Abstract
Background
The COVID-19 pandemic has demonstrated the need for efficient and comprehensive, simultaneous assessment of multiple combined novel therapies for viral infection across the range of illness severity. Randomized Controlled Trials (RCT) are the gold standard by which efficacy of therapeutic agents is demonstrated. However, they rarely are designed to assess treatment combinations across all relevant subgroups. A big data approach to analyzing real-world impacts of therapies may confirm or supplement RCT evidence to further assess effectiveness of therapeutic options for rapidly evolving diseases such as COVID-19.
Methods
Gradient Boosted Decision Tree, Deep and Convolutional Neural Network classifiers were implemented and trained on the National COVID Cohort Collaborative (N3C) data repository to predict the patients’ outcome of death or discharge. Models leveraged the patients’ characteristics, the severity of COVID-19 at diagnosis, and the calculated proportion of days on different treatment combinations after diagnosis as features to predict the outcome. Then, the most accurate model is utilized by eXplainable Artificial Intelligence (XAI) algorithms to provide insights about the learned treatment combination impacts on the model’s final outcome prediction.
Results
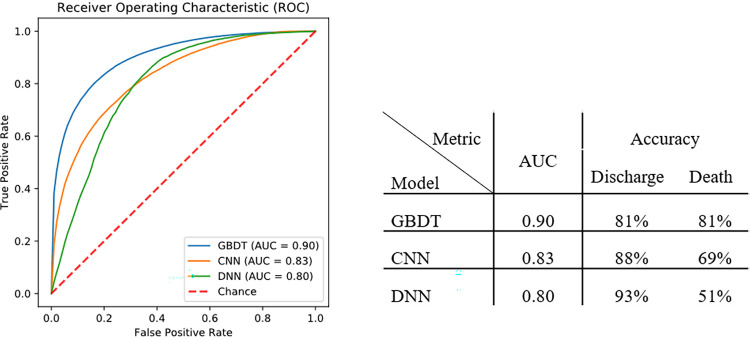
Gradient Boosted Decision Tree classifiers present the highest prediction accuracy in identifying patient outcomes with area under the receiver operator characteristic curve of 0.90 and accuracy of 0.81 for the outcomes of death or sufficient improvement to be discharged. The resulting model predicts the treatment combinations of anticoagulants and steroids are associated with the highest probability of improvement, followed by combined anticoagulants and targeted antivirals. In contrast, monotherapies of single drugs, including use of anticoagulants without steroid or antivirals are associated with poorer outcomes.
Conclusions
This machine learning model by accurately predicting the mortality provides insights about the treatment combinations associated with clinical improvement in COVID-19 patients. Analysis of the model’s components suggests benefit to treatment with combination of steroids, antivirals, and anticoagulant medication. The approach also provides a framework for simultaneously evaluating multiple real-world therapeutic combinations in future research studies.
Introduction
At the time of this writing, 8,029 completed or ongoing clinical trials for COVID-19 have been listed in ClinicalTrials.gov [1]. A majority of these trials are prospective randomized controlled trials (RCTs) or similarly designed clinical trials. These approaches offer the benefit of directly comparing therapeutic arms and control groups and can minimize bias. Notably, RCTs necessarily have inclusion and exclusion criteria that can limit the generalizability of the conclusions drawn from them. Further, RCTs, due to economic interests, required sample sizes, and the relative complexity of factorial designs, are rarely designed to explicitly address the optimal therapeutic combination(s) as a function of severity of illness.
Due to variable host-virus interactions, patients with SARS-CoV-2 infection may have a range of manifestations ranging from asymptomatic infection to critical illness [2]. Some studies of COVID-19 have described an initial viral stage of illness that can progress to a hyperinflammatory pulmonary stage, which can further evolve to a hypercoagulable phase or a late hyperinflammatory phase, as well as a chronic illness, referred to as the Post-Acute Sequelae of COVID-19 (PASC) or long-COVID [3]. Approaches to managing each of these phases are likely to require combinations of therapies directed at the respective underlying mechanisms and severity of illness. For instance, directly acting antiviral therapies would be anticipated to have the largest impact in the viral phase of the illness while anti-inflammatory treatments may be counterproductive as the body is mounting an antiviral immune response. By contrast, anti-inflammatory therapies would be expected to have the most beneficial effect in patients who have transitioned from the viral phase to a hyperinflammatory phase of illness. Antiviral agents may be less effective in this later phase. Especially in the early days of the pandemic, RCTs necessarily and largely evaluated individual therapeutic agents in critically ill patients, given the more favorable potential risk-benefit ratio. This approach may miss the impact of effective treatment combinations across the spectrum of COVID-19 illness severity.
Treatments have largely been studied individually in RCTs; for example, RCTs demonstrated that steroids benefit most patients requiring oxygen therapy [4], and the antiviral drug, Remdesivir, has been used successfully during the viral phase [5]. While one study recently reported benefit from the combination of Remdesivir and dexamethasone [6], data are lacking on the optimal combination of therapies for individual patients at various stages of the illness.
Review of patient outcomes from large, real-world data (RWD) sources offers the opportunity to assess the effect of therapies and their combinations not directly or adequately evaluated by RCTs, potentially augmenting our understanding of this increasingly complex therapeutic landscape [7]. Therefore, in this study, we explore the patient and treatment factors, particularly therapeutic agent combinations, associated with better outcomes using machine learning models (ML). The present study addresses key gaps in the extant literature by adopting ML models to evaluate the effect of therapeutic agents, singly and in combination, on patient outcomes using the large N3C cohort of patients.
Methods
Overall setting and study design
The National COVID Cohort Collaborative (N3C) is a high-granularity electronic health record (EHR) data repository containing harmonized, patient-level data from 72 sites across the United States (US). They are primarily tertiary care centers but also include data from health information exchanges and community hospitals. N3C data partners contribute data to N3C regularly. As of May 4, 2022 (Release 75), N3C contains data on more than 10 million patients, including more than 4.9 million COVID-19 SARS-CoV-2 infected persons.
N3C design, data ingestion and harmonization, and sampling approach have been detailed previously [8, 9]. In brief, N3C contributing sites provide the central repository EHR data, including demographics, healthcare visits, vital signs, medications, laboratory results, and diagnoses which are then harmonized into the Observational Medical Outcomes Partnership (OMOP) common data model. Participating sites submit EHR data on all patients with a positive SARS-CoV-2 lab test (Polymerase Chain Reaction, Antigen, or Antibody) or a COVID-19 diagnosis and a demographically matched comparison group of SARS-CoV-2 uninfected persons (1:2 matching positive: negative). For this study, we modify the N3C COVID-19 positivity definition [10] to exclude those with antibody-only positive results after December 10, 2020, the date when vaccinations became publicly available in the US [11].
To account for differences in data availability at the site level, we excluded sites with low medication reporting (<2 standard deviations below mean reporting for all sites). This approach excluded 17 of the 72 sites in N3C at the time of our data extraction.
Institutional Review Board (IRB) approval for this retrospective cohort study is obtained from the University of Mississippi Medical Center (IRB2020V0280, 3/31/2021), Johns Hopkins University (IRB00249128, 9/18/2020), Christiana Health (IRB604959, 5/07/2021), West Virginia University (IRB2012192778, 12/17/2020), University of Nebraska Medical Center (IRB050-21-EP, 2/9/2021), Nemour’s Children’s Health (IRB1700991, 2/17/2022), and Maine Medical Center (IRB1697848-2, 3/5/2021). Further approval by the N3C Data Access Committee (RP-504BA5) is granted that operates under the authority of the National Institute of Health IRB with Johns Hopkins University School of Medicine serving as the central IRB. A limited dataset was available for this project, however, zip codes were not used for the analyses described in the paper. No informed consent was obtained as the study utilizes a limited dataset.
Cohort identification
For the purpose of this study, we selected COVID-positive patients with at least one day of hospitalization during the 28 days after their initial COVID-19 diagnosis. The cohort under study includes patients in the United States who tested positive for COVID-19 and were hospitalized between January 1, 2020, and July 1, 2021.
