Abstract
The Development of reliable and field-compatible detection methods is essential to monitoring and controlling the spread of any global pandemic. We herein report a novel anti-RNA:DNA hybrid (anti-RDH) antibody-based biosensor for visual, colorimetric lateral flow assay, using gold nanoparticles, coupled with transcription-mediated-isothermal-RNA-amplification (TMIRA) for specific and sensitive detection of viral RNA. We have demonstrated its utility for SARS-CoV-2 RNA detection. This technique, which we have named RDH-LFA (anti-RNA:DNA hybrid antibody-based lateral flow assay), exploits anti-RDH antibody for immunocapture of viral RNA hybridized with specific DNA probes in lateral flow assay. This method uses biotinylated-oligonucleotides (DNAB) specific to SARS-CoV-2 RNA (vRNA) to generate a vRNA-DNAB hybrid. The biotin-tagged vRNA-DNAB hybrid molecules bind to streptavidin conjugated with gold nanoparticles. This hybrid complex is trapped by the anti-RDH antibody immobilized on the nitrocellulose membrane resulting in pink color signal leading to visual naked-eye detection in 1 minute. Combining RDH-LFA with isothermal RNA amplification (TMIRA) significantly improves the sensitivity (LOD:10 copies/µl) with a total turnaround time of an hour. More importantly, RDH-LFA coupled with the TMIRA method showed 96.6% sensitivity and 100% specificity for clinical samples when compared to a commercial gold standard reverse-transcription quantitative polymerase-chain-reaction assay. Thus, the present study reports a rapid, sensitive, specific, and simple method for visual detection of viral RNA, which can be used at the point-of-care without requiring sophisticated instrumentation.
Keywords: visual, detection, transcription-mediated isothermal RNA amplification, RNA:DNA hybrid, antibody, SARS-CoV 2, viral, lateral flow assay
Significance Statement.
We herein report an anti-RNA:DNA hybrid (anti-RDH) antibody-based biosensor for visual, colorimetric lateral flow assay (RDH-LFA) for viral RNA detection. RDH-LFA exploits the high affinity and specificity of anti-RDH antibody for immunocapture of viral RNA in lateral flow assay. Combining RDH-LFA with one-pot transcription-mediated-isothermal-RNA-amplification significantly improves the sensitivity (LOD:10 copies/µl) with a turnaround time of an hour. Though we demonstrate the use of this sensor for SARS-CoV-2 RNA detection, it can be used for the detection of any RNA, which makes it a diverse diagnostic approach. Our method is 96.6% sensitive and 100% specific when evaluated using clinical samples; is isothermal, user-friendly, and can be used at the point of care without sophisticated instrumentation.
Introduction
Three pillars to control any new infectious disease are testing, contact tracing, and quarantine at the right time. This is also true for the COVID-19 pandemic. As the symptoms of common flu and COVID-19 overlap, testing remains the only way to diagnose COVID-19 and curb its spread. With more than two years since its inception, the disease is still spreading fast, and the battle is far from over. Though vaccines have been developed, there is still limited evidence of long-term immunity and protection from new variants. With SARS-CoV-2 accumulating mutations, periodic seasonal waves are a challenge to the mankind. Moreover, there is a persistent threat of new deadly RNA viruses invading our immune system. Viral RNA detection by qRT-PCR is highly sensitive, however, it is time-taking, requires technically trained staff, and costly infrastructure. Use of detection devices at the primary care level would be a key step for the rapid testing and identification of the infection, thereby preventing subsequent community transmission. Therefore, continuous efforts are needed to develop accurate, fast, sensitive and field-compatible detection methods which can be used at the Point of Care (POC) for viral diagnostics. A POC device is portable to avoid transport of samples to the central laboratory saving transportation time, does not require trained personnel, needs minimal instrumentation and sample processing for testing. The development of POC devices would make it possible to acquire testing results in out-of-laboratory settings in a minimum turnaround time. Recently, a number of approaches have been developed and explored which have the potential to be used as POC devices for pathogen detection. The most promising one is CRISPR technology using either Cas13a or Cas12a or Cas 9 enzymes with fluorescence or colorimetric detection format (1). However, there are still challenges associated with the use of CRISPR technology, such as, a limited number of possible detectable target sequences due to the requirement of protospacer adjacent motifs (PAM) specific sequences, mismatch tolerance leading to off-target effects, the requirement of guide RNA instead of DNA probes makes it susceptible to RNase degradation, and need for specialized bioinformatics tools to design the best guide RNA.
The methods using CAS proteins alone have lower sensitivity; therefore, it needs to be combined with different amplification approaches to improve the overall sensitivity of detection methods. The isothermal amplification approaches, such as Recombinase Polymerase amplification (RPA), Reverse-transcription loop-mediated-isothermal-amplification (RT-LAMP), etc. (2–4), have a significant advantage over conventional PCR amplification as specialized instrumentation is not needed. RT-LAMP uses two enzymes, RT and DNA polymerase having strand displacement activity with a set of four to six specially designed primers. However, the use of multiple primers in LAMP increases the chances of primer–dimer interactions, which may hinder the overall amplification efficiency. Further, the RPA amplification approach used in SHERLOCK (specific high-sensitivity enzymatic reporter unlocking)-based CRISPR detection system (5) requires the use of multiple enzymes (reverse transcriptase, RPA: recombinase/polymerase/strand displacement proteins, T7 RNA polymerase, and Cas13a) and synthetic fluorescent reporter constructs for detection, making it complicated and difficult to implement outside laboratory settings. In both RT-LAMP and RPA approaches, the resultant product is amplified DNA, therefore, a separate step may be needed for conversion to RNA using T7 RP, which makes it tedious to implement for applications where RNA is needed as a final product for detection.
An alternative approach for direct amplification of RNA is to use Transcription-mediated-isothermal-RNA-amplification (TMIRA) for increasing RNA copy number by exploiting only two enzymes, Reverse Transcriptase (RT) and T7 RNA polymerase (T7-RP), in a single tube under constant temperature (6). In this, RNA-dependent DNA polymerase activity of RT synthesizes cDNA from sense RNA, which remains bound together in form of RNA:DNA hybrid. Subsequently, RNA moiety in the hybrid is degraded and second strand of DNA is synthesized by exploiting RNase H endonuclease and DNA-dependent DNA polymerase activities of RT in sequential manner in the presence of two primers, one of them having T7 promoter sequence at the 5'-end. This dsDNA with T7 promoter sequence is utilized by T7-RP for transcription, where multiple RNA copies are produced. This whole reaction in a single tube undergoes multiple cycles in isothermal conditions giving rise to 107 times amplification of RNA. The resultant products in this reaction include dsDNA, ssRNA, and RNA:DNA hybrid molecules.
