(s) (GRS) could enable POAG risk stratification. Despite significantly higher POAG burden among individuals of African ancestry (AFR), GRS are limited in this population. A recent large-scale, multi-ancestry meta-analysis identified 127 POAG-associated loci and calculated cross-ancestry and ancestry-specific effect estimates, including in EUR and AFR individuals. We assessed the utility of the 127-variant GRS for POAG risk stratification in EUR and AFR Veterans in the Million Veteran Program (MVP). We also explored the association between the GRS and documented invasive glaucoma surgery (IGS).
Design:
Cross-sectional study.
Participants:
5830 POAG cases (445 with IGS documented in electronic health record) and 64476 controls with imputed genetic data from the MVP.
Methods:
We tested unweighted and weighted GRS of 127 published risk variants in EUR (3382 cases, 58811 controls) and AFR (2448 cases, 5665 controls) Veterans in the MVP. Weighted GRS were calculated using effect estimates from the most recently published report of cross-ancestry and ancestry-specific meta-analyses. We also evaluated GRS in POAG cases with documented IGS.
Main Outcome Measures:
Performance of 127-variant GRS in EUR and AFR Veterans for POAG risk stratification and association with documented IGS.
Results:
GRS were significantly associated with POAG (p<5×10−5) in both groups; a higher proportion of EUR compared to AFR cases were consistently categorized in the top GRS decile (21.9–23.6% and 12.9–14.5%, respectively). Only GRS weighted by ancestry-specific effect estimates were associated with IGS documentation in AFR cases; all GRS types were associated with IGS in EUR cases.
Conclusions:
Varied performance of the GRS for POAG risk stratification and documented IGS association in EUR and AFR Veterans highlights (i) the complex risk architecture of POAG, (ii) the importance of diverse representation in genomics studies that inform GRS construction and evaluation, and (iii) the necessity of expanding diverse POAG-related genomic data so that GRS can equitably aid in screening individuals at high risk for POAG and who may require more aggressive treatment.
Primary open-angle glaucoma (POAG) is an age-related eye disease characterized by progressive damage to the optic nerve which typically manifests as loss of peripheral vision.1 POAG is the most common type of glaucoma and the leading cause of irreversible blindness worldwide.2,3 Because POAG is an age-related condition, the prevalence is expected to rise with the increase in size of the population over age 65 such that, by 2040, nearly 80 million individuals around the world are predicted to be affected by POAG.3,4 POAG is highly heritable,5,6 and its susceptibility is shaped by both genetic and environmental risk factors.7 To date, more than 127 loci have been identified as associated with POAG risk through genetic studies including genome-wide association studies (GWAS) and meta-analyses.8 While more recently discovered POAG-associated loci were identified through studies of multi-ancestry cohorts,8–14 most studies have been conducted within populations of non-Hispanic European ancestry (EUR)15–23 even though the prevalence of POAG is higher in other ancestral groups, especially in individuals of non-Hispanic African ancestry (AFR).24
POAG often develops earlier and results in more severe disease outcomes such as irreversible blindness in AFR individuals compared to individuals from other ancestral groups (reviewed in25). Such severe outcomes may be mitigated through earlier detection and treatment interventions aimed at lowering the elevated intraocular pressure (IOP), the most common risk factor for POAG.26 When pharmacologic and/or laser treatment methods fail to meet treatment goals, micro invasive surgical options are implemented in mild or moderate cases while invasive glaucoma surgery (IGS), such as trabeculectomies and glaucoma drainage implants (GDI) like tube shunts, are utilized in those with a more severe or refractory disease course.27 Therefore, improved POAG management would result from risk stratification methods that identify those who might develop POAG as well as those at risk for more aggressive disease progression resulting in need for IGS.
Genetic risk score(s) (GRS) and polygenic risk score(s) (PRS), which aggregate the additive effects of particular genetic variants, have been evaluated for prediction of many complex traits28 including age-related macular degeneration,29,30 breast cancer,31 cardiovascular disease,32 and Alzheimer’s disease.33 Typically, GRS aggregate effects of variants detected at genome-wide levels of significance in one or multiple studies; whereas, PRS extend to include additional genetic variation depending on preferred significance and level of linkage disequilibrium with surrounding genomic regions.34 Recent evidence indicates the utility of GRS and PRS for POAG prediction and risk stratification.15,35–40 For example, in EUR individuals, GRS and PRS were associated with POAG age of diagnosis and disease-relevant clinical parameters.35,36 Therefore, in the future, GRS may serve as a possible surrogate measure to assess POAG risk and inform recommendations for increased screening to facilitate early intervention.4,27
GRS and PRS are often calculated within a single genetic ancestry; therefore, they are limited in their potential for clinical implementation across different ancestries.41,42 Broader representation in genomics studies by including data from populations that have been historically underrepresented in research will inform more precise prediction models.43–45 One way to increase diversity in genetic studies is to access large-scale multi-ancestry biobanks linked to electronic health records (EHR), which have facilitated the discovery of risk loci for several complex traits.46,47 The Million Veteran Program (MVP) was established by the Department of Veterans Affairs (VA) Office of Research and Development (ORD) and has thus far enrolled over 800000 U.S. Veterans.48 Because the Veteran population includes older individuals at risk for age-related eye conditions, the Veterans Health Administration has experienced an increasing number of ocular disease diagnoses (i.e. age-related macular degeneration, cataract, and POAG) and use of eye care services in recent years.49,50 Previous analysis of the genetic ancestry of the MVP study population demonstrated that about 29% of MVP-enrolled Veterans are of ancestries that have been historically underrepresented in GWAS.51 Here, we leveraged the MVP EHR and genetic data to study the utility of previously identified POAG risk variants for predicting POAG across at-risk, demographically diverse populations.
Methods
Study Participants
The MVP is a national research program comprised of health, genetic, and lifestyle information for more than 800000 US military Veterans who provided informed consent to participate in this observational cohort study and mega-biobank (https://www.research.va.gov/mvp/). Over 450000 Veterans have genome-wide data available.51 Genetic ancestry of MVP participants was determined using the Harmonized Ancestry and Race/Ethnicity (HARE) algorithm.52 We used a previously validated computable phenotyping algorithm53 based on structured diagnosis data (ICD-CM) codes, Current Procedural Terminology (CPT) codes, and prescriptions to identify POAG cases and controls in the VA Computerized Patient Record System.54 Cases and controls were required to have ≥2 eye clinic procedural codes; exclusion codes indicative of other ocular conditions that could interfere with POAG case/control classification were used to refine phenotype definitions. Cases were required to be ≥30 years old, have ≥6 POAG diagnosis codes, and have semi-structured medication data for ≥2 separate IOP-lowering prescriptions. Controls were required to be ≥65 years old and have records free of POAG inclusion codes. This algorithm extracted data for Veterans of non-Hispanic European ancestry (EUR) and non-Hispanic African ancestry (AFR) and classified 5830 POAG cases (3382 EUR and 2448 AFR) and 64476 controls (58811 EUR and 5665 AFR) with imputed genetic data available.51 This study adhered to the tenets of the Declaration of Helsinki and was approved by the VA ORD Central Institutional Review Board.
POAG-associated Variant GRS Definition
We calculated GRS based on 127 POAG-associated variants (Table S1, available at www.aaojournal.org) that were identified in the largest to-date cross-ancestry GWAS meta-analysis and for which cross-ancestry and ancestry-specific effect estimates were generated. Published cross-ancestry effect estimates were available for all 127 variants in the study. However, published ancestry-specific weights for the AFR group were only available for 118 of the 127 variants due to unavailability or low minor allele frequency in the published meta-analysis. The published effect sizes varied by ancestral group for these variants;8 therefore, we examined heterogeneity between AFR and EUR Veterans in the MVP using a fixed-effects inverse-variance weighted approach and summarized them by Cochran’s Q and I2 measures. A p-value of less than 0.1 was used for Cochran’s Q, while I2 values of greater than 50% indicated moderate to very large heterogeneity.
Within each ancestral group (AFR and EUR), two models were considered: an unweighted additive model, which summed risk allele dosage for the K = 127 variants:
and a weighted additive model of the K = 127 variants, which weights each variant’s risk allele dosage by its published effect estimate8 before summing:
Odds ratios (ORs) were converted to log OR (betas). Unweighted GRS used risk alleles defined by OR>1 in cross-ancestry analyses. For the weighted models, we applied (i) cross-ancestry and (ii) ancestry-specific effect estimates from the most recently published multi-ancestry POAG meta-analysis.8 All GRS were standardized with mean zero and standard deviation of one for the ease of comparison.
Evaluation of GRS for POAG Risk Stratification
We performed logistic regression-based association analyses of GRS against POAG case-control status in EUR and AFR Veterans (Fig 1). We evaluated an unadjusted model as well as a model adjusting for age, sex, and 10 sample-specific principal components (PCs). We also tested for association between deciles of GRS and POAG case-control status using the middle 20% (percentiles 40–60%) as the reference. To evaluate model performance, we constructed receiver operator characteristic (ROC) curves for both unadjusted and adjusted models and estimated the area under the ROC curve (AUC) for each model using the pROC R Package (https://cran.r-project.org/web/packages/pROC/). We statistically compared GRS across ancestral groups using an extension of DeLong’s test for independent ROC curves. We also calculated case proportion by GRS decile, which was the number of cases in each decile divided by the total number of cases in the study. We calculated precision-recall curves (PRC) and area under the PRC (AUC-PRC), which may provide more information when there is imbalance between sample sizes of cases and controls.55 To examine robustness of AUC and AUC-PRC against case imbalance, we down-sampled controls by randomly selecting controls to equal the number of cases in 1000 iterations using the caret R package.56
Figure 1. Schematic overview of study design.
