Abstract
A common cause of frontotemporal dementia (FTD) are nonsense mutations in the progranulin (GRN) gene. Because nonsense mutations activate the nonsense-mediated RNA decay (NMD) pathway, we sought to inhibit this RNA turnover pathway as a means to increase progranulin levels. Using a knock-in mouse model harboring a common patient mutation, we tested whether either pharmacological or genetic inhibition of NMD upregulates progranulin in these GrnR493X mice. We first examined antisense oligonucleotides (ASOs) targeting an exonic region in GrnR493X mRNA predicted to block its degradation by NMD. As we previously reported, these ASOs effectively increased GrnR493X mRNA levels in fibroblasts in vitro. However, following CNS delivery, we found that none of the 8 ASOs we tested increased Grn mRNA levels in the brains of GrnR493X mice. This result was obtained despite broad ASO distribution in the brain. An ASO targeting a different mRNA was effective when administered in parallel to wild-type mice. As an independent approach to inhibit NMD, we examined the effect of loss of an NMD factor not required for embryonic viability: UPF3b. We found that while Upf3b deletion effectively perturbed NMD, it did not increase Grn mRNA levels in Grn+/R493X mouse brains. Together, our results suggest that the NMD-inhibition approaches that we used are likely not viable for increasing progranulin levels in individuals with FTD caused by nonsense GRN mutations. Thus, alternative approaches should be pursued.
Introduction
Progranulin is a lysosomal and secreted protein with pleiotropic effects, including promoting neuronal survival, neurite outgrowth, wound healing, tumor cell growth, and modulating inflammation [1, 2]. In humans, heterozygous GRN mutations cause frontotemporal dementia (FTD) due to progranulin haploinsufficiency [3, 4]. Therefore, increasing progranulin levels is a major therapeutic goal [5, 6]. Gene therapy studies in mice provide proof of concept that restoring progranulin levels in heterozygous Grn mice improves FTD-associated neuropathology and behavioral deficits [7]. Current therapeutic efforts are focused on small molecules that increase progranulin expression [8–11], gene therapies [7, 12], monoclonal antibodies that modulate progranulin trafficking [13], and protein replacement [14]. However, there are currently no approved therapies for progranulin-deficient FTD.
The vast majority (>80%) of FTD-associated GRN mutations are nonsense or frameshift mutations which introduce a premature termination codon (PTC) [15]. As a result, for many of these mutations, the mutant mRNA has been shown to be [3, 4, 16, 17], or is expected to be, degraded through the nonsense-mediated RNA decay (NMD) pathway [18]. Because the progranulin protein contains 7.5 conserved granulin domains, which are believed to be the bioactive units that are produced following proteolytic cleavage, stabilizing mutant GRN mRNAs would likely increase the levels of functional granulins. Together, this suggests that inhibiting NMD mechanisms may be feasible therapeutic strategies for increasing levels of progranulin mRNA and functional protein in the context of progranulin-deficient FTD.
NMD can be inhibited by several pharmacological and genetic methods. A number of compounds have been identified which broadly inhibit NMD; these include NMDI1, NMDI9, NMDI14, 5-azacytidine (5AzaC), thapsigargin, and others [19–22]. Another reported strategy for blocking NMD uses antisense oligonucleotides (ASOs), short synthetic oligonucleotides used to modulate target RNAs, to inhibit degradation of a specific PTC-containing transcript [23]. In a cell-based reporter system, Nomakuchi et al. demonstrated that ASOs targeting the exon-junction complex (EJC) at the 3’ end of the exon harboring the PTC can prevent binding of key EJC proteins that are required for NMD, thereby enabling the PTC-containing mutant mRNA to escape NMD-mediated degradation [23]. Most recently, ASO-mediated suppression of the NMD factor UPF3b has been suggested as a potential approach for diseases caused by nonsense mutations [24]. Notably, UPF3b depletion experiments have revealed that UPF3b is a branch-specific NMD factor that regulates a subset of NMD targets [24, 25].
We previously developed a GrnR493X mouse model that harbors the common GRNR493X patient nonsense mutation [17], as well as a panel of ASOs that block NMD-mediated degradation of the mutant GrnR493X mRNA in cultured mouse fibroblast cells [17]. Here, we tested two NMD-targeting strategies for increasing progranulin levels in the brains of GrnR493X mice. Specifically, we tested the ASOs in a pharmacological approach and Upf3b deletion in a genetic approach.
