Abstract
The stability of simple repetitive DNA sequences (microsatellites) is a sensitive indicator of the ability of a cell to repair DNA mismatches. In a genetic screen for yeast mutants with elevated microsatellite instability, we identified strains containing point mutations in the yeast mismatch repair genes, MSH2, MSH3, MLH1, and PMS1. Some of these mutations conferred phenotypes significantly different from those of null mutations in these genes. One semidominant MSH2 mutation was identified. Finally we showed that strains heterozygous for null mutations of mismatch repair genes in diploid strains in yeast confer subtle defects in the repair of small DNA loops.
Eukaryotic genomes contain many regions in which a single base or a small number of bases are repeated (microsatellites). Additions and deletions within microsatellites occur at a much higher frequency than that observed for nonrepetitive DNA sequences (43). In Saccharomyces cerevisiae, mutations in several genes involved in DNA replication result in decreased microsatellite stability. Mutations in the yeast DNA polymerases epsilon and delta increase the instability of microsatellites (24, 53). Some mutations of the POL30 gene (encoding the yeast homolog of the polymerase processivity factor PCNA) and mutations in RFC1, the large subunit of the yeast clamp loader, also destabilize dinucleotide tracts (21, 55, 57). In addition, mutations in the RAD27 gene, which encodes the yeast homolog of the human FEN1 gene, destabilize repetitive tracts (23, 41). The RAD27 gene product is required for processing of the Okazaki fragments during replication.
Mutations in genes affecting DNA mismatch repair (MMR) dramatically reduce microsatellite stability (43). Three yeast homologs of the prokaryotic mutS gene, MSH2, MSH3, and MSH6, have been shown to be involved in mismatch repair in the nuclear genome (25, 43). Mutations in MSH2 and MSH3 decrease the stability of repetitive tracts (repeat units of from 1 to 14 bp), while mutations in MSH6 decrease the stability of repetitive tracts with repeat units of 1 or 2 bp (22, 29, 44). Unlike mutations in the DNA replication machinery, loss of MMR activity does not affect the stability of repetitive tracts with repeat units of 16 bp or more (44). Msh2p and Msh6p, but not Msh3p, are involved in the repair of base-base mismatches (25).
Several yeast homologs of the prokaryotic mutL mismatch repair gene have been identified, and the products of four of these genes, Mlh1p, Mlh2p, Mlh3p, and Pms1p, have been implicated in the yeast postreplication DNA mismatch repair (13, 25, 43). Mutations in the genes MLH1 and PMS1 increase the instability of repetitive tracts with repeat units of 1 to 14 bp (44, 47). Mutations of MLH2 and MLH3 result in modest increases in the rate of 1-bp insertions and deletions in naturally occurring homopolymeric sequences (13). Mutations in MLH1 and PMS1, but not MLH3, result in elevated frequencies of base substitution mutations (13, 43).
The microsatellite-destabilizing effects of mutations affecting DNA mismatch repair are a consequence of two factors: (i) the high level of DNA polymerase slippage on repetitive DNA sequences, resulting in formation of DNA loops involving one or more displaced repeats, and (ii) a role of DNA mismatch repair in the recognition and repair of small DNA loops (43). The genetic data described above, as well as supporting biochemical data, have been interpreted as indicating that yeast cells have at least three mismatch repair complexes: (i) a complex containing Mlh1p, Pms1p, Msh2p, and Msh6p, required for the repair of base-base mismatches and DNA loops of 2 bp or less, (ii) a complex containing Mlh1p, Pms1p, Msh2p, and Msh3p that has the major role in the repair of DNA loops of between 1 and 14 bp, and (iii) a complex containing Mlh1p, Mlh3p, Msh2p, and Msh3p that has a minor role in the repair of small DNA loops (17, 25).
The microsatellite instability observed in yeast cells with mutations in the mismatch repair genes is also observed in mammalian cells with comparable mutations (25). Tumors from patients with hereditary nonpolyposis colorectal cancer (HNPCC), as well as many sporadic colorectal, pancreatic, and gastric tumors, are often associated with mutations of hMLH1 and hMSH2 (7, 12, 27, 33, 36). Mutations in hPMS2 (the human homologue of yeast PMS1), although rare, have also been identified in the germline of HNPCC patients (30, 32, 33). Although there is a high degree of conservation between the yeast and human mismatch repair systems, there may be significant differences. A biochemical study using human cell extracts indicates that the human Msh2p-Msh6p-containing complexes may recognize and repair larger-sized single-stranded loops (14). It is unlikely that the observed differences in the genetic and biochemical data reflect genetic redundancy between Msh3p and Msh6p for DNA loop repair in the genetic studies done with yeast, since the inferred repair of large DNA loops is identical in msh3 single-mutant and msh3 msh6 double-mutant strains (44).
Most of the early studies on microsatellite instability in yeast were done with null mutations of the DNA mismatch repair genes. More recently the effects of point mutations in these genes have been examined. Point mutations of yeast MSH2, PMS1, and MLH1, some of which mimic hMSH2 mutations in HNPCC patients, often elevate the frequency of frameshift mutations (9, 10, 35, 48). Point mutations that result in a dominant-negative phenotype (when the genes carrying the mutations are overexpressed) were identified for MSH2 (48), MLH1 (35), PMS1 (35), and MSH6 (5, 8, 49). Most of these mutations were not dominant or had only a subtle semidominant phenotype when expressed at normal levels. The interpretation of these data is somewhat complicated by the observation that overexpression of wild-type mismatch repair genes sometimes results in a mutator phenotype (42).
In most of the studies described above, point mutations in DNA mismatch repair genes were generated by in vitro mutagenesis, followed by an in vivo assay of the effects of the altered gene. In our study, we used a genetic screen to look for novel mutations affecting microsatellite stability. All of the mutant strains obtained in this screen, however, had mutations in MSH2, MLH1, MSH3, or PMS1. Some of the mutations resulted in phenotypes different from those of null mutant alleles. We also found a semidominant MSH2 mutation. In addition, we examined microsatellite stability in strains heterozygous for null mutations in mismatch repair genes.
MATERIALS AND METHODS
Plasmid constructions.
Plasmids (pEAS10 and pEAS18) with out-of-frame insertions of microsatellites within the coding sequence of ADE2 were constructed in several steps. First, the ADE2-associated DNA sequences between −563 to +671 were PCR amplified from wild-type yeast cells using the primers 5′ CTAGCGCACTACCAGTATATCATC and 5′ CGGACTCCGGAACTCTAGCAGGC. The resulting fragment was ligated into the TA-cloning vector pCR2.1 (Invitrogen) to generate pEAS7. The ADE2 fragment was excised from pEAS7 by treatment with BamHI and NotI, and the resulting fragment was ligated to BamHI-NotI-treated pRS306 (45), generating pEAS8. Microsatellite sequences were inserted into pEAS8 by digestion of the plasmid with XbaI and ligation with the annealed oligonucleotides 5′ CTAGT(GT)14GC and 5′CTAGC(AC)14A (pEAS10) or 5′ CTA(G)17 and 5′ CTA(G)16 (pEAS18). These manipulations result in the insertion of the microsatellite 7 bp after the initiating codon. The repetitive sequences in these plasmids were verified by sequence analysis. The poly(GT) tract in pEAS10 was in the +2 reading frame, and the poly(G) tract in pEAS18 was in the +1 reading frame.
Sequence analysis of the wild-type MSH2 gene in S. cerevisiae AMY125 showed that this strain has a single difference from the MSH2 sequence in the Stanford Genome Database, A to G at position 1105; this alteration does not change the amino acid sequence. One of the mutant MSH2 alleles (msh2-D621Y) was cloned into the integrating vector pRS306. Primers flanking the mutation (5′CAGTAAACGAACTGGTCCGCTCC and 5′ CTTGCGATATGTTCAGCAATTGCCC) were used to amplify sequences containing the mutation, and the resulting 1-kb fragment was inserted into the TA-cloning vector pCR2.1, generating the plasmid pEAS44. The pEAS44 plasmid was treated with BamHI and XbaI, and the resulting fragment was inserted into BamHI-XbaI-treated pRS306 to generate pEAS45. The plasmids used in the quantitative measurement of microsatellite stability have been described previously (19, 44). These plasmids have the following microsatellites (sequence of repeats in parentheses, number of repeats as subscripts): pMD28, (G)18; pSH44, (GT)16.5; pBK1, (CAGT)16; pBK10, (CAATCGGT)10; pEAS20, (CAACGCAATGCGTTGGATCT)3.
Yeast strains.
Strains used in this study were isogenic with AMY125 (MATα ade5-1 leu2-3 trp1-289 ura3-52 his7-2; obtained from A. Morrison and A. Sugino, Osaka University, Osaka, Japan) except for alterations introduced by transformation. The genotypes of these strains are described in Table 1. The yeast strains EAS63 and EAS154 contain out-of-frame insertions of poly(GT) (EAS63) or poly(G) (EAS154) within the ADE2 gene. These strains were constructed in several steps. First, we switched the mating type of MS71, a LEU2 derivative of AMY125 (46), to MATa by using the plasmid pGAL-HO (20), resulting in strain EAS18. Second, we selected a derivative of EAS18 (EAS28) in which the ade5-1 mutation was reverted to ADE5. To determine whether the Ade+ phenotype reflected an intragenic event (rather than an extragenic suppressor), we crossed EAS28 with an ADE5 strain of opposite mating type and dissected 20 tetrads from the resulting diploid. Since all spore colonies were Ade+, we conclude that the reversion was intragenic. To construct EAS63, we performed a two-step transplacement of EAS18 with BglII-treated pEAS10; EAS154 was constructed in the same way with BglII-treated pEAS18. Both EAS63 and EAS154 were Ade− strains that formed red colonies. White sectors within the colony arose as a consequence of frameshifts within the microsatellites that restored the correct reading frame.