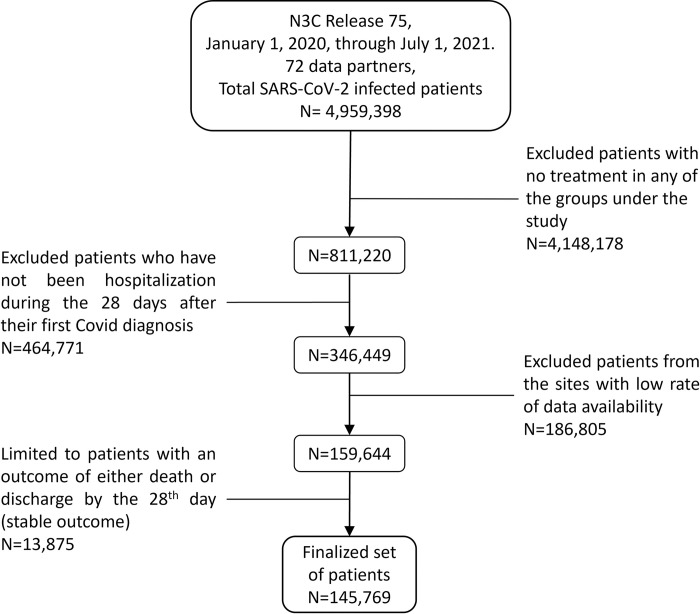
Selection is then further limited to patients with an outcome of either death or discharge by the 28th day (n = 145,769) after COVID-19 diagnosis. Patients with any other outcome at the end of the 28-day period are not considered as they are still being treated, and our interest is limited to those who have completed treatments [12–14]. This selected cohort is hereafter referred to as patients with a stable outcome, as treatment duration is completed and the final outcome of either death or discharge has been achieved. Fig 1 presents the information flow diagram for the final cohort under the study.
Fig 1. Information flow diagram for the cohort under the study.
Data extraction
Data were extracted on May 4, 2022 (N3C release 75) for the previously defined cohort with a stable outcome before July 1, 2021. The lag between the observation window cutoff and data extraction ensured that data from reporting sites was as complete as possible and placed the observation window before the rapid rise of the Delta variant. We developed concept sets for all conditions, drugs, and procedures used in this study, which include OMOP concept identifiers (derived from SNOMED CT, RxNorm, and other standardized vocabularies) contained with a patient’s EHR. Concept sets in use, available in Table 1, define computable phenotypes to programmatically identify patient health status at a point-in-time. All concept sets in use received review by three clinicians and one informatician during curation and implementation.
Table 1. Medications in each treatment category.
| Class | Medication | Concept Sets |
|---|---|---|
| Anticoagulants (Coag) | Apixaban | 259221776 |
| Betrixaban | 568693141 | |
| Dabigatran | 23600781 | |
| Enoxaparin | 858278110 | |
| Heparin | 357794478 | |
| Rivaroxaban | 544420473 | |
| Warfarin | 441951686 | |
| Targeted Antivirals (ViralTrgt) | Remdesivir | 719693192 |
| Nirmatrelvir/ritonavir (Paxlovid) | 285332632 | |
| Molnupiravir | 643666235 | |
| Macrolide and Quinolone Antibiotics (BiotMQ) | Azithromycin | 359938251 |
| Doxycycline | 950251876 | |
| Ciprofloxacin | 369973585 | |
| Moxifloxacin | 609610642 | |
| Gemifloxacin | 382925247 | |
| Delafloxacin | 103404439 | |
| Gatifloxacin | 932126058 | |
| Ofloxacin | 931604126 | |
| Norfloxacin | 292248378 | |
| Erythromycin | 4697796 | |
| Clarithromycin | 4697796 | |
| Levofloxacin | 4697796 | |
| Spike Protein Monoclonal Antibodies (MonoSP) | Bamlanivimab | 804283782 |
| Casirivimab/Imdevimab | 204936358 | |
| Etesevimab | 985547691 | |
| Sotrovimab | 550646109 | |
| Tixagevimab/Cilgavimab | 809722294 | |
| Bamlanivimab-Etesevimab combo | String search | |
| Bebtelovimab | String search | |
| Steroids Preparations (Ster) | Dexamethasone | 213873961 |
| Hydrocortisone | 932266800, 422007021 | |
| Methylprednisolone | 640520004 | |
| Prednisone | 783588396 | |
| Monoclonal Antibody Immunomodulators (MonoI) | Tocilizumab | 276204116 |
| Baricitinib | 394764748 | |
| Tofacitinib | 391595378 | |
| Sarilumab | 807728943 | |
| Unproven Antiviral Therapies (ViralUnp) | Hydroxychloroquine | 807281242 |
| Chloroquine | 818210864 | |
| Ivermectin | 980395214 | |
| Lopinavir/ritonavir | 165611849 | |
| Tenofovir | 563211602, 568417090 | |
| Interferon | 359012050, 531467540 | |
| Miscellaneous (Misc) | Vitamin D | 689338842 |
| Fluvoxamine | 424477820 |
Feature engineering
For the identified cohort, we have considered demographics, body mass index (BMI), comorbidities [15], treatment with pressors, the quarter of COVID-19 diagnosis, patient severity at the time of diagnosis, and prescribed treatments as input features for model development.
To measure patient severity, we used an Ordinal Scale (OS) developed for use with EHR data [16]. Specifically, this was a 6-point ordinal scale assigned with odd integers from 1 to 11, devised explicitly for patients diagnosed with COVID-19 based on discrete EHR data elements. In this context, a level of 1 represents an outpatient or patient discharged from the hospital, level 3 indicates hospitalization, while being hospitalized on Oxygen or Mechanical Ventilator is an indicator of levels 5 and 7, respectively, with level 9 representing patients hospitalized on ECMO and level 11 representing death.
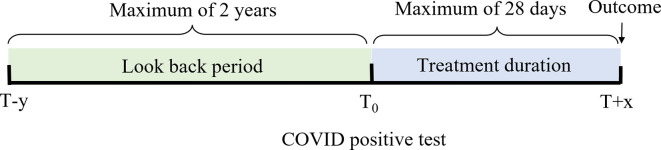
Fig 2 shows the lookback period used for determining the patient’s comorbidities in green with a minimum of 2 years, while highlighted in blue are the considered treatments’ duration within up to 28 days after the diagnosis, followed by the recorded patient’s outcome as of the last day of treatment.
Fig 2. Time windows for treatment, comorbidities, and outcome.
Prescribed therapeutics on each day after the diagnosis were categorized and considered in eight distinct groups, defined as anticoagulants (Coag), steroid preparations (Ster), unproven antiviral therapies (ViralUnp), targeted antivirals (ViralTrgt), spike protein monoclonal antibodies (MonoSP), monoclonal antibody Immunomodulators (MonoI), macrolide and quinolone antibiotics (BiotMQ), and a miscellaneous treatments (Misc) category that included other treatments presumed to be administered for treatment of COVID-19. Medications in each category are shown in Table 1.
The model considered the proportion of days on treatment combinations, any direct correlations between the treatment values and duration of treatment are removed, preventing the ML algorithm from leveraging this information directly for prediction. By using the proportion of days on treatment combinations, the modeling algorithm is forced to find the effect of different treatment distributions rather than attributing days on treatments to the outcome of interest.
Modeling
We implemented three models to predict the final patient outcomes at the end of the 28-day observation window. The first was a Gradient Boosted Decision Tree (GBDT) classifier based on an additive model that tunes a weak learner into a strong one by training on residuals in boosting rounds; GBDT combines the results of previous learners along the way, thus learning from the errors of previous iterations to improve accuracy [17]. Two Neural Network models were also implemented, the first was based on a Deep fully-connected Neural Network (DNN) with a self-attention mechanism to increase the attention of the model to key features. The second was a multi-layer Convolutional Neural Network (CNN), convolving over the features to provide levels of generalization and extract treatment patterns and their effects. For the CNN model, multiple convolution structures based on VGG-16 [18], Inception [19], and DenseNet [20] blocks were evaluated and results for the best model is reported.
For the ML models, the input features considered as predictors of outcome are demographics, BMI, quarter of diagnosis, comorbidities, the severity of the patient at the time of diagnosis, being treated with pressors, and prescribed treatment combinations after diagnosis. Due to the sensitivity of ML models to hyper-parameters and to make the study repeatable, we used HyperOpt [21], an open-source Bayesian optimization library, to increase the model’s Area Under receiver operating Characteristic (AUC) curve by fine-tuning the parameters. Hyper-parameter tuning is performed on stratified random train, validation, and test splits of 60%, 20%, and 20% respectively, with random over-sampling of the training dataset using the SMOTE [22] library to address the data imbalance in the training set. Then, given the discovered hyper-parameters, model evaluation is conducted by 5-fold cross-validation to report the models’ AUC and accuracy.
Model interpretability
Generally, machine learning models are considered black-box procedures, with limited insights and interpretability other than outcome prediction. However, recent years have seen many improvements in the ability to generate robust and interpretable insights from complex ML models [23]. Use of SHapley Additive exPlanation (SHAP) [24] values as an eXplainable Artificial Intelligence (XAI) algorithm can provide insightful interpretations of a complex machine learning model with high accuracy and robustness, similar to human interpretations.
