Abstract
ER-phagy is a form of autophagy that is mediated by ER-phagy receptors and selectively degrades endoplasmic reticulum (ER). Coronaviruses have been shown to use the ER as a membrane source to establish their double-membrane vesicles (DMVs). However, whether viruses modulate ER-phagy to drive viral DMV formation and its underlying molecular mechanisms remains largely unknown. Here, we demonstrate that coronavirus subverts ER-phagy by hijacking the ER-phagy receptors FAM134B and ATL3 into p62 condensates, resulting in increased viral replication. Mechanistically, we show that viral protein ORF8 binds to and undergoes condensation with p62. FAM134B and ATL3 interact with homodimer of ORF8 and are aggregated into ORF8/p62 liquid droplets, leading to ER-phagy inhibition. ORF8/p62 condensates disrupt ER-phagy to facilitate viral DMV formation and activate ER stress. Together, our data highlight how coronavirus modulates ER-phagy to drive viral replication by hijacking ER-phagy receptors.
Keywords: ER-phagy, p62 condendation, SARS-CoV-2, ORF8, double membrane vesicles, ER stress
Graphical abstract
Tan et al. describe an important mechanism by which SARS-CoV-2 protein ORF8 inhibits ER-phagy by hijacking the receptors FAM134B and ATL3 into p62 condensates, facilitating the production of viral replication organelle double-membrane vesicles.
Introduction
The endoplasmic reticulum (ER) is the largest dynamic organelle that enables protein folding, secretory protein biogenesis, and organelle communication and so on.1 The autophagic degradation of the ER, a selective form of autophagy which is termed “ER-phagy,” is a degradative process that controls the quality and abundance of ER to maintain normal homeostasis. Nutrient starvation or the unfold protein response (UPR) induces ER-phagy.2 The ER-phagy receptors connect domains of ER sheets or tubules to the autophagosome biogenesis machinery by binding to their partner LC3 to mediate the recognition of the discrete foci on the ER by autophagy.3 Several mammalian ER membrane proteins, including FAM134B, RTN3L, CCPG1, SEC62, TEX264, and ATL3, have been identified as ER-phagy receptors.4 , 5 , 6 , 7 , 8 , 9 , 10
Viruses use the ER as a source of membranes to establish their replication vesicular invaginations.11 , 12 The dengue virus (DENV), Zika virus (ZIKV), and West Nile virus (WNV) use their viral encoded NS3 proteases to directly cleave FAM134B at a single site, which blocks the formation of ER-phagy and sequestration of viral proteins within these structures to promote viral replication.13 Coronaviruses arrogate intracellular membranes to form double-membrane vesicles (DMVs) which serve as viral replication organelles.14 , 15 The ER is probably the main membrane donor for DMV formation and the membrane rearrangements can be an invagination toward the lumen of the ER or an extrusion of the ER membrane.16 However, relatively little is known about the relationship between DMV formation during SARS-CoV-2 infection and ER-phagy.
Among the accessory proteins of SARS-CoV-2, protein encode by ORF8 have drawn particular attention. ORF8 antibodies have been identified as a major marker of acute, convalescent, and long-term antibody response to SARS-CoV-2 infection because of its immunodominance and specificity.17 The SARS-CoV-2 ORF8 protein is a 121 amino acid (aa) long protein comprising an N-terminal hydrophobic signal peptide (1–17 aa) and an ORF8 chain (18–121 aa),18 displays various biological functions. It inhibits type I interferon signaling pathway,19 interacts with MHC-1 molecules and mediates their downregulation via autophagy, leading to an impairment of the antigen presentation system.20 ORF8 was also found to interact with many ER proteins21; however, the function of SARS-CoV-2 ORF8 in the ER remains unknown.
Phase separation, a general phenomenon in physics and chemistry, is a much more recent development in biology.22 Accumulating evidences have revealed that many proteins possess the phase transition properties, which regulate biochemical reactions, signal transduction, and stress resistance by increasing local protein concentration or decreasing molecular motion.23 , 24 , 25 Recently, several studies have examined various aspects of the phase separation behavior of the SARS-CoV-2 N protein, including its role in viral RNA packaging, stress granule modulation, regulation of host cell innate immune pathways, and regulation by kinases.26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 However, whether other SARS-CoV-2 viral proteins can undergo phase separation is poorly understood.
p62 was identified as the autophagic cargo receptor. It plays a key role in mediating the formation and autophagic clearance of intracellular protein aggregates.35 , 36 p62 also binds to ubiquitin (Ub)-coated viral particles, which are subsequently delivered to the autophagic machinery.37 p62 exists in two populations: the membrane-bound form interacts with autophagosomes and the cytosolic pool, while the membrane-less form has been shown to undergo phase separation, driven by polyubiquitin to segregate the autophagic cargos.38 , 39 , 40
In this study, we showed that the SARS-CoV-2 ORF8 protein undergoes phase separation in vitro and in vivo. It interacts and condenses with p62. ORF8/p62 condensates hijack two ER-phagy adaptors, FAM134B and ATL3 to impede ER-phagy, resulting in increased viral DMV biogenesis and severe ER stress. In addition, ORF8 homodimerization is required for ER-phagy receptors binding and ER-phagy inhibition, but not for the interaction and phase separation formation with p62.
Results
SARS-CoV-2 ORF8 interacts with p62 to undergo condensation
To investigate whether SARS-CoV-2 ORF8 has liquid-like properties, we first examined the distribution of ORF8. ORF8-Strep was transiently expressed in HeLa cells, small ORF8 puncta were observed in cultured HeLa cells (Figure S1A), and these puncta could undergo fusion and fission (Figure S1B). Fluorescence recovery after photo bleaching (FRAP) revealed that the fluorescent signal was recovered after bleaching of GFP-ORF8 puncta (Figure S1C). Purified GFP and recombinant GFP-tagged ORF8 were used for in vitro phase separation assays with or without 10% polyethylene glycol 8000 (PEG8000), which increases macromolecular crowding and promotes phase separation. We found that GFP protein remained clear in both buffer with or without PEG8000 (Figures S1D and S1E). Interestingly, although purified GFP-ORF8 exhibited puncta formation in 10% PEG8000 solution, it failed to undergo condensation without PEG8000 (Figures S1D and S1E). These data suggest that ORF8 alone cannot undergo phase separation and ORF8 forms biomolecular condensates only in PEG8000 solution.
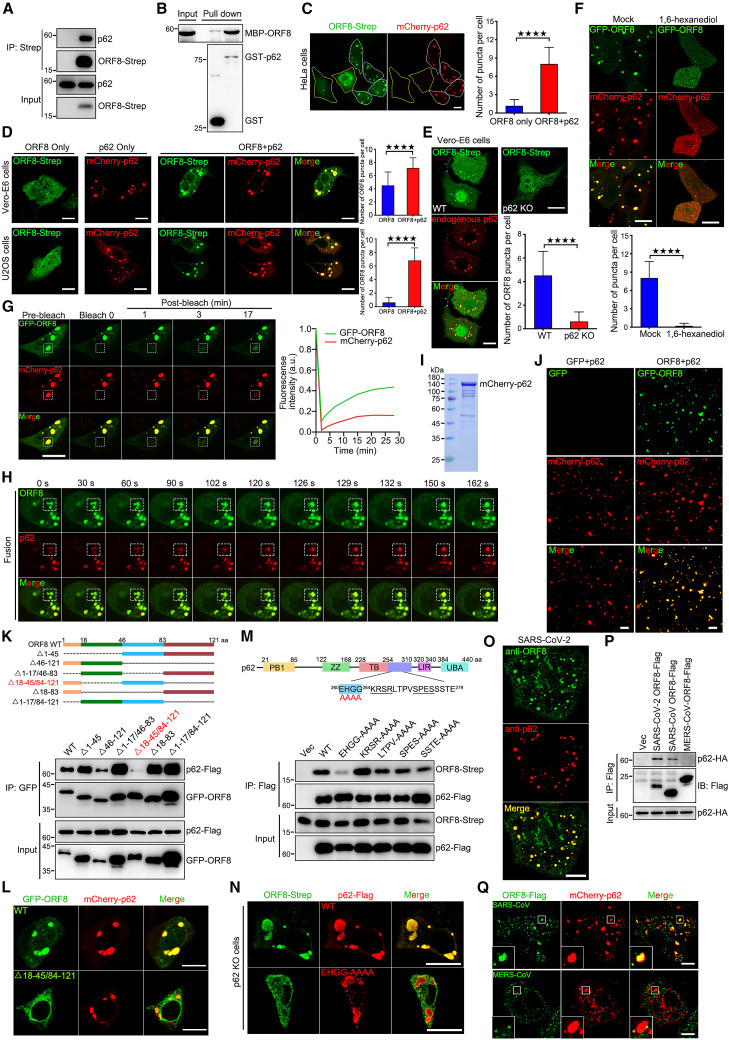
To investigate the functions of SARS-CoV-2 ORF8 condensates, we first performed an immunoprecipitation/mass spectrometry (IP/MS) screening with ORF8 as a bait to identify ORF8-interacting proteins. We found that p62, the autophagic cargo receptor in mammalian cells, is one of the candidates for ORF8 binding (Figure S1F). Several studies have suggested that the membrane-less accumulated p62 bodies undergo phase separation.38 , 39 , 40 , 41 , 42 We first confirmed that p62 was a positive candidate for ORF8 through in vivo co-immunoprecipitation (coIP) and in vitro GST pull-down assays (Figures 1A and 1B). Interestingly, we found that all ORF8 puncta were frequently co-localized with mCherry-p62 bodies in the cytoplasm (Figure S1G). ORF8 puncta with p62 always exhibited stronger signals and larger sizes compared with those without p62 expression (Figures 1C and 1D). Consistent with previous study,39 we observed huge intracellular droplet formation of exogenous p62 expression, and ORF8 co-expression did not significantly change the size or number of p62 bodies (Figures 1C and 1D). The number of ORF8 puncta decreased in p62-knockout (p62 KO) cells (Figure 1E). Treatment with 1,6-hexanediol, which can disrupt condensates, disrupted the ORF8/p62 puncta (Figure 1F). FRAP and fusion/fission assays revealed that GFP-ORF8/mCherry-p62 puncta undergo phase separation (Figures 1G, 1H, and S1H). Furthermore, purified GFP-ORF8, mCherry-p62, and poly-Ub were used for in vitro phase separation assays in PEG8000 free buffer. We found that GFP-ORF8, but not GFP protein was condensed into p62/poly-Ub condensates (Figures 1I and 1J). Taken together, these data suggest that SARS-CoV-2 ORF8 was condensed into p62 bodies.
Figure 1.
SARS-CoV-2 ORF8 interacts p62 to form condensates
(A) Co-precipitation analysis of endogenous p62 with ORF8-Strep in HEK293T.
(B) Purified MBP-ORF8 was incubated with purified GST or GST-p62, and analysis of the interaction between ORF8 and p62 by in vitro GST pull-down was performed.
(C and D) Cells were co-transfected with ORF8-Strep and mCherry-p62 for 24 h, then stained with anti-Strep antibody and imaged using confocal microscopy. Scale bar, 10 μm. The number of ORF8 puncta in each cell was counted from 50 cells of three independent experiments. Two-tailed unpaired Student’s t test, ∗∗∗∗p < 0.0001.
(E) p62 WT and KO Vero-E6 cells were transfected with ORF8-Strep for 24 h, then cells were stained with anti-Strep and anti-p62 antibodies, and imaged using confocal microscopy. Scale bar, 10 μm. The number of ORF8 puncta in each cell was counted from 50 cells of three independent experiments. Two-tailed unpaired Student’s t test, ∗∗∗∗p < 0.0001.
(F) HeLa cells were co-transfected with GFP-ORF8 and mCherry-p62 for 24 h, then cells were mock treated or treated with 1,6-hexanediol for 1 min and imaged using confocal microscopy. Scale bar, 10 μm. The number of ORF8/p62 puncta in each cell was counted from 50 cells of three independent experiments. Two-tailed unpaired Student’s t test, ∗∗∗∗p < 0.0001.
(G) HeLa cells with exogenous expression of GFP-ORF8 and mCherry-p62 undergo liquid-like behavior; fluorescence recovery after photobleaching (FRAP) images are shown before and at indicated time points after bleaching. Time 0 indicates the time of photobleaching. Scale bar, 10 μm. Quantification of fluorescence intensity recovery of GFP-ORF8 and mCherry-p62 in the bleached droplet.
(H) HeLa cells with exogenous expression of GFP-ORF8 and mCherry-p62 undergo fusion, images are shown at indicated time points after imaging.
(I) mCherry-p62 were purified from Escherichia coli BL21 and analyzed via Coomassie blue.
(J) In vitro phase separation assay of GFP and mCherry-p62/His6-UBx8 or GFP-ORF8 and mCherry-p62/His6-UBx8. Fluorescence images of 10 μM each protein in phase separation assay buffer without PEG8000. Representative fluorescence images of three independent experiments are shown. Scale bar, 10 μm.