TABLE 1.
S. cerevisiae strain names, strain constructions, and relevant genotypes
| Strain name | Strain construction or reference | Relevant genotypea |
|---|---|---|
| AMY125 | 47 | Wild-type |
| MS71 | 46 | LEU2 |
| GCY140 | 46 | LEU2 msh3Δ |
| EAS18 | Described in Materials and Methods | MATa |
| EAS28 | Selected revertant of ade5-1 mutation in EAS18 | MATaADE5 |
| EAS38 | 44 | msh6::LEU2 |
| EAS59 | 44 | msh2Δ msh3Δ msh6::LEU2 |
| EAS74 | 44 | LEU2 msh2Δ |
| AMY101 | 47 | pms1::LEU2 |
| AMY125 Δmlh1 | 47 | mlh1::LEU2 |
| AMY101 Δmlh1 | 47 | pms1::LEU2 mlh1::URA3 |
| EAS250 | 5-FOAR derivative of AMY101 Δmlh1 | pms1::LEU2 mlh1::ura3 |
| EAS63 | Described in Materials and Methods | MATaLEU2 ADE5 ade2::polyGT |
| EAS154 | Described in Materials and Methods | MATaLEU2 ADE5 ade2::polyG |
| MD47 | Haploid segregant of cross between AMY125 and EAS63 | MATaADE5 ade2::polyGT |
| MD48 | Haploid segregant of cross between AMY125 and EAS63 | ADE5 ade2::polyGT |
| MD81 | Two-step transplacement of MD47 with BglII-treated pEAS45 | MATaADE5 ade2::polyGT msh2-D621Y |
| EAS498 | Diploid formed by cross of MD47 and MD48, followed by transformation with pSH44 | MATa/α ADE5/ADE5 ade2::polyGT/ade2-polyGT + pSH44 |
| EAS528 | Haploid segregant of cross between MD47 and EAS59 | msh2Δ ADE5 ade2::polyGT |
| EAS529 | Haploid segregant of cross between MD47 and EAS59 | msh3Δ ADE5 ade2::polyGT |
| EAS530 | Haploid segregant of cross between MD47 and EAS59 | msh6::LEU2 ADE5 ade2::polyGT |
| EAS531 | Haploid segregant of cross between MD47 and EAS250 | mlh1::ura3 ADE5 ade2::polyGT |
| EAS532 | Haploid segregant of cross between MD47 and EAS250 | pms1::LEU2 ADE5 ade2::polyGT |
| EAS533 | Haploid segregant of cross between MD47 and EAS59 | MATamsh2Δ ADE5 ade2::polyGT |
| EAS534 | Haploid segregant of cross between MD47 and EAS59 | MATamsh3Δ ADE5 ade2::polyGT |
| EAS535 | Haploid segregant of cross between MD47 and EAS59 | MATamsh6::LEU2 ADE5 ade2::polyGT |
| EAS536 | Haploid segregant of cross between MD47 and EAS250 | MATamlh1::ura3 ADE5 ade2::polyGT |
| EAS537 | Haploid segregant of cross between MD47 and EAS250 | MATapms1::LEU2 ADE5 ade2::polyGT |
All strains in the study are isogenic (except for changes introduced by transformation) with AMY125 (MATα ade5-1 his7-2 leu2-3, 112 trp1-289 ura3-52). Only alleles that differ from those of the progenitor genotype are listed. The names of strains isogenic with EAS63, except for point mutations generated during this study, are listed separately in the other tables.
Quantitative analysis of microsatellite stability.
Microsatellite stability was measured using plasmids that contained in-frame insertions of various microsatellites into the coding region of URA3 (44). Strains with these plasmids are phenotypically Ura+ and, therefore, sensitive to 5-fluoro-orotate (5-FOA) (4). Alterations in the length of the microsatellite that alter the reading frame result in cells that are resistant to 5-FOA. Thus, to determine the rate of microsatellite instability, we measured the frequency of 5-FOAr colonies in 5 to 20 cultures, as described previously (19). We used the method of the median developed by Lea and Coulson (26) to calculate rates from the frequency data.
Screen for microsatellite-unstable mutants.
EAS63 and EAS154 were plated on rich growth medium (YPD) and mutagenized with ultraviolet light to 10% viability. The plates were incubated at 25°C for 5 to 6 days. Most of the colonies were red and had 0 to 2 white sectors. We screened for those with increased levels of sectoring, indicating an increase in the rate of frameshift mutations within the microsatellite within the ADE2 gene. Three hundred thousand colonies were screened for EAS63, and 200,000 colonies were screened for EAS154. Colonies with increased sectoring were purified to verify the sectoring phenotype. These strains were retested using the quantitative assay for microsatellite stability (described above) involving the plasmids pSH44 (EAS63-derived mutants) and pMD28 (EAS154-derived mutants).
All mutant strains (with reporter plasmids) were mated to the wild-type MD48 strain to determine whether the mutations were recessive. Recessive mutant strains (containing either pSH44 or pMD28) were tested for the ability to complement mutations in the known DNA mismatch repair genes by crossing them to EAS250 (pms1 mlh1) or EAS59 (msh2 msh3 msh6). Complementation was measured by monitoring the frequency of 5-FOAr derivatives in two independent diploids of each genotype. Strains that failed to complement the mutations in EAS250 were tested for complementation in strains with single mutations in mlh1 (AMY125 Δmlh1) and pms1 (AMY101) (47). Strains that failed to complement the mutations in EAS59 were further tested by mating with EAS74 (msh2), GCY140 (msh3), and EAS38 (msh6).
Mutant strains with rates of microsatellite instability that appeared to differ significantly from strains with a null mutation in the mismatch repair gene (strains with the mutations msh2-K893*, msh2-L574S, and pms1-R188T) were backcrossed twice to MD47, the presence of the mutation was verified by sequencing, and the rate of microsatellite stability was measured in 20 additional independent cultures. The msh2-D621Y mutation was tested by reintroducing it into MD47 by two-step replacement with the plasmid pEAS45 to yield the strain MD81 (Table 1).
Screen for dominant mutations affecting microsatellite stability.
A screen for dominant mutations was done using the diploid EAS498, a strain that was homozygous for the ade2::polyGT allele and that contained the plasmid pSH44. EAS498 cells were plated on YPD medium and exposed to ultraviolet light until the survival rate was approximately 15%. Two hundred thousand survivors were screened for the colony sectoring phenotype. Those strains with increased sectoring were purified. We examined microsatellite stability (using the pSH44 assay) with five independent colonies of these strains. The strains were sporulated, and haploid spore colonies that retained the sectoring phenotype were selected. These haploids were backcrossed to either MD47 or MD48 (depending on the mating type of the strain with the mutation), and the dominance of the mutations was verified.
To determine whether the dominant mutations were linked to any of the yeast mismatch repair genes, we crossed haploid strains containing the dominant mutations to strains with single mutations in genes affecting mismatch repair and the ade2::polyGT reporter gene; testers EAS528 to EAS532 were of the α mating type, and testers EAS533 to EAS537 were of the a mating type (Table 1). The resulting diploids were sporulated, and we looked for linkage between the new mutation and the mutation in the known DNA mismatch repair gene. For example, if all spores derived from a cross with an msh2 tester had multiple white sectors in the red spore colonies, we concluded that the new mutation was within the msh2 gene. This conclusion was then confirmed by sequencing the MSH2 gene.
Sequencing of the mismatch repair mutants.
The mismatch repair genes were amplified by PCR in approximately four equal fragments, and the sequences of these fragments were determined with automated sequencing. The primers used for amplification and sequencing are available on request.
Western blot analysis.
Whole-cell extracts were prepared by the method of cell disruption using glass beads (2). The quantity of protein present in these lysates was determined using the Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, Calif.) by the microassay method (supplied with the reagent). Fifty micrograms of total protein from each strain was subjected to sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis through an SDS–8% polyacrylamide gel and blotted using standard methods (2). The primary antibody used for detection of Msh2p was a rabbit polyclonal antibody obtained from E. Alani (Cornell University) (48). Reaction of Msh2p with the antibody was detected using the ECL Western kit (Amersham Pharmacia Biotech, Piscataway, N.J.).
Statistical analyses.
For comparisons of the rates of instability between two different strains, we used a method described previously (56). Briefly, the frequencies of 5-FOAR cells were determined for about 20 independent colonies per strain. The rates of microsatellite alterations were calculated as by the method of the median (26). To determine whether the rates were significantly different between two strains, we converted the frequencies of 5-FOAR cells in each single culture to a rate measurement (26). All rates for both strains were ranked in order from the lowest to the highest. We then determined by chi-square analysis whether one strain had significantly more colonies in the top half of the rate values. P values of <0.05 were considered to be significant.
RESULTS
Screen for yeast mutants with elevated levels of microsatellite instability.
