Abstract
To date, little is known about the effect of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus responsible for the coronavirus disease 2019 (COVID-19) pandemic, on the upper respiratory tract (URT) microbiota over time. To fill this knowledge gap, we used 16S ribosomal RNA gene sequencing to characterize the URT microbiota in 48 adults, including (1) 24 participants with mild-to-moderate COVID-19 who had serial mid-turbinate swabs collected up to 21 days after enrolment and (2) 24 asymptomatic, uninfected controls who had mid-turbinate swabs collected at enrolment only. To compare the URT microbiota between groups in a comprehensive manner, different types of statistical analyses that are frequently employed in microbial ecology were used, including ⍺-diversity, β-diversity and differential abundance analyses. Final statistical models included age, sex and the presence of at least one comorbidity as covariates. The median age of all participants was 34.00 (interquartile range=28.75–46.50) years. In comparison to samples from controls, those from participants with COVID-19 had a lower observed species index at day 21 (linear regression coefficient=−13.30; 95 % CI=−21.72 to −4.88; q=0.02). In addition, the Jaccard index was significantly different between samples from participants with COVID-19 and those from controls at all study time points (PERMANOVA q<0.05 for all comparisons). The abundance of three amplicon sequence variants (ASVs) (one Corynebacterium ASV, Frederiksenia canicola , and one Lactobacillus ASV) were decreased in samples from participants with COVID-19 at all seven study time points, whereas the abundance of one ASV (from the family Neisseriaceae ) was increased in samples from participants with COVID-19 at five (71.43 %) of the seven study time points. Our results suggest that mild-to-moderate COVID-19 can lead to alterations of the URT microbiota that persist for several weeks after the initial infection.
Keywords: airway, coronavirus, COVID-19, microbiota, nasal, respiratory, SARS-CoV-2
Data Summary
The authors confirm all supporting data and protocols have provided within the article or through supplementary data files.
Impact Statement.
The microbial communities that inhabit the upper respiratory tract (URT) (i.e. the URT microbiota) are key determinants of respiratory health. The URT microbiota can be easily disrupted by multiple environmental factors, such as acute respiratory infections by common respiratory viruses. However, little is known about the effect of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the virus responsible for the coronavirus disease 2019 (COVID-19) pandemic, on the URT microbiota over time. In our study, we show differences in multiple URT microbial ecology metrics between participants with mild-to-moderate COVID-19 and asymptomatic, uninfected controls. These differences could be found up to 21 days after study enrolment. Our results suggest that mild-to-moderate COVID-19 can lead to alterations of the URT microbiota that persists for several weeks after the initial infection.
Introduction
Unlike many other key determinants of respiratory health, the upper respiratory tract (URT) microbiota is dynamic and can be shaped by multiple environmental factors throughout life [1]. In particular, common respiratory viruses have been shown to disrupt the homeostasis, structure and function of the URT microbial communities [2, 3]. This is supported by the results of multiple prior studies from our group and others showing substantial differences in the URT microbiota between subjects with and without acute respiratory infections (ARIs) due to respiratory syncytial virus (RSV), human rhinovirus, or influenza virus [4–7]. Furthermore, current evidence suggests that the URT microbial disturbances caused by these common respiratory viruses can in turn impact on disease severity, the host immune response and even the development of chronic lung diseases following the initial ARI (such as asthma) [8–13].
To date, little is known about the resilience and recovery of the URT microbiota following ARI with severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the respiratory virus responsible for the ongoing coronavirus disease 2019 (COVID-19) pandemic. Through characterization of the URT microbiota in serial samples collected from participants with mild-to-moderate COVID-19 using 16S ribosomal RNA (rRNA) gene sequencing, our objectives were (1) to examine whether SARS-CoV-2 leads to specific URT microbiota changes over time and (2) to evaluate whether, among participants with COVID-19, SARS-CoV-2 viral load is associated with longitudinal changes of the URT microbiota.
Methods
Full details of the material and methods are available in the eMethods section of the Supplementary Material.
Overview of the study design
For the current study, we first included participants with confirmed COVID-19 enrolled as part of a randomized controlled trial (RCT) examining the effect of several types of nasal irrigations on COVID-19-related outcomes. The detailed methods for this RCT have been reported previously [14–16]. Inclusion criteria included age ≥18 years, a qualitative PCR test positive for SARS-CoV-2 performed at Vanderbilt University Medical Center (VUMC) or one of its affiliated centres, planned self-quarantine after being diagnosed with COVID-19, and residence within a 30 mile radius of the main VUMC campus in Nashville, Tennessee. Exclusion criteria included current use of nasal saline irrigations or other nasal medications (such as nasal steroids), inability to perform nasal irrigations or to collect URT samples in a separate house bathroom or away from household contacts and the need for hospitalization related to COVID-19. Thus, only participants with symptomatic, mild-to-moderate COVID-19 (based on criteria from the World Health Organization [17]) were included in the RCT. Eligible participants were contacted and enrolled in the RCT within 24 h of the initial diagnosis of COVID-19. Only participants who were not randomized to one of the RCT’s intervention arms –thus, were not assigned to any specific interventions and did not use one of the nasal irrigations being tested– were included in the current study (n=24).
In parallel to conducting the aforementioned RCT, we also enrolled asymptomatic participants within the VUMC community (including employees, students and faculty, among others) to serve as controls (n=24). Inclusion criteria included age ≥18 years and absence of COVID-19-related symptoms (e.g. runny nose, cough, or fever). Exclusion criteria included current use of nasal saline irrigations or other nasal medications (such as nasal steroids) and current or prior diagnosis of COVID-19.
Following adequate training, all of the above participants (n=48) were asked to collect a mid-turbinate swab at enrolment (day 1) using a self-collection kit (FLOQSwabs, Copan Diagnostics, Inc.). Those with COVID-19 were also asked to collect serial samples using the same method on follow-up days 3, 5, 7, 10, 14 and 21. The collection of all samples included in the current study occurred in the spring and summer of 2020. Each subject provided informed consent for his/her participation. The VUMC Institutional Review Board and Biosafety Committee approved this study.
SARS-CoV-2 testing by quantitative reverse transcription PCR
To measure viral load in participants with COVID-19 and rule out asymptomatic infection in controls, quantitative reverse transcription PCR in all mid-turbinate swabs collected at enrolment using United States Centers for Disease Control and Prevention primers and probes designed for the detection of SARS-CoV-2 was performed [16]. High viral load was defined as a cycle threshold value for the detection of the coronavirus nucleocapsid gene region 1 below the median of all samples collected at enrolment (day 1) from SARS-CoV-2-infected participants.
