Abstract
Sex determination (SD) is not conserved among teleost fishes and can even differ between populations of the same species. Across the outstandingly species-rich fish family Cichlidae, more and more SD systems are being discovered. Still, the picture of SD evolution in this group is far from being complete. Lake Tanganyika and its affluent rivers are home to Astatotilapia burtoni, which belongs to the extremely successful East African cichlid lineage Haplochromini. Previously, in different families of an A. burtoni laboratory strain, an XYW system and an XY system have been described. The latter was also found in a second laboratory strain. In a laboratory-reared family descending from a population of the species’ southern distribution, a second XY system was discovered. Yet, an analysis of sex chromosomes for the whole species distribution is missing. Here, we examined the genomes of 11 natural populations of A. burtoni, encompassing a wide range of its distribution, for sex-linked regions. We did not detect signs of differentiated sex chromosomes and also not the previously described sex chromosomal systems present in laboratory lines, suggesting different SD systems in the same species under natural and (long-term) artificial conditions. We suggest that SD in A. burtoni is more labile than previously assumed and consists of a combination of non-genetic, polygenic, or poorly differentiated sex chromosomes.
Keywords: sex determination, genome, GWAS, genome sequencing, cichlid fish, sex chromosomes
Introduction
Sex determination (SD) is the process of a sexually reproducing organism defining its sex (Capel 2017). Albeit serving the unifying goal of establishing sex, SD systems and their molecular mechanisms are not conserved across the tree of life (Cutting et al. 2013; Bachtrog et al. 2014) and can even differ among populations of the same species (Anderson et al. 2012; Yamamoto et al. 2014; Li et al. 2018; Pennell et al. 2018; Taslima et al. 2020). For example, in the European common frog (Rana temporaria), a gradient of sex chromosome differentiation exists along a geographical range (Rodrigues et al. 2014); sex chromosomes exhibit higher genetic differentiation in the northern-boreal population of the common frog compared to the southernmost population, while populations in between show intermediate levels of sex chromosome differentiation at the sequence level. In the latter, Rodrigues et al. (2014) noted some mismatches between genotypic and phenotypic sex, suggesting that other mechanisms or factors could be driving SD or overriding the otherwise strong genetic sex determiner. Likewise, the Japanese wrinkled frog (Rana rugosa) has four types of SD systems depending on its region of origin (Miura 2008). In the North of Japan, R. rugosa has a female ZW heterogametic system with heteromorphic sex chromosomes, while the three southern/western forms feature a male XY heterogametic system, but only one of them has differentiated sex chromosomes. Finally, in a stickleback (Gasterosteus aculeatus) population from the Japan Sea, a neo-Y sex chromosome was identified that seems to be coupled to reproductive isolation (Kitano et al. 2009).
Interestingly, previous studies revealed that laboratory model species can show discrepancies in their sex chromosomal systems compared to those present in their wild counterparts: Laboratory strains of zebrafish (Danio rerio) typically lack cytogenetically detectable heteromorphic sex chromosomes and have been assumed to use a polygenic SD system (Amores and Postlethwait 1998; Phillips et al. 2006). However, a study performed on natural zebrafish populations identified a heteromorphic ZW sex chromosomal system (Sharma et al. 1998). Within some laboratory strains, although lacking signs of sex chromosome differentiation, crosses also suggested dominant female sex determiners (Tong et al. 2010). Furthermore, SD in zebrafish is sensitive to harsh environmental conditions and different studies revealed different sex-associated loci (Anderson et al. 2012; Liew et al. 2012; Howe et al. 2013; Liew and Orbán 2014), suggesting SD to be polygenic (Bradley et al. 2011; Liew et al. 2012). Finally, Wilson et al. (2014) showed that the ZW system found in natural zebrafish populations got “accidentally” lost, or selected against, during the establishment of laboratory lines when selecting against mildly deleterious mutations.
In Siamese fighting fish (Betta splendens), laboratory strains show higher penetrance for an XY system than their wild counterparts, probably due to selection during domestication in favor of more predictable sex ratios. Other sex determination systems might be at play in the wild (Kwon et al. 2022).
Observations mainly of laboratory-reared fish have revealed polygenic SD in Lake Malawi cichlids with different co-existing sex chromosomes and hence several male and female genotypes with epistasis between alleles at the different loci determining sex (e.g. Moore et al. 2022).
Here, we set out to investigate genomic signatures of sex linkage in natural populations of the cichlid fish Astatotilapia burtoni (Günther 1893). A. burtoni has served as model species in numerous research fields, including neurobiology (Hofmann 2003; Maruska and Fernald 2018), developmental biology (Robison et al. 2001; Juntti et al. 2016), behavior (Hofmann 2003; Theis et al. 2017), genetics and genomics (Lang et al. 2006; Egger et al. 2017; El Taher et al. 2019), and speciation (Weber et al. 2021). A. burtoni is phylogenetically placed within the “modern haplochromines” (Salzburger et al. 2005; Meyer et al. 2015). The modern haplochromines form the most species-rich cichlid tribe comprising the Lake Tanganyika (LT) endemic Tropheini and the entire species flocks of the Lake Malawi and Victoria radiations, among others (Verheyen 2003; Salzburger et al. 2005, 2014). While A. burtoni is present in LT, it is not part of the endemic LT radiation (Salzburger 2018; Ronco et al. 2020, 2021). A. burtoni occurs in LT, tributary rivers and swamps (De Vos et al. 2001; Kullander and Roberts 2011), with a high degree of population structure between the northern and southern part of the lake detected by nuclear markers, as well as a deep divergence between southwestern populations and populations from the north and southeastern part of the lake (Pauquet et al. 2018; Weber et al. 2021).
Sex-determining regions of A. burtoni have been identified on three different chromosomes (Table 1). Following the cichlid community convention, the naming of A. burtoni chromosomes refers to linkage groups of the genome assembly of another cichlid species, the Nile tilapia (Oreochromis niloticus). Cichlids show high degrees of large-scale chromosomal synteny (Mazzuchelli et al. 2012). The Nile tilapia is often used as a reference due to the quality of its reference genome (Conte et al. 2017), its karyotype of 2n = 44 [the modal karyotype of African cichlids, (Majtánová et al. 2019)], as well as the fact that it is an outgroup to all cichlid species of the great lake radiations (Ronco et al. 2021, Extended Data Fig. 1, divergence of Oreochromini ∼16 Mya).
Table 1.
Astatotilapia burtoni sex determination systems previously described in laboratory-reared specimens.
| Reference | SD system type | Corresponding LG in O. niloticus genome | Genomic location on O. niloticus LG | SD candidate genes |
|---|---|---|---|---|
| Roberts et al. 2016 | XY | LG05-14 (fused in A. burtoni) | LG05 0–25 Mb LG14 0–18 Mb |
wnt4
wnt7a |
| XYW | LG13 | 3.2–7.7 Mb |
cyp17a1
bmpr1a |
|
| Böhne et al. 2016 | XY | LG05-14 (fused in A. burtoni) | LG05 6.8–20.3 Mb LG14 0–15 Mb |
wnt4
wnt7a |
| XY | LG18 | 3.9–19 Mb | — |
Fig. 1.
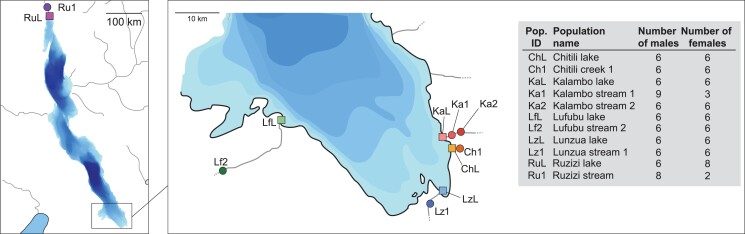
Map of Lake Tanganyika showing the 11 sampling locations of populations investigated. Squares represent lake, and circles represent stream populations. Bathymetric colors indicate lake depth (increasing color darkness represents increasing depth). Modified from Theis et al. (2017) with permission.
In two A. burtoni laboratory strains, an XY system is located on LGs 05 and 14, supporting a chromosomal fusion of these LGs compared to the Nile tilapia karyotype and in line with the A. burtoni karyotype (Böhne et al. 2016; Roberts et al. 2016). In families of one of the laboratory strains, an XYW system was detected on LG13 (Roberts et al. 2016). In a laboratory-reared family derived from wild-caught individuals of the south of LT, an XY system was found on LG18.
The reported XYW region on LG13 spans 4.6 Mb at the beginning of LG13 with cytochrome P450 17alpha-hydroxylase (cyp17a1) and type I bone morphogenetic protein receptor (bmpr1a) as prime candidate genes for SD; The XY region on LG05/14 has the highest density of and most significantly sex-associated SNPs located on LG05 between 6.8 and 20.3 Mb, including wingless-type MMTV integration site family member 4 (wnt4) as the top candidate gene for SD. The XY system on LG18 encompasses ∼16 Mb (Böhne et al. 2016). Until now, the interaction and evolutionary history of these three sex-linked systems remain unstudied, in particular in wild A. burtoni populations.
Here, we investigated signatures of sex-linkage based on genomic data of specimens of 11 natural populations of A. burtoni (Fig. 1). We searched the genomes of 61 females and 71 males for molecular traces of the three sex chromosomal systems identified previously (Böhne et al. 2016; Roberts et al. 2016). To this end, we performed calculations of intersex Fst and a genome-wide association (GWAS) test with sex. Next, we generated de novo genome assemblies for each sex of the 11 populations and performed coverage analyses to trace down differences between sexes. We further applied a k-mer-based approach to detect sex-associated regions across the genome and used those to confirm and narrow down previous genome-wide results. Finally, we tested the prevalence of previously described male-specific markers in the natural populations.