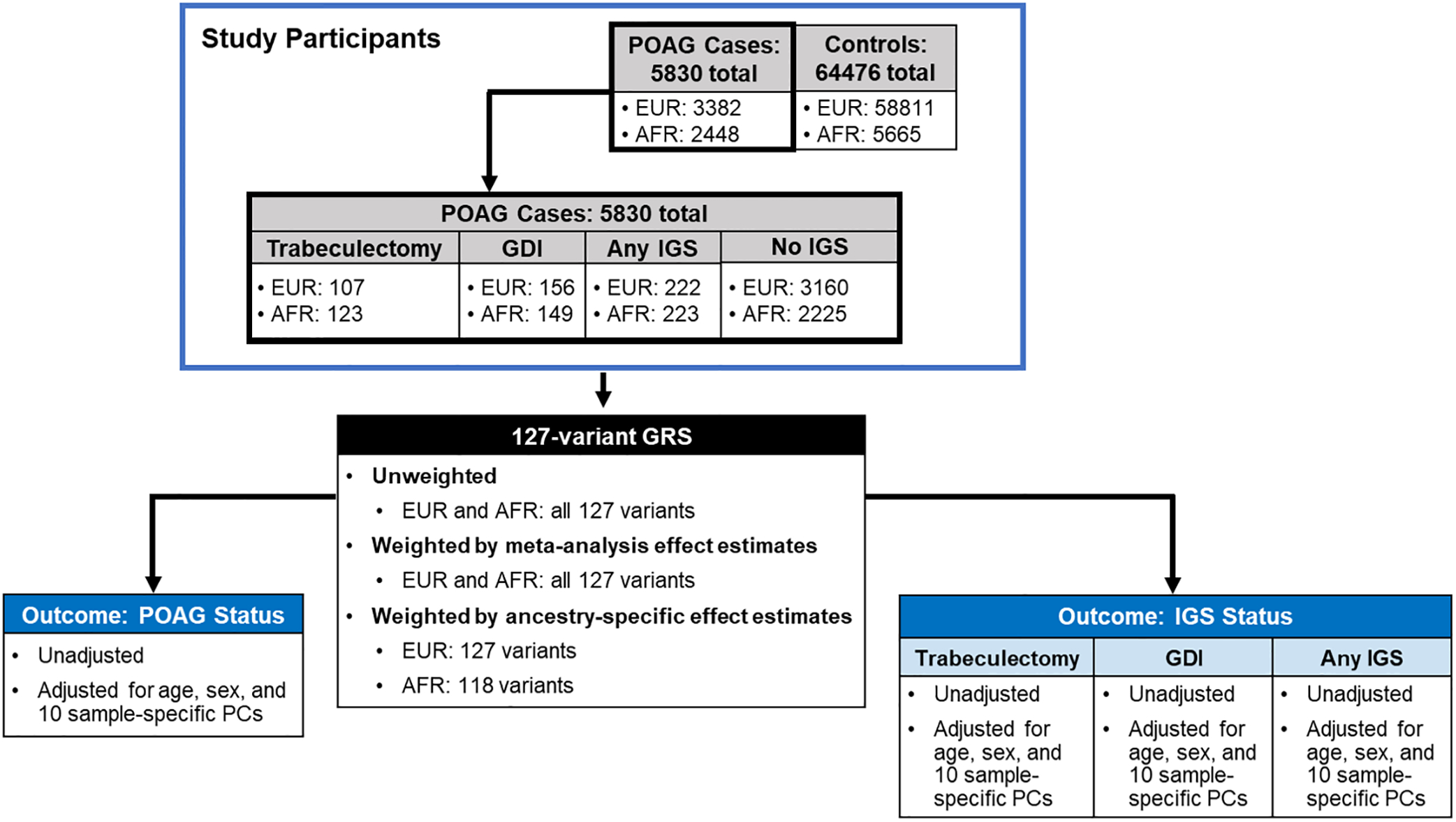
GRS were calculated for POAG cases and controls in the MVP with imputed genetic data and harmonized ancestry and race/ethnicity data. We performed logistic regression-based association analyses of the weighted and unweighted 127-variant GRS with POAG case-control status as well as IGS status. Receiver operator characteristic (ROC) curves for both unadjusted and adjusted models were constructed to evaluate GRS performance. Abbreviations: POAG = primary open-angle glaucoma, AFR = non-Hispanic African ancestry, EUR = non-Hispanic European ancestry, GRS = genetic risk score, IGS = invasive glaucoma surgeries, GDI = glaucoma drainage implants, PCs = principal components.
To further explore the estimated proportion of variance of POAG explained by each GRS, we calculated coefficients of determination (R2) on the observed scale (Nagelkerke’s)57 and the liability scale58 using a fixed disease prevalence of 2.4%.59 While disease prevalence varies between ancestral groups, we used a single value for the sake of comparison. The liability scale accounts for differences in ascertainment, which are apparent in the MVP groups: the case proportions are 0.302 and 0.054 in AFR and EUR Veterans, respectively. To partition the variance explained, we included the following variables in our models (i) age and sex, (ii) age, sex, and 10 PCs, and (iii) age, sex, 10 PCs, and each 127-variant GRS (unweighted, cross-ancestry meta-weighted, and ancestry-specific weighted). We calculated increases in R2 with the addition of each model variable. We also calculated the variance explained by the inclusion of GRS based on the single variant that achieved genome-wide significance in the published AFR-specific analysis (IQGAP1 variant rs16944405) with the 127-variant GRS in addition to our model variables: age, sex, 10 PCs. Differences in R2 and AUC were calculated with the inclusion of the IQGAP1 variant.
Association between GRS and Documented IGS
To further explore risk stratification among POAG cases, we evaluated GRS association with documented IGS (Fig 1). Three IGS case groups were considered for these analyses based on the CPT codes contained in the VA EHR: (i) trabeculectomy (CPT 66180 or 66179), (ii) GDI (CPT 66172 or 66170), and (iii) either trabeculectomies or GDI (any of the aforementioned CPT codes). Controls consisted of Veterans diagnosed with POAG who had not undergone trabeculectomies nor had GDI, as indicated by their lack of CPT codes for these procedures in their VA EHR.
We performed logistic regression-based association analyses of the unweighted and weighted GRS and binary IGS phenotypes, evaluating unadjusted models as well as models adjusting for age, sex, and 10 sample-specific PCs. We also performed logistic regression-based analyses to test for association between deciles of GRS and each IGS case group. The top decile (top 10%) was compared to the bottom 9 deciles (bottom 90%), which were used as the reference in these analyses. We also examined case proportion by decile for our comparisons. To evaluate GRS performance, we created and statistically compared ROC curves for our models. Comparisons across ancestral groups for each IGS status used an extension of DeLong’s test for independent ROC curves. Weighted versus unweighted GRS were statistically compared using DeLong’s test for correlated ROC curves. We also calculated PRC and AUC-PRC for these models.
Results
Study Demographics for POAG Cases and Controls
Our study population (Table 1) consisted of 5830 POAG cases (2448 AFR and 3382 EUR) and 64476 controls (5665 AFR and 58811 EUR) with imputed genetic data from the MVP. The majority of the sample was male (n = 68581; 97.5%). The average age for AFR cases (66.82±9.46 years) was significantly lower than the average age for AFR controls (70.99±6.70 years) in this dataset (p < 2 × 10−16) (Table 1). By contrast, the average ages among EUR cases and controls (73.32±9.55 and 73.11±7.30 years, respectively) were not significantly different (p = 0.20) (Table 1).
Table 1. POAG case-control demographics in the MVP.
Genetic ancestry of Veterans in the MVP was determined based on the Harmonized Ancestry and Race/Ethnicity (HARE) algorithm. Case-control status for POAG was determined using a phenotyping algorithm, which accessed electronic health records in the VA Computerized Patient Record System.
| African Ancestry | European Ancestry | |||||||
|---|---|---|---|---|---|---|---|---|
| POAG Cases | Controls | Total | p-valuea | POAG Cases | Controls | Total | p-valuea | |
| N (% total) | 2448 (30.17) | 5665 (69.83) | 8113 (100) | 3382 (5.44) | 58811 (94.56) | 62193 (100) | ||
| Average Age (SD) | 66.82 (9.46) | 70.99 (6.70) | 69.73 (7.88) | <2 × 10−16 | 73.32 (9.55) | 73.11 (7.3) | 73.12 (7.44) | 0.2021 |
| N Males (% total) | 2328 (95.1) | 5494 (96.98) | 7822 (96.41) | 3.75 × 10−5 | 3263 (96.48) | 57496 (97.76) | 60759 (97.69) | 1.8 × 10−6 |
Abbreviations: POAG = primary open-angle glaucoma, MVP = Million Veteran Program, SD = standard deviation.
p-values from Welch’s t-test for age, and chi-square test for sex.
