Abstract
The closely linked H19 and Igf2 genes show highly similar patterns of gene expression but are reciprocally imprinted. H19 is expressed almost exclusively from the maternally inherited chromosome, while Igf2 expression is mostly from the paternal chromosome. In humans, loss of imprinting at this locus is associated with tumors and with developmental disorders. Monoallelic expression at the imprinted Igf2/H19 locus occurs by at least two distinct mechanisms: a developmentally regulated silencing of the paternal H19 promoter, and transcriptional insulation of the maternal Igf2 promoters. Both mechanisms of allele-specific silencing are ultimately dependent on a common cis-acting element located just upstream of the H19 promoter. The coordinated expression patterns and some experimental data support the idea that positive regulatory elements are also shared by the two genes. To clarify the organization and function of positive and negative regulatory elements at the H19/Igf2 locus, we analyzed two mouse mutations. First, we generated a deletion allele to localize enhancers used in vivo for expression of both H19 and Igf2 in mesodermal tissues to sequences downstream of the H19 gene. Coincidentally, we demonstrated that some expression of Igf2 is independent of the shared enhancer element. Second, we used this new information to further characterize an ectopic H19 differentially regulated region and the associated insulator. We demonstrated that its activity is parent-of-origin dependent. In contrast to recent results from Drosophila model systems; we showed that this duplication of a mammalian insulator does not interfere with its normal function. Implications of these findings for current models for monoallelic gene expression at this locus are discussed.
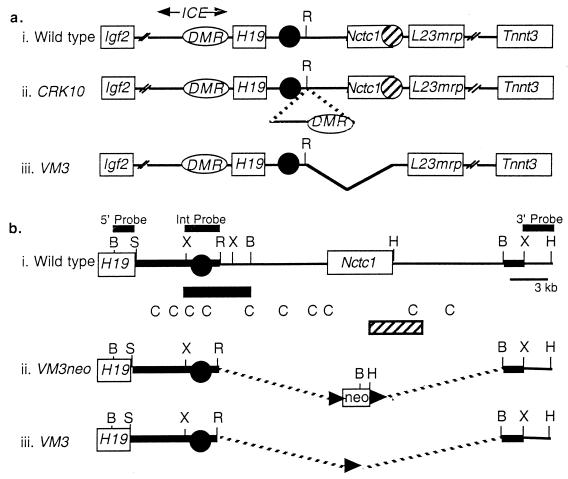
Igf2 and H19 are closely linked and reciprocally imprinted genes located on the distal end of mouse chromosome 7 (48) (Fig. 1a). The regulation of the two genes has been intensively studied both as a model system for understanding mechanisms of genomic imprinting and because dysregulation of IGF2 in humans is associated with the developmental disorder Beckwith-Wiedemann syndrome and with many types of tumors (33). The two genes are part of a large cluster of imprinted genes whose organization and monoallelic expression patterns are well conserved between mice and humans (30, 31). H19 represents one limit of this imprinted cluster, as the next known genes, Nctc1 and L23mrp, are both biallelically expressed (19, 49).
FIG. 1.
Diagram of the wild-type, CRK10, and VM3 chromosomes in the Igf2/H19 region. (a) Summary of the wild-type (i), CRK10 (ii), and VM3 (iii) alleles, depicting the relative organization of the Igf2, H19, Nctc1, L23mrp, and Tnnt3 genes (open rectangles). Endodermal and skeletal-muscle-specific enhancers for H19 and Igf2 expression are represented by closed and hatched circles, respectively. The mapped locations of these enhancers are based on in vivo analyses of chromosomal deletions (reference 26 and this study) and also on in vitro transfection experiments (20, 46). The ICE, for imprinting control element, is defined genetically as the minimal upstream sequence required to imprint single-copy H19 transgenes (20) and as a sequence whose deletion results in the loss of imprinting of H19 and Igf2 (20, 36, 42). Within the ICE is the 2-kb DMR (for differentially methylated region) (oval) that is hypermethylated specifically on the paternal chromosome (4, 8, 14, 29, 43, 44). Mapping to the DMR are domains of maternal-chromosome-specific nuclease hypersensitivity. The CRK10 allele (ii) carries a 9.2-kb insertion at kb +10.7 (relative to the H19 transcriptional start site). The inserted DNA includes the H19DMR, all the maternal-chromosome-specific nuclease-hypersensitive domains, and additional 5′ sequences that encompass all of the 5′ sequence elements required to imprint single-copy H19 transgenes (20). The inserted element is the minimal DNA sequence that is known to carry a parent-of-origin-specific transcriptional insulator (reference 20 and this study). The VM3 allele (iii) deletes sequences showing muscle-specific enhancer activity in vitro and in transgenic mice (18, 20). This deletion encompasses the Nctc1 gene body. (b) Strategy for construction of the VM3 allele by deleting from kb +10.7 to +34.7 downstream of the H19 gene. The wild-type chromosome (i), the targeted VM3neo chromosome (ii), and the targeted VM3 chromosome after Cre recombinase-mediated excision of the neomycin-selectable marker (iii) are depicted. The endodermal enhancers, as defined by in vitro transfection studies, are represented by a closed circle (46). The deletion confirming the role of these enhancers for in vivo expression of both H19 and Igf2 is indicated by the closed rectangle below line i (26). Also depicted below line i are sequences conserved between human and mouse (C) (18) as well as sequences showing enhancer activity specifically in myoblast cell lines (hatched rectangle) (20). The flanking sequences used to direct homologous recombination are represented by the thickened lines. Probes used to detect the correctly targeted clones are depicted above line i. B, BamHI; S, SalI; R, EcoRI; H, HindIII; X, XbaI.
