Abstract
We compared transcriptomic profiles of cerebral organoids differentiated from induced pluripotent stem cells of eight schizophrenia and eight bipolar disorder patients to identify genes that were differentially expressed in cerebral organoids between two disorders. Gene ontology analysis showed relative up-regulation in schizophrenia organoids of genes related to response to cytokines, antigen binding and clathrin-coated vesicles, while showing up-regulation in bipolar disorder of genes involved in calcium binding. Gene set enrichment analysis revealed enrichment in schizophrenia of genes involved in mitochondrial and oxidative phosphorylation while showing enrichment in bipolar disorder of genes involved in long term potentiation and neuro-transporters. We compared mitochondrial function in cerebral organoids from schizophrenia and bipolar disorder subjects and found that while schizophrenia organoids showed deficits in basal oxygen consumption rate and ATP production when compared to healthy control organoids, while bipolar disorder organoids did not show these deficits. Gene ontology analyses also revealed enrichment in bipolar disorder of genes in ion binding and regulation of transport. Experiments examining the interaction between mitochondria and endoplasmic reticulum in cortical neurons from bipolar disorder subjects showed a significantly lower number of contact sites between mitochondria and endoplasmic reticulum when compared to cortical neurons from schizophrenia patients. These results point to disease-specific deficits in mitochondrial respiration in schizophrenia and in mitochondrial-endoplasmic reticulum interactions in bipolar disorder.
Supplementary Information
The online version contains supplementary material available at 10.1007/s44192-023-00031-8.
Introduction
Schizophrenia (SCZ) and bipolar I disorder (BPI) are amongst the most severe psychiatric disorders, with significant disease burden in the global context. In its prototypical presentation, schizophrenia is characterized as a thought disorder, with disturbances in the realms of perception and cognition, including positive symptoms (i.e. hallucinations, delusions), negative symptoms (i.e. apathy, avolition, affective flattening) and cognitive deficits (i.e. working memory, executive functioning) and is a chronic condition. Bipolar disorder, on the other hand, is primarily a mood disorder, characterized by the two poles of mania and depression and usually recurrent. Clinical, neuropathological, neuroimaging and genetic studies show that there are shared facets as well as distinct differences between the two disorders [1, 2]. Genome-wide association studies (GWAS) of subjects with schizophrenia and bipolar disorder have revealed shared genetic susceptibilities though there are distinct risk variants for each of the disorders as well [3, 4]. With regards to treatment, antipsychotic medications are the mainstay for the treatment of schizophrenia but a number of antipsychotic medications also have high efficacy in the treatment for manic and depressive phases of bipolar disorder [5]. However, the converse does not hold true—mood stabilizers do not have high efficacy in the treatment of schizophrenia, especially in monotherapy [6]. Research aimed at delineating the shared and distinct biological features of these two disorders can provide insights into the neurobiological underpinnings of these disorders.
Schizophrenia and bipolar disorder have historically been treated as disorders with distinct symptoms and clinical courses, starting with the designation of dementia praecox and manic-depressive insanity by Emil Kraepelin over a century ago [2]. Dementia praecox was later coined as schizophrenia by Eugen Bleuler while manic-depressive insanity is now more commonly referred to as bipolar disorder. However, there is convergent evidence from genetic, clinical and neuropathological studies that indicate some shared biology and some unique facets between the two disorders [1, 2]. Studies examining gene expression in post-mortem brains had also revealed both shared features as well as distinct differences between schizophrenia and bipolar disorder [7]. Transcriptome analysis of post-mortem tissue from the anterior cingulate cortex of schizophrenia and bipolar disorder subjects showed some correlation in gene expression differences in schizophrenia and bipolar disorder that indicated shared biology related to dysregulation of cytoskeleton remodelling and lysosomal function as well as shared genes involved in synaptic biology, adherens junctions, mitochondrial biology and biosynthetic metabolic processes [8]. However, gene expression data from cortical brain tissues showed that while the transcriptome profiles schizophrenia and autism are significantly correlated, there was no statistically significant correlation between the schizophrenia and bipolar disorder transcriptome profiles [9].
Induced pluripotent stem cells (iPSCs) that can be reprogrammed from readily accessible tissue from human subjects have enabled new ways to study human brain development and provided methods to generate and study human neuronal cells in the laboratory [10–13]. Human iPSCs can be differentiated to generate a range of neuronal and glial cells relevant to disease biology of schizophrenia and bipolar disorder [14–17]. In addition to two-dimensional neuronal cultures, advances in tissue engineering now enable generation of human three-dimensional cerebral organoids from the iPSCs [18]. These cerebral organoids recapitulate human cortical development and contain many neuronal and glial subtypes, including cortical neuron subtypes, synapses and exhibiting electrophysiological activity [19].
We had previously undertaken RNA-seq experiments to compare gene expression profiles between cerebral organoids from iPSCs of schizophrenia subjects and healthy subjects as well as between cerebral organoids from iPSCs of bipolar disorder subjects and healthy subjects [20, 21]. Analyses of the transcriptomic data showed that both schizophrenia and bipolar disorder cerebral organoids have gene expression differences in pathways involved in neurodevelopment and in tissue generation, when compared to the gene expression profiles of cerebral organoids from healthy subjects [20, 21]. Here, we used the raw RNA-seq data from these experiments to undertake a head-to-head comparison of the transcriptome profiles of cerebral organoids from eight schizophrenia subjects and eight bipolar disorder subjects, with an eye towards identifying cellular pathways that were differentially regulated between these two disorders. In the head-to-head comparison, we found that cerebral organoids from schizophrenia subjects showed relative up-regulation, compared to bipolar disorder organoids, of genes involved in response to immune system and in mitochondrial function while cerebral organoids from bipolar disorder subjects showed relative up-regulation of genes involved in calcium signaling and in neuro-transporters. As a follow-up to the transcriptomic data, we had previously undertaken mitochondrial function studies in schizophrenia organoids and examined mitochondria-endoplasmic reticulum (ER) interactions in iPSC-derived cortical neurons in bipolar disorder [20, 21]. Here, report on additional experiments investigating mitochondrial function in bipolar disorder organoids and new data generated to analyze mitochondria-ER interactions in schizophrenia iPSC-derived cortical neurons. We compared our new data with the previously published findings so that we could delineate which cellular features are distinct in the two disorders. We found that schizophrenia organoids, but not bipolar disorder organoids, exhibit dysfunction in basal respiration rate and ATP production. Conversely, experiments measuring mitochondria-endoplasmic reticulum (ER) interactions in iPSC-derived cortical neurons showed a relative decrease in ER-mitochondria contact sites in bipolar disorder neurons when compared to schizophrenia and healthy control neurons.
Materials and methods
Differentiation of excitatory cortical neurons
The iPSC cultures were maintained in NutriStem hPSC XF Medium (Biological Industries, 01-0005). When iPSCs reached confluency, the cells were differentiated into excitatory neurons on Geltrex (ThermoFisher, A1413202) coated plates. Cells were maintained in N2/B27 media containing 485 mL Neurobasal medium (Life Technologies), 5 mL N2 (Gibco, 17502001), 5 mL Glutamax (ThermoFisher Scientific), 5 mL penicillin–streptomycin), 10 mL B27 supplement (Gibco, 17504044). For the first 7 days, the medium was supplemented with 10 μM SB431542 (Sigma-Aldrich, S4317), 1 μM dorsomorphin (Sigma-Aldrich, P5499) and 100 nM LDN193189 (Sigma-Aldrich, SML0559). On days 8–29, the N2/B27 media was changed daily, without supplementing the media with SB431542, dorsomorphin or LDN193189. On day 30, cells were transitioned in Brainphys Neuronal Medium (Stemcell Technologies, 05790) supplemented with B-27, with media change twice weekly. Methods for differentiation of cerebral organoids and extraction of RNA had been described in our previous studies [19, 20].