Variant Properties
From our heterogeneity analyses (full results in Table S1, available at www.aaojournal.org), we found that 20 of the 127 variants had Cochran’s Q p-value less than 0.10. I2 values ranged from 0–92.66%, with IQR of 0–38.10%. We also found that 25 of the variants had I2 values greater than 50%, indicating moderate to very large heterogeneity.
Evaluation of GRS for POAG Risk Stratification
We plotted the distributions of GRS by POAG case-control status for each ancestral group and model combination (Fig S1, available at www.aaojournal.org). All GRS were significantly associated (p < 0.05) with POAG status in both unadjusted and adjusted models in both ancestral groups (Table S2, available at www.aaojournal.org). We calculated log odds ratios (beta) with 95% confidence intervals for deciles of GRS using the middle 20% as the reference (Fig 2A–C; Table S3, available at www.aaojournal.org). We found that beta estimates were higher for GRS calculated for EUR compared to AFR Veterans (unweighted: 1.04 vs. 0.54, cross-ancestry meta-weighted: 1.14 vs. 0.59, and ancestry-specific: 1.15 vs. 0.29) (Table S3, available at www.aaojournal.org). While the beta estimates generally increased across deciles, the trend is stronger in EUR compared to AFR individuals (Fig 2A–C; Table S3, available at www.aaojournal.org). This is echoed in the ROC analysis (Fig 2D–F) which showed significant differences between the AFR and EUR AUCs in the unadjusted models (Table 2). In the adjusted models, we did not detect significant differences between AFR and EUR AUCs for the meta-weighted and ancestry-specific GRS (p = 0.053 and 0.43, respectively) (Table 2). Moreover, POAG case proportions consistently increased for each GRS type calculated for EUR individuals, with over 20% present in the highest risk deciles (Fig 3). By contrast, POAG case proportions only consistently increased across cross-ancestry meta-weighted GRS deciles in AFR individuals (Fig 3).
Figure 2. Comparison of performance of 127-variant POAG GRS in AFR and EUR Veterans.
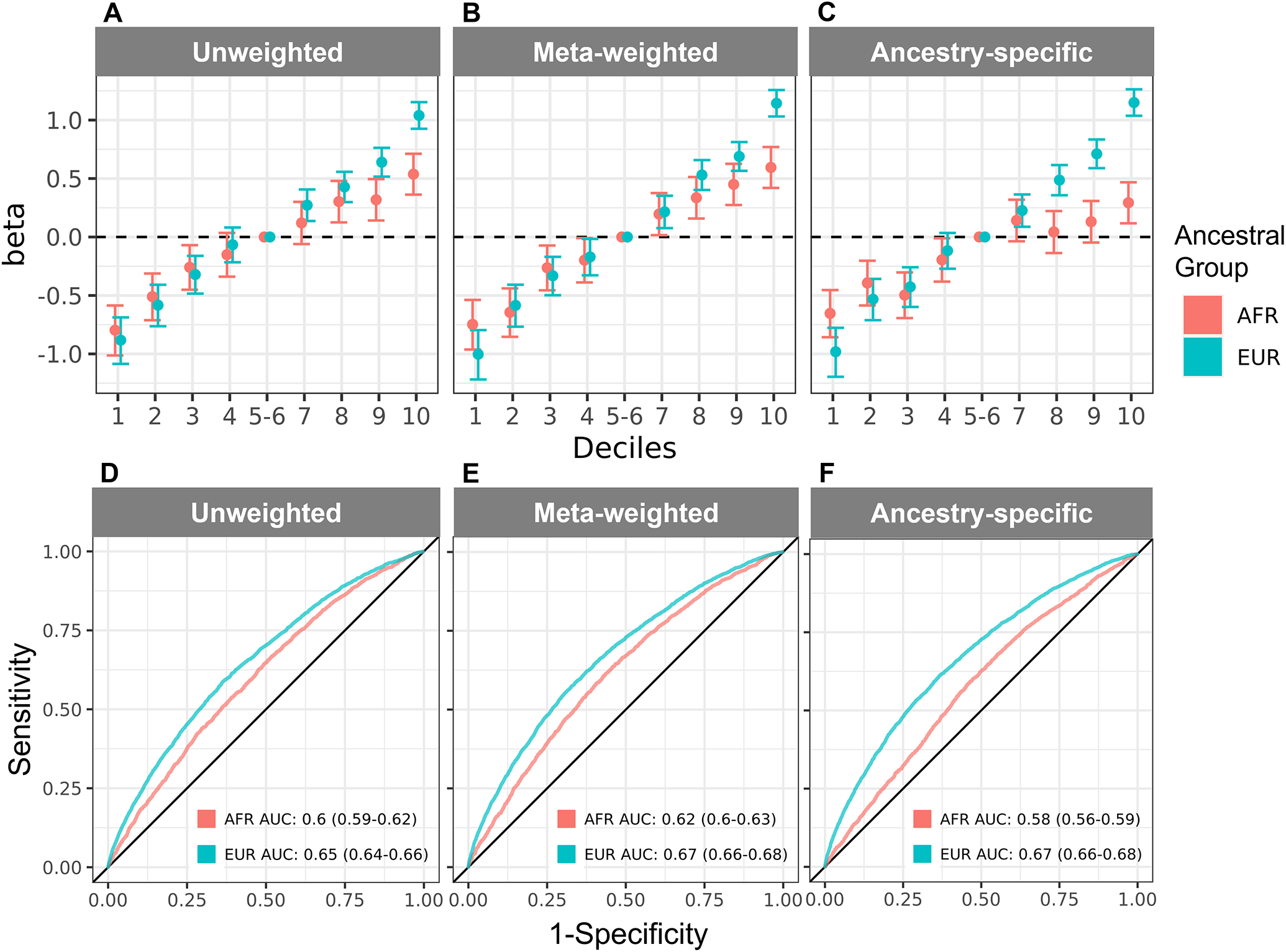
Log odds ratios (betas) for unadjusted logistic regression-based association analyses are shown across increasing deciles of (A) unweighted, (B) cross-ancestry meta-weighted, and (C) ancestry-specific GRS for AFR and EUR Veterans. In panels A-C, deciles 5–6 (40–60%) are used as the reference and denoted by the dashed line. A full description of log odds ratios for panels A-C are shown in Table S3 (available at www.aaojournal.org). ROC curves are shown for (D) unweighted, (E) cross-ancestry meta-weighted, and (F) ancestry-specific GRS performance in AFR and EUR Veterans. AFR and EUR AUCs are listed in the inset with their 95% confidence intervals in parentheses. Abbreviations: AFR = non-Hispanic African ancestry, EUR = non-Hispanic European ancestry, GRS = genetic risk scores, AUC = area under the curve, ROC = receiver operator characteristic.
Table 2. Performance of the 127-variant POAG GRS for predicting POAG case-control status in EUR and AFR individuals in the MVP.
We evaluated unadjusted models as well as models adjusting for age, sex, and 10 sample-specific PCs. GRS were unweighted or weighted by published cross-ancestry meta-analysis or ancestry-specific effect estimates. P-values represent the statistical comparisons we computed across ancestral groups using an extension of DeLong’s test for independent ROC curves.
| Model | GRS Type | AFR AUC (95% CI) | EUR AUC (95% CI) | AFR vs. EUR p-value |
|---|---|---|---|---|
| Unadjusted | Unweighted | 0.60 (0.59–0.62) | 0.65 (0.64–0.66) | 1.93 × 10−8 |
| Cross-ancestry meta-analysis | 0.62 (0.60–0.63) | 0.67 (0.66–0.68) | 8.35 × 10−10 | |
| Ancestry-specific | 0.58 (0.56–0.59) | 0.67 (0.66–0.68) | 2.14 ×10−26 | |
| Adjusted | Unweighted | 0.68 (0.67–0.70) | 0.65 (0.64–0.66) | 0.00068 |
| Cross-ancestry meta-analysis | 0.69 (0.67–0.70) | 0.67 (0.66–0.68) | 0.053 | |
| Ancestry-specific | 0.68 (0.66–0.69) | 0.67 (0.66–0.68) | 0.43 |
Abbreviations: GRS = genetic risk score, AFR = non-Hispanic African ancestry, EUR = non-Hispanic European ancestry, AUC = area under the curve, CI = confidence interval.
Figure 3. Comparison of POAG case classifications based on 127-variant POAG GRS in AFR and EUR Veterans.