Both H19 and Igf2 are highly expressed during fetal and early postnatal development and show essentially identical spatial and temporal specificities. In fact, as suggested by their close linkage, their reciprocal imprinting, and their overlapping expression patterns, the two genes share transcriptional regulatory elements. The paternal silencing of H19 and maternal repression of Igf2 both depend on a common cis-acting element, here called the H19ICE (for H19 imprinting control element), located upstream of the H19 promoter (Fig. 1a) (20, 42). The boundaries of the ICE are not entirely precise, as they differ depending on the specific assay used. Sequences to kb −7 (relative to the start site of the H19 mRNA transcript) are sufficient to imprint single-copy H19 transgenes (20). If sequences to only kb −3.8 are used, only transgenes inserted as multiple copies are imprinted, and even then paternally inherited H19 transgenes are expressed in occasional pups (32). Deletion of endogenous sequences between kb −3.8 and 2.0 (or between kb −7 and −0.8) on the maternal chromosome results in the loss of imprinting of the paternal H19 allele. Significantly, the same deletion inherited through the maternal germline results in activation of the normally silent maternal Igf2 allele (36, 42). The ICE includes DNA sequence domains, centered at kb −2.4 and −3.8, that show nuclease hypersensitivity specifically on the maternal chromosome (16, 23, 39). These regions of hypersensitivity were identified in somatic tissue and lie within a 2-kb domain of DNA showing hypermethylation specific to the paternal chromosome (4, 8, 14, 29, 43, 44). This differentially methylated region (H19DMR) is hypermethylated on the paternal chromosome at all stages of development and has been demonstrated to have both silencing and insulating activities (7, 9, 17, 20, 21). On the unmethylated maternal chromosome, the H19ICE functions as a transcriptional insulator and blocks activation of the Igf2 promoters by distal enhancer elements. On the paternal chromosome, the methylation imprint at the H19DMR acts to disrupt insulator function and thereby allows expression of Igf2 on that chromosome. Several labs have recently demonstrated that CTCF, a known enhancer-blocking protein, selectively binds to nonmethylated CpG-containing sequences within the H19DMR, thus providing a molecular basis for parent-of-origin-specific activity of the insulator (7, 17, 22, 38). Other studies, however, indicate that this model does not explain all aspects of Igf2 monoallelism and that elements in addition to the H19DMR are required for imprinting of Igf2 (1, 12).
The insulator model for Igf2 monoallelic expression demands that all H19 and Igf2 enhancers be located downstream of the H19DMR or its associated insulator, while the coexpression of Igf2 and H19 suggests that these enhancer elements are shared. In fact, much of the endoderm-specific expression of both genes is dependent on shared enhancers that must lie between 7.2 and 13 kb downstream of the H19 promoter and about 80 kb downstream of the Igf2 promoter (26). However, the location of regulatory elements required to drive expression in mesodermal tissues is less clear. Transgenic mouse and in vitro transfection studies together indicate that enhancers capable of directing expression of H19 in skeletal muscle are located at between kb +25 and +28 (18, 20). Transgenic studies also suggest that additional enhancer elements that can direct H19 expression in several tissues, including cardiac muscle, may lie downstream of kb +35 (2, 20). However, neither the necessity of these elements for expression of the H19 gene in its normal chromosomal location nor their role in the expression of Igf2 has yet been tested.
In addition to its role in regulating activity of the insulator element, the paternal imprint at the H19DMR has a second, genetically distinct function of blocking transcription of the paternal H19 allele by directing CpG methylation of the paternal H19 promoter and gene body sequences. Once established, this promoter methylation and associated epigenetic changes are capable of silencing the H19 promoter independently of the H19DMR (36). We have previously generated a mutant allele, CRK10, in which the sequences from kb −10 to −0.8 that encompass the H19DMR were inserted at the kb +10.7 position (Fig. 1b) (20). As predicted by the insulator model, transcription of H19 in skeletal muscle was blocked while expression in liver was unaffected. We also demonstrated that CpGs in the inserted DMR element were methylated only when paternally inherited. However, it could not be determined whether this methylation was capable of blocking insulator activity or whether the hypermethylation from the exogenous H19DMR could spread to neighboring sequences.
In this study, we generated a 24-kb chromosomal deletion of sequences that included the presumptive mesodermal enhancers, and we further analyzed the CRK10 insertion mutation to clarify the organization and function of regulatory elements at the Igf2/H19 locus. First, by deletion of the putative mesodermal enhancer elements, we demonstrated that normal expression of H19 and Igf2 in both the tongue and limb muscle is dependent on these sequences. However, expression in mesodermal cells in other organs is dependent on additional enhancers downstream of kb +36. We demonstrated the latter point by using the CRK10 allele of chromosome 7 in which the H19ICE has been moved to kb +10.7. In a second set of experiments, we further analyzed the function of this exogenously located ICE/H19DMR/Igf2-insulator. We showed that while the insulator function remained parent-of-origin dependent, in its new location it could not direct methylation of neighboring sequences as it does the H19 promoter. We also demonstrated that insertion of this second H19DMR element had no effect on expression of Igf2 from the maternal chromosome. Thus, the insertion of a second active insulator element did not interfere with silencing of the maternal Igf2 gene. Implications of this finding for possible mechanisms of transcriptional insulation are discussed. Finally and parenthetically, we demonstrated that the mouse Nctc1 gene is not required for normal mouse development or fertility.
MATERIALS AND METHODS
Generation of VM3 mutant mice.
For the structure of the flanking sequences used to direct homologous recombination, see Fig. 1b. The targeting vector was linearized and transfected into RI mouse embryonic stem cells. To identify VM3neo clones, DNAs from G418-resistant colonies were digested with BamHI and probed with a 1.2-kb BamHI-SalI fragment from just outside the 5′ flank to identify a 11.3-kb band in wild-type cells and an additional 10-kb band in targeted lines. Likewise, digestion with HindIII and probing with a 4-kb HindIII-XbaI fragment from just outside the 3′ flank identified a 15-kb band in wild-type cells and an additional 5.5-kb band in correctly targeted clones. Two independently derived lines were injected into C57/BL6-J blastocysts to generate chimeric founders. Founder males were mated with mice carrying the EIIa-cre transgene (25) to excise neomycin resistance-encoding sequences. Successful excision of the neomycin resistance gene was assayed by digestion with XbaI and hybridization to a 2.0-kb internal XbaI-EcoRI fragment which displays 3.1-, 2.4-, and 4.3-kb bands for the VM3, wild-type, and VM3neo alleles, respectively.
RNA analysis.
