Abstract
Snail family genes encode DNA binding zinc finger proteins that act as transcriptional repressors. Mouse embryos deficient for the Snail (Sna) gene exhibit defects in the formation of the mesoderm germ layer. In Sna−/− mutant embryos, a mesoderm layer forms and mesodermal marker genes are induced but the mutant mesoderm is morphologically abnormal. Lacunae form within the mesoderm layer of the mutant embryos, and cells lining these lacunae retain epithelial characteristics. These cells resemble a columnar epithelium and have apical-basal polarity, with microvilli along the apical surface and intercellular electron-dense adhesive junctions that resemble adherens junctions. E-cadherin expression is retained in the mesoderm of the Sna−/− embryos. These defects are strikingly similar to the gastrulation defects observed in snail-deficient Drosophila embryos, suggesting that the mechanism of repression of E-cadherin transcription by Snail family proteins may have been present in the metazoan ancestor of the arthropod and mammalian lineages.
Genes of the Snail family encode zinc finger proteins that function as transcriptional repressors in a variety of experimental systems (8, 10, 12, 16, 17, 21; reviewed in reference 11). The first gene of this family studied was the Drosophila snail gene, which is one of two genes required zygotically for mesoderm formation during Drosophila embryogenesis (1, 5, 9, 13, 24; reviewed in reference 19). Embryos homozygous for null mutations of snail exhibit defects in mesoderm formation, gastrulation movements, and germ band retraction (9, 24). The snail protein is a transcriptional repressor which acts to maintain proper germ layer boundaries by repressing the expression within the mesoderm of regulatory genes involved in ectodermal development (18). Snail family genes are evolutionarily conserved, and studies have implicated Snail family proteins in the regulation of epithelial-mesenchymal transitions in tissue culture systems and in both vertebrate and invertebrate embryos (3, 7, 17, 19, 20, 23, 26, 27).
Two mouse homologs of snail, termed Sna and Slug, have been cloned (15, 22, 28, 30). It has been previously demonstrated that mice homozygous for a null mutation of the Slug gene are viable, although they exhibit postnatal growth deficiency (15). We describe here the construction and analysis of a targeted mutation of the Sna gene. During gastrulation, Sna is expressed in the primitive streak and the mesoderm germ layer (22, 30). Sna-deficient mouse embryos die early in gestation, exhibiting defects in gastrulation and mesoderm formation.
MATERIALS AND METHODS
Gene targeting.
The Sna targeting vector was constructed from an 18-kb genomic clone containing the entire Sna gene (14). The 5′ arm was a 2.5-kb SalI-NruI genomic fragment subcloned upstream of a PGK-neo expression cassette. The 3′ arm was a 1.2-kb XbaI-EcoRI fragment. This resulted in the deletion of a 1.6-kb genomic fragment containing exons 1 and 2 of the Sna gene, which deletes the translation initiation site and amino acids 1 to 203 of the Sna protein, including degenerate zinc finger 1 and zinc fingers 2 and 3 of the DNA binding domain. A herpes simplex virus (HSV)-tk cassette was introduced for negative selection. Embryonic stem (ES) cell electroporation and selection and blastocyst injections were performed as previously described (31). DNAs from individual ES cell colonies were prescreened by PCR, and positive colonies were then screened by Southern blotting, using a 0.5-kb EcoRI-SphI genomic fragment as a probe on SphI-digested genomic DNA. Germ line transmission of the Sna mutant allele was obtained for two independently targeted clones. The official nomenclature for this mutant allele is Snatm1Grid.
Histology, in situ hybridization, and immunofluorescence.
Embryos were dissected at embryonic day 7.5 (E7.5) from timed matings of Sna+/− heterozygotes. Mutant homozygotes were identified by allele-specific PCR or by their characteristic morphology. A strict correlation was observed between genotype and the characteristic Sna−/− mutant phenotype. Some embryos were sectioned in their decidua for histological analysis. Decidua and isolated embryos were fixed in Bouin's fixative for histological analysis. Fixed embryos were dehydrated through graded alcohols, embedded in paraffin, sectioned, and stained with hematoxylin and eosin. Embryos for in situ hybridization were fixed overnight at 4°C in 4% paraformaldehyde in phosphate-buffered saline. Whole-mount in situ hybridization was performed as previously described (15). At least three Sna−/− mutant embryos were analyzed for each of the probes. For analysis of E-cadherin RNA expression, embryos were embedded in plastic resin after whole-mount in situ hybridization and sectioned.