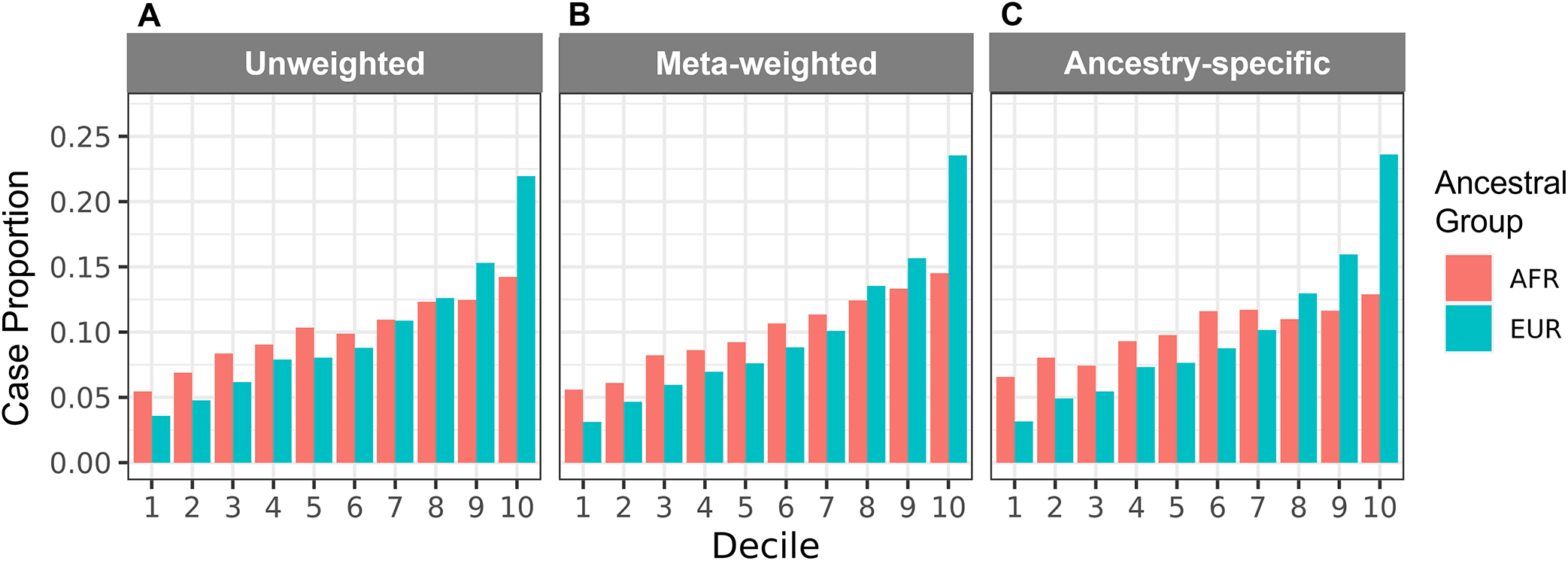
Case proportions are shown for each decile of the (A) unweighted, (B) meta-weighted, and (C) ancestry-specific weighted GRS by ancestry group. Case proportion by decile indicates the number of POAG cases in each decile divided by the total number of POAG cases in the MVP. Abbreviations: AFR = non-Hispanic African ancestry, EUR = non-Hispanic European ancestry.
In our unadjusted models, the EUR AUCs were higher than the AFR AUCs, but in our adjusted models, the AFR AUCs were higher than the EUR AUCs (Table 2). Because of these trends, we explored the estimated proportion of POAG variance explained by covariates and each GRS in our adjusted models (Table S4, available at www.aaojournal.org). We found that covariates alone (age, sex, and 10 sample-specific PCs) explained a higher proportion of POAG variance (calculated as coefficients of determination [R2] on both the observed scale [Nagelkerke’s] and liability scale) in AFR Veterans than EUR Veterans (Table S4, available at www.aaojournal.org). Moreover, the addition of the different GRS types (unweighted, cross-ancestry meta-weighted, and ancestry-specific weighted) resulted in a larger increase in the R2 values calculated for EUR Veterans than AFR Veterans. These observations suggest that the addition of covariates alone improved AUC estimates in AFR Veterans while GRS types were more informative for EUR AUCs in our adjusted models (Table S4, available at www.aaojournal.org). The addition of the IQGAP1 variant (rs16944405) resulted in small increases in R2 for unweighted and ancestry-specific weighted GRS and a decrease in R2 for cross-ancestry meta-weighted GRS (Table S4, available at www.aaojournal.org). We found no significant differences in AUC with the addition of the IQGAP1 variant (Table S5, available at www.aaojournal.org). We also tested the robustness of AUC (Fig 2D–F) and AUC-PRC (Fig S2, available at www.aaojournal.org) to case-control imbalance using down-sampling. While AUC-PRC was consistently a 0.01 lower than AUC, the results were comparable and preserve differences between AFR and EUR analyses (Fig S3, available at www.aaojournal.org). Additionally, we found that mean AUCs from down-sampling (Fig S3, available at www.aaojournal.org) were comparable to those calculated in the full dataset (Fig 2D–F; Table 2).
Association between GRS and Documented IGS
We calculated unweighted and weighted GRS for POAG cases who had or had not undergone IGS (Table S6, available at www.aaojournal.org). In total, 222 EUR and 223 AFR individuals had at least one type of IGS documented in their EHR, and 3160 EUR and 2225 AFR individuals had POAG but did not have any type of IGS documented in their EHR (Table S6, available at www.aaojournal.org). Among the individuals with documented IGS, 107 EUR and 123 AFR had trabeculectomies, and 156 EUR and 149 AFR had GDI (Table S6, available at www.aaojournal.org).
We evaluated the association of the 127-variant GRS with IGS types documented in the VA EHR and observed significant associations in both AFR and EUR individuals. The GRS we calculated (unweighted, cross-ancestry meta-analysis weighted, and ancestry-specific weighted) were significantly associated (p < 0.05) with having any type of IGS documented in the VA EHR for EUR individuals (Table 3). In AFR individuals, an association between GRS and IGS status was only detected with the ancestry-specific GRS (p = 0.0024 and 0.0018 in the unadjusted and covariate-adjusted models, respectively) (Table 3). Upon closer examination of associations with specific IGS types (trabeculectomies or GDI), we observed differences between GRS calculated for EUR and AFR individuals (Table S7, available at www.aaojournal.org). In AFR individuals, cross-ancestry meta-analysis and ancestry-specific GRS were associated with documented trabeculectomy (Table S7, available at www.aaojournal.org). Whereas, in EUR individuals, all GRS approaches were associated with documented GDI (Table S7, available at www.aaojournal.org).
Table 3. Association analysis results for 127-variant GRS and any type of IGS in the MVP.
We evaluated GRS approaches in MVP POAG cases who had undergone any type of IGS, including trabeculectomies or GDI. In our logistic regression-based association analyses, we used unadjusted models as well as models adjusting for age, sex, and 10 sample-specific PCs. GRS were unweighted or weighted by published cross-ancestry meta-analysis or ancestry-specific effect estimates.
| African Ancestry | European Ancestry | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Model | GRS Type | Estimate | Std. Error | z value | P | Estimate | Std. Error | z value | P |
| Unadjusted | Unweighted | 0.123 | 0.071 | 1.743 | 0.0814 | 0.248 | 0.07 | 3.563 | 0.000367 |
| Cross-ancestry meta-analysis | 0.129 | 0.07 | 1.838 | 0.066 | 0.265 | 0.068 | 3.927 | 8.61 × 10−5 | |
| Ancestry-specific | 0.216 | 0.071 | 3.036 | 0.0024 | 0.276 | 0.067 | 4.14 | 3.48 × 10−5 | |
| Adjusted | Unweighted | 0.109 | 0.073 | 1.495 | 0.1349 | 0.244 | 0.07 | 3.473 | 0.000515 |
| Cross-ancestry meta-analysis | 0.114 | 0.073 | 1.561 | 0.1185 | 0.265 | 0.068 | 3.909 | 9.27 × 10−5 | |
| Ancestry-specific | 0.226 | 0.072 | 3.121 | 0.0018 | 0.261 | 0.0646 | 4.045 | 4.09 × 10−5 | |
Abbreviations: GRS = genetic risk score, MVP = Million Veteran Program, IGS = invasive glaucoma surgeries.
A higher proportion of EUR versus AFR Veterans with any documented IGS fell within the highest GRS deciles, regardless of GRS type (Fig S4G–I, available at www.aaojournal.org). However, we only observed a significant difference between the top decile vs. the bottom 9 deciles for the ancestry-weighted GRS (Fig 4C; Table S8, available at www.aaojournal.org). We did not observe significant differences in GRS performance for identifying EUR and AFR Veterans with any IGS documented in the MVP, but some AUCs were higher in a particular ancestral group for a particular IGS type (Table S9; Figs S5 and S6, available at www.aaojournal.org).
Figure 4. Comparison of GRS deciles for any IGS in POAG patients in the MVP.