RNA-seq experiments and data processing
We used the Illumina RiboZero TruSeq Stranded Total RNA Library Prep Kit (Illumina) to construct the RNA-seq library and used the Illumina NovaSeq6000 platform for sequencing in the 100 nt, paired-end configuration, as described previously [20, 21]. We obtained an average of 60 million reads for each sample. The reads were trimmed with Cutadapt and aligned to the reference genome (hg38 UCSC assembly) to analyze gene expression using TopHat v2.0.14 and Bowtie v2.10 with default parameters and RefSeq annotation (genome-build GRCh38.p9) [22]. We used Cufflinks v2.2.1 to analyze distribution of alignments and quantile normalized FPKM (fragments per kilobase of exon model per million reads mapped) values [23, 24]. We utilized Cuffdiff v2.2.1 to perform differential expression testing. We did not consider sex differences for this analysis. The false discovery rate (FDR) was 0.05. The raw data analysis is included in Supplementary file 1, in sheet 3 titled significant genes. Tables 1, 2 and 3 show q values represent FDR-adjusted p-value of the test statistic. RT-PCR was used to validate a number of key relevant genes.
Table 1.
Gene set enrichment analysis
| Size | ES | NES | NOM p-val | FDR q-val | FWER p-val | Rank at max | Leading edge | |
|---|---|---|---|---|---|---|---|---|
| Reactome | ||||||||
| Reactome_Influenza_Infection | 154 | − 0.59132 | − 2.80241 | 0 | 0 | 0 | 10,503 | tags = 73%, list = 27%, signal = 99% |
| Reactome_Mitochondrial_Translation | 93 | − 0.60753 | − 2.62088 | 0 | 0 | 0 | 13,391 | tags = 86%, list = 34%, signal = 131% |
| Reactome_Respiratory_Electron_Transport | 89 | − 0.60034 | − 2.61611 | 0 | 0 | 0 | 12,856 | tags = 84%, list = 33%, signal = 125% |
| Reactome_Infectious_Disease | 371 | − 0.49008 | − 2.60519 | 0 | 0 | 0 | 13,579 | tags = 68%, list = 35%, signal = 103% |
| Reactome_Cell_Cycle_Checkpoints | 282 | − 0.51048 | − 2.60441 | 0 | 0 | 0 | 15,024 | tags = 74%, list = 39%, signal = 120% |
| Reactome_Naplus_Cl_Dependent_Neurotransmitter_Transporters | 19 | 0.515474 | 1.541765 | 0.038388 | 0.66119 | 1 | 1466 | tags = 32%, list = 4%, signal = 33% |
| Reactome_Role_Of_Phospholipids_In_Phagocytosis | 33 | 0.452627 | 1.529319 | 0.032136 | 0.623584 | 1 | 4477 | tags = 24%, list = 11%, signal = 27% |
| Reactome_Plasma_Lipoprotein_Assembly | 19 | 0.504823 | 1.504998 | 0.043222 | 0.652722 | 1 | 2605 | tags = 32%, list = 7%, signal = 34% |
| Reactome_Xenobiotics | 24 | 0.466743 | 1.485898 | 0.049904 | 0.666401 | 1 | 8655 | tags = 50%, list = 22%, signal = 64% |
| Reactome_Long_Term_Potentiation | 23 | 0.474369 | 1.470371 | 0.046693 | 0.608513 | 1 | 8104 | tags = 48%, list = 21%, signal = 60% |
| Hallmark | ||||||||
| Hallmark_Myogenesis | 199 | − 0.55399 | − 2.73616 | 0 | 0 | 0 | 5986 | tags = 43%, list = 15%, signal = 51% |
| Hallmark_E2F_Targets | 198 | − 0.51894 | − 2.56449 | 0 | 0 | 0 | 13,107 | tags = 65%, list = 34%, signal = 98% |
| Hallmark_Oxidative_Phosphorylation | 185 | − 0.52698 | − 2.53241 | 0 | 0 | 0 | 13,844 | tags = 71%, list = 36%, signal = 109% |
| Hallmark_Unfolded_Protein_Response | 111 | − 0.43549 | − 1.96044 | 0 | 0 | 0 | 14,339 | tags = 65%, list = 37%, signal = 102% |
| Hallmark_Glycolysis | 198 | − 0.38222 | − 1.88766 | 0 | 0 | 0 | 13,694 | tags = 57%, list = 35%, signal = 87% |
| Hallmark_Pancreas_Beta_Cells | 40 | 0.268541 | 0.960266 | 0.515539 | 1 | 1 | 2039 | tags = 10%, list = 5%, signal = 11% |
| Hallmark_Spermatogenesis | 132 | 0.17472 | 0.771267 | 0.937394 | 1 | 1 | 9809 | tags = 27%, list = 25%, signal = 36% |
| Hallmark_Xenobiotic_Metabolism | 200 | 0.164059 | 0.76698 | 0.98087 | 0.93739 | 1 | 5051 | tags = 12%, list = 13%, signal = 13% |
| KEGG | ||||||||
| KEGG_Ribosome | 86 | − 0.66314 | − 2.80034 | 0 | 0 | 0 | 10,890 | tags = 93%, list = 28%, signal = 129% |
| KEGG_Parkinsons_Disease | 99 | − 0.50716 | − 2.26377 | 0 | 0 | 0 | 13,694 | tags = 75%, list = 35%, signal = 115% |
| KEGG_Oxidative_Phosphorylation | 100 | − 0.51326 | − 2.26148 | 0 | 0 | 0 | 13,763 | tags = 71%, list = 35%, signal = 109% |
| KEGG_Small_Cell_Lung_Cancer | 84 | − 0.50382 | − 2.18709 | 0 | 0 | 0 | 8917 | tags = 50%, list = 23%, signal = 65% |
| KEGG_P53_Signaling_Pathway | 67 | − 0.52866 | − 2.14187 | 0 | 9.17E−04 | 0.003 | 12,063 | tags = 64%, list = 31%, signal = 93% |
| KEGG_Taste_Transduction | 51 | 0.457469 | 1.688169 | 0.001692 | 0.234029 | 0.361 | 7615 | tags = 49%, list = 20%, signal = 61% |
| KEGG_Linoleic_Acid_Metabolism | 28 | 0.510943 | 1.677292 | 0.011858 | 0.129439 | 0.388 | 8626 | tags = 43%, list = 22%, signal = 55% |
| KEGG_Type_I_Diabetes_Mellitus | 42 | 0.433637 | 1.558308 | 0.012797 | 0.188571 | 0.766 | 8960 | tags = 43%, list = 23%, signal = 56% |
| KEGG_Asthma | 29 | 0.486376 | 1.629667 | 0.013619 | 0.132661 | 0.538 | 8960 | tags = 52%, list = 23%, signal = 67% |
| KEGG_Retinol_Metabolism | 64 | 0.358277 | 1.385493 | 0.043956 | 0.410179 | 0.994 | 7681 | tags = 28%, list = 20%, signal = 35% |
Gene set enrichment analysis (GSEA) data for DEGs using three different databases (Hallmark, KEGG, Reactome) tabulated according to normalized enrichment score (NES) and False discovery rate (FDR) q-value. Positive correlation indicates relative association with gene expression in bipolar disorder and a negative correlation indicates relative association with gene expression in schizophrenia
Table 2.