Sna−/− mutant embryos and control littermates were stained at E7.5 in whole mount with a monoclonal anti-E-cadherin antibody (Zymed). Antibody staining was detected with fluorescein-conjugated anti-rat immunoglobulin G (Jackson ImmunoResearch). Whole-mount embryos were cut into thick sections using electrolytically sharpened tungsten needles, mounted in Vectashield mounting medium (Vector Laboratories), and examined with a fluorescent microscope.
Transmission electron microscopy.
Embryos were fixed overnight in 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.2). After being washed, embryos were postfixed in 1% osmium tetroxide in 0.1 M phosphate buffer. Embryos were washed, dehydrated in an ethanol series, and treated with propylene oxide. Embryos were infiltrated with Epon-araldite, and ultrathin sections were cut. Specimens were imaged on a JEOL 100CXII transmission electron microscope.
RESULTS
Disruption of the mouse Sna gene.
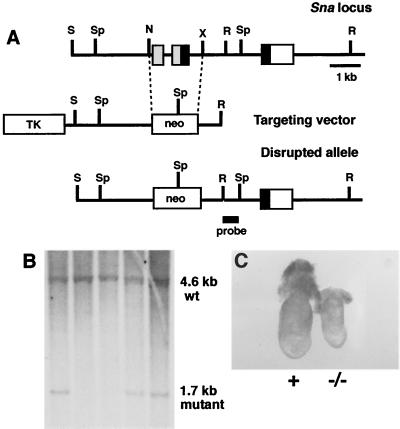
To analyze the role of the Sna gene during embryogenesis in mice, we used gene targeting to construct a mutant allele from which exons 1 and 2 of the Sna gene had been deleted (Fig. 1A). This deletion removes the exons encoding the translation initiation site and amino acids 1 to 203 of the 264-amino-acid Sna protein, including degenerate zinc finger 1 and zinc fingers 2 and 3 of the DNA binding domain. Germ line transmission of the Sna mutant allele was obtained for two independently targeted clones (Fig. 1B). Heterozygous Sna+/− mice appeared normal.
FIG. 1.
Targeted disruption of the mouse Sna gene. (A) Targeting scheme. The upper line shows the genomic organization of the Sna gene (14). The three exons are indicated by boxes. The region encoding the amino terminus of the Sna protein is indicated by gray boxes, the region encoding the zinc fingers is indicated by black boxes, and the 3′ untranslated region is indicated by a white box. The middle line represents the structure of the targeting vector. The lower line represents the predicted structure of the Sna locus following homologous recombination of the targeting vector. The probe used for Southern blot analysis is indicated. N, NruI; R, EcoRI; S, SalI; Sp, SphI; X, XbaI; TK, thymidine kinase. (B) DNAs isolated from targeted ES cells were digested with SphI, blotted, and hybridized with the indicated probe. Wild-type (wt) and mutant hybridization bands are indicated. Three independently targeted ES cell clones are shown. (C) Whole-mount morphology of a Sna−/− embryo (right) and a control littermate embryo (left) at E7.5. In all figures, normal littermate embryos were either Sna+/− or Sna+/+ and are indicated with a plus sign.
No homozygous Sna−/− mice were found among the progeny of the intercross of heterozygous Sna+/− mice, indicating that our Sna mutant allele is a recessive lethal mutation. To determine when homozygous mutant embryos were dying, embryos were isolated from timed matings. At E6.5, Sna−/− mutant embryos were not distinguishable from the embryos of heterozygous and wild-type littermates. At E7.5, however, the homozygous mutant embryos were smaller than the embryos of their littermates (Fig. 1C). By E8.5, the Sna−/− embryos were severely retarded compared to those of littermates and were being resorbed (data not shown).
Sna−/− mutant embryos form a mesoderm cell layer.