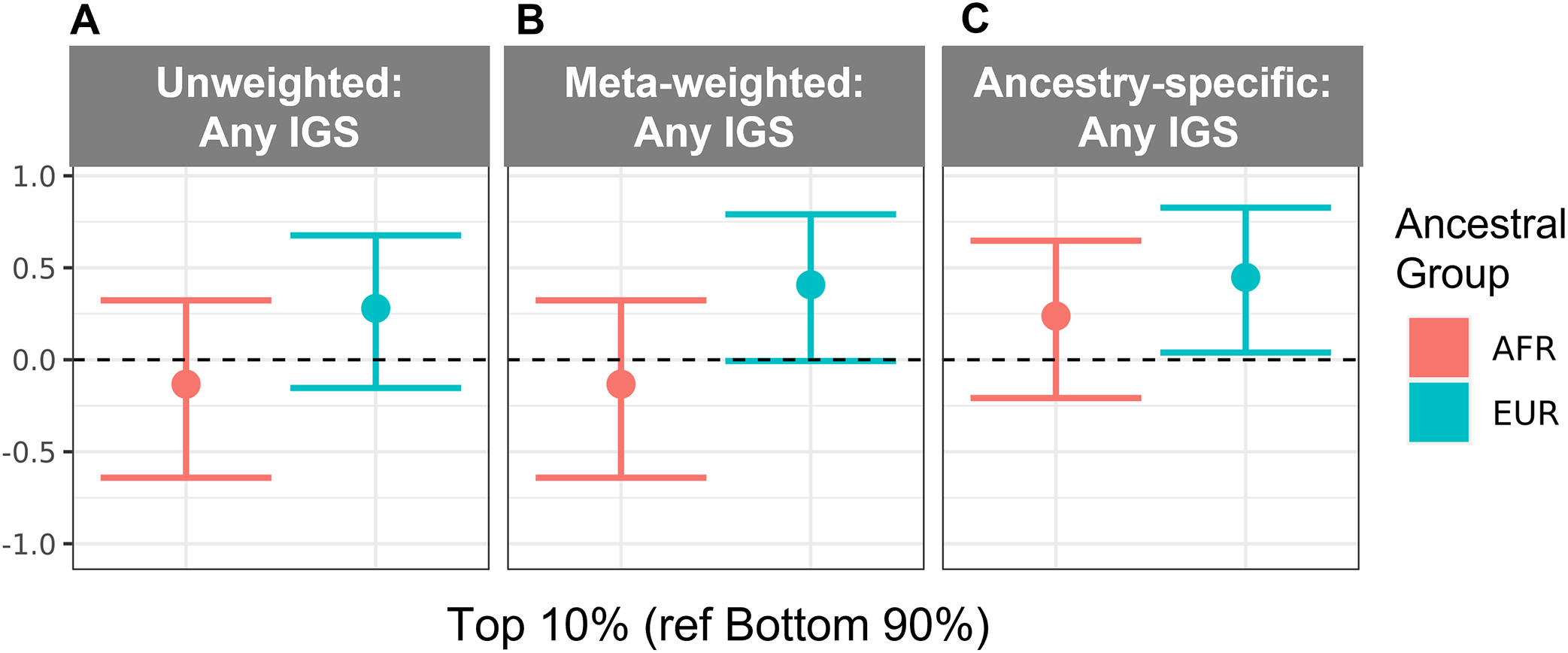
Log odds ratios (betas) for unadjusted logistic regression-based association analyses are shown for deciles of (A) unweighted, (B) cross-ancestry meta-weighted, and (C) ancestry-specific GRS for AFR and EUR Veterans. In all panels, deciles 1–9 (bottom 90%) were compared to decile 10 (top 10%). A full description of log odds ratios (betas) for panels A-C are shown in Table S8 (available at www.aaojournal.org). Abbreviations: AFR = non-Hispanic African ancestry, EUR = non-Hispanic European ancestry, GRS = genetic risk score, IGS = invasive glaucoma surgery.
Discussion
We evaluated the predictive ability of GRS for POAG and IGS based on 127 POAG-associated variants identified through the most recently published large-scale, multi-ancestry GWAS.8 This included evaluations of unweighted GRS and weighted GRS using published cross-ancestry and ancestry-specific effect estimates.8 Our findings demonstrated that the 127-variant GRS was significantly associated with POAG in EUR and AFR Veterans (p < 5×10−5) and better classified POAG status in EUR as compared to AFR cases in the MVP. In the subset of MVP POAG cases, we also detected association (p < 0.05) between the 127-variant GRS and documented IGS. We further determined that the highest GRS deciles, regardless of effect estimate weights, categorized a higher proportion of EUR Veterans with IGS as compared to AFR Veterans but still only categorized at most about 15% of IGS documentations in the VA EHR.
In the top GRS decile in each of our analyses, the 127-variant GRS identified a higher proportion of EUR POAG cases than AFR POAG cases (21.9–23.6% and 12.9–14.5%, respectively) (Fig 3). We previously evaluated GRS based on 83 POAG-associated variants; this GRS also identified EUR POAG cases more consistently than AFR POAG cases (top deciles: 13.2% and 9.6%, respectively).60 In our present study, we demonstrated that, even with the addition of 44 POAG risk variants newly identified from a cross-ancestry meta-analysis,8 GRS still performed better in EUR individuals as compared to AFR individuals. Moreover, the addition of the only genome-wide significant variant from the published AFR-specific analysis (rs16944405 in IQGAP1)8 to the GRS we evaluated in this study did not result in significant change in Nagelkerke’s R2 (Table S4, available at www.aaojournal.org) or AUC (Table S5, available at www.aaojournal.org). These observations emphasize the need for more diverse participant inclusion in POAG genetics research to facilitate the discovery of additional AFR-specific POAG-associated variants,8,12,13 which may improve our understanding of the complex genetic architecture of POAG and contribute to more informative POAG GRS.
Other GRS and PRS approaches have been applied to POAG in AFR individuals, albeit typically in studies with smaller sample sizes and often with variants identified in EUR GWAS. GRS of 10 POAG-associated variants identified in EUR GWAS were significantly associated (p = 0.00086) in African American prevalent cases (n = 658) compared to controls (n = 6067) but were not associated (p = 0.23) in 1062 African American incident cases.61 GRS comprised of 23 POAG risk variants from the NHGRI-EBI GWAS Catalog were significantly associated (p < 0.001) with POAG in the Primary Open-Angle African American Glaucoma Genetics (POAAGG) study, which included 3830 and 2135 individuals in their discovery and replication cohorts, respectively.11,62 A 400-variant PRS based on the results of a GWAS performed in 1875 POAG cases and 1709 controls of African ancestry ascertained by the African Descent and Glaucoma Evaluation Study (ADAGES) III and at Wake Forest School of Medicine achieved an AUC of 0.94.63 The predictive ability of this particular GRS may be inflated due to overfitting, as it was calculated in the sample used to select variants and estimate their effect sizes. Nevertheless, these observations emphasize the importance of having multiple independent studies with diverse ancestral representation and developing ancestry-specific risk scores based on genetic data. This point is particularly relevant for individuals of admixed genetic ancestry for whom ancestry-specific GRS based on a single continental genetic ancestry may insufficiently represent their genomic profile. For AFR Veterans in the MVP, the median proportion of AFR global genetic ancestry (calculated using ADMIXTURE 1.364 and reference populations in the 1000 Genomes Project65) was 0.83; whereas the median proportion of EUR global genetic ancestry for EUR Veterans in the MVP was 0.97 (Table S10, available at www.aaojournal.org). Therefore, future studies need to prioritize diverse representation, especially individuals with different levels of admixed genetic ancestry.
Several prior studies have evaluated the predictive capabilities of GRS and PRS for POAG endophenotypes (e.g. vertical cup-to-disc ratio (VCDR) and IOP) and have generally predicted glaucoma with AUCs ranging from 0.7 to 0.8.15,35–38 Quantitative POAG endophenotypes, including IOP and VCDR, are not currently available for analysis as a part of the MVP, so we were unable to evaluate the performance of the 127-variant GRS for variation in these clinical measures.
The goal of IGS is to reduce elevated IOP in patients for whom less invasive POAG treatment options (e.g. IOP-lowering drops, laser trabeculoplasty, and microinvasive glaucoma surgery) have not been effective despite adequate treatment compliance.27 Previous studies have reported associations between risk scores and the need for incisional surgery as well as other clinical POAG outcomes including maximum IOP, POAG severity, and number of POAG-affected family members.35,36 However, neither of these studies examined this phenomenon in AFR individuals, for whom POAG risk is higher. We posit that predictive tools like GRS could be used to stratify POAG patients based on whether they might advance to severe POAG and require IGS. We examined the ability of the 127-variant GRS to stratify POAG cases by documented IGS. We found that the top GRS deciles consistently categorized a higher proportion of EUR Veterans who had IGS documented in their VA EHR compared to AFR Veterans (Fig S4G–I, available at www.aaojournal.org). However, it is important to note both the poor overall performance of these models based on AUC (Table S9 and Fig S5, available at www.aaojournal.org) and AUC-PRC (Fig S7, available at www.aaojournal.org) as well as the influence of non-genetic risk factors, including access to eye care and treatment adherence, that are important risk factors that contribute to disease progression and the need for IGS.66 Therefore, considerations about GRS for IGS prediction warrant more comprehensive risk model assessments before they can be appropriately and equitably introduced to the clinic.
We detected associations between the GRS and specific IGS types (Table S7, available at www.aaojournal.org); however, it is important to note that the type of surgery performed is provider-dependent. Historically, trabeculectomies were preferred over tube-shunt procedures except in refractory cases or those at high risk for filtration failure.67 However, the number of tube-shunt implantations (e.g. GDI) in individuals with medically uncontrolled POAG has increased in recent years due to (i) the concern over long-term trabeculectomy-related complications, (ii) improved efficacy of tube shunts, and (iii) the higher rate of surgical success of tube shunts demonstrated by the Tube Versus Trabeculectomy (TVT) study.68 Therefore, from a clinical perspective, associations found with GRS and any type of IGS provide the most clinically useful information compared to associations for a particular IGS (trabeculectomy vs. GDI). Additional research is needed to evaluate the utility and implementation of predictive methods like GRS for risk stratification and clinical decisions in glaucoma care.