Materials and methods
ASOs
ASOs used in these studies were 18-mer ASOs, except the Malat1-targeting ASO was a 20-mer. The ASO sequences are provided in S1 Table. ASOs used for in vitro and cell-based studies were dissolved in water and stored at -20°C. For in vivo studies, lyophilized ASOs were dissolved in sterile PBS without calcium or magnesium (Gibco, 14190–250) and sterilized by passing through a 0.2 μm filter.
Cell culture
GrnR493X MEF cells [17] and HeLa cells were cultured in DMEM (Dulbecco’s Modified Eagle Medium, high-glucose) (Gibco, 11995–073) supplemented with 10% fetal bovine serum (FBS) (Gibco, 26140–095), 10 U/ml penicillin, and 10 μg/ml of streptomycin. For ASO treatments, GrnR493X MEF cells were seeded in 6-well plates, and then transfected as indicated on the following day with 100 nM ASO using 6 μl of Lipofectamine 2000 (Invitrogen). For progranulin expression, HeLa cells were seeded 6-well plates, and then transfected with 1 μg of the indicated plasmid on the following day using 3.75 μl of Lipofectamine 3000 (Invitrogen).
Mouse studies
Mice were housed in a pathogen-free barrier facility with a 12-h light/12-h dark cycle and provided food and water ad libitum. GrnR493X knock-in mice [17] and Upf3b knockout mice [26] were on the C57BL/6J background and were genotyped by real-time PCR (Transnetyx). For intracerebroventricular (ICV) ASO delivery, 200–500 μg ASO was administered by bolus injection into the right lateral ventricle of mice anesthetized with isoflurane, as previously described [27]. After 2–3 weeks, mice were sacrificed and brain tissues were collected for RNA and protein analyses, as described below. For immunofluorescence, mice were transcardially perfused with PBS followed by 4% paraformaldehyde. For intraperitoneal (IP) ASO delivery, 50 mg/kg of ASO was administered every other day for a total of 4 injections. One day after the final injection, mice were sacrificed and tissues were collected for qPCR analysis.
Animal procedures were approved by the Institutional Animal Care and Use Committee of Saint Louis University (protocol #2764) and followed NIH guidelines. For ICV administration, mice were anesthetized with isoflurane and also provided bupivacaine and buprenorphine. For perfusion, mice were anesthetized with a ketamine/xylazine cocktail followed by transcardial perfusion. For tissue collection, mice were anesthetized with ketamine/xylazine cocktail followed by rapid decapitation.
RNA analysis
Total RNA was isolated from cultured cells using the RNeasy Mini kit (Qiagen, 74106) with on-column DNase digestion (Qiagen, 79256). RNA was reverse-transcribed to obtain cDNA using the iScript cDNA synthesis kit (Bio-Rad, 1708891), and qPCR was performed using PowerUp SYBR Green Master Mix (ThermoFisher, A25777) with a Bio-Rad CFX384 Real-Time System. Primers sequences are provided in S2 Table. Results for qPCR were normalized to the housekeeping gene 36B4 and evaluated by the comparative CT method.
Western blot analysis
Mouse cortex samples were lysed in RIPA buffer containing protease inhibitors (Roche, cOmplete Mini EDTA-free Protease Inhibitor Cocktail). Cleared lysates were transferred to new tubes, and protein concentrations were determined using the Bio-Rad DC Protein Assay Kit II. For experiments analyzing secreted progranulin, conditioned media was collected from transfected HeLa cells and cleared at 10,000 x g for 10 min at 4° C. Sample buffer was added to the lysates or conditioned media, and the samples were heated at 95°C for 10 min. Equal amounts of protein lysates (100 μg) or equal volumes of conditioned media (30 μl) were separated on SDS–PAGE gels. Proteins were transferred to nitrocellulose membranes using the Bio-Rad Turbo-Blot transfer system. After blocking and antibody incubations, membranes were incubated with SuperSignal West or SuperSignal Femto chemiluminescent HRP substrate (ThermoFisher) and visualized using a Chemi-Doc system (Bio-Rad). The primary antibodies used for immunoblot analysis include: an anti-mouse progranulin polyclonal antibody (R&D Systems, AF2557, 1:200 dilution) and an anti-α-tubulin monoclonal antibody (Sigma, T9026, 1:2000 dilution). The HRP-conjugated secondary antibodies used were donkey anti-sheep IgG (H+L) (Jackson Immuno Research Labs, 713035147) and donkey anti-mouse IgG (H+L) (Jackson Immuno Research Labs, 715035150).