Histological analysis of Sna−/− mutant embryos at E7.5 demonstrated the presence of three germ layers (see below), indicating that a mesoderm layer had formed in the mutants. This was confirmed by analysis of the expression of several marker genes (Fig. 2). In wild-type embryos, the Brachyury (T) gene is expressed in the primitive streak and in the rostral axial mesoderm (32). In the Sna−/− embryos, expression levels of the T gene were reduced compared to those of the controls and expression did not extend as far rostrally in the embryo (Fig. 2A and B). The Lim1 gene in wild-type embryos is expressed in the primitive streak and the mesodermal wings (2). Lim1 was expressed in both of these structures in the Sna−/− embryos (Fig. 2C and D). In wild-type embryos at the prestreak and early streak stages, the Otx2 gene is expressed throughout the epiblast but its expression gradually becomes restricted to the anterior neuroectoderm (29). In Sna−/− mutants, Otx2 expression did not become anteriorly restricted (Fig. 2E and F). The Cer1 gene is expressed in wild-type embryos in the anterior visceral endoderm and the definitive endoderm (4). Cer1 was expressed in these tissues in Sna−/− embryos, but expression levels were reduced compared to those of the controls (Fig. 2G and H). These marker studies confirm that the mesoderm, ectoderm, and endoderm all differentiate in Sna−/− mutant embryos, although some differences in expression levels or details of the expression patterns were detected in the mutants.
FIG. 2.
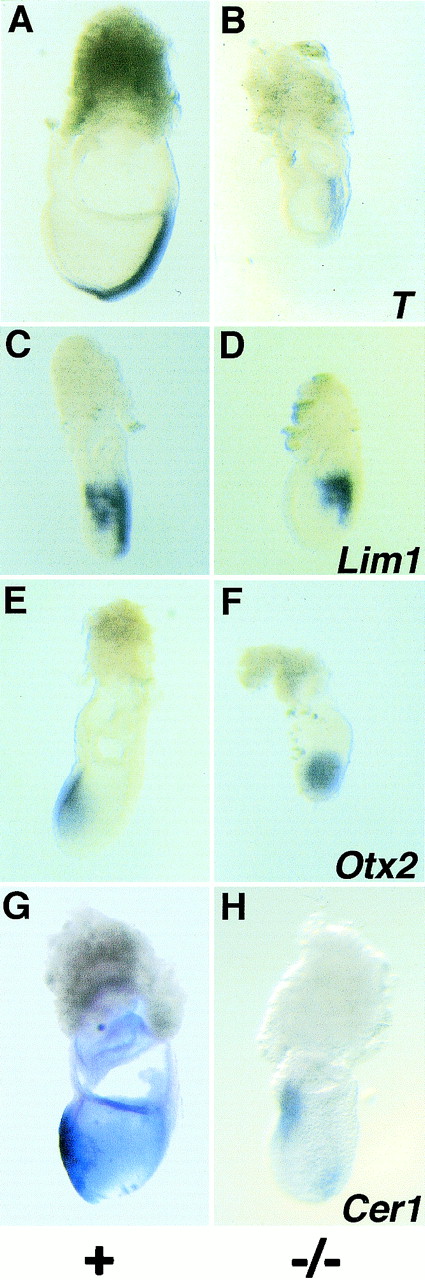
Analysis of marker gene expression in Sna−/− mutant embryos at E7.5. (A and B) T expression. T is expressed in axial mesoderm cells, and expression extends rostrally from the node (A). In the Sna−/− embryo, T is expressed, but at lower levels than in the control embryos, and does not extend as far rostrally in the embryo (B). (C and D) Lim1 expression. Lim1 is expressed in the primitive streak and the mesodermal wings (C). Lim1 is expressed in these tissues in the Sna−/− embryo (D). (E and F) Otx2 expression. Otx2 is expressed in the visceral endoderm and epiblast, and expression is gradually restricted to the anterior third of the embryo as the primitive streak extends (E). In the Sna−/− mutant, Otx2 expression is not restricted to the anterior portion of the embryo (F). (G and H) Cer1 expression. Cer1 is expressed in the anterior visceral endoderm and the definitive endoderm (G). In the Sna−/− embryos, Cer1 expression is reduced (H). All embryos are oriented with the anterior side towards the left.
The Sna−/− mutant mesoderm retains epithelial characteristics.