Tissues were homogenized in TRIZOL reagent (Life Technologies) by using a power homogenizer, and the RNA was isolated according to the manufacturer's protocol. Expression levels were analyzed by Northern blotting. Probes used to quantitate H19, Igf2, and EF2a RNAs were described previously (20). cDNA from p6 skeletal muscle was amplified using L23mrp-specific primers (5′-ATGTGTTGTACCCCCTTTACC-3′ and 5′-TGGTCCAAATCCTGCTGTC-3′) to obtain a 455-bp probe or using Tnnt3-specific primers (5′-AGAAGAGAGGAGGAGGATG-3′ and 5′-GTGCAGAGCTGGAATAAG-3′) to obtain a 475-bp probe. Expression levels were quantified with the Molecular Dynamics Storm PhosphorImaging System.
Allele-specific expression of Igf2.
Mice carrying the CRK10 mutation were crossed with Dis7CAS mice as described in the legend to Fig. 5. Dis7CAS mice are mostly Mus domesticus but are homozygous M. castaneous across the H19/Igf2 locus (15). Allele-specific expression of Igf2 was analyzed using the single-nucleotide primer extension (SNuPE) assay (36, 40).
FIG. 5.
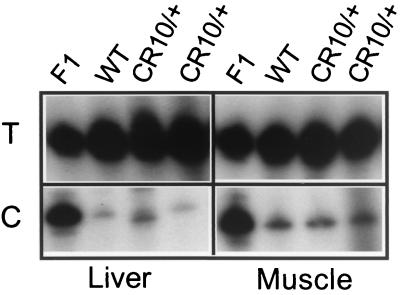
Effect of maternal-chromosome-based inheritance of the CRK10 mutation on Igf2 expression. Skeletal muscle and liver RNA samples were treated with DNase I, amplified for Igf2 by reverse transcription-PCR, and analyzed for allele-specific expression by SNuPE. Expression was analyzed in neonates inheriting the CRK10 mutation with an M. domesticus Igf2 allele via the maternal chromosome and inheriting a wild-type chromosome with an M. castaneous Igf2 allele via the paternal chromosome (CRK10/+). Their control littermates likewise have maternal-chromosome-derived domesticus and paternal-chromosome-derived M. castaneous alleles of Igf2, but both chromosomes are wild type (+/+). Lane F1 is a control representing a 50:50 mix of M. castaneous and M. domesticus substrates. M. domesticus (or maternal) RNA results in incorporation of cytosine (C), and M. casteneous (or paternal) RNA results in incorporation of thymidine (T).
RESULTS
A shared skeletal muscle enhancer for H19 and Igf2.
In vivo transgenic analyses indicated that enhancers capable of driving transcription of H19 in skeletal muscle lie between kb +13 and +36 (3, 10, 18, 20, 32). This region includes six short sequence elements that are well conserved between mice and humans and therefore are potentially important regulatory sites (18). In vitro transfection studies confirmed the presence of at least one set of skeletal muscle-specific enhancers in sequences centered between kb +22 and +28 (20). This 6-kb region includes one of the conserved sequence elements which had also been assayed by transient transgenesis and shown to drive expression of reporter constructs in midgestation embryos (18). To examine directly the requirement of these enhancer sequences for directing in vivo expression of H19 and Igf2 in skeletal muscle and in other mesodermal tissues, a 24-kb targeted deletion of sequences between kb 10.7 and 34.7 was constructed as shown in Fig. 1b. This VM3 allele was constructed in two steps so that the selection marker encoding resistance to neomycin sulfate was removed prior to quantitation of H19 and Igf2 expression levels.
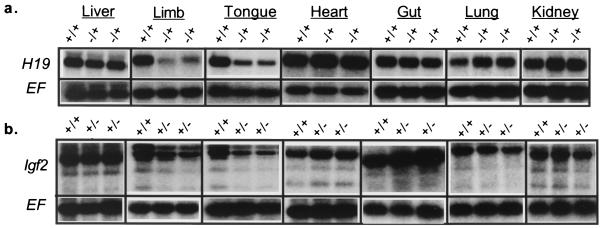
To determine whether the deleted elements are required for expression of H19, RNAs from pups inheriting the deletion via the maternal chromosome were analyzed by Northern blotting (Fig. 2a). Multiple samples from two mutant strains generated from independently derived cells lines were analyzed. H19 levels in liver, heart, lung, gut, and kidney tissues were unaffected. In contrast, expression of H19 in skeletal muscle and tongue tissue was reduced five- to sixfold compared with that of wild-type littermates. Thus, we concluded that the enhancer elements identified in transgene and transfection studies are, in fact, functional and necessary for expression of the H19 gene in its normal chromosomal context. These enhancers are not required for all mesodermal expression but are specific to skeletal muscle.
FIG. 2.
Northern analysis demonstrating the effect of the VM3 enhancer deletion on H19 and Igf2 expression. (a) H19 RNA levels were examined in p2 pups inheriting the VM3 mutation via the maternal chromosome (−/+). (b) Igf2 RNA levels were examined in p2 pups inheriting the VM3 mutation via the paternal chromosome (+/−). In both cases, mutants were compared to wild-type littermates (+/+). Blots were probed for EF2a to verify equal loading. For each tissue, RNA levels of at least two wild-type and two experimental animals from multiple independent litters were quantitated.
To determine whether mesodermal expression of H19 and Igf2 is likely driven through a common set of enhancers, the effect of paternal-chromosome-based inheritance of the VM3 deletion on Igf2 expression was examined by Northern blotting (Fig. 2b). Qualitatively, the effects of the mutation were identical to those noted for H19 on maternal-chromosome inheritance: Igf2 RNA levels were normal in liver, heart, lung, gut, and kidney tissues, while expression in skeletal muscle and tongue tissue was reduced. Quantitatively, expression of Igf2 in VM3 mutants was 3.5 times lower than that in wild-type littermates. These results provided experimental evidence that H19 and Igf2 do have a common set of mesodermal enhancers. The location of these enhancers downstream of the H19DMR is consistent with the demands of the insulator model.