Immunofluorescence
Fixed brains were frozen in O.C.T. solution (Tissue-Tek) and sectioned at 40 μm using a cryostat. Free floating sections were blocked and then incubated with a previously described pan-ASO antibody that recognizes the ASO backbone [28] at 1:2000 dilution overnight. After washing, sections were incubated with Alexa Fluor Plus 647 goat anti-rabbit IgG (Invitrogen, A32733, 1:300 dilution) for 1 h, followed by incubation with DAPI (Invitrogen, D1306). After washing, sections were mounted onto slides with Fluoromount-G mounting media (Invitrogen, 00-4958-02). Images were acquired on an Olympus FV1000 confocal microscope with a 20x objective.
Statistical analyses
Data are presented as means ± SD or means ± SEM, as indicated in the figure legends. Data were analyzed with GraphPad Prism software using the statistical tests described in the figure legends. P values < 0.05 were considered significant.
Results
We previously developed ASOs that inhibit NMD-mediated degradation of the GrnR493X mutant mRNA and reported that they increase progranulin mRNA and protein levels in mouse fibroblast cells [17]. These ASOs were designed to block the binding of NMD proteins to the EJC, thereby enabling the GrnR493X mutant mRNA to escape NMD-mediated degradation. The R493X nonsense mutation is located in exon 12 of the mouse Grn mRNA, 159 nucleotides upstream of the next intron; the ASOs target the EJC of exon 12, specifically 17–44 nucleotides upstream of the 3’ end of the exon 12.
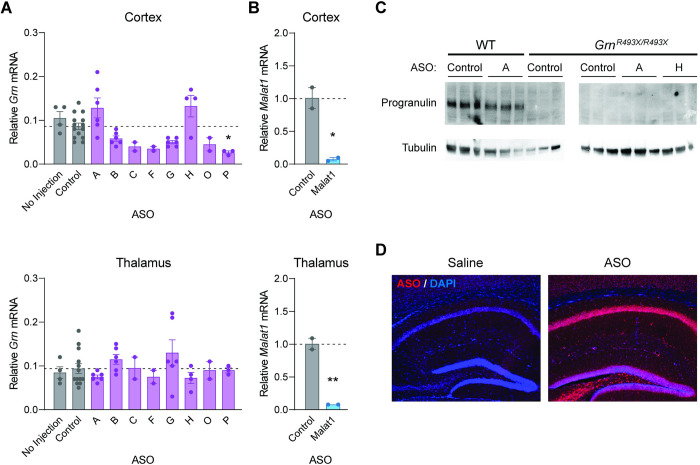
Here, we report in vivo testing of these 8 NMD-targeting Grn ASOs in the GrnR493X knock-in mouse model [17]. In contrast to our findings in cells, we failed to detect any significant increase in Grn mRNA levels in the cortex or thalamus of GrnR493X/R493X mice at 2–3 weeks following ICV administration of 200–500 μg ASO (Fig 1A). Surprisingly, in the cortex, we noted a trend toward decreased Grn mRNA levels with multiple ASOs, possibly due to effects on mRNA stability. As a positive control, we administered a validated Malat1-targeting ASO that is designed to decrease Malat1 mRNA levels [29]. As expected, we observed markedly decreased Malat1 mRNA in the cortex and thalamus (Fig 1B). With the NMD-targeting Grn ASOs, we also did not detect any increase in progranulin protein in the cortex (Fig 1C). Importantly, we confirmed that this polyclonal antibody is able to detect the truncated progranulin R493X protein (S1 Fig). Immunofluorescence staining confirmed broad distribution of the ASO throughout the brain in these studies (Fig 1D).
Fig 1. ICV administration of ASOs targeting NMD of the GrnR493X mRNA does not increase progranulin mRNA or protein levels in the brains of GrnR493X/R493X mice.
qPCR results from brains of GrnR493X/R493X mice at 2–3 weeks after ICV administration of 200–500 μg ASO. (A) Grn mRNA levels are presented relative to levels in tissues of wild-type mice that received control ASO. (B) Malat1 mRNA levels in wild-type mice. (C) Western blot of mouse progranulin levels in cortex of GrnR493X/R493X mice at 2 weeks after ICV administration of 500 μg ASO. (D) At 3 weeks after ICV administration of saline or ASO B (200 μg), brains were fixed and sections were stained with an ASO-antibody (red) and counterstained with nuclear stain DAPI (blue). Data are presented as means ± SEM; * indicates p<0.05 and ** indicates p<0.01, as determined by one-way ANOVA with Tukey post hoc test in (A) and by t-test in (B). WT, wild-type.