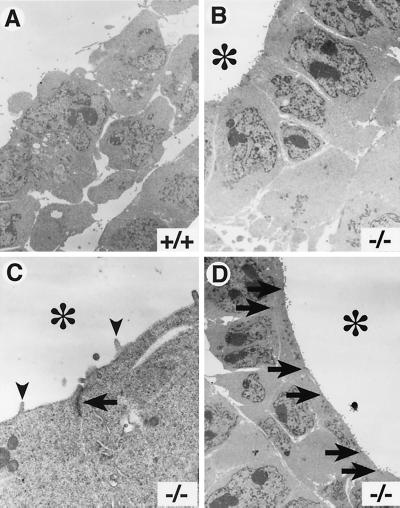
While histological and marker analyses clearly indicated that a mesoderm cell layer differentiated in the Sna−/− mutant embryos, morphological abnormalities were apparent in the mutant mesoderm. In wild-type and heterozygous embryos by E7.5, the mesoderm cell layer had delaminated from the primitive streak and had migrated anteriorly between the embryonic ectoderm and the visceral endoderm to form the mesodermal wings (Fig. 3A, C, and E). Cells in the mesoderm layer of these embryos had a morphology characteristic of that of mesenchymal cells. In Sna−/− mutant embryos, a primitive streak and a mesoderm layer formed and the cells migrated anteriorly to form the mesodermal wings. However, many of the mutant mesoderm cells did not have a characteristic mesenchymal morphology (Fig. 3B, D, and F). In most Sna−/− mutant embryos, cavities or lacunae formed in the mesoderm layer (Fig. 3D and F) and the mesoderm cells abutting these lacunae exhibited an epithelial morphology. The cells lining these lacunae had the appearance of a columnar epithelium (Fig. 3F and 4B). Transmission electron microscopic analysis revealed that the mutant mesoderm exhibited apical-basal polarity, which is typically observed in an epithelial cell layer. These cells contained microvilli along the apical surface (i.e., the surface facing into the lacunae) (Fig. 4C) and contained electron-dense adhesive junctions that resembled adherens junctions (Fig. 4C and D).
FIG. 3.
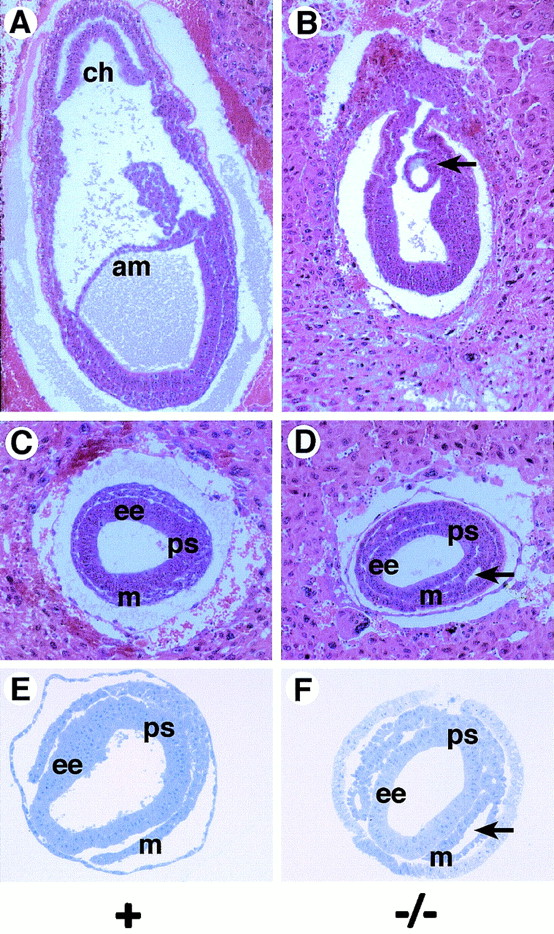
Morphological abnormalities in the Sna−/− mutant mesoderm. (A and B) Sagittal sections of embryos at E7.5. In the Sna−/− mutant embryo (B), a posterior amniotic fold forms (arrow) but no amnion or chorion is formed. (C to F) Transverse sections of embryos at E7.5. In Sna−/− mutant embryos, lacunae form within the mesoderm layers (arrows in D and F). Mesoderm cells lining these lacunae exhibit an epithelial morphology. (A to D) Hematoxylin-and eosin-stained paraffin sections. (E and F) Toluidine blue-stained plastic sections. Abbreviations: am, amnion; ch, chorion; ee, embryonic ectoderm; m, mesoderm; ps, primitive streak.
FIG. 4.