Lateral flow assay (LFA) is a popular paper-based POC diagnostic platform that can be easily performed without specific equipment, skills, or experience. LFA for viral RNA detection has been described earlier, where DNA oligos have been used to capture target RNA (7). However, the capture DNA oligos need to be modified with expensive chemical modifications to improve the immobilization efficiency. Moreover, secondary structures in the target RNA pose additional problem by hindering the hybridization process. The immobilization of protein-based biological material-like antibody on the lateral flow membrane takes place by direct physical absorption due to multiple hydrophobic and dipolar interactions. Here, we have developed a lateral flow assay using anti-RNA:DNA hybrid (anti-RDH) antibody, which binds to RNA:DNA hybrid (RDH) with very high affinity and specificity (8, 9). This technique, which we have named RDH-LFA (anti-RNA:DNA hybrid antibody-based lateral flow assay), exploits anti-RDH antibody for immunocapture of viral RNA in complex with specific biotinylated-DNA probes and streptavidin conjugated gold nanoparticles (strep-GNP) for visual, colorimetric lateral flow assay detection. For improved sensitivity, RDH-LFA is combined with TMIRA for viral RNA amplification. TMIRA uses complementary biotinylated-oligonucleotide primer specific to the viral RNA to give rise to a biotin-tagged-RDH TMIRA amplicons, which, when bound to anti-RDH antibody spotted on the LFA strip, can be visually detected by strep-GNP. We have demonstrated the use of this method for SARS-CoV-2 detection. Thus, the current study reports a simple, sensitive, specific, visual colorimetric, lateral flow-based biosensor for the viral RNA detection, without the requirement of sophisticated instrumentation.
Results and discussion
Scheme of RDH-LFA coupled with TMIRA
Lateral flow assays for viral RNA detection have been described earlier, where DNA oligos have been used to capture RNA (7). Anti-RNA:DNA hybrid antibody (anti-RDH) binds to RNA:DNA hybrid (RDH) with very high affinity and specificity (8, 9). Here, we have developed a simple, fast, visual colorimetric, anti-RDH antibody-based biosensor for lateral flow assay (RDH-LFA) coupled with TMIRA for the detection of viral RNA. This is an immunocapture-based approach that exploits the anti-RDH antibody for RDH capture in lateral flow assay (RDH-LFA) (Fig. 1A and 1B). We have demonstrated the use of this method for the detection of SARS-CoV-2, which is an RNA virus.
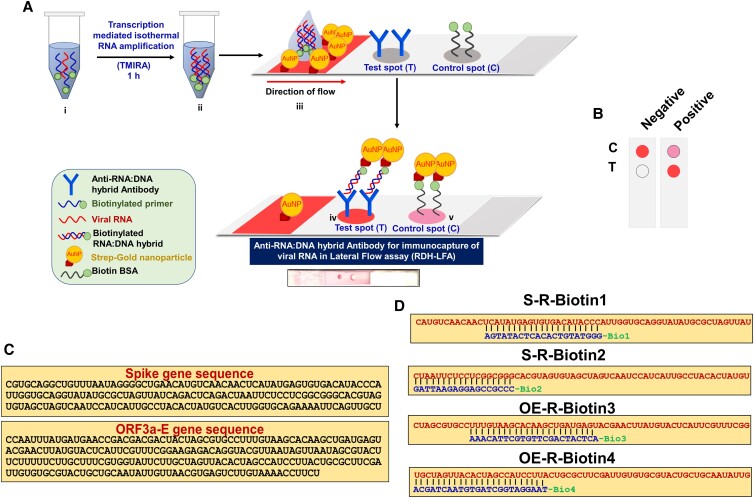
Fig. 1.
Scheme of RDH-LFA coupled with TMIRA. (A) Schematic representation showing workflow of anti-RDH antibody-based lateral flow assay (RDH-LFA) coupled with transcription-mediated-isothermal-RNA-amplification (TMIRA) for visual colorimetric detection of viral RNA. RDH-LFA exploits high affinity and specificity of anti-RDH antibody for immunocapture of viral RNA hybridized with specific DNA probes in lateral flow assay. To improve detection sensitivity, viral RNA is amplified (i) in a self-sustained isothermal amplification (TMIRA) for one hour in a one-pot reaction giving rise to biotin-tagged-RDH (ii), which is used in RDH-LFA (iii) and is concentrated at the test spot (T) where anti-RDH antibody is immobilized (iv). The result can be visualized in form of a colorimetric signal developed by aggregation of strep-GNP bound to the biotin moieties in RDH at the test spot (T) within one minute (iv). At the control spot, binding of spotted biotin BSA to strep-GNP will result in development of pink color irrespective of the presence or the absence of RNA (v). Refer to Figure S3 for detailed principle of TMIRA. (B) Interpretation of LFA: Development of pink color both at the test spot and control spot is an indication of positive signal while pink color only at the control spot represents a negative signal. (C) Specific target sequences in the genome of SARS-CoV-2 spanning the S (Spike), Orf3a and E genes, selected using in silico Blast analysis for specific detection of the viral RNA. (D) Sequences of four synthetic viral target RNA (Table S1) specific to SARS-CoV-2 RNA (in red) hybridized with corresponding complementary biotinylated-DNA probes (in blue) used for proof of principle experiments.
LFA strips used in this assay are assembled using a conjugate fiber, a nitrocellulose membrane, and an adsorbent pad. The anti-RDH antibody and Biotin-BSA are spotted on the lateral flow membrane at test spot (Spot-T, Fig. 1A) and control spot (Spot-C, Fig. 1A), respectively. For colorimetric detection, the conjugate fiber is coated with Streptavidin-linked-gold nanoparticles (strep-GNP) before the strip assembly. Refer to Methods section for complete details regarding fabrication of lateral flow strip.