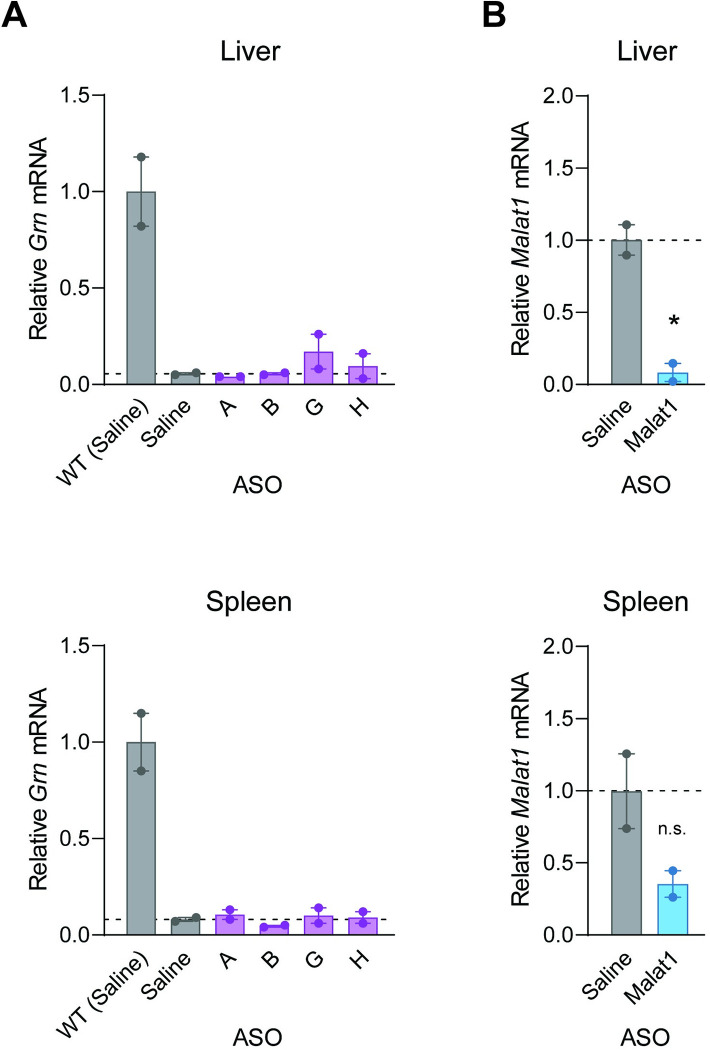
We also performed intraperitoneal (IP) administration and similarly found these ASOs failed to increase Grn mRNA levels in the liver and spleen of GrnR493X/R493X mice (Fig 2A). As a control, the Malat1-targeting ASO strongly decreased Malat1 mRNA in the liver and spleen (Fig 2B). Together, these results demonstrate that the ASOs targeting NMD of the GrnR493X mRNA failed to increase progranulin levels in vivo, despite showing efficacy in cells (S2 Fig) [17].
Fig 2. IP administration of ASOs targeting NMD of the GrnR493X mRNA does not increase Grn mRNA levels in the livers and spleens.
qPCR results from livers and spleens of GrnR493X/R493X mice following a series of four IP administrations of 50 mg/kg ASO. (A) Grn mRNA levels are presented relative to levels in tissues of wild-type mice that received control ASO. (B) Malat1 mRNA levels in wild-type mice. Data are presented as means ± SEM; * indicates p<0.05, as determined by one-way ANOVA with Tukey post hoc test in (A) and by t-test in (B). n.s., not significant.