Materials and methods
DNA samples
We analyzed whole-genome sequencing data from Weber et al. (2021), accessible under the NCBI BioProject accession number PRJNA485198, from 11 different A. burtoni populations, comprising a total of 132 individuals. Genome data were available for typically six females and six males per population; for exact numbers, geographic locations, names, and acronyms of the populations used in this study, see Fig. 1.
Variant calling
DNA-sequencing data per individual were quality filtered and adapters removed with Trimmomatic V0.36 (Bolger et al. 2014) in PE mode with the settings adapterfile:2:30:12:8:true MINLEN:30. Reads were mapped against the Nile tilapia (Oreochromis niloticus) genome assembly version 2 (RefSeq accession number GCF_001858045.1_ASM185804v2), which was the only cichlid genome assembly on chromosomal level available to us at the time. Prior to mapping, unplaced scaffolds of this genome assembly were concatenated lexicographically into an “UNPLACED” super chromosome. This customized reference was indexed with BWA V0.7.13 and individual DNA reads were aligned against it with bwa-mem under default parameters (Li and Durbin 2009). Alignments were coordinate-sorted and indexed with SAMtools 1.3.1 (Li et al. 2009). Variants were called with GATK’s V3.7 (McKenna et al. 2010) HaplotypeCaller (per individual and per chromosome), GenotypeGVCFs (per chromosome) and CatVariants (to merge all obtained VCF files). The final variants were filtered with GATK's VariantFiltration with settings “QD < 2.0”, “FS > 200.0”, “ReadPosRankSum < −20.0”, “SOR > 10.0”, “DP < 200” and “DP > 4,000” for indels and “MQ < 40.0”, “FS > 60.0”, “QD < 2.0”, “DP < 200”, “DP > 4,000”, ‘SOR > 7.5”, “MQRankSum < −12.5”, and “ReadPosRankSum < −10.0” for SNPs.
Genome-wide association test for sex
From the initially filtered vcf file with all populations combined, we removed sites with more than 20% missing data and sites that had more than two alleles, retaining only SNPs. The obtained vcf was phased and genotypes were imputed with beagle V5 (Browning and Browning 2016) and transformed into bed format with PLINK V1.9 (Purcell et al. 2007; Chang et al. 2015). We then ran an association test for sex using the univariate linear mixed model LMM integrated in GEMMA V0.97 (gemma -notsnp -lmm 4) accounting for population stratification with a relatedness matrix calculated within GEMMA (-k option) (Zhou and Stephens 2012). Genotypes of potential sex-linked sites were visualized with the R package Pheatmap V1.0.12 in R V3.6.0 (R Core Team 2018) using vcfR V1.8.0 (Knaus and Grünwald 2017).
Population genetic statistics
SNPs per population were subsetted from the filtered vcf file described above. We then calculated intersex Fst and male and female nucleotide diversity in windows of 10 kb with VCFtools V0.1.14 (Danecek et al. 2011).
Per population de novo genome assemblies
In order to analyze per-population coverage differences between the sexes, we generated de novo draft genome assemblies for each sex and each population. To guarantee sufficient coverage, sequencing reads of two randomly selected individuals per sex for the 11 A. burtoni populations were combined as input for these 22 de novo draft genome assemblies. Assemblies were generated as previously described (Malmstrøm et al. 2017; Böhne et al. 2019) using Celera Assembler V8.3 (Myers et al. 2000) and indexed with BWA V0.7.13 (Li and Durbin 2009). We aligned the draft assemblies against the Nile tilapia reference genome (refseq accession number GCF_001858045.2_O_niloticus_UMD_NMBU) using LAST V861 and lastal (Kiełbasa et al. 2011) to infer chromosomal locations of draft genome scaffolds.
Sequence coverage analyses per population
Following the strategy described by Böhne et al. (2019), we assessed differences in sequence coverage between males and females of a population to identify sex-linked genomic regions under the expectation that Y- and W-specific regions are not present in females and males, respectively, and that coverage on X and Z in sex-differentiated regions are reduced in males and females, respectively. To this aim, reads of each individual were trimmed and quality filtered prior to aligning them to the two de novo assemblies of the corresponding population, as described above (see variant calling section). Sequencing depth from all mapped reads per individual was calculated with SAMtools for all positions (i.e. samtools depth -aa). We next calculated the median of coverage per sex per population for each de novo assembly in R V3.5 (R Core Team 2018).
To visualize global patterns of coverage difference between males and females of a given population based on the method described in Böhne et al. (2019), we ordered scaffolds of each de novo assembly based on the coordinates retrieved from LAST alignments against the Nile tilapia reference genome. In addition, to extracting coverage from all alignments (which represents mixed coverage from both gametologs in the heterogametic sex if the reference genome only contains one sex chromosome and/or gametologs are poorly differentiated), we also retained only alignments with no mismatch (i.e. keep reads with flag “NM:i:0”) from reads mapped in proper pairs of the BAM files for each sex and population with SAMtools (i.e. samtools view –f 2). We estimated sequencing depth for those files in the same way described above, per sex and per population and as previously done in Böhne et al. (2019). Depth for males and females was visualized in 10 kb windows in R, as log2[(median values per sex per site)/(median of depth for the whole genome)]. To obtain genomic windows with significant differences in sequencing depth between sexes within linkage groups, we calculated the median male-female coverage ratio and median male-female coverage difference for each window, after depth normalization (i.e. median values per sex per site/median of depth for the whole genome). We identified windows with median-normalized coverage ratios outside the 99% quantile and a median-normalized coverage difference between the sexes >0.25 (i.e. the minimum value of coverage twice as high as the coverage in the opposite sex). Subsequently, we assessed statistical significance for each of the retained windows with a two-sided Wilcoxon signed-rank test for the median-normalized coverage values between sexes, with the corresponding 95% confidence interval. Finally, to test for differences in the number of significant windows with reduced coverage found for each assembly across chromosomes, a one-sided Fisher's exact test and Bonferroni correction for multiple testing were performed in R.
To identify sex-limited regions (i.e. Y- or W-chromosome sequences) in the de novo assembled scaffolds, we identified regions having (i) zero mapping coverage in the sex contrasting the one used in the assembly and (ii) mapping coverage of zero in no more than one individual of the same sex as used for the assembly.
We retained all regions with such a coverage pattern and a minimal length of 500 bp. Sex-specific median-normalized coverage of scaffolds containing one or more of these 500 bp regions were extracted, and coverage was visualized in 100 bp windows with the R package tidyverse (Wickham et al. 2019). Sequence similarities among scaffolds containing the so-identified sex-specific regions across all populations within each sex were assessed using the CAP3 sequence assembly program (Huang and Madan 1999) with default parameters, which collapses contiguous sequences. Next, we assigned sex-specific scaffolds to genomic regions of the Nile tilapia reference genome based on the LAST alignments described above. Finally, to identify protein-coding sequences in sex-specific regions, we extended the sequence of the sex-specific regions into their flanking regions to a total length of 5 kb (whenever contig length permitted) and blasted the obtained sequences to the NR database (version 2018-10-23) with blastx BLAST + V2.7.1 (Camacho et al. 2009). We retained the top 20 alignments with an e-value cutoff of <0.001.
Heterogametic system detection with k-mers
We applied a second approach to identify sex-specific sequences and their location in the genome as described in Akagi et al. (2014) and Böhne et al. (2019) for 10 of the 11 populations. We excluded the Ruzizi River population due to the low number of female samples (Fig. 1). Starting from trimmed sequencing reads (see above), we generated k-mer catalogs per population of all possible k-mers starting with “AG” and a length of 37 bp present in at least five specimens per population using a Python script provided in Akagi et al. (2014). We divided k-mer catalogs into four categories: Y-k-mers = male-specific, Z-k-mers = male-biased, X-k-mers = female-biased, and W-k-mers = female-specific. To this end, we applied a linear regression to the k-mer counts of each population and retained outliers from the general distribution by calculating studentized residuals from a linear model (i.e. jack-knifed residuals). Outliers were defined as all k-mers with an absolute studentized residual value equal to or bigger than 3, as an observation with an absolute value of 3 is deemed to be an outlier (Belsley et al. 1980; Hettmansperger 1987; Atkinson 1994). Subsequently, sex-specific k-mers (i.e. Y- or W-k-mers) were defined as k-mers having zero counts in one sex but not in the opposite sex. Sex-biased k-mers were obtained based on the ratio of counts between males and females, expecting larger counts for the homogametic sex (e.g. X-k-mers = female count/male count > 4, depending on the population analyzed). In summary, we retained outlier k-mers from the linear regression and from there we took (i) sex-specific k-mers (either Y- or W-k-mers) and (ii) sex-biased k-mers with ratios bigger than four for all populations but not in Kalambo River 1, (i.e. ratio threshold set to 12 for Z-k-mers due to the lower number of female samples for this population, see Fig. 1). Next, we tested for an increased amount of sex-specific k-mers per population with a Wilcoxon test, aiming to detect the heterogametic sex of the population. Additionally, we identified k-mers shared among populations in each category with UpSetR (Conway et al. 2017) in R.