For lateral flow assay, the viral RNA sample is mixed with complimentary biotinylated-oligonucleotides (DNAB) probes specific to SARS-CoV-2 RNA in a hybridization buffer to generate RNA:DNAB hybrid, which is used as an input in the assay. Once this mix is deposited at the conjugate fiber in the LFA strip, the RNA:DNAB hybrid is complexed with strep-GNP and moves along with buffer by capillary action towards the test spot where anti-RDH antibody is immobilized. The RNA:DNAB hybrid-strep-GNP complex is captured and is concentrated by lateral flow at the test spot, giving rise to a pink colorimetric signal within one minute developed by aggregation of a number of strep-GNP molecules (Strep–biotin interaction) bound to the biotin moieties in RDH. Biotin conjugated BSA at the control spot gives rise to a pink color spot (Spot-C, Fig. 1A) due to the direct binding of strep-GNP irrespective of viral RNA being present. Since the DNAB probe is carefully designed in such a way that it specifically binds to SARS-CoV2 RNA only and not any other target RNA, samples with the SARS-CoV2 RNA only will give rise to a positive signal, ensuring the high specificity of this method. Further, to improve the sensitivity, we have coupled RDH-LFA with a single tube amplification method, TMIRA (6), to increase the target RNA copy number. The principle of TMIRA is explained in detail in Figure S3. This method uses two thermostable enzymes (RT and T7-RP) in a single tube for target RNA amplification under isothermal conditions without the need of a costly real-time PCR machine. The resultant products in this reaction include dsDNA, RNA and RNA:DNA hybrid (RDH) molecules where RDH can be directly detected by the RDH-LFA method as explained above. The use of DNA gold-nano probes for pathogen detection has already been extensively explored (7, 10); however, this study exploits immunocapture of RDH by anti-RDH antibody and visualization by gold-nanoparticles in lateral flow assay (RDH-LFA) coupled with TMIRA, for viral RNA detection.
Probe designing and functionalization
In order to ensure high specificity of SARS-CoV-2 viral RNA detection, it is essential to design the probe sequences which bind to the viral RNA specifically. While designing the probes, we have taken utmost care to select the region which does not show significant similarity to the reference RNA from humans, other SARS viruses or common human pathogens (H1N1, Mycobacterium tuberculosis) to avoid false positive signal due to non-specific hybridization of probes with human RNA or RNA from other pathogens. In silico BLASTn analysis, queries of the individual regions (∼200 bp long) covering the entire SARS-CoV-2 RNA genome (GenBank accession no. MN908947) were performed against public domain nucleotide sequences in NCBI (National Center for Biotechnology Information) reference RNA sequences (refseq_rna) using default parameters with word size 7. Regions showing high similarity to human RNA, other SARS viruses or common human pathogens were screened out. Using these stringent criteria, we came across two specific target regions in the entire genome of SARS-CoV-2, which spanned the S, Orf3a and E genes (Fig. 1C). Accordingly, four different DNA probes complementary to these target regions were designed; two were specific to the S gene (S-R-Biotin1, S-R-Biotin2), the third one spans both Orf3a (OE-R-Biotin3) and E gene and the fourth one spans E gene (OE-R-Biotin4) sequences (Fig. 1D and Table S1). In addition, series of four viral RNA target sequences were selected spanning this specific region for the synthesis of synthetic viral RNA for testing proof of concept of RDH-LFA (Fig. 1D and Table S1). All the probes were functionalized with the biotin moiety at the 5'-end with a spacer arm [triethyleneglycol (TEG)] to avoid steric hindrance during target probe hybridization. The biotin moiety is required for binding to strep-GNP for signal development (Fig. 1A). For TMIRA, the amplification primers were designed from the selected specific regions (Table S1 and Fig. 1C). These TMIRA primers were again analyzed by in silico BLASTn analysis against public domain nucleotide sequences in NCBI (refseq_rna) using default parameters for similar small sequences to check for any significant similarity with human RNA, SARS-CoV virus or common human pathogens and no significant similarity was found.
Characterization of anti-RDH antibody binding specificity
Anti-RDH antibody binds to RDH with very high affinity (8, 9). For this study, it is important to test the binding specificity of the anti-RDH antibody for RDH. Therefore, we performed dot-blot assays using preformed RDH, ssDNA, only RNA, and dsDNA spotted on nitrocellulose membrane (Table S1 and methods). The dot blots with spotted samples were immuno-labeled with anti-RDH antibody and a secondary antibody conjugated with an IR dye. The development of a fluorescent signal is an indication of anti-RDH antibody binding to the spotted substrate. The fluorescent signal was obtained only for RDH, and serial dilution of RDH showed a proportionate decrease in the signal (Figs. 2A and B). These results demonstrated that anti-RDH antibody binds specifically to RDH and can potentially be used for specific RDH immunocapture in RDH-LFA.
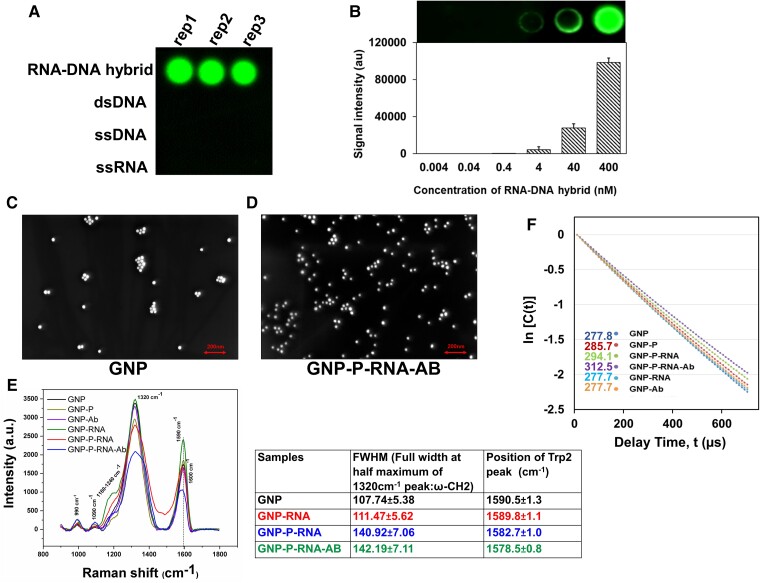
Fig. 2.
Validation of anti-RDH antibody. Dot-blot assays probed with anti-RDH antibody using (A) different nucleic acid substrates (B) different serial dilution of RDH and corresponding bar diagram. FESEM images of GNP in (C) absence and (D) presence of vRNA:DNAB:anti-RDH antibody complex (GNP-P-RNA-Ab) acquired at a magnification of 100.00 K X. (E) SERS data including graph (intensity vs Raman shift, cm−1) and table of FWHM (full width at half maximum) value of 1320 cm−1 peak and position of trp2 peak indicating Raman peak shift (cm−1) for different samples. (F) A scatterplot between Intensity autocorrelation function {ln [C(t)]} and delay time t(µs) using DLS. GNP-Ab and GNP-RNA were used as negative controls. The average relaxation time is mentioned on left hand side of the legend.The sample abbreviations used for (D) and (E) are: “strep-GNP” (GNP), “strep-GNP + biotinylated-DNAB probe” (GNP-P), “strep-GNP + biotinylated-DNAB probe + viral RNA” (GNP-P-RNA), “strep-GNP + biotinylated-DNAB probe + viral RNA + anti-RDH antibody” (GNP-P-RNA-Ab), “strep-GNP + anti-RDH antibody” (GNP-Ab), “strep-GNP + synthetic viral RNA” (GNP-RNA). Also see Figure S1.