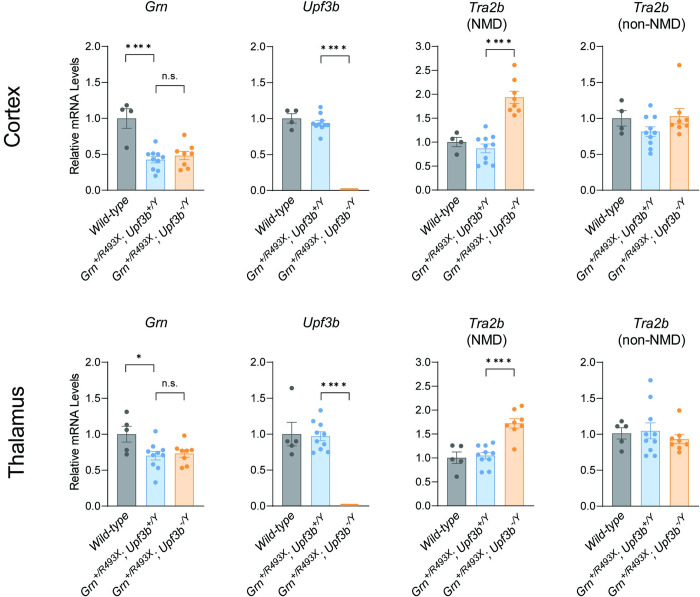
There are also recent efforts to block NMD through UPF3b inhibition [24]. Several studies have shown that UPF3b regulates degradation of a subset of NMD transcripts [24, 25]. To determine if the GrnR493X mRNA is regulated in a UPF3b-dependent manner, we crossed GrnR493X mice with Upf3b-null mice [26] and assessed Grn mRNA levels in the brain. Because the Upf3b gene is x-linked, we used UPF3b-expressing male mice (Upf3b+/Y) and UPF3b-deficient male mice (Upf3b–/Y) for comparisons. As expected, Grn+/R493X mice have ~50% Grn mRNA levels compared to wild-type mice, and male Upf3b–/Y mice do not express Upf3b mRNA (Fig 3). In the cortex and thalamus of age-matched mice, Upf3b deletion did not increase Grn mRNA levels; this was in contrast to the established NMD-sensitive isoform of Tra2b mRNA [21], which is significantly increased by UPF3b deficiency. Together, these results suggest that the GrnR493X mRNA is not subject to UPF3b-mediated degradation and therefore not amenable to UPF3b inhibition strategies.
Fig 3. Upf3b deletion does not increase Grn mRNA levels in the brains of Grn+/R493X mice.
qPCR results from cortex and thalamus of 10–12 week old male mice. Grn mRNA levels are presented relative to levels in tissues of wild-type mice. Data are presented as means ± SEM; * indicates p<0.05 and **** indicates p<0.0001, as determined by one-way ANOVA with Tukey post hoc test. n.s., not significant.
Discussion
In the current studies, we tested two NMD-targeting strategies for increasing progranulin levels in GrnR493X mice. In the pharmacological approach, it is unclear why the ASOs targeting NMD-mediated degradation of the GrnR493X mutant mRNA failed to increase progranulin levels in vivo. To rule out delivery issues, we used a validated Malat1-targeting ASO as a positive control and observed the expected effect of lowering Malat1 mRNA levels. We also confirmed broad ASO distribution in the brain by immunostaining. Lastly, after completion of our in vivo studies, we confirmed that these ASO stock solutions are inherently active in preventing NMD of the mutant GrnR493X mRNA when the ASOs were delivered to cultured cells via lipid-based transfection. Together, these results suggest that the NMD-targeting ASOs that are active in vitro may not necessarily be active in vivo.
The reason(s) for the lack of efficacy of the NMD-targeting ASOs in vivo are unclear, but possible reasons include ASO uptake in vivo and that the subcellular distribution of these ASOs might not be optimal for them to be efficacious in vivo. While there are several reports in cell-based studies [17, 23, 30, 31], to our knowledge there is no demonstration yet of in vivo use of ASOs to block NMD by targeting an EJC. Additionally, it is possible that ASO length could be important for targeting NMD in vivo; it is worth noting that the NMD-targeting ASOs used in this study are 18-mer ASOs, whereas the positive control Matat1-targeting ASO is a 20-mer. This is unlikely to account for the negative results with the NMD-targeting ASOs, because other studies have shown in vivo efficacy of centrally administered 18-mer ASOs that sterically block splicing or regulatory factors. For example, an 18-mer ASO targeting an intronic splicing silencer increased the inclusion of exon 7 of SMN2 in a humanized mouse model of spinal muscular atrophy [32]. Additionally, we recently showed that an 18-mer ASO that blocks a miR binding site in the GRN mRNA increases progranulin protein levels in a humanized mouse model [33]. Nonetheless, future studies could test ASOs of different lengths, such as 20-mers, targeting this same EJC region of the GrnR493X mRNA. Finally, because NMD targeting of an mRNA can vary between cells and tissues [34], we cannot rule out the possibility that the GrnR493X mRNA is not efficiently targeted to NMD in the brain, despite our previous findings that the mRNA is regulated by NMD in cultured cells and in peripheral tissues [17].
The other major finding, from our genetic approach, is that the GrnR493X mutant mRNA is not degraded through the UPF3b-dependent branch of NMD. These results further suggest that alternative strategies should be pursued for increasing progranulin levels in the context of progranulin-deficient FTD. One such potential strategy is to use ASOs to block miR binding sites, such as miR-659 and miR-29b [35–38], in the 3’ UTR of the GRN mRNA [33]. A notable advantage of this miR-targeting strategy is that it is agnostic to the specific disease mutation and could be used in the context of any of the >70 FTD-associated GRN mutations that have been identified [15]. On the other hand, the NMD-targeting strategy would require development of patient-specific ASOs to target the particular exon harboring the nonsense GRN mutation.