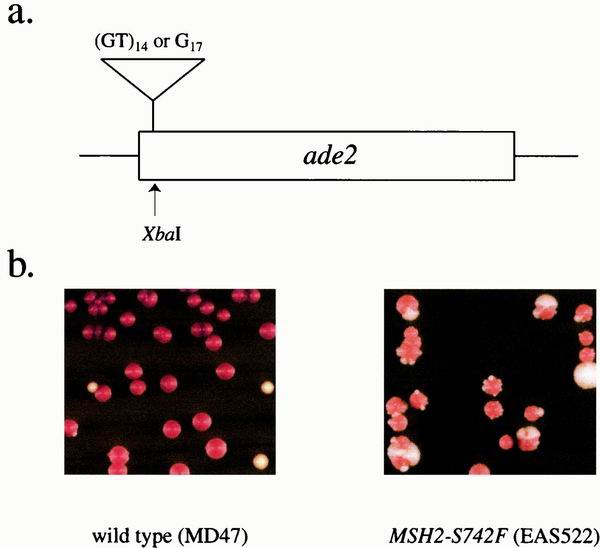
We constructed reporter strains that allowed us to screen yeast colonies for mutants with increased microsatellite instability using a frameshift assay. These strains contain an out-of-frame insertion of microsatellite sequences, either a 29-bp poly(GT) sequence (strain EAS63) or a 17-bp poly(G) (strain EAS154), inserted into the coding sequence of the ADE2 gene (Fig. 1a). Failure of these cells to express ADE2 results in red colonies. A change in microsatellite length, occurring during the growth of a colony, that restores the correct reading frame results in a white sector within the red colony. Similar screens for yeast mutants affecting microsatellite stability have also been done by Xie et al. (58), using an out-of-frame microsatellite insertion within lacZ, and by Tran et al. (53), using an out-of-frame homopolymeric tract in the LYS2 gene.
FIG. 1.
The screen used for microsatellite-unstable mutants. (a) A reporter gene was constructed in which an out-of-frame microsatellite (either 14 copies of a poly(GT) sequence or a tract of 17 guanines) was inserted into the ADE2 coding sequence at an Xbal site located 7 nucleotides 3′ of the start site. The resulting ade2 mutant genes were in the +2 (ade2::polyGT) and +1 (ade2::polyG) reading frames. These reporters were transplaced into the chromosome, replacing the wild-type ADE2 gene. (b) Both strains have the ade2::polyGT reporter. In the DNA mismatch repair-proficient strain (MD47), most of the colonies are red and only a few have sectors. In the mismatch repair-deficient strain (EAS522 with the MSH2-S742F mutation), most of the red colonies have white sectors, and there are numerous white colonies. Since the red pigment that accumulates in ade2 cells reduces growth rates, the white colonies are usually larger than red colonies.
We subjected EAS63 and EAS154 to mutagenesis with ultraviolet light and screened the survivors for increased sectoring (Fig. 1b). Mutant strains with increased sectoring were further screened with a second frameshift reporter. We introduced the plasmid pSH44 into the strains containing the ade2::polyGT reporter and pMD28 into the strains containing the ade2::polyG reporter. These plasmids contain repetitive tracts inserted in frame with the URA3 coding sequence. Plasmid pSH44 contains a 33-bp poly(GT) sequence, and pMD28 contains an 18bp poly(G) sequence. Cells containing these plasmids are phenotypically Ura+. Alterations in the repetitive tracts that result in a frameshift will generate cells that are resistant to 5-FOA, a drug that is toxic to cells expressing URA3 (4). The mutants with increased sectoring were screened for increased rates of mutation to 5-FOAR. We obtained 17 strains with recessive mutations that increased microsatellite instability in both assays. All 17 of these strains were found to contain mutations in known DNA mismatch repair genes as described below. We also constructed a diploid strain containing two copies of the ade2::polyGT reporter that was used to select for dominant microsatellite-unstable mutations. This strain was subjected to mutagenesis with ultraviolet light, and the survivors were screened for increased sectoring on rich medium. One strain with a semidominant mutation in MSH2 was identified.
Microsatellite stability in MSH2 mutants.
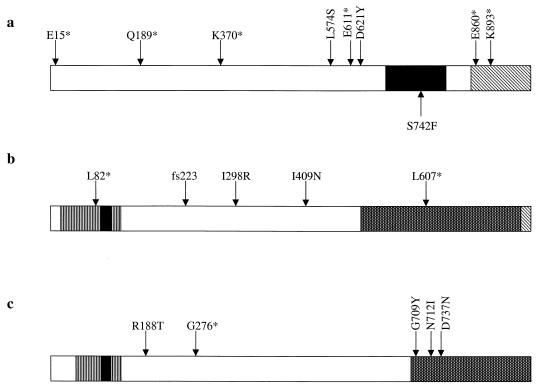
In our screening for microsatellite-unstable mutants with the haploid strain, we obtained eight strains with msh2 mutations (Fig. 2a). Of these eight, six were nonsense mutations and two were missense mutations. The D621Y change is within an amino acid conserved between yeast and human Msh2p; the equivalent position in hMsh2 is position 603 (48). The L574S alteration is also at a conserved amino acid (position 556 in hMsh2). The identical L574S mutation was identified independently in a screen for dominant-negative mutations of MSH2 (48). Although this mutation behaved as a dominant-negative mutation in this screen, in our study the mutation was recessive. This difference is likely to reflect the fact that the msh2-L574S allele was transcribed from a strong promoter (GAL10) on a high-copy-number plasmid (2μm-based) in the study of Studamire et al. (48) and was transcribed from its own promoter as a single-copy gene in our study. Neither the D621Y nor L574S mutation lies in a domain of Msh2p with a known function, and neither is at a position that has been observed with HNPCC patients (website: http://www.nfdht.nl).
FIG. 2.
Locations of mutations relative to functional domains of the DNA mismatch repair proteins. (a) Msh2p. The striped area at the C terminus of the protein indicates the region that has been shown to be required for interaction with Msh6p (1). The black region indicates the location of the ATP-binding domain (39). The arrows above the figure indicate the locations of the recessive mutations found in the MSH2 gene, and the arrow below the figure indicates the dominant mutation. Asterisks indicate nonsense mutations. (b) Mlh1p. The diagonally striped region at the C terminus indicates a sequence of 13 amino acids that is identical to those found at the C terminus of human Mlh1p (35). The stippled area indicates the region of Mlh1 that is required for the interaction with Pms1p as demonstrated by two-hybrid interactions (35). The vertically striped area at the N terminus indicates the region of the protein that is highly conserved among the MutL homologs; within this region is the sequence GFRGEAL, indicated by the black area, which has been termed the MutL box (35). (c) Pms1p. The stippled area indicates the region of Pms1p that is required for the interaction with Mlh1p as demonstrated by two-hybrid interactions (35). As above, the vertically striped area at the N terminus indicates the region of the protein that is highly conserved among the MutL homologs, and the GFRGEAL sequence is indicated by the black area (35).
To quantitatively measure the effects of the recessive msh2 mutations on the stability of microsatellites, we transformed each of the mutants with plasmids that contain in-frame insertions of microsatellites of various repeat unit sizes in the coding sequence of URA3. The repeat units (in base pairs) in the various reporter plasmids were the following: 1 (pMD28), 2 (pSH44), 8 (pBK10), and 20 (pEAS20); a plasmid containing a 4-bp repeat (pBK1) was also used in some studies. The rate of alterations within the microsatellite can be monitored by measuring the rate of occurrence of 5-FOAR derivatives within each strain (described in Materials and Methods). In our previous studies (44, 46, 47), we found that mono- or dinucleotide microsatellites were destabilized by null mutations in MSH2, MSH3, MSH6, MLH1, and PMS1. The same mutations, with the exception of msh6, destabilized microsatellites with 8-bp repeats. None of these mutations affected the stability of the reporter with 20-bp repeats (44).
Rate measurements (based initially on measuring the frequency of 5-FOAR in five independent cultures) are shown in Table 2. From similar, previous studies, we found that differences smaller than threefold, in experiments where no more than five independent cultures are used, are unlikely to be significant. By this criterion, three of the mutations had effects that were suggestively different from that of the null msh2 mutation. To confirm these differences and to more accurately measure the rate of alterations, we repeated the rate measurements using 20 independent cultures. Both the K893* nonsense mutation (resulting in a C-terminal deletion of 72 amino acids) and the D621Y missense mutation had a significantly (P < 0.05) smaller effect on the stability of the mononucleotide microsatellite than the null mutation, although the effects of these mutations on microsatellites with larger repeat units were similar to that observed with the null mutation (Table 2). In contrast, the L574S substitution affected the homopolymeric microsatellite to approximately the same extent as the null msh2 mutation, but it had a significantly smaller effect than the null mutation on the stability of the dinucleotide microsatellite.
TABLE 2.