Characterization of biotinylated-RDH binding to strep-GNP and anti-RDH antibody
RDH-LFA works on the principle of hybridization of specific biotinylated-DNA probes (DNAB) to the viral RNA (vRNA) for generation of vRNA:DNAB hybrid, the capture of this hybrid by the anti-RDH antibody, and then detection by binding of biotin moiety to strep-GNP. Here, vRNA:DNAB hybrid molecules were generated by binding of synthetic viral RNA (set of four, 60 b each) to corresponding biotinylated-DNA probes (Table S1) as described above. First, Field emission scanning electron microscope (FESEM) was used to assess the size distribution of strep-GNP with and without vRNA:DNAB:anti-RDH antibody complex. FESEM is a type of SEM that enables capturing of topographical and elemental information at very high magnification (up to 300,000x) and field depth. FESEM produces higher-resolution images with more clarity and lesser electrostatic distortion than conventional SEM. The FESEM (SEM, JEOL, Japan) (Fig. 2C and D, Figures S1A and S1B) images showed that both strep-GNP alone and strep-GNP-probe-RNA-antibody complex exhibited dispersed as well as agglomerated forms. Individual GNP molecules showed a uniform size distribution with an average diameter of ∼35 nm. Further, using Surface-Enhanced Raman Scattering (SERS) and Dynamic Light Scattering (DLS), we confirmed the generation of vRNA:DNAB hybrid molecules and binding of the hybrid molecules to strep-GNP (through strep–biotin interaction) and anti-RDH antibody.
Surface-enhanced Raman scattering
SERS is a surface-sensitive technique that reveals molecule-specific vibrational information generating a molecular fingerprint and offers information regarding molecular adsorption driving forces at nanoparticle surfaces and intermolecular interactions (11). The Raman spectra of Strep-GNP showed a few peaks, including two major peaks at 1320cm−1 (ω-CH2), and 1590 cm−1 (Trp2) (Fig. 2E and Figure S1C) (12). Upon addition of biotinylated-DNA probes (DNAB), additional peaks were observed corresponding to the functional groups in DNA molecules (Figure S1D) (13). Notably, upon addition of viral RNA (vRNA) to DNAB-strep-GNP, the Raman spectra showed a broadening of the peak at 1320 cm−1 and broadening as well as downshift of the peak at 1590 cm−1 (Fig. 2E and Figure S1E). This could be attributed to molecular conformation changes associated with the hybridization of vRNA with DNAB bound to strep-GNP. Addition of anti-RDH antibody to vRNA:DNAB-strep-GNP complex caused a further downshift of 1590 cm−1 peak indicating antibody interaction to the vRNA:DNAB-strep-GNP complex. The absence of these changes in negative controls, strep-GNP with antibody or RNA only (without DNAB probe) (Fig. 2E) indicates that these effects are due to specific hybridization of RNA to the complementary biotinylated-DNA bound to strep-GNP and further binding of anti-RDH antibody to the RDH molecules.
Dynamic light scattering
The binding of vRNA:DNAB hybrid with Strep-GNP and specific antibody was further confirmed by DLS. DLS is used to determine the diffusion coefficient (DC) and the size distribution profile (polydispersity index, PI) of small particles in suspension or polymers in solution (Methods) (14). The average relaxation time (indication of DC) for Strep-GNP was found to be 277.8 microseconds (PI:0.046) which increased to about 285.7 microseconds (PI:0.049) upon adding DNAB probe (Fig. 2F). Further sequential addition of vRNA:DNAB hybrid followed by anti-RDH antibody lead to an increase in relaxation time (294.1 and 312.5 microseconds, respectively). This increase in the relaxation time is an indication of slow diffusion of the particles due to its binding with RDH and specific antibody. On the other hand, in the presence of only RNA or antibody, both the decay (relaxation) time and PI remain unchanged (277.7 and 0.046), indicating that the increase is due to the specific binding of the vRNA to DNAB-strep-GNP and anti-RDH antibody to the RDH. Further, the average hydrodynamic diameter of strep-GNP showed a slight increase after addition of probe with RNA which further increased in the presence of anti-RDH antibody (Figure S1F), suggesting interaction of these ligands on the surface of strep-GNP.
These results confirmed the binding of vRNA-DNAB hybrid with strep-GNP and anti-RDH antibody, which are essential requirements for RDH-LFA to work successfully.
Proof of principle of RDH-LFA biosensor
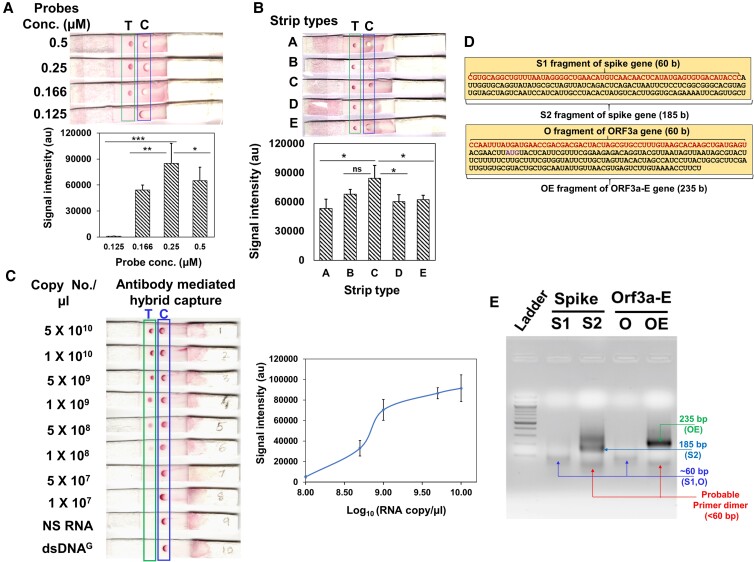
For optimization of RDH-LFA, we used a mix of the four synthetic viral RNAs and corresponding biotin DNA probes (Fig. 1D, Table S1). Four probes were used as multiple probes are known to give better sensitivity. Among the conditions assessed, probe concentration and the membrane wicking time were important for obtaining an efficient LFA signal (Figs. 3A and B, Figure S2A-C, Table S2). Using the optimized conditions (methods), we evaluated RDH-LFA sensitivity using serial dilutions of the four synthetic RNAs with a constant amount of corresponding probes in a 1X Hybridization buffer. The limit of detection (LoD), defined as the lowest conc. with a distinct higher signal than the negative control, was found to be 1×108 copies/µl (Fig. 3C). No signal was obtained with biotinylated-dsDNA, indicating that the antibody is specific to RDH, not dsDNA. Also, the signal was absent for non-specific human RNA ruling out non-specific hybridization of DNA probes with human RNA (Fig. 3C). Therefore, it was concluded that anti-RDH antibody can detect viral RNA as low as 1×108 copies/µl. Thus, these results show proof of principle for anti-RDH antibody-based lateral flow assay biosensor for colorimetric detection. We also compared the sensitivity of RDH-LFA with the capture DNA oligos approach, where DNA oligos are used to capture the RNA:DNA hybrid. For this, we spotted a mix of four capture oligos (Table S1) at the T-spot instead of anti-RDH antibody. Our results for synthetic viral RNA (concentration ranging from 1X109 to 5X1010 copies/μl) showed that RDH-LFA (LOD: 108 copies/µl, Figure S2D) is 100 times more sensitive than the oligo capture method (LOD: 1010 copies/µl, Figure S2D). The lower sensitivity of the oligo capture method could be due to inefficient binding of oligos to the membrane and secondary structures in the target RNA, which may hinder the hybridization process. Hence, RDH-LFA is a simple, more efficient, easy, and rapid (5 min) approach. However, the sensitivity is not sufficient for the diagnosis of poor viral loads; therefore, it can be used for testing of high viral load patients, e.g. at airport setup, where fast screening of asymptomatic super-spreader individuals (with high viral load) is essential.