For the Kalambo River (Ka2) and Chitili River (Ch1) populations, we extracted sequencing reads and their mates containing Y-k-mers of each population. Next, we assembled the extracted reads with MEGAHIT (Li et al. 2015) with –k-max 12. We also placed the resulting contigs onto the Nile tilapia reference genome with BWA and compared the contig data sets using blastX to the NR database in Blast2GO (Gotz et al. 2008) to retrieve functional annotations.
Population phylogenies using k-mers
To investigate if there are k-mers that would group individuals within or across populations by sex rather than reflecting the population structure, we computed phylogenies per population, for all samples, and for all samples within each chromosome using specimen k-mer counts generated with the alignment and assembly-free (AAF) software (Fan et al. 2015). We performed this calculation with two k-mer sizes, the default k-mer size of 25 bp and more specific 31 bp k-mers.
Comparing Y-specific sequences from a laboratory population
Using previously obtained and located (LG05) male-specific sequences from a restriction site associated DNA-sequencing experiment (Böhne et al. 2016), we tested for sequence similarities of these Y-specific RAD tags to the de novo assemblies with BlastN (Camacho et al. 2009).
Further sex chromosome identification approaches
We implemented SEX-DETector in a population through a Bayesian approach (SDpop) (Käfer et al. 2021) and Sex Assignment Through Coverage (SATC) (Nursyifa et al. 2022) per population and across all samples to detect sex chromosomes. These methods have been used to detect young and mildly degenerated sex chromosomes across different taxa.
Results
Intersex population genetics, sex association and previously identified sex-linked sites in A. burtoni natural populations
Similar to other studies (Wilson et al. 2014; Conte et al. 2017; Bergero et al. 2018; Pan et al. 2019), we searched for an accumulation of sex-specific alleles with the expectation that a sex chromosome would show increased sequence differences between males and females. Based on intersex Fst and comparisons of male-female nucleotide diversity within each population, we did not detect any sex-differentiated chromosome [i.e. expected increase of intersex Fst to 0.5 (Bhatia et al. 2013)] on the population level (Supplementary Fig. 1 File 1).
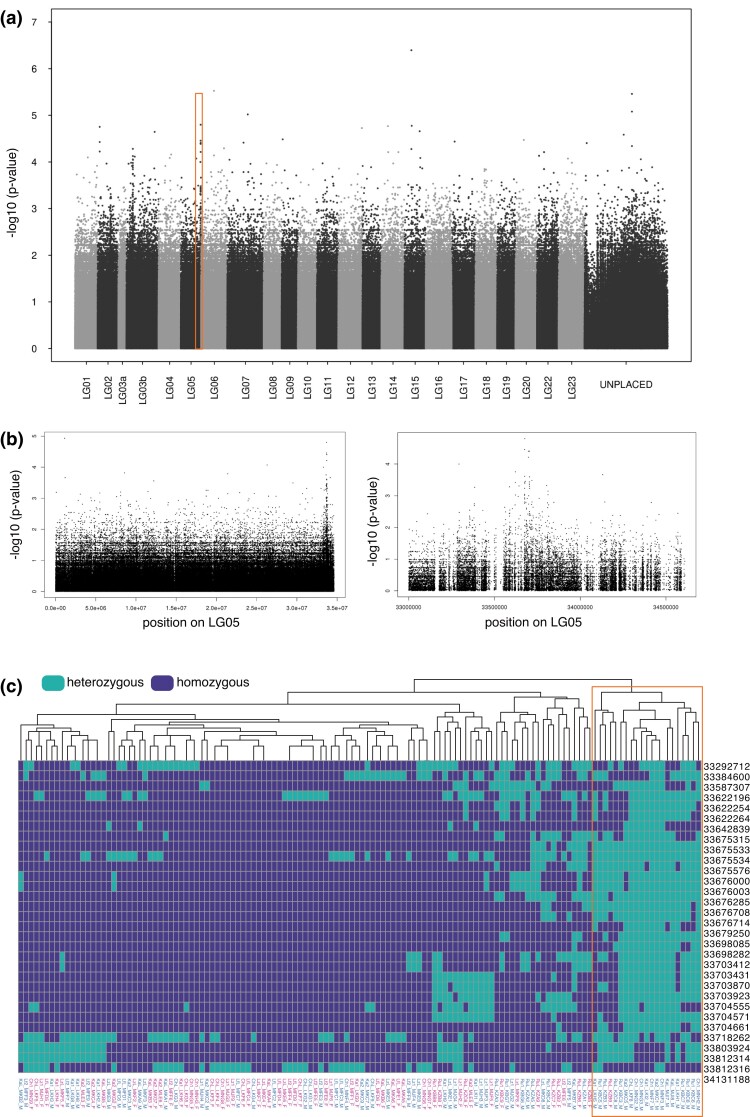
A GWAS analysis based on a dataset combining SNPs of all specimens of all populations did not identify a region of elevated, significant sex association. Concerning the previously identified sex-linked LGs in laboratory strains (Böhne et al. 2016; Roberts et al. 2016), visual inspection revealed only a narrow and not drastically elevated peak of SNPs with association to sex at the end of LG05 (Fig. 2a). Since this chromosome has previously been implicated in sex determination not only in A. burtoni but also other haplochromine cichlids (Böhne et al. 2016; Böhne et al. 2019; El Taher et al. 2021), we inspected the genotypes of the SNPs in this region. This revealed an excess of SNPs specific almost exclusively to a group of male samples from different populations (one female was included in this group as well as 20 males), matching an XY-patterning however only in a subset of samples and not reaching usual significant levels applied in GWAS (Fig. 2b and c, males from Ch1, Ru1, RuL, KaL, Ka1, and Ka2 population).
Fig. 2.
a) Manhattan plots of a GWAS analysis for an association with sex using the reference genome of the Nile tilapia (Oreochromis niloticus). Interchanging black and gray colors delimit chromosomes. Unplaced scaffolds were concatenated into an “UNPLACED” chromosome for visualization. The overlayed box indicates a region on LG05 with concentrated and increased association to sex. b) Magnifications of the region on chromosome 5 depicted with a box in panel a. c) The heatmap shows individual genotypes for SNPs with an elevated association to sex in a narrow peak region detected on LG05. A group of male individuals is largely heterozygous in this region (green squares) and groups by sex and not population (males from Ch1, Ru1, RuL, KaL, Ka1, and Ka2 populations; highlighted with a box). Chromosomal coordinates are shown on the right side; the sample clustering tree is shown on top; samples are indicated below the plot: _M for males and _F for females. Green: heterozygous genotypes, dark blue: homozygous genotypes.
Furthermore, while the chromosomal association would be consistent with previous results from A. burtoni, the exact position of these potentially male-specific SNPs differed from previous studies with a sex-determining region located at the opposite end of LG05 (Böhne et al. 2016).
We next inspected male and female genotypes of all individuals of genomic regions previously shown to be associated with sex (Böhne et al. 2016; Roberts et al. 2016). We did not observe any obvious clustering of sample genotypes by sex, which would have been indicative of an excess of heterozygous sites in one sex (e.g. males in the XY regions or females in the ZW region). Instead, we recovered the phylogenetic split between the southern and northern populations and mostly among the southern populations (Supplementary File 2).
We next investigated the presence of previously identified male-limited Y-specific sequences from a RAD-sequencing experiment of an A. burtoni laboratory strain in the population genomic data. Y-specific RAD markers identified from a laboratory strain were present in almost all male and female genome assemblies (Supplementary Table 1). Only one of the markers was not present in the female assemblies of two southern populations, ChL and Ka2 (Supplementary Table 1). Using a projection of our de novo assembled scaffolds to the Nile tilapia genome, we confirmed that the RAD tag sequences were located on scaffolds matching to LG05. For four of the assemblies, however, the scaffolds presented multiple mapping matches of similar quality also to other chromosomes (Lf2 female assembly to LG06, in ChL female assembly to LG03, in Ka1 male assembly to LG16 and in LfL female assembly to LG18). This suggests possible sequence duplications in the genome.
Identification of sex differences in coverage across the genome
We next targeted differences in coverage between sexes based on newly generated sex-specific de novo assemblies of each population. The quality analysis of the generated de novo assemblies revealed consistent and thus comparable quality across assemblies (Supplementary Table 2). We additionally tested for differences in the back-mapping quality within populations for the de novo assemblies (Supplementary Fig. 2 File 1 and Table 3), revealing a high average back-mapping rate (96.4–97.6%, Supplementary Table 3). There were no significant global differences in mapping and coverage between males and females within the same population, supporting no sex-specific skew in sequencing nor mapping (Supplementary Fig. 2 File 1). We did not detect sex-specific, chromosome-wide patterns of low sequence coverage in any of the populations (Supplementary File 2), which once more supports the result that there is no (strongly) differentiated sex chromosome in these populations.
We next identified 10 kb windows on each chromosome in each assembly with reduced sequence coverage in the opposite sex of the reference assembly (Supplementary Table 4) and tested if the number of such windows differed among chromosomes in each assembly (Fig. 3 and Supplementary Table 5). Assuming an XY system with sequence differentiation between X and Y, we would expect a reduced coverage in males for X-linked regions using the female assemblies, and vice versa a reduced coverage in females for Z-linked regions when using the male assemblies (Fig. 3). In poorly differentiated sex chromosome systems, we would, however, expect some read mapping to the Y chromosome from X-chromosome-originated reads as well as to the W-chromosome from Z-linked reads. Hence, we might not be able to unambiguously determine the type of heterogamety with this approach.