The generated SHAP values for input features of ML models can be used to characterize the effect of the inputs on the final model’s prediction. In this study, to communicate the effects of treatment combinations as features of patients’ outcomes, we first trained an accurate ML model on the patients’ data. Then, the trained model is utilized for generating the SHAP values of input features, providing insights into the features’ importance on the probability of a patient discharge prediction. For the analysis, a feature has a positive impact if the feature increases the probability of the discharge prediction, while the negative impact of a feature translates to a decrease in the probability of discharge prediction.
After model hyper-parameter optimization, training, and evaluation, the model was retrained on the entire dataset, using the same parameters, to learn all existing interactions within the dataset. Then for SHAP value calculations, two required inputs are generated, background samples as a base of comparisons and input samples for evaluation of the effects. Following SHAP’s best practices for calculating the required background samples in large datasets, we applied the K-Nearest Neighbor clustering algorithm (K = 50) to patients in each class of outcome (death and discharge), providing us with a total of 100 cluster centroids to be used for the SHAP analysis.
For input samples, we noticed, however, that in a highly imbalanced dataset, averaging the SHAP values for each feature to provide a holistic view of the effect can be biased by the class containing the larger sample size (which in this case was discharge), diminishing the impact of learned interactions within the smaller set (defined by death as the outcome). To overcome any unwanted effects that data imbalance may pose on the results, we used 1:1 matched sets of patients, matching on the demographics (age, sex, race, ethnicity), BMI, comorbidities (specified in Table 2, under comorbidities), quarter of the year, pressor status (presence or absence), and OS level at diagnosis as inputs for SHAP calculation, resulting in a more balanced set of patients, preserving the effects and discriminating factors learned from the smaller set.
Table 2. Patients’ characteristics.
| Characteristics | n = 145,769 |
|---|---|
| Gender (%) | |
| Female | 71,891 (49.3) |
| Male | 73,855 (50.7) |
| Other/ Unknown | <25 (0.0) |
| Age (mean (SD)) | 59.2 y (19.5) |
| Race (%) | |
| Asian | 5,305 (3.6) |
| Black | 32,467 (22.3) |
| Native Haw./Pac. Islander | 318 (0.2) |
| White | 75,658 (51.9) |
| Other/ Unknown | 30,883 (21.2) |
| Ethnicity (%) | |
| Hispanic/Latino | 29,419 (20.2) |
| Not Hispanic/Latino | 104,200 (71.5) |
| Other/ Unknown | 12,068 (8.3) |
| OS at day 1 (%) | |
| OS 1—outpatient | 35,328 (24.2) |
| OS 3—hospitalized | 94,601 (64.9) |
| OS 5—hospitalized on Oxygen | 9,326 (6.4) |
| OS 7—hospitalized on Mechanical Ventilator | 6,252 (4.3) |
| OS 9—hospitalized on ECMO | 262 (0.2) |
| OS 11—death | 0 (0) |
| BMI (mean (SD)) | 31.0 (7.1) |
| Comorbidities (%) | |
| Hypertension | 92,767 (63.6) |
| Diabetes Mellitus | 33,808 (23.2) |
| Myocardial Infarction | 19,197 (13.2) |
| Congestive Heart Failure | 31,629 (21.7) |
| Peripheral Vascular Disease | 24,011 (16.5) |
| Stroke | 24,168 (16.6) |
| Dementia | 13,037 (8.9) |
| Chronic Pulmonary Disease | 40,545 (27.8) |
| Rheumatologic Disease | 8,998 (6.2) |
| Mild Liver Disease | 15,014 (10.3) |
| Severe Liver Disease | 4,972 (3.4) |
| Upper GI bleed | 4,791 (3.3) |
| Renal Disease | 33,848 (23.2) |
| Peptic Ulcer Disease | 4,496 (3.1) |
| Paralysis | 5,116 (3.5) |
| Cancer | 14,397 (9.9) |
| Diabetes with chronic complications |
27,186 (18.7) |
| Metastatic solid tumor | 5,376 (3.7) |
| HIV/AIDS | 1,527 (1.0) |
| Quarter of Diagnosis (%) | |
| Jan-Mar 2020 | 6,956 (4.8) |
| Apr-Jun 2020 | 28,837 (19.8) |
| Jul-Sep 2020 | 17,111 (11.7) |
| Oct-Dec 2020 | 44,210 (30.3) |
| Jan-Mar 2021 | 33,468 (23.0) |
| Apr-Jun 2021 | 15,173 (10.4) |
| Outcomes (%, IQR) | |
| Discharged | 128,063 (87.9, 8) |
| Death | 17,706 (12.1, 13) |
Results
Study population
The dataset included 145,769 hospitalized patients (Table 2). Most patients (128,063; 87.9%) were discharged alive from the hospital within 28 days of COVID-19 diagnosis while the remaining 17,706 (12.1%) were deceased. Although 24.2% of patients were not hospitalized on day 1 of their diagnosis (OS level 1), they subsequently were hospitalized after day 1 as this study assessed only hospitalized patients.
Prescribed treatment combinations
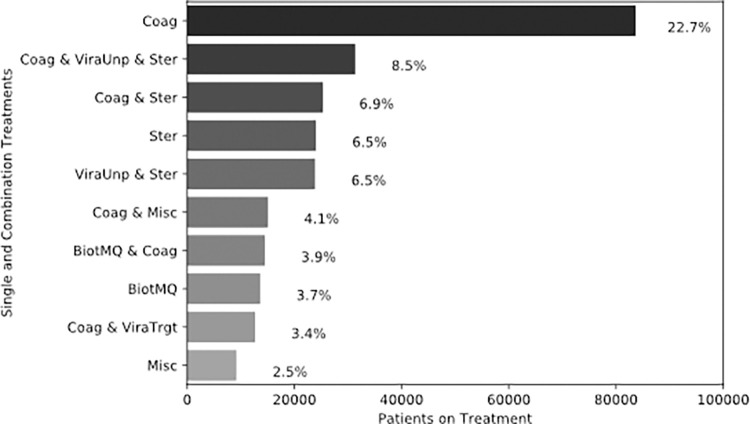
Among single agent treatments, anticoagulants (Coag), steroids (Ster), and macrolide and quinolone antibiotics (BiotMQ) are the top three most commonly prescribed to patients; 22.7% (n = 83,665), 6.5% (n = 24,026), and 3.7% (n = 13,625), respectively (Fig 3). The three most frequent treatment combinations prescribed were: 1) anticoagulants and steroids with unproven antivirals (ViraUnp) 2) anticoagulants and steroids, and 3) steroids with unproven antivirals (ViraUnp) (Fig 3).
Fig 3. Top 10 prescribed treatments to patients.
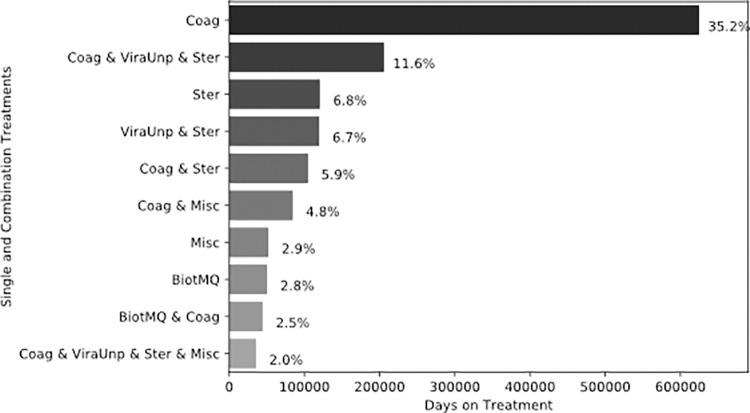
The top two prescribed treatments were also the treatments that patients received for the greatest number of days with 35.2% for anticoagulants and 11.6% for the combination therapy of anticoagulants with unproven antivirals (ViraUnp) and steroids. While steroids alone were the third most used therapeutic agent, patients spent roughly the same days on steroids in combination with anticoagulants (5.9%) and on steroid single therapy alone (6.8%). Fig 4 presents the top 10 therapeutics based on the cumulative number of days they were prescribed to patients.
Fig 4. Top 10 treatments based on the number of days prescribed.