Phenotype definition differences between large POAG studies are important to consider when using previously published effect estimates and risk variants in GRS analyses. Because POAG is phenotypically heterogeneous, consistency in phenotype definitions for POAG may not be feasible across samples comprising meta-analyses8 or of biobank mega-analyses;69 this lack of consistency can contribute to diminished statistical power to detect true association signals. In our study, we defined POAG case-control status based on a computable phenotyping algorithm,53 which we previously developed and validated in conjunction with eye clinic staff (optometrists and ophthalmologists with access to gold-standard clinical diagnosis data).53 Further, we ensured that phenotyping performance was consistent between EUR and AFR Veterans in the MVP.53
The EHR-linked MVP mega-biobank represents an important opportunity to independently evaluate POAG GRS in diverse populations using previously discovered POAG-associated variants and loci. Other GRS and PRS studies can suffer from performance overestimation when they use the same data to determine GWAS-based effect estimates and evaluate risk score performances.70,71 Additionally, the MVP database has greater ancestral diversity compared to other large biobanks. Historically, the majority of complex disease genetics research has consisted of data from EUR individuals,45 and the information gleaned from these studies has generally not replicated well in other ancestral groups,72–75 especially for AFR and admixed individuals.41,76 This disparity is particularly pertinent for POAG risk prediction in AFR individuals because POAG is more common,24 presents earlier,77 and progresses faster in AFR populations.25,78–80 As almost 30% of the Veterans enrolled in the MVP are of non-European ancestry,81 we were able to include a large number of AFR individuals (n = 2448 cases and 5665 controls) in this study. Despite having a sufficient sample size to power our calculations, we found that the GRS underperformed for AFR individuals (Fig 3). Based on previously described GRS limitations74,75,82,83 and the findings from this study, it is clear that greater ancestral diversity in genetics research is needed so that (i) health disparities are not further exacerbated in already medically underserved populations and (ii) the clinical utility of risk models based on genetic data are not limited in applicability across groups.
We recognize that there are limitations to this study. Due to the historical predominance of males in the U.S. military, the MVP consists mostly of males (92%);48 97.5% of the participants in this study were male due to the age criteria we used to define POAG cases and controls. Therefore, additional studies should be conducted to evaluate the performance of POAG GRS in females. Moreover, because the MVP is an EHR-linked biobank for Veterans who sought care through the VA healthcare system, we do not have access to information regarding diagnoses and procedures Veterans may have received from non-VA facilities. Although we calculated and evaluated GRS in EUR and AFR individuals, we did not examine the POAG risk architecture in other ancestral groups. Future work should expand GRS approaches to additional ancestral groups. Additionally, we limited our GRS to include only the 127 POAG-associated variants described in the largest-to-date cross-ancestry POAG meta-analysis, and ancestry-specific weights for the AFR group were unavailable for 4 variants on the X chromosome. Therefore, future work should consider the potential contributions of genetic variation beyond these 127 variants, especially for the sex chromosomes.
In summary, we leveraged the MVP, a large, multi-ancestry biobank linked to EHR data, to evaluate genetic risk for POAG. While GRS have clinical potential, inclusion of diverse ancestral groups in genomics research is needed to ensure that GRS are equitable and actionable.75,84 The results from this study further emphasize the necessity of including data from historically underrepresented populations in biobanks and biomedical research databases. Employing this strategy would improve understanding of POAG risk and disease management across diverse populations, including populations with high POAG burden.
Supplementary Material
Financial Support
This research is based on data from the Million Veteran Program, Office of Research and Development, Veterans Health Administration, and was supported by award I01 BX004557. This publication does not represent the views of the Department of Veteran Affairs or the United States Government. We are grateful to the VINCI and GENISIS support teams as well as the MVP Core Statistical Analysis team for their contributions to this study. We also appreciate the Veterans who enrolled in the MVP. Without them, this work would not be possible. This work was also funded by the Cleveland Institute for Computational Biology, NIH Core Grants (P30 EY025585, P30 EY011373), and unrestricted grants from Research to Prevent Blindness to Case Western Reserve University (CWRU), Cleveland Clinic Lerner College of Medicine of CWRU, and the University of Buffalo. A.R.W. was supported by the CWRU Visual Sciences Training Program (T32 EY 7157-19) and the CWRU Clinical and Translational Scientist Training Program (TL1 TR 002549-04). L.A.C. was supported by the National Heart, Lung, and Blood Institute (T32 HL 0075-67). This publication was made possible by the Clinical and Translational Science Collaborative of Cleveland (UL1TR0002548) from the National Center for Advancing Translational Sciences (NCATS) component of the National Institutes of Health and NIH roadmap for Medical Research. This work made use of the High Performance Computing Resource in the Core Facility for Advanced Research Computing at Case Western Reserve University.
Abbreviations and acronyms
- 1KGP3
1000 Genomes Phase 3
- ACB
African Caribbean in Barbados
- ADAGES
African Descent and Glaucoma Evaluation Study
- AFR
African ancestry
- ASW
African Ancestry in Southwest USA
- AUC
area under the curve
- AUC-PRC
area under the precision-recall curve
- CEU
Utah residents with Northern and Western European ancestry
- CI
confidence interval
- CPT
Current Procedural Terminology
- EHR
electronic health record
- ESN
Esan in Nigeria
- EUR
European ancestry
- FIN
Finnish in Finland
- GBR
British in England and Scotland
- GDI
glaucoma drainage implants
- GRS
genetic risk score(s)
- GWAS
genome-wide association study
- GWD
Gambian in Western Division, The Gambia
- HARE
Harmonized Ancestry and Race/Ethnicity
- HTG
high-tension glaucoma
- IBS
Iberian populations in Spain
- IGS
invasive glaucoma surgery
- IOP
intraocular pressure
- IQR
interquartile range
- LWK
Luhya in Webuye, Kenya
- MSL
Mende in Sierra Leone
- MVP
Million Veteran Program
- NTG
normal-tension glaucoma
- OR
odds ratio
- ORD
Office of Research and Development
- PC
principal component
- POAAGG
Primary Open-Angle African American Glaucoma Genetics
- POAG
primary open-angle glaucoma
- PRC
precision-recall curves
- PRS
polygenic risk score(s)
- ROC
receiver operator characteristic
- SE
standard error
- TSI
Toscani in Italy
- TVT
Tube Versus Trabeculectomy
- VA
Veterans Affairs
- VCDR
vertical cup-to-disc ratio
- YRI
Yoruba in Ibadan, Nigeria
Footnotes
Meeting Presentation
Association for Research in Vision and Ophthalmology Annual Meeting, 2021
American Society of Human Genetics Annual Meeting, 2021
American Academy of Optometry Annual Meeting, 2021
This article contains additional online-only material. The following should appear online-only: Figures S1–7 and Tables S1–10.
Conflict of Interest
No conflicting relationship exists for any author.