Fig. 3.
Proof of principle of RDH-LFA and TMIRA optimization. (A–C): Proof of principle of RDH-LFA:5×109 copies/µl of synthetic viral RNA with four biotinylated-DNA probes (12.5 nM) in 20 µl 1X hybridization buffer, unless otherwise indicated, were used for RDH-LFA; images showing results for different (A) probe conc. (B) strip type; the membrane wicking time in sec (membrane thickness in mm) are 150(110), 200(110), 100(105), 170(100), 120(105), for A, B, C, D, E type, respectively. (C) Image showing colorimetric signal developed in RDH-LFA for different conc. of the synthetic viral RNAs mixed with the four biotinylated-DNA probes (Table S1). NS RNA is non-specific RNA from human source. The signal intensity values from test spot are plotted against synthetic RNA conc. Also refer to Figure S2. (D) Target sequences to be amplified using TMIRA: different target lengths were tested for amplification efficiency [S1 (60 bp) and S2 (185 bp) for spike-gene, and O (61 bp) and OE (235 bp) for Orf3a-e gene]. (e) Agarose gel showing TMIRA products: lane 1 is 100 bp ladder, lane 2 and 3 are 60 bp (S1) and 185 bp (S2) amplicons using spike gene RNA template (S) with forward primer (T7-S-F) and reverse primers (S1-R for S1 and S2-R for S2, respectively), lane 4 and 5 are 61 bp (O) and 235 bp (OE) amplicons using Orf-3a-E gene RNA template with forward primer (T7-OE-F) and reverse primers (O-R for O and OE-R for OE, respectively). Different bands in the lanes are labeled with their respective sizes for better clarity.
Amplification of viral target RNA by TMIRA
Since the lowest limit of detection for direct detection by RDH-LFA was found to be 1×108 copies/µl, we used TMIRA for the amplification of target RNA, which amplifies RNA directly under isothermal conditions using reverse transcriptase (RT) and T7 RNA polymerase (T7-RP) in a single tube (Figure S3) (6) The principle of TMIRA is distinct from “RT-LAMP”, “Recombinase polymerase-based amplification” or “RTF-EXPAR” (4, 5, 15–17). We have specifically used the TMIRA method as we need RDH as a final product for RDH-LFA. We selected the two regions screened earlier (Fig. 1C) as amplification target sequences in the viral genome, one in the spike gene and the other in the Orf3a-E gene (Fig. 3D, Table S1). Also, different target lengths were tested for amplification efficiency [S1 (60 bp) and S2 (185 bp) for Spike-gene, and O (61 bp) and OE (235 bp) for Orf3a-E gene]. We used thermostable RT and T7-RP to facilitate amplification at a higher temperature for better specificity. We used an RT (Roche) that has RNA-directed DNA polymerase, DNA-dependent DNA polymerase, and RNaseH activities, which are required for TMIRA to work successfully (Figure S3). Here, the forward amplification primer contained T7-RP promoter sequence (29 b at 5'-end) as an overhang for transcription reaction, with the other half (at 3'-end) specific and complimentary to the viral RNA sequence (Table S3). As described in the figure (Figure S3), the T7 promoter sequence is required for transcription of the target sequence by T7-RP. The final species obtained after complete amplification are dsDNA, ssRNA, and RDH (Figure S3). The starting conditions for amplification were decided based on multiple published reports (6, 18); however, further extensive optimization was needed for specific amplification of SARS-CoV-2 RNA targets to obtain improved yield. Synthetic viral RNA targets (109 copies/µl or ∼130 pg) and appropriate forward and reverse primers, were used for amplification.
The results showed that longer-length amplicons (S2 and OE) were amplified with better efficiency. Among S2 and OE, the efficiency of the OE target was better (Fig. 3E); therefore, we used this target for further optimization of TMIRA with respect to different NTP conc., buffer composition, incubation conditions, etc. It was concluded that the optimum conc. of primers, MgCl2, DTT, and NTP are critical for a successful TMIRA (Figures S4A–S4D, Table S3). Though previous reports (6) recommend addition of RNaseH to the amplification reaction, it did not make any difference to the amplification yield (Figure S4E), indicating that the inherent RNaseH activity of RT is sufficient for amplification.
Among the incubation conditions tested, 42°C for 60 minutes of incubation gave the highest amplification yield (Figure S4F, Table S3). The absence of any visible product in reactions without T7-RP, RT, or T7 Forward primer (Figure S4G) as against the one with all the components present, confirmed that the amplification is indeed mediated by the combined action of both enzymes. Finally, amplification of OE with optimized conditions showed significantly better amplification (Figure S4H) compared to the initial conditions (Fig. 3E). Final conditions used for TMIRA are described in detail in the materials and methods.