(K) GFP-ORF8 truncations were expressed as indicated with p62-FLAG. Cell lysates were subjected to immunoprecipitation with GFP antibody and analyzed using western blotting (WB).
(L) Cells were transfected with GFP-ORF8 or mutant with mCherry-p62 for 24 h, and imaged using confocal microscopy. Scale bar, 10 μm.
(M) p62-FLAG mutations were expressed as indicated with ORF8-Strep. Cell lysates were subjected to immunoprecipitation with FLAG antibody and analyzed using WB.
(N) p62 KO Vero-E6 cells were transfected with p62-FLAG or mutant with ORF8-Strep for 24 h, then stained with anti-FLAG and anti-Strep antibodies and imaged using confocal microscopy. Scale bar, 10 μm.
(O) SARS-CoV-2 infected Vero-E6 cells were stained with anti-ORF8 and anti-p62 antibodies and imaged using confocal microscopy. Scale bar, 10 μm.
(P) Co-precipitation analysis of p62-HA with SARS-CoV-2 ORF8-FLAG or SARS-CoV ORF8-FLAG or MERS-CoV ORF8-FLAG in HEK293T.
(Q) Cells were co-transfected with mCherry-p62 with SARS-CoV ORF8-FLAG or MERS-CoV ORF8-FLAG for 24 h, then stained with anti-FLAG antibody and imaged using confocal microscopy. Scale bar, 10 μm.
See also Figures S1 and S2.
Next, we determined whether the ORF8-p62 interaction was critical for the formation of ORF8/p62 condensates. Treatment with 1,6-hexanediol weakened the interaction between p62 and ORF8 (Figure S1I). A series of progressively truncated ORF8 mutants were constructed and used for coIP (Figure 1K, upper panel). Deletion of 18–45/84–121 aa of ORF8 (ORF8Δ18–45/84–121) resulted in a failure to bind to p62 (Figure 1K, bottom panel) or co-localize with p62 (Figures 1L and S1J). Truncations and point mutations of p62 were also generated and we found that the mutation of amino acids 260–264 EHGG to AAAA (p62EHGG-AAAA) resulted in a failure to bind to ORF8 (Figures S1K, S1L, and 1M). Consistently, p62EHGG-AAAA failed to recruit ORF8 to p62 bodies in p62 KO cells (Figures 1N, S1M, and S1N). In native SARS-CoV-2-infected cells, endogenous ORF8, tracked by ORF8 antibody, exhibited significant colocalization with p62 bodies (Figure 1O). We further found that ORF8 protein of SARS-CoV also interacts with p62 and was condensed into p62 bodies, but not MERS-CoV ORF8 (Figures 1P and 1Q). Taken together, these data demonstrate that the ORF8-p62 interaction is critical for the formation of ORF8/p62 condensates.
ORF8 inhibits the autophagic degradation of p62
p62 bodies are engulfed by autophagosomes for degradation via the interaction of p62 with LC3.38 , 39 Overexpression of ORF8 inhibited the starvation-induced autophagic degradation of p62 by disrupting the interaction of p62 with LC3 (Figures S2A and S2B). Starvation enhanced the protein level of ORF8 (Figure S2C), the interaction between ORF8 and p62 (Figure S2D), and also the number of ORF8/p62 liquid droplets (Figure S2E). These results suggest that p62 was more stable in ORF8/p62 condensates than expression alone. Consistent with previous study,43 we further found that higher levels of endogenous p62 and p62 bodies were accumulated in SARS-CoV-2-infected cells than in mock-infected cells (Figures S2F and S2G). The kinetics of exogenous p62 expression paralleled the increase in its condensates (Figures S2H–S2K). These data suggest that SARS-CoV-2 infection enhances the protein level of p62 to promote p62/ORF8 condensation.
We have showed that the overexpression of ORF8 did not affect p62 condensates (Figure 1D). Next, we determine whether ORF8 affects the other functions of p62. Overexpression of ORF8 slightly enhanced the interaction between p62 and ubiquitin (Figure S2L) but had no effect on its self-interaction (Figure S2M) and the interaction between p62 and Keap1 (Figure S2N).
Taken together, these data indicate that ORF8 inhibits the autophagic degradation of p62 and SARS-CoV-2 infection results in the accumulation of p62 for ORF8/p62 condensation.
ORF8/p62 condensation subverts ER-phagy
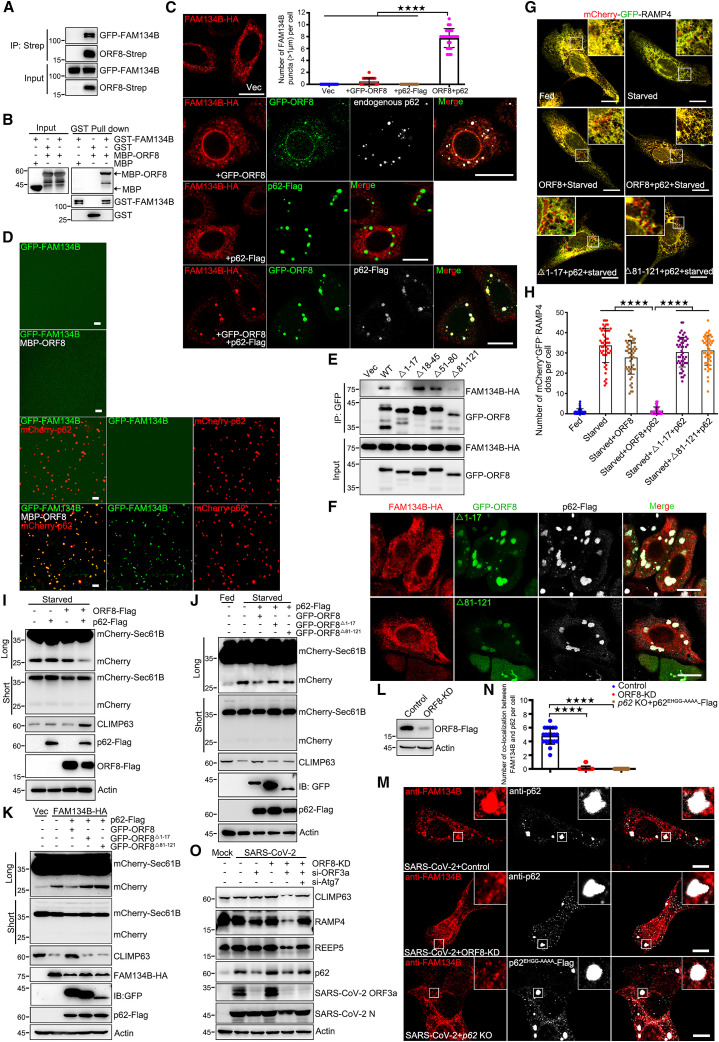
Next, we sought to determine the function of ORF8/p62 condensation. ORF8 was found to interact with many ER proteins (Figure S1F).21 We first used RAMP4, a subunit of the ER translocon complex, to track the ER. Compared with control cells, ER morphology was significantly changed and ER puncta were observed in ORF8/p62 co-expression cells (Figure S3A). These data indicate that ORF8/p62 co-expression caused ER damage which may explain why the ER lumen protein ORF8 can bind p62. ER-phagy receptor FAM134B is one of candidate from our ORF8 IP/MS result (Figure S1F). We then asked whether ORF8 could modulate ER-phagy. We first confirmed that ORF8 directly interacted with FAM134B in vivo coIP (Figure 2 A) and in vitro GST pull-down assays (Figure 2B). Intriguingly, FAM134B was recruited into ORF8/p62 liquid droplets, and the co-localization of the three was observed regardless of exogenous or endogenous p62 expression (Figure 2C). Notably, p62 bodies without ORF8 co-expression failed to recruit FAM134B (Figure 2C). In order to examine whether membrane protein FAM134B was condensed into ORF8/p62 condensates as a solubilized form or together with (ER-)membranes, we generated the internal reticulon domain (RTND) deleted mutant (Δ84–233).44 We found that Δ84–233 mutant still interacted with ORF8 and was condensed into ORF8/p62 condensates (Figures S3B and S3C). In order to further demonstrate that FAM134B was condensed into pre-formed p62/poly-Ub condensates by interacting with ORF8 in vitro, purified GFP-FAM134B, MBP-ORF8, His-UBx8 and mCherry-p62 were analyzed using in vitro phase separation assay in PEG8000 free buffer. We found that GFP-FAM134B were only condensed into ORF8/p62/poly-Ub condensates, but failed to form droplets when incubated alone, or incubated with ORF8, or incubated with p62/poly-Ub (Figures 2D and S3D). Two ORF8 mutants, deletion of amino acids 1–17 (ORF8Δ1–17) or 84–121 (ORF8Δ84–121), failed to interact with FAM134B and lost their ability to recruit FAM134B to ORF8/p62 liquid droplets (Figures 2E and 2F). These data confirm that FAM134B directly interacts with ORF8 and is recruited to ORF8/p62 condensates.
Figure 2.
ORF8/p62 condensates subvert ER-phagy
(A) Co-precipitation analysis of GFP-FAM134B with ORF8-Strep in HEK293T.
(B) Purified MBP or MBP-ORF8 was incubated with purified GST or GST-FAM134B, and analyzed the interaction between ORF8 and FAM134B by GST pull-down.
(C) Immunofluorescence of cells expressing FAM134B-HA or FAM134B-HA and GFP-ORF8 or FAM134B-HA, p62-FLAG and GFP-ORF8 with anti-HA and anti-FLAG antibodies. Scale bar, 10 μm. The number of FAM134B puncta (>1 μm) in each cell was counted from 50 cells of three independent experiments. Two-tailed unpaired Student’s t test, ∗∗∗∗p < 0.0001.
(D) In vitro phase separation assay of GFP-FAM134B alone or GFP-FAM134B and MBP-ORF8 or GFP-FAM134B and mCherry-p62/His-UBx8 or GFP-FAM134B, MBP-ORF8 and mCherry-p62/His-UBx8. Fluorescence images of 10 μM each protein in phase separation assay buffer without PEG8000. Representative images of three independent experiments are shown. Scale bar, 10 μm.
(E) GFP-ORF8 truncations were expressed as indicated with FAM134B-HA. Cell lysates were subjected to immunoprecipitation with GFP antibody and analyzed using WB.
(F) HeLa cells were transfected with GFP-ORF8 truncations with FAM134B-HA for 24 h, then stained with anti-HA antibody and imaged using confocal microscopy. Scale bar, 10 μm.
(G and H) U2OS cells expressing mCherry-GFP-RAMP4 were transfected with p62-FLAG with or without ORF8-Strep or mutants for 24 h, then cells were starved in EBSS for 12 h and imaged using confocal microscopy (G). Scale bar, 10 μm. The number of red puncta in each cell (H) was counted from 50 cells of three independent experiments. Two-tailed unpaired Student’s t test, ∗∗∗∗p < 0.0001.
(I and J) U2OS cells expressing mCherry-Sec61B were transfected with indicated plasmids for 24 h and then starved in EBSS for 12 h. Cell lysates were analyzed using WB.
(K) U2OS cells expressing mCherry-Sec61B were transfected with indicated plasmids for 24 h. Cell lysates were analyzed using WB.
(L) Vero-E6 cells were transfected with control or ORF8-KD plasmid for 36 h and analyzed the knockdown efficiency using WB.
(M and N) Vero-E6 WT cells were transfected with control or ORF8-KD plasmid, p62 KO Vero-E6 cells were transfected with p62EHGG-AAAA-FLAG for 12 h and then infected with SARS-CoV-2 for 24 h, then cells were stained with anti-FAM134B and anti-p62 or anti-FLAG antibodies and imaged using confocal microscopy. Scale bar, 10 μm. The number of colocalization between FAM134B and p62 droplets in each cell was counted from 20 cells of two independent experiments. Two-tailed unpaired Student’s t test, ∗∗∗∗p < 0.0001.
(O) Vero-E6 cells were transfected with ORF8-KD plasmid with or without si-ORF3a or si-Atg7 for 24 h and then infected with SARS-CoV-2 for another 24 h. Cell lysates were analyzed using WB.
See also Figure S3.