Apical-basal polarity and adhesive junctions in the Sna−/− mutant mesoderm. Transmission electron microscopic images for analysis of wild-type (A) and Sna−/− (B to D) embryos at E7.5. are shown. (A) Mesoderm cells in wild-type embryos exhibit a typical mesenchymal morphology. (B) In Sna−/− embryos, mesoderm cells lining the lacunae exhibit an ordered, columnar morphology. (B to D) The lumens of the lacunae are indicated with asterisks. (C) Mesoderm cells in the mutant embryos have microvilli (arrowheads) at the apical surface and exhibit electron-dense adhesive junctions (arrow) between the cells. (D) Adhesive junctions are numerous in the Sna−/− mutant mesoderm. The positions of intercellular adhesive junctions are indicated by arrows. Approximate magnifications: (A and B) ×4,000, (C) ×20,000, (D) ×3,000.
E-cadherin expression is not downregulated in the Sna−/− mutant mesoderm.
Recent work has shown that Sna expression represses E-cadherin transcription in cultured epithelial cell lines by binding to E boxes present in the E-cadherin promoter region and that Sna overexpression causes epithelial cell lines to adopt a fibroblast-like morphology and to acquire tumorigenic and invasive properties (3, 7). E-cadherin protein is a component of adherens junctions, and downregulation of E-cadherin expression in cells in the primitive streak is believed to be important for gastrulation in vertebrates (6). We therefore analyzed expression of the E-cadherin gene by in situ hybridization of Sna−/− mutant embryos and littermate controls (Fig. 5A to C). In the control embryos, E-cadherin expression was downregulated in the mesoderm (Fig. 5A and B). In Sna−/− embryos, E-cadherin RNA expression was maintained in the mesoderm layer (Fig. 5A and C). However, the levels of E-cadherin RNA observed in the mutant mesoderm were lower than those observed in the embryonic ectoderm. We also examined whether E-cadherin protein expression was maintained in the mesoderm of Sna−/− embryos. This analysis revealed that, as we observed with E-cadherin RNA, expression of E-cadherin protein was retained in the mesoderm of the Sna−/− embryos but at lower levels than were observed in the embryonic ectoderm (Fig. 5D to G).
FIG. 5.
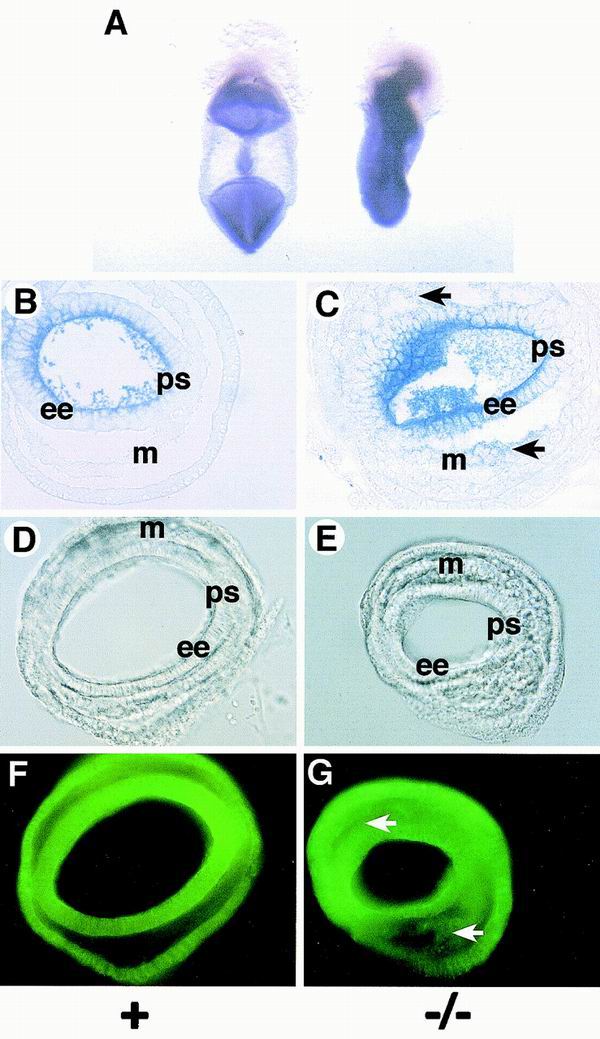
E-cadherin expression is retained in the mesoderm of Sna−/− mutant embryos. (A) Whole-mount in situ hybridization with E-cadherin antisense riboprobes of a control littermate (left) and a Sna−/− embryo (right). (B and C) Plastic sections of embryos treated as described for panel A. E-cadherin RNA expression is downregulated in the mesoderm of the control littermate embryo (B), but expression is retained (arrows) in the mesoderm of the Sna−/− embryo (C). (D to G) Immunofluorescence with anti-E-cadherin monoclonal antibody. (D and E) Nomarski optics. (F and G) Fluorescence optics. In the Sna−/− embryo (G), E-cadherin protein expression is retained in the mesoderm layer (arrows). Abbreviations: ee, embryonic ectoderm; m, mesoderm; ps, primitive streak.