Rates of microsatellite instability in strains with msh2 mutationsa
| Mutation (strain name)b | Length of repeat unit (bp) | Rate of 5-FOAR/ div.c | Rate relative to wild-typed | Rate relative to msh2Δe |
|---|---|---|---|---|
| msh2-E15* (ESY1) | 1 | 7.0 × 10−2 | 8,400 | 1.7 |
| 2 | 1.2 × 10−3 | 390 | 0.75 | |
| 8 | 7.7 × 10−5 | 9.1 | 0.55 | |
| 20 | 8.0 × 10−5 | 1.1 | 0.72 | |
| msh2-Q189* (ESY2) | 1 | 3.1 × 10−2 | 3,700 | 0.74 |
| 2 | 1.8 × 10−3 | 580 | 1.1 | |
| 8 | 8.2 × 10−5 | 10 | 0.58 | |
| 20 | 6.2 × 10−5 | 0.86 | 0.56 | |
| msh2-K370* (ESY3) | 1 | 4.2 × 10−2 | 5,100 | 1.0 |
| 2 | 1.1 × 10−3 | 360 | 0.69 | |
| 8 | 6.4 × 10−5 | 7.5 | 0.46 | |
| 20 | 3.7 × 10−5 | 0.51 | 0.34 | |
| msh2-L574S (EAS640) | 1 | 2.5 × 10−2 | 3,000 | 0.60 |
| 2 | 4.0 × 10−4 | 130 | 0.25∗∗ | |
| 8 | 8.2 × 10−5 | 9.6 | 0.59 | |
| 20 | 5.6 × 10−5 | 0.78 | 0.81 | |
| msh2-E611* (ESY4) | 1 | 2.2 × 10−2 | 2,700 | 0.52 |
| 2 | 1.1 × 10−3 | 360 | 0.69 | |
| 8 | 9.1 × 10−5 | 11 | 0.83 | |
| 20 | 1.2 × 10−4 | 1.7 | 1.7 | |
| msh2-D621Y (MD81) | 1 | 6.5 × 10−3 | 780 | 0.15∗∗ |
| 2 | 7.3 × 10−4 | 240 | 0.46 | |
| 8 | 8.2 × 10−5 | 9.6 | 0.59 | |
| 20 | 1.1 × 10−4 | 1.5 | 1.0 | |
| msh2-E860* (ESY5) | 1 | 2.2 × 10−2 | 2,700 | 0.52 |
| 2 | 1.4 × 10−3 | 450 | 0.88 | |
| 8 | 9.2 × 10−5 | 11 | 0.65 | |
| 20 | 8.4 × 10−5 | 1.2 | 0.76 | |
| msh2-K893* (MD82) | 1 | 6.9 × 10−3 | 830 | 0.16∗∗ |
| 2 | 1.0 × 10−3 | 320 | 0.6 | |
| 8 | 5.4 × 10−5 | 6.3 | 0.40 | |
| 20 | 7.9 × 10−5 | 1.1 | 0.72 | |
| MSH2-S742F (EAS522) | 1 | 1.6 × 10−2 | 1,900 | 0.38 |
| 2 | 1.7 × 10−3 | 550 | 1.1 | |
| 8 | 6.3 × 10−5 | 7.4 | 0.45 | |
| 20 | 8.0 × 10−5 | 1.1 | 0.73 |
Rates of microsatellite instability were monitored using plasmids with in-frame insertions of microsatellites into the URA3 coding sequence. Alterations in tract length resulting in loss of the wild-type reading frame resulting in a Ura− (5-FOAR) derivative. The following plasmids were used in the analysis: pMD28 (1-bp repeat), pSH44 (2-bp repeat), pBK10 (8-bp repeat), and pEAS20 (20-bp repeat).
The positions of the mutations are shown in Fig. 2a. All strains are ADE5 LEU2 derivatives of AMY125 except for the presence of the msh2 mutation and an ade2::polyGT or ade2::polyG frameshift reporter gene. A dominant mutant allele is shown in capital letters. Single asterisks indicate nonsense mutations.
Rates of mutation to 5-FOAR were determined using fluctuation analysis (described in Materials and Methods) with five independent cultures. When rates differed from the rates in the comparable wild-type strain, the rate estimates were repeated using 20 independent cultures. div., cell division.
These relative rates (mutant/wild-type) are shown to two significant figures. Rates of alterations (95% confidence limits in parentheses) for the isogenic wild-type strain were the following: 8.3 × 10−6 (6.9 to 15), 1-bp repeat; 3.1 × 10−6 (2.6 to 4.1), 2-bp repeat; 8.5 × 10−6 (4.9 to 1.1), 8-bp repeat; and 7.2 × 10−5 (6.2 to 8.3), 20-bp repeat.
The rates for the tested msh2 alleles were divided by the rates determined for the null msh2 mutation. Rates in the haploid msh2Δ strains (95% confidence limits in parentheses) were the following 4.2 × 10−2 (1.2 to 6.0), 1-bp repeat; 1.6 × 10−3 (1.7 to 3.8), 2-bp repeat; 1.4 × 10−4 (0.94 to 2.3), 8-bp repeat; 1.1 × 10−4 (0.98 to 1.8), 20-bp repeat (44). Rates significantly (P < 0.05) different from those for the msh2Δ strain (determined by chi-square analysis) are indicated by double asterisks.
In addition to the msh2 mutation, the stability of homopolymeric and dinucleotide microsatellites is strongly affected by the msh3 mutation and weakly affected by the msh6 mutation. The stability of microsatellites with repeat units greater than 3 bp is affected by the msh2 and msh3 mutations but not by the msh6 mutation (44). One interpretation of the effects of K893* and D621Y is that these mutations primarily affect interactions of Msh2p with Msh3p. Thus, one would expect stronger effects on the stability of microsatellites with repeat units greater than 1 bp. Consistent with this hypothesis, the D621Y substitution is in a region of yeast Msh2p that, in the human Msh2p, is required for interaction with hMsh3p but not hMsh6p (16). The effect of the D621Y substitution, however, is not solely a consequence of a lack of interaction with Msh3p, because this mutation (unlike the null msh3 mutation [44]) substantially elevated the rate of forward mutation at the CAN1 locus. The rate of can1 mutations in a strain with this substitution was 3.2 × 10−6/division. The rates in wild-type, null msh3, and null msh2 strains are 3.1 × 10−7, 3.7 × 10−7, and 1 × 10−5, respectively (44).
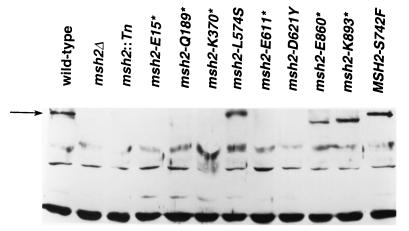
An alternative possibility is that the level of Msh2p is reduced in strains with the K893* and D621Y mutations and this reduction affects formation of the Msh2p/Msh3p complex more severely than formation of the Msh2p/Msh6p complex. To test this hypothesis, we performed Western blot analysis with strains with msh2 mutations (Fig. 3). The D621Y substitution resulted in a substantial decrease in the amount of Msh2p. Although no Msh2p is apparent in the gel shown in Fig. 3, a low level of Msh2p was observed when a larger amount of cellular proteins was loaded on the gel (data not shown).
FIG. 3.
Msh2p levels in the msh2 mutant strains. Protein samples (50 μg) of whole-cell extracts were separated by SDS-polyacrylamide gel electrophoresis. Western blot analysis was performed using polyclonal antibodies raised against Msh2p (48). In lane 3, msh2::Tn indicates a strain (EAS74) that carries a Tn10-LUK insertion near the 5′ end of the MSH2 gene, msh2::Tn10-LUK(7-7) (40). The arrow to the left indicates full-length Msh2p.
Approximately wild-type levels of the truncated K893* protein were produced. The nonsense mutation in msh2-K893* is an ochre mutation. In a previous study in strains with the same genetic background, we found low-frequency readthrough of an ochre mutation in DNA polymerase δ (24). This suppression was a consequence of the prion-like [PSI+] factor. When the strain was treated with guanidine hydrochloride, the [PSI+] factor was lost and the readthrough was suppressed (24). To determine whether [PSI+]-mediated suppression of the ochre mutation of msh2-K893* might be responsible for the non-null phenotype resulting from this mutation, we treated the strains containing this mutation with guanidine hydrochloride and then repeated the microsatellite instability assays. The rates of instability (95% confidence limits shown in parentheses) for the treated strains for microsatellites of repeat units of 1, 2, 8, and 20 bp, respectively, were 2.2 × 10−2 (1.7 × 10−2 to 3.2 × 10−2), 1.3 × 10−3 (1.0 × 10−3 to 2.1 × 10−3), 1.3 × 10−4 (0.9 × 10−4 to 1.7 × 10−4), and 1.0 × 10−4 (0.9 × 10−4 to 1.8 × 10−4). Since these rates are not significantly different from the rates found for strains with null msh2 mutations, we conclude that the non-null phenotype observed in strains with msh2-K893* reflects [PSI+]-mediated suppression. Since this type of suppression is very inefficient (usually less than 1%), the failure to observe Msh2p of wild-type length in the Western analysis (Fig. 3) is not surprising.
The strain with the L574S mutation was partly proficient for the repair of 2-bp loops, but not for the repair of larger or smaller loops (Table 2). This same mutation retains partial activity in the Msh2p-dependent removal of nonhomologous ends during mitotic recombination events (48). These effects cannot be explained as a consequence of a specific defect in either Msh2p-Msh3p or Msh2p-Msh6p interactions. It is likely that this substitution affects the binding and/or subsequent signaling events of the Msh2p-containing complexes in a manner dependent on the size of the loop in the mismatch repair substrate. This mutation maps to a position comparable to an amino acid near the DNA recognition domain IV of MutS (34).
Microsatellite instability in MLH1 mutants.