TMIRA improves sensitivity of RDH-LFA method
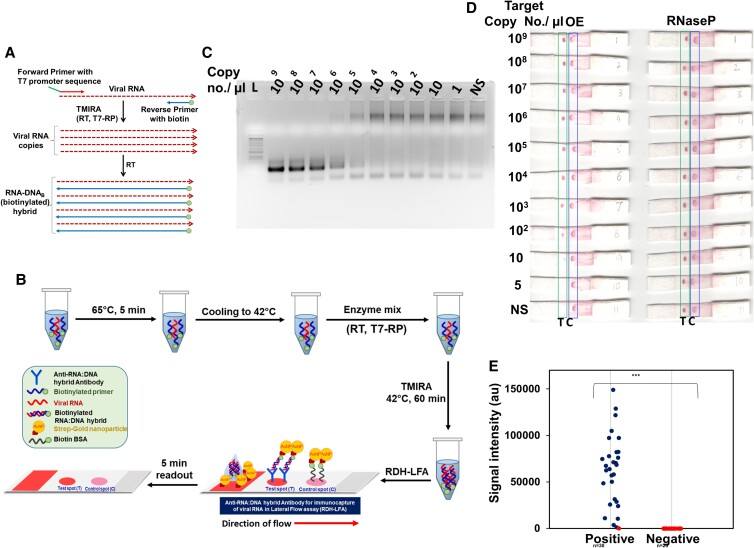
The use of 5'-biotin-tagged reverse primer during amplification results in the incorporation of Biotin primer in the RDH, giving rise to biotin-tagged-RDH, which can be detected by RDH-LFA using strep-GNP (Figs. 4A and B, Figure S3). TMIRA reaction products corresponding to 10-fold serial dilutions of synthetic viral RNA target (OE) mixed with a constant level of RNase P-RNA (internal positive control) were used for evaluating the performance of the TMIRA on RDH-LFA strips. Separate TMIRA reactions were set up for OE and RNase P. Agarose gel electrophoresis of OE showed a continuous decrease in the final product with decreasing RNA conc. (Fig. 4C). We observed a non-specific higher-sized band with no or low template, which disappeared in the absence of T7-RP primer or T7-RP (Figure S4I), suggesting that it may be due to the shift of T7-RP promoter primer DNA after binding T7-RP. Subsequently, for LFA detection, the amplicons mixed in hybridization buffer were dispensed on the RDH-LFA strips, separately for both OE and RNase P reactions (Fig. 4D). The signal at the test spot showed a concomitant decrease with decreasing OE target RNA copy number while the signal from RNase P reaction remained constant. The sample containing non-specific (NS) RNA did not result in any signal at the test spot, indicating that there is no non-specific signal from a human source (Fig. 4D). Using this method, we were able to detect the target RNA with LOD at 10 copies/µl in the sample. These results showed that combining RDH-LFA with TMIRA improves the sensitivity of detection by ∼107-fold and can be potentially used for ultra-sensitive viral RNA detection. This method is simple to perform and can be used at the point-of-care, with minimal technical expertise, and without sophisticated instrumentation; the only equipment required would be a heating block.
Fig. 4.
TMIRA improves sensitivity of RDH-LFA method. (A) Schematic representation of TMIRA, leading to amplification of target RNA in presence of forward primer having 5'-overhang containing T7-RP promoter sequence and reverse primer tagged with biotin at 5'-end giving rise to biotin-tagged-RDH (for details see Figure S3). (B) Experimental workflow of RDH-LFA coupled with TMIRA: Target RNA, specific primers and buffers are incubated at 65°C for 5 min for denaturation followed by annealing at 42°C. The thermostable enzymes mix (RT and T7-RP) along with NTPs are added to the reaction tube and incubated at 42°C for 60 min for a self-sustained isothermal amplification. The amplified product is then used for RDH-LFA and signal is read after 5 minutes. (C) Agarose gel showing TMIRA products by using 10-fold serial dilutions of the synthetic target RNA (OE) as template with forward primer (T7-OE-F) and reverse primer tagged with biotin at 5'-end (OE-R-Biotin4). (d) RDH-LFA Colorimetric signal developed by using TMIRA OE amplicons derived from (C); RNaseP (Human) RNA was used as an internal positive control which was spiked at constant level in the target RNA (See text for details). (E) Evaluation of sensitivity and specificity of RDH-LFA coupled with TMIRA using clinical samples (see Supp figure S5): Scatterplot showing signal intensities of test spot in RDH-LFA with TMIRA for samples from both Covid-19 patients (positive) and healthy individuals (negative) as previously determined by standard commercial qRT-PCR method. Among 30 RNA samples previously tested positive with qRT-PCR, 29 samples were found to be positive using RDH-LFA. In the scatterplot, the blue dot refers to a positive result and red dot refers to a negative result, using RDH-LFA coupled with TMIRA.
Evaluation of sensitivity and specificity of RDH-LFA coupled with TMIRA in Saliva spiked with RNA
RDH-LFA uses extracted RNA samples to proceed further. However, for a POC device, it may be useful to directly use the clinical nasopharyngeal and oropharyngeal swab samples. Therefore, we evaluated RDH-LFA-TMIRA efficiency using saliva spiked with synthetic RNA samples (Fig. S5A). Our results showed that LOD of RDH-LFA using saliva spiked with synthetic RNA is 100 copies/µl, which is still good. Therefore, while direct saliva can be used for low viral load, we recommend using pure RNA for RDH-LFA detection for very low viral load.
Evaluation of sensitivity and specificity of RDH-LFA coupled with TMIRA using clinical samples
For any method to be useful and promising for commercial use, it is essential to test it using clinical samples. Since qRT-PCR is the gold standard assay for SARS-CoV-2 RNA detection, we used a commercial qRT-PCR method (Truenat™) (19) for determining the clinical sensitivity and specificity of our method using clinical nasopharyngeal and oropharyngeal swab RNA samples. Using RDH-LFA coupled with the TMIRA method, we found 29 samples as positive among 30 COVID-19 patient RNA samples tested positive with qRT-PCR (ct value ≤ 32) (Figure S5B, Fig. 4E). Further, we did not find any detectable signal (Figure S5C, Fig. 4E, spot-T) for all the RNA samples (#25) tested as negative with qRT-PCR for COVID-19. Therefore, we conclude that RDH-LFA coupled with the TMIRA method offers 96.6% sensitivity and 100% specificity in clinical samples.
Conclusion
Taken together, we have developed a simple, sensitive, specific, fast, visual colorimetric, anti-RDH antibody-based biosensor for lateral flow assay (RDH-LFA) coupled with TMIRA for SARS-CoV-2 RNA detection. Anti-RNA:DNA hybrid antibody (anti-RDH), binds to RNA:DNA hybrid (RDH) with very high affinity and specificity (8, 9). Lateral flow assays for viral RNA detection using capture DNA oligos (instead of antibody) have been described earlier (7); however, we found that RDH-LFA works better with ∼100-fold higher analytical sensitivity compared to the oligo capture approach. The sensitivity of RDH-LFA increases considerably (LOD: 10 copies/µl) after it is combined with TMIRA having a total turnaround time of 1 h. While the cost of instrumentation, infrastructure, manpower requirements and basic set-up is low, the running reagent cost of our method is moderate (∼6 USD), which can be significantly reduced when it is manufactured at a bulk level.