Reference:
- 1.Wiggs JL, Pasquale LR. Genetics of glaucoma. Hum Mol Genet. 2017;26(R1):R21–R27. doi: 10.1093/hmg/ddx184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90(3):262–267. doi: 10.1136/bjo.2005.081224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tham YC, Li X, Wong TY, Quigley HA, Aung T, Cheng CY. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology. 2014;121(11):2081–2090. doi: 10.1016/j.ophtha.2014.05.013 [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization. World Report on Vision. World Health Organization; 2019. [Google Scholar]
- 5.Wang K, Gaitsch H, Poon H, Cox NJ, Rzhetsky A. Classification of common human diseases derived from shared genetic and environmental determinants. Nat Genet. 2017;49(9):1319–1325. doi: 10.1038/ng.3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanfilippo PG, Hewitt AW, Hammond CJ, Mackey DA. The heritability of ocular traits. Surv Ophthalmol. 2010;55(6):561–583. doi: 10.1016/j.survophthal.2010.07.003 [DOI] [PubMed] [Google Scholar]
- 7.Doucette LP, Rasnitsyn A, Seifi M, Walter MA. The interactions of genes, age, and environment in glaucoma pathogenesis. Surv Ophthalmol. 2015;60(4):310–326. doi: 10.1016/j.survophthal.2015.01.004 [DOI] [PubMed] [Google Scholar]
- 8.Gharahkhani P, Jorgenson E, Hysi P, et al. Genome-wide meta-analysis identifies 127 open-angle glaucoma loci with consistent effect across ancestries. Nature Communications. 2021;12(1):1258. doi: 10.1038/s41467-020-20851-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choquet H, Paylakhi S, Kneeland SC, et al. A multiethnic genome-wide association study of primary open-angle glaucoma identifies novel risk loci. Nature Communications. 2018;9(1):2278. doi: 10.1038/s41467-018-04555-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hysi PG, Cheng CY, Springelkamp H, et al. Genome-wide analysis of multi-ancestry cohorts identifies new loci influencing intraocular pressure and susceptibility to glaucoma. Nat Genet. 2014;46(10):1126–1130. doi: 10.1038/ng.3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gudiseva H v., Verma SS, Chavali VRM, et al. Genome wide-association study identifies novel loci in the Primary Open-Angle African American Glaucoma Genetics (POAAGG) study. bioRxiv. Published online March 12, 2020:2020.02.27.968156. doi: 10.1101/2020.02.27.968156 [DOI] [Google Scholar]
- 12.Bonnemaijer PWM, Iglesias AI, Nadkarni GN, et al. Genome-wide association study of primary open-angle glaucoma in continental and admixed African populations. Human Genetics. 2018;137(10):847–862. doi: 10.1007/s00439-018-1943-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The Genetics of Glaucoma in People of African Descent (GGLAD) Consortium, Hauser MA, Allingham RR, et al. Association of Genetic Variants With Primary Open-Angle Glaucoma Among Individuals With African Ancestry. JAMA. 2019;322(17):1682–1691. doi: 10.1001/jama.2019.16161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Allingham RR, Nakano M, et al. A common variant near TGFBR3 is associated with primary open angle glaucoma. Hum Mol Genet. 2015;24(13):3880–3892. doi: 10.1093/hmg/ddv128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khawaja AP, Bailey JNC, Wareham NJ, et al. Genome-wide analyses identify 68 new loci associated with intraocular pressure and improve risk prediction for primary open-angle glaucoma. Nature Genetics. 2018;50(6):778-+. doi: 10.1038/s41588-018-0126-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gharahkhani P, Burdon KP, Fogarty R, et al. Common variants near ABCA1, AFAP1 and GMDS confer risk of primary open-angle glaucoma. Nat Genet. 2014;46(10):1120–1125. doi: 10.1038/ng.3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Springelkamp H, Hohn R, Mishra A, et al. Meta-analysis of genome-wide association studies identifies novel loci that influence cupping and the glaucomatous process. Nature Communications. 2014;5:4883. doi: 10.1038/ncomms5883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Springelkamp H, Iglesias AI, Mishra A, et al. New insights into the genetics of primary open-angle glaucoma based on meta-analyses of intraocular pressure and optic disc characteristics. Human Molecular Genetics. 2017;26(2):438–453. doi: 10.1093/hmg/ddw399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheetz TE, Fingert JH, Wang K, et al. A Genome-Wide Association Study for Primary Open Angle Glaucoma and Macular Degeneration Reveals Novel Loci. PLoS ONE. 2013;8(3):e58657. doi: 10.1371/journal.pone.0058657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thorleifsson G, Walters GB, Hewitt AW, et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat Genet. 2010;42(10):906–909. doi: 10.1038/ng.661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibson J, Griffiths H, de Salvo G, et al. Genome-wide association study of primary open angle glaucoma risk and quantitative traits. Molecular Vision. 2012;18:1083. Accessed July 27, 2021. /pmc/articles/PMC3351427/ [PMC free article] [PubMed] [Google Scholar]
- 22.Ulmer M, Li J, Yaspan BL, et al. Genome-Wide Analysis of Central Corneal Thickness in Primary Open-Angle Glaucoma Cases in the NEIGHBOR and GLAUGEN Consortia. Investigative Opthalmology & Visual Science. 2012;53(8):4468–4474. doi: 10.1167/iovs.12-9784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooke Bailey JN, Loomis SJ, Kang JH, et al. Genome-wide association analysis identifies TXNRD2, ATXN2 and FOXC1 as susceptibility loci for primary open-angle glaucoma. Nature Genetics. 2016;48(2):189–194. doi: 10.1038/ng.3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choquet H, Wiggs JL, Khawaja AP. Clinical implications of recent advances in primary open-angle glaucoma genetics. Eye (Lond). 2020;34(1):29–39. doi: 10.1038/s41433-019-0632-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Restrepo NA, Bailey JNC. Primary Open-Angle Glaucoma Genetics in African Americans. Curr Genet Med Rep. 2017;5(4):167. doi: 10.1007/S40142-017-0131-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caprioli J Glaucoma: a disease of early cellular senescence. Invest Ophthalmol Vis Sci. 2013;54(14):ORSF60–ORSF67. doi: 10.1167/iovs.13-12716 [DOI] [PubMed] [Google Scholar]
- 27.Gedde SJ, Vinod K, Wright MM, et al. Primary Open-Angle Glaucoma Preferred Practice Pattern(R). Ophthalmology. Published online 2020. doi: 10.1016/j.ophtha.2020.10.022 [DOI] [PubMed] [Google Scholar]
- 28.Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nature Reviews Genetics 2018 19:9. 2018;19(9):581–590. doi: 10.1038/s41576-018-0018-x [DOI] [PubMed] [Google Scholar]
- 29.Cooke Bailey JN, Hoffman JD, Sardell RJ, Scott WK, Pericak-Vance MA, Haines JL. The Application of Genetic Risk Scores in Age-Related Macular Degeneration: A Review. J Clin Med. 2016;5(3):31. doi: 10.3390/jcm5030031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heesterbeek TJ, de Jong EK, Acar IE, et al. Genetic risk score has added value over initial clinical grading stage in predicting disease progression in age-related macular degeneration. Sci Rep. 2019;9(1):6611. doi: 10.1038/s41598-019-43144-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siu AL US Preventive Services Task Force. Screening for Breast Cancer: U.S. Preventive Services Task Force Recommendation Statement. Ann Intern Med. 2016;164(4):279–296. doi: 10.7326/M15-2886 [DOI] [PubMed] [Google Scholar]
- 32.Damen J, Hooft L, Schuit E, et al. Prediction models for cardiovascular disease risk in the general population: systematic review. BMJ. 2016;353:i2416. doi: 10.1136/BMJ.I2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daunt P, Ballard C, Creese B, et al. Polygenic Risk Scoring is an Effective Approach to Predict Those Individuals Most Likely to Decline Cognitively Due to Alzheimer’s Disease. J Prev Alzheimers Dis. 2021;8(1):78–83. doi: 10.14283/JPAD.2020.64 [DOI] [PubMed] [Google Scholar]
- 34.Osterman MD, Kinzy TG, Bailey JNC. Polygenic Risk Scores. Current Protocols. 2021;1(5). doi: 10.1002/cpz1.126 [DOI] [PubMed] [Google Scholar]
- 35.Craig JE, Han X, Qassim A, et al. Multitrait analysis of glaucoma identifies new risk loci and enables polygenic prediction of disease susceptibility and progression. Nature Genetics. 2020;52(2):160–166. doi: 10.1038/s41588-019-0556-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qassim A, Souzeau E, Siggs OM, et al. An Intraocular Pressure Polygenic Risk Score Stratifies Multiple Primary Open-Angle Glaucoma Parameters Including Treatment Intensity. Ophthalmology. 2020;127(7):901–907. doi: 10.1016/j.ophtha.2019.12.025 [DOI] [PubMed] [Google Scholar]
- 37.MacGregor S, Ong JS, An J, et al. Genome-wide association study of intraocular pressure uncovers new pathways to glaucoma. Nat Genet. 2018;50(8):1067–1071. doi: 10.1038/s41588-018-0176-y [DOI] [PubMed] [Google Scholar]