Mitochondrial genes upregulated in schizophrenia compared to bipolar disorder
| Gene | Log2(fold_change) | P_value | Q_value |
|---|---|---|---|
| B2M | inf | 0.00005 | 0.0168915 |
| SFTPB | 5.47445 | 0.00005 | 0.0168915 |
| P2RX3 | 4.09903 | 0.00015 | 0.041525 |
| PKHD1L1 | 4.00139 | 0.00005 | 0.0168915 |
| THSD7B | 3.9018 | 0.00005 | 0.0168915 |
| EBF2 | 3.73444 | 0.00005 | 0.0168915 |
| ACTA1 | 3.71003 | 0.00005 | 0.0168915 |
| SHOX2 | 3.54632 | 0.00005 | 0.0168915 |
| BCL6B | 3.40011 | 0.00005 | 0.0168915 |
| MUC5B | 3.27599 | 0.00005 | 0.0168915 |
| CD5 | 3.27526 | 0.00005 | 0.0168915 |
| CASQ2 | 3.27468 | 0.00005 | 0.0168915 |
| MYBPC2 | 3.20693 | 0.00005 | 0.0168915 |
| NPTX2 | 3.17192 | 0.0001 | 0.0297493 |
| NHLH1 | 3.15736 | 0.00005 | 0.0168915 |
| MYBPH | 3.05216 | 0.00005 | 0.0168915 |
| ASB4 | 3.01621 | 0.0001 | 0.0297493 |
| MUC12 | 2.87732 | 0.00005 | 0.0168915 |
| SHD | 2.83455 | 0.00005 | 0.0168915 |
| TRIM55 | 2.73832 | 0.00005 | 0.0168915 |
| ACTC1 | 2.72941 | 0.00005 | 0.0168915 |
| MYH3 | 2.66443 | 0.00005 | 0.0168915 |
| EBF1 | 2.6039 | 0.00005 | 0.0168915 |
| CHRND | 2.60343 | 0.00005 | 0.0168915 |
| APLN | 2.54672 | 0.00005 | 0.0168915 |
| GRID2 | 2.50442 | 0.0002 | 0.0480289 |
| MYL4 | 2.42199 | 0.0001 | 0.0297493 |
| KRT1 | 2.38926 | 0.0002 | 0.0480289 |
| TNNT2 | 2.26791 | 0.00005 | 0.0168915 |
| NEFM | 2.25293 | 0.00005 | 0.0168915 |
| AFAP1L1 | 2.2312 | 0.00005 | 0.0168915 |
| EYA1 | 2.10989 | 0.0002 | 0.0480289 |
| RBM24 | 2.05014 | 0.0002 | 0.0480289 |
| RYR1 | 2.00653 | 0.00005 | 0.0168915 |
| C7 | 1.92159 | 0.00005 | 0.0168915 |
| COL19A1 | 1.87283 | 0.0001 | 0.0297493 |
| DUOX2 | 1.86384 | 0.0001 | 0.0297493 |
| SERPINA3 | 1.79983 | 0.00005 | 0.0168915 |
| TNNC1 | 1.798 | 0.0002 | 0.0480289 |
| NTRK2 | 1.73311 | 0.00005 | 0.0168915 |
| CAPN6 | 1.60497 | 0.00005 | 0.0168915 |
| NEUROD1 | 1.53609 | 0.00005 | 0.0168915 |
| MCAM | 1.52598 | 0.00005 | 0.0168915 |
| SPOCK2 | 1.43396 | 0.00005 | 0.0168915 |
| ARHGAP29 | 1.29965 | 0.00005 | 0.0168915 |
| CDR1 | 1.0788 | 0.0001 | 0.0297493 |
| PEG10 | 1.07848 | 0.0002 | 0.0480289 |
| FREM2 | 1.00581 | 0.0002 | 0.0480289 |
List of mitochondria-associated genes in the MitoCarta 2.0 database that were upregulated in SCZ organoids vis-à-vis BPI organoids and in BPI organoids vis-à-vis SCZ organoids
Table 3.
Mitochondrial genes upregulated in bipolar disorder compared to schizophrenia
| Gene | Log2(fold_change) | P_value | Q_value |
|---|---|---|---|
| CCL25 | 4.42404 | 0.00005 | 0.0168915 |
| GIP | 4.04306 | 5.00E−05 | 0.0168915 |
| HLA-DRB1 | 3.66592 | 0.00005 | 0.0168915 |
| OPRK1 | 3.58192 | 0.00005 | 0.0168915 |
| CX3CR1 | 3.33744 | 0.00005 | 0.0168915 |
| ASAH2 | 3.32305 | 0.00005 | 0.0168915 |
| LCT | 3.24615 | 0.00005 | 0.0168915 |
| APOC3 | 3.05208 | 0.00005 | 0.0168915 |
| SLC2A2 | 2.78622 | 0.0001 | 0.0297493 |
| FOLH1 | 2.75393 | 0.00005 | 0.0168915 |
| GATA4 | 2.37677 | 0.0002 | 0.0480289 |
| ANXA13 | 2.13002 | 0.00005 | 0.0168915 |
| COL2A1 | 1.98566 | 0.00005 | 0.0168915 |
| SI | 1.92321 | 0.00005 | 0.0168915 |
| PIGR | 1.76004 | 0.00005 | 0.0168915 |
| SLC5A1 | 1.70496 | 0.0001 | 0.0297493 |
| APOB | 1.69844 | 0.00005 | 0.0168915 |
| SULT2A1 | 1.69654 | 0.0002 | 0.0480289 |
| MTTP | 1.66128 | 0.00005 | 0.0168915 |
| MALRD1 | 1.6595 | 0.00005 | 0.0168915 |
| OLFM4 | 1.61724 | 0.00005 | 0.0168915 |
| GSTA1 | 1.3839 | 0.00005 | 0.0168915 |
| OAT | 1.24558 | 0.00015 | 0.041525 |
| FOS | 1.2401 | 0.00005 | 0.0168915 |
| PRLR | 1.21405 | 0.00015 | 0.041525 |
| XDH | 1.21051 | 0.00005 | 0.0168915 |
| MME | 1.07592 | 0.00005 | 0.0168915 |
Gene ontology and gene set enrichment analyses
We used Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis on all differentially regulated genes, utilizing the Functional Enrichment Analysis unit of HOMER v.3. [25]. The reference list for the GO analysis is from Clarivate-MetaCore + MetaDrug™ version 19.1 build 69600. In the figures, we have listed the genes that reached significance (p < 0.05). We performed gene set enrichment analysis (GSEA) with default parameters using the GSEA software for all expressed genes with FPKM values calculated by Cufflinks against following data sets v6.2: Hallmark, REACTOME and KEGG [26]. We used Human MitoCarta 2.0 (https://www.broadinstitute.org/files/shared/metabolism/mitocarta/human.mitocarta2.0.html), was used to identify DEGs implicated in human mitochondrial pathways, as shown in Tables 2 and 3.
Mitochondrial respiration—seahorse mito stress test assay
We used the Seahorse XF Cell Mito Stress Test (Agilent, 103015–100) to assess mitochondrial respiration in cerebral organoids that had been grown for 9 months, as described previously for the comparison between healthy control and schizophrenia organoids [20]. We undertook the Seahorse assay for the bipolar disorder organoids as well using the same assay in order to compare the schizophrenia and bipolar disorder organoids. Cerebral organoids were cut into three sections and plated on Seahorse XFe96 Spheroid microplates (Agilent, 102978–100) coated with 10 μg/mL laminin (Sigma-Aldrich, L2020). Effects of the following perturbations were recorded sequentially: 2 mM oligomycin, 2 mM FCCP (Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone), and 2 mM rotenone and 2 mM antimycin + 2 mM rotenone. Data were analyzed with Wave software 2.4 (Agilent). All organoids used in this assay were generated at the same time.
Proximity ligation assay
The proximity ligation assay (PLA) was used to quantify contact points between the endoplasmic reticulum (ER) and mitochondria in the human iPSC-derived cortical neurons, using the Duolink In Situ PLA Probe (Sigma-Aldrich DUO92002, DUO92004) [27]. We had described the experimental protocol previously in comparing PLA in cortical neurons from bipolar disorder iPSCs and healthy control iPSCs [21]. We collected additional data on PLA in cortical neurons from schizophrenia iPSCs as well to compare with the previously reported PLA data in cortical neurons differentiated from bipolar disorder and healthy control cortical iPSCs. Cortical neurons that had been differentiated for 90 days were fixed with 4% PFA for 20 min, washed in PBS three times and then permeabilized in 0.1% Triton-X100 for 15 min. The samples were then incubated in the blocking solution for 30 min at 37 °C. The following primary antibodies, diluted in PBS, were added to the cells and incubated overnight at 4 °C: anti-VDAC1 antibody [20B12AF2] (Abcam, ab14734, diluted 1:100) and anti-IP3R1 antibody (Abcam, ab5804, diluted 1:500) [27]. The antibodies were then removed and the cells washed with PBS + 0.01% Triton-X100. The PLA probes, which were diluted 1:5 in the provided diluent, were added to the cells and incubated at 37 °C for 1 h. Ligation and amplification were undertaken using the Duolink® In Situ Detection Reagent, with ligation stock diluted 1:5 in high-purity water and the ligase diluted 1:40 and vortexed before being added to the samples. After 30 min incubation at 37 °C, the samples were washed twice with TBS-T. The amplification stock solution was diluted 1:5 in high-purity water and polymerase was diluted 1:80 in the amplification solution and vortexed before being added to the samples and incubated for 100 min at 37 °C. The samples were washed in 1 × saline sodium citrate (SSC) washing buffer for 2 min, followed by a wash in 0.01 × SSC washing buffer for another 2 min. Prolong Gold Antifade Mount with DAPI (Life Technologies, P36931) was then added and neurons which were imaged at 60× resolution with the PerkinElmer Opera Phoenix High-Content Screening System. The Opera Phoenix and Harmony software (Perkin Elmer) were used to quantify the number of PLA positive spots.