The analytical sensitivity of RDH-LFA combined with the TMIRA method (LOD:10 copies/µl) is comparable to CRISPR-based LFA for SARS-CoV-2 detection, like SHERLOCK, DETECTR, etc. where the sensitivity ranges from 10 to 100 copies/µl (5, 15) (Table S4). The CRISPR-based methods are always combined with different amplification approaches such as RPA or RT-LAMP to improve the overall sensitivity of detection methods. For clinical samples for SARS-CoV-2 detection, the sensitivity of CRISPR-based methods combined with RNA amplification has been found to be in the range of 87.6–96.6% and specificity 95.5–100% (15, 20). The 96.6% sensitivity and 100% specificity for clinical samples using RDH-LFA coupled with TMIRA raises the hope for commercial use of our method. The isothermal RNA amplification technique used for RDH-LFA (TMIRA) is compatible with our method as RDH is one of the species produced in this reaction, which can be directly used for RDH-LFA detection. The high specificity of our method is due to the carefully designed specific primers, which will give rise to a TMIRA amplification product only in the presence of SARS-CoV-2 RNA. Additionally, TMIRA is simple as the whole reaction takes place in a single tube with only two enzymes and two primers at a constant temperature, which can be carried out by minimally skilled personnel. In contrast, the presence of off-target effects and the need for many enzymes (#5 or 6), use of many primers (#4 to 6), specialized guide RNAs with highly specific PAM sequence, fluorescent reporter constructs and multiple separate steps in CRISPR-based assays with RT-LAMP or RPA make them more complicated and difficult to implement outside laboratory settings.
In the future, RDH-LFA can also be combined with Smartphone-based Lateral Flow Device Reader apps (21), which can be used for obtaining results in quantitative information, digitization of results and subsequent sharing with stakeholders. CNT-based electrochemical detection for quantitative information or digitization of results can also be explored (22). Furthermore, though GNPs are very popular for colorimetric detection, other type of nanomaterials such as quantum dots, UCNP, and magnetic nanoparticles (23–25) with unique chemical and optical properties are also being explored at the laboratory scale by researchers for LFA detection. While these techniques are very promising because of their high sensitivity, they are still in their initial stages of development. Further research is needed to address the associated challenges and accelerate the clinical use of these nanomaterials. Simplified large-scale synthesis of nanomaterials with high sensitivity, reproducibility and long-term storage stability remains a significant challenge. In most of these approaches, a specialized instrument for fluorescent/magnetic/electrical signal may be required depending on the detection format, which needs to be simplified before they can be used as part of the POC device. Nonetheless, in the future, the gold nanoparticles can be combined or substituted by such nanomaterials to improve sensitivity, and add versatility or for multiplexing in the system. RDH-LFA can also be combined with Microfluidics-based automation/Artificial intelligence-based diagnostics approach using fully integrated microfluidic cartridges compatible with the point of care to perform multiplex measurements and for scaling up to increase the throughput (26, 27).
Overall, the significant unique advantages of our method are 1) its simplicity, 2) its compatibility with a simplified LFA detection approach makes it possible to be used as a POC device, 3) low capital cost as it does not require specialized instrumentation, highly trained personnel or costly infrastructure, 4) turnaround time of 1 h, being a POC testing device it will save on transportation time as well, 5) high analytical sensitivity, clinical sensitivity and specificity, and 6) adaptability to any RNA sequence, e.g. detection of any other RNA virus, cancer RNA biomarkers, etc. and therefore, has a long-term utility. As dsDNA is also one of the reaction intermediates in TMIRA, with minor modifications to the protocol, this method can also be applied for DNA amplification and detection, which makes it an adaptable diagnostics method with broad applications. The present goal is to make this method available at resource-limited locations in near-patient settings for disease diagnosis where sophisticated instrumentation may not be available.
Methods
Detailed materials and methods are provided as SI Appendix.
Supplementary Material
Acknowledgments
The authors are thankful to Dr. Tapan K. Ghanty and Dr. Hema Rajaram for the support and encouragement for this work. The authors are thankful to Archana Joshi Saha and R. Shashidhar for the useful discussions during this work. We are also thankful to K. V. Ravikanth, BARC for assistance with SEM microcsopy. This work is supported by Bhabha Atomic Research Centre, Mumbai, India.
Contributor Information
Anusree Dey, Molecular Biology Division, Bhabha Atomic Research Centre, Trombay, Mumbai, 400085, India; Homi Bhabha National Institute, Anushakti Nagar, Mumbai, 400094, India.
Jyoti Prakash, Materials Group, Bhabha Atomic Research Centre, Trombay, Mumbai, 400085, India; Homi Bhabha National Institute, Anushakti Nagar, Mumbai, 400094, India.
Rituparna Das, Molecular Biology Division, Bhabha Atomic Research Centre, Trombay, Mumbai, 400085, India; Homi Bhabha National Institute, Anushakti Nagar, Mumbai, 400094, India.
Sandeep Shelar, Chemistry Division, Bhabha Atomic Research Centre, Trombay, Mumbai, 400085, India.
Ajay Saini, Molecular Biology Division, Bhabha Atomic Research Centre, Trombay, Mumbai, 400085, India; Homi Bhabha National Institute, Anushakti Nagar, Mumbai, 400094, India.
Susan Cherian, Medical Division, Bhabha Atomic Research Centre, Anushakti Nagar, Mumbai, 400094, India.
Sofia C Patel, Medical Division, Bhabha Atomic Research Centre, Anushakti Nagar, Mumbai, 400094, India.
Puthusserickal A Hassan, Chemistry Division, Bhabha Atomic Research Centre, Trombay, Mumbai, 400085, India; Homi Bhabha National Institute, Anushakti Nagar, Mumbai, 400094, India.
Ashwini Khandekar, Medical Division, Bhabha Atomic Research Centre, Anushakti Nagar, Mumbai, 400094, India.
Kinshuk Dasgupta, Materials Group, Bhabha Atomic Research Centre, Trombay, Mumbai, 400085, India; Homi Bhabha National Institute, Anushakti Nagar, Mumbai, 400094, India.
Hari Sharan Misra, Molecular Biology Division, Bhabha Atomic Research Centre, Trombay, Mumbai, 400085, India; Homi Bhabha National Institute, Anushakti Nagar, Mumbai, 400094, India.