Characterization of the URT microbiota
The methods used to characterize the URT microbiota have been previously described in detail [16, 18–21]. In brief, following bacterial DNA extraction from the mid-turbinate swabs, the V4 region of the 16S rRNA gene was amplified using universal primers. The pool was then sequenced on an Illumina MiSeq platform with 2×250 base pair reads. For quality control, negative controls were amplified and sequenced concurrently with participant samples. Next, the 16S rRNA gene sequences were processed using the R package dada2 by applying its standard operating procedure (available at: https://benjjneb.github.io/dada2/tutorial.html) [22]. To this end, sequences were grouped into amplicon sequence variants (ASVs), and taxonomy was assigned using the silva reference database version 132 [23]. Low-quality sequences, chimaeras and non-bacterial sequences were discarded as part of the dada2 pipeline. To remove any suspected contaminants that were found in the negative controls, the remaining sequences were processed using the R package decontam [24]. The full taxonomy of contaminant ASVs identified by decontam are listed in Table S1 (available in the online version of this article). Those ASVs that were present in >1 sample were then retained and samples with <1000 sequences (n=33) were discarded using the R package phyloseq [25, 26]. Last, the relative abundances of individual ASVs were calculated using simple proportions.
Statistical analyses
Statistical analyses were conducted at the ASV level in R version 3.1.10 [27]. To compare the URT microbiota between groups in a comprehensive manner, different types of statistical analyses that are frequently employed in microbial ecology were used, including ⍺-diversity (richness ±evenness), β-diversity (overall composition) and differential abundance (individual taxa) analyses. For ⍺- and β-diversity analyses only, the processed dataset was rarified to the lowest library size of all samples (n=1,154). This rarefaction process was repeated multiple times (n=400) and the results were averaged. Common ⍺-diversity (observed species, Shannon and inverse Simpson indices) and β-diversity (Bray–Curtis and Jaccard indices) metrics were then calculated. For differential abundance analyses, a non-rarified dataset was used and, to minimize the impact of rare taxa, only ASVs with a relative abundance across all samples >0.01 % were included. To examine non-longitudinal associations, linear regression (for ⍺-diversity analyses), permutational multivariate analysis of variance (PERMANOVA) (for β-diversity analyses) [28] and differential gene expression analysis based on the negative binomial distribution (DESeq2) (for differential abundance analyses) [29] were used. Based on pre-established criteria, ASVs were considered differentially abundant only if, in addition to statistical significance, they (1) had an absolute fold change ≥2 (equivalent to an absolute fold change ≥1 in the log2 scale) and (2) were differentially abundant in ≥3 of the 7 individual comparisons. To examine longitudinal associations, generalized estimating equations (GEEs) (for ⍺-diversity analyses), PERMANOVA (for β-diversity analyses) [28] and permuted spline-based statistical analyses and GEEs (for differential abundance analyses) were used [30]. All statistical models included age, sex and the presence of at least one comorbidity (i.e. obesity, diabetes, hypertension, lung disease, or heart disease, coded as yes vs no or not reported) as covariates. The statistical models for ⍺-diversity and differential abundance analyses using GEEs also included the study time point and a multiplicative interaction between SARS-CoV-2 viral load and the study time point as additional covariates. When appropriate, P-values were controlled for multiple testing using the Benjamini–Hochberg procedure and the resulting q-values are reported [31]. Statistical significance was defined as a P- or q-value <0.05.
Results
Baseline characteristics of the study population and samples collected
The baseline characteristics of the 48 participants included in the current study are shown in Table 1. The median age of all participants was 34.00 (interquartile range=28.75–46.50) years. The majority of participants were male, white non-Hispanic and non-obese. There were no significant differences in baseline characteristics between those with and without COVID-19. However, those with COVID-19 were less likely to currently smoke or have hypertension, and more likely to have at least one other comorbidity. Based on eligibility criteria, none of the participants were using nasal medications at study enrolment. Furthermore, none of them had used antibiotics in the 2 weeks prior to study enrolment.
Table 1.
Baseline characteristics of participants*†
|
Baseline characteristic |
SARS-CoV-2 |
All (n=48) |
p-value |
|
|---|---|---|---|---|
|
Uninfected (n=24) |
Infected (n=24) |
|||
|
Age (years) |
33.00 (29.00–45.00) |
35.50 (26.75–49.25) |
34.00 (28.75–46.50) |
0.8 |
|
Male sex |
13 (54.17 %) |
14 (58.33 %) |
27 (56.25 %) |
1.0 |
|
White non-Hispanic |
18 (75.00 %) |
18 (75.00 %) |
36 (75.00 %) |
1.0 |
|
Obesity |
4 (21.05 %) |
7 (31.82 %) |
11 (26.83 %) |
0.5 |
|
Diabetes |
1 (4.55 %) |
3 (12.50 %) |
4 (8.70 %) |
0.6 |
|
Hypertension |
4 (18.18 %) |
4 (16.67 %) |
8 (17.39 %) |
1.0 |
|
Lung Disease |
0 (0.00 %) |
4 (16.67 %) |
4 (8.70 %) |
0.1 |
|
Heart Disease |
1 (4.55 %) |
2 (8.33 %) |
3 (6.52 %) |
1.0 |
|
Current Smoker |
1 (4.55 %) |
0 (0.00 %) |
1 (2.17 %) |
0.5 |
|
Current use of nasal medications |
0 (0.00 %) |
0 (0.00 %) |
0 (0.00 %) |
– |
|
Use of antibiotics in the last 2 weeks |
0 (0.00 %) |
0 (0.00 %) |
0 (0.00 %) |
– |
*Data are presented as median (interquartile range) for age and number (%) for all other variables. Percentages were calculated for participants with complete data.
†P-values for the comparisons between SARS-CoV-2-infected and -uninfected participants were calculated using Mann–Whitney U or Fisher’s exact tests, as appropriate.