We did note some residual expression of H19 and of Igf2 in mutant mice. We noted this same level of expression even in pups that were homozygous for the VM3 deletion allele (data not shown). Thus, we did not see any evidence for transvection at this locus. Rather, the residual expression is likely due to the activity of enhancer elements outside the VM3 deletion. In situ hybridization experiments did not reveal any cell type-specific loss of expression in tongue tissue and skeletal muscle but rather showed a generalized decrease in transcript levels throughout these tissues (data not shown).
Nctc1 is not essential for normal mouse development.
Nctc1 is a skeletal-muscle-specific transcript that like H19 is unlikely to encode a peptide product (19). The VM3 mutation deletes the entire mRNA coding region for Nctc1. Since mice homozygous for the VM3 mutation are viable as well as fertile and appear and behave normally, we concluded that Nctc1 gene activity is not essential for normal mouse development. Although we consider it unlikely, we cannot rule out the possibility of a requirement for Nctc1 transcription for activation of H19 and Igf2 in cis.
VM3 allele and L23mrp expression.
The gene encoding the L23 (mitochondrion)-related protein (L23mrp or Rpl23), located 40 kb from the imprinted H19 gene (Fig. 1a), is biallelically and ubiquitously expressed at low levels relative to embryonic H19. Targeted deletion of the endodermal enhancers located downstream of H19 has no effect on L23mrp expression, suggesting that the latter gene is functionally insulated from H19 (49). Likewise, we saw that mice homozygous for the VM3 mesodermal enhancer deletion exhibited no loss of L23mrp expression in skeletal muscle, nor was there any increase in L23mrp transcription in fetal liver tissue (data not shown). Thus, the VM3 deletion, which removes sequences to within 4 kb of the L23mrp transcriptional start site, has no effect on the mechanism insulating L23mrp from the H19/Ig2 endodermal enhancers or on the mechanisms driving expression of L23mrp in muscle tissue.
VM3 and TNNT3 expression.
The gene encoding the faster-migrating isoform of skeletal muscle troponin-T (TNNT3) is located 55 kb from L23MRP (47). The linkage of Tnnt3 to H19 is conserved in mice (27). Tnnt3 is biallelically expressed and shows expression parallel to that of H19 in different adult skeletal muscle types, which led to the suggestion that H19 and TNNT3 have common enhancer elements (47). However, mice homozygous for the VM3 enhancer deletion exhibited no change in Tnnt3 expression levels (data not shown).
Additional mesodermal enhancer sequences downstream of kb +35.
Endoderm-specific enhancer elements shared by H19 and Igf2 were identified by deletion of sequences between kb +7.2 and +13 (26), while experiments described above demonstrated that some shared mesoderm-specific enhancer elements map to sequences centered at kb +26. Comparison of expression patterns of mice carrying YAC (3) and BAC (20) H19 transgenes extending to about kb +36 and +140, respectively, suggested that additional enhancers capable of driving high levels of mesoderm-specific expression of H19 in several tissues, including the gut, heart, lung, and kidney, are likely to lie downstream of kb +35. To test directly whether such enhancer elements play a role in expression of H19 in its normal chromosomal context, we examined the effect of maternal-chromosome-based inheritance of the CRK10 allele. The CRK10 mutation is an insertion of a 9.2-kb element, including the H19DMR/Igf2-insulator, +10.7 kb downstream of the H19 promoter (Fig. 1a).
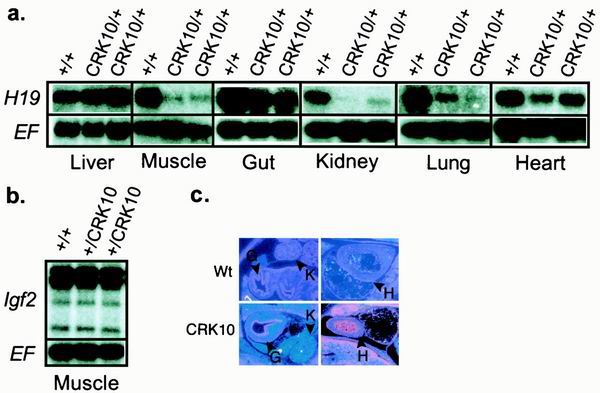
As previously demonstrated, there was no loss of expression in the liver, consistent with the location of the endodermal enhancers upstream of the inserted insulator (Fig. 3a). As previously demonstrated for limb muscle, expression in the tongue was almost completely lost (Fig. 3a). This loss of expression in skeletal muscle is consistent with our above-described demonstration that transcription of H19 in these tissues is dependent on enhancer elements downstream of the insulator insertion site as defined by the VM3 deletion.
FIG. 3.
Effect of the CRK10 mutation on H19 and Igf2 expression. (a) H19 RNA levels in embryonic day 18.5 pups inheriting the CRK10 mutation via the maternal chromosome (CRK10/+) were analyzed by Northern blotting. (b) Igf2 RNA levels in skeletal muscle of pups inheriting the mutation via the paternal chromosome (+/CRK10) were also examined by Northern blotting. Expression of wild type littermates (+/+) was also quantitated. Blots were probed with EF2c to verify equal loading. For each tissue, RNA levels of at least two wild-type and two experimental animals from multiple independent litters were quantitated. (c) In situ analysis of H19 expression patterns in embryonic day 14 embryos. Wt, wild type; H, heart; G, gut; K, kidney.
We next looked at tissues with significant contributions of cells of mesodermal origin but which were unaffected by the VM3 deletion. Any effect unique to CRK10 would presumably be due to the insulator-blocking enhancer elements that are downstream of kb +35. We noted a 50% reduction in H19 RNA levels in the gut (Fig. 3a). By in situ hybridization, we determined that the loss of expression was specific to the smooth muscle layer, with continued high levels being noted in the epithelium (Fig. 3c). This is essentially reciprocal to the pattern seen on deletion of the endodermal enhancers (26). The pattern is also consistent with previous transgenic studies in which the endodermal enhancers were demonstrated to be capable of driving expression only in the endodermally derived endothelium (9).
We also noted a modest decrease in total mRNA levels in the heart (Fig. 3a). Since only about 50% of cardiac expression is blocked by the CRK10 insulator insertion, and because expression in the heart is not altered by the VM3 deletion or by the deletion of the enhancers centered at kb +8, the location of most cardiac enhancer activity remains puzzling.