Next, to determine whether ORF8/p62 condensates modulate the ER-phagy by hijacking FAM134B under SARS-CoV-2 infection, we applied the mCherry-GFP tandem tagging strategy, mCherry-GFP-RAMP4.45 When autolysosomes digest the ER membrane fragments, the GFP of this tandem reporter is attenuated by lysosomal degradation, resulting in the loss of yellow fluorescence, leaving only the red fluorescence of RFP. We used prolonged amino acid starvation (12 h) with Earl’s buffered saline solution (EBSS) to induce ER-phagy. By comparing the level of p62 condensation between SARS-CoV-2 infection and exogenous p62 expression, we found that exogenous p62 showed similar condensation level with endogenous p62 in SARS-CoV-2-infected cells (Figures S2G, S2I, and S2K). We then used this physiologically relevant ORF8 and p62 co-expression system for the further investigation. We observed that ORF8/p62 co-expression, but not ORF8 expression alone, inhibited starvation-induced ER-phagy (Figures 2G and 2H), suggesting that huge droplets (ORF8 and p62 co-expression) were required for recruiting enough FAM134B for ER-phagy inhibition. Consistently, ORF8Δ1–17 and ORF8Δ84–121 failed to inhibit ER-phagy (Figures 2G and 2H). We further used a reporter construct mCherry-Sec61B, an ER sheet resident protein, which produces free mCherry visualized by western analysis when ER-phagy was induced because of the partial digestion of Sec61B. We first verified that starvation-induced ER-phagy produced mCherry fragments and degraded the ER membrane protein CLIMP-63 (Figure S3E). As expected, only ORF8/p62 co-expression decreased starvation-induced mCherry fragment production and blocked the degradation of CLIMP-63, but not ORF8 alone, p62 alone, mutant ORF8Δ1–17, or mutant ORF8Δ84–121 (Figures 2I and 2J). FAM134B expression revised ORF8/p62-inhibited ER-phagy (Figure S3F, lanes 3 and 5). Furthermore, ORF8/p62 also inhibited FAM134B overexpression-induced ER-phagy, but not ORF8 alone, mutant ORF8Δ1–17, or mutant ORF8Δ84–121 (Figure 2K). Similar results were also obtained using another reporter construct mCherry-RAMP4 (Figure S3G). Furthermore, SARS-CoV ORF8/p62 bodies also hijacked FAM134B and inhibited ER-phagy (Figures S3H and S3I).
Next, we used a verified construct, specifically degraded ORF8 protein by engineered E3 ubiquitin-protein ligase system,20 to knock down ORF8 (ORF8-KD) (Figure 2L). As expected, in native SARS-CoV-2-infected cells, endogenous FAM134B exhibited no colocalization with p62 bodies without ORF8 in WT cells or ORF8/p62EHGG-AAAA liquid droplets in p62 KO cells (Figures 2M and 2N). We further used siRNA to knock down ORF3a in SARS-CoV-2-infected cells to ensure the autophagic degradation,43 and further knock down ORF8 to subvert the recruitment of FAM134B into condensates, and then investigated the ER-phagy by examining ER membrane proteins CLIMP63, REEP5, and RAMP4. As expected, our data showed that SARS-CoV-2 induced autophagic degradation of CLIMP63, REEP5, and RAMP4, and failed to inhibit ER-phagy only under ORF3a and ORF8 double-knockdown condition (Figure 2O). Taken together, these results suggest that ORF8/p62 condensates subvert ER-phagy by hijacking FAM134B through the interaction between ORF8 and FAM134B.
We further used p62 mutants that underwent weaker or stronger condensation than the WT to test their ability of ER-phagy inhibition.38 Compared with the WT, the p62S403E mutant (with a phosphorylation defect) formed larger bodies and showed slightly stronger ER-phagy inhibition while the deletion of PB1 domain (p62ΔPB1, with a p62 polymerization defect) resulted in weaker condensation and a failure to affect ER-phagy (Figures S3J and S3K).
Next, we determined whether ORF8 affects the other functions of FAM134B. Starvation enhanced the interaction between ORF8 and FAM134B (Figure S3L). ORF8/p62 co-expression weakened the interaction between FAM134B and LC3 (Figure S3M) but had no effect on FAM134B self-interaction (Figure S3N)44 or FAM134B digestion (Figure S3O). These data indicate that ORF8/p62 condensates hijack FAM134B, disrupting the interaction between FAM134B and LC3, thus inhibiting ER-phagy.
Interestingly, under starvation plus FAM134B overexpression, ORF8/p62 still modulated ER-phagy (Figures S3F and S3G, lanes 4 and 5), suggesting that other ER-phagy receptors may be involved. CoIP assays showed that ORF8 also interacted with ATL3, Sec62, and CCPG1, but not RTN3 or TEX264 (Figure S3P). Surprisingly, only ATL3 was hijacked into ORF8/p62 condensates (Figure S3Q), but not Sec62, TEX264, RTN3L, or CCPG1, even under thapsigargin-induced ER stress or starvation-induced ER-phagy conditions (Figures S3R and S3S). ORF8 mutant, deletion of amino acids 1–17 (ORF8Δ1–17), failed to interact with ATL3 and lost their ability to recruit ATL3 to ORF8/p62 condensates (Figures S3T and S3U). Therefore, FAM134B and ATL3 are the two critical ER-phagy receptors involved in ORF8/p62 condensate-induced ER-phagy dysfunction.
ORF8/p62 condensates facilitate viral DMV biogenesis by inhibiting ER-phagy
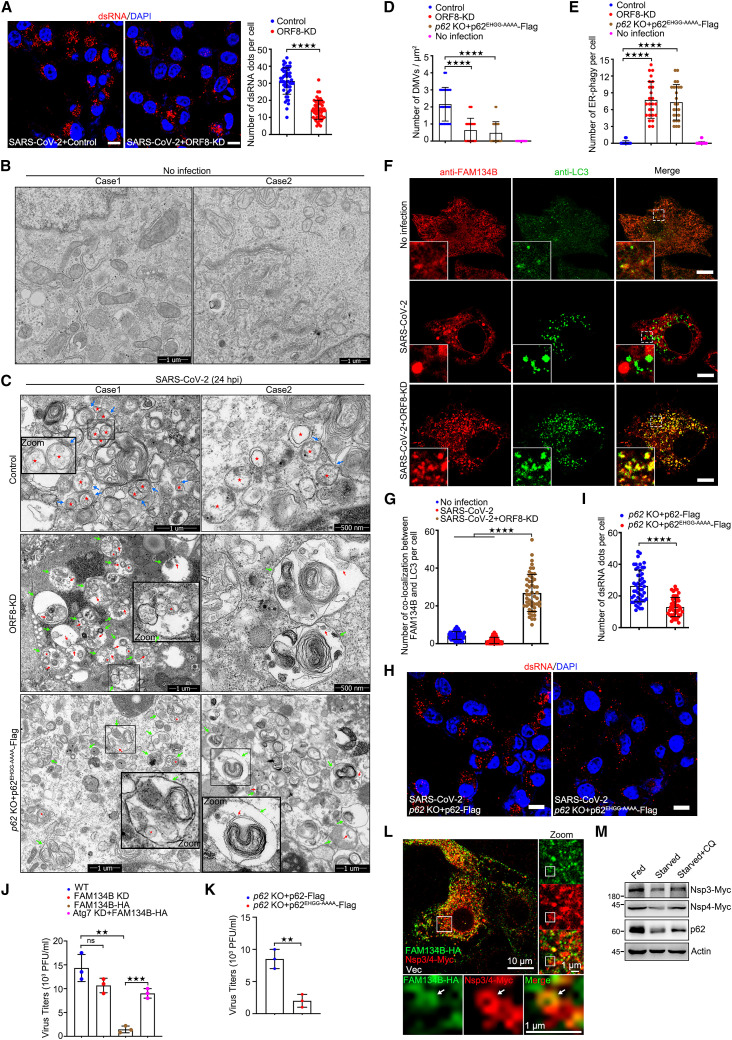
The ER has been shown to serve as the main membrane donor for SARS-CoV-2 DMVs.16 , 46 , 47 Next, we determined whether ORF8/p62 condensates could drive DMV biogenesis by inhibiting ER-phagy. The interior of DMVs contains double-stranded RNA (dsRNA),48 which was first used to track DMVs in SARS-CoV-2-infected cells. Consistent with previous studies,49 dsRNA showed dots in the cytosol, indicating the position and number of the DMVs. Intriguingly, knockdown of ORF8 decreased the number of dsRNA dots (Figure 3 A). Compared with non-infected cells, ultrastructural analysis using transmission electron microscopy (TEM) revealed the appearance of DMVs in SARS-CoV-2 infected Vero-E6 cells, some of which were associated with ER membranes in WT cells (Figures 3B and 3C). Similar DMVs from TEM assay were also observed in SARS-CoV-2-infected Calu-3 cells.49 In SARS-CoV-2-infected ORF8 knockdown cells, autophagosomes are larger than DMVs and contain ER fragmentation and/or other membrane structures (Figure 3C). We further observed a consistent reduction in DMV formation in SARS-CoV-2-infected ORF8-depleted cells (Figure 3C, middle panel; and 3D). In the control-infected Vero-E6 cells, ER-phagy was rarely observed (Figures 3B and 3E). In contrast, similar to the ER-phagy morphology in U2OS and HCT116 cells,4 , 9 the accumulation of numerous autophagic vacuoles containing ER fragments was observed in SARS-CoV-2 infected ORF8-depleted cells (Figure 3C, middle panel; and 3E). Consistent with TEM result, we confirmed that knockdown of ORF8 lead to the colocalization between FAM134B and LC3 under SARS-CoV-2 infection by confocal microcopy using FAM134B and LC3 antibodies, and FAM134B bodies, which did not co-localize with LC3, were observed in SARS-CoV-2 infected wild-type cells (Figures 3F and 3G). These data suggest that ORF8 inhibits ER-phagy in SARS-CoV-2-infected cells. Next, we determined whether ORF8/p62 condensates modulate ER-phagy in SARS-CoV-2-infected cells. We performed rescue experiments in p62 KO cells by transiently expressing p62EHGG-AAAA, which failed to bind to and recruit ORF8 into p62 bodies to form ORF8/p62 condensates. Compared with the WT cells, we observed frequent ER-phagy and fewer DMVs in the p62EHGG-AAAA-rescued p62 KO cells (Figure 3C, bottom panel, and Figures 3D and 3E). Consistent with TEM result, using dsRNA antibody tracked DMVs, we found that fewer DMVs in the p62EHGG-AAAA-rescued p62 KO cells than p62 wild-type-rescued p62 KO cells under SARS-CoV-2 infection (Figures 3H and 3I). These results indicate that ORF8/p62 condensates inhibit ER-phagy to facilitate SARS-CoV-2 DMVs biogenesis.
Figure 3.
ORF8/p62 condensates facilitate DMV formation
(A) Vero-E6 cells were transfected with control or ORF8-KD plasmid for 12 h and infected with SARS-CoV-2 for 24 h. Cells were stained with dsRNA antibody (red). Nuclear DNA was stained with DAPI (blue). Cells were imaged using confocal microscopy. Scale bar, 10 μm. The number of dsRNA puncta in each cell was counted from 50 cells of two independent experiments. Two-tailed unpaired Student’s t test, ∗∗∗∗p < 0.0001.
(B and C) Transmission electron microscopy images of p62 WT and KO Vero-E6 cells transfected with p62EHGG-AAAA with control or ORF8-KD plasmid for 12 h and then infected with or without SARS-CoV-2 for 24 h. The DMV structures (red pentagram), ER-DMVs interaction (blue arrowhead), autophagosomes (green arrowhead), ER in autophagosomes (red arrowhead) are shown.
(D and E) The number of DMV (D) and ER-phagy (E) in each cell was counted from 25 cells of two independent experiments. Two-tailed unpaired Student’s t test, ∗∗∗∗p < 0.0001.
(F) Vero-E6 cells were transfected with control or ORF8-KD plasmid for 12 h and then infected with or without SARS-CoV-2 for 24 h, then cells were stained with anti-FAM134B and anti-LC3 antibodies and imaged using confocal microscopy. Scale bar, 10 μm.
(G) The number of co-localizations between FAM134B and LC3 in each cell from (F) was counted from 25 cells of two independent experiments. Two-tailed unpaired Student’s t test, ∗∗∗∗p < 0.0001.
(H) p62 KO Vero-E6 cells transfected with WT or p62EHGG-AAAA for 12 h and then infected with SARS-CoV-2 for 24 h. Cells were stained with dsRNA antibody (red). Nuclear DNA was stained with DAPI (blue). Cells were imaged using confocal microscopy. Scale bar, 10 μm.
(I) The number of dsRNA puncta in each cell from (H) was counted from 50 cells of two independent experiments. Two-tailed unpaired Student’s t test, ∗∗∗∗p < 0.0001.
(J) Vero-E6 cells were transfected with sh-FAM134B or FAM134B-HA or si-Atg7 for 12 h, cells were infected with SARS-CoV-2 for 24 h, and media were collected and analyzed using plaque assay. Error bars, mean ± SD of three independent experiments. Two-tailed unpaired Student’s t test, ∗∗p < 0.01, ∗∗∗p < 0.001.
(K) p62 KO Vero-E6 cells were transfected with p62-FLAG or p62EHGG-AAAA-FLAG for 12 h and infected with SARS-CoV-2 for 24 h, and media were collected and analyzed using plaque assay. Error bars, mean ± SD of three independent experiments Two-tailed unpaired Student’s t test, ∗∗p < 0.01.