Model accuracy
Developing an ML model to leverage the aforementioned curated data and provide an accurate prediction, can be used not only as a predictive measure for taking therapeutic actions, but also as a means to evaluate the effect of patients’ characteristics and prescribed treatment combinations on the final patient outcomes. The devised models have been trained and evaluated using 5-fold cross-validation. Fig 5 shows the Receiver Operating Characteristic (ROC) curve, and accuracy of the models. Our results indicate that the Gradient Boosted Decision Tree (GBDT) classifier has superior (AUC = 0.90) and balanced accuracy (81% for both death and discharge classes) in identifying and discriminating patient outcomes compared to both Deep Neural Network (DNN) and Convolutional Neural Network (CNN) models.
Fig 5. ML models performance in predicting patient outcomes.
Feature importance
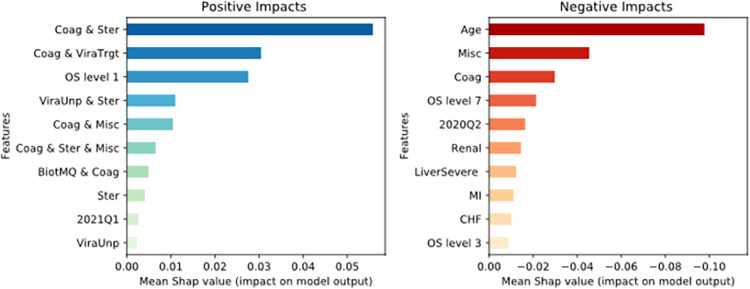
Given the accuracy and discriminative ability of the GBDT model, SHAP values were calculated to evaluate the impact of a feature on the model’s prediction. Specifically, positive SHAP values in this context indicate a positive impact on the predicted probability of classifying a patient as discharged while negative SHAP values indicate impact on the predicted probability of death. Fig 6 presents the top 10 features with the highest positive and negative impacts on the model’s predictive ability. It shows that six treatment combinations are among the top ten features with the highest positive impact underlining the importance of combination therapies; the steroid and anticoagulant combination provides the highest positive effect on model prediction. Monotherapies of both steroids and unproven antiviral therapies (ViralUnp) are ranked eighth and tenth after combination therapies. The other two features with high positive impact are COVID diagnosis in the first quarter of 2021 and OS severity level 1 (outpatient status) at the time of diagnosis. Among the features with the most negative impact, age is associated with the strongest negative impact on the model’s classification, followed by two of the single therapies: miscellaneous (Misc) and anticoagulants (Coag) alone. Among the comorbidities, renal disease (Renal), severe liver disease (LiverSevere), Myocardial Infarction (MI), and Congestive Heart Failure (CHF) are most highly associated with negative effects, in decreasing order of importance. In addition, OS levels 7 and 3 at the time of diagnosis are each associated with the negative outcome (death).
Fig 6. Top 10 features with the highest impact on final model prediction.
Discussion
We developed accurate machine learning models with high accuracy for predicting death and discharge outcomes from COVID-19. By examing factors contributing to these predictions we can better understand the impact of treatment combinations on outcomes. Specifically, our findings suggest that combination therapy with different classes of drugs is more effective than therapy with only a single agent. These models also demonstrate that patient characteristics and comorbidities such as age, kidney, liver, heart disease, and severity of illness at diagnosis have a large impact on disease outcome, confirming previous literature [25–29]. Indeed, the models suggest that for COVID-19 outcomes, patient characteristics are not surprisingly often as influential as the treatments administered. Of note, pre-existing renal, liver, and heart diseases were strongly associated with poor prognosis. However, several combinations of treatments appear to be associated with better or worse outcomes. Specifically, our models support the efficacy of steroids, antiviral drugs, and anticoagulation while raising the possibility of harm from miscellaneous category therapies of vitamin D and fluvoxamine. Data from clinical trials of fluvoxamine in COVID-19 is mixed; however, our findings support guidelines recommending against its routine use at this time [30, 31]. Similarly, our negative findings regarding vitamin D are consistent with a clinical trial showing lack of efficacy of vitamin D in reducing the length of stay in hospitalized COVID-19 patients [32]. Steroids have a well established therapeutic benefit in COVID-19 patients requiring oxygen but have not been shown to benefit patients not requiring oxygen [33]. With electronic health record data, our models also observe the association between steroid treatment and higher likelihood of recovery among COVID-19 patients requiring oxygen therapy.
COVID-19 is associated with micro and macrovascular thrombosis [34–36], and COVID-19 patients have high risk of thrombotic complications such as pulmonary embolism. Therefore, various doses of anticoagulation have been proposed as part of standard COVID-19 treatment. Intermediate dose anticoagulation in ICU patients failed to show benefit [37]. However, among hospitalized patients not requiring ICU care, full dose anticoagulation has been reported to have benefits [38, 39]. If and when to use higher dose anticoagulation remains controversial [40, 41]. Our models suggest the possibility of benefit from the addition of anticoagulation to steroids. The combination of two potentially beneficial therapies, steroids and anticoagulants, being associated with an increased likelihood of recovery may reinforce the need to consider therapeutic combinations when attempting to define the optimal treatment of COVID-19.
The association with poor outcome of use of anticoagulants alone without steroids or antivirals is intriguing. Perhaps the use of anticoagulation alone is a marker for patients who were not treated aggressively for COVID-19 or who had comorbidities such as poorly controlled diabetes which might cause clinicians to withhold steroids or renal/hepatic failure that might give clinicians pause regarding the use of Remdesivir. However, this hypothesis cannot be tested in our dataset.
Many experts have suggested that combining steroids with antivirals may be beneficial because of the potential immunosuppressive effect of steroids [42, 43]. We expected to see a benefit of the combination of steroids and antivirals with efficacy against COVID-19. However, the overall positive effect of steroids combined with antivirals of unproven efficacy was surprising. It may be that the decision to use the combination of steroids/antiviral drug before proven antiviral drugs were available may have been a marker of other aspects of care (for example, excellent supportive care such as proning) that may have been associated with better outcomes. Since many patients in the dataset were treated before the availability of proven antivirals with efficacy against COVID, this may have led to an association between use of unproven (and likely ineffective) antivirals and reduced mortality.
Due to anti-inflammatory effects and proposed antiviral effects, macrolides were applied as possibly effective treatments for COVID-19 early in the pandemic [44]. Similarly, fluoroquinolone antibiotics were also suggested as COVID-19 treatments [45]. This was the rationale for including the antibiotics in our analysis. As enthusiasm for use of these medications for specific treatment of COVID-19 per se has declined, the positive associations found by our machine learning algorithms are perhaps unexpected. Severely ill COVID-19 patients are known to be at high risk of secondary infections [46]. It is possible that macrolides and quinolones treated secondary infections or prevented the development of such infections. Alternatively, the association between antibiotics treatment and improved outcome may be confounded by serving as a marker of more aggressive treatment. Further study of the mechanisms responsible for this association are needed.
While this study demonstrates a generally applicable machine learning model (ML) approach to explore treatment factors, particularly therapeutic agent combinations for COVID-19, ML models have been successfully applied to other aspects of the COVID-19 pandemic. More specifically for COVID-19, ML models have been developed and validated to predict the outcomes of COVID-19 patients using metrics collected at the time of admission [47]. Another study using ML evaluated risk factors associated with increased mortality for COVID-19 patients [48]. ML has also been used to show the predictive effect of comorbidities and risk factors on progression of illness in COVID-19 patients [49, 50]. ML models generally demonstrate improved prediction of patient outcomes when compared to conventional statistical approaches [51–53].
Our study has several limitations. First, information on the doses of medications used is not available in the dataset. Similarly, the impact of steroid dose is unknown. However, the results of our study support the need for clinical trials to explore the efficacy of different doses of therapeutic combinations and single therapies. An additional limitation is that we have no knowledge as to why clinicians choose to administer or not administer certain therapeutic agents. Patients with treatment limitations, such as DNR orders, are more likely to die than those without such limitations [54]. It is possible that such care limitations or contraindications, especially early in the pandemic, influenced the decision to use or not use certain treatments. It is also possible that some patients were incidentally positive for COVID-19 but hospitalized for other serious illnesses, although this cannot be determined from the database. Another limitation of our study is the lack of full control over the diagnosis criteria that treating clinicians used and the possibility of false negatives or false positives, however, we followed the best practices provided by the NIH experts to define inclusion criteria for COVID-19 positivity.