DISCUSSION
We describe here the construction and analysis of a targeted null mutation of the mouse Sna gene. Although previous work had demonstrated that the related Slug gene is not essential for embryogenesis in mice (15), Sna−/− mutant embryos die early in gestation. The mutant embryos exhibit defects in gastrulation and in the epithelial-mesenchymal transition required for generation of the mesoderm cell layer. Our data indicate that formation of the mesoderm cell layer can occur despite the retention of E-cadherin expression. However, many cells in the mesoderm of Sna−/− mutant embryos retain apical-basal polarity and an epithelial morphology, presumably due to the retention of adherens junctions between the mesoderm cells in the mutant embryos. The phenotypic defects we observe in the Sna−/− mutant mouse embryos are strikingly similar to the gastrulation defects observed in snail mutant Drosophila embryos (25). The Drosophila E-cadherin gene is normally expressed in the epithelial cells of the cellular-blastoderm-stage embryo but is then downregulated in mesoderm precursor cells prior to invagination. In Drosophila embryos homozygous for a snail null mutation, E-cadherin downregulation does not occur and mesoderm precursors in the ventral region of the embryo retain adherens junctions and apical-basal polarity (25). As noted by Wolpert (33), the morphogenetic movements of gastrulation are more highly conserved than the establishment of the body plan during evolution. The similarity of the gastrulation defects in mutant Snail genes of both Drosophila and mice indicates that the molecules regulating mesoderm formation and gastrulation movements are conserved over an extremely wide evolutionary distance. This observation suggests that repression of E-cadherin transcription by Snail family proteins may have been an ancestral condition in the metazoan precursor to the arthropod and mammalian lineages.
Our studies provide the first genetic evidence that the Sna gene functions as a key regulator of the epithelial-mesenchymal transition in mice. Our data show that, as was found in cultured cells and in metastatic carcinomas (3, 7), the E-cadherin gene is a target for repression by the Sna protein. However, the level of expression of E-cadherin in the mesoderm of Sna−/− mutant embryos is considerably less than the level of E-cadherin RNA expression in the embryonic ectoderm of these embryos. This finding suggests that other regulators of E-cadherin transcription may not be maintained in the mesoderm of Sna−/− mutant embryos. For example, the embryonic ectoderm may express a positive regulator of E-cadherin transcription and this positive regulator might not be expressed in the mesoderm of the Sna−/− mutant embryos.
It is intriguing that despite the retention of E-cadherin expression and intercellular adherens junctions, a primitive streak forms and the mesoderm layer delaminates in Sna−/− mutant embryos. This may be due to the fact that these regions express distinctly lower levels of E-cadherin RNA than are expressed in the embryonic ectoderm. It would be interesting to overexpress E-cadherin in the primitive streak and the mesoderm to test whether higher levels of E-cadherin expression would entirely prevent streak formation and mesoderm delamination.
ACKNOWLEDGMENTS
We thank B. Holdener, J. Mercer, and M. Shen for helpful discussions; L. Bechtold and P. Finger for transmission electron microscopic analysis and plastic sectioning; G. Martin for help with fluorescent microscopy; C. Norton for technical assistance; S. Ang, R. Behringer, B. Hermann, and R. Kemler for in situ probes; and S. Ackerman and T. O'Brien for reading the manuscript.
This work was supported by a grant (HD34883) from the NIH to T.G. and a subcontract to T.G. under NIH Project Center grant DE13078 from Johns Hopkins University. This work was also supported by a training grant (CA09217) (E.A.C. and Y.L.) and a Core grant (CA34196) from the National Cancer Institute to the Jackson Laboratory.
E.A.C. and R.J. contributed equally to this work.