There were 154 samples collected from the 24 participants with COVID-19 out of the 168 samples expected (i.e. 24 subjects×7 study time points): 18 were collected at enrolment (day 1) and 20, 20, 14, 17, 15 and 17 were collected on follow-up days 3, 5, 7, 10, 14 and 21, respectively. Thus, the overall sample return rate among those with COVID-19 was 91.66 %. In total, 121 (78.57 %) of the 154 samples from participants with COVID-19 passed quality control, whereas all 24 (100 %) samples obtained from participants without COVID-19 passed quality control. The median high-quality sequence count per sample across all 145 samples included in statistical analyses was 7219 (interquartile range=3142–24 771), with a maximum high-quality sequence count of 139 348.
Comparisons between SARS-CoV-2-infected and -uninfected participants
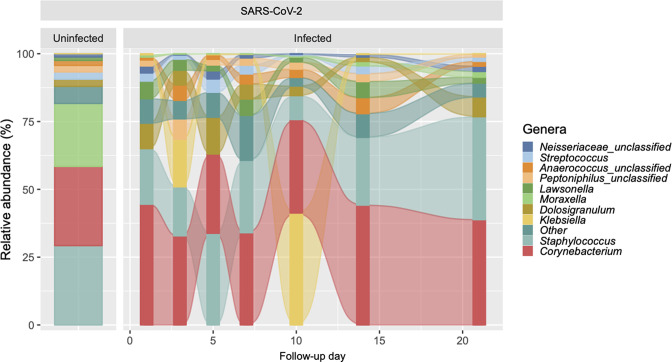
The genera with the highest total relative abundances across all 121 samples from participants with COVID-19 included Corynebacterium (35.47 %), Staphylococcus (24.38 %), Klebsiella (8.98 %), Dolosigranulum (7.32 %) and Lawsonella (3.94 %), whereas the genera with the highest total relative abundances across the 24 samples from controls included Staphylococcus (29.25 %), Corynebacterium (29.19 %), Moraxella (23.21 %), Dolosigranulum (2.65 %) and Streptococcus (2.64 %) (Fig. 1).
Fig. 1.
Predominant genera of the upper respiratory tract microbiota in SARS-CoV-2-uninfected and -infected participants. The figure shows alluvial plots of the relative abundance (%) of the top 10 most abundant genera (y-axis) per group and, for SARS-CoV-2-infected participants, by follow-up day (x-axis). The relative abundances of other genera were collapsed into the ‘other’ category.
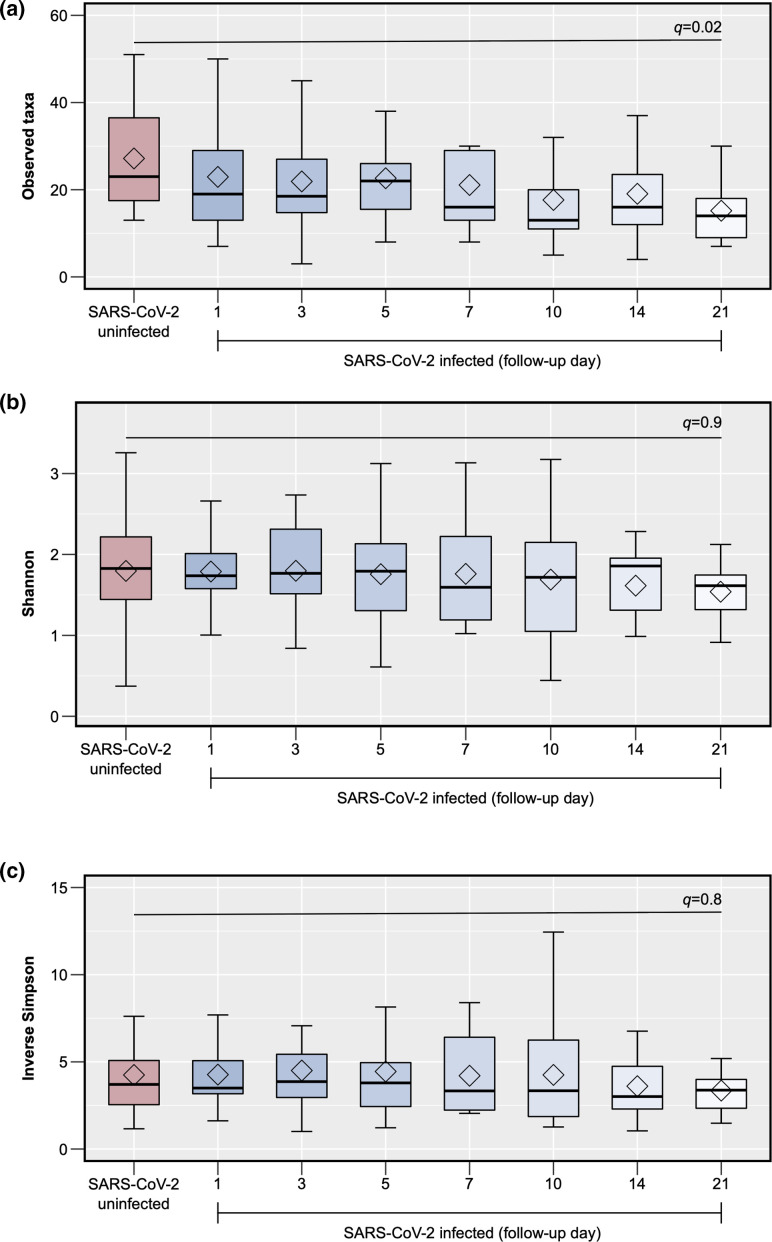
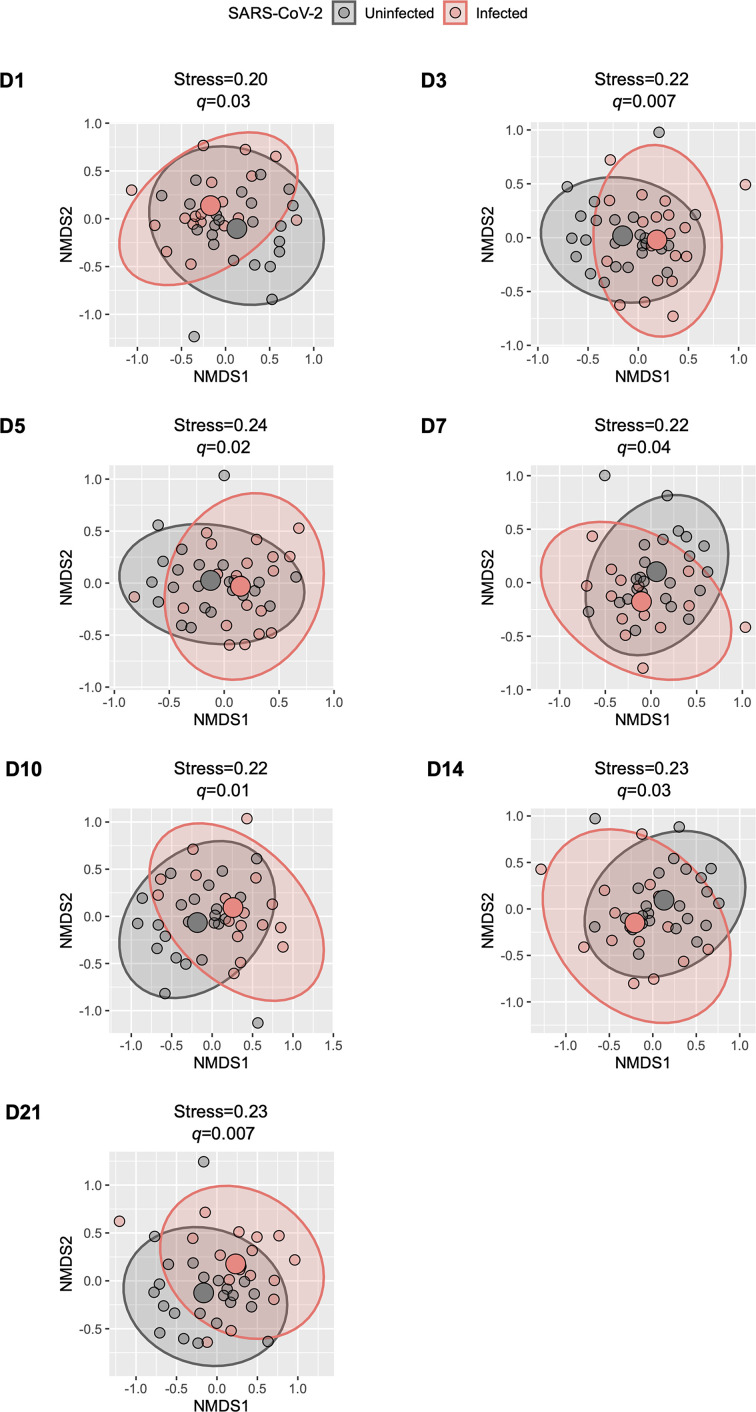
In comparison to samples from controls, those from participants with COVID-19 had an overall lower ⍺-diversity at most of the study time points, although only the observed species index from samples collected at day 21 was significantly different (linear regression coefficient=−13.30; 95 % CI=−21.72 to −4.88; q=0.02) (Fig. 2a–c). In β-diversity analyses, there were no significant differences in the Bray–Curtis index (based on taxa abundance) between groups (PERMANOVA q>0.05 for all comparisons); however, the Jaccard index (based on taxa presence vs absence) was significantly different between samples from participants with COVID-19 and those from controls at all study time points (PERMANOVA q<0.05 for all comparisons) (Fig. 3).
Fig. 2.