Conclusions
Machine learning algorithms can predict mortality in hospitalized COVID-19 patients with a high degree of accuracy. Future work may allow use of such algorithms to identify high risk patients needing more aggressive therapies. In the meantime, our analyses of a large multicenter cohort of COVID-19 patients using machine learning algorithms supports use of steroids, anti-virals, and anticoagulant medications in combination. Further study is needed on the associations of macrolide and fluoroquinolone antibiotics with survival in COVID-19. In addition to the beneficial observed effects of specific treatments and, in particular, their combinations, patient characteristics such as age and comorbidities are strong predictors of increased likelihood of death as expected, perhaps serving as negative controls suggesting validity of the models. More generally, this study demonstrates use of a machine learning model (ML) approach to explore treatment factors, particularly therapeutic agent combinations, associated with outcomes across comorbidity profiles and initial severity of illness. It potentially provides useful evidence, particularly with regard to therapeutic combinations, to supplement evidence from RCTs.
Supporting information
(DOCX)
(TIF)
(TIF)
Acknowledgments
National COVID Cohort Collaborative (N3C) Consortium membership includes:
Christopher G. Chute, DrPH–Schools of Medicine, Public Health, and Nursing, Johns Hopkins University, Baltimore, MD
Melissa A. Haendel, PhD–Center for Health AI, University of Colorado Anschutz Medical Campus, Aurora, CO
Richard Moffitt, PhD–Department of Biomedical Informatics, Stony Brook University, Stony Brook, NY
N3C Attribution
The analyses described in this publication were conducted with data or tools accessed through the NCATS N3C Data Enclave covid.cd2h.org/enclave and supported by NCATS U24 TR002306. This research was possible because of the patients whose information is included within the data from participating organizations (covid.cd2h.org/dtas) and the organizations and scientists (covid.cd2h.org/duas) who have contributed to the on-going development of this community resource.
Individual Acknowledgments for Core Contributors
We gratefully acknowledge contributions from the following N3C core teams:
(Asterisks indicate leads)
Principal Investigators: Melissa A. Haendel*, Christopher G. Chute*, Kenneth R. Gersing, Anita Walden
Workstream, subgroup and administrative leaders: Melissa A. Haendel*, Tellen D. Bennett, Christopher G. Chute, David A. Eichmann, Justin Guinney, Warren A. Kibbe, Hongfang Liu, Philip R.O. Payne, Emily R. Pfaff, Peter N. Robinson, Joel H. Saltz, Heidi Spratt, Justin Starren, Christine Suver, Adam B. Wilcox, Andrew E. Williams, Chunlei Wu
Key liaisons at data partner sites
Regulatory staff at data partner sites
Individuals at the sites who are responsible for creating the datasets and submitting data to N3C
Data Ingest and Harmonization Team: Christopher G. Chute*, Emily R. Pfaff*, Davera Gabriel, Stephanie S. Hong, Kristin Kostka, Harold P. Lehmann, Richard A. Moffitt, Michele Morris, Matvey B. Palchuk, Xiaohan Tanner Zhang, Richard L. Zhu
Phenotype Team (Individuals who create the scripts that the sites use to submit their data, based on the COVID and Long COVID definitions): Emily R. Pfaff*, Benjamin Amor, Mark M. Bissell, Marshall Clark, Andrew T. Girvin, Stephanie S. Hong, Kristin Kostka, Adam M. Lee, Robert T. Miller, Michele Morris, Matvey B. Palchuk, Kellie M. Walters
Project Management and Operations Team: Anita Walden*, Yooree Chae, Connor Cook, Alexandra Dest, Thomas Dillon, Patricia A. Francis, Rafael Fuentes, Alexis Graves, Julie A. McMurry, Andrew J. Neumann, Shawn T. O’Neil, Usman Sheikh, Elizabeth Zampino
Partners from NIH and other federal agencies: Christopher P. Austin*, Kenneth R. Gersing*, Samuel Bozzette, Mariam Deacy, Nicole Garbarini, Michael G. Kurilla, Sam G. Michael, Joni L. Rutter, Meredith Temple-O’Connor
Analytics Team (Individuals who build the Enclave infrastructure, help create codesets, variables, and help Domain Teams and project teams with their datasets): Benjamin Amor*, Mark M. Bissell, Katie Rebecca Bradwell, Andrew T. Girvin, Amin Manna, Nabeel Qureshi
Publication Committee Management Team: Mary Morrison Saltz*, Christine Suver*, Christopher G. Chute, Melissa A. Haendel, Julie A. McMurry, Anita Walden
Publication Committee Review Team: Carolyn Bramante, Jeremy Richard Harper, Wenndy Hernandez, Farrukh M Koraishy, Federico Mariona, Amit Saha, Satyanarayana Vedula
Data Partners with Released Data
Advocate Health Care Network—UL1TR002389: The Institute for Translational Medicine (ITM) • Boston University Medical Campus—UL1TR001430: Boston University Clinical and Translational Science Institute • Brown University—U54GM115677: Advance Clinical Translational Research (Advance-CTR) • Carilion Clinic—UL1TR003015: iTHRIV Integrated Translational health Research Institute of Virginia • Charleston Area Medical Center—U54GM104942: West Virginia Clinical and Translational Science Institute (WVCTSI) • Children’s Hospital Colorado—UL1TR002535: Colorado Clinical and Translational Sciences Institute • Columbia University Irving Medical Center—UL1TR001873: Irving Institute for Clinical and Translational Research • Duke University—UL1TR002553: Duke Clinical and Translational Science Institute • George Washington Children’s Research Institute—UL1TR001876: Clinical and Translational Science Institute at Children’s National (CTSA-CN) • George Washington University—UL1TR001876: Clinical and Translational Science Institute at Children’s National (CTSA-CN) • Indiana University School of Medicine—UL1TR002529: Indiana Clinical and Translational Science Institute • Johns Hopkins University—UL1TR003098: Johns Hopkins Institute for Clinical and Translational Research • Loyola Medicine—Loyola University Medical Center • Loyola University Medical Center—UL1TR002389: The Institute for Translational Medicine (ITM) • Maine Medical Center—U54GM115516: Northern New England Clinical & Translational Research (NNE-CTR) Network • Massachusetts General Brigham—UL1TR002541: Harvard Catalyst • Mayo Clinic Rochester—UL1TR002377: Mayo Clinic Center for Clinical and Translational Science (CCaTS) • Medical University of South Carolina—UL1TR001450: South Carolina Clinical & Translational Research Institute (SCTR) • Montefiore Medical Center—UL1TR002556: Institute for Clinical and Translational Research at Einstein and Montefiore • Nemours—U54GM104941: Delaware CTR ACCEL Program • NorthShore University HealthSystem—UL1TR002389: The Institute for Translational Medicine (ITM) • Northwestern University at Chicago—UL1TR001422: Northwestern University Clinical and Translational Science Institute (NUCATS) • OCHIN—INV-018455: Bill and Melinda Gates Foundation grant to Sage Bionetworks • Oregon Health & Science University—UL1TR002369: Oregon Clinical and Translational Research Institute • Penn State Health Milton S. Hershey Medical Center—UL1TR002014: Penn State Clinical and Translational Science Institute • Rush University Medical Center—UL1TR002389: The Institute for Translational Medicine (ITM) • Rutgers, The State University of New Jersey—UL1TR003017: New Jersey Alliance for Clinical and Translational Science • Stony Brook University—U24TR002306 • The Ohio State University—UL1TR002733: Center for Clinical and Translational Science • The State University of New York at Buffalo—UL1TR001412: Clinical and Translational Science Institute • The University of Chicago—UL1TR002389: The Institute for Translational Medicine (ITM) • The University of Iowa—UL1TR002537: Institute for Clinical and Translational Science • The University of Miami Leonard M. Miller School of Medicine—UL1TR002736: University of Miami Clinical and Translational Science Institute • The University of Michigan at Ann Arbor—UL1TR002240: Michigan Institute for Clinical and Health Research • The University of Texas Health Science Center at Houston—UL1TR003167: Center for Clinical and Translational Sciences (CCTS) • The University of Texas Medical Branch at Galveston—UL1TR001439: The Institute for Translational Sciences • The University of Utah—UL1TR002538: Uhealth Center for Clinical and Translational Science • Tufts Medical Center—UL1TR002544: Tufts Clinical and Translational Science Institute • Tulane University—UL1TR003096: Center for Clinical and Translational Science • University Medical Center New Orleans—U54GM104940: Louisiana Clinical and Translational Science (LA CaTS) Center • University of Alabama at Birmingham—UL1TR003096: Center for Clinical and Translational Science • University of Arkansas for Medical Sciences—UL1TR003107: UAMS Translational Research Institute • University of Cincinnati—UL1TR001425: Center for Clinical and Translational Science and Training • University of Colorado Denver, Anschutz Medical Campus—UL1TR002535: Colorado Clinical and Translational Sciences Institute • University of Illinois at Chicago—UL1TR002003: UIC Center for Clinical and Translational Science • University of Kansas Medical Center—UL1TR002366: Frontiers: University of Kansas Clinical and Translational Science Institute • University of Kentucky—UL1TR001998: UK Center for Clinical and Translational Science • University of Massachusetts Medical School