We obtained five strains with mlh1 mutations (Fig. 2b). Two of these mutations are nonsense mutations, and one is a complicated frameshift mutation at amino acid 223. In addition, we obtained two missense MLH1 (I298R and I409N) mutants. The effects of each of these mutations on microsatellite stability are shown in Table 3. Each of these mutations destabilized the repetitive tracts to approximately the same extent as the null mlh1 mutation (44). The I298R mutation changes an amino acid conserved in many MutL homologues, and an ATPase domain has been identified in this region in the E. coli MutL protein (3). The I409N substitution is in an Mlh1p region outside of both the highly conserved N terminus and the C-terminal region required for interaction with Pms1p (35). This mutation may define a novel domain required for the enzymatic activities of Mlh1p. Alternatively, it is possible that the I409N mutation destabilizes Mlh1p.
TABLE 3.
Rates of microsatellite instability in strains with mlh1 mutationsa
| Mutation (strain name)b | Length of repeat unit (bp) | Rate of 5-FOAR /div. | Rate relative to wild-typec | Rate relative to mlh1Δd |
|---|---|---|---|---|
| mlh1-L82* (ESY6) | 1 | 1.6 × 10−2 | 2,000 | 0.57 |
| 2 | 1.1 × 10−3 | 360 | 1.1 | |
| 8 | 6.9 × 10−5 | 8.1 | 0.53 | |
| 20 | 4.9 × 10−5 | 0.65 | 0.63 | |
| mlh1-223fse (ESY7) | 1 | 1.9 × 10−2 | 2,400 | 0.67 |
| 2 | 1.1 × 10−3 | 360 | 1.1 | |
| 8 | 8.6 × 10−5 | 10 | 0.66 | |
| 20 | 7.3 × 10−5 | 0.97 | 0.94 | |
| mlh1-1298R (EAS368) | 1 | 1.2 × 10−2 | 1,500 | 0.43 |
| 2 | 1.6 × 10−3 | 520 | 1.6 | |
| 8 | 1.1 × 10−4 | 13 | 0.85 | |
| 20 | 1.2 × 10−4 | 1.7 | 1.5 | |
| mlh1-1409N (ESY8) | 1 | 1.8 × 10−2 | 2,300 | 0.64 |
| 2 | 8.3 × 10−4 | 270 | 0.84 | |
| 8 | 9.2 × 10−5 | 11 | 0.71 | |
| 20 | 5.7 × 10−5 | 0.76 | 0.73 | |
| mlh1-L607* (EAS527) | 1 | 1.7 × 10−2 | 2,100 | 0.61 |
| 2 | 1.3 × 10−3 | 420 | 1.3 | |
| 8 | 5.7 × 10−5 | 6.7 | 0.44 | |
| 20 | 5.6 × 10−5 | 0.75 | 0.72 |
Methods of measuring microsatellite instability are described in the Table 2 legend. div., cell division.
The positions of the mutations are shown in Fig. 2b. Strains were identical to those described in Table 2 except for the substitution of the mlh1 mutation for the msh2 mutation. Single asterisks indicate nonsense mutations.
The rates of alteration in the wild-type strain can be found in the legend to Table 2.
The rates of alteration in the mlh1Δ strain (95% confidence limits shown in parentheses) were the following: 2.8 × 10−2 (2.2 to 3.9), 1-bp repeat; 9.9 × 10−4 (8.2 to 14), 2-bp repeat; 1.3 × 10−4 (0.92 to 1.6), 8-bp repeat; 7.8 × 10−5 (6.5 to 11), 20-bp repeat.
This mutation results in an alteration of the DNA sequence that includes codon 223 from TCGAAT to TCTATTT.
The strain containing the I298R mutation was found to contain two mutations that affected microsatellite stability in addition to mlh1. The rates shown in Table 3 are those of a strain derived from backcrosses with the wild-type strain, in which only the mlh1 mutation is present. One of the additional mutations in the original mutant strain was a frameshift mutation (a deletion of one A in a run of 10 A's near the 5′ end of the gene) at amino acid 152 of the MSH3 gene. Since frameshifts in homopolymeric tracts are greatly elevated in mlh1 strains (15, 44, 53), it is likely that the msh3 mutation arose after the mlh1 mutation; interestingly, a mammalian cell line with the comparable double mutations in hMLH1 and hMSH3 has also been observed (28). The second additional mutation in the original mutant isolate conferred sensitivity to UV light and had a modest but significant effect on the rate of alteration of tracts with repeat units of 2 bases (data not shown). This gene has not yet been identified.
Microsatellite instability in the PMS1 mutants.
We identified four strains with mutations in PMS1, one with a nonsense mutation (G276*) and three with missense mutations (R188T, N712I, and G709Y/D737N) (Fig. 2c). In the strain with the double-amino-acid substitution, we did not determine the individual effects of each of these mutations. Both of these substitutions and a third amino acid substitution (N712I) lie within the region that is required for the interaction of Pms1p and Mlh1p in yeast (35). The R188T substitution is at a position that is invariant among MutL homologues in prokaryotes, yeast, and humans (33, 38).
Strains with the G276*, N712I, and G709Y/D737N mutations had approximately the same microsatellite instability as strains with the null pms1 mutation (Table 4). It seems likely that these mutations eliminate the ability of Pms1p to function in repair by inhibiting the interaction of Pms1p with Mlh1p or by resulting in an unstable Pms1p protein. The R188T mutation had the same phenotype as the null mutation with the mononucleotide microsatellite, indicating a lack of ability to repair 1-base loops, but had a hypomorphic phenotype with the dinucleotide and octanucleotide microsatellites (Table 4). This result suggests that the repair defect in this strain is predominantly a deficiency in repair events that involve the MSH6-containing complex. It is possible that this region of Pms1p is required for stable interaction with Msh6p in an Msh2p-Msh6p heterodimer.
TABLE 4.
Rate of microsatellite instability in strains with pms1 mutationsa
| Mutationb | Length of repeat unit (bp) | Rate of 5-FOAR /div. | Rate relative to wild-typec | Rate relative to pms1Δd |
|---|---|---|---|---|
| pms1-R188T (EAS585) | 1 | 2.5 × 10−2 | 3,100 | 0.93 |
| 2 | 4.2 × 10−4 | 140 | 0.25∗∗ | |
| 8 | 2.5 × 10−5 | 2.9 | 0.18∗∗ | |
| 20 | 6.2 × 10−5 | 0.82 | 0.52 | |
| pms1-G276* (ESY9) | 1 | 1.8 × 10−2 | 2,300 | 0.67 |
| 2 | 1.5 × 10−3 | 480 | 0.88 | |
| 8 | 8.2 × 10−5 | 9.6 | 0.59 | |
| 20 | 4.1 × 10−5 | 0.55 | 0.34 | |
| pms1-GDe (ESY10) | 1 | 1.2 × 10−2 | 1,500 | 0.44 |
| 2 | 1.2 × 10−3 | 390 | 0.71 | |
| 8 | 1.3 × 10−4 | 15 | 0.93 | |
| 20 | 4.6 × 10−5 | 0.61 | 0.38 | |
| pms1-N7121 (ESY11) | 1 | 2.1 × 10−2 | 2,600 | 0.78 |
| 2 | 9.5 × 10−4 | 310 | 0.56 | |
| 8 | 7.0 × 10−5 | 8.2 | 0.50 | |
| 20 | 1.2 × 10−4 | 1.6 | 1.0 |
Methods of measuring the rates are described in Table 2. div., cell division.
The positions of the mutations are shown in Fig. 2c. The strains are isogenic to those described in Tables 2 and 3, except for the substitution of pms1 for the other mutations affecting DNA mismatch repair. Single asterisks indicate nonsense mutations.
The rates of alteration in the wild-type strain are in the legend to Table 2.
The rates of alteration (95% confidence limits in parentheses) in the pms1Δ strain are the following: 2.7 × 10−2 (2.2 to 3.9), 1-bp repeat; 1.7 × 10−3 (1.3 to 2.0), 2-bp repeat; 1.4 × 10−4 (1.2 to 1.5), 8-bp repeat; 1.2 × 10−4 (0.8 to 1.7), 20-bp repeat. The double asterisks show rates significantly different (P < 0.05) from those in the pms1Δ strain.
In this mutant strain, Pms1p has two substitutions, G709Y and D737N.
Mutator phenotypes of strains heterozygous for null mutations in DNA mismatch repair genes.
We investigated whether strains heterozygous for null mutations of the DNA mismatch repair genes had a mutator phenotype. In previous studies, a very subtle repair defect was found in strains heterozygous for null mutations of either MSH2 (10) or MLH1 (42). For example, in one assay, strains homozygous for an msh2 null mutation elevated the mutation rate 6,500-fold over that of the wild type; the mutator phenotype of the heterozygous null mutation was threefold (10). This effect did not reflect reduced repair capacity in the heterozygous strains but was a consequence of loss of the wild-type MSH2 or MLH1 allele in a small fraction of the cells in the initially heterozygous strain (11, 42). Thus, no evidence for haploinsufficiency for DNA mismatch repair genes has been previously reported.
To investigate this issue in more detail, we constructed diploid strains heterozygous for single or multiple null mutations of DNA mismatch repair genes that had reporter plasmids allowing us to monitor the stability of mono-, di-, and tetranucleotide microsatellites. Each rate estimate in these experiments was based on examination of 20 independent cultures. To determine the statistical significance of the data, we used a ranking method developed previously (56). In brief, the frequencies of 5-FOAR cells in each single culture were converted to a rate measurement. The rates from both strains being compared were ranked in order from the lowest to the highest. Chi-square analysis was used to determine whether one strain had significantly more cultures with rates in the top half of the ranking; P values of <0.05 were considered to be significant. The results from this analysis are shown in Table 5.