A limitation of our studies is that they relied heavily on the GrnR493X knock-in mouse model of progranulin-deficient FTD. While we have previously shown that NMD inhibition similarly increases progranulin mRNA levels in fibroblast cells derived from GrnR493X knock-in mice and in human fibroblast cells harboring the GRNR493X mutation [17], we cannot exclude the possibility that species differences may exist with respect to the current findings.
In conclusion, we found that pharmacological inhibition of NMD for the GrnR493X mRNA and genetic inhibition of the UPF3b-dependent branch of NMD do not increase progranulin levels in the Grn R493X mouse model. Our results suggest that these NMD-inhibition approaches are likely not viable for increasing progranulin levels in individuals with FTD caused by nonsense GRN mutations. Thus, alternative approaches should be pursued.
Supporting information
Immunoblot analysis of progranulin using conditioned medium from HeLa cells transfected with plasmids encoding GFP, wild-type (WT) progranulin, or R493X truncation mutant. mPGRN, mouse progranulin.
(PDF)
Cells were transfected with 100 nM ASO using Lipofectamine 2000. After 24 hours, RNA was isolated for qPCR. Grn mRNA levels are presented relative to levels in wild-type cells transfected with control ASO and presented as means ± SD; * indicates p<0.05, ** indicates p<0.01, *** indicates p<0.001, **** indicates p<0.0001, as determined by one-way ANOVA with Dunnett post hoc test.
(PDF)
(PDF)
(PDF)
Acknowledgments
We thank Laura Mitic, Robert Farese, Jr., Tobias Walther, Thi Nguyen, John Morley, and members of the lab for helpful discussions. We also thank Matthew Kim for his technical support with the Upf3b-null mice.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from the National Institutes of Health [AG064069 and AG047339] to ADN; National Institutes of Health/National Center for Advancing Translational Sciences (NCATS) [UL1TR002345] to ADN; and Bluefield Project to Cure FTD to ADN. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Ionis Pharmaceuticals provided support in the form of salaries for authors [K.L., P.J.-N., and F.R.], provided research materials, and contributed to the study design, data collection, and analysis, but did not have additional roles in the decision to publish and preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1.Kao AW, McKay A, Singh PP, Brunet A, Huang EJ. Progranulin, lysosomal regulation and neurodegenerative disease. Nat Rev Neurosci. 2017;18(6):325–33. doi: 10.1038/nrn.2017.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paushter DH, Du H, Feng T, Hu F. The lysosomal function of progranulin, a guardian against neurodegeneration. Acta Neuropathol. 2018;136(1):1–17. doi: 10.1007/s00401-018-1861-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, Pirici D, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442(7105):920–4. doi: 10.1038/nature05017 [DOI] [PubMed] [Google Scholar]
- 4.Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442(7105):916–9. doi: 10.1038/nature05016 [DOI] [PubMed] [Google Scholar]
- 5.Terryn J, Verfaillie CM, Van Damme P. Tweaking progranulin expression: therapeutic avenues and opportunities. Front Mol Neurosci. 2021;14:713031. doi: 10.3389/fnmol.2021.713031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amin S, Carling G, Gan L. New insights and therapeutic opportunities for progranulin-deficient frontotemporal dementia. Curr Opin Neurobiol. 2021;72:131–9. doi: 10.1016/j.conb.2021.10.001 [DOI] [PubMed] [Google Scholar]
- 7.Arrant AE, Filiano AJ, Unger DE, Young AH, Roberson ED. Restoring neuronal progranulin reverses deficits in a mouse model of frontotemporal dementia. Brain. 2017;140(5):1447–65. doi: 10.1093/brain/awx060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cenik B, Sephton C, Dewey C, Xian X, Wei S, Yu K, et al. Suberoylanilide hydroxamic acid (vorinostat) up-regulates progranulin transcription: rational therapeutic approach to frontotemporal dementia. J Biol Chem. 2011;286(18):16101–8. doi: 10.1074/jbc.M110.193433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holler CJ, Taylor G, McEachin ZT, Deng Q, Watkins WJ, Hudson K, et al. Trehalose upregulates progranulin expression in human and mouse models of GRN haploinsufficiency: a novel therapeutic lead to treat frontotemporal dementia. Mol Neurodegener. 2016;11(1):46. doi: 10.1186/s13024-016-0114-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sha SJ, Miller ZA, Min SW, Zhou Y, Brown J, Mitic LL, et al. An 8-week, open-label, dose-finding study of nimodipine for the treatment of progranulin insufficiency from GRN gene mutations. Alzheimers Dement (N Y). 2017;3(4):507–12. doi: 10.1016/j.trci.2017.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ljubenkov PA, Edwards L, Iaccarino L, La Joie R, Rojas JC, Koestler M, et al. Effect of the histone deacetylase inhibitor FRM-0334 on progranulin levels in patients with progranulin gene haploinsufficiency: a randomized clinical trial. JAMA Netw Open. 2021;4(9):e2125584. doi: 10.1001/jamanetworkopen.2021.25584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hinderer C, Miller R, Dyer C, Johansson J, Bell P, Buza E, et al. Adeno-associated virus serotype 1-based gene therapy for FTD caused by GRN mutations. Ann Clin Transl Neurol. 2020;7(10):1843–53. doi: 10.1002/acn3.51165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alzforum. AL001 boosts progranulin. Does it slow frontotemporal dementia? 2021 [cited 2022 February 16]. Available from: https://www.alzforum.org/news/conference-coverage/al001-boosts-progranulin-does-it-slow-frontotemporal-dementia.