Worcester—UL1TR001453: The UMass Center for Clinical and Translational Science (UMCCTS) • University of Minnesota—UL1TR002494: Clinical and Translational Science Institute • University of Mississippi Medical Center—U54GM115428: Mississippi Center for Clinical and Translational Research (CCTR) • University of Nebraska Medical Center—U54GM115458: Great Plains IDeA-Clinical & Translational Research • University of North Carolina at Chapel Hill—UL1TR002489: North Carolina Translational and Clinical Science Institute • University of Oklahoma Health Sciences Center—U54GM104938: Oklahoma Clinical and Translational Science Institute (OCTSI) • University of Rochester—UL1TR002001: UR Clinical & Translational Science Institute • University of Southern California—UL1TR001855: The Southern California Clinical and Translational Science Institute (SC CTSI) • University of Vermont—U54GM115516: Northern New England Clinical & Translational Research (NNE-CTR) Network • University of Virginia—UL1TR003015: iTHRIV Integrated Translational health Research Institute of Virginia • University of Washington—UL1TR002319: Institute of Translational Health Sciences • University of Wisconsin-Madison—UL1TR002373: UW Institute for Clinical and Translational Research • Vanderbilt University Medical Center—UL1TR002243: Vanderbilt Institute for Clinical and Translational Research • Virginia Commonwealth University—UL1TR002649: C. Kenneth and Dianne Wright Center for Clinical and Translational Research • Wake Forest University Health Sciences—UL1TR001420: Wake Forest Clinical and Translational Science Institute • Washington University in St. Louis—UL1TR002345: Institute of Clinical and Translational Sciences • Weill Medical College of Cornell University—UL1TR002384: Weill Cornell Medicine Clinical and Translational Science Center • West Virginia University—U54GM104942: West Virginia Clinical and Translational Science Institute (WVCTSI)
Additional Data Partners Who Have Signed a DTA and Whose Data Submitted
Icahn School of Medicine at Mount Sinai—UL1TR001433: ConduITS Institute for Translational Sciences • The University of Texas Health Science Center at Tyler—UL1TR003167: Center for Clinical and Translational Sciences (CCTS) • University of California, Davis—UL1TR001860: UCDavis Health Clinical and Translational Science Center • University of California, Irvine—UL1TR001414: The UC Irvine Institute for Clinical and Translational Science (ICTS) • University of California, Los Angeles—UL1TR001881: UCLA Clinical Translational Science Institute • University of California, San Diego—UL1TR001442: Altman Clinical and Translational Research Institute • University of California, San Francisco—UL1TR001872: UCSF Clinical and Translational Science Institute
Additional Data Partners Who Have Signed a DTA and Whose Data Release is Pending
Arkansas Children’s Hospital—UL1TR003107: UAMS Translational Research Institute • Baylor College of Medicine—None (Voluntary) • Children’s Hospital of Philadelphia—UL1TR001878: Institute for Translational Medicine and Therapeutics • Cincinnati Children’s Hospital Medical Center—UL1TR001425: Center for Clinical and Translational Science and Training • Emory University—UL1TR002378: Georgia Clinical and Translational Science Alliance • HonorHealth—None (Voluntary) • Loyola University Chicago—UL1TR002389: The Institute for Translational Medicine (ITM) • Medical College of Wisconsin—UL1TR001436: Clinical and Translational Science Institute of Southeast Wisconsin • MedStar Health Research Institute—UL1TR001409: The Georgetown-Howard Universities Center for Clinical and Translational Science (GHUCCTS) • MetroHealth—None (Voluntary) • Montana State University—U54GM115371: American Indian/Alaska Native CTR • NYU Langone Medical Center—UL1TR001445: Langone Health’s Clinical and Translational Science Institute • Ochsner Medical Center—U54GM104940: Louisiana Clinical and Translational Science (LA CaTS) Center • Regenstrief Institute—UL1TR002529: Indiana Clinical and Translational Science Institute • Sanford Research—None (Voluntary) • Stanford University—UL1TR003142: Spectrum: The Stanford Center for Clinical and Translational Research and Education • The Rockefeller University—UL1TR001866: Center for Clinical and Translational Science • The Scripps Research Institute—UL1TR002550: Scripps Research Translational Institute • University of Florida—UL1TR001427: UF Clinical and Translational Science Institute • University of New Mexico Health Sciences Center—UL1TR001449: University of New Mexico Clinical and Translational Science Center • University of Texas Health Science Center at San Antonio—UL1TR002645: Institute for Integration of Medicine and Science • Yale New Haven Hospital—UL1TR001863: Yale Center for Clinical Investigation
Additional Data Partners Who Have Signed a DTA and Whose Data Release is Pending
The Rockefeller University—UL1TR001866: Center for Clinical and Translational Science • The Scripps Research Institute—UL1TR002550: Scripps Research Translational Institute • University of Texas Health Science Center at San Antonio—UL1TR002645: Institute for Integration of Medicine and Science • The University of Texas Health Science Center at Houston—UL1TR003167: Center for Clinical and Translational Sciences (CCTS) • NorthShore University HealthSystem—UL1TR002389: The Institute for Translational Medicine (ITM) • Yale New Haven Hospital—UL1TR001863: Yale Center for Clinical Investigation • Emory University—UL1TR002378: Georgia Clinical and Translational Science Alliance • Weill Medical College of Cornell University—UL1TR002384: Weill Cornell Medicine Clinical and Translational Science Center • Montefiore Medical Center—UL1TR002556: Institute for Clinical and Translational Research at Einstein and Montefiore • Medical College of Wisconsin—UL1TR001436: Clinical and Translational Science Institute of Southeast Wisconsin • University of New Mexico Health Sciences Center—UL1TR001449: University of New Mexico Clinical and Translational Science Center • George Washington University—UL1TR001876: Clinical and Translational Science Institute at Children’s National (CTSA-CN) • Stanford University—UL1TR003142: Spectrum: The Stanford Center for Clinical and Translational Research and Education • Regenstrief Institute—UL1TR002529: Indiana Clinical and Translational Science Institute • Cincinnati Children’s Hospital Medical Center—UL1TR001425: Center for Clinical and Translational Science and Training • Boston University Medical Campus—UL1TR001430: Boston University Clinical and Translational Science Institute • The State University of New York at Buffalo—UL1TR001412: Clinical and Translational Science Institute • Aurora Health Care—UL1TR002373: Wisconsin Network For Health Research • Brown University—U54GM115677: Advance Clinical Translational Research (Advance-CTR) • Rutgers, The State University of New Jersey—UL1TR003017: New Jersey Alliance for Clinical and Translational Science • Loyola University Chicago—UL1TR002389: The Institute for Translational Medicine (ITM) • #N/A—UL1TR001445: Langone Health’s Clinical and Translational Science Institute • Children’s Hospital of Philadelphia—UL1TR001878: Institute for Translational Medicine and Therapeutics • University of Kansas Medical Center—UL1TR002366: Frontiers: University of Kansas Clinical and Translational Science Institute • Massachusetts General Brigham—UL1TR002541: Harvard Catalyst • Icahn School of Medicine at Mount Sinai—UL1TR001433: ConduITS Institute for Translational Sciences • Ochsner Medical Center—U54GM104940: Louisiana Clinical and Translational Science (LA CaTS) Center • HonorHealth—None (Voluntary) • University of California, Irvine—UL1TR001414: The UC Irvine Institute for Clinical and Translational Science (ICTS) • University of California, San Diego—UL1TR001442: Altman Clinical and Translational Research Institute • University of California, Davis—UL1TR001860: UCDavis Health Clinical and Translational Science Center • University of California, San Francisco—UL1TR001872: UCSF Clinical and Translational Science Institute • University of California, Los Angeles—UL1TR001881: UCLA Clinical Translational Science Institute • University of Vermont—U54GM115516: Northern New England Clinical & Translational Research (NNE-CTR) Network • Arkansas Children’s Hospital—UL1TR003107: UAMS Translational Research Institute.
Review board approvals and consent to participate
National Institute of Health’s (NIH) National COVID Cohort Collaborative (N3C) Data Utilization Request Approval committee approved the data utilization request of this project (RP-B3442B), which is approved under the authority of the National Institutes of Health Institutional Review Board and with Johns Hopkins University School of Medicine serving as a central institutional review board. The study protocol was reviewed by the University of Mississippi Medical Center (IRB2020V0280) and Johns Hopkins University’s (IRB00309495) IRBs. The N3C data transfer to NCATS is performed under a Johns Hopkins University Reliance Protocol # IRB00249128 or individual site agreements with NIH. The N3C Data Enclave is managed under the authority of the NIH; information can be found at https://ncats.nih.gov/n3c/resources. No informed consent was obtained because the study used a limited data set.