Results
Gene expression analysis of cerebral organoids from schizophrenia and bipolar disorder subjects
We used bulk RNA-seq data from iPSC-derived cerebral organoids from iPSCs of eight patients each with bipolar disorder and schizophrenia, as described in earlier studies [20, 21]. These cerebral organoids developed along the telencephalic lineage to continue to ~ 5 mm after 7 months. Cerebral organoids generated from iPSC lines different groups showed no gross differences between them and all groups showed a similar proportion of different cell types and expression of a range of neuronal and glial markers, including MAP2, Ctip2, Satb2, Pax6, TBR2, Cux1, LHX6, glutamine synthetase (GS), GFAP, oligodendrocytes-specific protein/claudin11 (OSP), myelin basic protein (MBP) and IBA1 [20, 21]. Overall, these results show overall consistency in the cellular composition of organoids generated from different lines.
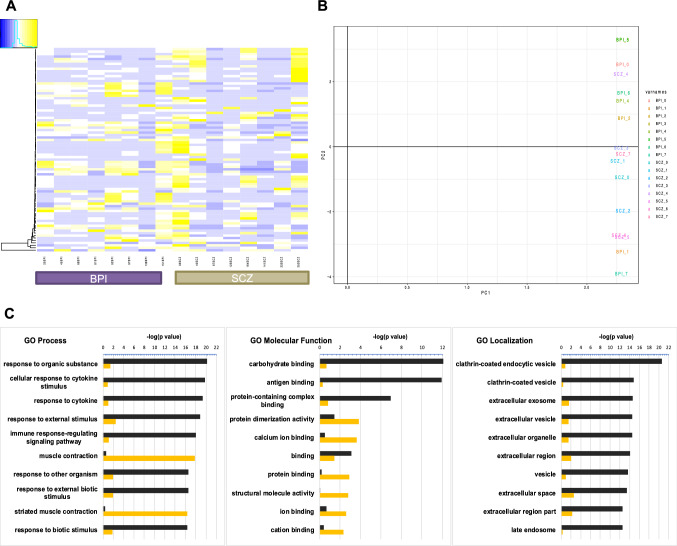
The heatmap depicting the differentially expressed genes (DEGs) in the RNA-seq experiments showed similar gene expression patterns between the two disease groups. (Fig. 1A). With principal component analysis, we assessed line-to-line and group-to-group variability and found that the gene expression data revealed overlap between schizophrenia and bipolar disorder (Fig. 1B). There were 83 significant DEGs between schizophrenia and bipolar disorder, out of which 53 genes were up-regulated in schizophrenia compared to bipolar disorder and 30 genes that were up-regulated in bipolar disorder compared to schizophrenia. Gene ontology analysis on these genes (Fig. 1C) revealed that schizophrenia organoids show relative up-regulation of genes involved in antigen binding, cytokine response and response to organic substances while the bipolar disorder organoids show relative up-regulation of genes involved in calcium binding, ion binding and muscle contraction. These analyses show that pathways representing genes that are up-regulated in schizophrenia organoids are enriched immune signaling processes while pathways representing genes that are up-regulated in bipolar disorder organoids are enriched for ion channel biology.
Fig. 1.
Comparative gene expression analysis of cerebral organoids differentiated from iPSCs of subjects with schizophrenia and bipolar disorder. A Heatmaps of differentially expressed genes (DEGs) of 6-month-old cerebral organoids generated from iPSCs of eight SCZ and eight BPI subjects. B Principal Component Analysis (PCA) of DEGs of SCZ and BPI cerebral organoids. C Gene ontology (GO) analysis of DEGs for SCZ vs BPI. GO analyses for biological process, molecular function and cellular component/protein localization are rank-ordered according to significance (p value) for the top 10 categories—categories up-regulated in SCZ are depicted in black and categories up-regulated in BPI are depicted in orange
Gene ontology
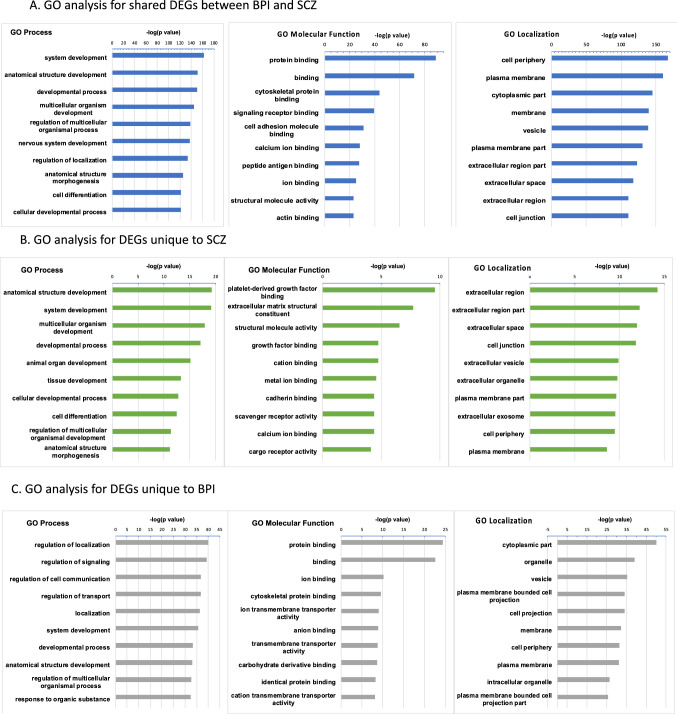
We categorized DEGs into the following three categories: (i) genes that were differentially regulated in the same manner in both schizophrenia and bipolar disorder when compared to healthy control organoids, (ii) genes that were differentially regulated in schizophrenia compared to healthy control but not in bipolar disorder and (iii) genes that were differentially regulated in bipolar disorder relative to healthy control but not in schizophrenia. We sought to delineate biological pathways that were similarly affected in the two disorders as well as pathways that were specific for the particular disease. We performed gene ontology (GO) analyses after rank-ordering the top hits according to significance (p-value) (Fig. 2A–C). Among significant GO: Biological Process categories that were differentially regulated in both schizophrenia and bipolar disorder were pathways involved in development, including nervous system development (Fig. 2A). GO: Localization analysis showed shared differences in cellular component/protein localization categories of cell periphery, plasma membrane cytoplasmic part and vesicles (Fig. 2A). The GO: Molecular Function analysis showed shared differences in schizophrenia and bipolar disorder in categories involving cytoskeletal protein binding, cell adhesion and peptide antigen binding (Fig. 2A).
Fig. 2.
Gene ontology analysis. A GO analysis of biological process, molecular function and cellular component/protein localization for DEGs that were shared between SCZ and BPI cerebral organoids when compared to CON cerebral organoids, rank-ordered according to significance (p value) for the top 10 categories. B GO analysis of biological process, molecular function and cellular component/protein localization for DEGs that were unique to SCZ cerebral organoids, rank-ordered according to significance (p value) for the top 10 categories. C GO analysis of biological process, molecular function and cellular component/protein localization for DEGs that were unique to BPI, rank-ordered according to significance (p value) for the top 10 categories
When gene ontology analyses were undertaken with DEGs unique to schizophrenia, the most significant GO: Biological Process categories included developmental processes while the significant GO: Localization categories showed differences in extracellular regions (Fig. 2B). The significant GO: Molecular Function categories indicated differences in schizophrenia in biology involving platelet-derived growth factor binding, extracellular matrix and cadherin binding (Fig. 2B). Similar analysis of DEGs unique to bipolar disorder for the significant GO: Biological Process categories showed differences in regulation of localization and signaling while the significant GO: Localization analysis shows significant differences in categories related to cytoplasm, organelles and vesicles (Fig. 2C). The significant GO: Molecular Function categories in bipolar disorder included a number of protein and ion binding as well as categories involving transporter activity (Fig. 2C).