REFERENCES
- 1.Alberga A, Boulay J-L, Kempe E, Dennefeld C, Haenlin M. The snail gene required for mesoderm formation in Drosophila is expressed dynamically in derivatives of all three germ layers. Development. 1991;111:983–992. doi: 10.1242/dev.111.4.983. [DOI] [PubMed] [Google Scholar]
- 2.Barnes J D, Crosby J L, Jones C M, Wright C V E, Hogan B L M. Embryonic expression of Lim-1, the mouse homolog of Xlim-1, suggests a role in lateral mesoderm differentiation and neurogenesis. Dev Biol. 1994;161:168–178. doi: 10.1006/dbio.1994.1018. [DOI] [PubMed] [Google Scholar]
- 3.Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia de Herreros A. The transcription factor Snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 4.Belo J A, Bouwmeester T, Leyns L, Kertesz N, Gallo M, Follettie M, De Robertis E M. Cerberus-like is a secreted factor with neutralizing activity expressed in the anterior primitive endoderm of the mouse gastrula. Mech Dev. 1997;68:45–57. doi: 10.1016/s0925-4773(97)00125-1. [DOI] [PubMed] [Google Scholar]
- 5.Boulay J L, Dennefeld C, Alberga A. The Drosophila developmental gene snail encodes a protein with nucleic acid binding fingers. Nature. 1987;330:395–398. doi: 10.1038/330395a0. [DOI] [PubMed] [Google Scholar]
- 6.Burdsal C A, Damsky C H, Pedersen R A. The role of E-cadherin and integrins in mesoderm differentiation and migration at the mammalian primitive streak. Development. 1993;118:829–844. doi: 10.1242/dev.118.3.829. [DOI] [PubMed] [Google Scholar]
- 7.Cano A, Pérez-Moreno M A, Rodrigo I, Locascio A, Blanco M J, del Barrio M G, Portillo F, Nieto M A. The transcription factor Snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 8.Fujiwara S, Corbo J C, Levine M. The snail repressor establishes a muscle/notochord boundary in the Ciona embryo. Development. 1998;125:2511–2520. doi: 10.1242/dev.125.13.2511. [DOI] [PubMed] [Google Scholar]
- 9.Grau Y, Carteret C, Simpson P. Mutations and chromosomal rearrangements affecting the expression of snail, a gene involved in embryonic patterning in Drosophila melanogaster. Genetics. 1984;108:347–360. doi: 10.1093/genetics/108.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray S, Szymanski P, Levine M. Short-range repression permits multiple enhancers to function autonomously within a complex promoter. Genes Dev. 1994;8:1829–1838. doi: 10.1101/gad.8.15.1829. [DOI] [PubMed] [Google Scholar]
- 11.Hemavathy K, Ashraf S I, Ip Y T. Snail/Slug family of repressors: slowly going into the fast lane of development and cancer. Gene. 2000;257:1–12. doi: 10.1016/s0378-1119(00)00371-1. [DOI] [PubMed] [Google Scholar]
- 12.Hemavathy K, Guru S C, Harris J, Chen J D, Ip Y T. Human Slug is a repressor that localizes to sites of active transcription. Mol Cell Biol. 2000;20:5087–5095. doi: 10.1128/mcb.20.14.5087-5095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ip Y T, Park R E, Kosman D, Bier E, Levine M. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev. 1992;6:1728–1739. doi: 10.1101/gad.6.9.1728. [DOI] [PubMed] [Google Scholar]
- 14.Jiang R, Copeland N G, Gilbert D J, Jenkins N A, Gridley T. Genomic organization and chromosomal localization of the mouse Snail (Sna) gene. Mamm Genome. 1997;8:686–688. doi: 10.1007/s003359900537. [DOI] [PubMed] [Google Scholar]
- 15.Jiang R, Lan Y, Norton C R, Sundberg J P, Gridley T. The Slug gene is not essential for mesoderm or neural crest development in mice. Dev Biol. 1998;198:277–285. [PubMed] [Google Scholar]
- 16.Kataoka H, Murayama T, Yokode M, Mori S, Sano H, Ozaki H, Yokota Y, Nishikawa S-I, Kita T. A novel Snail-related transcription faction Smuc regulates basic helix-loop-helix transcription factor activities via specific E-box motifs. Nucleic Acids Res. 2000;28:626–633. doi: 10.1093/nar/28.2.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LaBonne C, Bronner-Fraser M. Snail-related transcriptional repressors are required in Xenopus for both the induction of the neural crest and its subsequent migration. Dev Biol. 2000;221:195–205. doi: 10.1006/dbio.2000.9609. [DOI] [PubMed] [Google Scholar]