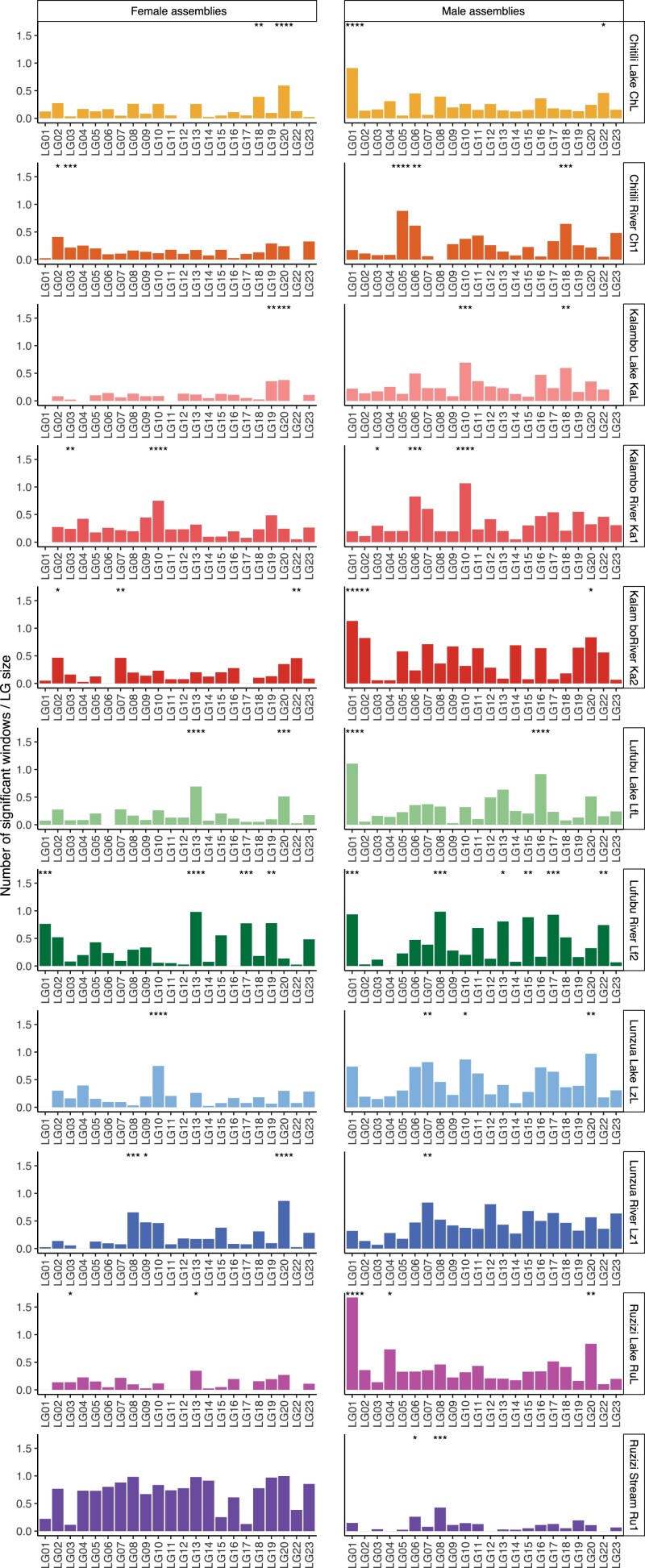
Fig. 3.
Reduced coverage based on mapping to sex-specific genome assemblies per chromosome (LGs). Bar plots show the number of 10 kb outlier windows of reduced coverage in males when mapped to female assemblies (left side) and in females when mapped to male assemblies (right side) by population and normalized by chromosomal length. Populations are color-coded as shown in Fig. 1. Significance levels after Bonferroni correction of a one-sided Fisher's exact test comparing the number of significant windows detected per each chromosome within a population and sex are indicated with *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
Concerning the previously described sex chromosomal system, we found little to no support for sex chromosome differentiation with this approach (Fig. 3 and Supplementary Table 5). For the two known XY systems, solely LG18 showed significantly reduced male coverage in the female assembly of ChL (which would support an XY system) but even more so on LG20. LG13 showed decreased female coverage only in Lf2 for male assemblies (supportive of a ZW system); however, several other LGs did as well. Interestingly, in the Ch1 male assembly, we detected significantly reduced female coverage on both LG05 and LG18. On LG05, most of the windows were located at the beginning of the chromosome (Supplementary File 3). As indicated above, while this might support loci with a ZW pattern, it could also be caused by Y-linked regions that still show some similarity to their X-linked alleles and, hence, have a poorer mapping in females. Out of all chromosomes that had previously not been described as sex-linked in A. burtoni, particularly, LG01 showed significantly reduced female mapping-coverage in five populations (ChL, Ka2, LfL, Lf2, RuL) and LG20 showed reduced male mapping-coverage in female assemblies in four populations (ChL, KaL, LfL, Lz1).
Based on the female assemblies, the two Lufubu populations (LfL and Lf2) showed a high number of windows of reduced male coverage that could support X-chromosome-like windows on LG13. The same pattern was true for the RuL population, albeit with a lower significance level (Fig. 3 and Supplementary Table 5). Overall, cichlid sex chromosomes are not strongly degenerated and hence the power to detect sex chromosomal sequences over coverage differences is limited, potential signals might be increased by increasing sample sizes but still be obscured if several SD systems coexist in A. burtoni.
Identification of sex-limited sequences
We next identified genomic regions from the de novo assemblies with regions of at least 500 bp length with zero read mapping in the sex not used for the assembly (Table 2 and Supplementary File 4). The number of these presumably sex-specific regions within the populations was rather low (0 to 56, average = 6.5) (Table 2). Some sex-specific regions were located within the same scaffold (Table 2 and Supplementary Fig. 3a & b File 1). The population that showed the largest number of female- and male-specific regions was Ru1, while Ka2 also showed an increase in male-specific regions; this suggests that Ru1 and Ka2 could have male sex-determining regions that are differentiated to some extent. However, it is important to keep in mind that sampling is sex-biased in the Ruzizi River population. We did not find a shared sex-specific region across populations within assemblies, supporting the hypothesis that there is no general conservation of sex chromosomes in A. burtoni. The number of sex-specific regions could lend some support for a ZW system in ChL and Lz1.
Table 2.
De novo assembled male and female scaffolds containing sex-specific regions (SSRs).
| Assembly sex | Population | Nr. of scaffolds in assembly | Nr. of SSR larger than 500 bp | Nr. of scaffolds with SSRs larger than 500 bp | 95% confidence intervals for the scaffolds with SSRs larger than 500 bp |
|---|---|---|---|---|---|
| Female | ChL | 46,282 | 6 | 5 | 1.62–11.67 |
| Ch1 | 47,268 | 2 | 2 | 0.24–7.22 | |
| KaL | 49,070 | 1 | 1 | 0.03–5.57 | |
| Ka1 | 46,494 | 5 | 4 | 1.09–10.24 | |
| Ka2 | 48,912 | 2 | 1 | 0.03–5.57 | |
| LfL | 46,346 | 3 | 3 | 0.62–8.77 | |
| Lf2 | 48,402 | 1 | 1 | 0.03–5.57 | |
| LzL | 47,122 | 1 | 1 | 0.03–5.57 | |
| Lz1 | 51,131 | 10 | 6 | 2.2–13.06 | |
| RuL | 47,758 | 0 | 0 | 0–3.69 | |
| Ru1 | 46,890 | 18 | 15 | 8.4–24.72 | |
| Male | ChL | 47,835 | 0 | 0 | 0–3.69 |
| Ch1 | 47,714 | 2 | 2 | 0.24–7.22 | |
| KaL | 47,992 | 0 | 0 | 0–3.69 | |
| Ka1 | 46,952 | 2 | 2 | 0.24–7.22 | |
| Ka2 | 48,541 | 24 | 14 | 7.66–23.48 | |
| LfL | 46,613 | 1 | 1 | 0.03–5.57 | |
| Lf2 | 48,243 | 1 | 1 | 0.03–5.57 | |
| LzL | 47,983 | 2 | 2 | 0.24–7.22 | |
| Lz1 | 49,422 | 4 | 1 | 0.03–5.57 | |
| RuL | 47,334 | 2 | 2 | 0.24–7.22 | |
| Ru1 | 45,710 | 56 | 39 | 27.74–53.3 |
Next, we inferred the coding potential for the scaffolds carrying the sex-limited regions and projected the scaffolds onto the Nile tilapia reference genome (Supplementary Fig. 3 File 1 and Supplementary Table 6). LG03 was the chromosome with the largest number of scaffolds with male-and female-limited regions (Supplementary Fig. 3 File 1 and Supplementary Table 6). This might be due to the fact that LG03 is the largest chromosome in the Nile tilapia genome. Note, however, that LG03 has also been reported as the sex chromosome in Oreochromini (Gammerdinger and Kocher 2018) and is rich in repetitive element content (Conte et al. 2017; Tao et al. 2020). Concerning the previously identified A. burtoni sex chromosomes, solely a female-specific region on a scaffold of Lf2 was located on LG13. However, we could not detect a coding gene in this region. Ka2 had one male-specific scaffold located on LG14 (Supplementary Fig. 3b File 1, Supplementary Table 6), yet, this scaffold also had secondary matches to different chromosomes (as LG05) and unplaced genomic regions (Supplementary Table 6). This scaffold had a protein sequence description associated with signaling receptor binding and regulation of immune response functions. Overall, no clear dominance on a single LG of the few sex-linked scaffolds was identified (Supplementary Fig. 3b File 1).