The loss of H19 expression from the CRK10 chromosome in the lung and the kidney was almost complete (Fig. 3a). In contrast to the patterns seen in the heart and gut, the effect does not appear to be cell type specific; rather, the reduction in RNA levels appears to be uniform. Curiously, these results are very similar to what is seen on deletion of the endodermal enhancer (26).
Parent-of-origin-specific activity of the CRK10 insertion.
Having demonstrated directly that Igf2 shares the downstream mesoderm-specific enhancers necessary for expression in skeletal muscle, the effect of a paternal-chromosome-based inheritance of the CRK10 insertion could be discerned by examining Igf2 expression. The ability of the inserted insulator to block activation of Igf2 by the distal skeletal muscle enhancers was studied. In contrast to its effect on H19 upon maternal inheritance, the insulator did not have the ability to block transcription of the paternal Igf2 allele (Fig. 3b). Rather, the paternal Igf2 promoters continued to be expressed at high levels in skeletal muscle despite the presence in cis of the CRK10 insertion separating them from the skeletal muscle enhancers. Thus, we concluded that the inserted insulator is not active on the paternal chromosome and therefore must be subject to the same parent-of-origin-specific modulation of its insulator function as is postulated in its normal location. These results are consistent with and perhaps were predicted by our previous finding that the ectopic insulator is specifically hypermethylated when inherited via the paternal chromosome (20).
Methylation activity of the CRK10 insertion.
In its normal location, the H19DMR not only functions as a transcriptional insulator that blocks activation of the maternal Igf2 promoter but also acts to direct methylation of cytosine residues in the paternal H19 promoter. These epigenetic modifications are presumed to then be directly responsible for monoallelic repression of H19. To determine whether the inserted sequences act as a nucleation center for developmentally regulated modification of adjacent sequences, the methylation status of DNA located immediately downstream of the CRK10 insertion was examined. Genomic DNAs were prepared from pups inheriting the CRK10 insertion and from their wild-type littermates. These DNAs were subjected to restriction digestion with enzymes sensitive to methylation. Whether inherited via the maternal or paternal chromosome, endogenous sequences adjacent to the inserted H19DMR remained unaffected; there was no apparent increase in CpG methylation of the downstream sequences (Fig. 4). Rather, the sequences in this region appear to be largely unmethylated.
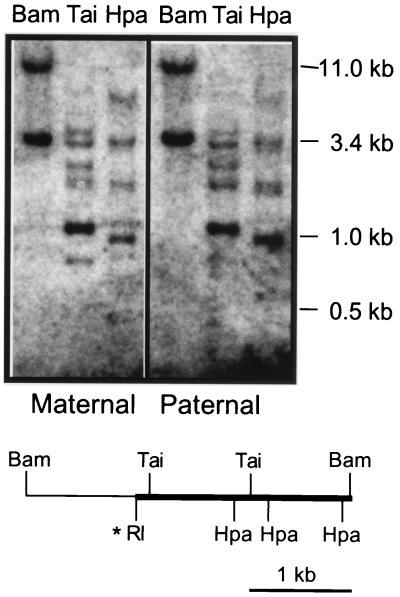
FIG. 4.
Methylation analysis of sequences downstream of the CRK10 insertion mutation. DNAs from heterozygous pups inheriting the CRK10 mutation via the maternal or the paternal chromosome were digested with BamHI and hybridized with an 2.4-kb EcoRI-BamHI fragment from sequences immediately 3′ of the CRK10 insertion. This hybridization reveals an 11-kb band specific to the wild-type chromosome and a 3.4-kb band specific to the CRK10 chromosome. The 3.4-kb band is thus a junction fragment including 1 kb of sequences from the H19DMR (thin line on the diagram) and 2.4 kb of 3′-flanking sequences (thick line on the diagram). Topologically, these flanking sequences are in the same position relative to the inserted H19DMR as the endogenous H19 promoter and gene body are to the endogenous H19DMR. To determine whether these flanking sequences are differentially methylated in a parent-of-origin-specific manner, aliquots of the BamHI-digested DNA were also digested with methylation-sensitive enzymes, including TaiI (Tai) and HpaII (Hpa). A restriction map of the 3.4-kb BamHI fragment specific to the CRK10 chromosome is depicted, with the probe shown as a thickened line. The thin line represents the sequences that are part of the H19DMR that was inserted at the EcoRI (RI) site.
Effect of insulator activity on Igf2 regulation.
In light of recent data suggesting that insulators work in tandem to “loop out” genetic loci into regulatory domains (see Discussion), we next examined the effect of the CRK10 mutation on maternal transcriptional regulation of Igf2 in the liver and also in skeletal muscle. The relative contribution of the maternal Igf2 allele was quantitated by SNuPE assay (24, 35). On the CRK10 chromosome, the enhancer responsible for expression in the liver is flanked by insulator elements and is separated from the Igf2 promoter by a single insulator, just as it is on the wild-type chromosome. As expected, we saw no expression of maternal Igf2 from the CRK10 chromosome in liver tissue (Fig. 5). However, we also did not detect any activation of maternal Igf2 from the CRK10 chromosome in skeletal muscle (Fig. 5), in which the relevant enhancer is separated from the promoter by a paired set of functional insulators. Thus, as discussed below, pairing of insulators did not result in their inactivation in this mammalian system.
DISCUSSION
Transcriptional regulation of the Igf2 and H19 genes has been intensively studied because of the association of dysregulation at this locus with several human diseases and also as a primary model system for investigating mechanisms of genomic imprinting. The currently favored model for imprinting of the two genes posits that monoallelic expression of each gene depends on a common cis-acting element, the ICE, located just upstream of the H19 promoter (Fig. 1) (34, 41). Sequences within this region are hypermethylated specifically on the paternal chromosome. This DNA methylation is postulated to enable expression of the Igf2 gene by inactivating a transcriptional insulator. The paternal-chromosome-specific DNA methylation also induces developmentally programmed epigenetic changes at the H19 promoter which silence it. This popular model requires that the enhancers for both H19 and Igf2 must be located downstream of the H19 gene, while the coordinate expression patterns of the two genes suggest that these enhancers are likely to be shared.