Review board approval and consent
National Institute of Health’s (NIH) National COVID Cohort Collaborative (N3C) Data Utilization Request (DUR) approval committee approved the data utilization request of this project (RP-504BA5). Each author’s home Institutional Review Board approved the study protocol (HM and WH # 2020V0280; TB #1700991; MK, BP, WK, and SH #2012192778; JA and JH #050-21-EP; SLS #1697848–2, MV #604959). The N3C data transfer to NCATS is performed under a Johns Hopkins University Reliance Protocol # IRB00249128 or individual site agreements with NIH. The N3C Data Enclave is managed under the authority of the NIH; information can be found at https://ncats.nih.gov/n3c/resources.
Data Availability
Data cannot be shared publicly because it contains sensitive patient data. Interested individuals can directly apply for access to the dataset through NIH/N3C by providing an approved IRB and signing a Data Use Agreement (DUA). Requirements for access to the dataset can be found on the N3C Enclave website (https://covid.cd2h.org/enclave). However, the source code utilized for the study is publicly available and can be found in our GitHub repository (https://github.com/hrmoradi/Therapeutics).
Funding Statement
The project described was supported by the National Institute of General Medical Sciences, U54GM104942-05S2, U54GM115458, U54GM104940, U54GM104938, U54GM115516, U54GM115677, U54GM115428, and U54GM104941. The analyses described in this publication were conducted with data or tools accessed through the NCATS N3C Data Enclave covid.cd2h.org/enclave and supported by CD2H - The National COVID Cohort Collaborative (N3C) IDeA CTR Collaboration 3U24TR002306-04S2 NCATS U24 TR002306. This research was possible because of the patients whose information is included within the data from participating organizations (covid.cd2h.org/dtas) and the organizations and scientists (covid.cd2h.org/duas) who have contributed to the on-going development of this community resource. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the N3C program.
References
- 1.Covid trials n.d. https://clinicaltrials.gov/ct2/results?cond=COVID-19.
- 2.Pereira NL, Ahmad F, Byku M, Cummins NW, Morris AA, Owens A, et al. COVID-19: Understanding Inter-Individual Variability and Implications for Precision Medicine. Mayo Clin Proc 2021;96:446–63. doi: 10.1016/j.mayocp.2020.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siddiqi HK, Mehra MR. COVID-19 illness in native and immunosuppressed states: A clinical–therapeutic staging proposal. J Heart Lung Transplant 2020;39:405–7. doi: 10.1016/j.healun.2020.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sterne JA, Murthy S, Diaz JV, Slutsky AS, Villar J, Angus DC, et al. Association between administration of systemic corticosteroids and mortality among critically ill patients with COVID-19: a meta-analysis. Jama 2020;324:1330–41. doi: 10.1001/jama.2020.17023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gottlieb RL, Vaca CE, Paredes R, Mera J, Webb BJ, Perez G, et al. Early remdesivir to prevent progression to severe covid-19 in outpatients. N Engl J Med 2021. doi: 10.1056/NEJMoa2116846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marrone A, Nevola R, Sellitto A, Cozzolino D, Romano C, Cuomo G, et al. Remdesivir plus dexamethasone versus dexamethasone alone for the treatment of COVID-19 patients requiring supplemental O2 therapy: a prospective controlled non-randomized study. Clin Infect Dis 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dreyer NA, Hall M, Christian JB. Modernizing Regulatory Evidence with Trials and Real-World Studies. Ther Innov Regul Sci 2020;54:1112–5. doi: 10.1007/s43441-020-00131-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett TD, Moffitt RA, Hajagos JG, Amor B, Anand A, Bissell MM, et al. Clinical Characterization and Prediction of Clinical Severity of SARS-CoV-2 Infection Among US Adults Using Data From the US National COVID Cohort Collaborative. JAMA Netw Open 2021;4:e2116901. doi: 10.1001/jamanetworkopen.2021.16901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haendel MA, Chute CG, Bennett TD, Eichmann DA, Guinney J, Kibbe WA, et al. The National COVID Cohort Collaborative (N3C): Rationale, design, infrastructure, and deployment. J Am Med Inform Assoc 2021;28:427–43. doi: 10.1093/jamia/ocaa196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Latest Phenotype · National-COVID-Cohort-Collaborative/Phenotype_Data_Acquisition Wiki. GitHub https://github.com/National-COVID-Cohort-Collaborative/Phenotype_Data_Acquisition (accessed December 29, 2022).
- 11.Browne SK, Beeler JA, Roberts JN. Summary of the Vaccines and Related Biological Products Advisory Committee meeting held to consider evaluation of vaccine candidates for the prevention of respiratory syncytial virus disease in RSV-naïve infants. Vaccine 2020;38:101–6. 10.1016/j.vaccine.2019.10.048. [DOI] [PubMed] [Google Scholar]
- 12.Desautels T, Das R, Calvert J, Trivedi M, Summers C, Wales DJ, et al. Prediction of early unplanned intensive care unit readmission in a UK tertiary care hospital: a cross-sectional machine learning approach. BMJ Open 2017;7:e017199. doi: 10.1136/bmjopen-2017-017199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heldt FS, Vizcaychipi MP, Peacock S, Cinelli M, McLachlan L, Andreotti F, et al. Early risk assessment for COVID-19 patients from emergency department data using machine learning. Sci Rep 2021;11:4200. doi: 10.1038/s41598-021-83784-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan L, Zhang H-T, Xiao Y, Wang M, Guo Y, Sun C, et al. Prediction of criticality in patients with severe Covid-19 infection using three clinical features: a machine learning-based prognostic model with clinical data in Wuhan 2020:2020.02.27.20028027. 10.1101/2020.02.27.20028027. [DOI] [Google Scholar]
- 15.Quan H, Li B, Couris CM, Fushimi K, Graham P, Hider P, et al. Updating and validating the Charlson comorbidity index and score for risk adjustment in hospital discharge abstracts using data from 6 countries. Am J Epidemiol 2011;173:676–82. doi: 10.1093/aje/kwq433 [DOI] [PubMed] [Google Scholar]
- 16.Khodaverdi M, Price BS, Porterfield JZ, Bunnell HT, Vest MT, Anzalone AJ, et al. An Ordinal Severity Scale for COVID-19 Retrospective Studies Using Electronic Health Record Data. JAMIA Open 2022:ooac066. doi: 10.1093/jamiaopen/ooac066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman JH. Greedy function approximation: a gradient boosting machine. Ann Stat 2001:1189–232. [Google Scholar]
- 18.Liu S, Deng W. Very deep convolutional neural network based image classification using small training sample size. 2015 3rd IAPR Asian Conf. Pattern Recognit. ACPR, 2015, p. 730–4. 10.1109/ACPR.2015.7486599. [DOI] [Google Scholar]
- 19.Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, et al. Going deeper with convolutions. 2015 IEEE Conf. Comput. Vis. Pattern Recognit. CVPR, 2015, p. 1–9. 10.1109/CVPR.2015.7298594. [DOI] [Google Scholar]
- 20.Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely Connected Convolutional Networks. 2017 IEEE Conf. Comput. Vis. Pattern Recognit. CVPR, 2017, p. 2261–9. 10.1109/CVPR.2017.243. [DOI] [Google Scholar]
- 21.Bergstra James, Yamins Daniel, Cox David. Making a Science of Model Search: Hyperparameter Optimization in Hundreds of Dimensions for Vision Architectures. In: Dasgupta Sanjoy, McAllester David, editors. Proc. 30th Int. Conf. Mach. Learn., vol. 28, PMLR; 2013, p. 115–23. [Google Scholar]
- 22.Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: synthetic minority over-sampling technique. J Artif Intell Res 2002;16:321–57. [Google Scholar]
- 23.Adadi A, Berrada M. Peeking inside the black-box: a survey on explainable artificial intelligence (XAI). IEEE Access 2018;6:52138–60. [Google Scholar]
- 24.Lundberg SM, Lee S-I. A Unified Approach to Interpreting Model Predictions. In: Guyon I, Luxburg UV, Bengio S, Wallach H, Fergus R, Vishwanathan S, et al., editors. Adv. Neural Inf. Process. Syst., vol. 30, Curran Associates, Inc.; 2017. [Google Scholar]