- 38.Gao XR, Huang H, Kim H. Polygenic Risk Score Is Associated With Intraocular Pressure and Improves Glaucoma Prediction in the UK Biobank Cohort. Translational Vision Science & Technology. 2019;8(2):10-10. doi: 10.1167/tvst.8.2.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zebardast N, Sekimitsu S, Wang J, et al. Characteristics of Gln368Ter Myocilin Variant and Influence of Polygenic Risk on Glaucoma Penetrance in the UK Biobank. Ophthalmology. 2021;S0161–6420(21):00189–5. doi: 10.1016/j.ophtha.2021.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan BJ, Bailey JC, Igo RP, et al. Association of a Primary Open-Angle Glaucoma Genetic Risk Score With Earlier Age at Diagnosis. JAMA Ophthalmology. 2019;137(10):1190–1194. doi: 10.1001/jamaophthalmol.2019.3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51(4):584–591. doi: 10.1038/s41588-019-0379-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cavazos TB, Witte JS. Inclusion of variants discovered from diverse populations improves polygenic risk score transferability. HGG Adv. 2021;2(1):100017. doi: 10.1016/j.xhgg.2020.100017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marquez-Luna C, Loh PR, South Asian Type 2 Diabetes C, Consortium ST 2 D, Price AL. Multiethnic polygenic risk scores improve risk prediction in diverse populations. Genet Epidemiol. 2017;41(8):811–823. doi: 10.1002/gepi.22083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sirugo G, Williams SM, Tishkoff SA. The Missing Diversity in Human Genetic Studies. Cell. 2019;177(1):26–31. doi: 10.1016/j.cell.2019.02.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duncan L, Shen H, Gelaye B, et al. Analysis of polygenic risk score usage and performance in diverse human populations. Nature Communications. 2019;10(1):1–9. doi: 10.1038/s41467-019-11112-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolford B, Willer C, Surakka I. Electronic health records: the next wave of complex disease genetics. Hum Mol Genet. 2018;27(R1):R14–R21. doi: 10.1093/HMG/DDY081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li R, Chen Y, Ritchie MD, Moore JH. Electronic health records and polygenic risk scores for predicting disease risk. Nat Rev Genet. 2020;21(8):493–502. doi: 10.1038/s41576-020-0224-1 [DOI] [PubMed] [Google Scholar]
- 48.Gaziano JM, Concato J, Brophy M, et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214–223. doi: 10.1016/j.jclinepi.2015.09.016 [DOI] [PubMed] [Google Scholar]
- 49.Saeedi O, Ashraf H, Slade EP, et al. Trends in Prevalence of Diagnosed Ocular Disease and Utilization of Eye Care Services in American Veterans. Am J Ophthalmol. 2017;173:70–75. doi: 10.1016/j.ajo.2016.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maa AY, Evans C, Delaune W, Lynch MG. Veteran eye disease after eligibility reform: prevalence and characteristics. Mil Med. 2013;178(7):811–815. doi: 10.7205/MILMED-D-12-00537 [DOI] [PubMed] [Google Scholar]
- 51.Hunter-Zinck H, Shi Y, Li M, et al. Genotyping Array Design and Data Quality Control in the Million Veteran Program. The American Journal of Human Genetics. 2020;106(4):535–548. doi: 10.1016/j.ajhg.2020.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fang H, Hui Q, Lynch J, et al. Harmonizing Genetic Ancestry and Self-identified Race/Ethnicity in Genome-wide Association Studies. Am J Hum Genet. 2019;105(4):763–772. doi: 10.1016/j.ajhg.2019.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nealon CL, Halladay CW, Kinzy TG, et al. Development and Evaluation of a Rules-based Algorithm for Primary Open-Angle Glaucoma in the VA Million Veteran Program. Ophthalmic Epidemiology. Published online November 25, 2021:1–9. doi: 10.1080/09286586.2021.1992784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fihn SD, Francis J, Clancy C, et al. Insights From Advanced Analytics At The Veterans Health Administration. Health Affairs. 2014;33(7):1203–1211. doi: 10.1377/hlthaff.2014.0054 [DOI] [PubMed] [Google Scholar]
- 55.Saito T, Rehmsmeier M. The Precision-Recall Plot Is More Informative than the ROC Plot When Evaluating Binary Classifiers on Imbalanced Datasets. PLOS ONE. 2015;10(3):e0118432. doi: 10.1371/JOURNAL.PONE.0118432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuhn M Building Predictive Models in R Using the caret Package. Journal of Statistical Software. 2008;28(5):1–26. doi: 10.18637/JSS.V028.I0527774042 [DOI] [Google Scholar]
- 57.Nagelkerke NJ. A note on a general definition of the coefficient of determination. Biometrika. 1991;78(3):691–692. doi: 10.1093/biomet/78.3.691 [DOI] [Google Scholar]
- 58.Lee SH, Goddard ME, Wray NR, Visscher PM. A Better Coefficient of Determination for Genetic Profile Analysis. Genetic Epidemiology. 2012;36(3):214–224. doi: 10.1002/gepi.21614 [DOI] [PubMed] [Google Scholar]
- 59.Zhang N, Wang J, Li Y, Jiang B. Prevalence of primary open angle glaucoma in the last 20 years: a meta-analysis and systematic review. Scientific Reports. 2021;11(1):13762. doi: 10.1038/s41598-021-92971-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cooke Bailey J, Kinzy TG, Nealon C, et al. POAG genetic risk score performs worse in African-descent than European-descent samples, highlighting need for expanded genetic studies in diverse populations. Investigative Ophthalmology & Visual Science. 2021;62(8):1513–1513. [Google Scholar]
- 61.Hoffmann TJ, Tang H, Thornton TA, et al. Genome-wide association and admixture analysis of glaucoma in the Women’s Health Initiative. Human Molecular Genetics. 2014;23(24):6634–6643. doi: 10.1093/hmg/ddu364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cole BS, Gudiseva H v., Pistilli M, et al. The Role of Genetic Ancestry as a Risk Factor for Primary Open-angle Glaucoma in African Americans. Investigative Opthalmology & Visual Science. 2021;62(2). doi: 10.1167/iovs.62.2.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taylor KD, Guo X, Zangwill LM, et al. Genetic Architecture of Primary Open-Angle Glaucoma in Individuals of African Descent. Ophthalmology. 2019;126(1):38–48. doi: 10.1016/j.ophtha.2018.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Research. 2009;19(9):1655–1664. doi: 10.1101/GR.094052.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.1000 Genomes Project Consortium, Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dreer L, Girkin C, Mansberger S. Determinants of medication adherence to topical glaucoma therapy. J Glaucoma. 2012;21(4):234–240. doi: 10.1097/IJG.0B013E31821DAC86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gedde S, Heuer D, Parrish R. Review of results from the Tube Versus Trabeculectomy Study. Curr Opin Ophthalmol. 2010;21(2):123–128. doi: 10.1097/ICU.0B013E3283360B68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gedde S, Singh K, Schiffman J, Feuer W. The Tube Versus Trabeculectomy Study: interpretation of results and application to clinical practice. Curr Opin Ophthalmol. 2012;23(2):118–126. doi: 10.1097/ICU.0B013E32834FF2D1 [DOI] [PubMed] [Google Scholar]
- 69.Zhou W, Kanai M, Wu KHH, et al. Global Biobank Meta-analysis Initiative: powering genetic discovery across human diseases. medRxiv. 2021;27(5):2021.11.19.21266436. doi: 10.1101/2021.11.19.21266436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shi J, Park JH, Duan J, et al. Winner’s Curse Correction and Variable Thresholding Improve Performance of Polygenic Risk Modeling Based on Genome-Wide Association Study Summary-Level Data. PLOS Genetics. 2016;12(12):e1006493. doi: 10.1371/journal.pgen.1006493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Janssens ACJW. Validity of polygenic risk scores: are we measuring what we think we are? Human Molecular Genetics. 2019;28(R2):R143–R150. doi: 10.1093/hmg/ddz205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Petrovski S, Goldstein DB. Unequal representation of genetic variation across ancestry groups creates healthcare inequality in the application of precision medicine. Genome Biology 2016 17:1. 2016;17(1):1–3. doi: 10.1186/S13059-016-1016-Y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bustamante CD, de La Vega FM, Burchard EG. Genomics for the world. Nature 2011 475:7355. 2011;475(7355):163–165. doi: 10.1038/475163a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Need AC, Goldstein DB. Next generation disparities in human genomics: concerns and remedies. Trends in Genetics. 2009;25(11):489–494. doi: 10.1016/J.TIG.2009.09.012 [DOI] [PubMed] [Google Scholar]
- 75.Adeyemo A, Balaconis MK, Darnes DR, et al. Responsible use of polygenic risk scores in the clinic: potential benefits, risks and gaps. Nature Medicine 2021. Published online November 15, 2021:1–9. doi: 10.1038/s41591-021-01549-6 [DOI] [PubMed] [Google Scholar]
- 76.Wang Y, Guo J, Ni G, Yang J, Visscher PM, Yengo L. Theoretical and empirical quantification of the accuracy of polygenic scores in ancestry divergent populations. Nature Communications. 2020;11(1):3865. doi: 10.1038/s41467-020-17719-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tielsch JM, Katz J, Sommer A, Quigley HA, Javitt JC. Family history and risk of primary open angle glaucoma. The Baltimore Eye Survey. Arch Ophthalmol. 1994;112(1):69–73. doi: 10.1001/archopht.1994.01090130079022 [DOI] [PubMed] [Google Scholar]
- 78.Grant W, Burke J. Why do some people go blind from glaucoma? Ophthalmology. 1982;89(9):991–998. doi: 10.1016/S0161-6420(82)34675-8 [DOI] [PubMed] [Google Scholar]
- 79.Martin M, Sommer A, Gold E, Diamond E. Race and primary open-angle glaucoma. Am J Ophthalmol. 1985;99(4):383–387. doi: 10.1016/0002-9394(85)90001-7 [DOI] [PubMed] [Google Scholar]
- 80.Bonnemaijer P, lo Faro V, Sanyiwa A, et al. Differences in clinical presentation of primary open-angle glaucoma between African and European populations. Acta Ophthalmol. 2021;99(7):e1118–e1126. doi: 10.1111/AOS.14772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hunter-Zinck H, Shi Y, Li M, et al. Measuring genetic variation in the multi-ethnic Million Veteran Program (MVP). bioRxiv. Published online January 7, 2020:2020.01.06.896613. doi: 10.1101/2020.01.06.896613 [DOI] [Google Scholar]
- 82.Manrai AK, Funke BH, Rehm HL, et al. Genetic Misdiagnoses and the Potential for Health Disparities. https://doi.org/101056/NEJMsa1507092. 2016;375(7):655–665. doi: 10.1056/NEJMSA1507092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hollitt GL, Siggs OM, Ridge B, et al. Attitudes towards polygenic risk testing in individuals with glaucoma. Ophthalmology Glaucoma. Published online November 11, 2021. doi: 10.1016/J.OGLA.2021.11.002 [DOI] [PubMed] [Google Scholar]
- 84.Han X, Hewitt AW, MacGregor S. Predicting the Future of Genetic Risk Profiling of Glaucoma: A Narrative Review. JAMA Ophthalmology. 2021;139(2):224–231. doi: 10.1001/JAMAOPHTHALMOL.2020.5404 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