Previous studies have demonstrated the corequirement of a common set of endodermal enhancer elements located downstream of H19 (26). Here we examined a mouse with a targeted deletion of a region downstream of the H19 gene which had previously been shown to have enhancer activity in vitro (20) and in transgenic lines (18, 20), and we directly demonstrated that the deleted sequences included the primary enhancer element important for skeletal muscle expression of both H19 and Igf2. Northern analysis demonstrated that both the H19 and Igf2 RNA levels in the liver, heart, gut, lung, and kidney were unaffected by the deletion. In contrast, the H19 and Igf2 RNA levels were reduced 5.5- and 3.5-fold, respectively, in skeletal muscle. The nonequivalent reduction in expression of the two genes suggests that expression of Igf2 may also be driven by other mesodermal enhancers which are not utilized by H19. This hypothesis could explain recent reports demonstrating that monoallelic expression of Igf2 requires sequences in addition to the Igf2 insulator at the ICE/H19DMR (1, 12).
With verification that H19 and Igf2 both utilize skeletal muscle enhancers downstream of the insertion site at +10.7 kb, the parent-of-origin activity of the CRK10 allele could be defined further. We first looked at paternal-chromosome inheritance and demonstrated that the CRK10 insertion mutation has no effect on transcriptional regulation of Igf2. Just as in its normal location upstream of the H19 gene, the insulator activity is parent-of-origin specific. Consistent with this finding, we had previously shown that the ectopic insulator remains unmethylated on the maternal chromosome but is specifically hypermethylated on paternal inheritance (20). Our results are in contrast with those obtained in a recent study by Hark and coworkers, in which the insulator activity of an ectopic H19ICE element was determined not to be parent-of-origin specific (17). In the latter study, the activity of a smaller insert encompassing only sequences between kb −3.8 and −0.8 was tested by using randomly integrated transgenic constructs. We are presently testing whether we were able to see maternal-chromosome-specific transcriptional insulation in our system because the genetic context of mouse distal chromosome 7 supplies crucial information or because of the additional sequences included in the CRK10 insertion (i.e., sequences in addition to the H19DMR).
Further analysis of the CRK10 allele revealed that the inserted DMR lacks the ability to affect the methylation status of adjacent sequences. Our experiments cannot distinguish between a requirement for particular target sequences, such as the relatively CpG-rich H19 promoter, and a deficiency in the ectopic element's function.
Insertion of the H19DMR between the liver and skeletal-muscle enhancer elements located downstream of H19 has confirmed the presence of downstream enhancer elements required for transcription of H19 in the mesoderm. Interestingly, the CRK10 mutation almost completely abolished H19 expression in the lung and kidney. Previous transgenic and deletion analyses have shown that endodermal enhancers required for both genes are located around kb 8 (10, 26). Deletion of this region caused a complete loss of H19 expression in the lung and kidney. Similarly, the insertion of an insulator at kb +10 caused an almost complete loss of H19 expression in those tissues. Together these results suggest that there are at least two required sets of enhancer elements necessary for the transcription of H19 in these tissues. Although these enhancers are very distant on the chromosome, their effects are extremely synergistic. This model explains the puzzling inability of the proximal endodermal enhancers to direct high levels of expression in kidney and lung tissues in transgenic mice (10, 32, 45) even while their necessity in these same tissues was demonstrated through deletion mutations. Future studies will examine the nature of the synergism to determine whether this is a mechanism for specifically directing the enhancers to H19 and Igf2 to the exclusion of other genes within striking distance of the enhancers.
Insulators are thought to play an important role in gene regulation. At least two models for insulator action have been proposed. In one case, the insulator is predicted to block the progress of a positive signal that moves from the enhancer toward the promoter. In a second model, insulators regulate gene activity by organizing loci into transcriptionally active and transcriptionally inactive domains (5, 37). Very recent studies examining the effect of paired insulator elements in Drosophila support the notion that insulators regulate enhancer-promoter interactions through the formation of chromatin loops (11, 28). In these experiments, an enhancer flanked by the Su(Hw) insulator was unable to interact with its promoter. However, when an enhancer was separated from its promoter by a paired set of insulators, the enhancer retained the ability to activate that promoter. It was proposed that two insulators work in tandem to loop out any intervening DNA between them and leave the enhancer and promoter elements, which remain in a single active domain, thus allowing the enhancer-promoter interactions. (See also a review by Bell et al. [6].)
To examine this model in a mammalian system, we returned to the CRK10 chromosome, whose organization relative to the Igf2 promoter mirrors that of the transgenic constructs used in the Drosophila experiments (Fig. 1a). The analysis was designed not to determine the minimal sequences required for insulator function but rather to determine a possible role for functional insulators in organizing the locus into independent arrays. On a maternal CRK10 chromosome, the endodermal enhancer sequences that drive expression of Igf2 and H19 in the liver are flanked by two functional insulator elements, but only a single insulator lies between the enhancer sequences and the Igf2 promoter. However, the skeletal-muscle-specific enhancer is separated from the Igf2 promoter by paired insulator elements. We examined expression of Igf2 specifically from the maternal CRK10 chromosome because we already knew from examination of H19 expression that the enhancers and the insulators were all functioning. In contrast to results demonstrated in Drosophila for the Su(Hw) insulator, the maternal Igf2 promoter remained silent not only in the liver but also in skeletal muscle. Thus, in the mammalian Igf2-insulator system, we do not see evidence for looping; instead, the very large distances over which the insulators and enhancers work in this system may be more supportive of alternate models for insulator function (5, 13).