(L) HeLa cells were transfected with FAM134B-HA, Nsp3/4-Myc for 24 h, then stained with anti-HA and anti-Myc, and imaged using confocal microscopy. Scale bar, 10 μm.
(M) HeLa cells were transfected with Nsp3/4-Myc for 24 h and starved for another 12 h with or without 2 h CQ treatment. Cell lysates were analyzed using WB.
See also Figure S4.
Intriguingly, we also observed that DMVs were engulfed in autophagosomes within the ER (Figure 3C). Hence, we speculated that the ER-phagy machinery could modulate the replication of SARS-CoV-2. To verify this hypothesis, we first examined SARS-CoV-2 replication in Vero-E6 cells using a plaque assay. Overexpression of FAM134B caused a reduction in SARS-CoV-2 replication but failed to inhibit viral replication in ER-phagy defect (Atg7 knockdown) cells but knockdown of FAM134B only had a minor effect on viral replication (Figure 3J). Meanwhile, compared with the control cells, viral production was lower in the p62EHGG-AAAA-rescued p62 KO cells (Figure 3K). Taken together, these data suggest that ORF8/p62 condensates inhibit ER-phagy to facilitate viral replication.
Several studies have suggested that co-expression of Nsp3 and Nsp4 is required and sufficient to induce DMVs biogenesis.50 , 51 , 52 Using TEM, we transiently co-expressed Nsp3 and Nsp4 of SARS-CoV-2 to directly visualize DMVs in p62 KO cells with rescuing indicate p62 and/or ORF8 mutants. Consistent with the conclusion from the native virus infection assay, p62 KO cells with rescuing the WT ORF8 and p62 increased the production of DMVs. In contrast, p62 KO cells with rescuing p62EHGG-AAAA (fail to bind to ORF8) plus ORF8 expression or rescuing WT p62 plus ORF8Δ18–45/84–121 (fail to bind to p62) expression resulted in reduction in DMV formation because of the activation of ER-phagy (Figures S4A and S4B), suggesting that ER-phagy inhibition caused by the recruitment of FAM134B into ORF8/p62 condensates is required for DMV accumulation.
A previous study indicated that DMVs originated from the rough ER and smooth ER connectors were seen linking DMVs to the rough ER.49 Meanwhile, we found that ER-phagy was inhibited during DMV formation in SARS-CoV-2-infected cells. Thus, we sought to determine whether ER-phagy could degrade the DMVs. For this purpose, we transiently co-expressed Nsp3 and Nsp4 of SARS-CoV-2 to examine the intracellular localization of ER-phagy receptor and Nsp3/4 tracked DMVs. Nsp3-Nsp4 co-expression in HeLa cells showed hollow structures, indicating the formation of DMVs, and that these structures appeared to co-localize with FAM134B (Figure 3L). We then examined whether ER-phagy could degrade Nsp3-Nsp4 tracked DMVs. Starvation-induced ER-phagy resulted in a decrease in the protein levels of Nsp3 and Nsp4, and which was prevented by chloroquine (CQ; which blocks autophagic degradation) (Figure 3M). Taken together, our results indicate that SARS-CoV-2 infection inhibits ER-phagy, thus increasing viral DMV formation and viral replication.
ORF8/p62 condensates activate ER stress
Previous studies have shown that coronaviruses induce ER stress because of the synthesis of viral proteins within the ER.53 , 54 ER-phagy was proposed to be the mechanism that eliminates ER stress and prevents apoptosis.55 Hence, we sought to determine whether SARS-CoV-2 infection or ORF8 expression could activate ER stress via ER-phagy inhibition. We treated cells with cyclopiazonic acid (CPA, a reversible inhibitor of the ER calcium pump) as the positive control for ER stress activation10 and evaluated ER stress by analyzing the protein level of CHOP (Figure S4C).56 ORF8 expression triggered slight ER stress, and higher protein levels of CHOP accumulated in ORF8-expressing cells than in control cells upon CPA treatment (Figure S4C), indicating that ORF8 expression could activate ER stress. In addition, we observed that co-expression of ORF8/p62, but not ORF8/p62EHGG-AAAA, induced more ER stress than ORF8 expression alone (Figures S4D and S4E). Knockdown of p62 abolished the ER stress induced by ORF8 expression (Figure S4F). Besides, expression of ORF8Δ18–45/84–121 (fail to bind to p62) failed to induce ER stress (Figure S4G). Similar results were obtained in p62 KO cells using rescue mutants: compared with vector, only p62 KO cells with rescuing the WT ORF8 and p62 induced ER stress, but not p62 KO cells with rescuing p62EHGG-AAAA (fail to bind to ORF8) plus ORF8 expression or rescuing WT p62 plus ORF8Δ18–45/84–121 (fail to bind to p62) expression (Figure S4H). These data indicate that the interaction between ORF8 and p62 is required for ER stress activation.
Next, to determine the association between ER stress and ER-phagy, we first proved that CPA-induced ER stress was attenuated upon starvation or FAM134B-activated ER-phagy (Figures S4I and S4J), indicating that ER-phagy may degrade the damaged ER to alleviate ER stress. Similarly, overexpression of FAM134B also abolished ORF8-induced ER stress (Figure S4K), and ORF8 expression triggered more ER stress in Atg7 knockdown cells than control cells (Figure S4L), suggesting that ORF8 expression activates ER stress by inhibiting ER-phagy. Taken together, our results demonstrate that ORF8/p62 condensates subvert ER-phagy and activates ER stress.
ORF8 homodimerization is required for ER-phagy inhibition
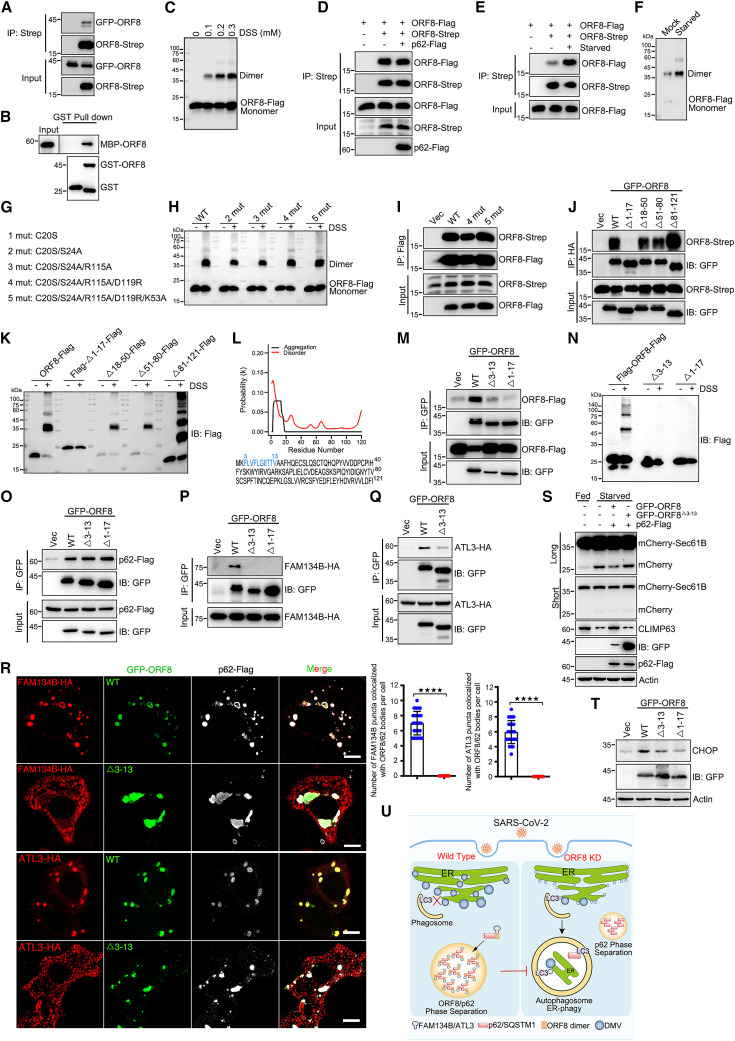
A previous study revealed the structure of SARS-CoV-2 ORF8 fragment comprising amino acids 18–121, which was crystallized as a covalent dimer.57 Next, we determined whether the homodimerization of ORF8 could be important for ORF8/p62 condensation or ER-phagy inhibition. We first confirmed the self-binding of ORF8 using coIP and GST pull-down assays (Figures 4A and 4B). In the presence of the cross-linker disuccinimidyl suberate (DSS), we further confirmed that ORF8 exists both in monomeric and dimeric states (Figure 4C). p62 expression had no effect on the self-binding of ORF8 (Figure 4D). Interestingly, starvation enhanced the homodimerization of ORF8 (Figures 4E and 4F), suggesting that the homodimerization of ORF8 could be important for ER-phagy inhibition. We then generated several point mutants based on the structure of ORF8 (Figure 4G).57 Surprisingly, these mutants still dimerize (Figures 4H and 4I). The reported structure of SARS-CoV-2 ORF8 was based on the fragment containing amino acids 18–121, not the full length of the protein. We generated several truncations and analyzed their dimerization in vivo. We found that the fragment containing amino acids 1–17 was responsible for the self-interaction and homodimerization of ORF8 in vivo (Figures 4J and 4K). On the basis of bioinformatics analysis, ORF8 contained an N-terminal aggregation domain (amino acids 3–13) (Figure 4L). Deleting these amino acids (ORF8Δ3–13) abolished the self-interaction and homodimerization of ORF8 in vivo (Figures 4M and 4N). Furthermore, ORF8Δ3–13 still interacted with p62 (Figure 4O). But it failed to bind to FAM134B or ATL3 (Figures 4P and 4Q), nor hijack FAM134B or ATL3 into ORF8/p62 liquid droplets (Figure 4R). As expected, ORF8Δ3–13 failed to inhibit ER-phagy or activate ER stress (Figures 4S and 4T). Taken together, these data confirm that ORF8 homodimerization is essential for its interaction with and recruitment of FAM134B and ATL3 into condensates to subvert ER-phagy.
Figure 4.
ORF8 homo-dimerization is important for ER-phagy inhibition
(A) Co-precipitation analysis of GFP-ORF8 with ORF8-Strep in HEK293T.
(B) Purified MBP-ORF8 was incubated with purified GST or GST-ORF8, and analysis of the self-interaction by GST pull-down.
(C) Analysis of the homo-dimerization of ORF8 by cross-linking with DSS. Cells were transfected with ORF8-FLAG for 24 h and treated with DSS as indicated for 30 min. Cell lysates were analyzed using WB.
(D) Analysis of the self-interaction of ORF8 with p62 overexpression by coIP assay.
(E) Analysis of the self-interaction of ORF8 under starvation treatment by coIP assay.
(F) Analysis of the homo-dimerization of ORF8 by cross-linking with DSS under starvation treatment. Cells stable expressing ORF8-FLAG were starved for 12 h. Cell lysates were analyzed using WB.
(G) Schematic illustration of ORF8 point mutations.
(H and I) Cross-linking with DSS assay (H) and coIP (I) analyzed ORF8 homo-dimerization. Cells were transfected with indicated plasmids for 24 h.
(J) Interaction between Strep-tagged WT ORF8 and mutants with GFP-ORF8-HA observed in a coIP assay.
(K) Analysis of the homodimerization of ORF8 deletion mutants by cross-linking with DSS and analyzed using WB.
(L) Prediction of ORF8 aggregation domain.
(M and N) Analysis of the homodimerization of ORF8 deletion mutants by coIP (M) and DSS assay (N).
(O) Interaction between GFP-ORF8 and mutants with p62-FLAG observed in a coIP assay.
(P and Q) Interaction between GFP-ORF8 and mutants with FAM134B-HA (P) or ATL3-HA (Q) observed in a coIP assay.
(R) Analysis of the colocalization of GFP-ORF8 and deletion mutant with FAM134B-HA or ATL3-HA with anti-HA or anti-FLAG antibodies using confocal microscopy. Scale bar, 10 μm. The number of colocalization of ORF8/p62 bodies with FAM134B or ATL3 in each cell was counted from 25 cells of three independent experiments. Two-tailed unpaired Student’s t test, ∗∗∗∗p < 0.0001.
(S) U2OS cells expressing mCherry-Sec61B were transfected with indicated plasmids for 24 h and then starved in EBSS for 12 h. Lysates were analyzed using WB.
(T) HEK293T cells were transfected with indicated plasmids for 24 h. Lysates were analyzed using WB.