TABLE 5.
Rates of microsatellite instability in a wild-type diploid strain and in strains heterozygous for mutations of mismatch repair genesa
| Relevant genotypeb (strain name) | Length of repeat unit (bp) | Rate of 5-FOAR/div. | 95% Confidence limits on rates | Rate relative to wild-typec |
|---|---|---|---|---|
| Wild-type (EAS309) | 1 | 4.0 × 10−6 | 3.1 × 10−6–4.8 × 10−6 | 1 |
| 7.1 × 10−6 | 4.7 × 10−6–11 × 10−6 | |||
| 2 | 4.0 × 10−6 | 2.4 × 10−6–5.9 × 10−6 | 1 | |
| 3.3 × 10−6 | 2.0 × 10−6–6.0 × 10−6 | |||
| 4 | 7.8 × 10−6 | 6.1 × 10−6–13 × 10−6 | 1 | |
| 8 | 1.1 × 10−5 | 0.71 × 10−5–1.6 × 10−5 | 1 | |
| 20 | 7.1 × 10−5 | 0.60 × 10−4–1.0 × 10−4 | 1 | |
| MSH2/msh2Δ (EAS278) | 1 | 1.7 × 10−5 | .88 × 10−5–4.3 × 10−5 | 2.9∗∗ |
| 1.5 × 10−5 | 1.1 × 10−5–2.9 × 10−5 | |||
| 2 | 9.1 × 10−6 | 3.1 × 10−6–14 × 10−6 | 2.7∗∗ | |
| 1.1 × 10−5 | 0.60 × 10−5–1.7 × 10−5 | |||
| 4 | 8.6 × 10−6 | 6.6 × 10−6–14 × 10−6 | 1.1 | |
| 8 | 1.2 × 10−5 | 1.1 × 10−5–2.2 × 10−5 | 1.1 | |
| MSH3/msh3Δ (EAS279) | 1 | 1.8 × 10−5 | 1.1 × 10−5–3.0 × 10−5 | 2.7∗∗ |
| 1.2 × 10−5 | 0.65 × 10−5–1.7 × 10−5 | |||
| 2 | 2.0 × 10−5 | 1.4 × 10−5–2.7 × 10−5 | 5.2∗∗ | |
| 1.8 × 10−5 | 1.3 × 10−5–2.9 × 10−5 | |||
| 4 | 1.3 × 10−5 | 0.77 × 10−5–2.6 × 10−5 | 1.7 | |
| MSH6/msh6Δ (EAS277) | 1 | 1.2 × 10−5 | 0.96 × 10−5–1.6 × 10−5 | 2.2 |
| 2 | 6.8 × 10−6 | 2.8 × 10−6–22 × 10−6 | 1.9 | |
| 4 | 5.8 × 10−6 | 4.6 × 10−6–8.4 × 10−6 | 0.74 | |
| MSH2/msh2Δ MSH3/msh3Δ MSH6/msh6Δ (EAS365) | 1 | 4.0 × 10−5 | 2.9 × 10−5–6.6 × 10−5 | 5.8∗∗ |
| 2.4 × 10−5 | 2.1 × 10−5–4.0 × 10−5 | |||
| 2 | 4.7 × 10−5 | 3.5 × 10−5–5.4 × 10−5 | 12∗∗ | |
| 4.0 × 10−5 | 3.0 × 10−5–5.9 × 10−5 | |||
| 4 | 2.5 × 10−5 | 1.2 × 10−5–6.9 × 10−5 | 3.2∗∗ | |
| MLH1/mlh1Δ (EAS433) | 1 | 9.0 × 10−6 | 0.60 × 10−5–2.1 × 10−5 | 2.1 |
| 1.4 × 10−5 | 0.71 × 10−5–3.2 × 10−5 | |||
| 2 | 6.4 × 10−6 | 0.35 × 10−5–1.5 × 10−5 | 1.8 | |
| PMS1/pms1Δ (EAS432) | 1 | 2.4 × 10−5 | 1.4 × 10−5–7.7 × 10−5 | 4.5∗∗ |
| 2.5 × 10−5 | 1.6 × 10−5–5.4 × 10−5 | |||
| 2 | 6.1 × 10−6 | 4.4 × 10−6–9.0 × 10−6 | 1.7 | |
| MLH1/mlh1Δ PMS1/pms1Δ (EAS631) | 1 | 4.6 × 10−5 | 1.2 × 10−5–7.5 × 10−5 | 7.9∗∗ |
| 4.2 × 10−5 | 2.8 × 10−5–5.4 × 10−5 | |||
| 2 | 6.6 × 10−6 | 0.46 × 10−5–2.2 × 10−5 | 1.8 | |
| MSH2/MSH2-S742F (EAS632) | 1 | 6.3 × 10−5 | 4.9 × 10−5–16 × 10−5 | 11∗∗ |
| 2 | 3.3 × 10−5 | 2.2 × 10−5–6.6 × 10−5 | 9.0∗∗ | |
| 8 | 4.3 × 10−5 | 3.6 × 10−5–5.8 × 10−5 | 3.9∗∗ | |
| 20 | 6.1 × 10−5 | 4.7 × 10−5–12 × 10−5 | 0.86 |
Rates of microsatellite instability were measured as described in the Table 2 legend, except that all experiments were performed using 20 independent cultures. Each rate value represents an independent experiment. div., cell division.
All null mutant alleles are shown in small letters. MSH2-S742F is a semi-dominant mutation, as described in the text. All diploids were constructed by crossing EAS63 (wild type for DNA mismatch repair genes; complete genotype in Table 1) with isogenic haploids containing mismatch repair mutations and the various reporter plasmids; the strain names and mutations introduced by the non-EAS63 haploids were the following: AMY125 (wild-type), EAS74 (msh2Δ), GCY140 (msh3Δ), EAS38 (msh6Δ), EAS59 (msh2Δ msh3Δ msh6Δ), AMY125 mlh1 (mlh1Δ), AMY101 (pms1Δ), AMY101 Δmlh1 (mlh1Δ pms1Δ), and EAS522 (MSH2-S742F).
The heterozygous diploid rates were divided by the wild-type diploid rates. When two experiments were performed, the rates were averaged to determine the relative rates. All rates marked with double asterisks were significantly (P < 0.05) elevated relative to the wild-type rate as determined using the chi-square test described in Materials and Methods.
We observed a small (approximately threefold), but significant, increase in the rate of instability of dinucleotide tracts in the msh2 and msh3 heterozygous strains, but not for those strains heterozygous for msh6 mutations. The msh2 msh3 msh6 triply heterozygous strain had significant repair defects with the mono-, di-, and tetranucleotide reporters. A strain heterozygous for a null mutation of pms1 displayed a significant increase in instability of the mononucleotide repeat, while a strain heterozygous for mlh1 mutation did not. Consistent with this result, in a strain heterozygous for both pms1 and mlh1 mutations, a significant effect was observed with the mononucleotide reporter (Table 5).
As described above, in previous similar studies the putative haploinsufficiency of the DNA mismatch repair mutations reflected an elevated level of mutations in a small fraction of the cells in which the wild-type repair gene had been lost (11, 42). If this mechanism is also responsible for the elevated microsatellite instability observed in our experiment, then we expect that many of the 5-FOAR derivatives should have two properties: (i) they should have mutation rates characteristic of strains with the null mismatch repair mutations, and (ii) they should no longer contain the DNA sequences characteristic of the wild-type allele of the originally heterozygous mismatch repair gene. We purified 16 independent 5-FOAR derivatives from MSH2/msh2Δ and MSH2/msh2Δ MSH3/msh3Δ MSH6/msh6Δ strains carrying the pSH44 dinucleotide reporter plasmid. If the 5-FOAR Ura− derivatives containing this plasmid have a mutator phenotype, one should observe reversion to the 5-FOAS Ura+ phenotype at high frequency. None of the 32 strains had this property. Since the wild-type and mutant alleles of the mismatch repair genes were readily distinguishable by PCR analysis, we also examined these 32 strains for the presence of the wild-type alleles. All 16 strains derived from the MSH2/msh2Δ strains retained the MSH2 allele, and all 16 strains derived from the triple heterozygote retained the wild-type MSH2, MSH3, and MSH6 alleles.
In summary, we conclude that strains heterozygous for mutations in some combinations of DNA mismatch repair genes have a subtle mutator phenotype. The differences between our results and those of others (11, 42) are likely to reflect properties of the assays (relative sensitivities or sequence-specific effects) or the different genetic backgrounds. In addition, we performed a different type of test to assess statistical significance.
A semidominant mutation of MSH2.
By mutagenizing a diploid strain with the ade2::polyGT reporter, we also identified a strain with a semidominant MSH2 mutation, MSH2-S742F (Fig. 2a). This mutation lies in the second of four highly conserved nucleotide binding motifs and, in a haploid, resulted in a repair defect similar to that observed for null msh2 mutants (Table 2). In the heterozygous diploid, this mutation destabilized microsatellites with repeat units of 1, 2, or 8 bp about fivefold compared to the wild type (Table 5). No significant destabilization was observed for the minisatellite (20-bp repeat unit).