REFERENCES
- 1.Ainscough J F, John R, Barton S, Surani M. A skeletal muscle-specific mouse Igf2 repressor lies 40 kb downstream of the gene. Development. 2000;127:3923–3930. doi: 10.1242/dev.127.18.3923. [DOI] [PubMed] [Google Scholar]
- 2.Ainscough J F, Dandola L, Surani M A. Appropriate expression of the mouse H19 gene utilises three or more distinct enhancer regions spread over more than 130 kb. Mech Dev. 2000;91:365–368. doi: 10.1016/s0925-4773(99)00289-0. [DOI] [PubMed] [Google Scholar]
- 3.Ainscough J F, Koide T, Tada M, Barton S, Surani M A. Imprinting of Igf2 and H19 from a 130 kb YAC transgene. Development. 1997;124:3621–3632. doi: 10.1242/dev.124.18.3621. [DOI] [PubMed] [Google Scholar]
- 4.Bartolomei M S, Webber A L, Brunkow M E, Tilghman S M. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 1993;7:1663–1673. doi: 10.1101/gad.7.9.1663. [DOI] [PubMed] [Google Scholar]
- 5.Bell A, Felsenfeld G. Stopped at the border: boundaries and insulators. Curr Opin Genet Dev. 1999;9:191–198. doi: 10.1016/S0959-437X(99)80029-X. [DOI] [PubMed] [Google Scholar]
- 6.Bell A, West A, Felsenfeld G. Insulators and boundaries: versatile regulatory elements in the eukaryotic genome. Science. 2001;291:447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]
- 7.Bell A C, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 8.Brandeis M, Kafri T, Ariel M, Chaillet J R, McCarrey J, Razin A, Cedar H. The ontogeny of allele-specific methylation associated with imprinted genes in the mouse. EMBO J. 1993;12:3669–3677. doi: 10.1002/j.1460-2075.1993.tb06041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brenton J D, Drewell R A, Viville S, Hilton K J, Barton S C, Ainscough J F, Surani M A. A silencer element identified in Drosophila is required for imprinting of H19 reporter transgenes in mice. Proc Natl Acad Sci USA. 1999;96:9242–9247. doi: 10.1073/pnas.96.16.9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunkow M E, Tilghman S M. Ectopic expression of the H19 gene in mice causes prenatal lethality. Genes Dev. 1991;5:1092–1101. doi: 10.1101/gad.5.6.1092. [DOI] [PubMed] [Google Scholar]
- 11.Cai H, Shen P. Effects of cis arrangements of chromatin insulators on enhancer-blocking activity. Science. 2001;291:493–495. doi: 10.1126/science.291.5503.493. [DOI] [PubMed] [Google Scholar]
- 12.Constancia M, Dean W, Lopes S, Moore T, Kelsey G, Reik W. Deletion of a silencer element in Igf2 results in loss of imprinting independent of H19. Nat Genet. 2000;26:203–206. doi: 10.1038/79930. [DOI] [PubMed] [Google Scholar]
- 13.Dorsett D. Distant liaisons: long-range enhancer-promoter interactions in Drosophila. Curr Opin Genet Dev. 1999;9:505–514. doi: 10.1016/s0959-437x(99)00002-7. [DOI] [PubMed] [Google Scholar]
- 14.Ferguson-Smith A C, Sasaki H, Cattanach B M, Surani M A. Parental-origin-specific epigenetic modifications of the mouse H19 gene. Nature. 1993;362:751–755. doi: 10.1038/362751a0. [DOI] [PubMed] [Google Scholar]
- 15.Gould T D, Pfeifer K. Imprinting of mouse Kvlqt1 is developmentally regulated. Hum Mol Genet. 1998;7:483–487. doi: 10.1093/hmg/7.3.483. [DOI] [PubMed] [Google Scholar]
- 16.Hark A T, Tilghman S M. Chromatin conformation of the H19 epigenetic mark. Hum Mol Genet. 1998;7:1979–1985. doi: 10.1093/hmg/7.12.1979. [DOI] [PubMed] [Google Scholar]
- 17.Hark A T, Schoenherr C J, Katz D J, Ingram R S, Levorse J M, Tilghman S M. CTCF mediates methylation-sensitive enhancer blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 18.Ishihara K, Hatano N, Furuumi H, Kato R, Iwaki T, Miura K, Jinno Y, Sasaki H. Comparative genomic sequencing identifies novel tissue-specific enhancers and sequence elements for methylation-sensitive factors implicated in Igf2/H19 imprinting. Genome Res. 2000;10:664–671. doi: 10.1101/gr.10.5.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishihara K, Kato R, Furuumi H, Zubair M, Sasaki H. Sequence of a 42-kb mouse region containing the imprinted H19 locus: identification of a novel muscle-specific transcription unit showing biallelic expression. Mamm Genome. 1998;9:775–777. doi: 10.1007/s003359900863. [DOI] [PubMed] [Google Scholar]
- 20.Kaffer C R, Srivastava M, Park K, Ives E, Hsieh S, Batlle J, Grinberg A, Huang S P, Pfeifer K. A transcriptional insulator at the imprinted H19/Igf2 Locus. Genes Dev. 2000;14:1908–1919. [PMC free article] [PubMed] [Google Scholar]
- 21.Kanduri C, Holmgren C, Pilartz M, Franklin G, Kanduri M, Liu L, Ginjala V, Ulleras E, Mattsson R, Ohlsson R. The 5′-flanking of the murine H19 gene in unusual chromatin conformation unidirectionally blocks enhancer-promoter communication. Curr Biol. 2000;10:449–457. doi: 10.1016/s0960-9822(00)00442-5. [DOI] [PubMed] [Google Scholar]
- 22.Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi C, Wolffe A, Ohlsson R, Lobanenkov V. Functional association of CTCF with the insulator upstream of the H19 gene is parent-of-origin specific and methylation-sensitive. Curr Biol. 2000;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- 23.Khosla S, Aitchison A, Gregory R, Allen N D, Feil R. Parental allele-specific chromatin configuration in a boundary–imprinting-control element upstream of the mouse H19 gene. Mol Cell Biol. 1999;19:2556–2566. doi: 10.1128/mcb.19.4.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuppuswamy M N, Hoffmann J W, Kasper C K, Spitzer S G, Groce S L, Bajaj S P. Single nucleotide primer extension to detect genetic diseases: experimental application to hemophilia B (factor IX) and cystic fibrosis. Proc Natl Acad Sci USA. 1991;88:1143–1147. doi: 10.1073/pnas.88.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lasko M, Picher J, Gorman J, Sauer B, Okamoto Y, Lee E, Alt F, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci USA. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leighton P A, Saam J R, Ingram R S, Stewart C L, Tilghman S M. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev. 1995;9:2079–2089. doi: 10.1101/gad.9.17.2079. [DOI] [PubMed] [Google Scholar]