Comparison of the ⍺-diversity of the upper respiratory tract microbiota between SARS-CoV-2-uninfected and -infected participants. The figure shows box plots of the observed species (1A), Shannon (1B) and inverse Simpson indices (1C) (y-axes) per group and, for SARS-CoV-2-infected participants, by follow-up day (x-axes). The P-values for the comparisons of follow-up day 21 for SARS-CoV-2-infected participants vs day 1 (enrolment) for uninfected participants using linear regression models adjusting for potential confounders (see text) and controlling for multiple testing (q-values) are shown. The sample sizes for each group ( n ) are also shown.
Fig. 3.
Comparison of the β-diversity of the upper respiratory tract microbiota between SARS-CoV-2-uninfected and -infected participants. The figure shows scatter plots representing the overall composition (small circles) of each participant’s upper respiratory tract microbiota per group. Each scatter plot depicts the comparisons of a particular follow-up day (d) for SARS-CoV-2-infected participants vs day 1 (enrolment) for uninfected participants. The scatter plots were generated using NMDS based on the Jaccard index and using two dimensions (x- and y-axes). The group’s centroid (large circles) and corresponding 95% CI (ellipses), as well as the NMDS stress values, are shown. The P-values for the comparisons between groups using permutational multivariate analysis of variance models adjusting for potential confounders (see text) and controlling for multiple testing (q-values) are also shown. NMDS, non-metric multidimensional scaling; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
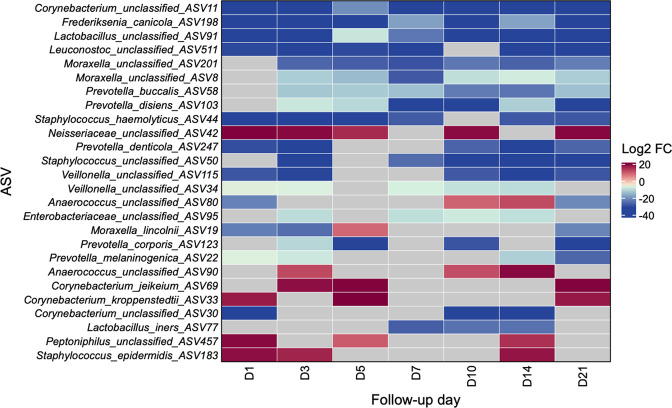
In differential abundance analyses, a total of 26 ASVs from 12 different genera (including 5 Prevotella , 4 Corynebacterium , 3 Moraxella , 3 Staphylococcus , 2 Lactobacillus , 2 Veillonella , 2 Anaerococcus , 1 Frederiksenia , 1 Leuconostoc , 1 Neisseriaceae , 1 Enterobacteriaceae and 1 Peptoniphilus ASV) met the pre-specified criteria to be considered differentially abundant between groups. Eighteen (69.23%) of these 26 ASVs were still differentially abundant in samples collected at the last available study time point. Furthermore, the abundance of three ASVs [one Corynebacterium ASV (potentially C. segmentosum by the Basic Local Alignment Search Tool (blast) database [32]), Frederiksenia canicola , and one Lactobacillus ASV (likely L. jensenii by the blast database)] were decreased in samples from participants with COVID-19 at all seven study time points, whereas the abundance of one ASV [from the family Neisseriaceae (not precisely classified by the blast database)] was increased in samples from participants with COVID-19 at five (71.43 %) of the seven study time points (Fig. 4). For many of the differential abundant taxa, the differences between groups were mainly driven by a few participants (Fig. S1).