As a summary of the coding potential function detected, nine of the inferred protein-coding sequences showed similarities to mobile elements (6 scaffolds in females and 14 scaffolds in male assemblies), which are indeed a characteristic feature of sex chromosomes (Supplementary Fig. 3c File 1, Supplementary Table 6). Besides, several sex-limited sequences had a common protein-coding description between males and females. Most of the sequences were related to transposable elements as RNA-directed DNA polymerase, others were associated to small structural molecules such as zinc finger, and others were related to the immune system such as NACHT, LRR, and PYD domains or GTPase IMAP family members, among other associations (Supplementary Fig. 3c File 1, Supplementary Table 6). Finally, we did not find similarities to proteins that have previously been associated with SD or sex differentiation.
K-mer approaches to detect heterogametic sex and sex-specific sequences
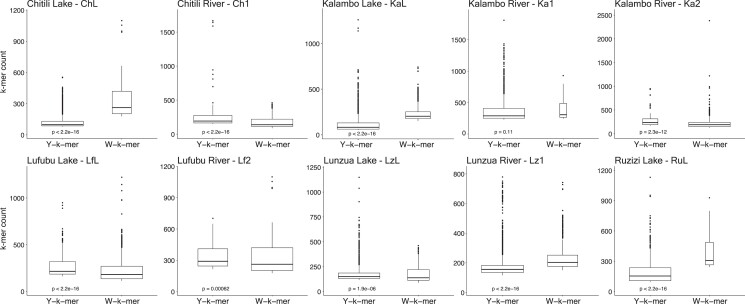
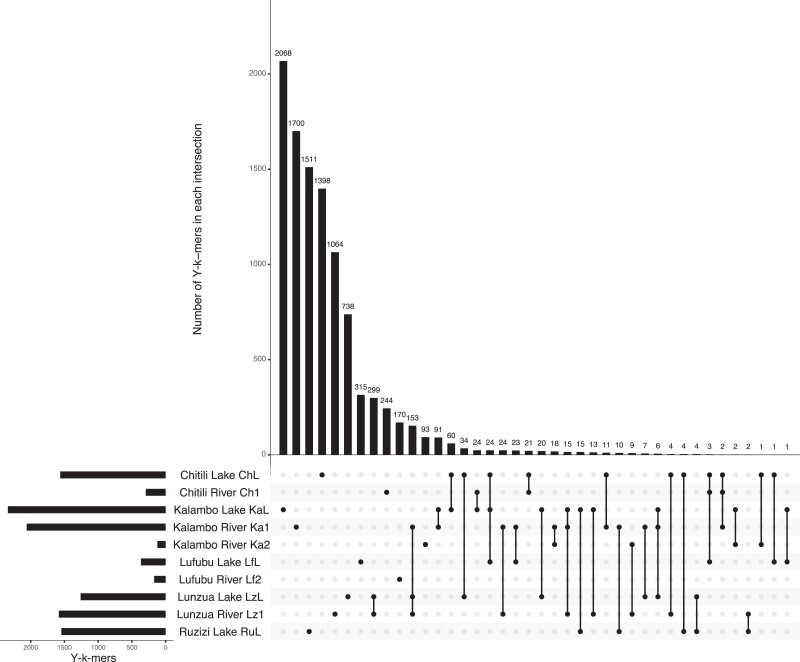
Since we did not detect major global sex differences along LGs, we next implemented a reference-genome-free approach by establishing k-mer catalogs for females and males of each population (Akagi et al. 2014; Böhne et al. 2019). Note that we excluded the Ru1 population due to the low number of female specimens available. We divided the k-mer counts of each population into categories, in either sex-specific (i.e. k-mers with zero counts in one sex, Y-mers or W-mers), or sex-biased (X- or Z-k-mers, see Supplementary Fig. 4 File 1 for a representation). We tested for a difference in Y-mer and W-mer coverage within each population as an indicator of a prevailing heterogametic system. Populations Ch1, Ka2, LfL and Lf2, and LzL had significantly higher coverage of Y-k-mers than W-k-mers (Fig. 4), supportive of male XY heterogametic systems. Conversely, ChL, KaL, Lz1, and RuL showed significantly higher coverage of W-k-mers than Y-k-mers, while having fewer k-mers overall (Fig. 4), suggesting a female ZW heterogametic system. Finally, we tested whether there were sex-specific and sex-biased k-mers shared across populations. Within each k-mer category, we found some degree of overlap between the k-mers of closely related populations; however, most k-mers were specific to a given population (Fig. 5 and Supplementary Fig. 5 File 1). Likewise, intersecting sex-specific and sex-biased k-mers showed that some k-mers belonged to opposite categories depending on the population investigated (e.g. 231 W-k-mers in KaL and Ka2 shared with Y-k-mers in LzL and Lz1, Supplementary Fig. 6 File 1). Finally, due to the fact that several analyses could support the presence of an XY male heterogametic system in Ka2, we further extracted reads containing Y-k-mers, assembled them and positioned the obtained contigs onto the Nile tilapia reference genome. We assembled 16 contigs, of which only two contigs had a location in a previously described sex-linked region, located in intergenic regions of LG13.
Fig. 4.
Boxplots of sex-specific k-mer coverage (Y- and W-k-mers) in the A. burtoni natural populations. Box width indicates the number of k-mers identified and the Y-axis their coverage. Wilcoxon test P-values are shown below for each population.
Fig. 5.
Comparison of Y-mers across populations. Number of male-specific k-mers per population is depicted by left-side bars. The compared populations are shown with dots (lower right) and the number of Y-k-mers found in each comparison is shown with bars. Note that there is a large proportion of k-mers specific to each population.
Phylogenetic reconstructions using k-mer differences
Next, based on the presence and coverage of k-mers across samples, we tested whether specimens within and between populations would group by sex rather than following the species tree, which would be indicative of shared/ancestral sex chromosome sequences (Böhne et al. 2019). As presumed, the k-mer-based phylogeny largely recovered the species/population topology (Supplementary Fig. 7a, b File 1), previously described by Egger et al. (2017); Pauquet et al. (2018); Weber et al. (2021), since most k-mers are not expected to be located on sex chromosomes. Exploring the tree topologies in more detail within each population revealed that the topologies are consistent with using all specimens together. In none of the cases, specimens grouped unambiguously by sex (Supplementary Fig. 8 File 1). These results would confirm a small number of sex-specific k-mers across the whole genome in each A. burtoni population.
We further investigated whether, for any chromosome, including the previously described sex chromosomes, specimens would group by sex. To this aim, we divided the k-mer catalog into the corresponding chromosome in Nile tilapia (Supplementary Fig. 9 File 1). As in the genome-wide analysis, we did not find any clustering by sex based on chromosome-specific k-mers but rather a similar general phylogenetic grouping as using genome-wide k-mers.
Further sex chromosome identification approaches
Additionally, we explored two recently published methods for SD detection that have successfully identified (young and mildly degenerated) sex chromosomes in other species: SATC (Nursyifa et al. 2022) and SDpop (Käfer et al. 2021). However, none of these novel methods detected any sex chromosome signal across all samples, nor within populations.
Discussion
In this study, using a suite of genomic tools, we investigated potentially sex-linked genomic regions of wild A. burtoni populations of LT and its surroundings and compared novel genomic sequences to previous results obtained from laboratory and wild strains. Our analyses with respect to potential sex-linked regions provide little support to the findings of previous studies on the sex-linked regions of A. burtoni laboratory strains and a wild population (Böhne et al. 2016; Roberts et al. 2016). Overall, we did not detect a shared sex chromosome system across all populations, nor strong population-specific sex chromosome signals.
On the per population level, our diverse set of approaches might support a male heterogametic XY system in the Ch1, Ka1, and Ka2 populations; however, we could not unambiguously locate this signal to chromosome level. With the samples at hand, we suspect the presence of female heterogametic systems in ChL and Lz1. However, a more extensive sampling per population or focusing on single families derived from these populations is needed to further disentangle if and which genomic regions are truly sex-linked in A. burtoni.
Across populations, we detected some signal of a potential XY system on LG05 (in populations ChL, Ch1, KaL, Ka1, Ka2, RuL, Ru1) that does, however, not overlap with the previously identified sex-differentiated region of A. burtoni laboratory strains on this chromosome. LG05 is among the chromosomes that emerged multiple times independently as a sex chromosome in different African cichlid lineages (El Taher et al. 2021). In the future, the inclusion of more samples per population, family-based data, and a detailed analysis of the sex-linked regions on this chromosome might shed light on the underlying genes driving the convergent evolution of the same chromosome as the sex chromosome in several cichlid species.
Applying similar methods as we did here to other (cichlid) species (Akagi et al. 2014; Böhne et al. 2019; El Taher et al. 2021), the detection and characterization of sex-linked regions are possible. Consequently, potential sex chromosomal system(s) in A. burtoni natural populations seem to show little, if any gametolog differentiation, rendering the applied approaches powerless even with a total of 132 individuals. This can either be attributed to the fact that sex chromosomes, if at all present, are of young age and/or show ongoing recombination (Charlesworth 2017). “Old” sex chromosomes might appear undifferentiated under continuous recombination as postulated under the “fountain-of-youth” model (Perrin 2009), which is based on the recombination of sex chromosomes in sex reversals. However, an assumption of this hypothesis is strong heterochiasmy between the sexes, for which we lack evidence in cichlids. Still, we did not find evidence for suppressed recombination on any chromosome in one sex, and thus continued recombination among most of the A. burtoni genome in both sexes seems to be in place.