- 18.Leptin M. twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes Dev. 1991;5:1568–1576. doi: 10.1101/gad.5.9.1568. [DOI] [PubMed] [Google Scholar]
- 19.Leptin M, Casal J, Grunewald B, Reuter R. Mechanisms of early Drosophila mesoderm formation. Dev Suppl. 1992;1992:23–31. [PubMed] [Google Scholar]
- 20.Locascio A, Nieto M A. Cell movements during vertebrate development: integrated tissue behaviour versus individual cell migration. Curr Opin Genet Dev. 2001;11:464–469. doi: 10.1016/s0959-437x(00)00218-5. [DOI] [PubMed] [Google Scholar]
- 21.Nakayama H, Scott I C, Cross J C. The transition to endoreduplication in trophoblast giant cells is regulated by the mSna zinc finger transcription factor. Dev Biol. 1998;199:150–163. doi: 10.1006/dbio.1998.8914. [DOI] [PubMed] [Google Scholar]
- 22.Nieto M A, Bennett M F, Sargent M G, Wilkinson D G. Cloning and developmental expression of Sna, the murine homolog of the Drosophila snail gene. Development. 1992;116:227–237. doi: 10.1242/dev.116.1.227. [DOI] [PubMed] [Google Scholar]
- 23.Nieto M A, Sargent M G, Wilkinson D G, Cooke J. Control of cell behavior during vertebrate development by Slug, a zinc finger gene. Science. 1994;264:835–839. doi: 10.1126/science.7513443. [DOI] [PubMed] [Google Scholar]
- 24.Nüsslein-Volhard C, Wieschaus E, Kluding H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. I Zygotic loci on the second chromosome. Roux's Arch Dev Biol. 1984;193:267–282. doi: 10.1007/BF00848156. [DOI] [PubMed] [Google Scholar]
- 25.Oda H, Tsukita S, Takeichi M. Dynamic behavior of the cadherin-based cell-cell adhesion system during Drosophila gastrulation. Dev Biol. 1998;203:435–450. doi: 10.1006/dbio.1998.9047. [DOI] [PubMed] [Google Scholar]
- 26.Romano L A, Runyan R B. Slug is a mediator of epithelial-mesenchymal cell transformation in the developing chicken heart. Dev Biol. 1999;212:243–254. doi: 10.1006/dbio.1999.9339. [DOI] [PubMed] [Google Scholar]
- 27.Savagner P, Yamada K M, Thiery J P. The zinc finger protein Slug causes desmosome dissociation, an initial and necessary step in growth factor-induced epithelial-mesenchymal transition. J Cell Biol. 1997;137:1403–1419. doi: 10.1083/jcb.137.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sefton M, Sánchez S, Nieto M A. Conserved and divergent roles for members of the Snail family of transcription factors in the chick and mouse embryo. Development. 1998;125:3111–3121. doi: 10.1242/dev.125.16.3111. [DOI] [PubMed] [Google Scholar]
- 29.Simeone A, Acampora D, Mallamaci A, Stornaiuolo A, D'Apice M R, Nigro V, Boncinelli E. A vertebrate gene related to orthodenticle contains a homeodomain of the bicoid class and demarcates anterior neuroectoderm in the gastrulating mouse embryo. EMBO J. 1993;12:2735–2747. doi: 10.1002/j.1460-2075.1993.tb05935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith D E, Franco del Amo F, Gridley T. Isolation of Sna, a mouse gene homologous to the Drosophila genes snail and escargot: its expression pattern suggests multiple roles during postimplantation development. Development. 1992;116:1033–1039. doi: 10.1242/dev.116.4.1033. [DOI] [PubMed] [Google Scholar]
- 31.Swiatek P, Gridley T. Perinatal lethality and defects in hindbrain development in mice homozygous for a targeted mutation of the zinc finger gene Krox20. Genes Dev. 1993;7:2071–2084. doi: 10.1101/gad.7.11.2071. [DOI] [PubMed] [Google Scholar]
- 32.Wilkinson D G, Bhatt S, Herrmann B G. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature. 1990;343:657–659. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- 33.Wolpert L. Gastrulation and the evolution of development. Dev Suppl. 1992;1990:7–13. [PubMed] [Google Scholar]