Gene set enrichment analysis
We carried out gene set enrichment analysis (GSEA) using three datasets (Hallmark, KEGG, Reactome) to identify gene-enriched pathways in DEGs for one disorder vis-à-vis the other. GSEA is an analytical method that allows us to examine gene expression data and determine whether a collated set of genes, annotated based on their common roles in various biological processes, show significant differences between two groups [26]. We tabulated positive and negative correlation with schizophrenia and bipolar disorder according to the normalized enrichment scores (Table 1). Analysis of DEGS with relative enrichment in schizophrenia cerebral organoids showed involvement of pathways involved in inflammatory response (Reactome), oxidative phosphorylation (KEGG, Hallmark) and mitochondrial function (Reactome) in all three pathway databases (Table 1). Analysis of pathways for DEGs with relative enrichment in bipolar disorder cerebral organoids showed involvement of categories including neuro-transporters (Reactome) and long-term potentiation (Reactome) (Table 1). These results point to disease-specific differences related to a number of pathways, including in mitochondrial function in schizophrenia.
Functional analysis of schizophrenia and bipolar disorder organoids
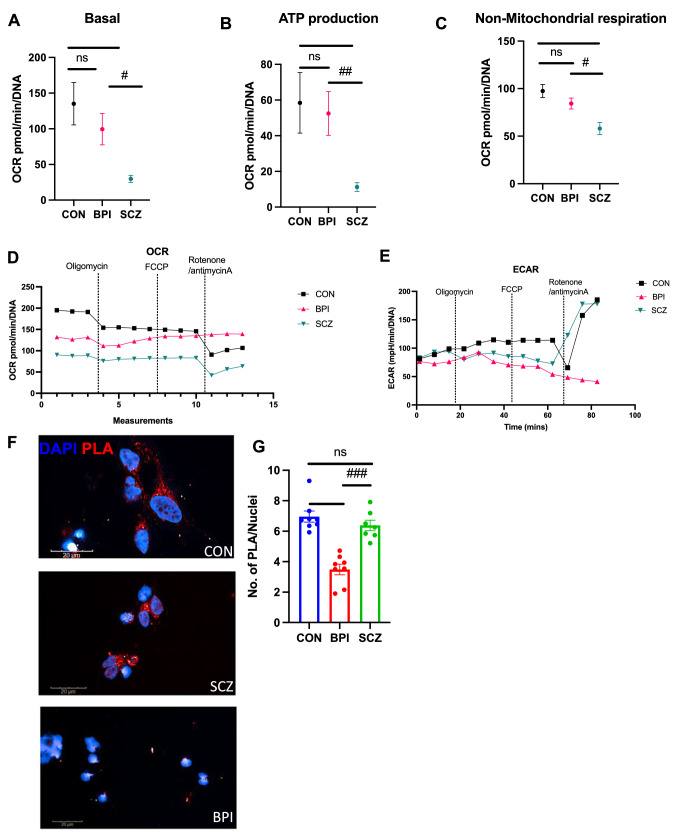
The GSEA analysis revealed significant differences in schizophrenia and bipolar disorder in categories of mitochondrial translation, respiratory electron transport and oxidative phosphorylation. Furthermore, we compared the DEGs from the bipolar disorder vs schizophrenia analysis to Human MitoCarta 2.0 database and found that 75 genes among the 83 DEGS were implicated in human mitochondrial pathways—genes that had relative up-regulation in schizophrenia are shown in Table 2 while genes that had relative up-regulation in bipolar disorder are shown in Table 3. To follow up on these observations, we undertook mitochondrial function experiments with the Seahorse Cell Mito Stress Test in cerebral organoids from bipolar disorder lines and compared them with the Seahorse Cell Mito Stress Test in cerebral organoids from schizophrenia lines that we had reported on previously [20]. These experiments involved live-cell metabolic experiments with 9-month-old cerebral organoids, in the setting of exposure to specific annotated perturbations in the assay. While the schizophrenia organoids had significantly lower basal oxygen consumption rate, ATP production and non-mitochondrial oxygen consumption when compared to healthy control organoids as we had reported earlier [20], the bipolar disorder organoids showed no significant differences in this regard (Fig. 3A–C). There was significant difference in the oxygen consumption rate (OCR) between schizophrenia and healthy control organoids in the setting of different perturbations, but these differences were also not observed in bipolar disorder organoids (Fig. 3D). However, bipolar disorder organoids showed lower ECAR rate which suggests lower rate of glycolysis when compared to healthy control and schizophrenia organoids (Fig. 3E).
Fig. 3.
Functional analysis of mitochondrial respiration in iPSC-derived cerebral organoids and mitochondrial-ER interactions in iPSC-derived cortical neurons. A–C Seahorse Cell Mito Stress Test assay conducted with 9-month-old cerebral organoids. Data collected from cerebral organoids differentiated from eight BPI iPSC lines and compared with data from SCZ and CON organoids reported previously [20]. Oxygen consumption rate (OCR) results for basal rate, ATP production, and non-mitochondrial oxygen consumption rate are shown, along with values for extracellular acidification rate (ECAR). Data collected from three sections in each condition and values shown as mean ± SEM. Normality was tested via Kolmogorov–Smirnov, but since all data were not normally distributed, Mann–Whitney test was performed. Basal: CON vs SCZ ***p = 0.0001, SCZ vs BPI #p = 0.0249. ATP production: CON vs SCZ **p = 0.0012, SCZ vs BPI ##p = 0.0016. Non-mitochondrial oxygen consumption rate: CON vs SCZ **p = 0.0003, SCZ vs BPI #p = 0.0237. CON vs BPI was insignificant in all categories. D Mitochondrial OCR (oxygen consumption rate) graph. Šídák's multiple comparisons test: CON vs SCZ ***p = 0.0001, CON vs BPI p = ns, SCZ vs BPI ***p = 0.0001. E ECAR (extracellular acid reflux rate) graph. Šídák's multiple comparisons test, performed CON vs SCZ, p = ns, CON vs BPI ***p = , CON vs BPI ***p = 0.0010 and SCZ vs BPI *p = 0.0102. F Representative images of proximity ligation assay (PLA) in CON, SCZ and BPI cortical neurons. Scale bar: 20 μM. F Quantification of PLA positive objects/nuclei. Data shown from cortical neurons generated from eight SCZ iPSC lines and compared with PLA data from eight CON and eight BPI iPSC lines reported previously [21]. Values are shown as mean ± SEM. Two-tailed Mann–Whitney U test: CON vs BPI ***p = 0.0002, SCZ vs BPI ###p = 0.0003, CON vs SCZ was insignificant
Mitochondria–endoplasmic reticulum interactions have been hypothesized to be abnormal in mood disorders [28, 29]. The contact sites between endoplasmic reticulum (ER) and mitochondria, called mitochondria-associated membranes (MAMs), facilitate Ca2+ exchange between the two organelles and play important roles in mitochondrial fission and fusion, intracellular trafficking, calcium homeostasis, ER stress and unfolded proteins, phospholipid exchange as well as autophagy and inflammation [30]. Since the GO analysis had indicated enrichment in bipolar disorder of genes involved in ion binding and regulation of transport (Figs. 1C and 2C), we sought to investigate if there were disease-specific differences in MAMs between schizophrenia and bipolar disorder. In a previous study, we had shown that bipolar disorder cortical neurons showed a significant reduction in MAMs when compared to healthy control cortical neurons, with data obtained using the proximity ligation assay to quantify the number of contact sites between the mitochondria and ER [21]. We collected additional data on MAMs in cortical neurons from schizophrenia iPSCs and compared them with the previous data from bipolar neurons. We found that the control and schizophrenia neurons were similar in terms of MAMs in the cortical neurons but only the bipolar disorder cortical neurons showed a significant reduction in MAMs (Fig. 3F and G).