Fig. 4.
Differential abundance of taxa of the upper respiratory tract microbiota between SARS-CoV-2-uninfected and -infected participants. The figure shows a heatmap of ASVs (rows) that were differentially abundant between groups when comparing a particular follow-up day (columns) for SARS-CoV-2-infected participants vs day 1 (enrolment) for uninfected participants. Each cell indicates the log2 FC in the normalized counts of an ASV between groups as shown by the colour scale (red, increased in SARS-CoV-2-infected participants; blue, increased in controls; grey, no significant association). These estimates were obtained from DESeq2 models adjusting for potential confounders and controlling for multiple testing (see text). ASV, amplicon sequence variant; FC, fold change; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Longitudinal effect of SARS-CoV-2 viral load on the URT microbiota
The median cycle threshold value for the detection of the coronavirus nucleocapsid gene region 1 across all samples collected at study enrolment (day 1) from the 24 participants with COVID-19 was 24.88 (interquartile range=21.24–33.37). Of the 121 samples from participants with COVID-19 included in statistical analyses, 64 (52.89 %) belonged to the 12 SARS-CoV-2-infected participants with high viral load and 57 (47.11 %) belonged to the 12 SARS-CoV-2-infected participants with low viral load.
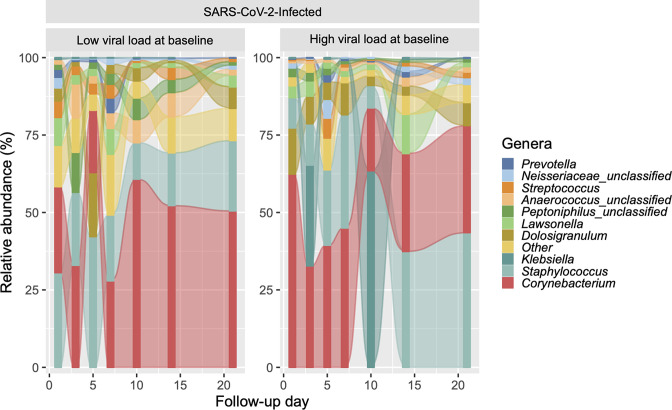
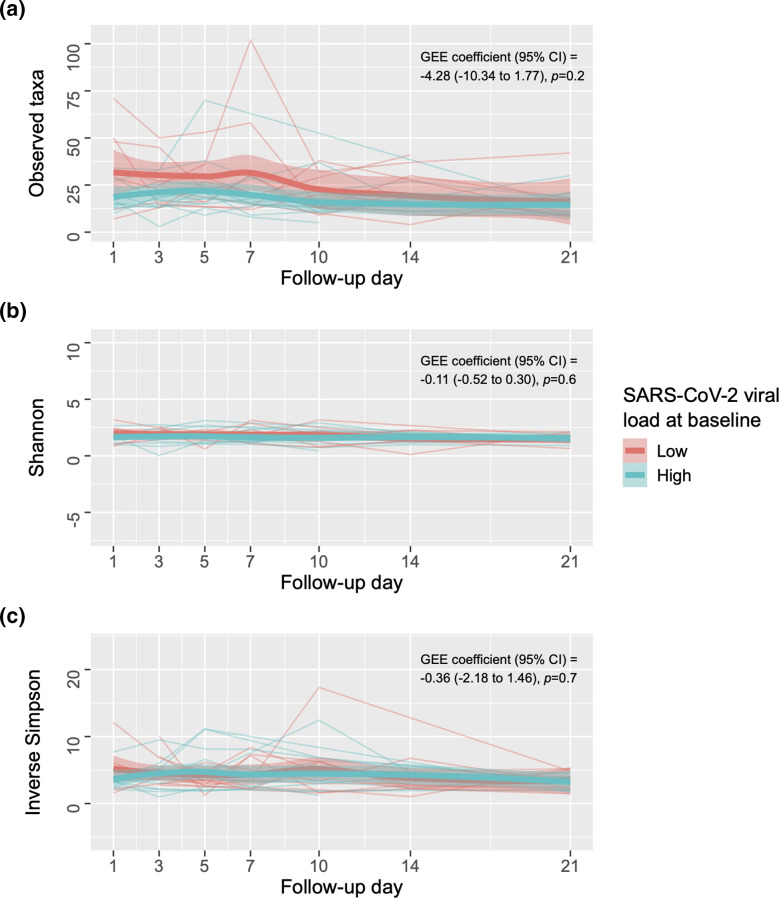
The genera with the highest total relative abundance across all 64 samples from SARS-CoV-2-infected participants with high viral load included Corynebacterium (37.15 %), Staphylococcus (22.28 %), Klebsiella (17.30 %), Dolosigranulum (7.70 %) and Lawsonella (3.40 %), whereas the genera with the highest total relative abundance across all 57 samples from those with low viral load included Corynebacterium (33.70 %), Staphylococcus (26.59 %), Dolosigranulum (6.94 %), Anaerococcus unclassified (5.50 %) and Peptoniphilus unclassified (5.11 %) (Fig. 5). However, in longitudinal analyses, high SARS-CoV-2 viral load was not associated with any of the ⍺-diversity (Fig. 6a–c) or β-diversity indices (q>0.05 for all comparisons). Likewise, high SARS-CoV-2 viral load did not impact on the abundance of the top 25 most predominant ASVs (which accounted for 90.34 % of all ASVs) over time (q>0.05 for all comparisons).
Fig. 5.
Longitudinal changes in the predominant genera of the upper respiratory tract microbiota in SARS-CoV-2-infected participants with and without high viral load at baseline. The figure shows alluvial plots of the relative abundance (%) of the top 10 most abundant genera (y-axis) per group and by follow-up day (x-axis). The relative abundances of other genera were collapsed into the ‘other’ category.
Fig. 6.
Longitudinal effect of SARS-CoV-2 viral load on the ⍺-diversity of the upper respiratory tract microbiota in participants with coronavirus disease 2019. The figure shows line plots of the observed species (1A), Shannon (1B) and inverse Simpson indices (1C) (y-axes) for each participant per group and follow-up day (x-axes). The coefficient, 95% CI, and P-value for the comparison between groups using a GEE model adjusting for potential confounders (see text) are shown. Each group’s fitted locally estimated scatterplot smoothing lines (bold lines) and corresponding 95 % CIs (shaded bands) are also shown. CI, confidence interval, GEE, generalized estimating equation; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Discussion
The URT microbiota has been shown to be a key determinant of respiratory health, but there is a paucity of evidence on the effect of SARS-CoV-2 on the URT microbiota over time. Our results suggest that (1) mild-to-moderate COVID-19 can lead to alterations in the URT microbiota that may continue for several weeks after the initial ARI and (2) that the longitudinal changes in the URT microbiota associated with COVID-19 are independent of viral load.