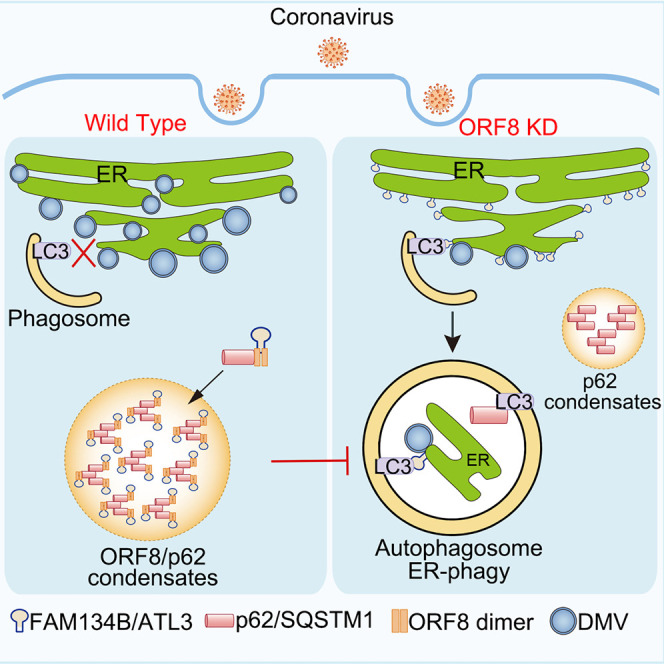
(U) Proposed model for the role of ORF8/p62 condensates in viral replication through ER-phagy inhibition.
Discussion
p62 exists in two populations: the membrane-bound form that functions in selective autophagy, and the membrane-less ubiquitinated p62 that undergoes condensates. p62 droplets wet autophagosomal membranes in WT cells which ensures piecemeal or complete droplet sequestration and droplet autophagy is important for the prevention of p62 accumulation. However, very little is known about the interactions between p62 droplets and ER-phagy. In this study, we uncovered an important mechanism that SARS-CoV-2 ORF8 protein interacted and formed condensates with p62 to facilitate viral DMV formation by subverting ER-phagy. The mechanism underlying ORF8/p62 condensate-induced suppression of ER-phagy is very intriguing. We found that the ER-phagy receptors, FAM134B and ATL3 were aggregated into ORF8/p62 liquid droplets via interaction with homodimer of ORF8 (Figure 4U).
The selective degradation of ER fragments containing viral proteins via ER-phagy is a reasonable coping strategy to limit viral replication, limit viral access to resources from the ER and prevent viral amplification. ER-phagy is also thought to dissolve ER fragments that are severely stressed by infection, thereby rescuing cells from death.58 , 59 , 60 Viruses, such as DENVs and ZIKVs, have developed a strategy to antagonize ER-phagy and impede the formation of ER- and viral protein-containing autophagosomes, thereby creating a favorable environment for their replication.61 Knockdown of RTN3 or ATL3 significantly reduced the replication of DENVs, ZIKVs, and WNVs.62 , 63 DENVs and ZIKVs were also found to exploit host BPIFB3 to inhibit FAM134B-dependent ER-phagy.64 Although we showed here that knockdown of FAM134B had minor effect on the replication of SARS-CoV-2, we further found that overexpression of FAM134B resulted in a reduction in the replication of SARS-CoV-2, and knockdown of Atg7 completely cut off the effect of FAM134B overexpression on viral production. Our results indicate that overexpression of FAM134B inhibits viral replication through ER-phagy activation.
Recent studies have shown that viral DMVs appeared at the early stage of SARS-CoV-2 infection, becoming detectable at 6 h after infection.49 SARS-CoV-2 exploits class III phosphatidylinositol 3-kinase (PI3K) and the lipid phosphatidic acid (PA) by acylglycerolphosphate acyltransferase (AGPAT) 1 and 2 in the ER for DMVs biogenesis.51 , 65 Our results showed that DMVs co-localize with FAM134B, ER-phagy can degrade Nsp3/4-tracked DMVs; and ORF8/p62 condensates inhibit the ER-phagic degradation of DMVs. Consistently, similar conclusions were obtained from native SARS-CoV-2 infection experiments, suggesting that ER-phagy was inhibited at the early stage of viral infection to facilitate DMV formation. In addition, we showed that ORF8/p62 inhibits ER-phagy by hijacking FAM134B and ATL3 into the condensates. In biology, condensation usually refers to a reversible biophysical process. ER-phagy was proposed to eliminate ER stress and prevents cellular apoptosis.55 Although we observed that ORF8/p62 inhibited ER-phagy during DMVs biogenesis, it will be of interest to determine whether in the very late stage of viral infected tissues, FAM134B and ATL3 could be released from ORF8/p62 condensates to mediate ER-phagy, clearing damaged ER fragments and alleviating severe ER stress, thereby preventing cell death.
ER morphology was significantly changed, and ER puncta were observed and ER stress was induced in ORF8/p62 co-expression cells. These results indicate that ORF8/p62 co-expression may lead to ER damage, which partly explain why the ER lumen protein ORF8 can bind p62: ORF8 may be released from damaged ER lumen to cytoplasm. The structure of SARS-CoV-2 ORF8 containing amino acids 18–121 was crystallized as a disulfide-linked homodimer via X-ray crystallography.57 However, in our in vivo coIP and DSS cross-linking assays, we were surprised to find that point mutants, based on the structure of ORF8, were still able to homo-dimerize. In contrast, our data suggest that the N-terminal amino acids 3–13 of ORF8 is responsible for its self-interaction and homodimerization. We speculated that this unexpected and apparent contradiction might be due to the different experimental systems used in the in vivo and in vitro assays. Further structural analyses of the full-length ORF8 and the different point mutants should be performed to elucidate and confirm the details of the mechanisms by which ORF8 homo-dimerizes. Nonetheless, the reported structure of SARS-CoV-2 ORF8 and our in vivo results provide important insights into the homo-dimerization of ORF8, which is critical for its interactions with ER-phagy receptors FAM134B or ATL3 and ER-phagy inhibition.
In summary, our study proposes an interesting and important model in which viral protein forms condensates with p62 to disrupt ER-phagy for viral DMV formation and viral replication. Moreover, our data also suggest that the development of therapeutics that could inhibit the ability of viral protein condensates and homodimerization could be an efficient approach to suppress viral replication.
Limitations of the study
This study has several limitations. Although we could distinguish between DMVs and autophagosomes by diameter and comparing our TEM images with the reported results, we failed to obtain the immune-EM by using LC3 antibody to track autophagosomes and dsRNA antibody to track DMVs. Furthermore, although we have showed that endogenous FAM134B can be recruited into p62 condensates and exhibited no colocalization with p62 bodies without ORF8 in SARS-CoV-2-infected cells, for obtaining most conclusions that ORF8/p62 condensates inhibit ER-phagy in this study, we relied on overexpression system rather than wild-type viruses. Further experiments, such as those involving the use of the real recombinant virus (ORF8Δ18–45/84–121), should be performed to confirm that SARS-CoV-2 infection induces formation of ORF8/p62 condensates, which sequester FAM134B, inhibit ER-phagy, and promote DMV formation.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-HA mAb | Sigma-Aldrich | Cat#: H9658; RRID: AB_260092 |
| Rabbit anti-HA pAb | Proteintech | Cat#: 51064-2-AP; RRID: AB_11042321 |
| Mouse anti-FLAG® M2 mAb | Sigma-Aldrich | Cat#: F1804; RRID: AB_262044 |
| Rabbit anti-Flag mAb | Abclonal | Cat#: AE092 |
| Mouse anti-Myc mAb | Cell Signaling Technology | Cat#: 2276; RRID: AB_331783 |
| Mouse anti-β-actin | Sigma-Aldrich | Cat#: A5441; RRID: AB_476744 |
| Mouse anti-Strep-tag II [GT661] | Abcam | Cat#: ab184224 |
| Mouse anti-SQSTM1 (clone: 2C11) | Abnova | Cat#: H00008878-M01; RRID: AB_437085 |
| Mouse anti-CHOP (L63F7) | Cell Signaling Technology | Cat#: 2895; RRID: AB_2089254 |
| Rabbit anti-Atg7 mAb | Cell Signaling Technology | Cat#: 8558; RRID: AB_10831194 |
| Mouse anti-mCherry mAb | Abclonal | Cat#: AE002; RRID: AB_2770407 |
| Mouse anti-GFP mAb | Abclonal | Cat#: AE012; RRID: AB_2770402 |
| Mouse anti-GST mAb | Abclonal | Cat#: AE001; RRID: AB_2770403 |
| Mouse anti-MBP mAb | Abclonal | Cat#: AE016; RRID: AB_2770406 |
| Mouse anti-His mAb | Abclonal | Cat#: AE003; RRID: AB_2728734 |
| Mouse anti-double stranded RNA (J2) | Scicons | Cat#: 10010200; RRID: AB_2651015 |
| Rabbit anti-SERP1 (RAMP4) pAb | Proteintech | Cat#: 17807-1-AP; RRID: AB_10597394 |
| Rabbit anti-REEP5 pAb | Proteintech | Cat#: 14643-1-AP; RRID: AB_2178440 |
| Rabbit anti-FAM134B pAb | Proteintech | Cat#: 21537-1-AP; RRID: AB_2878879 |
| Rabbit anti-CKAP4 (CLIMP63) | Proteintech | Cat#: 16686-1-AP; RRID: AB_2276275 |
| HRP-conjugated Goat Anti-Mouse IgG (H+L) | Abclonal | Cat#: AS003; RRID: AB_2769851 |
| HRP-conjugated Goat Anti- Rabbit IgG (H+L) | Abclonal | Cat#: AS014; RRID: AB_2769854 |
| HRP-conjugated Goat Anti-Mouse IgG Heavy Chain | Abclonal | Cat#: AS064; RRID: AB_2864058 |
| HRP-conjugated AffiniPure Goat Anti-Mouse IgG Light Chain | Abclonal | Cat#: AS062; RRID: AB_2864056 |
| Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat#: A32723; RRID: AB_1549585 |
| Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 488 | Thermo Fisher Scientific | Cat#: A32731; RRID: AB_2633280 |
| Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Thermo Fisher Scientific | Cat#: A-11031; RRID: AB_144696 |
| Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Thermo Fisher Scientific | Cat#: A-11036; RRID: AB_10563566 |
| Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat#: A-21236; RRID: AB_2535805 |
| Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat#: A-21244; RRID: AB_2535812 |
| Bacterial and virus strains | ||
| E. coli DH5α (DE3) | This paper | N/A |
| E. coli BL21 | This paper | N/A |
| SARS-CoV-2 (WBP) | This paper | Hubei Provincial Center for Disease Control and Prevention |
| Chemicals, peptides, and recombinant proteins | ||
| 1,6-hexanediol | Macklin | Cat#: H810887 |
| Chloroquine | Sigma-Aldrich | Cat#: C6628 |
| DSS | Thermo Fisher Scientific | Cat#: 21655 |
| Cyclopiazonic acid (CPA) | GLPBIO | Cat#: GC10268 |
| Thapsigargin (Tg) | Cell Signaling Technology | Cat#: 12758 |
| Polyethylenimine (PEI) | Yeasen | Cat#: 40815ES03 |
| Lipofectamine 2000 transfection reagent | Invitrogen | Cat#: 11668019 |
| Phosphate buffered saline (PBS) | Hyclone | Cat#:SH30256.01 |
| Dulbecco’s modified eagle’s medium (DMEM) | Gibco | Cat#: 11995065 |
| Fetal bovine serum (FBS) | Sigma-Aldrich | Cat#: 12303C |
| Penicillin-Streptomycin | Gibco | Cat#: 15140163 |
| 0.25% Trypsin-EDTA | Gibco | Cat#: 25200056 |
| OPTI-MEM | Gibco | Cat#: 31985088 |
| Protease inhibitor cocktail | Biosharp | Cat#: BL612A |
| Earle’s balanced salt solution (EBSS) | Sigma-Aldrich | Cat#: E2888 |
| Anti-Flag M2 affinity gel | Sigma-Aldrich | Cat#: A2220 |
| Anti-HA magnetic beads | Bimake | Cat#: B26202 |
| Streptactin beads 4FF | Smart Lifesciences | Cat#: SA053005 |
| MBPSep Dextrin Agarose Resin 6FF | Yeasen | Cat#: 20515ES08 |
| Glutathione Resin | Genscript | Cat#: L00206 |
| L-Glutathione reduced (GSH) | Biofrox | Cat#: 1392GR005 |
| DAPI | Servicebio | Cat#: G1012-10ML |
| Paraformaldehyde | Servicebio | Cat#: G1101-500ML |