- 14.Logan T, Simon MJ, Rana A, Cherf GM, Srivastava A, Davis SS, et al. Rescue of a lysosomal storage disorder caused by Grn loss of function with a brain penetrant progranulin biologic. Cell. 2021;184(18):4651–68.e25. doi: 10.1016/j.cell.2021.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cruts M, Theuns J, Van Broeckhoven C. Locus-specific mutation databases for neurodegenerative brain diseases. Hum Mutat. 2012;33(9):1340–4. doi: 10.1002/humu.22117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gass J, Cannon A, Mackenzie IR, Boeve B, Baker M, Adamson J, et al. Mutations in progranulin are a major cause of ubiquitin-positive frontotemporal lobar degeneration. Hum Mol Genet. 2006;15(20):2988–3001. doi: 10.1093/hmg/ddl241 [DOI] [PubMed] [Google Scholar]
- 17.Nguyen AD, Nguyen TA, Zhang J, Devireddy S, Zhou P, Xu X, et al. Murine knockin model for progranulin-deficient frontotemporal dementia with nonsense-mediated mRNA decay. Proc Natl Acad Sci U S A. 2018;115(12):E2849–E58. doi: 10.1073/pnas.1722344115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurosaki T, Popp MW, Maquat LE. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat Rev Mol Cell Biol. 2019;20(7):406–20. doi: 10.1038/s41580-019-0126-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin L, Grigoryan A, Wang D, Wang J, Breda L, Rivella S, et al. Identification and characterization of small molecules that inhibit nonsense-mediated RNA decay and suppress nonsense p53 mutations. Cancer Res. 2014;74(11):3104–13. doi: 10.1158/0008-5472.CAN-13-2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durand S, Cougot N, Mahuteau-Betzer F, Nguyen CH, Grierson DS, Bertrand E, et al. Inhibition of nonsense-mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P-bodies. J Cell Biol. 2007;178(7):1145–60. doi: 10.1083/jcb.200611086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z, Vuong JK, Zhang M, Stork C, Zheng S. Inhibition of nonsense-mediated RNA decay by ER stress. RNA. 2017;23(3):378–94. doi: 10.1261/rna.058040.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhuvanagiri M, Lewis J, Putzker K, Becker JP, Leicht S, Krijgsveld J, et al. 5-azacytidine inhibits nonsense-mediated decay in a MYC-dependent fashion. EMBO Mol Med. 2014;6(12):1593–609. doi: 10.15252/emmm.201404461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nomakuchi T, Rigo F, Aznarez I, Krainer A. Antisense oligonucleotide-directed inhibition of nonsense-mediated mRNA decay. Nat Biotechnol. 2016;34(2):164–6. doi: 10.1038/nbt.3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang L, Low A, Damle SS, Keenan MM, Kuntz S, Murray SF, et al. Antisense suppression of the nonsense mediated decay factor Upf3b as a potential treatment for diseases caused by nonsense mutations. Genome Biol. 2018;19(1):4. doi: 10.1186/s13059-017-1386-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan WK, Huang L, Gudikote JP, Chang YF, Imam JS, MacLean JA, 2nd, et al. An alternative branch of the nonsense-mediated decay pathway. EMBO J. 2007;26(7):1820–30. doi: 10.1038/sj.emboj.7601628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang L, Shum EY, Jones SH, Lou CH, Dumdie J, Kim H, et al. A Upf3b-mutant mouse model with behavioral and neurogenesis defects. Mol Psychiatry. 2018;23(8):1773–86. doi: 10.1038/mp.2017.