Another complementary explanation to our results is that SD in A. burtoni is a complex polygenic feature as previously proposed for an A. burtoni laboratory strain with different SD systems and alleles thereof in different families: XY on LG05/14 and XYW on LG13 (Roberts et al. 2016). Within some of the populations, we found support for both types of heterogamety. To understand if this is a sampling or sequencing artifact or indeed an indication of complex SD, an even more extended sampling of A. burtoni will be needed. Currently, since we could not unambiguously identify sex-linked loci, we cannot resolve the genetic architecture of SD in A. burtoni. Yet, if previously identified, on the population level co-existing two sex chromosomal systems (XYW on LG13 and XY on LG05/14) were indeed the sole possibilities, we should have picked up those regions when analyzing all populations together and probably also with the population-specific analyses given the 132 samples included here, which were sequenced to an average coverage per individual of 9.8× to 24.5× (Weber et al. 2021). Certainly, on the per population level, our power to detect a polygenic SD system is limited. While polygenic SD mechanism might occur in A. burtoni as it has also been reported for other cichlids (Ser et al. 2010; Liew et al. 2012; Moore et al. 2022), it is also possible that different A. burtoni laboratory strains display genetic mechanisms to determine sex not common in the wild, as supported by the previous finding of an XY system on LG18 in a wild population (Böhne et al. 2016) not found in laboratory strains. Our findings of multiple small regions that show sex association or are sex-limited to some degree may also suggest a polygenic SD system in A. burtoni, which may differ among strains and localities.
Our population data might support the hypothesis that SD in A. burtoni is indeed polygenic and probably involves more loci than previously thought. To resolve this scenario, further analyses of an even broader set of samples, including families would be needed. Analyses on the family or single population level, however, might fail to reveal the full complexity of SD of A. burtoni. Our data could also lend some support to the expectation that a polygenic system is unstable past a couple of generations (Rice 1986); polygenic SD is debated to represent a stable state (van Doorn and Kirkpatrick 2010) and could rather reflect a particular situation of sex chromosome turnover (Schartl et al. 2016).
It is also possible that SD in A. burtoni is leaky and dependent on geographic or ecological gradients, as we have shown for another haplochromine cichlid, Pseudocrenilabrus philander (Böhne et al. 2019). Similar scenarios have been described in the European frog (Rodrigues et al. 2014), the Japanese frog (Miura 2008), and to some extent in the Japanese stickleback (Kitano et al. 2009), which show an association between the SD system and the geographical location within a species. Nevertheless, we did not detect any sex-associated patterning in correlation to the A. burtoni habitat range.
As in many teleosts (and other species), the sex of A. burtoni can be reversed by interference with sex hormones (Heule et al. 2014). It is thus also possible that under natural conditions, SD in A. burtoni is less dependent on genetic factors but more so on the environment.
It is also possible that A. burtoni populations are in a transitioning state from one SD system to another. In general, sex chromosomes evolve rapidly in cichlids (Roberts et al. 2009; Ser et al. 2010) and indeed show frequent turnovers (El Taher et al. 2021).
Our results could further suggest that the easily identifiable and strongly sex-linked regions in the laboratory strains might result from an initial unintended sampling bias/founder effect or some inadvertent environmental modifications during laboratory handling and probably have been further selected for as a byproduct of selection under artificial breeding conditions (e.g. selection for early breeding, breeding of individuals at much smaller body size, early development of male-specific markings, among others).
Interestingly, data from fighting fish show a similar pattern as we identified, with a stronger prevalence of a simple sex chromosomal system in domesticated strains and less penetrance of this system in the wild (Kwon et al. 2022). Previous studies that characterized the SD system of zebrafish (Danio rerio) and the variation of SD mechanisms between natural and laboratory strains (Tong et al. 2010; Bradley et al. 2011; Wilson et al. 2014) revealed results in the opposite direction. Zebrafish wildtype populations have a ZZ/ZW system that lab strains have seemingly lost (Tong et al. 2010; Wilson et al. 2014).
The samples available to us cover the geographical distribution range of A. burtoni with representatives from 11 different populations across LT. While for two populations (i.e. Ka1 and Ru1), we had fewer female specimens than for the other populations that dataset was overall balanced; we further excluded biased populations in some analyses to minimize an effect of uneven sample sizes. Generally, the data include more representatives of the southern basin than the northern basin, a factor that could have an impact on the observed patterns and, therefore, might ask for a more in-depth analysis of more populations and specimens of the northern part of the LT basin, especially since the laboratory populations used in previous studies are derived from northern Ruzizi populations (Pauquet et al. 2018).
Our (laboratory) male-specific RAD tag marker analyses confirmed the genetic complexity in A. burtoni SD, as in some populations the location of the LG05 RAD-tags differed, suggesting different mechanisms of SD in those populations. We could not detect a distinctive, unique pattern of a simple sex-linkage in A. burtoni natural populations, while RAD data of laboratory strains identified clearly differentiated regions between gametologs. We suggest that A. burtoni SD in wild populations has not faced the same selective pressures as SD of specimens under laboratory rearing conditions; and while there could be a polygenic SD system in some laboratory strains and also in natural populations (though probably involving other or more loci), it is also possible that non-genetic factors affect SD or that sex chromosomes in A. burtoni are only very little differentiated. This opens the window to more in-depth studies of sex chromosomes in A. burtoni natural populations. For instance, assessing the sex ratio among several families might reveal the strength of the genetic component in A. burtoni SD (Liew et al. 2012); the more in-depth study of additional northern populations might show similarities in the SD system for laboratory stocks that were actually derived from these (Roberts et al. 2016; El Taher et al. 2019); genetic linkage mapping and association studies with larger numbers of specimens might identify particularly small sex differences as has been shown with a single fixed SNP difference between males and females of the pufferfish (Kamiya et al. 2012); and finally might reveal if there are instances of a mismatch between genotypic and phenotypic sex in A. burtoni natural populations indicative of sex reversal resembling the pattern observed in the European common frog (Rana temporaria) (Rodrigues et al. 2014).
Our shortcoming in detecting a sex-linked region in A. burtoni natural populations suggests that the SD system in A. burtoni is more unstable and prone to change and might involve more modifier loci than previously anticipated. Furthermore, we propose that the A. burtoni laboratory strains have undergone particular selection pressures under captive (in)breeding that might have given rise to the stronger differentiation of sex chromosomes. In general, sex chromosomal systems in teleost fish are labile (Kikuchi and Hamaguchi 2013; The Tree of Sex Consortium 2014), which might be perfectly exemplified in A. burtoni.
Supplementary Material
Acknowledgments
We thank the scientific computing center at the University of Basel (sciCORE http://scicore.unibas.ch/) where calculations were performed, with support by the SIB—Swiss Institute of Bioinformatics.
Contributor Information
Nicolás Lichilín, Zoological Institute, Department of Environmental Sciences, University of Basel, Vesalgasse 1, 4051 Basel, Switzerland; Department of Neuroscience and Developmental Biology, University of Vienna, Djerassiplatz 1, 1030 Vienna, Austria.
Walter Salzburger, Zoological Institute, Department of Environmental Sciences, University of Basel, Vesalgasse 1, 4051 Basel, Switzerland.
Astrid Böhne, Zoological Institute, Department of Environmental Sciences, University of Basel, Vesalgasse 1, 4051 Basel, Switzerland; Leibniz Institute for the Analysis of Biodiversity Change, Museum Koenig Bonn, Adenauerallee 127, 53113 Bonn, Germany.
Data availability
The raw data used in this study are derived from (Weber et al. 2021), accessible from the NCBI under the BioProject accession number PRJNA485198. All data generated in this manuscript are submitted with the main document and its Supplementary material uploaded to GSA figshare: https://doi.org/10.25387/g3.21623121.
Funding
This work was funded by the Swiss National Science Foundation (SNSF, Ambizione grant PZ00P3_161462) to AB and the Freiwillige Akademische Gesellschaft Basel (FAG) to NL.
Author contributions
AB and WS designed the study, NL and AB analyzed data, NL drafted the manuscript, all authors contributed to result interpretation and manuscript writing. All authors approved the final manuscript version.