| Bovine serum albumin (BSA) | Fisher Scientific | Cat#: BP1600-100 |
| Triton X-100 | Solarbio | Cat#: T8200 |
| Critical commercial assays | ||
| Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#: 23225 |
| Experimental models: Cell lines | ||
| HEK293T cells | CCTCC (China Center for Type Culture Collection) | Cat#: GDC187 |
| HeLa cells | ATCC | Cat#: CCL-2 |
| Vero E6 cells | Hubei Provincial Center for Disease Control and Prevention, China | N/A |
| U2OS cells | ATCC | Cat#: HTB-96 |
| Oligonucleotides | ||
| siRNA Targeting sequences: p62: GCATTGAAGTTGATATCGAT | This paper | N/A |
| siRNA Targeting sequences: Atg7 #1: GCCAACAUCCCUGGAUACAAG | This paper | N/A |
| siRNA Targeting sequences: Atg7 #2: CUGUGAACUUCUCUGACGU | This paper | N/A |
| shRNA Targeting sequences for FAM134B: GAGGTATCCTGGACT GATAAT |
This paper | N/A |
| sgRNA Targeting sequences for p62: GTGCGCCTGGAAGCCGC | This paper | N/A |
| Recombinant DNA | ||
| pLVX-EF1a-ORF8-Strep | Nevan J. Krogan Lab | Gordon et al., 202021 |
| pEGFP-C1-FAM134B | Qiming Sun Lab | Jiang et al., 202044 |
| pEGFP-C1-FAM134B-Δ84-233 | Qiming Sun Lab | Jiang et al., 202044 |
| mCherry-GFP-FAM134B | Qiming Sun Lab | Jiang et al., 202044 |
| pCDNA5-RTN3L-Flag | Qiming Sun Lab | N/A |
| pGEX-4T-GST-p62 | This paper | N/A |
| pEGFP-C3-p62 | Li Yu Lab | Sun et al., 201838 |
| pEGFP-C3-p62-ΔPB1 | Li Yu Lab | Sun et al., 201838 |
| pEGFP-C3-p62-S403E | Li Yu Lab | Sun et al., 201838 |
| pEGFP-C3-p62-M404V | Li Yu Lab | Sun et al., 201838 |
| MBP-mCherry-p62 | Li Yu Lab | Sun et al., 201838 |
| Flag-p62 | Wei Liu Lab | You et al., 201941 |
| Flag-p62-Δ1-85 | Wei Liu Lab | You et al., 201941 |
| Flag-p62-Δ86-121 | Wei Liu Lab | You et al., 201941 |
| Flag-p62-Δ122-168 | Wei Liu Lab | You et al., 201941 |
| Flag-p62-Δ169-253 | Wei Liu Lab | You et al., 201941 |
| Flag-p62-Δ254-310 | Wei Liu Lab | You et al., 201941 |
| Flag-p62-Δ311-384 | Wei Liu Lab | You et al., 201941 |
| Flag-p62-Δ385-440 | Wei Liu Lab | You et al., 201941 |
| pCDNA5-CCPG1-Flag | This paper | N/A |
| pCDNA3.1-scFv-ORF8-VIF-1/-2 | Hui Zhang Lab | Zhang et al., 202120 |
| pGEX-4T-GST-GFP-ORF8 | This paper | N/A |
| MBP-ORF8 | Yan Li Lab | N/A |
| pCDNA4-ORF8-Flag | This paper | N/A |
| pCDNA4-GFP-ORF8 | This paper | N/A |
| pCDNA4-GFP-ORF8-HA | This paper | N/A |
| pGEX-4T-GST-FAM134B | This paper | N/A |
| pCDNA4-FAM134B-HA | This paper | N/A |
| pCDNA4-ATL3-HA | This paper | N/A |
| pCDNA4-TEX264-HA | This paper | N/A |
| pCDNA4-SEC62-HA | This paper | N/A |
| pCDNA4-p62-Flag | This paper | N/A |
| pCDNA4-p62-Δ250-270-Flag | This paper | N/A |
| pCDNA4-p62-Δ260-280-Flag | This paper | N/A |
| pCDNA4-p62-Δ270-290-Flag | This paper | N/A |
| pCDNA4-p62-Δ280-300-Flag | This paper | N/A |
| pCDNA4-p62-Δ290-310-Flag | This paper | N/A |
| pCDNA4-p62-EHGG-AAAA-Flag | This paper | N/A |
| pCDNA4-p62-KRSR-AAAA-Flag | This paper | N/A |
| pCDNA4-p62-LTPV-AAAA-Flag | This paper | N/A |
| pCDNA4-p62-SPES-AAAA-Flag | This paper | N/A |
| pCDNA4-p62-SSTE-AAAA-Flag | This paper | N/A |
| mCherry-sec61β | This paper | N/A |
| pCDNA4-mCherry-RAMP4-HA | This paper | N/A |
| pCDNA4-mCherry-GFP-RAMP4-HA | This paper | N/A |
| pCDNA4-M-Flag | This paper | N/A |
| HA-Ub | This paper | N/A |
| pCDNA4-p62-HA | This paper | N/A |
| pEGFP-LC3 | This paper | N/A |
| pCDNA4-Keap-HA | This paper | N/A |
| pCDNA4-SARS-CoV-ORF8-Flag | This paper | N/A |
| pCDNA4-MERS-CoV-ORF8-Flag | This paper | N/A |
| NSP3-Myc | This paper | N/A |
| NSP4-Myc | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism 8 | GraphPad Software | N/A |
| Image J | NIH | https://imagej.en.softonic.com/ |
| IUPred2A | N/A | https://iupred2a.elte.hu/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Binbin Ding (dingbinbin@hust.edu.cn).
Materials availability
Plasmids and cell lines generated in this study will be available upon request with a completed Materials Transfer Agreement.
Experimental model and subject details
Cell cultures
HEK293T, HeLa, U2OS, Vero-E6, and p62 KO Vero-E6 cells were cultured in Dulbecco’s modified Eagle’s medium (11995065, Gibco) supplemented with 10% fetal bovine serum (12303C, Sigma-Aldrich) and 1% penicillin-streptomycin (15140163, Gibco) at 37°C with 5% CO2. CRISPR-Cas9 was used to generate p62 KO Vero cell line. For starvation treatment, cells were washed twice with phosphate-buffered saline (SH30256.01, Hyclone) and cultured in Earle’s balanced salt solution (E2888, Sigma-Aldrich) for 12 h. To induce ER stress, CPA (GC10268, GLPBIO) and thapsigargin (12758, Cell Signaling Technology) were used to treat cells for 12h or 6h, respectively. For chloroquine treatment, cells were incubated with 100 μM chloroquine (HY-17589A, MedChemExpress) for different times. For 1,6-hexanediol treatment, cells were washed with PBS for twice and incubated with 3% 1,6- hexanediol (H810887, Macklin) for 1 min, and then fixed in 4% PFA for immunofluorescence or lysed in TAP lysis buffer for subsequent immunoblotting. All cells were tested for mycoplasma negative.
Method details
Plasmids construction
Flag-p62 and deleted mutants were gifts from Wei Liu (Zhejiang University), pEGFP-C3-p62 and its point mutants, mCherry-p62, MBP-mCherry-p62, His6-UBx8 were gifts from Li Yu (Tsinghua University). ORF8-KD (namely pCDNA3.1-scFv-ORF8-VIF) was a gift from Hui Zhang (Sun Yat-Sen University). pLVX-EF1a-ORF8-Strep was a gift from Nevan J. Krogan Lab (UCSF). GFP-FAM134B, mCherry-GFP-FAM134B and pCDNA5-RTN3L-Flag were gifts from Qiming Sun (Zhejiang University). mCherry-Sec61B was obtained from Addgene (#172445). pCDNA4-ORF8-Flag, pCDNA4-p62-Flag were generated by respectively cloning SARS-CoV-2 ORF8 coding sequence or human p62 into pCDNA4-Flag vector. pCDNA4-EGFP-ORF8 and pCDNA4-EGFP-ORF8-HA were generated by inserting ORF8 ORF into pCDNA4 vector. pCDNA4-FAM134B-HA, pCDNA4-ATL3-HA, pCDNA4-SEC62-HA, pCDNA4-TEX264-HA pCDNA4-Keap1-HA were generated by cloning respective ORF into pCDNA4-HA vector. pCDNA4-mCherry-RAMP4-HA was cloned into pCDNA4-mCherry-HA vector and GFP was inserted into N-terminal of RAMP4 to get pCDNA4-mCherry-GFP-RAMP4-HA. MBP-ORF8 was a gift from Yan Li (Huazhong University of Science and Technology). GST-p62, GST-ORF8, GST-GFP-FAM134B were generated by respectively cloning human p62, ORF8 or GFP-FAM134B into pGEX-2T vector. GST-GFP-ORF8 was generated by inserting GFP coding sequence into C-terminal of GST according to GST-ORF8. All mutant constructs in this study were generated by PCR-mediated site-directed mutagenesis on the basis of their respective wide type constructs. All constructs were confirmed by DNA sequencing.
Antibodies and reagents
Mouse monoclonal anti-Flag (F1804), mouse anti-HA (H9658) and mouse anti-β-actin (A5441) were obtained from Sigma-Aldrich. Mouse anti-p62 (H00008878-M01) was obtained from Abnova. Mouse anti-Myc (2276), mouse anti-CHOP (2895) and rabbit anti-Atg7 (8558) were obtained from Cell Signaling Technology. Mouse anti-LC3 (M152-3) was obtained from MBL. Mouse anti-GFP (AE012), anti-mCherry (AE002), anti-GST (AE001), anti-MBP (AE016), anti-His (AE003), rabbit anti-Flag (AE092), rabbit anti-SARS-CoV-2 ORF3a (A20234), rabbit anti-SARS-CoV-2 ORF8 (A20235) were obtained from ABclonal. Mouse anti-Strep-tag-II (ab184224) was obtained from Abcam. Mouse anti-dsRNA (J2, 10010200) was obtained from Scicons. Rabbit anti-HA (51064-2-AP), anti-RAMP4 (17807-1-AP), anti-FAM134B (21537-1-AP), anti-CLIMP63 (16686-1-AP) and anti-REEP5 (14643-1-AP) were obtained from Proteintech. Anti-SARS-CoV-2 Nucleocapsid (N) (40143-MM05) was obtained from Sino Biological Inc. For immunoblotting, HRP-conjugated goat anti-mouse IgG (H+L) (AS003, Abclonal), HRP-conjugated goat anti-rabbit IgG (H+L) (AS014, Abclonal), HRP-conjugated goat anti-mouse IgG heavy chain (AS064, Abclonal) and HRP-conjugated AffiniPure goat anti-mouse IgG light chain (AS062, Abclonal) were used as secondary antibodies. For immunostaining, Alexa Fluor 488-conjugated goat anti-mouse IgG (H+L) (A32723), goat anti-rabbit IgG (H+L) (A32731), Alexa Fluor 568-conjugated goat anti-mouse IgG (H+L) (A-11031), goat anti-Rabbit IgG (H+L) (A-11036), Alexa Fluor 647-conjugated goat anti-mouse IgG (H+L) (A-21236), goat anti-rabbit IgG (H+L) (A-21244) were purchased from Thermo Fisher Scientific. Anti-HA magnetic beads (B26202) were obtained from Bimake. Anti-Flag M2 Affinity Gel (A2220) was obtained from Sigma-Aldrich. Streptactin beads 4FF was purchased from Smart Lifesciences.
RNA interference
siRNA oligonucleotides were obtained from GenePharma. siRNA sequences targeting specified genes are following: Atg7#1, GCCAACAUCCCUGGAUACAAG; Atg7#2, CUGUGAACUUCUCUGACGU p62, GCATTGAAGTTGATATCGAT; ORF3a, GAGAATCTTCACAATTGGAACTGTA. siRNAs were transfected into cells by using reagent according to the manufacturer’s instructions. Cells were harvested 36 h after transfection.
shRNA sequences targeting FAM134B: GAGGTATCCTGGACTGATAAT. The FAM134B annealed oligonucleotides were inserted into pLKO vector. The plasmid was transfected together with lentiviral package plasmids (phelper, VSV-G and TAT) into HEK293T cells to produce recombinant lentivirus. Then FAM134B KD Vero-E6 cells were obtained by lentivirus infection, 2 μg/mL puromycin selection and western blot verification.
Generation of stable cell line
The ORF8 coding sequence was cloned into plenti-puro-Flag vector. HeLa cells were infected with lentivirus collected from HEK293T cells transfected with lentiviral package plasmids (phelper, VSV-G and TAT) and plenti-ORF8-Flag. After 24 h, the stable transformants were cultured and selected in fresh DMEM containing 1 μg/mL puromycin.
Establishment of p62 knockdown Vero E6 cells
To generate p62 KO Vero-E6 cell line, p62 sgRNA targeting sequence (GTGCGCCTGGAAGCCGC) was inserted into pLKO-cas9 vector. HEK293T cells were transfected with pLKO-cas9-p62 sgRNA and lentiviral package plasmids (phelper, VSV-G and TAT). After 2 days, the supernatant was collected and used to infect Vero E6 cells. Twenty-four hours later, the medium was replaced with fresh medium containing with 2 μg/mL puromycin. Following 3 days screening, cells were seeded in 96 well plate and cultured. After two weeks, single clones were selected and seeded in 6 well plate. p62 KO cells were confirmed by WB and sequencing.