DISCUSSION
Our study represents one of three direct screenings for mutations affecting microsatellite stability in yeast (53, 58). In previous studies, mutations in genes affecting various components of the DNA replication (POL3, POL30, RAD27, and RFC1) and DNA mismatch repair (MSH2, MSH3, MLH1, MLH2, MLH3, MLH1, EXO1, and PMS1) systems were observed to destabilize nuclear microsatellites (17, 18, 25, 43, 57, 58). In our mutant hunt, however, we found only mutations affecting a subset of the DNA mismatch repair proteins. There are several likely explanations for the limited number of genes identified. First, since most of the DNA replication proteins (with the exception of Rad27p) are essential, the available mutational target within these proteins is smaller than for the nonessential DNA mismatch repair proteins. Second, since our ade2 reporter was in the +2 reading frame for the dinucleotide tract and the +1 reading frame for the mononucleotide tract, loss of a single repeat would restore the correct reading frame, whereas addition of a single repeat would not. Thus, our screening is more sensitive for detecting mutants that elevate the frequency of microsatellite deletions rather than additions. This factor may explain why we failed to detect rad27 mutants, since these mutations specifically elevate the frequency of additions rather than of deletions (23, 51, 58). In another mutant hunt in which the reporter gene was biased toward detection of additions to microsatellites, the strains with the rad27 mutation were readily detected (58). Third, our mutant hunt was presumably biased for detecting mutations that had the strongest effects on the stability of mono- and dinucleotide microsatellites. The effects of msh2, pms1, and mlh1 on these types of microsatellites are considerably stronger than those of exo1, msh3, msh6, mlh2, or mlh3 (13, 18, 44, 50). Our study failed to identify new genes that strongly affect microsatellite stability. As discussed above, it may be that such genes exist but are essential and, therefore, are small targets for mutation. Alternatively, these genes may be functionally redundant.
We found nine missense mutations in MMR genes that strongly destabilized microsatellites, one of which was semidominant (Tables 2 to 4). It is likely that the semidominant mutation affects the ATPase function of Msh2p, since it is a substitution of a highly conserved amino acid in this domain. This mutation may be semidominant because the protein is forming nonfunctional complexes with other components of the DNA mismatch repair machinery. It should be emphasized that MSH2-S742F represents a “classical” semidominant mutation, one that has a partial mutant phenotype when present in one copy in a diploid. Although many mutant alleles of mismatch repair genes result in a dominant-negative phenotype when overexpressed in yeast (9, 48), only one other dominant mutation of MSH2 has been reported (G693A) (10). Since different assays were used to examine the mutator phenotypes in our study and that of Drotschmann et al. (10), it is difficult to compare the relative effects of MSH2-S742F and MSH2-G693A.
Three dominant mutations have been found in patients with mismatch repair defects: a nonsense mutation at codon 134 in hPMS2 (equivalent to the yeast PMS1 gene), a missense mutation at codon 605 in hPMS2 (30), and a deletion of codon 618 in hMLH1 (37). A subsequent study of the mutation in hMLH1, however, suggests that it may not be dominant (31).
Based on biochemical studies of eukaryotic mismatch repair proteins (25), the msh2 mutations could affect interactions with Msh3p and/or Msh6p, binding to the mismatch, ATPase activity, formation of the ternary complex with Mlh1p/Pms1p, or subsequent interactions with other components of the DNA mismatch repair system (for example, PCNA [6]). The L574S and D621Y substitutions are in regions of Msh2p that are thought to be involved in maintaining the structural integrity of Msh2p, and in fact, there is a significant reduction in the steady-state levels of Msh2p in a strain carrying the D621Y, but not the L574S, mutation. The P361L substitution is in a position expected to be involved in interdomain interactions (34). In addition, the L574S mutation is near a domain important in mismatch recognition (34). Similarly, the mutant Mlh1p and Pms1p could be defective in formation of the Mlh1p/Pms1p heterodimer, interactions with the mismatch-bound Msh2p/Msh3p or Msh2p/Msh6p heterodimer, ATPase activity, or subsequent interactions with the MMR machinery (54). We also cannot rule out the possibility that the mutant substitutions in the mutL homologs affect protein stability. Finally, although all of the mutant strains discussed above, with the exception of D621Y (which was regenerated in a wild-type background by two-step transplacement), were backcrossed to the wild-type strain several times to eliminate additional mutations, it is possible that additional, closely linked mutations exist that affect the phenotypes of these mutant strains.
As discussed in Results, some of the MMR mutations specifically affected the stability of one type of microsatellite. Such mutations may differentially affect the formation of one class of MutS heterodimer (for msh2 mutations) or the interaction with one class of MutS heterodimer (for mlh1 or pms1 mutations). Alternatively, it is possible that certain mutational changes can specifically affect interaction of the MMR proteins with specific types of DNA mismatched substrates without affecting formation of the heterodimers or the ternary complex. Resolution of these issues will probably require biochemical characterization of the mutant proteins.
ACKNOWLEDGMENTS
The research was supported by a National Institutes of Health Grant, GM52319 (T.D.P.). E.A.S. is a recipient of a Burroughs Wellcome Fund Career Award in the Biomedical Sciences.
We thank R. M. Liskay and E. Alani for comments on the manuscript. We also thank E. Alani for generously supplying the anti-Msh2p antibody.
REFERENCES
- 1.Alani E. The Saccharomyces cerevisiae Msh2 and Msh6 proteins form a complex that specifically binds to duplex oligonucleotides containing mismatched DNA base pairs. Mol Cell Biol. 1996;16:5604–5615. doi: 10.1128/mcb.16.10.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons; 1994. [Google Scholar]
- 3.Ban C, Yang W. Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell. 1998;95:541–552. doi: 10.1016/s0092-8674(00)81621-9. [DOI] [PubMed] [Google Scholar]
- 4.Boeke J D, Lacroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoroorotic resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 5.Bowers J, Sokolsky T, Quach T, Alani E. A mutation in the MSH6 subunit of the Saccharomyces cerevisiae MSH2-MSH6 complex disrupts mismatch recognition. J Biol Chem. 1999;274:16115–16125. doi: 10.1074/jbc.274.23.16115. [DOI] [PubMed] [Google Scholar]
- 6.Bowers J, Tran P T, Joshi A, Liskay R M, Alani E. MSH-MLH complexes formed at a DNA mismatch are disrupted by the PCNA sliding clamp. J Mol Biol. 2001;306:957–968. doi: 10.1006/jmbi.2001.4467. [DOI] [PubMed] [Google Scholar]
- 7.Bronner C E, Baker S M, Morrison P T, Warren G, Smith L G, Lescoe M K, Kane M, Earabino C, Lipford J, Lindlom A, Tannergard P, Bollag R J, Godwin A R, Ward D C, Nordenskjold M, Fishel R, Kolodner R, Liskay R M. Mutation in the DNA mismatch repair gene homologue hMSH2 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 8.Das Gupta R, Kolodner R D. Novel dominant mutations in Saccharomyces cerevisiae MSH6. Nat Genet. 2000;24:53–56. doi: 10.1038/71684. [DOI] [PubMed] [Google Scholar]
- 9.Drotschmann K, Clark A B, Kunkel T A. Mutator phenotypes of common polymorphisms and missense mutations in MSH2. Curr Biol. 1999;9:907–910. doi: 10.1016/s0960-9822(99)80396-0. [DOI] [PubMed] [Google Scholar]
- 10.Drotschmann K, Clark A B, Tran H T, Resnick M A, Gordenin D A, Kunkel T A. Mutator phenotypes of yeast strains heterozygous for mutations in the MSH2 gene. Proc Natl Acad Sci USA. 1999;96:2970–2975. doi: 10.1073/pnas.96.6.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drotschmann K, Shcherbakova P V, Kunkel T A. Mutator phenotype due to loss of heterozygosity in diploid yeast strains with mutations in MSH2 and MLH1. Toxicol Lett. 2000;112:239–244. doi: 10.1016/s0378-4274(99)00276-3. [DOI] [PubMed] [Google Scholar]
- 12.Fishel R, Lescoe M K, Rao M R S, Copeland N G, Jenkins N A, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 13.Flores-Rozas H, Kolodner R D. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc Natl Acad Sci USA. 1998;95:12404–12409. doi: 10.1073/pnas.95.21.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genschel J, Littman S J, Drummand J T, Modrich P. Isolation of MutS(beta) from human cells and comparison of the mismatch repair specificities of MutS(beta) and MutSα. J Biol Chem. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 15.Greene C N, Jinks-Robertson S. Frameshift intermediates in homopolymeric runs are efficiently removed by yeast mismatch repair proteins. Mol Cell Biol. 1997;17:2844–2850. doi: 10.1128/mcb.17.5.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerrette S, Wilson T, Gradia S, Fishel R. Interactions of human hMSH2 with hMSH3 and hMSH2 with hMSH6: examination of mutations found in hereditary nonpolyposis colorectal cancer. Mol Cell Biol. 1998;18:6616–6623. doi: 10.1128/mcb.18.11.6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harfe B D, Jinks-Robertson S. DNA mismatch repair and genetic instability. Annu Rev Genet. 2000;34:359–399. doi: 10.1146/annurev.genet.34.1.359. [DOI] [PubMed] [Google Scholar]