Literature cited
- R Core Team . R: A Language and Environment for Statistical Computing. Vienna; R Foundation for Statistical Computing; 2018. [Google Scholar]
- Akagi T, Henry IM, Tao R, Comai L. A Y-chromosome-encoded small RNA acts as a sex determinant in persimmons. Science. 2014;346(6209):646–650. doi: 10.1126/science.1257225. [DOI] [PubMed] [Google Scholar]
- Amores A, Postlethwait JH. Chapter 18 Banded chromosomes and the zebrafish karyotype. In: William Detrich H, Westerfield M, Zon LI, editors. Methods in cell biology, Volume 60. San Diego (CA): Academic Press; 1998:323–338. doi: 10.1016/S0091-679X(08)61908-1. [DOI] [PubMed] [Google Scholar]
- Anderson JL, Marí A, Braasch I, Amores A, Hohenlohe P, Batzel P, Postlethwait JH. Multiple sex-associated regions and a putative sex chromosome in zebrafish revealed by RAD mapping and population genomics. PLoS One. 2012;7(7):e40701. doi: 10.1371/journal.pone.0040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson AC. Fast very robust methods for the detection of multiple outliers. J Am Stat Assoc. 1994;89(428):1329–1339. doi: 10.1080/01621459.1994.10476872. [DOI] [Google Scholar]
- Bachtrog D, Mank JE, Peichel CL, Kirkpatrick M, Otto SP, Ashman TL, Hahn MW, Kitano J, Mayrose I, Ming R, et al. Sex determination: why so many ways of doing it? PLoS Biol. 2014;12(7):1–13. doi: 10.1371/journal.pbio.1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsley DA, Kuh E, Welsch RE. Regression Diagnostics: Identifying Influential Data and Sources of Collinearity (Wiley Series in Probability and Mathematical Statistics). Hoboken: (NJ: ): Wiley-interscience; 1980. [Google Scholar]
- Bergero R, Gardner J, Bader B, Yong L, Charlesworth D. Sexually dimorphic recombination can facilitate the establishment of sexually antagonistic polymorphisms in guppies. bioRxiv. 365114. doi: 10.1101/365114, 09 July 2018, preprint: not peer reviewed. [DOI] [Google Scholar]
- Bhatia G, Patterson N, Sankararaman S, Price AL. Estimating and interpreting FST: the impact of rare variants. Genome Res. 2013;23(9):1514–1521. doi: 10.1101/gr.154831.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhne A, Weber AAT, Rajkov J, Rechsteiner M, Riss A, Egger B, Salzburger W. Repeated evolution versus common ancestry: sex chromosome evolution in the haplochromine cichlid Pseudocrenilabrus philander. Genome Biol Evol. 2019;11(2):439–458. doi: 10.1093/gbe/evz003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhne A, Wilson CA, Postlethwait JH, Salzburger W. Variations on a theme: genomics of sex determination in the cichlid fish Astatotilapia burtoni. BMC Genomics. 2016;17(1):883. doi: 10.1186/s12864-016-3178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley KM, Breyer JP, Melville DB, Broman KW, Knapik EW, Smith JR. An SNP-based linkage map for zebrafish reveals sex determination loci. G3 (Bethesda). 2011;1(1):3–9. doi: 10.1534/g3.111.000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR. Genotype imputation with millions of reference samples. Am J Hum Genet. 2016;98(1):116–126. doi: 10.1016/j.ajhg.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinf. 2009;10(1):421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capel B. Vertebrate sex determination: evolutionary plasticity of a fundamental switch. Nat Rev Genet. 2017;18(11):675–689. doi: 10.1038/nrg.2017.60. [DOI] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4(1):7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D. Evolution of recombination rates between sex chromosomes. Philos Trans R Soc Lond B Biol Sci. 2017;372(1736):20160456. doi: 10.1098/rstb.2016.0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte MA, Gammerdinger WJ, Bartie KL, Penman DJ, Kocher TD. A high quality assembly of the Nile Tilapia (Oreochromis niloticus) genome reveals the structure of two sex determination regions. BMC Genomics. 2017;18(1):1–19. doi: 10.1186/s12864-017-3723-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JR, Lex A, Gehlenborg N. Upsetr: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33(18):2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutting A, Chue J, Smith CA. Just how conserved is vertebrate sex determination? Dev Dyn. 2013;242(4):380–387. doi: 10.1002/dvdy.23944. [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos L, Snoeks J, van den Audenaerde DT. An annotated checklist of the fishes of Rwanda (East Central Africa), with historical data on introductions of commercially important species. J East African Nat Hist. 2001;90(1):41. doi: 10.2982/0012-8317(2001)90[41:AACOTF]2.0.CO;2. [DOI] [Google Scholar]
- Egger B, Roesti M, Böhne A, Roth O, Salzburger W. Demography and genome divergence of lake and stream populations of an East African cichlid fish. Mol Ecol. 2017;26(19):5016–5030. doi: 10.1111/mec.14248. [DOI] [PubMed] [Google Scholar]
- El Taher A, Lichilín N, Salzburger W, Böhne A. Time matters! developmental shift in gene expression between the head and the trunk region of the cichlid fish Astatotilapia burtoni. BMC Genomics. 2019;20(1):39. doi: 10.1186/s12864-018-5321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Taher A, Ronco F, Matschiner M, Salzburger W, Böhne A. Dynamics of sex chromosome evolution in a rapid radiation of cichlid fishes. Sci Adv. 2021;7(36):eabe8215. doi: 10.1126/sciadv.abe8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Ives AR, Surget-Groba Y, Cannon CH. An assembly and alignment-free method of phylogeny reconstruction from next-generation sequencing data. BMC Genomics. 2015;16(1):1–18. doi: 10.1186/s12864-015-1647-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammerdinger WJ, Kocher TD. Unusual diversity of sex chromosomes in African cichlid fishes. Genes. 2018;9(10):480. doi: 10.3390/genes9100480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talon M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nuc Acids Res. 2008;36(10):3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther ACLG. Second report on the reptiles, batrachians, and fishes transmitted by Mr. H. H. Johnston, C. B, from British Central Africa. Proc Zool Soc. 1893;1893(4):616–628. https://biostor.org/reference/70273. [Google Scholar]
- Hettmansperger TP. Why not try a robust regression? Austr J Stat. 1987;29(1):1–18. doi: 10.1111/j.1467-842X.1987.tb00716.x. [DOI] [Google Scholar]
- Heule C, Göppert C, Salzburger W, Böhne A. Genetics and timing of sex determination in the East African cichlid fish Astatotilapia burtoni. BMC Genet. 2014;15(1):1–17. doi: 10.1186/s12863-014-0140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann HA. Functional genomics of neural and behavioral plasticity. J Neurobiol. 2003;54(1):272–282. doi: 10.1002/neu.10172. [DOI] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496(7446):498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9(9):868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntti SA, Hilliard AT, Kent KR, Kumar A, Nguyen A, Jimenez MA, Loveland JL, Mourrain P, Fernald RD. A neural basis for control of cichlid female reproductive behavior by prostaglandin F2α. Curr Biol. 2016;26(7):943–949. doi: 10.1016/j.cub.2016.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käfer J, Lartillot N, Marais GAB, Picard F. Detecting sex-linked genes using genotyped individuals sampled in natural populations. Genetics. 2021;218(2):iyab053. doi: 10.1093/genetics/iyab053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya T, Kai W, Tasumi S, Oka A, Matsunaga T, Mizuno N, Fujita M, Suetake H, Suzuki S, Hosoya S, et al. A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger Pufferfish, Takifugu rubripes (Fugu). PLoS Genet. 2012;8(7):e1002798. doi: 10.1371/journal.pgen.1002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011;21(3):487–493. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Hamaguchi S. Novel sex-determining genes in fish and sex chromosome evolution. Dev Dyn. 2013;242(4):339–353. doi: 10.1002/dvdy.23927. [DOI] [PubMed] [Google Scholar]
- Kitano J, Ross JA, Mori S, Kume M, Jones FC, Chan YC, Absher DM, Grimwood J, Schmutz J, Myers RM, et al. A role for a neo-sex chromosome in stickleback speciation. Nature. 2009;461(7267):1079–1083. doi: 10.1038/nature08441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus BJ, Grünwald NJ. Vcfr : a package to manipulate and visualize variant call format data in R. Mol Ecol Res. 2017;17(1):44–53. doi: 10.1111/1755-0998.12549. [DOI] [PubMed] [Google Scholar]
- Kullander SO, Roberts TR. Out of Lake Tanganyika: endemic Lake fishes inhabit rapids of the Lukuga River. Ichthyol Explor Freshw. 2011;22(4):355–376. [Google Scholar]
- Kwon YM, Vranken N, Hoge C, Lichak MR, Norovich AL, Francis KX, Camacho-Garcia J, Bista I, Wood J, McCarthy S, et al. Genomic consequences of domestication of the Siamese fighting fish. Sci Adv. 2022;8(10):eabm4950. doi: 10.1126/sciadv.abm4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang M, Miyake T, Braasch I, Tinnemore D, Siegel N, Salzburger W, Amemiya CT, Meyer A. A BAC library of the East African haplochromine cichlid fish Astatotilapia burtoni. J Exp Zool B Mol Dev Evol. 2006;306B(1):35–44. doi: 10.1002/jez.b.21068. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DC, Liu CM, Luo R, Sadakane K, Lam TW. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31(10):1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- Li XY, Liu XL, Zhu YJ, Zhang J, Ding M, Wang MT, Wang ZW, Li Z, Zhang XJ, Zhou L, et al. Origin and transition of sex determination mechanisms in a gynogenetic hexaploid fish. Heredity 2018;121(1):64–74. doi: 10.1038/s41437-017-0049-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew WC, Bartfai R, Lim Z, Sreenivasan R, Siegfried KR, Orban L. Polygenic sex determination system in zebrafish. PLoS One. 2012;7(4):e34397. doi: 10.1371/journal.pone.0034397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew WC, Orbán L. Zebrafish sex: a complicated affair. Brief Funct Genomics. 2014;13(2):172–187. doi: 10.1093/bfgp/elt041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majtánová Z, Indermaur A, Nyom ARB, Ráb P, Musilova Z. Adaptive radiation from a chromosomal perspective: evidence of chromosome set stability in cichlid fishes (Cichlidae: Teleostei) from the Barombi Mbo Lake, Cameroon. Int J Mol Sci. 2019;20(20):4994. doi: 10.3390/ijms20204994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrøm M, Matschiner M, Tørresen OK, Jakobsen KS, Jentoft S. Whole genome sequencing data and de novo draft assemblies for 66 teleost species. Sci Data. 2017;4(1):1–13. doi: 10.1038/sdata.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruska KP, Fernald RD. Astatotilapia burtoni: a model system for analyzing the neurobiology of behavior. ACS Chem Neuro. 2018;9(8):1951–1962. doi: 10.1021/acschemneuro.7b00496. [DOI] [PubMed] [Google Scholar]