Sheetal Uppal, Molecular Biology Division, Bhabha Atomic Research Centre, Trombay, Mumbai, 400085, India; Homi Bhabha National Institute, Anushakti Nagar, Mumbai, 400094, India.
Ethics Approval Statement
This project was approved by the Medical Ethics committee, Bhabha Atomic Research Centre Hospital, Mumbai. Since there was no direct contact between the researcher and the patients, an exemption for consent was granted.
Supplementary Material
Supplementary material is available at PNAS Nexus online.
Funding
The authors declare no funding.
Author Contributions
A.d. conducted experiments, analyzed data and wrote the manuscript; J.P. carried out SEM and Raman experiments, analyzed data, and wrote the manuscript; R.D. conducted experiments and analyzed data; S.S. and P.A.H. provided Gold-nanoparticles for initial optimizations, helped with DLS experiments and analyzed data; A.S. designed the probes and wrote the manuscript; S.C. and S.C.P. collected samples did RNA isolation, qRT-PCR experiments; P.A.H. reviewed the manuscript and gave critical comments; A.K. recruited the patients; K.D. analyzed Raman data; H.S.M. reviewed the manuscript; S.U. conceived the study, designed the probes and amplification primers, conducted experiments, analyzed data, and wrote the manuscript; All the authors approved the final version.
Data Availability
All data associated with this study are present in the paper or the Supporting information.
References
- 1. Kumar P, et al. 2020. CRISPR-Cas System: an approach with potentials for COVID-19 diagnosis and therapeutics. Front Cell Infect Microbiol. 10:576875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. James AS, Alawneh JI. 2020. COVID-19 Infection diagnosis: potential impact of isothermal amplification technology to reduce community transmission of SARS-CoV-2. Diagnostics (Basel). 10(6):399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang WS, et al. 2021. Reverse transcription recombinase polymerase amplification coupled with CRISPR-Cas12a for Facile and highly sensitive colorimetric SARS-CoV-2 detection. Anal Chem. 93(8):4126–4133. [DOI] [PubMed] [Google Scholar]
- 4. Rabe BA, Cepko C. 2020. SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc Natl Acad Sci U S A. 117(39):24450–24458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Patchsung M, et al. 2020. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat Biomed Eng. 4(12):1140–1149. [DOI] [PubMed] [Google Scholar]
- 6. Fahy E, Kwoh DY, Gingeras TR. 1991. Self-sustained sequence replication (3SR): an isothermal transcription-based amplification system alternative to PCR. PCR Methods Appl. 1(1):25–33. [DOI] [PubMed] [Google Scholar]
- 7. Rohrman BA, Leautaud V, Molyneux E, Richards-Kortum RR. 2012. A lateral flow assay for quantitative detection of amplified HIV-1 RNA. PLoS One. 7(9):e45611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bou-Nader C, Bothra A, Garboczi DN, Leppla SH, Zhang J. 2022. Structural basis of R-loop recognition by the S9.6 monoclonal antibody. Nat Commun. 13(1):1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boguslawski SJ, et al. 1986. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J Immunol Methods. 89(1):123–130. [DOI] [PubMed] [Google Scholar]
- 10. Moitra P, Alafeef M, Dighe K, Frieman MB, Pan D. 2020. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 14(6):7617–7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Phan HT, Haes AJ. 2018. Impacts of pH and intermolecular interactions on surface-enhanced Raman scattering chemical enhancements. J Phys Chem C. 122(26):14846–14856. [Google Scholar]
- 12. Galarreta BC, Norton PR, Lagugné-Labarthet F. 2011. SERS Detection of streptavidin/biotin monolayer assemblies. Langmuir. 27(4):1494–1498. [DOI] [PubMed] [Google Scholar]
- 13. Langlet M, et al. 2012. Elaboration of an Ag°/TiO2 platform for DNA detection by surface enhanced Raman spectroscopy. Surface Sci. 606(23–24):2067–2072. [Google Scholar]
- 14. Hassan PA, Rana S, Verma G. 2015. Making sense of brownian motion: colloid characterization by dynamic light scattering. Langmuir. 31(1):3–12. 10.1021/la501789z. [DOI] [PubMed] [Google Scholar]
- 15. Broughton JP, et al. 2020. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 38(7):870–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ganguli A, et al. 2020. Rapid isothermal amplification and portable detection system for SARS-CoV-2. Proc Natl Acad Sci U S A. 117(37):22727–22735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carter JG, et al. 2021. Ultrarapid detection of SARS-CoV-2 RNA using a reverse transcription-free exponential amplification reaction, RTF-EXPAR. Proc Natl Acad Sci U S A. 118(35):e2100347118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guatelli JC, et al. 1990. Isothermal, in vitro amplification of nucleic acids by a multienzyme reaction modeled after retroviral replication. Proc Natl Acad Sci U S A. 87(5):1874–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gupta N, Rana S, Singh H. 2020. Innovative point-of-care molecular diagnostic test for COVID-19 in India. Lancet Microbe. 1(7):e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Selvam K, et al. 2021. RT-LAMP CRISPR-Cas12/13-Based SARS-CoV-2 detection methods. Diagnostics (Basel). 11(9):1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kadam R, et al. 2020. Target product profile for a mobile app to read rapid diagnostic tests to strengthen infectious disease surveillance. PLoS One. 15(1):e0228311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Prakash J, et al. 2021. Label-free rapid electrochemical detection of DNA hybridization using ultrasensitive standalone CNT aerogel biosensor. Biosens Bioelectron. 1(191):113480. [DOI] [PubMed] [Google Scholar]
- 23. Zhang Q, et al. 2022. SARS-CoV-2 detection using quantum dot fluorescence immunochromatography combined with isothermal amplification and CRISPR/Cas13a. Biosens Bioelectron. 202:113978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu K, et al. 2020. Magnetic-nanosensor-based virus and pathogen detection strategies before and during COVID-19. ACS Appl Nano Mater. 3(10):9560–9580. [DOI] [PubMed] [Google Scholar]
- 25. Alexaki K, et al. 2022. A SARS-cov-2 sensor based on upconversion nanoparticles and graphene oxide. RSC Adv. 12(29):18445–18449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kaushik AK, et al. 2020. Electrochemical SARS-CoV-2 sensing at point-of-care and artificial intelligence for intelligent COVID-19 management. ACS Appl Bio Mater. 3(11):7306–7325. [DOI] [PubMed] [Google Scholar]
- 27. Alexander R, et al. 2022. Machine learning approach for label-free rapid detection and identification of virus using Raman spectra. Intelligent Medicine. In Press. 10.1016/j.imed.2022.10.001 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are present in the paper or the Supporting information.