Immunoprecipitation and western blot
HEK293T cells were harvested and lysed with TAP lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% NP-40, 1 mM EDTA, Protease cocktail) for 30 min on ice. After centrifugation at 13000 rpm for 20 min at 4°C, the supernatants were collected and subjected to immunoprecipitation. For Flag and HA tag immunoprecipitation, tag affinity gel beads or magnetic beads were added in supernatants and incubated together overnight at 4°C. Beads with precipitated immunocomplexes were washed three times with TAP lysis buffer and boiled at 100°C for 10 min and subsequently analyzed by WB. Protein concentration was determined with BCA protein assay kit (23225; Thermo Fisher) according to manufacturer’s instructions. Equal amounts of protein were loaded and separated by appropriate concentration SDS-PAGE and electrophoretically transferred onto nitrocellulose membrane. After blocking with 5% nonfat milk in PBST for 1 h, membrane was incubated with the primary antibodies, followed by HRP-conjugated secondary antibodies.
IP/MS
HEK293T cells were transfected with ORF8-Flag for 36 hr and harvested and lysed with TAP lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% NP-40, 1 mM EDTA, Protease cocktail) for 30 min on ice. The supernatants were collected via centrifugation at 10,000 rpm for 30 min at 4°C and coimmunoprecipitated with Flag beads. The immunoprecipitated fractions were separated via 12% SDS polyacrylamide gel electrophoresis (PAGE) and analyzed via mass spectrometry using 30 min LC-MSMS (Shanghai Sangon Biotech Co., Ltd.).
Protein expression and purification
Escherichia coli strain BL21 (DE3) was transformed with plasmids (GST-GFP, MBP-mCherry-p62, GST-p62, GST-GFP-FAM134B, MBP-ORF8, GST-GFP-ORF8, His-UBx8) and Rosseta was selected to express the plasmid GST-GFP-FAM134B. Cells were cultured overnight in LB at 37°C. Then the overnight cultured bacteria were subcultured in fresh LB medium. When the optical density at 600 nm (OD600) reached 0.6–0.8, protein expression was induced with 0.5mM IPTG at 16°C for 16h. Cells pellet were suspended in lysis buffer (50mM Tris-HCl pH7.5, 150mM NaCl, 0.1%NP-40, 1mM DTT) and sonicated (15 cycles of 10 seconds on, 10 seconds off) on ice. After centrifugation at 12,000g for 20 mins at 4°C, the supernatants were incubated with corresponding agarose beads overnight at 4°C. The beads were washed with lysis buffer for three times and eluted with glutathione (GST-tag) or maltose (MBP-tag) or imidazole (His-tag) in 50mM Tris (pH 8.0). The elutes were further purified by chromatography. Protein expression and size were confirmed by SDS-PAGE and protein concentration was measured by Nanodrop.
GST pull-down assay
Escherichia coli strain BL21 (DE3) was selected to express recombinant protein. After the OD value reached 0.6–0.8, E. coli cells transformed with different plasmids were induced with 0.5mM IPTG at 16°C for 16h. Cell pellets were collected, lysed in lysis buffer (50mM Tris-HCl pH7.5, 150mM NaCl, 0.1%NP-40, 1mM DTT), and sonication on ice (15 cycles of 10 seconds on, 10 seconds off). The lysate was cleared by centrifugation at 12,000g for 20 mins at 4°C, and the supernatants were added to agarose beads to incubate overnight at 4°C. Then the beads were collected and resuspended in 50 μL SDS loading buffer, boiled at 100°C for 10min, and subjected to SDS-PAGE.
In vitro phase separation
For ORF8 in vitro phase separation assay, 10 μM recombinant proteins GST-GFP or GST-GFP-ORF8 were added in PEG8000 free buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mg/mL BSA) or added in PEG8000 buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mg/mL BSA, 10% PEG8000); for ORF8/p62 in vitro phase separation assay, 10 μM recombinant proteins GST-GFP with MBP-mCherry-p62/His-UBx8 or GST-GFP-ORF8 with MBP-mCherry-p62/His-UBx8 were added in phase separation buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mg/mL BSA); for FAM134B in vitro phase separation assay, 10 μM recombinant proteins GST-GFP-FAM134B or GST-GFP-FAM134B with MBP-ORF8 or GST-GFP-FAM134B with MBP-mCherry-p62/His-UBx8 or GST-GFP-FAM134B with MBP-ORF8, MBP-mCherry-p62/His-UBx8 were added in phase separation buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mg/mL BSA). After a reaction for 30 min at room temperature, 10 μL solution was loaded onto a chamber consist of a glass slide attached by two parallel strips of double-sided tape and coverslips, then observed and imaged by Zeiss LSM 780.
FRAP
Cells were transfected with indicated plasmids by using Lipofectamine 2000 (11668019, Thermo Fisher Scientific) according to the manufacturer’s instructions. Fluorescence recovery after photo bleaching (FRAP) assays were performed on an Olympus FV3000 confocal microscope with cell culture system. After 12 h, ORF8/p62 bodies were bleached for 10 s at intensity of 90% at 488 nm or 561 nm laser. Fluorescence recovery was recorded within a period of time. The focal plane was monitored and addressed by the Z drift compensator system. Images were acquired using a 60x/1.4 oil objective with Z series and time-series. Time Lapses was performed with 60s interval for 120 cycles. Mean fluorescence intensity was quantified by Image J and the values were normalized to background.
Fusion and fission
HeLa cells were seeded on confocal dishes and transfected with indicated plasmids. After 12 h, cells were imaged and captured by Olympus FV3000 Confocal Microscope with cell culture system. The focal plane was monitored and addressed by the Z drift compensator system. Images were acquired using a 60x/1.4 oil objective with Z series and time-series. Time Lapses was performed with 10 s interval for 100 cycles. Acquired images were processed with software FV31S-DT to analyze the fusion and fission events of ORF8/p62 puncta.
SARS-CoV-2 virus infection
All work with live SARS-CoV-2 virus were performed inside biosafety cabinets in the biosafety level 3 facility at Hubei Provincial Center for Disease Control and Prevention. Vero-E6 cells seeded in 6-well plates were infected with SARS-CoV-2 WBP at a MOI of 0.05 PFU/cell for 1 h at 37°C with 5% CO2, then infection medium was removed and replaced with fresh DMEM medium with 2% FBS.
Plaque assay
SARS-CoV-2-containing culture medium was serially 10-fold diluted. Vero-E6 cells in 6-well plates were grown to 60 to 70% confluency and infected with 100 μL of the dilutions. Plates were Incubated for 2 h at 37°C with 5% CO2, and then washed with PBS, the infection medium was replaced with methylcellulose, and plates were incubated at 37°C with 5% CO2 for another 3 to 4 days until visible viral plaques were detected. Plates were stained with 0.5% crystal violet for 4 h at room temperature and washed; then the plaques were counted and the viral titers were calculated.
Immunofluorescence analysis
Cells were seeded on coverslips within 12-well plates and transfected with indicated plasmids or treated with various stimuli as indicated. Twenty-four hours later, cells were washed with PBS and fixed with 4% paraformaldehyde for 15 min at room temperature, followed by permeabilized with 0.1% Triton X-100 for 10 min. After washing with PBS for three times, cells were blocked with 1% BSA for 30 min. Specific primary antibodies were diluted and incubated with cells overnight at 4°C. Then cells were washed with PBS for three times, followed by incubation with Alexa Fluor-conjugated goat anti-mouse IgG or goat anti-rabbit IgG secondary antibodies for 1 h at room temperature. Cells were then washed with PBS for three times and preserved with anti-quenching reagent and imaged by Zeiss LSM 780.
Quantification and statistical analysis
GraphPad Prism 8 software was used for statistical analysis. Statistical tests and sample sizes are indicated in the figure legends. A p value of <0.05 was considered statistically significant and a p value of >0.05 was considered statistically non-significant (ns).
Acknowledgments
We deeply thank Prof. Nevan J. Krogan (University of California, San Francisco [UCSF]) for providing SARS-CoV-2 viral proteins expression plasmids; Prof. Li Yu (Tsinghua University) and Prof. Wei Liu (Zhejiang University) for p62 expression plasmids; Prof. Qiming Sun (Zhejiang University) for EGFP-SEC61B, EGFP-FAM134B, and RTN3L expression plasmids and critical suggestions on ER-phagy assays; Prof. Hui Zhang (Sun Yat-Sen University) for pCDNA3.0-scFv-ORF8-VIF; Prof. Yan Li (Huazhong University of Science and Technology [HUST]) for MBP-ORF8 expression plasmid and helpful suggestions on protein purification; Prof. Shuguo Sun (HUST) for helpful suggestions on phase separation assays; and Pei Zhang (Wuhan Institute of Virology) for TEM assistance. We would like to thank Editage (www.editage.cn) for English language editing. This work was supported mainly by the Major Research Plan of the National Natural Science Foundation of China (92054107), the National Natural Science Foundation of China (U22A20337), and Open Research Fund Program of the State Key Laboratory of Virology of China (2022KF006).
Author contributions
X.T., J.L., and Z.Y. performed the experiments. R.C. contributed to protein purification. H.X. contributed to cloning. Y.Q., K.C., and B.H. contributed to SARS-CoV-2 infection experiments. C.X. provided critical reagents and analyzed the data. B.D. and Y.Q. conceived the project, designed the experiments, and analyzed the data. B.D., X.T., and J.L. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: March 10, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112286.