- Mazzuchelli J, Kocher TD, Yang F, Martins C. Integrating cytogenetics and genomics in comparative evolutionary studies of cichlid fish. BMC Genomics. 2012;13(1):463. doi: 10.1186/1471-2164-13-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1297. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BS, Matschiner M, Salzburger W. A tribal level phylogeny of Lake Tanganyika cichlid fishes based on a genomic multi-marker approach. Mol Phylo Evol. 2015;83:56–71. doi: 10.1016/j.ympev.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura I. An evolutionary witness: the frog Rana rugosa underwent change of heterogametic sex from XY male to ZW female. Sex Dev. 2008;1(6):323–331. doi: 10.1159/000111764. [DOI] [PubMed] [Google Scholar]
- Moore EC, Ciccotto PJ, Peterson EN, Lamm MS, Albertson RC, Roberts RB. Polygenic sex determination produces modular sex polymorphism in an African cichlid fish. Proc Natl Acad Sci U S A. 2022;119(14):e2118574119. doi: 10.1073/pnas.2118574119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, Kravitz SA, Mobarry CM, Reinert KH, Remington KA, et al. A whole-genome assembly of Drosophila. Science. 2000;287(5461):2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- Nursyifa C, Brüniche-Olsen A, Garcia-Erill G, Heller R, Albrechtsen A. Joint identification of sex and sex-linked scaffolds in non-model organisms using low depth sequencing data. Mol Ecol Res. 2022;22(2):458–467. doi: 10.1111/1755-0998.13491. [DOI] [PubMed] [Google Scholar]
- Pan Q, Feron R, Yano A, Guyomard R, Jouanno E, Vigouroux E, Wen M, Busnel JM, Bobe J, Concordet JP, et al. Identification of the master sex determining gene in Northern pike (Esox lucius) reveals restricted sex chromosome differentiation. PLoS Genet. 2019;15(8):e1008013. doi: 10.1371/journal.pgen.1008013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauquet G, Salzburger W, Egger B. The puzzling phylogeography of the haplochromine cichlid fish Astatotilapia burtoni. Ecol Evol. 2018;8(11):5637–5648. doi: 10.1002/ece3.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell MW, Mank JE, Peichel CL. Transitions in sex determination and sex chromosomes across vertebrate species. Mol Ecol. 2018;27(19):3950–3963. doi: 10.1111/mec.14540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin N. Sex reversal: a fountain of youth for sex chromosomes? Evolution. 2009;63(12):3043–3049. doi: 10.1111/j.1558-5646.2009.00837.x. [DOI] [PubMed] [Google Scholar]
- Phillips RB, Amores CA, Morasch MR, Wilson C, Postlethwait JH. Assignment of zebrafish genetic linkage groups to chromosomes. Cyto Gen Res. 2006;114(2):155–162. doi: 10.1159/000093332. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR. On the instability of polygenic sex determination: the effect of sex-specific selection. Evolution. 1986;40(3):633. doi: 10.1111/j.1558-5646.1986.tb00514.x. [DOI] [PubMed] [Google Scholar]
- Roberts NB, Juntti SA, Coyle KP, Dumont BL, Stanley MK, Ryan AQ, Fernald RD, Roberts RB. Polygenic sex determination in the cichlid fish Astatotilapia burtoni. BMC Genomics. 2016;17(1):1–13. doi: 10.1186/s12864-016-3177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RB, Ser JR, Kocher TD. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science. 2009;326(5955):998–1001. doi: 10.1126/science.1174705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison RR, White RB, Illing N, Troskie BE, Morley M, Millar RP, Fernald RD. Gonadotropin-releasing hormone receptor in the teleost Haplochromis burtoni: structure, location, and function. Endocrinology. 2001;142(5):1737–1743. doi: 10.1210/endo.142.5.8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues N, Merilä J, Patrelle C, Perrin N. Geographic variation in sex-chromosome differentiation in the common frog (Rana temporaria). Mol Ecol. 2014;23(14):3409–3418. doi: 10.1111/mec.12829. [DOI] [PubMed] [Google Scholar]
- Ronco F, Büscher HH, Indermaur A, Salzburger W. The taxonomic diversity of the cichlid fish fauna of ancient Lake Tanganyika, East Africa. J Great Lakes Res. 2020;46(5):1067–1078. doi: 10.1016/j.jglr.2019.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronco F, Matschiner M, Böhne A, Boila A, Büscher HH, El Taher A, Indermaur A, Malinsky M, Ricci V, Kahmen A, et al. Drivers and dynamics of a massive adaptive radiation in cichlid fishes. Nature. 2021;589(7840):76–81. doi: 10.1038/s41586-020-2930-4. [DOI] [PubMed] [Google Scholar]
- Salzburger W. Understanding explosive diversification through cichlid fish genomics. Nat Rev Genet. 2018;19(11):705–717. doi: 10.1038/s41576-018-0043-9. [DOI] [PubMed] [Google Scholar]
- Salzburger W, Mack T, Verheyen E, Meyer A. Out of Tanganyika: genesis, explosive speciation, key-innovations and phylogeography of the haplochromine cichlid fishes. BMC Evol Biol. 2005;5(1):1–15. doi: 10.1186/1471-2148-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzburger W, Van Bocxlaer B, Cohen AS. Ecology and evolution of the African great lakes and their faunas. Annu Rev Ecol Evol Syst. 2014;45(1):519–545. doi: 10.1146/annurev-ecolsys-120213-091804. [DOI] [Google Scholar]
- Schartl M, Schmid M, Nanda I. Dynamics of vertebrate sex chromosome evolution: from equal size to giants and dwarfs. Chromosoma. 2016;125(3):553–571. doi: 10.1007/s00412-015-0569-y. [DOI] [PubMed] [Google Scholar]
- Ser JR, Roberts RB, Kocher TD. Multiple interacting loci control sex determination in Lake Malawi cichlid fish. Evolution. 2010;64(2):486–501. doi: 10.1111/j.1558-5646.2009.00871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma KK, Sharma OP, Tripathi NK. Female heterogamety in Danio rerio (Cypriniformes: Cyprinidae). Proc Natl Acad Sci India Sect B. 1998;68(2):123–126. [Google Scholar]
- Tao W, Xu L, Zhao L, Zhu Z, Wu X, Min Q, Wang D, Zhou Q. High-quality chromosome-level genomes of two tilapia species reveal their evolution of repeat sequences and sex chromosomes. Mol Ecol Resour. 2020;21(2):543–560. doi: 10.1111/1755-0998.13273. [DOI] [PubMed] [Google Scholar]
- Taslima K, Wehner S, Taggart JB, de Verdal H, Benzie JAH, Bekaert M, McAndrew BJ, Penman DJ. Sex determination in the GIFT strain of tilapia is controlled by a locus in linkage group 23. BMC Genet. 2020;21(1):49. doi: 10.1186/s12863-020-00853-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis A, Roth O, Cortesi F, Ronco F, Salzburger W, Egger B. Variation of anal fin egg-spots along an environmental gradient in a haplochromine cichlid fish. Evolution. 2017;71(3):766–777. doi: 10.1111/evo.13166. [DOI] [PubMed] [Google Scholar]
- Tong SK, Hsu HJ, Chung BC. Zebrafish monosex population reveals female dominance in sex determination and earliest events of gonad differentiation. Dev Biol. 2010;344(2):849–856. doi: 10.1016/j.ydbio.2010.05.515. [DOI] [PubMed] [Google Scholar]
- The Tree of Sex Consortium . Tree of sex: a database of sexual systems. Sci Dat. 2014;1(140015):1–8. doi: 10.1038/sdata.2014.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn GS, Kirkpatrick M. Transitions between male and female heterogamety caused by sex-antagonistic selection. Genetics. 2010;186(2):629–645. doi: 10.1534/genetics.110.118596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheyen E. Origin of the superflock of cichlid fishes from Lake Victoria, East Africa. Science. 2003;300(5617):325–329. doi: 10.1126/science.1080699. [DOI] [PubMed] [Google Scholar]
- Weber AAT, Rajkov J, Smailus K, Egger B, Salzburger W. Speciation dynamics and extent of parallel evolution along a lake-stream environmental contrast in African cichlid fishes. Sci Adv. 2021;7(45):eabg5391. doi: 10.1126/sciadv.abg5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H, Averick M, Bryan J, Chang W, McGowan L, François R, Grolemund G, Hayes A, Henry L, Hester J, et al. Welcome to the Tidyverse. J Open Source Softw. 2019;4(43):1686. doi: 10.21105/joss.01686. [DOI] [Google Scholar]
- Wilson CA, High SK, McCluskey BM, Amores A, Yan YL, Titus TA, Anderson JL, Batzel P, Carvan MJ, Schartl M, et al. Wild sex in zebrafish: loss of the natural sex determinant in domesticated strains. Genetics. 2014;198(3):1291–1308. doi: 10.1534/genetics.114.169284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Zhang Y, Sarida M, Hattori RS, Strüssmann CA. Coexistence of genotypic and temperature-dependent sex determination in Pejerrey Odontesthes bonariensis. PLoS One. 2014;9(7):1–8. doi: 10.1371/journal.pone.0102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44(7):821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data used in this study are derived from (Weber et al. 2021), accessible from the NCBI under the BioProject accession number PRJNA485198. All data generated in this manuscript are submitted with the main document and its Supplementary material uploaded to GSA figshare: https://doi.org/10.25387/g3.21623121.