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farr SA, Erickson MA, Niehoff ML, Banks WA, Morley JE. Central and peripheral administration of antisense oligonucleotide targeting amyloid-β protein precursor improves learning and memory and reduces neuroinflammatory cytokines in Tg2576 (AβPPswe) mice. J Alzheimers Dis. 2014;40(4):1005–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeVos SL, Miller RL, Schoch KM, Holmes BB, Kebodeaux CS, Wegener AJ, et al. Tau reduction prevents neuronal loss and reverses pathological tau deposition and seeding in mice with tauopathy. Sci Transl Med. 2017;9(374):eaag0481. doi: 10.1126/scitranslmed.aag0481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jafar-Nejad P, Powers B, Soriano A, Zhao H, Norris DA, Matson J, et al. The atlas of RNase H antisense oligonucleotide distribution and activity in the CNS of rodents and non-human primates following central administration. Nucleic Acids Res. 2021;49(2):657–73. doi: 10.1093/nar/gkaa1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong Q, Stump MR, Zhou Z. Inhibition of nonsense-mediated mRNA decay by antisense morpholino oligonucleotides restores functional expression of hERG nonsense and frameshift mutations in long-QT syndrome. J Mol Cell Cardiol. 2011;50(1):223–9. doi: 10.1016/j.yjmcc.2010.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim YJ, Nomakuchi T, Papaleonidopoulou F, Yang L, Zhang Q, Krainer AR. Gene-specific nonsense-mediated mRNA decay targeting for cystic fibrosis therapy. Nat Commun. 2022;13(1):2978. doi: 10.1038/s41467-022-30668-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hua Y, Sahashi K, Hung G, Rigo F, Passini MA, Bennett CF, et al. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24(15):1634–44. doi: 10.1101/gad.1941310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aggarwal G, Banerjee S, Jones SA, Benchaar Y, Bélanger J, Sévigny M, et al. Antisense oligonucleotides targeting the miR-29b binding site in the GRN mRNA increase progranulin translation. BioRxiv [Preprint]. September 30, 2022. Available from: www.biorxiv.org/content/10.1101/2022.01.12.476053v3.full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zetoune AB, Fontanière S, Magnin D, Anczuków O, Buisson M, Zhang CX, et al. Comparison of nonsense-mediated mRNA decay efficiency in various murine tissues. BMC Genet. 2008;9:83. doi: 10.1186/1471-2156-9-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rademakers R, Eriksen JL, Baker M, Robinson T, Ahmed Z, Lincoln SJ, et al. Common variation in the miR-659 binding-site of GRN is a major risk factor for TDP43-positive frontotemporal dementia. Hum Mol Genet. 2008;17(23):3631–42. doi: 10.1093/hmg/ddn257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiao J, Herl LD, Farese RV, Gao FB. MicroRNA-29b regulates the expression level of human progranulin, a secreted glycoprotein implicated in frontotemporal dementia. PLoS One. 2010;5(5):e10551. doi: 10.1371/journal.pone.0010551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piscopo P, Grasso M, Fontana F, Crestini A, Puopolo M, Del Vescovo V, et al. Reduced miR-659-3p levels correlate with progranulin increase in hypoxic conditions: implications for frontotemporal dementia. Front Mol Neurosci. 2016;9:31. doi: 10.3389/fnmol.2016.00031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen L, Li Q, Wang J, Jin S, Zheng H, Lin J, et al. MiR-29b-3p promotes chondrocyte apoptosis and facilitates the occurrence and development of osteoarthritis by targeting PGRN. J Cell Mol Med. 2017;21(12):3347–59. doi: 10.1111/jcmm.13237 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunoblot analysis of progranulin using conditioned medium from HeLa cells transfected with plasmids encoding GFP, wild-type (WT) progranulin, or R493X truncation mutant. mPGRN, mouse progranulin.
(PDF)
Cells were transfected with 100 nM ASO using Lipofectamine 2000. After 24 hours, RNA was isolated for qPCR. Grn mRNA levels are presented relative to levels in wild-type cells transfected with control ASO and presented as means ± SD; * indicates p<0.05, ** indicates p<0.01, *** indicates p<0.001, **** indicates p<0.0001, as determined by one-way ANOVA with Dunnett post hoc test.
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.