In our study, we show differences in multiple URT microbial ecology metrics between participants with mild-to-moderate COVID-19 and asymptomatic, uninfected controls. These dissimilarities were more evident at the microbial community-level (i.e. in ⍺- and β-diversity analyses) than at the individual taxa level (i.e. in differential abundance analyses). Indeed, although we found multiple ASVs to be differentially abundant between groups, this was mainly explained by changes in a few participants and there was no evidence of a common URT microbiota signature related to SARS-CoV-2 infection. This could be a reflection of our small sample size but could also be seen if, even in less severe cases of COVID-19, SARS-CoV-2 leads to a broad instability of the URT microbiota and stochastic effects in its structure, rather than consistent changes in specific taxa [21, 33]. Interestingly, the differences between groups were still evident up to 21 days after enrolment (the last available follow-up). Because viral shedding in immunocompetent adults with non-severe COVID-19 (such as those included in our study) is likely to occur 1 to 2 weeks after the onset of disease [34, 35], this suggests that the modifications in the URT microbiota associated with SARS-CoV-2 can persist for several days after the virus is cleared. These findings are in line with the ample literature demonstrating that other respiratory viruses (such as RSV, human rhinovirus, or influenza virus) can lead to extensive alterations of the URT microbiota [4–7]. Of note, we only collected samples during a single time point from controls, and it is possible that there could be individual day-to-day variations in the URT microbiota, which could have biased our results. However, albeit limited, current evidence suggests that, in the absence of disease, the URT microbiota is relatively stable over short periods of time and mainly changes with seasons [36–39].
To date, the precise mechanisms by which SARS-CoV-2 can modify the URT microbiota are unknown, but –based on what is known for other respiratory viruses– these are likely multifaceted, complex, and related to changes in the local milieu that can result from viral-induced epithelial injury, influx of inflammatory cells and production of numerous immune mediators as part of the anti-viral immune response, impaired ciliary function, increased mucous secretions, fluctuations in the production of metabolites and availability of resources, and up- or downregulation of specific host cell receptors, among others [40]. These all, in turn, lead to a failure in the homeostasis of the microbial ecosystem that disturb inter-bacterial interactions, promoting the establishment or replication of certain taxa [41].
Our results add to the accumulating evidence showing an association between SARS-CoV-2 and the URT microbiota in adults [16, 21, 42–45]. However, the large majority of previous studies in this field have had a cross-sectional design and/or have included participants infected with other respiratory viruses as controls. In two prior studies that did include longitudinal analyses and true controls [43, 44], differences between groups were noted up to approximately 1 month after the diagnosis of COVID-19, which supports our findings of an incomplete restoration of the URT microbial communities several weeks following the initial exposure. Unlike ours, those two studies only included participants who were admitted to the hospital due to COVID-19 and mainly used β-diversity analyses. In addition, most of their statistical analyses did not account for potential confounders of the associations. The latter is an important limitation addressed by our study, as previous research has shown that several factors related to hospitalization or disease severity (such as underlying chronic lung disease, antibiotic use and/or respiratory support) can directly influence the URT microbiota, thus introducing bias [45]. To our knowledge, only one prior study has examined whether SARS-CoV-2 viral load can longitudinally impact on the URT microbiota [45]. In contrast to ours, that study showed a mild (albeit significant) inverse relationship between SARS-CoV-2 viral load and the ⍺-diversity of the URT microbiota over time in adjusted analyses. However, all of the participants with COVID-19 in that study were admitted to the intensive care unit, which may have led to some residual confounding. Furthermore, no significant associations were found in β-diversity or differential abundance analyses, suggesting that the effect of SARS-CoV-2 viral load on the URT microbiota is non-existent or minimal.
The results of several other published studies suggest that specific upper and lower respiratory tract microbiota signatures can determine COVID-19 severity [21, 42, 43, 46]. Our findings indicate that SARS-CoV-2 decreases the abundance of several URT taxa that are usually commensal or beneficial (e.g. certain members of the genera Corynebacterium and Lactobacillus ), while also increasing the abundance of other taxa that can be pathogenic or detrimental (e.g. some members of the genera Staphylococcus and Peptoniphilus ), which could be one of the mechanisms underlying COVID-19 severity. In the same context, we and others have previously shown that the URT microbiota associated with other common respiratory viruses (e.g. RSV) can influence their viral load, host transcriptome patterns and the local immune response, ultimately impacting on disease severity and the development of chronic lung diseases following the initial ARI (such as asthma) [8–11]. However, because most of the available data are from observational studies, whether the changes in the URT microbiota related to SARS-CoV-2 truly mediate short- or long-term clinical outcomes following COVID-19 remains largely unknown and merits further investigation.
Despite our study’s multiple strengths, we should acknowledge several other limitations. First, as noted above, our sample size was small, so we may have been underpowered to detect certain associations. Second, due to the inherent limitations of 16S rRNA gene sequencing, we were unable to accurately classify some taxa below the genus level or identify those that are transcriptionally active. Third, as suggested by our prior studies, URT viral–bacterial associations are likely complex and bi-directional [10], and thus are difficult to examine. Fourth, whether the URT microbiota patterns associated with SARS-CoV-2 are unique to this respiratory virus is unknown. However, this is suggested by the results of prior studies demonstrating substantial differences in the URT microbial communities of participants with COVID-19 and those with other respiratory viruses [42]. Furthermore, we have previously shown that respiratory viruses other than SARS-CoV-2 (e.g. RSV) are associated with distinct URT microbiota profiles [5]. Fifth, we did not have data on SARS-CoV-2 variants, and it is possible that individual strains of this respiratory virus could also cause specific changes in the URT microbiota. However, the samples included in our study were all collected in the spring and summer of 2020, when the wild-type SARS-CoV-2 was likely the dominant circulating strain. Sixth, we only included participants with mild-to-moderate COVID-19, so our results cannot be extended to those with asymptomatic, severe, or critical COVID-19. Last, as is the case with all observational studies, there is a possibility of residual confounding or introduction of other sources of bias. For example, although all of our SARS-CoV-2-infected participants were relatively young, had only mild-to-moderate COVID-19, and had no recent use of nasal medications or antibiotics, they may have been taking other medications during the study that were not considered in statistical analyses. Likewise, frequent sampling could result is some subtle changes of the URT microbiota, although the effect of this is unknown.