Discussion
Patient-derived cells provide live biological material that can be utilized to interrogate disease biology relevant to schizophrenia and bipolar disorder [13, 31, 32]. Recently iPSC-derived neuronal and glial cells have been utilized to examine gene expression differences in schizophrenia and bipolar disorder [33, 34]. While most of the early gene expression studies had focused on the use of neural progenitor cells and two-dimensional neuronal cultures, three-dimensional cerebral organoids are also being utilized for the study of disease biology, including examination of transcriptomic profiles [35, 36]. In our study comparing the similarities and differences in transcriptomic profiles of schizophrenia and bipolar disorder organoids, we found shared categories related to nervous system development in these two disorders. While there is significant evidence lending support to neurodevelopmental abnormalities in schizophrenia, the evidence for neurodevelopmental processes in bipolar disorder is not as well established [37]. In the ex vivo models that we studied which recapitulate neurodevelopmental processes, the findings suggest a shared feature related to deviation from normal development as captured by the gene expression patterns.
While we had previously examined transcriptomic differences in schizophrenia and bipolar disorder when compared to healthy control organoids, we undertook this additional study to examine what genes and pathways are differentially regulated in schizophrenia vs. bipolar disorder. While some of the biological and clinical features are shared between schizophrenia and bipolar disorder, it is hypothesized there are distinct differences in the neurobiology of the two disorders as well. By undertaking a head-to-head comparison, we hoped to delineate genes and pathways that distinguish the underlying biology in these two disorders. When examining the gene ontology patterns of genes that were differentially regulated in schizophrenia organoids vis-a-vis bipolar disorder organoids, we found that the differentially expressed genes in schizophrenia were enriched for pathways involved in immune signaling. This is intriguing given the convergent evidence for the role of the immune system dysregulation in schizophrenia [38]. Brain organoids generated from iPSCs generate a range of neuronal and glial cell types, including microglia, and some of the differences in schizophrenia organoids may result from differences in neuro-immune processes in the context of the developing brain [21]. Another category that showed distinct enrichment in schizophrenia cerebral organoids vis-à-vis bipolar disorder organoids related to genes involved in oxidative phosphorylation and mitochondrial function. This was borne out in the functional Seahorse assays as well that showed significantly lower basal oxygen consumption rate, ATP production and non-mitochondrial oxygen consumption in schizophrenia organoids but not in bipolar disorder organoids. There have been a number of studies examining mitochondrial function in schizophrenia and bipolar disorder, many involving post-mortem brain studies, peripheral cells and neuroimaging [39–43]. Previous studies in bipolar disorder have shown decreased expression of electron transport chain components in the cortex [44, 45] as well as decreased mitochondrial respiration and increased glycolytic capacity [46, 47]. Our gene expression results as well as the Seahorse experiments point to disease-specific deficits of mitochondrial function in cerebral organoids from schizophrenia, possibly pointing to unique differences in mitochondrial function in the context of neurodevelopment in schizophrenia. Our findings of differences in mitochondrial respiration in schizophrenia cerebral organoids is consistent with a large body of literature suggesting deficits in the oxidative phosphorylation system in schizophrenia [48].
The pathways that were enriched in the differentially regulated genes in bipolar disorder organoids vis-à-vis schizophrenia organoids involved categories involved in ion binding and regulation of transport. Further examination of mitochondria–ER interactions showed that cortical neurons from bipolar disorder subjects showed a significant reduction MAMs compared to control cortical neurons but the MAMs in schizophrenia cortical neurons were in the same range as the control cortical neurons. ER stress has been hypothesized to play in important role in bipolar disorder [49, 50]. Studies have also indicated that the mood stabilizers lithium and valproic acid may mediate their therapeutic effects through modulation of pathways involved in ER stress [51, 52]. While various aspects of mitochondrial biology have been hypothesized to play a role in a schizophrenia and bipolar disorder [53, 54], our findings of disease-specific differences in ER–mitochondria interactions in bipolar disorder vis-à-vis schizophrenia is of interest in light of these previous studies. Altered ER–mitochondrial interactions may be an underlying mechanism that leads to the abnormal form and distribution of the mitochondrial network observed in cells from patients with bipolar disorder [39].
Our findings are also of interest in the context of a recent study analyzing transcriptional data for mitochondria-related gene expression the dorsolateral prefrontal cortex in post-mortem brain specimens from subjects with schizophrenia and bipolar disorder [55]. This study showed that schizophrenia and bipolar disorder had distinct differences in the nature of mitochondrial dysfunction, with a greater subset of genes that were differentially expressed in schizophrenia compared to bipolar disorder. Moreover, they found that the schizophrenia-related DEGs were enriched for mitochondrial function pathways involved in energy production while there was no such enrichment for bipolar disorder-related DEGs [55]. Our findings of mitochondrial function differences in schizophrenia organoids but not in bipolar disorder organoids are consistent with those post-mortem findings. Moreover, our experiments with the cortical neurons suggest that the mitochondria-related deficits in bipolar disorder may primarily result from aberrant mitochondria–endoplasmic reticulum interactions rather than mitochondrial pathways involved in energy production.
Among other pathways implicated by the DEGs in our transcriptomic analyses were those related to ion channel biology and neuro-transporters. Ion channel genes have been implicated in the disease biology of bipolar disorder and some therapeutic approaches have been tried based on modulation of ion channels [56, 57]. Genetic studies of rare variants in bipolar disorder have also suggested that these risk variants are enriched for genes that are related to the regulation of neuronal excitability [58]. Studies have suggested a role of excitatory/inhibitory (E/I) synaptic imbalance in bipolar disorder as well as in schizophrenia [59, 60]. In our functional studies neuronal firing using multi-electrode arrays that we have previously published, we found no differences in spontaneous neuronal firing in bipolar disorder or schizophrenia organoids when compared to healthy control organoids [20, 21]. However, these experiments had also shown that there was a significant reduction in the response to electrical stimulation and to KCl-induced depolarization in both bipolar disorder and schizophrenia organoids, suggesting some shared functional deficits in relation to neuronal excitability [20, 21].
While our studies add to the body of literature examining biological features that are shared and distinct between the disorders, there are a number of caveats to the results described in our study. The generation and culturing of organoids for 6–9 month periods introduces the possibility of stochastic variations that may affect the differentiation process. We have attempted to minimize such confounding factors by undertaking the organoid generation and culturing of the different groups in parallel and managed by the same researcher to avoid user-dependent and batch effects. Since schizophrenia and bipolar disorder are idiopathic disorders with complex genetics, we would ideally like to have a much larger sample size. However, given the time, labor and resource intensiveness of the cerebral organoid generation process makes it difficult at this time. Generating and studying a total of 24 cerebral organoids over the course of 9 months was a significant undertaking. While cerebral organoids capture some elements of the human brain biology, there are many limitations in our attempts to recapitulate the biology in the brain. While cerebral organoids contain many neuronal and glial cell types, including mature neurons and synaptic connections, they lack a vasculature in the way cerebral organoids are currently generated. In addition, the transcriptomic patterns of cerebral organoids most closely resemble the gene expression patterns observed in the fetal cortex. Hence, there are limitations in the study of psychiatric disorders that usually have their onset in young adulthood using cellular models that resemble the biology of the fetal brain. Hence, these models may have more relevance in understanding some of the neurodevelopmental factors or vulnerabilities that may be present long before the clinical manifestation of the illnesses. Along those lines, these cellular models may also enable the testing of specific perturbations that may intersect with the underlying complex genetic risk to unmask aberrant pathway as related to the disease biology during neurodevelopment [31, 32].
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 (XLSX 34629 KB) Cuffdiff analysis for the bipolar disorder vs schizophrenia.
Supplementary file2 (DOCX 61 KB) Tables.
Acknowledgements
This work was supported by a National Institute of Mental Health Biobehavioral Research Awards for Innovative New Scientists (BRAINS) Award R01MH113858 (RK), National Institute of Mental Health Clinical Scientist Development Award K08MH086846 (RK), the Doris Duke Charitable Foundation Clinical Scientist Development Award (2015088) (to RK), the Ryan Licht Sang Bipolar Foundation (RK), the Jeanne Marie Lee Investigator Award-Osterhaus Family Foundation (AK), NARSAD Young Investigator Award from the Brain & Behavior Research Foundation (AK), the Phyllis & Jerome Lyle Rappaport Foundation (to RK), the Harvard Stem Cell Institute (RK) and by Steve Willis and Elissa Freud (RK). We would like to thank Joanne Huang for preparing fibroblasts for the reprogramming process, Steve Sheridan for logistical assistance in obtaining a quote and shipping materials to a contract research organization for the iPSC reprogramming process, Smita Jagtap for technical assistance with the Seahorse assays and Magdalena Vater for technical advice for the PLA experiments.