- 27.Misener V, Wielowieyski A, Brennan L, Beebakhee G, Jongstra J. The mouse Lsp1 and Tnnt3 genes are 4.3 kb apart on distal mouse chromosome 7. Mamm Genome. 1998;9:846–848. doi: 10.1007/s003359900880. [DOI] [PubMed] [Google Scholar]
- 28.Muravyova E, Golovnin A, Gracheva E, Parshikov A, Belenkaya T, Pirrotta V, Georgiev P. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science. 2001;291:495–498. doi: 10.1126/science.291.5503.495. [DOI] [PubMed] [Google Scholar]
- 29.Olek A, Walter J, Horsthemke B, Dittrich B, Buiting K. The pre-implantation ontogeny of the H19 methylation imprint. Nat Genet. 1997;17:275–276. doi: 10.1038/ng1197-275. [DOI] [PubMed] [Google Scholar]
- 30.Onyango P, Miller W, Lehoczky J, Leung C, Birren B, Wheelan S, Dewar K, Feinberg A. Sequence and comparative analysis of the mouse 1-megabase region orthologous to the human 11p15.5 imprinted domain. Genome Res. 2000;10:1697–1710. doi: 10.1101/gr.161800. [DOI] [PubMed] [Google Scholar]
- 31.Paulsen M, Davies K R, Bowden L M, Villar A J, Franck O, Fuermann M, Dean W L, Moore K R, Rodrigues N, Davies K E, Hu R-J, Feinberg A P, Maher E R, Reik W, Walter J. Syntenic organization of the mouse distal chromosome 7 imprinting cluster and the Beckwith-Wiedemann syndrome region in chromosome 11p15.5 Hum. Mol Genet. 1998;7:1149–1159. doi: 10.1093/hmg/7.7.1149. [DOI] [PubMed] [Google Scholar]
- 32.Pfeifer K, Leighton P A, Tilghman S M. The structural H19 gene is required for transgene imprinting. Proc Natl Acad Sci USA. 1996;93:13876–13883. doi: 10.1073/pnas.93.24.13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reik W, Constancia M, Dean W, Davies K, Bowden L, Murrell A, Feil R, Walter J, Kelsey G. Igf2 imprinting in development and disease. Int J Dev Biol. 2000;44:145–150. [PubMed] [Google Scholar]
- 34.Reik W, Murrell A. Genomic imprinting. Silence across the border. Nature. 2000;405:408–409. doi: 10.1038/35013178. [DOI] [PubMed] [Google Scholar]
- 35.Singer-Sam J, Chapman V, LeBon J M, Riggs A D. Parental imprinting studies by allele-specific primer extension after PCR: paternal X chromosome-linked genes are transcribed prior to preferential paternal X chromosome inactivation. Proc Natl Acad Sci USA. 1992;89:10469–10473. doi: 10.1073/pnas.89.21.10469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Srivastava M, Hsieh S, Grinberg A, Williams-Simon L, Huang S-P, Pfeifer K. H19 and Igf2 monoallelic expression is regulated in two distinct ways by a shared cis acting element. Genes Dev. 2000;14:1186–1195. [PMC free article] [PubMed] [Google Scholar]
- 37.Sun F, Elgin S. Putting boundaries on silence. Cell. 1999;99:459–462. doi: 10.1016/s0092-8674(00)81534-2. [DOI] [PubMed] [Google Scholar]
- 38.Szabo P, Tang S, Rentsendorj A, Pfeifer G, Mann J R. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr Biol. 2000;10:607–610. doi: 10.1016/s0960-9822(00)00489-9. [DOI] [PubMed] [Google Scholar]
- 39.Szabo P E, Pfeifer G P, Mann J R. Characterization of novel parent-specific epigenetic modifications upstream of the imprinted mouse H19 gene. Mol Cell Biol. 1998;18:6767–6776. doi: 10.1128/mcb.18.11.6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szabo P E, Mann J R. Biallelic expression of imprinted genes in the mouse germ line: implications for erasure, establishment, and mechanisms of genomic imprinting. Genes Dev. 1995;9:1857–1868. doi: 10.1101/gad.9.15.1857. [DOI] [PubMed] [Google Scholar]
- 41.Thorvaldsen J, Bartolomei M S. Mothers setting boundaries. Science. 2000;288:2145–2146. doi: 10.1126/science.288.5474.2145. [DOI] [PubMed] [Google Scholar]
- 42.Thorvaldsen J L, Duran K L, Bartolomei M S. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tremblay K D, Duran K L, Bartolomei M S. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol. 1997;17:4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tremblay K D, Saam J R, Ingram R S, Tilghman S M, Bartolomei M S. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat Genet. 1995;9:407–413. doi: 10.1038/ng0495-407. [DOI] [PubMed] [Google Scholar]
- 45.Webber A, Ingram R I, Levorse J, Tilghman S M. Location of enhancers is essential for imprinting of H19 and Igf2. Nature. 1998;391:711–715. doi: 10.1038/35655. [DOI] [PubMed] [Google Scholar]
- 46.Yoo-Warren H, Pachnis V, Ingram R S, Tilghman S M. Two regulatory domains flank the mouse H19 gene. Mol Cell Biol. 1988;8:4707–4715. doi: 10.1128/mcb.8.11.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan L, Qian N, Tycko B. An extended region of biallelic gene expression and rodent-human synteny downstream of the imprinted H19 gene on chromosome 11p15.5. Hum Mol Genet. 1996;5:1931–1937. doi: 10.1093/hmg/5.12.1931. [DOI] [PubMed] [Google Scholar]
- 48.Zemel S, Bartolomei M S, Tilghman S M. Physical linkage of two mammalian imprinted genes. Nat Genet. 1992;2:61–65. doi: 10.1038/ng0992-61. [DOI] [PubMed] [Google Scholar]
- 49.Zubair M, Hilton K, Saam J, Surani M, Tilghman S, Sasaki H. Structure and expression of the mouse L23mrp gene downstream of the imprinted H19 gene: biallelic expression and lack of interaction with H19 enhancers. Genomics. 1997;45:290–296. doi: 10.1006/geno.1997.4961. [DOI] [PubMed] [Google Scholar]