In summary, our results suggest that mild-to-moderate COVID-19 can lead to alterations of the URT microbiota that persists for several weeks after the initial infection. Our study sheds light on the dynamics of the URT microbial communities following ARI with SARS-CoV-2 and on the diverse consequences of COVID-19 on the human body. Future studies with larger sample sizes, using other next-generation sequencing technologies (e.g. metagenomics or metatranscriptomics), and capturing longer follow-up periods, are needed to better define whether the URT microbiota fully recovers after COVID-19, and the timeline of this recovery. In addition, mechanistic studies should explore the precise pathways through which SARS-CoV-2 can modify the URT microbiota. Ultimately, understanding whether URT microbial disturbances induced by SARS-CoV-2 can mediate COVID-19 severity is imperative to design interventions that could decrease the morbidity and mortality of this disease and benefit public health.
Supplementary Data
Funding information
This work was supported by funds from the Centers For Disease Control and Prevention (under award number 75D3012110094); the National Institute of Allergy and Infectious Diseases (under award numbers R21AI142321, R21AI154016, R21AI149262); the National Heart, Lung, and Blood Institute (under award numbers K23HL148638 and R01HL146401); and the Vanderbilt Technologies for Advanced Genomics Core (grant support from the National Institutes of Health under award numbers UL1RR024975, P30CA68485, P30EY08126, G20RR030956). The contents are solely the responsibility of the authors and do not necessarily represent official views of the funding agencies.
Author contribution
C.R.S., K.S.K., M.H.S., M.H.F., S.V.R., J.H.T., and S.R.D. contributed to the study design. K.S.K., M.H.S., B.C.W., M.H.F., V.G., and J.H.T. contributed to the sample collection. M.H.S., B.A.S., H.M.B., H.H.B., and S.R.D. contributed to the sample processing. C.R.S. and M.H.S. contributed to the bioinformatic processing and statistical analyses. J.H.T. and S.R.D. obtained the research funding supporting the current study. C.R.S., M.H.S., J.H.T., and S.R.D. wrote the initial version of the manuscript and all authors reviewed and approved the final version.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
The parent clinical trial for this study was registered on clinicaltrials.gov (NCT 04347538). This study was approved by the Vanderbilt University Medical Center Institutional Review Board and Biosafety Committee. All subjects consented to participate in this study and written consent was received prior to registration in the study.
Footnotes
Abbreviations: ARI, acute respiratory infection; ASV, amplicon sequence variant; COVID-19, coronavirus disease 2019; GEE, generalized estimating equation; PERMANOVA, permutational multivariate analysis of variance; RCT, randomized controlled trial; rRNA, ribosomal RNA; RSV, respiratory syncytial virus; SARS-CoV-2, severe acute respiratory syndrome coronavirus-2; URT, upper respiratory tract.
All supporting data, code and protocols have been provided within the article or through supplementary data files.
One supplementary figure and one supplementary table are available with the online version of this article.
References
- 1.Man WH, de Steenhuijsen Piters WAA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol. 2017;15:259–270. doi: 10.1038/nrmicro.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosch A, de Steenhuijsen Piters WAA, van Houten MA, Chu M, Biesbroek G, et al. Maturation of the infant respiratory microbiota, environmental drivers, and health consequences. A prospective cohort study. Am J Respir Crit Care Med. 2017;196:1582–1590. doi: 10.1164/rccm.201703-0554OC. [DOI] [PubMed] [Google Scholar]
- 3.Petersen C, Round JL. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol. 2014;16:1024–1033. doi: 10.1111/cmi.12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosas-Salazar C, Shilts MH, Tovchigrechko A, Chappell JD, Larkin EK, et al. Nasopharyngeal microbiome in respiratory syncytial virus resembles profile associated with increased childhood Asthma risk. Am J Respir Crit Care Med. 2016;193:1180–1183. doi: 10.1164/rccm.201512-2350LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosas-Salazar C, Shilts MH, Tovchigrechko A, Schobel S, Chappell JD, et al. Differences in the nasopharyngeal microbiome during acute respiratory tract infection with human rhinovirus and respiratory syncytial virus in infancy. J Infect Dis. 2016;214:1924–1928. doi: 10.1093/infdis/jiw456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen EK, Koeppel AF, Hendley JO, Turner SD, Winther B, et al. Characterization of the nasopharyngeal microbiota in health and during rhinovirus challenge. Microbiome. 2014;2:22. doi: 10.1186/2049-2618-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaul D, Rathnasinghe R, Ferres M, Tan GS, Barrera A, et al. Microbiome disturbance and resilience dynamics of the upper respiratory tract during influenza A virus infection. Nat Commun. 2020;11:2537. doi: 10.1038/s41467-020-16429-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ederveen THA, Ferwerda G, Ahout IM, Vissers M, de Groot R, et al. Haemophilus is overrepresented in the nasopharynx of infants hospitalized with RSV infection and associated with increased viral load and enhanced mucosal CXCL8 responses. Microbiome. 2018;6:10. doi: 10.1186/s40168-017-0395-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Steenhuijsen Piters WAA, Heinonen S, Hasrat R, Bunsow E, Smith B, et al. Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med. 2016;194:1104–1115. doi: 10.1164/rccm.201602-0220OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosas-Salazar C, Tang Z-Z, Shilts MH, Turi KN, Hong Q, et al. Upper respiratory tract bacterial-immune interactions during respiratory syncytial virus infection in infancy. J Allergy Clin Immunol. 2022;149:966–976. doi: 10.1016/j.jaci.2021.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosas-Salazar C, Shilts MH, Tovchigrechko A, Schobel S, Chappell JD, et al. Nasopharyngeal Lactobacillus is associated with a reduced risk of childhood wheezing illnesses following acute respiratory syncytial virus infection in infancy. J Allergy Clin Immunol. 2018;142:1447–1456. doi: 10.1016/j.jaci.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dumas O, Hasegawa K, Mansbach JM, Sullivan AF, Piedra PA, et al. Severe bronchiolitis profiles and risk of recurrent wheeze by age 3 years. J Allergy Clin Immunol. 2019;143:1371–1379. doi: 10.1016/j.jaci.2018.