Author contributions
AK and RK conceived the scientific approach and experimental plan. KL and AK generated the organoids and undertook the experiments. DM and BMC obtained fibroblasts from the subjects. AK and RK wrote the manuscript with input from KL, DM and BMC. All authors reviewed the manuscript. All authors read and approved the final manuscript.
Data availability
The data are available in NCBI's Gene Expression Omnibus had previously been deposited [20, 21] and are accessible through GEO Series accession number GSE134497 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE134497) and GSE133534 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE133534).
Declarations
Ethics approval and consent to participate
The study was approved undertaken with approval from the institutional review boards of Massachusetts General Hospital and McLean Hospital and performed in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Consent for publication
All human subjects had given informed consent to provide biological samples used in this study.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Maggioni E, Crespo-Facorro B, Nenadic I, Benedetti F, Gaser C, Sauer H, et al. Common and distinct structural features of schizophrenia and bipolar disorder: The European Network on Psychosis, Affective disorders and Cognitive Trajectory (ENPACT) study. PLoS ONE. 2017;12(11):e0188000. doi: 10.1371/journal.pone.0188000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fischer BA, Carpenter WT. Will the Kraepelinian dichotomy survive DSM-V? Neuropsychopharmacology. 2009;34(9):2081–2087. doi: 10.1038/npp.2009.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cross-Disorder Group of the Psychiatric Genomics Consortium Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381(9875):1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prata DP, Costa-Neves B, Cosme G, Vassos E. Unravelling the genetic basis of schizophrenia and bipolar disorder with GWAS: a systematic review. J Psychiatr Res. 2019;114:178–207. doi: 10.1016/j.jpsychires.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 5.López-Muñoz F, Shen WW, D’Ocon P, Romero A, Álamo C. A history of the pharmacological treatment of bipolar disorder. Int J Mol Sci. 2018;19(7):2143. doi: 10.3390/ijms19072143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berle J, Spigset O. Are mood stabilizers beneficial in the treatment of schizophrenia? Tidsskr Nor Laegeforen. 2005;125(13):1809–1812. [PubMed] [Google Scholar]
- 7.Ramaker RC, Bowling KM, Lasseigne BN, Hagenauer MH, Hardigan AA, Davis NS, et al. Post-mortem molecular profiling of three psychiatric disorders. Genome Med. 2017;9(1):72. doi: 10.1186/s13073-017-0458-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan J, Cai JJ, Ji G, Sham PC. Commonality in dysregulated expression of gene sets in cortical brains of individuals with autism, schizophrenia, and bipolar disorder. Transl Psychiatry. 2019;9(1):152. doi: 10.1038/s41398-019-0488-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis SE, Panitch R, West AB, Arking DE. Transcriptome analysis of cortical tissue reveals shared sets of downregulated genes in autism and schizophrenia. Transl Psychiatry. 2016;6:e817. doi: 10.1038/tp.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi K, Yamanaka S. Induced pluripotent stem cells in medicine and biology. Development. 2013;140(12):2457–2461. doi: 10.1242/dev.092551. [DOI] [PubMed] [Google Scholar]
- 11.Karmacharya R, Haggarty SJ. Stem cell models of neuropsychiatric disorders. Mol Cell Neurosci. 2016;73:1–2. doi: 10.1016/j.mcn.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Karmacharya R, Kieling C, Mondelli V. Integrating stem cell-based experiments in clinical research. Eur Psychiatry. 2020;63(1):e62. doi: 10.1192/j.eurpsy.2020.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kathuria A, Lopez-Lengowski K, Roffman JL, Karmacharya R. Distinct effects of interleukin-6 and interferon-gamma on differentiating human cortical neurons. Brain Behav Immun. 2022;103:97–108. doi: 10.1016/j.bbi.2022.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watmuff B, Berkovitch SS, Huang JH, Iaconelli J, Toffel S, Karmacharya R. Disease signatures for schizophrenia and bipolar disorder using patient-derived induced pluripotent stem cells. Mol Cell Neurosci. 2016;73:96–103. doi: 10.1016/j.mcn.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watmuff B, Liu B, Karmacharya R. Stem cell-derived neurons in the development of targeted treatment for schizophrenia and bipolar disorder. Pharmacogenomics. 2017;18(5):471–479. doi: 10.2217/pgs-2016-0187. [DOI] [PubMed] [Google Scholar]
- 16.Kathuria A, Lopez-Lengowski K, Watmuff B, McPhie D, Cohen BM, Karmacharya R. Synaptic deficits in iPSC-derived cortical interneurons in schizophrenia are mediated by NLGN2 and rescued by N-acetylcysteine. Transl Psychiatry. 2019;9(1):321. doi: 10.1038/s41398-019-0660-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez-Lengowski K, Kathuria A, Gerlovin K, Karmacharya R. Co-culturing microglia and cortical neurons differentiated from human induced pluripotent stem cells. J Vis Exp. 2021 doi: 10.3791/62480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Renner M, Lancaster MA, Bian S, Choi H, Ku T, Peer A, et al. Self-organized developmental patterning and differentiation in cerebral organoids. EMBO J. 2017;36(10):1316–1329. doi: 10.15252/embj.201694700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yakoub AM. Cerebral organoids exhibit mature neurons and astrocytes and recapitulate electrophysiological activity of the human brain. Neural Regen Res. 2019;14(5):757–761. doi: 10.4103/1673-5374.249283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kathuria A, Lopez-Lengowski K, Jagtap SS, McPhie D, Perlis RH, Cohen BM, et al. Transcriptomic landscape and functional characterization of induced pluripotent stem cell-derived cerebral organoids in schizophrenia. JAMA Psychiat. 2020;77(7):745–754. doi: 10.1001/jamapsychiatry.2020.0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kathuria A, Lopez-Lengowski K, Vater M, McPhie D, Cohen BM, Karmacharya R. Transcriptome analysis and functional characterization of cerebral organoids in bipolar disorder. Genome Med. 2020;12(1):34. doi: 10.1186/s13073-020-00733-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. 2011;17(1):10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 23.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tubbs E, Rieusset J. Study of endoplasmic reticulum and mitochondria interactions by in situ proximity ligation assay in fixed cells. J Vis Exp. 2016 doi: 10.3791/54899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pereira AC, Resende R, Morais S, Madeira N, Pereira CF. The ups and downs of cellular stress: the "MAM hypothesis" for bipolar disorder pathophysiology. Int J Clin Neurosci Ment Health. 2017;4:S04. doi: 10.21035/ijcnmh.2017.4(Suppl.3).S04. [DOI] [Google Scholar]
- 29.Resende R, Fernandes T, Pereira AC, De Pascale J, Marques AP, Oliveira P, et al. Mitochondria, endoplasmic reticulum and innate immune dysfunction in mood disorders: do mitochondria-associated membranes (MAMs) play a role? Biochim Biophys Acta Mol Basis Dis. 2020;1866(6):165752. doi: 10.1016/j.bbadis.2020.165752. [DOI] [PubMed] [Google Scholar]
- 30.Vance JE. MAM (mitochondria-associated membranes) in mammalian cells: lipids and beyond. Biochim Biophys Acta. 2014;1841(4):595–609. doi: 10.1016/j.bbalip.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 31.Huang JH, Berkovitch SS, Iaconelli J, Watmuff B, Park H, Chattopadhyay S, et al. Perturbational profiling of metabolites in patient fibroblasts implicates α-aminoadipate as a potential biomarker for bipolar disorder. Mol Neuropsychiatry. 2016;2(2):97–106. doi: 10.1159/000446654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang JH, Park H, Iaconelli J, Berkovitch SS, Watmuff B, McPhie D, et al. Unbiased metabolite profiling of schizophrenia fibroblasts under stressful perturbations reveals dysregulation of plasmalogens and phosphatidylcholines. J Proteome Res. 2017;16(2):481–493. doi: 10.1021/acs.jproteome.6b00628. [DOI] [PubMed] [Google Scholar]