- 25.Albitar O, Ballouze R, Ooi JP, Ghadzi SMS. Risk factors for mortality among COVID-19 patients. Diabetes Res Clin Pract 2020;166. doi: 10.1016/j.diabres.2020.108293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mikami T, Miyashita H, Yamada T, Harrington M, Steinberg D, Dunn A, et al. Risk Factors for Mortality in Patients with COVID-19 in New York City. J Gen Intern Med 2021;36:17–26. doi: 10.1007/s11606-020-05983-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vest MT, Caplan R, Fawcett M, Deitchman AR, Valentino D, Gajera M, et al. Intubation Timing in COVID-19 Based on ROX Index and Association With Patient Outcomes. Respir Care 2022:respcare.09937. doi: 10.4187/respcare.09937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwok S, Adam S, Ho JH, Iqbal Z, Turkington P, Razvi S, et al. Obesity: A critical risk factor in the COVID-19 pandemic. Clin Obes 2020;10:e12403. doi: 10.1111/cob.12403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge E, Li Y, Wu S, Candido E, Wei X. Association of pre-existing comorbidities with mortality and disease severity among 167,500 individuals with COVID-19 in Canada: A population-based cohort study. PLOS ONE 2021;16:e0258154. doi: 10.1371/journal.pone.0258154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reis G, Moreira-Silva EA dos S, Silva DCM, Thabane L, Milagres AC, Ferreira TS, et al. Effect of early treatment with fluvoxamine on risk of emergency care and hospitalisation among patients with COVID-19: the TOGETHER randomised, platform clinical trial. Lancet Glob Health 2022;10:e42–51. doi: 10.1016/S2214-109X(21)00448-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lenze EJ, Mattar C, Zorumski CF, Stevens A, Schweiger J, Nicol GE, et al. Fluvoxamine vs Placebo and Clinical Deterioration in Outpatients With Symptomatic COVID-19: A Randomized Clinical Trial. JAMA 2020;324:2292–300. doi: 10.1001/jama.2020.22760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murai IH, Fernandes AL, Sales LP, Pinto AJ, Goessler KF, Duran CSC, et al. Effect of a Single High Dose of Vitamin D3 on Hospital Length of Stay in Patients With Moderate to Severe COVID-19: A Randomized Clinical Trial. JAMA 2021;325:1053–60. doi: 10.1001/jama.2020.26848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dexamethasone in Hospitalized Patients with Covid-19. N Engl J Med 2021;384:693–704. doi: 10.1056/NEJMoa2021436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Helms J, Tacquard C, Severac F, Leonard-Lorant I, Ohana M, Delabranche X, et al. High risk of thrombosis in patients with severe SARS-CoV-2 infection: a multicenter prospective cohort study. Intensive Care Med 2020;46:1089–98. doi: 10.1007/s00134-020-06062-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19 | NEJM n.d. https://www.nejm.org/doi/full/10.1056/nejmoa2015432 (accessed July 22, 2022). [DOI] [PMC free article] [PubMed]
- 36.Kichloo A, Dettloff K, Aljadah M, Albosta M, Jamal S, Singh J, et al. COVID-19 and Hypercoagulability: A Review. Clin Appl Thromb Off J Int Acad Clin Appl Thromb 2020;26:1076029620962853. doi: 10.1177/1076029620962853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Investigators INSPIRATION. Effect of Intermediate-Dose vs Standard-Dose Prophylactic Anticoagulation on Thrombotic Events, Extracorporeal Membrane Oxygenation Treatment, or Mortality Among Patients With COVID-19 Admitted to the Intensive Care Unit: The INSPIRATION Randomized Clinical Trial. JAMA 2021;325:1620–30. doi: 10.1001/jama.2021.4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Therapeutic Anticoagulation with Heparin in Noncritically Ill Patients with Covid-19. N Engl J Med 2021;385:790–802. doi: 10.1056/NEJMoa2105911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Therapeutic Anticoagulation with Heparin in Critically Ill Patients with Covid-19. N Engl J Med 2021;385:777–89. doi: 10.1056/NEJMoa2103417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jimenez D, Rali P, Doerschug K. COUNTERPOINT: Should Therapeutic Heparin Be Administered to Acutely Ill Hospitalized Patients With COVID-19? No. Chest 2022;161:1448–51. doi: 10.1016/j.chest.2022.01.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tritschler T, Le Gal G, Brosnahan S, Carrier M. POINT: Should Therapeutic Heparin Be Administered to Acutely Ill Hospitalized Patients With COVID-19? Yes. Chest 2022;161:1446–8. doi: 10.1016/j.chest.2022.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hospitalized Adults: Therapeutic Management. COVID-19 Treat Guidel n.d. https://www.covid19treatmentguidelines.nih.gov/management/clinical-management/hospitalized-adults—therapeutic-management/ (accessed July 22, 2022).
- 43.Ho KS, Narasimhan B, Difabrizio L, Rogers L, Bose S, Li L, et al. Impact of corticosteroids in hospitalised COVID-19 patients. BMJ Open Respir Res 2021;8:e000766. doi: 10.1136/bmjresp-2020-000766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pani A, Lauriola M, Romandini A, Scaglione F. Macrolides and viral infections: focus on azithromycin in COVID-19 pathology. Int J Antimicrob Agents 2020;56:106053. doi: 10.1016/j.ijantimicag.2020.106053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karampela I, Dalamaga M. Could Respiratory Fluoroquinolones, Levofloxacin and Moxifloxacin, Prove to be Beneficial as an Adjunct Treatment in COVID-19? Arch Med Res 2020;51:741–2. doi: 10.1016/j.arcmed.2020.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adelman MW, Bhamidipati DR, Hernandez-Romieu AC, Babiker A, Woodworth MH, Robichaux C, et al. Secondary Bacterial Pneumonias and Bloodstream Infections in Patients Hospitalized with COVID-19. Ann Am Thorac Soc 2021;18:1584–7. doi: 10.1513/AnnalsATS.202009-1093RL [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaid A, Somani S, Russak AJ, De Freitas JK, Chaudhry FF, Paranjpe I, et al. Machine Learning to Predict Mortality and Critical Events in a Cohort of Patients With COVID-19 in New York City: Model Development and Validation. J Med Internet Res 2020;22:e24018. doi: 10.2196/24018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jia L, Wei Z, Zhang H, Wang J, Jia R, Zhou M, et al. An interpretable machine learning model based on a quick pre-screening system enables accurate deterioration risk prediction for COVID-19. Sci Rep 2021;11:23127. doi: 10.1038/s41598-021-02370-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snider B, Patel B, McBean E. Insights Into Co-Morbidity and Other Risk Factors Related to COVID-19 Within Ontario, Canada. Front Artif Intell 2021;4:684609. doi: 10.3389/frai.2021.684609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou K, Sun Y, Li L, Zang Z, Wang J, Li J, et al. Eleven routine clinical features predict COVID-19 severity uncovered by machine learning of longitudinal measurements. Comput Struct Biotechnol J 2021;19:3640–9. doi: 10.1016/j.csbj.2021.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Snider B, McBean EA, Yawney J, Gadsden SA, Patel B. Identification of Variable Importance for Predictions of Mortality From COVID-19 Using AI Models for Ontario, Canada. Front Public Health 2021;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhatia S, Makhija Y, Jayaswal S, Singh S, Malik PS, Venigalla SK, et al. Severity and mortality prediction models to triage Indian COVID-19 patients. PLOS Digit Health 2022;1:e0000020. doi: 10.1371/journal.pdig.0000020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jung C, Mamandipoor B, Fjølner J, Bruno RR, Wernly B, Artigas A, et al. Disease-Course Adapting Machine Learning Prognostication Models in Elderly Patients Critically Ill With COVID-19: Multicenter Cohort Study With External Validation. JMIR Med Inform 2022;10:e32949. doi: 10.2196/32949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walkey AJ, Weinberg J, Wiener RS, Cooke CR, Lindenauer PK. Association of Do-Not-Resuscitate Orders and Hospital Mortality Rate Among Patients With Pneumonia. JAMA Intern Med 2016;176:97–104. doi: 10.1001/jamainternmed.2015.6324 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(TIF)
(TIF)
Data Availability Statement
Data cannot be shared publicly because it contains sensitive patient data. Interested individuals can directly apply for access to the dataset through NIH/N3C by providing an approved IRB and signing a Data Use Agreement (DUA). Requirements for access to the dataset can be found on the N3C Enclave website (https://covid.cd2h.org/enclave). However, the source code utilized for the study is publicly available and can be found in our GitHub repository (https://github.com/hrmoradi/Therapeutics).