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raita Y, Pérez-Losada M, Freishtat RJ, Harmon B, Mansbach JM, et al. Integrated omics endotyping of infants with respiratory syncytial virus bronchiolitis and risk of childhood asthma. Nat Commun. 2021;12:3601. doi: 10.1038/s41467-021-23859-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimura KS, Freeman MH, Wessinger BC, Gupta V, Sheng Q, et al. Interim analysis of an open-label randomized controlled trial evaluating nasal irrigations in non-hospitalized patients with coronavirus disease 2019. Int Forum Allergy Rhinol. 2020;10:1325–1328. doi: 10.1002/alr.22703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esther CR, Kimura KS, Mikami Y, Edwards CE, Das SR, et al. Pharmacokinetic-based failure of a detergent virucidal for severe acute respiratory syndrome-coronavirus-2 (SARS-COV-2) nasal infections: a preclinical study and randomized controlled trial. Int Forum Allergy Rhinol. 2022;12:1137–1147. doi: 10.1002/alr.22975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosas-Salazar C, Kimura KS, Shilts MH, Strickland BA, Freeman MH, et al. SARS-CoV-2 infection and viral load are associated with the upper respiratory tract microbiome. J Allergy Clin Immunol. 2021;147:1226–1233. doi: 10.1016/j.jaci.2021.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.World Health Organization Operational considerations for case management of COVID-19 in health facility and community. Interim guidance. Pediatr Med Rodz. 2020;16:27–32. doi: 10.15557/PiMR.2020.0004. [DOI] [Google Scholar]
- 18.Singh K, Gobert AP, Coburn LA, Barry DP, Allaman M, et al. Dietary arginine regulates severity of experimental colitis and affects the colonic microbiome. Front Cell Infect Microbiol. 2019;9:66. doi: 10.3389/fcimb.2019.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiremath G, Shilts MH, Boone HH, Correa H, Acra S, et al. The salivary microbiome is altered in children with Eosinophilic Esophagitis and correlates with disease activity. Clin Transl Gastroenterol. 2019;10:e00039. doi: 10.14309/ctg.0000000000000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shilts MH, Rosas-Salazar C, Strickland BA, Kimura KS, Asad M, et al. Severe COVID-19 is associated with an altered upper respiratory tract microbiome. Front Cell Infect Microbiol. 2021;11:781968. doi: 10.3389/fcimb.2021.781968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:226. doi: 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 2017;5:27. doi: 10.1186/s40168-017-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.R Development Core Team Vienna, Austria: R Foundation for Statistical Computing; 2006. R: A language and environment for statistical computing.http://www.R-project.org/ [Google Scholar]
- 28.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x. [DOI] [Google Scholar]
- 29.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shields-Cutler RR, Al-Ghalith GA, Yassour M, Knights D. SplinectomeR enables group comparisons in longitudinal microbiome studies. Front Microbiol. 2018;9:785. doi: 10.3389/fmicb.2018.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc: Series B. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 32.McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–5. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaneveld JR, McMinds R, Vega Thurber R. Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat Microbiol. 2017;2:17121. doi: 10.1038/nmicrobiol.2017.121. [DOI] [PubMed] [Google Scholar]
- 34.Qi L, Yang Y, Jiang D, Tu C, Wan L, et al. Factors associated with the duration of viral shedding in adults with COVID-19 outside of Wuhan, China: a retrospective cohort study. Int J Infect Dis. 2020;96:531–537. doi: 10.1016/j.ijid.2020.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.World Health Organization Criteria for releasing COVID-19 patients from isolation: scientific brief. World Health Organization; 2020. [Google Scholar]
- 36.Wagner Mackenzie B, Chang K, Zoing M, Jain R, Hoggard M, et al. Longitudinal study of the bacterial and fungal microbiota in the human sinuses reveals seasonal and annual changes in diversity. Sci Rep. 2019;9:17416. doi: 10.1038/s41598-019-53975-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frank DN, Feazel LM, Bessesen MT, Price CS, Janoff EN, et al. The human nasal microbiota and Staphylococcus aureus carriage. PLoS One. 2010;5:e10598. doi: 10.1371/journal.pone.0010598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Camarinha-Silva A, Jáuregui R, Pieper DH, Wos-Oxley ML. The temporal dynamics of bacterial communities across human anterior nares. Environ Microbiol Rep. 2012;4:126–132. doi: 10.1111/j.1758-2229.2011.00313.x. [DOI] [PubMed] [Google Scholar]
- 40.McCullers JA. Insights into the interaction between influenza virus and pneumococcus. Clin Microbiol Rev. 2006;19:571–582. doi: 10.1128/CMR.00058-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanada S, Pirzadeh M, Carver KY, Deng JC. Respiratory viral infection-induced microbiome alterations and secondary bacterial Pneumonia. Front Immunol. 2018;9:2640. doi: 10.3389/fimmu.2018.02640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merenstein C, Liang G, Whiteside SA, Cobián-Güemes AG, Merlino MS, et al. Signatures of COVID-19 severity and immune response in the respiratory tract microbiome. mBio. 2021;12:e0177721. doi: 10.1128/mBio.01777-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren L, Wang Y, Zhong J, Li X, Xiao Y, et al. Dynamics of the upper respiratory tract microbiota and its association with mortality in COVID-19. Am J Respir Crit Care Med. 2021;204:1379–1390. doi: 10.1164/rccm.202103-0814OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu R, Lu R, Zhang T, Wu Q, Cai W, et al. Temporal association between human upper respiratory and gut bacterial microbiomes during the course of COVID-19 in adults. Commun Biol. 2021;4:240. doi: 10.1038/s42003-021-01796-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lloréns-Rico V, Gregory AC, Van Weyenbergh J, Jansen S, Van Buyten T, et al. Clinical practices underlie COVID-19 patient respiratory microbiome composition and its interactions with the host. Nat Commun. 2021;12:6243. doi: 10.1038/s41467-021-26500-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sulaiman I, Chung M, Angel L, Tsay J-CJ, Wu BG, et al. Microbial signatures in the lower airways of mechanically ventilated COVID-19 patients associated with poor clinical outcome. Nat Microbiol. 2021;6:1245–1258. doi: 10.1038/s41564-021-00961-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.