Supplemental information
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Borgese N., Francolini M., Snapp E. Endoplasmic reticulum architecture: structures in flux. Curr. Opin. Cell Biol. 2006;18:358–364. doi: 10.1016/j.ceb.2006.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferro-Novick S., Reggiori F., Brodsky J.L. ER-phagy, ER homeostasis, and ER quality control: implications for disease. Trends Biochem. Sci. 2021;46:630–639. doi: 10.1016/j.tibs.2020.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chino H., Mizushima N. ER-phagy: quality control and turnover of endoplasmic reticulum. Trends Cell Biol. 2020;30:384–398. doi: 10.1016/j.tcb.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Khaminets A., Heinrich T., Mari M., Grumati P., Huebner A.K., Akutsu M., Liebmann L., Stolz A., Nietzsche S., Koch N., et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522:354–358. doi: 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- 5.Chen Q., Xiao Y., Chai P., Zheng P., Teng J., Chen J. ATL3 is a tubular ER-phagy receptor for GABARAP-mediated selective autophagy. Curr. Biol. 2019;29:846–855.e6. doi: 10.1016/j.cub.2019.01.041. [DOI] [PubMed] [Google Scholar]
- 6.Smith M.D., Harley M.E., Kemp A.J., Wills J., Lee M., Arends M., von Kriegsheim A., Behrends C., Wilkinson S. CCPG1 is a non-canonical autophagy cargo receptor essential for ER-phagy and pancreatic ER proteostasis. Dev. Cell. 2018;44:217–232.e11. doi: 10.1016/j.devcel.2017.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grumati P., Morozzi G., Hölper S., Mari M., Harwardt M.L.I., Yan R., Müller S., Reggiori F., Heilemann M., Dikic I. Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. Elife. 2017;6 doi: 10.7554/eLife.25555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chino H., Hatta T., Natsume T., Mizushima N. Intrinsically disordered protein TEX264 mediates ER-phagy. Mol. Cell. 2019;74:909–921.e6. doi: 10.1016/j.molcel.2019.03.033. [DOI] [PubMed] [Google Scholar]
- 9.An H., Ordureau A., Paulo J.A., Shoemaker C.J., Denic V., Harper J.W. TEX264 is an endoplasmic reticulum-resident ATG8-interacting protein critical for ER remodeling during nutrient stress. Mol. Cell. 2019;74:891–908.e10. doi: 10.1016/j.molcel.2019.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fumagalli F., Noack J., Bergmann T.J., Cebollero E., Pisoni G.B., Fasana E., Fregno I., Galli C., Loi M., Soldà T., et al. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat. Cell Biol. 2016;18:1173–1184. doi: 10.1038/ncb3423. [DOI] [PubMed] [Google Scholar]
- 11.Junjhon J., Pennington J.G., Edwards T.J., Perera R., Lanman J., Kuhn R.J. Ultrastructural characterization and three-dimensional architecture of replication sites in dengue virus-infected mosquito cells. J. Virol. 2014;88:4687–4697. doi: 10.1128/jvi.00118-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Welsch S., Miller S., Romero-Brey I., Merz A., Bleck C.K.E., Walther P., Fuller S.D., Antony C., Krijnse-Locker J., Bartenschlager R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lennemann N.J., Coyne C.B. Dengue and Zika viruses subvert reticulophagy by NS2B3-mediated cleavage of FAM134B. Autophagy. 2017;13:322–332. doi: 10.1080/15548627.2016.1265192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knoops K., Kikkert M., Worm S.H.E.v.d., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Wilde A.H., Raj V.S., Oudshoorn D., Bestebroer T.M., van Nieuwkoop S., Limpens R.W.A.L., Posthuma C.C., van der Meer Y., Bárcena M., Haagmans B.L., et al. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-α treatment. J. Gen. Virol. 2013;94:1749–1760. doi: 10.1099/vir.0.052910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santerre M., Arjona S.P., Allen C.N., Shcherbik N., Sawaya B.E. Why do SARS-CoV-2 NSPs rush to the ER? J. Neurol. 2021;268:2013–2022. doi: 10.1007/s00415-020-10197-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hachim A., Kavian N., Cohen C.A., Chin A.W.H., Chu D.K.W., Mok C.K.P., Tsang O.T.Y., Yeung Y.C., Perera R.A.P.M., Poon L.L.M., et al. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat. Immunol. 2020;21:1293–1301. doi: 10.1038/s41590-020-0773-7. [DOI] [PubMed] [Google Scholar]
- 18.Alam I., Kamau A.A., Kulmanov M., Jaremko Ł., Arold S.T., Pain A., Gojobori T., Duarte C.M. Functional pangenome analysis shows key Features of E protein are preserved in SARS and SARS-CoV-2. Front. Cell. Infect. Microbiol. 2020;10:405. doi: 10.3389/fcimb.2020.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J.Y., Liao C.H., Wang Q., Tan Y.J., Luo R., Qiu Y., Ge X.Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286 doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Chen Y., Li Y., Huang F., Luo B., Yuan Y., Xia B., Ma X., Yang T., Yu F., et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Ι. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2024202118. e2024202118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hyman A.A., Weber C.A., Jülicher F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- 23.Boeynaems S., Alberti S., Fawzi N.L., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., Van Den Bosch L., et al. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 2018;28:420–435. doi: 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin Y., Brangwynne C.P. Liquid phase condensation in cell physiology and disease. Science. 2017;357 doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 25.Li P., Banjade S., Cheng H.C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J.V., King D.S., Banani S.F., et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu S., Ye Q., Singh D., Cao Y., Diedrich J.K., Yates J.R., 3rd, Villa E., Cleveland D.W., Corbett K.D. The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nat. Commun. 2021;12:502. doi: 10.1038/s41467-020-20768-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iserman C., Roden C.A., Boerneke M.A., Sealfon R.S.G., McLaughlin G.A., Jungreis I., Fritch E.J., Hou Y.J., Ekena J., Weidmann C.A., et al. Genomic RNA elements drive phase separation of the SARS-CoV-2 nucleocapsid. Mol. Cell. 2020;80:1078–1091.e6. doi: 10.1016/j.molcel.2020.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cubuk J., Alston J.J., Incicco J.J., Singh S., Stuchell-Brereton M.D., Ward M.D., Zimmerman M.I., Vithani N., Griffith D., Wagoner J.A., et al. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat. Commun. 2021;12:1936. doi: 10.1038/s41467-021-21953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S., Dai T., Qin Z., Pan T., Chu F., Lou L., Zhang L., Yang B., Huang H., Lu H., Zhou F. Targeting liquid-liquid phase separation of SARS-CoV-2 nucleocapsid protein promotes innate antiviral immunity by elevating MAVS activity. Nat. Cell Biol. 2021;23:718–732. doi: 10.1038/s41556-021-00710-0. [DOI] [PubMed] [Google Scholar]
- 30.Savastano A., Ibáñez de Opakua A., Rankovic M., Zweckstetter M. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat. Commun. 2020;11:6041. doi: 10.1038/s41467-020-19843-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carlson C.R., Asfaha J.B., Ghent C.M., Howard C.J., Hartooni N., Safari M., Frankel A.D., Morgan D.O. Phosphoregulation of phase separation by the SARS-CoV-2 N protein suggests a biophysical basis for its dual functions. Mol. Cell. 2020;80:1092–1103.e4. doi: 10.1016/j.molcel.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perdikari T.M., Murthy A.C., Ryan V.H., Watters S., Naik M.T., Fawzi N.L. SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs. EMBO J. 2020;39 doi: 10.15252/embj.2020106478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen H., Cui Y., Han X., Hu W., Sun M., Zhang Y., Wang P.H., Song G., Chen W., Lou J. Liquid-liquid phase separation by SARS-CoV-2 nucleocapsid protein and RNA. Cell Res. 2020;30:1143–1145. doi: 10.1038/s41422-020-00408-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Y., Ma L., Cai S., Zhuang Z., Zhao Z., Jin S., Xie W., Zhou L., Zhang L., Zhao J., Cui J. RNA-induced liquid phase separation of SARS-CoV-2 nucleocapsid protein facilitates NF-κB hyper-activation and inflammation. Signal Transduct. Target. Ther. 2021;6:167. doi: 10.1038/s41392-021-00575-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Komatsu M., Waguri S., Koike M., Sou Y.S., Ueno T., Hara T., Mizushima N., Iwata J.I., Ezaki J., Murata S., et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 37.Choi Y., Bowman J.W., Jung J.U. Autophagy during viral infection - a double-edged sword. Nat. Rev. Microbiol. 2018;16:341–354. doi: 10.1038/s41579-018-0003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun D., Wu R., Zheng J., Li P., Yu L. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 2018;28:405–415. doi: 10.1038/s41422-018-0017-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agudo-Canalejo J., Schultz S.W., Chino H., Migliano S.M., Saito C., Koyama-Honda I., Stenmark H., Brech A., May A.I., Mizushima N., Knorr R.L. Wetting regulates autophagy of phase-separated compartments and the cytosol. Nature. 2021;591:142–146. doi: 10.1038/s41586-020-2992-3. [DOI] [PubMed] [Google Scholar]
- 40.Zaffagnini G., Savova A., Danieli A., Romanov J., Tremel S., Ebner M., Peterbauer T., Sztacho M., Trapannone R., Tarafder A.K., et al. p62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J. 2018;37 doi: 10.15252/embj.201798308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.You Z., Jiang W.X., Qin L.Y., Gong Z., Wan W., Li J., Wang Y., Zhang H., Peng C., Zhou T., et al. Requirement for p62 acetylation in the aggregation of ubiquitylated proteins under nutrient stress. Nat. Commun. 2019;10:5792. doi: 10.1038/s41467-019-13718-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y., Willis T.L., Button R.W., Strang C.J., Fu Y., Wen X., Grayson P.R.C., Evans T., Sipthorpe R.J., Roberts S.L., et al. Cytoplasmic DAXX drives SQSTM1/p62 phase condensation to activate Nrf2-mediated stress response. Nat. Commun. 2019;10:3759. doi: 10.1038/s41467-019-11671-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miao G., Zhao H., Li Y., Ji M., Chen Y., Shi Y., Bi Y., Wang P., Zhang H. ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the SNARE complex required for autolysosome formation. Dev. Cell. 2021;56:427–442.e5. doi: 10.1016/j.devcel.2020.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang X., Wang X., Ding X., Du M., Li B., Weng X., Zhang J., Li L., Tian R., Zhu Q., et al. FAM134B oligomerization drives endoplasmic reticulum membrane scission for ER-phagy. EMBO J. 2020;39 doi: 10.15252/embj.2019102608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang J.R., Lingeman E., Luong T., Ahmed S., Muhar M., Nguyen T., Olzmann J.A., Corn J.E. A genome-wide ER-phagy screen highlights key roles of mitochondrial metabolism and ER-resident UFMylation. Cell. 2020;180:1160–1177.e20. doi: 10.1016/j.cell.2020.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klein S., Cortese M., Winter S.L., Wachsmuth-Melm M., Neufeldt C.J., Cerikan B., Stanifer M.L., Boulant S., Bartenschlager R., Chlanda P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020;11:5885. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snijder E.J., Limpens R.W.A.L., de Wilde A.H., de Jong A.W.M., Zevenhoven-Dobbe J.C., Maier H.J., Faas F.F.G.A., Koster A.J., Bárcena M. A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolff G., Melia C.E., Snijder E.J., Bárcena M. Double-membrane vesicles as platforms for viral replication. Trends Microbiol. 2020;28:1022–1033. doi: 10.1016/j.tim.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cortese M., Lee J.Y., Cerikan B., Neufeldt C.J., Oorschot V.M.J., Köhrer S., Hennies J., Schieber N.L., Ronchi P., Mizzon G., et al. Integrative imaging reveals SARS-CoV-2-induced reshaping of subcellular morphologies. Cell Host Microbe. 2020;28:853–866.e5. doi: 10.1016/j.chom.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oudshoorn D., Rijs K., Limpens R.W.A.L., Groen K., Koster A.J., Snijder E.J., Kikkert M., Bárcena M. Expression and cleavage of middle east respiratory syndrome coronavirus nsp3-4 polyprotein induce the formation of double-membrane vesicles that mimic those associated with coronaviral RNA replication. mBio. 2017;8 doi: 10.1128/mBio.01658-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Twu W.I., Lee J.Y., Kim H., Prasad V., Cerikan B., Haselmann U., Tabata K., Bartenschlager R. Contribution of autophagy machinery factors to HCV and SARS-CoV-2 replication organelle formation. Cell Rep. 2021;37 doi: 10.1016/j.celrep.2021.110049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Angelini M.M., Akhlaghpour M., Neuman B.W., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013;4 doi: 10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang X.S., Tang L.Y., Dai J., Zhou H., Li S.J., Xia Q.C., Wu J.R., Zeng R. Quantitative analysis of severe acute respiratory syndrome (SARS)-associated coronavirus-infected cells using proteomic approaches: implications for cellular responses to virus infection. Mol. Cell. Proteomics. 2005;4:902–913. doi: 10.1074/mcp.M400112-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yeung Y.S., Yip C.W., Hon C.C., Chow K.Y.C., Ma I.C.M., Zeng F., Leung F.C.C. Transcriptional profiling of Vero E6 cells over-expressing SARS-CoV S2 subunit: insights on viral regulation of apoptosis and proliferation. Virology. 2008;371:32–43. doi: 10.1016/j.virol.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Loi M., Raimondi A., Morone D., Molinari M. ESCRT-III-driven piecemeal micro-ER-phagy remodels the ER during recovery from ER stress. Nat. Commun. 2019;10:5058. doi: 10.1038/s41467-019-12991-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ke P.Y., Chen S.S.L. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J. Clin. Invest. 2011;121:37–56. doi: 10.1172/jci41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Flower T.G., Buffalo C.Z., Hooy R.M., Allaire M., Ren X., Hurley J.H. Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2021785118. e2021785118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moretti J., Roy S., Bozec D., Martinez J., Chapman J.R., Ueberheide B., Lamming D.W., Chen Z.J., Horng T., Yeretssian G., et al. STING senses microbial viability to orchestrate stress-mediated autophagy of the endoplasmic reticulum. Cell. 2017;171:809–823.e13. doi: 10.1016/j.cell.2017.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liang S., Wang F., Bao C., Han J., Guo Y., Liu F., Zhang Y. BAG2 ameliorates endoplasmic reticulum stress-induced cell apoptosis in Mycobacterium tuberculosis-infected macrophages through selective autophagy. Autophagy. 2020;16:1453–1467. doi: 10.1080/15548627.2019.1687214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chiramel A.I., Dougherty J.D., Nair V., Robertson S.J., Best S.M. FAM134B, the selective autophagy receptor for endoplasmic reticulum turnover, inhibits replication of Ebola virus strains Makona and Mayinga. J. Infect. Dis. 2016;214:S319–S325. doi: 10.1093/infdis/jiw270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li J., Gao E., Xu C., Wang H., Wei Y. ER-phagy and microbial infection. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.771353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aktepe T.E., Liebscher S., Prier J.E., Simmons C.P., Mackenzie J.M. The host protein reticulon 3.1A is utilized by Flaviviruses to facilitate membrane remodelling. Cell Rep. 2017;21:1639–1654. doi: 10.1016/j.celrep.2017.10.055. [DOI] [PubMed] [Google Scholar]
- 63.Neufeldt C.J., Cortese M., Scaturro P., Cerikan B., Wideman J.G., Tabata K., Moraes T., Oleksiuk O., Pichlmair A., Bartenschlager R. ER-shaping atlastin proteins act as central hubs to promote flavivirus replication and virion assembly. Nat. Microbiol. 2019;4:2416–2429. doi: 10.1038/s41564-019-0586-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Evans A.S., Lennemann N.J., Coyne C.B. BPIFB3 regulates endoplasmic reticulum morphology to facilitate flavivirus replication. J. Virol. 2020;94 doi: 10.1128/jvi.00029-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tabata K., Prasad V., Paul D., Lee J.Y., Pham M.T., Twu W.I., Neufeldt C.J., Cortese M., Cerikan B., Stahl Y., et al. Convergent use of phosphatidic acid for hepatitis C virus and SARS-CoV-2 replication organelle formation. Nat. Commun. 2021;12:7276. doi: 10.1038/s41467-021-27511-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.