- 18.Harfe B D, Minesinger B K, Jinks-Robertson S. Discrete in vivo roles for the MutL homologs Mlh2p and Mlh3p in the removal of frameshift intermediates in budding yeast. Curr Biol. 2000;10:145–148. doi: 10.1016/s0960-9822(00)00314-6. [DOI] [PubMed] [Google Scholar]
- 19.Henderson S T, Petes T D. Instability of simple sequence DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2749–2757. doi: 10.1128/mcb.12.6.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herskowitz I, Jensen R E. Putting the HO gene to work: practical uses for mating-type switching. Methods Enzymol. 1991;194:132–146. doi: 10.1016/0076-6879(91)94011-z. [DOI] [PubMed] [Google Scholar]
- 21.Johnson R E, Kovvali G K, Guzder S N, Amin N C, Holm C, Habraken Y, Sung P, Prakash L, Prakash S. Evidence for involvement of yeast proliferating cell nuclear antigen in DNA mismatch repair. J Biol Chem. 1996;271:27987–27990. doi: 10.1074/jbc.271.45.27987. [DOI] [PubMed] [Google Scholar]
- 22.Johnson R E, Kovvali G K, Prakash L, Prakash S. Requirement of the yeast MSH3 and MSH6 genes for MSH2-dependent genomic stability. J Biol Chem. 1996;271:7285–7288. doi: 10.1074/jbc.271.13.7285. [DOI] [PubMed] [Google Scholar]
- 23.Johnson R E, Kovvali G K, Prakash L, Prakash S. Requirement of the yeast RTH1 5′ to 3′ exonuclease for the stability of simple repetitive DNA. Science. 1995;269:238–240. doi: 10.1126/science.7618086. [DOI] [PubMed] [Google Scholar]
- 24.Kokoska R J, Stefanovic L, DeMai J, Petes T D. Increased rates of genomic deletions generated by mutations in the yeast gene encoding DNA polymerase δ or by decreases in the cellular levels of DNA polymerase δ. Mol Cell Biol. 2000;20:7490–7504. doi: 10.1128/mcb.20.20.7490-7504.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolodner R D, Marsischky G. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 26.Lea D E, Coulson C A. The distribution of the number of mutants in bacterial populations. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 27.Leach F S, Nicolaides N C, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen L A, Nystrom-Lahti M, Guan X-Y, Zhang J, Meltzer P S, Yu J-W, Kao F-T, Chen D J, Cerosaletti K M, Fournier R E K, Todd S, Lewis T, Leach R J, Naylor S L, Wissenbach J, Mecklin J-P, Jarvinen H, Petersen G M, Hamilton S R, Green J, Jass J, Watson P, Lynch H T, Trent J M, de la Chapelle A, Kinzler K W, Vogelstein B. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 28.Malkhosyan S, Rampino N, Yamamoto H, Perucho M. Frameshift mutator mutations. Nature. 1996;382:499–500. doi: 10.1038/382499a0. [DOI] [PubMed] [Google Scholar]
- 29.Marsischky G T, Filosi N, Kane M F, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 30.Miyaki M, Nishio J, Konishi M, Kikuchi-Yanoshita R, Tanaka K, Muraoka M, Nagato M, Chong J M, Koike M, Terada T, Kawahara Y, Fukutome A, Tomiyama J, Chuganji Y, Momoi M, Utsunomiya J. Drastic genetic instability of tumors and normal tissues in Turcot syndrome. Oncogene. 1997;15:2877–2881. doi: 10.1038/sj.onc.1201668. [DOI] [PubMed] [Google Scholar]
- 31.Moliaka Y K, Cella M, Chudina A P, Kolesnikova T N, Terracciano L, Cathomas G, Bochkov N P, Buerstedde J-M. Mechanisms underlying mismatch repair deficiencies in normal cells. Genes Chromosomes Cancer. 1997;20:305–309. [PubMed] [Google Scholar]
- 32.Nicolaides N C, Littman S J, Modrich P, Kinzler K W, Vogelstein B. A naturally occurring hPMS2 mutation can confer a dominant negative mutator phenotype. Mol Cell Biol. 1998;18:1635–1641. doi: 10.1128/mcb.18.3.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicolaides N C, Papadopoulos N, Liu B, Wei Y F, Carter K C, Ruben S M, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 34.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 35.Pang Q, Prolla T A, Liskay R M. Functional domains of the Saccharomyces cerevisiae Mlh1p and Pms1p DNA mismatch repair proteins and their relevance to human hereditary nonpolyposis colorectal cancer-associated mutations. Mol Cell Biol. 1997;17:4465–4473. doi: 10.1128/mcb.17.8.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papadopoulos N, Nicolaides N C, Wei Y-F, Ruben S M, Carter K C, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M, Adams M D, Venter J C, Hamilton S R, Petersen G M, Watson P, Lynch H T, Peltomaki P, Mecklin J-P, de la Chapelle A, Kinzler K W, Vogelstein B. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 37.Parsons R, Li G-M, Longley M, Modrich P, Liu B, Berk T, Hamilton S R, Kinzler K W, Vogelstein B. Mismatch repair deficiency in phenotypically normal human cells. Science. 1995;268:738–740. doi: 10.1126/science.7632227. [DOI] [PubMed] [Google Scholar]
- 38.Prolla T A, Pang Q, Christie D M, Liskay R M. Dual requirement in yeast DNA mismatch repair for MLH1 and PMS1, two homologs of the bacterial mutL gene. Mol Cell Biol. 1994;14:407–415. doi: 10.1128/mcb.14.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reenan R A G, Kolodner R D. Isolation and characterization of two Saccharomyces cerevisiae genes encoding homologs of the bacterial HexA and MutS mismatch repair proteins. Genetics. 1992;132:963–973. doi: 10.1093/genetics/132.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reenan R A G, Kolodner R D. Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: evidence for separate mitochondrial and nuclear functions. Genetics. 1992;132:975–985. doi: 10.1093/genetics/132.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schweitzer J K, Livingston D M. Expansions of CAG repeat tracts are frequent in a yeast mutant defective in Okazaki fragment maturation. Hum Mol Genet. 1998;7:69–74. doi: 10.1093/hmg/7.1.69. [DOI] [PubMed] [Google Scholar]
- 42.Shcherbakova P V, Kunkel T A. Mutator phenotypes conferred by MLH1 overexpression and by heterozygosity for mlh1 mutations. Mol Cell Biol. 1999;19:3177–3183. doi: 10.1128/mcb.19.4.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sia E A, Jinks-Robertson S, Petes T D. Genetic control of microsatellite instability. Mutat Res. 1997;383:61–70. doi: 10.1016/s0921-8777(96)00046-8. [DOI] [PubMed] [Google Scholar]
- 44.Sia E A, Kokoska R J, Dominska M, Greenwell P, Petes T D. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol Cell Biol. 1997;17:2851–2858. doi: 10.1128/mcb.17.5.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strand M, Earley M C, Crouse G F, Petes T D. Mutations in the MSH3 gene preferentially lead to deletions within tracts of simple repetitive DNA in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1995;92:10418–10421. doi: 10.1073/pnas.92.22.10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strand M, Prolla T A, Liskay R M, Petes T D. Destabilization of tract of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 48.Studamire B, Price G, Sugawara N, Haber J E, Alani E. Separation-of-function mutations in Saccharomyces cerevisiae MSH2 that confer mismatch repair defects but do not affect nonhomologous-tail removal during recombination. Mol Cell Biol. 1999;19:7558–7567. doi: 10.1128/mcb.19.11.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Studamire B, Quach T, Alani E. Saccharomyces cerevisiae Msh2p and Msh6p ATPase activities are both required during mismatch repair. Mol Cell Biol. 1998;18:7590–7601. doi: 10.1128/mcb.18.12.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tishkoff D X, Boerger A L, Bertrand P, Filosi N, Gaida G M, Kane M F, Kolodner R D. Isolation and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci USA. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tishkoff D X, Filosi N, Gaida G M, Kolodner R D. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 52.Tran H T, Keen J D, Kricker M, Resnick M A, Gordenin D A. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol Cell Biol. 1997;17:2859–2865. doi: 10.1128/mcb.17.5.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tran H T, Gordenin D A, Resnick M A. The 3′→5′ exonucleases of DNA polymerases δ and ɛ and the 5′→3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2000–2007. doi: 10.1128/mcb.19.3.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tran P T, Liskay R M. Functional studies on the candidate ATPase domains of Saccharomyces cerevisiae MutLα. Mol Cell Biol. 2000;20:6390–6398. doi: 10.1128/mcb.20.17.6390-6398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Umar A, Buermeyer A B, Simon J A, Thomas D C, Clark A B, Liskay R M, Kunkel T A. Requirement for PCNA in DNA mismatch repair at a step preceding DNA synthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 56.Wierdl M, Greene C N, Datta A, Jinks-Robertson S, Petes T D. Destabilization of simple repetitive DNA sequences by transcription in yeast. Genetics. 1996;143:713–721. doi: 10.1093/genetics/143.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie Y, Counter C, Alani E. Characterization of the repeat-tract instability and mutator phenotypes conferred by a Tn3 insertion in RFC1, the large subunit of the yeast clamp loader. Genetics. 1999;151:499–509. doi: 10.1093/genetics/151.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xie Y, Liu Y, Argueso J L, Hendricksen L A, Kao H-I, Bambara R A, Alani E. Identification of rad27 mutations that confer differential defects in mutation avoidance, repeat-tract instability, and flap cleavage. Mol Cell Biol. 2001;21:4889–4899. doi: 10.1128/MCB.21.15.4889-4899.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]