- 33.Akkouh IA, Hughes T, Steen VM, Glover JC, Andreassen OA, Djurovic S, et al. Transcriptome analysis reveals disparate expression of inflammation-related miRNAs and their gene targets in iPSC-astrocytes from people with schizophrenia. Brain Behav Immun. 2021;94:235–244. doi: 10.1016/j.bbi.2021.01.037. [DOI] [PubMed] [Google Scholar]
- 34.Lin M, Lachman HM, Zheng D. Transcriptomics analysis of iPSC-derived neurons and modeling of neuropsychiatric disorders. Mol Cell Neurosci. 2016;73:32–42. doi: 10.1016/j.mcn.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stachowiak EK, Benson CA, Narla ST, Dimitri A, Chuye LEB, Dhiman S, et al. Cerebral organoids reveal early cortical maldevelopment in schizophrenia-computational anatomy and genomics, role of FGFR1. Transl Psychiatry. 2017;7(11):6. doi: 10.1038/s41398-017-0054-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Notaras M, Lodhi A, Fang H, Greening D, Colak D. The proteomic architecture of schizophrenia iPSC-derived cerebral organoids reveals alterations in GWAS and neuronal development factors. Transl Psychiatry. 2021;11(1):541. doi: 10.1038/s41398-021-01664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parellada M, Gomez-Vallejo S, Burdeus M, Arango C. Developmental differences between schizophrenia and bipolar disorder. Schizophr Bull. 2017;43(6):1176–1189. doi: 10.1093/schbul/sbx126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller BJ, Goldsmith DR. Towards an immunophenotype of schizophrenia: progress, potential mechanisms, and future directions. Neuropsychopharmacology. 2017;42(1):299–317. doi: 10.1038/npp.2016.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cataldo AM, McPhie DL, Lange NT, Punzell S, Elmiligy S, Ye NZ, et al. Abnormalities in mitochondrial structure in cells from patients with bipolar disorder. Am J Pathol. 2010;177(2):575–585. doi: 10.2353/ajpath.2010.081068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Da Silva T, Wu A, Laksono I, Prce I, Maheandiran M, Kiang M, et al. Mitochondrial function in individuals at clinical high risk for psychosis. Sci Rep. 2018;8(1):6216. doi: 10.1038/s41598-018-24355-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hjelm BE, Rollins B, Mamdani F, Lauterborn JC, Kirov G, Lynch G, et al. Evidence of mitochondrial dysfunction within the complex genetic etiology of schizophrenia. Mol Neuropsychiatry. 2015;1(4):201–219. doi: 10.1159/000441252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holper L, Ben-Shachar D, Mann JJ. Multivariate meta-analyses of mitochondrial complex I and IV in major depressive disorder, bipolar disorder, schizophrenia, Alzheimer disease, and Parkinson disease. Neuropsychopharmacolog. 2019;44(5):837–849. doi: 10.1038/s41386-018-0090-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ben-Shachar D. Mitochondrial multifaceted dysfunction in schizophrenia; complex I as a possible pathological target. Schizophr Res. 2017;187:3–10. doi: 10.1016/j.schres.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 44.Sun X, Wang JF, Tseng M, Young LT. Downregulation in components of the mitochondrial electron transport chain in the postmortem frontal cortex of subjects with bipolar disorder. J Psychiatry Neurosci. 2006;31(3):189–196. [PMC free article] [PubMed] [Google Scholar]
- 45.Andreazza AC, Shao L, Wang JF, Young LT. Mitochondrial complex I activity and oxidative damage to mitochondrial proteins in the prefrontal cortex of patients with bipolar disorder. Arch Gen Psychiatry. 2010;67(4):360–368. doi: 10.1001/archgenpsychiatry.2010.22. [DOI] [PubMed] [Google Scholar]
- 46.Marques AP, Resende R, Silva DF, Batista M, Pereira D, Wildenberg B, et al. Mitochondrial alterations in fibroblasts of early stage bipolar disorder patients. Biomedicines. 2021;9(5):522. doi: 10.3390/biomedicines9050522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Regenold WT, Phatak P, Marano CM, Sassan A, Conley RR, Kling MA. Elevated cerebrospinal fluid lactate concentrations in patients with bipolar disorder and schizophrenia: implications for the mitochondrial dysfunction hypothesis. Biol Psychiatry. 2009;65(6):489–494. doi: 10.1016/j.biopsych.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bergman O, Ben-Shachar D. Mitochondrial oxidative phosphorylation system (OXPHOS) deficits in schizophrenia: possible interactions with cellular processes. Can J Psychiatry. 2016;61(8):457–469. doi: 10.1177/0706743716648290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pfaffenseller B, Wollenhaupt-Aguiar B, Fries GR, Colpo GD, Burque RK, Bristot G, et al. Impaired endoplasmic reticulum stress response in bipolar disorder: cellular evidence of illness progression. Int J Neuropsychopharmacol. 2014;17(9):1453–1463. doi: 10.1017/S1461145714000443. [DOI] [PubMed] [Google Scholar]
- 50.Hayashi A, Kasahara T, Kametani M, Toyota T, Yoshikawa T, Kato T. Aberrant endoplasmic reticulum stress response in lymphoblastoid cells from patients with bipolar disorder. Int J Neuropsychopharmacol. 2009;12(1):33–43. doi: 10.1017/S1461145708009358. [DOI] [PubMed] [Google Scholar]
- 51.Kakiuchi C, Ishigaki S, Oslowski CM, Fonseca SG, Kato T, Urano F. Valproate, a mood stabilizer, induces WFS1 expression and modulates its interaction with ER stress protein GRP94. PLoS ONE. 2009;4(1):e4134. doi: 10.1371/journal.pone.0004134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Breen MS, White CH, Shekhtman T, Lin K, Looney D, Woelk CH, et al. Lithium-responsive genes and gene networks in bipolar disorder patient-derived lymphoblastoid cell lines. Pharmacogenomics J. 2016;16(5):446–453. doi: 10.1038/tpj.2016.50. [DOI] [PubMed] [Google Scholar]
- 53.Dogan AE, Yuksel C, Du F, Chouinard VA, Öngür D. Brain lactate and pH in schizophrenia and bipolar disorder: a systematic review of findings from magnetic resonance studies. Neuropsychopharmacology. 2018;43(8):1681–1690. doi: 10.1038/s41386-018-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Machado AK, Pan AY, da Silva TM, Duong A, Andreazza AC. Upstream pathways controlling mitochondrial function in major psychosis: a focus on bipolar disorder. Can J Psychiatry. 2016;61(8):446–456. doi: 10.1177/0706743716648297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Glausier JR, Enwright JF, 3rd, Lewis DA. Diagnosis- and cell type-specific mitochondrial functional pathway signatures in schizophrenia and bipolar disorder. Am J Psychiatry. 2020;177(12):1140–1150. doi: 10.1176/appi.ajp.2020.19111210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Balaraman Y, Lahiri DK, Nurnberger JI. Variants in ion channel genes link phenotypic features of bipolar illness to specific neurobiological process domains. Mol Neuropsychiatry. 2015;1(1):23–35. doi: 10.1159/000371886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andreazza AC, Young LT. The neurobiology of bipolar disorder: identifying targets for specific agents and synergies for combination treatment. Int J Neuropsychopharmacol. 2014;17(7):1039–1052. doi: 10.1017/S1461145713000096. [DOI] [PubMed] [Google Scholar]
- 58.Ament SA, Szelinger S, Glusman G, Ashworth J, Hou L, Akula N, et al. Rare variants in neuronal excitability genes influence risk for bipolar disorder. Proc Natl Acad Sci USA. 2015;112(11):3576–3581. doi: 10.1073/pnas.1424958112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee Y, Zhang Y, Kim S, Han K. Excitatory and inhibitory synaptic dysfunction in mania: an emerging hypothesis from animal model studies. Exp Mol Med. 2018;50(4):12. doi: 10.1038/s12276-018-0028-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao R, Penzes P. Common mechanisms of excitatory and inhibitory imbalance in schizophrenia and autism spectrum disorders. Curr Mol Med. 2015;15(2):146–167. doi: 10.2174/1566524015666150303003028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1 (XLSX 34629 KB) Cuffdiff analysis for the bipolar disorder vs schizophrenia.
Supplementary file2 (DOCX 61 KB) Tables.
Data Availability Statement
The data are available in NCBI's Gene Expression Omnibus had previously been deposited [20, 21] and are accessible through GEO Series accession number GSE134497 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE134497) and GSE133534 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE133534).